Abstract
Exposure to environmental hazards occurs at different stages of our lifetime–infant, child, adult. This study integrates recently published toxicogenomics data to examine how exposure to a known rat chemical carcinogen (pentabrominated diphenyl ether (PBDE)) upregulated liver transcriptomic changes at different life cycle stages (PND 4, PND 22, adult). We found that at all three life cycle stages PBDE exposure induced hepatocellular transcriptomic changes in disease pathways including cancer, metabolic, membrane function, and Nrf2 antioxidant pathways, pathways all characteristics of chemical carcinogens. In addition, in the adult rat after a 5-day exposure to the chemical carcinogen, there was upregulation of members of the Ras oncogenic pathway, a specific pathway found to be activated in the PBDE-induced tumors in rats in a previous hazard identification cancer study. The findings of liver transcript changes characteristic of carcinogenic activity after early life exposures and after short-term adult exposures provides data to support the use of transcriptomic data to predict the apical cancer endpoints in model studies. Using data from gene expression profiling studies after neonatal, young, or adult short-term chemical exposure helps to meet the 21st century toxicology goal of developing study designs to reduce, refine, and replace the use of traditional 2-year rodent cancer studies to provide hazard identification information. The studies reported here find that key transcripts associated with carcinogenesis were elevated in neonate (PND 4), young (PND 22) and adult animals after short-term exposure to PBDE, a known experimental chemical carcinogen in model systems.
Keywords: pentabrominated diphenyl ether (PBDE), liver transcripts, PND 4, PND 22, adult life stage changes
1 Introduction
In the 21st century, the toxicology community seeks to develop hazard identification information using “modern tools”, including data from gene expression profiling studies (toxicogenomic (TGMX) studies). The U.S. National Academy of Sciences discusses this strategy in the 2007 report Toxicity Testing in the 21st Century: A Vision and a Strategy (U.S. National Research Councilo.t.N.A, 2007). This paper evaluates liver toxicogenomic changes after exposure to a model chemical carcinogen (pentabrominated diphenyl ether (Dunnick et al., 2018a; Dunnick et al., 2018b)) in male rats at PND 4 (after in utero exposure), PND 22 (after in utero/postnatal exposure), and in adult male rats (after 5-day exposures). The hypothesis for this study was that exposure to pentabrominated diphenyl ether at different life cycle stages would activate disease pathways that could be used to predict longer term toxic and carcinogenic effects, thereby reducing the number of animals needed to obtain toxic and cancer hazard identification information used in risk assessment evaluations (Dunnick et al., 2018c; Shockley et al., 2020a; Dunnick et al., 2020).
For the model carcinogen (pentabrominated diphenyl ether (PBDE)) used in this study, world-wide exposure continues to be of concern because PBDE containing products remain in the home and in the environment (Maddela et al., 2020; Simonetti et al., 2020; Jin et al., 2021), and PBDE exposures may occur during e-waste recycling (Wei et al., 2020; Zhou et al., 2020; Wannomai et al., 2021). As fat-soluble organic pollutants, PBDEs persist in human and animal tissues with long tissue half-lives (Möller et al., 2011; Abbasi et al., 2019; Maddela et al., 2020). The CDC NHANES program (National Health and Nutrition Examination Survey) has collected data on occurrence of these chemicals in human tissues, and exposures to these chemicals continue to be widespread in the United States (Sjödin et al., 2019; Vuong et al., 2020). In humans, exposure to these chemicals has been associated with various health hazards including alteration of thyroid function and developmental changes (Chen et al., 2014; Makey et al., 2016; Sjödin et al., 2020).
Three recent toxicogenomics studies evaluated the gene expression response to PBDE exposure in male rats at PND 4, PND 22, and adult animal evaluations (Dunnick et al., 2018c; Shockley et al., 2020a; Dunnick et al., 2020). Each of these studies had a separate research interest (i.e., the response of male rats to PBDE at either PND4, PND22, or adult). Accordingly, each investigation produced a distinctive dataset derived from the unique study design underlying its own particular scientific focus. These previous analyses were conducted independently and served to expand the field of PBDE research by characterizing the toxicological response to the carcinogenic agent at a specific point in male rat development. This study analyzes and compares the PBDE-induced liver transcriptomic changes at PND 4, PND 22, or in the adult to explore the gene expression response to PBDE in the male rat at different life cycle stages. The new information resulting from this analysis can be used in risk assessment and may help to lay a framework in predicting long-term toxic effects following exposure to environmental agents at different life cycle stages (Harrill et al., 2019).
2 Materials and methods
2.1 Chemicals
As described in previously reported studies (Dunnick et al., 2018c; Shockley et al., 2020a; Dunnick et al., 2020), the chemical pentabromodiphenyl ether-47 (PBDE-47, 2,2′,4,4′-tetrabromodiphenyl ether; CAS# 5436-43-1) was obtained from Cerillant Corp (Round Rock, TX; Lot ER081208-02). Chemical identity was confirmed by mass spectrometry and nuclear magnetic resonance spectroscopy. The purity for PBDE-47 was 99.6% by gas chromatography (GC) analysis using flame ionization detection. There were no quantifiable pentabrominated dibenzodioxins or furans present in the PBDE-47 sample. PBDE-mixture (DE-71, technical pentabromodiphenyl; CAS# 32534-81-9) was obtained from Great Lakes Corporation (West Lafayette, IN; Lot 2550OA30A). The identity and purity were determined as described previously. The DE-71 composition was: PBDE-99 (41.7%), PBDE-47 (35.7%), PBDE-100 (10.4%), PBDE-154 (3.6%), PBDE-153 (3.3%), and PBDE-85 (2%); low levels of pentabrominated dibenzodioxins and furans were also identified (approximately 7 × 10–6% by weight).
2.2 Animals and exposures
The animals used were either male Wistar Han rat or Harlan Sprague Dawley rats which induced transcripts encoding metabolic enzymes, members of the Nrf2 antioxidant pathway, and ABC transporters upon exposure to PBDE-47 and the PBDE-47 mixture as described in previous publications (Dunnick et al., 2018c; Shockley et al., 2020a; Shockley et al., 2020b; Dunnick et al., 2020). In this paper, the transcriptomic data from different life stages was analyzed by groups (Group 1–5, see Table 1). Group One examined rat liver toxicogenomic changes after PBDE-47 exposure for 5 days. In this study adult Harlan Sprague Dawley male rats were exposed to the PBDE mixture by oral gavage in corn oil for five consecutive days; on the sixth day liver samples were taken for toxicogenomic analysis (Shockley et al., 2020a; Shockley et al., 2020b). Group Two and Group Four examined rat liver toxicogenomic changes in PND 4 pups after exposure of dams to PBDE-47 or the PBDE mixture (DE-71), respectively (Dunnick et al., 2020). In these studies, Wistar Han rat dams were dosed with a dosing volume of 5 ml/kg/day from GD 6 through PND 4, 7 days per week. Controls received corn oil vehicles. Liver samples were taken for toxicogenomic analysis in male pups at PND 4. Group Three and Group Five examined liver toxicogenomic changes after exposure of dams and pups to PBDE-47 or the PBDE mixture, respectively (Dunnick et al., 2018c). In these studies, Wistar Han rat dams were dosed with a dosing volume of 5 ml/kg/day from GD 6 through PND 21, 7 days per week. Controls received corn oil vehicles. Pups received daily the PBDE mixture dosing from PND12-21. At PND 22 liver samples from pups were taken for toxicogenomic analysis.
TABLE 1.
Summary of thyroid hormone levels and liver pathology findings after exposure to PBDE 47 and/or PBDE mixture (DE-71) at different life cycle stages.
| Group 1 (Shockley et al., 2020a) | Group 2 (Dunnick et al., 2020) | Group 3 (Dunnick et al., 2018c) | Group 4 (Dunnick et al., 2020) | Group 5 (Dunnick et al., 2018c) | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chemical exposure | PBDE 47 | PBDE 47 | PBDE 47 | PBDE mixture (DE-71) | PBDE mixture (DE-71) | |||||||||||||||||
| Dosing Schedule | 5 days of consecutive dosing in seven-week-old Harlan Sprague Dawley Rats | Pups from Wistar Han rat dams exposed from GD 6 through PND 4 (no direct pup dosing) | Pups from Wistar Han rat dams exposed GD 6 through PND 21; pups received daily dosing from PND 12–21 | Pups from Wistar Han rat dams exposed from GD 6 through PND 4 (no direct pup dosing) | Pups from Wistar Han rat dams exposed GD 6 through PND 21; pups received daily dosing from PND 12–21 | |||||||||||||||||
| TGMX time point | After 5 Day Exposure | PND 4 | PND 22 | PND 4 | PND 22 | |||||||||||||||||
| Oral gavage (μmol/kg) | 0 | 0.1 | 1.0 | 10 | 100 | 1000 | 0 | 0.2 | 31 | 103 | 0 | 0.2 | 31 | 103 | 0 | 0.2 | 27 | 89 | 0 | 0.2 | 27 | 89 |
| Total Thyroxine TT4 (Ig/dL) | 5.77 ± 0.63 | 5.77 ± 0.35 | 5.00 ± 0.27 | 5.94 ± 0.28 | 1.48 ± 0.10** | 0.80 ± 0.08** | 1.14 ± 0.16 | 1.18 ± 0.13 | 0** | 0** | 5.08 ± 0.21 | 5.05 ± 0.26 | 3.20 ± 0.08** | 3.00 ± 0.10** | 0.96 ± 0.15 | 1.5 ± 0.15 | 0.59 ± 0.11 | 0** | 5.67 ± 0.36 | 5.67 ± 0.31 | 3.66 ± 0.19** | 2.69 ± 0.23** |
| T4 (% control) | - | 100 | 86 | 102 | 25 | 14 | - | 104 | 0 | 0 | - | 99 | 63 | 59 | - | 157 | 61 | 0 | - | 100 | 65 | 48 |
| Treatment related liver lesions | + | ++ | + | + | + | + | + | + | + | |||||||||||||
p = < 0.01 (treated group vs. control group).
Pathology Designation: + (minimal hepatocyte hypertrophy and/or central vacuolization); ++ (moderate or marked hepatocyte hypertrophy and/or central vacuolization).
2.3 Microarray, RNA extraction method, and statistical analysis
The methods for liver transcriptomics were similar across the different life cycle exposure scenarios. In all cases the left lateral lobe of the liver was processed for RNA isolation. Liver RNA was isolated from the liver tissue, amplified and biotin labeled, and hybridized to Affymetrix Rat Genome 230 2.0 arrays (Affymetrix, Santa Clara, CA) as described previously (Dunnick et al., 2018c; Shockley et al., 2020a; Dunnick et al., 2020). Each GeneChip® array was scanned using an Affymetrix GeneChip® Scanner 3000 7G to generate raw expression level data (.CEL files). Probe intensity data from all Rat Genome 230 version two Affymetrix GeneChip® arrays were read into the R software environment (http://www.R-project.org) directly from. CEL files using the R/affy package (Gautier et al., 2004). Normalization was carried out using the robust multi-array average (RMA) method separately for each Group.
