Abstract
The murine embryonic–trophoblast–extra-embryonic endoderm (ETX) model is an integrated stem cell–based model to study early postimplantation development. It is based on the self-assembly potential of embryonic, trophoblast, and hypoblast/primitive/visceral endoderm-type stem cell lines (ESC, TSC, and XEN, respectively) to arrange into postimplantation egg cylinder–like embryoids. Here, we provide an optimized method for reliable and efficient generation of ETX embryoids that develop into late gastrulation in static culture conditions. It is based on transgenic Gata6-overproducing ESCs and modified assembly and culture conditions. Using this method, up to 43% of assembled ETX embryoids exhibited a correct spatial distribution of the three stem cell derivatives at day 4 of culture. Of those, 40% progressed into ETX embryoids that both transcriptionally and morphologically faithfully mimicked in vivo postimplantation mouse development between E5.5 and E7.5. The ETX model system offers the opportunity to study the murine postimplantation egg cylinder stages and could serve as a source of various cell lineage precursors.
An optimized protocol results in the reliable and efficient generation of developmentally advanced ETX embryoids.
INTRODUCTION
Understanding the mechanisms that drive development of a mammalian zygote into all specialized cell types in the primitive body plan, laid down by gastrulation, and earliest organogenesis, is a major endeavor in developmental biology. The mouse embryo and fetus has always been the most accessible model to study mammalian development and has provided invaluable insights into the genetic control of pre- and postimplantation development. Many developmental principles, molecular mechanisms, and gene regulatory networks, however, still need to be documented in detail.
Mammalian zygotes create multicellularity first by several cleavage divisions, followed by the formation of the blastocyst, a structure displaying the first three lineages specified during early development (1). The blastocyst is composed of an outer layer called the trophectoderm (TE) that will establish the embryonic portion of the placenta; the naïve epiblast, which will form the fetus; and the primitive endoderm (PrE), which will be instrumental for embryo patterning and contributes to the yolk sac. After implantation in the uterine wall at embryonic day 4.5 (E4.5), this mouse blastocyst will develop into an egg cylinder stage embryo (2). Egg cylinder stage embryos (E5.5 to E7.5) display a cup-shape structure called the primed epiblast developing from the naïve epiblast and on the opposing side the extra-embryonic ectoderm (ExEc) that emerged from the polar TE of the blastocyst (3). Both the ExEc and primed epiblast are surrounded by a PrE-descendent cell layer called the extra-embryonic endoderm (ExEn) and the visceral endoderm (VE), respectively (4, 5). Subsequent to the development of the egg cylinder, the first three compartments undergo notable differentiations and their reciprocal interactions are crucial for proper development. The formation of an axis defining the anterior and posterior side of the E6 embryo is a defining step in embryogenesis progression and precedes gastrulation. The onset of gastrulation at E6.5 is marked by the formation of mesoderm and definitive endoderm (DE) in the primitive streak at the posterior side of the epiblast. By E7.5, DE and different types of mesoderm have formed, while primordial germ cells (PGCs) and hematoendothelial precursors have emerged at the posterior side of the primitive streak.
A better understanding on how these different early lineages coordinate each other’s differentiation to establish the body plan requires tools to study cellular, genetic, and molecular mechanisms of pre-, peri-, and early postimplantation embryogenesis. Artificially assembled murine postimplantation embryo-like structures, initially as ETS (embryonic–trophoblast stem) and later ETX (embryonic–trophoblast–extra-embryonic endoderm) (6–9) embryoids, are animal-free stem cell–based embryo models that allow to study these in vivo near-inaccessible early stages of development. These models rely on the self-assembly potential of embryonic stem cell (ESC) (10, 11), trophoblast stem cell (TSC) (12), and extra-embryonic endoderm cell (XEN) lines (13) derived from the naïve epiblast, TE, and PrE, respectively, of the mouse blastocyst. ETX models not only provide the opportunity to assess artificial development in vitro but also allow animal-free studies of genetic, mechanobiological, and physiological determinants of proper development. An additional critical advantage is that they enable functional studies of otherwise embryonic lethal genes during gametogenesis, fertilization itself, or the early cleavage stages. Published protocols to generate murine ETX embryoids, however, are not always as robust, and ETX development hardly progresses beyond early gastrulation in static culture conditions (6, 9). The use of special culture devices and adapted culture media was recently reported to support ETX development beyond late gastrulation (14–16), albeit at a low efficiency. To use the ETX model to study murine postimplantation events, the efficiency to produce ETX embryoids with a good developmental potential needed to be improved. Here, we report that the use of transgenic Gata6-overproducing ESCs instead of XEN cells, with delayed addition of TSCs as well as alterations to the culture medium composition, results in efficient production of ETX embryoids that reliably mimic in vivo embryonic development until E7.5 in static culture conditions.
RESULTS
PrE-ESCs and delayed addition of TSCs during the ETX assembly generate ETX embryoids with better developmental potential
The ETX model is based on the self-assembly potential of ESC, TSC, and XEN cells. To trace the ESC lineage easily during ETX assembly, a male Actin-GFP (green fluorescent protein) mouse ESC line was used, facilitating the microscopic selection of ETX embryoids with an apparent epiblast (17). As a source for ExEn, wild-type XEN cells or male ESCs expressing transgenic Fgfr2-E2A-Gata6 (from cDNA) as well as DsRed, under the control of a bidirectional Tet-inducible promoter (PrE-ESCs), were used (fig. S1A). Although forced expression of Gata4, Gata6, or Sox17 can convert mouse ESCs into PrE-like cells (18, 19), we opted to use an Fgfr2-E2A-Gata6 cDNA-based construct to transdifferentiate ESCs into PrE-like cells. Because PrE emerges during preimplantation development from Gata6-expressing cells, which subsequently start to express Gata4 and Sox17 following stimulation of the fibroblast growth factor (FGF)/extracellular signal–regulated kinase (ERK) pathway (20–22), we believed a forced up-regulation of an Fgfr2-E2A-Gata6–encoding construct in particular would more reliably mimic normal embryonic development. PrE-ESCs exposed to doxycycline (Dox; 1 μg/ml) up-regulated clearly Gata6 production after an exposure of 12 hours, reaching a maximal level after 24 hours (fig. S1B). This resulted ultimately in the emergence of a stable XEN cell–like cell type and line (fig. S1, C and D).
A female X-GFP TSC line was used as a source for ExEc. Because all but the X-GFP TSC line were male cell lines, the X-GFP TSC line could be transcriptionally traced by the expression of Xist, the master regulator of X chromosome inactivation in female mammalian (placental) cells (23).
Three different aggregation methods were compared (Fig. 1, A to C). The first method (referred to as A) relied on the simultaneous assembly of ESCs, TSCs, and XEN cells according to a previously described protocol (9). The second (B) also relied on this simultaneous assembly but used PrE-ESCs instead of XEN cells, similar to a previous protocol using Gata4-inducible ESCs (6). For the last method (C), PrE-ESCs were also used, but TSCs were added one day later in the aggregation protocol and the culture conditions were modified. Aggregations could be made in both AggreWell (800-μm) plates and 384-well low adhesion plates.
Fig. 1. Overview of methods and efficiencies of various ETX methods applied in this study.
ESCs and their derivatives presented a robust GFP signal (Actin-GFP, in green; XEN cells, PrE-ESCs, and their derivatives, in red), although only PrE-ESCs presented strong DSRED during live imaging at days 4 and 5. TSCs and their derivatives are also depicted (in blue), although during live imaging they modestly displayed EGFP stemming from their X-GFP transgene. (A) Method A corresponds to the original ETX embryoid procedure (9) with the representative pictures of the obtained ETX embryoids at days 4 to 6. (B) Method B consists of an adapted ETX procedure using Fgfr2-Gata6 transgenic ESCs instead of XEN cells and the obtained ETX embryoids on days 4 to 6. (C) Method C is the adapted ETX procedure described in this study, where TSCs are added the day following the aggregation of ESCs and PrE-ESCs (post-assembly). The obtained ETX embryoids on days 4 to 6 are shown. All scale bars correspond to 100 μm. (D) Efficiencies of correct cell allocation, epithelialization, observed anterior-posterior axis, and exocoelom are depicted for each method at different time points (chi-square test, n = number of studied structures). (E) Size (width and length) for ETX embryoids from each method at different time points (Mann-Whitney test). P values were corrected for multiple testing using the Benjamini-Hochberg procedure (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant).
The three-dimensional (3D) cell distribution encompassed ESCs with a strong GFP signal (Actin-GFP) and TSCs with a light GFP signal (X-GFP) forming two joined cell masses on opposite poles with XEN cells or DSRED presenting PrE-ESCs enveloping both of these masses. This correct 3D cell distribution could be observed from day 2 onward with an efficiency of 7, 30, and 43% on day 4 using methods A, B, and C, respectively (Fig. 1D and fig. S2). In ETX embryoids, using regular XEN cells (method A), the epithelialization of ESCs with the formation of a pro-amniotic cavity (a characteristic comarking the formation of the epiblast) was never observed by live imaging. This was in contrast to the protocols that used our PrE-ESCs (methods B/C). XEN cells applied in method A did generally not form a cohesive entity with either TSCs or ESCs, and they also had a preference to adhere to the TSCs (Fig. 1A). Replacing regular XEN cells by our PrE-ESCs treated with Dox (1 μg/ml) for 4 to 6 hours before the assembly and during the first 24 hours of the assembly (methods B/C) improved the developmental potential of the ETX embryoids substantially. The resulting PrE-like cells were assimilated within the ETX structures much more cohesively, and PrE-ESCs and ESCs seemed to support each other’s development (Fig. 1, B and C). Here, ETX embryoids increased in size, and epithelialization of the epiblast was observed from day 4 post-assembly onward, in 1 and 9% of ETX structures obtained with methods B and C, respectively (Fig. 1D and fig. S2). The use of knockout serum replacement (KSR) in IVC2 medium, when implementing method A, has been reported to be important for cavity formation in the ETX embryoids (9). The use of KSR in IVC2 medium for ETX embryoids created with PrE-ESCs (methods B/C) resulted, however, in ETX embryoids that tended to form very large pre-amniotic cavities. Therefore, KSR was replaced with normal fetal bovine serum (FBS) in IVC2 medium when PrE-ESCs were used instead of XEN cells. On day 5 post-assembly, 2 and 9% of ETX embryoids from methods B and C, respectively, developed a distinct anterior-posterior axis along the mesoderm (Fig. 1D and fig. S2). On day 6, ETX embryoids produced using regular XEN cells (method A) never became large (Fig. 1, A and E) and epithelialization of the epiblast was rarely observed in live-imaged ETX embryoids (method A). In contrast, ETX embryoids produced using PrE-ESCs produced larger epithelialized structures, comparable in size to E7.5 in vivo embryos (Fig. 1, B, C, and E). A fraction of day 6 ETX embryoids using PrE-ESCs (methods B and C) also succeeded to develop an apparent exocoelom (1 and 5%, respectively; Fig. 1D). Whereas the quality of the best ETX embryoids were comparable when using either method B or C, method C was significantly more robust and efficient (Fig. 1D).
