Abstract
Pasteurella multocida is the causative agent of a wide range of diseases in avian and mammalian hosts. Gene expression in response to low iron conditions was analyzed in P. multocida using whole-genome microarrays. The analysis shows that the expression of genes involved in energy metabolism and electron transport generally decreased 2.1- to 6-fold while that of genes used for iron binding and transport increased 2.1- to 7.7-fold in P. multocida during the first 2 h of growth under iron-limiting conditions compared with controls. Notably, 27% of the genes with significantly altered expression had no known function, illustrating the limitations of using publicly available databases to identify genes involved in microbial metabolism and pathogenesis. Taken together, the results of our investigations demonstrate the utility of whole-genome microarray analyses for the identification of genes with altered expression profiles during varying growth conditions and provide a framework for the detailed analysis of the molecular mechanisms of iron acquisition and metabolism in P. multocida and other gram-negative bacteria.
Pasteurella multocida is a gram-negative, nonmotile, rod-shaped, facultative anaerobe that has been isolated from a wide range of mammals and birds throughout the world. This organism is the etiologic agent of a variety of economically significant diseases, including fowl cholera in poultry, hemorrhagic septicemia in cattle and buffalo, atrophic rhinitis in swine, and snuffles in rabbits (9, 12). The global distribution, severity of disease caused, and the wide variety of livestock affected by P. multocida account for considerable economic losses due to this pathogen worldwide (12).
Iron is an essential nutrient for most organisms due to its important role in metabolic electron transport chains. Due to the presence of specialized protein carriers such as transferrin and lactoferrin in body fluids, the concentration of free iron normally present in mammalian and avian hosts is not enough to support the in vivo growth of bacteria (1, 2). Successful pathogens must therefore possess an effective response to the limited iron conditions encountered upon entry into a host. Previous studies illustrate the fact that the identities of genes and pathways involved in the acquisition, transport, and utilization of iron in P. multocida are poorly understood (5, 6, 10, 11, 13, 16). Here we have utilized whole-genome microarray analysis to identify genes with altered expression patterns when P. multocida is grown under iron-limiting conditions.
Bacterial growth and RNA isolation.
P. multocida PM70 was grown to log phase in a flask of brain-heart infusion (BHI) medium (Becton Dickinson) at 37°C. The culture was split into two 180-ml volumes, briefly centrifuged at 4°C, washed with 1× phosphate-buffered saline (pH 7.0), and centrifuged again. One pellet was resuspended in 180 ml of BHI medium, and the other was resuspended in 180 ml of BHI medium containing the iron chelator 2,2′-dipyridyl (200 μM) (Sigma, St. Louis, Mo.). The resuspended cultures were incubated on a rotary shaker at 37°C, and 30-ml volumes were removed 15, 30, 60, and 120 min after resuspension. These samples were briefly centrifuged at 4°C, and the pellets were flash frozen in dry ice and ethanol. Total RNA extractions were performed with RNeasy Maxi columns (Qiagen, Chatsworth, Calif.), with DNase digestions done on the column by adding 82 Kunitz units of enzyme (Qiagen) and incubating at room temperature for 15 min.
Microarray analysis.
Gene expression analysis with DNA microarrays was performed as described elsewhere (http: //www.cbc.umn.edu/ResearchProjects/AGAC/Pm/Pmarraydata.html) (8a). In brief, a library of targets representing all 2,014 open reading frames (ORFs) from P. multocida PM70 (AE004439) was constructed with primers designed to amplify fragments of ≤500 bp from each ORF from genomic DNA. Two successive rounds of PCR were performed to minimize genomic DNA contamination in the products of amplification, and the final 100-μl reactions were checked for quality on agarose gels and purified with MultiScreen PCR plates (Millipore, Bedford, Mass.). The 1,936 (96%) ORF segments that were successfully amplified were printed using a Total Array System robot (BioRobotics, Boston, Mass.). RNA from P. multocida grown in BHI medium alone or BHI medium containing 2,2′-dipyridyl were labeled with Cy3 and Cy5, respectively, and competitively hybridized with the printed microarrays. Images of the hybridized arrays were obtained with a Scanarray 5000 microarray scanner (GSI Lumonics, Watertown, Mass.). Two independent hybridizations using independent RNA extractions were performed for each time point. Fluorescent intensities for individual spots were normalized based on the total intensity of fluorescence in the Cy3 and Cy5 channels. Hierarchical clustering and analysis were performed using the publicly available programs Cluster and Treeview (M. Eisen; http://www.microarrays.org/software).
