PURPOSE
Poly ADP-ribose polymerase inhibitors (PARPi) are used for patients with advanced prostate cancer bearing alterations in homologous recombination repair (HRR) genes. We sought to characterize HRR gene variants and describe real-world outcomes for patients on PARPi.
METHODS
The US Department of Veterans Affairs’ National Precision Oncology Program’s database was reviewed to identify patients who underwent somatic DNA sequencing and were prescribed a PARPi before May 15, 2020. Somatic and germline variants within HRR genes were reported, and pathogenicity was reviewed via OncoKB. In patients treated with PARPi for > 4 weeks, the rate of those achieving a 30% decrease in prostate-specific antigen (PSA30) and composite progression-free survival (PFS) were compared between patients bearing pathogenic variants of BRCA2 and patients without these variants using Mann-Whitney and log-rank tests, respectively.
RESULTS
Forty-eight patients bearing 67 total HRR gene variants were prescribed PARPi for prostate cancer. Twenty-one patients (43.8%) were found to have at least one pathogenic HRR gene variant. Eight (16.6%) were referred to genetic counseling, and five (10.4%) were ultimately confirmed with germline variants. The median PFS was 4.0 months, and PSA30 was 25.6% (11 of 43) for all 43 evaluable patients. Patients with pathogenic BRCA2 variants (n = 13) had higher PSA30 (69.2% v 4.0%; P < .001) and longer PFS (7.2 v 2.8 months; P = .0291) than those without.
CONCLUSION
In a real-world setting, heavily pretreated patients with prostate cancer and pathogenic BRCA2 variants have a significant PSA response rate and a PFS > 7 months with PARPi. This work emphasizes the importance of determining pathogenicity and origin of HRR alterations to better inform clinical treatment decisions and highlights the need for provider education and other decision support tools.
BACKGROUND
In the United States, prostate cancer is the second most common malignancy among men, with an annual incidence of approximately 250,000.1 Although most patients with early-stage prostate cancer share a favorable prognosis, patients with metastatic castrate-resistant prostate cancer (mCRPC) frequently have poor outcomes, with a median overall survival ranging from 13 months2 to 32 months.3,4 Unfortunately, despite an overall decline in the incidence of prostate cancer in the United States, the incidence of metastatic prostate cancer continues to rise.5 Thus, efforts to improve treatment options for men with mCRPC are critical.
CONTEXT
Key Objective
Poly ADP-ribose polymerase inhibitors (PARPi) are approved for patients with advanced prostate cancer bearing pathogenic/likely pathogenic mutations in specific homologous recombination repair (HRR) genes. We sought to review the gene variants and outcomes among Veterans who were treated with PARPi and had underwent tumor DNA sequencing through the US Department of Veterans Affairs' National Precision Oncology Program.
Knowledge Generated
Approximately one half of patients with prostate cancer are prescribed PARPi without harboring a pathogenic/likely pathogenic HRR gene variant. Patients bearing pathogenic/likely pathogenic BRCA2 mutations more frequently have longer progression-free survival and are more likely to achieve a significant prostate-specific antigen reduction compared with others receiving PARPi.
Clinical Relevance
Patients with advanced prostate cancer bearing BRCA2 pathogenic mutations have better outcomes with PARPi than those with other HRR gene variants. Efforts should be made to improve provider education regarding gene variant pathogenicity and relationship to clinical decision making.
Poly ADP-ribose polymerase inhibitors (PARPi), a family of drugs that inhibit the repair of DNA single-strand breaks, have recently become a therapy of interest given the relatively high frequency of alterations in DNA repair genes among patients with mCRPC. Germline alterations in homologous recombination repair (HRR) genes have been found in 7%-12% of men with mCRPC,6 and somatic alterations in HRR genes have been reported in 19% (The Cancer Genome Atlas) to 27%7 of both primary and metastatic samples of prostate cancer. Within these HRR genes, variants in BRCA2 tend to be most frequently observed followed by those in ATM and BRCA1.8
Several completed and ongoing trials to determine the efficacy of PARPi in patients with mCRPC have shown promising results for populations with certain HRR alterations with differential responses on the basis of specific HRR gene alterations.9-11 Of these, the PROfound study was the first clinical trial to assess the efficacy of olaparib by specific cohorts of HRR genes.9 This study showed a more favorable response to olaparib among patients with BRCA1, BRCA2, and ATM deleterious alterations compared with those with alterations in other HRR genes, with exploratory analyses demonstrating longer progression-free survival (PFS) for patients with BRCA2 alterations (10.8 months). Notably, a recent report from the TOPARP-B trial identified biallelic BRCA2 deletions as one of the biomarkers associated with a significantly improved response to PARPi.12 Currently, reports of PARPi administration and treatment outcomes on the basis of the genotype have been primarily limited to clinical trial settings. For these reasons, a study assessing real-world outcomes of patients with mCRPC treated with PARPi on the basis of the HRR genotype is warranted.
Consequently, the first aim of the current study was to review the HRR gene variants and overall treatment outcomes for Veterans who were prescribed PARPi for prostate cancer. The second aim of this study was to compare outcomes between patients bearing pathogenic/likely pathogenic BRCA2 variants and those without. We hypothesized that in a real-world context, patients with a wide array of HRR gene alterations would be prescribed PARPi and those with pathogenic BRCA2 alterations would have better outcomes than patients with non-BRCA2 alterations.
METHODS
Included Patients
Patients who (1) had been diagnosed with prostate cancer; (2) had successful genomic sequencing of tumor tissue or plasma; and (3) were prescribed olaparib, rucaparib, niraparib, or talazoparib for prostate cancer before May 2020 were selected for evaluation. All included patients had confirmed mCRPC. Patients who received a PARPi for a malignancy other than prostate cancer were excluded. Patients were required to receive a PARPi for at least 4 weeks to be included in outcomes assessments. No patient received a PARPi as part of a clinical trial.
