Abstract
In late 2020, after circulating for almost a year in the human population, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) exhibited a major step change in its adaptation to humans. These highly mutated forms of SARS-CoV-2 had enhanced rates of transmission relative to previous variants and were termed ‘variants of concern’ (VOCs). Designated Alpha, Beta, Gamma, Delta and Omicron, the VOCs emerged independently from one another, and in turn each rapidly became dominant, regionally or globally, outcompeting previous variants. The success of each VOC relative to the previously dominant variant was enabled by altered intrinsic functional properties of the virus and, to various degrees, changes to virus antigenicity conferring the ability to evade a primed immune response. The increased virus fitness associated with VOCs is the result of a complex interplay of virus biology in the context of changing human immunity due to both vaccination and prior infection. In this Review, we summarize the literature on the relative transmissibility and antigenicity of SARS-CoV-2 variants, the role of mutations at the furin spike cleavage site and of non-spike proteins, the potential importance of recombination to virus success, and SARS-CoV-2 evolution in the context of T cells, innate immunity and population immunity. SARS-CoV-2 shows a complicated relationship among virus antigenicity, transmission and virulence, which has unpredictable implications for the future trajectory and disease burden of COVID-19.
Subject terms: SARS-CoV-2, Mutation, Viral genetics
In this Review, the authors summarize the mutations harboured by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern. They describe the impact of mutations on virus infectivity and transmissibility, and discuss SARS-CoV-2 evolution in the context of T cells, innate immunity and population immunity.
Introduction
Since its initial emergence in Wuhan in December 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused more than 641 million cases of COVID-19 and more than 6.6 million deaths as of December 2022 (ref. 1). SARS-CoV-2 (along with SARS-CoV, the cause of SARS) is a member of the species Severe acute respiratory syndrome-related coronavirus, the sole member of a subgenus of viruses, Sarbecovirus, primarily found in horseshoe bats2. Like other coronaviruses, SARS-CoV-2 possesses a large RNA genome, comprising ~30,000 nucleotides, whose replication is mediated by RNA-dependent RNA polymerase (RdRP) and an associated proofreading enzyme exoribonuclease (ExoN). This, combined with the discontinuous nature of coronavirus transcription, has resulted in coronaviruses with high rates of recombination, insertions and deletions, and point mutations (although the rates are lower than for other RNA viruses due to the proofreading), as previously reviewed3. The success of novel genetic variants generated, although prone to stochastic sampling processes, will be very dependent on natural selection; in particular, positive selection associated with mutations that are beneficial to the virus in which they occur.
SARS-CoV-2 has proven to be a highly capable human pathogen, but also a generalist in terms of host tropism, establishing infections in a variety of mammalian species, including infections in farmed mink4, a stable reservoir in white-tailed deer5,6 and incidental infections of many other animal species7. Once SARS-CoV-2 was in humans, the first months of SARS-CoV-2 evolution were characterized by limited adaptation and phenotypic change relative to its later evolution8. The first notable change, a single spike substitution (D614G), arose early in the pandemic and conferred an ~20% growth advantage relative to preceding variants9. A lineage defined by D614G (PANGO lineage10 B.1) quickly became dominant in Europe, giving an early indication of the potential for SARS-CoV-2 to increase its transmissibility in humans. As we described previously3,11, from October 2020 onwards, novel, more heavily mutated SARS-CoV-2 variants began to emerge. These variants were distinguished by higher numbers of non-synonymous mutations principally in the spike protein — particularly the case for Omicron — and distinct phenotypic properties, including altered transmissibility and antigenicity. To date, five SARS-CoV-2 variants have been declared variants of concern (VOCs) by the World Health Organization (and national public health agencies) on the basis that they exhibit substantially altered transmissibility or immune escape, warranting close monitoring. Each VOC showed transmission advantages over preceding variants and became dominant, either regionally in the cases of Alpha (PANGO lineage10 B.1.1.7), Beta (B.1.351) and Gamma (P.1) — in Europe, southern Africa and South America, respectively — or globally, in the cases of Delta (B.1.617.2/AY sublineages) and the many Omicron sublineages (B.1.1.529/BA sublineages, such as BA.1, BA.2 and BA.5).
In contrast to the expectation that viruses undergo rapid host adaptation following spillover12,13, selection analysis indicates that SARS-CoV-2 lacked notable levels of observable adaptation early in the pandemic14. It subsequently became clear that SARS-CoV-2 is a generalist virus capable of using a variety of mammalian angiotensin-converting enzyme 2 (ACE2) membrane proteins for cell entry15, enabling infection of a wide range of mammals14,16. The sarbecoviruses are transmitted frequently between different horseshoe bat species17 and non-bat species with ACE2-binding capability (the inferred ancestral trait in sarbecoviruses18), which happens to include humans. The SARS-CoV-2 spike protein contains important properties that are responsible and required for efficient human-to-human transmission, in particular: human ACE2 binding and the polybasic furin cleavage site (FCS) at the S1–S2 junction19,20. At present the SARS-CoV-2 S1–S2 FCS is unique among sarbecoviruses, although analogous sequences are observed in other betacoronaviruses.
The entry of SARS-CoV-2 into airway cells requires furin-mediated cleavage at the FCS, enabling membrane fusion. The FCS is therefore a key determinant of the high transmission rates of SARS-CoV-2, contributing to its efficient spread in humans19,21. Further optimization of the wild-type FCS during the course of the pandemic has resulted in enhanced furin cleavage of the Alpha and Delta spike proteins22–25. In concert with other mutations, notably those enhancing ACE2 binding26,27, mutations optimizing furin cleavage are thought to have contributed to enhanced transmissibility, and thus fitness, of the Alpha and Delta VOCs, with 65% and 55% higher relative transmissibility compared with the variants they replaced, respectively28–30. In contrast to Alpha and Delta, the evolutionary success of the Omicron variant is not linked to optimization of furin cleavage. Rather, Omicron is characterized by an altered entry phenotype31,32, coupled with significant immune escape31,33,34, enabling efficient infection of vaccinated or previously infected individuals. Although transmissibility in a naive population is largely determined by intrinsic viral properties, the increasingly complex immune landscape in which SARS-CoV-2 now circulates means that antibody escape (as opposed to transmissibility being enhanced by virus biology alone, a trait that might be difficult to optimize further than that achieved by Omicron) is becoming the prime driver of variant success. Before Omicron’s emergence (Box 1), each of the dominant variants had evolved from pre-VOC progenitors, rather than evolving from one another. By contrast, successive waves are now being caused by Omicron sublineages (for example, BA.5, one of its sublineages BQ.1 and BA.2.75, a sublineage of BA.2). Of note, it is possible that an undetected variant, potentially a recombinant (Box 2), could emerge with high transmissibility linked to intrinsic biology and novel antigenic properties.
Whether an entirely novel variant emerges, or future viruses evolve from the Omicron sublineages with novel antigenic changes (increasingly probable as previous VOCs are no longer circulating), it is clear that novel SARS-CoV-2 variants possessing unique combinations of mutations will continue to emerge and those with a fitness advantage will dominate relative to previous variants. To date, successful variants have also exhibited variation in clinically relevant traits, including disease severity, immune evasion and sensitivity to therapeutics (particularly monoclonal antibodies). It is therefore of public health and clinical importance to understand the drivers of SARS-CoV-2 fitness. Variant fitness — a virus’s reproductive success — depends on a variety of factors that determine its ability to infect, replicate within and spread between hosts. In this Review, we provide an overview of observed mutations that are described as impacting SARS-CoV-2 infectivity and transmissibility, and discuss the viral capacity to escape T cell-mediated, innate or humoral immunity11. See our previous review11 for more details on spike-mediated humoral immunity.
Box 1 The origin of SARS-CoV-2 variants of concern and the Omicron complex.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOCs) exhibit a number of distinct properties, one of the most intriguing being a relatively long phylogenetic branch length often with a lack of genetic intermediates before their detection67,193,194. Alpha was first detected in the UK during a time of low virus circulation and unprecedented SARS-CoV-2 surveillance67,131. Delta was first identified in India in April 2021 (ref. 195), and then was associated with a new epidemic wave in the UK196, generating several sublineages, some of which were even more transmissible than parental Delta, for example AY.4.2 (ref. 197). Similarly, the Omicron complex of variants has a long branch length and even more mutations than prior VOCs43,44.
The lack of intermediate sequences has led to several hypotheses of VOC origins198: (1) circulation in geographically undersampled regions; (2) cryptic circulation within an animal reservoir following reverse zoonosis of an early variant; (3) evolution within a chronic infection in an immunosuppressed host or multiple human hosts. Although there is evidence for establishment of stable animal reservoirs for SARS-CoV-2, particularly in farmed mink and white-tailed deer4,5,186, at present the ‘chronic infection hypothesis’ of variant emergence is the best supported of these scenarios67,183. Chronic infections are known to drive mutational profiles that are highly similar to those of VOCs113,184,199–201. There is also evidence for limited onwards transmission of chronic infections causing localized outbreaks113,182. Long-term evolution in the presence of constant subneutralizing antibody pressure might explain VOC antigenic distance, particularly for Omicron.
Unlike other VOCs, Omicron had evolved remarkable diversity before being first detected, and is currently divided into five major lineages (BA.1, BA.2, BA.3, BA.4 and BA.5 (ref. 43); see the figure), with many more sublineages being detected that are accumulating further antigenic changes157. BA.1 caused a global wave of infection at the end of 2021, but by early 2022, it had been replaced by BA.2. In April 2022, further BA sublineages, BA.4 and BA.5, were recognized, and, as of September 2022, BA.5 has driven a new Omicron wave internationally43. The Omicron lineages show a complex relatedness to each other, which likely included multiple intra-VOC recombination events before detection43,44. For example, BA.4 and BA.5 contain near identical 5′ ends of the genome until the M gene, but show large divergence after this point, suggesting a recent recombination event. Similarly, BA.3 could conceivably be the recombinant progeny of ancestral BA.1 and BA.2 viruses44. Hypothetical phylogenetic origins of the Omicron complex showing one potential pattern of recombination among sublineages BA.1, BA.2, BA.3, BA.4 and BA.5 are shown in the figure. Protein names indicate approximate (~) potential break points.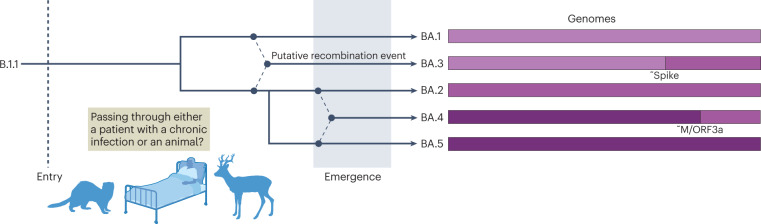
M, membrane protein.
Box 2 SARS-CoV-2 recombinants.
When two RNA viruses co-infect the same cell within an individual, there is a high chance during genome replication that the polymerase will switch from one genome sequence template to the heterologous genome. This results in a recombinant virus with part of its genome from one ‘parent’ and the remaining genomic sequence from the other (see the figure, part a). Several recombinant lineages have been identified in different locations and designated by the PANGO classification system since 2020 (ref. 10), identifiable by the ‘X-’ lineage prefix. Recombinants have been unambiguously identified when genetically distinct variants, such as two variants of concern, have transiently co-circulated leading to co-infections. For example the first assigned recombinant lineage, XA, was a recombinant between Alpha (B.1.1.7) and the lineage previously circulating in the UK, B.1.177 (ref. 202). XC was a recombinant between Alpha and Delta found in Japan203.
From the start of 2022, the number of recombinants identified rapidly increased, which was likely due to high levels of co-circulation between Delta and BA.1, or BA.1 and BA.2, in many countries at a time of phasing out of COVID-19 restrictions. Another explanation is better confidence in identifying recombinants, owing to higher sequence divergence among the genomes of Delta, BA.1 and BA.2. One example recombinant is XD — a Delta × BA.1 recombinant first found in January 2022 in France204. XD has two genome breakpoints, with a backbone and part of the spike protein amino-terminal domain from Delta and the remainder of the spike protein from BA.1 (see the figure, part b). Functionally, XD has been shown to have an intermediate pathogenicity phenotype between BA.1 and Delta in transgenic mice expressing human angiotensin-converting enzyme 2 (ACE2) under the control of the keratin 18 promoter, causing moderate weight loss, suggesting part of the differential rodent pathogenicity phenotype of Delta and BA.1 maps outside the spike protein204.
