Abstract
Deciphering the mechanisms by which Borrelia burgdorferi controls the synthesis of proteins associated with mammalian infection will be an important step toward understanding the pathogenic properties of Lyme disease-causing bacteria. We present results of studies indicating that B. burgdorferi senses a wide variety of environmental stimuli, including soluble chemicals, which enables it to independently control synthesis of the Erp and OspC proteins. Regulation of OspC and Erp expression appears to occur at the level of transcription. In this regard, we observed that one or more DNA-binding proteins interact specifically with erp promoter DNA but not with the ospC promoter.
To persist in nature, the Lyme disease spirochete Borrelia burgdorferi must efficiently infect both warm-blooded animals and ticks and must be competent for transmission between these hosts. In this cycle of B. burgdorferi infection, ticks ingest bacteria from infected animals along with the blood meal. Within the tick, spirochetes colonize the midgut and persist in this organ through the molt. As the tick feeds on its next vertebrate host, the bacteria penetrate the midgut lining, migrate through the hemolymph to the salivary glands, and are transmitted to the new host via the tick's saliva. The bacteria then disseminate throughout the host's body and reside in its tissues until they are transmitted to another tick feeding on the infected animal, continuing the cycle (37). This complex series of processes undoubtedly requires that B. burgdorferi sense its environment and synthesize proteins appropriate for interactions with the various tick and vertebrate tissues encountered. Consistent with this hypothesis, a recent study indicated that precise regulation of gene expression by B. burgdorferi is essential for the bacteria to disseminate in warm-blooded hosts and cause disease (4). The mechanisms by which B. burgdorferi senses its environment or regulates its genes are poorly understood at this time. Elucidation of the regulatory pathways employed by B. burgdorferi to control protein synthesis will doubtless provide important insights into the biology of these bacteria and the pathogenesis of Lyme disease, in addition to identifying targets for development of improved therapeutic treatments. Recombinant genetic methods for use in B. burgdorferi are sorely lacking (49), preventing studies of regulation through gene fusions or other techniques that might be performed with Escherichia coli or other, more tractable bacteria. However, insight into the mechanisms by which Lyme disease spirochetes regulate gene expression can be gained by more traditional methods.
The B. burgdorferi OspC protein is rarely detected on bacteria in the midguts of unfed ticks but is made by spirochetes in the tick after initiation of feeding and during the early stages of mammalian infection (15, 18, 19, 25, 38, 39, 53). OspC is a surface-exposed lipoprotein (16, 19, 29, 35, 52), which could therefore interact with host cells, extracellular matrices, or other substances. Regulation of OspC synthesis by cultured B. burgdorferi has been observed in response to culture conditions (10, 33, 39). Changes in OspC protein levels are accompanied by similar changes in ospC mRNA levels (48), suggesting that regulation occurs at the level of transcription. One well-studied phenomenon involves regulation of synthesis in response to culture temperature, with bacteria grown at 23°C producing very little OspC, while bacteria shifted from 23 to 34°C produce greater quantities of the protein (39). These temperatures mimic the environments before and during transmission from the tick vector, corresponding with air and blood temperatures, respectively. Thus B. burgdorferi likely uses temperature as a cue that the tick is feeding on a warm-blooded animal, indicating the need to express proteins required for transmission.
Synthesis of the B. burgdorferi Erp (OspEF-related) proteins is similarly regulated in response to culture temperature (1, 40, 43). As with OspC, Erp synthesis appears to be regulated at the level of transcription (40). Erp proteins are also expressed by B. burgdorferi during the initial stages of mammalian infection, as evidenced by reverse transcriptase PCR analyses (4, 14) and the appearance of Erp-directed antibodies during the first 2 to 4 weeks of infection (3, 14, 31, 32, 40, 43, 46, 47, 50). All Lyme disease spirochetes contain erp genes (45), but these genes have not been found in other species of the genus Borrelia (42), indicating that Erp proteins perform a function unique to the biology of Lyme disease borreliae. Like OspC, Erp proteins are surface-exposed lipoproteins (3, 16, 24, 50). At least some Erp proteins can bind complement factor H (21), suggesting that these proteins aid the bacteria during mammalian infection by actively inhibiting complement activation. While each B. burgdorferi bacterium contains a single ospC gene, located on the circular plasmid cp26 (26, 36, 48), individual bacteria carry multiple erp operons, each located on a different plasmid of the cp32 family (2, 13, 44, 45). Clonal B. burgdorferi with as many as nine different erp loci per cell have been characterized (2, 12, 13, 17, 40, 44). However, our previous studies and those presented in this report indicate that all erp genes of the B. burgdorferi type strain, B31, appear to be regulated in an identical manner (16, 40). The erp loci of strain B31 are named erpAB, erpCD, erpG, erpHY, erpIJ, erpK, erpLM, erpNO, erpPQ, and erpX (12, 13, 40, 44). The coding regions of the erpAB, erpIJ, and erpNO loci are identical, so their encoded proteins are indistinguishable and are collectively referred to as ErpA/I/N and ErpB/J/O (31, 40, 41, 45).
As discussed above, both OspC and Erp proteins are expressed by B. burgdorferi during the initial stages of mammalian infection, suggesting that these surface proteins are involved with transmission from the tick and/or enable interactions with tissues of the warm-blooded host. Since production of Erp proteins and production of OspC are similarly regulated by temperature in vitro, a model has been proposed in which these proteins are coexpressed in response to the same environmental clues during the B. burgdorferi infectious cycle (40, 43, 54). Further studies described below indicate that this model is in need of revision.
