Abstract
Subfamily Caesalpinioideae with ca. 4,600 species in 152 genera is the second-largest subfamily of legumes (Leguminosae) and forms an ecologically and economically important group of trees, shrubs and lianas with a pantropical distribution. Despite major advances in the last few decades towards aligning genera with clades across Caesalpinioideae, generic delimitation remains in a state of considerable flux, especially across the mimosoid clade. We test the monophyly of genera across Caesalpinioideae via phylogenomic analysis of 997 nuclear genes sequenced via targeted enrichment (Hybseq) for 420 species and 147 of the 152 genera currently recognised in the subfamily. We show that 22 genera are non-monophyletic or nested in other genera and that non-monophyly is concentrated in the mimosoid clade where ca. 25% of the 90 genera are found to be non-monophyletic. We suggest two main reasons for this pervasive generic non-monophyly: (i) extensive morphological homoplasy that we document here for a handful of important traits and, particularly, the repeated evolution of distinctive fruit types that were historically emphasised in delimiting genera and (ii) this is an artefact of the lack of pantropical taxonomic syntheses and sampling in previous phylogenies and the consequent failure to identify clades that span the Old World and New World or conversely amphi-Atlantic genera that are non-monophyletic, both of which are critical for delimiting genera across this large pantropical clade. Finally, we discuss taxon delimitation in the phylogenomic era and especially how assessing patterns of gene tree conflict can provide additional insights into generic delimitation. This new phylogenomic framework provides the foundations for a series of papers reclassifying genera that are presented here in Advances in Legume Systematics (ALS) 14 Part 1, for establishing a new higher-level phylogenetic tribal and clade-based classification of Caesalpinioideae that is the focus of ALS14 Part 2 and for downstream analyses of evolutionary diversification and biogeography of this important group of legumes which are presented elsewhere.
Keywords: Fabaceae, generic delimitation, mimosoid clade, monophyly, morphological homoplasy, phylogenomics
Introduction
In 2017, the Legume Phylogeny Working Group established a new subfamily classification of the Leguminosae (LPWG 2017), which dealt with the longstanding problem of the paraphyly of old sense subfamily Caesalpinioideae DC. by formally dividing the family into six subfamilies: Cercidoideae LPWG, Detarioideae Burmeist., Duparquetioideae LPWG, Dialioideae LPWG, Caesalpinioideae and Papilionoideae DC. Subfamily Caesalpinioideae was especially impacted by this new classification because several large clades previously included within it were afforded subfamily rank, while at the same time the former subfamily Mimosoideae DC., which is nested within Caesalpinioideae, was subsumed within the re-circumscribed Caesalpinioideae and is now simply referred to as the mimosoid clade (LPWG 2017). The idea that Leguminosae comprises six main lineages has since been amply confirmed by phylogenomic analyses of large nuclear gene and plastome DNA sequence datasets (Koenen et al. 2020a; Zhang et al. 2020; Zhao et al. 2021) providing robust support for the six subfamilies. Establishment of this new classification has shifted the focus of current legume systematics research to development of phylogenetically-based tribal (e.g. de la Estrella et al. 2018 for Detarioideae) and clade-based (e.g. Sinou et al. 2020 for Cercidoideae) higher-level classifications and, especially, towards establishment of robust generic systems for each subfamily. Here, we present a phylogenomic backbone for the re-circumscribed subfamily Caesalpinioideae as the basis for a new higher-level and generic classification of that subfamily.
Caesalpinioideae sensu LPWG (2017) is the second largest subfamily of legumes with ca. 4,600 species currently placed in 152 genera (LPWG 2017 plus additions, see below). Within this subfamily, ca. 3,400 species and 90 genera are placed in the mimosoid clade corresponding to the former subfamily Mimosoideae, which is nested within new sense Caesalpinioideae (LPWG 2017). Caesalpinioideae has a pantropical distribution and many of its lineages form ecologically abundant or dominant elements across each of the major lowland tropical biomes – seasonally dry tropical forests (“the succulent biome” sensu Schrire et al. 2005 and Ringelberg et al. 2020), savannas and tropical rain forests – thus spanning the full lowland tropical rainfall spectrum from arid to hyper-wet, with just a small fraction of species extending into the warm temperate zone, a subset of which are frost tolerant. Caesalpinioideae species are infrequent above 2500 m elevation in the tropics and are notably absent from mid- and high-elevation tropical montane forests, with only a few exceptions (e.g. some Inga Mill. spp., Paraseriantheslophantha(Vent.)I.C. Nielsensubsp.montana (Jungh.) I.C. Nielsen). The ecological versatility of the subfamily across the lowland tropical moisture availability spectrum is matched by its great diversity of life-history strategies, from massive canopy-emergent rainforest trees to small desert shrubs, and functionally-herbaceous savanna geoxyles to woody lianas and aquatic plants (Lewis et al. 2005; LPWG 2013, 2017; Koenen et al. 2020b; Ringelberg et al. 2022). Many species are economically important because of their highly-nutritious fruits, valuable wood, nitrogen-rich leaves and other products (Lewis et al. 2005) and are especially prominent as multipurpose trees in tropical silvo-pastoral and other agroforestry systems. Several other species constitute some of the world’s most serious invasive weeds (e.g. Leucaenaleucocephala (Lam.) de Wit, several Mimosa L. spp. and Acacia Mill. spp., Prosopisjuliflora (Sw.) DC.). Generic diversity is highest in the Neotropics and Africa and there are important centres of species diversity in Mexico and Central America, lowland South America, Africa, Madagascar, parts of S.E. Asia and Australia. Caesalpinioideae includes some of the largest genera in the legume family, such as Acacia with > 1,000 species concentrated in dry parts of Australia and Mimosa with > 500 species mostly in the Neotropics, as well as Chamaecrista Moench and Senna Mill., each with 300+ species distributed pantropically, Inga Mill. with ca. 300 species restricted to the Neotropics, almost entirely in rainforests and Vachellia Wight & Arn. (ca. 160 species) and Senegalia Raf. (ca. 220 species), two pantropical genera concentrated in drier environments, within which the iconic umbrella-crown trees of African savannas are found.
Numbers of genera across Caesalpinioideae have increased progressively through the last 270 years, but are difficult to track, because of the altered delimitation of the subfamily. However, the history of generic delimitation in mimosoids illustrates the overall trajectory of numbers of genera. Von Linnaeus (1753) placed all known mimosoids in a single genus Mimosa, which was later subdivided by Willdenow (1805) into five genera: Inga, Mimosa, Schrankia Willd., Desmanthus Willd. and Acacia. In 1825, de Candolle added five more genera, but the real foundations for all subsequent work were established by Bentham (1842, 1875) notably in his ‘Revision of suborder Mimoseae’ in 1875, which recognised six tribes and 46 genera, based on examination of 1,200 species known at that time.
The legacy of Bentham’s generic system has been long-lasting. At the heart of Bentham’s system were a set of large, geographically widespread genera, including Acacia, Calliandra Benth., Pithecellobium Mart. and Prosopis L., all of which, with the advent of molecular phylogenetics, have been shown to be non-monophyletic. The disintegration of Acacia into (currently) seven segregate genera (Acacia, Acaciella Britton & Rose, Mariosousa Seigler & Ebinger, Parasenegalia Seigler & Ebinger, Pseudosenegalia Seigler & Ebinger, Senegalia and Vachellia), based on 20 years of molecular phylogenetic studies (Clarke et al. 2000; Miller and Bayer 2000, 2001, 2003; Robinson and Harris 2000; Luckow et al. 2003; Miller et al. 2003, 2013, 2017; Murphy et al. 2003; Seigler et al. 2006a, b; Brown et al. 2008; Bouchenak-Khelladi et al. 2010; Gómez-Acevedo et al. 2010; Miller and Seigler 2012; Kyalangalilwa et al. 2013; Mishler et al. 2014; Boatwright et al. 2015; Terra et al. 2017; Koenen et al. 2020b) (Figs 1 and 6–8) has been the most prominent example in legumes of the dissolution of one of Bentham’s broadly circumscribed pantropical genera. Pithecellobium and Calliandra have suffered similar fates (Barneby and Grimes 1996, 1997; Barneby 1998; de Souza et al. 2013, 2016). In contrast, although Bentham (1875) had restricted his concept of the genus Albizia Durazz. to just Old World species, Nielsen (1981) expanded the genus pantropically, creating the last big ‘dustbin genus’ of mimosoids (Koenen et al. 2020b). By far the most persistent generic delimitation problems surround those of former tribe Ingeae, where starkly contrasting generic systems and numerous generic transfers have caused much on-going confusion (reviewed by Brown 2008).
Figure 1.
Phylogeny of Caesalpinioideae with clade names as inferred by Koenen et al. (2020b), the starting point for this study.
Figure 6.
Phylogeny of Caesalpinioideae (continued). See Fig. 2 for caption.
Figure 8.
Phylogeny of Caesalpinioideae (continued). See Fig. 2 for caption.
By 1981, the number of mimosoid genera had risen to 62 in Advances in Legume Systematics Part 1 (Elias 1981), 78 in Legumes of the World (Lewis et al. 2005) and in the most recent census (LPWG 2017) to 84, with 148 genera recognised in Caesalpinioideae as a whole.
Across the non-mimosoid Caesalpinioideae generic delimitation has also seen many changes. The most complex problems have been, without doubt, in the Caesalpinia Group and, especially, the genus Caesalpinia L. s.l. (Polhill and Vidal 1981; Lewis 1998; Gagnon et al. 2016), but these have now largely been resolved with the phylogenetically-based generic system of Gagnon et al. (2016), which recognised 26 genera, leaving just one residual generic problem in that group (see Clark et al. 2022).
Since LPWG (2017), two genera of Caesalpinioideae have been synonymised (i.e. Cathormion Hassk. within Albizia (Koenen et al. 2020b) and Lemuropisum H. Perrier within Delonix Raf. (Babineau and Bruneau 2017)) and six new genera have been segregated or resurrected (i.e. Lachesiodendron P.G. Ribeiro, L.P. Queiroz & Luckow (Ribeiro et al. 2018), Parasenegalia and Pseudosenegalia (Seigler et al. 2017), Jupunba Britton & Rose and Punjuba Britton & Rose (Soares et al. 2021) and Robrichia (Barneby & J.W. Grimes) A.R.M. Luz & E.R. Souza (de Souza et al. 2022a)), bringing the current tally of Caesalpinioideae genera to 152, of which 90 are mimosoids.
Despite this rapid on-going progress to align genera with clades in recent years, generic delimitation across Caesalpinioideae and, especially, the mimosoid clade, remains in a state of considerable flux and there is evidence to suggest that several more genera are non-monophyletic: Prosopis (Catalano et al. 2008), Dichrostachys (DC.) Wight & Arn. (Hughes et al. 2003; Luckow et al. 2005), Balizia Barneby & J.W. Grimes (Iganci et al. 2016; Koenen et al. 2020b), Zygia P. Browne (Ferm et al. 2019), Entada Adans. (Luckow et al. 2003), Caesalpinia (Gagnon et al. 2016), Albizia, Senegalia and Leucochloron Barneby & J.W. Grimes (Koenen et al. 2020b; Fig. 1). One factor that has undoubtedly contributed significantly to this widespread generic non-monophyly is the potentially pervasive homoplasy of multiple morphological characters previously used for generic delimitation, as well as reliance on only a few characters for delimiting taxa. This has led to tribes defined solely on stamen number and fusion into a staminal tube (Bentham 1875) and ‘fruit genera’, such as Calliandra, which was defined by Bentham (1875), based on its characteristic elastically dehiscent fruit. All mimosoid tribes and the genus Calliandra have since been shown to be non-monophyletic and their defining characters shown to have evolved multiple times across the subfamily (e.g. LPWG 2013; Barneby 1998). Such over-reliance on a small number of potentially homoplasious morphological characters, such as fruit type, connation and number of stamens and floral heteromorphy have likely repeatedly misled classification and resulted in widespread generic non-monophyly.
Another issue has been delimitation of the mimosoid clade with on-going uncertainties surrounding the inclusion or not of certain genera (Luckow et al. 2000, 2003; Manzanilla and Bruneau 2012). Although lacking valvate petals in bud (the putative synapomorphy of mimosoids), morphologically some members of the informal Dimorphandra group of Polhill and Vidal (1981) and Polhill (1994) show many similarities to mimosoids, with small, often numerous, regular flowers arranged in spikes or spiciform racemes, the hypanthium contracted, the anthers sagittate and introrse, the stamens becoming the most conspicuous and attractive part of the flower and pollen in tetrads in a few genera (Diptychandra Tul. and Dinizia Ducke) with possible affinities to the polyads that characterise many mimosoid lineages (Banks et al. 2010). These mimosoid-like features have prompted inclusion of some genera such as Dinizia in the mimosoid clade in the past (e.g. Burkart 1943; Luckow et al. 2000). Although none of these mimosoid-like genera has flowers with petals valvate in bud, previous molecular phylogenetic analyses have unexpectedly placed two Dimorphandra group genera in the mimosoid clade: Chidlowia Hoyle and Sympetalandra Stapf. The monospecific west African genus Chidlowia was placed with high support within the mimosoid clade in analyses based on few genetic markers (Manzanilla and Bruneau 2012; LPWG 2017), a result which was confirmed by the phylogenomic analyses of Koenen et al. (2020b; Fig. 1). The small Asian genus Sympetalandra was also recovered in the mimosoid clade in the matK tree of LPWG (2017), but was not sampled by Koenen et al. (2020b). Although support for the mimosoid clade is robust and the branch subtending that clade is long (Koenen et al. 2020b; Fig. 1), such that the monophyly of mimosoids is not in doubt, not all Caesalpinioideae genera have been included in phylogenomic analyses. By sampling widely and densely across Caesalpinioideae as a whole, we aim to further resolve which genera are placed in the mimosoid clade.
Several other issues have hindered a more complete understanding of the phylogeny and tribal / generic classification of subfamily Caesalpinioideae. First, the legacy of the traditional subfamily classification meant that taxon sampling in previous phylogenetic studies focused primarily on either old sense Caesalpinioideae (i.e. the grade subtending mimosoids (the ‘Caesalpinieae grade’ of Manzanilla and Bruneau 2012) of new sense Caesalpinioideae (Bruneau et al. 2008; Manzanilla and Bruneau 2012)), or on the mimosoid clade (e.g. Luckow et al. 2003, 2005; Koenen et al. 2020b). Few studies, apart from the family-wide analysis of plastid matK sequences (LPWG 2017), have sampled densely and widely across Caesalpinioideae as a whole. Second, several parts of the Caesalpinioideae phylogeny have been recalcitrant to phylogenetic resolution using traditional DNA sequence loci, most notably along the backbone of the grade subtending the mimosoid clade (Bruneau et al. 2008; Manzanilla and Bruneau 2012; LPWG 2017) and across the large ingoid clade sensu Koenen et al. (2020b). Third, lack of dense pantropical sampling of taxa in previous phylogenies means that the monophyly of several key genera with wide pantropical distributions, such as the ‘dustbin genus’ Albizia, has not been adequately tested and that possible sister-group relationships between New and Old World groups that are relevant to delimitation of genera may have been missed.