A second notable recombinant in the UK is the BA.1 × BA.2 recombinant XE, which was first detected in England on 19 January 2022. Before it was outcompeted by BA.5, XE comprised more than 2,500 genomes, mostly from the UK, and preliminary data suggest a modest increase in growth rate compared with BA.2 (ref. 205). XE contains the spike and structural proteins from BA.2 and part of ORF1ab from BA.1.
A recent notable lineage is XAY, a BA.2 × Delta recombinant, which was first discovered in South Africa in May 2022 alongside a smaller sister recombinant, XBA206. These recombinants have a much larger number of breakpoints than any prior known cluster, as well as many unique mutations not found in the parental lineages. One possible explanation for the appearance of these recombinants is a chronic Delta infection with BA.2, iterative recombination (alongside gradual gaining of unique mutations) before multiple emergences back into the general population. XBB is a recently emerged and rapidly growing recombinant between two second-generation BA.2 lineages — BJ.1 (also known as BA.2.10.1.1) and BA.2.75 — and it contains a large number of antigenic receptor-binding domain mutations and has been shown to be poorly neutralized by previous Omicron breakthrough antisera207.
As we have described, variant-specific properties can both map within and outside the spike protein. Recombinants, such as XD, demonstrate it is possible for recombination to generate viruses with phenotypic properties from both parental viruses204. Before the rapidly growing XBB, recombinants had not had a large impact on the course of the pandemic, usually appearing around the time new variants were replacing previously dominant lineages. It should be noted that such recombination is commonplace among the sarbecoviruses infecting horseshoe bats, as exemplified by the high levels of recombinant genomes detected and the tendency for the spike protein to be swapped, indicative of frequent antigenic shift events208. A recombination process occurring in an individual with Omicron and Delta infection (see the figure, part a). Genome maps of some notable contemporary recombinants (XD, XE, XAY and XBB) (see the figure, part b).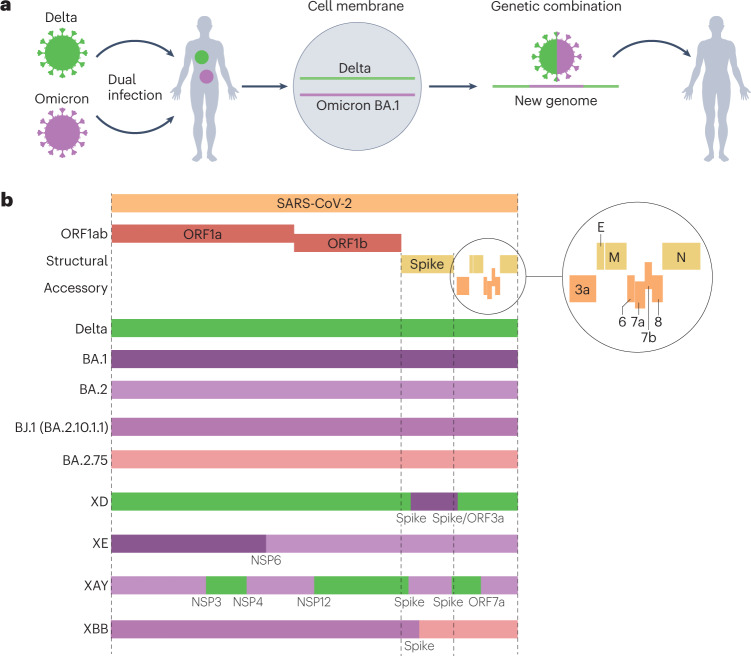
E, envelope protein; M, membrane protein; N, nucleocapsid protein; NSP, non-structural protein; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Antigenic escape and SARS-CoV-2 variants
An early concern regarding SARS-CoV-2 evolution was the potential emergence of antigenically distinct variants with the ability to evade vaccine- or infection-acquired immunity, as exemplified by the N439K spike substitution35. Before their update in late 2022, all widely used COVID-19 vaccines were based on the spike antigen of early variants, most using the reference sequence Wuhan-Hu-1, sampled from a lineage B infection at the Huanan Seafood Market, often with mutations that stabilize the spike protein in a prefusion conformation36. Although limited antigenic change was reported for Alpha25,37,38, moderate escape from vaccine-derived antibodies and convalescent sera was observed for Beta, Gamma and Delta in laboratory experiments23,37–39. Nonetheless, epidemiological studies provided evidence that vaccine effectiveness against Delta and Beta was largely preserved40–42. Thus, despite a design based on a very early spike sequence, first-generation SARS-CoV-2 vaccines conferred remarkable protection against severe disease and allowed much of the world to return to a semblance of normality.
The Omicron ‘complex’ — comprising the distinct sublineages BA.1, BA.2, BA.3, BA.4 and BA.5 (Box 1) — is capable of infecting the vaccinated and previously infected, bringing about the challenge of vaccine-sequence selection and universal vaccines to the fore of SARS-CoV-2 control strategy discussions. With more than 15 spike receptor-binding domain (RBD) mutations and a number of antigenic deletions and substitutions in the amino-terminal domain (NTD)43,44, BA.1, BA.2, BA.4 and BA.5 are very poorly neutralized with first-generation vaccines and by pre-Omicron infection-derived antibodies (Fig. 1). In addition, escape from the vast majority of current therapeutic monoclonal antibodies has been demonstrated; at present, only bebtelovimab — a monoclonal antibody targeting the RBD of the spike protein — has been reported to retain its efficacy against all SARS-CoV-2 variants31,33,34,38,45–50. This large antigenic ‘shift’ has led some to propose that Omicron lineages should be considered a separate strain or serotype compared with pre-Omicron lineages51,52. Importantly, the magnitude of this antigenic change is reflected in data on real-world vaccine effectiveness against infections and symptomatic disease41,53–57. Booster doses are required to maintain any vaccine effectiveness against Omicron, which reduces as antibody titres wane41,55. Indeed, vaccine effectiveness against severe disease for Omicron remained high 4 months after a booster dose and then decreased quickly, although the reduction was less rapid than that seen after primary vaccination58. Due to the short duration of protective immunity against Omicron infection with current vaccines, many vaccine manufacturers and academics are focusing on second-generation vaccines, such as monovalent or bivalent Omicron-specific boosters59 (which are being deployed at present), nasal vaccine delivery to stimulate greater mucosal immunity60 or universal vaccine approaches61. In common with seasonal human coronaviruses, the extent to which long-term acquired immunity can prevent reinfection by SARS-CoV-2 is limited due to a combination of antibody waning and virus antigenic drift — the incremental acquisition of mutations that permit immune evasion62,63. In addition to step changes in antigenicity, virus evolution during persistent infections has enabled SARS-CoV-2 to accumulate multiple mutations in the context of a single or a few long-term infections, contributing to antigenic shift events when these variants go on to infect others (Box 1).
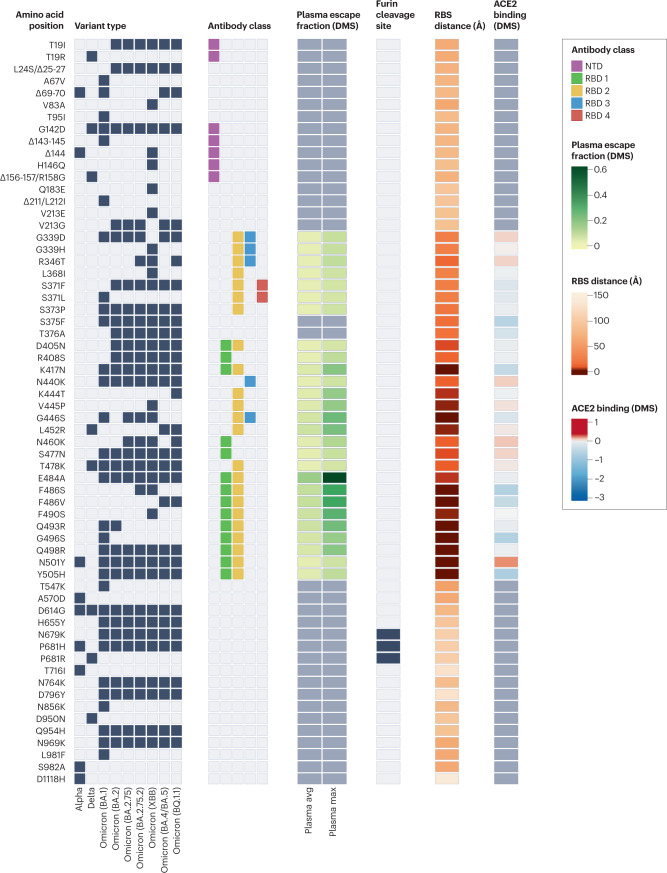
Fig. 1. Properties of amino acid substitutions or deletions in selected SARS-CoV-2 variants of concern.
Black boxes denote the presence of each mutation in the variant of concern. Epitope residues are coloured to indicate the amino-terminal domain (NTD) supersite187 or the receptor-binding domain (RBD) class188. For RBD residues, the results of deep mutational scanning (DMS) studies show the escape fraction (that is, a quantitative measure of the extent to which a mutation reduced polyclonal antibody binding) for each mutant averaged across plasma (‘plasma avg’) and for the most sensitive plasma (‘plasma max’)189, illustrating consistency or variation in the effect of a mutation depending on differences in the antibody repertoire of individuals. Mutations in the furin cleavage site are highlighted. Orange shading indicates the distance to angiotensin-converting enzyme 2 (ACE2)-contacting residues that form the receptor-binding site (RBS). Note that the RBS is defined as residues with an atom <4 Å from an ACE2 atom in the structure of the RBD bound to ACE2 (RCSB Protein Data Bank ID 6M0J190). Finally, ACE2-binding scores representing the binding constant (Δlog10 KD) relative to the wild-type reference amino acid from DMS experiments are shown in shades of red or blue26.
The FCS in SARS-CoV-2 variant emergence
Compared with other known sarbecoviruses, a unique feature of SARS-CoV-2 is the presence of an FCS within its spike protein that can be cleaved by furin. FCSs are nonetheless present in many other betacoronaviruses of the subgenera Embecovirus and Merbecovirus, such as human coronavirus OC43, human coronavirus HKU1 and Middle East respiratory syndrome coronavirus (Fig. 2a). The SARS-CoV-2 FCS has been shown to be vital for optimal virus replication in human airway cells64, transmissibility19,21 and pathogenicity65. Furin, a host protease, is most abundant in the Golgi apparatus, allowing cleavage during trafficking of the virus to the cell surface66. It is now clear, however, that the FCS of early SARS-CoV-2 variants was suboptimal and not efficiently cleaved by furin19,24,65. Interestingly, one of the roles described for the early substitution mutation, spike D614G, was modestly enhanced spike cleavage (reviewed in detail previously3). A number of subsequent SARS-CoV-2 variants harbour mutations adjacent to the FCS, which increase the number of basic amino acid residues — the known recognition site for furin; for example, Alpha, Mu and Omicron contain the FCS mutation P681H44,67,68, which is predicted to increase cleavage activity. Moreover, a different mutation at the same position, P681R, enhances the replication and pathogenicity of the Delta VOC23,30,69 (Fig. 2b). Of note, Omicron contains P681H, as well as the further mutation N679K44, which together result in an optimized FCS19,23–25,32,70. Importantly, however, furin site optimization alone does not enhance the transmissibility or replication of SARS-CoV-2 and might be detrimental to efficient virus transmission27, indicating that additional mutations seen in these variants are required for optimized replication and transmissibility.
Fig. 2. Furin cleavage site and variant success.
