Abstract
The cellular attachment and entry of pathogenic microorganisms can be facilitated by the expression of microbial adhesins that bind fibronectin. We have previously described a Borrelia burgdorferi gene, bbk32, that encodes a 47-kDa fibronectin-binding protein. In this study, the ligand-binding region of BBK32 from B. burgdorferi isolate B31 was localized to 32 amino acids. The bbk32 gene was cloned and sequenced from three additional B. burgdorferi isolates representing different genospecies of B. burgdorferi sensu lato. All four bbk32 genes encoded proteins having fibronectin-binding activity when expressed in Escherichia coli, and the deduced proteins shared 81 to 91% amino acid sequence identity within the ligand-binding domain. In addition, the ligand-binding region of BBK32 was found to share sequence homology with a fibronectin-binding peptide defined for protein F1 of Streptococcus pyogenes. The structural and functional similarity between the ligand-binding region of BBK32 and the UR region of protein F1 suggests a common mechanism of cellular adhesion and entry for B. burgdorferi and S. pyogenes.
Lyme disease remains the most prevalent vector-borne infectious disease in North America (17). The spirochete Borrelia burgdorferi was identified as the etiologic agent of Lyme disease in 1982 (1). Since then, at least 10 genospecies representing the complex B. burgdorferi sensu lato have been described (29). Only the genospecies B. burgdorferi sensu stricto, B. garinii, and B. afzelii are well established in causing disease in humans. Recently, however, B. burgdorferi strains resembling the newly described genospecies B. bissettii were isolated from Lyme disease patients in Slovenia (27). Human infections with B. burgdorferi are transmitted by ticks of the Ixodes subgenus. Once transmitted by tick bite, B. burgdorferi establishes a localized infection at the site of tick attachment. The spirochetes migrate in the skin, producing an oval rash termed erythema migrans in 80% of Lyme disease patients (6). Days to weeks later, the spirochetes enter the vasculature and disseminate to multiple tissue and organ sites. At this stage, the patient enters an early disseminated form of Lyme disease and may present with carditis, lymphadenopathy, meningitis, and migratory joint and muscle pain. Despite the presence of a strong host immune response, B. burgdorferi may persist in the host, and spirochetes may be isolated from the patient months to years after transmission. In this late stage of Lyme disease, the patient's clinical manifestations may include cutaneous, musculoskeletal, and neurologic involvement. While early intervention with antibiotics is generally efficacious, this late form of Lyme disease may be more refractory to treatment (26).
The mechanisms by which B. burgdorferi invades and colonizes the host are poorly understood. For many bacterial pathogens, the initial step in host colonization involves the expression of adhesive molecules that mediate bacterial adherence to cells or to the extracellular matrix (20, 30). The capacity of B. burgdorferi to bind a wide variety of cells and extracellular matrix components indicates that these organisms may also express adhesive molecules (24). A common feature of several pathogenic bacteria, most notably Staphylococcus aureus and streptococci, is the expression of adhesins that bind fibronectin (10). Fibronectin is a large, dimeric glycoprotein that is produced by a broad range of cell types (22). It exists as a soluble molecule in body fluids and as an insoluble component of cell membranes and the extracellular matrix. Structurally, fibronectin is a mosaic protein composed of three types of protein modules that are organized into distinct functional domains. Through these functional domains, fibronectin can interact with a variety of macromolecules including fibrin, heparin, collagen, and integrins. Many of these functional domains are also targeted by adhesins expressed by pathogenic microorganisms.
Borrelia species also express adhesins that bind fibronectin (7, 13, 21, 28). We have previously reported on the identification of a gene, bbk32, which encodes a 47-kDa fibronectin-binding adhesin expressed by B. burgdorferi isolate B31 (21). BBK32 was localized to the outer surface of isolate B31, and the adhesin was found to interact specifically with the collagen-binding domain of fibronectin. The ability of recombinant BBK32 to inhibit binding of isolate B31 to immobilized fibronectin was also demonstrated. These results indicated that BBK32 is the primary fibronectin-binding adhesin expressed by B. burgdorferi.
In this study, we extended these earlier observations by mapping the ligand-binding region of BBK32 from B. burgdorferi isolate B31. In addition, we cloned the bbk32 gene from four isolates representing different B. burgdorferi genospecies into Escherichia coli, and determined the degree of sequence conservation and functional activity of the expressed proteins.
