PURPOSE
Clinical utility of up-front multigene panel testing (MGPT) is directly related to the frequency of pathogenic variants (PVs) in the population screened and how genetic findings can be used to guide treatment decision making and cancer prevention efforts. The benefit of MGPT for many common malignancies remains to be determined. In this study, we evaluated up-front MGPT in unselected patients with endometrial cancer (EC) to determine the frequency of PVs in cancer susceptibility genes.
METHODS
Patients with EC were prospectively enrolled at nine Ohio institutions from October 1, 2017, to December 31, 2020. Nine hundred and sixty-one patients with newly diagnosed EC underwent clinical germline MGPT for 47 cancer susceptibility genes. In addition to estimating the prevalence of germline PVs, the number of individuals identified with Lynch syndrome (LS) was compared between MGPT and tumor-based screening.
RESULTS
Likely pathogenic variants or PVs were identified in 97 of 961 women (10.1%). LS was diagnosed in 29 of 961 patients (3%; 95% CI, 2.1 to 4.3), with PVs in PMS2 most frequent. MGPT revealed nine patients with LS in addition to the 20 identified through routine tumor-based screening. BRCA1 and BRCA2 PVs were found in 1% (10 of 961; 95% CI, 0.6 to 1.9) of patients and that group was significantly enriched for type II ECs.
CONCLUSION
This prospective, multicenter study revealed potentially actionable germline variants in 10% of unselected women with newly diagnosed EC, supporting the use of up-front MGPT for all EC patients. The discovery that BRCA1 or BRCA2 heterozygotes frequently had type II cancers points to therapeutic opportunities for women with aggressive histologic EC subtypes.
INTRODUCTION
Endometrial cancer (EC) is the most common gynecologic malignancy in developed countries.1 Risk factors include age, obesity, and inherited cancer susceptibility.2 Lynch syndrome (LS), which is caused by germline pathogenic variants (PVs) in the mismatch repair (MMR) genes MLH1, MSH2 (EPCAM), MSH6, and PMS2, accounts for 2%-5% of all ECs.3-5
CONTEXT
Key Objective
We sought to determine the true burden of cancer susceptibility in women with endometrial cancer using up-front multigene panel testing (MGPT) in a prospective multicenter study. The study compares tumor immunohistochemistry screening for Lynch syndrome (LS) and up-front gene testing findings. We document the frequency and types of cancer susceptibility alleles identified in what is the largest prospective series reported to date.
Knowledge Generated
PMS2 pathogenic variants are as frequent as MSH6 pathogenic variants in unselected patients with endometrial cancer. Up-front MGPT identifies women carrying LS and hereditary breast and ovarian cancer pathogenic or likely pathogenic gene variants that do not meet National Comprehensive Cancer Network criteria for genetic counseling and testing.
Relevance
Up-front MGPT is feasible for LS screening and obviates the need for follow-up germline testing when there is a tumor immunohistochemistry abnormality. It also identifies women who carry cancer susceptibility alleles that would otherwise go undetected.
There is substantial variability in the penetrance and expressivity of the different LS genes. MLH1 and MSH2 confer similar high risk for EC (21%-57%) and colorectal cancer (33%-61%), as well as for a variety of other malignancies.6 Early-onset cancers, synchronous and metachronous tumors, and strong family histories of cancer are hallmarks of LS associated with MLH1 and MSH2. LS families segregating MSH6 mutations have later-onset disease and fewer cancers overall.7 The incidence of pathogenic germline MSH6 variants in unselected patients with EC is higher than other LS genes.8 PMS2, like MSH6, has lower penetrance than MLH1 and MSH2.9
Identification of LS in patients with EC affords cancer prevention opportunities for both index cases and their family members. Universal tumor screening for patients with EC using immunohistochemistry (IHC) and reflex MLH1 promoter hypermethylation testing has helped identify women at risk of LS, especially when family history is noncontributory. However, not all centers have adopted universal tumor testing. Furthermore, even with a positive tumor screen result, confirming the diagnosis of LS depends on successful referral for genetic counseling and that patients follow through with germline testing.5,10 There is evidence of significant drop-off between finding an abnormal IHC result and completion of genetic testing.10,11 A recent countrywide initiative in Canada found that despite universal tumor testing, LS was underdiagnosed.12
Mutations in cancer susceptibility genes (CSGs) other than those associated with LS are thought to play a smaller role in EC risk.13 Although multigene panel testing (MGPT) studies performed for women with EC have identified PVs in other CSGs,14-16 the true burden of PVs in other CSGs remains unknown. The modest number of cases investigated (381 in the study by Ring et al,14 156 in Cadoo et al,15 and 98 in Samadder et al16), coupled with the fact that the cohorts overrepresent women with nonendometrioid, high-grade, and higher-stage tumors, limits our understanding of how frequent PVs in other CSGs are in the general EC population.
There is a growing body of evidence that incidental findings that come with MGPT are clinically relevant and can be used to guide both treatment and prevention strategies for patients with cancer and their relatives.17,18 Determining the molecular underpinnings of cancer is a cornerstone of precision oncology. Genetic testing for patients with cancer has become central to approaches to prevent second malignancies, treatment, and risk stratification, and for guiding the care of unaffected relatives. This is particularly important when the link between inherited factors and cancer risk have been well-established, and screening and prevention strategies exist or could be reasonably developed.
This study aimed to prospectively determine the frequency and spectrum of PVs causing LS and PVs in other CSGs by using up-front germline MGPT in a large and unselected series of patients with EC recruited from multiple gynecologic oncology practices. Secondary objectives included determining whether MGPT increases LS diagnoses and evaluating the relationship between PVs and clinicopathologic features.
METHODS
Study Design and Participants
The Ohio Prevention and Treatment of Endometrial Cancer (OPTEC) Initiative is a prospective collaborative study led by The Ohio State University (OSU) Comprehensive Cancer Center (NCT03460483).19 Collaborating centers are high-volume centers where women diagnosed with EC are cared for by board-certified gynecologic oncologists (Appendix Table A1). Two institutions are National Cancer Institute (NCI)–designated Comprehensive Cancer Centers. The study was approved by the Institutional Review Boards at each participating center with OSU serving as the Institutional Review Board of record. Written informed consent was obtained. All study-related services and/or procedures provided were at no cost to participants.
Women who had a hysterectomy or diagnostic biopsy proving a newly diagnosed EC from October 1, 2017, to December 31, 2020, were eligible. Clinical and family history data, pathology reports, tumor block, and blood and/or saliva specimen were collected and relevant demographic and clinicopathologic data were extracted from those records.
Genetic Testing and LS Risk Prediction
Germline testing was completed at Invitae Corporation using the 47-gene Common Hereditary Cancers Panel (Appendix Table A2). Patients with likely pathogenic (LP) variants or PVs received genetic counseling as part of the study. For the purposes of clinical management, patients with LP variants are counseled the same as those with PVs. Variants in CSGs other than LS-related genes were classified as high, moderate, or low penetrance or autosomal recessive on the basis of National Comprehensive Cancer Network (NCCN) designations20 and expert opinion.
