Abstract
Background
Neutralising antibodies against SARS-CoV-2 are a vital component in the fight against COVID-19 pandemic, having the potential of both therapeutic and prophylactic applications. Bispecific antibodies (BsAbs) against SARS-CoV-2 are particularly promising, given their ability to bind simultaneously to two distinct sites of the receptor-binding domain (RBD) of the viral spike protein. Such antibodies are complex molecules associated with multi-faceted mechanisms of action that require appropriate bioassays to ensure product quality and manufacturing consistency.
Methods
We developed procedures for biolayer interferometry (BLI) and a cell-based virus neutralisation assay, the focus reduction neutralisation test (FRNT). Using both assays, we tested a panel of five BsAbs against different spike variants (Ancestral, Delta and Omicron) to evaluate the use of these analytical methods in assessing binding and neutralisation activities of anti-SARS-CoV-2 therapeutics.
Results
We found comparable trends between BLI-derived binding affinity and FRNT-based virus neutralisation activity. Antibodies that displayed high binding affinity against a variant were often followed by potent neutralisation at lower concentrations, whereas those with low binding affinity also demonstrated reduced neutralisation activity.
Conclusion
The results support the utility of BLI and FRNT assays in measuring variant-specific binding and virus neutralisation activity of anti-SARS-CoV-2 antibodies.
Keywords: SARS-CoV-2, COVID-19, bispecific antibody, bioassay, product quality
Statement of significance: Bispecific antibodies are a promising class of therapeutics against SARS-CoV-2. BLI coupled with FRNT assays can be used to characterise the binding and neutralisation activity of these antibodies against specific SARS-CoV-2 variants.
INTRODUCTION
Antibody-based therapies are promising in the fight against COVID-19, having the potential for both therapeutic and prophylactic applications. These antibodies are primarily directed to bind specific sites on the receptor-binding domain (RBD) of SARS-CoV-2 spike protein, which the virus utilises to bind to the ACE2 receptor, thereby blocking viral entry into human cells or providing prophylactic immunity prior to infection [1, 2]. Interim clinical data suggested a therapeutic value for several monoclonal antibodies (mAbs) in COVID-19 patients when administered as a single agent or a cocktail of two mAbs. The US Food and Drug Administration (FDA) has issued Emergency Use Authorization (EUA) for five investigational anti-SARS-CoV-2 mAbs to treat COVID-19 in adult and pediatric patients; sotrovimab or bebtelovimab as a monotherapy and casirivimab together with imdevimab or bamlanivimab with etesevimab or tixagevimab plus cilgavimab as a combination therapy [3]. A major challenge remains, however, because SARS-CoV-2 is constantly mutating, resulting in genetic variants of concern such as Alpha (B.1.1.7), Beta (B.1.351), Delta (B.1.617), Gamma (P.1) and Omicron, and its subvariants (BA.1, BA.2, BA.3, BA.4, BA.5) [4]. These variants harbor mutations in the spike protein or directly in the RBD, thus decreasing the efficacy of certain antibody and vaccine formulations, underscoring the urgent need of developing new antibody-based therapies that potently neutralise the new variants of concern [5]. In this context, bispecific antibodies (BsAbs) against SARS-CoV-2 spike represent a promising and emerging therapeutic area for COVID-19. While sharing the epitope specificity and manufacturability of a conventional mAb, BsAbs can simultaneously bind to two distinct sites on the viral spike protein using a single therapeutic, thereby enhancing neutralisation potential against a wide range of genetic variants. This is supported by recent reports for several anti-SARS-CoV-2 BsAbs, including COV-X2 [6], CV503_521_GS [7], BsAb15 [8] and RBT-0813 [9].
Both mAbs and BsAbs against SARS-CoV-2 are complex molecules with multi-faceted mechanisms of action, including single or dual target binding, virus neutralisation and potential Fc effector functions [10–12]. For these complex products, the development and implementation of a potency assay(s) is critical to ensure that each lot is consistently produced with the potency necessary to achieve clinical efficacy and that such potency is maintained over the shelf life of the product (ICH Q6B) [13]. Compared with physicochemical methods, which are either compendial or platform methods, bioassays are product specific that require special considerations to satisfy regulatory requirements. Amid the COVID-19 pandemic, the FDA issued a guidance on potency assays for manufacturers of mAbs and other therapeutic proteins to treat COVID-19 [14]. The guidance document recommends a comprehensive assessment of anti-SARS-CoV-2 antibodies, including assays for measuring spike protein binding affinity, virus neutralisation and potential Fc effector functions. In surveying sponsor submissions of developmental therapeutics to the FDA, we found that surface plasmon resonance (SPR) is widely used to analyse viral spike-antibody interactions with respect to binding kinetics and affinity as well as binding specificity (Table 1). The plaque reduction neutralisation test (PRNT) remains a gold standard for measuring neutralisation titers of antibodies for many viral diseases. However, PRNT has relatively low throughput and is difficult to automate due to the necessity of visualising and counting individual plaques. As an alternative, enzyme-linked immunosorbent assays (ELISA) or pseudo-typed virus neutralisation assays are commonly used as bioassays for lot release or characterisation testing of anti-SARS-CoV-2 antibodies (Table 1). These assays measure the ability of an antibody to block RBD-ACE2 interactions, utilising recombinant RBD or full-length spike protein as partnering analytes. The selection of the right analyte is crucial for bioassay development because the RBD may not fully replicate the folding and conformation of the full-length S protein trimer. This is especially true for the new variants of concern, such as Omicron variants harboring several mutations outside of the RBD that can also impact how the virus interacts with ACE2. When surrogate bioassays are employed, it is important to demonstrate that a surrogate measurement (e.g., ELISA readout) correlates with the neutralisation activity derived from the cell-based assays utilising live viruses.