All of the studies reported below used infectious bacteria of the B. burgdorferi type strain, B31 (8, 11, 39, 40). B. burgdorferi was cultivated in several formulations of growth media. Barbour-Stoener-Kelly II (BSK-II) was prepared in our laboratory according to the recipe of A. G. Barbour (5), which includes 6% (vol/vol) rabbit serum (Sigma, St. Louis, Mo.). Two modifications of this medium were also prepared, either lacking Yeastolate (Difco, Detroit, Mich.) or lacking both Yeastolate and rabbit serum. Three different lots, including lot 38H8425, of a commercially prepared medium, BSK-H (34), were obtained from Sigma.
For immunoblot analyses, bacterial lysates containing equivalent amounts of total proteins (as determined by Coomassie brilliant blue staining and immunoblot analysis using the anti-FlaB monoclonal antibody [MAb] H9724) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were electrotransferred to Trans-blot nitrocellulose membranes (Bio-Rad, Hercules, Calif.) (31). Membranes were blocked for at least 1 h with 5% nonfat dry milk in Tris-buffered saline–Tween 20 (TBS-T) (20 mM Tris [pH 7.5], 150 mM NaCl, 0.05% [vol/vol] Tween 20). The membranes were then incubated for 1 h with an appropriate antibody, washed with TBS-T, and incubated for an additional hour with a protein A-horseradish peroxidase conjugate (Amersham, Piscataway, N.J.). Bound antibodies were detected by enhanced chemiluminescence (Amersham). MAbs B11 and B5, directed against the strain B31 ErpA/I/N and OspC proteins, respectively, were produced by hybridomas derived from mice infected with this strain by tick bite (16, 20, 30). Other Erp proteins were detected with polyclonal antibodies produced from rabbits vaccinated with recombinant Erp proteins (16, 31). MAb H9724, directed against the flagellar FlaB subunit (6), was provided by Tom Schwan (Rocky Mountain Laboratories, Hamilton, Mont.).
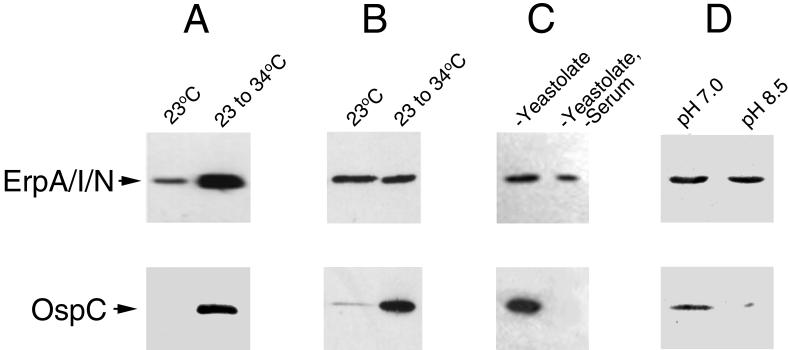
We and others have observed that B. burgdorferi strain B31 differentially expresses the OspC and Erp proteins in response to culture temperature, with significantly greater quantities of these proteins being made by bacteria shifted from 23 to 34°C relative to those maintained at 23°C (Fig. 1A) (33, 38–40, 43). These results have been consistently obtained from bacteria grown in either BSK-II made in our laboratories or commercially obtained BSK-H. Thus, we were surprised when aberrant results were obtained with BSK-H from lot 38H8425, purchased from Sigma Chemical Co. during autumn 1998. A normal temperature-regulated synthesis pattern was observed for OspC, while the levels of all Erp proteins were equally high in bacteria grown at a constant temperature of 23°C and bacteria shifted from 23 to 34°C (Fig. 1B and data not shown). The effect of this particular medium was consistently observed using B31 bacteria grown from different aliquots of frozen cells and different bottles of medium. We conclude that the constitutive synthesis of Erp proteins when bacteria were grown in this lot of BSK-H was a consequence of an unknown component of the medium, indicating that Erp synthesis is regulated by B. burgdorferi in response to a chemical signal(s) in addition to a temperature signal (Fig. 2). This signal has no apparent effect on OspC synthesis. Due to the fastidiousness of B. burgdorferi, BSK-II and BSK-H are very complex, undefined media, containing a multitude of amino acids, cofactors, and other nutrients, as well as a crude yeast extract, proteolysed crude protein, rabbit serum, and bovine serum albumin (5, 23, 34). Due to this overwhelming complexity, we have not been able to isolate the factor(s) responsible for constitutive Erp expression.
FIG. 1.
Representative immunoblots showing effects of culture medium composition or pH on Erp and OspC protein levels. Identical results were obtained for all the strain B31 Erp proteins (A) Bacteria grown in BSK-II medium at a constant temperature of 23°C or shifted from 23 to 34°C. (B) Bacteria grown in BSK-H medium, lot 38H8425, at a constant 23°C or shifted from 23 to 34°C. (C) Bacteria shifted from 23 to 34°C into BSK-II medium lacking either Yeastolate or both Yeastolate and rabbit serum. (D) Bacteria grown at 34°C in BSK-II medium buffered to remain at either pH 7.0 or 8.5.
