Abstract
In humans, the gut microbiome is strongly implicated in numerous sex-specific physiological processes and diseases. Given this, it is important to understand how sex differentiation of the gut microbiome occurs and how these differences contribute to host health and disease. While it is commonly believed that the gut microbiome stabilizes after 3 years of age, our review of the literature found considerable evidence that the gut microbiome continues to mature during and after puberty in a sex-dependent manner. We also review the intriguing, though sparse, literature on potential mechanisms by which host sex may influence the gut microbiome, and vice versa, via sex steroids, bile acids, and the immune system. We conclude that the evidence for the existence of a sex-specific gut microbiome is strong but that there is a dearth of research on how host-microbe interactions lead to this differentiation. Finally, we discuss the types of future studies needed to understand the processes driving the maturation of sex-specific microbial communities and the interplay between gut microbiota, host sex, and human health.
Keywords: steroid hormones, bile acid, puberty, gut microbiome
In Brief
Sex differences in the gut microbiome may impact multiple aspects of human health and disease. We review the evidence for microbial sex differences in puberty and adulthood and discuss potential mechanisms driving differentiation of the sex-specific gut microbiome.
Introduction
Next-generation sequencing (NGS) has revolutionized our understanding of the human gut microbiome. The combination of NGS, multiplex-library construction, and bioinformatics has revealed gut microbial diversity to be temporally dynamic and highly variable among both populations and individuals. Moreover, the gut microbial community collectively contains two orders of magnitude more genes than the human genome (Zhu, Wang, and Li 2010; Falony et al. 2016; Huttenhower et al. 2012; Levy et al. 2020). Variation in abiotic (pH, oxygen levels, nutrition) and biotic factors (immune surveillance, signaling molecules) create numerous niches along the alimentary tract that select for highly differentiated communities of microorganisms (Barlow, Bogatyrev, and Ismagilov 2020). Conversely, the microbes in the mammalian gut influence a broad range of host physiological parameters ranging from metabolism (Acharya et al. 2019; Baars et al. 2018; Colldén et al. 2019), to immunity (Vatanen et al. 2016; Candon et al. 2015; Markle et al. 2014), to the nervous system (Desbonnet et al. 2015; Heinzel et al. 2021; Murray et al. 2020), and to the reproductive system (Arroyo et al. 2019; Acharya et al. 2019).
The human gut microbiome develops rapidly after birth, when the alimentary canal is seeded by the mother and environmental microbiomes (Breitbart et al. 2008; Helve et al. 2019; Bäckhed et al. 2015; Ferretti et al. 2018; Dominguez-Bello et al. 2016; Song et al. 2021). Studies showed that the infant gut microbiome rapidly increases in diversity and begins to resemble adult gut microbiome taxonomic composition and functional pathways by 2 to 3 years of age (Koenig et al. 2011; Bokulich et al. 2016; Yatsunenko et al. 2012; Kurokawa et al. 2007). Based on these initial studies, the gut microbiome of young children was thought to be fully mature by age three. However, the gut microbiomes of children can be readily distinguished from adults (Agans et al. 2011; Yatsunenko et al. 2012; Ringel-Kulka et al. 2013; Hollister et al. 2015; Dominianni et al. 2015).
Many important and profound changes in human physiology occur during puberty, which may also be the case with the gut microbiome. In humans, sex-specific processes have been linked to the gut microbiome, including cognition and anxiety (Desbonnet et al. 2015; Murray et al. 2019), liver metabolism (Bhat et al. 2021; van Keulen et al. 2020), immunity (Wilharm et al. 2021; Shepherd et al. 2021), and menstrual regularity (Wang, Zhang, et al. 2020). In addition, diseases which emerge during and after puberty have been linked to changes in gut microbial composition, often in a sex-specific manner. These include inflammatory bowel disease (IBD) (Franzosa et al. 2019; Ferguson and Sedgwick 1994), type I diabetes (T1D) (Markle et al. 2013; Yurkovetskiy et al. 2013), lupus (Young et al. 2014; He et al. 2016), obesity (Joriba et al. 2019), polycystic ovary syndrome (PCOS) (Torres et al. 2018; van Hooff et al. 1999; Jobira et al. 2020), and endometriosis (Svensson et al. 2021; Yuan et al. 2018). In addition to playing a role in the etiology and pathophysiology of various diseases, emerging differences in gut microbiota during puberty may lead to variable responses to drug treatments. Understanding the development of sex differences in the gut microbiome may be critical for designing and optimizing therapeutic approaches to treat gut microbiome-related diseases. Given the number of sex-specific diseases related to the gut and the potential for these sex differences to impact drug therapies, it is worth reviewing what is currently known about sex differentiation of the gut microbiome and the processes that drive maturation of the sex-specific gut microbial community.
Sex Differences and the Gut Microbiome
A survey of the literature investigating the relationship of sex and the gut microbiome strongly supports the hypothesis that human and rodent gut microbiomes differentiate during puberty and that this differentiation results in sex-specific communities by adulthood. While early data from the Human Microbiome Project (HMP) did not detect sex differences in the gut microbiome (Huttenhower et al. 2012), subsequent studies have consistently identified distinct gut microbial communities between males and females. Numerous studies in both mice and humans have found sex differences in gut microbiome beta-diversity (between-sample taxonomic composition) (Fig 1) (Takagi et al. 2019; Markle et al. 2013; Sinha et al. 2019; Dominianni et al. 2015; Mayneris-Perxachs et al. 2020; Org et al. 2016; Sheng et al. 2017; Ding and Schloss 2014; Li et al. 2008; Falony et al. 2016; Gao et al. 2018; Borgo et al. 2018; Santos-Marcos et al. 2018; Mueller et al. 2006; Cui et al. 2021; Peters et al. 2022). Multiple studies have also shown that alpha diversity (within-sample taxonomic diversity) tends to be higher in adult females than adult males, though these differences are less pronounced in older adults (Takagi et al. 2019; de la Cuesta-Zuluaga et al. 2019; Sinha et al. 2019; Falony et al. 2016; Borgo et al. 2018; Gao et al. 2018; Peters et al. 2022). Sex was also among the ten factors that most explained variability in human gut microbial composition (Falony et al. 2016).
Figure 1.
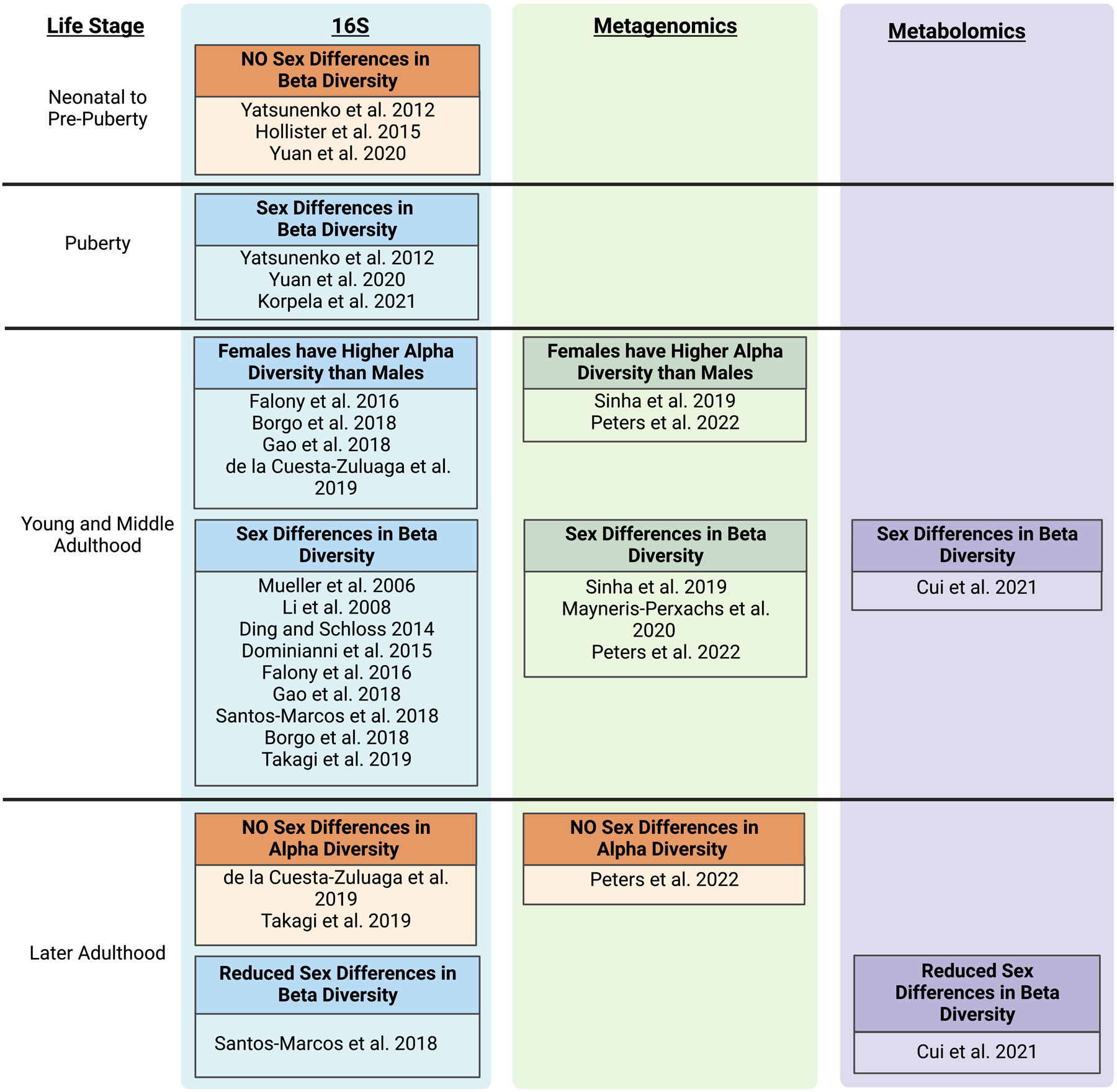
Sex differences in the human gut microbiome arise during puberty. Studies providing evidence of sex differences are categorized by developmental stage, type of data analysis (eg. 16S rRNA gene sequencing, metagenomics, or untargeted metabolomics) and observed effect on the gut microbiome. Created with BioRender.com.
Since multiple studies support the idea that there are sex differences in the adult gut microbiome, it follows that there must be a point during development where the gut microbiome differentiates by sex. While initial studies reported that the microbial community in young children (1–3 years of age) resembled that of adults (Yatsunenko et al. 2012; Kurokawa et al. 2007), follow-up studies showed significant differentiation of beta diversity between child and adult microbiomes (Ringel-Kulka et al. 2013; Hollister et al. 2015; Agans et al. 2011). A study of human dizygotic twins found no differentiation in gut microbial beta diversity between male and female infant twins, but did find differentiation between male and female pubertal twins (Yatsunenko et al. 2012). Similarly, another study revealed no sex differentiation in gut microbial beta diversity in pre-pubertal children, but did show differentiation by sex in pubertal subjects (Yuan et al. 2020). Moreover, beta diversity of pubertal male and female microbiomes was more similar to adult microbiomes of the same sex the further along the child was in puberty (Korpela et al. 2021). Similar patterns occurred in mouse studies: sex differences in alpha and beta diversity in mouse gut microbiomes were not observed before puberty but became evident after puberty (Markle et al. 2013; Yurkovetskiy et al. 2013). While some taxonomic differences between sexes in puberty were noted (Yuan et al. 2020; Korpela et al. 2021), there have been too few studies with small sample sizes to make strong conclusions about pubertal-specific taxonomic differences. Collectively, these studies support the idea that the mammalian gut microbiome differentiates during puberty, resulting in sex differences in adulthood.
Despite a consistent pattern of community-level sex differentiation in the gut microbiome, the specific taxa differentiated by sex varies among studies. For example, early work suggested that the ratio of the common gut phyla, Bacteroidetes and Firmicutes, differed between male and female gut microbiomes (Huttenhower et al. 2012; Gomez et al. 2012). However, follow-up studies found no difference in the Bacteroidetes to Firmicutes ratio, indicating that it is not a reliable indicator of sex differences (Takagi et al. 2019; Santos-Marcos et al. 2018; Singh and Manning 2016; Elderman et al. 2018). The abundances of specific bacterial genera or species have also not been reliable markers of sex differentiation. While individual studies identified bacterial taxa associated with microbiome sex-differentiation, the same taxa did not change consistently in all studies. The reasons for the lack of consistency are unclear, but may partly be due to external factors influencing the microbial gut composition in the various study populations (e.g., geography, age, and diet) (Valeri and Endres 2021; Kim et al. 2019; Jaggar et al. 2020). This inconsistency may also be a result of functional redundancy in the gut microbiome. So far, most studies of gut microbiome sex differentiation have employed 16S rRNA bacterial gene sequencing which cannot directly analyze gene function (Fig. 1). Future studies applying techniques such as metagenomics, transcriptomics or metabolomics to study gut microbiome function could be useful in determining the precise nature of gut microbiome sex differentiation.
Sex steroids and the Gut Microbiome
Sex steroids are produced by the gonads during puberty in response to activation of the hypothalamic-pituitary-gonadal axis (Fig 2). Observational studies in humans and studies in rodent models collectively suggest a linkage between sex steroids and the gut microbiome. For example, several studies manipulating sex steroid levels in mice indicated that estrogen and testosterone influenced gut microbial diversity and function. Gonadectomy of male and female mice shifted the gut community composition in both sexes and resulted in lower levels of sex differentiation as measured by beta diversity (Org et al. 2016; Kaliannan et al. 2018; Harada et al. 2016; Gao et al. 2021; Choi et al. 2017). Since gonad removal affects more than just the production of sex steroids, gonadectomy plus steroid replacement studies are necessary to discern whether sex steroids are responsible for a certain phenotype. One such study showed that treatment of ovariectomized female mice with estradiol shifted gut microbiome beta diversity to be more like intact females (Kaliannan et al. 2018). Another study demonstrated that treating castrated male mice with dihydrotestosterone shifted the gut microbiome beta diversity to be more like intact males (Gao et al. 2021).
Figure 2.