Data Source and Ethical Considerations
This retrospective study was approved by the Institutional Review Board of the Durham Veterans Affairs' (VA) Medical Center, which provided a waiver of patient informed consent.
Eligible patients were initially selected from the US Department of Veterans Affairs' National Precision Oncology Program's (NPOP) database. The NPOP database includes all Veterans who underwent tumor DNA sequencing via next generation sequencing (NGS) through an external commercial laboratory contracted with NPOP from July 2015 to the present time. Eight of the NGS panels are as follows: Personalis(ACE CancerPlus Test), PGDx(CancerSELECT 88, CancerSELECT 125, CancerSELECT 203, or PlasmaSELECT), and Foundation Medicine(FoundationOne CDx, FoundationOne Liquid, and FoundationOne Liquid CDx). To determine pathogenicity, each HRR gene variant was reviewed in OncoKB, an online precision oncology knowledge database, in May 2021.13 Apart from the NPOP database, the VA Corporate Data Warehouse,14 a repository comprising data from VA clinical and administrative systems, and Joint Legacy Viewer,15 which provides read-only access to all Veterans Health Administration electronic health records, were additional data sources.
Data Collection
The results of tumor DNA sequencing and dates of PARPi prescriptions and discontinuations were retrieved from NPOP. The Corporate Data Warehouse was accessed to obtain prostate-specific antigen (PSA) levels from 6 months before the initial date of PARPi prescription to up to 6 months after the last prescription date. Demographic data were then extracted via manual review of Joint Legacy Viewer. Imaging reports and notes from oncology providers and genetic counseling specialists were also reviewed. From oncology provider notes, dates of initial diagnoses, previous therapies received, duration of PARPi administration, provider comments on PSA levels, and PARPi-associated toxicities were recorded. From the genetic counseling specialists' notes, the results of germline mutational testing and reasons for germline testing were obtained. Medical records were reviewed up to January 15, 2021.
Data Analysis
Baseline demographics, clinical and disease characteristics, PARPi prescriptions, drug toxicities, HRR somatic gene variants, and available germline mutational testing results were reported for all selected patients. HRR gene variants annotated as pathogenic/likely pathogenic per OncoKB and with an allele fraction of > 30% were identified as those most likely to be germline variants, with the knowledge that the National Comprehensive Cancer Network16 recommends germline testing for all patients with metastatic prostate cancer.
Patients were then categorized on the basis of the presence of pathogenic/likely pathogenic BRCA2 gene variants and those without (including those with BRCA2 variants classified as variants of uncertain significance [VUS]). Furthermore, to eliminate concerns for patients likely bearing clonal hematopoiesis of indeterminate potential (CHIP), only BRCA2 variants discovered from plasma sequencing with an allele fraction > 1.0% were included in the BRCA2 arm.17-21 Clinical outcomes that were compared between these two patient cohorts included: PSA30, defined as the percentage of patients achieving a reduction in PSA by 30% or more after PARPi initiation,22 and a composite PFS end point, defined as the interval between drug initiation and the earliest of drug discontinuation, clinical or radiographic progression, or death. In addition, an exploratory analysis of patients with BRCA2 variants classified as VUS was completed. For the purposes of assessing radiographic progression, two authors (M.P. and V.V.) reviewed radiology reports from at least 6 months before PARPi administration and up to 1 year after PARPi discontinuation. The presence of increasing size of tumors and/or new tumor(s) was used to identify radiographic progression as per RECIST criteria.23 Only patients who were administered PARPi for 4 weeks or longer were included in the analysis of outcomes.
Descriptive statistics were used to summarize patient demographic data and clinical and disease characteristics. Specifically, medians and interquartile ranges were used for continuous variables, and the absolute number and frequency distributions were used for categorical variables. Regarding outcomes, PSA30 was compared using the Mann-Whitney test, and survival curves of composite PFS were drawn by the Kaplan-Meier method and compared by the log-rank test for patients with and without BRCA2 variants. Patients who discontinued PARPi because of toxicities were censored for PFS analyses. Follow-up was evaluated until January 15, 2021, at which time patients without progression were censored. All analyses were performed using Prism 8 software (GraphPad Software Corporation, version 8.3.1 (332)). The level of statistical significance was set at P < .05 for all analyses.
RESULTS
Selected Patients
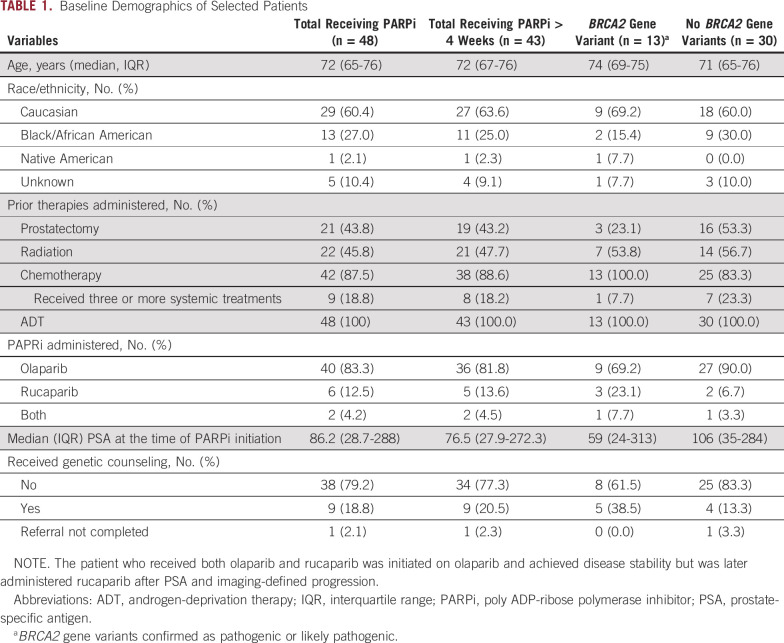
Of the 98 patients who had tumor DNA sequencing available in NPOP and received PARPi before May 15, 2020, 48 (49.0%) had a primary diagnosis of prostate cancer (Fig 1). Of these, 43 (91.7%) were administered a PARPi for > 4 weeks. The median age (interquartile range) of the cohort was 72 (65-76) years, and a slight majority of patients (60.4%) were Caucasian. At the time of PARPi initiation, all patients had metastatic prostate cancer; 21 (43.8%) and 22 (45.3%) patients had undergone prior prostatectomy or local radiation therapy, respectively (Table 1). All patients had disease progression with at least one prior line of systemic therapy, with nine patients (18.8%) having received > 3 lines of systemic treatments before PARPi administration.