The Dana-Farber Cancer Institute online calculator21 was used to determine PREMM5 scores.
Tumor Studies
Most participating centers performed MMR IHC in Clinical Laboratory Improvement Amendments–certified laboratories as part of universal screening. One center did not have routine screening, and one screened a subset of cases. All four MMR proteins were evaluated per the local institutions' IHC methods and interpretation guidelines.22 Reflex MLH1 methylation testing was performed for tumors lacking MLH1 and PMS2 expression.17 For the LS cases that did not have universal IHC performed, blinded IHC was performed in the Clinical Laboratory Improvement Amendments–certified laboratory at OSU. In addition, for a subset of cases with equivocal findings, tumor IHC was repeated (interpretation by A.S.) and/or microsatellite instability typing was performed using the Promega v1.2 panel.17
Statistical Analyses
Fisher's exact test was used to test for differences in proportions and Student's t test for between-group comparisons of continuous variables. The rates of mutations in PMS2 and BRCA genes were compared between the present OPTEC cohort and previously published population-based genetic testing studies using Fisher's exact test and Clopper-Pearson 95% CIs. These are illustrated with forest plots (Appendix Fig A1). All tests of statistical significance were two-sided, and statistical significance was considered as P value < .05.
RESULTS
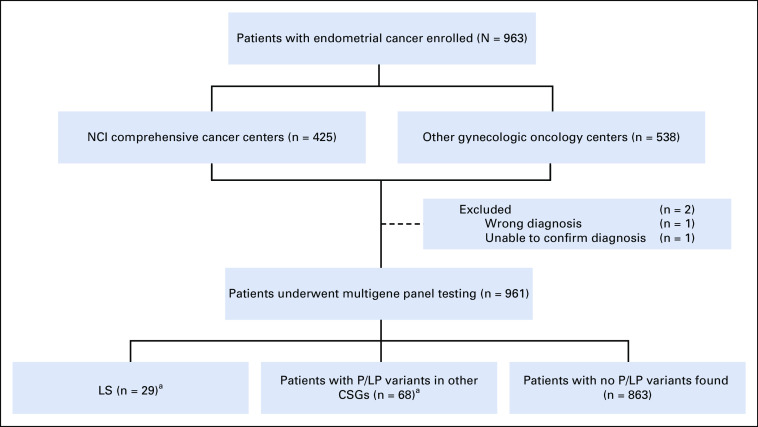
Nine hundred sixty-three patients were recruited at nine centers and their affiliated sites (Appendix Table A1). The cohort represents an unselected subset of patients treated during the enrollment period. Two patients were excluded (one wrong diagnosis and inability to confirm the EC for the second; Fig 1). The 961 patients with EC represent approximately 12% of the estimated total number of EC cases in Ohio during the study period. Baseline patient and tumor characteristics are presented in Table 1.
FIG 1.
Study profile. aThere were 100 P/LP variants identified in 97 patients; three patients were found to carry two P/LP variants each. One PMS2 LS patient was also heterozygous for a BRCA2 PV. Two patients were each found to have two PVs in other CSGs. One additional patient was found to have a TP53 likely mosaic PV (not included in any category). CSGs, cancer susceptibility genes; LS, Lynch syndrome; NCI, National Cancer Institute; P/LP, pathogenic/likely pathogenic; PVs, pathogenic variants.
TABLE 1.
Clinicopathologic Data (n = 961)
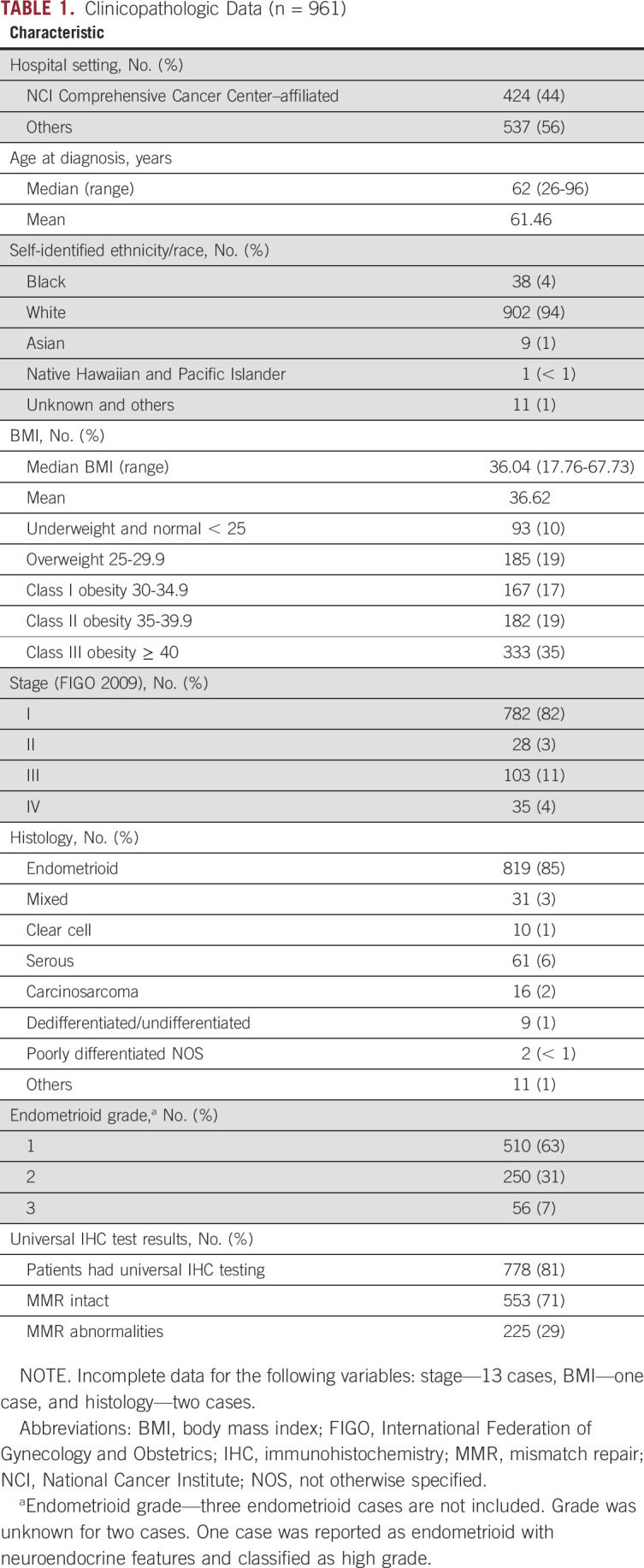
The clinical and demographic features (age, body mass index [BMI], histology, stage, and grade) are largely consistent with those for the United States overall.23 A noteworthy exception is the racial makeup of our study population is only 4% Black compared with the estimated 9% for the state of Ohio and 13% for the United States overall. Among the 778 women who had universal LS screening, 29% had IHC abnormalities.