P. multocida genes with altered expression profiles.
A complete set of all results is presented on our website at http: //www.cbc.umn.edu/ResearchProjects/AGAC/Pm/Pmarraydata.html. Of the 1,936 ORFs represented on the array, 135 had changes in expression of at least twofold over the course of the experiment (Table 1 and Fig. 1). The remaining ORFs either were not well measured or did not have detectably altered expression levels. A twofold change in the level of expression was used as an indication of significance based on the reproducibility of results obtained in our lab. These genes can be functionally classified based on homology with previously described proteins in public databases (Fig. 2). The classification strategy we used was based on the system developed for the Haemophilus influenzae genome (4). Several genes involved in energy metabolism had altered expression levels. For example, three genes encoding glycolysis enzymes (gapdh, pgk, and eno) had, on average, 2.8-fold decreases in expression in response to low iron conditions, while lactate dehydrogenase expression increased nearly 8-fold. The gene encoding Fnr, a transcriptional activator of genes involved in anaerobic metabolism, was expressed at 3.6-fold higher levels. Interestingly, some genes that are normally transcriptionally activated by Fnr, such as fumarate reductase and formate dehydrogenase (frdABCD and fdxGHI), had an over fourfold decrease in average expression. Additionally, the hypothetical protein encoded by ORF 0064 decreased 3.8-fold and is homologous to YfiD, a pyruvate formate lyase also regulated by Fnr. These discrepancies in Fnr regulation may be due to the disruption of FeS cluster cofactors that Fnr has been proposed to utilize for oxygen sensing (8). The expression of arcA, a transcriptional repressor of aerobic metabolism genes, decreased nearly threefold. In contrast, four genes involved in amino acid biosynthesis (aroA, trpG, hisH, and ilvM) had an average increase in expression of 2.6-fold. Together, these data show that in general the expression levels of genes involved in energy metabolism decreased 2.1- to 6-fold while those of genes involved in DNA and central intermediary metabolism, as well as those involved in amino acid biosynthesis, generally increased (Fig. 2).
TABLE 1.
P. multocida genes with alterations in expression level of at least twofold in response to low iron conditions
| ORF | Gene | Description | Fold changea in expression level |