FIG 1.
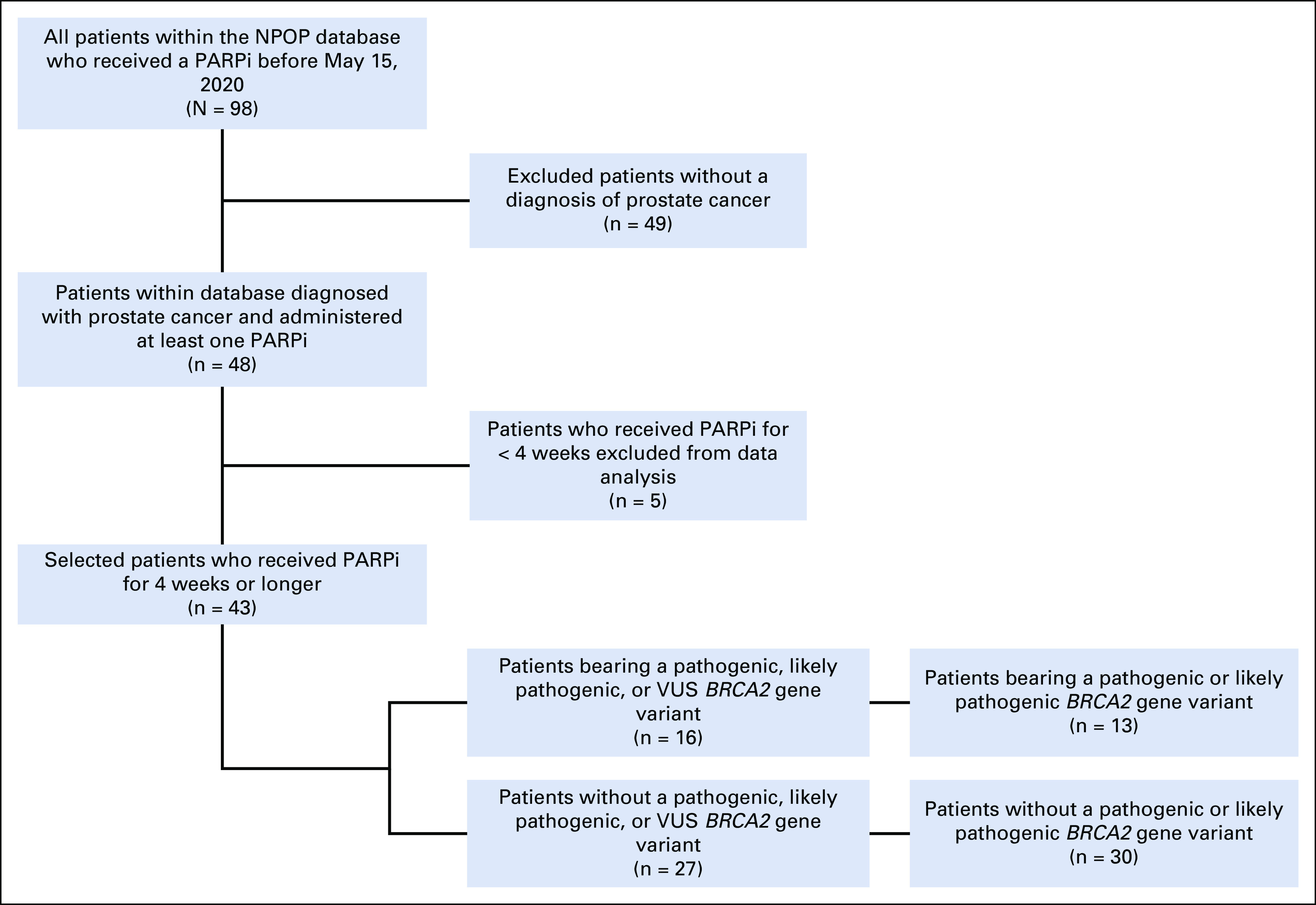
CONSORT diagram. NPOP, National Precision Oncology Program; PARPi, poly ADP-ribose polymerase inhibitor; VUS, variants of uncertain significance.
TABLE 1.
Baseline Demographics of Selected Patients
Variants Within HRR Genes
The HRR genes most commonly containing variants among patients receiving PARPi were BRCA2 (36 variants, 25 patients, 52.1%), ATM (13 variants, 10 patients, 20.8%), and BRCA1 (eight variants, eight patients, 16.7%; Fig 2). Of the BRCA2 alterations, 17 were classified as pathogenic/likely pathogenic alterations, 13 were VUS, two were likely benign, and three were likely CHIP (allele fraction < 1% from plasma sequencing) variants. A total of 16 patients had at least one BRCA2 variant classified as pathogenic/likely pathogenic (13 of whom received a PARPi for > 4 weeks). The remaining variants were within the following DNA repair genes: BAP1, CHEK2, FANCA, PALB2, and RAD51C. Of the 67 total reported HRR gene variants, 27 variants from 22 patients (45.8%) were determined to be pathogenic/likely pathogenic after review in OncoKB (Data Supplement). Nine (13.4%) variants were likely CHIP, with allele fractions of 1.0% or less from plasma sequencing. Twenty-five patients (52.1%) were prescribed PARPi without evidence of a pathogenic/likely pathogenic variant. Six of these patients had only likely CHIP HRR gene variants, and five did not have any US Food and Drug Administration label HRR gene variants.