Statistical contrasts were used to find pairwise gene expression differences between the control group and each dose group using the R/maanova package (Wu et al., 2003). For each flame retardant, the model
| (1) |
was used to fit the log2 transformed gene expression measures Yi, where µ is the mean for each array and εi captures random error for probe set i. All statistical tests were performed using Fs, a modified F-statistic incorporating shrinkage estimates of variance components (Cui et al., 2005). To reduce the number of false positives, p-values were adjusted for multiple hypothesis testing corresponding to all probe sets on the array using the Benjamin-Hochberg false discovery rate (FDR) procedure implemented using the p.adjust() function in R. This correction controls the expected proportion of errors among the significant results (Benjamini and Hochberg, 1995). Unless otherwise noted, an FDR threshold of 0.05 was used for statistical significance. Log2 fold changes for each exposure group were calculated by subtracting the control (0 μmol/kg) and dose treated relative expression values obtained from model (1) above (Churchill, 2004).
Overrepresented canonical pathways were determined from each gene list obtained above by testing for association with gene pathway relationships (www.ingenuity.com). Pathway enrichment was determined based on the one-tailed Fisher exact test, where p-values were adjusted for multiple testing using the FDR approach described above.
The transcriptomic data for these studies can be found on the National Library of Medicine’s GEO database web site. The Affymetrix data for each dataset can be accessed from GEO using GSE153366 for Group One, GSE154914 for Group Two and Group Four, and GSE124431 for Group Three and Group Five.
The benchmark dose (BMD) is defined as the dose corresponding to a stimulated change in response referred to as the benchmark response (BMR). Liver transcriptomic data were used to calculate the BMD and the lower bound of the 95% confidence interval of the BMD using BMDExpress version 2.0 (Phillips et al., 2019). All BMD calculations were performed within the BMDExpress framework separately for each combination of chemical and Group.
The BMD analysis followed the criteria described in the Genomic Dose Response recommendations (National Toxicology Program, 2018). First, we determined whether there was adequate signal in each dataset. Control-AFFX probe sets were first removed from each dataset, and a classical one-way ANOVA was used to filter the remaining RMA-normalized probe set intensities to find transcripts that were differentially expressed across experimental groups with a p-value <0.05. The presence of differentially expressed transcripts indicated that the dataset contained adequate signal. Next, for datasets with adequate signal, we used a bootstrap version of Williams’ trend test to find probe sets with dose group changes relative to the control group using a p-value <0.05 and a fold change of at least 1.5 computed with 10,000 permutations of group labels. Next, the Hill, power, linear, second-degree polynomial, and a set of four exponential models were fit to the data for each remaining probe set. The BMR level was set to 1 standard deviation above or below the control group. The lowest Akaike information criterion (AIC) was used to select the best fitting model. Hill model fits were not selected if the estimated dose at half maximal response was less than 1/3 of the lowest positive dose, and the next best model was selected instead.
The calculated BMD values are used as input data for Gene Ontology (GO) analyses. When more than one probe set mapped to the same Entrez ID, the BMD values were averaged across probe sets to obtain a single expression value for each Entrez ID. Probe sets that mapped to more than one Entrez ID were removed from the analysis. The resulting Entrez IDs were matched to Biological Process GO terms as a basis for gene set definitions. The output consists of a range of summary exposure levels (mmol/kg/day) for BMD and BMDL for each category computed from the BMD and BMDL values for the genes in a category. Probes were removed if BMDU/BMDL >40 with a fit p-value <0.1 and if BMD > highest dose level. The final GO Biological Process results were further processed to have ≥3 genes and ≥5% of the genes in the category with genes that passed the filtering scheme described above.
3 Results
Exposure of the male rat to PBDE-47 or PBDE mixture at different life cycle stages (PND 4, PND 22, and adult) caused treatment-related rat liver transcript changes (Table 2). Generally, there were more transcripts upregulated than downregulated for each of the exposure groups. At approximately 100 mmol/kg treatment, the number of significant transcripts was greatest in the adult animal, followed by PND 22 animals, and finally in PND 4 animals. While this study uses a false discovery rate significance threshold of 0.05, the trends described above were consistent across a wide range of significance thresholds (Table 2). In each age group, the number of significant transcripts increased with increasing exposure to PBDE. At PND 4 and PND 22, there were more significant transcripts after PBDE 47 exposure than after exposure to the PBDE mixture at the same exposure levels, except at PND 4 at 100 mmol/kg treatment in which there was a comparable number of changes.
TABLE 2.
Number of significant transcripts induced in the liver by pentabrominated diphenyl ether (PBDE) or PBDE mixture (DE-71).
| False discovery rate (FDR) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Group | Animals | Chemical | TGMX time point | Dose (μmol/kg) | 0.2 | 0.05 | 0.01 | 0.001 | 10–10 |
| 1 | HSD Rats | PBDE 47 | After 5 Days | 0.1 | 280 | 32 | 5 | 0 | 0 |
| 1 | HSD Rats | PBDE 47 | After 5 Days | 1.0 | 247 | 22 | 3 | 0 | 0 |
| 1 | HSD Rats | PBDE 47 | After 5 Days | 10 | 479 | 99 | 24 | 7 | 2 |
| 1 | HSD Rats | PBDE 47 | After 5 Days | 100 | 2162 | 948 | 446 | 195 | 33 |
| 1 | HSD Rats | PBDE 47 | After 5 Days | 1000 | 4670 | 2937 | 2016 | 1268 | 496 |
| 2 | WH pups | PBDE 47 | PND4 | 0.2 | 139 | 2 | 0 | 0 | 0 |
| 2 | WH pups | PBDE 47 | PND4 | 31 | 342 | 40 | 11 | 3 | 2 |
| 2 | WH pups | PBDE 47 | PND4 | 103 | 655 | 140 | 50 | 20 | 3 |
| 3 | WH pups | PBDE mixture | PND4 | 0.2 | 2 | 0 | 0 | 0 | 0 |
| 3 | WH pups | PBDE mixture | PND4 | 31 | 103 | 13 | 1 | 0 | 0 |
| 3 | WH pups | PBDE mixture | PND4 | 103 | 709 | 161 | 24 | 0 | 0 |
| 4 | WH pups | PBDE 47 | PND22 | 0.2 | 255 | 13 | 1 | 0 | 0 |
| 4 | WH pups | PBDE 47 | PND22 | 27 | 999 | 263 | 92 | 32 | 13 |
| 4 | WH pups | PBDE 47 | PND22 | 89 | 1675 | 590 | 240 | 82 | 16 |
| 5 | WH pups | PBDE mixture | PND22 | 0.2 | 33 | 1 | 0 | 0 | 0 |
| 5 | WH pups | PBDE mixture | PND22 | 27 | 318 | 101 | 43 | 6 | 3 |
| 5 | WH pups | PBDE mixture | PND22 | 89 | 832 | 232 | 91 | 10 | 5 |
The significant liver transcripts at all life cycle stages included upregulation of transcripts for liver metabolic phase one and phase two enzymes, NrF2 antioxidant pathway transcripts, and membrane transport protein transcripts (Table 3 and Figure 1). Some of the transcripts with the greatest fold change (compared to controls) were changes in metabolic function transcripts including upregulation of cytochrome activity transcripts for Cyp2b6, Cyp1a1, Cyp3a5, Por (Table 3). The upregulation of UGT transcripts (Table 3) was accompanied by lower levels of thyroid hormone level (T4) after PBDE exposure (Table 1). T4 levels were decreased with increasing PBDE exposure levels in rats at PND4, PND22, and in adult animals, and there were treatment-related liver lesions at the higher exposure levels in rats at all three life cycle stages (Table 1). The liver lesions were most severe in the PBDE adult exposed rats (Shockley et al., 2020a).
TABLE 3.
Selected PBDE-induced liver transcript alterations associated with metabolic, membrane, NRF2 pathway, and lipid function and cancer pathways.
