Abstract
Background
The genus Zingiber of the Zingiberaceae is distributed in tropical, subtropical, and in Far East Asia. This genus contains about 100–150 species, with many species valued as important agricultural, medicinal and horticultural resources. However, genomic resources and suitable molecular markers for species identification are currently sparse.
Results
We conducted comparative genomics and phylogenetic analyses on Zingiber species. The Zingiber chloroplast genome (size range 162,507–163,711 bp) possess typical quadripartite structures that consist of a large single copy (LSC, 86,986–88,200 bp), a small single copy (SSC, 15,498–15,891 bp) and a pair of inverted repeats (IRs, 29,765–29,934 bp). The genomes contain 113 unique genes, including 79 protein coding genes, 30 tRNA and 4 rRNA genes. The genome structures, gene contents, amino acid frequencies, codon usage patterns, RNA editing sites, simple sequence repeats and long repeats are conservative in the genomes of Zingiber. The analysis of sequence divergence indicates that the following genes undergo positive selection (ccsA, ndhA, ndhB, petD, psbA, psbB, psbC, rbcL, rpl12, rpl20, rpl23, rpl33, rpoC2, rps7, rps12 and ycf3). Eight highly variable regions are identified including seven intergenic regions (petA-pabJ, rbcL-accD, rpl32-trnL-UAG, rps16-trnQ-UUG, trnC-GCA-psbM, psbC-trnS-UGA and ndhF-rpl32) and one genic regions (ycf1). The phylogenetic analysis revealed that the sect. Zingiber was sister to sect. Cryptanthium rather than sect. Pleuranthesis.
Conclusions
This study reports 14 complete chloroplast genomes of Zingiber species. Overall, this study provided a solid backbone phylogeny of Zingiber. The polymorphisms we have uncovered in the sequencing of the genome offer a rare possibility (for Zingiber) of the generation of DNA markers. These results provide a foundation for future studies that seek to understand the molecular evolutionary dynamics or individual population variation in the genus Zingiber.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-023-09115-9.
Keywords: Zingiber, Chloroplast Genome, Phylogeny, Comparative genomics
Background
Zingiber Boehm. is a diverse genus of the family Zingiberaceae and consists of approximately 100–150 species that are widely distributed in the tropical and subtropical regions of Asia and Far East Asia [1, 2]. Zingiber contains many economically important species. Some species have long-lasting inflorescences and an assemblage of tightly clasped, brightly colored bracts and floral that often highly showy. They are widely used as landscaping and cut-flower in floral arrangements including chocolate pinecone ginger (Z. montanum) and Chiang Mai Princess (Z. citriodorum) [1–3]. In addition, some Zingiber species are widely cultivated as edible crop and among the best-known nonprescription drugs in traditional medicinal systems such as myoga ginger (Z. mioga), shampoo ginger (Z. zerumbet) and ginger (Z. officinale) [4–6]. Ginger have the pharmacological and biological potential effects of analgesic and anti-inflammatory, antibacterial, antitumor and antidiabetic [7–9]. In recent years, ginger was even considered as an alternative therapeutic agent for COVID-19 treatment based on its anti-viral activity [10–12].
The genus Zingiber could be distinguished based on nutritional and floral characteristics [1, 2]. Previous studies have shown that, species of Zingber can be divided into four groups, namely sect. Zingiber, sect. Dymczewiczia, sect. Pleuranthesis and sect. Cryptanthium based on the habit of inflorescences [13–15]. However, sect. Dymczewiczia was amalgamated with Sects. Zingiber and resolved as sister to sect. Pleuranthesis with weak support value according to the phylogenetic analysis of internal transcribed spacer (ITS) sequence of 23 Zingiber species and pollen morphology [16]. Zingiber species share similar characteristics of leaves and other vegetative organs, which makes it extremely difficult to identify species in the non-flowering stage [1–3]. Recently years, efforts have been made to explore the phylogenetic relationships among Zingiber species based on molecular data [16–19]. Kerss, et al. [17] found low resolution in identifying six Zingiber species using ITS and chloroplast matK regions. According to the analyses of amplified fragment length polymorphism (AFLP) DNA markers, Z. montanum was closely related to Z. zerumbet other than to Z. officinale [18]. These results were also revealed by Li, et al. [19] based on the complete chloroplast genome data. Overall, these previous studies have succeeded in clarifying the phylogenetic relationships of some Zingiber species, however, only small number of samples were used and the relationships among many species within the genus Zingiber are still unclear.
Chloroplast genomes have been used to address the chloroplast genome evolution, patterns and rates of nucleotide substitutions and phylogenetic relationships among land plants [20]. Chloroplast is a kind of vital organelle that can transform light energy into chemical energy in green plants [21, 22]. The chloroplast genome usually has a typical quadripartite structure consisting of a large single copy (LSC) region, a small single copy (SSC) region, and two copies of inverted repeats (IRs) shows and encodes 110–130 genes with a size range of 120–180 kb and [23–25]. In compare with mitochondrial and nuclear genome, chloroplast genome is typically inherited maternally and non-recombining [26]. Although the chloroplast genome structure is usually conserved in angiosperms, variations in genome size, genome structure, and gene substitution rate have been identified [27, 28]. In recent years, more than 40 complete chloroplast genomes have been sequenced in the family Zingiberaceae and divergent hotspots, which could be used for phylogenetic analyses, have been identified [25, 29–31]. However, only seven chloroplast genomes of Zingiber have been reported, which hindering the molecular plant identification and phylogenetic relationship clarification of Zingiber species. High throughput sequencing technology has made obtaining chloroplast genome sequences more practical and provides a unique opportunity to study the evolution of the chloroplast genome and the phylogeny of the genus Zingiber.
In this study, to characterize the genome structures, gene content, phylogeny and other characteristics of Zingiber, we sequenced chloroplast genomes of fourteen Zingiber species (Table 1). Then, we explored the molecular features of each genome and compared them with six other published chloroplast genomes within the Zingiber. Finally, we determined the chloroplast genome sequence variation, molecular evolution and phylogenetic relationships among 20 within the Zingiber.
Table 1.
Summary features of complete chloroplast genomes of Zingiber species
| Genome feature | Zingiber cochleariforme | Zingiber densissimum |
Zingiber ellipticum |
Zingiber flavomaculosum |
Zingiber koshunense |
Zingiber leptorrhizum |
Zingiber neotruncatum |
|---|---|---|---|---|---|---|---|
| Genome size (bp) | 163,665 | 163,607 | 163,455 | 163,298 | 163,394 | 162,956 | 162,484 |
| LSC length (bp) | 88,167 | 87,981 | 87,946 | 88,124 | 87,785 | 87,430 | 86,712 |
| SSC length (bp) | 15,788 | 15,846 | 15,771 | 15,644 | 15,835 | 15,722 | 15,812 |
| IR length (bp) | 29,855 | 29,890 | 29,869 | 29,765 | 29,887 | 29,902 | 29,957 |
| GC content (%) | |||||||
| Total genome | 36.04% | 36.08% | 36.16% | 36.12% | 36.07% | 36.18% | 36.13% |
| LSC | 33.78% | 33.88% | 33.92% | 33.88% | 33.84% | 34.02% | 33.95% |
| SSC | 29.57% | 29.40% | 29.73% | 29.68% | 29.49% | 29.61% | 29.29% |
| IR | 41.09% | 41.07% | 41.16% | 41.14% | 41.07% | 41.07% | 41.13% |
| Genes (total/different) | 133/113 | 133/113 | 133/113 | 133/113 | 133/113 | 133/113 | 133/113 |