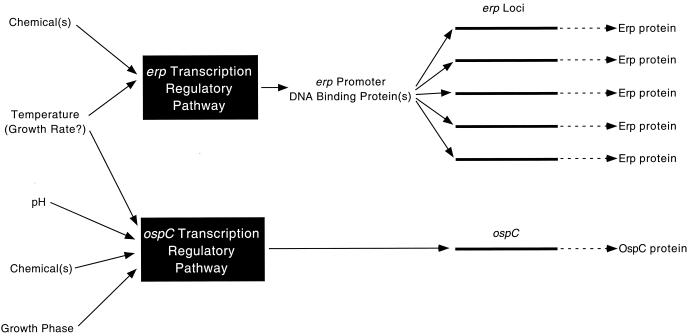
FIG. 2.
A model of the pathways by which B. burgdorferi regulates synthesis of OspC and Erp proteins. Synthesis of Erp proteins is affected by both temperature and one or more chemicals, while synthesis of OspC is modulated by temperature, pH, culture growth phase, and a different chemical signal(s). Since bacteria grow faster at the higher temperatures tested, it cannot yet be determined whether it is temperature, growth rate, or both that influence expression of these genes.
The effects of medium composition on Erp and OspC synthesis were further examined by culturing strain B31 organisms in medium that lacked one or more component of BSK-II. Bacteria were grown in complete BSK-II at 23°C, diluted 1:100 into BSK-II lacking either Difco Yeastolate (a yeast extract) or both Yeastolate and rabbit serum, and cultured to mid-logarithmic phase at 34°C. Those bacteria cultured in Yeastolate-deficient medium expressed Erp and OspC proteins, while those grown in the absence of both Yeastolate and serum produced all the Erp proteins but failed to synthesize OspC (Fig. 1C and data not shown). These results further indicate that synthesis of Erp and synthesis of OspC can be regulated independently, and suggest that OspC production is also controlled in response to an environmental chemical signal (Fig. 2).
B. burgdorferi grown in media with a pH at or below 7 produce significantly greater amounts of OspC than do bacteria grown at pH 8 or higher (Fig. 1D) (10). To examine the effects of environmental pH on Erp protein synthesis, bacteria were grown at a constant 34°C in media buffered by addition of 25 mM HEPES to remain at either pH 7 or 8.5 during bacterial growth (9, 10). No effects on the level of any Erp protein were detected (Fig. 1D and data not shown), indicating that synthesis of this protein family is unaffected by culture medium pH (Fig. 2). Similar results were obtained regardless of the culture medium used in the experiment.
It has been reported that synthesis of OspC is affected by bacterial growth phase, with bacteria in late-logarithmic or stationary phase producing greater amounts of this protein than bacteria in early growth phases (35). With this in mind, we assessed the influence of culture phase on Erp protein levels. Bacteria were inoculated at an initial density of 1 × 106 bacteria per ml and grown at 34°C, with aliquots harvested at the following time points and cell densities: 23 h (6.0 × 106 bacteria/ml), 44 h (1.5 × 107 bacteria/ml), 64 h (6.0 × 107 bacteria/ml), and 83 h (1.7 × 108 bacteria/ml). Increases in cell density as the culture passed through logarithmic growth phase and into stationary phase had no detectable effect on Erp synthesis (data not shown).
The differences described above indicate that expression of OspC and Erp proteins is controlled through distinct regulatory pathways. Since both protein types appear to be regulated at the level of transcription (40, 48), and transcription is often controlled through the binding of regulatory proteins to specific DNA sequences, we compared the promoter DNA sequences of the strain B31 erp and ospC loci, revealing essentially no similarity (Fig. 3). The strain B31 ospC promoter contains two large inverted repeats (IRs), neither of which have homologs in the erp 5′ noncoding region. In many European Lyme disease borreliae, the larger of the B31 ospC promoter IRs is disrupted by as many as seven copies of an 11-bp direct repeat (48), which also has no homology with the erp 5′ noncoding regions. These comparisons suggested that it is unlikely that erp and ospC promoters could interact with the same DNA binding protein, as was borne out by the studies described below.
FIG. 3.
(A) Comparison of the DNA sequences located 5′ of the strain B31 erp loci, from the initiation codon of the divergently transcribed gene (CAT) to the initiation codon of the first gene in the erp locus (ATG). The extensive similarities of these promoter sequences suggests that transcription of all the strain B31 erp genes is regulated similarly, as has been consistently observed in this and other studies (16, 40). The complete sequence of the DNA located 5′ of the erpIJ operon has not yet been determined, as the plasmid containing this locus (cp32-5) is absent from the recently sequenced culture of strain B31 (12, 13). (B) DNA sequence 5′ of the strain B31 ospC gene, from the divergently transcribed guaA gene (CAT) to the ospC initiation codon (ATG). Inverted repeats found in each 5′ noncoding region are indicated by arrows under the DNA sequence.
The abilities of B. burgdorferi proteins to bind to the 5′ noncoding regions of the erp and ospC loci were analyzed by electrophoretic mobility shift assays (EMSA). A divergently transcribed gene, guaA, is located 5′ of the B. burgdorferi ospC gene (27, 48). We reasoned that DNA sequences important in the regulation of ospC transcription would most likely be located between these two genes, so a DNA fragment spanning the region between ospC and guaA was PCR amplified for EMSA. Similarly, all erp loci are bordered on the 5′ end by a divergently transcribed gene (2, 12, 44). The DNA sequences preceding the B31 erp loci are all similar (Fig. 3), suggesting that all might have comparable affinities for any DNA binding protein. Thus we randomly chose the erpG, erpK, and erpLM 5′ noncoding regions for these studies. Fragments consisting of the DNA between the start codons of these erp loci and their upstream genes were synthesized by PCR. The B. burgdorferi flaB gene is constitutively expressed (7), and approximately 500 bp of 5′ noncoding sequence (7, 51) was synthesized by PCR. Oligonucleotides used in this PCR are listed in Table 1. DNA fragments were further purified from template DNA by agarose gel electrophoresis followed by extraction with Amicon Ultrafree-DA spin columns (Millipore, Bedford, Mass.). PCR-generated fragments of the erpG or ospC promoter regions were dephosphorylated with calf intestinal alkaline phosphatase and gel purified. Approximately 50 ng of the PCR product was end labeled using [γ-32P]dATP (Amersham) and T4 polynucleotide kinase (New England Biolabs, Beverly, Mass.).