| Gene | Group 1 (Shockley et al., 2020a) | Group 2 (Dunnick et al., 2020) | Group 3 (Dunnick et al., 2018c) | Group 4 (Dunnick et al., 2020) | Group 5 (Dunnick et al., 2018c) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chemical Exposure | PBDE 47 | PBDE 47 | PBDE 47 | PBDE mixture (DE-71) | PBDE mixture (DE-71) | ||||||||||||
| Dosing Schedule | 5 days of consecutive dosing in seven-week-old Harlan Sprague Dawley Rats | Pups from Wistar Han rat dams exposed from GD 6 through PND 4 (no direct pup dosing) | Pups from Wistar Han rat dams exposed GD 6 through PND 21; pups received daily dosing from PND 12–21 | Pups from Wistar Han rat dams exposed from GD 6 through PND 4 (no direct pup dosing) | Pups from Wistar Han rat dams exposed GD 6 through PND 21; pups received daily dosing from PND 12–21 | ||||||||||||
| TGMX Time Point | After 5-day exposure | PND4 | PND22 | PND4 | PND22 | ||||||||||||
| Oral Gavage (μmol/kg) | 0.1 | 1 | 10 | 100 | 1000 | 0.2 | 31 | 103 | 0.2 | 31 | 103 | 0.2 | 27 | 89 | 0.2 | 27 | 89 |
| Liver Metabolism | |||||||||||||||||
| 1370269_at Cyp1a1 cytochrome P450 family 1 subfamily A member 1 | 6.11*** | 65.54*** | 5.66** | 10.98*** | 4.28* | 59.68* | 91.3* | 5.13 | 60.47*** | 77.64*** | |||||||
| 1387243_at Cyp1a2 cytochrome P450 family 1 subfamily A member 2 | 1.23* | 2.26 | 2.09 | −1.27 | 13.88 | 53.83* | 2* | 2.49* | |||||||||
| 1369136_at Cyp2a6 (includes others) cytochrome P450 family 2 subfamily A member 6 | 5.33* | 7.17** | 14.54*** | 23.38*** | 6.99* | 16.93* | 14.92* | 14.31* | |||||||||
| 1371076_at Cyp2Bb cytochrome P450 family 2 subfamily B member 6 | 4.51*** | 10.94*** | 12.51*** | 3.98*** | 4.4*** | 4.25*** | 4.52*** | 3.41* | 4.03* | 4.89** | 4.7** | ||||||
| 1370241_at Cyp2c8 cytochrome P450 family 2 subfamily C member 8 | 1.28* | 5.73* | 9.26** | 24.91*** | 31.68*** | 5.53* | 11.15* | 46.27** | 55.8** | ||||||||
| 1370580_a_at Cyp2c19 cytochrome P450 family 2 subfamily C member 19 | 1.2** | 1.34*** | 1.41*** | 3.37*** | 3.49*** | 1.64*** | 1.69*** | 4.34* | 4.89* | 1.69* | 1.72* | ||||||
| 1387118_at Cyp3a5 cytochrome P450 family 3 subfamily A member 5 | 1.37* | 2.21*** | 2.84*** | 1.62* | 1.71* | −1.23 | 1.68** | 1.84** | 1.6* | 1.78* | 1.96*** | 1.97*** | |||||
| 1387109_at Por cytochrome p450 oxidoreductase | 2.24* | 3.85*** | 1.48 | 1.76* | 1.76* | 1.92* | 2.49* | 2.1* | 2.52* | ||||||||
| 1368607_at Cyp4a22 cytochrome P450 family 4 subfamily A member 22 | 3.32** | 4.36*** | 2.82* | 2.57 | 3.85* | 3.46* | |||||||||||
| 1368905_at Ces2c (includes others) carboxylesterase 2C | 1.68** | 6*** | 18.53*** | 2.15 | 3.29* | 10.86*** | 14.86*** | 1.96 | 3.37* | 9.29*** | 7.19*** | ||||||
| 1387328_at Cyp2c9 cytochrome P450 family 2 subfamily C member 9 | 1.66* | 1.73* | 1.73 | 3.66** | 3.94** | 2.56 | 3.6 | 3.54 | |||||||||
| 1387022_at Aldh1a1 aldehyde dehydrogenase 1 family member A1 | 1.2 | 1.47** | 2.49*** | 4.04*** | 1.96 | 3.96** | 2.95*** | 3.72*** | 2.42* | 2.65* | |||||||
| 1368718_at Aldh1a7 aldehyde dehydrogenase family 1, subfamily A7 | 9.7 | 15.45 | 1.29 | 2.91* | 6.16** | 3.01* | 4.76* | ||||||||||
| 1370067_at Me1 malic enzyme 1 | 1.7** | 3.12*** | 1.78* | 2.77* | 4.08** | 2.33* | 3.83* | 4.67* | |||||||||
| 1370870_at Me1 malic enzyme 1 | 2.04** | 3.83*** | 1.8* | 2.7* | 3.94* | 2.12* | 3.12 | 3.83* | |||||||||
| Conjugating Enzymes | |||||||||||||||||
| 1369850_at Ugt2a1 | 1.24* | 1.32* | 2.06*** | 1.46 | 1.63* | 1.72* | 2.14* | 1.5 | 1.98* | 2.33* | 2.3* | ||||||
| UDP glucuronosyltransferase family 2 member A1 complex locus | |||||||||||||||||
| 1370698_at Ugt2b17 | 1.7** | 2.38*** | 2.73*** | 1.78* | 1.74** | 2.05*** | 2*** | 1.93* | 2.05* | 2.15* | 2.34* | ||||||
| UDP glucuronosyltransferase family 2 member B17 | |||||||||||||||||
| 1,387,955_at Ugt2b17 | 1.35* | 1.67* | 1.64* | 1.33 | 1.81* | 2.06* | 1.63* | ||||||||||
| UDP glucuronosyltransferase 2 family, polypeptide B15 | |||||||||||||||||
| 1,381,852_at Ugt2b11 | -1.2 | 1.44** | 3.26*** | 1.56 | 2.47* | ||||||||||||
| UDP glucuronosyltransferase family 2 member B11 | |||||||||||||||||
| 1,368,397_at Ugt2b7 | 1.07 | 1.13** | 1.2*** | 1.27* | 1.25 | 1.28* | 1.39* | 1.35* | 1.41* | 1.31* | 1.29* | ||||||
| UDP glucuronosyltransferase family 2 member B7 | |||||||||||||||||
| 1,387,825_at Ugt2b28 | 3.57* | 4.01* | 6.15 | ||||||||||||||
| UDP glucuronosyltransferase family 2 member B28 | |||||||||||||||||
| 1,387,314_at Sult1b1 sulfotransferase family 1B member 1 | 1.11 | 1.11 | 1.29** | 3.74* | 5.17** | 2.83 | 4.22* | ||||||||||
| NrF2 antioxidant | |||||||||||||||||
| 1,367,843_at Akr7a2 aldo-keto reductase family 7 member A2 | 1.11 | 1.21* | 1.39*** | 1.44* | 1.44* | 1.59** | 1.36 | 1.6* | |||||||||
| 1,378,392_at Dnaja3 | 1.19* | 1.25* | 1.22 | 1.28* | 1.54 | ||||||||||||
| DnaJ heat shock protein family (Hsp40) member A3 | |||||||||||||||||
| 1,387,669_a_at Ephx1 epoxide hydrolase 1 | 1.26* | 1.65*** | 2.13*** | 1.78* | 2.28** | 2.4* | 2.94* | 2.77* | 3.46* | 3.35* | |||||||
| 1,368,180_s_at Gsta1 glutathione S-transferase alpha 1 | 1.26 | 1.56** | 2.11*** | 1.8 | 2.27* | 2.05* | 2.45* | 3.38* | 2.99* | 3.61* | |||||||
| 1,371,089_at Gsta3 glutathione S-transferase alpha 3 | 1.39 | 1.67* | 3.98*** | 13.87*** | 1.46 | 1.76* | 1.9* | 1.77* | 1.91* | ||||||||
| 1,386,985_at Gstm5 glutathione S-transferase mu 5 | 1.13 | 1.44*** | 1.86*** | 1.7* | 1.46* | 1.56* | 2.44* | 1.83* | 1.92* | ||||||||
| 1,368,409_at Gstt2/Gstt2B glutathione S-transferase theta 2 (gene/pseudogene) | 1.11 | 1.31 | 1.45* | 1.74* | |||||||||||||
| 1,367,613_at Prdx1 peroxiredoxin 1 | 1.08* | 1.27*** | 1.26** | 1.19 | 1.21* | 1.21* | |||||||||||
| 1,387,599_a_at Nqo1 | 1.39* | 2.64*** | 6.62*** | 3.16 | 7.19* | 2.6* | 2.93* | ||||||||||
| NAD(P)H quinone dehydrogenase 1 | |||||||||||||||||
| 1,374,070_at Gpx2 glutathione peroxidase 2 | 1.65* | 1.84 | 4.32* | 6.38* | 4.92 | ||||||||||||
| 1,368,121_at Akr7a3 aldo-keto reductase family 7 member A3 | 1.7* | 3.21*** | 1.93 | 3 | 4.9* | 6.92* | |||||||||||
| 1,367,982_at Alas1 | 2.73** | 4.04*** | 1.78* | 2.88** | 2.38* | 2.68* | 1.76 | 2.85* | 2.75* | 2.06* | |||||||
| 5′-aminolevulinate synthase 1 | |||||||||||||||||
| Membrane Function | |||||||||||||||||
| 1,369,455_at Abcg5 | -1.89* | -3.99*** | -3.45* | -6.29** | -3.05 | ||||||||||||
| ATP binding cassette subfamily G member 5 | |||||||||||||||||
| 1,369,440_at Abcg8 | -1.95* | -4.66*** | -5.15** | -7.72*** | -3.5* | ||||||||||||
| ATP binding cassette subfamily G member 8 | |||||||||||||||||
| 1,369,698_at Abcc3 | 1.35 | 2.09*** | 9.62*** | 23.41*** | 2.36* | 2.83** | 2.99* | 3.42* | 2.32 | 3.76* | 3.4** | 3.37** | |||||
| ATP binding cassette subfamily C member 3 | |||||||||||||||||
| 1,370,464_at Abcb1 | 1.87** | 3.45*** | 1.86 | 2.1** | 2.2** | 2.28 | 3.1* | 1.99 | |||||||||
| ATP binding cassette subfamily B member 1 | |||||||||||||||||
| 1,389,391_at Slc35e3 solute carrier family 35 member E3 | 1.25 | 2.03*** | 1.38 | 1.6* | 1.55 | ||||||||||||
| 1,372,479_at Slc4a4 solute carrier family 4 member 4 | -1.43 | 1.77* | |||||||||||||||
| 1,368,191_a_at Slc22a1 solute carrier family 22 member 1 | 1.12 | 1.27** | 1.49*** | 1.53* | 1.68* | 1.38 | 1.44 | 1.56 | 2.13* | 2.01* | |||||||
| 1,368,600_at Slc26a1 solute carrier family 26 member 1 | 1.6* | ||||||||||||||||
| 1,395,325_s_at Mmgt1 membrane magnesium transporter 1 | 1.26* | 1.95*** | 2.54*** | 1.74* | 2.28** | 1.96* | 2.22** | 1.91 | 2.37* | 1.91* | 1.9* | ||||||
| 1,376,168_at Mmgt1 membrane magnesium transporter 1 | 1.2 | 1.81*** | 2.43*** | 1.66* | 2.09** | 2** | 2.23** | 1.89 | 2.13* | 1.89* | 1.82 | ||||||