FIG 2.
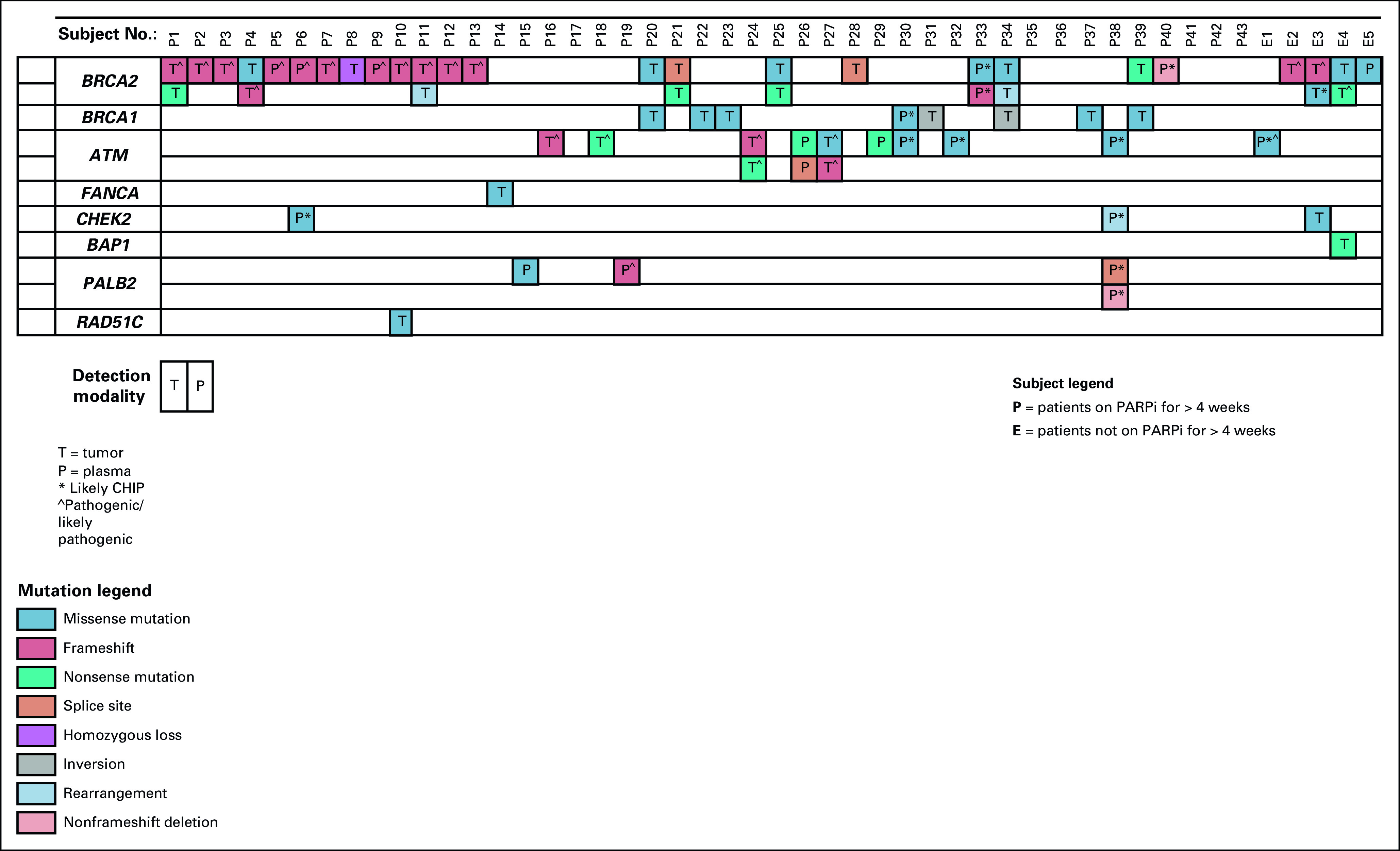
Oncoplot of HRR gene variants detected in patients who received PARPi delineated by those who received PARPi for > 4 weeks and those who received PARPi for < 4 weeks. CHIP, clonal hematopoiesis of indeterminate potential; HRR, homologous recombination repair; PARPi, poly ADP-ribose polymerase inhibitor.
Per the criteria of having at least one pathogenic HRR alteration with an allele fraction of > 30%, a total of 20 gene variants from 17 (35.4%) patients were considered to potentially represent germline mutations.24 Three (17.6%) of these 17 patients were referred to genetic counseling. Overall, eight patients (16.7%) of the selected 48 were referred to genetic counseling (Data Supplement). Three of these eight patients had pathogenic/likely pathogenic somatic BRCA2 variants with allele fractions > 30%, and all three were found to have germline mutations. Two patients had somatic BRCA2 pathogenic/likely pathogenic variants with fewer allele fractions, and one was found to have a germline variant. Two patients had BRCA2 VUS per OncoKB classifications, and one was found to have a germline variant. One patient was referred solely for likely CHIP variants and ultimately did not undergo germline testing.