TABLE 1.
Oligonucleotides used in these studies
| Primer | Sequence (5′→3′) | Use for PCR |
|---|---|---|
| E-8 | CATAAGTTACTCC | erpG promoter region |
| E-43 | AAAATTTTAGTCAAATTTGGAGTG | erpG promoter region |
| PC-29 | TCTCTAATTCTTCTTGCAATTAGTTG | ospC promoter region |
| PC-40 | TCCTGAATTATTACAAGATATAAATA | ospC promoter region |
| FLA-1 | CAAGAAAATACATTAAAGGC | flaB promoter region |
| FLA-14 | AGAACCTCTGTCTGCATCTG | flaB promoter region |
| E-8 | CATAAGTTACTCC | erpLM promoter region |
| E-62 | AAAATTTTAGTCAAATTTTG | erpLM promoter region |
| E-8 | CATAAGTTACTCC | erpK promoter region |
| E-62 | AAAATTTTAGTCAAATTTTG | erpK promoter region |
Cell extracts were prepared from B. burgdorferi grown in several different media. For erpG promoter fragment studies, bacteria were grown at 34°C in BSK-H lot 38H8425, in which Erp proteins are constitutively synthesized. For ospC promoter DNA binding studies, bacteria were harvested from cultures grown at 34°C in BSK-II with the pH adjusted to either 7.0 or 8.5, conditions under which OspC synthesis is either switched on or off, respectively. For each cell extract, 500-ml cultures of B. burgdorferi B31 were grown to mid-logarithmic phase. An aliquot of the culture was assayed by immunoblotting to confirm the presence or absence of Erp or OspC proteins. The cultures were then harvested by centrifugation, washed twice with phosphate-buffered saline, resuspended in 50 mM Tris (pH 8.0)–10 mM EDTA–10% (w/vol) sucrose, and frozen in a dry ice-ethanol bath. The bacteria were thawed in an icewater bath, and NaCl, dithiothreitol, and lysozyme were added to final concentrations of 140 mM, 1 mM, and 0.4 mg/ml, respectively. Bacteria were incubated on ice for 45 min with gentle mixing, then subjected to 4 freeze-thaw cycles using dry ice-ethanol and icewater. Cellular debris was removed from the bacterial lysates by centrifugation in a fixed-angle rotor at 43,000 × g for 1 h. The resultant protein extracts were aliquoted and frozen at −80°C.
Ten micrograms of B. burgdorferi cell-free protein extract, 2 μg of poly (dI-dC), 2 μl of 10 mM dithiothreitol, and 6 μl of 5× buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 50 mM KCl, 1 mM EGTA, 20% [vol/vol] glycerol, 0.5 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, 2 mM sodium orthovanadate) were mixed together, and water was added to a final volume of 30 μl. After incubation for 15 min at 4°C [to permit binding of nonspecific DNA binding proteins with the poly (dI-dC)], 2 ng of labeled DNA was added and incubated for an additional 30 min at 4°C. DNA and protein-DNA complexes were separated by electrophoresis at 4°C in 4% nondenaturing polyacrylamide gels. Gels were then dried, and DNA bands were visualized by autoradiography.
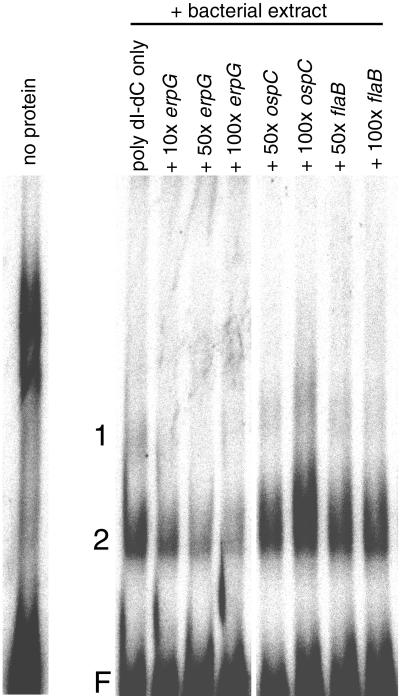
When the labeled erpG DNA fragment was incubated with a cell extract from B. burgdorferi expressing Erp proteins, two distinct bands were consistently seen (Fig. 4). For competition studies, unlabeled erpG, erpLM, erpK, ospC, or flaB promoter DNAs were added prior to the addition of labeled promoter fragment. After a 15-min incubation at 4°C, labeled DNA was added and processed as above. Addition of increasing amounts of unlabeled erpG promoter DNA significantly decreased the levels of both complexes (Fig. 4). Addition of either unlabeled erpLM or erpK promoter DNA fragments had similar effects (data not shown). In contrast, addition of even a 100-fold excess of either ospC or flaB promoter DNAs had no detectable effect on the erpG promoter-protein complexes. As a control, proteinase K (Boehringer Mannheim, Indianapolis, Ind.) was added at a final concentration of 2 ng/ml to cell extracts prior to labeled DNA addition, and incubated at 4°C for 30 min. Labeled promoter fragment was then added and processed as described above. Addition of protease completely eliminated all EMSA bands (data not shown). These results indicate that a protein(s) in the B. burgdorferi cell extract bound specifically to all the tested erp promoter DNAs but did not bind to either ospC or flaB promoter DNAs.