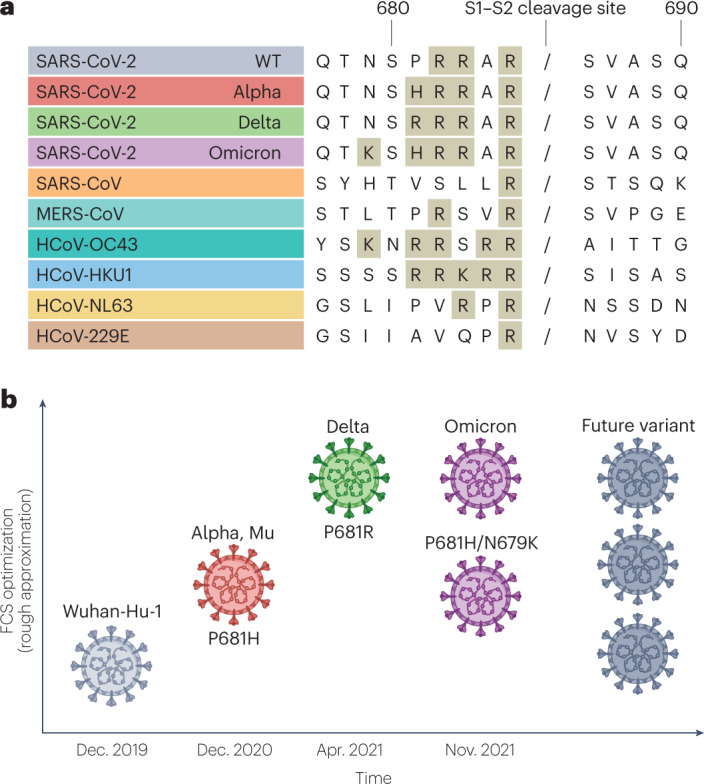
a, Comparison of S1–S2 cleavage site sequences in wild-type (WT) severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and Alpha, Delta and Omicron variants of concern compared with other coronaviruses: SARS-CoV, Middle East respiratory syndrome coronavirus (MERS-CoV), human coronavirus OC43 (HCoV-OC43), HCoV-HKU1, HCoV-NL63 and HCoV-229E. A slash indicates the putative furin/serine protease cleavage site. Amino acids contributing to monobasic or polybasic cleavages sites are shaded. b, Diagrammatic representation of estimated relative optimization of the S1–S2 furin cleavage site (FCS) for the variants of concern. The mutations affecting FCS function are indicated. Note that non-concordant results have been observed for Omicron, as indicated. The level of FCS optimization for future variants is uncertain. Data on Alpha from refs. 19,24,25, data on Delta from refs. 22,23,69 and data on Omicron from refs. 32,75,76.
The exact mechanism by which the observed FCS mutations enhance furin cleavage remains a topic of debate. Although there is fairly strong evidence that P681R directly enhances furin engagement and cleavage of the S1–S2 site19,24,69, the functional consequence of P681H71 is less clear. Mechanistically, it is likely relevant that position T678 of the spike protein, located close to the residue P681, can be post-translationally modified with an O-linked glycosylation site72–74. Downstream prolines are known to promote O-linked glycosylation; therefore, an alternative explanation could be that the removal of P681, rather than the addition of the histidine per se, leads to loss of the potentially furin-obstructing glycosylation site and associated enhancement of cleavage.
Recent insights into Omicron biology have challenged the hypothesis that enhancing spike cleavage is essential for increased viral transmission. All Omicron lineages contain P681H and N679K43,44, which alone, or together, enhance cleavage of the S1–S2 site in the wild-type spike protein32,70. However, in the context of the full Omicron spike protein, evidence of such improved cleavage is less clear, with some studies finding that the S1–S2 junction appears more poorly cleaved than in previous VOCs50,75, whereas others show cleavage efficiency comparable to that of Delta32,76,77. Regardless of the cleavage phenotype, many groups have described how Omicron is able to efficiently utilize an alternative cell entry pathway. Indeed, whereas previous VOCs such as Delta are highly reliant on fusion priming by transmembrane protease serine 2 (TMPRSS2) at the cell surface, Omicron is also able to be efficiently primed by endosomal proteases, such as cathepsins, in a manner similar to SARS-CoV31,32,50,75,76,78,79. This alternative mechanism is hypothesized to be partly responsible for the reduced severity of Omicron, at least in rodent models32,75,78,80,81, due to lower fusogenicity and potentially altered tissue tropism, with a bias towards infection of the upper respiratory tract over the lower respiratory tract. Several studies have suggested molecular mechanisms for this reduced fusogenicity and altered entry route, including mutations in the spike RBD of Omicron31,82, H655Y83 or mutations in the S2 domain31,32,76, specifically N969K32, although it is contentious whether this trait is conserved across all Omicron lineages32,84. Nonetheless, Omicron lineages continue to also show high transmissibility, at least equivalent to that of Delta85,86, implying a potential decoupling between efficiency of furin cleavage and fusogenicity and their contributions to virus transmissibility.
Beyond the S1–S2 site, betacoronaviruses also require cleavage of a second protease cleavage site, known as the S2′ site. Following cleavage of the S1–S2 site and cognate receptor binding, the S2′ site becomes exposed in the S2 domain of the spike protein87. Cleavage of S2′ directly liberates the fusion peptide, leading to virus–host membrane fusion88–90. For SARS-CoV-2 entry into airway cells, this site is preferentially cleaved by host serine proteases, such as TMPRSS2, but it can alternatively be cleaved by endolyosomal cathepsins19,91. A number of recent articles have suggested that variation in the NTD of SARS-CoV-2, particularly via remodelling of external loops through the acquisition of deletions or insertions, can allosterically influence both S1–S2 cleavage and S2′ cleavage, and therefore fusion92–94.
Overall, the relationship between protease usage, S1–S2 cleavage efficiency, tropism, pathogenicity and transmissibility of SARS-CoV-2 is highly complex and, at times, results among studies have been inconsistent. Further work is, thus, required to bridge these gaps in knowledge and to fully understand this system.
Other structural and non-structural proteins and infectivity
Several recent studies have investigated the consequence of mutations in structural proteins other than the spike protein, including membrane (M), envelope (E) and nucleocapsid (N) proteins. The B.1.1 lineage is defined by a pair of substitutions in the N protein — R203K and G204R. Variants derived from B.1.1 (such as Alpha, Gamma and Omicron) inherited the same mutations, whereas Delta and Beta independently evolved R203M and T205I, respectively, with convergent functional properties. Specifically, these mutations have been shown to increase viral infectivity95–97, although the exact mechanism of action remains disputed. On the one hand, data from a virus-like particle-based reporter-based assay95 suggested that these mutations directly increase virus particle formation, whereas another report suggested a role for phosphorylation of N protein allowing escape restriction by the kinase GSK3 (ref. 97). An alternative explanation is that the R203K and G204R mutations introduce a novel transcriptional regulation site into the middle of the N gene, allowing the expression of a truncated form of N protein (termed ‘N*’ or ‘N.iORF3’), which might enhance virus infectivity through enhanced interferon antagonism98,99. Interestingly, there are several further examples of SARS-CoV-2 lineages evolving novel transcriptional regulation site sequences that could result in expression of truncated protein products, either in frame or out of frame, most prominently in non-structural protein 16 (NSP16)98.
Alongside N protein, mutations in M and E proteins have also been implicated in modulating SARS-CoV-2 infectivity. Substitutions in the M and E proteins of BA.1 (Omicron) have been shown to reduce cell entry of virus-like particles, although these mutations are compensated for by further substitutions in S and N proteins100. Coronavirus E proteins have several functions, one of which is to act as a cation channel, potentially within the endoplasmic reticulum (ER) and Golgi compartments to regulate multiple stages of the viral life cycle101. The T9I mutation found in Omicron E protein has been shown to attenuate this ion channel activity in vitro102, although its functional consequences are unclear.
Although ORF1ab makes up two-thirds of the SARS-CoV-2 genome, it remains the region where the impact of variant mutations is least understood. One exception is a deletion at positions 106–108 within NSP6, a mutation that is conserved among all the VOCs except Delta. NSP6 is a multipass transmembrane protein associated with the formation of the coronavirus replication organelle — the ER-derived membranous structure that is produced during infection, providing a compartment for viral RNA replication shielded from innate immunity103. A recent study showed that NSP6 forms homodimers and mediates the formation of a ‘zippered ER’ — narrow and exclusive membranous channels that connect ‘double-membrane vesicles’, which are the primary site of viral genome replication103. It was found that the 106–108 deletion in NSP6 specifically enhances the formation of this zippered ER, indicating a potential host-specific adaptation. The exact mechanism of this enhancement remains to be elucidated, although the study authors postulated that the deletion removes a putative O-linked glycosylation site. At present it remains unclear why Delta and several other variants were never observed to have gained this adaptation despite the ease with which this deletion can occur.
Beyond the structural and ORF1ab proteins, there is some very limited evidence showing adaptive changes in accessory proteins, which is covered later in this Review. Unfortunately, experimental characterization of non-spike mutations and any associated adaptations to humans in SARS-CoV-2 variants remains far behind that of the spike protein. This is due to a number of factors, including the ubiquitousness of pseudovirus technology used to study spike phenotypes compared with the technical complexity of reverse genetics (usually required for virological studies of non-spike mutations) and the scarcity of in vitro systems for investigating non-spike proteins. It is clear from current work that non-spike adaptations contribute largely to virus fitness and pathogenicity, and the continued development of systems to study these regions is vital to ongoing research.
T cell responses and antigenic escape
T cells are a major part of the adaptive immune response to SARS-CoV-2 infection, with a profound CD4+ T cell and CD8+ T cell response observed in most infected individuals104. Several studies suggest an important role for T cell immunity in protection from severe COVID-19, although this is likely to be more nuanced and complex than the well-characterized correlate of protection from infection of neutralizing antibody responses105,106. Although CD4+ helper T cell responses are likely to be broadly important for antibody generation, the importance of T cells in reducing disease severity might be relatively more important in scenarios where neutralizing antibody responses are diminished or not yet detectable. Early induction of SARS-CoV-2-specific T cells is seen more frequently in mild infections than in severe infections107, and CD8+ T cells may be particularly important in reducing severe outcomes in patients with B cell deficiency108. Furthermore, a fully functional CD8+ T cell response is mobilized 1 week after the first dose of mRNA BNT162b2 vaccine (Pfizer–BioNTech), at a time when neutralizing antibodies are not fully induced, raising the possibility that early vaccine-induced protection may be mostly reliant on T cells109.
Given the integral role of T cells in SARS-CoV-2 immunity, the potential exists for selective pressure to lead to T cell escape, although the extent to which SARS-CoV-2 mutations affect T cells is currently poorly understood. Functional T cell responses are directed against multiple virus proteins, with the magnitude of response correlating with viral protein expression levels. Responses to spike protein, N protein and M protein dominate, with appreciable responses also seen against ORF3a and the non-structural proteins NSP3 and NSP12 (ref. 110). As the T cell response targets epitopes across the SARS-CoV-2 genome, the footprints of T cell escape are more broadly distributed than antibody-driven changes, which are concentrated within dominant epitopes of the spike protein (Fig. 3). Few studies have documented intrahost evolution within T cell epitopes, which would serve as direct evidence of T cell escape. Mutations within CD8+ epitopes in N protein (M322I and L331F), M protein (L90F) and the spike protein (L270F) were noted within minority variants in one study during the course of acute infections, resulting in loss of epitope-specific responses111. Prolonged SARS-CoV-2 infections in immunocompromised hosts may offer greater opportunities for T cell escape, akin to the extensively described examples in HIV-1 infection112. Emergence of the NSP3 T504P mutation resulting in loss of a CD8+ epitope response has been reported in multiple individuals with impaired humoral immune deficiency but preserved T cell responses in the context of chronic SARS-CoV-2 infection113,114. These findings are limited to a few cases, demonstrating the need for more prospective cohort studies systematically evaluating the risk of T cell escape in certain patient populations.
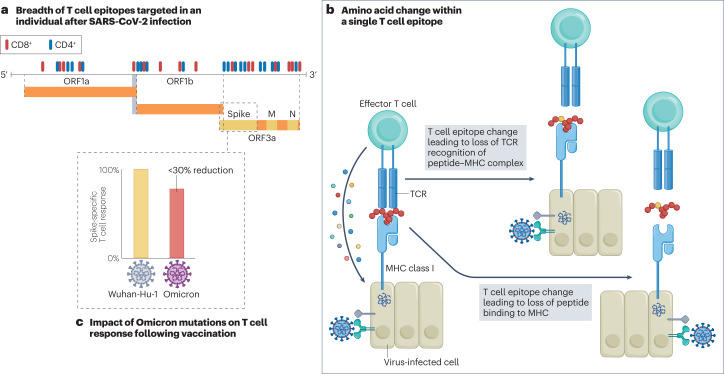
Fig. 3. Potential impact of SARS-CoV-2 variants on T cell responses and innate immunity.
a, Following severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, CD4+ T cell and CD8+ T cell responses are generated against 30–40 epitopes across the virus genome110 (epitopes are shown in red and blue). b, An example of how an amino acid change within one epitope might impact epitope-specific cytotoxic T cell responses, thereby inhibiting the elimination of virus-infected cells115. T cell evasion of SARS-CoV-2 has been shown to be the consequence of impaired peptide binding to the major histocompatibility complex (MHC) or poor binding of the T cell receptor (TCR) to the peptide–MHC complex. c, Although the T cell response to vaccination is focused on the spike protein alone, even the multiple spike mutations in the Omicron variant of concern reduce the vaccine-induced spike-specific T cell response only by less than 30%, with considerable interindividual variability120,191. M, membrane protein; N, nucleocapsid protein.