More robust foundations to overcome these difficulties were established by Koenen et al. (2020b) in a phylogenomic study of the mimosoid clade. By developing a clade-specific bait set (Mimobaits) for targeted enrichment of 964 nuclear genes, Koenen et al. (2020b) opened the way for generating DNA sequence datasets orders of magnitude larger than those used previously, thereby providing much enhanced phylogenetic resolution. Using these new data, Koenen et al. (2020b) established a new phylogenomic framework and recognised three large informally named higher-level clades each successively nested within Caesalpinioideae (Fig. 1). The mimosoid clade, core mimosoid clade and ingoid clade were all strongly supported by high proportions of gene trees and subtended by long branches. In addition, a set of 15 smaller informally named subclades across mimosoids were proposed by Koenen et al. (2020b) (Fig. 1) to replace the previously defined tribes and informal groups and alliances, almost all of which have been shown by numerous studies to be non-monophyletic (Luckow et al. 2003; LPWG 2013, 2017; Koenen et al. 2020b). Furthermore, although the Mimobaits bait set was designed based on RNA-seq data from species of four mimosoid genera and used initially for the mimosoid clade, the results of Koenen et al. (2020b) suggested that they work well across the non-mimosoid Caesalpinioideae, opening the way to potentially sequence these genes across the subfamily as a whole. The Koenen et al. (2020b) study also further revealed or confirmed the non-monophyly of several genera, but it lacked sufficient taxon sampling to fully test generic monophyly and sampling was largely restricted to the mimosoid clade. Here, we capitalise on these foundations using a slightly modified version of the Mimobaits gene set covering 997 nuclear genes to extend taxon sampling to 420 species from 147 of the 152 genera and establish a robust phylogenomic hypothesis for subfamily Caesalpinioideae as a whole.
This new phylogeny provides the basis for testing the monophyly of genera (the main focus of this paper and of this Special Issue Advances in Legume Systematics (ALS) 14, Part 1), establishing a new higher-level classification of the subfamily (the focus of ALS 14, Part 2) and for downstream analyses of biogeography, trait evolution and diversification (de Faria et al. 2022; Ringelberg et al. 2022). Caesalpinioideae provides an excellent clade for investigating evolutionary diversification and phylogenetic turnover across the lowland tropics (Lavin et al. 2004; Gagnon et al. 2019; Ringelberg et al. 2020, 2022), as well as the evolution of several prominent plant functional traits including compound leaves, armature, extrafloral nectaries and ant associations (Marazzi et al. 2019), agglomeration of pollen into polyads, plant growth forms (Gagnon et al. 2019), floral morphology and pollination syndromes, fruit morphology and seed dispersal syndromes and the ability to form nitrogen-fixing root nodule symbiosis (Sprent et al. 2017; de Faria et al. 2022). However, all of these opportunities require a robust and well-sampled subfamily-wide phylogeny of Caesalpinioideae. In turn, some of these traits have been used for generic delimitation in the past and, in this paper, we also evaluate a handful of such traits in a preliminary way by mapping them on to the phylogeny.
Methods
Phylogeny: taxon and gene sampling, and tree building
To test generic monophyly as thoroughly as possible, we sampled taxa to encompass known or suspected cases of generic non-monophyly, as well as sets of representative species spanning the root nodes of larger genera in Caesalpinioideae (Suppl. material 1). The final phylogenomic dataset comprised 420 Caesalpinioideae taxa covering 147 of the 152 genera. The five missing genera are: Stenodrepanum Harms, the monospecific sister genus of Hoffmannseggia Cav. in the Caesalpinia Group (Gagnon et al. 2016); Hultholia Gagnon & G.P. Lewis, another monospecific genus in the Caesalpinia Group (Gagnon et al. 2016); Microlobius C. Presl, which is also monospecific and nested within the mimosoid genus Stryphnodendron Mart. (Simon et al. 2016; Ribeiro et al. 2018; Lima et al. 2022); Vouacapoua Aubl., a genus of three species, whose phylogenetic placement is uncertain, but most likely falls into the Cassia clade (Bruneau et al. 2008; LPWG 2017); and Pterogyne Tul., another monospecific genus whose placement has been uncertain (Manzanilla and Bruneau 2012; Zhang et al. 2020), but which is probably sister to all Caesalpinioideae, excluding the Arcoa and Umtiza clades (Zhao et al. 2021). In total, 89 of 90 mimosoid genera and 58 of the 62 non-mimosoid Caesalpinioideae genera were sampled.
We sequenced a set of 997 nuclear genes specifically selected for phylogenomic analyses of the mimosoid clade (Koenen et al. 2020b) via targeted enrichment and hybrid capture. This Hybseq approach has quickly become the method of choice to generate phylogenomic data because of its versatility and relatively low cost (e.g. Nicholls et al. 2015; Barrett et al. 2016; Hart et al. 2016; Dodsworth et al. 2019; Johnson et al. 2019; Koenen et al. 2020b). Library preparation, hybrid capture, enrichment and sequencing were performed by Arbor Biosciences (previously MYcroarray; Ann Arbor, USA). Full details about how the new Caesalpinioideae phylogeny was inferred are presented by Ringelberg et al. (2022), but briefly, HybPiper (Johnson et al. 2016) was used to assemble the loci and the pipeline of Yang and Smith (2014) was used for data cleaning and orthology assessment. Various phylogenetic methods, including the multi-species coalescent approach using individual gene trees with ASTRAL (Zhang et al. 2018), Maximum Likelihood based on concatenated alignments with RAxML (Stamatakis 2014) and Bayesian gene jack-knifing with PhyloBayes (Lartillot et al. 2013), were used to infer ten nuclear species trees, which also differ in whether nucleotide or amino acid sequences were used and in the way orthology was assessed (Ringelberg et al. 2022). In addition, a chloroplast phylogeny was inferred using off-target plastid sequences, bringing the total number of phylogenies to eleven. Topological congruence between these eleven different phylogenies was assessed. Support for relationships was expressed in numbers of supporting and conflicting gene trees using PhyParts (Smith et al. 2015) and QuartetScores (Zhou et al. 2020) (Figs 2–12), rather than conventional bootstrap or posterior support values that are known to be inflated in large phylogenomic datasets (Rokas and Carroll 2006; Pease et al. 2018).
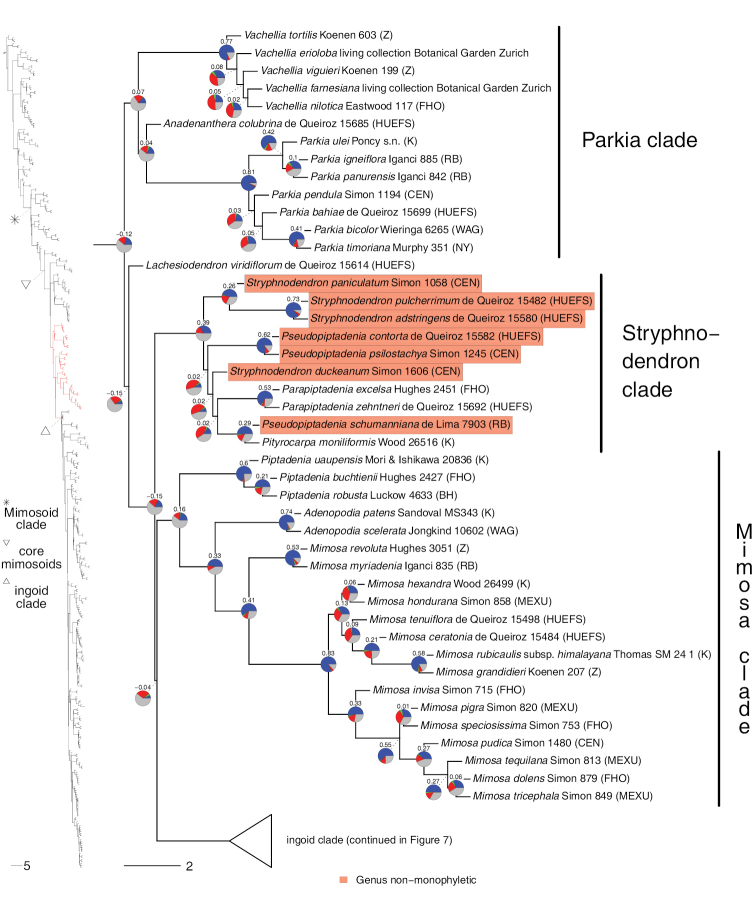
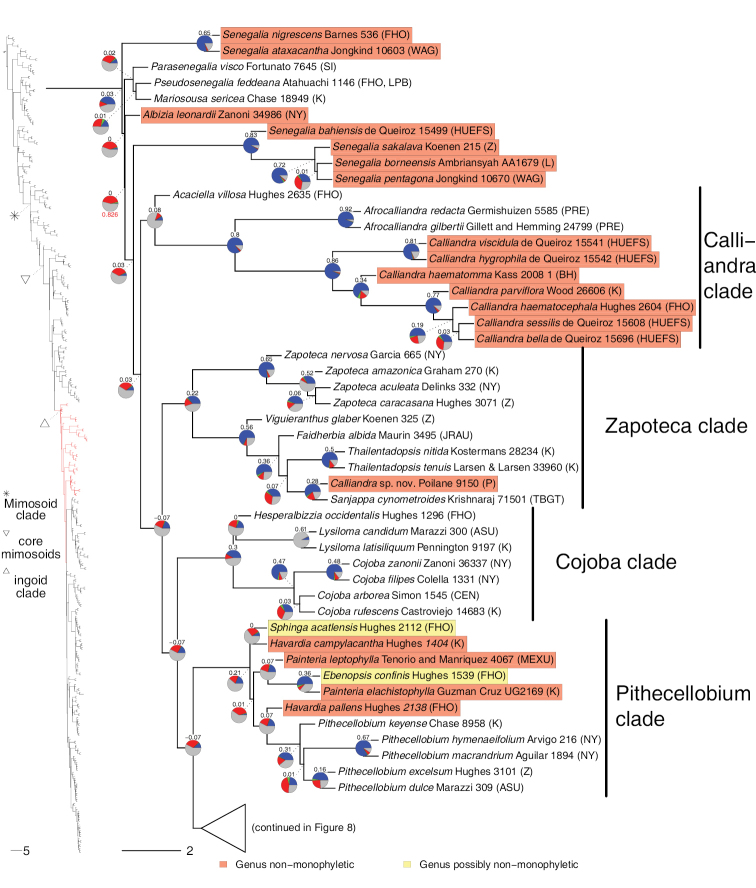
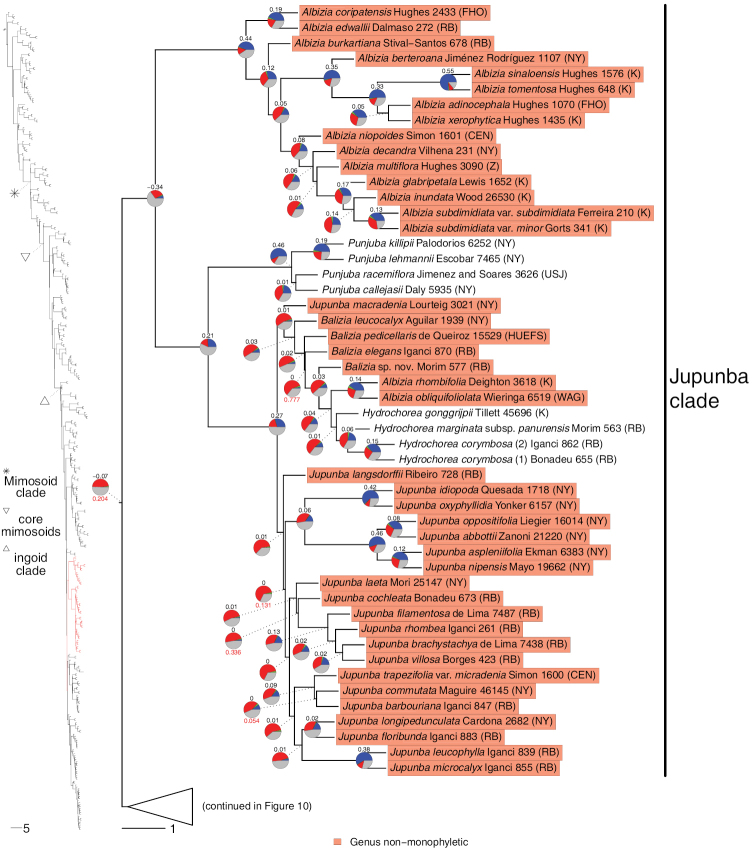
Figure 2.
Phylogeny of Caesalpinioideae, part 1 (continued in Figs 3–12). Left part of figure shows complete Caesalpinioideae phylogeny with highlighted in red the part shown in detail on the right. Depicted phylogeny is the ASTRAL (Zhang et al. 2018) phylogeny based on 821 single-copy nuclear gene trees, with branch lengths expressed in coalescent units and terminal branches assigned an arbitrary uniform length for visual clarity. Genera resolved as (potentially) non-monophyletic are highlighted and clades recognised by Koenen et al. (2020b) are labelled. Support for relationships is based on gene tree conflict: pie charts show the fractions of supporting and conflicting gene trees per node calculated using PhyParts (Smith et al. 2015), with blue representing supporting gene trees, green gene trees supporting the most common alternative topology, red gene trees supporting further alternative topologies and grey gene trees uninformative for this node. Numbers above nodes are Extended Quadripartition Internode Certainty scores calculated with QuartetScores (Zhou et al. 2020). Numbers below nodes are the outcome of ASTRAL’s polytomy test (Sayyari and Mirarab 2018), which tests for each node whether the polytomy null model can be rejected. Only non-significant (i.e. > 0.05) scores are shown, i.e. only for nodes that are better regarded as polytomies according to the test.
Figure 12.
Phylogeny of Caesalpinioideae (continued). See Fig. 2 for caption.
Character evolution
To explore evolution of morphological traits that have been important for generic delimitation, we scored variation in armature, aspects of floral heteromorphy and mode of fruit dehiscence and mapped their distribution across the Caesalpinioideae phylogeny. Our goal was to highlight how an over-reliance on broadly-defined character complexes or functional traits may have misled classification in the past, rather than to perform detailed reconstructions of character evolution through time or to thoroughly assess the homology of various character states.