Mapping the fibronectin-binding Region of BBK32.
The minimal region of BBK32 required to bind fibronectin was localized by creating a bbk32 gene fragment library using the Novatope system (Novagen, Madison, Wis.) and screening the library by ligand blotting. The bbk32 gene was amplified from B. burgdorferi isolate B31 as previously described (21). The amplicon was purified using QIAquick spin columns (Qiagen, Valencia, Calif.), and 5 μg of bbk32 was digested with bovine pancreatic DNase I in 50 mM Tris-HCl (pH 7.5)–0.05 mg of bovine serum albumin/ml–10 mM MnCl2. The DNA fragments were separated on a 2% NuSieve agarose gel (FMC Bioproducts, Rockland, Maine); the 50- to 150-bp fragments were excised from the gel and purified using QIAquick spin columns. Blunt ends were created by treatment of the DNA fragments with T4 DNA polymerase. A 3′ adenosine overhang was then added by Tth polymerase, and the DNA fragments were ligated into the Novagen pScreen T vector, a pET vector derivative possessing a T-cloning site upstream of a T7 gene 10 fusion partner. The bbk32 gene fragment library was constructed by transforming Novablue (DE3) competent E. coli (Novagen) with the ligation mixture and selecting for transformants by plating the bacteria on Luria-Bertani (LB) agar plates containing 50 μg of carbenicillin/ml. Transformants were transferred to nitrocellulose membranes, lysed with chloroform vapors, and denatured with 20 mM Tris (pH 8.0)–6 M urea–0.5 M NaCl. For ligand blotting, the membranes were washed extensively with 25 mM Tris (pH 7.5)–150 mM NaCl–0.05% Tween 20 (TTBS), blocked with 3% bovine serum albumin, and probed for 1 h with a 1:30,000 dilution of alkaline phosphatase-labeled human fibronectin as previously described (21). After the membranes were washed with TTBS, bound fibronectin was detected by immersion of the membranes in a solution of 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium (Kirkegaard & Perry Laboratories, Gaithersburg, Md.). Following this initial screening of the bbk32 gene fragment library, the fibronectin-binding activity of each positive colony was confirmed by a second round of ligand blotting (data not shown).
Seventeen colonies expressing fibronectin-binding activity by ligand blotting were selected for DNA sequence analysis. Plasmid DNA was purified from each of the 17 clones using QIAquick spin columns, and the insert DNA was sequenced by dye terminator cycle sequencing on an ABI 373 sequencer (PE Biosystems, Foster City, Calif.). Vector-specific primers, T7 terminator, and STAG (Novagen) were used to sequence both strands of the insert DNA. Among the 17 clones expressing fibronectin-binding activity, the smallest bbk32 insert encoded amino acids 131 to 167 (AA 131-167 construct). Nucleic acid sequence from a second clone encompassed amino acids 105 to 162 of BBK32 (AA 105-162 construct). The sequence overlap between these two clones suggested that amino acids 131 to 162 of BBK32 were sufficient to mediate fibronectin binding. All 17 clones possessed nucleic acid sequence encoding this segment of BBK32, indicating that this region is likely the principal fibronectin-binding domain of BBK32.
To further delimit the ligand-binding region of BBK32, we amplified, cloned, and expressed the segments of bbk32 encoding amino acids 131 to 162, 136 to 162, and 131 to 157 (AA 131-162, AA 136-162, and AA 131-157 constructs) and evaluated the fibronectin-binding activities of these clones by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and ligand blotting. The primers used to amplify these fragments of bbk32 are listed in Table 1. The amplification products were ligated into the pScreen T vector, and Novablue (DE3) competent E. coli were transformed with each construct. Transformants were selected on LB agar plates supplemented with carbenicillin (50 μg/ml), and E. coli clones possessing the desired fragment of bbk32 were verified by DNA sequencing as described above. The clones were subcultured to LB broth for 8 h, and recombinant protein expression was induced for 2 h by the addition of isopropylthiogalactoside to final concentration of 0.3 mM. The bacteria were harvested by centrifugation and lysed in SDS-PAGE sample buffer, and proteins were resolved on a 12% polyacrylamide gel. Following electrophoretic transfer of the proteins to a nitrocellulose membrane, ligand blotting was performed as described earlier.
TABLE 1.