|---|---|---|---|
| 576 | hemR | Heme-hemopexin utilization protein C | 10.6 |
| 639 | rpL25 | Ribosomal protein L25 | 8.2 |
| 288 | lldD | l-Lactate dehydrogenase | 7.8 |
| 42 | Hypothetical HI0854 protein | 7.7 | |
| 336 | Putative TonB-dependent receptor | 7.7 | |
| 399 | yfeB | Iron(III) dicitrate transport ATP-binding protein | 7.2 |
| 299 | Hypothetical HI0854 protein | 6.2 | |
| 300 | Putative TonB-dependent receptor | 6.0 | |
| 456 | ponB | Penicillin-binding protein 1B | 5.6 |
| 779 | hexC | HexC, capsule biosynthetic locus | 5.4 |
| 1078 | Hemin binding receptor | 5.2 | |
| 51 | fbpA | Iron binding protein FbpA precursor | 5.0 |
| 400 | yfeA | Mn transport protein, ABC transporter | 5.0 |
| 1459 | pgtC | Phosphoglycerate transport regulatory protein | 4.9 |
| 803 | Hypothetical HI1369 protein | 4.8 | |
| 1186 | exbB | Biopolymer transport protein homolog | 4.7 |
| 121 | hpt | Hypoxanthine phosphoribosyltransferase | 4.6 |
| 131 | fecB | Iron(III) dicitrate-binding periplasmic protein | 4.3 |
| 122 | gmhA | Phosphoheptose isomerase | 4.0 |
| 398 | yfeC | Iron (chelated) ABC transporter | 4.0 |
| 129 | fecD | Iron(III) dicitrate transport system permease | 3.6 |
| 1079 | FecCD family membrane transport | 3.6 | |
| 668 | fnr | Fumarate (and nitrate) reduction regulatory protein | 3.6 |
| 1802 | Hypothetical Hl0847 protein | 3.4 | |
| 130 | fecC | Iron(III) dicitrate transport system permease | 3.4 |
| 1460 | Phosphoglycerate transport system activator protein | 3.3 | |
| 1205 | hisH | Amidotransferase | 3.3 |
| 397 | yfeD | Iron (chelated) transporter, permease protein | 3.1 |
| 357 | menE | o-Succinylbenzoate–CoA ligase | 3.1 |
| 1748 | hslV | Putative heat shock protein | 3.0 |
| 212 | Hypothetical HI1601 protein | 2.9 | |
| 1842 | Unknown | 2.9 | |
| 424 | Hypothetical Hl1736 protein | 2.9 | |
| 583 | trpG | Anthranilate synthase component II | 2.9 |
| 1639 | tal | Transaldolase | 2.9 |
| 1481 | Unknown | 2.8 | |
| 1456 | Putative ATP-binding transport protein | 2.8 | |
| 1095 | Unknown | 2.8 | |
| 1457 | Putative periplasmic iron-binding protein | 2.7 | |
| 452 | Hypothetical Yersinia pestis protein | 2.6 | |
| 1816 | recX | Regulatory protein, RecX homolog | 2.6 |
| 1474 | cydD | Transport ATP-binding protein | 2.6 |
| 741 | Hemoglobin receptor precursor | 2.5 | |
| 1188 | tonB | TonB protein | 2.5 |
| 1697 | folB | Dihydroneopterin aldolase | 2.5 |
| 804 | pqqL | Zinc protease | 2.5 |
