Abstract
Ethiopian wolves, a canid species endemic to the Ethiopian Highlands, have been steadily declining in numbers for decades. Currently, out of 35 extant species, it is now one of the world's most endangered canids. Most conservation efforts have focused on preventing disease, monitoring movements and behavior, and assessing the geographic ranges of sub-populations. Here, we add an essential layer by determining the Ethiopian wolf's demographic and evolutionary history using high-coverage (∼40×) whole-genome sequencing from 10 Ethiopian wolves from the Bale Mountains. We observe exceptionally low diversity and enrichment of weakly deleterious variants in the Ethiopian wolves in comparison with two North American gray wolf populations and four dog breeds. These patterns are consequences of long-term small population size, rather than recent inbreeding. We infer the demographic history of the Ethiopian wolf and find it to be concordant with historic records and previous genetic analyses, suggesting Ethiopian wolves experienced a series of both ancient and recent bottlenecks, resulting in a census population size of fewer than 500 individuals and an estimated effective population size of approximately 100 individuals. Additionally, long-term small population size may have limited the accumulation of strongly deleterious recessive mutations. Finally, as the Ethiopian wolves have inhabited high-altitude areas for thousands of years, we searched for evidence of high-altitude adaptation, finding evidence of positive selection at a transcription factor in a hypoxia-response pathway [CREB-binding protein (CREBBP)]. Our findings are pertinent to continuing conservation efforts and understanding how demography influences the persistence of deleterious variation in small populations.
Keywords: conservation, adaptation to high altitude, demography, Ethiopian wolf
Introduction
Conservation biology has long been concerned with the consequences of small effective population sizes in endangered species and how it may lead to extinction (Gilpin and Soulé 1986 ; Lande and Barrowdough 1987; Lande 1993; Caughley 1994; Lynch, Conery, and Bürger 1995a; Lynch, Conery, and Bürger 1995b; Frankham 2005). Time to extinction can be accelerated in small populations due to the accumulation of deleterious mutations, inbreeding depression, or loss of adaptive potential, all of which can result in a large reduction in fitness (Lande 1988). Since the demographic history of a population impacts the distribution and prevalence of deleterious variation (Ohta 1973; Lande 1988; Lohmueller et al. 2008) elucidating the interaction between demography and genetics is a key component of effective conservation. Furthermore, recent research has shown that the small-population paradigm (Gilpin and Soulé 1986) may not fit all small populations (Robinson et al. 2018), and that some small populations have the ability to avoid extinction presumably through the purging of strongly deleterious recessive alleles.
The Ethiopian wolf (Canis simensis) is endemic to the Ethiopian Highlands, where it is restricted to the Afroalpine habitat due to its specialized diet of burrowing mole rats (Sillero-Zubiri and Gottelli 1995). The Ethiopian wolf population has presumably been declining since the Holocene (∼10–15,000 years ago) when the Afroalpine habitat began to retract (Gottelli et al. 2004), and has experienced recent crashes caused by anthropogenic activity. The expansion of human agriculture into the Highlands has resulted in increased habitat fragmentation, and range restriction, in addition to rabies and canine distemper virus outbreaks from increased proximity to domestic dogs (Sillero-Zubiri et al. 1996a; Whitby et al. 1997; Laurenson et al. 1998; Randall et al. 2004; Randall et al. 2006; Gordon et al. 2015). Two populations have become locally extinct in the last 25 years (Gottelli et al. 2013) and today, there remain fewer than 500 individuals (Group ICS 2011; Marino et al. 2013) sub-divided between six small isolated populations, having between 15 and 200 wolves each. These numbers are particularly concerning given the smaller proportion of mature wolves, and the social breeding system, whereby only a single dominant female and a few males breed per pack, which results in a very small effective population size (Sillero-Zubiri et al. 1996b; Randall et al. 2004; Marino and Sillero-Zubiri 2011). The largest remaining population is found in the Bale Mountains, where approximately 250 wolves reside in three genetically differentiated sub-populations (Randall et al. 2010). Gene flow occurs between these sub-populations through both male and female dispersal, and it is believed these mating behaviors allow for inbreeding avoidance despite this small population size (Randall et al. 2007; Randall et al. 2010). Previous surveys of genetic diversity have been conducted using either mitochondrial DNA (Gottelli et al. 1994) or microsatellites (Randall et al. 2007; Randall et al. 2010). These studies have shown that diversity in the remaining Ethiopian wolf populations is low and have suggested that long-term effective population sizes (Ne) are small across sub-populations from the Bale Mountains (Randall et al. 2010). However, this research was limited to a handful of putatively neutral loci, providing little information about loci under selection.
In sum, conservation efforts have provided a wealth of information about the behavior, disease, diet, and range of the Ethiopian wolf, but there is limited knowledge about genetic diversity and historical demography. Here, we determine how both ancient and recent demography has affected the genetic diversity of Ethiopian wolves. We generated high-coverage (∼40×) whole-genome sequence data of 10 contemporary Ethiopian wolves from the Bale Mountains. To examine the effect of long-term small population sizes on deleterious mutations, we compare the Ethiopian wolf with two gray wolves (Canis lupus) populations: 1) the Arctic wolf, a large extant gray wolf population and 2) the Isle Royale wolf, an isolated island population which was founded in the 1940s by 2–3 wolves. The Isle Royale population has since remained at a low population size averaging fewer than 25 wolves and exhibits several features of inbreeding depression (Peterson et al. 2014; Robinson et al. 2019). We also contrast these wild canids to four domestic dogs (Canis lupus familiaris) breeds: labrador retriever, pug, border collie, and Tibetan mastiff.
We find that the Ethiopian wolf exhibits remarkably low diversity relative to both gray wolves and breed dogs, as well as an enrichment of derived putatively deleterious variation. The inferred demography of the Ethiopian wolf includes multiple bottlenecks and suggests that the current effective size (Ne) is quite low. Despite the low Ne, the distribution of runs of homozygosity (ROHs) in the Ethiopian wolf does not suggest that there has been recent inbreeding in the population, demonstrating that small populations can avoid inbreeding under specific social and mating structures. We also find evidence of adaptation to high altitude through positive selection at the transcription factor CREB-binding protein (CREBBP).
Results and Discussion
Genetic Diversity of the Ethiopian Wolf
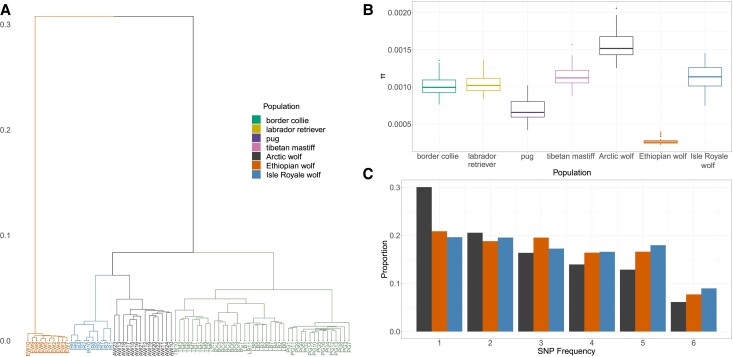
We used multiple approaches to compare the genetic diversity of the Ethiopian wolf with gray wolves and dogs (fig. 1). First, we assessed the phylogenetic relationships among canids, and performed hierarchical clustering on the shared identity-by-state (IBS) loci between individuals. The Ethiopian wolf has the deepest divergence, as it formed a separate clade from both wolves and dogs (fig. 1A), which is concordant with recent phylogenetic work showing Ethiopian wolves as being closer to the root of the genus Canis (Gopalakrishnan et al. 2018; Chavez et al. 2019). When we compute weighted pairwise-FST between the seven sampled groups, the Ethiopian wolf is most differentiated from the gray wolves and dogs, with an average pairwise-FST of approximately 0.8 (supplementary fig. S1A, Supplementary Material online). For example, when compared with other wolves and dogs, the Arctic wolves have FST of 0.38 and the Tibetan Mastiffs have an FST of 0.35. Unweighted average FST values are smaller in magnitude but reveal the same qualitative patterns of differentiation (supplementary fig. S1B, Supplementary Material online).
Fig. 1.
Summaries of genetic variation in the Ethiopian wolf compared with other canids. (A) Hierarchical clustering based on shared IBS loci between individuals, where the dendrogram is cut into K = 4 groups. Note that the Ethiopian wolves are an outgroup compared with other canids. (B) Diversity in dogs and wolves measured using the average number of pairwise differences between sequences, π. Ethiopian wolves have exceptionally low genetic diversity. Boxes represent the distribution over each chromosome. (C) The folded site frequency spectrum (SFS) for each wolf population. The full-unfolded SFS can be found in supplementary fig. S2, Supplementary Material online.