Oligonucleotide primers used for amplification of bbk32 gene fragments
| BBK32 construct | Forward primer | Reverse primer |
|---|---|---|
| AA 131-162 | 5′-CAAGGAAGTTTAAATTCCCT | 5′-CCTTAAATCAGAATCTATAG |
| AA 131-157 | 5′-CAAGGAAGTTTAAATTCCCT | 5′-TATAGTAAGATCAATTTC |
| AA 136-162 | 5′-TCCCTTAGCGGTGAAAGT | 5′-CCTTAAATCAGAATCTATAG |
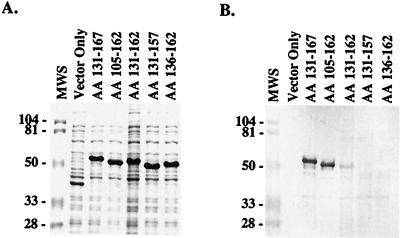
Using the pScreen T-vector expression system, the cloned DNA is expressed as a peptide fused to a 37-kDa vector-derived peptide. As shown in Fig. 1A, expression of the various pScreen/bbk32 constructs in E. coli resulted in the production of recombinant proteins ranging in size from 48 to 54 kDa. With the exception of the AA 105-162 construct, the apparent molecular weight of the recombinant protein exceeded the molecular weight predicted for each construct. A similar observation has been made for recombinant proteins derived from different streptococcal and staphylococcal fibronectin-binding proteins (11). The AA 131-162 construct produced a 50-kDa protein that bound fibronectin, as demonstrated by SDS-PAGE and ligand blotting (Fig. 1). However, the level of fibronectin-binding activity displayed by this clone was much lower than those of the AA 131-167 and AA 105-162 constructs. Furthermore, this fibronectin-binding band appears to have migrated slightly faster than would be anticipated from the corresponding Coomassie blue-stained gel. Deletion of five amino acids from either the N-terminal (AA 136-162 construct) or C-terminal (AA 131-157 construct) end of amino acids 131 to 162 completely abolished fibronectin-binding activity (Fig. 1). Little or no fibronectin-binding activity was associated with an E. coli clone transformed with pScreen containing no insert DNA (Fig. 1, Vector Only lane). These results indicate that amino acids 131 to 162, QGSLNSLSGESGELEEPIESNEIDLTIDSDLR, defined the fibronectin-binding region of BBK32. A striking feature of this sequence is the relatively high number of acidic amino acid residues. This sequence characteristic has been noted for other bacterial fibronectin-binding proteins (11).
FIG. 1.
Mapping the ligand-binding region of BBK32. Fragments of the bbk32 gene from B. burgdorferi isolate B31 were cloned into the pScreen T vector and expressed as recombinant fusion proteins in E. coli. Proteins from E. coli lysates were separated on a 12% polyacrylamide gel and stained with Coomassie blue (A) or transferred to a nitrocellulose membrane and probed with alkaline phosphatase-labeled human fibronectin (B). The BBK32 amino acid sequence expressed by each clone is indicated above each lane. A clone expressing the vector-derived peptide of 37 kDa is labeled Vector Only. Sizes of the molecular weight standards (MWS) are provided in kilodaltons.
Functional activity and sequence analysis of BBK32 from different genospecies of B. burgdorferi sensu lato.