PARPi Prescriptions and Toxicities
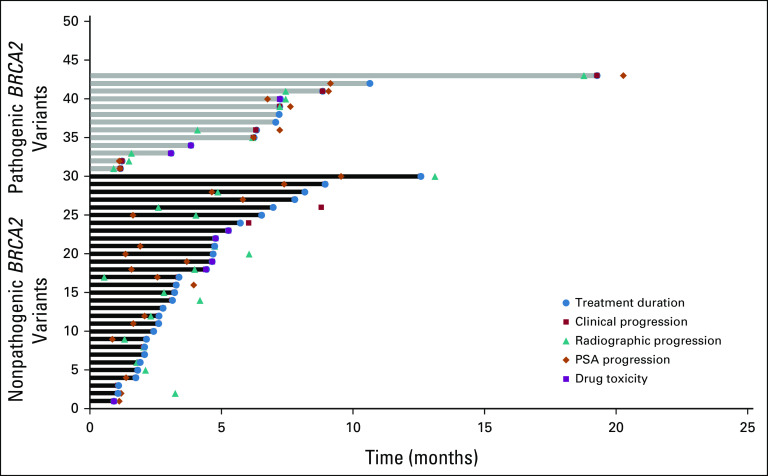
Patients received olaparib (83.3%), rucaparib (12.5%), or both (4.5%). Of the two patients receiving both drugs, one patient was transitioned to rucaparib after progression after initial response to olaparib. The second patient was initially discontinued from olaparib because of toxicities, and after progression on other systemic regimens, the patient was treated with rucaparib, which was ultimately discontinued because of fatigue. Of the five patients who were prescribed a PARPi for < 4 weeks, two experienced significant toxicities including fatigue and repeated falls, one progressed rapidly, one was hospitalized and transitioned to hospice, and one passed away before initiating the prescribed drug. Of the 43 patients who were administered a PARPi for 4 weeks or longer, 10 had documented drug-related toxicities, with anemia being most common followed by weight loss and fatigue (Table 2), which resulted in drug discontinuation in nine patients (Fig 5).
TABLE 2.
Toxicities Among All Patients Who Were Prescribed PARPi by Drug
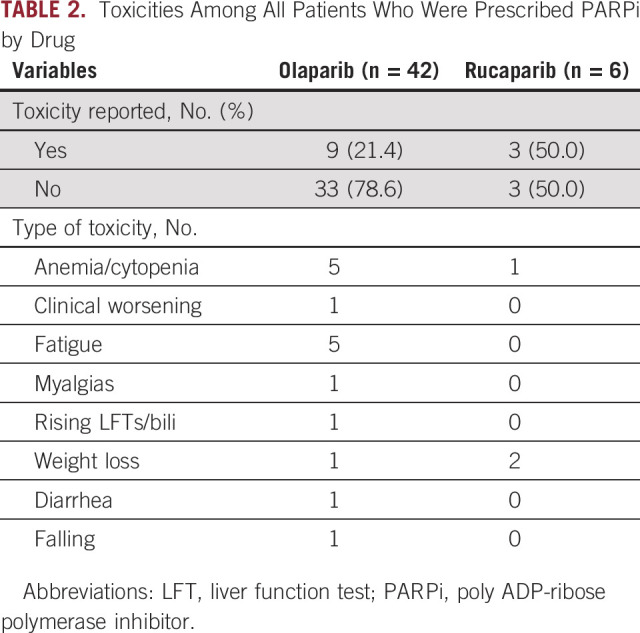
FIG 5.
Swimmer plot of time on treatment for each patient, indicating the time of clinical progression, radiographic progression, drug toxicity, and PSA progression on the basis of the presence of pathogenic/likely pathogenic BRCA2 gene variants. PSA, prostate-specific antigen.
Clinical Outcomes Among Patients With Prostate Cancer Treated With PARPi
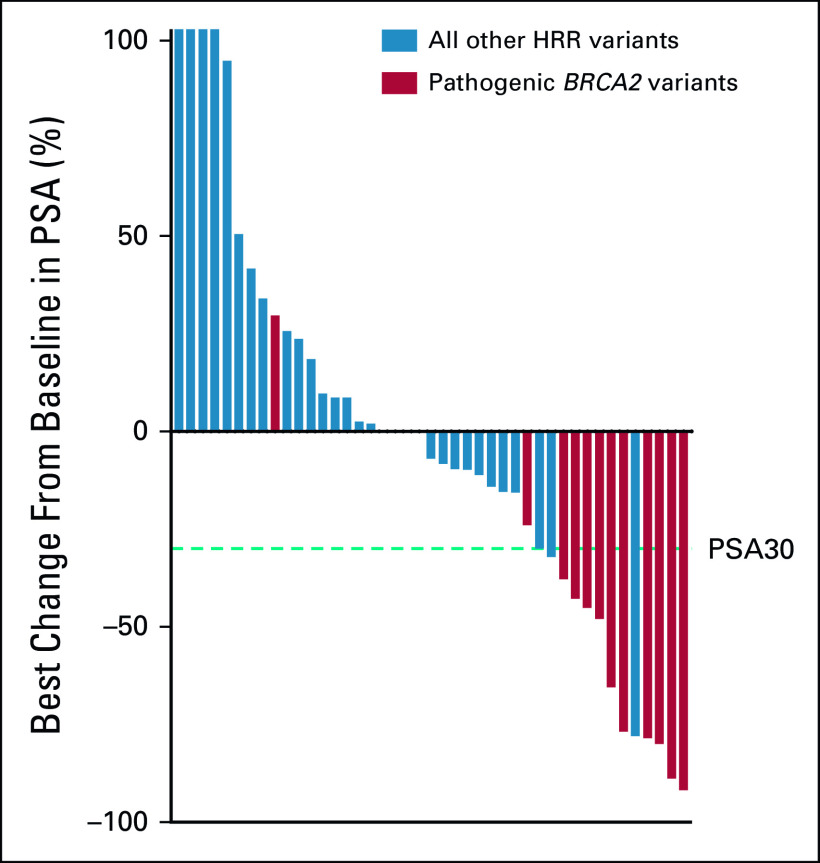
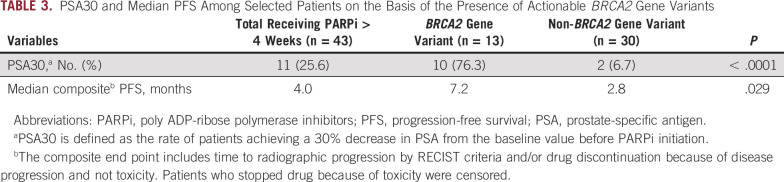
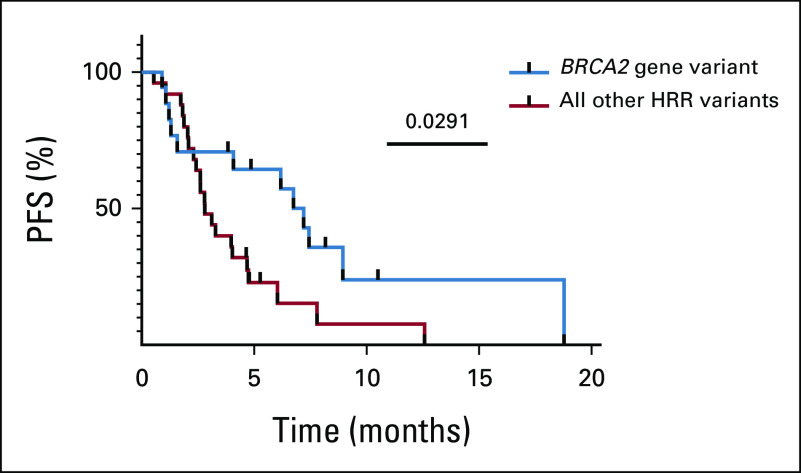
For all 43 patients who received PARPi for 4 weeks or longer, PSA30 was 27.9% (12 of 43; Fig 3). The 13 patients with at least one BRCA2 variant classified as pathogenic/likely pathogenic had a higher PSA30 rate than patients without a pathogenic/likely pathogenic BRCA2 gene variant (76.3% v 6.7%, P < .0001; Table 3; Fig 3). For all treated patients, the median composite PFS was 4.0 months. Patients with pathogenic/likely pathogenic BRCA2 variants had a significantly longer median PFS compared with those without (7.2 months v 2.8 months, P = .0291; hazard ratio 0.486; 95% CI, 0.25 to 0.95; Fig 4). Among the patients without pathogenic/likely pathogenic BRCA2 variants, there were five patients with BRCA2 VUS alterations; they had a median PFS of 5.1 months, and only one achieved PSA30. The median PFS for those who had a pathogenic, likely pathogenic, or VUS alteration not meeting criteria for likely CHIP in an HRR gene other than BRCA2 was 3.9 months, and only one achieved PSA30 (Fig 3). Detailed data regarding radiographic, clinical, and PSA progression along with drug toxicity timelines are presented in Figure 5.
