Abstract
For the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, clinical manifestations are broad and highly heterogeneous for both sexes. We aimed to determine how biological sex and age impact immune gene expression, particularly influencing the humoral neutralizing antibody (NAb) response and the cytokine production in coronavirus disease 2019 (COVID-19) subjects. The immune gene expression, according to biological sex and age, was assessed using the genome wide expression profile of blood proteins from healthy individuals using the Genotype Tissue Expression (GTEx) database. Moreover, anti-SARS-CoV-2 neutralizing antibody titers and cytokine levels were determined in blood samples from 141 COVID-19 individuals from Medellín, Colombia. Among subjects with COVID-19, males had statistically significantly higher median NAb titers and serum concentrations of interleukin-6 and CC chemokine ligand 3 than females. Overall, our findings point out a more robust innate immune response in women that could help recognize and restrain the virus faster than in men.
Keywords: Gene, SARS-CoV-2, Immunology, Male, Virus, Sex
Highlights
-
•
SARS-CoV-2 infection are influenced by sex differences in the immune gene.
-
•
There were more male-biased than female-biased genes.
-
•
The number of differentially expressed genes was higher in the youngest cohort studied.
1. Introduction
Viral infectious diseases affect humans worldwide and typically behave differently in each sex, influenced by the disease-causing virus and the host's immune response. In most infections, the exposure to a specific virus is usually considered the same between males and females; however, the vulnerability to the infection and its clinical course can vary in line with endogenous and exogenous factors such as the biological sex, gender, and its associated behavioral practices, immune-endocrine, and host-microbiome interactions [1]. Interestingly, females exhibit intensified innate and adaptive inflammatory responses that contribute to reducing the prevalence of many viral infections as compared with the infection rates among males, including dengue fever, hepatitis B, hepatitis C, human immunodeficiency virus (HIV), and West Nile fever [2, 3, 4, 5, 6].
For the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), infection rates are similar in men and women [7]. However, older males are more vulnerable to worse clinical outcomes, as highlighted in the sex-disaggregated data from several countries [8, 9, 10], and the report of the significantly high male-to-female ratio of intensive care unit (ICU) admission and in-hospital mortality [7]. Even though there is a male predominance in the severity of the disease, the clinical spectrum of COVID-19 is broad for both sexes and ranges from asymptomatic infection to more severe and life-threatening presentations [11].
These clinically diverse features are mainly attributed to the expression and distribution of the SARS-CoV-2 entry receptor, the angiotensin-converting enzyme 2 (ACE2) [12], and the immune response upon the infection. Of particular interest, disparities in the immune phenotypes of infected subjects may trigger the heterogeneous clinical traits and distinct disease courses displayed in both sexes. While females with COVID-19 show a more robust T cell response, males with the progressive disease show a lower proportion of activated T cells [13]. Moreover, the antibody response to viral proteins and neutralizing antibody titers to SARS-CoV-2 also follow a sex-specific pattern. Females show higher antibody titers against the protein S2 fragment of the virus [14], whereas neutralizing antibodies tend to decline faster in men [15].
Such sexual dimorphism in baseline cellular and humoral mechanisms of the innate and adaptive immune responses has been broadly documented under the primary hypothesis that sex chromosome-linked genes along with sex hormonal differences and fluctuations drive the heightened immune status in women [16, 17]. Notably, due to the X chromosome inactivation escape, many immunological factors encoded on the X chromosome are expressed in higher levels in women, granting them more advantage to face immune challenges [18, 19]. Regarding sex hormones, estrogen has an immune enhancing effect as it boosts the expression of several viral receptors and the production of interferon (IFN) [20, 21], whereas androgens generally promote an anti-inflammatory environment by diminishing the production of proinflammatory cytokines [22].
Notwithstanding the aforementioned evidence, the underlying molecular mechanisms that might impact the immune response to COVID-19 remain a subject of investigation. In this study, we evaluated genetic differences in immune regulation using the whole-genome expression profile of blood proteins from healthy individuals, employing the Genotype-Tissue Expression (GTEx) database. In addition, the neutralizing antibody response and cytokine production in individuals with COVID-19 from Medellín, Colombia were analyzed to determine how sex- and age-stratified effects of genetic variations could bias the inflammation phenomena during SARS-CoV-2 infection.
2. Materials and methods
2.1. Differential expression analysis of RNA-Seq data
We downloaded the GTEx RNA-seq dataset of blood samples from males and females by age groups as follows: 20–29, 30–39, 40–49, 50–59, and 60–69 years. The 70–79 age group was not included because there were not enough female specimens to run the comparison analysis. A total of 402 samples (261 males and 141 females) were included, and none of the samples were affected by a disease, under any medication treatment, or altered by a gene perturbation.
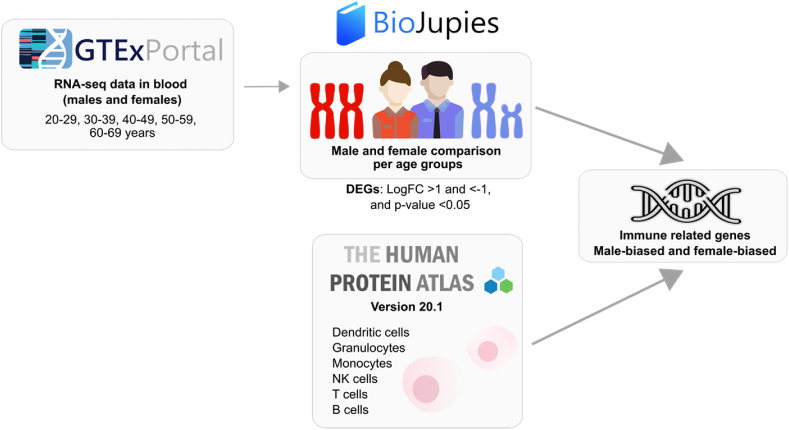
We then used the BioJupies platform (https://amp.pharm.mssm.edu/biojupies/) [23] to compare male and female RNA-seq data in the same age ranges and identify differentially expressed genes (DEGs) in whole blood. Genes with an absolute fold change ≥2 and p-value <0.05 were considered DEGs (Fig. 1).