Table 1.
Common bioassays for anti-SARS-CoV-2 therapeutics
| Type | Binding and Neutralisation | ||
|---|---|---|---|
| Release | Characterisation | ||
| 1 | IgG1 | Virus neutralisation | ELISA SPR |
| 2 | IgG1 | ELISA | SPR Flow cytometry Virus neutralisation Pseudovirus neutralisation |
| 3 | IgG1 | ELISA | SPR BLI PRNT |
| 4 | IgG1 | ELISA | ELISA FACS BLI Virus neutralisation Pseudovirus neutralisation |
| 5 | IgG1κ | ELISA | SPR Blocking ELISA Virus neutralisation |
| 6 | IgG1κ | ELISA | SPR Pseudovirus neutralisation Virus neutralisation |
| 7 | IgG1 | ELISA | Virus neutralisation |
| 8 | IgG1 | ELISA | Virus neutralisation |
| 9 | IgG1 | Micro-neutralisation assay | ELISA |
| 10 | IgG1κ | ELISA | BLI Flow cytometry Pseudovirus neutralisation Virus neutralisation PRNT |
| 11 | IgG1 | ELISA | Pseudovirus neutralisation SPR |
| 12 | IgG1κ | ELISA Pseudovirus neutralisation bioassay |
Virus neutralisation BLI |
| 13 | IgG1 | SPR | Pseudovirus neutralisation |
| 14 | IgG1 | ELISA | Virus neutralisation |
Common methods used to assess the binding and neutralisation. A survey was performed of INDs that have been submitted to the FDA that target SARS-CoV-2. The most common method for determination of potency at drug substance/drug product release testing was found to be ELISA binding assay.
Biolayer interferometry, BLI; enzyme-linked immunosorbent assay, ELISA; fluorescence activated cell sorting, FACS; immunoglobin G, IgG; plaque reduction neutralisation test, PRNT; surface plasmon resonance, SPR
To address the full range of a product’s biological activities adequately, it is often necessary to apply more than one analytical procedure to evaluate the same quality attribute. Methods that use different physicochemical or biological principles to assess the same attribute are especially valuable because they provide independent data to support the quality of that attribute (e.g., orthogonal methods to assess RBD-binding kinetics). In addition, the use of complementary analytical methods in series, such as binding assays and cell-based virus neutralisation assays, should provide a meaningful and sensitive method for comparing products in the same class. As SARS-CoV-2 continues to circulate and new variants emerge, there is a heightened need to rapidly and adequately assess attributes that demonstrate the effectiveness of developmental anti-SARS-CoV-2 therapeutics. Here, we developed procedures for the biolayer interferometry (BLI) and a focus reduction neutralisation test (FRNT) with the aim of creating additional protocols useful in characterising BsAbs against SARS-CoV-2. BLI is a label-free biosensing technique that enables direct measurement of biomolecular interactions in real time. This technique has been used in several recent publications to characterise anti-SARS-CoV-2 mAbs and assess their binding affinities [15, 16]. The FRNT assay is proven to be capable of analysing neutralising antibodies with a wide range of SARS-CoV-2 variants [17]. After testing a panel of five distinct BsAbs, we found comparable trends between BLI-derived binding affinity and FRNT-based virus neutralisation activity. The BLI system, coupled with the FRNT assays, complements the current toolbox (Table 1), thus enhancing the analytical capacity for quality assessment of anti-SARS-CoV-2 antibodies.
MATERIALS AND METHODS
Viral protein sources
All spike RBD and trimer proteins were obtained from Sinobiological. The catalog numbers for RBDs are as follows: Ancestral: 40592-V08H, Delta (B.1.617.2): 40592-V08H90 and Omicron (B.1.1.529): 40592-V08H121. The catalog numbers for trimers are as follows: Ancestral: 40589-V08H4, Delta (B.1.617.2): 40589-V08H10 and Omicron (B.1.1.529): 40589-V08H26.
Bispecific antibodies
Bispecific antibodies, CoV-X2-WT and CoV-X2-LALA, were generated and provided by Dr Luca Varani (Institute for Research in Biomedicine, University of Switzerland) [6]. CV503_664_EL, CV503_521_GS and CV503_993_EL were produced and provided by Dr Joshua Tan (National Institute of Allergy and Infectious Diseases, National Institutes of Health) [7].
Cell culture
H1299-hACE2 cells were generated in-house at the FDA and maintained in DMEM supplemented with 5% penicillin and streptomycin, and 10% fetal bovine serum (FBS) at 37 °C with 5% CO2. Vero E6 cells (Cat # CRL-1586) were purchased from American Type Culture Collection and cultured in Dulbecco’s minimal essential medium supplemented with 10% fetal bovine serum (Invitrogen) and 1% penicillin/streptomycin and L-glutamine.
SARS-CoV-2 isolates
SARS-CoV-2 isolate WA1/2020 (NR-52281, lot 70033175) was obtained from BEI Resources, NIAID, NIH and passaged three times in Vero cells prior to acquisition. It was passaged one more time in Vero E6 cells in our lab to generate viral stocks. SARS-CoV-2 Delta variants (Pango lineage B.1.617.2) hCoV-19/MD/05647/2021 (NR-55672), used for the FRNT50 assays, were obtained from BEI resources, NIAID, NIH. Delta variants were passaged once more in H1299-hACE2 cells in our lab to generate viral stocks. Omicron (Pango lineage B.1.1.529) hCoV-19/USA/HI-CDC-4359259-001/2021 was obtained from BEI resources (NR-56475) and used directly in experiments. Passaged viruses were deep sequenced to confirm identity.