Day 4 ETX embryoids (method C) display epithelialization reflective of E5.5 to E6.25 in vivo embryos
ETX embryoids produced according to our method C were further characterized to assess their resemblance to in vivo embryos. ETX embryoids with a good developmental potential displayed a pro-amniotic cavity, ExEc, epiblast, and VE/ExEn on day 4 post-assembly (Fig. 2A). The molecular signature of these compartments was verified by immunostaining and single-cell RNA sequencing (scRNA-seq). The TSC-derived ExEc displayed CDX2 and ELF5 staining (24, 25). The cell layer surrounding both the epiblast and ExEc stained positive for the VE and ExEn marker FOXA2 (26). POU5F1 with OTX2 was observed in the epithelialized ESC-derived compartment, indicating that the epiblast was advancing to its primed pluripotency state (Fig. 2B, fig. S3, and table S1). Morphologically, an anterior-posterior axis could not be discriminated yet at this stage (Fig. 2A), and therefore, day 4 ETX embryoids correlated best with in vivo E5.5 to E6.25 pregastrula stage embryos (Fig. 2C).
Fig. 2. Day 4 ETX embryoids display epithelialization reflective of E5.5 to E6.25 in vivo embryos.
(A) Live imaging of post-assembly day 4 ETX embryoids; bright-field overlaid with detected fluorescences (strong EGFP+++, ESC → epiblast; DSRED+, PrE-ESC → ExEn/VE, EGFP+, TSC → ExEc). Scale bar, 100 μm. (B) Confocal images of day 4 ETX embryoids stained for POU5F1, FOXA2, and CDX2 and for OTX2, FOXA2, and ELF5. Scale bar, 100μm. A schematic representation of these markers in E5.5 to E6.25 in vivo embryos is presented on top, and colored areas present the specified marker. (C) Schematic representation of E5.5 and E6.25 in vivo mouse embryos. (D) Cellular composition of pooled day 4 ETX embryoids resolved by scRNA-seq. UMAP plots displaying identified clusters and expression of Xist (specific to TSC cells), Dsred (in PrE-ESC only), and robust Egfp (ESC). (E) Heatmap of the expression of marker genes in day 4 ETX embryoids. (F) UMAP plots showing expression of Fgf5 (primed epiblast), Fgf8 (posterior epiblast), Nanog and Pou3F1 (anterior epiblast), and Brachyury (T; onset primitive streak formation) in day 4 ETX embryoids.
scRNA-seq analysis primarily confirmed these findings and facilitated a more detailed developmental assessment. Expression of Egfp (ESC), DsRed (PrE-ESC), or Xist (TSC) provided useful marker RNAs to allocate the developmental origin of the sequenced cells. Three distinct cell origins could be discerned (Fig. 2D) that had adopted the transcriptional profile from the lineage they were expected to specify (Fig. 2E). Cells within the ESC-derived population of day 4 ETX embryoids were still pluripotent based on expression of Pou5f1 and Nanog (Fig. 2, E and F). The ESC-derived cells could be subdivided into two primary cell clusters, a smaller population expressing naïve pluripotency marker genes such as Klf2 and Zfp42 and a larger population presenting markers of primed pluripotency such as Fgf5, Otx2, and Pou3f1 (Fig. 2, E and F) (27, 28). Although morphological discrimination of an anterior versus posterior axis was still lacking in the primed pluripotent cell population, two subregions within the formed epiblast were indicative for its emerging polarity. One subregion presented cells with high Fgf8 and Nanog and low Pou3f1 steady-state mRNA levels in comparison to the other epiblast subregion (Fig. 2F). As expected for in vivo pregastrula stage embryos (29), few cells of the epiblast of day 4 ETX embryoids seemed to express mesoderm marker genes. Some cells, however, within the posterior polarized epiblast region appeared to express Brachyury (T), highlighting that some cells within the epiblast started to form the primitive streak (Fig. 2F) (30, 31). The integration of the scRNA-seq data from our day 4 ETX embryoids with scRNA-seq data from in vivo E6.5 embryos (32) showed the extent of similarity between these two datasets (Fig. 3, A and B, and fig. S4, A to D) and that a majority of the differentially expressed genes (DEGs) were dataset specific rather than cell type specific (Fig. 3B and fig. S4D). The extra-embryonic cell populations (VE/ExEn/ExEc) from the day 4 ETX embryoids seemed to diverge the most from the E6.5 in vivo embryos. This may possibly be attributed to a subset of PrE-ESCs that still presented Pou5f1 expression and therefore appeared to differentiate slower than in in vivo embryos (Fig. 3C). Our integration also highlighted that day 4 ETX embryoids were developmentally a little less advanced than in vivo E6.5 embryos because day 4 ETX embryoids still presented cells residing in naïve pluripotency (Fig. 3A and fig. S4A).
Fig. 3. Day 4 ETX embryoids display distinct populations of visceral and ExEn.
(A) Integration of scRNA-seq data of pooled day 4 ETX embryoids with E6.5 in vivo embryos; color annotation in (B). (B) Number of DEGs between day 4 ETX embryoids and E6.5 in vivo embryos for each cell type separately. DEGs between all cells of day 4 ETX embryoids and in vivo embryos are indicated in darker shades. (C) UMAP plots showing expression of DsRed and Pou5f1 in day 4 ETX embryoids. (D) UMAP plots showing gene scores of VE and ExEn marker genes in day 4 ETX embryoids. (E) UMAP plot showing gene scores of distal and anterior VE (DVE and AVE) marker genes in day 4 ETX embryoids. (F) Confocal imaging of day 4 ETX embryoids stained with OTX2 and FOXA2; clear accumulation for OTX2 (shown with arrow) demarcates AVE in the top panel and DVE in the bottom panel; scale bar, 100 μm.
Day 4 ETX embryoids obtained in method C display distinct populations of VE and ExEn
In vivo, PrE-derived cells that envelop both the epiblast and ExEc have different expression profiles and are respectively defined as VE or ExEn according to this localization (32, 33). These differences were also observed within the PrE-ESC–derived cell populations of the ETX embryoids. These PrE-ESC–derived cell populations in scRNA-seq data of ETX embryoids were identified by tracing DsRed-positive cell populations. Because DsRed was along Fgfr2-E2A-Gata6 transcribed in a transient inducible manner in PrE-ESCs, however, DsRed transcripts diminished as the ETX embryoids aged, hampering assessment of DsRed expression in day 6 ETX embryoids. The DsRed-positive cell population in day 4 ETX embryoids contained a proportion of PrE-ESCs with delayed specification to endodermal fate as these cells were still positive for Pou5f1 (Fig. 3C). This observation is similar to the slow decrease in Pou5f1 expression in transdifferentiating PrE-ESCs into XEN cells (fig. S1B). From day 4 onward, this population of delayed-differentiated PrE-ESCs diminished in the ETX embryoids (fig. S5, A to C).
Two distinct DsRed-positive cell populations representing the ExEn (expressing Lgals2, Cubn, and Aqp8) and VE (Mest, Fgf5, Eomes, Dkk1, and Fgf8) could be distinguished in day 4 ETX embryoids (Fig. 3D) and, albeit to a lesser extent, in day 5 ETX embryoids (fig. S5, D and E) (32). In vivo, the VE has an important orchestrating role during embryogenesis and is composed of differing cell subpopulations (34). The VE cells at the distal tip of the egg cylinder of E5.5 in vivo embryos differentiate into the distal VE (DVE). After their formation, DVE cells migrate proximally on one side of the epiblast to become the anterior VE (AVE) (35, 36), which safeguards adjacent epiblast cells from posteriorizing cues and ectopic primitive streak formation. Molecularly, DVE/AVE is in in vivo embryos marked by expression of the transcription factor encoding genes Otx2, Hhex, Hesx1, Foxa2, and Lhx1 and the Wnt and/or Nodal antagonist-encoding genes Dkk1, Cer1, and Lefty1 (35, 37–43). scRNA-seq data of day 4 ETX embryoids highlighted that a subpopulation within the VE shared these characteristics (Fig. 3E). Immunostaining of day 4 ETX embryoids showed an accumulation of OTX2 either at the distal tip or more anteriorly located (Fig. 3F). These data, together with the columnar shape of VE cells observed on one side in day 5 ETX embryoids (Fig. 4, A and B, and fig. S6A), confirmed the presence of DVE/AVE.
Fig. 4. Day 5 ETX embryoids develop anterior-posterior polarity and undergo gastrulation.
(A and B) Live imaging of day 5 ETX embryoids with early [early day 5 ETX (A); for more details, see main text] and more pronounced [mid day 5 ETX (B)] signs of gastrulation; bright-field overlapped with observed fluorescence (ESC → epiblast, PrE-ESC → ExEn/VE, TSC → ExEc). Scale bar, 100 μm. (C) Confocal imaging of mid day 5 ETX embryoid stained for POU5F1, EOMES, and POU3F1. A schematic representation of these markers in E6.25 to E7 in vivo embryos is presented on top colored areas, which reflect the specified marker. Region “1” marks the anterior side, whereas region “2” marks the posterior side; these regions are magnified in the bottom two rows. Scale bar, 100 μm.
Day 5 ETX embryoids (method C) develop anterior-posterior polarity and undergo gastrulation
In vivo, in the mouse embryo, anterior-posterior axis formation precedes gastrulation (33, 44–48), which can be observed from E6 to E6.5 onward, respectively. A more prominent epiblast on the posterior side accompanied with columnar-shaped VE cells marking AVE on the anterior side (49) are key morphological features associated with the formation of the anterior-posterior axis. Both features could be discerned in day 5 ETX embryoids (Fig. 4, A and B, and fig. S6A). The apparent axis in day 5 ETX embryoids was subsequently validated by assessing its molecular determinants observed in in vivo embryos via immunostaining and scRNA-seq (33, 47, 48).