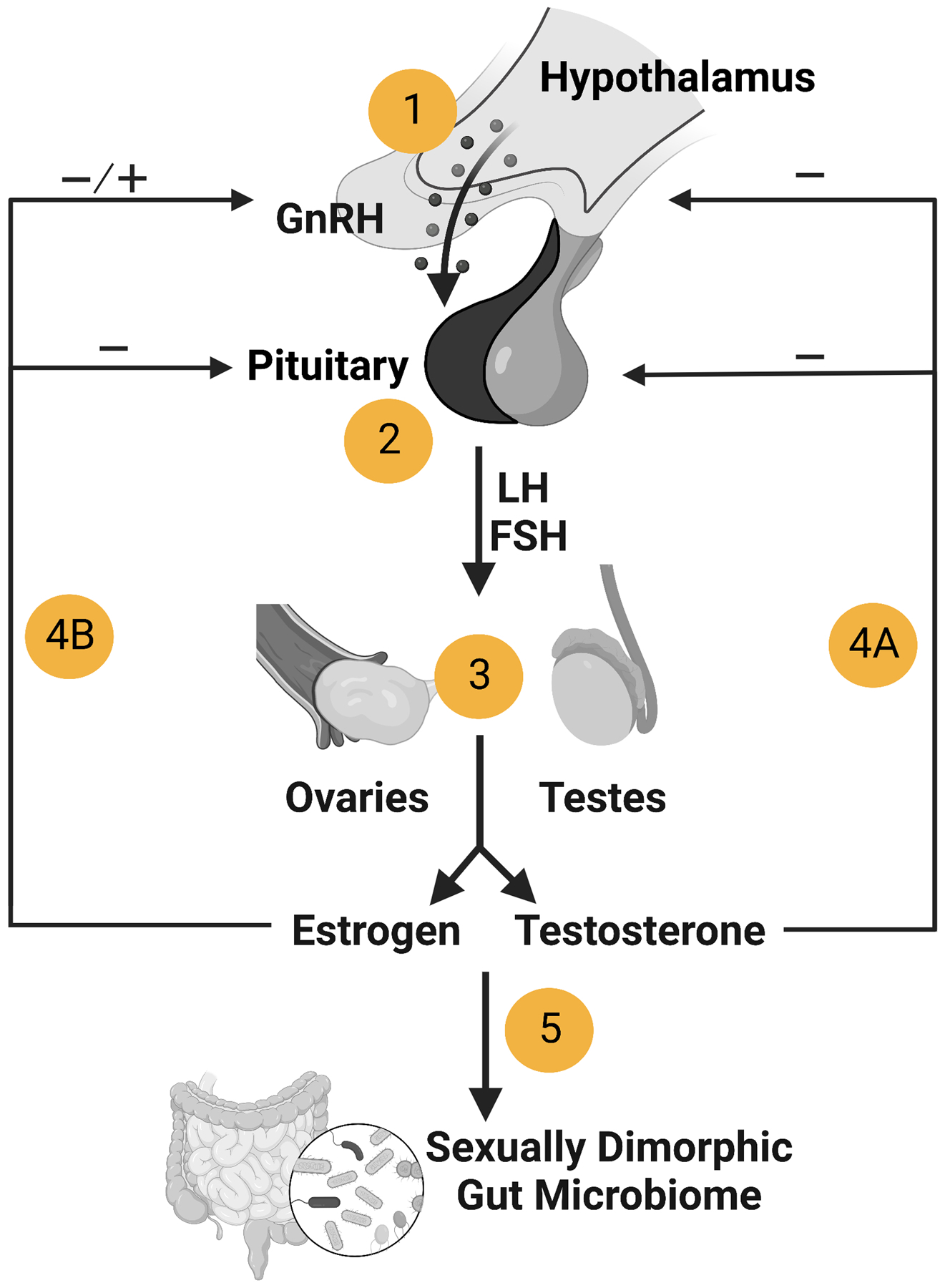
Activation of the hypothalamic–pituitary–gonadal axis during puberty. 1) Gonadotropin-releasing hormone (GnRH) production is stimulated by kisspeptin secreted from neurons in the hypothalamus. 2) GnRH induces pituitary gonadotrope cells to produce and secrete the gonadotropins, follicle stimulating hormone (FSH) and luteinizing hormone (LH). 3) In males, LH signaling in testicular Leydig cells results in testosterone production, while in females LH stimulates testosterone production in ovarian theca cells, which is then converted to estrogen in granulosa cells by aromatase. 4A) Testosterone exerts negative feedback in the hypothalamus and pituitary, while 4B) estrogen exerts both negative and positive feedback. 5) Sex steroids may directly or indirectly modulate the gut microbiome to be sexually dimorphic. Created with BioRender.com.
In addition to studies with gonadectomized mice, which results in steroid insufficiency, other studies have employed steroid excess to study the relationship between sex steroids and the gut microbiome. For example, studies have shown that treatment of pubertal female mice or rats with the aromatase inhibitor letrozole, which elevates testosterone and decreases estrogen (Kauffman et al. 2015) results in a shift in gut bacterial alpha and beta diversity (Kelley et al. 2016; Guo et al. 2016). Treatment of female rats with dihydrotestosterone (a non-aromatizable androgen) was also found to alter the gut microbiome (Zheng et al. 2021; Rodriguez Paris et al. 2022). Interestingly, letrozole treatment affected the gut microbiome and metabolic phenotype of pubertal females differently than adult females, indicating that puberty may be a sensitive period for effects of steroids on the gut microbiome as well as the host (Torres, Skarra, et al. 2019; Arroyo et al. 2019; Torres, Ho, et al. 2019). Moreover, cohousing letrozole- and placebo-treated mice, in which mice exchanged gut microbiota through coprophagy, reduced testosterone levels and ameliorated reproductive and metabolic phenotypes in letrozole-treated mice (Torres, Ho, et al. 2019; Ho et al. 2021). This potential bidirectional relationship between host sex steroids and the gut microbiome was also suggested by another study which showed that maternal high-fat diet induced metabolic dysregulation, early puberty, and infertility could be partially reversed in female offspring cohoused with healthy females (Wang, Zhang, et al. 2020).
In humans, the evidence for a relationship between sex steroids and the gut microbiome comes from various observational studies with postmenopausal women or women using oral contraceptives. Two gut microbiome studies of women using oral contraceptive pills (estrogen and progesterone) found they had lower alpha diversity, differentiated beta diversity, and distinct gut microbial metabolic pathways compared with women not using oral contraceptives (Mihajlovic et al. 2021; Hua et al. 2022). Other studies showed that urinary estrogens and their metabolites were negatively correlated with gut microbiota alpha diversity in postmenopausal females but not premenopausal females (Fuhrman et al. 2014; Flores et al. 2012). In postmenopausal women, estrone levels and the ratio of parent estrogens to estrogen metabolites were associated with an increased relative abundance of specific bacterial taxa (Fuhrman et al. 2014; Flores et al. 2012). A metagenomics study also found significant differences in the taxonomic and functional pathway beta diversity of postmenopausal women compared to premenopausal women (Zhao et al. 2019). Interestingly, the one study we found that investigated changes in the gut microbiome directly within the intestinal tract, namely the duodenum, also showed a relationship between sex steroids and the microbiome. Specifically, Leite et al. reported that the duodenal microbiome of postmenopausal women treated with hormone replacement therapy was more like premenopausal women than postmenopausal women not taking hormone replacement therapy (Leite et al. 2022).
Another link between sex steroids and the gut microbiome was identified in women with PCOS with elevated testosterone levels (hyperandrogenism). Studies of gut microbial diversity found that women and adolescent girls with PCOS had lower alpha diversity and differences in beta diversity compared to healthy women and adolescent girls (Torres et al. 2018; Lindheim et al. 2017; Qi et al. 2019; Liu et al. 2017; Jobira et al. 2020; Garcia-Beltran et al. 2021). Testosterone levels in women with PCOS also correlated with alpha diversity and the abundance of specific members of the gut microbial community [reviewed in (Rizk and Thackray 2020)]. Intriguingly, Qi et al. reported that female mice transplanted with stool from women with PCOS developed metabolic and reproductive dysregulation (Qi et al. 2019) suggesting that changes in the gut microbiome may be sufficient to induce PCOS-like phenotypes.
Microbial Modifications of Sex Steroids
Several key studies support the hypothesis that gut bacteria play a critical role in enterohepatic circulation of sex steroids (Fig 3). Studies in humans and rats demonstrated that antibiotic treatment dramatically increased the ratio of conjugated to deconjugated estrogens excreted in urine and feces (Adlercreutz et al. 1984; Goldin and Gorbach 1984; Flores et al. 2012). Moreover, Colldén et al. found negligible deconjugation of testosterone glucuronide in the intestinal tract of germ-free male mice (Colldén et al. 2019). This study also showed that the concentration of androgen glucuronide in normal mice was high in the small intestine and low in the cecum and colon, indicating that microbial deconjugation of androgen glucuronide (and presumably other steroids) occurs largely in the small intestine.
Figure 3.
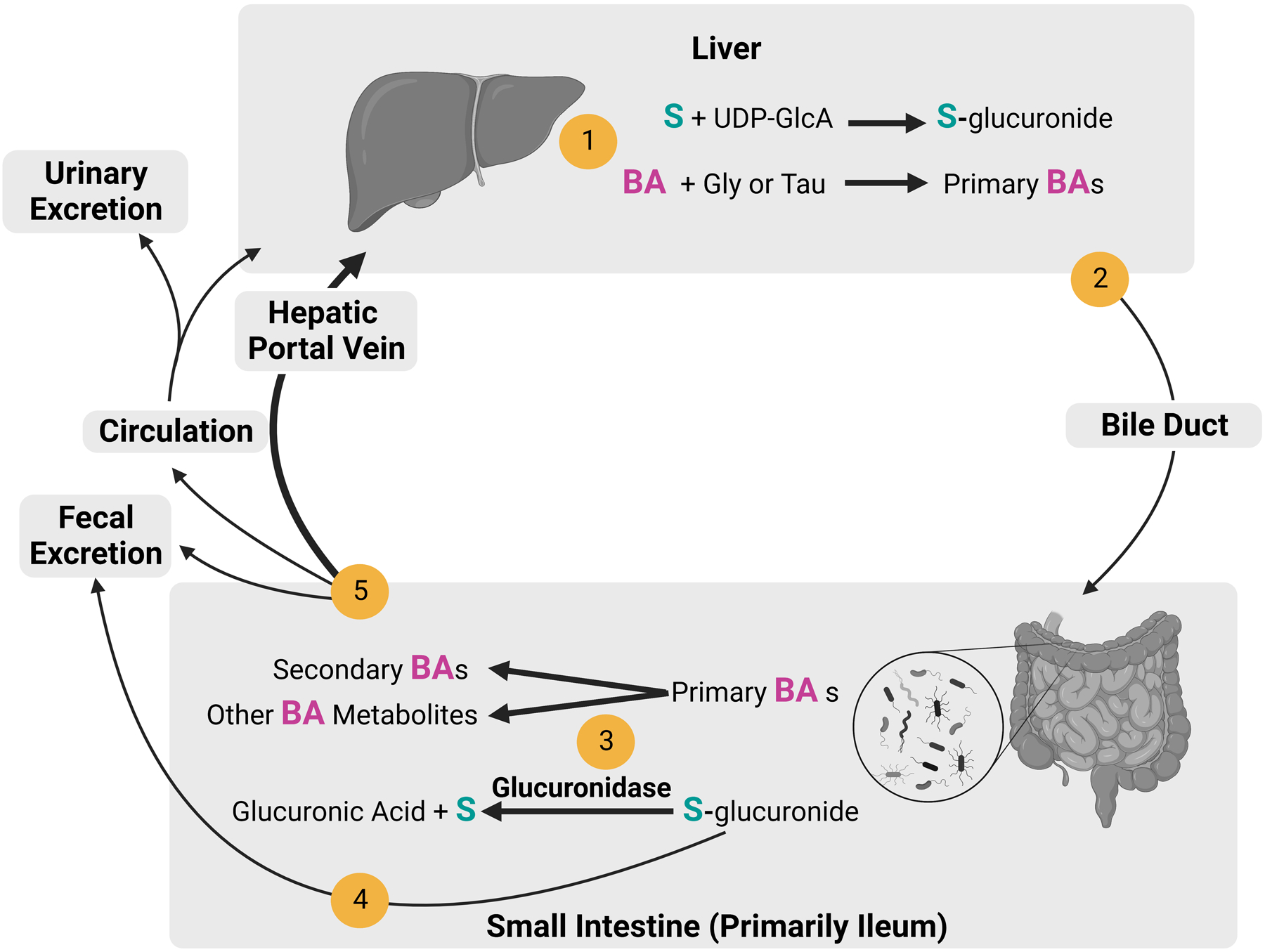
Enterohepatic circulation of sex steroids and bile acids. 1) Sex steroids (S) are conjugated with glucuronide from uridine 5’-diphosphoglucuronic acid (UDP-GlcA) by liver enzymes. Bile acids (BAs) are synthesized and conjugated with glycine (Gly) or taurine (Tau) in the liver. 2) Sex steroid glucuronides (S-glucuronide) and primary bile acids are transported to the gallbladder and excreted into the small intestine via the bile duct. 3) Gut bacteria deconjugate S-glucuronide and transform BAs into secondary BAs or other BA metabolites. 4) S-glucuronides that are not deconjugated are excreted through the feces. 5) Secondary BAs, other BAs metabolites, and deconjugated sex steroids are primarily reabsorbed through the hepatic portal vein to the liver. Alternatively, they are excreted through feces or returned to circulation, to be reabsorbed by the liver or excreted through urine. Created with BioRender.com.