Clinicopathologic features (age, race, BMI, stage, and grade) were similar for patients enrolled at the two NCI sites and the seven other practices, apart from a modest excess of nonendometrioid histologies at the NCI sites (16.8% v 12.2%; P = .05).
MGPT Findings
LP variants or PVs were identified in 97 women (10.1%). Three patients carried two PVs. An additional 321 women (33.4%) carried germline variants of uncertain significance (VUSs; Fig 1, Data Supplement).
LS genes.
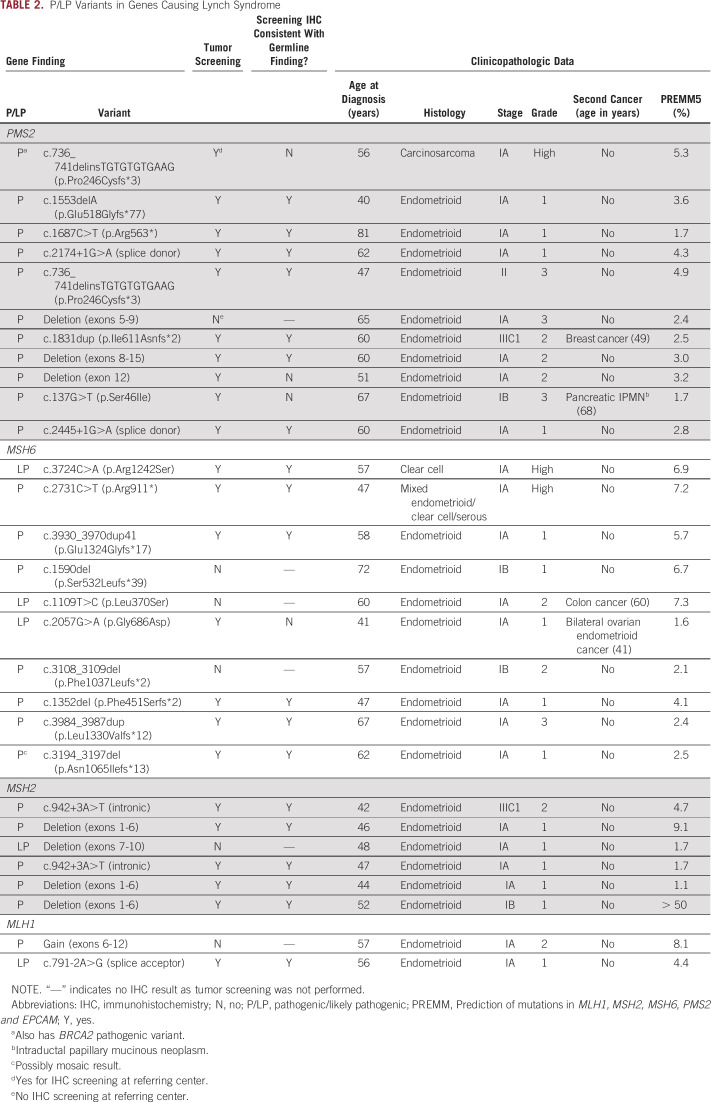
Three percent (29 of 961; 95% CI, 2.1 to 4.3) of patients were found to have LP variants or PVs in an MMR gene and consequently a LS diagnosis. The most common gene defects were in PMS2 (11) and MSH6 (10), followed by MSH2 (six) and MLH1 (two). Of the 29 LS variants, 24 were classified as PVs and five as LP variants (Table 2). Of note, one MSH6 PV (p.Asn1065Ilefs*13) was reported as possibly mosaic.
TABLE 2.
P/LP Variants in Genes Causing Lynch Syndrome
Clinicopathologic features for LS cases were similar to the study population as a whole. Most LS patients had early-stage endometrioid tumors. LS cases were significantly younger (mean 55.5 v 61.6 years; P = .002), and BMIs were lower (29.1 v 36.8; P ≤ .001) compared with non-LS patients. Although the LS cohort together represents younger women with lower BMIs, when considered by gene, there was a nonsignificant trend toward higher BMIs and older age among PMS2 heterozygotes.
MGPT identified 10 patients with LS (34.5% of all LS cases) that would not otherwise have been recognized: six patients did not have MMR IHC performed and four patients (one MSH6 and three PMS2) who had universal tumor screening had IHC and/or methylation findings that would not have triggered germline testing (Table 2).
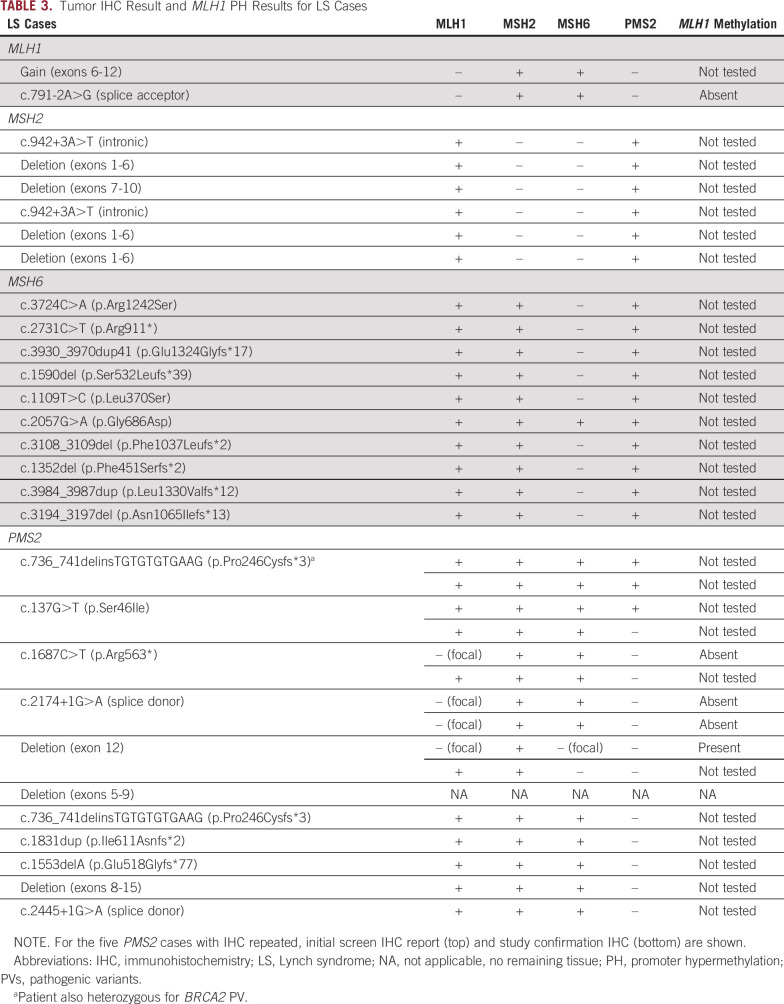
IHC was performed for LS cases that did not have prior testing (Table 3). IHC findings were largely consistent with the expected patterns of MMR expression for those women with LP variants or PVs involving MSH2, MLH1, and MSH6. All six MSH2 heterozygotes and both MLH1 heterozygotes had expected MMR expression patterns (absent MSH2 and MSH6 and absent MLH1 and PMS2, respectively). Nine of 10 MSH6 heterozygotes had isolated MSH6 loss, including the patient with the possibly mosaic result (p.Asn1065Ilefs*13). One patient with a LP missense MSH6 variant (p.Gly686Asp) had normal IHC for all four MMR proteins.