Several mutations within immunodominant ORF3a and N protein CD8+ T cell epitopes that result in complete loss of recognition have arisen independently in multiple SARS-CoV-2 lineages115. Among these is N protein P13L, present in Omicron within a B*27:05 restricted CD8+ epitope. Given the hypothesis that VOCs emerge in chronic infections, it is tempting to speculate that the presence of P13L in the Omicron VOC reflects selection due to T cell pressure during a chronic infection, in addition to the constellation of spike mutations that are likely driven by antibody pressure. L452R found in Delta, Epsilon, Kappa and the BA.4/BA.5 variant spike proteins results in loss of an A*24:02-restricted CD8+ response116. The spike P272L substitution has arisen in multiple lineages worldwide, and leads to loss of a dominant HLA A*02:01 restricted CD8+ epitope117. The role that T cells have in driving this change, in addition to antibody evasion and enhanced ACE2-binding affinity, is uncertain. Other spike mutations in VOCs that have been associated with loss of specific CD4+ responses include L18F, D80A and D215G in Beta, and D1118H in Alpha118,119. The degree to which these observations also represent incidental effects on T cell responses with mutations driven by other pressures is currently unknown.
Despite the loss of these specific responses, several studies show that the overall T cell response induced by infections and first-generation vaccines is preserved against most VOCs105,118–120. Even the extensive mutations in the Omicron spike protein result in only a modest less than 30% reduction in total CD4+ and CD8+ responses, with considerable interindividual variability119,120. Most high-frequency spike CD4+ epitope responses are focused on discrete regions of the NTD, carboxy terminus and fusion protein regions, with very few in the RBD110. There are no clear hotspots of spike CD8+ epitopes110. Mutations concentrated in the spike RBD and NTD found in many VOCs that are thought to be driven by antibody evasion and increased ACE2-binding affinity might therefore have limited impact on the overall T cell response. Thus, most T cell epitopes are conserved in different VOCs, and this is likely to contribute to the preserved vaccine effectiveness against hospitalization and death with Omicron seen after a second dose and after a third dose when compared with no vaccination121.
The other key reason for the modest impact of variants on T cell immunity is the breadth of response generated, with each individual mounting responses to 30–40 epitopes following infection110. At a population level, there is also far greater heterogeneity to the T cell response than antibody immunity due to a number of polymorphisms present in human HLA genes. At an individual level, however, significant reductions in spike-specific CD8+ responses to Omicron have been reported in ~15% of convalescent and vaccinated donors117, with another study noting more than 50% loss of CD4+ T cell and CD8+ T cell responses in ~20% of individuals tested122. Although the generalizability of these results is limited by small sample sizes and the HLA distribution of the populations studied, it nonetheless highlights the potential impact of VOCs on T cell responses in certain individuals who have spike-specific immunity generated only through vaccination.
It seems likely that antibody evasion and enhanced transmissibility will continue to be greater drivers of emerging VOCs than significant T cell escape. Whether we will see slow and sequential loss of CD8+ epitopes over time, similar to long-term adaptation in H3N2 influenza, is difficult to predict97. T cell escape can occur through several mechanisms98. Amino acid changes within epitopes or flanking regions can disrupt antigen processing, and changes to anchor residues can interfere with major histocompatibility complex (MHC) binding to epitopes123. Both these mechanisms can result in irreversible loss of T cell responsiveness to a particular epitope. Changes that impair T cell receptor binding to the peptide–MHC complex can, by contrast, result in partial or complete escape. This latter scenario might also be overcome by de novo T cell responses using alternative T cell receptor repertoires, as previously described in HIV-1 infection124.
As well as potential T cell escape, SARS-CoV-2, like many other viruses, directly downregulates MHC class I (MHC-I) expression on infected cells to evade T cell recognition, best described for the accessory protein ORF8 (refs. 125,126). ORF7a (as well as ORF3a and ORF6 (refs. 127,128)) has also been reported to downregulate MHC-I (refs. 128,129), although it is unclear whether this is a specific effect or merely the result of nonspecific Golgi fragmentation130. Recent work has shown that common substitutions seen in VOCs do not alter the ability of ORF8 to suppress MHC-I expression, with the exception of a premature stop codon at amino acid 27 in Alpha, resulting in the expression of a truncated, non-functional form of ORF8 (refs. 126,131). Despite the truncation of ORF8, in the context of infection, Alpha still downregulates MHC-I expression, implying that this variant, or possibly SARS-CoV-2 more generally, has evolved redundant mechanisms to inhibit this pathway126.
Innate immunity and SARS-CoV-2 variants
Innate immunity is an integral part of host defence against pathogens, with a key role in early viral control and tuning of adaptive immune responses132. Innate immune responses are particularly critical against novel viruses, such as zoonotic pathogens, for which there is usually no pre-existing adaptive immunity. It is surprising when novel zoonotic viruses are effective at spreading between humans through effective antagonism of innate host defences, despite recent successful replication and transmission in a distantly related host species, with its typically divergent innate immune system. A consequence of the robustness of the human innate immune system is that the proportion of zoonotic viruses that go on to cause pandemics is extremely low, and properties that evolved in the SARS-CoV-2 reservoir species, specifically generalist host tropism (which happened to include humans) coupled with acquisition of its FCS in the spike protein, possibly in an intermediate host species, were critical features for starting the COVID-19 pandemic. It is noteworthy that the related SARS virus SARS-CoV did not establish itself in the human population, despite also being highly transmissible, its eradication being attributed to less asymptomatic spread compared with SARS-CoV-2, making it easier identify infections. Importantly, as SARS-CoV-2 VOCs have emerged, it has become clear that they are acquiring adaptations to more efficiently infect humans, and this in part is through enhanced innate immune evasion. Indeed, Alpha133,134 and more recent VOCs77,135,136 have evolved reduced sensitivity to interferons, consistent with this being a key selective pressure for virus transmission in humans. Surprisingly, rather than adapting to specifically antagonize human proteins, Alpha and Omicron sublineages BA.4 and BA.5 have in part achieved this by upregulating expression of innate immunity suppressing viral proteins, particularly the protein ORF6 (ref. 77). ORF6 inhibits nuclear transport of transcription factors, including STAT1 and IRF3 (ref. 137), which control the expression of antiviral proteins and soluble pro-inflammatory mediators. Alpha has also enhanced expression of ORF9b (which inhibits signalling downstream of RNA sensing138) and N protein (which sequesters viral RNA to prevent activation of sensing mechanisms98), as well as de novo expression of N*/N.iORF3 – the amino-terminally truncated form of N protein, which displays some interferon antagonism but is expressed at low levels98. Increased levels of these proteins are likely the result of mutations in regulatory regions that modulate subgenomic RNA synthesis and protein expression. This highlights the importance of changes outside the spike protein in determining VOC properties, particularly in regulatory regions. Critically, mutations in the N protein Kozak sequence that are expected to influence expression of Alpha N protein and the overlapping protein ORF9b also appear in the dominant VOCs Delta and Omicron, but their full impact on innate immune antagonism in these VOCs remains to be determined. The relationship between SARS-CoV-2 and innate immunity is highly complex. For example, the viral antagonist ORF9b appears to be negatively regulated through phosphorylation by host kinases, suggesting its innate immune inhibition could be switched off at some point during infection, perhaps once host responses are triggered in infected cells133. Such an inflammatory switch mechanism, which essentially regulates host responses to infection, could drive symptomology and subsequent viral spread by altering cellular activation.
At least 15 SARS-CoV-2 proteins have been suggested to contribute to antagonism of innate responses to date (reviewed in detail elsewhere139,140). These proteins have typically been identified via reporter screens in which the SARS-CoV-2 protein of interest is expressed during a simulated in vitro innate immune response and its capacity to antagonize the response is evaluated. Such experiments do not typically provide mechanistic insight but are effective in discovering novel protein functions. Several of the VOCs exhibit amino acid changes in many of the proteins implicated as innate immune system antagonists, including NSP1, NSP3, NSP6, ORF3a, ORF6, ORF7b, ORF8 and N protein. Whether these coding mutations reflect adaptations to better antagonize human innate immunity is yet to be established in most cases. Moreover, the contribution of these proteins to the VOC phenotype is poorly understood, as this requires the painstaking generation of isogenic mutants using reverse genetics and assessment of their impact on replication, interferon production and sensitivity.
As well as reducing the induction of interferon, the Alpha and Omicron VOCs are more resistant to inhibition by its antiviral effects126,133–135,141. This has been best described as being related to spike adaptations which reduce sensitivity to interferon-induced transmembrane (IFITM) restriction factors76,134,142. The exact effect of IFITM proteins during SARS-CoV-2 replication is contentious, with some studies showing that IFITM proteins inhibit cellular entry of SARS-CoV-2, in a similar manner to influenza19,76,134,142, whereas others suggest that in some contexts they enhance infection, in a manner similar to that described for OC43 and HKU1 (refs. 76,134,143–145). The exact role of IFITM proteins is likely to be context specific, such as the particular entry pathway used by a particular virus in a given cell type or cell line, the use of live virus versus pseudovirus and the level of IFITM protein expressed. Members of the IFITM family are small, interferon-stimulated transmembrane proteins that are associated with different cell membranes; human IFITM1 is generally associated with cell surface membranes, whereas IFITM2 and IFITM3 are more localized to late and early endosomes, respectively76,146. The exact mechanism by which IFITM proteins impact viral entry is not fully resolved, but inhibition of viral glycoprotein fusion with host membranes is thought to be involved147,148. Several VOCs have been shown to have different degrees of sensitivity to IFITM protein inhibition or enhancement, quite often associated with specific entry pathways or furin cleavage phenotypes. For example, Omicron, which is more efficient at endosomal entry than early variants and other VOCs, appears to show greater inhibition (or in some cases enhancement) by endosomal IFITM proteins, although this does appear to be highly dependent on the cell system used32,76,145. This is possibly due to Omicron either having to compromise innate evasion due to adaptive immunity avoidance, particularly in the context of vaccine-induced spike antibodies, or having to adapt to use IFITM proteins as cofactors for entry.
It is not yet clear how enhanced antagonism of innate immunity might influence virus transmission. We hypothesize that the capacity of SARS-CoV-2 to spread efficiently is strongly linked to its ability to evade and antagonize innate immune responses in the first cells that encounter the virus in the airway. Indeed, the efficiency of infection events is expected to influence viral spread through the airway and thus the likelihood of seeding a productive infection at all. Type I interferon responses have been shown to be important in determining infection efficiency and outcome for other viruses, and have been well characterized for lentiviral infection of macaques149.
Finally, the fact that at present each VOC has evolved independently from an ancestral virus circulating early during the pandemic means that each VOC has taken a different mutational pathway to acquire distinct adaptations to humans. As a consequence, VOCs, or indeed other variants of SARS-CoV-2, could potentially recombine to unite independently encoded adaptations and thus phenotypic advantages from different variant genomes (Box 2).
Antigenic distance in determining transmission and fitness
Ever-changing population immunity creates a dynamic fitness landscape for virus variants because their fitness is as much dependent on acquired immunity as on their set of unique mutations. Although complexities such as the breadth and duration of immunity are important considerations150,151, the cumulative number of humans exposed to SARS-CoV-2, via infection and/or vaccination, results in a population much less susceptible to most circulating (and past) variants with an ever-decreasing number of immunologically naive, fully susceptible hosts (Fig. 4).