| 1377854_at Tmem62 transmembrane protein 62 | 1.21 | 1.29 | 1.34 | 1.61* | 1.3 | 1.38* | |||||||||||
| 1370807_at Vmp1 vacuole membrane protein 1 | 1.54* | 1.42* | |||||||||||||||
| 1368977_a_at Timm10b translocase of inner mitochondrial membrane 10B | 1.35** | 1.81*** | 1.34 | 1.37 | 1.52* | 1.51* | 1.92 | ||||||||||
| 1387013_at Cltrn collectrin, amino acid transport regulator | 11.24*** | 19.39*** | 10.7* | 14.91* | |||||||||||||
| Lipid/Protein Disease | |||||||||||||||||
| 1371572_at App amyloid beta precursor protein | 1.32* | 2.43*** | 3.05*** | 2.64*** | 3.37*** | 3.01* | 3.11* | ||||||||||
| 1369727_at Apoa2 apolipoprotein A2 | 1.78 | 3.68** | 5.14** | 3.15* | 5.5* | ||||||||||||
| Other | |||||||||||||||||
| 1383585_s_at Snx10 sorting nexin 10 | 3.53** | 6.35*** | 1.39 | 3.09* | 5.06* | 2.29* | 4.06 | 8.08* | |||||||||
| 1370371_a_at Ceacam4 CEA cell adhesion molecule 4 | -1.45 | 6.33* | 25.18*** | ||||||||||||||
| 1370349_a_at LOC100360095 (includes others) urinary protein 2 | 45.56* | ||||||||||||||||
| 1370828_at Zdhhc2 zinc finger DHHC-type containing 2 | 1.3 | 2.41*** | 7.17*** | 3.69*** | 6.17*** | 2.8* | 4.68*** | ||||||||||
| 1377662_at Pir pirin | 1.74** | 3.44*** | 1.43 | 1.94* | 1.83 | 2.36* | 2.41* | 2.02 | 2.77* | ||||||||
| Cancer-Related, Cancer Driver Genes, & Mitochondria function | |||||||||||||||||
| 1,369,957_at Rgs5 regulator of G protein signaling 5 | 1.78** | 1.75** | |||||||||||||||
| 1,383,288_at Mdm2 MDM2 proto-oncogene | 1.55** | 2.32*** | 1.55* | 1.66* | 1.2 | ||||||||||||
| 1,383,420_at Afg1l AFG1 like ATPase | 1.2 | 1.3* | 1.3* | ||||||||||||||
| 1,387,109_at Por cytochrome p450 oxidoreductase | 2.24* | 3.85*** | 1.48 | 1.76* | 1.76* | 1.92* | 2.49* | 2.1* | 2.52* | ||||||||
| 1,387,599_a_at Nqo1 NAD(P)H quinone dehydrogenase 1 | 1.39* | 2.64*** | 6.62*** | 3.16 | 7.19* | 2.6* | 2.93* | ||||||||||
| 1,367,613_at Prdx1 peroxiredoxin 1 | 1.08* | 1.27*** | 1.26** | 1.19 | 1.21* | 1.21* | |||||||||||
| 1,398,310_at Akr1d1 aldo-keto reductase family 1 member D1 | 1.2* | 1.38*** | |||||||||||||||
| 1,368,121_at Akr7a3 aldo-keto reductase family 7 member A3 | 1.7* | 3.21*** | 1.93 | 3 | 4.9* | 6.92* | |||||||||||
| 1,368,143_at Anxa7 annexin A7 | 1.64** | 3*** | 1.77* | 2.31** | 1.9* | 2.79* | |||||||||||
| 1,387,376_at Aox1 aldehyde oxidase 1 | 1.32** | 2.21*** | 1.73 | 1.45 | 1.66* | 1.47 | 1.56 | ||||||||||
| 1,370,964_at Ass1 argininosuccinate synthase 1 | -1.37** | -1.63*** | -1.84 | -1.88* | -1.94* | ||||||||||||
| 1,368,741_at C9 complement C9 | -1.16* | -1.2* | |||||||||||||||
| 1,367,733_at Ca2 carbonic anhydrase 2 | 1.8** | 3.46*** | 1.63* | 1.89** | |||||||||||||
| 1,382,603_at Cd274 CD274 molecule | -1.69* | -2.16** | |||||||||||||||
| 1,371,810_at Copg1 coatomer protein complex subunit gamma 1 | |||||||||||||||||
| 1,384,334_at Cps1 carbamoyl-phosphate synthase 1 | -1.15* | -1.23** | -1.2 | -1.21* | |||||||||||||
| 1,387,083_at Ctf1 cardiotrophin 1 | 1.27* | 1.67*** | |||||||||||||||
| 1,367,651_at Ctsd cathepsin D | 1.19* | 1.43*** | 1.58* | 1.55* | |||||||||||||
| 1,386,904_a_at Cyb5a cytochrome b5 type A | 1.13** | 1.27*** | 1.45*** | 1.15 | 1.23* | 1.21 | 1.27 | 1.33 | 1.42* | 1.32 | |||||||
| 1,370,475_at Cyp2b13/Cyp2b9 cytochrome P450, family 2, subfamily b, polypeptide 9 | -1.1* | -1.29*** | |||||||||||||||
| 1,368,607_at Cyp4a22 cytochrome P450 family 4 subfamily A member 22 | 3.32** | 4.36*** | 2.82* | 2.57 | 3.85* | 3.46* | |||||||||||
| 1,371,622_at Dph1 diphthamide biosynthesis 1 | 1.2* | 1.17 | |||||||||||||||
| 1,370,511_at Fgb fibrinogen beta chain | -1.1* | -1.24*** | 1.15 | ||||||||||||||
| 1,383,288_at Mdm2 MDM2 proto-oncogene | 1.55** | 2.32*** | 1.55* | 1.66* | 1.2 | ||||||||||||
| 1,370,831_at Mgll monoglyceride lipase | 1.34* | 2.26*** | 1.83 | ||||||||||||||
| 1,378,506_at Pik3c2a phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 alpha | |||||||||||||||||
| 1,398,814_at Rab11a RAB11A, member RAS oncogene family | 1.1* | 1.27*** | 1.2* | 1.31* | 1.39 | ||||||||||||
| 1,387,185_at Apbb3 amyloid beta precursor protein binding family B member 3 | -1.21 | -1.38* | |||||||||||||||
| 1,398,375_at Mta3 metastasis associated 1 family member 3 | 1.21 | 1.4** | 1.43** | 1.25 | 1.3 | 1.35* | |||||||||||
| 1,371,643_at Ccnd1 cyclin D1 | -1.45 | -2.13* | -2.14* | -2.7* | |||||||||||||
| 1,381,748_at Raph1 Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 | 1.55* | 1.78** | 1.52 | 1.58* | 1.96 | 1.53 | |||||||||||
| 1,383,709_at Rab8b RAB8B, member RAS oncogene family | 1.15 | 1.29** | 1.29* | ||||||||||||||
| 1,372,017_at Diablo/LOC100360940 diablo, IAP-binding mitochondrial protein | 1.19* | 1.35** | 1.22 | 1.24 | 1.46* | ||||||||||||
| 1,377,872_at Mals1 mitochondrial assembly of ribosomal large subunit 1 | 1.25** | 1.53*** | 1.21 | 1.41* | 1.51* | 1.36 | 1.66 | ||||||||||
| 1,373,074_at Timm21 translocase of inner mitochondrial membrane 21 | 1.22* | 1.36** | 1.41 | 1.63* | 1.3 | ||||||||||||
| 1,383,171_at Tfb2m transcription factor B2, mitochondrial | 1.17* | 1.28** | 1.33* | 1.32* | |||||||||||||
| 1,372,927_at Mrpl50 mitochondrial ribosomal protein L50 | 1.11 | 1.3** | 1.37*** | 1.42 | 1.45* | 1.54* | |||||||||||
| 1,368,977_a_at Timm10b translocase of inner mitochondrial membrane 10B | 1.35** | 1.81*** | 1.34 | 1.37 | 1.52* | 1.51* | 1.92 | ||||||||||
| 1,389,518_at Tomm5 translocase of outer mitochondrial membrane 5 | 1.25* | 1.4** | |||||||||||||||
| 1,369,023_at Mipep mitochondrial intermediate peptidase | 1.13 | 1.24* | 1.55*** | 1.31 | 1.28 | 1.42* | |||||||||||
| 1,372,105_at Micu2 mitochondrial calcium uptake 2 | 1.21 | 1.26* | 1.25 | 1.32* | |||||||||||||
| 1,372,456_at Mrps31 mitochondrial ribosomal protein S31 | 1.12 | 1.21* | 1.22 | 1.33* | |||||||||||||
| 1,373,982_at Tefm transcription elongation factor, mitochondrial | 1.23* | 1.28* | 1.25 | 1.33* | 1.31 | ||||||||||||
| Cancer Driver Genes | |||||||||||||||||
| 1,370,563_at Akr1c14 aldo-keto reductase family 1, member C14 | 1.16 | 1.23* | 1.54*** | 1.86*** | 1.3* | 1.31 | 1.44* | 1.65 | 1.64* | 1.63* | |||||||
| 1,387,077_at Arpp19 cAMP regulated phosphoprotein 19 | 1.42** | 2.09*** | 1.49* | 1.72* | 1.59* | 1.95* | |||||||||||
| 1,377,855_at Cep83 centrosomal protein 83 | 1.6** | 2.06*** | 1.48 | 1.93* | 2.21** | 1.3 | 1.57* | 1.56 | |||||||||
| 1,372,510_at Srxn1 sulfiredoxin 1 | 1.57* | 2.56*** | 1.91 | 2.6 | |||||||||||||
| 1,371,113_a_at Tfrc transferrin receptor | 1.66* | 2.09** | 6.02*** | ||||||||||||||
| 1,384,169_a_at Vav2 vav guanine nucleotide exchange factor 2 | 2.08*** | 3.02*** | 2.25* | 2.3* | 1.45 | 2.1* | 2.06* | ||||||||||
| 1,379,283_at Braf B-Raf proto-oncogene, serine/threonine kinase | 1.19 | 1.19 | 1.24* | ||||||||||||||
| 1,382,264_at Crebbp CREB binding protein | -1.25* | -1.2* | |||||||||||||||
| 1,372,774_at Entpd5 ectonucleoside triphosphate diphosphohydrolase 5 (inactive) | 1.19 | 1.52*** | 1.28 | 1.71* | 2.08* | 1.85* | 1.82 | ||||||||||
| 1,369,150_at Pdk4 pyruvate dehydrogenase kinase 4 | 1.44* | ||||||||||||||||
| 1,383,242_a_at Cebpa CCAAT enhancer binding protein alpha | -1.39 | -1.6* | -1.79 | ||||||||||||||
| 1,369,954_at Idh1 isocitrate dehydrogenase (NADP (+)) 1 | 1.2* | 1.23 | 1.14 | 1.23* | |||||||||||||
| 1,370,957_at Il6st interleukin 6 signal transducer | -1.41 | ||||||||||||||||
| 1,376,082_at Mecom MDS1 and EVI1 complex locus | -1.41* | ||||||||||||||||
Fold change values are present if they are significant for FDR <0.2. Asterisks indicate further levels of statistical significance (*FDR<0.05, **FDR< 0.001, ***FDR<10 < -10). Annotations based on Ingenuity Pathway Analysis.