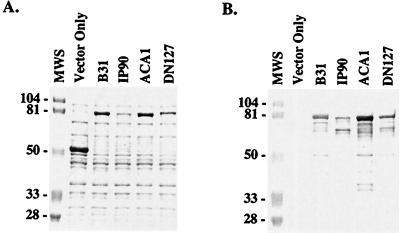
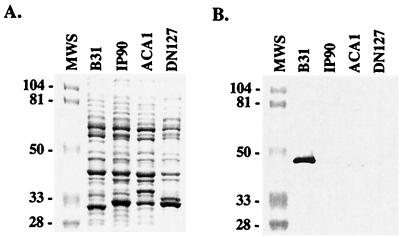
Having defined the ligand-binding region of BBK32, we sought to establish whether BBK32 is functionally and genetically conserved among isolates of B. burgdorferi sensu lato. To this end, we cloned and expressed bbk32 from four isolates representing different genospecies of B. burgdorferi: isolate B31, B. burgdorferi sensu stricto; isolate IP90, B. garinii; isolate ACA1, B. afzelii; and isolate DN127, B. bissettii. As previously described for isolate B31 (21), the bbk32 gene was amplified from each isolate and cloned into the pMalc2 expression vector (New England Biolabs, Inc., Beverly, Mass.), in which the gene of interest is expressed as a peptide fused to a 42-kDa vector-derived peptide. In each case, the transformation of E. coli with a bbk32 construct derived from either B31, IP90, ACA1, or DN127 resulted in the expression of an 80-kDa recombinant fusion protein that bound fibronectin, as determined by SDS-PAGE and ligand blot analysis (Fig. 2). The appearance of lower-molecular-weight bands having fibronectin-binding activity in Fig. 2 likely represents proteolysis of the overexpressed recombinant proteins. These results demonstrate that each B. burgdorferi isolate possesses a bbk32 gene that was capable of encoding a functional protein upon expression in E. coli. In contrast, when spirochetal lysates of B31, IP90, ACA1, and DN127 were tested by SDS-PAGE and ligand blotting for fibronectin-binding activity, only two of these four B. burgdorferi isolates expressed a fibronectin-binding protein (Fig. 3). As observed previously (21), isolate B31 expressed relatively high levels of a 47-kDa fibronectin-binding protein (BBK32), whereas ACA1 fibronectin-binding activity was visible as a faint band of 45 kDa. No fibronectin-binding activity was detected for the IP90 and DN127 lysates by ligand blotting. The lack of fibronectin-binding activity observed for isolates IP90 and DN127, despite the presence in both cases of a bbk32 gene capable of encoding a functional protein, suggest that bbk32 expression may be tightly regulated during in vitro cultivation of these isolates.
FIG. 2.
Fibronectin-binding activities of E. coli clones expressing the bbk32 gene from isolates representing different genospecies of B. burgdorferi. The bbk32 gene was amplified from each isolate and cloned into the pMalc2 vector. Expression of the cloned gene results in a recombinant fusion protein of 80 kDa. Lysates from each E. coli clone were subjected to SDS-PAGE on a 12% polyacrylamide gel and stained with Coomassie blue (A) or transferred to nitrocellulose for ligand blotting with alkaline phosphatase-labeled human fibronectin (B). The isolate from which bbk32 was derived is indicated above each lane. The genospecies designation for each isolate is as follows: B31, B. burgdorferi sensu stricto; IP90, B. garinii; ACA1, B. afzelii; and DN127, B. bissettii. Expression of the pMalc2 vector in E. coli produces a fusion protein of 52 kDa (Vector Only). Sizes of the molecular weight markers (MWS) are provided in kilodaltons.
FIG. 3.
Fibronectin-binding activities of isolates representing different B. burgdorferi genospecies. Proteins from the spirochete lysates were separated by SDS-PAGE on a 12% polyacrylamide gel. The proteins were detected by staining with Coomassie blue (A) or transferred to a nitrocellulose membrane for ligand blotting with alkaline phosphatase-labeled human fibronectin (B). Isolates B31, IP90, ACA1, and DN127 have been genotyped as B. burgdorferi sensu stricto, B. garinii, B. afzelii, and B. bissettii, respectively. The weak fibronectin-binding activity of strain ACA1 was evident in the original ligand blot but may not be clearly visible in photograph (B). Sizes of the molecular weight markers (MWS) are shown in kilodaltons.
Next, we evaluated the degree of sequence conservation in the ligand-binding region of BBK32 by sequencing the cloned gene from isolates IP90, ACA1, and DN127. For each pMa1c2/bbk32 construct, three representative clones were sequenced by dye terminator cycle sequencing using an ABI 373 DNA sequencer. Approximately 90% of the gene sequence predicted to encode the mature BBK32 protein was sequenced for each isolate, and the sequences were deposited in GenBank. The alignment by clustal analysis (Lasergene 99; DNASTAR, Inc., Madison, Wis.) of the predicted BBK32 amino acid sequences from these isolates and isolate B31 revealed sequence identity ranging from 69 to 90% (amino acids 35 to 337 [data not shown]). Slightly less variability was observed within the ligand-binding region of BBK32 (Fig. 4). Sequence identity for this segment of BBK32 ranged from 81 to 91% between isolates. Within the ligand-binding region of BBK32, the amino acid sequence motifs LSGESGEL and IESNEID were conserved among all four isolates.
FIG. 4.