Fig. 1.
Schematic representation of the in silico study.
2.2. Immune-related genes analysis
We filtered the DEGs coding for immune-related genes based on the lists available at The Human Protein Atlas (Supplementary Table 1). From the Immune Cells summary section (www.proteinatlas.org/humanproteome/immune+cell) [24], we selected protein-coding genes with the highest expression in blood or lymphoid tissues for each immune cell lineage, which included B, T and NK cells, monocytes, granulocytes, and dendritic cells, and sought the resultant genes in the DEGs list to obtain the genes with immune cell specificity across all age groups. The immune-related DEGs were classified in male-biased if upregulated in men (LogFC < -1, p < 0.05) and female-biased if upregulated in women (LogFC >1, p < 0.05) (Fig. 1).
The gene ontology enrichment analysis was generated by analyzing the female- and male-biased immune related genes using the Gene Ontology (GO) project (http://geneontology.org/), which also allowed to identify biological processes, molecular functions, and cellular components over-represented in the whole gene set. After applying Bonferroni's correction, significant designations were determined by using a cut-off of p-value <0.05 [25] (Fig. 1).
2.3. Clinical testing of subjects with COVID-19
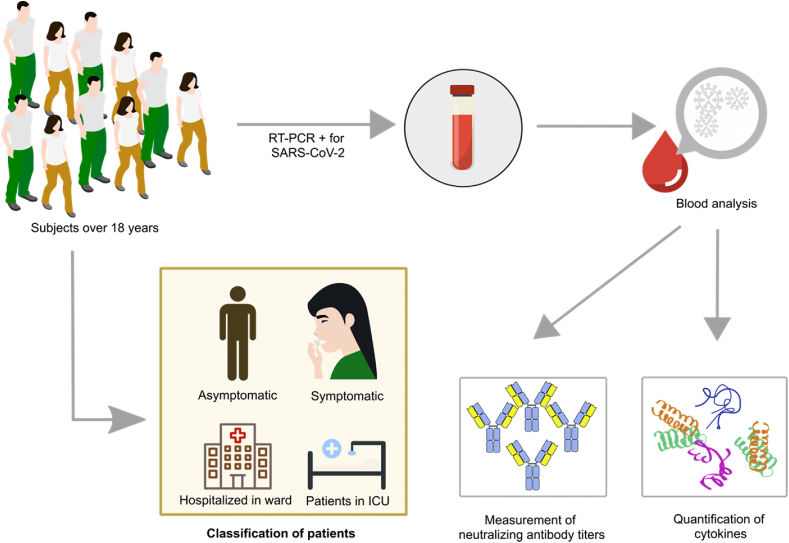
We studied a cohort of 141 individuals over 18 years from Medellín, Colombia, confirmed positive for SARS-CoV-2 infection by reverse transcription-polymerase chain reaction (RT-PCR) (Fig. 2). Individuals were recruited in Medellin, Colombia, at the IPS Universitaria, Hospital Universitario San Vicente Fundación and from the laboratories Grupo Inmunovirología and PECET from Universidad de Antioquia, authorized to carry out diagnostic tests for SARS-CoV-2.
Fig. 2.
Schematic representation of the in vitro study.
Pregnant women, patients with acute respiratory infection who did not meet the COVID-19 case criteria established by the Colombian Ministry of Health, subjects who did not accept participating, and those vaccinated against COVID-19 at the time of the enrollment were excluded from this study. The Bioethics Committee from the Faculty of Medicine of Universidad de Antioquia (F-017-0) approved the protocol, and all participants signed an informed consent form.
2.4. Plaque reduction neutralization test
A blood sample was obtained from each participant at the study enrollment. In asymptomatic participants, samples were taken with a maximum of 5 days after a positive RT-PCR SARS-CoV-2 test or after close contact with a COVID-19 positive case. In symptomatic participants, samples were collected around five days after the onset of symptoms, and preferably within 7 days since their onset. Finally, samples were taken within the first three days of hospital admission in hospitalized and ICU participants. Neutralizing antibody titers were determined by a plaque reduction neutralization test (PRNT) using a SARS-CoV-2 isolate (B.1 lineage), as previously described [26].
2.5. Quantification of cytokines
Quantifying interleukin (IL)-6 and CC chemokine ligand 3 (CCL3) were performed by BD Cytometric Bead Array (CBA), using Human Inflammatory Cytokines Kit, and Human Flex Set (Becton Dickinson, NJ, USA) in serum samples. The analysis was performed using FlowJo 10.8.1 (TreeStar, San Carlos, CA, USA).
2.6. Statistical analysis
We assessed sex differences in PRNT50 titers and cytokines levels using the Mann-Whitney test. Data are presented as median and interquartile ranges. Spearman's rank test analyzed correlations between PRNT50 titers and age. All the analyses were performed using GraphPad Prism version 9.0.1 (GraphPad Software Inc., San Diego, California, USA).