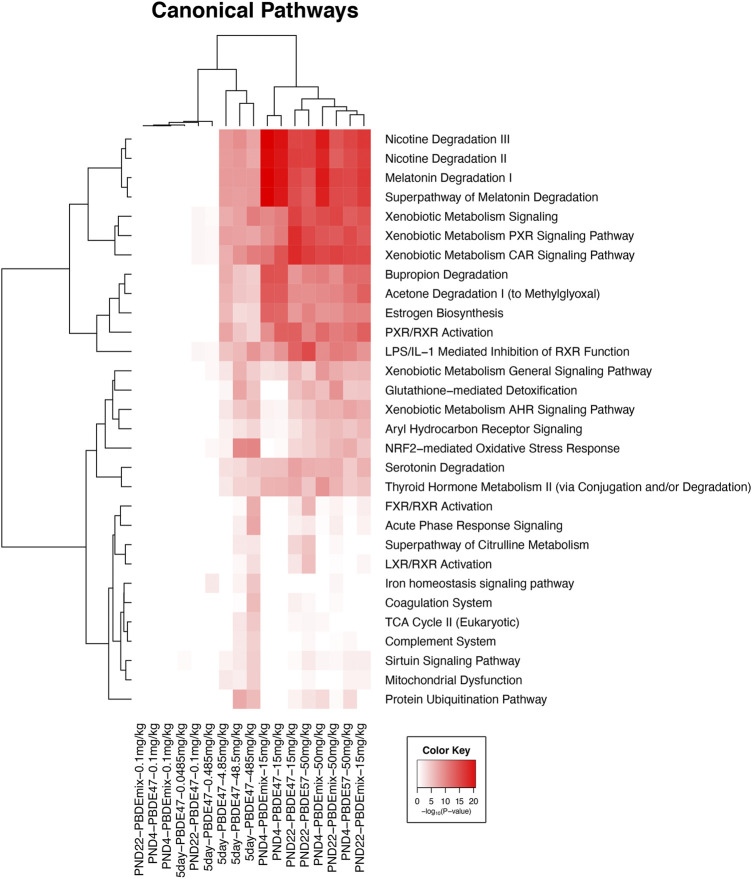
FIGURE 1.
Clustered heat map of log10 p-values associated with enriched Ingenuity Canonical Pathways associated with the changed transcripts from each experiment in all five groups in this study. Included pathways have at least one significant pathway after multiple test correction (FDR <0.01). The color key scale indicates the minus log10 p-value of pathway significance.
Of note was the upregulation of the Nrf2 antioxidant pathways, a pathway that is turned on as a defense when oxidative damage. This was particularly noted in the adult 5-day treatment group (Group One) including upregulation of Nqo1 (NAD(P)H quinone dehydrogenase 1), Akr7a3 (aldo-keto reductase family seven member A3), and Ephx1 (epoxide hydrolase 1) (Table 3).
Membrane protein transcripts were upregulated including the transporters Abcc3, Abcb1d, solute carriers, and other ion transporters. Abcg5 and Abcg8 were significantly downregulated at PND 22 and in adult animals but not in PND 4 animals.
The transcriptomic signals for cancer were upregulated particularly in Group one (the 5-day adult exposure group) and included upregulation of protooncogene and oncogene transcripts (Table 2). The hepatocellular disease markers indicated activation of Ras signaling transcripts (e.g., Rgs5, Rab11a, and Raph1). These Ras disease pathway transcripts were not upregulated in the PND 4 or PND 22 groups after PBDE-47 or PBDE mixture exposures. Other cancer transcripts for genes “characterized” as cancer driver genes (Bailey et al., 2018) were upregulated in PND 4, PND 22, and adult exposure groups. Mitochondria function transcripts were significantly upregulated particularly in the adult exposure group.
After PBDE-47 exposure there were significant transcriptional changes at PND 4 and in adult animals at the lowest exposure level (0.1–0.2 mmol/kg; FDR ≤0.05), changes that were more pronounced than after comparable PBDE mixture exposures. This suggests that PBDE-47 may be more toxic at lower exposure levels than the PBDE mixture which contained lower amounts of PBDE-47. PBDE mixture exposure at 27–31 mmol/kg and 89–103 mmol/kg also had fewer significantly changed liver transcripts than the PBDE-47 exposure groups at these exposure levels.
Male rat PBDE transcript data was used for Benchmark dose analysis (Figure 2). The BMDL (where there is estimated to be a 10% increase in toxicity over controls) was between ∼ 1 and 6 mmol/kg/day for PBDE exposures (for Groups 1–5). Lowest BMDL was based on GO categories with ≥3 genes (and ≥5% of the genes in the category being significant) that pass the selection criteria in BMDExpress 2.0 (p-value, ≥0.05 and |FC| > 1.5). Generally, these top GO categories were involved in various phases of metabolism including lipid metabolism.
FIGURE 2.

Toxicogenomic analysis of the five experimental datasets examined in this study. (A) Visualization of biological process gene ontology (GO) terms in concentration-response with the number of biological processes (y-axis) for a given level of the mean lower genomic benchmark dose (mean BMDL, x-axis). Group 1 (5 day PBDE-47 study) is shown in black, Group 2 (PBDE-47 at PND4) is shown in red, Group 3 (PBDE-47 at PND 22) is shown in blue, Group 4 (PBDE-mixture at PND4) is shown in green and Group 5 (PBDE-mixture at PND22) is shown in cyan. (B) The lowest mean BMDL for each group is shown in gray for each group for the most sensitive biological processes. An “x” indicates that a mean BMDL was not returned for a given biological process.
4 Discussion
T4 levels are decreased in humans and animals after PBDE exposure (National Toxicology Program, 2015; Allen et al., 2016; Makey et al., 2016). Human exposure and body burden levels of PBDEs have been reviewed by the Agency for Toxic Substances and Disease Registry, Center for Disease Control (Agency for Toxic Substances and Disease Control, 2017), and they also report that PBDE exposure is associated with decreased blood levels of T4 in both humans and animals. These studies show that higher levels of PBDEs are needed in animals than in humans to cause decreases in T4 levels. Thus, humans may be more sensitive to this PBDE toxic effect than rodent models.
Transcriptomic changes in model systems can be used to discover biomarkers of disease and in risk assessment, and these data can provide an understanding of the relationship between molecular pathways and pathology of disease. This information can be used in risk assessment for linking pathologies and molecular pathways (Merrick, 2019; Szabo and Devlin, 2019; Sprenger et al., 2022).
Our analysis of previously published liver gene expression profiling data provides evidence that key characteristics of chemical carcinogens (Smith et al., 2016) were upregulated at PND 4, PND 22, and in adult male rats after exposure to the rodent chemical carcinogen, PBDE. The significant liver transcript changes at all three life cycle stages included upregulation of transcripts for metabolic, membrane function, and the Nrf2 pathways, all characteristic of chemical carcinogens (Ma, 2013; Choi et al., 2021). These liver transcript changes after in utero exposure, neonatal exposure, or short-term adult exposures are biomarkers for upregulation of changes that can eventually lead to cancer.
The disruption of metabolic processes, as indicated by the PBDE-induced liver transcripts signals, is one of the key characteristics of chemical carcinogens (Smith et al., 2016; Guyton et al., 2018; La Merrill et al., 2020). This included upregulation of phase one and two liver metabolic enzymes (Table 3). Serum thyroxin (T4) levels were reduced in PBDE exposed groups, and this is attributed to PBDE-induced increase in levels of UGT transcripts, whose transcribed enzymes are used to catalyze the formation of thyroxine (T4) glucuronide which facilitates excretion of thyroid hormones in the bile (Richardson et al., 2008; Hoffman et al., 2017). Thyroid hormones were particularly low in the PND 4 rat because the development of thyroid production capacity is not yet fully developed at this life cycle stage (Dubois and Dussault, 1977; Choksi et al., 2003). If low levels of thyroid persist, this can promote development of hepatocellular cancer conditions (Frau et al., 2015; Lin et al., 2020).
PBDE metabolites can cause oxidative damage (Costa et al., 2014; Costa et al., 2015; National Toxicology Program, 2015). In this study, various Nrf2 antioxidant pathway gene transcripts (Ma, 2013) were activated in response to PBDE treatment, particularly in the adult rat. The metabolic capabilities of younger animals may not yet be fully developed to complete all the metabolic steps necessary (MacLeod et al., 1972; Moscovitz and Aleksunes, 2013; O'Hara et al., 2015) to form the PBDE metabolites capable of causing oxidative damage (National Toxicology Program, 2015) and, thus, this may have contributed to the observed lower activation of Nrf2 pathway transcripts in the younger animals.
Membrane transport protein transcripts were upregulated after PBDE exposure at all three life cycle stages. This included upregulation of xenobiotic efflux transporters and ion transporters. Upregulation of membrane transporters are characteristic of hepatocellular disease (Pedersen and Stock, 2013; Oosterwijk and Gillies, 2014; Prevarskaya et al., 2018), a disease that can become more severe with continued PBDE exposure (National Toxicology Program, 2015). For example, Abcc3 transcript, an efflux transporter, was upregulated after PBDE exposure in the liver, and upregulation of this membrane transcript is characteristic of preneoplastic and neoplastic hepatocellular lesions in humans and rodents (Carrasco-Torres et al., 2016). Upregulation of membrane transporters is also characteristic of carcinogenesis processes in other organ systems (Donepudi et al., 2016).
The acidic nature of cancer cells is governed by ion transport (Swietach et al., 2014), and in this study many ion membrane transport gene transcripts were upregulated in all three life cycle stages after PBDE exposures. However, there was downregulation of the lipid efflux membrane transporter (Abcg5/8) (Patel et al., 2018) at PND 22 and in adult animals but not in PND 4 pups. Liver lipids levels in young animals are generally lower than in adult rodents (Smith and Abraham, 1970), and this may explain in part why lipid efflux pump transcripts were not changed in the PND 4 pups. Downregulation of ABCG5/8 can alter sterol excretion from cells (Patel et al., 2018).
Cancer and mitochondria function disease gene transcripts were upregulated in the adult rats (Table 3). This included Ras pathway transcripts Rgs5, Rab11a, Rab8b, and Raph1 corresponding to human RGS5 (Umeno et al., 2018; Abe et al., 2019; Shioga et al., 2020; Zhang et al., 2020), human RAB11A (Abd El Gwad et al., 2018; Cao et al., 2019), Xenopus and zebrafish RAB8B (Demir et al., 2013), and human RAPH1 (Kurozumi et al., 2018). The RGS family is a group of multifunctional proteins that regulate cellular signaling events downstream of G-protein coupled receptors (Hurst and Hooks, 2009). RGS5 expression is increased in multiple cancers (e.g., breast, ovarian, acute myeloid leukemia, and liver) and expression of Rgs5 in rodents can also lead to other liver diseases (Bahrami et al., 2014). In addition, Rgs5 expression in rodents promotes portal vein invasion and intrahepatic metastasis from hepatocellular carcinoma (Umeno et al., 2018).
The Raph1 transcript was elevated after PBDE-47 exposure in the adult rat, a gene involved in cytoskeleton regulation (Kurozumi et al., 2018). High RAPH1 expression has been correlated with aggressive breast cancer phenotypes and provides independent prognostic value in invasive breast cancer (Batistela et al., 2013). Rab8b, another member of the RAS family was also upregulated after PBDE exposure, and has been reported to be an essential evolutionary conserved component of Wnt/β-catenin signaling (Demir et al., 2013; Das et al., 2015). The elevated Ras pathway transcripts found in this study correlated with the finding of Ras mutations in PBDE-induced liver tumors in a 2-year cancer study (Dunnick et al., 2018b).