Biolayer interferometry (BLI)
Binding kinetics were measured by BLI using an OctetRed96e. All experiments were performed at 30 °C shaking at a speed of 1000 rpm, using 10X kinetics buffer (Sartorius, CAT NO. 18-1105) diluted to 1X with PBS. Briefly, Anti-human Fab-CH1 2nd-generation (FAB2G) biosensors (Sartorius, CAT NO. 18-5126) were pre-hydrated in 1X Kinetics buffer for 10 minutes prior to initiation of experiments. CV503 and CoV-X2 BsAbs, diluted to 2 and 1.25 ug/mL, respectively, were immobilised on biosensors for 300 s. Biosensors containing immobilised BsAbs were immersed in wells containing buffer only for 180 s to establish a baseline, followed by association with 1:2 serially diluted SARS-CoV-2 RBD or trimer protein for 300 s. Analyte-free wells were used as reference to subtract background signals during data analysis. Biosensors containing the BsAb-viral protein complex were then dipped in wells containing buffer only to allow for dissociation for 600 s. The data were analysed using the Data Analysis Software HT 12.0 (ForteBio) using a global fitting model of 1:2 with chi-squared value less than 3 and R2 value greater than 0.95. Experiments were performed in duplicate.
Cell-based virus neutralisation assays
Antibody samples were diluted 2-fold in 5% FBS DMEM for a Focus-Reduction-Neutralisation-50% (FRNT50) assay that was stained as described previously [18] except a secondary Alexa-488 conjugated anti-Rabbit monoclonal antibody (Invitrogen) was used for fluorescent staining. Cell nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI), imaged using a Cytation7 (Agilent) and foci counted using Gen5 software. Neutralising titers were determined as described previously [19, 20]. Each neutralisation assay was run with n = 4 biological replicates at each antibody dilution with n = 12 antibody-free controls used to normalise percent neutralisation of each sample.
RESULTS
Antibody-RBD binding kinetics
We evaluated the BLI for measuring the binding kinetics between anti-SARS-CoV-2 antibodies and RBDs or full-length spike trimer proteins. To this end, we obtained a panel of five BsAbs against SARS-CoV-2, namely CV503_664_EL, CV503_521_GS, CV503_993_EL, CoV-X2-WT and CoV-X2-LALA. These samples represent a diverse array of BsAb formats, thus allowing a comprehensive evaluation of assay suitability. The CV503 subclass contains three BsAbs. CV503_664_EL and CV503_664_EL can be classified as RBD-recognising BsAbs in which by both arms are against epitopes within the RBD region, including CV503 that binds to the ACE2- RBS, as well as CV664 and CV993 that bind within the RBD but away from the RBS. The linkers, termed “EL”, are based on the elbow region between the variable region and the constant region. The CV503_521_GS BsAb can be classified as one that recognises the RBD and the N-terminal domain (NTD) due to the CV521 arm. The linker for this BsAb, termed “GS”, connects both heavy chains and both light chains. All CV503 BsAbs were generated using a dual variable domain-Ig format [7]. CoV-X2-WT and CoV-X2-LALA represent two BsAbs generated using a cross-mAb platform from well-studied parental mAbs, C121 and C135 [6]. C135 is known to target the RBD outside of the ACE2-RBS while the C121 RBD epitope overlaps with the ACE2-RBS [21]. The differing feature between the two CoV-X2 BsAbs is the presence of LALA-PG mutations to modulate potential effector functions, which was not expected to influence binding affinity with the virus proteins.
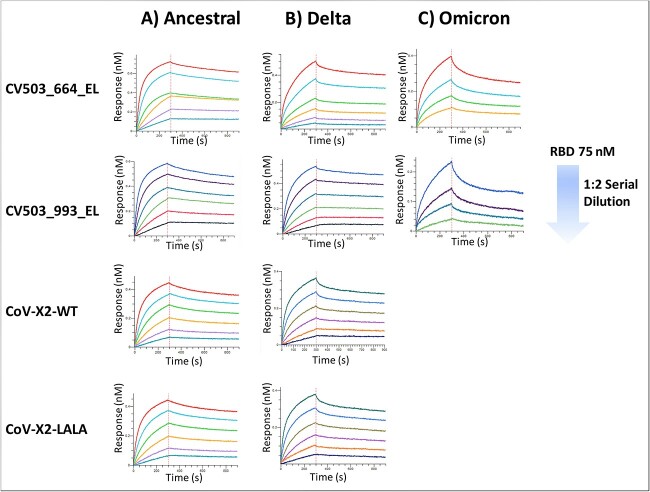
The BsAb samples were first tested in parallel with individual RBDs corresponding to Ancestral, Delta (B.167.2) and Omicron (B.1.1.529) variants. Briefly, an antibody sample was immobilised onto the biosensor tips, which were then immersed into 96-plate wells containing serial dilutions of RBD variants in kinetics buffer for 300 s to allow for association, followed by 600 s in kinetics buffer alone to allow for dissociation (Figure 1A–C). The recorded sensorgrams were fitted to a 1:2 bivalent analyte model (Supplementary Figures 1–2). The apparent binding affinity (KD), association (Kon) and dissociation (Koff) constants are summarised in Table 2. Each BsAb bound at a low nanomolar affinity to the Ancestral RBD, with KD values ranging from 2.2 to 4.4 nM. Binding affinity for most BsAbs when assessed with the Delta variant was not significantly impacted. Here, KD values were determined ranged from 1.4 nM for CV503_993_EL to 3.8 nM for CoV-X2-WT. Binding affinity was significantly diminished when analysed against the Omicron variant for most of the BsAbs. Affinity of CV503_664_EL and CV503_993_EL BsAbs reduced to 11 and 64 nM, respectively. With both CV503 BsAbs, the recorded Koff rates were significantly higher, contributing to the increases in KD values. When evaluated, the CoV-X2 antibodies displayed the most significant impact to binding as no signal was observed when analysed with the Omicron variant.
