ABSTRACT
There are limited studies on the antibiotic resistance patterns of slowly growing mycobacteria (SGM) species and their related gene mutations in Iran. This study aimed to elucidate the antibiotic susceptibility profiles and the mutations in some genes that are associated with the antibiotic resistance among SGM isolates from Iran. The SGM strains were isolated from sputum samples of suspected tuberculosis (TB) patients. SGM species were identified by standard phenotypic tests and were assigned to species level by amplification and sequencing of the dnaK gene. The minimum inhibitory concentration (MIC) of eight antibiotics was determined using broth microdilution method. The mutations in rrl, rpoB, gyrA, and gyrB genes were investigated in clarithromycin, rifampin, and moxifloxacin resistant isolates using sequencing method. A total of 77 SGM isolates including 46 (59.7%) Mycobacterium kansasii, 21 (27.3%) Mycbacterium simiae, and 10 (13%) Mycobacterium avium complex (MAC) were detected. The amikacin and linezolid with the susceptibility rates of 97.4% and 1.3% were the most and the least effective antibiotics, respectively. All MAC and M. simiae isolates, and 32 (69.6%) M. kansasii strains had multiple-drug resistance (MDR) profiles. The rrl, rpoB, gyrA, and gyrB genes showed various mutations in resistant isolates. Although the current study showed an association among resistance to the clarithromycin, rifampin, and moxifloxacin with mutations in the relevant genes, further research using the whole-genome sequencing is needed to provide a clearer insight into the molecular origins of drug resistance in SGM isolates.
KEYWORDS: Antibiotic resistance, nontuberculous mycobacteria, mutations, slowly growing mycobacteria, Iran
Introduction
Non-tuberculous mycobacteria (NTM) comprising more than 200 species which are found throughout the world. In routine diagnostic methods, such as acid-fast staining, NTM have a similar shape to Mycobacterium tuberculosis. Thus, preliminary differentiating them from each other has always been a serious challenge for health-care systems and medical diagnostic laboratories [1,2]. Iran is one of the countries where mycobacterial infections are endemic, therefore, the detection of different NTM in medical laboratories is necessary [3]. The results of one study in Iran showed that an unpredicted number of patients (15.1%) who were diagnosed and treated for TB were infected by NTM [4]. Many multi-drug resistants (MDR), extensively drug resistant (XDR), and extremely drug resistant TB (XXDR TB), may be a member of NTM [5]. One important factor that involved in the misdiagnosis of these bacteria is the lack of new and advanced diagnostic technologies in developing countries.
Microbiologically, NTM are categorized into rapidly growing mycobacteria (RGM) and slowly growing mycobacteria (SGM). In comparison to RGM, SGM are more frequent and show higher rates of drug resistance. Several species including M. kansasii, M. simiae, and M. avium complex (MAC) are belonged to SGM [6–8]. Despite the existence of detailed epidemiological information about TB in Iran, the prevalence and epidemiology of SGM in Iran remains largely unknown.
Although there is no standard treatment criterion for NTM infections at the moment, several antibiotics have been recommended for the treatment of these infections, including amikacin (AMK), moxifloxacin (MXF), clarithromycin (CLR), clofazimine (CFZ), ethambutol (EMB), isoniazid (INH), linezolid (LZD), and rifampin (RIF) [8,9]. The British Thoracic Society recently published a set of guidelines for the treatment of NTM pulmonary infections. The main consensus recommendation is to use a macrolide-based multidrug regimen to treat MAC-related pulmonary disease. Because surgical lung resection is frequently the only solution for patients who failed drug therapy due to antibiotic resistance, the prescription of multidrug therapy is critical to avoiding macrolide resistance and preventing unnecessary deaths in more severe cases. Rifamycins, like rifampin, complete the NTM treatment bases and are an important part of the M. kansasii treatment bases. Fluoroquinolones have significant activity against M. kansasii and are critical in cases of rifamycin-resistant M. kansasii. Clofazimine also has potential applications against NTM skin and soft tissue infections due to its high penetration in skin and soft tissue [9]. However, due to their extensive range of antibiotic resistance, the treatment of infections caused by these bacteria is a major challenge for health-care centers. Patient age, infecting species, and comorbidities determine the NTM infection remission rate [6].
Different mechanisms are contributed to wide range of antibiotic resistance in NTM species. While in many bacteria, acquired drug resistance is broadly obtained through horizontal gene transfer of mobile genetic elements, NTM species resist to various antibiotics by spontaneous mutations in chromosomal genes [7,10,11]. So far, the association of certain antibiotic resistance with mutation in different genes, such as rpoB (rifampin), katG and inhA (isoniazid), rrl (clarithromycin), embA and embB (ethambutol), gyrA and gyrB (fluoroquinolone), and Rv0678 (clofazimine) has been reported in mycobacteria [10,12].
There are limited studies on the antibiotic resistance patterns of SGM species and their related gene mutations, especially in Iran. Hence, this study aimed to elucidate the antibiotic susceptibility profiles and the mutations in some genes that are associated with this resistance among SGM isolates from Iran.
Material and methods
Ethics statement
This study was approved by the Ethics Committee of Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran (No: IR.AJUMS.MEDICINE.REC.1399.005). Written informed consent was obtained from all patients.
Mycobacteria isolation
In this cross-sectional study, the SGM isolates were collected from sputum samples of suspected TB patients (with symptoms of tuberculosis) referred to several Regional Tuberculosis Reference Laboratories of Iran, from May 2017 to August 2020. In this study, clinical, radiographic, and microbiological criteria were included for identifying NTM pulmonary infection on the basis of the American Thoracic Society (ATS)/Infectious Diseases Society of America (IDSA) guidelines [13]. Based on the systematic review and meta-analysis by Nasiri et al. [3], the prevalence of NTM species ranged from 5.6 to 19.2% after year 2000. Considering the minimum prevalence of 5.0%, 95% confidence level, and margin of error of 0.05%, the sample size was estimated approximately 70 isolates.
The demographic data were recorded and stored in archive of the TB centers. Early morning sputa were collected in a sterile container from each patient and stored at −20°C and later used for culture and molecular identification. After performing preliminary steps such as decontamination and homogenization, the specimens were cultured on Lowenstein–Jensen (LJ) medium (Sigma-Aldrich Co, USA). All mycobacterial positive cultures suspected to SGM were sent to the Ahvaz TB Reference Center for identification of species. For preparation of a fresh subculture, LJ medium was used and after incubation at 37°C, the colony was used for phenotypic and molecular tests.
Primary identification of SGM species
SGM were identified to species level according to the guidelines of the ATS/IDSA [13]. The phenotypic tests of acid-fast staining, growth rate, pigment production, clonal morphology, and conventional biochemical tests including nitrate reduction, Tween 80 hydrolysis, catalase production, and arylsulfatase test were used. M. avium ATCC® 700,898 was used as quality reference strains.
Confirmation and differentiation of SGM by dnaK gene sequencing
The SGM isolates were assigned to species level by amplification and sequencing of the dnaK gene. The DNA was extracted from the isolates by boiling method as previously described [14].