3. Results
3.1. Expression of immune-related genes in the whole blood of healthy subjects
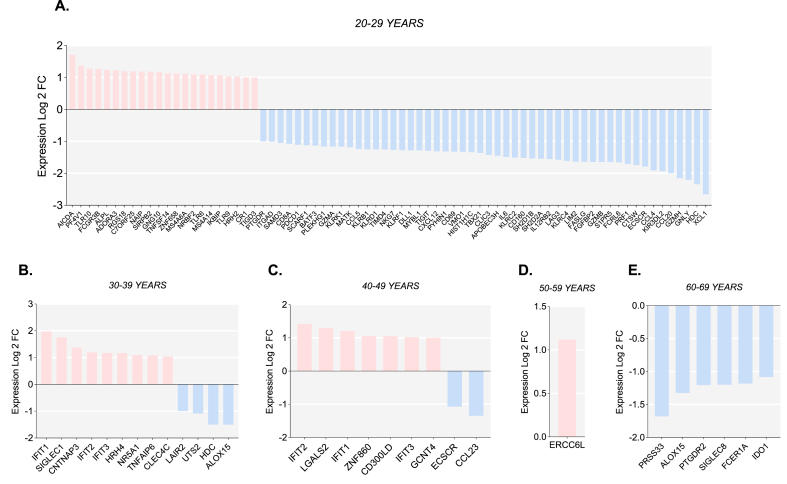
Across all the studied samples, we analyzed 103 DEGs, including common genes among the different age groups evaluated (Fig. 3 and Supplementary Table 2). Thirty-nine genes were female-biased, and a higher count was identified as male-biased (64 genes); both were mainly grouped within the 20-29-year category (Fig. 3A). In contrast to the youngest group, a higher proportion of female-biased genes as compared with male-biased genes was found in the 30–39 year and 40-49-year groups (Fig. 3B and C). No male-biased genes and female-biased genes were identified in the 50–59 year and the 60-69-year groups, respectively (Fig. 3D and E). Only ERCC6L, involved in the sister chromatid disjunction during mitosis, was significantly differentially expressed and female-biased in the 50-59-year group (Fig. 3D).
Fig. 3.
Sex differences in the expression of immune related genes in whole blood. Differentially expressed genes (Log2 fold change >1 or < -1 and p-value <0.05) in males and females aged 20–29 years (A), 30–39 years (B), 40–49 years (C), 50–59 years (D) and 60–69 years (E). Female-biased genes are shown as positive LogFC (in pink), and male biased genes are shown as negative LogFC (in blue). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Most upregulated genes in women were related in function to the innate immune system, followed by genes that participate in both innate and adaptive immunity and, in a smaller proportion, genes with a specific function in adaptive immunity. Among the resulting genes, microbe recognition (TLR6, TLR8, TLR9, TLR10, CLEC4C, IFIT1, and CD300LD), secretion of proinflammatory cytokines (TNFSF14 and TNFAIP6), and transcription factor (NR5A1) were the main associated functions. Only three genes were repeated in the whole women samples (30-39- and 40-49-year groups) and correspond to antiviral factors (IFIT1-3) (Fig. 3B and C).
Compared to the upregulated genes in women, most of the upregulated genes in men were associated predominantly with T and NK cell function. In the 20–29 years old group, we observed a pronounced expression of genes that participate in the regulation of the cytotoxic activity of NK and T cells (GNLY, GZMA, GZMB, GZMH, CTSW, PRF1, FCRL6, FASLG, KLRC2, FGFBP2, and CD8A), followed by genes that codify for proteins with chemotactic activity (CXCL1, CCL4, CCL5, CCL20, and CXCL12) and genes that regulate NK and T cells activation (LAG3, CD160, KLRC2, CD69, and BATF3) and inhibition (KIR3DL2, PCDC1, and KLRB1). Remarkably, we also noted in men aged 20–29 the overexpression of IL6 (Fig. 3A) that codifies for a pleiotropic cytokine with a wide spectrum of activities in the inflammatory response, but the upregulation was not observed in the rest of the age categories included. In older groups, male-biased genes were associated with enzymatic reactions of biological processes (HDC, ALOX15, and IDO1) and chemotaxis (CCL23, ECSCR, and PTGDR2) as well.
Moreover, a comparable amount of DEGs between men and women were involved in B lymphocyte activation. Noteworthy, the upregulation of BATF3 and CXCL12 was observed in males aged 20–29 years, whereas in women aged 20–29 years the upregulation of CR1, TLR9 and AICDA was found. No significant differences in the expression of genes implicated in B cell activation and immunoglobulin production were found in the remaining age categories.
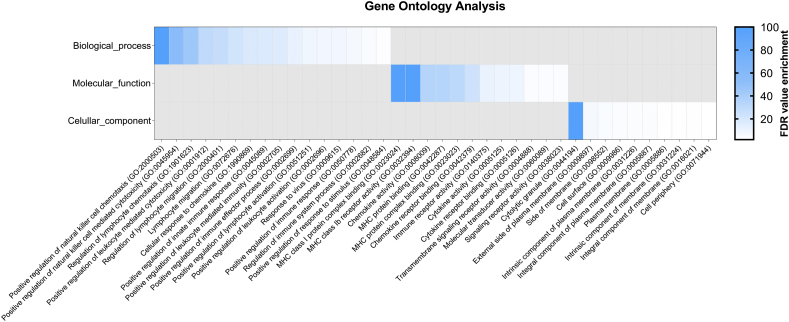
Using the Gene Ontology project (http://geneontology.org/), we further performed gene ontology (GO) enrichment analysis for the DEGs noted in all age groups. We identified several GO terms involved in immunological processes and positively enriched functions within the 20–29 years old subgroup data set. The most prominent biological process was the positive regulation of natural killer cell chemotaxis, followed by the positive regulation of natural killer cell mediated cytotoxicity and regulation of lymphocyte chemotaxis (Fig. 4). Other relevant biological processes included the positive regulation of leukocyte activation and its mediated cytotoxicity of the immune response to viruses, and another stimulus (Fig. 4). Furthermore, the most pronounced enriched GO molecular function involved the MHC complex (MHC class I protein complex binding, MHC class Ib receptor activity, MHC protein binding). The most significant enriched term for the cellular component ontology was cytolytic granule. No significantly enriched gene ontology term related to biological process, molecular function, or cellular component was observed in the rest of the age groups (data not shown).
Fig. 4.
Enrichment analysis of differentially expressed genes in subjects aged 20–29 years. FDR: False discovery rate.