Figure 1.
Binding kinetics of bispecific antibodies against Spike RBDs. (A–C) Kinetics experiments were performed using BLI with bispecific antibodies against SARS-CoV-2 RBDs. Representative sensorgrams of binding of BsAbs with (A) Ancestral, (B) Delta and (C) Omicron variants are shown. Due to the lack of binding signals, a sensorgram of CoV-X2 antibodies against Omicron is not displayed. Each experiment was run in duplicate.
Table 2.
Affinity measurements for bispecific antibodies against SARS-CoV-2
| Protein Analyte | Bispecific Antibody | Apparent K on (1/Ms) | Apparent K off (1/s) | Apparent KD (nM) |
|---|---|---|---|---|
| Ancestral RBD | CV503_664_EL | 1.6 × 105 ± 3.6 × 104 | 3.6 × 10−4 ± 1.2 × 10−4 | 2.2 ± 3.4 × 10−1 |
| CV503_993_EL | 2.2 × 105 ± 5.5 × 104 | 4.0 × 10−4 ± 1.3 × 10−4 | 2.2 ± 4.6 × 10−1 | |
| CoV-X2-WT | 8.9 × 104 ± 1.0 × 105 | 5.0 × 10−3 ± 6.4 × 10−3 | 4.4 ± 1.9 | |
| CoV-X2-LALA | 1.3 × 105 ± 2.1 × 104 | 3.5 × 10−4 ± 8.5 × 10−5 | 2.7 ± 2.1 × 10−1 | |
| Delta RBD | CV503_664_EL | 3.3 × 105 ± 1.4 × 105 | 4.4 × 10−4 ± 1.7 × 10−5 | 3.0 ± 1.7 × 10−1 |
| CV503_993_EL | 1.4 × 105 ± 1.4 × 103 | 2.0 × 10−4 ± 2.0 × 10−5 | 1.4 ± 1.8 × 10−1 | |
| CoV-X2-WT | 1.4 × 105 ± 4.2 × 104 | 5.3 × 10−4 ± 1.6 × 10−4 | 3.8 | |
| CoV-X2-LALA | 1.8 × 105 ± 6.4 × 104 | 5.3 × 10−4 ± 7.1 × 10−6 | 3.2 ± 1.1 | |
| Omicron RBD | CV503_664_EL | 1.5 × 105 ± 6.5 × 103 | 1.5 × 10−3 | 11 |
| CV503_993_EL | 7.1 × 104 ± 2.5 × 104 | 4.5 × 10−3 ± 1.2 × 10−3 | 64 ± 9.1 | |
| CoV-X2-WT | N/A * | N/A * | N/A * | |
| CoV-X2-LALA | N/A * | N/A * | N/A * | |
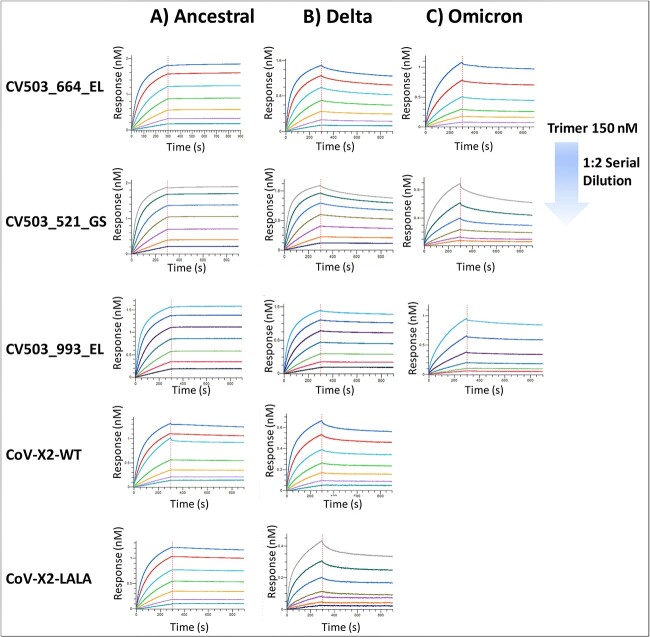
| Ancestral Trimer | CV503_664_EL | 4.8 × 104 ± 1.6 × 103 | <1 × 10−7 | <1 × 1012 |
| CV503_521_GS | 5.3 × 104 ± 3.7 × 103 | <1 × 10−7 | <1 × 1012 | |
| CV503_993_EL | 7.0 × 104 ± 6.4 × 103 | <1 × 10−7 | <1 × 1012 | |
| CoV-X2-WT | 5.4 × 104 ± 7.1 × 102 | 1.25 × 10−4 ± 3.5 × 10−5 | 2.4 ± 7.1 × 10−1 | |
| CoV-X2-LALA | 5.2 × 104 ± 4.2 × 103 | 1.2 × 10−4 ± 5.3 × 10−5 | 2.4 ± 9.2 × 10−1 | |
| Delta Trimer | CV503_664_EL | 9.2 × 104 ± 7.1 × 102 | 3.7 × 10−4 ± 1.4 × 10−5 | 4.2 ± 7.1 × 10−2 |
| CV503_521_GS | 8.8 × 104 ± 7.1 × 101 | 6.9 × 10−4 ± 6.0 × 10−5 | 8.0 ± 1.1 | |
| CV503_993_EL | 8.6 × 104 ± 6.4 × 103 | 1.3 × 10−4 ± 2.8 × 10−5 | 1.6 ± 2.1 × 10−1 | |
| CoV-X2-WT | 9.4 × 104 ± 1.4 × 103 | 3.3 × 10−4 ± 2.8 × 10−5 | 3.5 ± 2.8 × 10−1 | |
| CoV-X2-LALA | 9.1 × 104 ± 4.2 × 103 | 4.6 × 10−4 ± 5.0 × 10−5 | 5.1 ± 7.8 × 10−1 | |
| Omicron Trimer | CV503_664_EL | 5.1 × 104 ± 3.5 × 103 | 2.6 × 10−4 ± 5.7 × 10−5 | 5.2 ± 7.8 × 10−1 |
| CV503_521_GS | 6.5 × 104 ± 3.5 × 103 | 9.3 × 10−4 ± 2.4 × 10−4 | 14.5 ± 3.2 | |
| CV503_993_EL | 3.7 × 104 ± 7.1 × 102 | 2.2 × 10−4 ± 3.5 × 10−5 | 6.0 ± 7.8 × 10−1 | |
| CoV-X2-WT | N/A * | N/A * | N/A * | |
| CoV-X2-LALA | N/A * | N/A * | N/A * |
Apparent affinity constants recorded for each bispecific antibody against SARS-CoV-2 spike RBD or trimer protein, including association (Kon), dissociation (Koff) and equilibrium (KD) determined from duplicate experiments of each antibody. Affinity constants are mean ± standard deviation (SD).