TABLE 3.
Tumor IHC Result and MLH1 PH Results for LS Cases
Screening IHC findings for women with germline PMS2 PVs were highly variable and highlight the challenges associated with using IHC to identify PMS2-LS (Table 3). Only five of the 10 PMS2 heterozygotes for which tissues were available had isolated PMS2 absence on the basis of initial clinical reports. Two cases had normal staining for all four MMR proteins (p.Ser46Ile and p.Pro246Cysfs*3). The first case with intact MMR protein expression (p.Ser46Ile) was reclassified as having isolated loss of PMS2 on review requested by her gynecologic oncologist. The second case with intact MMR expression was found in a patient with carcinosarcoma who was heterozygous for PMS2 p.Pro246Cysfs*3 and BRCA2 PVs. Her tumor was microsatellite-stable (Appendix Fig A2). Together, MSI and IHC findings indicate that although this patient has a diagnosis of LS, her EC is not likely to be causally associated with PMS2.
The three remaining PMS2 PV cases were reported as having MLH1 abnormalities in addition to PMS2 loss. IHC was repeated for all three cases. One case (p.Arg563*), whose tumor was originally interpreted as having loss of both PMS2 and MLH1 with no MLH1 promoter methylation, revealed isolated loss of PMS2 on repeat IHC. The second (c.2174+1G>A [splice donor]) had focal weak staining of MLH1 (5%-20%) and loss of PMS2 on initial and repeat IHC using an independent block. This case had no MLH1 promoter methylation. The third (deletion exon 12) had absent PMS2, partial absent MLH1, partial absent MSH6, and normal MSH2 on initial IHC. Repeat IHC showed intact MLH1 and MSH2 and loss of MSH6 and PMS2. This case had MLH1 methylation.
Five of the 29 patients with LS in our series (two MLH1, two MSH2, and one PMS2) had prior knowledge of LS in the family before study enrollment. Of the remaining 24 patients, 21 ultimately had IHC results or a personal or family history that fulfilled NCCN criteria for germline testing.6 Family history did not predict nearly one third of patients with LS (see PREMM5 Model scores, Table 2).
In addition to the LP variants or PVs identified, there were 56 MMR VUSs, most of which were found in women who had intact MMR IHC. However, two patients had unexplained IHC abnormalities: one had MLH1 p.Val201Leu VUS with absent MLH1 and PMS2 and no MLH1 promoter methylation; the other had MSH6 p.Ala1162Asp VUS with specific loss of MSH6. The patient with the MSH6 VUS had colon cancer at age 38 years and a family history unremarkable for LS. The MLH1 VUS patient had no history suggestive of LS.
Other CSG findings.
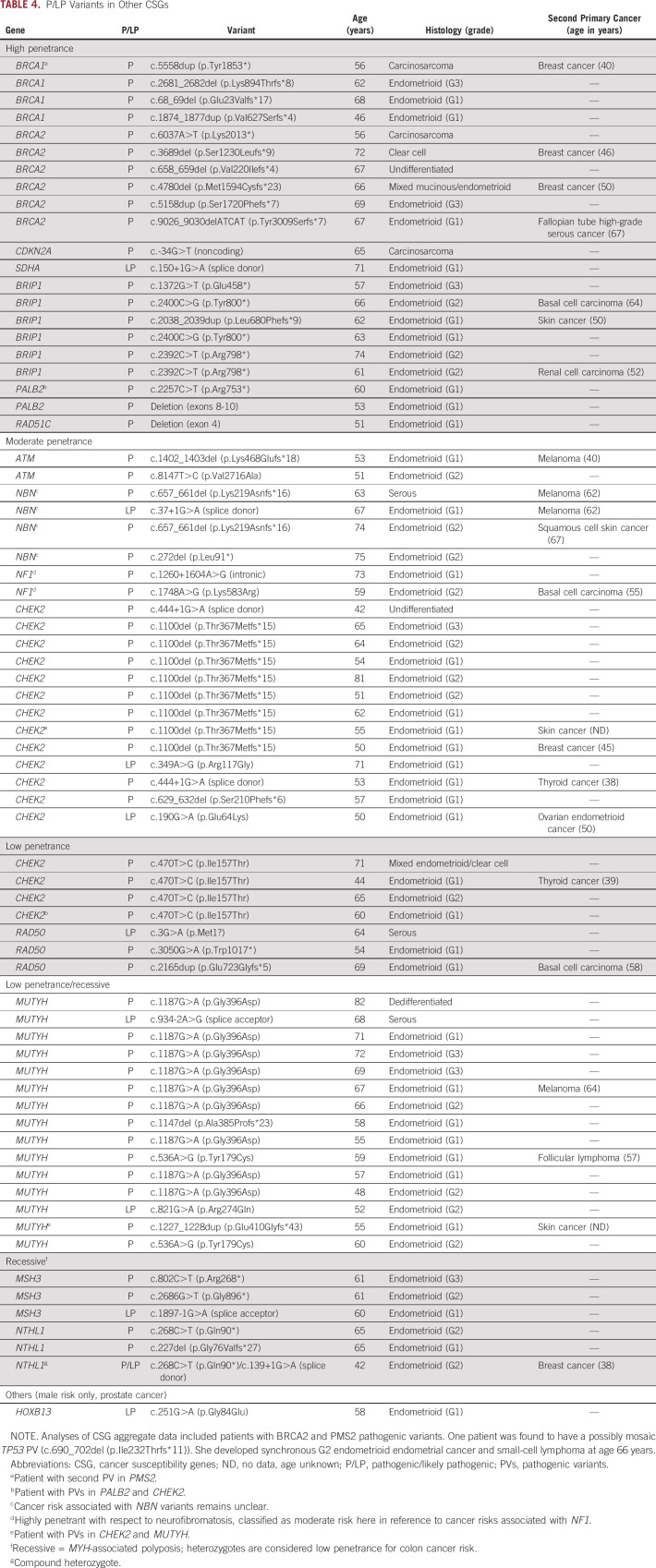
Sixty-eight women (7.1%) were found to have LP variants or PVs in 16 different CSGs (Table 4). Three women each carried two PVs. In aggregate, there were no differences in age, stage, grade, and histology for those 68 women compared with the rest of the cohort, LS excluded. Twenty-one patients (2.2% of study population) had LP variants or PVs in high-penetrance CSGs other than the LS genes. Nineteen of the 21 had LP variants or PVs in genes associated with breast and/or ovarian cancer.