FIG. 4.
EMSA of erp promoter DNA fragments. Assays used, labeled erpG promoter DNA alone or preincubated with B. burgdorferi bacterial extract either with poly (dI-dC) competitor only or with poly (dI-dC) plus a 10-, 50- or 100-fold excess of B. burgdorferi promoter DNA fragments. Competitor DNAs used were the 5′ noncoding regions of erpG, ospC, and flaB (indicated above each lane). The autoradiograph band corresponding with unbound (“free”) erpG promoter DNA is indicated by an “F,” and two DNA-protein complexes are labeled with the numbers 1 and 2.
Similar studies were also performed with labeled ospC promoter using extracts prepared from bacteria cultivated in medium buffered to remain at pH 7.0 or pH 8.5, conditions under which the bacteria do or do not express OspC, respectively. Both extracts yielded faint EMSA bands, yet addition of unlabeled ospC promoter DNA as a competitor had a negligible effect (data not shown). None of the other competitor promoter DNA fragments had any effect, either. Thus it appears that while a protein(s) in the cell extracts bound the ospC promoter region, none of these DNA-protein interactions were specific.
Based on previous observations that both OspC and Erp proteins are regulated in response to culture temperature, it has been hypothesized that these proteins are expressed at the same point in the B. burgdorferi infectious cycle. Since OspC appears to be involved in transmission from the tick vector, the Erp proteins might be, too (40, 43, 54). The data presented in the present work revise this hypothesis and demonstrate that expression of the OspC and Erp proteins is controlled through distinct mechanisms (diagrammed in Fig. 2). Both are regulated in vitro in response to temperature, although since the growth rates of bacteria cultivated at 23 and 34°C are different (40), this effect may be due to the bacteria sensing either temperature or change in growth rate (or both). Each locus type is controlled by a different chemical(s), which may be specific to host tissues in which Erp and OspC proteins function. The culture pH affected OspC expression but had no detectable effect on Erp production.
We also demonstrated that a B. burgdorferi protein(s) specifically binds to the 5′ noncoding DNA of erp loci but not to the 5′ noncoding region of ospC, suggesting that different activator or repressor proteins regulate transcription of these two gene families. Very little is known about how B. burgdorferi regulates gene expression. While the levels of several bacterial proteins have been shown to be regulated by B. burgdorferi, few studies have explored the underlying mechanisms (22, 28). Since we observed protein binding to the erp promoter DNAs in extracts of bacteria that expressed high levels of Erp proteins, this DNA binding protein(s) is more likely a transcriptional activator than a repressor. We are presently purifying this DNA binding activity to characterize its effect on erp expression. Analysis of the ospC 5′ noncoding region indicated that bacterial proteins bind to this DNA, although there was no convincing evidence of specificity for this locus. The results of our studies suggest that ospC transcription is regulated through a DNA binding protein(s) that either was unstable in the cell extracts, does not bind to DNA which is not supercoiled, or binds to a DNA sequence distant from the ospC-guaA intergenic region; or the expression of this gene is controlled through some mechanism other than DNA binding proteins.
The observations that culture medium composition can influence the synthesis of Erp proteins may account for differences among reports of Erp protein synthesis in vitro. We have reported that expression of every B31 erp gene and Erp protein can be detected in vitro (16, 40, 43), while some other researchers have been unable to detect Erp synthesis by cultured bacteria (3, 14, 46, 50). While production of Erp proteins by B. burgdorferi strain 297 was initially described as being undetectable in bacteria grown in vitro (3), further examination of cultured bacteria shifted from 23 to 34°C revealed transcription of all the strain 297 erp genes (1, 45; D. Akins, personal communication). Three additional reports, examining Erp proteins of B. burgdorferi strains N40 and ZS7, noted that certain Erp proteins of these bacteria could not be detected in cultured bacteria (14, 46, 50). Additionally, a recent study observed regulated expression of a strain 297 Erp protein in response to culture medium pH (54), while pH had no detectable effect on the strain B31 Erp proteins in our studies. These differences may have been a consequence of the culture medium used by those researchers, which did not permit synthesis of these proteins in quantities sufficient for detection. Genetic differences between the studied strains may also have influenced experimental results. Clearly, additional studies of these and other B. burgdorferi strains will be required to sort out these variations in results.
T. Schwan and colleagues have reported that B. burgdorferi bacteria within unfed ticks did not increase their synthesis of OspC when the ticks were placed at 34°C, indicating that temperature alone is insufficient to stimulate OspC synthesis (39). The observation that a chemical signal and pH also affect OspC production suggests that the absence of one of these cues may have contributed to the bacteria's inability to express OspC in their study. Since that experiment was performed with unfed ticks, where there are negligible nutrients for bacterial growth, the inability of the bacteria to increase their growth rate may also have suppressed the production of OspC.