While gut bacteria facilitate enterohepatic cycling of sex steroids, the role of specific bacterial species in this process is not well understood. Studies have shown that a bacterial enzyme, β-glucuronidase, is primarily responsible for the deconjugation of steroid glucuronides (Pellock and Redinbo 2017; Sui, Wu, and Chen 2021) and that β-glucuronidase homologs are present and active in many common gut bacteria in mice and humans (Lombardi et al. 1978; Dabek et al. 2008; McIntosh et al. 2012; Winter and Bokkenheuser 1987; Gloux et al. 2011; Gadelle, Raibaud, and Sacquet 1985). β-glucuronidase homologs were found in members of many common gut bacterial families, including Ruminococcaea, Lachnospiraceae, Clostridiaceae, Bacteroidaceae, and Tannerellaceae. Many studies have demonstrated E. coli deconjugation of various estrogen-glucuronides (Buehler, Katzman, and Doisy 1951; Graef, Furuya, and Nishikaze 1977; Lombardi et al. 1978; Legler et al. 2002), though studies on androgen glucuronides are lacking. Recently, Ervin et al. reported deconjugation of estrogen glucuronides by β-glucuronidases isolated from other gut bacteria, including Faecalibacterium prausnitzii, Roseburia species, and Clostridium species (Ervin et al. 2019). As β-glucuronidase genes are not present in all gut microbes, possession of this gene could provide a competitive advantage during puberty. Phylogenetic and structural diversity analyses of bacterial β-glucuronidase genes indicate that microbial β-glucuronidases are substrate specific and fall into functional classes (Biernat et al. 2019; Little et al. 2018; Ervin et al. 2019). The fact that many common gut bacteria possess β-glucuronidase activity with different substrate specificity provides a potential mechanisms by which sex steroids could differentiate the gut microbiome. In support of this idea, glucuronidase genes in gut metagenomes were reported to be higher in premenopausal women than in postmenopausal women and adult men (Peters et al. 2022).
Another mechanism by which sex steroids may impact bacterial communities is via bacterial metabolism of unconjugated (free) steroids. Metabolism of free androgens and estrogens, including redox reactions and partial cleavage of steroids, has been reported in human fecal cultures (Lombardi et al. 1978; Eriksson and Gustafsson 1971). These studies indicated that free steroids were not broken down completely by bacteria, so this activity may be for detoxifying steroids rather than for energy or anabolism. Indeed, free testosterone was shown to be toxic to gut bacteria such as E. faecalis and E. coli, suggesting that high concentrations of steroids may be toxic to some bacteria but not others (Plotkin et al. 2003). On the other hand, additional studies showed that estrogen and progesterone were completely degraded when added to fecal cultures (Coombes et al. 2020; Li et al. 2018). Collectively, these studies provide evidence that gut microbes metabolize sex steroids, and that this metabolism could potentially influence the composition of the gut microbial community in a sex-specific manner. However, the mechanisms by which this might occur are poorly understood and many more studies are needed to determine if and how these processes occur.
Bile acids and the Gut Microbiome
While direct interaction of sex steroids with gut microbiota may lead to a sexually dimorphic gut microbiome, this is not the only possible mechanism. Sex steroids may also indirectly influence the gut microbial community by regulating host production and secretion of bile acids, which may then modulate the gut microbiota. Bile acids are important for emulsifying fats during digestion and as signaling molecules in glucose and lipid metabolism (Perino et al. 2021). In humans, the liver produces cholic acid (CA), and chenodeoxycholic acid (CDCA) from cholesterol (Russell 2003). Muricholic acid (MCA) is produced instead of CDCA in mice. Bile acids are conjugated with either taurine or glycine, then secreted as primary bile acids through the bile duct into the small intestine. Like sex steroids, bile acids circulate between the liver and the intestine through enterohepatic circulation (Fig 3). Once secreted into the small intestine, microbial bile salt hydrolase (BSH) deconjugates bile acids with glycine or taurine. 70–90% of bile acids are reabsorbed from the ileum and transported to the liver through the hepatic portal vein (Angelin et al. 1982; Wahlström et al. 2016). The BSH gene is present in most major gut microbiome phyla (Firmicutes, Bacteroidetes, Actinobacteria, Fusobacteria, and Proteobacteria), and BSH enzymes in different phyla have different substrate specificity for glycine- or taurine-conjugated bile acids (Jones et al. 2008; Song et al. 2019). The widespread presence of BSH among members of the gut microbiota suggests that it could confer a competitive advantage by providing resistance to bile acid detergent toxicity (Jones et al. 2008). Another transformation of bile acids by gut microbiota is 7α-dehydroxylation, which produces the secondary bile acids deoxycholic acid (DCA) and lithocholic acid (LCA; MDCA in mice) (Lucas et al. 2021). 7α-dehydroxylase activity is less widespread among human gut bacteria than BSH activity, which may lead to niche specialization by species with this activity (Lucas et al. 2021). Less common transformations of bile acids include dehydroxylation, epimerization, isomerization, and oxidation (Eyssen et al. 1983; Degirolamo et al. 2014). As secondary bile acids have varying levels of toxicity to different microbes, producing more secondary bile acids may provide a competitive advantage for bacteria able to tolerate this toxicity and a mechanism for the host to influence the gut microbial community (Tian et al. 2020).
Multiple reviews indicate that microbial bile acid metabolism is widespread within the gut microbiota and that it creates a large diversity of bile-acid derived metabolites (Wahlström et al. 2016; Foley et al. 2019; Winston and Theriot 2020). However, apart from BSH activity, the prevalence of these functions within gut microbiota is unknown (Winston and Theriot 2020). While some effects of secondary bile acids on the host are known, such as anti-inflammatory properties, much remains to be discovered about how microbial transformation of bile acids affects host physiology (Ward et al. 2017; Lajczak-McGinley et al. 2020). A growing body of literature indicate that bile acids have a profound effect on host biology. For instance, both primary and secondary bile acids bind to host receptors such as farnesoid X receptor (FXR), G Protein-Coupled Bile Acid Receptor 1, pregnane X receptor, vitamin D receptor, and retinoid-related orphan receptor, and regulate metabolic and immune functions (Fiorucci et al. 2021; Chiang 2013). FXR, specifically, has been widely studied and implicated in glucose metabolism, lipid homeostasis, inflammation, and permeability of the intestinal barrier (Ding et al. 2015).
Sex differences and Bile Acids
In humans, the bile acid profiles of newborn infants (infants less than 48 hours old) were largely composed of primary bile acids and were markedly different from adult bile acid profiles (Wang, Chen, et al. 2020). In mice and rats, the gut microbiota and bile acid pool both diversify rapidly after birth. Interestingly, the diversity of secondary bile acids appears to increase until puberty after which time it stabilizes (van Best et al. 2020; Morris, Little, and Lester 1983). During and after puberty, studies have shown that primary and secondary bile acid levels are higher in female than male rodents (Morris, Little, and Lester 1983; Li-Hawkins et al. 2002; Schwarz et al. 2001; Turley et al. 1998; Jahnel et al. 2015; Baars et al. 2018). Ma et al. also identified differences in secondary bile acid levels between young and older adult mice, and determined that these differences were reduced by cohousing, indicating that the gut microbiome influences bile acid levels (Ma et al. 2020).
There is also evidence for sex differences in bile acids in humans. Multiple studies have determined that total bile acid concentrations and levels of bile acid synthesis are higher in males than females (Xiang et al. 2012; Frommherz et al. 2016; Steiner et al. 2012; Gälman, Angelin, and Rudling 2011; Bennion et al. 1978). Since mice show the opposite pattern, this difference is important to consider when using mice as a model for the effects of bile acids on physiology. While there are consistent sex differences in the total bile acid pool in humans, the reported sex differences in terms of the types of primary and secondary acids varies among studies (Wang, Chen, et al. 2020; Fisher and Yousef 1973; Bennion et al. 1978). In addition, little is known about how age interacts with sex differences in primary and secondary bile acid metabolism although one study indicated that sex steroids, which change in concentration throughout the lifespan, may indirectly affect secondary bile acid metabolism (Ridlon, Kang, and Hylemon 2010).
Interplay between Bile Acids, Gut Microbiome, and PCOS
Ho et al. found a relationship between gut microbiota and secondary bile acids in mice. In this study, letrozole treatment of female mice during puberty resulted in changes to secondary bile acid levels, gut microbiota abundances, metabolism, and the reproductive axis (Ho et al. 2021). In humans, women with PCOS were reported to have different levels of specific secondary bile acids compared to healthy controls, and higher levels of BSH genes in their gut microbial metagenomes (Yang, Wu, et al. 2021; Qi et al. 2019; Zhang et al. 2019). Transplantation of stool from women with PCOS into adult female mice resulted in impaired fertility and glucose and insulin sensitivity, potentially modulated by the secondary bile acids tauroursodeoxycholic acid and glycoursodeoxycholic acid (Qi et al. 2019). Another study with fecal microbiome transplantation from women with PCOS to adult female mice showed decreased expression of the FXR receptor in the ileum and impaired insulin sensitivity (Yang, Zhou, et al. 2021). These studies indicate a potential influence of the gut/liver axis on metabolic dysregulation and vice versa.
The Immune System, Sex, and the Gut Microbiome
The immune system differentiates in a sex-specific manner during puberty and remains sexually dimorphic in adulthood (Rizzetto et al. 2018; Hooper, Littman, and Macpherson 2012; Fuhler 2020). Since both innate and adaptive immunity shapes many of the interactions in the gut microbiome and vice versa (Hooper, Littman, and Macpherson 2012; Foster et al. 2017; Shi et al. 2017; Wiertsema et al. 2021), the sexually dimorphic nature of the immune system creates another possible mechanism for generating a sex-specific gut microbiome (Whitacre et al. 1999; Lamason et al. 2006; Sharma et al. 2018; Klein and Flanagan 2016). The immune system is known to influence the gut microbiome through a number of mechanisms, such as preventing infiltration of the gut microbiota into host tissues (Gallo and Hooper 2012), secreting mucins and antimicrobial compounds into the intestine, and propagating microbial signals to other immune cells (Abreu 2010; Peterson and Artis 2014). Adaptive immune cells also surveil the gut microbiota and alter community composition through immunoglobulin secretion (Zhang et al. 2015; Fransen et al. 2015). In turn, the gut microbiome affects differentiation of T and B cells and pro-and anti-inflammatory cytokine levels (Zhao and Elson 2018; Fujimura et al. 2016; Luu et al. 2018; Sampson et al. 2016; Li et al. 2019; Lajczak-McGinley et al. 2020).
Given the importance of the immune system in regulating the gut microbiome, sex-specific differences in the immune system could shape the gut microbiome in a sex-specific manner. This idea is supported by studies showing an association between the gut microbiome and autoimmune disorders. Ulcerative colitis, a form of IBD, is diagnosed more frequently in females, while Chron’s disease, another form of IBD, is diagnosed more often in males (Hayter and Cook 2012; Herzog et al. 2014; Henderson et al. 2011). Studies indicate that the gut microbiome is a major factor in IBD incidence and severity. Microbiome and metabolome composition in patients with IBD was different from healthy patients and associated with gut inflammation levels (Franzosa et al. 2019; Dong et al. 2019; Clooney et al. 2019). Gut microbiota may affect severity of IBD directly through activation of immune cells through pattern recognition receptors or immunoglobulin binding or indirectly through bile acid signaling (Ding et al. 2015; Schirmer et al. 2019). Additionally, fecal microbiome transplantation is an effective treatment for many patients, though differences in effectiveness by donor and recipient are not well understood (Zhou et al. 2021; Mańkowska-Wierzbicka et al. 2020; Cheng et al. 2021). While differences in gut microbiome composition between patients with ulcerative colitis or Crohn’s disease were reported, sex differences were not observed (Clooney et al. 2019; Dong et al. 2019). However, since sex differences in IBD patients or their microbiomes are often not analyzed or reported, it is possible that sex differentiation of the gut microbiome may contribute to differences in the development and pathology of the IBD subtypes and warrants future investigation. Given that sex-specific immune responses to bacterial metabolites are a candidate for how the gut microbiome contributes to sex differences in the incidence of IBD (Murray et al. 2019; Murray et al. 2020; Bhattarai et al. 2020; Spichak et al. 2021), it may also be important to determine if the sex of the donor and recipient needs to be considered in fecal microbiome transplantation for IBD treatment.
Another notable example of sex-specific differences in the immune system-gut microbiome relationship occurs in the non-obese diabetic (NOD) mouse model for the autoimmune disorder, T1D. In the NOD mouse model, T1D incidence after 30 weeks of age is ~80% in females and 20% in males, although this ratio varies depending on the mouse colony (Makino et al. 1980; Yurkovetskiy et al. 2013; Pozzilli et al. 1993). Intriguingly, castration of NOD males results in a T1D incidence similar to females, indicating that gonadal sex steroids may be important in protecting male mice from T1D (Makino et al. 1981). Additionally, Markle et al. found no sex differences in T1D in NOD mice raised in a germ-free environment, indicating that the gut microbiome was required for the sex differences in T1D incidence (Markle et al. 2013). This and other studies also showed that the male microbiome was protective against T1D and that delivery of male gut microbiota to female mice via fecal transplantation was protective against T1D (Markle et al. 2014; Markle et al. 2013; Yurkovetskiy et al. 2013). Altogether, this evidence suggests that host-microbe interactions may play a key role in modulating sex differences in autoimmunity.
Conclusions and Future Perspectives
Our review of the current literature on the relationship of sex to the gut microbiome in humans and in rodent models showed that a sex-specific gut microbiome develops during puberty and continues into adulthood. The majority of these studies relied on NGS of bacterial 16S rRNA gene amplicon libraries due to its cost-effectiveness and its ability to taxonomically identify both cultured and uncultured bacteria across all phyla. However, while the 16S gene provides useful information about gut bacterial diversity, it reveals little about the function of gut microbes. Metagenomic, metabolomic, and transcriptomic approaches can inform us about microbial gene content and expression, and metabolite production, and can identify changes in archaea, fungi, and viruses in addition to bacteria. Future studies employing these approaches will be needed to understand comprehensively how sex and sex steroids regulate the composition and function of gut microbial communities along the alimentary canal.
Understanding the role of gut microbiota in sex-specific human health and disease will require better knowledge of how sex steroids affect the gut microbiome, and vice versa. In humans, there is a lack of clinical studies investigating the relationship between sex steroids and the gut microbiome in men. Given that men commonly experience hypogonadism due to factors such as age, disease (e.g. obesity and type 2 diabetes), or drug treatment, e.g. GnRH agonist for prostate cancer (Zarotsky et al. 2014; Mittan et al. 2002), future studies are needed to discern the relationship between sex steroids and the gut microbiome in men. Since gender-affirming care for transgender individuals often involves hormone therapy, study of the gut microbiome in individuals receiving cross sex steroid treatment is also important.