*N/A indicates uncollected measurements due to a lack of binding to spike RBD or trimer.
Antibody-spike protein interactions
We also analysed the kinetics of each BsAb with the full-length trimer protein of each variant containing both the S1 and S2 subunits along with the extracellular domain (Figure 2A–C). Interestingly, we found each CV503 BsAb bound to Ancestral trimer at apparent subnanomolar affinity with minimal noticeable dissociation from the analyte. Dissociation from the spike trimer was more observable with the CoV-X2-WT and LALA BsAbs, resulting in apparent KD values of 2.4 nM for both. Among all BsAbs, binding affinity was observed to be in the low nanomolar range when evaluated with the Delta variant. As expected, the most drastic effect to binding affinity was observed with the Omicron variant. CoV-X2 antibodies displayed no binding signal, mirroring the results with the RBD of Omicron. However, affinity of all CV503 antibodies was observed at low nanomolar with the largest apparent KD value being 14.5 nM from the CV503_521_GS BsAb (Figure 2, Table 2).
Figure 2.
Binding kinetics of bispecific antibodies against Spike trimers. (A–C) Kinetics experiments were performed using BLI with bispecific antibodies against SARS-CoV-2 trimer proteins. Representative sensorgrams of binding of BsAbs with (A) Ancestral, (B) Delta and (C) Omicron variants are shown. Due to the lack of binding signals, a sensorgram of CoV-X2 antibodies against Omicron is not displayed. Experiments were run in duplicate.
Focus reduction neutralisation test (FRNT)
We developed procedures for an FRNT that was proven to be suitable for measuring antibody neutralising activity against a wide range of SARS-CoV-2 variants (Figure 3). The assay utilises H1299-hACE2 cells for infection (a human epithelial-like cell line derived from lung carcinoma, engineered to express human ACE2) in a 96-well based fluorescence assay format (see details in Methods and Materials). The FRNT50 results demonstrated differential neutralising activity of the BsAbs samples against Ancestral SARS-CoV-2 (WA1/2020), Delta B.1.617.2 (hCoV-19/MD/05647/2021) and Omicron B.1.1.529 (hCoV-19/USA/HI-CDC-4359259-001/2021) (Figure 4).
Figure 3.
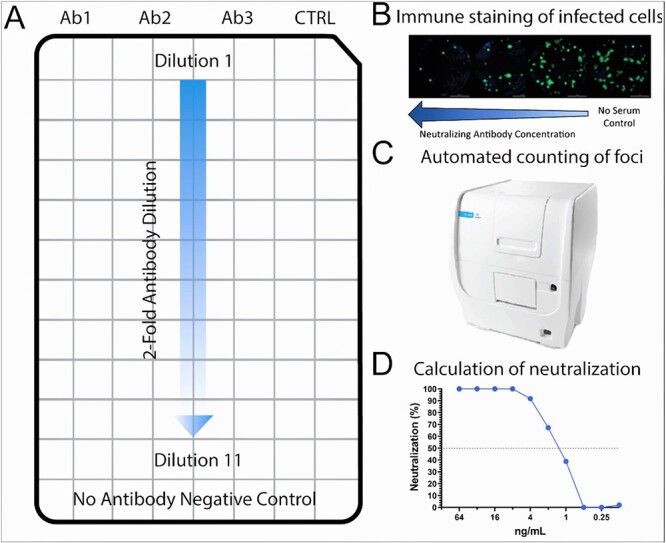
Schematic of fluorescent focus-reduction-neutralisation-test 50% using H1299-hACE2 cells. (A) Layout of a 96-well plate with H1299-hACE2 cells infected with known concentrations of SARS-CoV-2 isolates. Wells are treated with antibodies that are diluted 2-fold along with control antibodies and no-antibody negative controls. (B) Visualisation of infected cells by using fluorescent antibodies following overnight incubation under treatment conditions. (C) The Cytation7 machine can be used to count foci. (D) Resulting foci counts can be used to generate 50% end-point neutralisation titers.