3.2. Differences in the immune response between men and women with SARS-CoV-2 infection
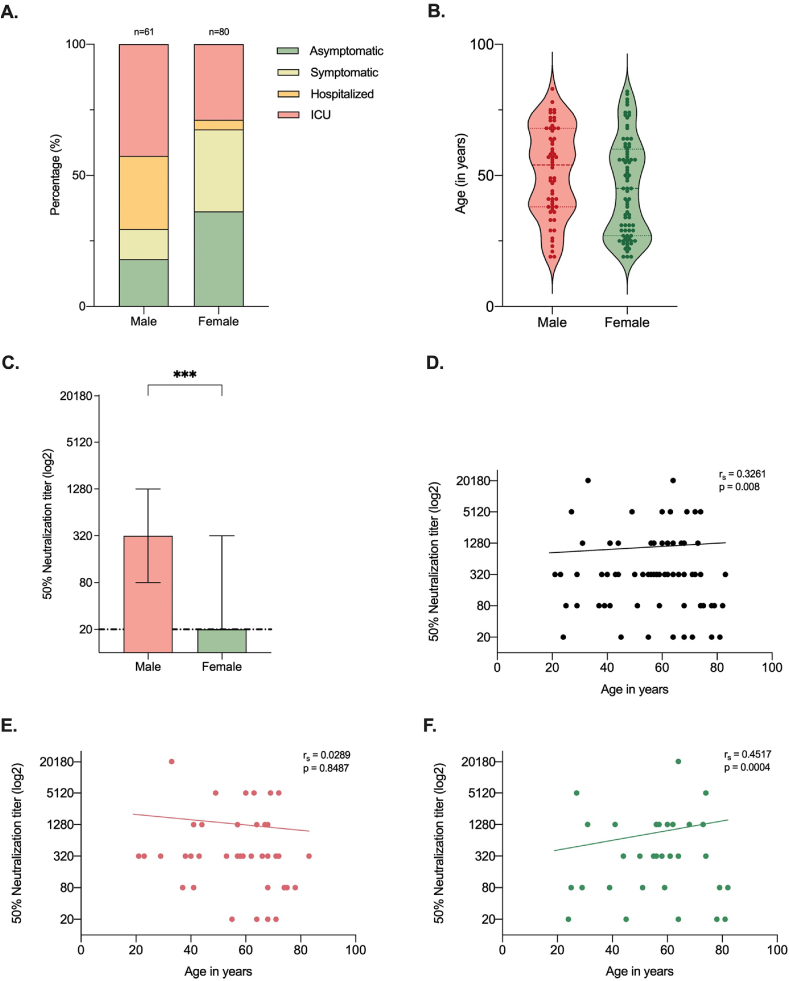
The present study included 141 Hispanic/Latino adults with a positive test result for SARS-CoV-2 infection. Of these participants, 80 (56.7%) were women and 61 (43.3%) were men (Fig. 5A). At the time of diagnosis, the median age of our participants was 49 (range 19–64) years; in males 55 (range 19–64) years, and females 44.5 (19–63) years (Fig. 5B). The complete baseline demographic and clinical characteristics are provided in Supplementary Table 3.
Fig. 5.
Disease severity and neutralizing antibody response to SARS-CoV-2 infection classified by sex and age. (A) Severity classification of SARS-CoV-2 infection by sex. (B) Age distribution of male and female patients. C. Neutralizing antibody titers in COVID-19 patients differentiated by sex. Median and interquartile ranges are reported for each sex. (D) Scatter plot of neutralization titers for the entire cohort of patients and the subgroup of men (E) and women (F). ***p ≤ 0.001.
All participants were categorized into asymptomatic, symptomatic, hospitalized or ICU groups based on the severity of their illness. A total of 36 individuals (25.5%) had no symptoms or nonspecific COVID-19 symptoms of less than 2 days duration (asymptomatic group), of which 7 (19.4%) were men and 29 (80.6%) were women (Fig. 5A). Eleven (18%) men and twenty-five (31.3%) women had mild to moderate symptoms related to SARS-CoV-2 infection but did not require hospitalization (symptomatic group). Moreover, sixty-nine patients had severe symptoms and required hospital admission (48.9%, 43 males and 26 females). Of the 43 (70.5%) men admitted to the hospital, 26 (60.5%) were admitted to ICU (ICU group), and the remaining 17 (39.5%) men were treated at a general medical ward (hospitalized group). Patients in the medical ward only had non-invasive oxygen support with high or low flow systems depending on the medical criteria, while all patients admitted to ICU required invasive mechanical ventilation. Furthermore, most female patients did not require hospital admission (54 women, 67.6%), but of those who were admitted, a higher proportion compared to males (23 females, 88.5%) required ICU admission, and only 3 women (11.5%) were confined to the general medical ward (Fig. 5A).
Of the whole analyzed serum samples, male participants had statistically significant higher median PNRT50 titers (1:320, IQR 1:80-1:1280) than women (1:20, IQR 0-1:320) (Fig. 5C); however, there was considerable variability in NAb titers among all participants. Sex- and severity-based comparisons showed that in patients with more severe disease (ICU group), male NAb titers were higher than females, although the difference was non-significant (data not shown).
In the linear simple regression model, we observed a weak positive correlation between age and NAb titers (rs = 0.3262, p = 0.008) (Fig. 5D), suggesting that older subjects tend to have higher neutralizing titers compared to younger individuals. We then stratified the data by sex (Fig. 5E and F) and found no statistically significant correlation between age and NAb titers in men (rs = 0.0289, p = 0.8487) (Fig. 5E). On the contrary, women's analysis did show a weak positive correlation between age and NAb titers (rs = 0.4517, p = 0.004) (Fig. 5F), suggesting that the positive correlation found between age and NAb titers in the entire cohort of participants is primarily due to female population.
