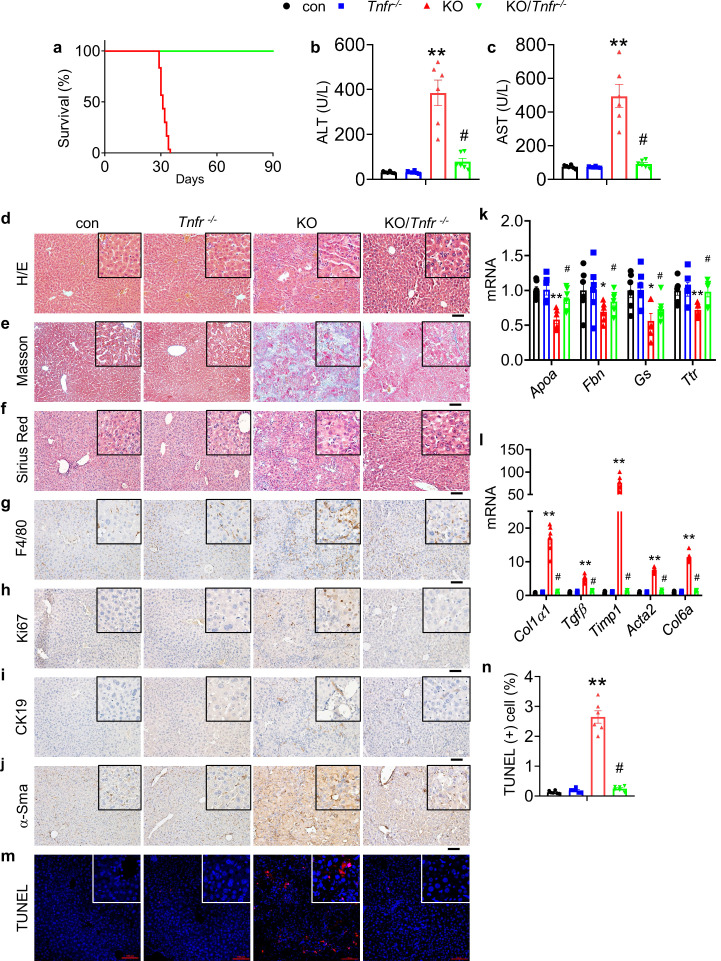
Figure 5. Global Tnfr genes deletion rescues the liver injury and lethality induced by Kindlin-2 loss.
(a) Survival curve of KO and KO/Tnfr-/- mice. N=30 mice in KO group, N=33 for KO/Tnfr-/- group. (b, c) Serum alanine transaminase (ALT) and aspartate transaminase (AST) levels in control, KO, Tnfr-/-, and KO/Tnfr-/- mice. N=6 mice/group. (d-f) Liver histology. Liver sections from mice of the indicated genotypes were subjected to hematoxylin and eosin (H/E) staining (d), Masson’s trichrome staining (e) and Sirius Red staining (f). Scale bars,100 μm. (g–j) Immunohistochemistry staining of liver sections from mice of the indicated genotypes for expression of F4/80 (g), Ki67 (h), CK19 (i), and α-Sma (j). Scale bars,100 μm. (k, l) Quantitative real-time RT-PCR (qRT-PCR) analysis. RNAs isolated from liver tissues of the indicated genotypes were subjected to qRT-PCR analysis for expression of the indicated genes. N=6 mice/group. (m, n) TUNEL staining of the indicated genotypes and quantification. N=6 mice/group. Scale bars,100 μm. The results are shown as means ± SEM. *p<0.05, **p<0.01, vs control. #p<0.05, vs KO.