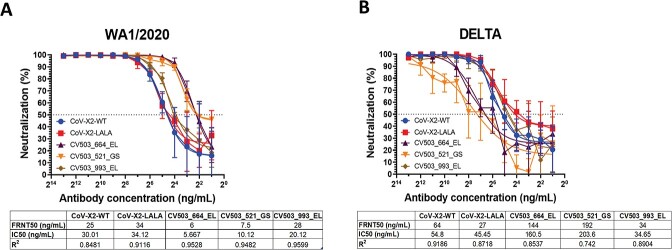
Figure 4.
Neutralisation of infectious Ancestral and Delta variant SARS-CoV-2 isolates using reduction of fluorescent foci in H1299-hACE2 cells. A panel of bispecific antibodies was tested by FRNT50 against Ancestral (A) and Delta (B) SARS-CoV-2 with n = 4 biological replicates per antibody. FRNT50, IC50 (4PL nonlinear regression) and R2 values are listed below the relevant panel.
CoV-X2-WT (25 ng/mL) and CoV-X2-LALA (34 ng/mL) had similar neutralising titers against Ancestral SARS-CoV-2 but did display some differences against delta, interestingly, neutralising at 64 and 27 ng/mL, respectively. CV503_993_EL had FRNT50s to Ancestral SARS-CoV-2 (WA1/2020) and Delta of 28 and 34 ng/mL, respectively. FRNT50 values for CV503_664_EL (144 ng/mL) and CV503_521_GS (192 ng/mL) against Delta were 22 times higher than anti-WA1/2020 titers. However, none of the tested BsAbs displayed neutralising activity against Omicron B.1.1.529.
DISCUSSION
Anti-SARS-CoV-2 antibodies are complex biomolecules which require the development and implementation of adequate bioassays to ensure product quality and manufacturing consistency. Such antibodies act primarily by blocking virus docking onto the ACE2 receptor of human cells; therefore, bioassay development is focused on assessing their binding and neutralisation activity against specific SARS-CoV-2 variants. Here, we developed procedures for the BLI to measure the binding kinetics between anti-SARS-CoV-2 antibodies and RBDs or the full-length spike trimers. We also developed an FRNT utilising live virus isolates to infect an engineered human lung cell line expressing human ACE2. BLI is high-throughput and quantitative, yielding results that correlate with those obtained from the FRNT virus neutralisation assays and allow the simultaneous evaluation of multiple spike protein variants. These assays complement the current toolbox (Table 1) and can be adopted for quality assessment in the development of antibody products against SARS-CoV-2 variants.
BLI is a label-free biosensing technology that is widely used for assessing biomolecular interactions. In a “dip-and-read” format, BLI offers a rapid and real-time monitoring of the binding kinetics to generate quantitative results regarding on- and off-rates, and equilibrium constant (KD values). Using BLI, we analysed a panel of five BsAbs in parallel against Ancestral, Delta and Omicron SARS-CoV-2 viral proteins. In general, trends in apparent binding affinity between antibodies with either RBD or trimer proteins appeared to correlate. We observed low nanomolar binding of most of the BsAbs to Ancestral and Delta viral proteins (Figures 1–2, Table 2), potentially indicating insignificant effect on antibody binding by the mutations acquired with the Delta variant, including those on the S protein reported to generate resistance to neutralising antibodies, such as L452R, T487Kand P618R [22]. Consistently, CV503_521_GS showed more noticeable reduction in affinity particularly against Delta and Omicron variants compared with the other CV503 antibodies (Figures 2, Table 2). The inclusion of CV503_521_GS on the panel of BsAbs indicates the importance of selecting the proper analyte (RBD or full-length spike trimer) during bioassay development for estimating binding affinity of potential therapeutics while taking into consideration the intended target of the antibody, in this case, both RBD and NTD. Of note, antibodies targeting the NTD can be more susceptible to variant mutations compared with the RBD. Compared with earlier variants, Delta and Omicron both contain several mutations in the NTD, which may aid in escape from neutralising antibodies against the NTD [23].
CoV-X2 BsAbs did not display significant differences in affinity between Ancestral and Delta proteins, which may be due to the lack of the E484K in B.1.617.2 specifically. This point mutation has been reported to promote escape from C121 as well as the retention of C135 binding epitopes, R346 and N440 [21]. However, the Omicron proteins tested here contain mutations at both E484, N440, which may provide some explanation as to the significant impact on binding of CoV-X2 BsAbs and Omicron (Figures 1–2). Supporting the potential use of BLI as an orthogonal method to SPR, which is commonly used by sponsors to assess binding affinity, we found the BLI results of CoV-X2 BsAbs to be similar to published SPR data. This includes an apparent KD value of 2.7 nM between CoV-X2-WT and Ancestral RBD, whereas the apparent KD was reported as 2.35 nM using SPR [6]; however, a direct comparison is limited due to variations in assay setup and viral proteins. The lack of testing with the RBDs and trimer proteins of Delta and Omicron variants in the previous work further limits our ability to draw comparison with the binding parameters we observed. Overall, the simplicity in the assay design, capability to perform parallel analysis and the mostly automated system makes the use of BLI a valuable technique in rapid analysis of anti-SARS-CoV-2 therapeutics.