TABLE 4.
P/LP Variants in Other CSGs
Ten patients were found to have PVs in BRCA1 or BRCA2 (1.04% of the study population; 95% CI, 0.6 to 1.9). Among the patients with BRCA PVs, there were significantly more type II (grade 3 endometrioid or nonendometrioid histologies) cancers compared with the rest of the cohort (6 of 10; P = .005; Table 4). Several had prior or synchronous malignancies with three patients having prior breast cancer diagnoses: two had histories of tamoxifen use. One patient had synchronous fallopian tube high-grade serous cancer at the time of her EC diagnosis. Half of the patients harboring BRCA1/2 PVs (5 of 10) had prior knowledge of the mutation at the time of OPTEC enrollment. Eight BRCA1/2 heterozygotes met NCCN criteria for genetic testing on the basis of either knowledge of a mutation in the family or family history.20
We identified one compound heterozygote for NTHL1 LP variants and PVs in a patient with a breast cancer (age 38 years) who had tamoxifen therapy. Another patient with active lymphoma was reported as possibly mosaic for a TP53 PV. Because we were unable to determine if the mosaicism in her blood DNA reflected somatic mosaicism, circulating tumor cell DNA, or clonal hematopoiesis, we chose not to include her among cases with LP variants or PVs.
MGPT identified 24 high- or moderate-penetrance variants in LS and/or hereditary breast and ovarian cancer genes in women who would not have been recommended for MGPT on the basis of NCCN family history and MMR IHC criteria. This represents an approximately 2.5% increase in yield with up-front germline testing.
Moderate- and low-penetrance PVs made up nearly half of all variants reported (Table 4). CHEK2 and MUTYH variants were most frequent, consistent with general population frequencies.
DISCUSSION
To our knowledge, this is the largest MGPT study for newly diagnosed EC to date. The prospective nature of the study, uniformity of testing, and the fact that incident cases were unselected for features suggestive of inherited cancer risk allow for reliable estimates of the incidence of germline LP variants or PVs. Furthermore, the multi-institutional recruitment, with more than half of the patients treated at hospitals that are not affiliated with Comprehensive Cancer Centers, improves the generalizability of findings from this cohort.
The 3% rate (95% CI, 2.1 to 4.3) of LS is consistent with prior estimates. The most common genetic cause of LS in this cohort was PMS2, and together MSH6 and PMS2 represent 72% of LS cases. The importance of MSH6 in the development of EC came to light nearly two decades ago,8 and the rate of MSH6 LP variants or PVs in OPTEC is consistent with prior estimates of incidence. Our finding that 1% of patients with EC harbored PMS2 PVs is somewhat unexpected and demonstrates the importance of PMS2 in EC risk.
Delayed development of clinical testing for PMS2 and lower penetrance of PMS2-associated LS contributed to the historical assumption that PMS2 is a rare cause of LS. MGPT revealed that PVs in PMS2 are not uncommon.24 As testing techniques evolved, it became apparent that deletions, often previously not tested, contributed to a substantial portion of LS-causing PMS2 variants.25 This becomes important when comparing our PMS2 PV rate with other cohorts. If the testing method used in other studies could not detect deletions, then the number of PMS2 variants would underestimate PV incidence. PMS2 deletions accounted for 3 of 11 (27%) of the PVs found in OPTEC, and this rate mirrors the frequency found in a recent large colorectal cohort.26
Although cancer risks associated with PMS2 are lower than other MMR genes, penetrance of PMS2 PVs remains to be determined.9,27,28 Comparison with three large population-based studies revealed the frequency of PMS2 PVs in OPTEC (excluding the three deletion cases) is significantly higher than expected (Appendix Fig A1).29-31 MSI and IHC confirmed that EC was causally associated with PMS2 PVs in 8 of 10 cases (the patient with both PMS2 and BRCA2 PVs being a clear exception and the patient with MLH1 promoter methylation not easily explained). Together these findings demonstrate the important role PMS2 plays in EC risk.
IHC screening to identify patients with PMS2-LS is particularly problematic.32 Only half of PMS2 heterozygotes had isolated loss of PMS2 in their tumors. The complex IHC patterns seen, including the three cases reported as having MLH1 abnormalities in addition to PMS2 loss, have been previously described as consistent with PMS2-related disease.32
Our study revealed that MGPT identified more patients with LS than universal LS tumor screening. This benefit may be particularly important for identifying the lower-penetrance MSH6 and PMS2 variants as family history is unlikely to trigger referral for germline testing and IHC can be problematic. Acknowledging that many centers do not routinely screen for tumor MMR defects, implementing MGPT in the up-front setting would be a more direct way to identify patients with EC with LS and other highly penetrant cancer syndromes. MMR IHC will, however, remain an integral component of determining candidacy for immune checkpoint blockade therapy.
More than 7% (68 of 961) of the OPTEC patients were heterozygous for LP variants or PVs in 16 different CSGs, other than the LS genes. The high frequency of BRCA1 and BRCA2 PVs in our cohort (1.04%; 95% CI, 0.6 to 1.9) is noteworthy. Comparing OPTEC findings with the unselected populations in United States, United Kingdom, and Australia, the prevalence of BRCA PVs was significantly higher (Appendix Fig A1).29-31
The specific association between germline BRCA PVs and type II ECs known to have worse outcomes is an important finding. Hysterectomy at the time of risk-reducing surgery for BRCA carriers would greatly reduce the chance of developing aggressive type II ECs and would eliminate the increased EC risk associated with the widespread tamoxifen use for prevention and treatment of BRCA-related breast cancer. Furthermore, if BRCA carrier ECs are deficient in homologous repair, poly (ADP-ribose) polymerase inhibition may be a therapeutic option.
MGPT revealed actionable results in an additional 58 women. Because of the small numbers of PVs seen in CSGs other than the LS and BRCA loci and the fact that we did not investigate patient tumors, we are unable to speculate as to whether those variants are causally associated with EC.
In summary, we found that 10.1% of patients with EC have germline LP variants or PVs in a CSG, which is similar to the rates found in patients with colorectal and breast cancer.18,33 We confirm that LS accounts for approximately 3% of all EC patients and demonstrate that approximately 1% of patients with EC carry BRCA1 or BRCA2 LP variants or PVs. Our data support the up-front use of MGPT for all EC patients to improve LS detection and to better understand the role genetic risk plays in this common malignancy.