FIG 3.
Waterfall plot of maximum percentage change from baseline in PSA during treatment in the presence or absence of the pathogenic/likely pathogenic BRCA2 variant. HRR, homologous recombination repair; PSA, prostate-specific antigen.
TABLE 3.
PSA30 and Median PFS Among Selected Patients on the Basis of the Presence of Actionable BRCA2 Gene Variants
FIG 4.
Composite PFS on the basis of the presence of pathogenic/likely pathogenic BRCA2 gene variants. HRR, homologous recombination repair; PFS, progression-free survival.
DISCUSSION
This study's first objective was to review the HRR gene variants and overall treatment outcomes among Veterans with mCRPC treated with PARPi. Clinical trials confirming the effectiveness of PARPi have treated tumors with a variety of pathogenic variants, most commonly, BRCA2, BRCA1, and ATM genes.7,10,25,26 Although variants were most frequently found within the same genes in our study, only 41.3% of the nonlikely CHIP variants were annotated as pathogenic/likely pathogenic when evaluated with OncoKB.
Various platforms including vendor laboratory reports and regularly updated online databases such as ClinVar and OncoKB are available to determine the pathogenicity of gene variants.13,27 In addition, health care systems with access to molecular pathologist expertise, such as the VA's NPOP, frequently review the relevance of discovered gene variants during molecular tumor boards or through consultation services. Despite the availability of such services within VA, we found that a substantial percentage of patients were prescribed PARPi on the basis of sequencing reports, which did not show pathogenic/likely pathogenic variants in HRR genes. The inappropriate prescription of PARPi was mostly based on misinterpretation of an alteration as pathogenic/likely pathogenic or reporting of alterations with very low allele fractions (< 1%), which likely represent CHIP.17-21 As multigene NGS panels are used more frequently, providers will be increasingly tasked with evaluating results and prescribing on the basis of approved indications. Further difficulties arise for settings such as PARPi in prostate cancer, in which drugs are approved for pathogenic/likely pathogenic variants across a wide spectrum of HRR genes. Given these concerns, efforts should be directed toward improvement of sequencing reports to more clearly indicate that alterations are not known to be pathogenic and improving provider education/accessibility to contemporary gene variant interpretations.