We developed an FRNT50 assay that can assess a wide range of SARS-CoV-2 variants, including Omicron variants. The FRNT assay allows for relatively short visualisation time of foci formed from virus infection of all tested variants. FRNT50 assays can also be adapted to use with 96-well plates, incubated overnight and read automatically using imaging machinery (Cytation7) and software, resulting in a rapid, high-throughput assay that can complement other cell-based neutralisation assays [24]. The results of our FRNT showed higher potency of BsAbs against Ancestral that decreased as the variant evolved, resulting in no neutralisation activity observed with Omicron.
This study demonstrated the suitability of BLI together with FRNT assays to evaluate the binding and neutralisation of anti-SARS-CoV-2 BsAbs. The results of each assay mostly complemented each other showing similar trends between binding and neutralisation activity, demonstrating the ability of BLI to detect sub- and low nanomolar binding which was echoed by low concentrations of antibodies required to neutralise the virus in the FRNT studies. As an exception, some BsAbs failed to neutralise Omicron variant despite their nanomolar binding affinity to the viral spike protein (Figures 2 and 4). The causes of this disagreement are not clear but may involve several factors: (1) mutations present in the Omicron variant that affect neutralisation activity primarily, perhaps by enhancing its internalisation capabilities; (2) recognition of additional receptors present on the cell surface that may aid in viral entry of the Omicron variant; (3) competition between ACE2 and BsAbs for binding to Omicron, given the known higher affinity of Omicron for the receptor compared with earlier variants [25]. This would only be evident when ACE2 is present in an assay system (e.g., ACE2-expressing cell lines), rather than evaluating protein–protein interactions between Omicron and BsAb alone. In this context, the BLI assay design can be expanded to investigate possible explanations for viral escape, including assessing binding affinity between Omicron and other potential co-receptors that have been suggested in recent publications [26, 27], as well as performing competition studies to determine the ability of the BsAbs to inhibit Omicron–ACE2 interactions. Both assays could aid in understanding the mechanism(s) in which Omicron is able to escape neutralisation by the BsAbs, even when RBD or spike protein binding seems to be unhindered.
As shown for several monoclonal antibodies granted EUA, antigenic drift of emerging SARS-CoV-2 variants of concern can render the monoclonal antibody ineffective as mutations in the spike protein confer resistance on successive pandemic waves. Bispecific antibodies allow for multivalent targeting of different SARS-CoV-2 isolates and an expanded effective range in neutralising activity to reduce infectivity. However, new genetic mutations on the S protein continue to pose a challenge, giving rise to variants with enhanced ability to evade neutralising antibodies, as we show here with the Omicron variant. The circulation of Omicron and its subvariants, along with the possibility of the emergence of new genetic variants with increased resistance and infectivity, heightens the need for technologies that can rapidly assess binding and neutralisation of antibody therapeutics against SARS-CoV-2. In this context, coupling BLI and FRNT assays may help generate data to guide the design of next generation antibody formulations to address the ongoing COVID-19 pandemic.
Supplementary Material
ACKNOWLEDGEMENTS
Production of the CV503 BsAbs (Tan’s laboratory) was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. Production of the CoV-X2 BsAbs (Varani’s laboratory) was supported by the Institute for Research in Biomedicine, University of Switzerland. H1299-hACE2 cells were a kind gift of Dr Shufeng Liu [OVRR, CBER, FDA]. The authors would like to thank Drs Jennifer Swisher [Office of Biotechnology Products (OBP), CDER, FDA], Shen Luo [OBP, CDER, FDA] and Serge Beaucage [OBP, CDER, FDA] for their critical review on the manuscript.
We would also like to acknowledge the reagents obtained through Biodefense and Emerging Infections Research Resources: The following reagent was deposited by the Centers for Diseases Control and Prevention and obtained through BEI Research Resources Repository, NIAID, NIH: SARS-Related Coronavirus 2, Isolate USA-WA1/2020, NR-52281. The following reagent was obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate hCoV-19/USA/MD-HP05647/2021 (Lineage B.1.617.2; Delta variant), NR-55672, contributed by Dr. Andrew S. Pekosz. The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate hCoV-19/USA/HI-CDC-4359259-001/2021 (Lineage B.1.1.529; Omicron Variant), NR-56475, contributed by Centers for Disease Control.
Disclaimer: This article reflects the views of the authors and should not be construed to represent FDA’s views or policies.
Contributor Information
Alexis Q Dean, Office of Biotechnology Products, Center for Drug Evaluation and Research, Food and Drug Administration, Silver Spring, MD 20993, USA.
Charles B Stauft, Laboratory of Vector-Borne Viral Diseases, Division of Viral Products, Office of Vaccine Research and Review, Food and Drug Administration, White Oak, MD 20993, USA.
Julianne D Twomey, Office of Biotechnology Products, Center for Drug Evaluation and Research, Food and Drug Administration, Silver Spring, MD 20993, USA.
Joshua Tan, Antibody Biology Unit, Laboratory of Immunogenetics, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Rockville, MD 20852, USA.
Luca Varani, Institute for Research in Biomedicine, University of Switzerland, CH-1015 Lausanne, Switzerland.
Tony T Wang, Laboratory of Vector-Borne Viral Diseases, Division of Viral Products, Office of Vaccine Research and Review, Food and Drug Administration, White Oak, MD 20993, USA.
Baolin Zhang, Office of Biotechnology Products, Center for Drug Evaluation and Research, Food and Drug Administration, Silver Spring, MD 20993, USA.
CONFLICT OF INTEREST STATEMENT
None declared.