To summarize patterns of genetic variation in the Ethiopian wolf, we compared the genome-wide average number of pairwise differences (π) with those of other canids (fig. 1B). The Ethiopian wolf had the lowest levels of diversity compared with the other wolf-like canids (mean π ≈ 0.000269) and the Arctic wolf had the highest level of diversity (mean π ≈ 0.00156). The Isle Royale wolf had levels of diversity close to breed dogs (mean π ≈ 0.00113). The pug had the lowest diversity in breed dogs (mean π ≈ 0.000679). To further assess this depletion of diversity in the Ethiopian wolf, we examined the proportional site frequency spectrum (SFS) in all wolf populations (fig. 1C). Similar to the pattern we observed with π, Arctic wolves had the highest proportion of singletons. The Ethiopian wolves and the Isle Royale wolves had markedly lower proportions of singletons. For the Ethiopian wolves, along with the reduction of singletons, there was a flattening of the SFS, suggesting a loss of rare alleles. This flattening in the Ethiopian wolf is likely a consequence of population contractions. The Isle Royale wolf SFS showed enrichment of intermediate frequency variants, relative to any other population, suggesting there was a recent contraction in size coupled with severe inbreeding in this island population (Robinson et al. 2019) (supplementary fig. S2, Supplementary Material online).
Measures of Inbreeding
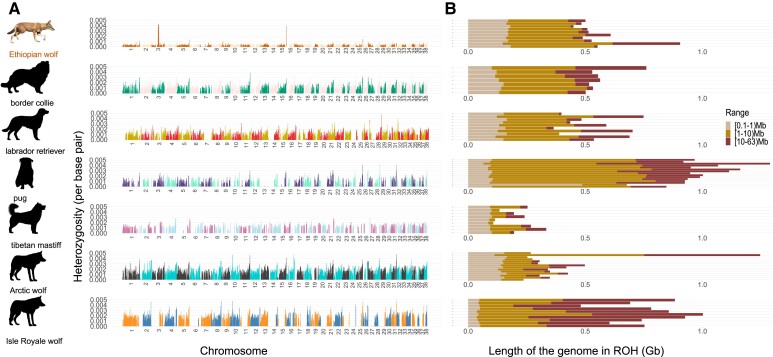
Next, we examined spatial patterns of heterozygosity and ROH across the genome to assess whether recent inbreeding has affected the Ethiopian wolf population (fig. 2). The Ethiopian wolf showed signs of genomic-flatlining, whereby individuals had very low diversity genome-wide and a few peaks of heterozygosity, alongside stretches of short (100 Kb up to 1 Mb) and intermediate-sized (1 Mb up to 10 Mb) ROH (supplementary Table S1, Supplementary Material online). As short ROH tend to be formed by common ancestry in the distant past, the increase in short and intermediate ROH along with the low heterozygosity in the genome suggest a long-term small population size in the Ethiopian wolf (Pemberton et al. 2012; Ceballos et al. 2018). Conversely, the Arctic wolf had high levels of heterozygosity across the genome and ROH were infrequent. These patterns are consistent with a demographic history of large population size (Musiani et al. 2007; Gray et al. 2009; Pollinger et al. 2011). Whereas, the Isle Royale wolf had a saw-tooth pattern of heterozygosity, stretches of high heterozygosity intertwined with long ROH (10 Mb and greater). Isle Royale wolf had the largest fraction of its genome in long ROH (population mean ∼430 Mb), even relative to dogs (largest population mean ∼289 Mb). The large amount of long ROH is well explained by the recent population crash and extensive inbreeding in the Isle Royale wolf population (Hedrick et al. 2014; Robinson et al. 2019).
Fig. 2.
Distribution of homozygosity across canid genomes. (A) Sliding-windows of heterozygosity across the genome from a single representative sample in each population. Chromosomes are ordered from 1 to 38. Note the low heterozygosity across the entire Ethiopian wolf genome. (B) The distribution of short (10 Kb–1 Mb), intermediate (1–10 Mb), and long ROH (10–63 Mb) in dogs and wolves. Each row of the ROH plot represents a single individual. Supplementary Table S1, Supplementary Material online reports the population averages per ROH range.
We found that breed dogs generally had very long ROH interspersed between regions with high heterozygosity (fig. 2). Long ROH were particularly prominent in the pug (population mean ∼288 Mb), which also had lower levels of heterozygosity relative to the other breed dogs. These patterns are likely consequences of extremely small effective population sizes in the recent history of the pug. All breed dogs had an enrichment of long ROH (dog mean ∼173 Mb) compared with the non-inbred Arctic and Ethiopian wolves (mean ∼85 Mb). The enrichment of long ROH is a consequence of the domestication bottleneck (common to all dogs) and bottlenecks of varying intensity experienced during breed formation (Boyko et al. 2010; Mooney et al. 2021) as well as inbreeding (Lindblad-Toh et al. 2005). These breed formation bottlenecks along with strong artificial selection for phenotypic traits of interest likely played the most significant role in generating long ROHs in dogs (Boyko et al. 2010).
In sum, unlike breed dogs and the Isle Royale wolf, overall the Ethiopian wolf population does not appear to carry long ROH, except for one outlier individual, which are characteristic of recent inbreeding. The outlier individual is a product of recent inbreeding and when excluded from analyses the average amount of the genome in a long ROH drops to ∼41 Mb in contrast to the ∼66 Mb reported in supplementary Table S1, Supplementary Material online. Nevertheless, overall, patterns of heterozygosity and shorter ROH suggest long-term small population size and extensive genetic drift.
Demographic History of the Ethiopian Wolf
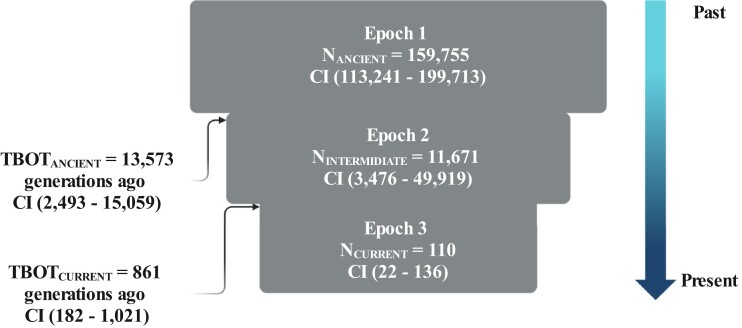
Our results are largely consistent with previous surveys of genetic diversity (Gottelli et al. 1994; Randall et al. 2007; Randall et al. 2010) and demographic analysis which suggested that the effective population size of Ethiopian wolves has been decreasing through time (Gottelli et al. 2004; Randall et al. 2010). However, none of the previous studies reconstructed the demographic history of the Ethiopian wolf. Given multiple genomes and high-coverage data, we were able to infer the demographic history of the Ethiopian wolf using approximate Bayesian computation (ABC). We inferred demographic parameters using the distribution of segregating sites (S) and π from a subset of four individuals (see Methods for details). We fit a three-epoch model (fig. 3) with a series of instantaneous population contractions, chosen based on population size change trajectories from running MSMC2 independently on four genomes (supplementary fig. S3, Supplementary Material online). The ABC inference suggested a large ancestral population size of approximatley 159,754 individuals and an ancient population contraction approximately 13,572 generations ago. The ancient contraction was severe and decreased population size to approximately 11,600 individuals, or 7% of the original population size. This contraction was followed by an additional recent severe contraction, which decreased the current effective population size to approximately 100 individuals (fig. 3). Our estimated current effective population size is on the same order of magnitude as estimates from field surveys, which place current census size as 197 mature individuals and decreasing (https://www.iucnredlist.org/species/3748/10051312). Importantly, we inferred this most recent contraction to 100 individuals occurred 861 generations ago, suggesting that the Ethiopian wolf has had a small population size for a very long time. Such a demographic history can explain the low levels of heterozygosity across the genome.
Fig. 3.
The Ethiopian wolf population has been at small size for an extended time. The demographic history of the Ethiopian wolf inferred using ABC and a three-epoch model. The inferred demographic history of the Ethiopian wolf, with point estimates (maximum a posteriori probability) and 95% credible intervals (CI) of the population size and bottleneck times (supplementary Table S2, Supplementary Material online). Demography schematic was created with BioRender.com.
Deleterious Variation
We next examined the prevalence of deleterious variation in the Ethiopian wolf. Understanding the prevalence of deleterious mutations is particularly relevant in the Ethiopian wolf because of their small population size, recent population bottlenecks due to disease, and recent sub-population extinctions. To compare across canids, we first polarized alleles using the wild dog (Lycaon pictus) as an outgroup. Then, GERP was used to annotate variants as putatively neutral or deleterious (see Methods for details). We implemented multiple approaches to count deleterious variants in the genome of an individual: 1) summing homozygous derived genotypes (counting homozygotes); 2) summing the total number of homozygous and heterozygous derived genotypes (counting variants); and 3) tallying twice the number of homozygous derived genotypes plus heterozygous genotypes (counting alleles). If deleterious alleles are recessive, counting derived homozygotes is most relevant. Conversely counting alleles is most relevant if deleterious alleles have additive effects on fitness (Lohmueller 2014; Simons and Sella 2016).