The next step –the clinical testing–, considering the differential expression in the in-silico analysis of some cytokines that participate in the cytokine dysregulation in COVID-19 and the presence of another acute respiratory distress syndrome (ARDS)-related cytokines in our results, was measuring serum levels of IL-6 and CCL-3 in our cohort of participants with SARS-CoV-2 infection. Interestingly, in agreement with the in-silico results, we observed statistically significant higher levels of the systemic inflammatory cytokine IL-6 in males than females (p = 0.0085) (Fig. 6A), coequal to the upregulation of IL6 gene observed in healthy young men in-silico (Fig. 3A). Additionally, we found significantly higher CCL-3 levels in males compared to females (p = 0.0261) (Fig. 6B). We finally examined correlations between age and serum levels of IL-6 and CCL-3, in a sex-disaggregated manner. In females, we observed a weak and a strong correlation between age, CCL3 and IL-6, respectively (Fig. 6C), while in the male population, the correlation between both cytokines evaluated and age was moderate (Fig. 6D).
Fig. 6.
Cytokine expression profile in patients with COVID-19. Serum levels (pg/mL) of IL-6 (A) and CCL3 (B) disaggregated by sex. Correlation between age and cytokine levels of IL-6 and CCL3 in female (C) and male (D) patients. Mann Whitney test was performed for statistical significance, shown as *p ≤ 0.05; ***p ≤ 0.001. Spearman's Rank correlation was used to calculate correlation coefficients (rs) and p-values, shown on the top left of each correlation plot.
4. Discussion
Clinical outcomes at all ages following SARS-CoV-2 infection are primarily influenced by sex differences in immune gene expression and the changes in physiological antiviral responses that occur throughout the aging process [1, 27]. Studies have demonstrated that males are more vulnerable to severe COVID-19 [7, 28]; however, understanding specific genetic traits that shape both cellular and humoral immunity in males and females across the lifespan contributes to the pathogenesis of this disease, and is still subject to investigation. Here, we assessed the expression of immune related genes in healthy subjects in a sex- and age-disaggregated manner using the GTEx database and evaluated the neutralizing antibody response, and cytokine profile of individuals tested positive for SARS-CoV-2 infection.
As expected, we observed disparities in the expression of several immune genes when analyzed by biological sex and age; in addition, we found varied immunological functions represented in the DEGs. It is noteworthy that in both sexes, most DEGs regulate antimicrobial and proinflammatory responses; however, women showed the highest number of upregulated innate immune genes. These results are congruent with previous data showing that adult females predominantly hold more robust innate immunological pathways than men, partly explained by the positive regulatory effects of estrogen and progesterone on various routes of inflammation [29]. In our study, IFIT1-3 genes were among the most striking female-biased genes as they encode potent anti-viral proteins that can restrict virus replication and even bind to viral RNA [30, 31], providing pivotal innate defense against pathogens. This observation is analogous to the interferon-driven upregulation of IFIT1-3 in response to SARS-CoV-2 infection observed by Lieberman and colleagues [32], and highlights the value of well-stablished innate immunity functions in fighting viral infections.
Additionally, we observed in young women a higher expression of genes codifying for viral nucleic acids receptors, such as TLR8 and TLR9, which might as well lead to faster SARS-CoV-2 recognition and clearance upon exposure. Regarding TLR9, although its role in SARS-CoV-2 recognition has not been discussed yet, the study of the SARS-CoV-2 genome shows that the coding region of the E protein and the ORF10 are enriched with CpG islands, regions that bind directly to TLR9, triggering strong early innate responses [33]. Our data suggest that genetics confer women an advantage in the first line of defense against SARS-CoV-2 since females mount more robust innate immune responses that could limit SARS-CoV-2 entry to human cells, promoting early viral clearance, and stimulating adaptive responses [34]. Therefore, women with COVID-19 manifest less severe or fatal outcomes than men, as confirmed by the lower hospitalization and ICU admission rates in females compared to males’ rates as seen in this study.
Interestingly, IL6 gene was upregulated in the youngest male population and cytokine levels were also increased in male COVID-19 subjects compared to female levels; results agree with estrogen's inhibitory effect on the IL6 gene [35]. Such increased expression in males, observed in silico and in the clinical testing of participants, supports the hypothesis that IL-6 overproduction in men contributes to the pathogenesis, progression and worsening of COVID-19 as it drives immune dysregulation leads to systemic inflammatory syndromes [36, 37], rather than promoting viral control. Moreover, several genes encoding chemokines were upregulated in young men, including CCL5 and CCL20. Regardless of their recognized antimicrobial properties, higher levels of CCL5 have been hypothesized to instigate immunopathogenesis during SARS-CoV-2 infection [38], while lower levels are believed to be protective [39]. Moreover, although males often show higher levels of CCL5 than females during the disease course, elevated levels have been correlated with worse disease progression only in women [13]. In our study, CCL3 levels were higher in men compared to females and might contribute to COVID-19 pathogenesis and severity of disease in men as it drives inflammatory signaling, enhances cytokine storm and leads to unfavorable outcomes [40].
Furthermore, we noted a significant number of DEGs playing a role in T lymphocyte and NK cell cytotoxic functions. Here, a significant part of those genes was upregulated in young male subjects, contrary to our expected higher frequency in women, given their strongest expression of cytotoxicity-related genes in other studies [41, 42]. In theory, our data suggest that the integrity of cytotoxic pathways and effector mechanisms to target and kill virus-infected cells might help reduce the estimates of severity during COVID-19 in young adult males and thus shorten male to female fatality ratios in younger individuals as compared with older subjects ratios [43]. Nevertheless, it is worth highlighting that in host defense against SARS-CoV-2, the expression of cytotoxic genes in young men may not be sufficient or guarantee the functioning of antiviral properties as even in young adults, the male gender is still predictive of severe disease [44].
Even though we found only a slight number of significant DEGs in males and females concerning B cell activation and antibody production, some relevant genes were differentially expressed in the youngest population. In men, BATF3, CXCL12, and BCL6, recognized for their role in the germinal center formation and B cell differentiation [45], were upregulated. At the same time, CR1, TLR9, and AICDA, also involved in B cell function, were upregulated in women. However, the overall number of DEGs does not reflect the superior adaptive immune response widely recognized in women [1], or explain the higher NAb titers in male than female subjects upon SARS-CoV-2 infection observed in this study.