Polymerase chain reaction (PCR) mixture was prepared in a final volume of 25 μL comprising 10X PCR buffer, 1.5 mmol of MgCl2, 0.2 mmol of each dNTPs, 1 U/μL of Taq DNA Polymerase, 1 μL of each primer (F:5′-CTGACCAAGGACAAGATGGC-3′ and R: 5′-TCGATCAGCTTGGTCATCAC −3′), which amplify a DNA fragment of 451 bp [15], 5 μL DNA template (50 ng), and 18 μl sterile deionized water.
The PCR program was carried out in a thermocycler (Eppendorf, Hamburg, Germany) as follows: initial denaturation for 1 min at 95°C, followed by 30 cycles of denaturation at 95°C for 30 sec, annealing at 55°C for 30 sec, extension at 72°C for 30 sec, and a final extension for 2 min at 72°C [15]. PCR products were separated by electrophoresis on 1% agarose gel and visualized by visualized under UV light using a gel documentation system (Protein Simple, USA). The size of PCR products was determined using a 100-bp molecular marker.
PCR products were sent for sequencing (Cardiogenetic Research Center, Tehran, Iran). The resulting sequences were searched in the GenBank (https://blast.ncbi.nlm.nih.gov/) database and the species identification was confirmed based on the percentage of similarity in comparison with the sequences in the database. The dendrogram (the maximum likelihood tree) was created by neighbor-joining (NJ) method with 1000 bootstrap replications using Mega X software [16].
Determination of antibiotic resistance patterns
Considering the importance of rifampin and MXF in the treatment of M. kansasii and physicians’ recommendation to macrolide-based diet for effective treatment of NTM infections, RIF (rpoB) and MXF (gyrA and gyrB) antibiotic resistance genes for dominant SGM (M. kansasii) and CLR (rrl) gene for all resistant isolates were evaluated using PCR and sequencing.
MIC of eight antibiotics was determined against SGM isolates using broth microdilution method according to the protocol provided by the Clinical and Laboratory Standards Institute (CLSI) guidelines [17]. As no defined breakpoint has mentioned for all NTMs by CLSI so far, and only there are breakpoints for few species of SGM in literature, the experiments of the susceptibility testing for other antibiotics that not mentioned by CLSI, were done and interpreted according to breakpoints for the M. kansasii and MAC isolates or based on the previously described studies [18,19]. The antibiotic powders (Sigma-Aldrich Co, USA) were as follows: AMK, CLR, CFZ, EMB, INH, LZD, MXF, and RIF. After preparing the stock solution of each antibiotic in its solvent as per provider’s instructions, and an appropriate amount was added to Middlebrook 7H9 broth medium (Liofilchem Srl, Italy) containing glycerol (2.0 ml/L) and oleic acid/dextrose/catalase (OADC; 100 ml/L). Serial double dilutions were prepared as follows: AMK, 0.5 to 16 μg/ml; MXF, 0.25 to 16 μg/ml; CLR, 0.5 to 32 μg/ml; LZD, 0.5 to 32 μg/ml; RIF, 0.125 to 16 μg/ml; EMB, 0.25 to 16 μg/ml; CFZ, 2 to 64 μg/ml; INH, 0.5 to 32 μg/ml.
The bacterial suspensions were prepared according to the CLSI guidelines [17], for each isolate a suspension in the broth was prepared from an agar medium with adequate growth of the isolate and their turbidity were adjusted to a 0.5 McFarland standard turbidity suspension to achieve a final concentration of 5 × 105 CFU/ml. Suspensions were then diluted to achieve a final concentration from 1 × 105 to 5 × 105 CFU/ml and were inoculated in each well of microtiter plates containing 100 μl of 7H9 broth supplemented with OADC and serial dilutions of antimicrobial agents, and incubated at 37 oC for 14 days. After incubation time, the MIC was read for each antibiotic, and the lowest antibiotic concentration that inhibits bacterial growth was considered as the MIC value. For each dilution series, one well containing bacterial suspension without antibiotics was considered as positive control and one well containing antibiotic without bacterial suspension was considered as negative control. MICs were determined 7–14 days after culture when sufficient bacterial growth in positive control was well observed. All experiments were performed in duplicate. MIC breakpoints, representing resistant, intermediate, and sensitive strains against eight studied antibiotics were interpreted based on the CLSI [17]. M. avium ATCC® 700,898 was used as quality reference strain.
Quality control (QC) of laboratory experiments
All experiments were performed in Tuberculosis Reference Laboratory in a special and self-contained room physically separated from other parts under the supervision of trained staff. A calibrated autoclave was available for decontaminating laboratory waste. Directional air flow was maintained by extracting room air. All procedures were undertook under a Class II biosafety cabinet. Gloves and masks were used during the experiments. In each run of culture medium preparation or antibiotic susceptibility experiment, the QC strains were used. Subcultures of working QC strains were performed weekly or whenever susceptibility testing was conducted. Storage of these stock cultures was carried out at –80°C in tryptic soy broth with 15% glycerol. When a new batch of test medium was prepared, QC strains were used. Also, there was a free antimicrobial medium with QC strains in each microdilution broth tray to verify viability of the test organisms.
Study of resistance genes mutations
The resistance genes related to CLR (rrl), RIF (rpoB), and MXF (gyrA and gyrB) antibiotics were evaluated only for phenotypically resistant isolates using PCR method and sequencing. The used primers are shown in Table 1. For rrl and rpoB genes, the PCR was performed by the newly designed primers using primer blast (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). For gyrA and gyrB genes, the previously described primers were used [20]. PCR mixture was prepared in a final volume of 25 μl comprising 10X PCR buffer, 1.5 mmol of MgCl2, 0.2 mmol of each dNTPs, 1 U/μl of Taq DNA Polymerase, 1 μl of each primer, 2 μl genomic DNA (20 ng), and 8.5 μl sterile deionized water. The PCR program was carried out in a thermocycler (Eppendorf, Hamburg, Germany) as follows: 95°C for 5 min, 35 cycles of denaturation at 95°C for 30 sec, annealing at optimal temperature of each primer for 30 sec, extension at 72°C for 40 sec (rpoB, gyrA, and gyrB) and 1 min (rrl), and a final extension at 72°C for 5 min [15]. The PCR products were then visualized by electrophoresis on 1% agarose gel. A 100 bp molecular size marker was used to determine the size of produced fragments. Amplified products were sent for sequencing to investigate possible mutations in genes that may be associated with antibiotic resistance. The obtained sequences were aligned with the homologous sequences of the reference strain of M. tuberculosis (H37Rv) and M. kansasii ATCC12478 in MEGA X software.
Table 1.