Overall, breed dogs had slightly more homozygous deleterious variants than the Arctic wolf, and the Isle Royale wolf carried deleterious variation at levels closer to dogs (Marsden et al. 2016; Robinson et al. 2019) (supplementary fig. S4, Supplementary Material online). This was expected given the recent breed formation bottleneck and inbreeding. Remarkably, we found that the Ethiopian wolf carried more neutral and deleterious derived homozygotes than dogs and wolves (supplementary fig. S4, Supplementary Material online). On average, we observed a 1.56-fold (Mann–Whitney U (MWU) P-value = 3.655e−06) and 1.76-fold (MWU P-value = 1.872e−05) increase of deleterious homozygotes in the Ethiopian wolf relative to dogs or wolves, respectively. When counting variants, all three wolf populations carried comparable numbers of deleterious derived variants, and there was a slight increase of synonymous variants relative to dog (1.13-fold and MWU P-value = 3.653e−06). The Ethiopian wolf also appeared to have a depletion of neutral segregating variants, suggesting that most derived alleles are fixed rather than segregating as heterozygotes. This result is concordant with our observations of low genome-wide diversity (fig. 1B). Finally, when counting alleles, we found the Ethiopian wolf carried a comparable number of neutral alleles as dogs and wolves, and a 1.29-fold (MWU P-value = 3.665e−06) and 1.24-fold (MWU P-value = 1.872e−05) increase of deleterious derived alleles relative to dogs and wolves, respectively. Carrying a similar number of neutral alleles demonstrated that the pipeline to process and merge the data was successful, as the number of derived neutral alleles per genome is predicted to be insensitive to demographic history (Simons and Sella 2016). The elevated levels of predicted deleterious homozygous genotypes and derived alleles in the Ethiopian wolf compared with gray wolf populations and breed dogs, and no phenotypic manifestation of inbreeding suggest an accumulation of weakly deleterious variation due to continued small population size.
Adaptation to High Altitude
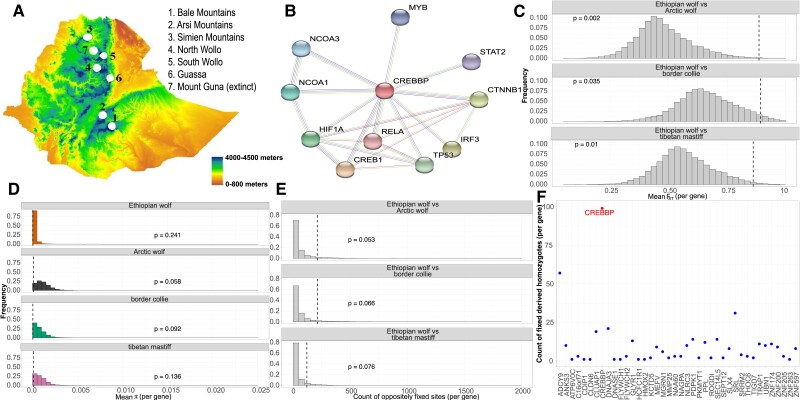
The Ethiopian wolf exclusively inhabits the high-altitude (3,000–4,500 m) regions of the Bale Mountains and has specialized to hunt mole rats native to this area (Gottelli and Sillero-Zubiri 1992; Sillero-Zubiri and Gottelli 1995) (fig. 4A). The Ethiopian wolf has been restricted to high-altitude for thousands of generations. Thus, we hypothesized that the species may have adapted to a hypoxic environment, much like the Tibetan wolves and Tibetan mastiffs had (Li et al. 2014; Miao et al. 2017; vonHoldt et al. 2017; Witt and Huerta-Sánchez 2019; Wang et al. 2020a). We performed a genome-wide scan for signatures of positive selection where we focused on high-altitude genes (see Methods). We compared the Ethiopian wolf with multiple breed dogs (Tibetan mastiff and border collie) and the Arctic wolf, and searched for genes showing unusually high average FST, low levels of diversity (π) and elevated counts of homozygous genotypes for derived alleles (see fig. 4). There was a single gene within the hypoxia inducible factor (HIF)-1α pathway (fig. 4B), CREBBP, that fell within the top 5% of all FST comparisons, regardless of which reference population was used (fig. 4C). Additionally, CREBBP had: 1) low values of π (fig. 4D); 2) an enrichment of sites fixed for the derived allele in the Ethiopian wolves relative to the three comparison groups, for example sites that are fixed as either ancestral in Ethiopian wolves and derived in the comparison population or derived in Ethiopian wolves and ancestral in the comparison population (fig. 4E); and 3) an enrichment of fixed derived homozygous sites (fig. 4F) in CREBBP relative to all other genes in the Ethiopian wolf genome.
Fig. 4.
Signals of high-altitude adaptation in the Ethiopian wolf. (A) Topography of the Ethiopian wolf habitat and sub-population locations. The lowest altitude begins at sea level and the highest altitude where the Bale Mountain population resides is approximately 4000 meters above sea level. Topography is modified from Gutema et al. 2018. (B) The primary protein-interaction network for CREBBP (represented by a red node) in Canis lupus familiaris from STRING (Szklarczyk et al. 2016). Each node represents a protein-coding gene, and the edges represent protein–protein associations that contribute to a shared function. The colors of each edge represent methods of validation of interactions (turquoise: curated database; purple: experimentally curated; green: text mining, black: co-expression). For (C)–(E), the vertical dashed line indicates where CREBBP falls in the distribution. P-values were computed as the proportion of genes with summary statistic values as or more extreme than what was observed for CREBBP. (C) The distribution of unweighted mean FST per gene when comparing the Ethiopian wolf to each reference population. (D) Distribution of π per gene for each population. (E) The distribution of oppositely fixed sites (fixed as ancestral in Ethiopian wolf and derived in comparison population or the converse) per gene when comparing the Ethiopian wolf with each reference population. (F) The count of fixed derived homozygous alleles in each gene within a 1 Mb window around CREBBP. CREBBP has the largest number of fixed derived sites.
Importantly, there were no other genes in the genome that were more extreme in all these patterns of genetic variation than CREBBP. Furthermore, to ensure that these patterns were not simply reflecting genomic phenomena that is potentially unusual in CREBBP that are unrelated to its role in altitude adaptation, we performed a control analysis. Here, we repeated the FST analysis, but now considering border collie as the focal population of interest instead of the Ethiopian wolf. We compared the border collie with the Arctic wolf and Tibetan mastiff. In these comparisons, CREBBP is not an outlier for FST (supplementary fig. S5, Supplementary Material online). Finally, we compared CREBBP-derived allele counts with other genes in the HIF-1α pathway that were identified as outliers in at least one scan (supplementary Table S3, Supplementary Material online). We observed that CREBBP contains more fixed derived alleles than any other gene in a 1Mb flanking region. This finding suggests that CREBBP, rather than another nearby gene, may be the target of selection. The three other genes in the HIF-1α pathway that showed elevated differentiation did not show elevated counts of derived alleles compared with the genes surrounding them (supplementary fig. S6, Supplementary Material online). Taken together, our results suggest that the patterns of genetic variation in CREBBP are unique with respect to the rest of the genome. Thus, this gene may contribute to the adaptation of the Ethiopian wolf to the high-altitude Bale Mountains environment.
CREB is a transcription factor that is critical to cellular processes, such as cell development, proliferation, differentiation, hypoxia-response, and circadian rhythm (Ginty et al. 1993; Arany et al. 1996; Kallio et al. 1998; Shaywitz and Greenberg 1999). One of the primary protein–protein interactions of CREBBP is with HIF-1α (fig. 4B), which has been validated with multiple methods shown above with STRING (Szklarczyk et al. 2016). The CREB transcriptional complex (CREB, CREBBP, and p300) interacts with multiple hypoxia-activated genes via HIF-1α in response to oxygen deprivation (Arany et al. 1996; Kallio et al. 1998). Since p300 directly interacts with DNA-bound HIF-1, the CREB transcriptional complex can physically modulate the cellular response to hypoxia (Arany et al. 1996). Cells can specifically adapt to hypoxic conditions via HIF-1-dependent induction of erythropoietin (Epo) which increases red blood cell production (Scholz et al. 1990; Arany et al. 1996; Kallio et al. 1998). Furthermore, previous work on pathways involved in response to hypoxia in human Ethiopian populations identified BHLHE41 as showing a strong signature of selection, and CREBBP directly interacts with BHLHE41 (Witt and Huerta-Sánchez 2019).