FUNDING
This work was funded by the US Food and Drug Administration. The funder had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
ETHICS AND CONSENT
Not applicable.
DATA AND MATERIALS AVAILABILITY
The data that support the findings of this study are available within the article and corresponding supplementary information. Additional data available upon request to corresponding author.
ANIMAL RESEARCH
Not applicable.
References
- 1. Twomey, JD, Luo, S, Dean, AQ et al. COVID-19 update: the race to therapeutic development. Drug Resist Updat 2020; 53: 100733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dean, AQ, Bozza, WP, Twomey, JD et al. The fight against COVID-19: striking a balance in the renin-angiotensin system. Drug Discov Today 2021; 26: 2214–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. US Food and Drug Administration . Emergency Use Authorization. 2020; 2020. https://www.fda.gov/emergency-preparedness-and-response/mcm-legal-regulatory-and-policy-framework/emergency-use-authorization. [Google Scholar]
- 4. Centers for Disease Control and Prevention (CDC), National Center for Immunization and Respiratory Diseases (NCRID), Division for Viral Diseases . SARS-CoV-2 Variant Classifications and Definitions. 2020; 2020. https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html. [Google Scholar]
- 5. Chen, P, Nirula, A, Heller, B et al. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with COVID-19. N Engl J Med 2021; 384: 229–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. De Gasparo, R, Pedotti, M, Simonelli, L et al. Bispecific IgG neutralizes SARS-CoV-2 variants and prevents escape in mice. Nature 2021; 593: 424–8. [DOI] [PubMed] [Google Scholar]
- 7. Cho, H, Gonzales-Wartz, KK, Huang, D et al. Bispecific antibodies targeting distinct regions of the spike protein potently neutralize SARS-CoV-2 variants of concern. Sci Transl Med 2021; 13: eabj5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li, Z, Li, S, Zhang, G et al. An engineered bispecific human monoclonal antibody against SARS-CoV-2. Nat Immunol 2022; 23: 423–30. [DOI] [PubMed] [Google Scholar]
- 9. Tom, YZ, Carolina, L, Valter, MS et al. A synthetic bispecific antibody capable of neutralizing SARS-CoV-2 Delta and Omicron bioRxiv. 2022; Preprint.
- 10. Lee, WS, Wheatley, AK, Kent, SJ et al. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat Microbiol 2020; 5: 1185–91. [DOI] [PubMed] [Google Scholar]
- 11. Winkler, ES, Gilchuk, P, Yu, J et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell 2021; 184: 1804–1820.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Iwasaki, A, Yang, Y. The potential danger of suboptimal antibody responses in COVID-19. Nat Rev Immunol 2020; 20: 339–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. US Food and Drug Administration, Center for Drug Evaluation and Research and Center for Biologics Evaluation and Research . Q6B Specifications: Test Procedures and Acceptance Criteria for Biotechnological/Biological Products. 1999; 1999. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/q6b-specifications-test-procedures-and-acceptance-criteria-biotechnologicalbiological-products. [Google Scholar]
- 14. US Food and Drug Administration, Center for Drug Evaluation and Research (CDER) . COVID-19: Potency Assay Considerations for Monoclonal Antibodies and Other Therapeutic Proteins Targeting SARS-CoV-2 Infectivity, Guidance for Industry. 2021; 2021. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/covid-19-potency-assay-considerations-monoclonal-antibodies-and-other-therapeutic-proteins-targeting. [Google Scholar]
- 15. Wang, Y, Yuan, M, Lv, H et al. A large-scale systematic survey reveals recurring molecular features of public antibody responses to SARS-CoV-2. Immunity 2022; 55: 1105–17.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fu, D, Zhang, G, Wang, Y et al. Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes. PLoS Biol 2021; 19: e3001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vanderheiden, A, Edara, VV, Floyd, K et al. Development of a rapid focus reduction neutralization test assay for measuring SARS-CoV-2 neutralizing antibodies. Curr Protoc Immunol 2020; 131: e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cele, S, Jackson, L, Khoury, DS et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature 2022; 602: 654–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reed, LJ, Muench, H. A simple method of estimating fifty per cent ENDPOINTS12. Am J Epidemiol 1938; 27: 493–7. [Google Scholar]
- 20. Stauft, CB, Selvaraj, P, Lien, CZ et al. Long-term immunity in convalescent Syrian hamsters provides protection against new-variant SARS-CoV-2 infection of the lower but not upper respiratory tract. J Med Virol 2022; 94: 2833–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Greaney, AJ, Starr, TN, Barnes, CO et al. Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat Commun 2021; 12: 4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McCallum, M, Walls, AC, Sprouse, KR et al. Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants. Science 2021; 374: 1621–6. [DOI] [PubMed] [Google Scholar]
- 23. Mittal, A, Khattri, A, Verma, V. Structural and antigenic variations in the spike protein of emerging SARS-CoV-2 variants. PLoS Pathog 2022; 18: e1010260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stauft, CB, Sangare, K, Wang, TT. Differences in new variant of concern replication at physiological temperatures in vitro. J Infect Dis 2022; jiac264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lupala, CS, Ye, Y, Chen, H et al. Mutations on RBD of SARS-CoV-2 Omicron variant result in stronger binding to human ACE2 receptor. Biochem Biophys Res Commun 2022; 590: 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang, K, Chen, W, Zhang, Z et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct Target Ther 2020; 5: 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Baindara, P, Roy, D, Mandal, SM et al. Conservation and enhanced binding of SARS-CoV-2 omicron spike protein to Coreceptor Neuropilin-1 predicted by docking analysis. Infect Dis Rep 2022; 14: 243–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.