The three character complexes and their states were defined as follows:
armature (six states): unarmed; nodal or internodal prickles on stem; stipular spines; nodal axillary thorns, including the axillary inflorescence axes which are modified into spines in Chloroleucon (Benth.) Britton & Rose; spinescent shoots.
floral heteromorphy (three states): homomorphic, i.e. with no conspicuous modification or variation amongst flowers within an inflorescence (here we include inflorescences that do not show any conspicuous phenotypic variation beyond the very common occurrence of variable proportions of male and bisexual flowers within inflorescences of many mimosoid genera); heteromorphic 1 = basal flowers of the inflorescence with showy staminodia; heteromorphic 2 = the central flower (or flowers) enlarged/sessile cf. the peripheral (sometimes pedicellate) flowers.
pod dehiscence (six states): indehiscent; inertly dehiscent along one or both sutures; explosively dehiscent, the woody valves twisting and splitting along both sutures along whole length of pod simultaneously; elastically dehiscent from the apex, the valves recurving, but not laterally twisting; craspedium, fruits breaking up into free-falling one-seeded articles leaving a persistent replum or whole valve breaking away intact from replum (valvately dehiscent); lomentiform fruit, the valves readily cracking between the seeds into one-seeded articles, taken here to include crypto-lomentiform fruits.
Data were assembled from taxonomic monographs, revisions and floras. Character evolution was simulated across the phylogeny using the ‘make.simmap’ function in the phytools (Revell 2012) R (R Core Team 2022) package, with 300 independent simulations and a ‘symmetrical rates’ (SYM) model. In each analysis, the character complex of interest (i.e. armature, floral heteromorphy and pod dehiscence) was treated as a single character with multiple states. A rooted phylogeny, without outgroups, was used for the analyses. The root character state was assigned an uninformed prior (i.e. each character state had the same initial probability of occurrence).
Data availability
A tree file of the ASTRAL phylogeny based on the single-copy genes (depicted in Figs 2–12) is included as online Suppl. material 4. In this tree file, all taxon names have been updated to reflect taxonomic changes made in all the entries in Advances in Legume Systematics 14 Part 1.
Results
Phylogenomics
For full results of the sequencing, orthology assembly and phylogenetic inference, see Ringelberg et al. (2022). Here a brief overview is provided.
Hybrid capture and sequencing yielded a large phylogenomic dataset with little missing data: the concatenated nucleotide alignment of the 821 single-copy nuclear genes (a subset of all 997 genes, see below) contains 944,871 sites, 824,713 alignment patterns (i.e. an indication of the phylogenetic informativeness of the alignment, determined by RAxML) and only 11.88% gaps. The ten nuclear species trees that were inferred using different phylogenetic methods are well-supported in terms of gene tree congruence measures (Figs 2–12) and largely congruent with each other. The few topological differences between different phylogenies typically involve only small numbers of species within relatively recent radiations, or deeper putative polytomies such as along the backbone of the ingoid clade, characterised by lack of phylogenetic signal across almost all genes (Koenen et al. 2020b), or the backbone of the Archidendron clade (Fig. 8), characterised by both lack of signal and high conflict amongst gene trees. These minor topological differences do not affect any of the findings of generic non-monophyly discussed below.
The plastid phylogeny (Suppl. material 3) differs more substantially from the nuclear species trees, reflecting the fact that nuclear and chloroplast genomes have unique and sometimes conflicting evolutionary histories (Bruun-Lund et al. 2017; Lee-Yaw et al. 2019; Rose et al. 2021). Cytonuclear discordance affects the monophyly of Senegalia Raf. (Terra et al. 2022), Archidendron F. Muell. (Brown et al. 2022), Dimorphandra Schott, the placement of Desmanthusbalsensis J.L. Contreras (Hughes et al. 2022b) and whether Zygiainundata (Ducke) H.C. Lima ex Barneby & J.W. Grimes and Z.sabatieri Barneby & J.W. Grimes form the sister clade of Inga or a grade subtending Inga.
Hereafter the ASTRAL phylogeny based on the subset of 821 single-copy nuclear gene trees is used as the ‘reference’ Caesalpinioideae backbone phylogeny (Figs 2–12). We use this particular tree over the plastome phylogeny because the nuclear dataset is based on hundreds of independent loci and contains considerably more sites, taxa and fewer gaps, while the plastome phylogeny is based on a single non-recombining locus. The nuclear trees, therefore, likely better represent an approximation of the true evolutionary history of Caesalpinioideae than the phylogeny based on maternally inherited plastid data. Of the various nuclear trees, we select the ASTRAL phylogeny because we find extensive conflict amongst individual gene trees in certain parts of the phylogeny (Figs 2–12), which violates the central assumption of the concatenation model (Jiang et al. 2020) and because the multi-species coalescent model has been shown to consistently outperform the concatenation model on a range of phylogenomic datasets (Jiang et al. 2020). Our analyses reveal that different approaches to orthology assessment have a very minor impact on the final Caesalpinioideae phylogeny, likely because the vast majority of nuclear genes in our dataset are single-copy (i.e. 821 of 997) (see Ringelberg et al. 2022 for details). Nevertheless, how to deal with multi-copy genes is a contentious topic in phylogenetics (Yang and Smith 2014; Moore et al. 2018; Karimi et al. 2019) and we, therefore, focus on the ASTRAL phylogeny based on just the 821 single-copy genes.
The resultant ASTRAL phylogeny is, in general, robustly supported across the majority of nodes using measures of gene tree support and conflict (Figs 2–12). However, there are also some specific parts of the phylogeny which show high levels of gene tree conflict and/or lack of phylogenetic signal across large fractions of genes, which appears to be a feature of most phylogenies based on large phylogenomic datasets (Salichos and Rokas 2013; Wang et al. 2019; Jiang et al. 2020; Koenen et al. 2020a,b; Yang et al. 2020). In most cases, the primary source of gene tree conflict is limited signal in individual gene trees rather than the presence of strongly-supported alternative topologies amongst the gene trees (Figs 2–12, Koenen et al. 2020b), suggesting that the conflict often has methodological rather than biological causes and implying that the presence of conflict per se is no reason for doubts about the recovered Caesalpinioideae topology. However, some parts of the phylogeny with high levels of gene tree conflict or lack of signal may be better viewed as potential polytomies, including the previously identified putative hard polytomy subtending a set of six or seven lineages along the backbone of the ingoid clade (Koenen et al. 2020b) and a putative polytomy across the backbone of the large Archidendron clade (see Appendix 1). These parts of the phylogeny showing high gene tree conflict affect only a few decisions about generic delimitation, most notably across the grade comprising Senegalia and allies (Fig. 7; Terra et al. 2022) and across the backbone of the Archidendron clade (Fig. 8; Brown et al. 2022).
Figure 7.
Phylogeny of Caesalpinioideae (continued). See Fig. 2 for caption.
All the informally named clades of Koenen et al. (2020b; Fig. 1) are here confirmed with robust support in this new phylogeny (Figs 2–12), including the mimosoid clade that is robustly supported and subtended by a long branch (Fig. 4). Our results confirm placement of Chidlowia and Sympetalandra within the mimosoid clade and Dinizia outside the mimosoid clade, with high support (Fig. 4). Higher-level relationships that form the basis for the clade- and tribal-based classification of Caesalpinioideae presented in “Advances in Legume Systematics 14, Part 2”, are not further discussed here.
Figure 4.
Phylogeny of Caesalpinioideae (continued). See Fig. 2 for caption.
Generic non-monophyly
Twenty-two genera were recovered as non-monophyletic or were nested within another genus and, therefore, likely require generic re-delimitation (Figs 2–12; Appendix 1). In addition, based on our results, the taxonomic status of Gagnebina Neck. ex DC., Sphinga Barneby & J.W. Grimes and Ebenopsis Britton & Rose, each represented here by a single taxon and nested in clades with complex generic relationships, require additional species sampling. Furthermore, although Archidendron species form a clade (Fig. 8), the genus is not supported as monophyletic in a substantial fraction of the individual gene trees (Fig. 8), nor in the plastid tree (Suppl. material 3) (see Brown et al. 2022). Overall, our results therefore show that 14(–17)% of the 152 Caesalpinioideae genera require re-delimitation and taxonomic updating. Only two of these genera are non-mimosoid Caesalpinioideae: Dimorphandra Schott and Caesalpinia. Almost all the non-monophyly issues are, therefore, in the mimosoid clade, where 22(–27)% of the 90 genera will require name changes.
Appendix 1 lists all (potentially) non-monophyletic genera with notes and pointers to papers in this Special Issue that discuss these genera and, in many cases, propose nomenclatural changes that resolve many of the non-monophyly issues revealed in our analyses. In some cases, it is clear that formal taxonomic re-circumscription must await more densely-sampled phylogenies and detailed morphological analyses. It is also important to note that, unless explicitly stated otherwise, the reported generic non-monophyly is recovered in all trees (i.e. the nuclear ASTRAL, RAxML and PhyloBayes species trees and chloroplast phylogeny) with high support values expressed and assessed in terms of numbers or fractions of supporting or conflicting genes.
Character evolution
Armature, types of inflorescence heteromorphy and pod dehiscence type each show high levels of homoplasy (Figs 13–15, Table S2) with all types of armature, floral heteromorphy and pod dehiscence hypothesised to have evolved multiple times.
Figure 13.
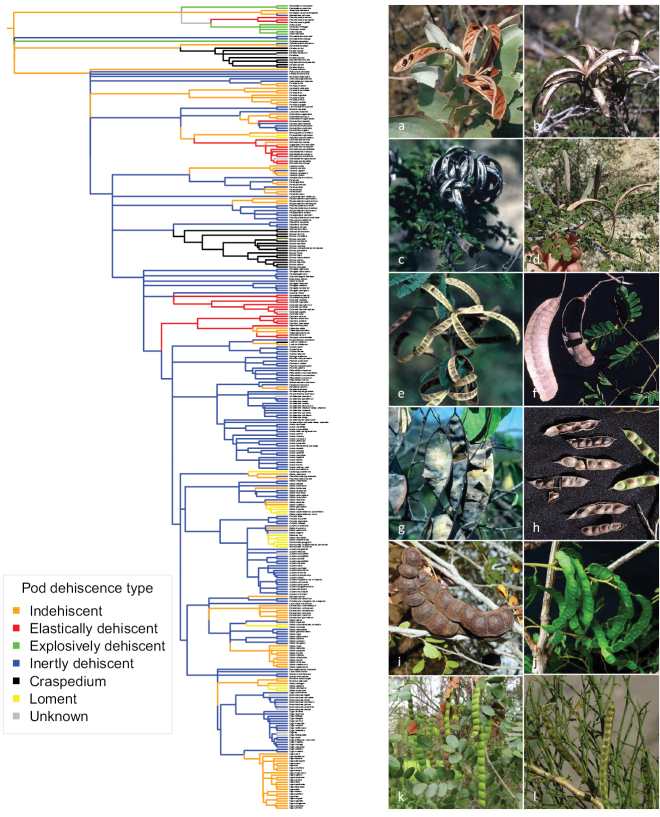
Evolution of fruit dehiscence types across the mimosoid clade. Character states were defined as: indehiscent; inertly dehiscent along one or both sutures; explosively dehiscent, whereby the woody valves twist and split along both sutures along whole length of pod simultaneously; elastically dehiscent from the apex, the valves recurving, but not laterally twisting; craspedium, i.e. fruits breaking up into free-falling one-seeded articles leaving a persistent replum or whole valve breaking away intact from replum (valvately dehiscent); lomentiform fruit, i.e. the valves readily cracking between the seeds into one-seeded articles, taken here to include crypto-lomentiform fruits. Branch lengths are not informative in this figure. Photos a–e elastically dehiscent aAcaciaargyraea Tindale bCalliandraprostrata Benth. cCalliandropsisnervosa (Britton & Rose) H.M. Hern. & P. Guinet dAlantsilodendronmahafalense (R. Vig.) Villiers eZapotecaportoricensis (Jacq.) H.M. Hern f–h craspedium fEntadapolystachya (L.) DC. gLysilomatergeminum Benth. hMimosamontanaKunth.var.sandemanii Barneby i–l lomentiform iAlbiziamoniliformis (DC.) F. Muell. jAlbiziasubdimidiata (Splitg.) Barneby & J.W. Grimes kAlbiziapistaciifolia (Willd.) Barneby & J.W. Grimes lProsopidastrumglobosum (Gillies ex Hook. & Arn.) Burkart. Photos a Bruce Maslin b, c, e–h Colin Hughes dhttp://clubbotatoliara.e-monsite.com/pages/posters-films-rapports/photos.htmli Garry Sankowsky http://www.rainforestmagic.com.auj Marcelo Simon k Xavier Cornejo lhttps://www.floramendocina.com.ar.
Figure 15.
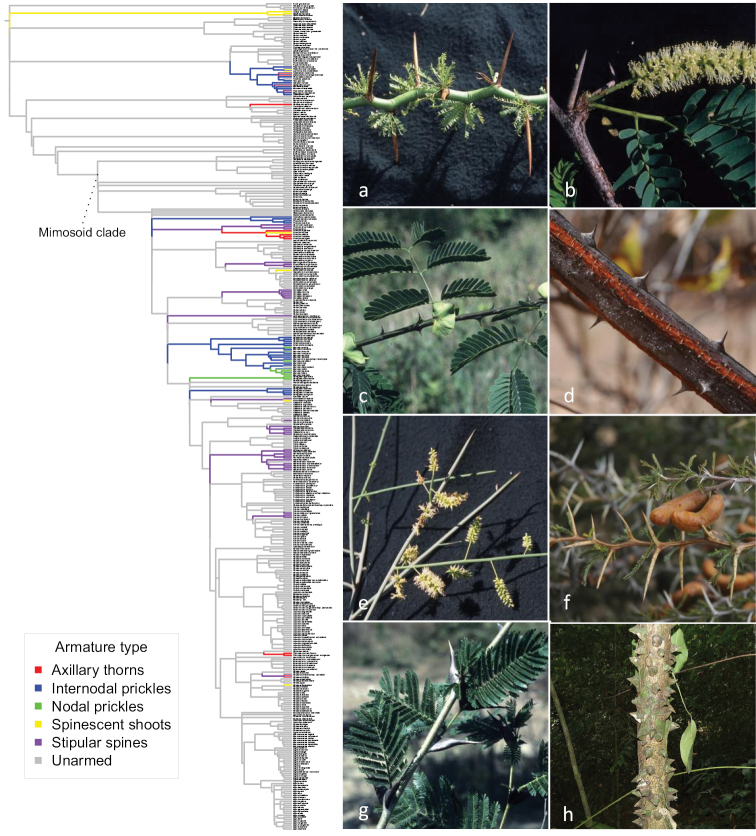
Evolution of different types of armature across Caesalpinioideae. Character states were defined as: unarmed; nodal or internodal prickles on stem; stipular spines; nodal axillary thorns including modified inflorescence axes of Chloroleucon; spinescent shoots. Branch lengths are not informative in this figure. Photos a and b axillary thorns aParkinsoniaandicola (Griseb.) Varjão & Mansano bProsopisjuliflora (Sw.) DC. c, d, h internodal prickles cSenegaliatamarindifolia (L.) Britton & Rose dMimosaophthalmocentra Mart. ex Benth. e spinescent shoots, Prosopiskuntzei Harms f and g stipular spines fProsopisferox Griseb. gVachelliacornigera (L.) Seigler & Ebinger hCylicodiscusgabunensis Harms. All photos Colin Hughes, except h William Hawthorne.
