The advent of immunotherapy, including immune checkpoint inhibitors (ICIs), has contributed to improved survival in patients with lung cancer.1 Three anti–programmed death-1 (PD-1)/programmed death ligand 1 (PD-L1) therapies, nivolumab, pembrolizumab, and atezolizumab, are approved by the US Food and Drug Administration (FDA) for the treatment of patients with advanced non–small-cell lung cancer (NSCLC) in the first-line setting.2-4
Despite the improvements in survival seen with ICIs, there remains a subset of patients who do not benefit from ICI monotherapy or combination treatments.5 As a result, many research efforts have been initiated to identify biomarkers predictive of response to treatment with ICIs. Predictive biomarkers that have entered clinical practice include PD-L1 expression on tumor and immune cells, and tumor mutational burden.2-4,6,7 However, the accuracy of predictions of patient response based on current biomarkers leaves ample room for improvement. Efforts to identify additional biomarkers of sufficient specificity and predictive power are critical for selecting patients for ICI monotherapy or combination regimens. Successful attempts will likely rely on integrated models combining multiple features.
To tackle such complex challenges, the scientific community has become increasingly interested in the possibilities offered by data sharing and crowdsourcing as a framework for mobilizing groups of people with shared interests around the world and bringing forward augmented solutions to the existing problems while encouraging collaboration. In this context, data sharing can be executed according to two paradigms.8 In the data-to-modeler paradigm, both training and validation data sets are provided to participants. However, in some circumstances, this is not compatible with the maintenance of confidentiality of individual patient data mandated by Institutional Review Boards. The model-to-data paradigm, where participants submit “containerized” models to organizers, allows validation data sets to remain hidden from participants, which both protects confidential data and helps ensure unbiased assessment of predictions. A container software such as Docker or RKT enables the packaging and transfer of a model and necessary software components in a platform-agnostic way. Such innovative approaches that improve access to clinical trial data and leverage worldwide modeling expertise must be embraced to accelerate the identification of biomarkers predictive of response. These approaches are instrumental in broadening our understanding of tumor biology and the tumor microenvironment, which will ultimately drive precision medicine with immunotherapy.
To enable such collaboration among scientists and physicians, Dialogue on Reverse Engineering and Assessment Methods (DREAM) Challenges engage experts from around the world and offer novel solutions to biological problems.9 Since the first DREAM Challenges were initiated in 2006, more than 60 have completed, with analysis results generated through virtual collaborative interactions.10 In oncology, crowdsourcing approaches have been used in multiple tumor types since 2012 for augmenting the work of teams generating data, developing protocols for randomized controlled trials, developing prognostic models, and assessing healthcare behaviors on a large scale.11 More specifically, three key DREAM Challenges in oncology have demonstrated the value of crowdsourcing for the development of prognosis models or the refinement of diagnostic methods. The Prostate Cancer DREAM Challenge, by benchmarking 50 models developed internationally, identified novel prognostic biomarkers previously under-reported and demonstrated that data sharing in combination with a crowdsourced challenge is a powerful framework to develop new prognostic models.12 The Multiple Myeloma DREAM Challenge assessed 171 models, resulting in the discovery of the epigenetic regulator PHF19 as a novel marker of aggressive disease and high risk of early progression.13 The Digital Mammography DREAM Challenge, the first large-scale benchmarking effort using a model-to-data paradigm on imaging data, suggested that integrated artificial intelligence and single-radiologist assessment can improve the interpretation of breast cancer–screening mammograms.14
Following on from these successful DREAM Challenges, experts in lung cancer have shown interest in accessing primary clinical trial data to explore alternative biomarkers to PD-L1 expression and tumor mutational burden. The Anti–PD-1 Response Prediction Challenge will be the first DREAM Challenge focused on the prediction of response to immuno-oncology therapies using data from a large phase III clinical trial, with the goal of addressing the need for additional predictive biomarkers to use with ICIs in patients with NSCLC. Moreover, this initiative will also enhance the understanding of tumor biology and provide further insights into the potential mechanisms underpinning treatment resistance to ICIs.