Alignment of BBK32 amino acid sequences from isolates representing different genospecies of B. burgdorferi with the ligand-binding region (amino acids 131 to 162) of BBK32 from isolate B31. The genospecies designations for the isolates are as follows: B31, B. burgdorferi sensu stricto; IP90, B. garinii; ACA1, B. afzelii; and DN127, B. bissettii. Sequence dissimilarity is indicated with a single-letter amino acid code. Sequence identity is shown as a period. Shaded amino acids residues represent sequence identity between the ligand-binding regions of BBK32 from isolate B31 and the UR region of protein F1 from S. pyogenes.
A BLAST search of GenBank for sequences homologous to the BBK32 ligand-binding region from isolate B31 produced a single match with an in vivo-expressed protein described for B. burgdorferi sensu stricto isolate N40 (3). The N40 amino acid sequence was identical to the BBK32 ligand-binding region of isolate B31, indicating that the N40 homolog may also interact with fibronectin. As was observed for isolates IP90 and DN127 in our study, N40 does not appear to express bbk32 during cultivation in vitro. However, bbk32 expression by N40 was detected following reverse transcription-PCR analyses of tissues from mice experimentally infected with this isolate (3). Expression of bbk32 has also been detected in skin and joint tissue biopsies from patients with Lyme disease (4). Despite variability in bbk32 expression levels among cultivated B. burgdorferi isolates, these studies indicate that bbk32 expression may be upregulated by B. burgdorferi during colonization of the mammalian host.
We also aligned the BBK32 ligand-binding region from isolate B31 with fibronectin-binding motifs described for other bacterial proteins including fibronectin attachment protein (23) and antigen 85b (16) from mycobacteria, FnbpA from Staphylococcus aureus (25), and protein F1 from Streptococcus pyogenes (18). Among these proteins, only protein F1 from S. pyogenes shared significant sequence homology with the ligand-binding region of BBK32. The BBK32 ligand-binding region of isolate B31 and the UR region of protein F1 shared sequence identity at 8 of 13 contiguous amino acids: LXGESGEXEXXXE, where X is a dissimilar amino acid (Fig. 4). The motif LXGESGE was also conserved among all B. burgdorferi sensu lato isolates sequenced in our study. Like BBK32, the UR region of protein F1 interacts with the collagen-binding domain of fibronectin (18, 21). The sequence homology shared between the ligand-binding region of BBK32 and the UR region of protein F1 may dictate the specificity of these proteins for the collagen-binding domain of fibronectin. A similar motif, LAGESGET, has also been recognized in the fibronectin-binding protein, FNZ, from Streptococcus equi subsp. zooepidemicus (14). The region of fibronectin bound by this segment of FNZ awaits localization.
The structural and functional resemblance of BBK32 to the UR region of protein F1 suggest that BBK32 may provide B. burgdorferi with a mechanism of host colonization similar to the role proposed for protein F1 during S. pyogenes infections. Protein F1 expression has been implicated in the cellular adherence and entry of S. pyogenes (9, 15, 19). Protein F1 may facilitate S. pyogenes adherence by binding fibronectin deposited on cell surfaces or by interacting with soluble fibronectin, which in turn binds to cell surface molecules such as integrins. Ozeri et al. (19) have demonstrated that the indirect interaction of protein F1 with β1 integrins via a fibronectin bridge can induce internalization of adherent S. pyogenes by the cell. The cellular uptake of S. pyogenes through the indirect interaction of protein F1 with integrins may provide this pathogen with a mechanism of penetrating tissue barriers and establishing deep tissue infections. Likewise, the ability of BBK32 to bind fibronectin, and indirectly integrins, may provide B. burgdorferi with a similar mechanism of cellular adherence and entry. The observation that β1 and β3 integrins may be involved in the adherence of B. burgdorferi to human cells lends support to this hypothesis (2). Furthermore, the ability of B. burgdorferi to invade cultured cells and survive intracellularly has been documented by electron microscopy, by confocal microscopy, and by antibiotic protection assays performed with ceftriaxone (5, 8, 12). Despite support from these in vitro studies, the in vivo capacity of B. burgdorferi to invade and persist within cells has proven difficult to establish. The potential intracellular existence of B. burgdorferi, however, may help explain the persistent nature of B. burgdorferi infections and the occasional failure of antibiotic therapy. We are currently investigating the role of BBK32 in cell adherence and invasion by expressing bbk32 in a heterologous host.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the bbk32 sequence from B. burgdorferi isolates B31, IP90, ACA1, DN127, and N40 are AE000788, AF213178, AF213179, AF213180, and U82107, respectively.