Discussion
Generic non-monophyly
The new Caesalpinioideae phylogeny (Figs 2–12) reveals extensive generic non-monophyly: 22 genera are non-monophyletic or nested within another genus and four other genera could likely also be non-monophyletic (Appendix 1). Notably, there are just two non-monophyletic genera (3% of the 62) across the non-mimosoid Caesalpinioideae, while 20 (to 24) mimosoid genera (i.e. 22(–27)% of 90 genera) are non-monophyletic. The discovery of such a high level of generic non-monophyly in the mimosoid clade is likely attributable to the denser taxon sampling in mimosoids than non-mimosoids in our analyses; the greater species-richness of mimosoids, which account for ca. 75% of the ca. 4,600 Caesalpinioideae species (LPWG 2017), but only 59% of the 152 genera, indicating that, on average, mimosoid genera are more species-rich and, therefore, more likely to have monophyly issues than non-mimosoid Caesalpinioideae genera; the fact that the Caesalpinia Group, the most problematic clade of non-mimosoid Caesalpinioideae in terms of generic delimitation, was already largely resolved by Gagnon et al. (2016), further reducing the likelihood of non-monophyly issues across non-mimosoid Caesalpinioideae; and finally, the continued legacy of Bentham’s broadly circumscribed mimosoid genera which has still not been fully resolved. For example, Acacia, which as indicated earlier, was once a pantropical genus with over 1,400 species (Miller and Seigler 2012) and now comprises seven genera, yet one of these genera, Senegalia, is here recovered as non-monophyletic (Fig. 7) and further subdivision of Senegalia seems likely (Terra et al. 2022). Similarly, Calliandra once had a pantropical distribution until Barneby (1998) restricted it to the New World (de Souza et al. 2013). However, not all Old World Calliandra species have yet been assigned to other genera and Calliandra, therefore, also remains non-monophyletic (Fig. 7). Finally, Albizia, the last mimosoid ‘dustbin genus’ (Barneby and Grimes 1996; Brown 2008; Koenen et al. 2020b) is here confirmed to be non-monophyletic in line with previous findings (Koenen et al. 2020b) (Figs 7–11), but with two previously unsampled Neotropical species each representing additional evolutionary lineages (Terra et al. 2022; Koenen 2022b). Nevertheless, most African, Madagascan and Asian Albizia species do form a single clade (Fig. 10; Koenen et al., unpublished data), while most Neotropical species are also in a single clade (Aviles et al. 2022) (Fig. 9, see Appendix 1).
Figure 11.
Phylogeny of Caesalpinioideae (continued). See Fig. 2 for caption.
Figure 10.
Phylogeny of Caesalpinioideae (continued). See Fig. 2 for caption.
Figure 9.
Phylogeny of Caesalpinioideae (continued). See Fig. 2 for caption.
Morphological homoplasy
Given the extensive re-arrangements of genera in Caesalpinioideae over the last two decades, the question arises why such a significant fraction of genera is still non-monophyletic in these new phylogenomic analyses. We identify two main reasons for this. First, extensive morphological homoplasy has misled generic delimitation and second, lack of pantropical taxonomic synthesis and phylogenetic sampling have resulted in failure to identify clades that span the Old World and New World or, conversely, amphi-Atlantic genera that are non-monophyletic, i.e. potential trans-continental connections and disconnects.
First, and most importantly, the likely extent of homoplasy of morphology and functional traits across Caesalpinioideae is only now starting to be revealed using this new phylogeny (Figs 13–15; de Faria et al. 2022). Here, we reconstructed hypotheses for the evolutionary trajectories of three trait syndromes – armature, mode of fruit dehiscence and aspects of floral heteromorphy – to demonstrate the extent of homoplasy and to show how the repeated evolution of distinctive types of, for example, fruit dehiscence has misled generic delimitation.
Fruits are highly diverse across Caesalpinioideae reflecting adaptations for hydrochory, anemochory, endozoochory, ornithochory, and myrmecochory, as well as several forms of mechanical seed dispersal via explosively, elastically and inertly dehiscent fruits. Here, we show that fruit dehiscence type shows extensive homoplasy across the mimosoid clade, with repeated evolution of, for example, pods elastically dehiscent from the apex, craspedia and lomentiform fruits (Fig. 13). It is now clear that repeated, potentially convergent evolution of fruit types has repeatedly misled generic delimitation and provided the basis for ‘fruit genera’ that have subsequently been shown to be non-monophyletic.
For example, as pointed out by Barneby (1998), the only character uniting Bentham’s (1875) broadly circumscribed pantropical Calliandra was the elastically dehiscent fruit, opening from the apex with the valves recurving, but not laterally twisting (Fig. 13a–e). Just how misplaced this reliance on fruit type as a generic synapomorphy was, is evident from the long parade of new genera segregated from Calliandra, most of them in the two decades after Barneby (1998) restricted the genus to just the New World species: Zapoteca H.M. Hern. (Hernández 1986), Viguieranthus Villiers (Du Puy et al. 2002), Thailentadopsis Kostermans (Lewis and Schrire 2003), Afrocalliandra E.R. Souza & L.P. Queiroz (de Souza et al. 2013) and Sanjappa E.R. Souza & M.V. Krishnaraj (de Souza et al. 2016). This procession is still incomplete given that Calliandra is still non-monophyletic (Fig. 7), pending phylogenetic placement of the Asian Calliandraumbrosa (Wall.) Benth. (see de Souza et al. 2016) and an, as yet, undescribed species (Fig. 7), the last remaining of the species excluded from Calliandra by Barneby (1998) that have not yet been placed in a segregate genus. It is clear that the distinctive ‘Calliandra pod’ has evolved at least six times independently across Caesalpinioideae (Fig. 13) and occurs in at least 12 phylogenetically scattered genera including Jaqueshuberia Ducke, Bussea Harms, Pseudoprosopis Harms, some species of Dichrostachys (DC.) Wight & Arn., Alantsilodendron Villiers, Calliandropsis H.M. Hern. & P. Guinet, Calliandra, Zapoteca, Viguieranthus, Sanjappa, Afrocalliandra and a small subset of species of Acacia. Of course, it is possible that more detailed anatomical investigation of these morphologically and functionally similar fruits will reveal anatomical differences that show that the homology of this fruit type is misplaced, but the structure of the pod valves and raised sutures of most of these are remarkably similar (Fig. 13a–e).
There are several other examples of classifications and especially genera being misled by parallel evolution of fruit types. For example, the polyphyly of the genus Enterolobium Mart. (de Souza et al. 2022a; Figs 10–11) was unexpected because the two clades of Enterolobium species share the distinctive indehiscent thickened and curled ‘ear pod’ fruit type. Similarly, it also seems clear that septate lomentiform fruits with valves readily cracking between the seeds and breaking up into one-seeded articles have also evolved multiple times (Fig. 13), often within genera (e.g. Capuron 1970; Aviles et al. 2022; Koenen 2022a; Soares et al. 2022) associated with hydrochory in species adapted to grow in seasonally inundated habitats and this has impacted on generic delimitation. For example, Barneby and Grimes (1996) separated their newly-segregated genera Balizia and Hydrochorea Barneby & J.W. Grimes on fruit types, yet it is clear that Hydrochorea is nested within a paraphyletic Balizia (Fig. 9; Soares et al. 2022) and that the distinctive lomentiform fruits of Hydrochorea are derived from non-lomentiform indehiscent or follicularly dehiscent pods within this clade (Aviles et al. 2022; Soares et al. 2022). This prevalence of homoplasy associated with fruit types across the mimosoid clade matches that seen across other legume clades (e.g. in subfamily Papilionoideae; Geesink 1984; Hu et al. 2000; Lavin et al. 2001) suggesting that the late developmental stages of the legume pod and associated legume seed dispersal syndromes are prone to convergent evolution, as previously suggested (Geesink 1984; Hu et al. 2000).
Of course, homoplasy per se in no way negates the value and importance of morphology for classification, but instead prompts re-evaluation of homology and the utility of specific morphological characters via reciprocal illumination with new molecular phylogenetic evidence. For example, armature is also homoplasious across Caesalpinioideae with repeated evolution of stipular spines, nodal and internodal prickles, axillary thorns and spinescent shoots (Fig. 15). While armature has been little used as the basis for defining genera because vegetative characters were generally downplayed compared to floral and fruit characters (e.g. Bentham 1875; Burkart 1976), the utility of armature for delimiting some groups within individual clades is increasingly apparent. For example, the four genera segregated from the non-monophyletic Prosopis s.l. by Hughes et al. (2022a) are diagnosed by different types of armature (Fig. 15). Similarly, armature is an important character distinguishing the segregates of Acacia s.l. (spinescent stipules in Vachellia, nodal and internodal prickles in Senegalia, unarmed in Acacia s.s., Parasenegalia, Pseudosenegalia, Mariosousa and Acaciella) and the distribution of prickles (nodal vs. internodal) is discussed in relation to the non-monophyly of Senegalia (Terra et al. 2022). Similarly, the two major clades of genera that make up the Caesalpinia Group (Figs 2 and 15) are separated by differences in armature.
Detailed phylogenetic reconstructions for other characters, based on more rigorous and detailed anatomical assessment of homology, will undoubtedly be worthwhile, but it is already clear that the three traits mapped here (Figs 13–15) are not exceptional in terms of their high levels of homoplasy. Leaves also show evolutionarily labile patterns with numerous repeated transitions from micro- to macrophyllidinous leaves within a large majority of Caesalpinioideae genera. Even the more prominent leaf type innovations of bipinnate vs. pinnate leaves, presence of phyllodes and presence or absence of extrafloral leaf nectaries (EFNs) are all hypothesised to be homoplasious. Multiple reversals to once-pinnate leaves within mimosoids (Inga, Calliandrahymenaeodes (Persoon) Benth., Sanjappacynometroides (Bedd.) E.R. Souza & M.V. Krishnaraj and Cojobarufescens (Benth.) Britton & Rose), multiple origins of phyllodes (in Acacia pro parte, species of Senna including S.phyllodinea (R. Br.) Symon and some varieties of S.artemisoides (Gaudich. ex DC.) Randell and Mimosa species including, for example, M.extranea Benth. and M.phyllodinea Benth. (Barneby 1991)), and multiple losses of EFNs (Marazzi et al. 2019) need to be hypothesised to account for the phylogenetic distributions of these traits. Floral traits show similar extensive homoplasy with multiple derivations of different types of floral heteromorphy (Fig. 14), numerous switches between spikes and capitula and repeated evolution of diverse compound inflorescence conformations (Grimes 1999), homoplasious occurrences of different types of anther glands (Luckow and Grimes 1997) and extremely diverse and evolutionarily labile shapes and sizes of polyads, even within some genera (e.g. Hughes 1997). As indicated above, number of stamens and their connation or not into a staminal tube, the two androecial traits that underpinned the tribal classification of mimosoids first established by Bentham (1875), are also homoplasious across mimosoids such that the tribal classification has not stood the test of time and molecular phylogenetics. Plant functional traits including nodulation (de Faria et al. 2022) and growth forms (Gagnon et al. 2019) also show high levels of homoplasy. Indeed, it appears that nearly all Caesalpinioideae morphological characters and functional traits are homoplasious, given that collectively we, as authors familiar with Caesalpinioideae, have been unable to come up with any morphological characters or functional traits that provide robust synapomorphies subtending larger subclades within Caesalpinioideae, due to either multiple evolutionary origins or repeated independent losses or reversals. Perhaps the one exception to this would be the aquatic habit in Neptunia Lour. spp., which is unique within Caesalpinioideae, although many mimosoids are rheophytes, tolerant of seasonal flooding. This is very much in line with the idea that vegetative, flower and fruit characters may be equally homoplasious, as found in other legume groups such as the dalbergioid clade in Papilionoideae (Lavin et al. 2001).
Figure 14.
Evolution of types of floral heteromorphy across the mimosoid clade. Character states were defined as: homomorphic, i.e. with no conspicuous modification or variation amongst flowers within an inflorescence (here we include inflorescences that can comprise proportions of male and bisexual flowers, but no other more conspicuous variation); heteromorphic 1 = basal flowers of the inflorescence with showy staminodia; heteromorphic 2 = flowers dimorphic within an inflorescence, the central flower (or flowers) enlarged/sessile cf. the peripheral (sometimes pedicellate) flowers. Branch lengths are not informative in this figure. Photos a–h heteromorphic 1 aNeptuniaplena (L.) Benth. bDichrostachyscinerea (L.) Wight & Arn. cDichrostachysmyriophylla Baker dGagnebinapterocarpa (Lam.) Baill. eDichrostachysbernieriana Baill. fDichrostachysakataensis Villiers gParkiabahiae H.C. Hopkins hParkianitida Miq. i–l heteromorphic 2 iPseudosamaneaguachapele (Kunth) Harms jAlbiziaobliquifoliolata De Wild. kHydrochoreacorymbosa (Rich.) Barneby & J.W. Grimes lAlbiziagrandibracteata Taub. Photos a, b, g, i Colin Hughes c, k, l Erik Koenen d Melissa Luckow e, f Dave Du Puy h Giacomo Sellan https://identify.plantnet.org/the-plant-list/observations/1012799991j Jan Wieringa.
Pre-eminence of certain morphological characters over others in classification of a group and the prevalence of ‘organogenera’ (sensu Nielsen 1981) united by just a single character, in situations where morphology is pervasively homoplasious, has been at the root of many of the disagreements about generic delimitation in mimosoids, as pointed out by Guinet (1981).
Trans-continental sampling
A second important reason for the extensive generic non-monophyly is the lack of pantropical synthesis and integration that has been the hallmark of much taxonomic work on Caesalpinioideae up to now and the lack of adequate pantropical sampling of taxa in previous phylogenies. In this light, it is notable that two of the most productive and influential mimosoid taxonomists of the twentieth century, both of whom significantly reshaped the generic classification – Rupert Barneby and Ivan Nielsen – worked largely independently in different geographical areas, especially on genera of the former tribe Ingeae. While both were very much aware of the wider pantropical dimensions and elements of their groups, Barneby focused primarily on New World mimosoids (e.g. Barneby 1991, 1998; Barneby and Grimes 1996, 1997), while Nielsen concentrated on Australasian mimosoids (e.g. Nielsen 1981, 1992) and neither was fully familiar with the details of species of the other (see e.g. Barneby and Grimes 1996), such that no pantropical synthesis across mimosoids was fully achieved and New World – Old World clades that span the Old World and New World or conversely, amphi-Atlantic genera that are non-monophyletic, although hypothesised by both authors, were not resolved.