The primers sequences used in this study.
| Antibiotic | Gene | Nucleotide sequence (5’-3’) | Annealing temperature | Product length (bp) | Reference |
|---|---|---|---|---|---|
| Clarithromycin | rrl | F: CGGGAWYCGGYCGCAGAAC R: CCAGGTCTGGCCTATCRAWC |
60 | 1110 | Designed in this study |
| Rifampin | rpoB | F: GGGAACGGATGACCACTCA R: GCGGTACGGCGTCTCGATGAAG |
53 | 350 | Designed in this study |
| Moxifloxacin | gyrA | F: ATTCTGCCGAACGGATCGAG R: CGACCGCGTTATCCGAATTG |
51 | 459 | [20] |
| gyrB | F: TGGGCAACACCGAGGTGAAG R: ACGGGTCCATGGTGGTTTCC |
70 | 762 |
Statistical analysis
Descriptive statistics were used to express the frequency and percentage of strains and resistance of each antibiotic using Statistical Package for the Social Sciences (SPSS) software version 22.0 (IBM Corporation, Armonk, NY, USA).
Results
SGM isolates identification
In total 77 SGM isolates were detected using phenotypic and standard biochemical methods. All of these isolates were positive for the 451 bp dnaK gene and confirmed as NTM. The demographic information for 40 patients who infected with SGM isolates are presented in Table 2. The isolates were recovered from samples belonged to 31 (77.5%) male patients and 9 (22.5%) female patients with the mean age of 54.5 years (ranged 28–81 years). Most patients had no underlying medical conditions and in 42% of them fever was the only symptom. Due to the confidentiality of information on the TB suspected patients in Iran, demographic data for some isolates was not accessible. The dnaK gene sequencing differentiated the isolates as follows: 46 (59.7%) M. kansasii, 21 (27.3%) M. simiae (Runyon Group I), and 10 (13%) M. avium complex (Runyon Group III) (Figure 1).
Table 2.
Demographic data of some patients infected with slowly growing mycobacteria.
| Characteristics | Identification by dnaK |
|
|---|---|---|
|
M. kansasii No. of strains (%) |
M. simiae No. of strains (%) |
|
| Age | ||
| 20–39 | 5 (16 %) | 2 (22 %) |
| 40–59 | 13 (41 %) | 5 (55 %) |
| ≥60 | 13 (41 %) | 2 (22 %) |
| Gender | ||
| Female | 7 (22 %) | 2 (22 %) |
| Male | 24 (77 %) | 7 (77 %) |
| Previous medical history | ||
| Treated tuberculosis | 3 (9 %) | 1 (11 %) |
| *HIV infection | 6 (9 %) | 0 (0%) |
| Open heart surgery | 2 (6 %) | 0 (0%) |
| COPD | 1 (3 %) | 0 (0%) |
| Immunocompromised | 1 (3 %) | 0 (0%) |
| Renal failure | 2 (6 %) | 0 (0%) |
| Diabetic | 2 (6 %) | 0 (0%) |
| Smoking | 1 (3 %) | 0 (0%) |
| Sarcoma | 1 (3 %) | 0 (0%) |
| Pemphigus Vulgaris | 1 (3 %) | 0 (0%) |
| Normal | 11 (35%) | 8 (88 %) |
| Main symptoms | ||
| Fever | 10 (32 %) | 7 (77 %) |
| Fever, cough | 10 (32 %) | 2 (22 %) |
| Fever, inflammation and tenderness in joint | 1 (3 %) | 0 (0%) |
| Productive cough, fever, body weight loss | 3 (9 %) | 0 (0%) |
| Productive cough | 2 (6 %) | 0 (0%) |
| Local pain, small pale nodule | 2 (6 %) | 0 (0%) |
| Productive cough, chest wall pain & weight loss | 1 (3 %) | 0 (0%) |
| Urinary tract infection | 1 (3.2%) | 0 (0%) |
| Local abscess and discharge | 1 (3.2%) | 0 (0%) |
* HIV, human immunodeficiency virus.
Figure 1.
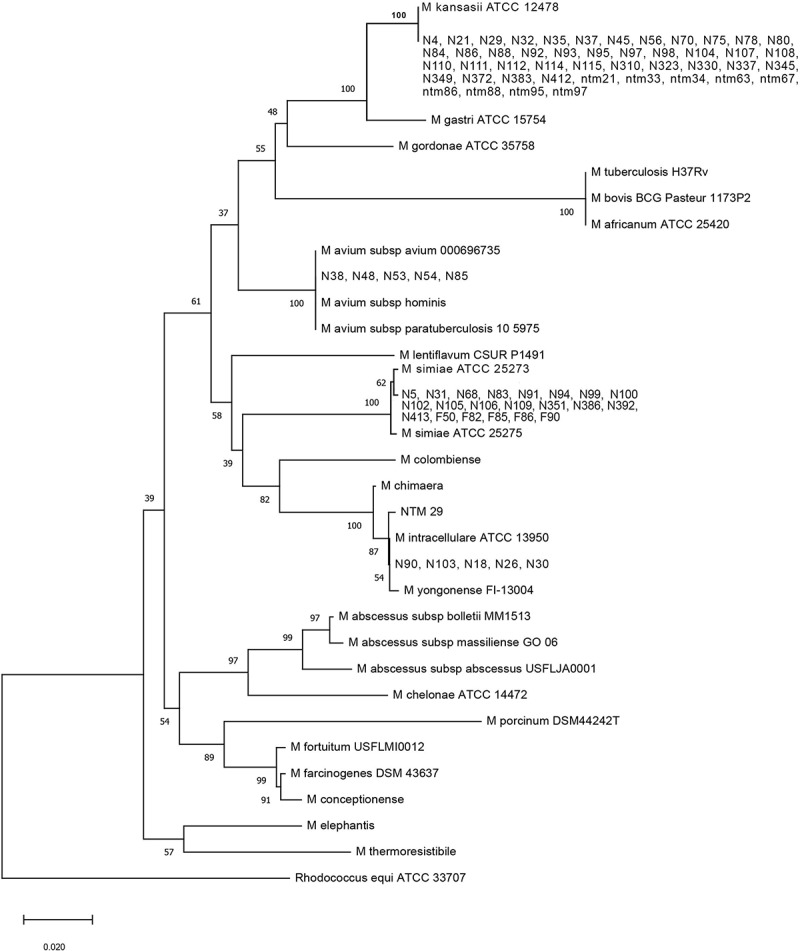
A phylogenetic tree based on dnaK gene sequences demonstrates the relationship between the 77 mycobacterium isolates and 1 out-group strain. The tree was created using the neighbor-joining method, and bootstrap analysis was performed from 100 replications.
Antibiotic resistance profiles
The antibiotic resistance rates of SGM isolates detected in present study, are shown in Table 3. According to the broth microdilution results, the AMK and LZD with the susceptibility rates of 97.4% and 1.3% were the most and the least effective antibiotics, respectively. All 77 isolates were resistant to at least one antibiotic, for instance, high resistance of 100% was observed in M. simiae strains against LZD, RIF, INH, and CFZ, and for MAC strains the full resistance against LZD, RIF, and INH was also noted (Table 3). High susceptibility to AMK was observed in all studied species with MIC range of 0.5–4 μg/ml (Table 4). In total, 81.8% of studied isolates were MDR. All MAC and M. simiae isolates, and 32 (69.56%) M. kansasii strains, comprised MDR profiles, indicating resistance to three or more antibiotic classes [10].