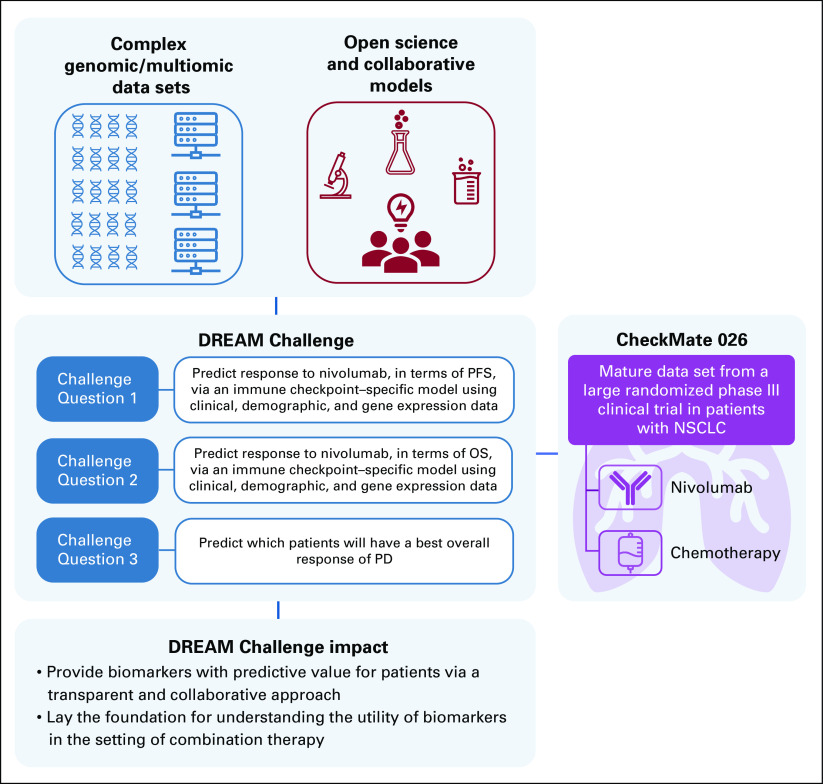
Challenge Objectives
The Anti–PD-1 Response Prediction Challenge will focus on developing models that are predictive of benefit from ICI treatment, specifically nivolumab monotherapy, using patient samples obtained during the conduct of a clinical trial. Clinical trial data related to nivolumab, including data related to PD-L1 expression, tumor mutational burden, and tumor transcriptomics, will be used to assess the performance of predictive models submitted in a crowdsourced challenge setting. The testing data set will use deidentified data from CheckMate 026 (ClinicalTrials.gov identifier: NCT02041533), a phase III trial that compared nivolumab with platinum-based chemotherapy in patients with metastatic or recurrent NSCLC and PD-L1 tumor expression ≥ 5%.15 Critically, the data set from CheckMate 026 is large, randomized, mature, and well annotated, allowing it to support robust testing of predictive models. Furthermore, the availability of a control arm allows the differentiation between predictive and prognostic models. As such, it is one of the first efforts to benchmark crowdsourced models of response to an ICI. This Challenge also lays the foundation for potential future challenges as the immuno-oncology landscape is rapidly evolving.
The Anti–PD-1 Response Prediction Challenge aims to assess the accuracy of models for predicting clinical outcomes after nivolumab monotherapy, including efficacy end points such as overall survival, progression-free survival, and best overall response, applying clinical and genomic data from CheckMate 026. The Challenge will comprise three subchallenges, and their objectives are summarized in Figure 1.
FIG 1.
Anti–PD-1 Response Prediction Challenge questions. NSCLC, non–small-cell lung cancer; OS, overall survival; PD, progressive disease; PD-1, programmed death-1; PFS, progression-free survival.
Challenge Design
The Challenge will rely on a partnership between Sage Bionetworks, the organization supporting the DREAM Challenge initiatives, and Bristol Myers Squibb and will comprise three phases: competitive, collaborative, and final. The Challenge framework has been designed to facilitate reproducibility and to ensure the protection of patient-level data. Specifically, in full adherence with Institutional Review Board approvals, the model-to-data process, as described above, will be used. The framework has been developed to receive and evaluate Dockerized models, which are in silico packages comprising the model itself plus necessary software components to run the model as developed by the participant team.