| CDS (total/different) | 87/79 | 87/79 | 87/79 | 87/79 | 87/79 | 87/79 | 87/79 |
| tRNA (total/different) | 38/30 | 38/30 | 38/30 | 38/30 | 38/30 | 38/30 | 38/30 |
| rRNA (total/different) | 8/4 | 8/4 | 8/4 | 8/4 | 8/4 | 8/4 | 8/4 |
| Genes with introns | 18 | 18 | 18 | 18 | 18 | 18 | 18 |
| Different CDS in LSC | 61 | 61 | 61 | 61 | 61 | 61 | 61 |
| Different CDS in SSC | 12 | 12 | 12 | 12 | 12 | 12 | 12 |
| Different CDS in IRB | 8 | 8 | 8 | 8 | 8 | 8 | 8 |
| Different CDS in IRA | 8 | 8 | 8 | 8 | 8 | 8 | 8 |
| GenBank accession | OP869986 | OP869975 | OP869976 | OP869987 | OP869977 | OP869984 | OP869978 |
| Genome feature |
Zingiber orbiculatum |
Zingiber purpureum |
Zingiber smilesianum |
Zingiber striolatum |
Zingiber xishuangbannaense |
Zingiber yingjiangense |
Zingiber Montanum |
|---|---|---|---|---|---|---|---|
| Genome size (bp) | 163,527 | 163,135 | 163,640 | 163,711 | 163,487 | 163,623 | 163,476 |
| LSC length (bp) | 88,032 | 87,730 | 88,142 | 88,026 | 88,199 | 87,957 | 87,797 |
| SSC length (bp) | 15,829 | 15,795 | 15,782 | 15,871 | 15,498 | 15,798 | 15,995 |
| IR length (bp) | 29,833 | 29,805 | 29,858 | 29,907 | 29,895 | 29,934 | 29,842 |
| GC content (%) | |||||||
| Total genome | 36.06% | 35.89% | 36.07% | 36.03% | 36.04% | 36.08% | 35.90% |
| LSC | 33.85% | 33.66% | 33.84% | 33.82% | 33.82% | 33.88% | 33.68% |
| SSC | 29.39% | 29.26% | 29.53% | 29.35% | 29.71% | 29.45% | 29.27% |
| IR | 41.10% | 40.95% | 41.09% | 41.06% | 40.95% | 41.05% | 40.93% |
| Genes (total/different) | 133/113 | 133/113 | 133/113 | 133/113 | 133/113 | 133/113 | 133/113 |
| CDS (total/different) | 87/79 | 87/79 | 87/79 | 87/79 | 87/79 | 87/79 | 87/79 |
| tRNA (total/different) | 38/30 | 38/30 | 38/30 | 38/30 | 38/30 | 38/30 | 38/30 |
| rRNA (total/different) | 8/4 | 8/4 | 8/4 | 8/4 | 8/4 | 8/4 | 8/4 |
| Genes with introns | 18 | 18 | 18 | 18 | 18 | 18 | 18 |
| Different CDS in LSC | 61 | 61 | 61 | 61 | 61 | 61 | 61 |
| Different CDS in SSC | 12 | 12 | 12 | 12 | 12 | 12 | 12 |
| Different CDS in IRB | 8 | 8 | 8 | 8 | 8 | 8 | 8 |
| Different CDS in IRA | 8 | 8 | 8 | 8 | 8 | 8 | 8 |
| GenBank accession | OP869979 | OP869980 | OP869981 | ON646165 | OP869982 | OP869983 | OP869985 |
LSC Lager single copy region, SSC Simple single copy region, IR Inverted repeat, CDS Protein coding gene
Results
Features of the Zingiber chloroplast genomes
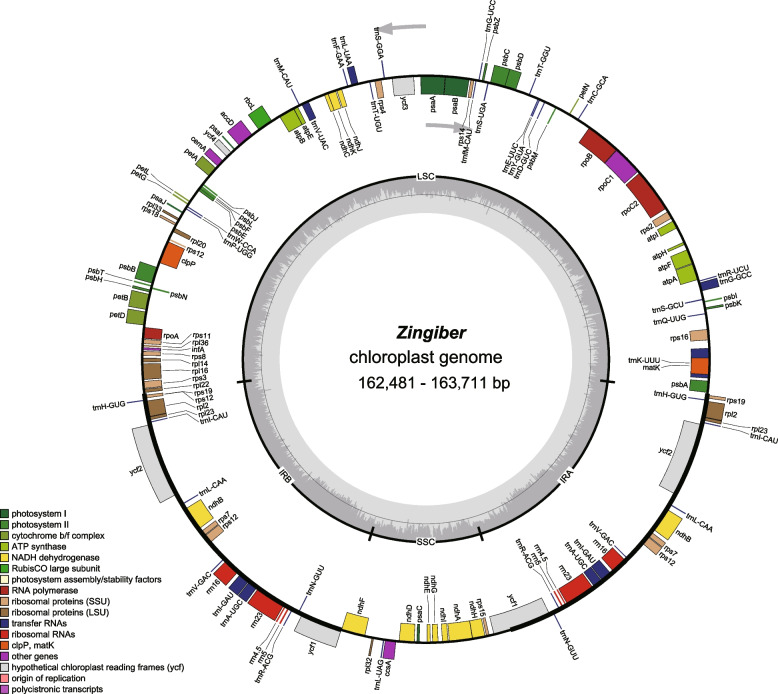
All fourteen sequenced chloroplast genomes of Zingiber have a typical quadripartite structure containing one large single copy (LSC), one small single copy (SSC) and two inverted repeat regions (IRA and IRB) (Fig. 1, Table 1). The chloroplast genomes size of them ranged from 162,481 bp (Z. neotruncatum) to 163,711 bp (Z. striolatum), with an LSC region (86,988–88,199 bp) and an SSC region (15,498–15,995 bp) separated by two inverted repeat (IR) regions (29,765–29,934 bp). All fourteen chloroplast genomes show similar total GC content (35.89–36.18%), and the IR regions (40.93–41.16%) were significantly higher than the other two regions (Table 1, Fig. 1). The 14 sequenced chloroplast genomes contain 133 predicted functional genes, of which 113 were unique genes, including 79 protein coding genes, 30 tRNA genes, and 4 rRNA genes (Tables 1 and 2). Among the different protein coding genes in our fourteen sequenced chloroplast genomes, 61 genes are located in the LSC regions, 12 genes are located in the SSC regions, and 8 genes are duplicated in the IR regions (Table 1). There were 18 genes containing introns, most of them have only a single intron, whereas ycf3 and clpP genes contain two introns (Table 2).
Fig. 1.
Chloroplast genome map of the genus Zingiber in this study. Genes belonging to different functional groups are shown in different colors in the outermost first ring. Genes shown on the outside of the outermost first ring are transcribed counter-clockwise and on the inside clockwise. The gray arrowheads indicate the direction of the genes. The tRNA genes are indicated by one letter code of amino acids with anticodons. LSC, large single copy region; IR, inverted repeat; SSC, small single copy region
Table 2.
Genes present in fourteen sequenced chloroplast genomes
| Category for genes | Group of genes | Name of genes |
|---|---|---|
| Photosynthesis | Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Subunits of photosystem II |
psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ |
|
| Subunits of cytochrome b/f complex | petA, petBa, petDa, petG, petL, petN, | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpFa, atpH, atpI | |
| Subunits of NADH dehydrogenase | ndhAa, ndhB(×2)a, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, | |
| ndhJ, ndhK | ||
| Subunit of rubisco | rbcL | |
| Self-replication | RNA polymerase | rpoA, rpoB, rpoC1a, rpoC2 |
| Large subunit of ribosomal proteins | rpl2(×2)a, rpl14, rpl16a, rpl20, rpl22, rpl23(× 2), rpl32, rpl33 | |
| rpl36 | ||
| Small subunit of ribosomal proteins | rps2, rps3, rps4, rps7(×2), rps8, rps11, rps12(× 2)a, rps14, | |
| rps15, rps16a, rps18, rps19(×2) | ||
| Ribosomal RNAs | rrn4.5(×2), rrn5(× 2), rrn16(× 2), rrn23(× 2) | |
| Transfer RNAs | trnA-UGC(×2)a, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-GCCa, trnG-UCC, trnH-GUG(× 2), trnI-CAU (× 2), trnI-GAU(× 2)a, trnK-UUUa, trnL-CAA(× 2), trnL-UAAa, trnL-UAG, trnM-CAU, trnN-GUU(× 2), trnP-UGG, trnQ-UUG, trnR-ACG(× 2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(× 2), trnV-UACa, trnW-CCA, trnY-GUA | |
| Other genes | Subunit of acetyl-coA-carboxylase | accD |
| c-type cytochrome synthesis gene | ccsA | |
| Envelop membrane protein | cemA | |
| Protease | clpPb | |
| Translational initiation factor | infA | |
| Maturase | matK | |
| Unknown function | Conserved open reading frames | ycf1(×2), ycf2(×2), ycf3b, ycf4 |
(×2): gene with two copies; agene containing one intron; bgene containing two introns;
Codon usage and RNA editing sites
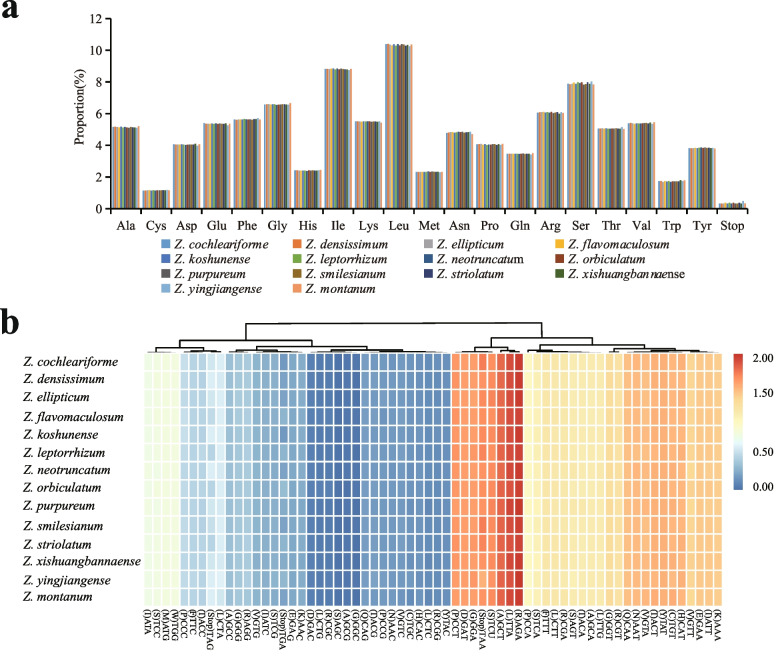
Codon usage patterns and nucleotide composition help to lay a theoretical foundation for genetic modifications of the chloroplast genome [32]. A total of 79 protein coding genes in all 14 sequenced chloroplast genomes in Zingiber are analyzed for codon usage frequency. They comprise 25,557 (Z. montanum) to 26,354 (Z. xishuangbannaense) codons. Of the 25,557–26,354 codons, leucine (Leu) is the most abundant amino acid, with a frequency of 10.25–10.40%, followed by isoleucine (Ile) with a frequency of 8.75–8.85%, while cysteine (Cys) is the least common, with a frequency of 1.14–1.18% (Fig. 2a). Because of the value of relative synonymous codon usage (RSCU) > 1.00, thirty codons show codon usage bias in protein coding genes of the 14 sequenced chloroplast genomes. Stop codon usage is biased toward TAA (RSCU > 1.00) (Fig. 2b). Both methionine (Met) and tryptophan (Trp) exhibit no codon bias and have RSCU values of 1.00 (Fig. 2b).
Fig. 2.
Codon content of all protein coding genes. a amino acids and stop codons proportion in protein coding sequences of fourteen sequenced chloroplast genomes and b heat map analysis for codon distribution of all protein coding genes of fourteen sequenced chloroplast genomes. Red colour indicates higher RSCU values and blue colour indicates lower RSCU values
Furthermore, 72–81 RNA editing sites were identified in 27 protein-coding genes of 14 chloroplast genomes, with the least in Z. montanum (72 sites) and Z. purpureum (72 sites), and the most in Z. orbiculatum (81 sites) (Table S1). In the 14 identified chloroplast genomes that we sequenced, the ndhB gene has the highest number of potential editing sites (11 sites), followed by the ndhD gene (7 sites) (Table S1). All of these editing sites are C-to-T transitions that occur at the first or second positions of the codons.