The overexpression of the Mdm2 oncoprotein transcript after PBDE exposure seen in the adult rat in these studies, has been observed to frequently occur in hepatocellular carcinoma (HCC) (Wang et al., 2019). MDM2 is an oncogene that is an inhibitor of the tumor suppressor, p53 (Ishizawa et al., 2018), and is an E3 ubiquitin ligase that directly binds to the N-terminal 1–42 aa of p53 to induce ubiquitin-mediated proteasomal degradation (Chen et al., 2019). Mdm2 inhibits p53 translocation from the nucleus to the cytoplasm and enhances p53 degradation via ubiquitin-proteasome pathway (Soussi and Kroemer, 2018). About half of all cancers retain wild type p53, however the p53 pathway may be inactivated due to the overexpression of endogenous negative regulators, including the murine double minute 2 (MDM2), as occurred in PBDE treated rats in this study.
Generating enough energy is essential for cancer processes to proceed and critical to this is mitochondria function (Sokol et al., 2014; Porporato et al., 2018). In this study, mitochondria transcripts were upregulated in the adult animals. This included upregulation of genes encoding TIMM proteins, which have been characterized as diagnostic markers of poor prognosis for surviving cancer (Sotgia and Lisanti, 2017). The TIMM proteins, which are involved in the mitochondria protein-important machinery, are necessary for import of proteins needed for mitochondria function, but coded for in the nucleus (Pfanner and Meijer, 1997).
In short, the 5-day transcriptomics study of adult animals exposed to PBDEs offered comparable results to changes found after exposure in earlier life stages (PND4 and PND22). Furthermore, the analysis of this transcriptomic data identified alterations in toxic gene expression pathways predictive of longer-term toxic and carcinogenic effects using fewer animals and shorter exposure times than used in traditional toxicology studies (e.g., 13-week studies or 2-year bioassays). Thus, the 5-day transcriptomics study design helps to meet the objective to reduce, refine, and replace (3Rs) the use of animals in toxicology testing, an important national and international goal (National Toxicology Program, 2022).
More work is needed with other chemical carcinogens to compare transcriptomic changes at different life cycle stages, other target organs, and different model systems in order to determine the most appropriate conditions for predicting carcinogenic outcomes after short term exposures. Moreover, in this study we compared gene expression response with thyroid hormone levels and liver pathology. Future studies should likewise incorporate appropriate hematological and biochemical parameters, pathology, or other supporting data to complement and validate the transcriptomic approach.
5 Conclusion
This study presents liver gene expression profiles for a model chemical carcinogen (PBDE), that caused a liver carcinogenic response in rats and mice (National Toxicology Program, 2015), and also associated with toxicity in humans including lower thyroid hormone levels and developmental toxicity (Lam et al., 2017). Our results based on a combined analysis of previously published studies demonstrated that disease pathways are activated at all life cycle stages after PBDE exposure.
We show that for PBDEs a 5-day exposure period with transcriptomic endpoints could reduce the amount of time needed to identify toxic and carcinogenic potential compared to the traditional 13-week study, while using a fewer number of animals. More work is needed to understand whether the results presented here could similarly be used to reduce the number of animals needed to obtain toxic and cancer hazard information for other chemicals or classes of chemicals. If so, a similar transcriptomic assessment approach could be used to provide information on the underlying mechanisms.
Humans may be exposed to brominated chemicals early in life is a concern for adverse health effects (Vuong et al., 2018; Margolis et al., 2020; Varshavsky et al., 2020; Vuong et al., 2020), because early exposures can lead to disease later in life (Ferguson et al., 2017; Center for Disease Control, 2021). Other rodent model studies showed that short-term exposure to brominated chemical can lead to cancer later in life (Dunnick et al., 1997), and that the resultant tumors are characterized by alterations in the Ras pathway (Ton et al., 2004). These and other DTT studies suggest that further work is needed to characterize cancer hazards from exposure to mixtures of brominated chemicals (Peng et al., 2017; Dhungana et al., 2019; Han et al., 2020).
Acknowledgments
We thank Dr. Scott Auerbach (Predictive Toxicology Branch, NIEHS) and Dr. John House (Biostatistics and Computational Biology Branch, NIEHS) for their review of the paper. However, the statements, opinions or conclusions contained therein do not necessarily represent the statements, opinions, or conclusions of NTP, NIEHS, NIH or the United States government.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: The datasets analyzed for this study can be found on the National Library of Medicine’s GEO database web site (https://www.ncbi.nlm.nih.gov/geo/). The Affymetrix data for each dataset can be accessed from GEO using GSE153366 for Group One, GSE154914 for Group Two and Group Four, and GSE124431 for Group Three and Group Five.
Ethics statement
The animal study was reviewed and approved by the care of animals on this study was according to NIH procedures as described in the U.S. Public Health Service Policy on Humane Care and Use of Laboratory Animals (available online at: https://olaw.nih.gov). The study protocols were approved by the institutional animal care and use committee.
Author contributions
KS and JD designed the study, analyzed the data, and wrote the manuscript.
Funding
This research was supported [in part] by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences, including Intramural Research projects ZIA ES103349 and ZIA ES103316.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer, DR, declared a shared affiliation with the authors to the handling editor at the time of review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Abbasi G., Li L., Breivik K. (2019). Global historical stocks and emissions of PBDEs. Environ. Sci. Technol. 53 (11), 6330–6340. 10.1021/acs.est.8b07032 [DOI] [PubMed] [Google Scholar]
- Abd El Gwad A., Matboli M., El-Tawdi A., Habib E. K., Shehata H., Ibrahim D., et al. (2018). Role of exosomal competing endogenous RNA in patients with hepatocellular carcinoma. J. Cell. Biochem. 119 (10), 8600–8610. 10.1002/jcb.27109 [DOI] [PubMed] [Google Scholar]
- Abe Y., Ogasawara S., Akiba J., Naito Y., Kondo R., Nakamura K., et al. (2019). Expression and role of regulator of G-protein signaling 5 in squamous cell carcinoma of the tongue. Clin. Exp. Dent. Res. 5 (2), 160–169. 10.1002/cre2.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agency for Toxic Substances and Disease Control (2017). C.f.D ctoxicological profile for polybrominated diphenyl ethers (PBDEs). Available at:https://www.atsdr.cdc.gov/toxprofiles/tp207.pdf (Accessed October 11, 2022). [PubMed]
- Allen J. G., Gale S., Zoeller R. T., Spengler J. D., Birnbaum L., McNeely E. (2016). PBDE flame retardants, thyroid disease, and menopausal status in U.S. women. Environ. Health 15 (1), 60. 10.1186/s12940-016-0141-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrami A. J., Gunaje J. J., Hayes B. J., Riehle K. J., Kenerson H. L., Yeung R. S., et al. (2014). Regulator of G-protein signaling-5 is a marker of hepatic stellate cells and expression mediates response to liver injury. PLoS One 9 (10), e108505. 10.1371/journal.pone.0108505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey M. H., Tokheim C., Porta-Pardo E., Sengupta S., Bertrand D., Weerasinghe A., et al. (2018). Comprehensive characterization of cancer driver genes and mutations. Cell 174 (4), 1034–1035. 10.1016/j.cell.2018.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistela M. S., Boberg D. R., Andrade F. A., Pecharki M., de S. F. R. E. M., Cavalli I. J., et al. (2013). Amplification and deletion of the RAPH1 gene in breast cancer patients. Mol. Biol. Rep. 40 (12), 6613–6617. 10.1007/s11033-013-2774-1 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57 (1), 289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Cao S. Q., Zheng H., Sun B. C., Wang Z. L., Liu T., Guo D. H., et al. (2019). Long non-coding RNA highly up-regulated in liver cancer promotes exosome secretion. World J. Gastroenterol. 25 (35), 5283–5299. 10.3748/wjg.v25.i35.5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco-Torres G., Fattel-Fazenda S., López-Alvarez G. S., García-Román R., Villa-Treviño S., Vásquez-Garzón V. R. (2016). The transmembrane transporter ABCC3 participates in liver cancer progression and is a potential biomarker. Tumour Biol. 37 (2), 2007–2014. 10.1007/s13277-015-3999-5 [DOI] [PubMed] [Google Scholar]
- Center for Disease Control (2021). N.I.f.O.S.a.H.N. (2021 (accessed)). Exposome and Exposomics. Available at:https://www.cdc.gov/niosh/topics/exposome/default.html (Accessed October 11, 2022).