Our new phylogeny with its near-complete generic sampling reveals several instances of Old World – New World connections and disconnects that have important implications for generic delimitation and which were not fully apparent before. First, the amphi-Atlantic genus Prosopis is shown to be non-monophyletic (Figs 4 and 5), confirming earlier evidence of Catalano et al. (2008). Prosopisafricana (Guill. & Perr.) Taub. forms a monospecific lineage unrelated to the rest of Prosopis, while the remaining three Old World species are sister to the Indo-Nepalese Indopiptadenia Brenan and New World Prosopis has the Namibian-Namaqualand monospecific Xerocladia Harv. nested within it (Fig. 5). It is, therefore, clear that Burkart’s (1976) broad trans-continental concept of Prosopis s.l., which followed Bentham’s (1842, 1875) circumscription, is not sustainable (see Hughes et al. 2022a). A second example of disconnection between Old and New World elements of a pantropical genus is Albizia, where species of New World section Arthrosamanea (Britton & Rose) Barneby & J.W. Grimes form a clade quite separate from Old World Albizia s.s. (Figs 9 and 10; Koenen et al. 2020b: see Aviles et al. 2022). Conversely, two previously poorly understood New World – Old World connections have been revealed. First, it is now clear that the African rainforest species Albiziaobliquifoliolata De Willd. and A.rhombifolia Benth. (previously often referred to the genus Cathormion) are nested within the New World Balizia / Hydrochorea clade (Fig. 9), which is the focus of generic re-delimitation by Soares et al. (2022). Similarly, the recently segregated Neotropical Robrichia (formerly EnterolobiumsectionRobrichia – see de Souza et al. 2022a) is sister to a clade of African mainly rainforest species (Albiziadinklagei (Harms) Harms / A.altissima Hook. f. / A.eriorhachis Harms / A.leptophylla Harms) whose generic placements in Albizia, Cathormion or Samanea (Benth.) Merr. have long been uncertain and neglected (Fig. 11), also prompting further generic re-arrangement in this Special Issue by Koenen (2022a). For the first time, the pantropical sampling employed here is more fully documenting these issues.
Figure 5.
Phylogeny of Caesalpinioideae (continued). See Fig. 2 for caption.
The mimosoid clade
We recover both Chidlowia and Sympetalandra as firmly nested in the mimosoid clade (Fig. 4), confirming previous molecular phylogenetic studies (Chidlowia: Manzanilla and Bruneau 2012; LPWG 2017; Koenen et al. 2020b; Sympetalandra: LPWG 2017). Of the ten genera previously included in the Dimorphandra group (sensu Polhill and Vidal 1981), Sympetalandra, comprising five species (van Steenis 1975; Hou 1996) in the forests of Malaya, Borneo, the Philippine Islands and the Lesser Sunda Islands, is unique in having its stamens shortly joined to the petals and Chidlowia Hoyle (Hoyle 1932) from West Africa (Sierra Leone to Ghana) stands out by having dorsifixed (rather than basifixed) anthers. These two genera are placed between the Xylia and Entada clades of the early-diverging lineages of the mimosoid clade (Fig. 4), outside the core mimosoid clade sensu Koenen et al. (2020b). For Chidlowia, once-pinnate leaves and relatively large flowers with showy red petals which are strongly imbricate in bud are more suggestive of placement outside the mimosoids. For example, Hoyle (1932) suggested an affinity with the detarioid genus Schotia Jacq., but the regular flowers with equally-sized petals, the showy red stamen filaments partly joined at the base (they were described as free in the genus protologue (Hoyle 1932)) and the small campanulate, gamosepalous calyces, support placement in the mimosoid clade. The placement of Sympetalandra in the mimosoid clade, based on molecular analyses, is supported by its racemose or paniculate inflorescences of small, essentially regular, flowers. Finally, the genus Dinizia, which on morphological grounds has sometimes been included in mimosoids in the past (Burkart 1943), is here placed in the grade of genera directly subtending the mimosoid clade, confirming the results of previous molecular phylogenetic studies (Luckow et al. 2005; Bouchenak-Khelladi et al. 2010; Marazzi and Sanderson 2010; Manzanilla and Bruneau 2012; Cardoso et al. 2013; Kyalangalilwa et al. 2013; LPWG 2017; Zhang et al. 2020).
The mimosoid clade, i.e. the subfamily formerly known as the mimosoideae, was traditionally diagnosed by petals valvate, as opposed to imbricate, in bud. Valvate petal aestivation is mostly a reflection of whether or not the flowers are actinomorphic vs. zygomorphic, i.e. as the flowers become radially symmetrical the petals become valvate in bud. Across the non-mimosoid grade of Caesalpinioideae subtending the mimosoid clade, taxa with imbricate and valvate aestivation are phylogenetically intermingled. Although the vast majority of mimosoids do, indeed, have valvate petal aestivation, three exceptions: Chidlowia (as indicated above), alongside Mimozyganthus Burkart and Parkia R.Br., both of which are deeply nested within the mimosoid clade, show imbricate petal aestivation, providing further evidence of the homoplasy of this character. Further work to characterise petal aestivation across all relevant genera of Caesalpinioideae is needed, but it is clear that valvate aestivation does not provide a unique diagnostic synapomorphy for the mimosoid clade.
All other aspects of higher-level relationships are discussed in ALS14 Part 2.
Taxonomy in the age of phylogenomics
Once purely the domain of morphological analyses (e.g. Barneby and Grimes 1996, 1997; Barneby 1998), decisions on delimiting and naming taxa have increasingly been based on genes rather than morphology (Muñoz-Rodríguez et al. 2019). Employing a large phylogenomic dataset and explicitly considering numbers of genes that support particular generic configurations contribute to naming taxa that are more likely to be robust to future sampling of additional species and genomic regions and, hence, to taxonomic stability (Orthia et al. 2005; Pfeil and Crisp 2005; Humphreys and Linder 2009). However, use of ever larger phylogenomic datasets also raises questions about how to delimit taxa and especially about how conflict amongst gene trees reflecting the widely different evolutionary histories of different parts of the genome (e.g. Salichos and Rokas 2013; Wang et al. 2019; Jiang et al. 2020; Koenen et al. 2020a, b) should inform delimitation of taxa. For example, what fraction of genes supporting a clade should be used as a cut-off for delimiting taxa? To what extent does it matter if there are alternative topologies that are supported by a substantial fraction of genes, even if that number is lower than the number of genes that supports the ‘main’ topology and what are the classificatory implications when only a small fraction of genes is informative for certain relationships (Shen et al. 2017)? Employing large numbers of genes is also enhancing our ability to identify putative hard polytomies on nodes where all, or almost all, genes lack phylogenetic signal (e.g. Koenen et al. 2020b), raising questions about whether it is justified to delimit multiple segregate genera when the relationships amongst them are unresolved and potentially form a polytomy. Large phylogenomic datasets also highlight cases of cytonuclear discordance even more starkly than before, raising questions about what is the best approach when different genomes (i.e. nuclear, plastid and mitochondrial) have different evolutionary histories, as is often the case (e.g. Bruun-Lund et al. 2017; Thielsch et al. 2017; Lee-Yaw et al. 2019; Rose et al. 2021; Debray et al. 2022)? Finally, we might also ask what, fundamentally, is now the role of morphology in delimiting taxa in the phylogenomic era (Muñoz-Rodríguez et al. 2019)?
The phylogeny of Caesalpinioideae presented here (Figs 2–12) poses many of these questions and provides some possible answers. First, the ubiquity of gene tree conflict found here and more generally in phylogenomics (Salichos and Rokas 2013; Wang et al. 2019; Jiang et al. 2020; Koenen et al. 2020b; Yang et al. 2020), suggests that the presence of conflicting topologies for a particular node alone is not sufficient reason to avoid naming the clade subtended by that node. If many conflicting topologies exist, but none of these occurs at a high frequency amongst the gene trees, low support values are indicative of lack of signal rather than true conflict (Koenen et al. 2020b) and do not need to affect classificatory decisions if there is support for the species tree topology amongst a sizable fraction of the gene trees. The nodes subtending Macrosamanea Britton & Rose, Zygia and Inga (Figs 11 and 12) are good examples of an abundance of conflicting topologies none of which is widespread and the monophyly of these genera is, therefore, not in question (except for a few outlier species of Zygia – see Appendix 1). However, if low support for a node in the species tree is caused by an alternative topology that is common across gene trees, the situation is more complex and the clade in question should probably not be named pending further study with additional accessions and genomic regions. The crown node of Archidendron (Fig. 8) provides an example of a node with a relatively abundant alternative topology, raising doubts about the monophyly of Archidendron (see Appendix 1; Brown et al. 2022). Second, in cases of cytonuclear discordance (as we see across several key nodes that affect decisions about generic delimitation), the smaller size of the plastid dataset and the fact that the chloroplast genome can be considered as a single, albeit large, uniparentally-inherited locus, suggest that, in most cases, nuclear phylogenies provide a more accurate approximation of the true species tree (see Terra et al. 2022).
Finally, despite providing the main (usually sole) source of information for classification for centuries, morphology was rapidly eclipsed as a source of data for phylogeny reconstruction with the advent of molecular data (e.g. Scotland et al. 2003). Nevertheless, despite the dominance of phylogenomic data for building accurate and robust trees, morphology continues to play a central role as a complementary source of evidence for delimiting taxa in the light of monophyly inferred from phylogenomic data (Humphreys and Linder 2009; Gagnon et al. 2016). For example, placement of Zygiasabatieri and Z.inundata not in a clade with the remainder of Zygia, but instead as the sister clade of Inga in the nuclear ASTRAL phylogeny (Fig. 12) or in a grade subtending Inga in the plastome phylogeny (Suppl. material 3; Ferm et al. 2019), presents several options for delimiting genera: transfer these two species to the genus Inga, place both species in a new segregate genus or place each species in separate segregate genera. All three options are valid from the perspective of monophyly, but not from a morphological standpoint, because Z.sabatieri and Z.inundata have dehiscent pods and Z.sabatieri has bipinnate leaves, in contrast to the once-pinnate leaves and indehiscent pods that are diagnostic of the genus Inga. From a morphological perspective, it will be preferable to assign Z.inundata and Z.sabatieri to a new segregate genus rather than to transfer them to Inga, thereby retaining the morphological integrity and diagnosability of the genus Inga (see Appendix 1). This example demonstrates the important role that morphology continues to play in the era of phylogenomics: not to determine relationships and infer monophyly, but to inform and guide decisions about how to partition a phylogeny into monophyletic taxa (see also Terra et al. 2022 for another example).
Conclusions and future work
Here, we present a series of phylogenomic analyses including detailed assessment of gene tree conflict and support that suggest that about one quarter of mimosoid genera are non-monophyletic (Figs 2–12). This new backbone phylogeny, building on the 122-taxon version of Koenen et al. (2020b), provides robust foundations for aligning genera with monophyletic groups across a clade where generic delimitation has long been contentious with starkly contrasting generic systems (Lewis et al. 2005; Brown 2008) and for the higher-level classification presented in Advances in Legume Systematics 14, Part 2. The limitations of previous work focused either just on the Old World (e.g. Nielsen 1981, 1992) or just on the New World (e.g. Barneby and Grimes 1996, 1997; Barneby 1998) have become more starkly apparent now that pantropical sampling has been achieved, revealing the non-monophyly of well-known pantropical genera, such as Albizia (Koenen et al. 2020b; Aviles et al. 2022) and Prosopis (Hughes et al. 2022a), as well as previously unrecognised clades with trans-Atlantic distributions (Soares et al. 2022; Koenen 2022a). Our analyses provide a glimpse of the likely extent of morphological homoplasy (Figs 13–15).
However, despite including 420 taxa in the current analyses, it is clear that additional taxon sampling will be needed to fully resolve all the possible non-monophyly issues within Caesalpinioideae. Several priorities for future research are apparent. First, denser taxon sampling across Senegalia and allies is needed to address the unusual dilemmas posed by extreme lack of resolution and cytonuclear discordance surrounding delimitation of the genera across the paraphyletic grade comprising Senegalia, Pseudosenegalia, Parasenegalia and Mariosousa (Fig. 7) that are explored here by Terra et al. (2022) who provided a list of priority taxa for future sampling with molecular data. Second, the likely non-monophyly of Archidendron (see Brown et al. 2022 and Appendix 1) also remains unresolved with a clear need for additional work, especially as many species are known from incomplete material. Archidendron and Senegalia are now the largest genera in Caesalpinioideae where doubts remain about their monophyly and delimitation. Third, a much more comprehensively sampled study is needed to address the longstanding non-monophyly of Dimorphandra Schott (Fig. 3). Fourth, the generic affinities of Calliandraumbrosa (Fig. 7; de Souza et al. 2016) and Calliandra sp. nov., the last species removed from Calliandra by Barneby (1998) yet to be placed in another genus, remain to be assessed. Finally, the taxonomic implications of the non-monophyly of Zygia revealed by Ferm et al. (2019) and confirmed here (Figs 11 and 12) have not yet been addressed. Like Archidendron, many species of Zygia remain poorly understood.
Figure 3.
Phylogeny of Caesalpinioideae (continued). See Figure 2 for caption.
Furthermore, although there is no evidence that any large clades in Caesalpinioideae are subtended by whole genome duplication (WGD) events (Koenen et al. 2020a), it is clear that polyploidisation events have happened many times more recently, scattered across the phylogeny of Caesalpinioideae, for example in Leucaena (Govindarajulu et al. 2011; Bailey et al., in prep.), Vachellia and Mimosa (Dahmer et al. 2011; Simon et al. 2011). Furthermore, high numbers of gene duplications detected on branches subtending, for example, Sympetalandra, Lemurodendron Villiers & P. Guinet and Schleinitzia Warb. point to possible additional WGDs (Ringelberg et al., unpublished data). More work is needed to understand all these possible polyploidisation events, whether they involved auto- or allopolyploidisation and how such events affect assessments of character evolution, homoplasy and generic delimitation.
Finally, our preliminary assessments of homoplasy (Figs 13–15) notwithstanding, there is a clear need for rigorous analysis and comparison of morphological traits across the subfamily, based on more detailed homology assessment of morphological, developmental and genomic data. Morphological diagnosability of taxa is centrally important, especially for the acceptance of novel taxonomy by the end-users of scientific names, a group that is much larger than that of the scientific taxonomic community. We hope that the new phylogeny presented here can provide the evolutionary framework for future morphological studies that assess character evolution and homoplasy in greater detail.