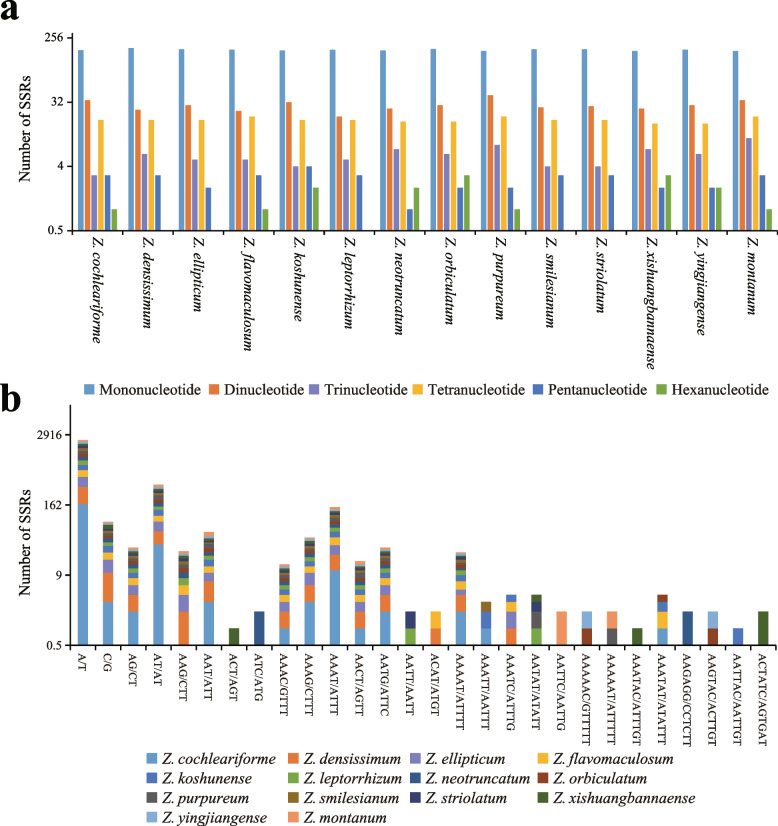
Features of simple sequence repeats (SSRs) and long repeats
A total number of 221 to 238 SSRs were identified in all sequenced chloroplast genome. (Fig. 3). Among each sequenced chloroplast genome, mononucleotide repeats were the most frequent, with numbers ranging from 167 to 184, which accounted for 70.18–79.09% of all SSRs, followed by dinucleotide, ranging from 24 to 40 (9.09–16.81%), tetranucleotide, ranging from 16 to 20 (6.96–8.77%), trinucleotide, ranging from 3 to 10 (1.30–4.26%), pentanucleotide, ranging from 1 to 4 (0.45–1.74%), and hexanucleotide, ranging from 0 to 3 (0–1.36%). The majority of the mononucleotide SSRs were A/T repeats, which accounted for 68.07–75.00% of all the repeat types among the fourteen sequenced chloroplast genomes, followed by AT/AT repeats, ranging from 8.18–15.97%, and the remaining repeat types below 6% (Fig. 3b).
Fig. 3.
Comparison of the simple sequence repeats (SSRs) among fourteen Zingiber species. a the number of different SSR types. b the frequency of the identified SSRs in different repeat class types
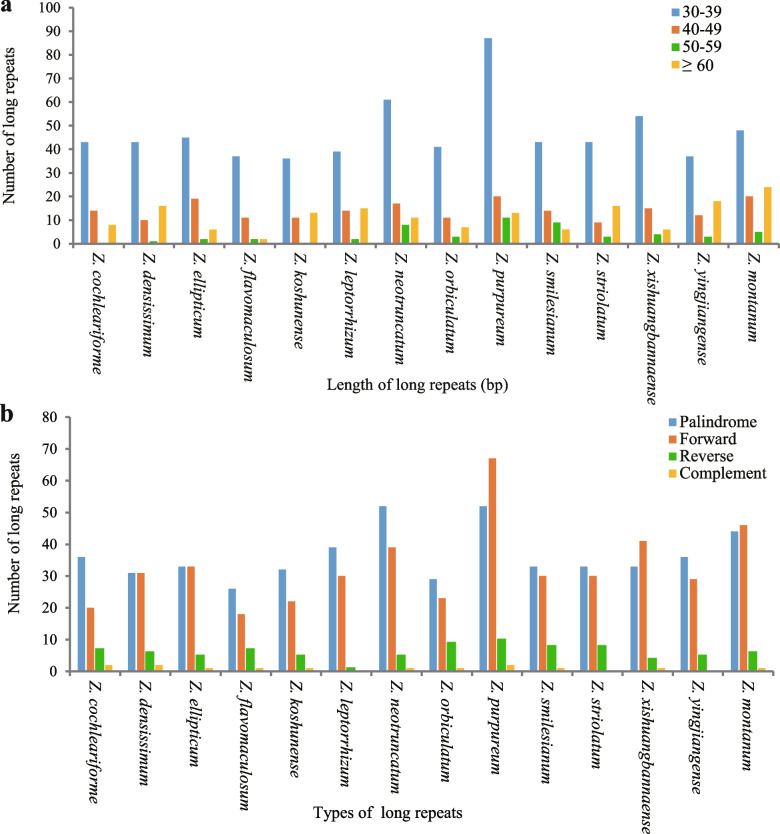
Long repeats that longer than 30 bp may have the function of promoting chloroplast genome rearrangement and increasing population genetic diversity, which has been a hotspot in genomic research [33]. In this study, 14 sequenced chloroplast genomes had 1068 long repeats that consisted of 509 palindromic repeats, 459 forward repeats, 86 reverse repeats, 14 complement repeats, 86 reverse repeats (Fig. 4a). Z. montanum had the largest number (131), and Z. flavomaculosum had the smallest number of long repeats (52) (Fig. 4a). In addition, the numbers of the four repeat types are quite different in Zingiber, with palindromic repeats and forward repeats a clear quantitative superiority (20–67), while complement repeats and reverse repeats are less abundant (0–10) (Fig. 4b). Moreover, among all long repeats, most sequences were between 30 and 39 bp (657) in length, followed by 40–49 bp (197), and 50–59 bp had the least number (53) (Fig. 4b).
Fig. 4.
Long repeat sequences among fourteen Zingiber species. a total of four long repeat types in fourteen chloroplast genomes and b numbers of long repeat sequences by length
Contraction and expansion of inverted repeats (IRs)
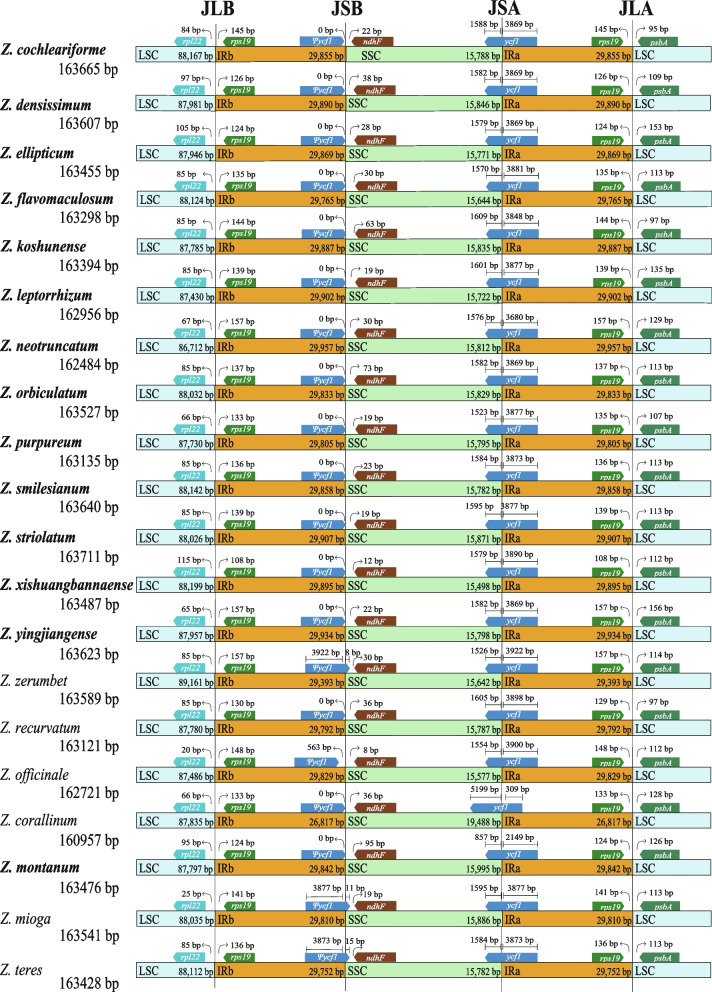
A comprehensive comparison at the LSC/IRs/SSC boundaries was performed among the 20 Zingiber species (Fig. 5). Although the inverted repeat regions (IRA and IRB) are the most conserved regions of the chloroplast genome, shrinkage and expansion of the IR boundaries are hypothesized to help explain size differences between chloroplast genomes beyond genus. The length of the IR region in the 14 chloroplast genomes exhibited a modest expansion, ranging from 29,765 bp to 29,957 bp. Within the 20 chloroplast genomes of Zingiber species, the rpl22 and rps19 genes were located in the boundaries of the LSC/ IRB regions (Fig. 5). There were 20–115 bp between rpl22 and the LSC/IRB borders, and the distance between rps19 and the LSC/IRB boundary ranged from 108 bp to 157 bp (Fig. 5).
Fig. 5.
Comparisons of LSC, SSC and IR regions boundaries among 20 chloroplast genomes. Ψ: pseudogenes
Ψycf1-ndhF genes were located at the boundaries of the IRB/SSC regions in all 20 Zingiber species. The IRB/SSC borders of 8 species (Z. cochleariforme, Z. densissimum, Z. ellipticum, Z. flavomaculosum, Z. orbiculatum, Z. yingjiangense, Z. recurvatum and Z. corallinum) were all situated adjacent to the end of Ψycf1. In addition, Ψycf1 expanded into the SSC regions in 8 species, namely, Z. koshunense, Z. purpureum, Z. smilesianum, Z. xishuangbannaense, Z. zerumbet, Z. montanum, Z. mioga and Z. teres, for 25 bp, 5 bp, 9 bp, 1 bp, 8 bp, 83 bp, 11 bp and 15 bp, respectively (Fig. 5). There were 63 bp, 19 bp, 23 bp, 12 bp, 30 bp, 95 bp, 19 bp and 15 bp between the ndhF and LSC/IRB borders in Z. koshunense, Z. purpureum, Z. smilesianum, Z. xishuangbannaense, Z. zerumbet, Z. montanum, Z. mioga and Z. teres, respectively (Fig. 5).
The SSC/IRA boundary was situated in the ycf1 coding region, which crossed into the IRA region in all 20 Zingiber species. However, the length of ycf1 in the IRA region varied among the 20 Zingiber species from 309 bp to 3922 bp (Fig. 5).
The rps19 and psbA genes were situated in the boundaries of the IRA/LSC regions in all 20 Zingiber species, in which the distances between rps19 and the IRA/LSC border ranged from 108 bp to 157 bp (Fig. 5). For all 20 Zingiber species, a 95–156 bp distance was observed between the psbA gene and the IRA/LSC border (Fig. 5).