The Challenge is open to anyone who will be able to construct models for submission, subject to specifications in the Challenge rules, including experts and innovators in genomics, computational biology, and translational biomarker development. Participants can sign up on the Synapse website.16 To facilitate data ingestion, participants will have access to a small synthetic data set with the same formatting as the validation data set. The full synthetic data set can be downloaded on the Synapse website.17
In the first phase, participants will construct models using any data available to them. The iAtlas platform,18 a database supporting the study of interactions between cancer and the immune microenvironment, hosts harmonized genomics data and associated clinical annotations from published immuno-oncology studies. Participants can use these as training data sets or to augment other training data already available to them.19 Participants could also use Professor Shirley Liu's lab's TIDE resources,20 a computational framework that integrated and modeled data from 189 human cancer studies, comprising 33,197 samples.21 More resources to consider are listed on the Anti–PD-1 Response Prediction Challenge website.16
Participants' Dockerized models will be applied to the CheckMate 026 data and evaluated during the competitive phase. Performance metrics, such as area under the receiver operating characteristic or precision-recall curves and Harrell's C-index, will be used to assess the predictive performance of participants' models in each arm of the CheckMate 026 trial.
Top-performing teams will be invited to participate in the collaborative model-development phase, during which data will be analyzed and interpreted to deepen the understanding of the submitted models. Eventually, opportunities for additional subchallenges and potential projects will be defined based on the learnings from the current Challenge, and teams submitting the best-performing models will have the opportunity to co-author the planned publication in accordance with medical publication standards and best practices.
Anticipated Outcome and Impact
The identified models are expected to identify potential biomarkers of response to nivolumab monotherapy and may then be applied to predict which patients are more likely to derive clinical benefit from ICIs. Ultimately, the identification of novel biomarkers may also contribute to the refinement of stratification criteria in future clinical trials and support the development of innovative treatments for patients. It is anticipated that some of the models may also have prognostic value for patients with NSCLC.
The Challenge results should also provide a deeper understanding of biological mechanisms resulting in tumor resistance to treatment and assist in understanding why patients with advanced NSCLC receiving nivolumab had similar efficacy outcomes to patients receiving chemotherapy in CheckMate 026. An improved understanding of the biological mechanisms behind resistance could pave the way to therapeutic interventions that immunologically activate tumors that do not display an immune-infiltrated phenotype, thereby increasing their responsiveness to ICI therapies.
Successful completion of this DREAM Challenge will provide a model for future collaborations among industry, academia, and citizen scientists to discover determinants of response or resistance to immunotherapy, facilitating rapid development of clinically actionable biomarkers. As the first DREAM Challenge in immunotherapy, this Challenge lays the foundation for future challenges as the immuno-oncology therapeutic landscape in NSCLC is rapidly evolving.
SUPPORT
Medical writing and editorial support for this commentary were provided by Thierry Deltheil, PhD, and Jay Rathi, MA, of Spark Medica Inc, funded by Bristol Myers Squibb.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO’s conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Benjamin G. Vincent
Leadership: Select Immunogenomics
Research Funding: Merck
Joseph D. Szustakowski
Employment: Bristol Myers Squibb
Stock and Other Ownership Interests: Bristol Myers Squibb
Research Funding: Bristol Myers Squibb
Patents, Royalties, Other Intellectual Property: I am an employee of Bristol Myers Squibb, and an inventor on one or more pending patent applications, including applications for TMB as a predictive biomarker of immunotherapy
Travel, Accommodations, Expenses: Bristol Myers Squibb
Parul Doshi
Employment: Bristol Myers Squibb
Stock and Other Ownership Interests: Bristol Myers Squibb
Travel, Accommodations, Expenses: Bristol Myers Squibb
Justin Guinney
Consulting or Advisory Role: AstraZeneca
Research Funding: Celgene, Genentech/Roche, Bristol Myers Squibb
David P. Carbone
Employment: James Cancer Center
Honoraria: AstraZeneca, Nexus Pharmaceuticals Inc
Consulting or Advisory Role: Bayer, Boehringer Ingelheim, Merck, Novartis, Pfizer, AstraZeneca, Helsinn Therapeutics, AbbVie, Inivata, Loxo, Incyte, Bristol Myers Squibb, Daiichi Sankyo, EMD Serono, GlaxoSmithKline, Inovio Pharmaceuticals, Janssen, Kyowa Hakko Kirin, Takeda
Research Funding: Bristol Myers Squibb
No other potential conflicts of interest were reported.