Table 3.
The resistance rates of 77 slowly growing mycobacteria isolates against eight antibiotics using the broth microdilution method.
| SGM species | Antibiotics | Antibiotics breakpoints |
MIC50 | MIC90 | Susceptibility profile |
||||
|---|---|---|---|---|---|---|---|---|---|
| Susceptible | Intermediate | Resistant | S (%) | I (%) | R (%) | ||||
|
M. kansasii (n = 46) |
RIF | ≤ 1 | - | ≥ 2 | 1.0 | 16.0 | 26 (56.5) | 0 (0.0) | 20 (43.5) |
| MXF | ≤ 1 | 2 | ≥ 4 | 0.25 | 4.0 | 30 (65.2) | 10 (21.7) | 6 (13.0) | |
| CLR | ≤ 8 | 16 | ≥ 32 | 16.0 | ≥ 32 | 18 (39.1) | 20 (43.5) | 8 (17.4) | |
| EMB | ≤ 2 | 4 | ≥ 8 | 4.0 | 16.0 | 24 (52.1) | 8 (17.4) | 14 (30.4) | |
| AMK | ≤ 16 | 32 | ≥ 64 | 1.0 | 4.0 | 46 (100) | 0 (0.0) | 0 (0.0) | |
| LZD | ≤ 8 | 16 | ≥ 32 | ≥ 32 | ≥ 32 | 1 (2.2) | 4 (8.6) | 41 (89.1) | |
| INH | ≤ 0.5 | - | ≥1 | ≥ 32 | ≥ 32 | 1 (2.2) | 0 (0.0) | 45 (97.8) | |
| CFZ | - | - | ≥1 | ≥ 64 | ≥ 64 | 12 (26.0) | 0 (0.0) | 34 (73.9) | |
|
M. simiae (n = 21) |
RIF | ≤ 1 | - | ≥ 2 | ≥ 16 | ≥ 16 | 0 (0.0) | 0 (0.0) | 21 (100) |
| MXF | ≤ 1 | 2 | ≥ 4 | 2.0 | 16.0 | 6 (28.5) | 5 (23.8) | 10 (47.6) | |
| CLR | ≤ 8 | 16 | ≥ 32 | 16.0 | ≥ 32 | 4 (19.0) | 12 (57.1) | 5 (23.8) | |
| EMB | ≤ 2 | 4 | ≥ 8 | 16.0 | 16.0 | 10 (47.6) | 0 (0.0) | 11 (52.4) | |
| AMK | ≤ 16 | 32 | ≥ 64 | 1.0 | 4.0 | 19 (90.4) | 0 (0.0) | 2 (9.5) | |
| LZD | ≤ 8 | 16 | ≥ 32 | ≥ 32 | ≥ 32 | 0 (0.0) | 0 (0.0) | 21 (100) | |
| INH | ≤ 0.5 | - | ≥1 | ≥ 32 | ≥ 32 | 0 (0.0) | 0 (0.0) | 21 (100) | |
| CFZ | - | - | ≥ 1 | ≥ 64 | ≥ 64 | 0 (0.0) | 0 (0.0) | 21 (100) | |
|
MAC (n = 10) |
RIF | ≤ 1 | - | ≥ 2 | 4.0 | 16.0 | 0 (0.0) | 0 (0.0) | 10 (100) |
| MXF | ≤ 1 | 2 | ≥ 4 | ≤ 0.25 | 2.0 | 7 (70.0) | 2 (20.0) | 1 (10.0) | |
| CLR | ≤ 8 | 16 | ≥ 32 | 8.0 | 16.0 | 7 (70.0) | 2 (20.0) | 1 (10.0) | |
| EMB | ≤ 2 | 4 | ≥ 8 | 2.0 | 8.0 | 6 (60.0) | 1 (10.0) | 3 (30.0) | |
| AMK | ≤ 16 | 32 | ≥ 64 | ≤ 0.5 | 4 | 10 (100) | 0 (0.0) | 0 (0.0) | |
| LZD | ≤ 8 | 16 | ≥ 32 | ≥ 32 | ≥ 32 | 0 (0.0) | 0 (0.0) | 10 (100) | |
| INH | ≤ 0.5 | - | ≥ 1 | ≥ 32 | ≥ 32 | 0 (0.0) | 0 (0.0) | 10 (100) | |
| CFZ | - | - | ≥ 1 | 16.0 | ≥ 64 | 3 (30) | 0 (0.0) | 7 (70) | |
|
Total resistant n (%) |
RIF | MXF | CLR | EMB | AMK | LZD | CFZ | INH | |
| 51 (66.2) | 17 (22.1) | 14 (18.2) | 26 (33.8) | 2 (2.6) | 76 (98.7) | 68 (88.3) | 58 (75.3) | ||
S: susceptible; I: intermediate; R: resistant; RIF: rifampin; MXF: moxifloxacin; CLR: clarithromycin; EMB: ethambutol; AMK: amikacin; LZD: linezolid; CFZ: clofazimine; INH: isoniazid.
Table 4.
The MIC (µg/ml) ranges of different antibiotics in various slowly growing mycobacterial species.
| Antibiotics | RIF | MXF | CLR | EMB | AMK | LZD | CFZ | INH |
|---|---|---|---|---|---|---|---|---|
|
M. kansasii MIC: n (%) |
0.125≥: 3 (6.5) 0.25: 7 (15.2) 1: 16 (34.7) 2: 12 (26.0) 4: 1 (2.1) 8: 1 (2.1) 16≤: 6 (13.0) |
0.25≥: 24 (52.1) 0.5: 2 (4.3) 1: 4 (8.6) 2: 10 (21.7) 4: 2 (4.3) 16: 4 (8.6) |
0.5≥: 11(23.9) 1: 1 (2.1) 4: 2 (4.3) 8: 4 (8.6) 16: 20 (43.4) 32≤: 8 (17.3) |
0.125 ≥: 2 (4.3) 0.25: 2 (4.3) 0.5: 2 (4.3) 1: 9 (19.5) 2: 7 (15.2) 4: 10 (21.7) 16: 14 (30.4) |
0.5≥: 14 (21.7) 1: 23 (50) 2: 2 (4.3) 4: 7 (15.2) |
0.5≥: 1 (2.1) 16≤:45(97.8) |
2≥:12(26.0) 8: 4 (8.6) 64≤:30(65.2) |
0.5≥: 1 (2.1) 1: 2 (4.3) 2: 11 (23.9) 4: 3 (6.5) 8: 4 (8.6) 16: 8 (17.3) 32≥: 17 (36.9) |
|
M. simiae MIC: n (%) |
16 | 0.25≥: 4 (19.0) 1: 2 (9.5) 2: 5 (23.8) 16: 10 (47.6) |
0.5≥:2(9.5) 8: 2 (9.5) 16: 12 (57.1) 32≤: 5 (23.8) |
0.125: 1 (4.7) 1: 2 (9.5) 2: 7 (33.3) 16: 11 (52.3) |
1: 18 (85.7) 4: 1 (4.7) 32≤: 2 (9.5) |
16< | 64: 6 (28.5) 64<: 15 (71.4) |
32< |
|
MAC MIC: n (%) |
2: 4 (40) 4: 1 (10) 8: 1 (10) 16≤: 4 (40) |
0.25≥: 6 (60) 1: 1 (10) 2: 2 (20) 16≤: 1 (10) |
0.5≥:2(20) 4: 2 (20) 8: 3 (30) 16: 2 (20) 32≤: 1 (10) |
0.125 ≥: 1 (10) 0.5: 1 (10) 2: 4 (40) 4: 1 (10) 8: 2 (20) 16: 1 (10) |
0.5≥: 6 (60) 1: 1 (10) 2: 1 (10) 4: 2 (20) |
16< | 2≥: 3 (30) 4: 1 (10) 16: 4 (40) 64≤: 2 (20) |
2: 1 (10) 4: 1 (10) 32≥: 8 (80) |
MIC: minimum inhibitory concentration; RIF: rifampin; MXF: moxifloxacin; CLR: clarithromycin; EMB: ethambutol; AMK: amikacin; LZD: linezolid; CFZ: clofazimine; INH: isoniazid; MAC: Mycobacterium avium complex.