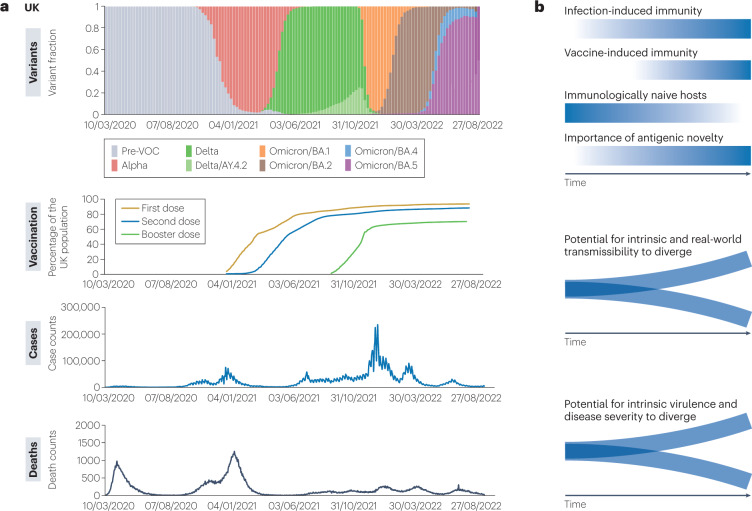
Fig. 4. Dominant SARS-CoV-2 variants, vaccinations, infections and deaths since early 2020 in the UK.
a, Waves of dominant variants in the UK (B.1, B.1.1.7/Alpha, B.1.617.2/Delta, AY.4.2/Delta and Omicron sublineages BA.1, BA.2, BA.4 and BA.5), proportion of the UK population with one or two doses and with one booster vaccination, number of COVID-19 cases and number of reported COVID-19-related deaths. Data from COG-UK Mutation Explorer192 and GOV.UK. b, Diagrammatic visualization of the dynamic relationship between variant transmissibility, antigenicity, virulence and fitness. As population immunity derived from infection and vaccination increases, the fraction of completely immunologically naive hosts declines (gradient blue lines). Consequently, the importance of antigenic novelty in determining variant fitness increases. Antigenic distance to previously circulating variants becomes an increasingly key determinant of variant transmissibility, increasing the potential for intrinsic and real-world transmissibility to diverge. Similarly, antigenic distance influences a variant’s potential to infect and cause disease in immune hosts, increasing the potential for a variant’s intrinsic virulence to diverge from its real-world clinical impact. VOC, variant of concern.
Early in the COVID-19 pandemic, when the fraction of naive hosts was greatest, there was little evolutionary benefit of antigenic novelty relative to wild-type variants. Instead, selection favoured variants capable of maximizing reproductive success through adaptation of intrinsic biological features such as the conformational change in the spike protein caused by D614G, the defining mutation of PANGO lineage B.1, or the enhanced furin cleavage phenotype coupled with increased ACE2 binding exhibited by Alpha27,29. As immunity in the host population increased, a variant’s antigenic novelty played an increasingly important role in its reproductive success, relative to intrinsic biological changes152. Subsequently, the Delta VOC became dominant globally, displacing previous variants in countries with partially immune populations with moderate-to-high vaccination coverage23,153. Virus neutralization data indicated moderate immune escape from neutralizing antibodies by the Delta VOC23,37–39, and vaccine effectiveness data indicated that antigenic novelty was not the primary driver of increased transmissibility40–42,154,155, indicating the high fitness of Delta was more the result of intrinsic viral properties such as optimization of spike furin cleavage22,23,69.
Compared with previous variants, Omicron showed an unprecedented degree of antigenic novelty31,33,34, arguably comparable to an influenza-like antigenic shift event51. The ‘shift’ here, the accumulation of mutations contributing to antigenic distance, probably occurs at least partly in the context of a chronic infection or chronic infections (Box 1). Comparison of transmission dynamics within vaccinated and unvaccinated households has indicated that immune escape was a critical component of the increased transmissibility of Omicron (BA.1) relative to Delta during their period of co-circulation86. The Omicron sublineage BA.2 has proved even more capable of infecting both unvaccinated and vaccinated individuals, potentially driven by immune evasiveness properties similar to those of BA.1 but with higher intrinsic transmissibility156. Most recently BA.4, BA.5, BA.2.75 and their sublineages have shown not only even greater evasion of immunity than pre-Omicron variants, but also escape from immunity generated from prior infection with Omicron, particularly BA.1 (refs. 46–49,157–159). This has been largely attributed to mutations at antigenically potent RBD positions, particularly L452R and F486V in the example of BA.4/BA.5 (ref. 160). Although there may remains the potential for further optimization of SARS-CoV-2 for transmission within humans, it now seems likely that the antigenic novelty and immune evasiveness of emerging variants will be the primary determinant of variant fitness and evolutionary success going forward. Consequently, understanding the complexities of cross-protection between variants is a major research priority.
Relative severity of SARS-CoV-2 variants
There is an imperative to understand how the virulence of SARS-CoV-2 variants might evolve in response to changing selection pressures. Pathogen virulence, alongside immunity, individual susceptibility, disease predisposition and other host factors, is a major contributor to disease severity and is defined in the evolutionary literature as the increased morbidity and mortality of individuals due to infection. Virulence does not necessarily decrease with time in the host population, rather, modelling data generally show a trade-off between transmission rate and virulence161,162. However, the predictability of virulence evolution is complicated by several mechanisms, including within-host competition, changing transmission routes and tropism, and interactions with the immune system162. For instance, repeated epidemic waves, characteristic of an antigenically evolving pathogen, can select for higher pathogen virulence163. Assessing the relative virulence of SARS-CoV-2 variants through disease severity in humans is challenging due to changing immune status and developments in medical interventions throughout the pandemic, although it is possible to compare disease severity of variants that infected the same population within a given period164. This approach suggests inconsistency in the direction of change in disease severity between successively dominant SARS-CoV-2 variants: the successful variants exhibited increased disease severity as Alpha replaced B.1.177, and as Delta replaced Alpha, correlating with relative changes in transmissibility164. By contrast, Omicron exhibited reduced disease severity in the period in which it co-existed with Delta, a decrease which appears to reflect a complex combination of factors including both higher infection rates of those with some degree of prior infection and an intrinsically lower virulence164–168. Proposed explanations for the lower disease severity of Omicron infections include reduced fusogenicity of the spike protein, leading to less tissue damage, and altered tropism restricted more to the upper respiratory tract (due to altered TMPRSS2 use)32,75,78,79. These previous studies compared the virulence of variants existing in the same population, although they could not determine the ‘intrinsic’ virulence of non-overlapping variants that circulated in populations with different immune statuses, such as Alpha and Omicron. Given that the immune status of an individual influences the severity of symptoms in addition to the likelihood of infection, the antigenic ‘distance’ between past exposure and a diverged variant has the potential to facilitate onset of disease in an otherwise protected individual, representing the potential for divergence between inherent potential to cause harm and actual virulence in infected people.
A complementary approach to measure severity of SARS-CoV-2 variants involves the use of animal models (reviewed in more depth previously169). Common animal models have included naive rodents, such as transgenic mice expressing human ACE2 under the control of the keratin 18 promoter (expressed abundantly in epithelial cells170) or hamsters, whose natively expressed ACE2 proteins are efficiently used by all current SARS-CoV-2 VOCs32,171. Pathogenicity in these rodent models is most commonly measured as a function of percentage weight loss, sometimes alongside use of survival curves and measures of pulmonary function172. These rodent models have largely recapitulated equivalent severity data from epidemiological studies in humans, such as Delta being more pathogenic than earlier variants and Omicron being less pathogenic than Delta32,69,75,173,174. However, the models have several limitations, as exemplified by recent epidemiological evidence from Hong Kong suggesting that Omicron sublineage BA.2 displays a disease severity similar to that of first-wave variants175, whereas rodent models generally show that Omicron is less severe than previous variants32,75,173,174,176. This inconsistency could potentially be explained by ongoing adaptation of SARS-CoV-2 to human hosts, resulting in concomitant adaptation away from rodents and other animal models32,80,177,178.
Disease severity following SARS-CoV-2 infection is correlated with several risk factors, including advanced age, being male and clinical comorbidities such as obesity and immunodeficiency, and several inflammatory markers179. As reviewed elsewhere, several recent genetic studies have focused on characteristics that might explain why some individuals are more susceptible to SARS-CoV-2 infections and others develop more severe symptoms180. However, there is an urgent need to combine these findings with data from specific variants and interventions (that is, vaccines, drugs and monoclonal antibodies) that might directly affect the observed phenotypes179. Furthermore, although the most common and easily measurable outcomes from acute infections are hospitalization or death, outcomes that are more difficult to measure also differ widely among variants, such as primary symptomatology181 or post-acute COVID-19 syndrome, although these likely differ greatly by prior immune status as well.
Conclusion
SARS-CoV-2 has been circulating for 3 years within the human population, infecting hundreds of millions of individuals. It remains, however, a relatively new human virus that continues to evolve and acquire adaptations to its new host species. Unprecedented SARS-CoV-2 genome sequence datasets generated globally have revealed evidence of beneficial mutations arising in real time and have guided laboratory experiments to better understand intrinsic properties of the interactions between the virus and the host. Although we have this extraordinary understanding of SARS-CoV-2 biology, virus fitness is highly dynamic and the ability of SARS-CoV-2 to infect, replicate within and spread among the human population has depended explicitly on the specific immune context at different periods of the pandemic. At present, Omicron is dominant worldwide, with infections being driven by emergent BA.2 and BA.5 sublineages. Although our understanding of SARS-CoV-2 is improving, virus evolution is inherently unpredictable, and a likely future scenario is the emergence of a new VOC that is antigenically and, potentially, phenotypically distinct from the early forms of Omicron. At the same time, population immunity against SARS-CoV-2 continues to accumulate and may well compensate in the case of a future variant emerging with higher severity, leading to milder acute disease.
All of the VOC progenitors have evolved from an ancestral pre-VOC virus present during the first wave of the pandemic, taking different but often convergent pathways to more efficiently infect and spread among humans, and to resist antibodies, T cell-driven immunity and innate immunity. The required adaptations, as we have discussed throughout this Review, are a mixture of changes to the host via intrinsic virus properties and escape of innate or adaptive immunity (Box 3). The prevailing hypothesis is that variants originate from chronic infections in immunocompromised individuals, in which the virus is able to establish a persistent infection due to impaired immune function67,113,182. This hypothesis explains the step change of seemingly rapid evolution seen before the emergence of new variants67,183,184. However, it should be noted that future variants will probably be directly derived from prior or contemporary VOCs, most recently exemplified by the spate of ‘second-generation’ Omicron variants derived from BA.2, such as BA.2.75, BJ.1 and BA.2.10.4 (ref. 185). While intralineage recombination serves as an opportunity for the virus to gain additive adaptations and phenotypic advantages from distantly related circulating variants, before the emergence of XBB, recombinants have exerted only a minor impact on the course of the pandemic at present (Box 2). Furthermore, although there is currently very limited evidence for the establishment of long-term circulation and evolution in animal reservoir species, intensive and active surveillance of susceptible species is needed as reverse zoonosis is being documented4,5,186. There are many countries with low sequencing capacity, or places with previously good surveillance that are decreasing or phasing out sequencing altogether. This is troublesome as a lack of genomic surveillance will mean future variants will be detected much later or could be circulating at low levels before eventual detection. There is, thus, a need for widespread and equitable surveillance coverage to rapidly detect potential new VOCs among these individuals and communities before they spread more widely.
Box 3 Fitness and antigenicity.
Darwinian fitness209, usually referred to as just ‘fitness’, is distinct from replicative fitness: the capacity of a virus to produce infectious progeny, measured experimentally in cultured cells, in tissue culture or within individual hosts210,211. By contrast, the broader definition of fitness is reproductive success so it is highly context dependent often varying over time and between locations. The fitness of a given severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant will depend on the changing immune profile of the host population in which it is circulating, and the success of an individual variant is relative to the properties of competing variants within the virus population and stochastic sampling processes. As immunity in the host population increases, the conditions can result in a previously highly transmissible variant, such as in an immune naive population, now being less fit relative to more evolved variants (Fig. 4b). Although intrinsic transmissibility is thus based on biological properties of the virus only, actual transmissibility depends on these properties in the context of population immunity, including the interplay of host immunity from past exposures, variant antigenicity and stochastic effects. Moreover, although it is possible that variants may differ in their relative antigenicity — the level of immune response they stimulate — variation in the degree of antigenic similarity to other variants is usually of greater impact. As immunity in the host population increases, the antigenic distance of a variant to previously circulating variants can determine its capacity to infect, replicate within and spread between hosts. The evolution of antigenic novelty therefore becomes the critical determinant of variant reproductive success and fitness.
Supplementary information
Acknowledgements
The COVID-19 Genomics UK Consortium is supported by funding from the UK Medical Research Council, which is part of UK Research and Innovation, the UK National Institute of Health and Care Research (grant code MC_PC_19027) and Genome Research Limited, operating as the Wellcome Sanger Institute. The authors acknowledge funding from the Medical Research Council (grant code MC_UU_12014/12) and UK Research and Innovation to the G2P-UK National Virology Consortium (MR/W005611/1).