Acknowledgements
The authors thank B. Adhikari, D. Lorence, B. Marazzi, É. de Souza, the G, K, JRAU, L, MEL, NY, FHO, P, RB, WAG and Z Herbaria, the Millennium Seed Bank, Kew, the South African National Botanical Institute, the Direction de Environment, New Caledonia and the National Tropical Botanical Garden, Hawaii, U.S.A. for provision of leaf or DNA samples; P. Ribeiro, É. de Souza, M. Morim, M. Simon and F. Bonadeu in Brazil and staff of the Kew Madagascar Conservation Centre and Botanical and Zoological Garden of Tsimbazaza, Madagascar for fieldwork support; the national regulatory authorities of Brazil for access to DNA of Brazilian plants (authorised through SISGEN n° A464716), and Madagascar (research permit 006/14/MEF/SG/DGF/DCB.SAP/SCB); U. Grossniklaus, V. Gagliardini, Dept Plant & Microbial Biology, Univ. Zurich for use of their TapeStation; the S3IT, Univ. Zurich for use of the ScienceCloud computational infrastructure; and the Club Botanique de Toliara, Madagascar, the Flora Mendocina project, Argentina, B. Maslin, G. Sankowsky, M. Simon, and X. Cornejo for permission to use photos included in Figs 13–15. This work was supported by the Swiss National Science Foundation through grants 310003A_156140 and 31003A_182453/1 to CEH and an Early.Postdoc.Mobility fellowship P2ZHP3_199693 to EJMK, the Claraz Schenkung Foundation, Switzerland to CEH, the Natural Sciences and Engineering Research Council of Canada, NSERC, to AB, FAPESB, Brazil (PTX0004 & APP0096 to LPdQ) and CNPq, Brazil (480530/2012-2 & 311847/2021-8 to JRI and PROTAX 440487 to LPdQ).
Appendix 1
Generic non-monophyly in Caesalpinioideae – towards a new generic system for the subfamily
Caesalpinia
Divergent circumscriptions of the genus Caesalpinia L. were largely resolved by Gagnon et al. (2016) who reduced Caesalpinia to ca. nine species and established a new generic system for the Caesalpinia Group as a whole, with 26 genera plus their ‘Ticanto clade’ (Caesalpiniacrista L. and allies) as a putative 27th genus. This 27th genus accounts for the non-monophyly of Caesalpinia in our analysis (Fig. 2) with Caesalpiniacrista representing the Ticanto clade that is re-instated as a genus in this Special Issue by Clark et al. (2022).
Dimorphandra
In line with previous studies (Luckow et al. 2005; LPWG 2017), Dimorphandra Schott is non-monophyletic in the nuclear phylogeny (Fig. 3), but robustly supported (99% bootstrap support (BS)) as monophyletic in the plastid tree (Suppl. material 3), indicating cytonuclear discordance. This implies either splitting Dimorphandra into two genera or sinking Mora Schomb. ex Benth., Stachyothyrsus Harms and Burkea Benth. into Dimorphandra (which predates these other three genera). Evidence suggests splitting Dimorphandra as the preferred option. First, the three Dimorphandra species sampled here represent the three morphologically delimited subgenera (da Silva 1986) with representatives of these subgenera intermingled with other genera rendering Dimorphandra polyphyletic in the legume-wide matK phylogeny (LPWG 2017) and Burkea and Mora are not closely related to Dimorphandra in the plastid phylogeny (Suppl. material 3; Stachyothyrsus is not included in the plastid analysis). Second, while Mora has been included in Dimorphandra based on morphological similarities (Sandwith 1932; van Steenis 1975), the two genera differ in floral, seed and pod morphology and have generally been treated as distinct (Sandwith 1932; van Steenis 1975; da Silva 1986). African Stachyothyrsus and Burkea are morphologically (van Steenis 1975) and geographically distinct from South American Dimorphandra and Mora. All of this suggests that Dimorphandra will need to be split into two genera or potentially three, although the robustly supported sister group relationship between D.davisii and D.macrostachya (internode certainty 0.77, subtended by a long branch) would perhaps favour two genera, rather than three. Additional taxon sampling, to test the monophyly of the three subgenera, is required before taxonomic re-arrangements can be made. If the genus is to be split, the name Dimorphandrawould remain attached tosubgenusDimorphandra, here represented by D.gardneriana Tul. Dimorphandraexaltata Schott is the type species of the genus. The names of the other two subgenera, Phaneropsia Tulasne and Pocillum Tulasne, would be available for the remaining species. Both names originate from the same publication (Tulasne 1844), but since Pocillum also refers to a genus of fungi (Kirk et al. 2008), Phaneropsia would be the more suitable generic name for the species not in Dimorphandra s.s. However, as taxon names have no priority at different rank (Turland et al. 2018), a new generic name may also be proposed.
Xylia and Calpocalyx
The non-monophyly of Xylia with Calpocalyx nested within it was documented using matK sequences (LPWG 2017) and is confirmed here (Fig. 4). This does not come as a great surprise, as these genera have always been considered closely related (Villiers 1984; Lewis et al. 2005). They have overlapping geographical and ecological distributions mainly in the tropical rainforests of central and western Africa (although Xylia has a wider distribution in Africa, Madagascar and Asia). The two genera also share a suite of morphological characteristics (Villiers 1984; Luckow et al. 2003), including robust woody sickle-shaped explosively dehiscent fruits (Fig. 13), a chromosome count of 2n = 12 (Goldblatt and Davidse 1977) and pollen grains in small-sized polyads (Jumah 1991). Since the name Xylia (Bentham 1841) predates Calpocalyx (Engler and Prantl 1897) and given the morphological and ecological similarities of the two genera, the most straightforward solution to the non-monophyly presented here would be the transfer of the species of Calpocalyx to Xylia. However, this apparently straightforward incorporation of Calpocalyx into Xylia is complicated by the name Esclerona Raf., an apparently valid name predating Xylia, raising the possibility of proposing conservation of the name Xylia prior to merging these two genera.
Entada and Elephantorrhiza
A close relationship between Entada Adans. and Elephantorrhiza Benth. has long been suggested in all molecular phylogenies that sampled these genera (e.g. Luckow et al. 2003; Koenen et al. 2020b). With denser sampling of species, it has become clear that Elephantorrhiza is nested within Entada (LPWG 2017), a result that is confirmed here (Fig. 4) and which provides the basis for re-circumscription of Entada to include Elephantorrhiza by O’Donnell et al. (2022) in this Special Issue.
Prosopis
One of the most striking and robustly supported examples of generic non-monophyly in our analyses is Prosopis s.l. whose species are placed in four separate lineages (Figs 4 and 5). The nodes supporting this non-monophyly are some of the most robustly supported across the Caesalpinioideae phylogeny as a whole (Fig. 5). This shows that P.africana is not closely related to the rest of Prosopis s.l., but is placed in a grade with other monospecific or species-poor genera subtending the core mimosoid clade (Fig. 4), confirming results from earlier studies (Catalano et al. 2008; LPWG 2017; Koenen et al. 2020b). The rest of Old World Prosopis (three species) is sister to the Indo-Nepalese genus Indopiptadenia and New World Prosopis has the Namibian – S. African Xerocladia nested within it (Fig. 5). A new generic classification of Prosopis s.l., accounting for this non-monophyly, is presented in this Special Issue by Hughes et al. (2022a).
Desmanthus
The non-monophyly of Desmanthus with the monospecific Hawaiian endemic Kanaloa Lorence & K.R. Wood nested within it (Fig. 5) mirrors earlier phylogenies (Hughes et al. 2003; Luckow et al. 2003, 2005) and is in line with the morphological distinctiveness of Desmanthusbalsensis J.L. Contreras from the remaining species of Desmanthus (Contreras Jiménez 1986; Luckow 1993). A new monospecific segregate genus to account for this non-monophyly is proposed in this Special Issue by Hughes et al. (2022b).
Dichrostachys , Gagnebina and Alantsilodendron
Dichrostachys (DC.) Wight & Arn. and Alantsilodendron Villiers are both recovered as non-monophyletic in our sparsely sampled analysis (Fig. 5), raising questions about the monophyly of Gagnebina Neck. ex DC., here represented by just a single species. The Malagasy members of these three genera (all species in our phylogeny, except D.cinerea R. Vig.) cluster together in a clade characterised by very short branches and extensive gene tree conflict (Fig. 5) suggestive of an early burst model of diversification typical of a rapid radiation on Madagascar (Aebli 2015). Previous molecular phylogenetic studies have also found at least some of these genera to be non-monophyletic (Hughes et al. 2003; Luckow et al. 2003, 2005; Aebli 2015) and some species have been transferred between genera based on morphology (Lewis and Guinet 1986). Each of these genera contains several other species from Madagascar not sampled here. While a parsimonious solution could be to merge the three genera into Gagnebina (de Candolle 1825) (a name predating Dichrostachys (Wight and Walker-Arnott 1834) and Alantsilodendron (Villiers 1994)), such a move would result in a highly variable genus, with no consistent morphological character to distinguish it. A forthcoming monograph (Luckow, unpublished data) will resolve the non-monophyly of these genera by transferring two species of Dichrostachys to Alantsilodendron and seven to a new genus (Phillipson et al. 2022). Additional sampling of non-Malagasy species of Dichrostachys would also be important, especially Australian D.spicata, as it has been placed as sister to the combined Dichrostachys / Gagnebina / Alantsilodendron + Calliandropsisnervosa (Britton & Rose) H.M. Hern. & Guinet clade in several studies (Hughes et al. 2003; Luckow et al. 2003, 2005; Aebli 2015). The African species D.dehiscens Balf. f. and D.kirkii Benth. also need to be sampled as they share a dehiscent fruit type with members of the new Madagascan genus.
Stryphnodendron and Pseudopiptadenia
Our analyses support the monophyly of the Stryphnodendron clade sensu Koenen et al. (2020b) comprising the genera Parapiptadenia Brenan, Pityrocarpa (Benth. & Hook.f.) Britton & Rose, Pseudopiptadenia Rauschert and Stryphnodendron Mart. (Fig. 6) and presumably Microlobius C. Presl., which, although not sampled here, has been shown to be nested within or sister to Stryphnodendron (Ribeiro et al. 2018; Simon et al. 2016; see also Lima et al. 2022). Of these genera, only Parapiptadenia is monophyletic in our analyses, although Pityrocarpa is here only represented by a single taxon (Fig. 6). Stryphnodendron is non-monophyletic as S.duckeanum Occhioni does not group with the rest of the genus (Fig. 6), in line with flower, fruit and branching characteristics that suggested transfer of S.duckeanum to another genus (Scalon 2007) and with previous molecular phylogenies showing S.duckeanum separated from the rest of Stryphnodendron (Jobson and Luckow 2007; Simon et al. 2016; Ribeiro et al. 2018; Sauter 2019). Similarly, Pseudopiptadenia is also non-monophyletic with P.schumanniana placed as sister to the single sampled species of Pityrocarpa, rather than forming a clade with Pseudopiptadeniacontorta (DC.) G.P. Lewis & M.P. Lima and P.psilostachya (DC.) G.P. Lewis & M.P. Lima (Fig. 6). Several previous molecular phylogenies also found Pseudopiptadenia to be non-monophyletic – however, those studies did not include P.schumanniana and found P.brenanii G.P. Lewis & M.P. Lima (not sampled here) to be the outlier instead (Simon et al. 2016; Ribeiro et al. 2018). The sparsely sampled backbone phylogeny of the Stryphnodendron clade presented here provides the foundations for more densely sampled analyses and re-delimitation of both Stryphnodendron (Lima et al. 2022) and Pseudopiptadenia / Pityrocarpa (Borges et al. 2022) in this Special Issue.
The remaining genera in the Stryphnodendron and Mimosa clades are all monophyletic (Fig. 6), confirming previous phylogenetic studies and taxonomic rearrangements, including segregation of Lachesiodendron P.G. Ribeiro, L.P. Queiroz & Luckow from Piptadenia (Ribeiro et al. 2018), as well as placement of amphi-Atlantic Adenopodia C. Presl as sister to Mimosa and the sister group relationships amongst the main clades of Mimosa (Simon et al. 2011).
Senegalia and allied genera
The striking cytonuclear discordance whereby Senegalia Raf. appears as non-monophyletic in the analyses of nuclear gene sequences, but as monophyletic in the analyses of plastomes, was first revealed by Koenen et al. (2020b), a result confirmed here by sampling more species of Senegalia, plus the closely related Mariosousa, Parasenegalia and Pseudosenegalia (Fig. 7). In the nuclear gene analyses, the two clades of Senegalia plus these three other genera and the incompletely known Albizialeonardii Britton & Rose ex Barneby & J.W. Grimes form a paraphyletic grade with very short and poorly supported or unsupported internal branches (Fig. 7). The complex and intriguing issues these features raise for delimitation of Senegalia are explored by Terra et al. (2022), who conclude that sequencing of more species is required.
Calliandra
Following reduction of Bentham’s (1875) broad trans-continental circumscription of Calliandra Benth. to just the New World species by Barneby (1998), five genera have been segregated to account for the majority of the Old World species. Now just a handful of Old World species remain to be resolved, including the Asian Calliandra sp. nov. (Poilane 9150), that, as expected, does not group together with the New World Calliandra s.s., but is instead sister to the Indian monospecific genus Sanjappa É.R. Souza & M.V. Krishnaraj in the Zapoteca clade (Fig. 7). Bentham (1875) included four Asian species in Calliandra (de Souza et al. 2013), which share the apically dehiscent pods of Calliandra (Fig. 13a–f), but in other respects present anomalies, especially in the configuration of their polyads. The identities of these Asian Calliandra species have long been considered ambiguous (Barneby 1998). Two of these Asian species have been assigned to different genera (C.cynometroides Bedd. to Sanjappa (de Souza et al. 2016) and C.geminata (Wight & Arn.) Benth. to Thailentadopsis Kosterm. (Lewis and Schrire 2003)), while the generic placement of the remaining species, C.umbrosa (Wall.) Benth., remains unknown. The fourth species, C.griffithii Baker ex Benth., is now considered a subspecies of C.umbrosa (Paul 1979). Calliandraumbrosa has never been included in a molecular phylogenetic analysis (de Souza et al. 2013, 2016) and, unfortunately, sequencing of C.umbrosa was unsuccessful in this study. However, polyad, leaf, corolla and pod morphology, plus the presence of facultatively spinescent stipules, distinguish C.umbrosa from other genera, suggesting that it should potentially be assigned to a new genus (de Souza et al. 2013, 2016). Until DNA sequences of C.umbrosa can be obtained to ascertain its relationship to Calliandra sp. nov., this residual non-monophyly of the genus Calliandra cannot be resolved.