While many studies in humans and rodents have established correlations between sex differences and factors such as sex steroids, bile acids and the immune system, few studies have investigated causal relationships and mechanisms. More studies are needed to investigate how microbiome manipulation affects sex steroid levels (e.g., via antibiotics, cohousing, or fecal transplants) and how sex steroid manipulation affects the gut microbiome (e.g., via gonadectomy and steroid replacement). Additionally, research should be undertaken to measure levels of both conjugated and unconjugated steroids in males and females, especially when paired with gut microbiome manipulation. The use of germ-free or antibiotic-depleted mice will be key in establishing which sex-specific diseases require the presence of the gut microbiome and which disease phenotypes can be recapitulated by fecal microbiome transplantation.
Since there is growing evidence that there are sex differences in bile acid production in the liver, future research should investigate when sex differences in primary and secondary bile acid levels emerge in humans and rodents and how the gut microbiome contributes to these differences. Gut microbiota manipulation may provide insight into how the gut microbiome affects sex-specific bile acid metabolism and signaling. Altering host bile acid levels through host genetic knock-outs in the bile acid pathway or through diet supplementation could potentially be used to determine how sex-specific bile acid levels affect gut microbiota composition. Deciphering this relationship may be essential to understand how gut microbiota contribute to sex-specific metabolic disorders like PCOS and type 2 diabetes.
Finally, given the number of sex-specific autoimmune diseases, understanding the relationship between the gut microbiome and the immune system is likely to be important for developing microbial-based preventative measures and therapeutics for these disorders. Studies manipulating the immune system are needed to understand how the immune system shapes the sex-specific gut microbiome. Likewise, more studies are required to provide insight into how gut microbiota manipulation affects development of sex-specific autoimmune diseases like IBD and type I diabetes. Understanding how sex steroids, bile acids, the immune system, and the gut microbiome interact to create and maintain sex differences in microbiome-related diseases may have far-reaching implications especially since the gut microbiome is increasingly being considered a therapeutic target.
Financial Support:
This work was funded by Grant R01 HD095412. (V.G.T.), F31 HD105403 (L.S-H.) and a SDSU ARCS Foundation Scholar Award (L.S-H.)
Footnotes
Disclosure Summary: The authors have nothing to disclose.
References
- Abreu Maria T. 2010. ‘Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function’, Nature Reviews Immunology, 10: 131–44. [DOI] [PubMed] [Google Scholar]
- Acharya Kalpana D., Gao Xing, Bless Elizabeth P., Chen Jun, and Tetel Marc J.. 2019. ‘Estradiol and high fat diet associate with changes in gut microbiota in female ob/ob mice’, Scientific reports, 9: 20192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adlercreutz H, Pulkkinen MO, Hämäläinen EK, and Korpela JT. 1984. ‘Studies on the role of intestinal bacteria in metabolism of synthetic and natural steroid hormones’, J Steroid Biochem, 20: 217–29. [DOI] [PubMed] [Google Scholar]
- Agans Richard, Rigsbee Laura, Kenche Harshavardhan, Michail Sonia, Khamis Harry J., and Paliy Oleg. 2011. ‘Distal gut microbiota of adolescent children is different from that of adults’, FEMS Microbiology Ecology, 77: 404–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelin Bo, Ingemar Björkhem Kurt Einarsson, and Ewerth Staffan. 1982. ‘Hepatic Uptake of Bile Acids in Man: FASTING AND POSTPRANDIAL CONCENTRATIONS OF INDIVIDUAL BILE ACIDS IN PORTAL VENOUS AND SYSTEMIC BLOOD SERUM’, The Journal of clinical investigation, 70: 724–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo Pablo, Ho Bryan S., Sau Lillian, Kelley Scott T., and Thackray Varykina G.. 2019. ‘Letrozole treatment of pubertal female mice results in activational effects on reproduction, metabolism and the gut microbiome’, PLOS ONE, 14: e0223274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baars Annemarie, Oosting Annemarie, Lohuis Mirjam, Koehorst Martijn, Sahar El Aidy Floor Hugenholtz, Smidt Hauke, Mischke Mona, Boekschoten Mark V., Verkade Henkjan J., Garssen Johan, van der Beek Eline M., Knol Jan, Vos Paul de, van Bergenhenegouwen Jeroen, and Fransen Floris. 2018. ‘Sex differences in lipid metabolism are affected by presence of the gut microbiota’, Scientific reports, 8: 13426–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, Khan MT, Zhang J, Li J, Xiao L, Al-Aama J, Zhang D, Lee YS, Kotowska D, Colding C, Tremaroli V, Yin Y, Bergman S, Xu X, Madsen L, Kristiansen K, Dahlgren J, and Wang J. 2015. ‘Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life’, Cell Host Microbe, 17: 690–703. [DOI] [PubMed] [Google Scholar]
- Barlow Jacob T., Bogatyrev Said R., and Ismagilov Rustem F.. 2020. ‘A quantitative sequencing framework for absolute abundance measurements of mucosal and lumenal microbial communities’, Nature Communications, 11: 2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennion Lynn J., Drobny Elaine, Knowler William C., Ginsberg Ronald L., Garnick Marc B., Adler Ronald D., and Duane William C.. 1978. ‘Sex differences in the size of bile acid pools’, Metabolism, 27: 961–69. [DOI] [PubMed] [Google Scholar]
- Bhat SF, Pinney SE, Kennedy KM, McCourt CR, Mundy MA, Surette MG, Sloboda DM, and Simmons RA. 2021. ‘Exposure to high fructose corn syrup during adolescence in the mouse alters hepatic metabolism and the microbiome in a sex-specific manner’, J Physiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai Yogesh, Jie Si, Linden David R., Ghatak Sayak, Mars Ruben A. T., Williams Brianna B., Pu Meng, Sonnenburg Justin L., Fischbach Michael A., Farrugia Gianrico, Sha Lei, and Kashyap Purna C.. 2020. ‘Bacterially Derived Tryptamine Increases Mucus Release by Activating a Host Receptor in a Mouse Model of Inflammatory Bowel Disease’, iScience, 23: 101798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biernat Kristen A., Pellock Samuel J., Bhatt Aadra P., Bivins Marissa M., Walton William G., Tran Bich Ngoc T., Wei Lianjie, Snider Michael C., Cesmat Andrew P., Tripathy Ashutosh, Erie Dorothy A., and Redinbo Matthew R.. 2019. ‘Structure, function, and inhibition of drug reactivating human gut microbial β-glucuronidases’, Scientific reports, 9: 825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich Nicholas A., Chung Jennifer, Battaglia Thomas, Henderson Nora, Jay Melanie, Li Huilin, Lieber Arnon D., Wu Fen, Perez-Perez Guillermo I., Chen Yu, Schweizer William, Zheng Xuhui, Contreras Monica, Dominguez-Bello Maria Gloria, and Blaser Martin J.. 2016. ‘Antibiotics, birth mode, and diet shape microbiome maturation during early life’, Science Translational Medicine, 8: 343ra82–43ra82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgo Francesca, Garbossa Stefania, Riva Alessandra, Severgnini Marco, Luigiano Carmelo, Benetti Albero, Pontiroli Antonio E., Morace Giulia, and Borghi Elisa. 2018. ‘Body Mass Index and Sex Affect Diverse Microbial Niches within the Gut’, Frontiers in Microbiology, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart Mya, Haynes Matthew, Kelley Scott, Angly Florent, Edwards Robert A., Felts Ben, Mahaffy Joseph M., Mueller Jennifer, Nulton James, Rayhawk Steve, Beltran Rodriguez-Brito Peter Salamon, and Rohwer Forest. 2008. ‘Viral diversity and dynamics in an infant gut’, Research in Microbiology, 159: 367–73. [DOI] [PubMed] [Google Scholar]
- Buehler HJ, Katzman PA, and Doisy EA. 1951. ‘Studies on beta-glucuronidase from E. coli’, Proc Soc Exp Biol Med, 76: 672–6. [DOI] [PubMed] [Google Scholar]
- Candon Sophie, Alicia Perez-Arroyo Cindy Marquet, Valette Fabrice, Foray Anne-Perrine, Pelletier Benjamin, Milani Cristian, Ventura Marco, Bach Jean-François, and Chatenoud Lucienne. 2015. ‘Antibiotics in Early Life Alter the Gut Microbiome and Increase Disease Incidence in a Spontaneous Mouse Model of Autoimmune Insulin-Dependent Diabetes’, PLOS ONE, 10: e0125448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Huang Z, Wei W, and Li Z. 2021. ‘Fecal microbiota transplantation for Crohn’s disease: a systematic review and meta-analysis’, Techniques in Coloproctology, 25: 495–504. [DOI] [PubMed] [Google Scholar]
- Chiang JY 2013. ‘Bile acid metabolism and signaling’, Compr Physiol, 3: 1191–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Hwang YJ, Shin MJ, and Yi H. 2017. ‘Difference in the Gut Microbiome between Ovariectomy-Induced Obesity and Diet-Induced Obesity’, J Microbiol Biotechnol, 27: 2228–36. [DOI] [PubMed] [Google Scholar]
- Clooney Adam G., Sutton Thomas D. S., Shkoporov Andrey N., Holohan Ross K., Daly Karen M., Orla O’Regan Feargal J. Ryan, Draper Lorraine A., Plevy Scott E., Ross R. Paul, and Hill Colin. 2019. ‘Whole-Virome Analysis Sheds Light on Viral Dark Matter in Inflammatory Bowel Disease’, Cell Host & Microbe, 26: 764–78.e5. [DOI] [PubMed] [Google Scholar]
- Colldén Hannah, Landin Andreas, Wallenius Ville, Elebring Erik, Lars Fändriks Maria E. Nilsson, Ryberg Henrik, Poutanen Matti, Klara Sjögren Liesbeth Vandenput, and Ohlsson Claes. 2019. ‘The gut microbiota is a major regulator of androgen metabolism in intestinal contents’, American journal of physiology. Endocrinology and metabolism, 317: E1182–E92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes Z, Yadav V, McCoubrey E Freire L,C, Basit W Conlan A,RS, and Gonzalez D. 2020. ‘Progestogens Are Metabolized by the Gut Microbiota: Implications for Colonic Drug Delivery’, Pharmaceutics, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M, Trimigno A, Aru V, Rasmussen MA, Khakimov B, and Engelsen SB. 2021. ‘Influence of Age, Sex, and Diet on the Human Fecal Metabolome Investigated by (1)H NMR Spectroscopy’, J Proteome Res. [DOI] [PubMed] [Google Scholar]
- Dabek M, McCrae SI, Stevens VJ, Duncan SH, and Louis P. 2008. ‘Distribution of beta-glucosidase and beta-glucuronidase activity and of beta-glucuronidase gene gus in human colonic bacteria’, FEMS Microbiol Ecol, 66: 487–95. [DOI] [PubMed] [Google Scholar]
- de la Cuesta-Zuluaga Jacobo, Kelley Scott T., Chen Yingfeng, Escobar Juan S., Mueller Noel T., Ley Ruth E., Daniel McDonald Shi Huang, Swafford Austin D., Knight Rob, and Thackray Varykina G.. 2019. ‘Age- and Sex-Dependent Patterns of Gut Microbial Diversity in Human Adults’, mSystems, 4: e00261–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degirolamo C, Rainaldi S, Bovenga F, Murzilli S, and Moschetta A. 2014. ‘Microbiota modification with probiotics induces hepatic bile acid synthesis via downregulation of the Fxr-Fgf15 axis in mice’, Cell Rep, 7: 12–8. [DOI] [PubMed] [Google Scholar]
- Desbonnet Lieve, Clarke Gerard, Traplin Alexander, Orla O’Sullivan Fiona Crispie, Moloney Rachel D., Cotter Paul D., Dinan Timothy G., and Cryan John F.. 2015. ‘Gut microbiota depletion from early adolescence in mice: Implications for brain and behaviour’, Brain, Behavior, and Immunity, 48: 165–73. [DOI] [PubMed] [Google Scholar]
- Ding L, Yang L, Wang Z, and Huang W. 2015. ‘Bile acid nuclear receptor FXR and digestive system diseases’, Acta Pharm Sin B, 5: 135–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Tao, and Schloss Patrick D.. 2014. ‘Dynamics and associations of microbial community types across the human body’, Nature, 509: 357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello Maria G., Jesus-Laboy Kassandra M. De, Shen Nan, Cox Laura M., Amir Amnon, Gonzalez Antonio, Bokulich Nicholas A., Song Se Jin, Hoashi Marina, Rivera-Vinas Juana I., Mendez Keimari, Knight Rob, and Clemente Jose C.. 2016. ‘Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer’, Nature Medicine, 22: 250–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominianni Christine, Sinha Rashmi, Goedert James J., Pei Zhiheng, Yang Liying, Hayes Richard B., and Ahn Jiyoung. 2015. ‘Sex, body mass index, and dietary fiber intake influence the human gut microbiome’, PLOS ONE, 10: e0124599–e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong LN, Wang M, Guo J, and Wang JP. 2019. ‘Role of intestinal microbiota and metabolites in inflammatory bowel disease’, Chin Med J (Engl), 132: 1610–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elderman Marlies, Hugenholtz Floor, Belzer Clara, Boekschoten Mark, van Beek Adriaan, de Haan Bart, Savelkoul Huub, de Vos Paul, and Faas Marijke. 2018. ‘Sex and strain dependent differences in mucosal immunology and microbiota composition in mice’, Biology of Sex Differences, 9: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson Håkan, and Gustafsson Jan-Åke. 1971. ‘Excretion of Steroid Hormones in Adults’, European Journal of Biochemistry, 18: 146–50. [DOI] [PubMed] [Google Scholar]