- Chen A., Yolton K., Rauch S. A., Webster G. M., Hornung R., Sjödin A., et al. (2014). Prenatal polybrominated diphenyl ether exposures and neurodevelopment in U.S. Children through 5 years of age: The HOME study. Environ. Health Perspect. 122 (8), 856–862. 10.1289/ehp.1307562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. L., Zhang C. Z., Liu L. L., Lu S. X., Pan Y. H., Wang C. H., et al. (2019). A GYS2/p53 negative feedback loop restricts tumor growth in HBV-related hepatocellular carcinoma. Cancer Res. 79 (3), 534–545. 10.1158/0008-5472.Can-18-2357 [DOI] [PubMed] [Google Scholar]
- Choi B. H., Kim J. M., Kwak M. K. (2021). The multifaceted role of NRF2 in cancer progression and cancer stem cells maintenance. Arch. Pharm. Res. 44 (3), 263–280. 10.1007/s12272-021-01316-8 [DOI] [PubMed] [Google Scholar]
- Choksi N. Y., Jahnke G. D., St Hilaire C., Shelby M. (2003). Role of thyroid hormones in human and laboratory animal reproductive health. Birth Defects Res. B Dev. Reprod. Toxicol. 68 (6), 479–491. 10.1002/bdrb.10045 [DOI] [PubMed] [Google Scholar]
- Churchill G. A. (2004). Using ANOVA to analyze microarray data. Biotechniques 37 (2), 173–175. 10.2144/04372TE01 [DOI] [PubMed] [Google Scholar]
- Costa L. G., de Laat R., Tagliaferri S., Pellacani C. (2014). A mechanistic view of polybrominated diphenyl ether (PBDE) developmental neurotoxicity. Toxicol. Lett. 230 (2), 282–294. 10.1016/j.toxlet.2013.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa L. G., Pellacani C., Dao K., Kavanagh T. J., Roque P. J. (2015). The brominated flame retardant BDE-47 causes oxidative stress and apoptotic cell death in vitro and in vivo in mice. Neurotoxicology 48, 68–76. 10.1016/j.neuro.2015.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X., Hwang J. T., Qiu J., Blades N. J., Churchill G. A. (2005). Improved statistical tests for differential gene expression by shrinking variance components estimates. Biostatistics 6 (1), 59–75. 10.1093/biostatistics/kxh018 [DOI] [PubMed] [Google Scholar]
- Das S., Yu S., Sakamori R., Vedula P., Feng Q., Flores J., et al. (2015). Rab8a vesicles regulate Wnt ligand delivery and Paneth cell maturation at the intestinal stem cell niche. Development 142 (12), 2147–2162. 10.1242/dev.121046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir K., Kirsch N., Beretta C. A., Erdmann G., Ingelfinger D., Moro E., et al. (2013). RAB8B is required for activity and caveolar endocytosis of LRP6. Cell Rep. 4 (6), 1224–1234. 10.1016/j.celrep.2013.08.008 [DOI] [PubMed] [Google Scholar]
- Dhungana B., Peng H., Kutarna S., Umbuzeiro G., Shrestha S., Liu J., et al. (2019). Abundances and concentrations of brominated azo dyes detected in indoor dust. Environ. Pollut. 252, 784–793. 10.1016/j.envpol.2019.05.153 [DOI] [PubMed] [Google Scholar]
- Donepudi A. C., Cheng Q., Lu Z. J., Cherrington N. J., Slitt A. L. (2016). Hepatic transporter expression in metabolic syndrome: Phenotype, serum metabolic hormones, and transcription factor expression. Drug Metab. Dispos. 44 (4), 518–526. 10.1124/dmd.115.066779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois J. D., Dussault J. H. (1977). Ontogenesis of thyroid function in the neonatal rat. Thyroxine (T4) and triiodothyronine (T3) production rates. Endocrinology 101 (2), 435–441. 10.1210/endo-101-2-435 [DOI] [PubMed] [Google Scholar]
- Dunnick J. K., Heath J. E., Farnell D. R., Prejean J. D., Haseman J. K., Elwell M. R. (1997). Carcinogenic activity of the flame retardant, 2, 2-bis(bromomethyl)-1, 3-propanediol in rodents, and comparison with the carcinogenicity of other NTP brominated chemicals. Toxicol. Pathol. 25 (6), 541–548. 10.1177/019262339702500602 [DOI] [PubMed] [Google Scholar]
- Dunnick J. K., Pandiri A. R., Merrick B. A., Kissling G. E., Cunny H., Mutlu E., et al. (2018a). Carcinogenic activity of pentabrominated diphenyl ether mixture (DE-71) in rats and mice. Toxicol. Rep. 5, 615–624. 10.1016/j.toxrep.2018.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnick J. K., Pandiri A. R., Merrick B. A., Kissling G. E., Cunny H., Mutlu E., et al. (2018b). Mutational analysis of pentabrominated diphenyl-induced hepatocellular tumors in rats and mice, tissue levels of PBDE congeners in rats and mice, and AhR genotyping of Wistar Han rats. Data Brief. 21, 2125–2128. 10.1016/j.dib.2018.10.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnick J. K., Shockley K. R., Morgan D. L., Travlos G. S., Gerrish K., Ton T. T., et al. (2020). Hepatic transcriptomic patterns in the neonatal rat after pentabromodiphenyl ether exposure. Toxicol. Pathol. 48 (2), 338–349. 10.1177/0192623319888433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnick J. K., Shockley K. R., Pandiri A. R., Kissling G. E., Gerrish K. E., Ton T. V., et al. (2018c). PBDE-47 and PBDE mixture (DE-71) toxicities and liver transcriptomic changes at PND 22 after in utero/postnatal exposure in the rat. Arch. Toxicol. 92 (11), 3415–3433. 10.1007/s00204-018-2292-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson A., Penney R., Solo-Gabriele H. (2017). A review of the field on children's exposure to environmental contaminants: A risk assessment approach. Int J environ res public health 14(3). 10.3390/ijerph14030265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frau C., Loi R., Petrelli A., Perra A., Menegon S., Kowalik M. A., et al. (2015). Local hypothyroidism favors the progression of preneoplastic lesions to hepatocellular carcinoma in rats. Hepatology 61 (1), 249–259. 10.1002/hep.27399 [DOI] [PubMed] [Google Scholar]
- Gautier L., Cope L., Bolstad B. M., Irizarry R. A. (2004). affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20 (3), 307–315. 10.1093/bioinformatics/btg405 [DOI] [PubMed] [Google Scholar]
- Guyton K. Z., Rusyn I., Chiu W. A., Corpet D. E., van den Berg M., Ross M. K., et al. (2018). Application of the key characteristics of carcinogens in cancer hazard identification. Carcinogenesis 39 (4), 614–622. 10.1093/carcin/bgy031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Yang D., Hall D. R., Liu J., Sun J., Gu W., et al. (2020). Toxicokinetics of brominated azo dyes in the early life stages of zebrafish (Danio rerio) is prone to aromatic substituent changes. Environ. Sci. Technol. 54 (7), 4421–4431. 10.1021/acs.est.9b07178 [DOI] [PubMed] [Google Scholar]
- Harrill J., Shah I., Setzer R. W., Haggard D., Auerbach S., Judson R., et al. (2019). Considerations for strategic use of high-throughput transcriptomics chemical screening data in regulatory decisions. Curr. Opin. Toxicol. 15, 64–75. 10.1016/j.cotox.2019.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman K., Sosa J. A., Stapleton H. M. (2017). Do flame retardant chemicals increase the risk for thyroid dysregulation and cancer? Curr. Opin. Oncol. 29 (1), 7–13. 10.1097/cco.0000000000000335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst J. H., Hooks S. B. (2009). Regulator of G-protein signaling (RGS) proteins in cancer biology. Biochem. Pharmacol. 78 (10), 1289–1297. 10.1016/j.bcp.2009.06.028 [DOI] [PubMed] [Google Scholar]
- Ishizawa J., Nakamaru K., Seki T., Tazaki K., Kojima K., Chachad D., et al. (2018). Predictive gene signatures determine tumor sensitivity to MDM2 inhibition. Cancer Res. 78 (10), 2721–2731. 10.1158/0008-5472.Can-17-0949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M., Zhang S., He J., Lu Z., Zhou S., Ye N. (2021). Polybrominated diphenyl ethers from automobile microenvironment: Occurrence, sources, and exposure assessment. Sci. Total Environ. 781, 146658. 10.1016/j.scitotenv.2021.146658 [DOI] [PubMed] [Google Scholar]
- Kurozumi S., Joseph C., Sonbul S., Aleskandarany M. A., Pigera M., Alsaleem M., et al. (2018). Clinicopathological and prognostic significance of Ras association and pleckstrin homology domains 1 (RAPH1) in breast cancer. Breast Cancer Res. Treat. 172 (1), 61–68. 10.1007/s10549-018-4891-y [DOI] [PubMed] [Google Scholar]
- La Merrill M. A., Vandenberg L. N., Smith M. T., Goodson W., Browne P., Patisaul H. B., et al. (2020). Consensus on the key characteristics of endocrine-disrupting chemicals as a basis for hazard identification. Nat. Rev. Endocrinol. 16 (1), 45–57. 10.1038/s41574-019-0273-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam J., Lanphear B. P., Bellinger D., Axelrad D. A., McPartland J., Sutton P., et al. (2017). Developmental PBDE exposure and IQ/ADHD in childhood: A systematic review and meta-analysis. Environ. Health Perspect. 125, 086001. 10.1289/ehp1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. H., Lin K. H., Yeh C. T. (2020). Thyroid hormone in hepatocellular carcinoma: Cancer risk, growth regulation, and anticancer drug resistance. Front. Med. 7, 174. 10.3389/fmed.2020.00174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q. (2013). Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 53, 401–426. 10.1146/annurev-pharmtox-011112-140320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod S. M., Renton K. W., Eade N. R. (1972). Development of hepatic microsomal drug-oxidizing enzymes in immature male and female rats. J. Pharmacol. Exp. Ther. 183 (3), 489–498. [PubMed] [Google Scholar]
- Maddela N. R., Venkateswarlu K., Kakarla D., Megharaj M. (2020). Inevitable human exposure to emissions of polybrominated diphenyl ethers: A perspective on potential health risks. Environ. Pollut. 266, 115240. 10.1016/j.envpol.2020.115240 [DOI] [PubMed] [Google Scholar]
- Makey C. M., McClean M. D., Braverman L. E., Pearce E. N., He X. M., Sjödin A., et al. (2016). Polybrominated diphenyl ether exposure and thyroid function tests in north American adults. Environ. Health Perspect. 124 (4), 420–425. 10.1289/ehp.1509755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis A. E., Banker S., Pagliaccio D., De Water E., Curtin P., Bonilla A., et al. (2020). Functional connectivity of the reading network is associated with prenatal polybrominated diphenyl ether concentrations in a community sample of 5 year-old children: A preliminary study. Environ. Int. 134, 105212. 10.1016/j.envint.2019.105212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick B. A. (2019). Next generation sequencing data for use in risk assessment. Curr. Opin. Toxicol. 18, 18–26. 10.1016/j.cotox.2019.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller A., Xie Z., Cai M., Zhong G., Huang P., Cai M., et al. (2011). Polybrominated diphenyl ethers vs alternate brominated flame retardants and Dechloranes from East Asia to the Arctic. Environ. Sci. Technol. 45 (16), 6793–6799. 10.1021/es201850n [DOI] [PubMed] [Google Scholar]
- Moscovitz J. E., Aleksunes L. M. (2013). Establishment of metabolism and transport pathways in the rodent and human fetal liver. Int. J. Mol. Sci. 14 (12), 23801–23827. 10.3390/ijms141223801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Toxicology Program (2022). (2022 (accessed)). Regulatory Applications of 3Rs. Available at:https://ntp.niehs.nih.gov/whatwestudy/niceatm/accept-methods/apps/index.html (Accessed October 11, 2022).
- National Toxicology Program (2018). National toxicology program approach to genomic dose-response modeling. Available at:https://ntp.niehs.nih.gov/ntp/results/pubs/rr/reports/rr05_508.pdf (Accessed October 11, 2022). [PubMed]
- National Toxicology Program (2015). Toxicology of a pentabromodiphenyl oxide mixture (DE71) (Cas no. 32534-81-9) in F344/N rats and B6C3F1/N mice and toxicology and carcinogenesis studies of a pentabromodiphenyl oxide mixture (DE71) in Wistar Han [Crl:WI(Han)] rats and B6C3F1/N mice (gavage and perinatal and postnatal gavage studies) NTP TR 589. Available at:https://ntp.niehs.nih.gov/publications/reports/tr/500s/tr589/index.html?utm_source=direct&utm_medium=prod&utm_campaign=ntpgolinks&utm_term=tr589abs (Accessed October 11, 2022).