Pithecellobium and allies
While the Pithecellobium alliance is the only one of the informal alliances of Barneby and Grimes (1996) whose monophyly has withstood the test of phylogenomic analysis (Koenen et al. 2020b), other than Pithecellobium Mart. itself, our sparsely sampled phylogeny of this clade suggests that the monophyly of the four other genera placed in the Pithecellobium clade (Painteria Britton & Rose, Havardia Small, Ebenopsis Britton & Rose and Sphinga Barneby & J.W. Grimes) is doubtful and needs to be further tested with more complete taxon sampling (Fig. 7). Even with our limited taxon sampling, Painteria and Havardia are clearly non-monophyletic (Fig. 7), raising significant doubts about the taxonomic status of Ebenopsis and Sphinga, which are both represented by only one species in our trees. Painteria is especially poorly distinguished from Havardia; Sphinga was originally described in Havardia and previous studies (Nielsen 1981; Polhill 1994) placed all four genera in a more broadly defined Havardia (Brown 2008). Such a solution might, therefore, seem sensible, but together they form a paraphyletic grade in our phylogenies (Fig. 7), suggesting that unless all four genera were to be sunk back into Pithecellobium (from which they were segregated (Barneby and Grimes 1996)), these four genera require at least three names, as they are divided over three (poorly-supported) lineages: one comprising Spingaacatlensis (Benth.) Barneby & J.W. Grimes and Havardiacampylacantha (L. Rico & M. Sousa) Barneby & J.W. Grimes, one Painterialeptophylla (DC.) Britton & Rose, Pa.elachistophylla (A. Gray ex S. Watson) Britton & Rose and Ebenopsisconfinis (Standl.) Britton & Rose and one H.pallens (Benth.) Britton & Rose, which is the type species of Havardia and sister to Pithecellobium. Clearly, taxon sampling in our phylogeny is too limited to draw firm taxonomic conclusions. A new phylogeny of the Pithecellobium clade, presented here in this Special Issue, is used as the basis for erecting two new genera to account for these generic non-monophyly issues (Tamayo-Cen et al. 2022). This new phylogeny, based on a small set of DNA sequence loci, but with denser taxon sampling than that encompassed here, is not fully congruent with the phylogenomic backbone presented in Fig. 7.
The Archidendron clade
The genera and lineages of the large Archidendron clade comprising Acacia Mill., Archidendron F. Muell. and six smaller genera (Fig. 8; Koenen et al. 2020b), together make up over one third of all mimosoid species and are restricted to Australasia. Relationships across the backbone of this clade are complex and generally poorly resolved with very short branches and high levels of gene tree conflict and lack of phylogenetic signal across a significant fraction of genes (Fig. 8), such that the topologies across different analytical approaches can differ. This suggests that some nodes across this backbone should better be viewed as putative polytomies. Three genera in this clade, Wallaceodendron Koord., Pararchidendron I.C. Nielsen and Paraserianthes I.C. Nielsen, are monospecific. Falcataria (I.C. Nielsen) Barneby & J.W. Grimes comprises three species but is represented by only one taxon in our phylogeny, so no conclusion can, therefore, be made about its monophyly, although our results support the segregation of this genus from Paraserianthes (Barneby and Grimes 1996; Brown et al. 2011). Three of the four remaining genera are monophyletic: Acacia, Archidendron and Serianthes Benth. (confirming the results of Demeulenaere et al. (2022) in this Special Issue). However, the monophyly of Archidendron remains doubtful as it is supported by few gene trees and opposed by many (Fig. 8) and the genus is not monophyletic in the plastid tree (Suppl. material 3). This is very much in line with previous findings of a non-monophyletic Archidendron (Brown et al. 2008, 2011; Iganci et al. 2016; LPWG 2017). The likely non-monophyly of Archidendron is explored in more detail in this Special Issue by Brown et al. (2022). It is notable that the two well-supported Archidendron subclades found here are replicated by Brown et al. (2022), where their morphological and geographical identities are discussed in detail. Finally, the non-monophyly of Archidendropsis I.C. Nielsen, documented and addressed in this Special Issue by Brown et al. (2022), is confirmed by the much larger phylogenomic dataset analysed here (Fig. 8).
Our results weakly support Paraseriantheslophantha as sister to Acacia (Fig. 8), in line with earlier findings (Brown et al. 2008, 2011; Koenen et al. 2020b) and shared morphological similarities including hard seeds that are stimulated to germinate by fire (Brown et al. 2011), minute anthers and numerous stamens (Barneby and Grimes 1996). As P.lophantha contains two geographically disjunct subspecies, P.lophanthasubsp.montana (Jungh.) I.C. Nielsen in Indonesia and P.lophanthasubsp.lophantha (the subspecies sequenced here) in southern Australia (Brown et al. 2011), sequencing the missing subspecies would be worthwhile to check that the two cluster together as sister to Acacia. However, it is important to note that this relationship is sensitive to the type of dataset and phylogenetic method: the ASTRAL trees (Fig. 8) recover P.lophantha as the sister of Acacia, whereas the nuclear RAxML phylogenies (Ringelberg et al. 2022) find a sister relationship between Acacia and Archidendron plus Archidendropsisxanthoxylon (C.T. White & W.D. Francis) I.C. Nielsen, the PhyloBayes gene jack-knifing phylogeny (Ringelberg et al. 2022) resolves the whole Archidendron clade as one large polytomy lacking a clear sister lineage to Acacia and the plastid tree (Suppl. material 3) recovers Archidendropsisxanthoxylon as sole sister of Acacia. Furthermore, P.lophantha and several species of Archidendron are also identified as species often changing positions across trees by RogueNarok (Aberer et al. 2013). The high levels of intergenic conflict, very short branches, extremely low bootstrap support values especially in the nucleotide RAxML phylogenies, lack of concordance and signal amongst the gene trees and failure to reject a polytomy by ASTRAL (Fig. 8), all suggest that the backbone of the Archidendron clade should perhaps best be viewed as one large polytomy, as depicted in the PhyloBayes consensus tree (Ringelberg et al. 2022). However, the number (eight in the PhyloBayes phylogeny) and precise identity of lineages arising from this tangle remain unclear and relationships amongst the genera of this clade remain highly uncertain pending additional taxon sampling and detailed investigation of the causes of gene tree conflict and possible evidence for introgression.
Albizia
At the start of this study, the genus Albizia was dubbed the last pantropical so-called ‘dustbin’ genus pending resolution (Koenen et al. 2020b). Here, we show that Albizia s.l. is rampantly non-monophyletic, most notably because the bulk of the Old and New World species are placed in separate clades (Figs 9 and 10). This Old World – New World split is remedied in this Special Issue by Aviles et al. (2022) who resurrect the genus Pseudalbizzia Britton & Rose for the majority of the New World species placed in Barneby’s AlbiziasectionArthrosamanea, with Albizia s.s. now restricted to just the Old World species, which still includes ca. 90 spp. (Koenen et al., unpubl. data). Furthermore, the disparate placements of several other species of Albizia across the phylogeny, viz: Albiziacarbonaria Britton (Fig. 8), the long-neglected African Albizia species previously often placed in Cathormion or Samanea (Benth.) Merr. (Figs 9 and 11) and Albizialeonardii (Fig. 7), are all accounted for with new generic placements and nomenclatural combinations (Koenen 2022b; Soares et al. 2022), one synonymisation (Terra et al. 2022) and a new segregate genus (Koenen 2022a), all of them being published in this Special Issue.
Abarema , Hydrochorea and Balizia
The recent re-circumscription of Abarema Pittier to include just two species and transfer of the remaining species to the re-instated Punjuba Britton & Rose and Jupunba Britton & Rose (Guerra et al. 2016, 2019; Iganci et al. 2016; Soares et al. 2021), is broadly supported here (Figs 9 and 11), except for the anomalous placement of Jupunbamacradenia (Pittier) M.V.B. Soares, M.P. Morim & Iganci which is sister to the Hydrochorea + Balizia clade (Fig. 9). This placement is unexpected and somewhat suspect considering J.macradenia is firmly placed in Jupunba in Soares et al. (2021). As found by Iganci et al. (2016), Koenen et al. (2020b) and Soares et al. (2021), Balizia is non-monophyletic with the genus Hydrochorea plus two African species of Albizia nested within it (Fig. 9). Hydrochorea is re-circumscribed to accommodate all these elements by Soares et al. (2022) in this Special Issue.
Leucochloron
Koenen et al. (2020b) showed that Leucochloron is polyphyletic and that result is confirmed here, split between the Albizia and Inga clades (Figs 10 and 11). A new segregate genus to account for this non-monophyly is proposed in this Special Issue by de Souza et al. (2022b).
Zygia , Macrosamanea and Inga
Alongside Archidendron, the large Neotropical, mainly rainforest genus Zygia remains one of the least well-documented genera of mimosoids, with many species known from incomplete material (Barneby and Grimes 1997). Previous work by Ferm et al. (2019) showed that, while the bulk of genus Zygia is monophyletic, a handful of outlier species have affinities to other genera: Zygiaocumarensis (Pittier) Barneby & J.W. Grimes is sister to Macrosamanea Britton & Rose ex Britton & Killip, Marmaroxylonmagdalenae Killip ex. L. Rico (treated as a synonym of Z.ocumarensis by Barneby and Grimes (1997)) is nested in Jupunba and Z.inundata and Z.sabatieri are together sister to Inga. With the exception of M.magdalenae, which is not included in this study, these placements are confirmed here with phylogenomic data (Figs 11 and 12) and reflect the morphological distinctiveness of these species from the rest of the genus (Barneby and Grimes 1997; Ferm et al. 2019) which prompted placements in their own separate monospecific sections of Zygia (Barneby and Grimes 1997). New nomenclatural combinations to deal with these outlier Zygia species are still pending. We suggest that Zygiaocumarensis should best be transferred to Macrosamanea, as it shares bipinnate leaves with multiple pairs of pinnae and an absence of cauli-/ramiflory (which is almost universal in Zygia) with several species of Macrosamanea (Barneby and Grimes 1996; Ferm et al. 2019). The identity of Marmaroxylonmagdalenae needs to be re-evaluated, but the evidence of Ferm et al. (2019), who sampled the type material, suggests it should be transferred to Jupunba. The generic placements of Z.inundata and Z.sabatieri are more contentious. Arguments can be made to transfer Z.inundata to Inga (Ferm et al. 2019): it was originally described in Inga and it shares once-pinnate leaves and absence of cauli-/ramiflory with Inga (Barneby and Grimes 1997; Ferm et al. 2019). However, Z.inundata was placed as the sole sister of Inga in the plastid tree (Suppl. material 3) and by Ferm et al. (2019), whereas the nuclear gene data suggest that Z.inundata is sister to Z.sabatieri and together these two species form the sister clade of Inga (Fig. 12). Zygiasabatieri has bipinnate leaves and both Z.sabatieri and Z.inundata have dehiscent pods, characteristics that distinguish these species from Inga with its uniformly once-pinnate leaves and indehiscent pods. In order to maintain a morphologically coherent and homogeneous Inga with respect to these diagnostic characters, segregating Z.inundata and Z.sabatieri as a new genus would appear to be advantageous. Ingopsis Barneby & J.W. Grimes and Pseudocojoba Barneby & J.W. Grimes, the names for the monospecific sections containing Z.inundata and Z.sabatieri, respectively (Barneby and Grimes 1997), are two available names, of which Ingopsis would be preferable given the morphological and phylogenetic proximity of this clade to Inga and the lack of a close relationship to Cojoba Britton & Rose. However, since these sectional names have no priority at generic rank (Turland et al. 2018), alternatively, a new name could equally be proposed. Finally, while Zygia s.s. was reasonably well sampled by Ferm et al. (2019) and also in the current study (Fig. 12), alongside further herbarium taxonomic work and field studies to clarify species, denser phylogenetic taxon sampling is desirable, in particular to include Z.eperuetorum (Sandwith) Barneby & J.W. Grimes. This species is known only from the Essequibo Valley in Guyana, was placed in its own section by Barneby and Grimes (1997), has an unusual combination of morphological characters not found elsewhere in Zygia and the fruit remains unknown. Zygiaeperuetorum may well, therefore, represent an additional separate lineage that could potentially merit recognition as a distinct genus.
Citation
Ringelberg JJ, Koenen EJM, Iganci JR, de Queiroz LP, Murphy DJ, Gaudeul M, Bruneau A, Luckow M, Lewis GP, Hughes CE (2022) Phylogenomic analysis of 997 nuclear genes reveals the need for extensive generic re-delimitation in Caesalpinioideae (Leguminosae). In: Hughes CE, de Queiroz LP, Lewis GP (Eds) Advances in Legume Systematics 14. Classification of Caesalpinioideae Part 1: New generic delimitations. PhytoKeys 205: 3–58. https://doi.org/10.3897/phytokeys.205.85866
Supplementary materials
Table S1
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Jens J. Ringelberg, Erik J.M. Koenen, João R. Iganci, Luciano P. de Queiroz, Daniel J. Murphy, Myriam Gaudeul, Anne Bruneau, Melissa Luckow, Gwilym P. Lewis, Colin E. Hughes
Data type
excel file.
Explanation note
Samples included in this study.
Table S2
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Jens J. Ringelberg, Erik J.M. Koenen, João R. Iganci, Luciano P. de Queiroz, Daniel J. Murphy, Myriam Gaudeul, Anne Bruneau, Melissa Luckow, Gwilym P. Lewis, Colin E. Hughes
Data type
excel file.
Explanation note
Trait data used for character evolution analyses.
Figure S1
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Jens J. Ringelberg, Erik J.M. Koenen, João R. Iganci, Luciano P. de Queiroz, Daniel J. Murphy, Myriam Gaudeul, Anne Bruneau, Melissa Luckow, Gwilym P. Lewis, Colin E. Hughes
Data type
Pdf file.
Explanation note
Chloroplast phylogeny of Caesalpinioideae. Only bootstrap support values lower than 100% are shown.
Supplementary tree file
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Jens J. Ringelberg, Erik J.M. Koenen, João R. Iganci, Luciano P. de Queiroz, Daniel J. Murphy, Myriam Gaudeul, Anne Bruneau, Melissa Luckow, Gwilym P. Lewis, Colin E. Hughes
Data type
Tree file (Newick format).
Explanation note
Tree file of the ASTRAL phylogeny based on the single-copy genes (depicted in Figs 2–12), in which taxon labels have been updated to reflect taxonomic changes made in all the entries in Advances in Legume Systematics 14 Part 1.
References
- Aberer AJ, Krompass D, Stamatakis A. (2013) Pruning rogue taxa improves phylogenetic accuracy: An efficient algorithm and webservice. Systematic Biology 62(1): 162–166. 10.1093/sysbio/sys078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebli A. (2015) Assembly of the Madagascan biota by replicated adaptive radiations: Case studies in Leguminosae-Mimosoideae. Unpublished MSc thesis, University of Zurich.