Genomic comparative and nucleotide diversity analyses
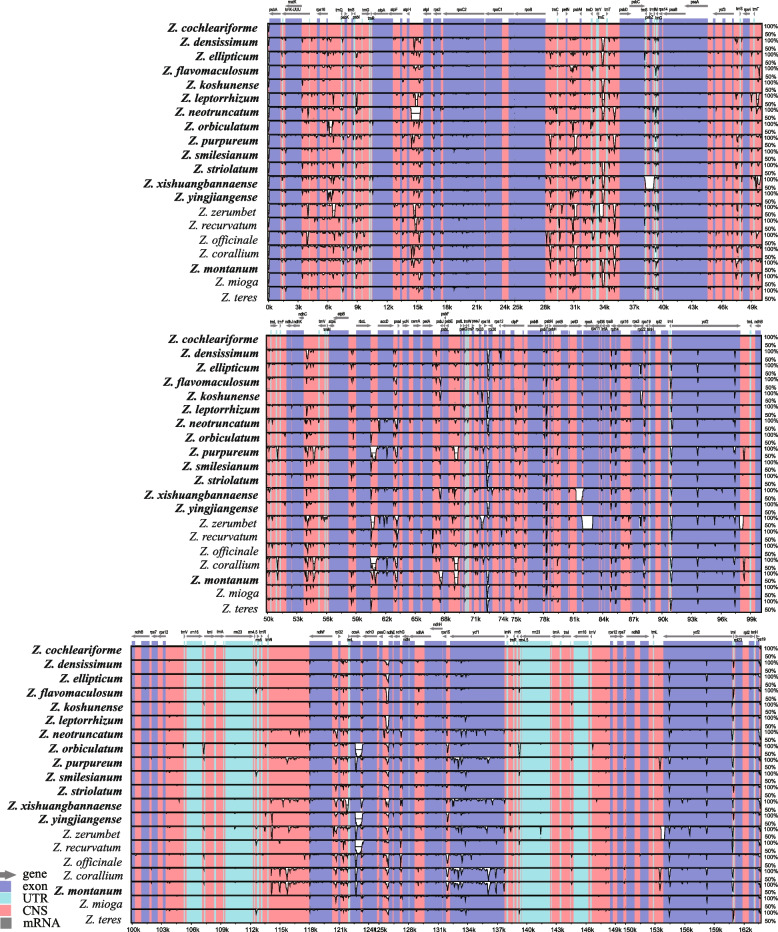
Multiple alignments of 20 Zingiber chloroplast genomes were compared by mVISTA, with the annotated Z. cochleariforme genome sequence as the reference (Fig. 6). The mVISTA comparison showed that the LSC and SSC regions were more divergent than the two IR regions. Moreover, the non-coding region exhibited more nucleotide divergence than the coding regions. The main divergences for the coding regions were located in the region of accD, ccsA, rpoC2 and ycf1. For the non-coding regions, strongly divergent regions were rbcL-accD, trnT-UGU-trnL-UAA, rps16-trnQ-UUG, atpI-atpH, petN-psbM, trnT-UGU-trnL-UAA, ndhF-rpl32, rpl32-trnL-UAG, trnN-ndhF and trnL-ycf1 (Fig. 6).
Fig. 6.
Comparative plots of percent sequence identity of 20 chloroplast genomes in Zingiber. Coarse species represent chloroplast genome obtained in this study
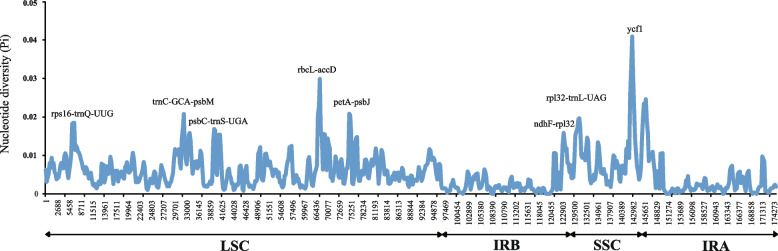
Furthermore, nucleotide diversity (Pi) values were calculated within 800 bp windows (Fig. 7) to identify sequence divergence hotspots. The results showed that the Pi value of the whole Zingiber chloroplast genome varied from 0 to 0.04088. Eight highly variable regions (Pi> 0.016) were detected: petA-pabJ, rbcL-accD, rpl32-trnL-UAG, rps16-trnQ-UUG, trnC-GCA-psbM, psbC-trnS-UGA and ndhF-rpl32 and ycf1. Among these, five regions (petA-pabJ, rbcL-accD, rps16-trnQ-UUG, trnC-GCA-psbM and psbC-trnS-UGA were located in the LSC region, and the remaining three were in the SSC region (Fig. 7). This is consistent with preceding results that the IR region is generally more conserved than the LSC and the SSC regions.
Fig. 7.
Nucleotide diversity (Pi) values of various regions in 20 chloroplast genomes
Characterization of substitution rates and positive selection analyses
The non-synonymous (dN) and synonymous (dS) substitution rates of all 79 protein coding genes were analyzed across 20 Zingiber species. Most of the genes were subjected to purifying selection. Using the likelihood ratio test, we found that 19 protein coding genes were under positive selection with posterior probability greater than 0.95 (Table 3). Among the 19 protein coding genes, ycf1 showed the highest number of positive amino acids sites (52), followed by ycf2 (24) and clpP (12) (Table 3). The other 16 protein coding genes, ccsA, ndhA, ndhB, petD, psbA, psbB, psbC, rbcL, rpl12, rpl20, rpl23, rpl33, rpoC2, rps7, rps12 and ycf3, presented 2, 5, 3, 1, 2, 1, 2, 11, 1, 5, 1, 5, 3, 1, 1 and 1 amino acids sites were truly under positive selection respectively (Table 3).
Table 3.
Positive selective amino acid loci and estimation of parameters
| Gene | Ln L | Estimates of parameters | Positively selected sites |
|---|---|---|---|
| ccsA | − 1688.900532 |
p0 = 0.99053p = 0.00799q = 0.00500 (p1 = 0.00947)ω = 122.48812 |
200 F 0.999**, 201 I 0.986* |
| clpP | − 1189.046814 |
p0 = 0.91790p = 0.02800q = 0.01097 (p1 = 0.08210)ω = 22.05525 |
22 E 0.966*, 24 Y 1.000**, 39 K 0.972*, 40 E 0.969*, 58 Q 0.979*, 72 W 0.979*, 75 S 0.998**, 78 A 0.960*, 120 V 1.000**, 153 T 0.975*, 192 L 0.961*, 204 L 0.953* |
| ndhA | − 1741.334638 |
p0 = 0.98308p = 12.72091q = 99.00000 (p1 = 0.01692)ω = 52.52860 |
132 S 0.996**, 189 R 1.000**, 190 V 1.000**, 191 I 0.998**, 192 L 0.998** |
| ndhB | − 2096.337425 |
p0 = 0.98968p = 0.00500q = 1.58164 (p1 = 0.01032)ω = 999.00000 |
133 I 0.995**, 145 G 0.951*, 181 T 0.995** |
| petD | − 753.029169 |
p0 = 0.96547p = 0.00500q = 1.79217 (p1 = 0.03453)ω = 7.43965 |
8 I 0.970* |
| psaA | − 3396.235357 |
p0 = 0.99301p = 0.00500q = 0.75911 (p1 = 0.00699)ω = 41.45919 |
261 F 0.999**, 292 A 0.981* |
| psbB | − 2254.262995 |
p0 = 0.99605p = 0.00500q = 1.52674 (p1 = 0.00395)ω = 373.49395 |
488 A 1.000** |
| psbC | − 2130.498976 |
p0 = 0.99356p = 0.00500q = 1.59626 (p1 = 0.00644)ω = 85.67788 |
24 T 0.970*, 280 S 0.978* |
| rbcL | − 2275.465641 |
p0 = 0.96928p = 0.48224q = 5.55236 (p1 = 0.03072)ω = 24.32347 |
19 G 0.992**, 30 T 0.999**, 176 L 0.988*, 232 L 1.000**, 233 F 1.000**, 254 C 0.981*, 269 T 0.970*, 289 H 0.998**, 333 I 0.998**, 431 L 0.987*, 456 S 1.000** |
| rpl12 | − 1180.117502 |
p0 = 0.99203p = 0.00500q = 3.98754 (p1 = 0.00797)ω = 77.23012 |
140 S 0.966* |
| rpl20 | − 745.824555 |
p0 = 0.95179p = 0.00871q = 0.02374 (p1 = 0.04821)ω = 35.61813 |
120 S 1.000**, 121 N 1.000**, 122 K 1.000**, 123 V 1.000**, 124 H 1.000** |
| rpl23 | −430.542995 |
p0 = 0.97858p = 0.00500q = 2.07543 (p1 = 0.02142)ω = 999.00000 |
57 E 0.976* |
| rpl33 | − 288.922817 |
p0 = 0.00001p = 0.00500q = 2.19296 (p1 = 0.99999)ω = 999.00000 |
3 K 0.988*, 6 D 0.988*, 29 G 0.988*, 42 M 0.988*, 43 P 0.988* |
| rpoC2 | − 6439.688674 |
p0 = 0.99492p = 0.04204q = 0.16546 (p1 = 0.00508)ω = 22.05755 |
593 D 0.978*, 743 P 0.985*, 1181 W 0.986* |
| rps7 | − 698.656393 |
p0 = 0.98065p = 0.00500q = 1.44246 (p1 = 0.01935)ω = 485.63912 |
81 G 0.966* |
| rps12 | −575.913647 |
p0 = 0.99188p = 0.01544q = 0.03483 (p1 = 0.00812)ω = 999.00000 |
116 Q 0.958* |
| ycf1 | −10,914.54603 |
p0 = 0.70387p = 0.00500q = 1.91338 (p1 = 0.29613)ω = 7.28992 |
14 S 0.996**, 16 I 0.990*, 48 R 0.970*, 65 I 0.994**, 315 R 0.958*, 353 R 0.977*, 384 S 0.961*, 404 L 0.994**, 464 E 0.952*, 588 N 0.997**, 598 F 0.994**, 622 E 0.990*, 682 L 0.953*, 684 A 0.994**, 699 H 0.994**, 705 Q 0.996**, 728 S 1.000**, 729 V 0.997**, 748 Q 0.996**, 753 R 1.000**, 861 I 0.998**, 869 L 0.961*, 883 L 0.965*, 900 Y 0.972*, 905 E 0.951*, 936 T 0.991**, 944 L 0.952*, 982 T 0.960*, 991 A 0.968*, 1027 P 0.991**, 1339 R 0.996**, 1449 R 0.997**, 1538 I 0.995**, 1539 S 0.996**, 1547 H 0.990**, 1564 S 1.000**, 1568 W 0.979*, 1569 S 0.977*, 1597 F 0.962*, 1657 T 0.991**, 1671 P 0.965*, 1672 L 0.956*, 1690 S 0.996**, 1695 L 0.953*, 1713 I 0.994**, 1715 H 0.993**, 1719 R 0.990*, 1748 L 0.996**, 1750 A 1.000**, 1758 T 0.954*, 1759 L 0.997**, 1780 G 0.999** |
| ycf2 | −10,302.30873 |
p0 = 0.96727p = 5.07671q = 3.06985 (p1 = 0.03273)ω = 65.06865 |
847 R 0.994**, 1008 D 1.000**, 1087 G 0.993**, 1353 L 1.000**, 1390 T 0.993**, 1417 E 0.993**, 1421 S 0.994**, 1461 P 0.994**, 1654 H 0.993**, 1677 I 0.993**, 2117 R 0.994**, 1997 T 0.993**, 2117 R 0.994**, 2231 Q 0.994**, 2247 R 1.000**, 2317 L 0.972*, 2319 H 0.993**, 2321 T 0.999**, 2322 G 0.995**, 2323 E 0.993**, 2324 R 0.993**, 2325 F 0.999**, 2327 I 0.993**, 2328 P 0.994** |
| ycf3 | − 735.294401 |
p0 = 0.98001p = 0.00500q = 1.68882 (p1 = 0.01999)ω = 32.38804 |
44 M 0.998** |
The degree of freedom for each gene was 38; * and ** indicate posterior probability higher than 0.95 and 0.99, respectively
Phylogenetic analyses
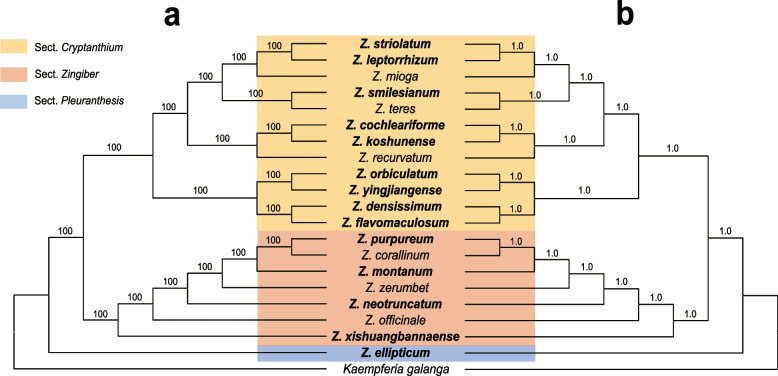
The phylogeny of 55 Zingiberaceae species were well resolved (Fig. S1). Zingiber is monophyletic (BS = 100%) and was well resolved as sister to Kaempferia with strong support (BS = 100%). Based on the chloroplast genome dataset, we generated a well-resolved phylogeny of Zingiber (Fig. 8). The support values of all the branches in both ML and BI trees were robust (BI = 1.0, BS = 100%). Thus, we will not include the support values in the text below. Zingiber was divided into three sections: sect. Crytanthium, sect. Zingiber, and sect. Pleuranthesis. Sect. Crytanthium is resolved as sister to sect. Zingiber. There are four major clades of the sect. Crytanthium. The first branch was well supported and comprised Z. flavomaculosum + Z. densissimum as sister to Z. yingjiangense + Z. orbiculatum. The second clade was Z. recurvatum + (Z. koshunense + Z. cochleariforme). Within the rest of the sect. Cryanthium, two subclades were recovered: Z. teres + Z. smilesianum and Z. mioga + (Z. leptorrhizum + Z. striolatum). In sect. Zingiber, Z. xishuangbannaense, subsequently followed by Z. officinale, Z. neotruncatum, Z. zerumbet, Z. montanum was sister to Z. corallinum + Z. purpureum. As for sect. Pleuranthesis, which contains only one species (Z. ellipticum).