A model was recently proposed (54) in which regulation of protein synthesis by B. burgdorferi can be described as being either (i) expressed during transmission from tick to mammal and stimulated by temperature increase and pH decrease or (ii) expressed only during tick infection and stimulated by lower temperatures and higher pH. The results presented here indicate that regulation of B. burgdorferi protein synthesis is far more complex than previously envisioned. The nature of the B. burgdorferi infectious cycle suggests that such complexity is to be expected. Transmission from tick to mammal involves penetration of the midgut lining, targeting to the salivary glands, penetration of the salivary glands, and entry into the salivary ducts, a series of processes that possibly requires different surface proteins at each step. Mammalian infection also is likely to require unique proteins at different steps, with some proteins facilitating interactions with host tissues as the bacteria disseminate while others permit persistent infection of the host. We therefore predict that B. burgdorferi possesses many different mechanisms for control of protein synthesis, each of which is responsive to a particular set of environmental stimuli. The model presented in Fig. 2 will undoubtedly grow in complexity as additional studies are performed on the regulation of OspC, Erp proteins, and other B. burgdorferi proteins. Definition of these regulatory mechanisms will be important milestones in understanding the natural history of B. burgdorferi and elucidating the bacterium's role in the pathology of Lyme disease.
Acknowledgments
This research was funded by United States Public Health Service grant RO1-AI44254 and University of Kentucky Chandler Medical Center Research Fund grant 949 to B. Stevenson.
We thank Julie Stewart for technical assistance; Adrian Centers, George Chaconas, and Martha Peterson for advice on DNA binding protein analysis studies; Tom Schwan, Robert Gilmore, Jr., and Lamine Mbow for providing monoclonal antibodies; and Tony Sinai and Wolf Zückert for helpful comments on the manuscript.
REFERENCES
- 1.Akins D R, Bourell K W, Caimano M J, Norgard M V, Radolf J D. A new animal model for studying Lyme disease spirochetes in a mammalian host-adapted state. J Clin Investig. 1998;101:2240–2250. doi: 10.1172/JCI2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akins D R, Caimano M J, Yang X, Cerna F, Norgard M V, Radolf J D. Molecular and evolutionary analysis of Borrelia burgdorferi 297 circular plasmid-encoded lipoproteins with OspE- and OspF-like leader peptides. Infect Immun. 1999;67:1526–1532. doi: 10.1128/iai.67.3.1526-1532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akins D R, Porcella S F, Popova T G, Shevchenko D, Baker S I, Li M, Norgard M V, Radolf J D. Evidence for in vivo but not in vitro expression of a Borrelia burgdorferi outer surface protein F (OspF) homologue. Mol Microbiol. 1995;18:507–520. doi: 10.1111/j.1365-2958.1995.mmi_18030507.x. [DOI] [PubMed] [Google Scholar]
- 4.Anguita J, Samanta S, Revilla B, Suk K, Das S, Barthold S W, Fikrig E. Borrelia burgdorferi gene expression in vivo and spirochete pathogenicity. Infect Immun. 2000;68:1222–1230. doi: 10.1128/iai.68.3.1222-1230.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbour A G. Isolation and cultivation of Lyme disease spirochetes. Yale J Biol Med. 1984;57:521–525. [PMC free article] [PubMed] [Google Scholar]
- 6.Barbour A G, Hayes S F, Heiland R A, Schrumpf M E, Tessier S L. A Borrelia-specific monoclonal antibody binds to a flagellar epitope. Infect Immun. 1986;52:549–554. doi: 10.1128/iai.52.2.549-554.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bono J L, Elias A F, Kupko J J, Stevenson B, Tilly K, Rosa P. Efficient targeted mutagenesis in Borrelia burgdorferi. J Bacteriol. 2000;182:2445–2452. doi: 10.1128/jb.182.9.2445-2452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burgdorfer W, Barbour A G, Hayes S F, Benach J L, Grunwaldt E, Davis J P. Lyme disease—a tick-borne spirochetosis? Science. 1982;216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- 9.Carroll J A, Cordova R M, Garon C F. Identification of eleven pH-regulated genes in Borrelia burgdorferi localized to linear plasmids. Infect Immun. 2000;68:6677–6684. doi: 10.1128/iai.68.12.6677-6684.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carroll J A, Garon C F, Schwan T G. Effects of environmental pH on membrane proteins in Borrelia burgdorferi. Infect Immun. 1999;67:3181–3187. doi: 10.1128/iai.67.7.3181-3187.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carroll J A, Gherardini F C. Membrane protein variations associated with in vitro passage of Borrelia burgdorferi. Infect Immun. 1996;64:392–398. doi: 10.1128/iai.64.2.392-398.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casjens S, Palmer N, van Vugt R, Huang W M, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson R J, Haft D, Hickey E, Gwinn M, White O, Fraser C. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs of an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- 13.Casjens S, van Vugt R, Tilly K, Rosa P A, Stevenson B. Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J Bacteriol. 1997;179:217–227. doi: 10.1128/jb.179.1.217-227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das S, Barthold S W, Stocker Giles S, Montgomery R R, Telford S R, Fikrig E. Temporal pattern of Borrelia burgdorferi p21 expression in ticks and the mammalian host. J Clin Investig. 1997;99:987–995. doi: 10.1172/JCI119264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Silva A M, Telford S R, Brunet L R, Barthold S W, Fikrig E. Borrelia burgdorferi OspA is an arthropod-specific transmission-blocking Lyme disease vaccine. J Exp Med. 1996;183:271–275. doi: 10.1084/jem.183.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Hage N, Babb K, Carroll J A, Lindstrom N, Fischer E R, Miller J C, Gilmore R D, Jr, Mbow M L, Stevenson B. Surface exposure and protease insensitivity of Borrelia burgdorferi Erp (OspEF-related) lipoproteins. Microbiology. 2001;147:821–830. doi: 10.1099/00221287-147-4-821. [DOI] [PubMed] [Google Scholar]