Mutation analysis related to antibiotic resistance
The results of mutation analysis are presented in Tables 5–8. All phenotypically resistant isolates comprised mutations in their resistance determinants simultaneously, except for 17 RIF-resistant M. kansasii isolates. Six mutation types were detected at positions 1249, 1356, 1407, 1479, 1533, and 1536 of the 350-bp fragment of rpoB gene except for 17 RIF-resistant M. kansasii isolates (Table 5). The mutations were detected in 350 bp fragment of rpoB gene in 3 RIF-resistant M. kansasii isolates (Table 6). Moreover, all MXF-resistant M. kansasii, had mutations in their gyrA and gyrB genes. The 459 bp fragment of gyrA gene showed mutations at positions 238, 239, 247,249, 257,258, 260,261, 2449, 2450, and 2452 in MXF-resistant M. kansasii. Also, 762 bp fragment of gyrB gene had mutations at positions 602,603, 950, 1332, 1339, 1390,1392, 1406, and 1594 in MXF-resistant M. kansasii isolates (Table 7). The eight CLR-resistant M. kansasii isolates showed mutations at positions 2058, 2059, and 2266 of 1110 bp fragment of rrl gene.
Table 5.
The frequency of genotypic resistance determinants in phenotypically resistance isolates.
| SGM species | Antibiotics | Phenotypic resistance n (%) |
Genotypic resistance determinants |
|
|---|---|---|---|---|
| Without mutation n (%) | With mutation n (%) | |||
| M. kansasii (n = 46) | RIF | 20 (43.5) | 17 (85.0) | 3 (15.0%) |
| MXF | 6 (13.0) | 0 (0.0) | 6 (100.0) | |
| CLR | 8 (17.4) | 0 (0.0) | 8 (100.0) | |
| M. simiae (n = 21) | CLR | 5 (23.8) | 0 (0.0) | 5 (100.0) |
| MAC (n = 10) | CLR | 1 (10.0) | 0 (0.0) | 1 (100.0) |
RIF: rifampin; MXF: moxifloxacin; CLR: clarithromycin; MAC: Mycobacterium avium complex; SGM: slowly growing mycobacteria.
Table 6.
Gene mutations in Rifampin-resistant isolates.
| Isolates | Gene | Nucleotide (codon) | Mutation | Amino acid change | N of mutants |
|---|---|---|---|---|---|
| M. kansasii | rpoB | 1249 (417) | A→C | Asparagine → Histidine | 1 |
| 1356 (452) | G→C | - | 1 | ||
| 1407 (469) | A→G | - | 1 | ||
| 1479 (493) | C→T | - | 1 | ||
| 1533 (511) | G→A | - | 3 | ||
| 1536 (512) | G→C | - | 2 |
Table 7.
Gene mutations in Moxifloxacin-resistant isolates.
| Isolates | Gene | Nucleotide (codon) | Mutation | Amino acid change | N of mutants |
|---|---|---|---|---|---|
| M. kansasii | gyrA | 238,239 (80) | ACG→GGG | Threonine →Glycine | 1 |
| 247,249 (83) | AAC→CAA | Asparagine →Glutamine | 2 | ||
| 257,258 (86) | CCG→CGA | Proline →Arginine | 1 | ||
| 260,261 (87) | CAC→CGG | Histidine → Arginine | 4 | ||
| gyrB | 2449,2450 (817) | GCG→TGG | Alanine →Tryptophan | 4 | |
| 2452 (818) | CTC→TTC | Leucine -→Phenylalanine | 2 | ||
| 602,603 (201) | AAC→ACG | Asparagine → Threonine | 2 | ||
| 950 (317) | GAA→CGA | Glutamic acid → Glycine | 3 | ||
| 1332 (444) | GAC→GAG | Aspartic acid→Glutamic acid | 2 | ||
| 1339 (447) | TCG→ACG | Serine → Theronine | 2 | ||
| 1390,1392 (464) | GGC→TGT | Glycine → Cysteine | 1 | ||
| 1406 (469) | AGC→AAC | Serine → Asparagine | 2 | ||
| 1594 (532) | GAC→AAC | Aspartic acid → Asparagine | 2 |
Table 8.
Gene mutations in clarithromycin-resistant isolates in rrl gene.
| Isolates | Nucleotide | Mutation | Amino acid change | N of mutants |
|---|---|---|---|---|
| M. kansasii | 2058 | A→C | - | 4 |
| 2058 | A→T | - | 3 | |
| 2059 | A→G | - | 1 | |
| 2266 | A→C | - | 8 | |
| MAC | 2058 | A→G | - | 1 |
| 2059 | A→G | - | 1 | |
| 2212 | C→A | - | 1 | |
| 2419 | A→C | - | 1 | |
| M. simiae | 22 | C→T | - | 2 |
| 25 | C→T | - | 3 | |
| 143 | T→C | - | 5 | |
| 250 | A→T | - | 5 | |
| 261 | G→T | - | 5 | |
| 293 | C→G | - | 5 | |
| 351 | T→C | - | 5 | |
| 477 | C→T | - | 5 | |
| 884 | C→T | - | 4 |
The only CLR-resistant MAC isolate in this study, showed four simultaneous mutations at positions 2058, 2059, 2212, and 2419 of rrl gene. Moreover, the five CLR-resistant M. simiae isolates had mutations at positions 22, 25, 143, 250, 261, 293, 351, 477, and 884 of rrl gene (Table 8).