Approximately 35% of selected patients in this study harbored pathogenic/likely pathogenic variants with allele fractions of > 30%, potentially representing germline variants,24 yet only 17.6% of these patients and < 20% of all selected patients were referred to genetic counseling for germline testing. Current data indicate that approximately one of every 10 patients with mCRPC carries a germline HRR alteration.6,8 With such a high rate of germline HRR gene alterations among patients with mCRPC, genetic counselors play an important role in identifying and managing those with hereditary cancer syndromes.28
This study's second objective was to compare outcomes between patients with pathogenic BRCA2 gene variants and those without. We observed a significant benefit in PSA30 for patients bearing pathogenic/likely pathogenic BRCA2 variants. Of note, the baseline PSA in this cohort of patients with pathogenic/likely pathogenic BRCA2 alterations was significantly lower than that of the cohort without, suggesting that clinicians may be more likely to prescribe PARPi earlier in the disease course for patients with BRCA2 gene variants. However, there was a similar amount of treatment before initiation between the two groups as reflected by the fraction of patients who received > 3 systemic regimens before PARPi initiation. The median PFS was longer among patients with pathogenic BRCA2 variants compared with those without (7.2 and 2.8 months, respectively). Collectively, these findings reinforce the concept that the presence of BRCA2 variants may serve as a better predictive biomarker for PARPi response than variants in other HRR genes.12
Compared with other studies evaluating PARPi in HRR-altered cancers,9-11,25,29,30 the overall PFS of 4 months observed in this retrospective analysis is quite low. This could potentially be attributed to two reasons. First, a significant number of patients included in our real-world outcomes study did not have true pathogenic/likely pathogenic HRR gene variants. Second, patients enrolled in these trials frequently received only 1-2 prior systemic therapy regimens, whereas many of our patients received > 3. Overall, the patients included in our study likely had more treatment-refractory disease when they were started on PARPi compared with those enrolled in trials. By contrast, patients in our study with pathogenic BRCA2 alterations had a significantly longer PFS than the whole group, consistent with results from multiple clinical trials.9,11,25 Given the less clear benefit among mCRPC patients with non-BRCA2 HRR gene variants, the decision to administer PARPi should be balanced against the adverse effects of PARPi, the financial toxicity associated with these drugs, and the efficacy of other approved systemic treatments for prostate cancer.31
Our study has several strengths. First, this study reflects real-world treatment practices and outcomes for patients with mCRPC receiving PARPi in a large integrated health care system with access to detailed data regarding outcomes. Second, because of the centralized NPOP sequencing program, we were able to review the pathogenicity of each HRR gene variant and the likelihood that an underlying germline variant is present for all selected patients.
Our study also has some limitations. First, our retrospective cohort of patients treated with PARPi is smaller than that in previously published clinical trials. Although we were able to discern differences in outcomes by genetic variations, more refined associations may be discernable in a larger patient population. Second, we primarily conducted our outcome analyses on the basis of the presence of HRR gene variants, yet less than half of the patients had pathogenic/likely pathogenic gene variants after review via OncoKB. Real-world outcomes with PARPi prescriptions would likely be more pronounced if only patients with pathogenic/likely pathogenic are selected for treatment. Third, the outcomes of our study specifically pertain to Veterans being served by oncology practices within the VA, who have a higher frequency of comorbidities. Consequently, patients with less comorbidities in other real-world settings might have better treatment responses to PARPi.
In conclusion, in a real-world setting, heavily pretreated patients with mCRPC with HRR gene variants achieve a PFS of 4 months with PARPi. Patients harboring pathogenic/likely pathogenic BRCA2 variants have substantially longer PFS and a higher PSA30 rate than non-BRCA2 alterations, consistent with clinical trials. More than half of patients prescribed PARPi had no pathogenic/likely pathogenic HRR gene variants. Only 18% of patients whose somatic sequencing suggested potential germline alterations had documentation of germline testing. This work emphasizes the importance of improving oncology provider education and health informatics tools to increase conformance of practice to recommended precision oncology care of men with metastatic prostate cancer.
Vishal Vashistha
Employment: UnitedHealthcare (I)
Research Funding: IBM Watson Health (Inst)
Other Relationship: IBM
Michael J. Kelley
Research Funding: Novartis (Inst), AstraZeneca (Inst), Bristol Myers Squibb (Inst), Regeneron (Inst), Genentech (Inst)
Other Relationship: IBM (Inst)
Bruce Montgomery
Research Funding: AstraZeneca (Inst), Janssen Oncology (Inst), Clovis Oncology (Inst), Astellas Pharma (Inst), BeiGene (Inst)
No other potential conflicts of interest were reported.
SUPPORT
Supported by a Prostate Cancer Foundation Valor Award (B.M.), the Pacific Northwest Prostate Cancer SPORE CA097186 (B.M.), the Department of Veterans Affairs (M.J.K.), and VA Big Data Scientist Enhancement Program (M.J.K.).
M.P. and V.V. contributed equally to this work.
AUTHOR CONTRIBUTIONS
Conception and design: Meghan Price, Vishal Vashistha, Michael J. Kelley, Bruce Montgomery
Collection and assembly of data: Meghan Price, Vishal Vashistha, David Winski, Bruce Montgomery
Data analysis and interpretation: Meghan Price, Vishal Vashistha, Michael J. Kelley, Rhonda L. Bitting, Bruce Montgomery
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Vishal Vashistha
Employment: UnitedHealthcare (I)
Research Funding: IBM Watson Health (Inst)
Other Relationship: IBM
Michael J. Kelley
Research Funding: Novartis (Inst), AstraZeneca (Inst), Bristol Myers Squibb (Inst), Regeneron (Inst), Genentech (Inst)
Other Relationship: IBM (Inst)
Bruce Montgomery
Research Funding: AstraZeneca (Inst), Janssen Oncology (Inst), Clovis Oncology (Inst), Astellas Pharma (Inst), BeiGene (Inst)
No other potential conflicts of interest were reported.