Author contributions
A.M.C., T.P.P., L.G.T., W.T.H., G.J.T. and T.I.d.S. researched data for the article. D.L.R., A.M.C., T.P.P., J.H., G.J.T. and T.I.d.S. contributed substantially to discussion of the content. D.L.R., A.M.C., T.P.P., L.G.T., W.T.H., J.H., G.J.T. and T.I.d.S. wrote the article. D.L.R., A.M.C., T.P.P., L.G.T., W.T.H., J.H., S.J.P., W.S.B., G.J.T. and T.I.d.S. reviewed and/or edited the manuscript before submission.
Peer review
Peer review information
Nature Reviews Microbiology thanks Frank Kirchhoff; Olivier Schwartz; Wendy Burgers, who co-reviewed with Akiko Suzuki; and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
COG-UK Mutation Explorer: https://sars2.cvr.gla.ac.uk/cog-uk/
GISAID: https://gisaid.org
GOV.UK: https://www.gov.uk/coronavirus
These authors contributed equally: Alessandro M. Carabelli, Thomas P. Peacock.
A list of authors and their affiliations appears at the end of the paper.
A full list of members and their affiliations appears in the Supplementary information.
Contributor Information
David L. Robertson, Email: david.l.robertson@glasgow.ac.uk
COVID-19 Genomics UK Consortium:
Supplementary information
The online version contains supplementary material available at 10.1038/s41579-022-00841-7.
References
- 1.World Health Organization. https://www.who.int/news/item/05-05-2022-14.9-million-excess-deaths-were-associated-with-the-covid-19-pandemic-in-2020-and-2021WHO (2022).
- 2.Gorbalenya AE, et al. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peacock TP, Penrice-Randal R, Hiscox JA, Barclay WS. SARS-CoV-2 one year on: evidence for ongoing viral adaptation. J. Gen. Virol. 2021 doi: 10.1099/jgv.0.001584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu L, et al. Adaptation, spread and transmission of SARS-CoV-2 in farmed minks and associated humans in the Netherlands. Nat. Commun. 2021;12:6802. doi: 10.1038/s41467-021-27096-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marques AD, et al. Multiple introductions of SARS-CoV-2 Alpha and Delta variants into white-tailed deer in Pennsylvania. mBio. 2022;13:e02101-22. doi: 10.1128/mbio.02101-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hale VL, et al. SARS-CoV-2 infection in free-ranging white-tailed deer. Nature. 2022;602:481–486. doi: 10.1038/s41586-021-04353-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bashor L, et al. SARS-CoV-2 evolution in animals suggests mechanisms for rapid variant selection. Proc. Natl Acad. Sci. USA. 2021;118:e2105253118. doi: 10.1073/pnas.2105253118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacLean OA, Orton RJ, Singer JB, Robertson DL. No evidence for distinct types in the evolution of SARS-CoV-2. Virus Evolut. 2020 doi: 10.1093/ve/veaa034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Volz E, et al. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2021;184:64–75.e11. doi: 10.1016/j.cell.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Toole Á, Pybus OG, Abram ME, Kelly EJ, Rambaut A. Pango lineage designation and assignment using SARS-CoV-2 spike gene nucleotide sequences. BMC Genom. 2022;23:121–121. doi: 10.1186/s12864-022-08358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harvey WT, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woolhouse ME, Taylor LH, Haydon DT. Population biology of multihost pathogens. Science. 2001;292:1109–1112. doi: 10.1126/science.1059026. [DOI] [PubMed] [Google Scholar]
- 13.Parrish CR, et al. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol. Mol. Biol. Rev. 2008;72:457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacLean OA, et al. Natural selection in the evolution of SARS-CoV-2 in bats created a generalist virus and highly capable human pathogen. PLoS Biol. 2021;19:e3001115. doi: 10.1371/journal.pbio.3001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conceicao C, et al. The SARS-CoV-2 spike protein has a broad tropism for mammalian ACE2 proteins. PLoS Biol. 2020;18:e3001016. doi: 10.1371/journal.pbio.3001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delaune D, et al. A novel SARS-CoV-2 related coronavirus in bats from Cambodia. Nat. Commun. 2021;12:6563. doi: 10.1038/s41467-021-26809-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin XD, et al. Extensive diversity of coronaviruses in bats from China. Virology. 2017;507:1–10. doi: 10.1016/j.virol.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Starr TN, et al. ACE2 binding is an ancestral and evolvable trait of sarbecoviruses. Nature. 2022;603:913–918. doi: 10.1038/s41586-022-04464-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peacock TP, et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021;6:899–909. doi: 10.1038/s41564-021-00908-w. [DOI] [PubMed] [Google Scholar]
- 20.Hoffmann M, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Y, et al. A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry. Nat. Commun. 2021;12:961. doi: 10.1038/s41467-021-21213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peacock, T. P. et al. The SARS-CoV-2 variants associated with infections in India, B.1.617, show enhanced spike cleavage by furin. Preprint at bioRxiv10.1101/2021.05.28.446163 (2021).
- 23.Mlcochova P, et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599:114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lubinski B, et al. Spike protein cleavage-activation in the context of the SARS-CoV-2 P681R mutation: an analysis from its first appearance in lineage A.23.1 identified in Uganda. Microbiol. Spectr. 2022;10:e0151422. doi: 10.1128/spectrum.01514-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown, J. C. et al. Increased transmission of SARS-CoV-2 lineage B.1.1.7 (VOC 2020212/01) is not accounted for by a replicative advantage in primary airway cells or antibody escape. Preprint at bioRxiv10.1101/2021.02.24.432576 (2021).
- 26.Starr TN, et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, et al. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature. 2022;602:294–299. doi: 10.1038/s41586-021-04245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell F, et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Eurosurveillance. 2021;26:2100509. doi: 10.2807/1560-7917.ES.2021.26.24.2100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies NG, et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372:eabg3055. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, et al. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. Cell Rep. 2022;39:110829. doi: 10.1016/j.celrep.2022.110829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willett BJ, et al. SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nat. Microbiol. 2022 doi: 10.1038/s41564-022-01143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peacock, T. P. et al. The altered entry pathway and antigenic distance of the SARS-CoV-2 Omicron variant map to separate domains of spike protein. Preprint at bioRxiv10.1101/2021.12.31.474653 (2022).
- 33.Cao Y, et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou, J. et al. Omicron breakthrough infections in vaccinated or previously infected hamsters. Preprint at bioRxiv10.1101/2022.05.20.492779 (2022).
- 35.Thomson EC, et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184:1171–1187.e20. doi: 10.1016/j.cell.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsieh C-L, et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia-Beltran WF, et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383.e9. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newman J, et al. Neutralizing antibody activity against 21 SARS-CoV-2 variants in older adults vaccinated with BNT162b2. Nat. Microbiol. 2022;7:1180–1188. doi: 10.1038/s41564-022-01163-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis C, et al. Reduced neutralisation of the Delta (B.1.617.2) SARS-CoV-2 variant of concern following vaccination. PLoS Pathog. 2021;17:e1010022. doi: 10.1371/journal.ppat.1010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lopez Bernal J, et al. Effectiveness of Covid-19 vaccines against the B.1.617.2 (Delta) variant. N. Engl. J. Med. 2021;385:585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andrews N, et al. Covid-19 Vaccine Effectiveness against the Omicron (B.1.1.529) Variant. N. Engl. J. Med. 2022 doi: 10.1056/NEJMoa2119451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abu-Raddad LJ, Chemaitelly H, Butt AA. Effectiveness of the BNT162b2 Covid-19 vaccine against the B.1.1.7 and B.1.351 variants. N. Engl. J. Med. 2021;385:187–189. doi: 10.1056/NEJMc2104974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tegally H, et al. Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat. Med. 2022 doi: 10.1038/s41591-022-01911-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Viana R, et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022 doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cele S, et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willett, B. J. et al. Distinct antigenic properties of the SARS-CoV-2 Omicron lineages BA.4 and BA.5. Preprint at bioRxiv10.1101/2022.05.25.493397 (2022).
- 47.Tuekprakhon A, et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell. 2022 doi: 10.1016/j.cell.2022.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao Y, et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022 doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khan K, et al. Omicron BA.4/BA.5 escape neutralizing immunity elicited by BA.1 infection. Nat. Commun. 2022;13:4686. doi: 10.1038/s41467-022-32396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meng B, et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts tropism and fusogenicity. Nature. 2022 doi: 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simon-Loriere E, Schwartz O. Towards SARS-CoV-2 serotypes? Nat. Rev. Microbiol. 2022;20:187–188. doi: 10.1038/s41579-022-00708-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van der Straten K, et al. Antigenic cartography using sera from sequence-confirmed SARS-CoV-2 variants of concern infections reveals antigenic divergence of Omicron. Immunity. 2022 doi: 10.1016/j.immuni.2022.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Accorsi EK, et al. Association between 3 doses of mRNA COVID-19 vaccine and symptomatic infection caused by the SARS-CoV-2 Omicron and Delta variants. JAMA. 2022;327:639–651. doi: 10.1001/jama.2022.0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kherabi Y, Launay O, Luong Nguyen LB. COVID-19 vaccines against Omicron variant: real-world data on effectiveness. Viruses. 2022;14:2086. doi: 10.3390/v14102086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kirsebom FCM, et al. COVID-19 vaccine effectiveness against the omicron (BA.2) variant in England. Lancet Infect. Dis. 2022 doi: 10.1016/S1473-3099(22)00309-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ferdinands, J. M. et al. Waning 2-dose and 3-dose effectiveness of mRNA vaccines against COVID-19-associated emergency department and urgent care encounters and hospitalizations among adults during periods of Delta and Omicron variant predominance - VISION Network, 10 states, August 2021-January 2022. MMWR Morb. Mortal. Wkly Rep. 71, 255–263 (2022). [DOI] [PMC free article] [PubMed]
- 57.Collie S, Champion J, Moultrie H, Bekker L-G, Gray G. Effectiveness of BNT162b2 vaccine against Omicron variant in South Africa. N. Engl. J. Med. 2021;386:494–496. doi: 10.1056/NEJMc2119270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Higdon MM, et al. Duration of effectiveness of vaccination against COVID-19 caused by the omicron variant. Lancet Infect. Dis. 2022;22:1114–1116. doi: 10.1016/S1473-3099(22)00409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fang Z, et al. Omicron-specific mRNA vaccination alone and as a heterologous booster against SARS-CoV-2. Nat. Commun. 2022;13:3250. doi: 10.1038/s41467-022-30878-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alu A, et al. Intranasal COVID-19 vaccines: from bench to bed. eBioMedicine. 2022;76:103841. doi: 10.1016/j.ebiom.2022.103841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dolgin E. Pan-coronavirus vaccine pipeline takes form. Nat. Rev. Drug Discov. 2022;21:324–326. doi: 10.1038/d41573-022-00074-6. [DOI] [PubMed] [Google Scholar]
- 62.Eguia RT, et al. A human coronavirus evolves antigenically to escape antibody immunity. PLoS Pathog. 2021;17:e1009453. doi: 10.1371/journal.ppat.1009453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edridge AWD, et al. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. 2020;26:1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- 64.Hoffmann M, Kleine-Weber H, Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson BA, et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature. 2021;591:293–299. doi: 10.1038/s41586-021-03237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Misumi Y, et al. Functional expression of furin demonstrating its intracellular localization and endoprotease activity for processing of proalbumin and complement pro-C3. J. Biol. Chem. 1991;266:16954–16959. doi: 10.1016/S0021-9258(18)55396-5. [DOI] [PubMed] [Google Scholar]
- 67.Hill V, et al. The origins and molecular evolution of SARS-CoV-2 lineage B.1.1.7 in the UK. Virus Evol. 2022 doi: 10.1093/ve/veac080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Laiton-Donato K, et al. Characterization of the emerging B.1.621 variant of interest of SARS-CoV-2. Infect. Genet. Evol. 2021;95:105038. doi: 10.1016/j.meegid.2021.105038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saito A, et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature. 2022;602:300–306. doi: 10.1038/s41586-021-04266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lubinski, B., Jaimes, J. A. & Whittaker, G. R. Intrinsic furin-mediated cleavability of the spike S1/S2 site from SARS-CoV-2 variant B.1.1.529 (Omicron). Preprint at bioRxiv10.1101/2022.04.20.488969 (2022).