Loss of PRDM9
Finally, we examined PRDM9, a gene known for its involvement in specific hot spots associated with meiotic recombination in humans and other mammalian species. PRDM9 contains loss-of-function mutations in dogs, coyotes, and golden jackals (Muñoz-Fuentes et al. 2011; Axelsson et al. 2012; Baker et al. 2017; Grey et al. 2018). Due to the loss of PRDM9 function, meiotic recombination is believed to occur by a different mechanism in canid species than in other mammals (Muñoz-Fuentes et al. 2011). Since the Ethiopian wolf is an outgroup on the canid phylogeny compared with dogs, coyotes, and golden jackals, whether PRDM9 also contains loss-of-function mutations in Ethiopian wolves can be used to date when the loss-of-function events occurred. Thus, we reconstructed the gene tree using amino acid (or nucleotide) sequences of PRDM9 (supplementary fig. S7, Supplementary Material online) using the domestic cat as the outgroup. We compared PRDM9 with GAPDH, a housekeeping gene that is involved with glycolysis and other cellular processes as a control (supplementary fig. S7, Supplementary Material online). The phylogenetic tree for GAPDH follows the expected species tree given inter-species evolutionary divergence. In comparison, for PRDM9 we observed that the human and macaque clustered together, as expected, and remained close to the other species that retained a functional copy of PRDM9, whereas canids formed a single divergent clade. We used GeneWise (Birney et al. 2004) to test whether PRDM9 was functional. We observed multiple frameshift mutations and stop codons in the zinc-finger region of PRDM9 in all the canid species (supplementary file, Supplementary Material online). Thus, our results indicated that the functionality of PRDM9 was lost prior to the speciation of the Ethiopian wolf, and likely has been lost across the entire grouping of wolf-like canids since the Dhole also retained a non-functional copy of PRDM9.
Conclusions
Our study is the first to use whole-genome sequence data to investigate genetic diversity and infer the demographic history of the Ethiopian wolf, a severely endangered canid. Despite showing molecular evidence for adaptation to its high-altitude environment, the species has been steadily declining for millennia and experienced recent sub-population extinction. We have shown that despite having a long-term small effective population size, low genetic diversity, and an enrichment of deleterious variation relative to breed dogs and gray wolves, the Ethiopian wolf has persisted for about 13,500 generations since an ancient contraction (or ∼27,000 or 40,500 years ago, assuming a 2- or 3-year generation time). Throughout the years, the population has continued declining and experiencing both recent crashes and sub-population extinction due to anthropogenic forces. The most recent crashes are a result of human agriculture in the Highlands and disease due to the introduction and close proximity to domestic dogs (Sillero-Zubiri et al. 1996a; Whitby et al. 1997; Laurenson et al. 1998; Randall et al. 2004; Randall et al. 2006; Gordon et al. 2015). Close proximity to the domestic dog has not only resulted in disease but also resulted in admixture between the two species. In some cases, admixture with dogs may actually enhance adaptation and disease resistance (vonHoldt et al. 2017; Cubaynes et al. 2022). However, we found no evidence of admixture with sampled domestic dogs in our dataset or village dogs (supplementary fig. S8–S10, Supplementary Material online) when we merge the data with previously published datasets (Hayward et al. 2016; Fitak et al. 2018). Our findings do not mean that a recent admixture has not occurred between these populations, but that this admixture is not detectable within our data. We believe this result is likely because we pre-screened our data with STRUCTURE to be unadmixed with domestic dogs (see Methods).
Given the recent population crashes, sub-population extinction, and long-term small population size of the Ethiopian wolf, the observed enrichment of derived deleterious homozygotes is expected (supplementary fig. S4, Supplementary Material online) and has been observed in other small populations. For example, the Ethiopian wolf has levels of heterozygosity (fig. 2) and levels of deleterious homozygous alleles (supplementary fig. S4, Supplementary Material online) comparable with that of the Island Fox population (Robinson et al. 2016; Robinson et al. 2018). Furthermore, we observe that the majority of sites in the genome are homozygous and that most deleterious mutations are carried as homozygotes rather than heterozygotes when comparing counts of variants with counts of homozygotes (supplementary fig. S4, Supplementary Material online). Despite this population accumulating weakly deleterious variation, there have not been any reported cases of apparent inbreeding depression (Sillero-Zubiri et al. 1996b; Randall et al. 2007; Randall et al. 2010), suggesting that purging has perhaps removed the most strongly deleterious recessive variants, as observed in simulations (Kyriazis et al. 2021) and empirical data from other small populations (Xue et al. 2015; Robinson et al. 2018; Grossen et al. 2020). Thus, our results combined with the literature mentioned above suggest that the long-term persistence of the Ethiopian wolf could be enabled through the purging of strongly deleterious recessive alleles and avoidance of inbreeding.
Overall, our results have illuminated unique interactions between demography and genetics in the Ethiopian wolf, which can better inform conservation efforts, that previously focused on addressing disease, monitoring movement,s and social behavior, and the ranges of extant sub-populations. Specifically, wild populations may be less susceptible to inbreeding depression as individuals possess mechanisms for inbreeding avoidance and our genomic analysis suggests an absence of appreciable recent inbreeding. Moreover, inbreeding depression is likely to be less severe in this species due to purging in the past and the long-term small population size (Kyriazis et al. 2021). Nonetheless, the genetic exchange should be re-established between now-isolated populations to maintain variation and enhance adaptation to climate change. Finally, there are no captive breeding populations of Ethiopian wolves. Captive reservoirs could be augmented by a frozen sperm or egg collection obtained from live or recently deceased individuals. Efforts to establish a reservoir in captivity are a pressing concern, especially given habitat loss, climate change, and expanding human populations (Gottelli and Sillero-Zubiri 1992; Gottelli et al. 1994; Marino et al. 2013).
Materials and Methods
Ethiopian Wolf Samples
The Ethiopian wolf samples for this study were selected from 85 previously collected tissue samples from the Bale Mountains population for which relatedness and microsatellite data were available (Randall et al. 2010). The Bale Mountains population has previously been shown to consist of three sub-populations, Sanetti Plateau, Morebawa, and Web Valley (Randall et al. 2010). These populations are genetically differentiated due to restricted gene flow between areas. Moreover, individual packs are primarily composed of close relatives. We used the microsatellite data to select individuals for sequencing that clustered with the Morebawa population and avoided sampling close relatives. All selected wolves had pairwise relatedness values <0.11. All but one selected wolf had a STRUCTURE (Pritchard et al. 2000) cluster assignment value for the Morebawa population of >0.88. The one exception, T431, had an assignment value of 0.489 and appeared to be admixed between Morebawa and Sanetti Plateau. STRUCTURE also allowed us to pre-screen the data to ensure there was no evidence of admixture with domestic dogs. The IDs of the wolves that were selected for sequencing included T699, T294, T279, T47, T392, T437, T431, T577, T852, T30. Tissue samples from these individuals were sent to MedGenome for extraction, library preparation, and sequencing to approximately 40 × coverage using Illumina HiSeq X Ten machines. The section below describes how the raw sequence data was processed. The data are available on SRA under project PRJNA889449.
Sequence Data Processing
We merged the newly generated Ethiopian wolf sequence data with several existing canid whole-genome sequencing datasets. These included: Arctic wolf (N = 15) (Robinson et al. 2019) with approximately 39 × coverage, Isle Royale wolf (N = 10) (Robinson et al. 2019) with approximately 24 × coverage, pug (N = 15) (Marchant et al. 2017) with approximately 47 × coverage, labrador retriever (N = 10) (Plassais et al. 2019) with approximately 30 × coverage, Tibetan mastiff (N = 10) (Plassais et al. 2019) with approximately 15 × coverage, border collie (N = 10) (Plassais et al. 2019) with approximately 24 × coverage. Data sets were chosen to be composed of the highest coverage sequencing data possible for publicly available data. Data from Robinson et al. are available on SRA under PRJNA512209; from Marchant et al. are available on European Nucleotide Archive (ENA) under PRJEB17926; and from Plassais et al. are available on SRA under PRJNA448733.
Raw whole-genome sequences were processed following GaTK best practices and with steps 1 through 9 in the NGS pipeline from Phung et al. 2019 (https://github.com/tanyaphung/NGS_pipeline). In brief, fastq files were first aligned to the dog genome (canFam3.1) with BWA (Li and Durbin 2010), duplicate reads were marked with Picard tools (http://broadinstitute.github.io/picard/), poor reads were removed using samtools (Li et al. 2009; Li 2011), and base quality scores were recalibrated with BQSR in GaTK v3.8 (Van der Auwera et al. 2013). We performed joint genotyping with Haplotype Caller and emitted all sites (variant and invariant). We need to output both variant and invariant sites to compute π and generate the site frequency spectra (SFS). To reduce bias in SNP calling accuracy between dogs, where we had many samples, and the wolves, where we had fewer samples, we conducted joint genotyping on each dog breed and each wild canid species separately. For example, joint genotyping was conducted on the Ethiopian wolves as a group, and jointly on the labrador retrievers as a group.
We then applied post hoc filtering to the seven populations VCFs. Specifically, we applied GaTK filtering recommendations for variant sites in non-model species: QD < 2.0, FS > 60.0, MQ < 40.0, MQRankSum < − 12.5, ReadPosRankSum < −8.0, and a minimum genotype quality (GQ) of 20 and we removed clustered SNPs (> 3 SNPs within 10 base pairs). For invariant sites, where no best practices were available, we removed sites with QUAL < 30, and RGQ < 1. For both variant and invariant sites, we applied a minimum depth filter of 10 for each genotype, as previous work has found heterozygous calls are unreliable below this depth (Marsden et al. 2016), and a maximum depth filter of 2.5 times the average genomic coverage (specific for each sample). We also removed any sites where all individuals were heterozygous, sites where fewer than 80% of individuals in a group had a genotype call after post hoc filtering was applied, and any sites within the UCSC repeat regions (http://hgdownload.soe.ucsc.edu/goldenPath/canFam3/database/rmsk.txt.gz).