Immunostainings demonstrated that the POU5F1-positive epiblast was enriched for POU3F1 on the presumptive anterior side and that EOMES, a marker of nascent mesoderm, localized to the posterior side (Fig. 4C, fig. S6B, and table S1). The numbers of EOMES-positive cells varied between ETX embryoids, possibly reflecting diverging speeds of development between the embryoids. Day 5 ETX embryoids were therefore classified as “early” (E6.75) or “mid” (E7) depending on whether they presented signs of early or progressed gastrulation, respectively (Fig. 4, A and B, and fig. S6A). Similar to in vivo embryos (33, 47, 48), POU3F1 additionally marked ExEc, and EOMES additionally labeled the ExEc and embryonal VE in ETX embryoids (Fig. 4C and fig. S6B). BRACHYURY (T) along POU5F1 staining of day 5 ETX embryoids was used to visualize the emerging mesoderm in the posterior epiblast (fig. S7 and table S1).
The anterior-posterior axis was also shown by virtue of scRNA-seq data (Fig. 5, A to F). Hence, the ESC-derived epiblast cluster marked by Otx2 and Fgf5 transcripts could be further subdivided in an anterior (Pou3f1) and a posterior epiblast cell population, the latter expressing the primitive streak marker Fgf8. Eomes and Brachyury (T) also marked the primitive streak and the emerging mesoderm cluster, whereas Fgf3 identified specified posterior mesoderm. According to those observations, day 5 ETX embryoids displayed anterior-posterior axis formation mimicking E6.5 to E7 in vivo embryonic development. To assess the extent of similarity of ETX development compared to in vivo development, scRNA-seq data from the day 5 ETX embryoids were integrated with existing data (32) from various stages of in vivo egg cylinder stage embryos. Early and mid day 5 ETX embryoids correlated the best with E6.75 and E7 in vivo embryos, respectively (Fig. 5G and figs. S8 to S11) (6, 32). Similarly to day 4 ETX embryoids, this comparison highlighted that the extra-embryonic tissues (VE/ExEn/ExEc) from day 5 ETX embryoids diverged mostly from their in vivo counterpart (figs. S8D and S10C). The number of cells undergoing gastrulation by assessing the percentage of Brachyury (T)– and/or Eomes-expressing cells in the scRNA-seq data from mid day 5 ETX embryoids and E7 in vivo embryos also seemed similar (Fig. 5H). Whereas Brachyury (T) and Eomes both mark emerging mesoderm from the epiblast, only Brachyury (T) exclusively marks emerging mesoderm as Eomes is also expressed in the ExEc and VE (33, 47, 48). The Brachyury (T) graph shows convincingly that mesoderm emerges solely from ESCs and not from PrE-ESCs because only Egfp-expressing cells express Brachyury (T).
Fig. 5. Day 5 ETX embryoids develop anterior-posterior polarity and undergo gastrulation.
(A) Schematic representation of E6.25 to E6.75 in vivo embryonic development. (B) Cellular composition of pooled early day 5 ETX embryoids resolved by scRNA-seq. UMAP plot displaying identified clusters. (C) Schematic representation of E6.5 to E7 in vivo embryonic development. (D) Cellular composition of mid day 5 ETX embryoids revealed by scRNA-seq. UMAP plot displaying identified clusters. (E and F) UMAP plots showing expression of various lineage markers in (E). Early day 5 ETX embryoids in (F). Mid day 5 ETX embryoids. (G) Integration of scRNA-seq data of mid day 5 ETX embryoids with E7 in vivo embryos. (H) Gastrulation domain in mid day 5 ETX embryoids and in vivo E7 embryos by assessing the percentage of T- and/or Eomes-expressing cells in scRNA-seq data from this study and (32), respectively.
Day 5 ETX embryoids (method C) develop DE and specify PGCs
In vivo, both progenitors of DE and PGCs emerge soon after formation of the egg cylinder. The first progenitors of DE in vivo can already be detected in the pregastrulation posterior epiblast at E5.5 and express Foxa2 (30, 31, 50–52). Day 4 ETX embryoids presented FOXA2 in a few epiblast cells, which was indicative that the primitive streak including DE progenitors was being formed (Fig. 6A and table S1). In day 5 ETX embryoids, as expected from E6.5 to E7 in vivo embryos (Fig. 5, A and C), a larger population of FOXA2-presenting cells could be detected in the epiblast (Fig. 6B, fig. S12A, and table S1). Soon after their emergence in vivo, these FOXA2-presenting cells in the epiblast progressively start to express other transcription factors crucial for DE specification. Following FOXA2, SOX17 is deemed one of the other early specifiers of the DE lineage (53–55). Because DE and VE cells are molecularly hard to distinguish from each other (52, 54, 56) and merely differ in their developmental origin, our ETX embryoids using stem cells with different cell markers provided here a unique property to identify the DE cell population. Our scRNA-seq data confirmed an Egfp-positive cell population with DE character clustering together with the DsRed-positive cell population (VE/ExEn) in day 5 and day 6 ETX embryoids (Fig. 6C and fig. S12, B and C). Using this system also allowed us to estimate the size of the DE cell population in ETX embryoids at various stages of development (Fig. 6D). From E7.25 onward, some DE cells intersperse in vivo with the VE (55), whereas the noninterspersed DE cells will translocate to the anterior side of the embryo along with the migrating axial mesoderm. DE cells at the anterior side of the embryo will subsequently also intercalate in the VE (57, 58). Because of their large size, the imaging of immunostained ETX embryoids beyond 5 days was difficult; nonetheless, live imaging displayed enhanced GFP (EGFP) ESC-derived cells that were found intercalated in the DSRED-positive VE (for those ETX embryoids that retained DsRed expression somewhat longer) (Fig. 6E).
Fig. 6. ETX embryoids develop DE and specify PGCs.
(A) Confocal imaging of day 4 ETX embryoids stained for CDX2, FOXA2, and OTX2; magnification shows FOXA2-positive cells in the epiblast. Scale bar, 100 μm. (B) Confocal imaging of day 5 ETX embryoids stained for FOXA2, POU5F1, and GATA6; a magnification displays an accumulation of FOXA2 in the epiblast region of the ETX embryoid, which is indicative of DE formation. Scale bar, 100 μm. (C) UMAP showing gene scores of DE markers in pooled mid day 5 ETX embryoids. (D) Percentage of DE cells in (per stage) ETX embryoids and in vivo embryos by assessing the percentage Foxa2/Pou5f1 (ETX day 4 embryoid and E6.5 in vivo embryos) or Foxa2/Sox17 (ETX mid day 5 and day 6 ETX embryoids and E7.0 and E7.5 in vivo embryos) in scRNA-seq data from this study and (32), respectively. (E) Live imaging of day 6 ETX embryoids; bright-field overlapped with observed fluorescence depicting the various stem cell–derived lineages; intercalated EGFP-positive cells (ESC-derived) into the DSRED-positive VE can be visualized. (F) Confocal imaging of day 5 ETX embryoids stained for POU5F1, EOMES, and BLIMP1; accumulation of EOMES on the posterior side marking mesoderm can be observed; dispersed BLIMP1-positive cells on the posterior side mark the emergence of primordial germ cells (PGCs). Scale bar, 100 μm.
Besides DE, PGC progenitors were also detected in ETX embryoids. The competence to form PGCs in mice is induced at E6.5 in some cells of the proximal epiblast in response to stimuli from extra-embryonic tissues (59–65) and is characterized by the expression of Ifitm1, Ifitm3 (27, 66), Dnd1, Prdm1 (Blimp1), Prdm14, and Tfap2c (67–74).The PGCs will further mature and specify around E7.5 with detectable expression of Stella (Dppa3) (75, 76). In day 5 and day 6 ETX embryoids, scRNA-seq identified a distinct cell population expressing Ifitm1, Ifitm3, Prdm1 (Blimp1), Prdm14, Tfap2c, and Dnd1 (figs. S13, A and B, and S14A). Immunostaining of day 5 ETX embryoids confirmed BLIMP1/PRDM1-positive cells dispersed within the most posteriorly forming mesoderm/primitive streak of the ETX embryoid, highlighting that PGC precursors are as expected emerging in the posterior epiblast (Fig. 6F and fig. S14B). A small cell population additionally expressed Stella (Dppa3) besides Prdm1 (Blimp1), Prdm14, and Tfap2c in both mid day 5 and day 6 ETX embryoids. The size of this population was comparable to their in vivo counterpart and was estimated following removal of the undifferentiated ESC clusters in ETX embryoids to prevent an overestimation of the PGC population size in ETX embryoids (because PGCs and ESCs share many characteristics; fig. S14C).