- Ervin SM, Li H, Lim L, Roberts LR, Liang X, Mani S, and Redinbo MR. 2019. ‘Gut microbial β-glucuronidases reactivate estrogens as components of the estrobolome that reactivate estrogens’, The Journal of biological chemistry, 294: 18586–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyssen H, De Pauw G, Stragier J, and Verhulst A. 1983. ‘Cooperative formation of omega-muricholic acid by intestinal microorganisms’, Appl Environ Microbiol, 45: 141–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, Kurilshikov A, Bonder MJ, Valles-Colomer M, Vandeputte D, Tito RY, Chaffron S, Rymenans L, Verspecht C, De Sutter L, Lima-Mendez G, D’Hoe K, Jonckheere K, Homola D, Garcia R, Tigchelaar EF, Eeckhaudt L, Fu J, Henckaerts L, Zhernakova A, Wijmenga C, and Raes J. 2016. ‘Population-level analysis of gut microbiome variation’, Science, 352: 560–4. [DOI] [PubMed] [Google Scholar]
- Ferguson A, and Sedgwick DM. 1994. ‘Juvenile onset inflammatory bowel disease: height and body mass index in adult life’, Bmj, 308: 1259–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti Pamela, Pasolli Edoardo, Tett Adrian, Asnicar Francesco, Gorfer Valentina, Fedi Sabina, Armanini Federica, Duy Tin Truong Serena Manara, Zolfo Moreno, Beghini Francesco, Bertorelli Roberto, Veronica De Sanctis Ilaria Bariletti, Canto Rosarita, Clementi Rosanna, Cologna Marina, Tiziana Crifò Giuseppina Cusumano, Gottardi Stefania, Innamorati Claudia, Caterina Masè Daniela Postai, Savoi Daniela, Duranti Sabrina, Gabriele Andrea Lugli Leonardo Mancabelli, Turroni Francesca, Ferrario Chiara, Milani Christian, Mangifesta Marta, Anzalone Rosaria, Viappiani Alice, Yassour Moran, Vlamakis Hera, Xavier Ramnik, Carmen Maria Collado Omry Koren, Tateo Saverio, Soffiati Massimo, Pedrotti Anna, Ventura Marco, Huttenhower Curtis, Bork Peer, and Segata Nicola. 2018. ‘Mother-to-Infant Microbial Transmission from Different Body Sites Shapes the Developing Infant Gut Microbiome’, Cell Host & Microbe, 24: 133–45.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorucci S, Carino A, Baldoni M, Santucci L, Costanzi E, Graziosi L, Distrutti E, and Biagioli M. 2021. ‘Bile Acid Signaling in Inflammatory Bowel Diseases’, Dig Dis Sci, 66: 674–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher MM, and Yousef IM. 1973. ‘Sex differences in the bile acid composition of human bile: studies in patients with and without gallstones’, Canadian Medical Association journal, 109: 190–93. [PMC free article] [PubMed] [Google Scholar]
- Flores Roberto, Shi Jianxin, Fuhrman Barbara, Xu Xia, Veenstra Timothy D., Gail Mitchell H., Gajer Pawel, Ravel Jacques, and Goedert James J.. 2012. ‘Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: a cross-sectional study’, Journal of Translational Medicine, 10: 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley Matthew H., Sarah O’Flaherty Rodolphe Barrangou, and Theriot Casey M.. 2019. ‘Bile salt hydrolases: Gatekeepers of bile acid metabolism and host-microbiome crosstalk in the gastrointestinal tract’, PLOS Pathogens, 15: e1007581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster Kevin R., Schluter Jonas, Coyte Katharine Z., and Rakoff-Nahoum Seth. 2017. ‘The evolution of the host microbiome as an ecosystem on a leash’, Nature, 548: 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen Floris, Zagato Elena, Mazzini Elisa, Fosso Bruno, Manzari Caterina, Aidy Sahar El, Chiavelli Andrea, D’Erchia Anna Maria, Sethi Maya K, Pabst Oliver, Marzano Marinella, Moretti Silvia, Romani Luigina, Penna Giuseppe, Pesole Graziano, and Rescigno Maria. 2015. ‘BALB/c and C57BL/6 Mice Differ in Polyreactive IgA Abundance, which Impacts the Generation of Antigen-Specific IgA and Microbiota Diversity’, Immunity, 43: 527–40. [DOI] [PubMed] [Google Scholar]
- Franzosa Eric A., Alexandra Sirota-Madi Julian Avila-Pacheco, Fornelos Nadine, Haiser Henry J., Reinker Stefan, Vatanen Tommi, Hall A. Brantley, Mallick Himel, McIver Lauren J., Sauk Jenny S., Wilson Robin G., Stevens Betsy W., Scott Justin M., Pierce Kerry, Deik Amy A., Bullock Kevin, Imhann Floris, Porter Jeffrey A., Zhernakova Alexandra, Fu Jingyuan, Weersma Rinse K., Wijmenga Cisca, Clish Clary B., Vlamakis Hera, Huttenhower Curtis, and Xavier Ramnik J.. 2019. ‘Gut microbiome structure and metabolic activity in inflammatory bowel disease’, Nature Microbiology, 4: 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommherz Lara, Bub Achim, Hummel Eva, Rist Manuela J., Roth Alexander, Watzl Bernhard, and Kulling Sabine E.. 2016. ‘Age-Related Changes of Plasma Bile Acid Concentrations in Healthy Adults—Results from the Cross-Sectional KarMeN Study’, PLOS ONE, 11: e0153959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhler GM 2020. ‘The immune system and microbiome in pregnancy’, Best Pract Res Clin Gastroenterol, 44–45: 101671. [DOI] [PubMed] [Google Scholar]
- Fuhrman Barbara J., Heather Spencer Feigelson Roberto Flores, Gail Mitchell H., Xu Xia, Ravel Jacques, and Goedert James J.. 2014. ‘Associations of the Fecal Microbiome With Urinary Estrogens and Estrogen Metabolites in Postmenopausal Women’, The Journal of Clinical Endocrinology & Metabolism, 99: 4632–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura Kei E., Sitarik Alexandra R., Havstad Suzanne, Lin Din L., Levan Sophia, Fadrosh Douglas, Panzer Ariane R., Brandon LaMere Elze Rackaityte, Lukacs Nicholas W., Wegienka Ganesa, Boushey Homer A., Ownby Dennis R., Zoratti Edward M., Levin Albert M., Johnson Christine C., and Lynch Susan V.. 2016. ‘Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation’, Nature Medicine, 22: 1187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadelle D, Raibaud P, and Sacquet E. 1985. ‘beta-Glucuronidase activities of intestinal bacteria determined both in vitro and in vivo in gnotobiotic rats’, Appl Environ Microbiol, 49: 682–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo Richard L., and Hooper Lora V.. 2012. ‘Epithelial antimicrobial defence of the skin and intestine’, Nature Reviews Immunology, 12: 503–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gälman C, Angelin B, and Rudling M. 2011. ‘Pronounced variation in bile acid synthesis in humans is related to gender, hypertriglyceridaemia and circulating levels of fibroblast growth factor 19’, J Intern Med, 270: 580–8. [DOI] [PubMed] [Google Scholar]
- Gao A, Su J, Liu R, Zhao S, Li W, Xu X, Li D, Shi J, Gu B, Zhang J, Li Q, Wang X, Zhang Y, Xu Y, Lu J, Ning G, Hong J, Bi Y, Gu W, Wang J, and Wang W. 2021. ‘Sexual dimorphism in glucose metabolism is shaped by androgen-driven gut microbiome’, Nat Commun, 12: 7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Zhang M, Xue J, Huang J, Zhuang R, Zhou X, Zhang H, Fu Q, and Hao Y. 2018. ‘Body Mass Index Differences in the Gut Microbiota Are Gender Specific’, Front Microbiol, 9: 1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran Cristina, Malpique Rita, Carbonetto Belen, Pedro González-Torres Desirée Henares, Brotons Pedro, Carmen Muñoz-Almagro Abel López-Bermejo, Zegher Francis de, and Ibáñez Lourdes. 2021. ‘Gut microbiota in adolescent girls with polycystic ovary syndrome: Effects of randomized treatments’, Pediatric Obesity, 16: e12734. [DOI] [PubMed] [Google Scholar]
- Gloux Karine, Berteau Olivier, Hanane El oumami Fabienne Béguet, Leclerc Marion, and Doré Joël. 2011. ‘A metagenomic β-glucuronidase uncovers a core adaptive function of the human intestinal microbiome’, Proceedings of the National Academy of Sciences, 108: 4539–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldin BR, and Gorbach SL. 1984. ‘Alterations of the intestinal microflora by diet, oral antibiotics, and Lactobacillus: decreased production of free amines from aromatic nitro compounds, azo dyes, and glucuronides’, J Natl Cancer Inst, 73: 689–95. [PubMed] [Google Scholar]
- Gomez Andres, Luckey David, Yeoman Carl J., Marietta Eric V., Berg Miller Margret E., Murray Joseph A., White Bryan A., and Taneja Veena. 2012. ‘Loss of Sex and Age Driven Differences in the Gut Microbiome Characterize Arthritis-Susceptible *0401 Mice but Not Arthritis-Resistant *0402 Mice’, PLOS ONE, 7: e36095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef V, Furuya E, and Nishikaze O. 1977. ‘Hydrolysis of steroid glucuronides with beta-glucuronidase preparations from bovine liver, Helix pomatia, and E. coli’, Clinical Chemistry, 23: 532–35. [PubMed] [Google Scholar]
- Guo Y, Qi Y, Yang X, Zhao L, Wen S, Liu Y, and Tang L. 2016. ‘Association between Polycystic Ovary Syndrome and Gut Microbiota’, PLOS ONE, 11: e0153196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada Naoki, Hanaoka Ryo, Horiuchi Hiroko, Kitakaze Tomoya, Mitani Takakazu, Inui Hiroshi, and Yamaji Ryoichi. 2016. ‘Castration influences intestinal microflora and induces abdominal obesity in high-fat diet-fed mice’, Scientific reports, 6: 23001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayter SM, and Cook MC. 2012. ‘Updated assessment of the prevalence, spectrum and case definition of autoimmune disease’, Autoimmun Rev, 11: 754–65. [DOI] [PubMed] [Google Scholar]
- He Zhixing, Shao Tiejuan, Li Haichang, Xie Zhijun, and Wen Chengping. 2016. ‘Alterations of the gut microbiome in Chinese patients with systemic lupus erythematosus’, Gut Pathogens, 8: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzel S, Aho VTE, Suenkel U, von Thaler AK, Schulte C, Deuschle C, Paulin L, Hantunen S, Brockmann K, Eschweiler GW, Maetzler W, Berg D, Auvinen P, and Scheperjans F. 2021. ‘Gut microbiome signatures of risk and prodromal markers of Parkinson’s disease’, Ann Neurol. [DOI] [PubMed] [Google Scholar]
- Helve Otto, Korpela Katri, Kolho Kaija-Leena, Saisto Terhi, Skogberg Kirsi, Dikareva Evgenia, Stefanovic Vedran, Salonen Anne, de Vos Willem M, and Andersson Sture. 2019. ‘2843. Maternal Fecal Transplantation to Infants Born by Cesarean Section: Safety and Feasibility’, Open Forum Infectious Diseases, 6: S68–S68. [Google Scholar]
- Henderson Paul, Hansen Richard, Cameron Fiona L., Gerasimidis Kostas, Rogers Pam, Bisset Michael W., Reynish Emma L., Drummond Hazel E., Anderson Niall H., Johan Van Limbergen Richard K. Russell, Satsangi Jack, and Wilson David C.. 2011. ‘Rising Incidence of Pediatric Inflammatory Bowel Disease in Scotland’, Inflammatory Bowel Diseases, 18: 999–1005. [DOI] [PubMed] [Google Scholar]
- Herzog D, Buehr P, Koller R, Rueger V, Heyland K, Nydegger A, Spalinger J, Schibli S, and Braegger CP. 2014. ‘Gender differences in paediatric patients of the swiss inflammatory bowel disease cohort study’, Pediatr Gastroenterol Hepatol Nutr, 17: 147–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Bryan, Ryback Daniel, Benson Basilin, Mason Cayla N., Torres Pedro J., Quinn Robert A., Thackray Varykina G., Kelley Scott T., and Metcalf Jessica L.. 2021. ‘Gut Metabolites Are More Predictive of Disease and Cohoused States than Gut Bacterial Features in a Polycystic Ovary Syndrome-Like Mouse Model’, mSystems, 6: e01149–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister Emily B., Riehle Kevin, Ruth Ann Luna Erica M. Weidler, Michelle Rubio-Gonzales Toni-Ann Mistretta, Raza Sabeen, Doddapaneni Harsha V., Metcalf Ginger A., Muzny Donna M., Gibbs Richard A., Petrosino Joseph F., Shulman Robert J., and Versalovic James. 2015. ‘Structure and function of the healthy pre-adolescent pediatric gut microbiome’, Microbiome, 3: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, and Macpherson AJ. 2012. ‘Interactions between the microbiota and the immune system’, Science, 336: 1268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Xinwei, Cao Yueming, Morgan David M., Miller Kaia, Chin Samantha M., Bellavance Danielle, and Khalili Hamed. 2022. ‘Longitudinal analysis of the impact of oral contraceptive use on the gut microbiome’, Journal of Medical Microbiology, 71. [DOI] [PubMed] [Google Scholar]