REFERENCES
- 1.American Cancer Society : Key Statistics for Prostate Cancer. Kennesaw, GA, American Cancer Society, 2021 [Google Scholar]
- 2.Tablazon IL, Howard LE, De Hoedt AM, et al. : Predictors of skeletal-related events and mortality in men with metastatic, castration-resistant prostate cancer: Results from the Shared Equal Access Regional Cancer Hospital (SEARCH) database. Cancer 125:4003-4010, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beer TM, Armstrong AJ, Rathkopf DE, et al. : Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med 371:424-433, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tannock IF, de Wit R, Berry WR, et al. : Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med 351:1502-1512, 2004 [DOI] [PubMed] [Google Scholar]
- 5.Broderick JM: Incidence of metastatic prostate cancer incidence on the rise. Oncology 34:460, 2020 [DOI] [PubMed] [Google Scholar]
- 6.Pritchard CC, Mateo J, Walsh MF, et al. : Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med 375:443-453, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armenia J, Wankowicz SAM, Liu D, et al. : The long tail of oncogenic drivers in prostate cancer. Nat Genet 50:645-651, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson D, Van Allen EM, Wu YM, et al. : Integrative clinical genomics of advanced prostate cancer. Cell 161:1215-1228, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hussain M, Mateo J, Fizazi K, et al. : Survival with olaparib in metastatic castration-resistant prostate cancer. N Engl J Med 383:2345-2357, 2020 [DOI] [PubMed] [Google Scholar]
- 10.Mateo J, Carreira S, Sandhu S, et al. : DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med 373:1697-1708, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abida W, Campbell D, Patnaik A, et al. : Preliminary results from the TRITON2 study of rucaparib in patients with DNA damage repair (DDR)-deficient metastatic castration-resistant prostate cancer (mCRPC): Updated analyses. Presented at ESMO Congress 2019, Barcelona, Spain, September 27, 2019-October 1, 2019
- 12.Carreira S, Porta N, Arce-Gallego S, et al. : Biomarkers associating with PARP inhibitor benefit in prostate cancer in the TOPARP-B trial. Cancer Discov 11:2812-2827, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakravarty D, Gao J, Phillips SM, et al. : OncoKB: A precision oncology knowledge base. JCO Precis Oncol 10.1200/PO.17.00011, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price LE, Shea K, Gephart S: The Veterans Affairs's Corporate Data Warehouse: Uses and implications for nursing research and practice. Nurs Adm Q 39:311-318, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Legler A, Price M, Parikh M, et al. : Effect on VA patient satisfaction of provider's use of an integrated viewer of multiple electronic health records. J Gen Intern Med 34:132-136, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.National Comprehensive Cancer Network : Prostate Cancer (Version 2.2021). Plymouth Meeting, PA, National Comprehensive Cancer Network, 2021 [Google Scholar]
- 17.Jensen K, Konnick EQ, Schweizer MT, et al. : Association of clonal hematopoiesis in DNA repair genes with prostate cancer plasma cell-free DNA testing interference. JAMA Oncol 7:107-110, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyatt AW, Annala M, Aggarwal R, et al. : Concordance of circulating tumor DNA and matched metastatic tissue biopsy in prostate cancer. J Natl Cancer Inst 109:djx118, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Razavi P, Li BT, Brown DN, et al. : High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med 25:1928-1937, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stetson D, Ahmed A, Xu X, et al. : Orthogonal comparison of four plasma NGS tests with tumor suggests technical factors are a major source of assay discordance. JCO Precis Oncol 3:1-9, 2019 [DOI] [PubMed] [Google Scholar]
- 21.Taavitsainen S, Annala M, Ledet E, et al. : Evaluation of commercial circulating tumor DNA test in metastatic prostate cancer. JCO Precis Oncol 3:1-9, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petrylak DP, Ankerst DP, Jiang CS, et al. : Evaluation of prostate-specific antigen declines for surrogacy in patients treated on SWOG 99-16. J Natl Cancer Inst 98:516-521, 2006 [DOI] [PubMed] [Google Scholar]
- 23.Eisenhauer EA, Therasse P, Bogaerts J, et al. : New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur J Cancer 45:228-247, 2009 [DOI] [PubMed] [Google Scholar]
- 24.Mandelker D, Donoghue M, Talukdar S, et al. : Germline-focussed analysis of tumour-only sequencing: Recommendations from the ESMO Precision Medicine Working Group. Ann Oncol 30:1221-1231, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mateo J, Porta N, Bianchini D, et al. : Olaparib in patients with metastatic castration-resistant prostate cancer with DNA repair gene aberrations (TOPARP-B): A multicentre, open-label, randomised, phase 2 trial. Lancet Oncol 21:162-174, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quigley DA, Dang HX, Zhao SG, et al. : Genomic hallmarks and structural variation in metastatic prostate cancer. Cell 174:758-769.e9, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landrum MJ, Lee JM, Benson M, et al. : ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 46:D1062-D1067, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pujol P, Barberis M, Beer P, et al. : Clinical practice guidelines for BRCA1 and BRCA2 genetic testing. Eur J Cancer 146:30-47, 2021 [DOI] [PubMed] [Google Scholar]
- 29.Smith MR, Sandhu SK, Kelly WK, et al. : Pre-specified interim analysis of GALAHAD: A phase II study of niraparib in patients (pts) with metastatic castration-resistant prostate cancer (mCRPC) and biallelic DNA-repair gene defects (DRD). Ann Oncol 30:V884-V885, 2019 [Google Scholar]
- 30.De Bono JS, Mehra N, Higano CS, et al. : TALAPRO-1: A phase II study of talazoparib (TALA) in men with DNA damage repair mutations (DDRmut) and metastatic castration-resistant prostate cancer (mCRPC)—first interim analysis (IA). J Clin Oncol 38, 2020. (suppl 6; abstr 119) [Google Scholar]
- 31.Schweizer MT, Cheng HH, Nelson PS, et al. : Two steps forward and one step back for precision in prostate cancer treatment. J Clin Oncol 38:3740-3742, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]