- 71.Lubinski B, et al. Functional evaluation of the P681H mutation on the proteolytic activation of the SARS-CoV-2 variant B.1.1.7 (Alpha) spike. iScience. 2022;25:103589. doi: 10.1016/j.isci.2021.103589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang L, et al. Furin cleavage of the SARS-CoV-2 spike is modulated by O-glycosylation. Proc. Natl Acad. Sci. USA. 2021;118:e2109905118. doi: 10.1073/pnas.2109905118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sanda M, Morrison L, Goldman R. N- and O-glycosylation of the SARS-CoV-2 spike protein. Anal. Chem. 2021;93:2003–2009. doi: 10.1021/acs.analchem.0c03173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gonzalez-Rodriguez, E. et al. O-linked sialoglycans modulate the proteolysis of SARS-CoV-2 spike and contribute to the mutational trajectory in variants of concern. Preprint at bioRxiv10.1101/2022.09.15.508093 (2022). [DOI] [PMC free article] [PubMed]
- 75.Suzuki R, et al. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature. 2022 doi: 10.1038/s41586-022-04462-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mesner, D. et al. SARS-CoV-2 Spike evolution influences GBP and IFITM sensitivity. Preprint at bioRxiv10.1101/2022.03.07.481785 (2022).
- 77.Reuschl, A.-K. et al. Enhanced innate immune suppression by SARS-CoV-2 Omicron subvariants BA.4 and BA.5. Preprint at bioRxiv10.1101/2022.07.12.499603 (2022).
- 78.Hui KPY, et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature. 2022 doi: 10.1038/s41586-022-04479-6. [DOI] [PubMed] [Google Scholar]
- 79.Aggarwal A, et al. Platform for isolation and characterization of SARS-CoV-2 variants enables rapid characterization of Omicron in Australia. Nat. Microbiol. 2022;7:896–908. doi: 10.1038/s41564-022-01135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barut, T. et al. The spike gene is a major determinant for the SARS-CoV-2 Omicron-BA.1 phenotype. Preprint at bioRxiv10.1101/2022.04.28.489537 (2022). [DOI] [PMC free article] [PubMed]
- 81.Bentley, E. G. et al. SARS-CoV-2 Omicron-B.1.1.529 Variant leads to less severe disease than Pango B and Delta variants strains in a mouse model of severe COVID-19. Preprint at bioRxiv10.1101/2021.12.26.474085 (2021).
- 82.Kimura, I. et al. SARS-CoV-2 spike S375F mutation characterizes the Omicron BA.1 variant. Preprint at bioRxiv10.1101/2022.04.03.486864 (2022). [DOI] [PMC free article] [PubMed]
- 83.Yamamoto, M. et al. SARS-CoV-2 Omicron spike H655Y mutation is responsible for enhancement of the endosomal entry pathway and reduction of cell surface entry pathways. Preprint at bioRxiv10.1101/2022.03.21.485084 (2022).
- 84.Kimura I, et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants including BA.4 and BA.5. Cell. 2022 doi: 10.1016/j.cell.2022.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Allen, H. et al. Comparative transmission of SARS-CoV-2 Omicron (B.1.1.529) and Delta (B.1.617.2) variants and the impact of vaccination: national cohort study, England. Preprint at medRxiv10.1101/2022.02.15.22271001 (2022). [DOI] [PMC free article] [PubMed]
- 86.Lyngse FP, et al. Household transmission of the SARS-CoV-2 Omicron variant in Denmark. Nat. Commun. 2022;13:5573. doi: 10.1038/s41467-022-33328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yu S, et al. SARS-CoV-2 spike engagement of ACE2 primes S2′ site cleavage and fusion initiation. Proc. Natl Acad. Sci. USA. 2022;119:e2111199119. doi: 10.1073/pnas.2111199119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Millet JK, Whittaker GR. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl Acad. Sci. USA. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Belouzard S, Chu VC, Whittaker GR. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl Acad. Sci. USA. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Benton DJ, et al. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 2020;588:327–330. doi: 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bestle D, et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance. 2020;3:e202000786. doi: 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Qing E, et al. Inter-domain communication in SARS-CoV-2 spike proteins controls protease-triggered cell entry. Cell Rep. 2022;39:110786. doi: 10.1016/j.celrep.2022.110786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Meng B, et al. SARS-CoV-2 spike N-terminal domain modulates TMPRSS2-dependent viral entry and fusogenicity. Cell Rep. 2022;40:111220. doi: 10.1016/j.celrep.2022.111220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cantoni D, et al. Evolutionary remodelling of N-terminal domain loops fine-tunes SARS-CoV-2 spike. EMBO Rep. 2022;23:e54322. doi: 10.15252/embr.202154322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Syed AM, et al. Rapid assessment of SARS-CoV-2-evolved variants using virus-like particles. Science. 2021;374:1626–1632. doi: 10.1126/science.abl6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu H, et al. Nucleocapsid mutations R203K/G204R increase the infectivity, fitness, and virulence of SARS-CoV-2. Cell Host Microbe. 2021;29:1788–1801.e6. doi: 10.1016/j.chom.2021.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Johnson BA, et al. Nucleocapsid mutations in SARS-CoV-2 augment replication and pathogenesis. PLoS Pathog. 2022;18:e1010627. doi: 10.1371/journal.ppat.1010627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mears, H. V. et al. Emergence of new subgenomic mRNAs in SARS-CoV-2. Preprint at bioRxiv10.1101/2022.04.20.488895 (2022).
- 99.Leary S, et al. Generation of a novel SARS-CoV-2 sub-genomic RNA due to the R203K/G204R variant in nucleocapsid: homologous recombination has potential to change SARS-CoV-2 at both protein and RNA level. Pathog. Immun. 2021;6:27–49. doi: 10.20411/pai.v6i2.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Syed AM, et al. Omicron mutations enhance infectivity and reduce antibody neutralization of SARS-CoV-2 virus-like particles. Proc. Natl Acad. Sci. USA. 2022;119:e2200592119. doi: 10.1073/pnas.2200592119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xia, B. et al. Why SARS-CoV-2 Omicron variant is milder? A single high-frequency mutation of structural envelope protein matters. Preprint at bioRxiv10.1101/2022.02.01.478647 (2022).
- 103.Ricciardi S, et al. The role of NSP6 in the biogenesis of the SARS-CoV-2 replication organelle. Nature. 2022 doi: 10.1038/s41586-022-04835-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Peng Y, et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Moss P. The T cell immune response against SARS-CoV-2. Nat. Immunol. 2022;23:186–193. doi: 10.1038/s41590-021-01122-w. [DOI] [PubMed] [Google Scholar]
- 106.Feng S, et al. Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:2032–2040. doi: 10.1038/s41591-021-01540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tan AT, et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021;34:108728. doi: 10.1016/j.celrep.2021.108728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bange EM, et al. CD8+ T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat. Med. 2021;27:1280–1289. doi: 10.1038/s41591-021-01386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Oberhardt V, et al. Rapid and stable mobilization of CD8+ T cells by SARS-CoV-2 mRNA vaccine. Nature. 2021;597:268–273. doi: 10.1038/s41586-021-03841-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tarke A, et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021;2:100204. doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Agerer B, et al. SARS-CoV-2 mutations in MHC-I-restricted epitopes evade CD8+ T cell responses. Sci. Immunol. 2021;6:eabg6461. doi: 10.1126/sciimmunol.abg6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Goonetilleke N, et al. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J. Exp. Med. 2009;206:1253–1272. doi: 10.1084/jem.20090365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wilkinson SAJ, et al. Recurrent SARS-CoV-2 mutations in immunodeficient patients. Virus Evol. 2022 doi: 10.1093/ve/veac050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stanevich, O. et al. SARS-CoV-2 escape from cytotoxic T cells during long-term COVID-19. Preprint at Res. Sq. 10.21203/rs.3.rs-750741/v1 (2021). [DOI] [PMC free article] [PubMed]
- 115.de Silva TI, et al. The impact of viral mutations on recognition by SARS-CoV-2 specific T cells. iScience. 2021;24:103353. doi: 10.1016/j.isci.2021.103353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Motozono C, et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe. 2021;29:1124–1136.e11. doi: 10.1016/j.chom.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dolton G, et al. Emergence of immune escape at dominant SARS-CoV-2 killer T cell epitope. Cell. 2022;185:2936–2951.e19. doi: 10.1016/j.cell.2022.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Riou C, et al. Escape from recognition of SARS-CoV-2 variant spike epitopes but overall preservation of T cell immunity. Sci. Transl Med. 2022;14:eabj6824. doi: 10.1126/scitranslmed.abj6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Reynolds CJ, et al. Prior SARS-CoV-2 infection rescues B and T cell responses to variants after first vaccine dose. Science. 2021;372:1418–1423. doi: 10.1126/science.abh1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tarke A, et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell. 2022;185:847–859.e11. doi: 10.1016/j.cell.2022.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.UKHSA. COVID-19 Vaccine Surveillance Report (UKHSA, 2022).
- 122.Naranbhai V, et al. T cell reactivity to the SARS-CoV-2 Omicron variant is preserved in most but not all individuals. Cell. 2022;185:1041–1051.e6. doi: 10.1016/j.cell.2022.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wellington, D. et al. SARS-CoV-2 mutations affect proteasome processing to alter CD8+ T cell responses. Preprint at bioRxiv10.1101/2022.04.08.487623 (2022).
- 124.Ladell K, et al. A molecular basis for the control of preimmune escape variants by HIV-specific CD8+ T cells. Immunity. 2013;38:425–436. doi: 10.1016/j.immuni.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 125.Zhang Y, et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-I. Proc. Natl Acad. Sci. USA. 2021;118:e2024202118. doi: 10.1073/pnas.2024202118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Moriyama, M., Lucas, C., Monteiro, V. S., Yale SARS-CoV-2 Genomic Surveillance Initiative & Iwasaki, A. SARS-CoV-2 Omicron subvariants evolved to promote further escape from MHC-I recognition. Preprint at bioRxiv10.1101/2022.05.04.490614 (2022).
- 127.Yoo J-S, et al. SARS-CoV-2 inhibits induction of the MHC class I pathway by targeting the STAT1-IRF1-NLRC5 axis. Nat. Commun. 2021;12:6602. doi: 10.1038/s41467-021-26910-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Arshad, N. et al. SARS-CoV-2 accessory proteins ORF7a and ORF3a use distinct mechanisms to downregulate MHC-I surface expression. Preprint at bioRxiv10.1101/2022.05.17.492198 (2022).
- 129.Zhang, F. et al. Inhibition of major histocompatibility complex-I antigen presentation by sarbecovirus ORF7a proteins. Preprint at bioRxiv10.1101/2022.05.25.493467 (2022). [DOI] [PMC free article] [PubMed]
- 130.Stewart, H. et al. Tetherin antagonism by SARS-CoV-2 enhances virus release: multiple mechanisms including ORF3a-mediated defective retrograde traffic. Preprint at bioRxiv10.1101/2021.01.06.425396 (2021).
- 131.Rambaut, A. et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Virological.orghttps://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 (2020).
- 132.Mandl JN, et al. Reservoir host immune responses to emerging zoonotic viruses. Cell. 2015;160:20–35. doi: 10.1016/j.cell.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Thorne LG, et al. Evolution of enhanced innate immune evasion by SARS-CoV-2. Nature. 2022;602:487–495. doi: 10.1038/s41586-021-04352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lista, M. J. et al. The P681H mutation in the spike glycoprotein confers type I interferon resistance in the SARS-CoV-2 alpha (B.1.1.7) variant. Preprint at bioRxiv10.1101/2021.11.09.467693 (2021).
- 135.Guo K, et al. Interferon resistance of emerging SARS-CoV-2 variants. Proc. Natl Acad. Sci. USA. 2022;119:e2203760119. doi: 10.1073/pnas.2203760119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bouhaddoum, M. et al. Global landscape of the host response to SARS-CoV-2 variants reveals viral evolutionary trajectories. Preprint at bioRxiv10.1101/2022.10.19.512927 (2022).