REFERENCES
- 1.Burgdorfer W, Barbour A G, Hayes S F, Benach J L, Grundwald E, Davis J P. Lyme disease: a tick-borne spirochetosis? Science. 1982;216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- 2.Coburn J, Magoun L, Bodary S C, Leong J M. Integrins αvβ3 and α5β1 mediate attachment of Lyme disease spirochetes to human cells. Infect Immun. 1998;66:1246–1252. doi: 10.1128/iai.66.5.1946-1952.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fikrig E, Barthold S W, Sun W, Feng W, Telford S R, Flavell R A. Borrelia burgdorferi P35 and P37 proteins, expressed in vivo, elicit protective immunity. Immunity. 1997;6:1–20. doi: 10.1016/s1074-7613(00)80341-6. [DOI] [PubMed] [Google Scholar]
- 4.Fikrig E, Feng W, Aversa J, Schoen R T, Flavell R A. Differential expression of Borrelia burgdorferi genes during erythema migrans and Lyme arthritis. J Infect Dis. 1998;178:1198–1201. doi: 10.1086/515684. [DOI] [PubMed] [Google Scholar]
- 5.Georgilis K, Peacocke M, Klempner M S. Fibroblasts protect the Lyme disease spirochete, Borrelia burgdorferi, from ceftriaxone in vitro. J Infect Dis. 1992;166:440–444. doi: 10.1093/infdis/166.2.440. [DOI] [PubMed] [Google Scholar]
- 6.Gerber M A, Shapiro E D, Burke G S, Parcells V J, Bell G L. Lyme disease in children in southeastern Connecticut. Pediatric Lyme disease study group. N Engl J Med. 1996;335:1270–1274. doi: 10.1056/NEJM199610243351703. [DOI] [PubMed] [Google Scholar]
- 7.Grab D J, Givens C, Kennedy R. Fibronectin-binding activity in Borrelia burgdorferi. Biochim Biophys Acta. 1998;1407:135–145. doi: 10.1016/s0925-4439(98)00038-6. [DOI] [PubMed] [Google Scholar]
- 8.Hechemy K E, Samsonoff W A, Harris H L, McKee M. Adherence and entry of Borrelia burgdorferi in Vero cells. J Med Microbiol. 1992;36:229–238. doi: 10.1099/00222615-36-4-229. [DOI] [PubMed] [Google Scholar]
- 9.Jadoun J, Ozeri V, Burstein E, Skutelsky E, Hanski E, Sela S. Protein F1 is required for efficient entry of Streptococcus pyogenes into epithelial cells. J Infect Dis. 1998;178:147–158. doi: 10.1086/515589. [DOI] [PubMed] [Google Scholar]
- 10.Joh D, Höök M. Role of fibronectin-binding MSCRAMMs in bacterial adherence and entry into mammalian cells. Matrix Biol. 1999;18:211–223. doi: 10.1016/s0945-053x(99)00025-6. [DOI] [PubMed] [Google Scholar]
- 11.Joh H J, House-Pompeo K, Patti J M, Gurusiddappa S, Höök M. Fibronectin receptors from Gram-positive bacteria: comparison of active sites. Biochemistry. 1994;33:6086–6092. doi: 10.1021/bi00186a007. [DOI] [PubMed] [Google Scholar]
- 12.Klempner M S, Noring R, Rogers R A. Invasion of human skin fibroblasts by the Lyme disease spirochete, Borrelia burgdorferi. J Infect Dis. 1993;167:1074–1081. doi: 10.1093/infdis/167.5.1074. [DOI] [PubMed] [Google Scholar]
- 13.Kopp P A, Schmitt M, Wellensiek H, Blobel H. Isolation and characterization of fibronectin-binding sites of Borrelia garinii N34. Infect Immun. 1995;63:3804–3808. doi: 10.1128/iai.63.10.3804-3808.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindmark H, Jacobsson K, Frykberg L, Guss B. Fibronectin-binding protein of Streptococcus equi subsp. zooepidemicus. Infect Immun. 1996;64:3993–3999. doi: 10.1128/iai.64.10.3993-3999.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molinari G, Talay S R, Valentin-Weigand P, Roche M, Chhatwal G S. The fibronectin-binding protein of Streptococcus pyogenes, SfbI, is involved in the internalization of group A streptococci by epithelial cells. Infect Immun. 1997;65:1357–1363. doi: 10.1128/iai.65.4.1357-1363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naito M, Ohara N, Matsumoto S, Yamada T. The novel fibronectin-binding motif and residues of Mycobacteria. J Biol Chem. 1998;273:2905–2909. doi: 10.1074/jbc.273.5.2905. [DOI] [PubMed] [Google Scholar]