ACKNOWLEDGMENT
We would like to thank the patients, as well as their families and caregivers, for their participation in the OPTEC study. We would also like to acknowledge the important contributions made by the many physicians and staff at all the participating institutions. We thank Dr Dan Jones, Director of Molecular Pathology for the Ohio State University Comprehensive Cancer Center, for assistance with study-specific tumor methylation studies. Finally, we thank Invitae Corporation for genetic testing services and Ms Jessica Gillespie and Mr Keith Brownewell for MSI typings.
APPENDIX
FIG A1.
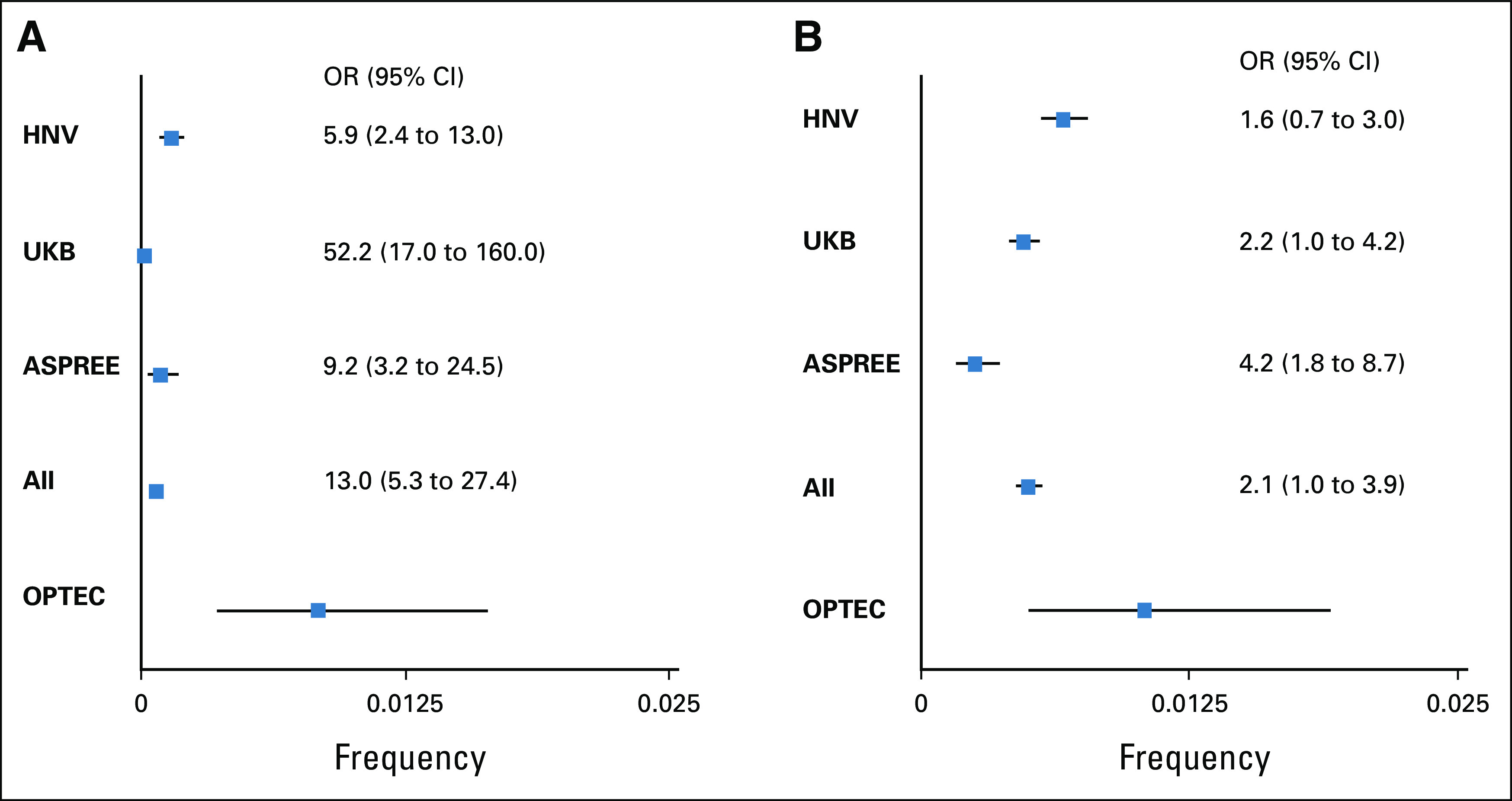
OPTEC PMS2 and BRCA1/2 PV frequency compared with general population. Forest plot demonstrating the increased frequency of (A) PMS2 and (B) BRCA1/2 PVs in the prospective OPTEC cohort. Frequencies and 95% CIs (Clopper-Pearson exact tests) for OPTEC, three large population studies and the three control populations combined are shown. The OR of the combined population studies is 13.0 (95% CI, 5.3 to 27.4; P = 5.14 × 10–7) for PMS2 and 2.1 (95% CI, 1.0 to 3.9; P = .03) for BRCA1/2. Because the population studies did not report deletion and insertion mutations, we did not include the cases with scale mutations in our calculation to avoid inflations of the ORs. All, HNV, UKB, and ASPREE combined; ASPREE, ASPirin in Reducing Events in the Elderly trial30; HNV, Healthy Nevada Project31; OPTEC, Ohio Prevention and Treatment of Endometrial Cancer; OR, odds ratio; PVs, pathogenic variants; UKB, UK Biobank.29
FIG A2.