- O'Hara K., Wright I. M., Schneider J. J., Jones A. L., Martin J. H. (2015). Pharmacokinetics in neonatal prescribing: Evidence base, paradigms and the future. Br. J. Clin. Pharmacol. 80 (6), 1281–1288. 10.1111/bcp.12741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterwijk E., Gillies R. J. (2014). Targeting ion transport in cancer. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369 (1638), 20130107. 10.1098/rstb.2013.0107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S. B., Graf G. A., Temel R. E. (2018). ABCG5 and ABCG8: More than a defense against xenosterols. J. Lipid Res. 59 (7), 1103–1113. 10.1194/jlr.R084244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen S. F., Stock C. (2013). Ion channels and transporters in cancer: Pathophysiology, regulation, and clinical potential. Cancer Res. 73 (6), 1658–1661. 10.1158/0008-5472.Can-12-4188 [DOI] [PubMed] [Google Scholar]
- Peng H., Saunders D. M., Jones P. D., Giesy J. P. (2017). Response to comment on "mutagenic azo dyes, rather than flame retardants, are the predominant brominated compounds in House dust. Environ. Sci. Technol. 51 (6), 3591–3592. 10.1021/acs.est.7b00675 [DOI] [PubMed] [Google Scholar]
- Pfanner N., Meijer M. (1997). The tom and tim machine. Curr. Biol. 7 (2), R100–R103. 10.1016/s0960-9822(06)00048-0 [DOI] [PubMed] [Google Scholar]
- Phillips J. R., Svoboda D. L., Tandon A., Patel S., Sedykh A., Mav D., et al. (2019). BMDExpress 2: Enhanced transcriptomic dose-response analysis workflow. Bioinformatics 35 (10), 1780–1782. 10.1093/bioinformatics/bty878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porporato P. E., Filigheddu N., Pedro J. M. B., Kroemer G., Galluzzi L. (2018). Mitochondrial metabolism and cancer. Cell Res. 28 (3), 265–280. 10.1038/cr.2017.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevarskaya N., Skryma R., Shuba Y. (2018). Ion channels in cancer: Are cancer hallmarks oncochannelopathies? Physiol. Rev. 98 (2), 559–621. 10.1152/physrev.00044.2016 [DOI] [PubMed] [Google Scholar]
- Richardson V. M., Staskal D. F., Ross D. G., Diliberto J. J., DeVito M. J., Birnbaum L. S. (2008). Possible mechanisms of thyroid hormone disruption in mice by BDE 47, a major polybrominated diphenyl ether congener. Toxicol. Appl. Pharmacol. 226 (3), 244–250. 10.1016/j.taap.2007.09.015 [DOI] [PubMed] [Google Scholar]
- Shioga T., Kondo R., Ogasawara S., Akiba J., Mizuochi S., Kusano H., et al. (2020). Usefulness of tumor tissue biopsy for predicting the biological behavior of hepatocellular carcinoma. Anticancer Res. 40 (7), 4105–4113. 10.21873/anticanres.14409 [DOI] [PubMed] [Google Scholar]
- Shockley K. R., Cora M. C., Malarkey D. E., Jackson-Humbles D., Vallant M., Collins B. J., et al. (2020a). Comparative toxicity and liver transcriptomics of legacy and emerging brominated flame retardants following 5-day exposure in the rat. Toxicol. Lett. 332, 222–234. 10.1016/j.toxlet.2020.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockley K. R., Cora M. C., Malarkey D. E., Jackson-Humbles D., Vallant M., Collins B. J., et al. (2020b). Transcriptomic data from the rat liver after five days of exposure to legacy or emerging brominated flame retardants. Data Brief. 32, 106136. 10.1016/j.dib.2020.106136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti G., Di Filippo P., Riccardi C., Pomata D., Sonego E., Buiarelli F. (2020). Occurrence of halogenated pollutants in domestic and occupational indoor dust. Int. J. Environ. Res. Public Health 17, 3813. 10.3390/ijerph17113813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjödin A., Jones R. S., Wong L. Y., Caudill S. P., Calafat A. M. (2019). Polybrominated diphenyl ethers and biphenyl in serum: Time trend study from the national health and nutrition examination Survey for years 2005/06 through 2013/14. Environ. Sci. Technol. 53 (10), 6018–6024. 10.1021/acs.est.9b00471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjödin A., Mueller J. F., Jones R., Schütze A., Wong L. Y., Caudill S. P., et al. (2020). Serum elimination half-lives adjusted for ongoing exposure of tri-to hexabrominated diphenyl ethers: Determined in persons moving from North America to Australia. Chemosphere 248, 125905. 10.1016/j.chemosphere.2020.125905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. T., Guyton K. Z., Gibbons C. F., Fritz J. M., Portier C. J., Rusyn I., et al. (2016). Key characteristics of carcinogens as a basis for organizing data on mechanisms of carcinogenesis. Environ. Health Perspect. 124 (6), 713–721. 10.1289/ehp.1509912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S., Abraham S. (1970). Fatty acid synthesis in developing mouse liver. Arch. Biochem. Biophys. 136 (1), 112–121. 10.1016/0003-9861(70)90333-4 [DOI] [PubMed] [Google Scholar]
- Sokol A. M., Sztolsztener M. E., Wasilewski M., Heinz E., Chacinska A. (2014). Mitochondrial protein translocases for survival and wellbeing. FEBS Lett. 588 (15), 2484–2495. 10.1016/j.febslet.2014.05.028 [DOI] [PubMed] [Google Scholar]
- Sotgia F., Lisanti M. P. (2017). Mitochondrial biomarkers predict tumor progression and poor overall survival in gastric cancers: Companion diagnostics for personalized medicine. Oncotarget 8 (40), 67117–67128. 10.18632/oncotarget.19962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soussi T., Kroemer G. (2018). MDM2-TP53 crossregulation: An underestimated target to promote loss of TP53 function and cell survival. Trends Cancer 4 (9), 602–605. 10.1016/j.trecan.2018.07.001 [DOI] [PubMed] [Google Scholar]
- Sprenger H., Kreuzer K., Alarcan J., Herrmann K., Buchmüller J., Marx-Stoelting P., et al. (2022). Use of transcriptomics in hazard identification and next generation risk assessment: A case study with clothianidin. Food Chem. Toxicol. 166, 113212. 10.1016/j.fct.2022.113212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swietach P., Vaughan-Jones R. D., Harris A. L., Hulikova A. (2014). The chemistry, physiology and pathology of pH in cancer. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369 (1638), 20130099. 10.1098/rstb.2013.0099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo D. T., Devlin A. A. (2019). Transcriptomic biomarkers in safety and risk assessment of chemicals. Biomarkers Toxicol. 63, 1125–1134. 10.1016/b978-0-12-814655-2.00063-3 [DOI] [Google Scholar]
- Ton T. V., Hong H. H., Anna C. H., Dunnick J. K., Devereux T. R., Sills R. C., et al. (2004). Predominant K-ras codon 12 G--> A transition in chemically induced lung neoplasms in B6C3F1 mice. Toxicol. Pathol. 32 (1), 16–21. 10.1080/01926230490260682 [DOI] [PubMed] [Google Scholar]
- Umeno Y., Ogasawara S., Akiba J., Hattori S., Kusano H., Nakashima O., et al. (2018). Regulator of G-protein signaling 5 enhances portal vein invasion in hepatocellular carcinoma. Oncol. Lett. 15 (2), 1763–1770. 10.3892/ol.2017.7474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- U. S. National Research Council, o.t.N.A (2007). Toxicity testing in the 21st century. Available at:https://www.nap.edu/catalog/11970/toxicity-testing-in-the-21st-century-a-vision-and-a (Accessed October 11, 2022).
- Varshavsky J. R., Sen S., Robinson J. F., Smith S. C., Frankenfield J., Wang Y., et al. (2020). Racial/ethnic and geographic differences in polybrominated diphenyl ether (PBDE) levels across maternal, placental, and fetal tissues during mid-gestation. Sci. Rep. 10 (1), 12247. 10.1038/s41598-020-69067-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong A. M., Braun J. M., Webster G. M., Thomas Zoeller R., Hoofnagle A. N., Sjödin A., et al. (2018). Polybrominated diphenyl ether (PBDE) exposures and thyroid hormones in children at age 3 years. Environ. Int. 117, 339–347. 10.1016/j.envint.2018.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong A. M., Xie C., Jandarov R., Dietrich K. N., Zhang H., Sjödin A., et al. (2020). Prenatal exposure to a mixture of persistent organic pollutants (POPs) and child reading skills at school age. Int. J. Hyg. Environ. Health 228, 113527. 10.1016/j.ijheh.2020.113527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Cheng J. W., Qin J. J., Hu B., Li X., Nijampatnam B., et al. (2019). MDM2-NFAT1 dual inhibitor, MA242: Effective against hepatocellular carcinoma, independent of p53. Cancer Lett. 459, 156–167. 10.1016/j.canlet.2019.114429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wannomai T., Matsukami H., Uchida N., Takahashi F., Tuyen L. H., Viet P. H., et al. (2021). Inhalation bioaccessibility and health risk assessment of flame retardants in indoor dust from Vietnamese e-waste-dismantling workshops. Sci. Total Environ. 760, 143862. 10.1016/j.scitotenv.2020.143862 [DOI] [PubMed] [Google Scholar]
- Wei B. K., Liu C., Wang Y., Jin J. (2020). Polybrominated diphenyl ether in E-waste dismantling sites in taizhou city, zhejiang province: Concentration, distribution, and migration trend. Huan Jing Ke Xue 41 (10), 4740–4748. 10.13227/j.hjkx.202003188 [DOI] [PubMed] [Google Scholar]
- Wu H., Kerr M., Cui X., Churchill G. A. (2003). Maanova: A software package for the analysis fo spotted cDNA mciroarray experiments. I.R. Parmgiani GGE. in The analysis of gene expression data: An overview of methods and software," in The analysis of gene expression data: An overview of methods and software, edZeger SL (ed). Editors Parmgiani G. G. E., Irizarry R. A., Zeger S. L. (New York, NY: Springer; ). [Google Scholar]
- Zhang S., Liu Z., Wu D., Chen L., Xie L. (2020). Single-Cell RNA-seq analysis reveals microenvironmental infiltration of plasma cells and hepatocytic prognostic markers in HCC with cirrhosis. Front. Oncol. 10, 596318. 10.3389/fonc.2020.596318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Sun J., Wang L., Zhu G., Li M., Liu J., et al. (2020). Multiple classes of chemical contaminants in soil from an e-waste disposal site in China: Occurrence and spatial distribution. Sci. Total Environ. 752, 141924. 10.1016/j.scitotenv.2020.141924 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: The datasets analyzed for this study can be found on the National Library of Medicine’s GEO database web site (https://www.ncbi.nlm.nih.gov/geo/). The Affymetrix data for each dataset can be accessed from GEO using GSE153366 for Group One, GSE154914 for Group Two and Group Four, and GSE124431 for Group Three and Group Five.