Fig. 8.
Molecular phylogenetic tree based on 20 chloroplast genomes within the genus Zingiber. a Maximum likelihood tree. b Bayesian tree. Coarse species represent chloroplast genome obtained in this study
Discussion
In this study, 14 Zingiber chloroplast genomes were newly reported. Their genome size (162,481 bp-163,711 bp), GC content (35.89–36.18%), genome quadripartite structure, gene composition, all of the protein-coding genes, tRNA and rRNA showed high similarity, which were in consistent with other Zingiberoideae chloroplast genomes [25, 29–31]. The conservation of plastomes had been observed in various angiosperms such as Malvaceae, Araceae in which the same gene content and gene order had been reported [34–37]. Nevertheless, plastome rearrangement, gene duplication, gene loss and intron loss are reported in a number of plant lineages [22, 25, 38]. Although structure variations occurred in some Zingiberoideae plants for example, both trnS-GGA and trnT-GGU were lost in the chloroplast genome of Globba schomburgkii. The lhbA gene were lost in both Hedychium coccineum and Hedychium neocarneum [25]. However, the chloroplast genomes of Zingiber species were highly conserved in current study, which is in agreement with previous studies at genus level Camellia [39], Sinosenecio [40], and Chrysosplenium [41]. Plastomes are very conservative which was maintained by multiple molecular mechanisms including uniparental inheritance, rarity of plastid fusion, and the presence of an active repair mechanism [21, 35]. Hence, the typically conservative nature of the Zingiber plastomes is linked to a certain molecular mechanism.
The expansion and contraction at the borders of the IR regions of chloroplast genomes is common in angiosperms, which may cause size variations, gene duplication or the reduction, and the origination of pseudogenes [20, 42, 43]. Abnormal expansion of IR regions had been observed in some taxon e.g., Pilea [44], Erodium [45] and Pelargonium [46], which transferred numerous genes from the SC regions into the IR. In this study, we found that expansion and contraction of IRs showed much similarity among the species of the genus Zingiber, and the distribution and locations of gene types in these regions were highly consistent. These results are in agreement with previous report of Zingiberoideae [25]. The IR/SSC boundary shifts always cause the increased length in the IR regions. Here, we found the IR/SC boundary of Zingiber is relatively stable. The pseudogene of ycf1 originated at the junction of IR in Zingiber plants which was also observed in other angiosperms [25, 35]. Compared with the chloroplast genomes of six Zingiber species published in NCBI, the length of IR region of all species assembled by ourselves was basically the same, and no gene loss was detected. Overall, the conservation of the IR of the Zingiber plants may be one of the reasons for its stability in length and structure.
Highly variable regions are always used as DNA barcode markers for the studies on species identification and phylogenetic analyses. The high similarity of the vegetative characteristics has made it extremely difficult to distinguish Zingiber plants [16]. Since some classical DNA barcodes are insufficient for species identification and phylogeny of Zingiber, it is very important to find more highly variable regions at genus level that could be developed as representing potential markers for future variety identification research. Based on the results of mVISTA and nucleotide diversity, eight highly variable regions among 20 Zingiber species are identified including seven intergenic regions (petA-pabJ, rbcL-accD, rpl32-trnL-UAG, rps16-trnQ-UUG, trnC-GCA-psbM, psbC-trnS-UGA and ndhF-rpl32) and one genic region (ycf1). These highly variable regions could be used as potential DNA barcode for species identification and phylogenetic analysis for the Zingiber species. Among them, ndhF-rpl32, rpl32-trnL-UAG and ycf1 were reported as suitable for species identification at subfamily and genus level in Zingiberoideae [25]. The ycf1 gene is the most variable site in the chloroplast genome, showing greater variability than existing chloroplast candidate barcodes such as matK and rbcL [47]. Other five intergenic regions that identified in the present study are also reported in other plants at species level. For example, petA-pabJ was demonstrated well utilization as DNA barcodes for Lindera plant [48] and rbcL-accD was identified to be an effective marker for Rumex species [49]. Sun, et al. [50] suggested that petA-psbJ, ndhF-rpl32 and rpl32-trnL potentially be used as molecular genetic markers for population genetics and phylogenetic studies of Magnolia polytepala. And rps16-trnQ-UUG, trnC-GCA-psbM, psbC-trnS-UGA are also reported in previous studies [51, 52]. Generally, although several candidate barcoding regions were identified, further research is still necessary to determine whether these highly divergent markers could be used in the identification and phylogenetic analyses of Zingiber species.
Positive selection is assumed to play key roles in the adaptation of organisms to diverse environments [53], while negative (purifying) selection is a ubiquitous evolutionary force responsible for genomic sequence conservation across long evolutionary timescales [54]. In this study, 19 genes with positive selection sites are identified in Zingiber. Among these genes containing amino acid positive sites, we found that ycf1 and ycf2 genes possess higher number (52, 24, respectively) of positive amino acid sites within Zingiber species, suggesting that the ycf1 gene may play important roles in the adaptive evolution of Zingiber species. Six genes (rpl12, rpl20, rpl23, rpl33, rps7 and rps12) encoding ribosomal subunit proteins are under positive selection, and these genes are considered to be essential for chloroplast biogenesis and function, suggesting that Zingiber plants may increase the adaptability of evolution by regulating encoding ribosomal subunit proteins in chloroplasts [55]. Moreover, eleven genes, namely ccsA, clpP, ndhA, ndhB, petD, psaA, psbB, psbC, rbcL, rpoC2 and ycf3, have also been identified with positive selection sites in current study. Recent studies have indicated that these nineteen genes with positive selection in some angiosperms are common. For examples, ccsA, rbcL, rpoC2 have been identified under positive selection in Orchidaceae, Euterpe, and Pterocarpus [24, 56, 57]; In Zingiberoideae, ccsA, ndhA, ndhB, psbJ, rbcL, rpl20, rpoC1, rpoC2, rps12, rps18, ycf1, ycf2 and ycf4 have also been identified under positive selection [25]. Zingiber species mainly inhabited warm, humid, semi-shaded environment and maintain a high level of plant diversity [1, 3]. Therefore, based on our analyses, we believe that positive selection of these chloroplast genes may be promote the adaptation of Zingiber plants to semi-shaded environment, but the detailed adaptation mechanism needs further in-depth research.