In line with other studies [46, 47], we found a modest correlation between age and NAb titers, but surprisingly, when disaggregated the data by sex, the correlation was no longer statistically significant in the male population, in contrast with the positive correlation stated in other studies [48, 49]. Yet, this may be due to our small sample size and the large variability in NAb titers among individuals. Furthermore, despite having a favorable antibody response, the greater severity of COVID-19 in men may have different explanations. First, neutralizing antibodies might appear later in severe than milder COVID-19 cases, when the hyperinflammatory state has already initiated [50]. In these cases, antibodies cannot correctly control SARS-CoV-2 or dampen the solid inflammatory response against the virus [50, 51]. On the other hand, this may merely denote an intent of the immune system to control the higher viral loads proven in patients with more severe disease [52], and in parallel be influenced by sex steroids, the expression of immune-related genes and other unknown factors incompletely understood so far.
One of the most intriguing observations in our study was that the number of DEGs found decreased with aging. As discussed above, older subjects are at increased risk for developing worse clinical features of COVID-19 [53], determined to a great degree by the interaction of key variables in the elderly; for instance, the existence of comorbidities that can widely heighten the chance of complications [54]. In such cases, the term immunosenescence is significant as it provokes immune dysfunction, altering gene expression of antimicrobial receptors and proinflammatory cytokines [55, 56, 57]. Altogether, these findings enlighten the crucialness of long-standing and balanced expression of immune-related genes during infectious diseases at any age.
There are several limitations in our study. First, the immune gene expression was analyzed with data from healthy individuals that may differ from the gene regulation induced by viral infectious diseases such as COVID-19. Moreover, genetic samples were only taken from elevated genes in the blood that may not reflect the entire immunological genome of other tissues where SARS-CoV-2 can be found, including the respiratory epithelium where the virus arouses hallmark manifestations. Second, the NAb response analysis and cytokine evaluation were done in a small sample from Colombia that did not allow comparisons per age group as it was done in-silico and could not reflect all the dynamics driven by SARS-CoV-2 infection in other races and ethnicities. Third, new variants of SARS-CoV-2 emerged during our enrolment period and NAb titers differ from wild-type, but we could not characterize them due to a lack of sufficient genotyping tests in our settings. However, throughout the recruitment timeframe of our participants B.1 was more predominant than variants and by the end Mu (B.1.621) was circulating the most; still, we cannot estimate the impact of these variants on our participants [58]. Finally, we did not follow up on subjects over time because it was out of our scope, and no other cofounders that could bias our results were measured.
In conclusion, the clinical outcome of SARS-CoV-2 infection varies among individuals. Many variables, including biological sex, age, and comorbidities, contribute to the observed differences. Collectively, our findings suggest that stronger innate responses in women, resulting from variations in immune gene expression may influence distinct outcomes observed between male and female subjects with COVID-19. Moreover, the expression of immune system genes is mainly regulated by the effects of the sex hormones estrogen and testosterone, promoting different shifts in the innate and adaptive immune responses. SARS-CoV-2 also induces changes in gene regulation that affect the kinetics of cellular and humoral responses by sex and age and should be explored in additional studies. Further, specific treatment of infections, not only caused by SARS-CoV-2, should include the effect of these variables on host immune responses.
Author contribution statement
Vicky Margarita Montaño Mendoza; Yorjagis Andres Mendez Cortina; Geysson Javier Fernandez; Ana Lucía Rodríguez-Perea; María Teresa Rugeles; Paula A. Velilla Hernandez; Walter D Cardona Maya: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This study was financed by Universidad de Antioquia and ICGEB Research Grant (CRP/20/015).
Data availability statement
Data will be made available on request.
Declaration of interest's statement
The authors declare that there are no conflicts of interest.
Acknowledgments
The authors acknowledge the valuable contributions of IPS Universitaria, Hospital San Vicente Fundación, and the volunteers. We also thank all the laboratory members for their support.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.heliyon.2023.e13045.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Klein S.L., Flanagan K.L. Sex differences in immune responses. Nat. Rev. Immunol. 2016;16(10):626–638. doi: 10.1038/nri.2016.90. 2016 1610. [DOI] [PubMed] [Google Scholar]
- 2.Guha-Sapir D., Schimmer B. Dengue fever: new paradigms for a changing epidemiology. Emerg. Themes Epidemiol. 2005;2:1. doi: 10.1186/1742-7622-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baig S. Gender disparity in infections of Hepatitis B virus. J. Coll. Phys. Surg. Pak. 2009;19(9):598–600. [PubMed] [Google Scholar]
- 4.Daniels D., Grytdal S., Wasley A. Surveillance for acute viral hepatitis — United States, 2007. Morb. Mortal. Wkly. Rep. - Surveillance Summ. 2009;58(3):1–27. http://www.jstor.org/stable/24805750 [PubMed] [Google Scholar]