- 17.El-Hage N, Lieto L D, Stevenson B. Stability of erp loci during Borrelia burgdorferi infection: recombination is not required for chronic infection of immunocompetent mice. Infect Immun. 1999;67:3146–3150. doi: 10.1128/iai.67.6.3146-3150.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fingerle V, Hauser U, Liegl G, Petko B, Preac-Mursic V, Wilske B. Expression of outer surface proteins A and C of Borrelia burgdorferi in Ixodes ricinus. J Clin Microbiol. 1995;33:1867–1869. doi: 10.1128/jcm.33.7.1867-1869.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuchs R, Jauris S, Lottspeich F, Preac-Mursic V, Wilske B, Soutschek E. Molecular analysis and expression of a Borrelia burgdorferi gene encoding a 22-kDa protein (pC) in Escherichia coli. Mol Microbiol. 1992;6:503–509. doi: 10.1111/j.1365-2958.1992.tb01495.x. [DOI] [PubMed] [Google Scholar]
- 20.Gilmore R D, Jr, Mbow M L. Conformational nature of the Borrelia burgdorferi B31 outer surface protein C protective epitope. Infect Immun. 1999;67:5463–5469. doi: 10.1128/iai.67.10.5463-5469.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hellwage J, Meri T, Heikkilä T, Alitalo A, Panelius J, Lahdenne P, Seppälä I J T, Meri S. The complement regulatory factor H binds to the surface protein OspE of Borrelia burgdorferi. J Biol Chem. 2001;276:8427–8435. doi: 10.1074/jbc.M007994200. [DOI] [PubMed] [Google Scholar]
- 22.Indest K J, Philipp M T. DNA-binding proteins possibly involved in regulation of the post-logarithmic-phase expression of lipoprotein P35 in Borrelia burgdorferi. J Bacteriol. 2000;182:522–525. doi: 10.1128/jb.182.2.522-525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly R. Cultivation of Borrelia hermsi. Science. 1971;173:443–444. doi: 10.1126/science.173.3995.443. [DOI] [PubMed] [Google Scholar]
- 24.Lam T T, Nguyen T-P K, Montgomery R R, Kantor F S, Fikrig E, Flavell R A. Outer surface proteins E and F of Borrelia burgdorferi, the agent of Lyme disease. Infect Immun. 1994;62:290–298. doi: 10.1128/iai.62.1.290-298.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leuba-Garcia S, Martinez R, Gern L. Expression of outer surface proteins A and C of Borrelia afzelii in Ixodes ricinus ticks and in the skin of mice. Zentbl Bakteriol. 1998;287:475–484. doi: 10.1016/s0934-8840(98)80187-4. [DOI] [PubMed] [Google Scholar]
- 26.Marconi R T, Samuels D S, Garon C F. Transcriptional analyses and mapping of the ospC gene in Lyme disease spirochetes. J Bacteriol. 1993;175:926–932. doi: 10.1128/jb.175.4.926-932.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Margolis N, Hogan D, Tilly K, Rosa P A. Plasmid location of Borrelia purine biosynthesis gene homologs. J Bacteriol. 1994;176:6427–6432. doi: 10.1128/jb.176.21.6427-6432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Margolis N, Samuels D S. Proteins binding to the promoter region of the operon encoding the major outer surface proteins OspA and OspB of Borrelia burgdorferi. Mol Biol Rep. 1995;21:159–164. doi: 10.1007/BF00997238. [DOI] [PubMed] [Google Scholar]
- 29.Mathiesen M J, Holm A, Christiansen M, Blom J, Hansen K, Østergaard S, Theisen M. The dominant epitope of Borrelia garinii outer surface protein C recognized by sera from patients with neuroborreliosis has a surface-exposed conserved structural motif. Infect Immun. 1998;66:4073–4079. doi: 10.1128/iai.66.9.4073-4079.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mbow M L, Gilmore R D, Jr, Titus R G. An OspC-specific monoclonal antibody passively protects mice from tick-transmitted infection by Borrelia burgdorferi B31. Infect Immun. 1999;67:5470–5472. doi: 10.1128/iai.67.10.5470-5472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller J C, El-Hage N, Babb K, Stevenson B. Borrelia burgdorferi B31 Erp proteins that are dominant immunoblot antigens of animals infected with isolate B31 are recognized by only a subset of human Lyme disease patient sera. J Clin Microbiol. 2000;38:1569–1574. doi: 10.1128/jcm.38.4.1569-1574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen T-P K, Lam T T, Barthold S W, Telford S R, Flavell R A, Fikrig E. Partial destruction of Borrelia burgdorferi within ticks that engorged on OspE- or OspF-immunized mice. Infect Immun. 1994;62:2079–2084. doi: 10.1128/iai.62.5.2079-2084.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Obonyo M, Munderloh U G, Fingerle V, Wilske B, Kurtti T J. Borrelia burgdorferi in tick cell culture modulates expression of outer surface proteins A and C in response to temperature. J Clin Microbiol. 1999;37:2137–2141. doi: 10.1128/jcm.37.7.2137-2141.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pollack R J, Telford S R, Spielman A. Standardization of medium for culturing Lyme disease spirochetes. J Clin Microbiol. 