Discussion
While in-vitro antibiotic susceptibility tests for most NTM species have not been validated, these tests can nevertheless be essential in the management of diseases associated with NTM [21]. The long-term antimicrobial therapy and its related toxicities may pose major problems for clinicians to choose the ideal treatment regimen for SGM-infected patients. There is not adequate information about SGM antibiotic susceptibility profiles in Iran. Because in developing countries such as Iran, it is difficult to obtain commercial kits and modern devices to perform laboratory experiments due to sanctions. Also, the purchase and supply of antibiotic powders, which are mostly imported, are so problematic and time-consuming process. These limitations have made TB laboratories still not prioritize the detection of NTM strains in Iran [22].
Antimicrobial susceptibility testing (AST) in-vitro is the first step in predicting the success or failure of a new antibiotic medication. These assays assess the responsiveness of isolated organisms to a specific antibiotic medication. They are generally inexpensive and rapid, as well as simple to duplicate and scale. The CLSI has developed the most commonly acknowledged technique for AST of NTMs, which has suggested the use of microdilution as the gold standard for determining antibacterial susceptibilities. Other techniques for assessing antimicrobial effectiveness against NTMs are not advised. For example, whereas the proportion method is regularly employed for M. tuberculosis, it frequently gives misleading findings for NTMs; while the epsilometer test is quick and easy, it suffers from lack of repeatability and exaggeration of drug sensitivity as measured by other procedures owing to ellipse tailing; moreover, the agar disk diffusion technique has an inherent problem in interpreting zones of inhibition, particularly when the quantity of drug in the disk is near the drug’s breakpoint [7]. In study of Krishnan et al. [23] MICs of CLR and RIF for 10 MAC strains using the MGIT system and BACTEC 460TB system methods were lower than present findings.
The role of in vitro susceptibility testing for NTM infections is debatable. This is primarily due to the unpredictability of the correlation between in vitro and clinical outcomes: correlation is especially poor for M. simiae, while it is reasonably satisfactory for M. kansasii, and for other species, such as MAC, the correlation holds valid only for particular medications like as macrolides, and not for others. This disparity is most likely the result of multiple causes, ranging from strain selection and testing settings to the absence of host effects in the assays [24]. There are also practical aspects to take into account. An essential characteristic of NTM is that their cell walls are intrinsically hydrophobic; Rather of remaining in the aqueous solution, they typically cling to the surface of the individual wells in the 96-well plates. As a result, just measuring the turbidity of cell suspension might produce inaccurate findings that differ from those obtained from cells present in surface-attached biofilms [25]. Although microdilution remains the most accepted in vitro technique, it does have important weaknesses: a lengthy testing period, the possibility of cross-contamination, a many reagents, the potential of false positives owing to extensive incubation durations, etc [26].
This study investigated the susceptibility patterns of 77 SGM isolates against eight most commonly used antibiotics. Among the studied antimicrobials, the AMK with susceptibility rate of 97.4% was the most effective antibiotic against SGM isolates. In a study by Park et al. [10] from Korea, although AMK was one of the most effective antibiotics against NTM, its resistance (23.3%) was much higher than the current study (2.6%). In this study, all M. kansasii and MAC isolates, and 90.4% of M. simiae species were susceptible to AMK. Almost close results were reported by Liu et al. [27] from China and Wetzstein et al. [28] from Germany.
Another finding of this study, was the resistance of more than 80% of the SGM isolates to each of the LZD and INH antibiotics. These results were in good agreement with the previous reports from China [27], Iran [22], and Ghana [29] that reported a high resistance rate for LZD and INH in some SGM isolates. In our study, all M. simiae and MAC isolates, and 89.1% of M. kansasii strains were resistant to LZD. Also, all M. simiae and MAC isolates, and 97.8% of M. kansasii, isolates were resistant to INH. In parallel with the current findings, Heidarieh et al. [30] from Iran, reported 100% and 90% resistance rates for INH and LZD in M. simiae isolates, respectively. However, the M. kansasii isolates of their study, were completely susceptible to LZD and INH that was differ from our results. Although the INH antibiotic is part of the treatment regimen for M. kansasii, the CLSI does not recommend the reporting the result of sensitivity test for this drug in NTM species. Because its MIC values are not commensurate with the findings observed in the clinical response [31].
CFZ is presently solely registered for leprosy treatment. However, it is also used for the treatment of MDR-TB and in some cases for NTM species [32]. The previous and new editions of CLSI sources do not specify a defined breakpoint for the CFZ. In the current research, most strains had MIC greater than 64 μg/ml, which was higher than expected. This finding was inconsistent with research conducted by Luo et al. [32] who revealed that CFZ had strong effects on most SGM species in vitro. One of the reasons for the discrepancy in these results, is due to lack of widely accepted method or breakpoint for susceptibility testing for various NTM species, and several investigations used different breakpoints for NTM strains [10,18,27].
Another drug of choice recommended for the treatment of MAC infections by ATS/IDSA [13], is EMB. The results showed that 60% of MAC, 52.1% of M. kansasii and 47.6% of M. simiae isolates were susceptible to EMB. Previous study by Zhou et al. [33] from China, reported more resistance rates for EMB against SGM isolates, showing the variability of resistance in different geographical regions.
In the current study, all M. simiae and MAC, and 43.5% of M. kansasii isolates were resistant to RIF, another recommended drug for combination therapy of NTM infections. In comparison with the current results, Heidarieh et al. [30] from Iran, reported lower (77%) and almost equal (50%) resistance rates for M. simiae and M. kansasii isolates, respectively. However, Park et al. [10] from Korea, reported a low resistance rate (12.7%) for RIF in NTM isolates that was inconsistent with the current study. This discrepancy may be due to the different sample sources of two studies.
The most remarkable result to emerge from the current study, was the high efficacy of MXF and CLR toward high proportion of SGM isolates. MXF and CLR with low resistance rates were the most effective antibiotics after AMK. These findings approved the argue of some specialists suggesting a macrolide-based regimen combined with quinolones (preferred MXF rather than ciprofloxacin) and AMK for effective therapy of MDR NTM infections particularly, M. simiae [27]. The low resistance rate of MXF concurred well with the previous report from Korea [10].
In this study, 81.8% of studied isolates were MDR. All MAC and M. simiae isolates, and 69.6% of M. kansasii strains showed these criteria. However, these findings need to be interpreted with caution, because some of the tested antibiotics did not have a defined breakpoint for SGM species by CLSI [17]. Previous reports from Korea [10], Iran [22], Ghana [29], and China [33], indicated the circulation of MDR-NTM isolates in environment and health-care systems. The comparison of resistance profiles of SGM isolates showed that the M. simiae had a more potential to resist the antibiotics than MAC and M. kansasii, as previous researches from Iran reported similar results [22,30]. One of the reasons that can justify this phenomenon, is the genetic changes that have made bacteria predispose to antibiotic resistance in the last decades, which should be revealed through modern technologies, such as whole-genome sequencing. The different antibiotic patterns of NTM in various geographic regions emphasize the necessity for AST prior to treatment. The correct NTM species identification and a proper therapy regimen can help a lot psychologically and reduce the cost of treatment for patients. Also, the NTM species determination may help in selecting the suitable treatment algorithm. This algorithm may vary depending on the clinical manifestations of the infection. For example, in the MAC pulmonary infection, the treatment criteria are different and depends on the presence or absence of cavitary disease. In patients with MAC cavitary disease, a daily treatment regimen of 3 drugs (azithromycin, ethambutol, and rifampin) is prescribed. Patients may also need 2 months of intravenous amikacin if the disease has progressed. However, studies have demonstrated that a thrice-weekly regimen containing three drugs is equally effective and better tolerated than daily treatment for patients without MAC cavitary infection [34].