- 137.Hall R, et al. SARS-CoV-2 ORF6 disrupts innate immune signalling by inhibiting cellular mRNA export. PLoS Pathog. 2022;18:e1010349. doi: 10.1371/journal.ppat.1010349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jiang H-w, et al. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell. Mol. Immunol. 2020;17:998–1000. doi: 10.1038/s41423-020-0514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Beyer DK, Forero A. Mechanisms of antiviral immune evasion of SARS-CoV-2. J. Mol. Biol. 2022;434:167265. doi: 10.1016/j.jmb.2021.167265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Xu D, Biswal M, Neal A, Hai R. Devil’s tools: SARS-CoV-2 antagonists against innate immunity. Curr. Res. Virol. Sci. 2021;2:100013. doi: 10.1016/j.crviro.2021.100013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shalamova L, et al. Omicron variant of SARS-CoV-2 exhibits an increased resilience to the antiviral type I interferon response. PNAS Nexus. 2022 doi: 10.1093/pnasnexus/pgac067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Winstone H, et al. The polybasic cleavage site in SARS-CoV-2 spike modulates viral sensitivity to type I interferon and IFITM2. J. Virol. 2021 doi: 10.1128/jvi.02422-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhao X, et al. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc. Natl Acad. Sci. USA. 2014;111:6756–6761. doi: 10.1073/pnas.1320856111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Prelli Bozzo C, et al. IFITM proteins promote SARS-CoV-2 infection and are targets for virus inhibition in vitro. Nat. Commun. 2021;12:4584. doi: 10.1038/s41467-021-24817-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nchioua R, et al. SARS-CoV-2 variants of concern hijack IFITM2 for efficient replication in human lung cells. J. Virol. 2022;96:e00594-22. doi: 10.1128/jvi.00594-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 2013;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Li K, et al. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 2013;9:e1003124. doi: 10.1371/journal.ppat.1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Klein, S. et al. IFITM3 blocks viral entry by sorting lipids and stabilizing hemifusion. Preprint at bioRxiv10.1101/2022.09.01.506185 (2022).
- 149.Sandler NG, Douek DC. Microbial translocation in HIV infection: causes, consequences and treatment opportunities. Nat. Rev. Microbiol. 2012;10:655–666. doi: 10.1038/nrmicro2848. [DOI] [PubMed] [Google Scholar]
- 150.Wang Z, et al. Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature. 2021;595:426–431. doi: 10.1038/s41586-021-03696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Levin EG, et al. Waning immune humoral response to BNT162b2 Covid-19 vaccine over 6 months. N. Engl. J. Med. 2021;385:e84. doi: 10.1056/NEJMoa2114583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Paton RS, Overton CE, Ward T. The rapid replacement of the Delta variant by Omicron (B.1.1.529) in England. Sci. Transl Med. 2022;14:eabo5395. doi: 10.1126/scitranslmed.abo5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Earnest R, et al. Comparative transmissibility of SARS-CoV-2 variants Delta and Alpha in New England, USA. Cell Rep. Med. 2022;3:100583. doi: 10.1016/j.xcrm.2022.100583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tang P, et al. BNT162b2 and mRNA-1273 COVID-19 vaccine effectiveness against the SARS-CoV-2 Delta variant in Qatar. Nat. Med. 2021;27:2136–2143. doi: 10.1038/s41591-021-01583-4. [DOI] [PubMed] [Google Scholar]
- 155.Bruxvoort KJ, et al. Effectiveness of mRNA-1273 against delta, mu, and other emerging variants of SARS-CoV-2: test negative case-control study. BMJ. 2021;375:e068848. doi: 10.1136/bmj-2021-068848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lyngse, F. P. et al. Transmission of SARS-CoV-2 Omicron VOC subvariants BA.1 and BA.2: evidence from Danish households. Preprint at medRxiv10.1101/2022.01.28.22270044 (2022).
- 157.Cao, Y. et al. Imprinted SARS-CoV-2 humoral immunity induces converging Omicron RBD evolution. Preprint at bioRxiv10.1101/2022.09.15.507787 (2022).
- 158.Sheward, D. J. et al. Omicron sublineage BA.2.75.2 exhibits extensive escape from neutralising antibodies. Preprint at bioRxiv10.1101/2022.09.16.508299 (2022). [DOI] [PMC free article] [PubMed]
- 159.Sheward DJ, et al. Evasion of neutralising antibodies by omicron sublineage BA.2.75. Lancet Infect. Dis. 2022 doi: 10.1016/S1473-3099(22)00524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang Q, et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature. 2022;608:603–608. doi: 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Anderson RM, May RM. Coevolution of hosts and parasites. Parasitology. 1982;85:411–426. doi: 10.1017/S0031182000055360. [DOI] [PubMed] [Google Scholar]
- 162.Alizon S, Hurford A, Mideo N, Van Baalen M. Virulence evolution and the trade-off hypothesis: history, current state of affairs and the future. J. Evol. Biol. 2009;22:245–259. doi: 10.1111/j.1420-9101.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- 163.Sasaki A, Lion S, Boots M. Antigenic escape selects for the evolution of higher pathogen transmission and virulence. Nat. Ecol. Evol. 2022;6:51–62. doi: 10.1038/s41559-021-01603-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pascall, D. J. et al. Inconsistent directions of change in case severity across successive SARS-CoV-2 variant waves suggests an unpredictable future. Preprint at medRxiv10.1101/2022.03.24.22272915 (2022).
- 165.Wolter N, et al. Early assessment of the clinical severity of the SARS-CoV-2 omicron variant in South Africa: a data linkage study. Lancet. 2022;399:437–446. doi: 10.1016/S0140-6736(22)00017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bager P, et al. Risk of hospitalisation associated with infection with SARS-CoV-2 omicron variant versus delta variant in Denmark: an observational cohort study. Lancet Infect. Dis. 2022 doi: 10.1016/s1473-3099(22)00154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Nyberg T, et al. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: a cohort study. Lancet. 2022 doi: 10.1016/S0140-6736(22)00462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lewnard JA, et al. Clinical outcomes associated with SARS-CoV-2 Omicron (B.1.1.529) variant and BA.1/BA.1.1 or BA.2 subvariant infection in southern California. Nat. Med. 2022 doi: 10.1038/s41591-022-01887-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lamers MM, Haagmans BL. SARS-CoV-2 pathogenesis. Nat. Rev. Microbiol. 2022;20:270–284. doi: 10.1038/s41579-022-00713-0. [DOI] [PubMed] [Google Scholar]
- 170.Dong W, et al. The K18-human ACE2 transgenic mouse model recapitulates non-severe and severe COVID-19 in response to an infectious dose of the SARS-CoV-2 virus. J. Virol. 2022;96:e0096421. doi: 10.1128/JVI.00964-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Thakur N, et al. SARS-CoV-2 variants of concern alpha, beta, gamma and delta have extended ACE2 receptor host ranges. J. Gen. Virol. 2022 doi: 10.1099/jgv.0.001735. [DOI] [PubMed] [Google Scholar]
- 172.Menachery VD, Gralinski LE, Baric RS, Ferris MT. New metrics for evaluating viral respiratory pathogenesis. PLoS ONE. 2015;10:e0131451. doi: 10.1371/journal.pone.0131451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Uraki R, et al. Characterization and antiviral susceptibility of SARS-CoV-2 Omicron/BA.2. Nature. 2022 doi: 10.1038/s41586-022-04856-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Halfmann PJ, et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 2022;603:687–692. doi: 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Mefsin, Y. et al. Epidemiology of infections with SARS-CoV-2 Omicron BA.2 variant in Hong Kong, January-March 2022. Preprint at medRxiv10.1101/2022.04.07.22273595 (2022). [DOI] [PMC free article] [PubMed]
- 176.Shuai H, et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature. 2022;603:693–699. doi: 10.1038/s41586-022-04442-5. [DOI] [PubMed] [Google Scholar]
- 177.van Doremalen, N. et al. SARS-CoV-2 Omicron BA.1 and BA.2 are attenuated in rhesus macaques as compared to Delta. Preprint at bioRxiv10.1101/2022.08.01.502390 (2022). [DOI] [PMC free article] [PubMed]
- 178.Su W, et al. Omicron BA.1 and BA.2 sub-lineages show reduced pathogenicity and transmission potential than the early SARS-CoV-2 D614G variant in Syrian hamsters. J. Infect. Dis. 2022 doi: 10.1093/infdis/jiac276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hu B, Guo H, Zhou P, Shi Z-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Niemi MEK, Daly MJ, Ganna A. The human genetic epidemiology of COVID-19. Nat. Rev. Genet. 2022 doi: 10.1038/s41576-022-00478-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Whitaker, M. et al. Variant-specific symptoms of COVID-19 among 1,542,510 people in England. Preprint at medRxiv10.1101/2022.05.21.22275368 (2022).
- 182.Gonzalez-Reiche, A. S. et al. SARS-CoV-2 variants in the making: Sequential intrahost evolution and forward transmissions in the context of persistent infections. Preprint at medRxiv10.1101/2022.05.25.22275533 (2022).
- 183.Ghafari M, Liu Q, Dhillon A, Katzourakis A, Weissman DB. Investigating the evolutionary origins of the first three SARS-CoV-2 variants of concern. Front. Virol. 2022 doi: 10.3389/fviro.2022.942555. [DOI] [Google Scholar]
- 184.Harari S, et al. Drivers of adaptive evolution during chronic SARS-CoV-2 infections. Nat. Med. 2022 doi: 10.1038/s41591-022-01882-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Roemer, C. Variant report 2022-08-31. https://github.com/neherlab/SARS-CoV-2_variant-reports/tree/main/reports (2022).
- 186.Pickering, B. et al. Highly divergent white-tailed deer SARS-CoV-2 with potential deer-to-human transmission. Preprint at bioRxiv10.1101/2022.02.22.481551 (2022).
- 187.McCallum M, et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347.e16. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Barnes CO, et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Greaney AJ, et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29:463–476.e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lan J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 191.Gao Y, et al. Ancestral SARS-CoV-2-specific T cells cross-recognize the Omicron variant. Nat. Med. 2022;28:472–476. doi: 10.1038/s41591-022-01700-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wright DW, et al. Tracking SARS-CoV-2 mutations and variants through the COG-UK-Mutation Explorer. Virus Evol. 2022 doi: 10.1093/ve/veac023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Faria NR, et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372:815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Tegally H, et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 195.Dhar MS, et al. Genomic characterization and epidemiology of an emerging SARS-CoV-2 variant in Delhi, India. Science. 2021;374:995–999. doi: 10.1126/science.abj9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.McCrone JT, et al. Context-specific emergence and growth of the SARS-CoV-2 Delta variant. Nature. 2022 doi: 10.1038/s41586-022-05200-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.UKHSA. SARS-CoV-2 Variants of Concern and Variants Under Investigation in England: Technical Briefing 25 (UKHSA, 2021).
- 198.Mallapaty S. Where did Omicron come from? Three key theories. Nature. 2022;602:26–28. doi: 10.1038/d41586-022-00215-2. [DOI] [PubMed] [Google Scholar]
- 199.Choi B, et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N. Engl. J. Med. 2020;383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chaguza, C. et al. Accelerated SARS-CoV-2 intrahost evolution leading to distinct genotypes during chronic infection. Preprint at medRxiv10.1101/2022.06.29.22276868 (2022). [DOI] [PMC free article] [PubMed]
- 201.Kemp SA, et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592:277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Jackson B, et al. Generation and transmission of interlineage recombinants in the SARS-CoV-2 pandemic. Cell. 2021;184:5179–5188.e8. doi: 10.1016/j.cell.2021.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Sekizuka T, et al. Genome recombination between Delta and Alpha variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Jpn J. Infect. Dis. 2022 doi: 10.7883/yoken.JJID.2021.844. [DOI] [PubMed] [Google Scholar]
- 204.Simon-Loriere, E. et al. Rapid characterization of a Delta-Omicron SARS-CoV-2 recombinant detected in Europe. Preprint at Res. Sq. 10.21203/rs.3.rs-1502293/v1 (2022).
- 205.UKHSA. SARS-CoV-2 Variants of Concern and Variants Under Investigation in England: Technical Briefing 40 (UKHSA, 2022).
- 206.Network for Surveillance-South Africa (NGS-SA). SARS-CoV-2 Sequencing Update 15 July 2022 (NGS-SA, 2022).
- 207.Cao, Y. et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Preprint at bioRxiv10.1101/2022.09.15.507787 (2022). [DOI] [PMC free article] [PubMed]
- 208.Lytras S, et al. Exploring the natural origins of SARS-CoV-2 in the light of recombination. Genome Biol. Evol. 2022 doi: 10.1093/gbe/evac018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Demetrius L, Ziehe M. Darwinian fitness. Theor. Popul. Biol. 2007;72:323–345. doi: 10.1016/j.tpb.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 210.Domingo E, Holland JJ. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 211.Wargo AR, Kurath G. Viral fitness: definitions, measurement, and current insights. Curr. Opin. Virol. 2012;2:538–545. doi: 10.1016/j.coviro.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.