- Huttenhower Curtis, Gevers Dirk, Knight Rob, Abubucker Sahar, Badger Jonathan H., Chinwalla Asif T., Creasy Heather H., Earl Ashlee M., FitzGerald Michael G., Fulton Robert S., Giglio Michelle G., Kymberlie Hallsworth-Pepin Elizabeth A. Lobos, Madupu Ramana, Magrini Vincent, Martin John C., Mitreva Makedonka, Muzny Donna M., Sodergren Erica J., Versalovic James, Wollam Aye M., Worley Kim C., Wortman Jennifer R., Young Sarah K., Zeng Qiandong, Aagaard Kjersti M., Abolude Olukemi O., Emma Allen-Vercoe Eric J. Alm, Alvarado Lucia, Andersen Gary L., Anderson Scott, Appelbaum Elizabeth, Arachchi Harindra M., Armitage Gary, Arze Cesar A., Ayvaz Tulin, Baker Carl C., Begg Lisa, Belachew Tsegahiwot, Bhonagiri Veena, Bihan Monika, Blaser Martin J., Bloom Toby, Bonazzi Vivien, Brooks J. Paul, Buck Gregory A., Buhay Christian J., Busam Dana A., Campbell Joseph L., Canon Shane R., Cantarel Brandi L., Chain Patrick S. G., Chen I. Min A., Chen Lei, Chhibba Shaila, Chu Ken, Ciulla Dawn M., Clemente Jose C., Clifton Sandra W., Conlan Sean, Crabtree Jonathan, Cutting Mary A., Davidovics Noam J., Davis Catherine C., DeSantis Todd Z., Deal Carolyn, Delehaunty Kimberley D., Dewhirst Floyd E., Deych Elena, Ding Yan, Dooling David J., Dugan Shannon P., Dunne Wm Michael, Durkin A. Scott, Edgar Robert C., Erlich Rachel L., Farmer Candace N., Farrell Ruth M., Faust Karoline, Feldgarden Michael, Felix Victor M., Fisher Sheila, Fodor Anthony A., Forney Larry J., Foster Leslie, Francesco Valentina Di, Friedman Jonathan, Friedrich Dennis C., Fronick Catrina C., Fulton Lucinda L., Gao Hongyu, Garcia Nathalia, Giannoukos Georgia, Giblin Christina, Giovanni Maria Y., Goldberg Jonathan M., Goll Johannes, Gonzalez Antonio, Griggs Allison, Gujja Sharvari, Susan Kinder Haake Brian J. Haas, Hamilton Holli A., Harris Emily L., Hepburn Theresa A., Herter Brandi, Hoffmann Diane E., Holder Michael E., Howarth Clinton, Huang Katherine H., Huse Susan M., Izard Jacques, Jansson Janet K., Jiang Huaiyang, Jordan Catherine, Joshi Vandita, Katancik James A., Keitel Wendy A., Kelley Scott T., Kells Cristyn, King Nicholas B., Knights Dan, Kong Heidi H., Koren Omry, Koren Sergey, Kota Karthik C., Kovar Christie L., Kyrpides Nikos C., La Rosa Patricio S., Lee Sandra L., Lemon Katherine P., Lennon Niall, Lewis Cecil M., Lewis Lora, Ley Ruth E., Li Kelvin, Liolios Konstantinos, Liu Bo, Liu Yue, Lo Chien-Chi, Lozupone Catherine A., Lunsford R. Dwayne, Madden Tessa, Mahurkar Anup A., Mannon Peter J., Mardis Elaine R., Markowitz Victor M., Mavromatis Konstantinos, McCorrison Jamison M., McDonald Daniel, McEwen Jean, McGuire Amy L., Pamela McInnes Teena Mehta, Mihindukulasuriya Kathie A., Miller Jason R., Minx Patrick J., Newsham Irene, Nusbaum Chad, Michelle O’Laughlin Joshua Orvis, Pagani Ioanna, Palaniappan Krishna, Patel Shital M., Pearson Matthew, Peterson Jane, Podar Mircea, Pohl Craig, Pollard Katherine S., Pop Mihai, Priest Margaret E., Proctor Lita M., Qin Xiang, Raes Jeroen, Ravel Jacques, Reid Jeffrey G., Rho Mina, Rhodes Rosamond, Riehle Kevin P., Rivera Maria C., Beltran Rodriguez-Mueller Yu-Hui Rogers, Ross Matthew C., Russ Carsten, Sanka Ravi K., Sankar Pamela, Sathirapongsasuti J. Fah, Schloss Jeffery A., Schloss Patrick D., Schmidt Thomas M., Scholz Matthew, Schriml Lynn, Schubert Alyxandria M., Segata Nicola, Segre Julia A., Shannon William D., Sharp Richard R., Sharpton Thomas J., Shenoy Narmada, Sheth Nihar U., Simone Gina A., Singh Indresh, Smillie Christopher S., Sobel Jack D., Sommer Daniel D., Spicer Paul, Sutton Granger G., Sykes Sean M., Tabbaa Diana G., Thiagarajan Mathangi, Tomlinson Chad M., Torralba Manolito, Treangen Todd J., Truty Rebecca M., Vishnivetskaya Tatiana A., Walker Jason, Wang Lu, Wang Zhengyuan, Ward Doyle V., Warren Wesley, Watson Mark A., Wellington Christopher, Wetterstrand Kris A., White James R., Katarzyna Wilczek-Boney YuanQing Wu, Wylie Kristine M., Wylie Todd, Yandava Chandri, Ye Liang, Ye Yuzhen, Yooseph Shibu, Youmans Bonnie P., Zhang Lan, Zhou Yanjiao, Zhu Yiming, Zoloth Laurie, Zucker Jeremy D., Birren Bruce W., Gibbs Richard A., Highlander Sarah K., Methé Barbara A., Nelson Karen E., Petrosino Joseph F., Weinstock George M., Wilson Richard K., White Owen, and Consortium The Human Microbiome Project. 2012. ‘Structure, function and diversity of the healthy human microbiome’, Nature, 486: 207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaggar Minal, Rea Kieran, Spichak Simon, Dinan Timothy G., and Cryan John F.. 2020. ‘You’ve got male: Sex and the microbiota-gut-brain axis across the lifespan’, Frontiers in Neuroendocrinology, 56: 100815. [DOI] [PubMed] [Google Scholar]
- Jahnel J, Zöhrer E, Scharnagl H, Erwa W, Fauler G, and Stojakovic T. 2015. ‘Reference ranges of serum bile acids in children and adolescents’, Clin Chem Lab Med, 53: 1807–13. [DOI] [PubMed] [Google Scholar]
- Jobira B, Frank DN, Pyle L, Silveira LJ, Kelsey MM, Garcia-Reyes Y, Robertson CE, Ir D, Nadeau KJ, and Cree-Green M. 2020. ‘Obese Adolescents With PCOS Have Altered Biodiversity and Relative Abundance in Gastrointestinal Microbiota’, The Journal of clinical endocrinology and metabolism, 105: e2134–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones Brian V., Begley Máire, Hill Colin, Gahan Cormac G. M., and Marchesi Julian R.. 2008. ‘Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome’, Proceedings of the National Academy of Sciences, 105: 13580–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joriba Beza, Frank Daniel N., Pyle Laura, Gross Susan, Diana IR Wesley Pendelton, Robertson Charles E., Naedeau Kristen J., and Kelsey Megan M.. 2019. ‘183-LB: The Gut Microbiome and Puberty in Girls at Risk for Type 2 Diabetes (T2D)’, Diabetes, 68: 183–LB. [Google Scholar]
- Kaliannan Kanakaraju, Robertson Ruairi C., Murphy Kiera, Stanton Catherine, Kang Chao, Wang Bin, Hao Lei, Bhan Atul K., and Kang Jing X.. 2018. ‘Estrogen-mediated gut microbiome alterations influence sexual dimorphism in metabolic syndrome in mice’, Microbiome, 6: 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman Alexander S., Thackray Varykina G., Ryan Genevieve E., Tolson Kristen P., Glidewell-Kenney Christine A., Semaan Sheila J., Poling Matthew C., Iwata Nahoko, Breen Kellie M., Duleba Antoni J., Stener-Victorin Elisabet, Shimasaki Shunichi, Webster Nicholas J., and Mellon Pamela L.. 2015. ‘A Novel Letrozole Model Recapitulates Both the Reproductive and Metabolic Phenotypes of Polycystic Ovary Syndrome in Female Mice1’, Biology of Reproduction, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley Scott T., Skarra Danalea V., Rivera Alissa J., and Thackray Varykina G.. 2016. ‘The Gut Microbiome Is Altered in a Letrozole-Induced Mouse Model of Polycystic Ovary Syndrome’, PLOS ONE, 11: e0146509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Yong Sung, Unno Tatsuya, Kim Byung-Yong, and Park Mi-Sung. 2019. ‘Sex Differences in Gut Microbiota’, wjmh, 38: 48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein Sabra L., and Flanagan Katie L.. 2016. ‘Sex differences in immune responses’, Nature Reviews Immunology, 16: 626–38. [DOI] [PubMed] [Google Scholar]
- Koenig Jeremy E., Spor Aymé, Scalfone Nicholas, Fricker Ashwana D., Stombaugh Jesse, Knight Rob, Angenent Largus T., and Ley Ruth E.. 2011. ‘Succession of microbial consortia in the developing infant gut microbiome’, Proceedings of the National Academy of Sciences, 108: 4578–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpela Katri, Kallio Sampo, Salonen Anne, Hero Matti, Anna Kaarina Kukkonen Päivi J. Miettinen, Savilahti Erkki, Kohva Ella, Kariola Laura, Suutela Maria, Tarkkanen Annika, de Vos Willem M., Raivio Taneli, and Kuitunen Mikael. 2021. ‘Gut microbiota develop towards an adult profile in a sex-specific manner during puberty’, Scientific reports, 11: 23297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa Ken, Itoh Takehiko, Kuwahara Tomomi, Oshima Kenshiro, Toh Hidehiro, Toyoda Atsushi, Takami Hideto, Morita Hidetoshi, Sharma Vineet K., Srivastava Tulika P., Taylor Todd D., Noguchi Hideki, Mori Hiroshi, Ogura Yoshitoshi, Ehrlich Dusko S., Itoh Kikuji, Takagi Toshihisa, Sakaki Yoshiyuki, Hayashi Tetsuya, and Hattori Masahira. 2007. ‘Comparative Metagenomics Revealed Commonly Enriched Gene Sets in Human Gut Microbiomes’, DNA Research, 14: 169–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajczak-McGinley Natalia K., Porru Emanule, Fallon Ciara M., Smyth Jessica, Curley Caitriona, McCarron Paul A., Tambuwala Murtaza M., Roda Aldo, and Keely Stephen J.. 2020. ‘The secondary bile acids, ursodeoxycholic acid and lithocholic acid, protect against intestinal inflammation by inhibition of epithelial apoptosis’, Physiological Reports, 8: e14456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamason Rebecca, Zhao Po, Rawat Rashmi, Davis Adrian, Hall John C., Jae Jin Chae Rajeev Agarwal, Cohen Phillip, Rosen Antony, Hoffman Eric P., and Nagaraju Kanneboyina. 2006. ‘Sexual dimorphism in immune response genes as a function of puberty’, BMC Immunology, 7: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legler Juliette, Jonas Arjen, Lahr Joost, Vethaak A. Dick, Brouwer Abraham, and Murk Albertinka J.. 2002. ‘Biological measurement of estrogenic activity in urine and bile conjugates with the in vitro ER-CALUX reporter gene assay’, Environmental Toxicology and Chemistry, 21: 473–79. [PubMed] [Google Scholar]
- Levy Roie, Magis Andrew T., Earls John C., Manor Ohad, Wilmanski Tomasz, Lovejoy Jennifer, Gibbons Sean M., Omenn Gilbert S., Hood Leroy, and Price Nathan D.. 2020. ‘Longitudinal analysis reveals transition barriers between dominant ecological states in the gut microbiome’, Proceedings of the National Academy of Sciences, 117: 13839–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Hawkins Jia, Mats Gåfvels Maria Olin, Lund Erik G., Andersson Ulla, Schuster Gertrud, Ingemar Björkhem David W. Russell, and Eggertsen Gosta. 2002. ‘Cholic acid mediates negative feedback regulation of bile acid synthesis in mice’, The Journal of clinical investigation, 110: 1191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Min, Wang Baohong, Zhang Menghui, Rantalainen Mattias, Wang Shengyue, Zhou Haokui, Zhang Yan, Shen Jian, Pang Xiaoyan, Zhang Meiling, Wei Hua, Chen Yu, Lu Haifeng, Zuo Jian, Su Mingming, Qiu Yunping, Jia Wei, Xiao Chaoni, Smith Leon M., Yang Shengli, Holmes Elaine, Tang Huiru, Zhao Guoping, Nicholson Jeremy K., Li Lanjuan, and Zhao Liping. 2008. ‘Symbiotic gut microbes modulate human metabolic phenotypes’, Proceedings of the National Academy of Sciences, 105: 2117–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Mingtang, Zhao Xingmin, Zhang Xiufang, Wu Di, and Leng Su. 2018. ‘Biodegradation of 17β-estradiol by Bacterial Co-culture Isolated from Manure’, Scientific reports, 8: 3787–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Wang Q, Wang Y, Sun A, Lin Y, Jin Y, and Li X. 2019. ‘Fecal microbiota transplantation from chronic unpredictable mild stress mice donors affects anxiety-like and depression-like behavior in recipient mice via the gut microbiota-inflammation-brain axis’, Stress, 22: 592–602. [DOI] [PubMed] [Google Scholar]
- Lindheim Lisa, Bashir Mina, Julia Münzker Christian Trummer, Zachhuber Verena, Leber Bettina, Horvath Angela, Pieber Thomas R., Gorkiewicz Gregor, Stadlbauer Vanessa, and Obermayer-Pietsch Barbara. 2017. ‘Alterations in Gut Microbiome Composition and Barrier Function Are Associated with Reproductive and Metabolic Defects in Women with Polycystic Ovary Syndrome (PCOS): A Pilot Study’, PLOS ONE, 12: e0168390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little Michael S., Pellock Samuel J., Walton William G., Tripathy Ashutosh, and Redinbo Matthew R.. 2018. ‘Structural basis for the regulation of β-glucuronidase expression by human gut Enterobacteriaceae’, Proceedings of the National Academy of Sciences, 115: E152–E61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Zhang C, Shi Y, Zhang F, Li L, Wang X, Ling Y, Fu H, Dong W, Shen J, Reeves A, Greenberg AS, Zhao L, Peng Y, and Ding X. 2017. ‘Dysbiosis of Gut Microbiota Associated with Clinical Parameters in Polycystic Ovary Syndrome’, Front Microbiol, 8: 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi Patricia, Goldin Barry, Boutin Eugenie, and Gorbach Sherwood L.. 1978. ‘Metabolism of androgens and estrogens by human fecal microorganisms’, Journal of Steroid Biochemistry, 9: 795–801. [DOI] [PubMed] [Google Scholar]