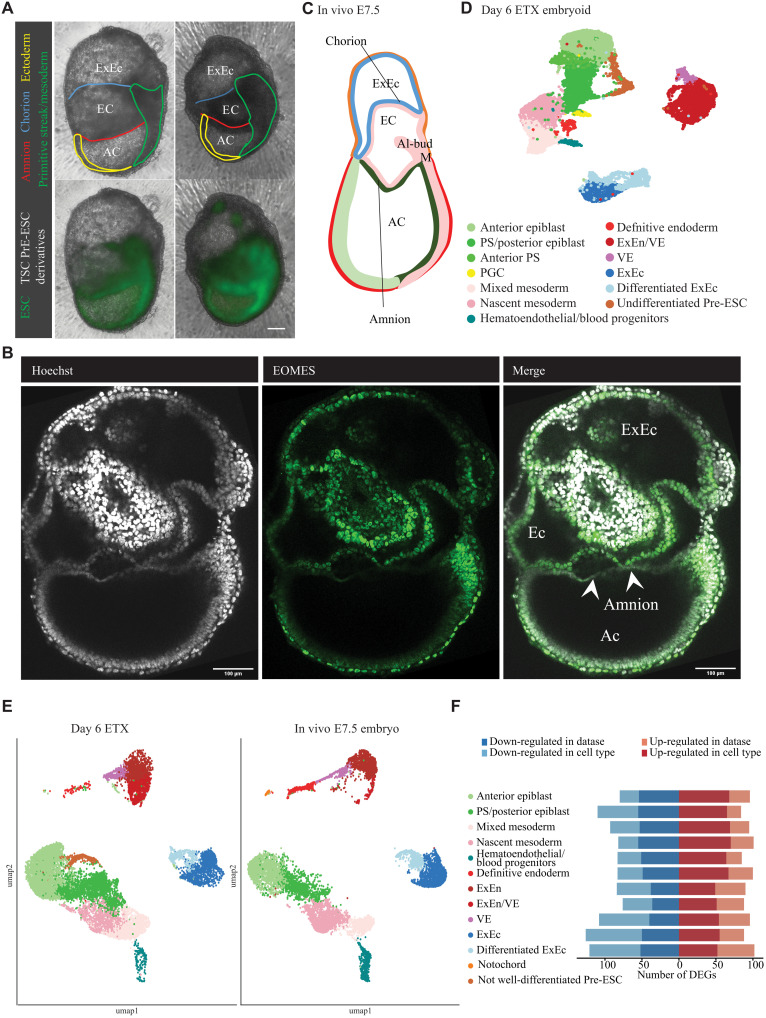
Day 6 ETX embryoids (method C) form amnion and chorion and develop hematoendothelial progenitors
The extra-embryonic mesoderm forms and delineates, following its expansion, a single cavity called the exocoelom (77). These mesodermal cells proximally line cells of the ExEc and form the chorion, whereas distally they line embryonic ectoderm cells and form the amnion. Therefore, both the chorion and amnion are composed of two different cell layers. Mid day 5 ETX embryoids and day 6 ETX embryoids could morphologically develop an exocoelom, amnion, and chorion (Fig. 7A and fig. S15A). Immunostainings confirmed the presence of an amnion-like bilayered membrane consisting of an EOMES-positive cell population (mesoderm origin) and an EOMES-negative cell population (Fig. 7B and table S1). As expected for E7.5 in vivo embryos (Fig. 7C), ETX embryoids that developed an exocoelom also presented, besides PGCs and DE, a hematoendothelial progenitor cell population expressing Lmo2, Tal1, Gata2, Runx1, and Fli1 (78). Hematoendothelial progenitors could also be detected in mid day 5 ETX embryoids but not at earlier stages (Figs. 5G and 7D). Visualization of these hematoendothelial progenitors was difficult due to the size of the ETX embryoids and difficulties to image whole-mount stained ETX embryoids. A faint TAL1-positive cell population could be discerned in whole-mount stainings of relative small day 6 ETX embryoids (fig. S15B and table S1). This should ideally be confirmed by sectioning of large day 6 ETX embryoids; however, controlling the section plane is difficult. scRNA-seq integration from our day 6 ETX embryoids with scRNA-seq data from in vivo E7.5 embryos published by others (32) again highlighted the similarity between these two datasets (Fig. 7, E and F, and figs. S16 and S17). Sporadically, ETX embryoids cultured until day 7 or day 8 seemed to display the formation of a headfold and a heart, but it is clear that culture conditions need further modification to efficiently support development beyond late gastrulation (fig. S18A).
Fig. 7. Day 6 ETX embryoids form exocoelom and develop hematoendothelial progenitors.
(A) Live imaging of day 6 ETX embryoids; bright-field overlapped with observed fluorescence depicting the various stem cell–derived lineages; amniotic coelom (AC) and exocoelom (EC) can be visualized. Scale bar, 100 μm. (B) Confocal imaging of day 6 ETX embryoids stained for EOMES and Hoechst shows the formation of a double cell layer characteristic for the amnion. Scale bar, 100 μm. (C) Schematic representation of E7.5 in vivo embryonic development, mesoderm (M), and allantois (Al-bud). (D) Cellular composition of pooled day 6 ETX embryoids resolved by scRNA-seq. UMAP plot displaying identified clusters. (E) Integration of scRNA-seq data of day 6 ETX embryoids with E7.5 in vivo embryos. (F) DEGs between day 6 ETX embryoids and E7.5 in vivo embryos for each cell type present in both datasets separately. DEGs between all cells of day 6 ETX embryoids and E7.5 in vivo embryos are indicated in darker shades.
DISCUSSION
The ETX model relies on the self-assembly potential of combined ESCs, TSCs, and XEN cells (6, 9) to mimic murine postimplantation development from E5.5 onward. This model is becoming a powerful animal-free alternative for descriptive and functional studies in early embryogenesis. Whereas ETX development hardly surpasses early gastrulation in static culture conditions (6, 9), recent reports have shown that the model could develop as far as forming a headfold and primitive heart when using complex culture devices and adapting culture media (14–16). However, this model is still hampered by its low efficiency to produce reliably developmentally advanced ETX embryoids. In addition, it is crucial to assess whether the used stem cell lineages remain developmentally restricted or display promiscuous behavior and contribute to different compartments of the ETX embryoid.
The current manuscript describes an adaptation of the ETX protocol of Zernicka-Goetz and co-workers (6, 9). A first critical modification that we introduced was the replacement of XEN cells by transgenic ESCs equipped to transdifferentiate into PrE-like cells (named here PrE-ESCs). These PrE-ESCs overexpressed an Fgfr2-E2A-Gata6 cDNA-based transgene in an inducible manner to facilitate PrE-like cell formation. Most likely, Fgfr2 is dispensable in this process because ESCs overexpressing solely Gata6 differentiated as rapidly to XEN cells (19). A similar PrE-like cell approach was recently described using Gata4-overexpressing ESCs (6, 14–16). Biologically, Gata6 is the earliest marker of the PrE lineage during embryonic development (20, 21), which motivated our choice for Gata6. Preaggregation of ESCs and PrE-ESCs in the presence of leukemia inhibitory factor (LIF) and Dox was a second critical modification to improve the correct 3D spatial distribution of the used stem cell lineages and their developmental potential. This preaggregation aimed to further trigger the transdifferentiation of PrE-ESCs into PrE, while ESCs would maintain pluripotency. Another modification was that TSCs were added only to the aggregates on the subsequent day. We reasoned that the delayed TSC addition biologically approached better the in vivo event. While TE emerges simultaneously with the ICM, TE cells do not instantly express all critical transcription factors of TSCs (e.g., Sox2) (79, 80). The TSC compartment emerges thus later than the pluripotent ICM. Last, stem cell lines with marker genes were used to test whether the stem cell lines remained developmentally confined.
This adapted protocol yielded up to 43% of ETX embryoids with correct spatial distribution of the three stem cell types on day 4. Of those embryoids, 40% progressed into ETX embryoids that morphologically resembled in vivo postimplantation development between E6 and E7.5. Immunostaining and single-cell transcriptomes confirmed that, on day 4, the embryonic compartment of ETX embryoids exit naïve pluripotency to establish epiblast; moreover, an ExEc compartment forms, and a VE subpopulation develops into DVE/AVE. On days 5 and 6 of ETX development, the three embryonic germ layers are present and their regionalization and patterning is like what is observed in vivo. This is further underscored by the presence of PGCs and hematoendothelial precursors. Beyond day 6, an apparent headfold and formation of the heart is sporadically observed, despite the static culture conditions, and without the use of complex culture devices and adapted culture media.
The ETX model remains, however, a delicate system in which minor perturbations during the early stages of development can affect later developmental potency. Hence, it remains imperative to use competent cell lines whose cell numbers need to be carefully titrated to simulate normal, dose-dependent effects of signaling factors that steer early embryogenesis and model murine embryonic development as accurately as possible. Whereas ETX embryoids sporadically developed beyond late gastrulation in our static system, future adaptations to support more robust later development may require more advanced culture systems that support oxygenation and nutrient delivery to the early gastrulating ETX embryoids, as was recently reported (14–16, 81, 82).
Our current ETX embryoid protocol provides opportunities to efficiently assess postimplantation developmental events that are difficult to study in vivo. These events include the formation of the epiblast, the definition of the anterior-posterior axis, the formation of germ layers, and even the emergence of both PGCs and hematoendothelial precursors. The observation that the used stem cell lines remain properly confined to their normal developmental origin and capacities as evaluated in chimeric embryos (19, 83–85) is crucial to study developmental events using the ETX model. Hence, it provides an opportunity to study early lineage interactions by using stem cell lines containing targeted mutations of key developmental genes. Last, the ETX model not only is an invaluable resource to study animal-free early mouse embryogenesis but also can equally serve as a source of various poorly accessible cell lineage precursors.
MATERIALS AND METHODS
Experimental design
The aim of this study was to improve the existing protocol (9) for generation of ETX embryoids. Because a major limitation of the original ETX protocol, in our hands, was the failure of most XEN cells to form a cohesive mass with TSCs and/or ESCs, we aimed in first instance to replace these cells. For this, transgenic ESC lines that express an Fgfr2-E2A-Gata6 cDNA construct in an inducible manner were generated and used in the ETX assembly protocol. All subsequent modifications to the ETX protocol were empirically determined. ETX embryoids generated using our best modified method were subsequently characterized to assess their capacity to mimic in vivo embryonic development.
Culture of XEN, ES, and TS cell lines
Derivation and culture of wild-type and transgenic (Actin-GFP and X-GFP) (17, 86) ES and TS cell lines from embryos from female 129S2/SvHsd (Envigo, the Netherlands) and male C57BL/6JOlaHsd (Envigo, the Netherlands) mice was as previously described (12, 87). Briefly, ESC lines were derived and cultured on cell culture plates coated with 0.2% gelatin and irradiated mouse embryonic fibroblasts (MEFs; density 3 × 104/cm2) in ESC medium [Dulbecco’s modified Eagle’s medium (DMEM; Gibco, 41966-029), 15% FBS, penicillin-streptomycin (100 U/ml), 100 μM nonessential amino acids (NEAAs), 0.1 mM β-mercaptoethanol, and LIF (20 ng/ml)] supplemented with 2i (1 μM ERK inhibitor PD0325901, 3.3 μM GSK3 inhibitor CHIR99021; Stemgent) and grown at 37°C and 5% CO2. Following establishment, ESC lines were intermittently cultured in ESC medium with or without 2i. TSC lines were derived and cultured onto 0.2% gelatin and irradiated MEF-coated (density 1.5 × 104/cm2) dishes in TSC medium [RPMI 1640, 20% FBS, 1 mM sodium pyruvate, 2 mM l-alanyl-l-glutamine, penicillin-streptomycin (100 U/ml), 0.1 mM β-mercaptoethanol, recombinant human FGF4 (rhFGF4; 25 ng/ml), and heparin] and grown at 37°C and 5% CO2. XEN cell lines were a gift from J. Rossant and were cultured as earlier described (13) in TSC medium devoid of rhFGF4 and heparin and grown at 37°C and 5% CO2. All cell lines were passaged using 0.25% trypsin-EDTA (5 to 7 min at 37°C) for dissociation of the cells. Trypsin-EDTA was subsequently inactivated with an equivalent amount of FBS, and the cell suspension was then centrifuged at 1000 rpm for 5 min and suspended in their respective cell culture medium. Cells were split in ratios 1:6 to 1:40 depending on the cell type.