As a final step, we merged all autosomal data into a single joint-VCF for our analyses that contained sites that were present in at least 90% of individuals. There was a total of 8,818,790 sites that were variable in at least one individual in the final merged data set. This data processing pipeline is necessary for us to confidently compare across species while retaining information about variant and invariant sites.
Relatedness
Relatedness between individuals was computed using VCFTools (Danecek et al. 2011), specifically the –relatedness2 option which incorporates the KING (Manichaikul et al. 2010) algorithm for computing pairwise kinship. The results were cross-checked using the PLINK (Chang et al. 2015) –genome command to estimate relatedness. First degree relatives were removed from each sampled group resulting in the following sample sizes used for subsequent analyses: Ethiopian wolf (N = 10), Arctic wolf (N = 14), Isle Royale wolf (N = 10), pug (N = 15), labrador retriever (N = 10), Tibetan mastiff (N = 9), border collie (N = 7).
Variant Annotation
The ancestral allele at each site was determined using the African wild dog (NCBI SRA: SAMN09924608) (Chavez et al. 2019) aligned to the canFam3.1 reference. For sites where the wild dog was homozygous, we used the wild dog allele as the ancestral allele. For sites where the wild dog was heterozygous and one of the alleles matched the canFam3.1 reference allele, we used that allele as the ancestral allele. Variant annotation was done using Ensembl VEP (version 94) with SIFT annotations enabled (McLaren et al. 2010; Kumar et al. 2018). Genomic Evolutionary Rate Profiling (GERP) scores (Davydov et al. 2010) were used alongside VEP to annotate variants as either neutral (synonymous with GERP Score <2) or deleterious (nonsynonymous with a GERP score >4). For details about how GERP scores were generated, see Marsden et al. (2016). Counts were compared using a Wilcoxon Rank Sum test.
Runs of Homozygosity
ROH were called in VCFTools version 0.1.16 (Danecek et al. 2011) with the –LROH command and BCFTools version 1.3.1 (Li 2011) using “bcftools roh” and converting positions to ranges with an open source perl script linked here. We used three approaches to call ROH: 1) using VCFTools and calling ROH separately for each population; 2) using BCFTools without a genetic map; and 3) using BCFTools with a genetic map from canFam3.1. Ultimately, we chose to use option one for our analyses because BCFTools roughly doubled (with a genetic map) or quadrupled (without a genetic map) the number of small and intermediate length ROH in the Ethiopians wolves, relative to VCFTools (supplementary fig. S11, Supplementary Material online). After calling ROH with VCFTools, we tallied the number of callable sites in each individual's genome that lie within each ROH. Our final set of ROH included those at least 10 Kb in length and where at least 40% of the run overlapped callable sites. FROH was calculated as the length of the genome within a ROH of at least 1 Mb divided by the total length of the canFam3.1 genome.
Genetic Diversity
We calculated π, the average number of pairwise differences per site, in a subsample of six individuals from each population. π was computed across the genome as:
where n is the total number of chromosomes sampled, p is the frequency of a given allele, and L is the length in base pairs of the sampled region.
Population Differentiation
Genome-wide unweighted (per site FST) and weighted FST (supplementary fig. S1, Supplementary Material online) between all pairs of populations was computed using Weir and Cockerham's formula (Weir and Cockerham 1984) as implemented in the –weir-fst-pop command from VCFTools (Danecek et al. 2011). Importantly, the weighted FST estimator computes the variance components in the numerator and denominator of FST separately for all SNPs across the genome first and then divides at the end to obtain FST (rather than computing FST for each SNP and then finding the mean over all SNPs).
We used the unsupervised learning model in ADMIXTURE (Alexander et al. 2009) to determine the number of distinct genetic clusters within the data. 780,150 LD-pruned were used after implementing the suggested PLINK (Chang et al. 2015) command from the manual. SNPs were ascertained in a 50-step window that advances 10 SNPs at a time, and only kept if r2 was less than 0.1 with any other SNP in the window. The number of source populations varied between K = 2 and K = 10, and the lowest cross-validation error was produced when K = 6 (supplementary fig. S8, Supplementary Material online). We saw no admixture between Ethiopian wolves and dogs in this analysis, but this is unsurprising given our sample selection.
PCA (supplementary fig. S9, Supplementary Material online) of the genetic variation data was conducted in R using a combination of SNPRelate (Zheng et al. 2012), PC-AiR (Conomos et al. 2015), and PC-Relate (Conomos et al. 2016). We first generated a genetic relatedness matrix using SNPRelate's implementation of KING (Manichaikul et al. 2010), to correct for ancestry, then PC-AiR and PC-Relate were used to perform PCA as these methods are robust to population structure, cryptic relatedness, and admixture. We also performed hierarchical clustering on the IBS matrix generated by SNPRelate's implementation of KING with the default settings.
Demographic Inference
We inferred the population history of the Ethiopian wolf using ABC (Tavaré et al. 1997). We took an approach similar to Robinson et al. (2016). First, we ran MSMC2 (Malaspinas et al. 2016; Schiffels and Wang 2020; Wang et al. 2020b) on each of the four individuals separately (EW2, EW3, EW9, EW10) to determine possible demographic history and inform priors for changes in population size and the bottleneck time (see supplementary fig. S3, Supplementary Material online). MSMC2 was run for 25 iterations with the parameters “-I 25 -t 4”. The results suggest a population contraction. Thus, we considered a three-epoch demographic model with serial contractions. We set priors for NANCIENT ranging from to 40,000 to 200,000 individuals, NINTERMIDIATE from 1,000–60,000 individuals, NCURRENT from 10–1,000 individuals, an ancient bottleneck (TBOTANCIENT) from 1,500–15,000 generations ago, and a recent bottleneck (TBOTRECENT) from 10–1,000 generations ago. We drew parameter values from a uniform prior distribution and performed 200,000 coalescent simulations. We assessed the fit of the model to the data using a joint statistic of π and segregating sites (S).
Coalescent simulations were performed with ms (Hudson 2002). We simulated data for n = 4 diploid individuals across 21,724 neutral regions of at least 1 Kb in length. As natural selection can confound demographic inference (Schrider et al. 2016; Pouyet et al. 2018), we identified neutral regions in canFam3.1 using the approach of Phung and colleagues (2019). Neutral regions are outside of genes, outside of phastCons (Siepel et al. 2005) conserved regions, and are at least 0.4 centimorgans (cM) away from the nearest gene. For each neutral region in the empirical data, S was tabulated with the VCFTools –snpdensity option and a window size of 10 base pairs and –site-pi option was used to compute π per site. Simulated neutral regions were length-matched to our remaining empirical data from four genomes that were not used to inform the prior (EW4, EW5, EW6, EW7). For all analyses, we assumed a mutation rate of 4.96e−09 (Phung et al. 2019) and a generation time of 3 years (Gottelli et al. 2004).
We used the following approach for the simulations. First, draw once from the uniform prior for each parameter to create a set of parameter values the simulation. Second, for each neutral region, generate a set of simulation values to be used in ms based on the drawn parameter values. Thus, each region will have a value population-scaled mutation rate (θ) for the population in the present, population recombination rate (ρ), length of the window (l), timing of the second epoch (scaled by 4 × NCURRENT), population size during the second epoch (scaled by NCURRENT), timing of the third epoch (scaled by 4 × NCURRENT), and finally, population size during the third epoch (scaled by NCURRENT). Similar to previous work (Robinson et al. 2016) for computing ρ, recombination rate, r, across regions varied and was assumed to be distributed following a gamma distribution with parameters (0.1,0.0000001). This is done to mimic the recombination rate map from dogs. Therefore, r had a mean of 1 × 10−8 and a standard deviation of 3.16 × 10−8. After we sampled one r value, we obtained ρ as 4 × Ne×r×l. Third, simulate each of the 21,724 neutral regions. Fourth, we used the summary statistics in ms to compute S and π. We repeated these steps for each simulation replicate.
From each of the 200,000 replicates, we obtained a distribution of both S and π across the 21,724 neutral regions that we compared with empirical data. In order to compare whole number counts for S and floats for π, each summary statistic was binned. We denoted the maximum number of bins as M, which was fixed to be 50. For S, a bin is defined as the number of SNPs within the neutral region. For π, we split the range, from the minimum of 0 to the maximum value of 0.0025, using increments of 0.0005. Thus, we partitioned π, into 50 bins and had a comparable number of bins with our SNP counts, S. In other words, the lowest value of π corresponded to the lowest value of S, and so on.
Next two distance metrics were used to create a joint statistic, Jπ, S, and decide which simulations to accept or reject. For π, we compute:
where i is the bin of interest and M is the maximum number of bins (fixed to be 50). Here, each value of i has corresponding range (e.g., i = 0 corresponds to [0, 0.0005], i = 1 corresponds to [0.0005, 0.001] … i = 50), Ei is the number of regions from the empirical data that have a value of π within range i, and Xi is the number of regions from the simulated data that have a value of π within bin i.