| 734 | htrA | Heat shock protein, periplasmic serine protease | 2.5 |
| 883 | Hypothetical Hl1293/94 protein | 2.5 | |
| 1529 | hpaG | 2-Hydroxyhepta-2,4-diene-1,7,-dioate | 2.5 |
| 1627 | ilvM | Acetolactate synthase small subunit | 2.5 |
| 719 | nrdB | Ribonucleoside diphosphate reductase, beta chain | 2.5 |
| 720 | Hypothetical Hl1309 | 2.5 | |
| 298 | Hypothetical Shigella dysenteriae protein | 2.4 | |
| 1722 | Hypothetical Hl0857 protein | 2.3 | |
| 839 | aroA | 3-Phosphoshikimate 1-carboxyvinyltransferase | 2.3 |
| 213 | Hypothetical Hl1602 protein | 2.3 | |
| 50 | fbpB | Putative transmembrane permease | 2.3 |
| 1348 | slyX | SlyX protein homolog | 2.3 |
| 611 | sbcB | Exonuclease I | 2.3 |
| 1187 | exbD | Biopolymer transport protein | 2.3 |
| 451 | Hypothetical Y. pestis protein | 2.3 | |
| 90 | Unknown | 2.2 | |
| 1730 | glmS | Glucosamine–fructose-6-phosphate aminotransferase | 2.2 |
| 875 | nagB | Glucosamine-6-phosphate isomerase | 2.2 |
| 1080 | Hypothetical S. dysenteriae protein | 2.2 | |
| 1226 | comD | Putative competence D protein | 2.2 |
| 656 | tehB | Tellurite resistance protein | 2.2 |
| 353 | fldA | Flavodoxin | 2.2 |
| 1483 | Hypothetical Hl0668 protein | 2.2 | |
| 49 | fbpC | ATPase, ABC-type transport protein | 2.2 |
| 1171 | Hypothetical Escherichia coli protein | 2.2 | |
| 1142 | rpL31 | Ribosomal protein L31 | 2.2 |
| 1163 | mpA | RNase P | 2.1 |
| 805 | Unknown | 2.1 | |
| 723 | ttrB | Tetrathionate reductase subunit B | 2.1 |
| 713 | Hypothetical HI1333 protein | 2.1 | |
| 1727 | ATP-binding protein, ABC transporter | 2.1 | |
| 1604 | Hypothetical E. coli protein | 2.1 | |
| 91 | Hypothetical HI0883 protein | 2.0 | |
| 317 | Hypothetical Hl0379 protein | 2.0 | |
| 1201 | Unknown | 2.0 | |
| 1473 | Putative ABC transporter, ATP-binding protein | 2.0 | |
| 1993 | skp | Skp, lipid A biosynthesis | 2.0 |
| 41 | Hypothetical ABC transporter, ATP-binding protein I | 2.0 | |
| 1761 | ORF-3, hypothetical Streptococcus pneumoniae protein | 2.0 | |
| 1792 | Pentahemic c-type cytochrome | −2.0 | |
| 1026 | sprT | SprT homolog | −2.0 |
| 1434 | dcuB | Anaerobic dicarboxylate transport protein homolog | −2.0 |
| 587 | Hypothetical E. coli protein | −2.0 | |
| 1331 | Putative NADH:ubiquinone oxidoreductase | −2.0 | |
| 823 | fumC | Fumarate hydratase class II | −2.1 |
| 1379 | Ribose ABC transporter, ATP-binding protein | −2.1 | |
| 832 | ptnD | Phosphotransferase system enzyme II | −2.1 |
| 667 | rsgA | Ferritin-like protein 2 | −2.1 |