The genetic origin of drug resistance in SGM has been very rarely investigated, and there is little evidence in this regard, particularly in Iran. In this research, we examined the association among the mutations in rrl, gyrA/B, and rpoB genes with CLR, MXF, and RIF resistance in M. kansasii isolates. Also, the association of rrl mutations with CLR resistance was investigated in MAC and M. simiae isolates.
The methylation of domain V of the 23S rRNA via erythromycin ribosomal methylase (erm) and the point mutations in the peptidyl transferase domain of the 23S rRNA (rrl) gene at certain locations are two main mechanisms of macrolides resistance in NTM species [10]. In CLR-resistant strains (≥32 µg/ml), 3 mutation types in M. kansasii and 4 mutation types in MAC isolates were found in different positions that may be related with resistance to this antibiotic (Table 8). The current observed mutation types including 2058, 2059, 2212, 2266, and 2419 in the rrl gene of M. kansasii and MAC isolates have also been reported in the previous studies from Korea [10], Poland [20], and Germany [28]. Our experiment confirmed that acquired CLR resistance may be related with mutation types at positions 2058/2059 of the rrl gene [35]. Also, the CLR-resistant M. simiae isolates showed various mutation types in rrl gene. To the best of our knowledge, there is no adequate information about the macrolide resistance mechanisms in M. simiae isolates in the world. The only previous study that investigated the association of mutation in some resistance genes and antibiotic resistance in M. simiae, was a case report by Lotfi et al. [36] from Iran. They reported a deletion of adenine at position 217 in the rrl gene of a MDR M. simiae isolate in a 65-year-old woman with respiratory infection.
The role of gyrA and/or gyrB mutations is unclear in fluoroquinolone (FQ) resistant NTM species and rare studies have investigated this association. FQ resistance may be related to mutations in the same codons that occurred in FQ-resistant M. tuberculosis (83 in gyrA, 447 and 464 in gyrB) [37]. In the present study, mutations in these three codons have also been observed in MXF-resistant M. kansasii (Table 7). In contrast to our findings, Bakula et al. [20] from Poland, reported no mutations in the gyrA and gyrB genes of 17 ciprofloxacin-resistant M. kansasii isolates indicating the existence of other mechanisms involved in FQ-resistance.
Six mutation types were detected at positions 1249, 1356, 1407, 1479, 1533, and 1536 of the 350-bp fragment of rpoB gene in 3 RIF-resistant M. kansasii isolates. Also, the rpoB gene sequencing revealed no mutations in 85% of RIF-resistant M. kansasii isolates that suggesting the contribution of other different factors in this resistance. Some of these factors including lipid-rich cell wall [37], enzymes involved in cell wall integrity (MurA, MurB, Ldt, PonA1, and PonA2) [11], and efflux pumps systems [20]. There were few studies in this field to be able to fully compare the results of this study with them. In a recent study by Huh et al. [12], silent mutations outside the RIF resistance-determining region (RRDR) in the rpoB gene were found in RIF-resistant NTM strains, suggesting that these regions should also be examined for RIF resistance.
Currently, unlike TB, there is no specific control program for NTM infections that is institutionalized in many countries including Iran. NTM diseases are frequently misdiagnosed as TB in many cases because mycobacterium species are often not identified due to the lack of inadequate laboratory facilities 38. Based on our findings, we believe that NTM infections may be a major factor in lung disease in our region, therefore, it is imperative that NTM infection be taken into account in TB control programs. Furthermore, due to the lack of a standard for defining and reporting NTM infections, they have not received adequate public health attention [39]. Hence, for assessing the efficacy of previous chemotherapy regimens and for detecting errors in past treatments, continual surveillance of the resistance patterns of NTM is recommended.
The main limitation of this study was the lack of complete demographic, clinical, and radiographic data for all patients. For this reason, we could not estimate the true prevalence of each isolate. Failure to perform sequencing for all resistant isolates due to financial constraints was another limitation of the present study.
Conclusions
This study revealed that AMK, MXF, and CLR are the most effective antibiotics against SGM isolates in vitro. The majority of SGM isolates showed MDR profiles and M. simiae comprised a more resistance capability compared to other SGM. Based on the results, there may be a need to perform the antibiotic susceptibility testing before starting treatment for NTM infections in Iran. Although the current study showed an association among resistance to the CLR, MXF, and RIF with mutations in the relevant genes, our knowledge of other antibiotic resistance mechanisms in SGM isolates is still insufficient. Further research, including the sequencing technology is needed to provide a clearer insight into the molecular origins of drug resistance in SGM isolates.
Acknowledgments
This work was part of M. Sc. thesis for Sousan Akrami, which was approved in Infectious and Tropical Diseases Research Center, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran. The authors would like to thank Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran, for providing financial support for this research.
Funding Statement
This work was supported by the research affairs of Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran under Grant number [OG-9906].
Author contributions
Azar Dokht Khosravi: Supervision; Writing – review & editing of manuscript, Data validation
Mohammad Hashemzadeh: Conceptualization; Formal analysis; Resources
Sousan Akrami: Data curation; Investigation; Methodology; Validation; Writing – original draft.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data Availability
All relevant data are within the manuscript.