For S, we computed the score:
where i corresponds to SNP counts for that bin. Then, Ei is the number of regions from the empirical data with a SNP count equal to i, and Xi is the number of regions from the simulated data with a SNP count equal to i.
Finally, our joint statistic was computed for all bins as
The top 200 simulations that minimized Jπ,S were kept and composed the posterior distribution. Supplementary figures S12 and S13, Supplementary Material online compare bins of π and S from the top 200 simulations against the empirical data, shows the posterior and priors for each parameter, and supplementary Table S2, Supplementary Material online contains the 95% credible interval for each parameter point estimate.
ABC was used for inference because it provides a more flexible and reliable framework than MSMC2 (Beichman et al. 2017; Beichman et al. 2018). We only use 8 of the 10 wolves when conducting inference to avoid overfitting and using all 10 wolves does not add any more variation to the higher frequency bins. We use four wolves for generating the priors from MSMC2, four wolves for training the demographic model, and drop the remaining two wolves.
Statistics to Detect Positive Selection
Since the Ethiopian wolf genome is highly homozygous, we did not use haplotype tests for positive selection. Instead, we used a combination of outlier methods (FST, π, and derived allele counts) to identify potential regions of the genome undergoing positive natural selection. We compared the Ethiopian wolf with several other groups: border collie, Tibetan mastiff, and Arctic wolf. We chose to include the border collie and Arctic wolf since neither has been shown to be adapted to high-altitude. Conversely the Tibetan mastiff has been shown to have adapted to high-altitude (Gou et al. 2014; Miao et al. 2017; Wang et al. 2020b; Witt and Huerta-Sánchez 2019). We do not use the Isle Royale wolf as its recent demographic history may confound analyses.
To perform the selection scan, first, we computed the value of FST per gene between the Ethiopian wolf and each comparison population with VCFTools using the unweighted values from –weir-fst-pop command. Here, FST is calculated for each SNP and then averaged over all the SNPs in each gene. We downloaded canFam3.1 gene coordinates from Ensembl, which included 14,970 gene annotations once we narrow down to single transcript per gene. We then summed the per site FST and divided by the total number of sites within each gene. Genes with fewer than 20 SNPs were not included, leaving us with 9,850 genes. The top 5% of genes with the highest FST values across all three population comparisons were retained. This resulted in a total of 82 genes for further analysis.
Then, candidate genes for high-altitude adaptation were identified. These candidate genes were either in the HIF-1α pathway, when using PANTHER (http://www.pantherdb.org/), or in a curated list of previously identified loci from high-altitude adaptation selection scans (Witt and Huerta-Sánchez 2019). CREBBP was the only candidate gene that was found as an outlier in all three FST comparisons (i.e., it was the only candidate in the set of 82 extreme genes).
Using CREBBP as the focal gene, we then calculated a number of other summary statistics that might be affected by positive selection. We wanted to assess whether other genes in the genome had more extreme values of the statistics than CREBBP. First, we examined π per gene in each comparison group, border collie, Ethiopian wolf, and Arctic wolf, where π per site was computed using the VCFTools –site-pi command. π per site was summed across all variants within each gene, then we and divided by the total number of sites within each gene. As with the FST analysis, genes with fewer than 20 SNPs were not included.
Second, we examined the number of sites fixed in opposite directions between the Ethiopian wolf and each of the other wolf-like canid populations. Third, we determined the derived allele (with respect to the Wild dog) count in each gene, and within genes in a 1-Mb window around it.
We determined whether any of the 82 genes identified as FST outliers overlapped with genes that also have a value of π less than or equal to CREBBP in Ethiopian wolves. There were 37 genes that had more extreme values of π (supplementary Table S4, Supplementary Material online). However, none of 37 genes fell within the tail of all fixed sites distribution comparisons as well.
Phylogenetic Tree Reconstruction and Multiple Sequence Alignment in PRDM9
Given that Ethiopian wolves are an outgroup to most canids (Gopalakrishnan et al. 2018), we wanted to test whether they have a functional version of PRDM9. PRDM9 is involved in recombination and is responsible for the positioning of recombination hotspots in the genome. We compared PRDM9 with another gene, GAPDH, which is a housekeeping gene in the glycolysis pathway. The PRDM9 and GAPDH coordinates of hg19, mm10, rheMac2, and canFam2 were identified using the ECR browser (https://ecrbrowser.dcode.org/) and a fasta file was generated. The data for felCat8 was pulled from UCSC. These fasta files were subsequently merged with a second fasta file containing the homologous regions of a single Arctic wolf (AW15), a single Ethiopian wolf (EW7), a single Tibetan mastiff (TM3), and a single Dhole (Dhole01) (Plassais et al. 2019). Then, multiple sequence alignment was conducted using an online version of MAFFT (https://mafft.cbrc.jp/alignment/server/). Finally, a neighbor-joining tree was constructed using the Jukes–Cantor substitution model (Jukes and Cantor 1969).
For PRDM9, GeneWise (Birney et al. 2004) was used to align the protein sequence of the Ethiopian wolf, Arctic wolf, and Tibetan mastiff relative to hg19 DNA sequence. GeneWise allowed us to identify intronic and frameshift errors, which would affect the functionality of the gene.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Annabel Beichman, Diego Ortega Del Vecchyo, Jacqueline Robinson, Eduardo Amorim, Xinjun Zhang, and Matilde Miranda for helpful scripts and productive discussions and Deb Randall for allowing to use the banked samples collected under permit (Randall et al. 2010). The authors would also like to acknowledge and remember Bob Wayne. Bob spent his career pioneering genomic methods to study demography and selection in populations of conservation concern. He will be greatly missed by his colleagues, students, and the conservation genomics community. This work was supported through the National Science Foundation Graduate Research Fellowship under Grant Number DGE-1650604 and the National Science Foundation Postdoctoral Research Fellowship in Biology awarded to J.A.M. K.E.L. was supported by National Institutes of Health grant R35GM119856.
Contributor Information
Jazlyn A Mooney, Department of Human Genetics, University of California Los Angeles, Los Angeles, CA 90095, USA; Department of Biology, Stanford University, Stanford, CA, USA; Department of Quantitative and Computational Biology, University of Southern California, Los Angeles, CA, USA.
Clare D Marsden, Department of Ecology & Evolutionary Biology, University of California Los Angeles, Los Angeles, CA, USA.
Abigail Yohannes, Department of Chemistry and Biochemistry, University of California Los Angeles, Los Angeles, CA, USA.
Robert K Wayne, Department of Ecology & Evolutionary Biology, University of California Los Angeles, Los Angeles, CA, USA.
Kirk E Lohmueller, Department of Human Genetics, University of California Los Angeles, Los Angeles, CA 90095, USA; Department of Ecology & Evolutionary Biology, University of California Los Angeles, Los Angeles, CA, USA.
Supplementary material
Supplementary data are available at Molecular Biology and Evolution online.