| 1696 | Hypothetical HI0266 protein | −2.2 | |
| 1211 | Hypothetical HI1048 protein | −2.2 | |
| 1592 | napF | Ferredoxin-type protein | −2.3 |
| 1491 | atpH | ATP synthase, delta chain | −2.3 |
| 1854 | Putative iron sulfur protein | −2.3 | |
| 1374 | Unknown | −2.3 | |
| 409 | fdxG | Formate dehydrogenase, alpha major subunit | −2.3 |
| 1286 | uspA | Universal stress protein, phosphotransferase | −2.4 |
| 897 | ptsl | Phosphoenolpyruvate protein, phosphotransferase | −2.4 |
| 834 | Phosphotransferase system enzyme II | −2.4 | |
| 1299 | Hypothetical HI0585 protein | −2.4 | |
| 81 | Hypothetical HI0902 protein | −2.4 | |
| 898 | ptsH | Phosphocarrier protein Hpr | −2.4 |
| 408 | fdxG | Formate dehydrogenase, alpha major subunit | −2.5 |
| 75 | pflB | Formate c-acetyltransferase | −2.5 |
| 1480 | Hypothetical E. coli protein | −2.5 | |
| 942 | Hypothetical Hl0081 protein | −2.5 | |
| 1372 | Putative sugar kinase | −2.6 | |
| 641 | bioD1 | Dethiobiotin synthase | −2.6 |
| 74 | Putative formate transporter | −2.6 | |
| 1860 | pgk | Phosphoglycerate kinase | −2.6 |
| 406 | fdxl | Formate dehydrogenase, gamma subunit | −2.6 |
| 1754 | dmsA | Anaerobic dimethyl sulfoxide reductase, chain A | −2.6 |
| 1285 | mazG | MazG protein | −2.8 |
| 1871 | eno | Phosphopyruvate hydratase (enolase) | −2.8 |
| 407 | fdxH | Formate dehydrogenase, beta subunit | −2.8 |
| 666 | rsgA | Ferritin-like protein 1 | −2.9 |
| 431 | phoR | Phosphate regulon sensor protein | −2.9 |
| 219 | arcA | Aerobic respiration control protein | −3.0 |
| 1852 | lctP | l-Lactate permease | −3.2 |
| 924 | gapdH | Glyceraldehyde-3-phosphate dehydrogenase | −3.3 |
| 1212 | merT | Putative mercuric ion transport protein | −3.3 |
| 1378 | Ribose ABC transporter, permease protein | −3.5 | |
| 198 | frdD | Fumarate reductase, 13-kDa hydrophobic protein | −3.7 |
| 1377 | rbsB | d-Ribose binding periplasmic protein precursor | −3.8 |
| 199 | frdC | Fumarate reductase, 15-kDa hydrophobic protein | −3.8 |
| 64 | Hypothetical Hl0017 protein | −3.8 | |
| 1453 | adh2 | Alcohol dehydrogenase 2 | −4.4 |
| 201 | frdA | Fumarate reductase, flavoprotein subunit | −5.5 |
| 200 | frdB | Fumarate reductase, iron sulfur protein | −6.0 |
| 331 | ompW | Outer membrane protein OmpW precursor | −6.5 |
Fold change is the maximum value observed over four time points. Negative values indicate decreases in the expression level.
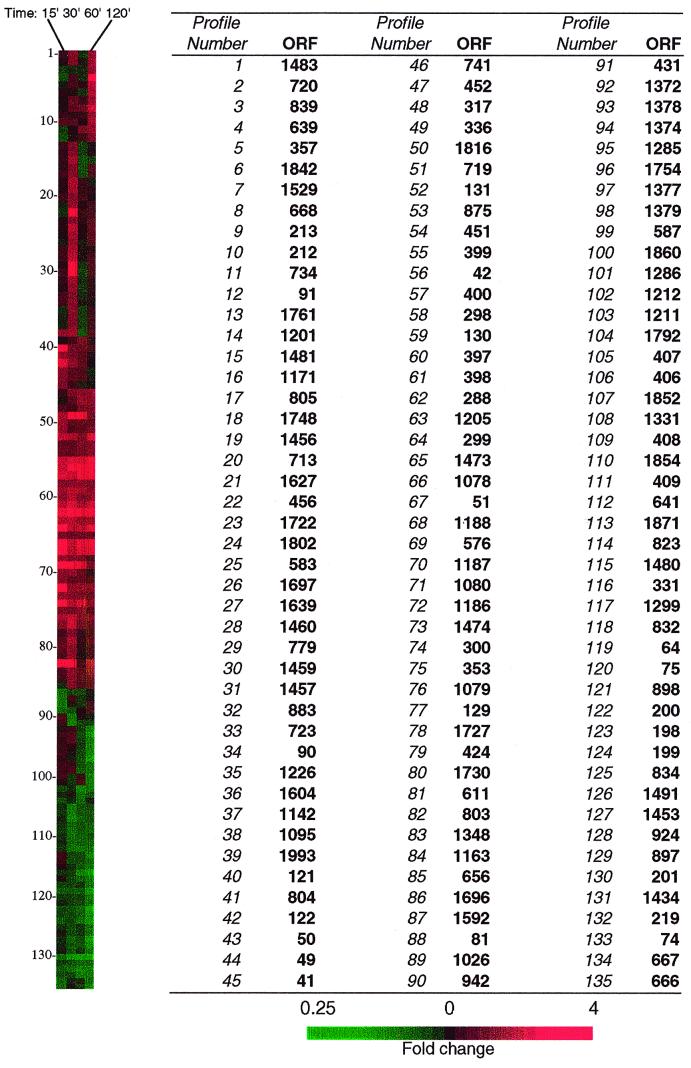
FIG. 1.