AUTHOR CONTRIBUTIONS
Conception and design: All authors
Provision of study materials or patients: Parul Doshi
Collection and assembly of data: Joseph D. Szustakowski, Parul Doshi, Justin Guinney
Data analysis and interpretation: Justin Guinney, David P. Carbone
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
REFERENCES
- 1.American Cancer Society : Facts & Figures 2020 Reports Largest One-Year Drop in Cancer Mortality. https://www.cancer.org/latest-news/facts-and-figures-2020.html [Google Scholar]
- 2.Bristol Myers Squibb : OPDIVO® (Nivolumab) [package insert]. https://packageinserts.bms.com/pi/pi_opdivo.pdf [Google Scholar]
- 3.Genentech Inc : TECENTRIQ® (Atezolizumab) [package insert]. https://www.gene.com/download/pdf/tecentriq_prescribing.pdf [Google Scholar]
- 4.Merck & Co Inc : KEYTRUDA® (Pembrolizumab) [package insert]. https://www.merck.com/product/usa/pi_circulars/k/keytruda/keytruda_pi.pdf [Google Scholar]
- 5.Sambi M, Bagheri L, Szewczuk MR: Current challenges in cancer immunotherapy: Multimodal approaches to improve efficacy and patient response rates. J Oncol 2019:4508794, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bristol Myers Squibb : YERVOY® (Ipilimumab) [package insert]. http://packageinserts.bms.com/pi/pi_yervoy.pdf [Google Scholar]
- 7.AstraZeneca : IMFINZI® (Durvalumab) [package insert]. https://www.azpicentral.com/imfinzi/imfinzi.pdf#page=1 [Google Scholar]
- 8.Guinney J, Saez-Rodriguez J: Alternative models for sharing confidential biomedical data. Nat Biotechnol 36:391-392, 2018 [DOI] [PubMed] [Google Scholar]
- 9.Saez-Rodriguez J, Costello JC, Friend SH, et al. : Crowdsourcing biomedical research: Leveraging communities as innovation engines. Nat Rev Genet 17:470-486, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DREAM Challenges : Closed Challenges. http://dreamchallenges.org/project-list/closed/ [Google Scholar]
- 11.Lee YJ, Arida JA, Donovan HS: The application of crowdsourcing approaches to cancer research: A systematic review. Cancer Med 6:2595-2605, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guinney J, Wang T, Laajala TD, et al. : Prediction of overall survival for patients with metastatic castration-resistant prostate cancer: Development of a prognostic model through a crowdsourced challenge with open clinical trial data. Lancet Oncol 18:132-142, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mason MJ, Schinke C, Eng CLP, et al. : Multiple myeloma DREAM Challenge reveals epigenetic regulator PHF19 as marker of aggressive disease. Leukemia 34:1866-1874, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaffter T, Buist DSM, Lee CI, et al. : Evaluation of combined artificial intelligence and radiologist assessment to interpret screening mammograms. JAMA Netw Open 3:e200265, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carbone DP, Reck M, Paz-Ares L, et al. : First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N Engl J Med 376:2415-2426, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Synapse : https://synapse.org/CheckmateChallenge
- 17.Synapse : synapse.org/#!Synapse:syn22360672
- 18.CRI iAtlas : https://www.cri-iatlas.org/
- 19.Thorsson V, Gibbs DL, Brown SD, et al. : The immune landscape of cancer. Immunity 48:812-830.e814, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.TIDE : http://tide.dfci.harvard.edu/login/
- 21.Jiang P, Gu S, Pan D, et al. : Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med 24:1550-1558, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]