Generation of PrE-ES cell lines
For the generation of PrE-ESCs that could transdifferentiate into a PrE-like state upon Dox exposure, two constructs were targeted to both safe harbor TIGRE loci of male 129S2/SvHsd ESCs using CRISPR-Cas9. One construct contained a CAG-driven rtTA cassette (88) and another construct presented a bidirectional tetracycline-inducible promoter expressing DsRed in one direction and Ffgr2-E2A-Gata6 in the opposite direction. Both constructs were targeted simultaneously. When ESC colonies had a considerable size, targeted ESCs were exposed to Dox (1 μg/ml). The following day, ESC clones that presented DSRED were selected for picking. Picked colonies were expanded in conventional ESC medium without Dox and screened using polymerase chain reaction for correct integration into the TIGRE loci.
Original assembly method A
ETX embryoids using standard XEN cells (>5 replicates) were aggregated as previously described aiming at assembling approximately 24 ESCs, 18 XEN cells, and 64 TSCs per microwell (9) in AggreWell plates (800 μm) or per well of a 384-well low adhesion plate. Actin-GFP ESCs, TSCs, and XEN cells were trypsinized as described. Following centrifugation at 1000 rpm during 5 min, cells were resuspended in ETX medium [DMEM (Gibco, 41966-029), 15% FBS, 100 μM NEAA, 1 mM sodium pyruvate, 2 mM l-alanyl-l-glutamine, penicillin-streptomycin (100 U/ml), and 0.1 mM β-mercaptoethanol]. Stem cells were assembled in ETX medium, and the day of assembly was annotated as day 0. On days 1 and 2, the aggregates were refreshed with ETX medium. On day 3, aggregates were refreshed with IVC1 [Advanced DMEM/F-12, 1× ITS-X, 2 mM l-alanyl-l-glutamine, and penicillin-streptomycin (25 U/ml)] supplemented with 20% FBS, 8 nM β-estradiol, progesterone (200 ng/ml), and 25 μM N-acetyl-l-cysteine and on days 4 and 5 with IVC2 (IVC1 with 30% KSR instead of 20% FBS). To assess the efficiency, rows of a 384-well plate with each well containing one ETX embryoid were randomly chosen to evaluate development of the ETX embryoids.
Adapted assembly method B
In method B (>5 replicates), XEN cells were replaced with PrE-ESCs instead of regular XEN cells. PrE-ESCs were treated with Dox (1 μg/ml) for 4 to 6 hours to initiate the expression of both Fgfr2 and Gata6 before the ETX assembly was initiated on ETX day 0. Stem cells were dissociated as described. Cell numbers were adapted aiming at assembling 20 to 24 ESCs, 5 to 10 PrE-ESCs, and 70 TSCs per microwell of an AggreWell plate (800 μm) or per well of a 384-well low adhesion plate. Dox (1 μg/ml) was added to the ETX medium the day of the aggregation (day 0) and refreshed with ETX medium devoid of Dox on days 1 and 2. On day 3, aggregates were refreshed with IVC1 on days 4 and 5 with IVC2 (IVC1 with 30% FBS instead of 20% FBS). To assess the efficiency, rows of a 384-well plate with each well containing one ETX embryoid were randomly chosen to evaluate development of the ETX embryoids.
Adapted assembly method C
In method C (>5 replicates), following a 4- to 6-hour treatment of PrE-ESCs with Dox (1 μg/ml), Actin-GFP ESCs and PrE-ESCs were dissociated as described. Cell numbers were adapted aiming at assembling on day 0, 20 to 24 ESCs and 5 to 10 PrE-ESCs per well of a 384-well low adhesion plate (final volume of 50 μl/well). Using a multichannel, 50 μl of this cell suspension was pipetted into each well of a 384-well low attachment plate and incubated overnight at 37°C and 5% CO2. Dox (1 μg/ml) and LIF (20 ng/ml) were added to the ETX medium on the day of the aggregation. The following day, the wells were rinsed twice with ETX medium devoid of LIF and Dox. TSCs were dissociated as described, and a cell suspension containing 70 TSCs per 60 μl of ETX medium supplemented with recombinant activin A (0.1 ng/ml; PeproTech) was added to each well containing approximately 20 μl of ETX medium. The medium of the cell aggregates was refreshed on day 3 with IVC1 (20% FBS) and with IVC2 on days 4 and 5 (30% FBS). ETX embryoids of interest were manually picked with a pipette. To assess the efficiency, rows of a 384-well plate with each well containing one ETX embryoid were randomly chosen to evaluate development of the ETX embryoids.
Immunostaining
ETX embryoids and slides with ESCs were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 to 30 min, washed in PBS (three times 10 min each), permeabilized with 0.25% Triton X-100 in PBS for 20 min, washed in PBS (three times 10 min each), and incubated overnight in blocking solution [0.5% bovine serum albumin (BSA) and 1% Tween 20 in PBS)] at 4°C (up to 1 week). Following incubation with the blocking solution, ETXs and slides with ESCs were placed in primary antibody dilutions in blocking solution (Table 1) for 2 days at 4°C. Subsequently, the ETXs and slides with ES cells were washed with blocking solution (three times 10 min each) before being incubated for 2 days at 4°C with secondary antibodies diluted in blocking solution. To remove the secondary antibodies, the ETXs and slides with ES cells were washed with blocking solution during three consecutive PBS washes (10 min each), and Hoechst 33342 (20 μg/ml) was added to the first wash. ETXs were embedded in Matrigel drops and visualized with a confocal microscope with a 20× water-immersion objective, whereas slides were mounted in antifade mounting medium (Vectashield) and visualized with a conventional confocal Leica microscope.
Table 1. Antibodies used.
| Antibody | Catalog | Dilution | Host |
|---|---|---|---|
| CDX2 | BioGenex MU392A-UC | 1:100 | Mouse |
| ELF5 | Santa Cruz Biotechnology sc376737 | 1:100 | Mouse |
| FOXA2 | Cell Signaling Technology 8186T | 1:200 | Rabbit |
| GATA6 | R&D Systems AF1700 | 1:500 | Goat |
| GATA4 | Santa Cruz Biotechnology sc9053 | 1:200 | Rabbit |
| POU5F1 | Santa Cruz Biotechnology sc8628 | 1:500 | Goat |
| POU5F1 | Santa Cruz Biotechnology sc5279 | 1:500 | Mouse |
| OTX2 | R&D Systems AF1979 | 1:100 | Goat |
| TBR2 | Abcam AB23345 | 1:200 | Rabbit |
| POU3F1 | Absea Biotechnology 060204E04 | 1:50 | Mouse |
| BLIMP1/PRDM1 | Santa Cruz Biotechnology sc130917 | 1:200 | Rat |
| SCL/TAL1 | Novus Biologicals NBP-33757 | 1:200 | Rabbit |
| T/BRACHYURY | Cell Signaling Technology 81694 | 1:200 | Rabbit |
| Rat Alexa Fluor 488 | A11006 | 1:500 | Goat |
| Mouse Alexa Fluor 633 | A21146 | 1:500 | Donkey |
| Rabbit Alexa Fluor 488 | A21206 | 1:500 | Donkey |
| Goat Alexa Fluor 555 | A21432 | 1:500 | Donkey |
Processing of ETXs for 10× sequencing
ETX embryoids were collected on days 4 to 6. ETX structures were selected on day 4 when they displayed a clear epiblast and a pro-amniotic cavity. Day 5 ETX embryoids were divided in a group displaying a morphology of early gastrulation and a group of ETX structures displaying signs of more advanced gastrulation. Selected day 6 ETX embryoids displayed a more developed phenotype with the apparent formation of an amnion, chorion, and exocoelom. Per selected stage, 10 ETX structures were processed for 10× sequencing as earlier described (32). Briefly, selected ETX embryoids were carefully removed from the wells and dissociated with TrypLE Express dissociation reagent (Life Technologies) at 37°C for 5 min and gently dissociated manually. Following inactivation with an equivalent amount of FBS, the single cells were filtered through the Flowmi Tip Strainer with 40-μm porosity (Thermo Fisher Scientific, no. 136800040), washed, and resuspended in PBS with 0.4% BSA.
Single-cell library preparation and sequencing
Single cells were processed on the 10× Genomics Chromium Platform using the Chromium Next GEM Single Cell 3′ Reagent kit v3.1 with dual indexing (10× Genomics) following the manufacturer’s protocol. Briefly, 15,000 cells were loaded in each channel of a chip to be partitioned into gel beads in emulsion (GEMs). Within GEMs, cells were lysed followed by barcoded reverse transcription of RNA. Breaking of the GEMs was followed by amplification, fragmentation, and addition of adapter and sample index. Libraries were pooled and sequenced 28-10-10-90 cycles on an Illumina NovaSeq6000 instrument aiming for a minimum coverage of 25,000 raw reads per cell.
Analysis of scRNA-seq data
10× Genomics Cell Ranger mkref v6.0.0 (89) was used to create a custom reference package containing the mm10 reference genome and the sequences of DsRed, GFP, and EGFP. The raw FASTQ files were processed on the basis of the custom reference package using cellranger count. Cells with fewer than 500 counts, 250 detected genes, and low complexity (log10 value of number of genes detected per count <0.8) were removed. Genes with zero expression in all cells and that are expressed in less than 10 cells were removed. Downstream data analysis was performed using the R package Seurat (version 4.0.3). Datasets were normalized by the “SCTransform” function. Cell cycle heterogeneity was scored, and differences between S and G2-M cell cycle phases were regressed out using the functions “CellCycleScoring” and “ScaleData.” Uniform manifold approximation and projection (UMAP) analysis was performed using the “RunUMAP” function including 30 principal components. Clusters were identified using the “FindClusters” function with a resolution of 0.6. Cluster marker genes were identified by differential expression analysis using the “FindAllMarkers” function with a minimum log fold change value of >0.25 and genes detected in a minimum of 25% cells in each cluster. Lineage-specific gene signatures were plotted using the “AddModuleScore” function.