Data Availability
Raw fastq files with for each of the Ethiopian wolf genomes are available on SRA under project PRJNA889449. Data for Isle Royale and Arctic wolves are available on SRA under PRJNA512209; Pug genomes are available on European Nucleotide Archive (ENA) under PRJEB17926; and Border collies, Labrador retriever and Tibetan mastiff data are on SRA under PRJNA448733. All code is freely available on Jazlyn Mooney's GitHub at https://github.com/jaam92/DogProject_Clare.git
References
- Alexander DH, Novembre J, Lange K. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arany Z, Huang LE, Eckner R, Bhattacharya S, Jiang C, Goldberg MA, Bunn HF, Livingston DM. 1996. An essential role for p300/CBP in the cellular response to hypoxia. Proc Natl Acad Sci USA. 93:12969–12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson E, Webster MT, Ratnakumar A, Ponting CP, Lindblad-Toh K, Consortium L. 2012. Death of PRDM9 coincides with stabilization of the recombination landscape in the dog genome. Genome Res. 22:51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker Z, Schumer M, Haba Y, Bashkirova L, Holland C, Rosenthal GG, Przeworski M. 2017. Repeated losses of PRDM9-directed recombination despite the conservation of PRDM9 across vertebrates. Elife. 6:e24133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beichman AC, Huerta-Sanchez E, Lohmueller KE. 2018. Using genomic data to infer historic population dynamics of nonmodel organisms. Annu Rev Ecol Evol Syst. 49:433–456. [Google Scholar]
- Beichman AC, Phung TN, Lohmueller KE. 2017. Comparison of single genome and allele frequency data reveals discordant demographic histories. G3 Genes. 7:3605–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R. 2004. Genewise and genomewise. Genome Res. 14:988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko AR, Quignon P, Li L, Schoenebeck JJ, Degenhardt JD, Lohmueller KE, Zhao K, Brisbin A, Parker HG, Cargill M. 2010. A simple genetic architecture underlies morphological variation in dogs. PLoS Biol. 8:e1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughley G. 1994. Directions in conservation biology. J Animal Ecol 63(2):215–244. [Google Scholar]
- Ceballos FC, Joshi PK, Clark DW, Ramsay M, Wilson JF. 2018. Runs of homozygosity: windows into population history and trait architecture. Nat Rev Genet. 19(4):220–234. [DOI] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ.. 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez DE, Gronau I, Hains T, Kliver S, Koepfli K-P, Wayne RK. 2019. Comparative genomics provides new insights into the remarkable adaptations of the African wild dog (Lycaon pictus). Sci Rep. 9:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conomos MP, Miller MB, Thornton TA. 2015. Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet Epidemiol. 39:276–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conomos MP, Reiner AP, Weir BS, Thornton TA. 2016. Model-free estimation of recent genetic relatedness. Am J Human Genetics. 98:127–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubaynes S, Brandell EE, Stahler DR, Smith DW, Almberg ES, Schindler S, Wayne RK, Dobson AP, vonHoldt BM, MacNulty DR, et al. 2022. Disease outbreaks select for mate choice and coat color in wolves. Science. 378:300–303. [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST. 2011. The variant call format and VCFtools. Bioinformatics. 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. 2010. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol. 6:e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitak RR, Rinkevich SE, Culver M. 2018. Genome-Wide analysis of SNPs is consistent with No domestic dog ancestry in the endangered Mexican wolf (Canis lupus baileyi). J Hered. 109:372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R. 2005. Genetics and extinction. Biol Conserv. 126:131–140. [Google Scholar]
- Gilpin ME, Soulé M. 1986. Minimum viable populations: processes of species extinctions. In: Soulé M, editor. Conservation biology: the science of scarcity and diversity. Sunderland, MA: Sinauer Associates [Google Scholar]
- Ginty DD, Kornhauser JM, Thompson MA, Bading H, Mayo KE, Takahashi JS, Greenberg ME. 1993. Regulation of CREB phosphorylation in the suprachiasmatic nucleus by light and a circadian clock. Science. 260:238–241. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan S, Sinding M-HS, Ramos-Madrigal J, Niemann J, Castruita JAS, Vieira FG, Carøe C, de Manuel Montero M, Kuderna L, Serres A. 2018. Interspecific gene flow shaped the evolution of the genus Canis. Curr Biol. 28:3441–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon CH, Banyard AC, Hussein A, Laurenson MK, Malcolm JR, Marino J, Regassa F, Stewart A-ME, Fooks AR, Sillero-Zubiri C. 2015. Canine distemper in endangered Ethiopian wolves. Emerg Infect Dis. 21:824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottelli D, Marino J, Sillero-Zubiri C, Funk SM. 2004. The effect of the last glacial age on speciation and population genetic structure of the endangered Ethiopian wolf (Canis simensis). Mol Ecol. 13:2275–2286. [DOI] [PubMed] [Google Scholar]
- Gottelli D, Sillero-Zubiri C. 1992. The Ethiopian wolf–an endangered endemic canid. Oryx. 26:205–214. [Google Scholar]
- Gottelli D, Sillero-Zubiri C, Applebaum GD, Roy MS, Girman DJ, Garcia-Moreno J, Ostrander EA, Wayne RK. 1994. Molecular genetics of the most endangered canid: the Ethiopian wolf Canis simensis. Mol Ecol. 3:301–312. [DOI] [PubMed] [Google Scholar]
- Gottelli D, Sillero-Zubiri C, Marino J, Funk SM, Wang J. 2013. Genetic structure and patterns of gene flow among populations of the endangered Ethiopian wolf. Anim Conserv. 16:234–247. [Google Scholar]
- Gou X, Wang Z, Li N, Qiu F, Xu Z, Yan D, Yang S, Jia J, Kong X, Wei Z. 2014. Whole-genome sequencing of six dog breeds from continuous altitudes reveals adaptation to high-altitude hypoxia. Genome Res. 24:1308–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MM, Granka JM, Bustamante CD, Sutter NB, Boyko AR, Zhu L, Ostrander EA, Wayne RK. 2009. Linkage disequilibrium and demographic history of wild and domestic canids. Genetics. 181:1493–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey C, Baudat F, de Massy B. 2018. PRDM9, A driver of the genetic map. PLoS Genet. 14(8):e1007479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossen C, Guillaume F, Keller LF, Croll D. 2020. Purging of highly deleterious mutations through severe bottlenecks in alpine ibex. Nat Commun. 11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutema TM, Atickem A, Bekele A, Sillero-Zubiri C, Kasso M, Tsegaye D, Venkataraman VV, Fashing PJ, Zinner D, Stenseth NC. 2018. Competition between sympatric wolf taxa: an example involving African and Ethiopian wolves. R Soc Open Sci. 5:172207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward JJ, Castelhano MG, Oliveira KC, Corey E, Balkman C, Baxter TL, Casal ML, Center SA, Fang M, Garrison SJ, et al. 2016. Complex disease and phenotype mapping in the domestic dog. Nat Commun. 7:10460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick PW, Peterson RO, Vucetich LM, Adams JR, Vucetich JA. 2014. Genetic rescue in isle royale wolves: genetic analysis and the collapse of the population. Conserv Genet. 15:1111–1121. [Google Scholar]
- Hudson RR. 2002. Generating samples under a Wright–Fisher neutral model of genetic variation. Bioinformatics. 18:337–338. [DOI] [PubMed] [Google Scholar]
- IUCN Species Survival Commission . 2011. Strategic plan for Ethiopian wolf conservation. UK: IUCN/SSC Canid Specialist Group Oxford. [Google Scholar]
- Jukes TH, Cantor CR. 1969. Evolution of protein molecules. Mammal Prot Metabol . 3:21–132. [Google Scholar]
- Kallio PJ, Okamoto K, O’Brien S, Carrero P, Makino Y, Tanaka H, Poellinger L. 1998. Signal transduction in hypoxic cells: inducible nuclear translocation and recruitment of theCBP/p300 coactivator by the hypoxia-induciblefactor-1α. EMBO J. 17:6573–6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriazis CC, Wayne RK, Lohmueller KE. 2021. Strongly deleterious mutations are a primary determinant of extinction risk due to inbreeding depression. Evolution letters. 5:33–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R. 1988. Genetics and demography in biological conservation. Science. 241:1455–1460. [DOI] [PubMed] [Google Scholar]
- Lande R. 1993. Risks of population extinction from demographic and environmental stochasticity and random catastrophes. Am Nat. 142:911–927. [DOI] [PubMed] [Google Scholar]
- Lande R, Barrowdough G. 1987. Effective population size, genetic variation, and their use in population. Viable Popul Conservat. 87:87–124. [Google Scholar]
- Laurenson K, Sillero-Zubiri C, Thompson H, Shiferaw F, Thirgood S, Malcolm J. 1998. Disease as a threat to endangered species: Ethiopian wolves, domestic dogs and canine pathogens. In: Animal conservation forum. Vol. 1. Cambridge, United Kingdom: Cambridge University Press. p. 273–280. [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 27:2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with burrows–wheeler transform. Bioinformatics. 26:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wu D-D, Boyko AR, Wang G-D, Wu S-F, Irwin DM, Zhang Y-P. 2014. Population variation revealed high-altitude adaptation of Tibetan Mastiffs. Mol Biol Evol . 31(5):1200–1205. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ, Zody MC. 2005. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 438:803–819. [DOI] [PubMed] [Google Scholar]
- Lohmueller KE. 2014. The distribution of deleterious genetic variation in human populations. Curr Opin Genet Dev. 29:139–146. [DOI] [PubMed] [Google Scholar]
- Lohmueller KE, Indap AR, Schmidt S, Boyko AR, Hernandez RD, Hubisz MJ, Sninsky JJ, White TJ, Sunyaev SR, Nielsen R. 2008. Proportionally more deleterious genetic variation in European than in African populations. Nature. 451:994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery J, Burger R. 1995a. Mutation accumulation and the extinction of small populations. Am Nat. 146:489–518. [Google Scholar]