- 5.Wang H., Wolock T.M., Carter A., et al. Estimates of global, regional, and national incidence, prevalence, and mortality of HIV, 1980–2015: the Global Burden of Disease Study 2015. Lancet HIV. 2016;3(8):e361–e387. doi: 10.1016/S2352-3018(16)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riccò M., Peruzzi S., Balzarini F. Epidemiology of West Nile virus infections in humans, Italy, 2012–2020: a summary of available evidences. Trav. Med. Infect. Dis. 2021;6(2) doi: 10.3390/TROPICALMED6020061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peckham H., de Gruijter N.M., Raine C., et al. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat. Commun. 2020;11(1):1–10. doi: 10.1038/s41467-020-19741-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capuano A., Rossi F., Paolisso G. Covid-19 kills more men than women: an overview of possible reasons. Front. Cardiovasc. Med. 2020;7:131. doi: 10.3389/fcvm.2020.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The sex, gender and COVID-19 project . Global Health 5050. https://globalhealth5050.org/the-sex-gender-and-covid-19-project/
- 10.Montaño Mendoza V.M., Mendez Cortina Y.A., Velilla Hernández P.A., Cardona Maya W.D. Understanding SARS-CoV-2 infection in males. J. Media Res. 2020;6(5):180–182. doi: 10.1016/j.rbmo.2020.04.009. [DOI] [Google Scholar]
- 11.Tsai P.H., Lai W.Y., Lin Y.Y., et al. Clinical manifestation and disease progression in COVID-19 infection. J. Chin. Med. Assoc. 2021;84(1):3–8. doi: 10.1097/JCMA.0000000000000463. [DOI] [PubMed] [Google Scholar]
- 12.Dong M., Zhang J., Ma X., et al. ACE2, TMPRSS2 distribution and extrapulmonary organ injury in patients with COVID-19. Biomed. Pharmacother. 2020;131 doi: 10.1016/J.BIOPHA.2020.110678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi T., Ellingson M.K., Wong P., et al. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature. 2020;588(315–320):1–6. doi: 10.1038/s41586-020-2700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsverava L., Chitadze N., Chanturia G., et al. Antibody profiling reveals gender differences in response to SARS-COVID-2 infection. medRxiv. 2021 doi: 10.1101/2021.06.01.21258175. Published online June 4, 2021.06.01.21258175. [DOI] [Google Scholar]
- 15.Grzelak L., Velay A., Madec Y., et al. Sex differences in the evolution of neutralizing antibodies to severe acute respiratory syndrome coronavirus 2. J. Infect. Dis. 2021;224(6):983–988. doi: 10.1093/INFDIS/JIAB127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schurz H., Salie M., Tromp G., Hoal E.G., Kinnear C.J., Möller M. The X chromosome and sex-specific effects in infectious disease susceptibility. Hum. Genom. 2019;13(1):2. doi: 10.1186/s40246-018-0185-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taneja V. Sex hormones determine immune response. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.01931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Libert C., Dejager L., Pinheiro I. The X chromosome in immune functions: when a chromosome makes the difference. Nat. Rev. Immunol. 2010;10(8):594–604. doi: 10.1038/nri2815. [DOI] [PubMed] [Google Scholar]
- 19.Markle J.G., Fish E.N. SeXX matters in immunity. Trends Immunol. 2014;35(3):97–104. doi: 10.1016/j.it.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Seillet C., Laffont S., Trémollières F., et al. The TLR-mediated response of plasmacytoid dendritic cells is positively regulated by estradiol in vivo through cell-intrinsic estrogen receptor α signaling. Blood. 2012;119(2):454–464. doi: 10.1182/blood-2011-08-371831. [DOI] [PubMed] [Google Scholar]
- 21.Bengtsson Å.K., Ryan E.J., Giordano D., Magaletti D.M., Clark E.A. 17β-estradiol (E2) modulates cytokine and chemokine expression in human monocyte-derived dendritic cells. Blood. 2004;104(5):1404–1410. doi: 10.1182/blood-2003-10-3380. [DOI] [PubMed] [Google Scholar]
- 22.Trigunaite A., Dimo J., Jørgensen T.N. Suppressive effects of androgens on the immune system. Cell. Immunol. 2015;294(2):87–94. doi: 10.1016/j.cellimm.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 23.Torre D., Lachmann A., Ma’ayan A. Cell Syst; 2018. BioJupies: Automated Generation of Interactive Notebooks for RNA-Seq Data Analysis in the Cloud. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The expression profiles in human immune cells. The Human Protein Atlas. https://www.proteinatlas.org/humanproteome/immune+cell
- 25.Gene ontology resource. http://geneontology.org/
- 26.Aguilar-Jiménez W., Flórez-Alvarez L., Rincón D.S., et al. Immune characterization of a Colombian family cluster with SARS-CoV-2 infection. Biomedica. 2021;41(Suppl 2):86. doi: 10.7705/BIOMEDICA.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lieberman N.A.P., Peddu V., Xie H., et al. In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age. PLoS Biol. 2020;18(9) doi: 10.1371/JOURNAL.PBIO.3000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pivonello R., Auriemma R.S., Pivonello C., et al. Sex Disparities in COVID-19 Severity and Outcome: Are Men Weaker or Women Stronger? Neuroendocrinology. 2021 doi: 10.1159/000513346. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shepherd R., Cheung A.S., Pang K., Saffery R., Novakovic B. Sexual dimorphism in innate immunity: the role of sex hormones and epigenetics. Front. Immunol. 2021;11:3559. doi: 10.3389/FIMMU.2020.604000/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou X., Michal J.J., Zhang L., et al. Interferon induced IFIT family genes in host antiviral defense. Int. J. Biol. Sci. 2013;9(2):200. doi: 10.7150/IJBS.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vladimer G.I., Górna M.W., Superti-Furga G. IFITs: emerging roles as key anti-viral proteins. Front. Immunol. 2014;5(MAR):94. doi: 10.3389/FIMMU.2014.00094/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lieberman N.A.P., Peddu V., Xie H., et al. In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age. PLoS Biol. 2020;18(9) doi: 10.1371/JOURNAL.PBIO.3000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y., Chen M., Cao H., Zhu Y., Zheng J., Zhou H. Extraordinary GU-rich single-strand RNA identified from SARS coronavirus contributes an excessive innate immune response. Microb. Infect. 2013;15(2):88. doi: 10.1016/J.MICINF.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diamond M.S., Kanneganti T.D. Innate immunity: the first line of defense against SARS-CoV-2. Nat. Immunol. 2022;23(2):165–176. doi: 10.1038/s41590-021-01091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu H., Liu K., Bodenner D.L. Estrogen receptor inhibits interleukin-6 gene expression by disruption of nuclear factor kappaB transactivation. Cytokine. 