Integration analysis on ETX and in vivo scRNA-seq data
To compare the ETX scRNA-seq data to scRNA-seq from in vivo mouse embryos, publicly available scRNA-seq from in vivo embryos was used (6, 32). On the basis of morphology and availability of in vivo data from specific time points, ETX day 4, early day 5, mid day 5, and day 6 embryoids were compared to E6.5, E6.75, E.7.0, and E7.5 in vivo embryos (32), respectively. Each ETX dataset was integrated with the cells from its corresponding in vivo data using Seurat v4.1.1 (90). For the ETX data, SoupX (91) was used to remove ambient background RNAs from the dataset. Only cells with at least 1000 features and less than 50,000 features and a mitochondrial percentage below 10% were used for the analysis. Moreover, genes present in less than three cells, DsRed, GFP, and EGFP were removed. The dataset was normalized using SCTransform v2 with the “glmGamPoi” method (92) and percentages of ribosomal and mitochondrial genes as vars.to.regress. For the in vivo datasets, the cells were also filtered for a number of features between 1000 and 50,000 and a mitochondrial percentage below 10%. Moreover, cells identified as doublet or lacking an annotated cell type were removed and only genes present in at least three cells were used. The cells were split into separate datasets for each sample separately (except for E6.5, which was analyzed as one dataset), after which they were normalized using SCTransform v2 (glmGamPoi; percentages of ribosomal and mitochondrial genes as vars.to.regress).
The integration of the ETX and embryo datasets was prepared using the “SelectIntegrationFeatures” (nfeatures = 3000), “PrepSCTIntegration,” and “RunPCA” functions. Integration anchors were found using “FindIntegrationAnchors” with SCT as normalization.method, 30 dimensions, “rpca” reduction, and 20 anchors, after which the cells were integrated using “IntegrateData.” UMAPs were calculated using “RunUMAP” based on the first 30 principal components from RunPCA. Last, “FindNeighbors” and “FindClusters” with a resolution of 0.5 were used to identify clusters. The clusters were annotated on the basis of the overlap with cell type annotation from the embryo cells (32), the cell type annotation from the ETX cells, and expression of known markers.
To evaluate the differences between embryoid and embryo, we identified DEGs using the corrected counts from “PrepSCTFindMarkers,” which are normalized for sequencing depth differences. DEGs between the datasets were selected using “FindMarkers” with “SCT” as assay, a minimum fold change of 1.5, and an adjusted P value of <0.05. Moreover, the cells were compared for each cluster separately when the cluster contained at least 10% cells from both datasets. The DEGs within each cluster were then divided into dataset-specific and cell type–specific DEGs based on the overlap with the DEGs between all cells from both datasets. The DE results were visualized using scatterplots, where the average counts per million (CPMs) of embryoid and embryo cells of each cluster were shown and genes that were significantly up- or down-regulated were highlighted. We performed Gene Ontology enrichment analysis on the DE genes that were either up- or down-regulated using the enrichr module of the GSEAPY package (93).
The percentage of cell types present in embryo and embryoid were assessed by selecting cells with a combination of marker genes expressed based on minimum CPM thresholds. The embryoid cells were split into an EGFP-positive and an EGFP-negative group (EGFP > 4 or EGFP < 4, respectively). For the day 4 ETX embryoids and E6.5 embryos, cells with Foxa2 > 2 and Pou5f1 > 5 were selected. Moreover, we extracted cells with T > 2 and Pou5f1 > 5, and cells with Pou5f1 > 5 and Eomes > 2. For the day 5 and day 6 ETX embryoids and E7.0 and E7.5 embryos, DE cells were filtered with Foxa2 > 2 and Sox17 > 2, and PGCs with Tfap2c > 2, Prdm1 > 2, and Prdm14 > 2. Hematoendothelial cells were selected using a combination of Lmo2 > 2, Tal1 > 2, and Gata2 > 2. Last, day 5 ETX and E7.0 embryo cells that were single positive or double positive for T and Eomes were extracted (T > 2, Eomes > 2).
Statistical methods
Statistical methods for identifying cell clusters and the integration are described in their respective section. For the comparison of efficiencies between methods of obtaining ETX embryoids, the number of embryoids with different phenotypic characteristics was scored and the proportion between the phenotypes was compared using a chi-square test and corrected for multiple testing using the Benjamini-Hochberg procedure. The size and width of the embryoids were compared using a Mann-Whitney test corrected using the Benjamini-Hochberg procedure.
Acknowledgments
We thank J. Rossant for male XEN cell lines. We also thank G.-J. Kremers, M. de Gruiter, G. van Cappellen, L. Koorndreef, and A. Francis for troubleshooting and technical support during ETX embryoid imaging.
Funding: This work has become possible through Erasmus University Rotterdam (EUR) funding to Erasmus University Medical Center for establishing new collaborations and investing in novel technologies within the departments of the Theme of Biomedical Sciences (BIG funding, incl. to J.G. and D.H.) and a ZonMw-PSIDER grant (10250022120002) awarded to J.G.
Author contributions: C.D. conceived and executed all experiments and wrote the original draft. E.M.B. prepared the single-cell library and performed the scRNA-seq. B.F.T., O.J.M.S., and C.D. processed and bioinformatically analyzed the scRNA-seq data. D.H., J.G., A.Z., S.M., O.J.M.S., B.F.T., and C.D. revised and edited this manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Raw sequencing data generated here have been deposited in the NCBI GEO repository (accession no. GSE206132) and are publicly available as of the date of publication. Reused data were retrieved from ArrayExpress (E-MTAB-6967). Stem cell lines and constructs can be provided by C.D. pending scientific review and a completed material transfer agreement. Requests for stem cell lines or constructs should be submitted to C.D.
Supplementary Materials
This PDF file includes:
Figs. S1 to S18
Table S1
REFERENCES AND NOTES
- 1.K. Cockburn, J. Rossant, Making the blastocyst: Lessons from the mouse. J. Clin. Invest. 120, 995–1003 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.P. P. Tam, R. R. Behringer, Mouse gastrulation: The formation of a mammalian body plan. Mech. Dev. 68, 3–25 (1997). [DOI] [PubMed] [Google Scholar]
- 3.D. J. Chavez, A. C. Enders, S. Schlafke, Trophectoderm cell subpopulations in the periimplantation mouse blastocyst. J. Exp. Zool. 231, 267–271 (1984). [DOI] [PubMed] [Google Scholar]
- 4.R. L. Gardner, Origin and differentiation of extraembryonic tissues in the mouse. Int. Rev. Exp. Pathol. 24, 63–133 (1983). [PubMed] [Google Scholar]
- 5.R. L. Gardner, J. Rossant, Investigation of the fate of 4-5 day post-coitum mouse inner cell mass cells by blastocyst injection. J. Embryol. Exp. Morphol. 52, 141–152 (1979). [PubMed] [Google Scholar]
- 6.G. Amadei, K. Y. C. Lau, J. De Jonghe, C. W. Gantner, B. Sozen, C. Chan, M. Zhu, C. Kyprianou, F. Hollfelder, M. Zernicka-Goetz, Inducible stem-cell-derived embryos capture mouse morphogenetic events in vitro. Dev. Cell 56, 366–382.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.S. Zhang, T. Chen, N. Chen, D. Gao, B. Shi, S. Kong, R. C. West, Y. Yuan, M. Zhi, Q. Wei, J. Xiang, H. Mu, L. Yue, X. Lei, X. Wang, L. Zhong, H. Liang, S. Cao, J. C. I. Belmonte, H. Wang, J. Han, Implantation initiation of self-assembled embryo-like structures generated using three types of mouse blastocyst-derived stem cells. Nat. Commun. 10, 496 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.S. E. Harrison, B. Sozen, M. Zernicka-Goetz, In vitro generation of mouse polarized embryo-like structures from embryonic and trophoblast stem cells. Nat. Protoc. 13, 1586–1602 (2018). [DOI] [PubMed] [Google Scholar]
- 9.B. Sozen, G. Amadei, A. Cox, R. Wang, E. Na, S. Czukiewska, L. Chappell, T. Voet, G. Michel, N. Jing, D. M. Glover, M. Zernicka-Goetz, Self-assembly of embryonic and two extra-embryonic stem cell types into gastrulating embryo-like structures. Nat. Cell Biol. 20, 979–989 (2018). [DOI] [PubMed] [Google Scholar]
- 10.G. R. Martin, Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U.S.A. 78, 7634–7638 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.M. J. Evans, M. H. Kaufman, Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 (1981). [DOI] [PubMed] [Google Scholar]
- 12.S. Tanaka, T. Kunath, A. K. Hadjantonakis, A. Nagy, J. Rossant, Promotion of trophoblast stem cell proliferation by FGF4. Science 282, 2072–2075 (1998). [DOI] [PubMed] [Google Scholar]
- 13.T. Kunath, D. Arnaud, G. D. Uy, I. Okamoto, C. Chureau, Y. Yamanaka, E. Heard, R. L. Gardner, P. Avner, J. Rossant, Imprinted X-inactivation in extra-embryonic endoderm cell lines from mouse blastocysts. Development 132, 1649–1661 (2005). [DOI] [PubMed] [Google Scholar]