Hierarchical clustering of 135 P. multocida ORFs that had significantly altered expression levels under low iron conditions. Clustering and visualization were performed using the software programs Cluster and Treeview. Red and green colors represent fold increase and decrease, respectively, in gene expression in response to low iron conditions. The gene expression profiles for the 15-, 30-, 60-, and 120-min time points are shown in the panel on the left, and corresponding ORF numbers in the P. multocida genome are also shown.
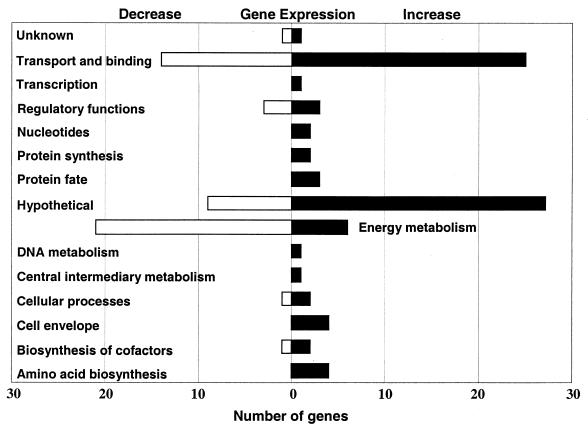
FIG. 2.
Functional classification of P. multocida genes that had altered expression levels, of twofold or more, in response to low iron conditions. Open and closed bars represent the numbers of genes that decreased and increased in expression, respectively.
As expected, many genes encoding proteins involved in iron transport increased their expression levels from 2.1- to 7.5-fold in response to low iron conditions (Table 1). These genes included yfeABCD, fbpABC, fecBCD, tonB, and exbBD. The Yfe, Fbp, and Fec systems are involved in the transport of iron into the cytoplasm, while TonB and ExbBD provide the energy for this to occur. Interestingly, five genes homologous to ABC transport proteins also had an average increase in expression of 2.3-fold. These may represent additional, uncharacterized bacterial transport systems that appear to be regulated by cellular iron content. Additionally, 10 genes involved in the transport of carbohydrates had significantly decreased expression (Table 1). This observation may be related to the decrease in the expression of genes involved in energy metabolism.
Several stress response genes had altered expression levels under low iron conditions. One of these, recX, increased its expression 2.6-fold and has been shown to play a role in the SOS response by binding and inactivating RecA, which normally triggers DNA repair and mutagenesis during an SOS response (17). RecA itself did not have altered expression, but transcription of the universal stress protein A gene (uspA) decreased 2.3-fold. Recent work has shown that uspA may be positively regulated in part by recA (3), and therefore it is possible that the relative excess of RecX may be contributing to a decrease in RecA activity. The expression of the heat shock proteins HslV and HtrA also increased an average of 2.8-fold in response to low iron. HtrA has been shown to respond to periplasmic stress (15), while HslV may be involved in the regulation of cell division protein SulA (14). Two ribosomal proteins, rpL25 and rpL31, had increases in expression of 8- and 2.1-fold, respectively, under low iron conditions. These changes may represent global responses to iron deprivation. Four genes involved in cell surface biosynthesis (gmhA, skp, hexC, and ponB) were also expressed at levels that were an average of fourfold higher than controls. The increase in expression of both stress-related and cell surface synthesis genes indicates that the iron chelator may have had a detrimental effect on the bacterial membrane or that the low iron conditions resulted in modifications and alterations in the bacterial cell surface.
Among the most noteworthy findings was the observation that 27% of the genes with significantly altered expression levels encode hypothetical proteins or have homology to hypothetical proteins from other bacteria (Fig. 2). These results reveal the limitations of our current understanding of genes involved in major processes in bacterial growth and metabolism. Interestingly, many of these hypothetical genes are physically located next to genes with homology to proteins with known functions that also had altered expression profiles (Table 2). This suggests the possibility that some of these hypothetical proteins are coregulated with these previously characterized genes and may possess similar or complementary functions. For example, the hypothetical protein encoded by ORF 0803 appears to have a TonB-dependent outer membrane receptor motif at the C-terminal end. Expression of ORF 0803 increased 4.8-fold, while ORF 0804 (a putative zinc protease) expression in P. multocida increased 2.5-fold. It is possible that these proteins may function together to bind and degrade iron-containing proteins such as transferrin, a hypothesis that remains to be directly tested (7).