- 17.Orloski K A, Hayes E B, Dennis D T. Surveillance for Lyme disease—United States, 1992–1998. Morbid Mortal Wkly Rep CDC Surveill Summ. 2000;49:1–9. [PubMed] [Google Scholar]
- 18.Ozeri V, Tovi A, Burstein I, Natanson-Yaron S, Caparon M G, Yamada K M, Akiyama S K, Vlodavsky I, Hanski E. A two-domain mechanism for group A streptococcal adherence through protein F to the extracellular matrix. EMBO J. 1996;15:989–998. [PMC free article] [PubMed] [Google Scholar]
- 19.Ozeri V, Rosenshine I, Mosher D F, Fassler R, Hanski E. Roles of integrins and fibronectin in the entry of Streptococcus pyogenes into cells via protein F1. Mol Microbiol. 1998;30:625–637. doi: 10.1046/j.1365-2958.1998.01097.x. [DOI] [PubMed] [Google Scholar]
- 20.Patti J M, Allen B L, McGavin M J, Höök M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol. 1994;48:585–617. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- 21.Probert W S, Johnson B J B. Identification of a 47 kDa fibronectin-binding protein expressed by Borrelia burgdorferi isolate B31. Mol Microbiol. 1998;30:1003–1015. doi: 10.1046/j.1365-2958.1998.01127.x. [DOI] [PubMed] [Google Scholar]
- 22.Proctor R A. Fibronectin: a brief overview of its structure, function, and physiology. Rev Infect Dis. 1987;9:S317–S321. doi: 10.1093/clinids/9.supplement_4.s317. [DOI] [PubMed] [Google Scholar]
- 23.Schorey J S, Holsti M A, Ratliff T L, Allen P M, Brown E J. Characterization of the fibronectin-attachment protein of Mycobacterium avium reveals a fibronectin-binding motif conserved among mycobacteria. Mol Microbiol. 1996;21:321–329. doi: 10.1046/j.1365-2958.1996.6381353.x. [DOI] [PubMed] [Google Scholar]
- 24.Sigal L H. Lyme disease: a review of aspects of its immunology and immunopathogenesis. Annu Rev Immunol. 1997;15:63–92. doi: 10.1146/annurev.immunol.15.1.63. [DOI] [PubMed] [Google Scholar]
- 25.Signas C, Raucci G, Jonsson K, Lindgren P E, Anantharamaiah G M, Höök M, Lindberg M. Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus: use of this peptide sequence in the synthesis of biologically active peptides. Proc Natl Acad Sci USA. 1989;86:699–703. doi: 10.1073/pnas.86.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steere A C. Lyme Disease. N Engl J Med. 1989;321:586–596. doi: 10.1056/NEJM198908313210906. [DOI] [PubMed] [Google Scholar]
- 27.Strle F, Picken R N, Cheng Y, Cimperman J, Maraspin V, Lotric-Furlan S, Ruzic-Sabljic E, Picken M M. Clinical findings for patients with Lyme borreliosis caused by Borrelia burgdorferi sensu lato with genotypic and phenotypic similarities to strain 25015. Clin Infect Dis. 1997;25:273–280. doi: 10.1086/514551. [DOI] [PubMed] [Google Scholar]
- 28.Szczepanski A, Furie M B, Benach J L, Lane B P, Fleit H B. Interaction between Borrelia burgdorferi and endothelium in vitro. J Clin Investig. 1990;85:1637–1647. doi: 10.1172/JCI114615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang G, van Dam A P, Schwartz I, Dankert J. Molecular typing of Borrelia burgdorferi sensu lato: taxonomic, epidemiological, and clinical implications. Clin Microbiol Rev. 1999;12:633–653. doi: 10.1128/cmr.12.4.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westerlund B, Korhonen T K. Bacterial proteins binding to the mammalian extracellular matrix. Mol Microbiol. 1993;9:687–694. doi: 10.1111/j.1365-2958.1993.tb01729.x. [DOI] [PubMed] [Google Scholar]