- 14.K. Y. C. Lau, H. Rubinstein, C. W. Gantner, R. Hadas, G. Amadei, Y. Stelzer, M. Zernicka-Goetz, Mouse embryo model derived exclusively from embryonic stem cells undergoes neurulation and heart development. Cell Stem Cell 29, 1445–1458.e8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.G. Amadei, C. E. Handford, C. Qiu, J. De Jonghe, H. Greenfeld, M. Tran, B. K. Martin, D.-Y. Chen, A. Aguilera-Castrejon, J. H. Hanna, M. B. Elowitz, F. Hollfelder, J. Shendure, D. M. Glover, M. Zernicka-Goetz, Embryo model completes gastrulation to neurulation and organogenesis. Nature 610, 143–153 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.S. Tarazi, A. Aguilera-Castrejon, C. Joubran, N. Ghanem, S. Ashouokhi, F. Roncato, E. Wildschutz, M. Haddad, B. Oldak, E. Gomez-Cesar, N. Livnat, S. Viukov, D. Lokshtanov, S. Naveh-Tassa, M. Rose, S. Hanna, C. Raanan, O. Brenner, M. Kedmi, H. Keren-Shaul, T. Lapidot, I. Maza, N. Novershtern, J. H. Hanna, Post-gastrulation synthetic embryos generated ex utero from mouse naive ESCs. Cell 185, 3290–3306.e25 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.M. Okabe, M. Ikawa, K. Kominami, T. Nakanishi, Y. Nishimune, “Green mice” as a source of ubiquitous green cells. FEBS Lett. 407, 313–319 (1997). [DOI] [PubMed] [Google Scholar]
- 18.A. C. McDonald, S. Biechele, J. Rossant, W. L. Stanford, Sox17-mediated XEN cell conversion identifies dynamic networks controlling cell-fate decisions in embryo-derived stem cells. Cell Rep. 9, 780–793 (2014). [DOI] [PubMed] [Google Scholar]
- 19.D. Shimosato, M. Shiki, H. Niwa, Extra-embryonic endoderm cells derived from ES cells induced by GATA factors acquire the character of XEN cells. BMC Dev. Biol. 7, 80 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.C. Chazaud, Y. Yamanaka, T. Pawson, J. Rossant, Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev. Cell 10, 615–624 (2006). [DOI] [PubMed] [Google Scholar]
- 21.S. Frankenberg, F. Gerbe, S. Bessonnard, C. Belville, P. Pouchin, O. Bardot, C. Chazaud, Primitive endoderm differentiates via a three-step mechanism involving Nanog and RTK signaling. Dev. Cell 21, 1005–1013 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Y. Yamanaka, F. Lanner, J. Rossant, FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development 137, 715–724 (2010). [DOI] [PubMed] [Google Scholar]
- 23.M. F. Lyon, X-chromosome inactivation. Curr. Biol. 9, R235–R237 (1999). [DOI] [PubMed] [Google Scholar]
- 24.F. Beck, T. Erler, A. Russell, R. James, Expression of Cdx-2 in the mouse embryo and placenta: Possible role in patterning of the extra-embryonic membranes. Dev. Dyn. 204, 219–227 (1995). [DOI] [PubMed] [Google Scholar]
- 25.M. Donnison, A. Beaton, H. W. Davey, R. Broadhurst, P. L'Huillier, P. L. Pfeffer, Loss of the extraembryonic ectoderm in Elf5 mutants leads to defects in embryonic patterning. Development 132, 2299–2308 (2005). [DOI] [PubMed] [Google Scholar]
- 26.H. Sasaki, B. L. Hogan, Differential expression of multiple fork head related genes during gastrulation and axial pattern formation in the mouse embryo. Development 118, 47–59 (1993). [DOI] [PubMed] [Google Scholar]
- 27.M. Saitou, S. C. Barton, M. A. Surani, A molecular programme for the specification of germ cell fate in mice. Nature 418, 293–300 (2002). [DOI] [PubMed] [Google Scholar]
- 28.P. H. Crossley, G. R. Martin, The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development 121, 439–451 (1995). [DOI] [PubMed] [Google Scholar]
- 29.G. Winnier, M. Blessing, P. A. Labosky, B. L. Hogan, Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 9, 2105–2116 (1995). [DOI] [PubMed] [Google Scholar]
- 30.D. G. Wilkinson, S. Bhatt, B. G. Herrmann, Expression pattern of the mouse T gene and its role in mesoderm formation. Nature 343, 657–659 (1990). [DOI] [PubMed] [Google Scholar]
- 31.I. Burtscher, H. Lickert, Foxa2 regulates polarity and epithelialization in the endoderm germ layer of the mouse embryo. Development 136, 1029–1038 (2009). [DOI] [PubMed] [Google Scholar]
- 32.B. Pijuan-Sala, J. A. Griffiths, C. Guibentif, T. W. Hiscock, W. Jawaid, F. J. Calero-Nieto, C. Mulas, X. Ibarra-Soria, R. C. V. Tyser, D. L. L. Ho, W. Reik, S. Srinivas, B. D. Simons, J. Nichols, J. C. Marioni, B. Göttgens, A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490–495 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.G. S. Kwon, A.-K. Hadjantonakis, Eomes::GFP—A tool for live imaging cells of the trophoblast, primitive streak, and telencephalon in the mouse embryo. Genesis 45, 208–217 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.H. Hoshino, G. Shioi, S. Aizawa, AVE protein expression and visceral endoderm cell behavior during anterior–posterior axis formation in mouse embryos: Asymmetry in OTX2 and DKK1 expression. Dev. Biol. 402, 175–191 (2015). [DOI] [PubMed] [Google Scholar]
- 35.P. Q. Thomas, A. Brown, R. S. Beddington, Hex: A homeobox gene revealing peri-implantation asymmetry in the mouse embryo and an early transient marker of endothelial cell precursors. Development 125, 85–94 (1998). [DOI] [PubMed] [Google Scholar]
- 36.P. Thomas, R. Beddington, Anterior primitive endoderm may be responsible for patterning the anterior neural plate in the mouse embryo. Curr. Biol. 6, 1487–1496 (1996). [DOI] [PubMed] [Google Scholar]
- 37.C. Kimura, K. Yoshinaga, E. Tian, M. Suzuki, S. Aizawa, I. Matsuo, Visceral endoderm mediates forebrain development by suppressing posteriorizing signals. Dev. Biol. 225, 304–321 (2000). [DOI] [PubMed] [Google Scholar]
- 38.A. Perea-Gomez, W. Shawlot, H. Sasaki, R. R. Behringer, S. Ang, HNF3beta and Lim1 interact in the visceral endoderm to regulate primitive streak formation and anterior-posterior polarity in the mouse embryo. Development 126, 4499–4511 (1999). [DOI] [PubMed] [Google Scholar]
- 39.D. Acampora, S. Mazan, Y. Lallemand, V. Avantaggiato, M. Maury, A. Simeone, P. Brulet, Forebrain and midbrain regions are deleted in Otx2−/− mutants due to a defective anterior neuroectoderm specification during gastrulation. Development 121, 3279–3290 (1995). [DOI] [PubMed] [Google Scholar]
- 40.J. A. Belo, T. Bouwmeester, L. Leyns, N. Kertesz, M. Gallo, M. Follettie, E. M. De Robertis, Cerberus-like is a secreted factor with neutralizing activity expressed in the anterior primitive endoderm of the mouse gastrula. Mech. Dev. 68, 45–57 (1997). [DOI] [PubMed] [Google Scholar]
- 41.C. Kimura-Yoshida, H. Nakano, D. Okamura, K. Nakao, S. Yonemura, J. A. Belo, S. Aizawa, Y. Matsui, I. Matsuo, Canonical Wnt signaling and its antagonist regulate anterior-posterior axis polarization by guiding cell migration in mouse visceral endoderm. Dev. Cell 9, 639–650 (2005). [DOI] [PubMed] [Google Scholar]
- 42.A. Perea-Gomez, F. D. Vella, W. Shawlot, M. Oulad-Abdelghani, C. Chazaud, C. Meno, V. Pfister, L. Chen, E. Robertson, H. Hamada, R. R. Behringer, S.-L. Ang, Nodal antagonists in the anterior visceral endoderm prevent the formation of multiple primitive streaks. Dev. Cell 3, 745–756 (2002). [DOI] [PubMed] [Google Scholar]
- 43.M. Yamamoto, Y. Saijoh, A. Perea-Gomez, W. Shawlot, R. R. Behringer, S.-L. Ang, H. Hamada, C. Meno, Nodal antagonists regulate formation of the anteroposterior axis of the mouse embryo. Nature 428, 387–392 (2004). [DOI] [PubMed] [Google Scholar]
- 44.S. J. Arnold, U. K. Hofmann, E. K. Bikoff, E. J. Robertson, Pivotal roles for eomesodermin during axis formation, epithelium-to-mesenchyme transition and endoderm specification in the mouse. Development 135, 501–511 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.A. P. Russ, S. Wattler, W. H. Colledge, S. A. Aparicio, M. B. Carlton, J. J. Pearce, S. C. Barton, M. A. Surani, K. Ryan, M. C. Nehls, V. Wilson, M. J. Evans, Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature 404, 95–99 (2000). [DOI] [PubMed] [Google Scholar]
- 46.I. Costello, I. M. Pimeisl, S. Drager, E. K. Bikoff, E. J. Robertson, S. J. Arnold, The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nat. Cell Biol. 13, 1084–1091 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.S. J. Arnold, E. J. Robertson, Making a commitment: Cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 10, 91–103 (2009). [DOI] [PubMed] [Google Scholar]
- 48.L. Li, C. Liu, S. Biechele, Q. Zhu, L. Song, F. Lanner, N. Jing, J. Rossant, Location of transient ectodermal progenitor potential in mouse development. Development 140, 4533–4543 (2013). [DOI] [PubMed] [Google Scholar]
- 49.J. A. Rivera-Pérez, J. Mager, T. Magnuson, Dynamic morphogenetic events characterize the mouse visceral endoderm. Dev. Biol. 261, 470–487 (2003). [DOI] [PubMed] [Google Scholar]
- 50.S. L. Ang, J. Rossant, HNF-3β is essential for node and notochord formation in mouse development. Cell 78, 561–574 (1994). [DOI] [PubMed] [Google Scholar]
- 51.D. C. Weinstein, A. R. i. Altaba, W. S. Chen, P. Hoodless, V. R. Prezioso, T. M. Jessell, J. E. Darnell Jr., The winged-helix transcription factor HNF-3β is required for notochord development in the mouse embryo. Cell 78, 575–588 (1994). [DOI] [PubMed] [Google Scholar]
- 52.S. L. Ang, A. Wierda, D. Wong, K. A. Stevens, S. Cascio, J. Rossant, K. S. Zaret, The formation and maintenance of the definitive endoderm lineage in the mouse: Involvement of HNF3/forkhead proteins. Development 119, 1301–1315 (1993). [DOI] [PubMed] [Google Scholar]
- 53.M. Viotti, S. Nowotschin, A.-K. Hadjantonakis, SOX17 links gut endoderm morphogenesis and germ layer segregation. Nat. Cell Biol. 16, 1146–1156 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.I. Burtscher, W. Barkey, M. Schwarzfischer, F. J. Theis, H. Lickert, The Sox17-mCherry fusion mouse line allows visualization of endoderm and vascular endothelial development. Genesis 50, 496–505 (2012). [DOI] [PubMed] [Google Scholar]
- 55.G. S. Kwon, M. Viotti, A.-K. Hadjantonakis, The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Dev. Cell 15, 509–520 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.M. Kanai-Azuma, Y. Kanai, J. M. Gad, Y. Tajima, C. Taya, M. Kurohmaru, Y. Sanai, H. Yonekawa, K. Yazaki, P. P. Tam, Y. Hayashi, Depletion of definitive gut endoderm in Sox17-null mutant mice. Development 129, 2367–2379 (2002). [DOI] [PubMed] [Google Scholar]