TABLE 2.
Hypothetical genes in close proximity to known genes that also had altered expression levelsa
| ORF cluster | Function of known gene(s) |
|---|---|
| 1211, 1212 | Mercuric ion transport |
| 212, 213 | Hypothetical |
| 298, 299, 300 | TonB-dependent receptor |
| 803, 804, 805 | Zinc protease |
| 1078, 1079, 1080 | Hemin binding receptor |
| 1456, 1457 | Putative ABC transporter |
Hypothetical genes are shown in bold.
Utility of the microarray-based approach for profiling of gene expression in bacteria.
A major advantage of the microarray approach is that it enables simultaneous profiling of the transcriptional activity of the entire bacterial genome in a time- and cost-efficient manner. This is especially important in ascribing possible functions to the many hypothetical genes discovered through the whole genome sequencing approach. Furthermore, knowledge of the transcriptional profile of the entire genome enables a holistic approach to the understanding of the metabolic state of the organism under various conditions. Apart from being relatively inexpensive to set up and perform, the use of two-color hybridizations permits comparative analyses of bacterial gene expression by obviating many of the sources of variation inherent in other methods, including single color- or radiolabel-based hybridization methods and many of the PCR-based approaches.
However, there are numerous limitations that must be considered when interpreting results of microarray-based expression analyses. For example, the analyses are limited in that they only index changes in the transcription of a gene and do not account for posttranscriptional regulation that may influence gene and protein expression. Furthermore, short-lived and unstable transcripts are often not well measured since microarrays are essentially a “snapshot” of the transcriptional activity at a fixed time point. The sensitivity of microarrays to detect small changes in gene expression is also currently unknown. Therefore, the results of microarray analyses need to be confirmed with more sensitive techniques such as quantitative PCR-based approaches both for validation purposes and to minimize the occurrence of “false positives.” Overall, it is important to note that these limitations do not detract from the overall utility of the microarray-based approach for global gene expression profiling in bacteria. Therefore, while microarrays do not provide the definitive answer to all questions of gene expression and regulation in bacterial pathogens, they serve as an excellent starting point for screening large numbers of genes to determine patterns of differential gene expression and to compare transcript profiles of bacterial cells of differing phenotypes or those that are subject to different environmental stimuli.
In summary, the results of our investigations show that microarray-based analysis of gene expression provides an effective tool for the identification of gene targets that are involved in major metabolic processes in bacterial pathogens as well as in the initial stages of infection and iron acquisition from hosts.
Acknowledgments
M.P. is supported by an NIH NIGMS Training for Future Biotechnology Development Grant (T32 GM08347). Funding for this project was provided by research grants from the Minnesota Turkey Growers Association, the Minnesota Agricultural Experiment Station, the University of Minnesota Academic Health Center, and the United States Department of Agriculture's National Research Initiative (to V.K.).