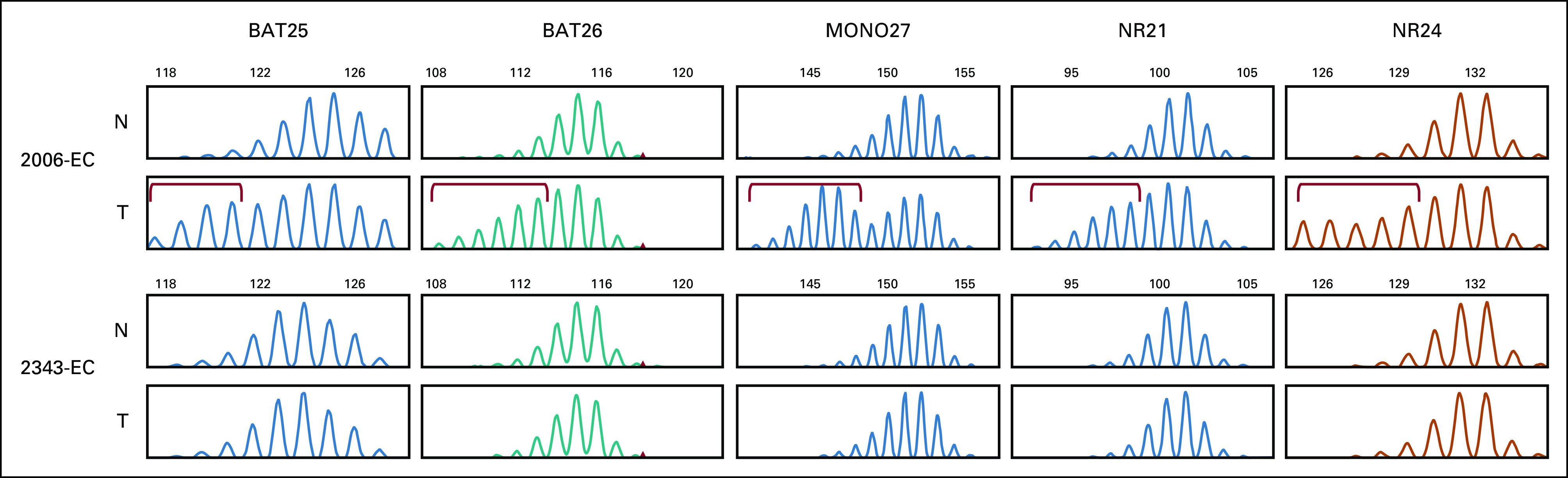
Promega v1.2 MSI typing for tumors from patients with germline pathogenic PMS2 variants. Tumor 2006-EC is a representative MSI-high case with MLH1 methylation and absent MLH1 and PMS2. Fragment sizes not evident in matched normal DNA are seen with five mononucleotide repeats. Tumor 2343-EC (PMS2 p.Pro246Cysfs*3) is microsatellite-stable. Aberrant fragment sizes are marked with red brackets. EC, endometrial cancer; MSI, microsatellite instability; N, normal DNA; T, tumor DNA.
TABLE A1.
OPTEC Collaborating Centers
TABLE A2.
Invitae Common Hereditary Cancers Panel—47 Genes
Heather Hampel
This author is a member of the JCO Precision Oncology Editorial Board. Journal policy recused the author from having any role in the peer review of this manuscript.
Stock and Other Ownership Interests: Genome Medical
Consulting or Advisory Role: Invitae, Genome Medical, Promega, 23andMe
David Cohn
Consulting or Advisory Role: Oncology Analytics
Research Funding: NRG Oncology, Advaxis, Agenus, Ajinomoto, Array BioPharma, AstraZeneca, Bristol Myers Squibb, Clovis Oncology, Exelixis, Genentech, GlaxoSmithKline, Gynecologic Oncology Group, ImmunoGen, INC Research, inVentiv Health, Janssen Research & Development, Ludwig Institute for Cancer Research, EMD Serono, Stemcentrx, Tesaro, AbbVie, Henry Jackson Foundation, PharmaMar, Sanofi, Eisai, Pfizer, Novartis, Regeneron, Tricon Pharmaceuticals
Other Relationship: Elsevier, UpToDate
Joseph P. McElroy
Employment: Pfizer (I)
Steven Waggoner
Consulting or Advisory Role: Regeneron
John Nakayama
Honoraria: ZoomRx, Medscape, M3, Curio Science
Consulting or Advisory Role: AstraZeneca, Clovis Oncology
Speakers' Bureau: Merck, Eisai
Research Funding: Xodus
Kim Resnick
Honoraria: Clovis Oncology
Sareena Singh
Speakers' Bureau: AstraZeneca, Merck
Aine Clements
Consulting or Advisory Role: AstraZeneca, Tesaro/GSK
Paul J. Goodfellow
Research Funding: Promega
No other potential conflicts of interest were reported.
PRIOR PRESENTATION
Presented in part as a poster at the virtual Society of Gynecologic Oncology Annual Meeting, March 19, 2021.
SUPPORT
Supported by an Ohio State University James Comprehensive Cancer Center Statewide Pelotonia Cancer Impact Award and R01CA223219.
DATA SHARING STATEMENT
A data sharing statement provided by the authors is available with this article at DOI https://doi.org/10.1200/PO.21.00249.
AUTHOR CONTRIBUTIONS
Conception and design: Heather Hampel, David E. Cohn, Sareena Singh, Paul J. Goodfellow
Financial support: Heather Hampel, Paul J. Goodfellow
Administrative support: Alexis Chassen
Provision of study materials or patients: David Cohn, Steven Waggoner, John Nakayama, Caroline Billingsley, Kim Resnick, Stephen Andrews, Eric Jenison, Aine Clements, Robert Neff
Collection and assembly of data: Monica D. Levine, Rachel Pearlman, Casey Cosgrove, Alexis Chassen, Adrian Suarez, David A. Barrington, Sareena Singh, Paul J. Goodfellow
Data analysis and interpretation: Monica D. Levine, Heather Hampel, Joseph P. McElroy, Paul J. Goodfellow
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Heather Hampel
This author is a member of the JCO Precision Oncology Editorial Board. Journal policy recused the author from having any role in the peer review of this manuscript.
Stock and Other Ownership Interests: Genome Medical
Consulting or Advisory Role: Invitae, Genome Medical, Promega, 23andMe
David Cohn
Consulting or Advisory Role: Oncology Analytics
Research Funding: NRG Oncology, Advaxis, Agenus, Ajinomoto, Array BioPharma, AstraZeneca, Bristol Myers Squibb, Clovis Oncology, Exelixis, Genentech, GlaxoSmithKline, Gynecologic Oncology Group, ImmunoGen, INC Research, inVentiv Health, Janssen Research & Development, Ludwig Institute for Cancer Research, EMD Serono, Stemcentrx, Tesaro, AbbVie, Henry Jackson Foundation, PharmaMar, Sanofi, Eisai, Pfizer, Novartis, Regeneron, Tricon Pharmaceuticals
Other Relationship: Elsevier, UpToDate
Joseph P. McElroy
Employment: Pfizer (I)
Steven Waggoner
Consulting or Advisory Role: Regeneron
John Nakayama
Honoraria: ZoomRx, Medscape, M3, Curio Science
Consulting or Advisory Role: AstraZeneca, Clovis Oncology
Speakers' Bureau: Merck, Eisai
Research Funding: Xodus
Kim Resnick
Honoraria: Clovis Oncology
Sareena Singh
Speakers' Bureau: AstraZeneca, Merck
Aine Clements
Consulting or Advisory Role: AstraZeneca, Tesaro/GSK
Paul J. Goodfellow
Research Funding: Promega
No other potential conflicts of interest were reported.