The phylogenetic analysis of 55 Zingiberaceae species showed that Zingiber was well resolved as sister to Kaempferia with strong support (Fig. S1), which is consistent with previous studies [17, 19, 25, 58–60]. Previously, the classification of Zingiber species was usually based on the type of inflorescence and pollen morphology, which generally solved the classification problems of Zingiber plants [61]. Zingiber was classified into three sections based on ITS sequences analyses together with similarity in pollen morphology and inflorescence habit [16, 17]. Our species-level phylogenetic tree of Zingiber showed that three traditionally accepted sections were monophyletic with strong support. In different with the result of Theerakulpisut [16] based on the ITS analyses, our results strongly supported sect. Cryptanthium as sister to sect Zingiber rather than sect. Pleuranthesis. Conflicts between phylogenetic trees delineated by chloroplast genomes and nuclear genes are also common in some angiosperms, such as Asteraceae and Zingiberaceae [62–68]. The conflict phenomenon may be due to reticulate evolution in the events of rapid diversification or uniparental inheritance of the plastome [35, 62]. However, the mechanism that leads to the conflict in Zingiber require further in-deep research. Additionally, the phylogeny indicated strong support for interspecies relationships. In sect. Zingiber, Z. purpureum was well resolved as sister to Z. corallinum. Z. xishuangbannaense, a species endemic to china, was resolved as the first lineage split from Zingiber in this study. The reminder Zingiber species formed a monophyletic clade with strong support, which is consistent with previous studies [16, 25]. The rest of the sect. Zingiber formed a strong supported clade. Although Theerakulpisut, et al. [16] recognized this clade, but the bootstrap value is below 50% and relationships among a number of lineages of this clade are uncertain. Our results demonstrated that Z. neotruncatum subsequently followed by Z. zerumbet was sister to Z. montanum + (Z. corallinum + Z. purpureum). For sect. Cryptanthium, 12 species, including 9 newly sequenced species in this study, were sampled, which is the mostly densest sampling to date. The relationships among lineages of sect. Cryptanthium were well resolved with robust support and provided a back bone for further classification at the infrageneric level and for investigating the biogeography of this group.
Conclusions
In this study, fourteen complete chloroplast genomes of Zingiber species have been sequenced, assembled and annotated for the first time. The structural characteristics of these fourteen chloroplast genomes are shown to be conservative, which are similar to those reported chloroplast genomes of Zingiberoideae species. Meanwhile, comparative analyses of 20 Zingiber chloroplast genomes have generated 8 highly variable regions, which may be used as a potential source of molecular markers for species identification. Based on whole chloroplast genomes data, phylogenetic relationships among 20 Zingiber species have been clearly resolved. We found sect. Cryptanthium as sister to sect Zingiber rather than to sect. Pleuranthesis. The conflict phenomenon may be due to reticulate evolution in the events of rapid diversification or uniparental inheritance of the plastome. In addition, 19 genes are under positive selection with high posterior probabilities, which may play important roles in Zingiber species adaption to semi-shaded environment. Overall, our research has greatly enriched the genome resources of Zingiber, which will help to further analyze the phylogeny of Zingiber and resolve the genetic relationships within Zingiber in the future.
Materials and methods
Plant material, DNA extraction, and sequencing
A total of 21 chloroplast genomes were used for this study, including seven chloroplast genomes obtained from GenBank (www.ncbi.nlm.nih.gov/genbank) and fourteen newly generated in this study (Table 1). Genomic DNA was isolated from silica-gel dried leaf tissue or herbarium specimens (Table S2) using Plant Genomic DNA Kit (TIANGEN, Beijing, China), The concentration and quantity of each isolated genomic DNA sample were determined with a NanoDrop 2000 micro spectrometer (Wilmington, DE, USA) and 1% agarose gel electrophoresis, respectively. DNA was used to construct PE libraries with insert sizes of 150 bp and sequenced by the MGI DNBSEQ-T7 platform (MGI-TECH, Shen Zhen, China).
Chloroplast genome assembly and annotation
For each accession, 5.0 Gb raw data were generated with pair-end 150 bp read length. Trimmomatic v0.39 [69] was used to remove low-quality and adapter-containing reads. The clean data were then assembled using GetOrganelle v1.7.5 [70]. The assembled chloroplast genomes were annotated in Geneious R11 with Z. officinal (MW602894), Z. teres (NC_062457), Z. mioga (NC_057615), Z. recurvatum (MT473712) and Z. zerumbet (MK262726) as references, and then manually checked for start/stop codons. Finally, the OGDRAW v1.3.1 program was used to draw the circular chloroplast genome maps of the Zingiber species with default settings.
Codon usage and RNA editing sites
Codon usage patterns and nucleotide composition could help to lay a theoretical foundation for genetic modifications of the chloroplast genome [32]. Here, to examine the deviation in synonymous codon usage, the relative synonymous codon usage (RSCU) was calculated using the software CodonW (University of Texas, Houston, TX, USA) with the RSCU value (Fig. 2a). When the RSCU value > 1.00, it means that the use of a codon is more frequent than expected, and vice versa. The clustered heat map of RSCU values of fourteen sequenced Zingiber chloroplast genomes was conducted by R v3.6.3 (https://www.R-project.org.) (Fig. 2b). To predict possible RNA editing sites in the twenty chloroplast genomes, protein coding genes were used to predict potential RNA editing sites using the online program Predictive RNA Editor for Plants (PREP) suite (http://prep.unl.edu/) with a cut of value of 0.8.
Analyses of SSRs and long repeats
Chloroplast SSR has high variation level within the same species and is an important source for developing molecular markers, which are widely used in phylogenetic and population genetic analysis [71]. MIcroSAtellite (MISA) (http://pgrc.ipk-gatersleben.de/misa/) was used to detect the simple sequence repeat (SSRs or microsatellites) motifs in fourteen sequenced chloroplast genomes with the settings as follows: 8 for mono-, 5 for di-, 4 for tri-, and 3 for tetra-, pena-, and hexa-nucleotide SSRs (Fig. 3). The REPuter software was employed to identify long repeats such as forward, palindrome, reverse and complement repeats. The criteria for determining long repeats were as follows: (1) a minimal repeat size of more than 30 bp; (2) a repeat identity of more than 90%; and (3) a hamming distance equal to 3 (Fig. 4).
Genome comparison and nucleotide variation analysis
To detect the contractions and expansions of the IR regions in the chloroplast genomes of the Zingiber, 20 whole genomes within Zingiber were compared (Fig. 5). The online software mVISTA tool with the Shufe-LAGAN mode [72] was used to make pairwise alignments among these 20 whole chloroplast genomes with the annotated chloroplast genome of Z. cochleariforme as reference (Fig. 6). The 20 chloroplast genomes of Zingiber were first aligned using MAFFT v7 [73] and then manually adjusted using BioEdit v7.0.9 [74]. DnaSP v5.10 software [75] was used to calculate the nucleotide variability (Pi) of the 20 chloroplast genomes within the Zingiber, with a sliding window analysis with the step size and window length set as 200 bp and 800 bp (Fig. 7).
Positive selection analysis
To identify the genes under selection, we scanned the chloroplast genomes of fourteen species within Zingiber using the software EasyCondeML [76]. The software was used for calculating the non-synonymous (dN) and synonymous (dS) substitution rates, along with their ratios (ω = dN/dS). The analyses of selective pressures were conducted along the ML tree of these fourteen species in Newick format. Each single-copy CDS sequences was aligned according to their amino acid sequence. The site-specific model with five site models (M0, M1a & M2a, M7 & M8) were employed to identify the signatures of adaptation across chloroplast genomes. This model allowed the ω ratio to vary among sites, with a fixed ω ratio in all the branches. The site-specific model, M1a (nearly neutral) vs. M2a (positive selection) and M7 (β) vs. M8 (β & ω) were calculated in order to detect positive selection [77]. Likelihood ratio test (LRT) of the comparison (M1a vs. M2a and M7 vs. M8) was used to evaluate of the selection strength respectively and the p value of Chi square (χ2) smaller than 0.05 is thought as significant. The Bayes Empirical Bayes (BEB) inference [78] was implemented in site models M2a and M8 to estimate the posterior probabilities and positive selection pressures of the selected genes.
Phylogenetic analyses
The phylogenetic analyses of 20 Zingiber species were performed based on chloroplast genomic data. The Maximum Likelihood (ML) method in Geneious R11 was used to construct the phylogenetic tree with default settings including 1000 bootstrap replications and the general time-reversible model with a gamma distribution of substitution rate among sites (GTR + G). In addition, Bayesian Inference (BI) was performed using MrBayes v3.2 [79], using the substitution model GTR and running parameters were as follows: the Markov Chain Monte Carlo algorithm was applied for 2 million generations with four Markov chains and sampled of trees every 100 generations, then the first 10% of trees were discarded as burn-in. The software Figtree v1.4 was used to edit and visualize the final BI tree and ML tree (Fig. 8). In addition, to clarify the phylogenetic position of Zingiber within the Zingiberaceae, we constructed a maximum likelihood tree based on chloroplast genome dataset of 55 Zingiberaceae species.
Supplementary Information
Additional file 1: Fig. S1. Molecular phylogenetic tree based on 55 chloroplast genomes of Zingiberaceae. Species name in red color represent chloroplast genome obtained in this study. Table S1. List of RNA editing sites in fourteen Zingiber species by PREP program. Table S2. List of 14 species of Zingiber sequenced in this study.
Acknowledgements
We thank Dr. Renbin Zhu (XTBG) and Qiang Zhang (GXIB) help collect materials. And we sincerely thank Dr. Zhiduan Chen from IBCAS for carefully reading an early draft of the manuscript.
Abbreviations
- bp
Base pairs
- ITS
Internal transcribed spacer
- BI
Bayesian Inference
- CNS
Conserved non coding sequence
- dN
Non synonymous
- DNA
Deoxyribonucleic acid
- dS
Synonymous
- IR
Inverted repeat
- LSC
Large single copy region
- SSC
Small single copy region
- ML
Maximum Likelihood
- RSCU
Relative synonymous codon usage
- SSR
Simple sequence repeats
Authors’ contributions
H.-L.L. and Y.L. conceived the study. D.J., X.C., M.G., H.-L.L., S.T. and H.X. performed the experiments. D.J., X.C., H.-L.L., S.D., M.X., S.T. and J.L. contributed reagents/materials/ analysis tools and analyzed the data. H.-L.L. and D.J. wrote the paper. H.-L.L., D.J., X.C., Y.L. and M.X. edited the paper. The author(s) read and approved the final manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (32270237), the Scientific and Technological Research Program of Chongqing Municipal Education Commission (KJZD-M202101301), the Natural Science Foundation of Chongqing (cstc2019jcyj-msxmX0300) and the Foundation for High-level Talents of Chongqing University of Arts and Science (R2022YS09, 2017RTZ21, P2018TZ05).