2005;31(4):251–257. doi: 10.1016/J.CYTO.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 36.Santa Cruz A., Mendes-Frias A., Oliveira A.I., et al. Interleukin-6 is a biomarker for the development of fatal severe acute respiratory syndrome coronavirus 2 pneumonia. Front. Immunol. 2021;12 doi: 10.3389/FIMMU.2021.613422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montaño Mendoza V.M. Interleukin-17: a potential therapeutic target in COVID-19. J. Infect. 2020;81(2):e136–e138. doi: 10.1016/j.jinf.2020.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patterson B.K., Seethamraju H., Dhody K., et al. CCR5 inhibition in critical COVID-19 patients decreases inflammatory cytokines, increases CD8 T-cells, and decreases SARS-CoV2 RNA in plasma by day 14. Int. J. Infect. Dis. 2021;103:25–32. doi: 10.1016/J.IJID.2020.10.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agresti N., Lalezari J.P., Amodeo P.P., et al. Disruption of CCR5 signaling to treat COVID-19-associated cytokine storm: case series of four critically ill patients treated with leronlimab. J. Transl. Autoimmun. 2021;4 doi: 10.1016/J.JTAUTO.2021.100083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coperchini F., Chiovato L., Ricci G., Croce L., Magri F., Rotondi M. The cytokine storm in COVID-19: further advances in our understanding the role of specific chemokines involved. Cytokine Growth Factor Rev. 2021;58:82. doi: 10.1016/J.CYTOGFR.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hewagama A., Patel D., Yarlagadda S., Strickland F.M., Richardson B.C. Stronger inflammatory/cytotoxic T cell response in women identified by microarray analysis. Gene Immun. 2009;10(5):509. doi: 10.1038/GENE.2009.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Libert C., Dejager L., Pinheiro I. The X chromosome in immune functions: when a chromosome makes the difference. Nat. Rev. Immunol. 2010;10(8):594–604. doi: 10.1038/NRI2815. [DOI] [PubMed] [Google Scholar]
- 43.Green M.S., Nitzan D., Schwartz N., Niv Y., Peer V. Sex differences in the case-fatality rates for COVID-19—a comparison of the age-related differences and consistency over seven countries. PLoS One. 2021;16(4) doi: 10.1371/JOURNAL.PONE.0250523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandoval M., Nguyen D.T., Vahidy F.S., Graviss E.A. Risk factors for severity of COVID-19 in hospital patients age 18–29 years. PLoS One. 2021;16(7) doi: 10.1371/JOURNAL.PONE.0255544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song S., Matthias P.D. The transcriptional regulation of germinal center formation. Front. Immunol. 2018;9(SEP):2026. doi: 10.3389/FIMMU.2018.02026/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karuna S., Li S.S., Grant S., et al. Neutralizing antibody responses over time in demographically and clinically diverse individuals recovered from SARS-CoV-2 infection in the United States and Peru: a cohort study. PLoS Med. 2021;18(12) doi: 10.1371/journal.pmed.1003868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lei Q., Hou H., Yu C., et al. Kinetics of neutralizing antibody response underscores clinical COVID-19 progression. J. Immunol. Res. 2021;2021:1–11. doi: 10.1155/2021/9822706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klein S.L., Pekosz A., Park H.S., et al. Sex, age, and hospitalization drive antibody responses in a COVID-19 convalescent plasma donor population. J. Clin. Invest. 2020;130(11):6141. doi: 10.1172/JCI142004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu L., To K.K.W., Chan K.H., et al. High neutralizing antibody titer in intensive care unit patients with COVID-19. Emerg. Microb. Infect. 2020;9(1):1664. doi: 10.1080/22221751.2020.1791738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawasuji H., Morinaga Y., Tani H., et al. Delayed neutralizing antibody response in the acute phase correlates with severe progression of COVID-19. Sci. Rep. 2021;11(1):1–5. doi: 10.1038/s41598-021-96143-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chvatal-Medina M., Mendez-Cortina Y., Patiño P.J., Velilla P.A., Rugeles M.T. Antibody responses in COVID-19: a review. Front. Immunol. 2021;12:1208. doi: 10.3389/FIMMU.2021.633184/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dadras O., Afsahi A.M., Pashaei Z., et al. The relationship between COVID-19 viral load and disease severity: a systematic review. Immun. Inflamm. Dis. 2022;10(3) doi: 10.1002/IID3.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singhal S., Kumar P., Singh S., Saha S., Dey A.B. Clinical features and outcomes of COVID-19 in older adults: a systematic review and meta-analysis. BMC Geriatr. 2021;21(1):1–9. doi: 10.1186/S12877-021-02261-3/TABLES/3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dai S.P., Zhao X., Wu J hui. Effects of comorbidities on the elderly patients with COVID-19: clinical characteristics of elderly patients infected with COVID-19 from sichuan, China. J. Nutr. Health Aging. 2021;25(1):18. doi: 10.1007/S12603-020-1486-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sidler C., Wóycicki R., Ilnytskyy Y., Metz G., Kovalchuk I., Kovalchuk O. Immunosenescence is associated with altered gene expression and epigenetic regulation in primary and secondary immune organs. Front. Genet. 2013;4 doi: 10.3389/FGENE.2013.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosenstiel P., Derer S., Till A., et al. Systematic expression profiling of innate immune genes defines a complex pattern of immunosenescence in peripheral and intestinal leukocytes. Gene Immun. 2008;9(2):103–114. doi: 10.1038/SJ.GENE.6364454. [DOI] [PubMed] [Google Scholar]
- 57.Huang Z., Chen B., Liu X., et al. Effects of sex and aging on the immune cell landscape as assessed by single-cell transcriptomic analysis. Proc. Natl. Acad. Sci. U. S. A. 2021;118(33) doi: 10.1073/PNAS.2023216118/-/DCSUPPLEMENTAL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tracking of hCoV-19 Variants GISAID. Published 22AD. https://gisaid.org/hcov19-variants/ Accessed.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.