1993;31:1251–1255. doi: 10.1128/jcm.31.5.1251-1255.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramamoorthy R, Philipp M T. Differential expression of Borrelia burgdorferi proteins during growth in vitro. Infect Immun. 1998;66:5119–5124. doi: 10.1128/iai.66.11.5119-5124.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sadziene A, Wilske B, Ferdows M S, Barbour A G. The cryptic ospC gene of Borrelia burgdorferi B31 is located on a circular plasmid. Infect Immun. 1993;61:2192–2195. doi: 10.1128/iai.61.5.2192-2195.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwan T G, Burgdorfer W, Rosa P A. Borrelia. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of Clinical Microbiology. 7th ed. Washington, D.C.: American Society for Microbiology; 1999. pp. 746–758. [Google Scholar]
- 38.Schwan T G, Piesman J. Temporal changes in outer surface proteins A and C of the Lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J Clin Microbiol. 2000;38:382–388. doi: 10.1128/jcm.38.1.382-388.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwan T G, Piesman J, Golde W T, Dolan M C, Rosa P A. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc Natl Acad Sci USA. 1995;92:2909–2913. doi: 10.1073/pnas.92.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stevenson B, Bono J L, Schwan T G, Rosa P. Borrelia burgdorferi Erp proteins are immunogenic in mammals infected by tick bite, and their synthesis is inducible in cultured bacteria. Infect Immun. 1998;66:2648–2654. doi: 10.1128/iai.66.6.2648-2654.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stevenson B, Casjens S, Rosa P. Evidence of past recombination events among the genes encoding the Erp antigens of Borrelia burgdorferi. Microbiology. 1998;144:1869–1879. doi: 10.1099/00221287-144-7-1869. [DOI] [PubMed] [Google Scholar]
- 42.Stevenson B, Porcella S F, Oie K L, Fitzpatrick C A, Raffel S J, Lubke L, Schrumpf M E, Schwan T G. The relapsing fever spirochete Borrelia hermsii contains multiple, antigen-encoding circular plasmids that are homologous to the cp32 plasmids of Lyme disease spirochetes. Infect Immun. 2000;68:3900–3908. doi: 10.1128/iai.68.7.3900-3908.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stevenson B, Schwan T G, Rosa P A. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect Immun. 1995;63:4535–4539. doi: 10.1128/iai.63.11.4535-4539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stevenson B, Tilly K, Rosa P A. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J Bacteriol. 1996;178:3508–3516. doi: 10.1128/jb.178.12.3508-3516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stevenson B, Zückert W R, Akins D R. Repetition, conservation, and variation: the multiple cp32 plasmids of Borrelia species. J Mol Microbiol Biotechnol. 2000;2:411–422. [PubMed] [Google Scholar]
- 46.Suk K, Das S, Sun W, Jwang B, Barthold S W, Flavell R A, Fikrig E. Borrelia burgdorferi genes selectively expressed in the infected host. Proc Natl Acad Sci USA. 1995;92:4269–4273. doi: 10.1073/pnas.92.10.4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sung S Y, McDowell J V, Carlyon J A, Marconi R T. Mutation and recombination in the upstream homology box-flanked ospE-related genes of the Lyme disease spirochetes result in the development of new antigenic variants during infection. Infect Immun. 2000;68:1319–1327. doi: 10.1128/iai.68.3.1319-1327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tilly K, Casjens S, Stevenson B, Bono J L, Samuels D S, Hogan D, Rosa P. The Borrelia burgdorferi circular plasmid cp26: conservation of plasmid structure and targeted inactivation of the ospC gene. Mol Microbiol. 1997;25:361–373. doi: 10.1046/j.1365-2958.1997.4711838.x. [DOI] [PubMed] [Google Scholar]
- 49.Tilly K, Elias A F, Bono J L, Stewart P, Rosa P. DNA exchange and insertional inactivation in spirochetes. J Mol Microbiol Biotechnol. 2000;2:433–442. [PubMed] [Google Scholar]
- 50.Wallich R, Brenner C, Kramer M D, Simon M M. Molecular cloning and immunological characterization of a novel linear-plasmid-encoded gene, pG, of Borrelia burgdorferi expressed only in vivo. Infect Immun. 1995;63:3327–3335. doi: 10.1128/iai.63.9.3327-3335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wallich R, Moter S E, Simon M M, Ebnet K, Heiberger A, Kramer M D. The Borrelia burgdorferi flagellum-associated 41-kilodalton antigen (flagellin): molecular cloning, expression, and amplification of the gene. Infect Immun. 1990;58:1711–1719. doi: 10.1128/iai.58.6.1711-1719.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilske B, Preac-Mursic V, Jauris S, Hofmann A, Pradel I, Soutschek E, Schwab E, Will G, Wanner G. Immunological and molecular polymorphisms of OspC, an immunodominant major outer surface protein of Borrelia burgdorferi. Infect Immun. 1993;61:2182–2191. doi: 10.1128/iai.61.5.2182-2191.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilske B, Preac-Mursic V, Schierz G, Busch K V. Immunochemical and immunological analysis of European Borrelia burgdorferi strains. Zentbl Bakteriol Hyg A. 1986;263:92–102. doi: 10.1016/s0176-6724(86)80108-0. [DOI] [PubMed] [Google Scholar]
- 54.Yang X, Goldberg M S, Popova T G, Schoeler G B, Wikel S K, Hagman K E, Norgard M V. Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi. Mol Microbiol. 2000;37:1470–1479. doi: 10.1046/j.1365-2958.2000.02104.x. [DOI] [PubMed] [Google Scholar]