Availability of data and materials
The complete chloroplast genomes generated during the current study were deposited in NCBI database (Accession number: OP869975, OP869976, OP869977, OP869978, OP869979, OP869980, OP869981, OP869982, OP869983, OP869984, OP869985, OP869986, OP869987, ON646165).
Declarations
Ethics approval and consent to participate
The authors declare that the collection of plant materials for this study complies with relevant institutional, national and international guidelines and legislation.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interest.
Footnotes
The original version of this article was revised: The second author's affiliation was wrong and Fig. 3 was not updated. This has been solved.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Dongzhu Jiang and Xiaodong Cai are co-first authors and contributed equally to this work.
Change history
7/14/2023
A Correction to this paper has been published: 10.1186/s12864-023-09393-3
Contributor Information
Yiqing Liu, Email: liung906@163.com.
Hong-Lei Li, Email: lihonglei215@163.com.
References
- 1.Wu D, Liu N, Ye Y. The zingiberaceous resources in china. Wuhan: Huazhong university of science and technology university press; 2016. p. 143. [Google Scholar]
- 2.Branney TM. Hardy gingers: Including hedychium, roscoea, and zingiber. Portland: Timber press, Inc.; 2005. pp. 44–55. [Google Scholar]
- 3.Gao J, Xia Y, Huang J, Li Q. Zhongguo jiangke huahui. Beijing: Science press; 2006. p. 40. [Google Scholar]
- 4.Sasidharan I, Nirmala MA. Comparative chemical composition and antimicrobial activity fresh & dry ginger oils (Zingiber officinale roscoe). J Int Pharm Res. 2010;2: 40-43.
- 5.Banerjee S, Mullick H, Banerjee J, Ghosh A. Zingiber officinale: ‘A natural gold’. Int J Pharmaceutical Bio-Sci. 2011;2:283–294. [Google Scholar]
- 6.Prasad S, Tyagi AK. Ginger and its constituents: Role in prevention and treatment of gastrointestinal cancer. Gastroent Res Pract. 2015;2015: 142979. [DOI] [PMC free article] [PubMed]
- 7.Kubra IR, Rao LJM. An impression on current developments in the technology, chemistry, and biological activities of ginger (Zingiber officinale roscoe) Crit Rev Food Sci Nutr. 2012;52:651–688. doi: 10.1080/10408398.2010.505689. [DOI] [PubMed] [Google Scholar]
- 8.Shareef HK, Muhammed HJ, Hussein HM, Hameed IH. Antibacterial effect of ginger (Zingiber officinale) roscoe and bioactive chemical analysis using gas chromatography mass spectrum. Orient J Chem. 2016;32:20–40. doi: 10.13005/ojc/320207. [DOI] [Google Scholar]
- 9.Li H-L, Wu L, Dong Z, Jiang Y, Jiang S, Xing H, et al. Haplotype-resolved genome of diploid ginger (Zingiber officinale) and its unique gingerol biosynthetic pathway. Hort Res. 2021;8: 1-1. [DOI] [PMC free article] [PubMed]
- 10.Jafarzadeh A, Jafarzadeh S, Nemati M. Therapeutic potential of ginger against COVID-19: Is there enough evidence? J Tradit Chinese Medical Sci. 2021;8:267–279. [Google Scholar]
- 11.San Chang J, Wang KC, Yeh CF, Shieh DE, Chiang LC. Fresh ginger (Zingiber officinale) has anti-viral activity against human respiratory syncytial virus in human respiratory tract cell lines. J Ethnopharmacol. 2013;145:146–151. doi: 10.1016/j.jep.2012.10.043. [DOI] [PubMed] [Google Scholar]
- 12.Thota SM, Balan V, Sivaramakrishnan V. Natural products as home-based prophylactic and symptom management agents in the setting of COVID-19. Phytother Res. 2020;34:3148–3167. doi: 10.1002/ptr.6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Theilade I. Revision of the genus Zingiber in peninsular Malaysia. The Gardens’ Bulletin Singapore. 1996;48:207–236. [Google Scholar]
- 14.Theilade I. A synopsis of the genus Zingiber (Zingiberaceae) in Thailand. Nord J Bot. 1999;19:389–410. doi: 10.1111/j.1756-1051.1999.tb01220.x. [DOI] [Google Scholar]
- 15.Theilade I, Mærsk-Møller M, Theilade J, Larsen K. Pollen morphology and structure of Zingiber (Zingiberaceae) Grana. 1993;32:338–342. doi: 10.1080/00173139309428961. [DOI] [Google Scholar]
- 16.Theerakulpisut P, Triboun P, Mahakham W, Maensiri D, Khampila J, Chantaranothai P. Phylogeny of the genus Zingiber (Zingiberaceae) based on nuclear its sequence data. Kew Bull. 2012;67:389–395. doi: 10.1007/s12225-012-9368-2. [DOI] [Google Scholar]
- 17.Kress WJ, Prince LM, Williams KJ. The phylogeny and a new classification of the gingers (Zingiberaceae): Evidence from molecular data. Am J Bot. 2002;89:1682–1696. doi: 10.3732/ajb.89.10.1682. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh S, Majumder P, Mandi SS. Species-specific AFLP markers for identification of Zingiber officinale, Z. montanum and Z. zerumbet (Zingiberaceae) Genet Mol Res. 2011;10:218–229. doi: 10.4238/vol10-1gmr1154. [DOI] [PubMed] [Google Scholar]
- 19.Li D-M, Ye Y-J, Xu Y-C, Liu J-M, Zhu G-F. Complete chloroplast genomes of Zingiber montanum and Zingiber zerumbet: Genome structure, comparative and phylogenetic analyses. PLoS One. 2020;15:e0236590. doi: 10.1371/journal.pone.0236590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo Y-Y, Yang J-X, Bai M-Z, Zhang G-Q, Liu Z-J. The chloroplast genome evolution of venus slipper (Paphiopedilum): Ir expansion, ssc contraction, and highly rearranged ssc regions. BMC Plant Biol. 2021;21:1–14. doi: 10.1186/s12870-021-03053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wicke S, Schneeweiss GM, Depamphilis CW, Müller KF, Quandt D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol Biol. 2011;76:273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daniell H, Lin C-S, Yu M, Chang W-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17:1–29. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Zuo Y, Zhu X, Liao S, Ma J. Complete chloroplast genomes and comparative analysis of sequences evolution among seven Aristolochia (Aristolochiaceae) medicinal species. Int J Mol Sci. 2019;20:1045. doi: 10.3390/ijms20051045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong Z, Wu Z, Zhao K, Yang Z, Zhang N, Guo J, Tembrock LR, Xu D. Comparative analyses of five complete chloroplast genomes from the genus Pterocarpus (Fabacaeae) Int J Mol Sci. 2020;21:3758. doi: 10.3390/ijms21113758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li D-M, Li J, Wang D-R, Xu Y-C, Zhu G-F. Molecular evolution of chloroplast genomes in subfamily Zingiberoideae (Zingiberaceae) BMC Plant Biol. 2021;21:1–24. doi: 10.1186/s12870-021-03315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsunewaki K, Ogihara Y. The molecular basis of genetic diversity among cytoplasms of Triticum and Aegilops species. Ii. On the origin of polyploid wheat cytoplasms as suggested by chloroplast DNA restriction fragment patterns. Genetics. 1983;104:155–171. doi: 10.1093/genetics/104.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrett CF, Wicke S, Sass C. Dense infraspecific sampling reveals rapid and independent trajectories of plastome degradation in a heterotrophic orchid complex. New Phytol. 2018;218:1192–1204. doi: 10.1111/nph.15072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrett CF, Sinn BT, Kennedy AH. Unprecedented parallel photosynthetic losses in a heterotrophic orchid genus. Mol Biol Evol. 2019;36:1884–1901. doi: 10.1093/molbev/msz111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui Y, Nie L, Sun W, Xu Z, Wang Y, Yu J, Song J, Yao H. Comparative and phylogenetic analyses of ginger (Zingiber officinale) in the family Zingiberaceae based on the complete chloroplast genome. Plants. 2019;8:283. doi: 10.3390/plants8080283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li D-M, Zhao C-Y, Liu X-F. Complete chloroplast genome sequences of Kaempferia galanga and Kaempferia elegans: Molecular structures and comparative analysis. Molecules. 2019;24:474. doi: 10.3390/molecules24030474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li D-M, Zhao C-Y, Zhu G-F, Xu Y-C. Complete chloroplast genome sequence of Hedychium coronarium. Mitochondrial DNA Part B. 2019;4:2806–2807. doi: 10.1080/23802359.2019.1659114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazumdar P, Binti Othman R, Mebus K, Ramakrishnan N, Ann HJ. Codon usage and codon pair patterns in non-grass monocot genomes. Ann Bot. 2017;120:893–909. doi: 10.1093/aob/mcx112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Timme RE, Kuehl JV, Boore JL, Jansen RK. A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes: Identification of divergent regions and categorization of shared repeats. Am J Bot. 2007;94:302–312. doi: 10.3732/ajb.94.3.302. [DOI] [PubMed] [Google Scholar]
- 34.Mehmood F, Shahzadi I, Ali Z, Islam M, Naeem M, Mirza B, Lockhart PJ, Ahmed I, Waheed MT. Correlations among oligonucleotide repeats, nucleotide substitutions, and insertion-deletion mutations in chloroplast genomes of plant family Malvaceae. J Syst Evol. 2021;59:388–402. doi: 10.1111/jse.12585. [DOI] [Google Scholar]
- 35.Mehmood F, Rahim A, Heidari P, Ahmed I, Poczai P. Comparative plastome analysis of Blumea, with implications for genome evolution and phylogeny of Asteroideae. Ecol Evol. 2021;11:7810–7826. doi: 10.1002/ece3.7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waseem S, Mirza B, Ahmed I, Waheed MT. Comparative analyses of chloroplast genomes of Theobroma cacao and Theobroma grandiflorum. Biologia. 2020;75:761–771. doi: 10.2478/s11756-019-00388-8. [DOI] [Google Scholar]
- 37.Henriquez CL, Mehmood F, Hayat A, Sammad A, Waseem S, Waheed MT, Matthews PJ, Croat TB, Poczai P, Ahmed I. Chloroplast genome evolution in the Dracunculus clade (Aroideae, Araceae) Genomics. 2021;113:183–192. doi: 10.1016/j.ygeno.2020.12.016. [DOI] [PubMed] [Google Scholar]