- Lucas LN, Barrett K, Kerby RL, Zhang Q, Cattaneo LE, Stevenson D, Rey FE, and Amador-Noguez D. 2021. ‘Dominant Bacterial Phyla from the Human Gut Show Widespread Ability To Transform and Conjugate Bile Acids’, mSystems: e0080521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu Maik, Weigand Katharina, Wedi Fatana, Breidenbend Carina, Leister Hanna, Pautz Sabine, Adhikary Till, and Visekruna Alexander. 2018. ‘Regulation of the effector function of CD8+ T cells by gut microbiota-derived metabolite butyrate’, Scientific reports, 8: 14430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Hong Y, Zheng N, Xie G, Lyu Y, Gu Y, Xi C, Chen L, Wu G, Li Y, Tao X, Zhong J, Huang Z, Wu W, Yuan L, Lin M, Lu X, Zhang W, Jia W, Sheng L, and Li H. 2020. ‘Gut microbiota remodeling reverses aging-associated inflammation and dysregulation of systemic bile acid homeostasis in mice sex-specifically’, Gut Microbes, 11: 1450–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Kunimoto K, Muraoka Y, and Katagiri K. 1981. ‘Effect of castration on the appearance of diabetes in NOD mouse’, Jikken Dobutsu, 30: 137–40. [DOI] [PubMed] [Google Scholar]
- Makino S, Kunimoto K, Muraoka Y, Mizushima Y, Katagiri K, and Tochino Y. 1980. ‘Breeding of a non-obese, diabetic strain of mice’, Jikken Dobutsu, 29: 1–13. [DOI] [PubMed] [Google Scholar]
- Mańkowska-Wierzbicka Dorota, Marta Stelmach-Mardas Marcin Gabryel, Tomczak Hanna, Marzena Skrzypczak-Zielińska Oliwia Zakerska-Banaszak, Anna Sowińska Dagmara Mahadea, Baturo Alina, Wolko Łukasz, Słomski Ryszard, and Dobrowolska Agnieszka. 2020. ‘The Effectiveness of Multi-Session FMT Treatment in Active Ulcerative Colitis Patients: A Pilot Study’, Biomedicines, 8: 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markle Janet G. M., Frank Daniel N., Adeli Khosrow, von Bergen Martin, and Danska Jayne S.. 2014. ‘Microbiome manipulation modifies sex-specific risk for autoimmunity’, Gut Microbes, 5: 485–93. [DOI] [PubMed] [Google Scholar]
- Markle Janet G. M., Frank Daniel N., Steven Mortin-Toth Charles E. Robertson, Feazel Leah M., Rolle-Kampczyk Ulrike, von Bergen Martin, McCoy Kathy D., Macpherson Andrew J., and Danska Jayne S.. 2013. ‘Sex Differences in the Gut Microbiome Drive Hormone-Dependent Regulation of Autoimmunity’, Science, 339: 1084–88. [DOI] [PubMed] [Google Scholar]
- Mayneris-Perxachs J, Arnoriaga-Rodríguez M, Luque-Córdoba D, Priego-Capote F, Pérez-Brocal V, Moya A, Burokas A, Maldonado R, and Fernández-Real JM. 2020. ‘Gut microbiota steroid sexual dimorphism and its impact on gonadal steroids: influences of obesity and menopausal status’, Microbiome, 8: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh FM, Maison N, Holtrop G, Young P, Stevens VJ, Ince J, Johnstone AM, Lobley GE, Flint HJ, and Louis P. 2012. ‘Phylogenetic distribution of genes encoding beta-glucuronidase activity in human colonic bacteria and the impact of diet on faecal glycosidase activities’, Environ Microbiol, 14: 1876–87. [DOI] [PubMed] [Google Scholar]
- Mihajlovic Jovana, Leutner Michael, Hausmann Bela, Kohl Gudrun, Schwarz Jasmin, Hannah Röver Nina Stimakovits, Wolf Peter, Maruszczak Katharina, Bastian Magdalena, Kautzky-Willer Alexandra, and Berry David. 2021. ‘Combined hormonal contraceptives are associated with minor changes in composition and diversity in gut microbiota of healthy women’, Environmental Microbiology, 23: 3037–47. [DOI] [PubMed] [Google Scholar]
- Mittan Daniella, Lee Shuko, Miller Elizabeth, Perez Reina C., Basler Joseph W., and Bruder Jan M.. 2002. ‘Bone Loss following Hypogonadism in Men with Prostate Cancer Treated with GnRH Analogs’, The Journal of Clinical Endocrinology & Metabolism, 87: 3656–61. [DOI] [PubMed] [Google Scholar]
- Morris AI, Little JM, and Lester R. 1983. ‘Development of the Bile Acid Pool in Rats from Neonatal Life through Puberty to Maturity’, Digestion, 28: 216–24. [DOI] [PubMed] [Google Scholar]
- Mueller Susanne, Saunier Katiana, Hanisch Christiana, Norin Elisabeth, Alm Livia, Midtvedt Tore, Cresci Alberto, Silvi Stefania, Orpianesi Carla, Maria Cristina Verdenelli Thomas Clavel, Koebnick Corinna, Hans-Joachim Franz Zunft Joël Doré, and Blaut Michael. 2006. ‘Differences in Fecal Microbiota in Different European Study Populations in Relation to Age, Gender, and Country: a Cross-Sectional Study’, Applied and Environmental Microbiology, 72: 1027–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray Emma, Sharma Rupali, Smith Kevin B., Mar Kendall D., Barve Rudra, Lukasik Matthew, Pirwani Atiqa F., Etienne Malette-Guyon Sanjeevani Lamba, Thomas Bronwen J., Homa Sadeghi-Emamchaie Jacky Liang, Mallet Jean-François, Matar Chantal, and Ismail Nafissa. 2019. ‘Probiotic consumption during puberty mitigates LPS-induced immune responses and protects against stress-induced depression- and anxiety-like behaviors in adulthood in a sex-specific manner’, Brain, Behavior, and Immunity, 81: 198–212. [DOI] [PubMed] [Google Scholar]
- Murray Emma, Smith Kevin B., Stoby Karlene S., Thomas Bronwen J., Swenson Michael J., Arber Lauren A., Frenette Emilie, and Ismail Nafissa. 2020. ‘Pubertal probiotic blocks LPS-induced anxiety and the associated neurochemical and microbial outcomes, in a sex dependent manner’, Psychoneuroendocrinology, 112: 104481. [DOI] [PubMed] [Google Scholar]
- Org Elin, Mehrabian Margarete, Parks Brian W., Shipkova Petia, Liu Xiaoqin, Drake Thomas A., and Lusis Aldons J.. 2016. ‘Sex differences and hormonal effects on gut microbiota composition in mice’, Gut Microbes, 7: 313–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellock Samuel J., and Redinbo Matthew R.. 2017. ‘Glucuronides in the gut: Sugar-driven symbioses between microbe and host’, The Journal of biological chemistry, 292: 8569–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perino Alessia, Demagny Hadrien, Velazquez-Villegas Laura, and Schoonjans Kristina. 2021. ‘Molecular physiology of bile acid signaling in health, disease, and aging’, Physiological Reviews, 101: 683–731. [DOI] [PubMed] [Google Scholar]
- Peters Brandilyn A., Lin Juan, Qi Qibin, Usyk Mykhaylo, Isasi Carmen R., Yasmin Mossavar-Rahmani Carol A. Derby, Santoro Nanette, Perreira Krista M., Daviglus Martha L., Kominiarek Michelle A., Cai Jianwen, Knight Rob, Burk Robert D., and Kaplan Robert C.. 2022. ‘Menopause Is Associated with an Altered Gut Microbiome and Estrobolome, with Implications for Adverse Cardiometabolic Risk in the Hispanic Community Health Study/Study of Latinos’, mSystems, 7: e00273–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson Lance W., and Artis David. 2014. ‘Intestinal epithelial cells: regulators of barrier function and immune homeostasis’, Nature Reviews Immunology, 14: 141–53. [DOI] [PubMed] [Google Scholar]
- Plotkin Balbina J., Roose Robert J., Erikson Quenby, and Viselli Susan M.. 2003. ‘Effect of Androgens and Glucocorticoids on Microbial Growth and Antimicrobial Susceptibility’, Current Microbiology, 47: 514–20. [DOI] [PubMed] [Google Scholar]
- Pozzilli P, Signore A, Williams AJ, and Beales PE. 1993. ‘NOD mouse colonies around the world--recent facts and figures’, Immunol Today, 14: 193–6. [DOI] [PubMed] [Google Scholar]
- Qi X, Yun C, Sun L, Xia J, Wu Q, Wang Y, Wang L, Zhang Y, Liang X, Wang L, Gonzalez FJ, Patterson AD, Liu H, Mu L, Zhou Z, Zhao Y, Li R, Liu P, Zhong C, Pang Y, Jiang C, and Qiao J. 2019. ‘Gut microbiota-bile acid-interleukin-22 axis orchestrates polycystic ovary syndrome’, Nat Med, 25: 1225–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon Jason M., Kang Dae-Joong, and Hylemon Phillip B.. 2010. ‘Isolation and characterization of a bile acid inducible 7α-dehydroxylating operon in Clostridium hylemonae TN271’, Anaerobe, 16: 137–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringel-Kulka Tamar, Cheng Jing, Ringel Yehuda, Jarkko Salojärvi Ian Carroll, Palva Airi, de Vos Willem M., and Satokari Reetta. 2013. ‘Intestinal Microbiota in Healthy U.S. Young Children and Adults—A High Throughput Microarray Analysis’, PLOS ONE, 8: e64315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk Maryan G, and Thackray Varykina G. 2020. ‘Intersection of Polycystic Ovary Syndrome and the Gut Microbiome’, Journal of the Endocrine Society, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzetto Lisa, Fava Francesca, Tuohy Kieran M., and Selmi Carlo. 2018. ‘Connecting the immune system, systemic chronic inflammation and the gut microbiome: The role of sex’, Journal of Autoimmunity, 92: 12–34. [DOI] [PubMed] [Google Scholar]
- Paris Rodriguez, Valentina Xin Yi Denise Wong, Solon-Biet Samantha M., Edwards Melissa C., Aflatounian Ali, Gilchrist Robert B., Simpson Stephen J., Handelsman David J., Kaakoush Nadeem O., and Walters Kirsty A.. 2022. ‘The interplay between PCOS pathology and diet on gut microbiota in a mouse model’, Gut Microbes, 14: 2085961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DW 2003. ‘The enzymes, regulation, and genetics of bile acid synthesis’, Annu Rev Biochem, 72: 137–74. [DOI] [PubMed] [Google Scholar]
- Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, Challis C, Schretter CE, Rocha S, Gradinaru V, Chesselet MF, Keshavarzian A, Shannon KM, Krajmalnik-Brown R, Wittung-Stafshede P, Knight R, and Mazmanian SK. 2016. ‘Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson’s Disease’, Cell, 167: 1469–80.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Marcos JA, Rangel-Zuñiga OA, Jimenez-Lucena R, Quintana-Navarro GM, Garcia-Carpintero S, Malagon MM, Landa BB, Tena-Sempere M, Perez-Martinez P, Lopez-Miranda J, Perez-Jimenez F, and Camargo A. 2018. ‘Influence of gender and menopausal status on gut microbiota’, Maturitas, 116: 43–53. [DOI] [PubMed] [Google Scholar]
- Schirmer M, Garner A, Vlamakis H, and Xavier RJ. 2019. ‘Microbial genes and pathways in inflammatory bowel disease’, Nat Rev Microbiol, 17: 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz M, Russell DW, Dietschy JM, and Turley SD. 2001. ‘Alternate pathways of bile acid synthesis in the cholesterol 7alpha-hydroxylase knockout mouse are not upregulated by either cholesterol or cholestyramine feeding’, J Lipid Res, 42: 1594–603. [PubMed] [Google Scholar]
- Sharma Rupali, Rooke Jasmine, Kolmogorova Daria, Melanson Brett, Mallet Jean-François, Matar Chantal, Schwarz Jaclyn, and Ismail Nafissa. 2018. ‘Sex differences in the peripheral and central immune responses following lipopolysaccharide treatment in pubertal and adult CD-1 mice’, International Journal of Developmental Neuroscience, 71: 94–104. [DOI] [PubMed] [Google Scholar]
- Sheng Lili, Prasant Kumar Jena Hui-Xin Liu, Kalanetra Karen M., Gonzalez Frank J., French Samuel W., Krishnan Viswanathan V., Mills David A., and Yvonne Wan Yu-Jui. 2017. ‘Gender Differences in Bile Acids and Microbiota in Relationship with Gender Dissimilarity in Steatosis Induced by Diet and FXR Inactivation’, Scientific reports, 7: 1748–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd Rebecca, Cheung Ada S., Pang Ken, Saffery Richard, and Novakovic Boris. 2021. ‘Sexual Dimorphism in Innate Immunity: The Role of Sex Hormones and Epigenetics’, Frontiers in Immunology, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi N, Li N, Duan X, and Niu H. 2017. ‘Interaction between the gut microbiome and mucosal immune system’, Mil Med Res, 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, and Manning SD. 2016. ‘Impact of age and sex on the composition and abundance of the intestinal microbiota in individuals with and without enteric infections’, Ann Epidemiol, 26: 380–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha Trishla, Arnau Vich Vila Sanzhima Garmaeva, Jankipersadsing Soesma A., Imhann Floris, Collij Valerie, Marc Jan Bonder Xiaofang Jiang, Gurry Thomas, Alm Eric J., Mauro D’Amato Rinse K. Weersma, Scherjon Sicco, Wijmenga Cisca, Fu Jingyuan, Kurilshikov Alexander, and Zhernakova Alexandra. 2019. ‘Analysis of 1135 gut metagenomes identifies sex-specific resistome profiles’, Gut Microbes, 10: 358–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Se Jin, Wang Jincheng, Martino Cameron, Jiang Lingjing, Thompson Wesley K., Shenhav Liat, Daniel McDonald Clarisse Marotz, Harris Paul R., Hernandez Caroll D., Henderson Nora, Ackley Elizabeth, Nardella Deanna, Gillihan Charles, Montacuti Valentina, Schweizer William, Jay Melanie, Combellick Joan, Sun Haipeng, Izaskun Garcia-Mantrana Fernando Gil Raga, Maria Carmen Collado Juana I. Rivera-Viñas, Maribel Campos-Rivera Jean F. Ruiz-Calderon, Knight Rob, and Dominguez-Bello Maria Gloria. 2021. ‘Naturalization of the microbiota developmental trajectory of Cesarean-born neonates after vaginal seeding’, Med, 2: 951–64.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Ziwei, Cai Yuanyuan, Lao Xingzhen, Wang Xue, Lin Xiaoxuan, Cui Yingyun, Praveen Kumar Kalavagunta Jun Liao, Jin Liang, Shang Jing, and Li Jing. 2019. ‘Taxonomic profiling and populational patterns of bacterial bile salt hydrolase (BSH) genes based on worldwide human gut microbiome’, Microbiome, 7: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spichak Simon, Donoso Francisco, Moloney Gerard M., Gunnigle Eoin, Brown Jillian M., Codagnone Martin, Dinan Timothy G., and Cryan John F.. 2021. ‘Microbially-derived short-chain fatty acids impact astrocyte gene expression in a sex-specific manner’, Brain, Behavior, & Immunity - Health, 16: 100318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner C, Holleboom AG, Karuna R, Motazacker MM, Kuivenhoven JA, Frikke-Schmidt R, Tybjaerg-Hansen A, Rohrer L, Rentsch KM, and Eckardstein Av. 2012. ‘Lipoprotein distribution and serum concentrations of 7α-hydroxy-4-cholesten-3-one and bile acids: effects of monogenic disturbances in high-density lipoprotein metabolism’, Clin Sci (Lond), 122: 385–96. [DOI] [PubMed] [Google Scholar]