- 57.S. Nowotschin, M. Setty, Y. Y. Kuo, V. Liu, V. Garg, R. Sharma, C. S. Simon, N. Saiz, R. Gardner, S. C. Boutet, D. M. Church, P. A. Hoodless, A. K. Hadjantonakis, D. Pe'er, The emergent landscape of the mouse gut endoderm at single-cell resolution. Nature 569, 361–367 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.M. Pour, A. S. Kumar, N. Farag, A. Bolondi, H. Kretzmer, M. Walther, L. Wittler, A. Meissner, I. Nachman, Emergence and patterning dynamics of mouse-definitive endoderm. iScience 25, 103556 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.S. M. de Sousa Lopes, B. A. Roelen, R. M. Monteiro, R. Emmens, H. Y. Lin, E. Li, K. A. Lawson, C. L. Mummery, BMP signaling mediated by ALK2 in the visceral endoderm is necessary for the generation of primordial germ cells in the mouse embryo. Genes Dev. 18, 1838–1849 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.K. A. Lawson, N. R. Dunn, B. A. Roelen, L. M. Zeinstra, A. M. Davis, C. V. E. Wright, J. P. W. F. M. Korving, B. L. M. Hogan, Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 13, 424–436 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.K. A. Lawson, W. J. Hage, Clonal analysis of the origin of primordial germ cells in the mouse. Ciba Found. Symp. 182, 68–84 (1994). [DOI] [PubMed] [Google Scholar]
- 62.Y. Ying, X.-M. Liu, A. Marble, K. A. Lawson, G.-Q. Zhao, Requirement of Bmp8b for the generation of primordial germ cells in the mouse. Mol. Endocrinol. 14, 1053–1063 (2000). [DOI] [PubMed] [Google Scholar]
- 63.A. McLaren, Primordial germ cells in the mouse. Dev. Biol. 262, 1–15 (2003). [DOI] [PubMed] [Google Scholar]
- 64.T. Yoshimizu, M. Obinata, Y. Matsui, Stage-specific tissue and cell interactions play key roles in mouse germ cell specification. Development 128, 481–490 (2001). [DOI] [PubMed] [Google Scholar]
- 65.P. P. Tam, S. X. Zhou, The allocation of epiblast cells to ectodermal and germ-line lineages is influenced by the position of the cells in the gastrulating mouse embryo. Dev. Biol. 178, 124–132 (1996). [DOI] [PubMed] [Google Scholar]
- 66.S. S. Tanaka, Y. Matsui, Developmentally regulated expression of mil-1 and mil-2, mouse interferon-induced transmembrane protein like genes, during formation and differentiation of primordial germ cells. Gene Expr. Patterns 2, 297–303 (2002). [DOI] [PubMed] [Google Scholar]
- 67.Y. Yabuta, K. Kurimoto, Y. Ohinata, Y. Seki, M. Saitou, Gene expression dynamics during germline specification in mice identified by quantitative single-cell gene expression profiling. Biol. Reprod. 75, 705–716 (2006). [DOI] [PubMed] [Google Scholar]
- 68.M. S. Cook, D. Coveney, I. Batchvarov, J. H. Nadeau, B. Capel, BAX-mediated cell death affects early germ cell loss and incidence of testicular teratomas in Dnd1Ter/Ter mice. Dev. Biol. 328, 377–383 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.K. K. Youngren, D. Coveney, X. Peng, C. Bhattacharya, L. S. Schmidt, M. L. Nickerson, B. T. Lamb, J. M. Deng, R. R. Behringer, B. Capel, E. M. Rubin, J. H. Nadeau, A. Matin, The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature 435, 360–364 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.T. Noguchi, M. Noguchi, A recessive mutation (ter) causing germ cell deficiency and a high incidence of congenital testicular teratomas in 129/Sv-ter mice. J. Natl. Cancer Inst. 75, 385–392 (1985). [PubMed] [Google Scholar]
- 71.S. Aramaki, K. Hayashi, K. Kurimoto, H. Ohta, Y. Yabuta, H. Iwanari, Y. Mochizuki, T. Hamakubo, Y. Kato, K. Shirahige, M. Saitou, A mesodermal factor, T, specifies mouse germ cell fate by directly activating germline determinants. Dev. Cell 27, 516–529 (2013). [DOI] [PubMed] [Google Scholar]
- 72.E. Magnúsdóttir, S. Dietmann, K. Murakami, U. Gunesdogan, F. Tang, S. Bao, E. Diamanti, K. Lao, B. Gottgens, M. Azim Surani, A tripartite transcription factor network regulates primordial germ cell specification in mice. Nat. Cell Biol. 15, 905–915 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.E. Magnúsdóttir, M. A. Surani, How to make a primordial germ cell. Development 141, 245–252 (2014). [DOI] [PubMed] [Google Scholar]
- 74.Y. Ohinata, B. Payer, D. O'Carroll, K. Ancelin, Y. Ono, M. Sano, S. C. Barton, T. Obukhanych, M. Nussenzweig, A. Tarakhovsky, M. Saitou, M. A. Surani, Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436, 207–213 (2005). [DOI] [PubMed] [Google Scholar]
- 75.M. Sato, T. Kimura, K. Kurokawa, Y. Fujita, K. Abe, M. Masuhara, T. Yasunaga, A. Ryo, M. Yamamoto, T. Nakano, Identification of PGC7, a new gene expressed specifically in preimplantation embryos and germ cells. Mech. Dev. 113, 91–94 (2002). [DOI] [PubMed] [Google Scholar]
- 76.A. Bortvin, M. Goodheart, M. Liao, D. C. Page, Dppa3/Pgc7/stella is a maternal factor and is not required for germ cell specification in mice. BMC Dev. Biol. 4, 2 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.P. N. Pereira, M. P. Dobreva, L. Graham, D. Huylebroeck, K. A. Lawson, A. N. Zwijsen, Amnion formation in the mouse embryo: The single amniochorionic fold model. BMC Dev. Biol. 11, 48 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.I. Bergiers, T. Andrews, O. Vargel Bolukbasi, A. Buness, E. Janosz, N. Lopez-Anguita, K. Ganter, K. Kosim, C. Celen, G. Itir Percin, P. Collier, B. Baying, V. Benes, M. Hemberg, C. Lancrin, Single-cell transcriptomics reveals a new dynamical function of transcription factors during embryonic hematopoiesis. eLife 7, e29312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.K. Adachi, I. Nikaido, H. Ohta, S. Ohtsuka, H. Ura, M. Kadota, T. Wakayama, H. R. Ueda, H. Niwa, Context-dependent wiring of Sox2 regulatory networks for self-renewal of embryonic and trophoblast stem cells. Mol. Cell 52, 380–392 (2013). [DOI] [PubMed] [Google Scholar]
- 80.E. Wicklow, S. Blij, T. Frum, Y. Hirate, R. A. Lang, H. Sasaki, A. Ralston, HIPPO pathway members restrict SOX2 to the inner cell mass where it promotes ICM fates in the mouse blastocyst. PLOS Genet. 10, e1004618 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.A. Aguilera-Castrejon, B. Oldak, T. Shani, N. Ghanem, C. Itzkovich, S. Slomovich, S. Tarazi, J. Bayerl, V. Chugaeva, M. Ayyash, S. Ashouokhi, D. Sheban, N. Livnat, L. Lasman, S. Viukov, M. Zerbib, Y. Addadi, Y. Rais, S. Cheng, Y. Stelzer, H. Keren-Shaul, R. Shlomo, R. Massarwa, N. Novershtern, I. Maza, J. H. Hanna, Ex utero mouse embryogenesis from pre-gastrulation to late organogenesis. Nature 593, 119–124 (2021). [DOI] [PubMed] [Google Scholar]
- 82.A. Aguilera-Castrejon, J. H. Hanna, Ex utero culture of mouse embryos from pregastrulation to advanced organogenesis. J. Vis. Exp. 176, e63160 (2021). [DOI] [PubMed] [Google Scholar]
- 83.A. Nagy, K. Nagy, M. Gertsenstein, Production of mouse chimeras by aggregating pluripotent stem cells with embryos. Methods Enzymol. 476, 123–149 (2010). [DOI] [PubMed] [Google Scholar]
- 84.J. Artus, A.-K. Hadjantonakis, Generation of chimeras by aggregation of embryonic stem cells with diploid or tetraploid mouse embryos. Methods Mol. Biol. 693, 37–56 (2011). [DOI] [PubMed] [Google Scholar]
- 85.Y. Ohinata, T. Tsukiyama, Establishment of trophoblast stem cells under defined culture conditions in mice. PLOS ONE 9, e107308 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.A.-K. Hadjantonakis, L. L. Cox, P. P. Tam, A. Nagy, An X-linked GFP transgene reveals unexpected paternal X-chromosome activity in trophoblastic giant cells of the mouse placenta. Genesis 29, 133–140 (2001). [DOI] [PubMed] [Google Scholar]
- 87.C. Dupont, F. Loos, J. Kong-A-San, J. Gribnau, FGF treatment of host embryos injected with ES cells increases rates of chimaerism. Transgenic Res. 26, 237–246 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.T. B. Nesterova, G. Wei, H. Coker, G. Pintacuda, J. S. Bowness, T. Zhang, M. Almeida, B. Bloechl, B. Moindrot, E. J. Carter, I. Alvarez Rodrigo, Q. Pan, Y. Bi, C. X. Song, N. Brockdorff, Systematic allelic analysis defines the interplay of key pathways in X chromosome inactivation. Nat. Commun. 10, 3129 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.G. X. Y. Zheng, J. M. Terry, P. Belgrader, P. Ryvkin, Z. W. Bent, R. Wilson, S. B. Ziraldo, T. D. Wheeler, G. P. McDermott, J. Zhu, M. T. Gregory, J. Shuga, L. Montesclaros, J. G. Underwood, D. A. Masquelier, S. Y. Nishimura, M. Schnall-Levin, P. W. Wyatt, C. M. Hindson, R. Bharadwaj, A. Wong, K. D. Ness, L. W. Beppu, H. J. Deeg, C. McFarland, K. R. Loeb, W. J. Valente, N. G. Ericson, E. A. Stevens, J. P. Radich, T. S. Mikkelsen, B. J. Hindson, J. H. Bielas, Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.T. Stuart, A. Butler, P. Hoffman, C. Hafemeister, E. Papalexi, W. M. Mauck III, Y. Hao, M. Stoeckius, P. Smibert, R. Satija, Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.M. D. Young, S. Behjati, SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. Gigascience 9, giaa151 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.C. Ahlmann-Eltze, W. Huber, glmGamPoi: Fitting gamma-poisson generalized linear models on single cell count data. Bioinformatics 36, 5701–5702 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.M. V. Kuleshov, M. R. Jones, A. D. Rouillard, N. F. Fernandez, Q. Duan, Z. Wang, S. Koplev, S. L. Jenkins, K. M. Jagodnik, A. Lachmann, M. G. McDermott, C. D. Monteiro, G. W. Gundersen, A. Ma'ayan, Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S18
Table S1