- Lynch M, Conery J, Bürger R. 1995b. Mutational meltdowns in sexual populations. Evolution. 49:1067–1080. [DOI] [PubMed] [Google Scholar]
- Malaspinas A-S, Westaway MC, Muller C, Sousa VC, Lao O, Alves I, Bergström A, Athanasiadis G, Cheng JY, Crawford JE. 2016. A genomic history of aboriginal Australia. Nature. 538:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen W-M. 2010. Robust relationship inference in genome-wide association studies. Bioinformatics. 26:2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant TW, Johnson EJ, McTeir L, Johnson CI, Gow A, Liuti T, Kuehn D, Svenson K, Bermingham ML, Drögemüller M. 2017. Canine brachycephaly is associated with a retrotransposon-mediated missplicing of SMOC2. Curr Biol. 27:1573–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino J, Bedin E, Sillero-Zubiri C, EWCP Team . 2017. Ethiopian Wolf Conservation Programme Annual Report 2017. Bale, Ethiopia Available from: https://www.ethiopianwolf.org/resources/EWCP%20Annual%20Report%20%20April%202017.pdf
- Marino J, Sillero-Zubiri C. 2011. Canis simensis. IUCN Red List of Threatened Species. 10:2011–2011. [Google Scholar]
- Marino J, Sillero-Zubiri C, Gottelli D, Johnson PJ, Macdonald DW. 2013. The fall and rise of Ethiopian wolves: lessons for conservation of long-lived, social predators. Anim Conserv. 16:621–632. [Google Scholar]
- Marsden CD, Ortega-Del Vecchyo D, O’Brien DP, Taylor JF, Ramirez O, Vilà C, Marques-Bonet T, Schnabel RD, Wayne RK, Lohmueller KE. 2016. Bottlenecks and selective sweeps during domestication have increased deleterious genetic variation in dogs. Proc Natl Acad Sci USA. 113:152–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. 2010. Deriving the consequences of genomic variants with the Ensembl API and SNP effect predictor. Bioinformatics. 26:2069–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao B, Wang Z, Li Y. 2017. Genomic analysis reveals hypoxia adaptation in the Tibetan mastiff by introgression of the gray wolf from the Tibetan plateau. Mol Biol Evol. 34:734–743. [DOI] [PubMed] [Google Scholar]
- Mooney JA, Yohannes A, Lohmueller KE. 2021. The impact of identity by descent on fitness and disease in dogs. Proc Natl Acad Sci USA. 118(16):e2019116118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Fuentes V, Di Rienzo A, Vilà C. 2011. Prdm9, a major determinant of meiotic recombination hotspots, is not functional in dogs and their wild relatives, wolves and coyotes. PloS One. 6:e25498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musiani M, Leonard JA, Cluff HD, Gates CC, Mariani S, Paquet PC, Vila C, Wayne RK. 2007. Differentiation of tundra/taiga and boreal coniferous forest wolves: genetics, coat colour and association with migratory caribou. Mol Ecol. 16:4149–4170. [DOI] [PubMed] [Google Scholar]
- Ohta T. 1973. Slightly deleterious mutant substitutions in evolution. Nature. 246:96. [DOI] [PubMed] [Google Scholar]
- Pemberton TJ, Absher D, Feldman MW, Myers RM, Rosenberg NA, Li JZ. 2012. Genomic patterns of homozygosity in worldwide human populations. Am J of Human Genetics. 91:275–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RO, Vucetich JA, Bump JM, Smith DW. 2014. Trophic cascades in a multicausal world: isle royale and Yellowstone. Ann Rev Ecol Evol Syst. 45:325–345. [Google Scholar]
- Phung TN, Wayne RK, Wilson MA, Lohmueller KE. 2019. Complex patterns of sex-biased demography in canines. Proc Roy Soc B. 286:20181976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plassais J, Kim J, Davis BW, Karyadi DM, Hogan AN, Harris AC, Decker B, Parker HG, Ostrander EA. 2019. Whole genome sequencing of canids reveals genomic regions under selection and variants influencing morphology. Nat Commun. 10(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollinger JP, Earl DA, Knowles JC, Boyko AR, Parker H, Geffen E, Pilot M, Jedrzejewski W, Jedrzejewska B, Sidorovich V. 2011. A genome-wide perspective on the evolutionary history of enigmatic wolf-like canids. Genome Res. 21(8):1294–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouyet F, Aeschbacher S, Thiéry A, Excoffier L. 2018. Background selection and biased gene conversion affect more than 95% of the human genome and bias demographic inferences. Elife. 7:e36317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics. 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall DA, Marino J, Haydon DT, Sillero-Zubiri C, Knobel DL, Tallents LA, Macdonald DW, Laurenson MK. 2006. An integrated disease management strategy for the control of rabies in Ethiopian wolves. Biol Conserv. 131:151–162. [Google Scholar]
- Randall DA, Pollinger JP, Argaw K, Macdonald DW, Wayne RK. 2010. Fine-scale genetic structure in Ethiopian wolves imposed by sociality, migration, and population bottlenecks. Conserv Genetics. 11:89–101. [Google Scholar]
- Randall DA, Pollinger JP, Wayne RK, Tallents LA, Johnson PJ, Macdonald DW. 2007. Inbreeding is reduced by female-biased dispersal and mating behavior in Ethiopian wolves. Behav Ecol. 18:579–589. [Google Scholar]
- Randall DA, Williams SD, Kuzmin IV, Rupprecht CE, Tallents LA, Tefera Z, Argaw K, Shiferaw F, Knobel DL, Sillero-Zubiri C. 2004. Rabies in endangered Ethiopian wolves. Emerg Infect Dis. 10:2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JA, Brown C, Kim BY, Lohmueller KE, Wayne RK. 2018. Purging of strongly deleterious mutations explains long-term persistence and absence of inbreeding depression in island foxes. Curr Biol. 28:3487–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JA, Ortega-Del Vecchyo D, Fan Z, Kim BY, Marsden CD, Lohmueller KE, Wayne RK. 2016. Genomic flatlining in the endangered island fox. Curr Biol. 26:1183–1189. [DOI] [PubMed] [Google Scholar]
- Robinson JA, Räikkönen J, Vucetich LM, Vucetich JA, Peterson RO, Lohmueller KE, Wayne RK. 2019. Genomic signatures of extensive inbreeding in isle royale wolves, a population on the threshold of extinction. Sci Adv. 5:eaau0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffels S, Wang K. 2020. MSMC And MSMC2: the multiple sequentially markovian coalescent. In: Statistical population genomics. New York, NY: Humana. p.147–166. [DOI] [PubMed] [Google Scholar]
- Scholz H, Schurek H-J, Eckardt K-U, Bauer C. 1990. Role of erythropoietin in adaptation to hypoxia. Experientia. 46:1197–1201. [DOI] [PubMed] [Google Scholar]
- Schrider DR, Shanku AG, Kern AD. 2016. Effects of linked selective sweeps on demographic inference and model selection. Genetics. 204:1207–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaywitz AJ, Greenberg ME. 1999. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 68:821–861. [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15:1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillero-Zubiri C, Gottelli D. 1995. Diet and feeding behavior of Ethiopian wolves (Canis simensis). J Mammal. 76:531–541. [Google Scholar]
- Sillero-Zubiri C, King AA, Macdonald DW. 1996a. Rabies and mortality in Ethiopian wolves (Canis simensis). J Wildl Dis. 32:80–86. [DOI] [PubMed] [Google Scholar]
- Sillero-Zubiri C, Gottelli D, Macdonald DW. 1996b. Male philopatry, extra-pack copulations and inbreeding avoidance in Ethiopian wolves (Canis simensis). Behav Ecol Sociobiol (Print). 38:331–340. [Google Scholar]
- Simons YB, Sella G. 2016. The impact of recent population history on the deleterious mutation load in humans and close evolutionary relatives. Curr Opin Genet Dev. 41:150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P. 2016. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 45(D1):D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavaré S, Balding DJ, Griffiths RC, Donnelly P. 1997. Inferring coalescence times from DNA sequence data. Genetics. 145:505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J. 2013. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. 43:11–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vonHoldt B, Fan Z, Ortega-Del Vecchyo D, Wayne RK. 2017. EPAS1 Variants in high altitude Tibetan wolves were selectively introgressed into highland dogs. PeerJ. 5:e3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M-S, Wang S, Li Y, Jhala Y, Thakur M, Otecko NO, Si J-F, Chen H-M, Shapiro B, Nielsen R. 2020a. Ancient hybridization with an unknown population facilitated high-altitude adaptation of canids. Mol Biol Evol. 37:2616–2629. [DOI] [PubMed] [Google Scholar]
- Wang K, Mathieson I, O’Connell J, Schiffels S. 2020b. Tracking human population structure through time from whole genome sequences. PLoS Genet. 16:e1008552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. 1984. Estimating F-statistics for the analysis of population structure. Evolution. 38:1358–1370. [DOI] [PubMed] [Google Scholar]
- Whitby JE, Johnstone P, Sillero-Zubiri C. 1997. Rabies virus in the decomposed brain of an Ethiopian wolf detected by nested reverse transcription-polymerase chain reaction. J Wildl Dis. 33:912–915. [DOI] [PubMed] [Google Scholar]
- Witt KE, Huerta-Sánchez E. 2019. Convergent evolution in human and domesticate adaptation to high-altitude environments. Philos Trans R Soc B. 374:20180235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Prado-Martinez J, Sudmant PH, Narasimhan V, Ayub Q, Szpak M, Frandsen P, Chen Y, Yngvadottir B, Cooper DN. 2015. Mountain gorilla genomes reveal the impact of long-term population decline and inbreeding. Science. 348:242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. 2012. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 28:3326–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw fastq files with for each of the Ethiopian wolf genomes are available on SRA under project PRJNA889449. Data for Isle Royale and Arctic wolves are available on SRA under PRJNA512209; Pug genomes are available on European Nucleotide Archive (ENA) under PRJEB17926; and Border collies, Labrador retriever and Tibetan mastiff data are on SRA under PRJNA448733. All code is freely available on Jazlyn Mooney's GitHub at https://github.com/jaam92/DogProject_Clare.git