REFERENCES
- 1.Lortet-Tieulent J, Ferlay J, Bray F, et al. : International patterns and trends in endometrial cancer incidence, 1978-2013. J Natl Cancer Inst 110:354-361, 2018 [DOI] [PubMed] [Google Scholar]
- 2.Arnold M, Pandeya N, Byrnes G, et al. : Global burden of cancer attributable to high body-mass index in 2012: A population-based study. Lancet Oncol 16:36-46, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodfellow PJ, Billingsley CC, Lankes HA, et al. : Combined microsatellite instability, MLH1 methylation analysis, and immunohistochemistry for Lynch syndrome screening in endometrial cancers from GOG210: An NRG Oncology and Gynecologic Oncology Group study. J Clin Oncol 33:4301-4308, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchanan DD, Tan YY, Walsh MD, et al. : Tumor mismatch repair immunohistochemistry and DNA MLH1 methylation testing of patients with endometrial cancer diagnosed at age younger than 60 years optimizes triage for population-level germline mismatch repair gene mutation testing. J Clin Oncol 32:90-100, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryan NAJ, Glaire MA, Blake D, et al. : The proportion of endometrial cancers associated with Lynch syndrome: A systematic review of the literature and meta-analysis. Genet Med 21:2167-2180, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.National Comprehensive Cancer Network : Genetic/Familial High-Risk Assessment: Colorectal, Version: 1.2021, 2020 [Google Scholar]
- 7.Baglietto L, Lindor NM, Dowty JG, et al. : Risks of Lynch syndrome cancers for MSH6 mutation carriers. J Natl Cancer Inst 102:193-201, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodfellow PJ, Buttin BM, Herzog TJ, et al. : Prevalence of defective DNA mismatch repair and MSH6 mutation in an unselected series of endometrial cancers. Proc Natl Acad Sci USA 100:5908-5913, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ten Broeke SW, van der Klift HM, Tops CMJ, et al. : Cancer risks for PMS2-associated Lynch syndrome. J Clin Oncol 36:2961-2968, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kahn RM, Gordhandas S, Maddy BP, et al. : Universal endometrial cancer tumor typing: How much has immunohistochemistry, microsatellite instability, and MLH1 methylation improved the diagnosis of Lynch syndrome across the population? Cancer 125:3172-3183, 2019 [DOI] [PubMed] [Google Scholar]
- 11.Tjalsma AS, Wagner A, Dinjens WNM, et al. : Evaluation of a nationwide Dutch guideline to detect Lynch syndrome in patients with endometrial cancer. Gynecol Oncol 160:771-776, 2021 [DOI] [PubMed] [Google Scholar]
- 12.Thompson E, Huvila J, Leung S, et al. : 4 Refining pathologic interpretation of endometrial carcinomas: Lessons learned from a nationwide study in a new era of molecular classification. Int J Gynecol Cancer 30:A3-A4, 2020 [Google Scholar]
- 13.Dork T, Hillemanns P, Tempfer C, et al. : Genetic susceptibility to endometrial cancer: Risk factors and clinical management. Cancers (Basel) 12:2407, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cadoo KA, Mandelker DL, Mukherjee S, et al. : Understanding inherited risk in unselected newly diagnosed patients with endometrial cancer. JCO Precis Oncol 3, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ring KL, Bruegl AS, Allen BA, et al. : Germline multi-gene hereditary cancer panel testing in an unselected endometrial cancer cohort. Mod Pathol 29:1381-1389, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samadder NJ, Riegert-Johnson D, Boardman L, et al. : Comparison of universal genetic testing vs guideline-directed targeted testing for patients with hereditary cancer syndrome. JAMA Oncol 7:230-237, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearlman R, Frankel WL, Swanson B, et al. : Prevalence and spectrum of germline cancer susceptibility gene mutations among patients with early-onset colorectal cancer. JAMA Oncol 3:464-471, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yurgelun MB, Allen B, Kaldate RR, et al. : Identification of a variety of mutations in cancer predisposition genes in patients with suspected Lynch syndrome. Gastroenterology 149:604-613.e20, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.ClinicalTrials.gov: Universal endometrial cancer DNA sequencing for detection of Lynch syndrome and personalized care, https://clinicaltrials.gov/ct2/show/NCT03460483
- 20.National Comprehensive Cancer Network : Genetic/Familial High-Risk Assessment: Breast, Ovarian and Pancreatic, Version: 1.2022, 2021 [Google Scholar]
- 21.Lynch syndrome prediction model: MLH1, MSH2, MSH6, PMS2, and EPCAM gene mutations, https://premm.dfci.harvard.edu/
- 22.Cosgrove CM, Cohn DE, Hampel H, et al. : Epigenetic silencing of MLH1 in endometrial cancers is associated with larger tumor volume, increased rate of lymph node positivity and reduced recurrence-free survival. Gynecol Oncol 146:588-595, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Felix AS, Brinton LA: Cancer progress and priorities: Uterine cancer. Cancer Epidemiol Biomarkers Prev 27:985-994, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Espenschied CR, LaDuca H, Li S, et al. : Multigene panel testing provides a new perspective on Lynch syndrome. J Clin Oncol 35:2568-2575, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Klift H, Wijnen J, Wagner A, et al. : Molecular characterization of the spectrum of genomic deletions in the mismatch repair genes MSH2, MLH1, MSH6, and PMS2 responsible for hereditary nonpolyposis colorectal cancer (HNPCC). Genes Chromosomes Cancer 44:123-138, 2005 [DOI] [PubMed] [Google Scholar]
- 26.Pearlman RFW, Swanson BJ, Jones D, et al. : A prospective Statewide study of universal screening for hereditary colorectal cancer: The Ohio Colorectal Cancer Prevention Initiative. JCO Precis Oncol 5:779-791, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moller P, Seppala TT, Bernstein I, et al. : Cancer risk and survival in path_MMR carriers by gene and gender up to 75 years of age: A report from the prospective Lynch syndrome database. Gut 67:1306-1316, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goodenberger ML, Thomas BC, Riegert-Johnson D, et al. : PMS2 monoallelic mutation carriers: The known unknown. Genet Med 18:13-19, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel AP, Wang M, Fahed AC, et al. : Association of rare pathogenic DNA variants for familial hypercholesterolemia, hereditary breast and ovarian cancer syndrome, and Lynch syndrome with disease risk in adults according to family history. JAMA Netw Open 3:e203959, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lacaze P, Sebra R, Riaz M, et al. : Medically actionable pathogenic variants in a population of 13,131 healthy elderly individuals. Genet Med 22:1883-1886, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grzymski JJ, Elhanan G, Morales Rosado JA, et al. : Population genetic screening efficiently identifies carriers of autosomal dominant diseases. Nat Med 26:1235-1239, 2020 [DOI] [PubMed] [Google Scholar]
- 32.van der Klift HM, Mensenkamp AR, Drost M, et al. : Comprehensive mutation analysis of PMS2 in a large cohort of probands suspected of Lynch syndrome or constitutional mismatch repair deficiency syndrome. Hum Mutat 37:1162-1179, 2016 [DOI] [PubMed] [Google Scholar]
- 33.Buys SS, Sandbach JF, Gammon A, et al. : A study of over 35,000 women with breast cancer tested with a 25-gene panel of hereditary cancer genes. Cancer 123:1721-1730, 2017 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
A data sharing statement provided by the authors is available with this article at DOI https://doi.org/10.1200/PO.21.00249.