- Sui Yue, Wu Jianming, and Chen Jianping. 2021. ‘The Role of Gut Microbial β-Glucuronidase in Estrogen Reactivation and Breast Cancer’, Frontiers in Cell and Developmental Biology, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson Agnes, Brunkwall Louise, Roth Bodil, Orho-Melander Marju, and Ohlsson Bodil. 2021. ‘Associations Between Endometriosis and Gut Microbiota’, Reproductive Sciences, 28: 2367–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi Tomohisa, Naito Yuji, Inoue Ryo, Kashiwagi Saori, Uchiyama Kazuhiko, Mizushima Katsura, Tsuchiya Saeko, Dohi Osamu, Yoshida Naohisa, Kamada Kazuhiro, Ishikawa Takeshi, Handa Osamu, Konishi Hideyuki, Okuda Kayo, Tsujimoto Yoshimasa, Ohnogi Hiromu, and Itoh Yoshito. 2019. ‘Differences in gut microbiota associated with age, sex, and stool consistency in healthy Japanese subjects’, Journal of Gastroenterology, 54: 53–63. [DOI] [PubMed] [Google Scholar]
- Tian Yuan, Gui Wei, Koo Imhoi, Smith Philip B., Allman Erik L., Nichols Robert G., Rimal Bipin, Cai Jingwei, Liu Qing, and Patterson Andrew D.. 2020. ‘The microbiome modulating activity of bile acids’, Gut Microbes, 11: 979–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres Pedro J, Ho Bryan S, Arroyo Pablo, Sau Lillian, Chen Annie, Kelley Scott T, and Thackray Varykina G. 2019. ‘Exposure to a Healthy Gut Microbiome Protects Against Reproductive and Metabolic Dysregulation in a PCOS Mouse Model’, Endocrinology, 160: 1193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres Pedro J., Siakowska Martyna, Banaszewska Beata, Pawelczyk Leszek, Duleba Antoni J., Kelley Scott T., and Thackray Varykina G.. 2018. ‘Gut Microbial Diversity in Women With Polycystic Ovary Syndrome Correlates With Hyperandrogenism’, The Journal of clinical endocrinology and metabolism, 103: 1502–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres Pedro J., Skarra Danalea V., Ho Bryan S., Sau Lillian, Anvar Arya R., Kelley Scott T., and Thackray Varykina G.. 2019. ‘Letrozole treatment of adult female mice results in a similar reproductive phenotype but distinct changes in metabolism and the gut microbiome compared to pubertal mice’, BMC Microbiology, 19: 57–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turley SD, Schwarz M, Spady DK, and Dietschy JM. 1998. ‘Gender-related differences in bile acid and sterol metabolism in outbred CD-1 mice fed low- and high-cholesterol diets’, Hepatology, 28: 1088–94. [DOI] [PubMed] [Google Scholar]
- Valeri Francesco, and Endres Kristina. 2021. ‘How biological sex of the host shapes its gut microbiota’, Frontiers in Neuroendocrinology, 61: 100912. [DOI] [PubMed] [Google Scholar]
- van Best N, Rolle-Kampczyk U, Schaap FG, Basic M, Olde Damink SWM, Bleich A, Savelkoul PHM, von Bergen M, Penders J, and Hornef MW. 2020. ‘Bile acids drive the newborn’s gut microbiota maturation’, Nat Commun, 11: 3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hooff MH, Voorhorst FJ, Kaptein MB, Hirasing RA, Koppenaal C, and Schoemaker J. 1999. ‘Endocrine features of polycystic ovary syndrome in a random population sample of 14–16 year old adolescents’, Hum Reprod, 14: 2223–9. [DOI] [PubMed] [Google Scholar]
- van Keulen Britt J, Dolan Conor V, van der Voorn Bibian, Andrew Ruth, Walker Brian R, Hulshoff Pol Hilleke, Boomsma Dorret I, Rotteveel Joost, and Finken Martijn J J. 2020. ‘Sexual dimorphism in cortisol metabolism throughout pubertal development: a longitudinal study’, Endocrine Connections, 9: 542–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatanen Tommi, Kostic Aleksandar D, d’Hennezel Eva, Siljander Heli, Franzosa Eric A, Yassour Moran, Kolde Raivo, Vlamakis Hera, Arthur Timothy D, Hämäläinen Anu-Maaria, Peet Aleksandr, Tillmann Vallo, Uibo Raivo, Mokurov Sergei, Dorshakova Natalya, Ilonen Jorma, Virtanen Suvi M, Szabo Susanne J, Porter Jeffrey A, Lähdesmäki Harri, Huttenhower Curtis, Gevers Dirk, Cullen Thomas W, Knip Mikael, and Xavier Ramnik J. 2016. ‘Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans’, Cell, 165: 842–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlström A, Sayin SI, Marschall HU, and Bäckhed F. 2016. ‘Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism’, Cell Metab, 24: 41–50. [DOI] [PubMed] [Google Scholar]
- Wang Mengjie, Zhang Youjie, Miller David, Naveen O Rehman Xi Cheng, Yeo Ji-Youn, Joe Bina, and Hill Jennifer W. 2020. ‘Microbial Reconstitution Reverses Early Female Puberty Induced by Maternal High-fat Diet During Lactation’, Endocrinology, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Wen-Xia, Chen Li, Wang Guo-Yu, Zhang Jin-Ling, Tan Xian-Wen, Lin Qiu-Hong, Chen Yu-Jie, Zhang Jian, Zhu Ping-Ping, Miao Jia, Su Ming-Ming, Liu Chang-Xiao, Jia Wei, and Lan Ke. 2020. ‘Urinary Bile Acid Profile of Newborns Born by Cesarean Section Is Characterized by Oxidative Metabolism of Primary Bile Acids: Limited Roles of Fetal-Specific CYP3A7 in Cholate Oxidations’, Drug Metabolism and Disposition, 48: 662–72. [DOI] [PubMed] [Google Scholar]
- Ward Joseph B. J., Lajczak Natalia K., Kelly Orlaith B., O’Dwyer Aoife M., Giddam Ashwini K., Gabhann Joan Ní, Franco Placido, Tambuwala Murtaza M., Jefferies Caroline A., Keely Simon, Roda Aldo, and Keely Stephen J.. 2017. ‘Ursodeoxycholic acid and lithocholic acid exert anti-inflammatory actions in the colon’, American Journal of Physiology-Gastrointestinal and Liver Physiology, 312: G550–G58. [DOI] [PubMed] [Google Scholar]
- Whitacre Caroline C., Reingold Stephen C., O’Looney Patricia A., Blankenhorn Elizabeth, Brinley Floyd, Collier Elaine, Duquette Pierre, Fox Howard, Giesser Barbara, Gilmore Wendy, Lahita Robert, Nelson J. Lee, Reiss Carol, Riskind Peter, and Voskuhl Rhonda. 1999. ‘A Gender Gap in Autoimmunity’, Science, 283: 1277–78. [DOI] [PubMed] [Google Scholar]
- Wiertsema SP, van Bergenhenegouwen J, Garssen J, and Knippels LMJ. 2021. ‘The Interplay between the Gut Microbiome and the Immune System in the Context of Infectious Diseases throughout Life and the Role of Nutrition in Optimizing Treatment Strategies’, Nutrients, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilharm Anneke, Brigas Helena C., Sandrock Inga, Ribeiro Miguel, Amado Tiago, Reinhardt Annika, Demera Abdi, Hoenicke Lisa, Strowig Till, Carvalho Tânia, Prinz Immo, and Ribot Julie C.. 2021. ‘Microbiota-dependent expansion of testicular IL-17-producing Vγ6+ γδ T cells upon puberty promotes local tissue immune surveillance’, Mucosal Immunology, 14: 242–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston Jenessa A., and Theriot Casey M.. 2020. ‘Diversification of host bile acids by members of the gut microbiota’, Gut Microbes, 11: 158–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter Jeanette, and Bokkenheuser Victor D.. 1987. ‘Bacterial metabolism of natural and synthetic sex hormones undergoing enterohepatic circulation’, Journal of Steroid Biochemistry, 27: 1145–49. [DOI] [PubMed] [Google Scholar]
- Xiang Xiaoqiang, Backman Janne T., Neuvonen Pertti J., and Niemi Mikko. 2012. ‘Gender, but not CYP7A1 or SLCO1B1 Polymorphism, Affects the Fasting Plasma Concentrations of Bile Acids in Human Beings’, Basic & Clinical Pharmacology & Toxicology, 110: 245–52. [DOI] [PubMed] [Google Scholar]
- Yang Xiao, Wu Richao, Qi Dan, Fu Linlin, Song Tian, Wang Ying, Bian Yuehong, and Shi Yuhua. 2021. ‘Profile of Bile Acid Metabolomics in the Follicular Fluid of PCOS Patients’, Metabolites, 11: 845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YL, Zhou WW, Wu S, Tang WL, Wang ZW, Zhou ZY, Li ZW, Huang QF, He Y, and Zhou HW. 2021. ‘Intestinal Flora is a Key Factor in Insulin Resistance and Contributes to the Development of Polycystic Ovary Syndrome’, Endocrinology, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko Tanya, Rey Federico E., Manary Mark J., Trehan Indi, Maria Gloria Dominguez-Bello Monica Contreras, Magris Magda, Hidalgo Glida, Baldassano Robert N., Anokhin Andrey P., Heath Andrew C., Warner Barbara, Reeder Jens, Kuczynski Justin, Caporaso J. Gregory, Lozupone Catherine A., Lauber Christian, Clemente Jose Carlos, Knights Dan, Knight Rob, and Gordon Jeffrey I.. 2012. ‘Human gut microbiome viewed across age and geography’, Nature, 486: 222–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young Nicholas A., Wu Lai-Chu, Burd Craig J., Friedman Alexandra K., Kaffenberger Benjamin H., Rajaram Murugesan V. S., Schlesinger Larry S., James Hayley, Shupnik Margaret A., and Jarjour Wael N.. 2014. ‘Estrogen modulation of endosome-associated toll-like receptor 8: An IFNα-independent mechanism of sex-bias in systemic lupus erythematosus’, Clinical Immunology, 151: 66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Ming, Li Dong, Zhang Zhe, Sun Huihui, An Min, and Wang Guoyun. 2018. ‘Endometriosis induces gut microbiota alterations in mice’, Human Reproduction, 33: 607–16. [DOI] [PubMed] [Google Scholar]
- Yuan Xin, Chen Ruimin, Zhang Ying, Lin Xiangquan, and Yang Xiaohong. 2020. ‘Sexual dimorphism of gut microbiota at different pubertal status’, Microbial Cell Factories, 19: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy Leonid, Burrows Michael, Khan Aly A, Graham Laura, Volchkov Pavel, Becker Lev, Antonopoulos Dionysios, Umesaki Yoshinori, and Chervonsky Alexander V. 2013. ‘Gender Bias in Autoimmunity Is Influenced by Microbiota’, Immunity, 39: 400–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarotsky V, Huang M-Y, Carman W, Morgentaler A, Singhal PK, Coffin D, and Jones TH. 2014. ‘Systematic literature review of the risk factors, comorbidities, and consequences of hypogonadism in men’, Andrology, 2: 819–34. [DOI] [PubMed] [Google Scholar]
- Zhang Bingjie, Shen Shanmei, Gu Tianwei, Hong Ting, Liu Jiayi, Sun Jie, Wang Hongdong, Bi Yan, and Zhu Dalong. 2019. ‘Increased circulating conjugated primary bile acids are associated with hyperandrogenism in women with polycystic ovary syndrome’, The Journal of Steroid Biochemistry and Molecular Biology, 189: 171–75. [DOI] [PubMed] [Google Scholar]
- Zhang Husen, Sparks Joshua B., Karyala Saikumar V., Settlage Robert, and Luo Xin M.. 2015. ‘Host adaptive immunity alters gut microbiota’, The ISME Journal, 9: 770–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Hui, Chen Juanjuan, Li Xiaoping, Sun Qiang, Qin Panpan, and Wang Qi. 2019. ‘Compositional and functional features of the female premenopausal and postmenopausal gut microbiota’, FEBS Letters, 593: 2655–64. [DOI] [PubMed] [Google Scholar]
- Zhao Qing, and Elson Charles O.. 2018. ‘Adaptive immune education by gut microbiota antigens’, Immunology, 154: 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Yu J, Liang C, Li S, Wen X, and Li Y. 2021. ‘Characterization on gut microbiome of PCOS rats and its further design by shifts in high-fat diet and dihydrotestosterone induction in PCOS rats’, Bioprocess Biosyst Eng, 44: 953–64. [DOI] [PubMed] [Google Scholar]
- Zhou Hao-Yue, Guo Biao, Lufumpa Eniya, Li Xiao-Mei, Chen Li-Hong, Meng Xiang, and Li Bao-Zhu. 2021. ‘Comparative of the Effectiveness and Safety of Biological Agents, Tofacitinib, and Fecal Microbiota Transplantation in Ulcerative Colitis: Systematic Review and Network Meta-Analysis’, Immunological Investigations, 50: 323–37. [DOI] [PubMed] [Google Scholar]
- Zhu Baoli, Wang Xin, and Li Lanjuan. 2010. ‘Human gut microbiome: the second genome of human body’, Protein & Cell, 1: 718–25. [DOI] [PMC free article] [PubMed] [Google Scholar]