- 38.Lee C, Ruhlman TA, Jansen RK. Unprecedented intraindividual structural heteroplasmy in Eleocharis (Cyperaceae, Poales) plastomes. Genome Biol Evol. 2020;12:641–655. doi: 10.1093/gbe/evaa076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li L, Hu Y, He M, Zhang B, Wu W, Cai P, Huo D, Hong Y. Comparative chloroplast genomes: Insights into the evolution of the chloroplast genome of Camellia sinensis and the phylogeny of Camellia. BMC Genomics. 2021;22:1–22. doi: 10.1186/s12864-021-07427-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng J-Y, Zhang X-S, Zhang D-G, Wang Y, Deng T, Huang X-H, Kuang T-H, Zhou Q. Newly reported chloroplast genome of Sinosenecio albonervius y. Liu & qe yang and comparative analyses with other Sinosenecio species. BMC Genomics. 2022;23:1–13. doi: 10.1186/s12864-022-08872-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Z, Liao R, Yang T, Dong X, Lan D, Qin R, Liu H. Analysis of six chloroplast genomes provides insight into the evolution of Chrysosplenium (Saxifragaceae) BMC Genomics. 2020;21:1–14. doi: 10.1186/s12864-020-07045-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu C-S, Chaw S-M. Large-scale comparative analysis reveals the mechanisms driving plastomic compaction, reduction, and inversions in conifers II (Cupressophytes) Genome Biol Evol. 2016;8:3740–3750. doi: 10.1093/gbe/evw278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu A, Guo W, Gupta S, Fan W, Mower JP. Evolutionary dynamics of the plastid inverted repeat: The effects of expansion, contraction, and loss on substitution rates. New Phytol. 2016;209:1747–1756. doi: 10.1111/nph.13743. [DOI] [PubMed] [Google Scholar]
- 44.Li J, Tang J, Zeng S, Han F, Yuan J, Yu J. Comparative plastid genomics of four Pilea (Urticaceae) species: Insight into interspecific plastid genome diversity in Pilea. BMC Plant Biol. 2021;21:1–13. doi: 10.1186/s12870-020-02793-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blazier JC, Jansen RK, Mower JP, Govindu M, Zhang J, Weng M-L, Ruhlman TA. Variable presence of the inverted repeat and plastome stability in Erodium. Ann Bot. 2016;117:1209–1220. doi: 10.1093/aob/mcw065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weng ML, Ruhlman TA, Jansen RK. Expansion of inverted repeat does not decrease substitution rates in Pelargonium plastid genomes. New Phytol. 2017;214:842–851. doi: 10.1111/nph.14375. [DOI] [PubMed] [Google Scholar]
- 47.Dong W, Xu C, Li C, Sun J, Zuo Y, Shi S, Cheng T, Guo J, Zhou S. Ycf1, the most promising plastid DNA barcode of land plants. Sci Rep. 2015;5:1–5. doi: 10.1038/srep08348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao M-L, Song Y, Ni J, Yao X, Tan Y-H, Xu Z-F. Comparative chloroplast genomics and phylogenetics of nine Lindera species (Lauraceae) Sci Rep. 2018;8:1–11. doi: 10.1038/s41598-018-27090-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhandari GS, Park C-W. Molecular evidence for natural hybridization between Rumex crispus and R. obtusifolius (Polygonaceae) in Korea. Sci Rep. 2022;12:1–12. doi: 10.1038/s41598-022-09292-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun L, Jiang Z, Wan X, Zou X, Yao X, Wang Y, Yin Z. The complete chloroplast genome of Magnolia polytepala: Comparative analyses offer implication for genetics and phylogeny of Yulania. Gene. 2020;736:144410. doi: 10.1016/j.gene.2020.144410. [DOI] [PubMed] [Google Scholar]
- 51.Amenu SG, Wei N, Wu L, Oyebanji O, Hu G, Zhou Y, Wang Q. Phylogenomic and comparative analyses of Coffeeae alliance (Rubiaceae): Deep insights into phylogenetic relationships and plastome evolution. BMC Plant Biol. 2022;22:1–13. doi: 10.1186/s12870-022-03480-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jo S, Kim Y-K, Cheon S-H, Fan Q, Kim K-J. Characterization of 20 complete plastomes from the tribe Laureae (Lauraceae) and distribution of small inversions. PLoS One. 2019;14:e0224622. doi: 10.1371/journal.pone.0224622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moseley RC, Mewalal R, Motta F, Tuskan GA, Haase S, Yang X. Conservation and diversification of circadian rhythmicity between a model crassulacean acid metabolism plant Kalanchoë edtschenkoi and a model c3 photosynthesis plant Arabidopsis thaliana. Front Plant Sci. 2018;9:1757. doi: 10.3389/fpls.2018.01757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cvijović I, Good BH, Desai MM. The effect of strong purifying selection on genetic diversity. Genetics. 2018;209:1235–1278. doi: 10.1534/genetics.118.301058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee K, Leister D, Kleine T. Arabidopsis mitochondrial transcription termination factor mterf2 promotes splicing of group iib introns. Cells. 2021;10:315. doi: 10.3390/cells10020315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dong W-L, Wang R-N, Zhang N-Y, Fan W-B, Fang M-F, Li Z-H. Molecular evolution of chloroplast genomes of orchid species: Insights into phylogenetic relationship and adaptive evolution. Int J Mol Sci. 2018;19:716. doi: 10.3390/ijms19030716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Santana LA, Gomes Pacheco T, Nascimento da Silva O, do Nascimento Vieira L, Guerra MP, Pacca Luna Mattar E, de Baura VA, Balsanelli E, Maltempi de Souza E, de Oliveira Pedrosa F, et al. Plastid genome evolution in Amazonian acaí palm (Euterpe oleracea mart.) and Atlantic forest açaí palm (Euterpe edulis mart.) Plant Mol Biol. 2021;105:559–574. doi: 10.1007/s11103-020-01109-5. [DOI] [PubMed] [Google Scholar]
- 58.Ngamriabsakul C, Newman M, Cronk Q. The phylogeny of tribe Zingibereae (Zingiberaceae) based on its (nrDNA) and trnL-F (cpdna) sequences. Edinb J Bot. 2003;60:483–507. doi: 10.1017/S0960428603000362. [DOI] [Google Scholar]
- 59.Wood T, Whitten W, Williams N. Phylogeny of Hedychium and Related genera (Zingiberaceae) based on its sequence data. Edinb J Bot. 2000;57:261–270. doi: 10.1017/S0960428600000196. [DOI] [Google Scholar]
- 60.Williams KJ, Kress WJ, Manos PS. The phylogeny, evolution, and classification of the genus Globba and tribe Globbeae (Zingiberaceae): Appendages do matter. Am J Bot. 2004;91:100–114. doi: 10.3732/ajb.91.1.100. [DOI] [PubMed] [Google Scholar]
- 61.Valeton T. New notes on the Zingiberaceae of Java and Malaya n archipelago. Bull Jard Bot Buitenzorg ser. 1918;27:1–166. [Google Scholar]
- 62.Watson LE, Siniscalchi CM, Mandel J. Phylogenomics of the hyperdiverse daisy tribes: Anthemideae, Astereae, Calenduleae, Gnaphalieae, and Senecioneae. J Syst Evol. 2020;58:841–852. doi: 10.1111/jse.12698. [DOI] [Google Scholar]
- 63.Vargas OM, Ortiz EM, Simpson BB. Conflicting phylogenomic signals reveal a pattern of reticulate evolution in a recent high-Andean diversification (Asteraceae: Astereae: Diplostephium) New Phytol. 2017;214:1736–1750. doi: 10.1111/nph.14530. [DOI] [PubMed] [Google Scholar]
- 64.Gao B, Yuan L, Tang T, Hou J, Pan K, Wei N. The complete chloroplast genome sequence of Alpinia oxyphylla miq. and comparison analysis within the Zingiberaceae family. PLoS One. 2019;14:e0218817. doi: 10.1371/journal.pone.0218817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gui L, Jiang S, Xie D, Yu L, Huang Y, Zhang Z, Liu Y. Analysis of complete chloroplast genomes of Curcuma and the contribution to phylogeny and adaptive evolution. Gene. 2020;732:144355. doi: 10.1016/j.gene.2020.144355. [DOI] [PubMed] [Google Scholar]
- 66.Liang H, Zhang Y, Deng J, Gao G, Ding C, Zhang L, Yang R. The complete chloroplast genome sequences of 14 Curcuma species: Insights into genome evolution and phylogenetic relationships within Zingiberales. Front Genet. 2020;11:802. doi: 10.3389/fgene.2020.00802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li D-M, Zhu G-F, Xu Y-C, Ye Y-J, Liu J-M. Complete chloroplast genomes of three medicinal Alpinia species: Genome organization, comparative analyses and phylogenetic relationships in family Zingiberaceae. Plants. 2020;9:286. doi: 10.3390/plants9020286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li D-M, Zhao C-Y, Zhu G-F, Xu Y-C. Complete chloroplast genome sequence of Amomum villosum. Mitochondrial DNA Part B. 2019;4:2673–2674. doi: 10.1080/23802359.2019.1644237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jin J-J, Yu W-B, Yang J-B, Song Y, DePamphilis CW, Yi T-S, Li D-Z. Getorganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020;21:1–31. doi: 10.1186/s13059-020-02154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Srivastava D, Shanker A. Identification of simple sequence repeats in chloroplast genomes of Magnoliids through bioinformatics approach. Interdiscip Sci. 2016;8:327–336. doi: 10.1007/s12539-015-0129-4. [DOI] [PubMed] [Google Scholar]
- 72.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. Vista: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–WW79. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Katoh K, Misawa K, Ki K, Miyata T. Mafft: A novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 75.Librado P, Rozas J. Dnasp v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 76.Gao F, Chen C, Arab DA, Du Z, He Y, Ho SY. Easycodeml: A visual tool for analysis of selection using codeml. Ecol Evol. 2019;9:3891–3898. doi: 10.1002/ece3.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang Z, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- 78.Yang Z, Wong WS, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 79.Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. Molecular phylogenetic tree based on 55 chloroplast genomes of Zingiberaceae. Species name in red color represent chloroplast genome obtained in this study. Table S1. List of RNA editing sites in fourteen Zingiber species by PREP program. Table S2. List of 14 species of Zingiber sequenced in this study.
Data Availability Statement
The complete chloroplast genomes generated during the current study were deposited in NCBI database (Accession number: OP869975, OP869976, OP869977, OP869978, OP869979, OP869980, OP869981, OP869982, OP869983, OP869984, OP869985, OP869986, OP869987, ON646165).