REFERENCES
- 1.Bullen J J. The significance of iron in infection. Rev Infect Dis. 1981;3:1127–1138. doi: 10.1093/clinids/3.6.1127. [DOI] [PubMed] [Google Scholar]
- 2.Bullen J J, Rogers H J, Griffiths E. Role of iron in bacterial infection. Curr Top Microbiol Immunol. 1978;80:1–35. doi: 10.1007/978-3-642-66956-9_1. [DOI] [PubMed] [Google Scholar]
- 3.Diez A, Gustavsson N, Nystrom T. The universal stress protein A of Escherichia coli is required for resistance to DNA damaging agents and is regulated by a RecA/FtsK-dependent regulatory pathway. Mol Microbiol. 2000;36:1494–1503. doi: 10.1046/j.1365-2958.2000.01979.x. [DOI] [PubMed] [Google Scholar]
- 4.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 5.Flossmann K D, Grajetzki C, Rosner H. Demonstration of iron transport activity in Pasteurella multocida cultures. J Basic Microbiol. 1985;25:559–567. doi: 10.1002/jobm.3620250902. [DOI] [PubMed] [Google Scholar]
- 6.Hu S P, Felice L J, Sivanandan V, Maheswaran S K. Siderophore production by Pasteurella multocida. Infect Immun. 1986;54:804–810. doi: 10.1128/iai.54.3.804-810.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.James B W, Mauchline W S, Dennis P J, Keevil C W. A study of iron acquisition mechanisms of Legionella pneumophila grown in chemostat culture. Curr Microbiol. 1997;34:238–243. doi: 10.1007/s002849900176. [DOI] [PubMed] [Google Scholar]
- 8.Khoroshilova N, Beinert H, Kiley P J. Association of a polynuclear iron-sulfur center with a mutant FNR protein enhances DNA binding. Proc Natl Acad Sci USA. 1995;92:2499–2503. doi: 10.1073/pnas.92.7.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.May B J, Zhang Q, Li L L, Paustian M L, Whittam T S, Kapur V. Complete genomic sequence of Pasteurella multocida, Pm70. Proc Natl Acad Sci USA. 2001;98:3460–3465. doi: 10.1073/pnas.051634598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morishita T Y, Lowenstine L J, Hirsh D C, Brooks D L. Pasteurella multocida in raptors: prevalence and characterization. Avian Dis. 1996;40:908–918. [PubMed] [Google Scholar]
- 10.Ogunnariwo J A, Alcantara J, Schryvers A B. Evidence for non-siderophore-mediated acquisition of transferrin-bound iron by Pasteurella multocida. Microb Pathog. 1991;11:47–56. doi: 10.1016/0882-4010(91)90093-p. [DOI] [PubMed] [Google Scholar]
- 11.Reissbrodt R, Erler W, Winkelmann G. Iron supply of Pasteurella multocida and Pasteurella haemolytica. J Basic Microbiol. 1994;34:61–63. doi: 10.1002/jobm.3620340114. [DOI] [PubMed] [Google Scholar]
- 12.Rimler R B, Angus R D, Phillips M. Evaluation of the specificity of Pasteurella multocida somatic antigen-typing antisera prepared in chickens, using ribosome-lipopolysaccharide complexes as inocula. Am J Vet Res. 1989;50:29–31. [PubMed] [Google Scholar]
- 13.Ruffolo C G, Jost B H, Adler B. Iron-regulated outer membrane proteins of Pasteurella multocida and their role in immunity. Vet Microbiol. 1998;59:123–137. doi: 10.1016/s0378-1135(97)00123-5. [DOI] [PubMed] [Google Scholar]
- 14.Seong I S, Oh J Y, Yoo S J, Seol J H, Chung C H. ATP-dependent degradation of SulA, a cell division inhibitor, by the HslVU protease in Escherichia coli. FEBS Lett. 1999;456:211–214. doi: 10.1016/s0014-5793(99)00935-7. [DOI] [PubMed] [Google Scholar]
- 15.Strauch K L, Johnson K, Beckwith J. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J Bacteriol. 1989;171:2689–2696. doi: 10.1128/jb.171.5.2689-2696.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veken J W, Shah N H, Klaasen P, Oudega B, de Graaf F K. Binding of host iron-binding proteins and expression of iron-regulated membrane proteins by different serotypes of Pasteurella multocida causing haemorrhagic septicaemia. Microb Pathog. 1996;21:59–64. doi: 10.1006/mpat.1996.0042. [DOI] [PubMed] [Google Scholar]
- 17.Vierling S, Weber T, Wohlleben W, Muth G. Transcriptional and mutational analyses of the Streptomyces lividans recX gene and its interference with RecA activity. J Bacteriol. 2000;182:4005–4011. doi: 10.1128/jb.182.14.4005-4011.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]