References
- [1].Lagune M, Petit C, Sotomayor FV, et al. Conserved and specialized functions of type VII secretion systems in non-tuberculous mycobacteria. Microbiology. 2021;167(7):001054. [DOI] [PubMed] [Google Scholar]
- [2].Gopalaswamy R, Shanmugam S, Mondal R, et al. Of tuberculosis and non-tuberculous mycobacterial infections–a comparative analysis of epidemiology, diagnosis and treatment. J Biomed Sci. 2020;27(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Nasiri MJ, Dabiri H, Darban-Sarokhalil D, et al. Prevalence of non-tuberculosis mycobacterial infections among tuberculosis suspects in Iran: systematic review and meta-analysis. PloS One. 2015;10(6):e0129073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Nasiri MJ, Dabiri H, Fooladi AAI, et al. High rates of nontuberculous mycobacteria isolation from patients with presumptive tuberculosis in Iran. New Microbes New Infect. 2018;21:12–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hashemi Shahraki A, Heidarieh P, Zaker Bostanabad S, et al. Multidrug-resistant tuberculosis may be nontuberculous mycobacteria. Eur J Intern Med. 2015;26:279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ratnatunga CN, Lutzky VP, Kupz A, et al. The rise of non-tuberculosis mycobacterial lung disease. Front Immunol. 2020;11:303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Saxena S, Spaink HP, Forn-Cuní G.. Drug resistance in nontuberculous mycobacteria: mechanisms and models. Biology (Basel). 2021;10(2):96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bento CM, Gomes MS, Silva T. Looking beyond typical treatments for atypical mycobacteria. Antibiotics (Basel). 2020;9(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Haworth CS, Banks J, Capstick T, et al. British Thoracic Society guidelines for the management of non-tuberculous mycobacterial pulmonary disease (NTM-PD). Thorax. 2017;72(Suppl 2):ii1–64. [DOI] [PubMed] [Google Scholar]
- [10].Park H-E, Kim S, Shim S, et al. 16S and 23S rRNA gene mutation independent multidrug resistance of non-tuberculous mycobacteria isolated from South Korean Soil. Microorganisms. 2020;8(8):1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nasiri MJ, Haeili M, Ghazi M, et al. New insights in to the intrinsic and acquired drug resistance mechanisms in mycobacteria. Front Microbiol. 2017;8:681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Huh HJ, Kim S-Y, Jhun BW, et al. Recent advances in molecular diagnostics and understanding mechanisms of drug resistance in nontuberculous mycobacterial diseases. Infect Genet Evol. 2019;72:169–182. [DOI] [PubMed] [Google Scholar]
- [13].Daley CL, Iaccarino JM, Lange C, et al. Treatment of nontuberculous mycobacterial pulmonary disease: an official ATS/ERS/ESCMID/IDSA clinical practice guideline. Clin Infect Dis. 2020;71(4):e1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mohammadi S, Esfahani BN, Moghim S, et al. Optimal DNA isolation method for detection of nontuberculous mycobacteria by polymerase chain reaction. Adv Biomed Res. 2017;6:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dai J, Chen Y, Dean S, et al. Multiple-genome comparison reveals new loci for mycobacterium species identification. J Clin Microbiol. 2011;49(1):144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Clinical and Laboratory Standards Institute . Susceptibility testing of mycobacteria, Nocardia spp., and other aerobic actinomycetes. In: CLSI standard document M24-A2. 2rd ed. Wayne PA: Clinical and Laboratory Standards Institute; 2011. 20–25. . [PubMed] [Google Scholar]
- [18].Pang H, Jiang Y, Wan K. Drug susceptibility of 33 reference strains of slowly growing mycobacteria to 19 antimicrobial agents. BioMed Res Int. 2017;2017:1584658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Araj GF, Baba OZ, Itani LY, et al. Non-tuberculous mycobacteria profiles and their anti-mycobacterial resistance at a major medical center in Lebanon. J Infect Dev Ctries. 2019;13(7):612–618. [DOI] [PubMed] [Google Scholar]
- [20].Bakuła Z, Modrzejewska M, Pennings L, et al. Drug susceptibility profiling and genetic determinants of drug resistance in mycobacterium kansasii. Antimicrob Agents Chemother. 2018;62(4):e01788–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shen Y, Wang X, Jin J, et al. In vitro susceptibility of mycobacterium abscessus and mycobacterium fortuitum isolates to 30 antibiotics. BioMed Res Int. 2018;2018:4902941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Karami-Zarandi M, Bahador A, Gizaw Feysia S, et al. Identification of Non-Tuberculosis Mycobacteria by Line Probe Assay and Determination of Drug Resistance Patterns of Isolates in Iranian Patients. Arch Razi Inst. 2019;74(4):375–384. [DOI] [PubMed] [Google Scholar]
- [23].Krishnan MY, Manning EJ, Collins MT. Comparison of three methods for susceptibility testing of mycobacterium avium subsp. paratuberculosis to 11 antimicrobial drugs. J Antimicrob Chemother. 2009;64(2):310–316. [DOI] [PubMed] [Google Scholar]
- [24].Griffith DE, Aksamit T, Brown-Elliott BA, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007;175(4):367–416. [DOI] [PubMed] [Google Scholar]
- [25].Falkinham JO. Challenges of NTM drug development. Front Microbiol. 2018;9:1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Khan ZA, Siddiqui MF, Park S. Current and emerging methods of antibiotic susceptibility testing. Diagnostics (Basel). 2019;9(2):49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Liu CF, Song YM, He WC, et al. Nontuberculous mycobacteria in China: incidence and antimicrobial resistance spectrum from a nationwide survey. Infect Dis Poverty. 2021;10:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wetzstein N, Kohl TA, Andres S, et al. Comparative analysis of phenotypic and genotypic antibiotic susceptibility patterns in mycobacterium avium complex. Int J Infect Dis. 2020;93:320–328. [DOI] [PubMed] [Google Scholar]
- [29].Otchere ID, Asante-Poku A, Osei-Wusu S, et al. Isolation and characterization of nontuberculous mycobacteria from patients with pulmonary tuberculosis in Ghana. Int J Mycobacteriol. 2017;6(1):70–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Heidarieh P, Mirsaeidi M, Hashemzadeh M, et al. In vitro antimicrobial susceptibility of nontuberculous mycobacteria in Iran. Microb Drug Resist. 2016;22(2):172–178. [DOI] [PubMed] [Google Scholar]
- [31].Brown-Elliott BA, Woods GL. Antimycobacterial susceptibility testing of nontuberculous mycobacteria. J Clin Microbiol. 2019;57(10):e00834–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Luo J, Yu X, Jiang G, et al. In vitro activity of CFZ against nontuberculous mycobacteria isolated in Beijing, China. Antimicrob Agents Chemother. 2018;62(7):e00072–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhou W, Zhao H, Yuan H, et al. Prevalence and antimicrobial susceptibility of non-tuberculous mycobacteria isolated from sputum samples of patients with pulmonary infections in China. Jundishapur J Microbiol. 2021;14(1):e109676. [Google Scholar]
- [34].Daley CL, Winthrop KL. Mycobacterium avium complex: addressing gaps in diagnosis and management. J Infect Dis. 2020;222(Supplement_4):S199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Huh HJ, Kim SY, Shim HJ, et al. GenoType NTM-DR performance evaluation for identification of mycobacterium avium complex and mycobacterium abscessus and determination of clarithromycin and amikacin resistance. J Clin Microbiol. 2019;57(8):e00516–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lotfi H, Aryan E, Sankian M, et al. A case of multidrug-resistant mycobacterium simiae in an elderly woman. Respirol Case Rep. 2021;9(3):e00715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].De Moura VC, Da Silva MG, Gomes KM, et al. Phenotypic and molecular characterization of quinolone resistance in mycobacterium abscessus subsp. bolletii recovered from postsurgical infections. J Med Microbiol. 2012;61(1):115–125. [DOI] [PubMed] [Google Scholar]
- [38].Rindi L. Efflux pump inhibitors against nontuberculous mycobacteria. Int J Mol Sci. 2020;21(12):4191 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hoza AS, Mfinanga SG, Rodloff AC, et al. Increased isolation of nontuberculous mycobacteria among TB suspects in Northeastern, Tanzania: public health and diagnostic implications for control programmes. BMC Res Notes. 2016;9:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the manuscript.