- Aviles GP, Koenen E, Riina R, Hughes CE, Ringelberg JJ, Fernández-Concha GC, Morillo IR, Itza LLC, Tamayo-Cen I, Prado JHR, Cornejo X, Mattapha S, de Stefano RD. (2022) Re-establishment of the genus Pseudalbizzia (Leguminosae, Caesalpinioideae: mimosoid clade): the New World species formerly placed in Albizia. In: Hughes CE, de Queiroz LP, Lewis GP. (Eds) Advances in Legume Systematics 14. Classification of Caesalpinioideae Part 1: New generic delimitations.PhytoKeys 205: 371–400. 10.3897/phytokeys.205.76821 [DOI] [PMC free article] [PubMed]
- Babineau M, Bruneau A. (2017) Phylogenetic and biogeographical history of the Afro-Madagascan genera Delonix, Colvillea and Lemuropisum (Fabaceae: Caesalpinioideae). Botanical Journal of the Linnean Society 184(1): 59–78. 10.1093/botlinnean/box009 [DOI] [Google Scholar]
- Banks H, Himanen I, Lewis GP. (2010) Evolution of pollen, stigmas and ovule numbers at the caesalpinioid-mimosoid interface (Fabaceae). Botanical Journal of the Linnean Society 162(4): 594–615. 10.1111/j.1095-8339.2010.01038.x [DOI] [Google Scholar]
- Barneby RC. (1991) Sensitivae Censitae - A description of the genus Mimosa Linnaeus (Mimosaceae) in the New World. Memoirs of the New York Botanical Garden 65: 1–835. [Google Scholar]
- Barneby RC. (1998) Silk tree, guanacaste, monkey’s earring. A generic system for the synandrous Mimosaceae of the Americas. Part III: Calliandra. Memoirs of the New York Botanical Garden 74: 1–223. [Google Scholar]
- Barneby RC, Grimes JW. (1996) Silk tree, guanacaste, monkey’s earring. A generic system for the synandrous Mimosaceae of the Americas. Part I: Abarema, Albizia, and allies. Memoirs of the New York Botanical Garden 74: 1–292. [Google Scholar]
- Barneby RC, Grimes JW. (1997) Silk tree, guanacaste, monkey’s earring. A generic system for the synandrous Mimosaceae of the Americas. Part II: Pithecellobium, Cojoba, and Zygia. Memoirs of the New York Botanical Garden 74: 1–149. [Google Scholar]
- Barrett CF, Bacon CD, Antonelli A, Cano Á, Hofmann T. (2016) An introduction to plant phylogenomics with a focus on palms. Botanical Journal of the Linnean Society 182(2): 234–255. 10.1111/boj.12399 [DOI] [Google Scholar]
- Bentham G. (1841) Notes on Mimoseae, with a short synopsis of species. Journal of Botany (Hooker) 4: 323–417. [Google Scholar]
- Bentham G. (1842) Notes on Mimoseae, with a synopsis of species. Tribe III. Acacieae (cont.). The London Journal of Botany 1: 318–392. [Google Scholar]
- Bentham G. (1875) Revision of the suborder Mimoseae. Transactions of the Linnean Society of London 30(3): 335–664. 10.1111/j.1096-3642.1875.tb00005.x [DOI] [Google Scholar]
- Boatwright JS, Maurin O, van der Bank M. (2015) Phylogenetic position of Madagascan species of Acacia s.l. and new combinations in Senegalia and Vachellia (Fabaceae, Mimosoideae, Acacieae). Botanical Journal of the Linnean Society 179(2): 288–294. 10.1111/boj.12320 [DOI] [Google Scholar]
- Borges L, Inglis PW, Simon M, Ribeiro P, de Queiroz L. (2022) Misleading fruits: The non-monophyly of Pseudopiptadenia and Pityrocarpa supports generic re-circumscriptions and a new genus within mimosoid legumes. In: Hughes CE, de Queiroz LP, Lewis GP. (Eds) Advances in Legume Systematics 14. Classification of Caesalpinioideae Part 1: New generic delimitations.PhytoKeys 205: 239–260. 10.3897/phytokeys.205.82275 [DOI] [PMC free article] [PubMed]
- Bouchenak-Khelladi Y, Maurin O, Hurter J, van der Bank M. (2010) The evolutionary history and biogeography of Mimosoideae (Leguminosae): An emphasis on African acacias. Molecular Phylogenetics and Evolution 57(2): 495–508. 10.1016/j.ympev.2010.07.019 [DOI] [PubMed] [Google Scholar]
- Brown GK. (2008) Systematics of the tribe Ingeae. Muelleria 26: 27–42. [Google Scholar]
- Brown GK, Murphy DJ, Miller JT, Ladiges PY. (2008) Acacia s.s. and its relationship among tropical legumes, tribe Ingeae (Leguminosae: Mimosoideae). Systematic Botany 33(4): 739–751. 10.1600/036364408786500136 [DOI] [Google Scholar]
- Brown GK, Murphy DJ, Ladiges PY. (2011) Relationships of the Australo-Malesian genus Paraserianthes (Mimosoideae: Leguminosae) identifies the sister group of Acacia sensu stricto and two biogeographical tracks. Cladistics 27(4): 380–390. 10.1111/j.1096-0031.2011.00349.x [DOI] [PubMed] [Google Scholar]
- Brown GK, Aju J, Bayly MJ, Murphy DJ, McLay TGB. (2022) Phylogeny and classification of the Australasian and Indomalayan mimosoid legumes Archidendron and Archidendropsis (Leguminosae, subfamily Caesalpinioideae, mimosoid clade). In: Hughes CE, de Queiroz LP, Lewis GP. (Eds) Advances in Legume Systematics 14. Classification of Caesalpinioideae Part 1: New generic delimitations.PhytoKeys 205: 299–334. 10.3897/phytokeys.205.79381 [DOI] [PMC free article] [PubMed]
- Bruneau A, Mercure M, Lewis GP, Herendeen PS. (2008) Phylogenetic patterns and diversification in the caesalpinioid legumes. Botany 86(7): 697–718. 10.1139/B08-058 [DOI] [Google Scholar]
- Bruun-Lund S, Clement WL, Kjellberg F, Rønsted N. (2017) First plastid phylogenomic study reveals potential cyto-nuclear discordance in the evolutionary history of Ficus L. (Moraceae). Molecular Phylogenetics and Evolution 109: 93–104. 10.1016/j.ympev.2016.12.031 [DOI] [PubMed] [Google Scholar]
- Burkart A. (1943) Las leguminosas argentinas, silvestres y cultivadas. Acme Agency, Buenos Aires, Argentina.
- Burkart A. (1976) A monograph of the genus Prosopis (Leguminosae subfam. Mimosoidae). Journal of the Arnold Arboretum 57(3): 219–525. 10.5962/p.185864 [DOI] [Google Scholar]
- de Candolle AP. (1825) Prodromus systematis naturalis regni vegetabilis - pars II.
- Capuron R. (1970) Contribution à l’étude de la florestière de Madagascar: Notes sur les Albizia Duraz. Adansonia 2: 357–382. [Google Scholar]
- Cardoso D, Pennington RT, de Queiroz LP, Boatwright JS, Van Wyk BE, Wojciechowski MF, Lavin M. (2013) Reconstructing the deep-branching relationships of the papilionoid legumes. South African Journal of Botany 89: 58–75. 10.1016/j.sajb.2013.05.001 [DOI] [Google Scholar]
- Catalano SA, Vilardi JC, Tosto D, Saidman BO. (2008) Molecular phylogeny and diversification history of Prosopis (Fabaceae: Mimosoideae). Biological Journal of the Linnean Society. Linnean Society of London 93(3): 621–640. 10.1111/j.1095-8312.2007.00907.x [DOI] [Google Scholar]
- Clark RP, Jiang K-W, Gagnon E. (2022) Reinstatement of Ticanto (Leguminosae-Caesalpinioideae) – the final piece in the Caesalpinia group puzzle. In: Hughes CE, de Queiroz LP, Lewis GP. (Eds) Advances in Legume Systematics 14. Classification of Caesalpinioideae Part 1: New generic delimitations.PhytoKeys 205: 59–98. 10.3897/phytokeys.205.82300 [DOI] [PMC free article] [PubMed]
- Clarke HD, Downie SR, Seigler DS. (2000) Implications of chloroplast DNA restriction site variation for systematics of Acacia (Fabaceae : Mimosoideae). Systematic Botany 25(4): 618–632. 10.2307/2666724 [DOI] [Google Scholar]
- Contreras Jiménez JL. (1986) Desmanthusbalsensis (Leguminosae: Mimosoideae), una especie nueva de la depresión del Río Balsas en Guerrero, Mexico. Phytologia 60: 89–92. 10.5962/bhl.part.3792 [DOI] [Google Scholar]
- da Silva MF. (1986) Dimorphandra (Caesalpiniaceae). Flora Neotropica 44: 1–127. [Google Scholar]
- Dahmer N, Simon MF, Schifino-Wittmann MT, Hughes CE, Miotto STS, Giuliani JC. (2011) Chromosome numbers in the genus Mimosa L.: Cytotaxonomic and evolutionary implications. Plant Systematics and Evolution 291(3–4): 211–220. 10.1007/s00606-010-0382-2 [DOI] [Google Scholar]
- de Faria SM, Ringelberg JJ, Gross E, Koenen EJM, Cardoso D, Ametsitsi GKD, Akomatey J, Maluk M, Tak N, Gehlot HS, Wright KM, Teaumroong N, Songwattana P, De Lima HC, Prin Y, Zartmann C, Sprent JI, Ardley J, Hughes CE, James EK. (2022) The innovation of the symbiosome has enhanced the evolutionary stability of nitrogen fixation in legumes. New Phytologist. 10.1111/nph.18321 [DOI] [PMC free article] [PubMed]
- de la Estrella M, Forest F, Klitgård B, Lewis GP, Mackinder BA, De Queiroz LP, Wieringa JJ, Bruneau A. (2018) A new phylogeny-based tribal classification of subfamily Detarioideae, an early branching clade of florally diverse tropical arborescent legumes. Scientific Reports 8(1): 1–14. 10.1038/s41598-018-24687-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza ÉR, Lewis GP, Forest F, Schnadelbach AS, van den Berg C, de Queiroz LP. (2013) Phylogeny of Calliandra (Leguminosae: Mimosoideae) based on nuclear and plastid molecular markers. Taxon 62(6): 1200–1219. 10.12705/626.2 [DOI] [Google Scholar]
- de Souza ÉR, Krishnaraj MV, de Queiroz LP. (2016) Sanjappa, a new genus in the tribe Ingeae (Leguminosae: Mimosoideae) from India. Rheedea 26: 1–12. [Google Scholar]
- de Souza ÉR, Luz ARM, Rocha L, Lewis GP. (2022a) Molecular and morphological analysis supports the separation of Robrichia as a genus distinct from Enterolobium (Leguminosae: Caesalpinioideae: Mimosoid Clade). Systematic Botany 47(1): 268–277. 10.1600/036364422X16442669847067 [DOI] [Google Scholar]
- de Souza ÉR, de Almeida PGC, Rocha L, Koenen EJM, Burgos MA, Lewis GP, Hughes CE. (2022b) Boliviadendron, a new segregate genus of mimosoid legume (Leguminosae, Caesalpinioideae, mimosoid clade) narrowly endemic to the interior Andean valleys of Bolivia. In: Hughes CE, de Queiroz LP, Lewis GP. (Eds) Advances in Legume Systematics 14. Classification of Caesalpinioideae Part 1: New generic delimitations.PhytoKeys 205: 439–452. 10.3897/phytokeys.205.82256 [DOI] [PMC free article] [PubMed]
- Debray K, Le Paslier M-C, Bérard A, Thouroude T, Michel G, Marie-Magdelaine J, Bruneau A, Foucher F, Malécot V. (2022) Unveiling the patterns of reticulated evolutionary processes with phylogenomics: Hybridization and polyploidy in the genus Rosa. Systematic Biology 71(3): 547–569. 10.1093/sysbio/syab064 [DOI] [PubMed] [Google Scholar]
- Demeulenaere E, Schils T, Burleigh JG, Ringelberg JJ, Koenen EJM, Ickert-Bond SM. (2022) Phylogenomic assessment prompts recognition of the Serianthes clade and confirms the monophyly of Serianthes and its relationship with Falcataria and Wallaceodendron in the wider ingoid clade (Leguminosae, Caesalpinioideae). In: Hughes CE, de Queiroz LP, Lewis GP. (Eds) Advances in Legume Systematics 14. Classification of Caesalpinioideae Part 1: New generic delimitations.PhytoKeys 205: 335–362. 10.3897/phytokeys.205.79144 [DOI] [PMC free article] [PubMed]
- Dodsworth S, Pokorny L, Johnson MG, Kim JT, Maurin O, Wickett NJ, Forest F, Baker WJ. (2019) Hyb-Seq for flowering plant systematics. Trends in Plant Science 24(10): 887–891. 10.1016/j.tplants.2019.07.011 [DOI] [PubMed] [Google Scholar]
- Du Puy DJ, Labat J-N, Rabevohitra R, Villiers J-F, Bosser J, Moat J. (2002) The Leguminosae of Madagascar. Royal Botanic Gardens, Kew, United Kingdom, 737 pp. [Google Scholar]
- Elias TS. (1981) Mimosoideae. In: Polhill RM, Raven PH. (Eds) Advances in Legume Systematics 1.Royal Botanic Gardens, Kew, United Kingdom, 143–152.
- Engler HGA, Prantl KAE. (1897) Die Natürlichen Pflanzenfamilien 2–4, Nachträge 1. Wilhelm Engelmann, Leipzig, Germany.
- Ferm J, Korall P, Lewis GP, Ståhl B. (2019) Phylogeny of the Neotropical legume genera Zygia and Marmaroxylon and close relatives. Taxon 68(4): 661–672. 10.1002/tax.12117 [DOI] [Google Scholar]
- Gagnon E, Bruneau A, Hughes CE, de Queiroz L, Lewis GP. (2016) A new generic system for the pantropical Caesalpinia group (Leguminosae). PhytoKeys 71: 1–160. 10.3897/phytokeys.71.9203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon E, Ringelberg JJ, Bruneau A, Lewis GP, Hughes CE. (2019) Global Succulent Biome phylogenetic conservatism across the pantropical Caesalpinia Group (Leguminosae). New Phytologist 222(4): 1994–2008. 10.1111/nph.15633 [DOI] [PubMed] [Google Scholar]
- Geesink R. (1984) Scala Milletiearum: a survey of the genera of the tribe Millettieae (Leguminosae-Papilionoideae). Leiden Botanical Series 1, EJ Brill & Leiden University Press, Leiden.
- Goldblatt P, Davidse G. (1977) Chromosome numbers in legumes. Annals of the Missouri Botanical Garden 64(1): 121–128. 10.2307/2395237 [DOI] [Google Scholar]
- Gómez-Acevedo S, Rico-Arce L, Delgado-Salinas A, Magallón S, Eguiarte LE. (2010) Neotropical mutualism between Acacia and Pseudomyrmex: Phylogeny and divergence times. Molecular Phylogenetics and Evolution 56(1): 393–408. 10.1016/j.ympev.2010.03.018 [DOI] [PubMed] [Google Scholar]
- Govindarajulu R, Hughes CE, Alexander PJ, Donovan Bailey C. (2011) The complex evolutionary dynamics of ancient and recent polyploidy in Leucaena (Leguminosae; Mimosoideae). American Journal of Botany 98(12): 2064–2076. 10.3732/ajb.1100260 [DOI] [PubMed] [Google Scholar]
- Grimes J. (1999) Inflorescence morphology, heterochrony, and phylogeny in the Mimosoid tribes Ingeae and Acacieae (Leguminosae: Mimosoideae). Botanical Review 65(4): 317–347. 10.1007/BF02857753 [DOI] [Google Scholar]
- Guerra E, Morim MP, Iganci J. (2016) A new species of Abarema (Fabaceae) from Brazil. Phytotaxa 289(1): 77–82. 10.11646/phytotaxa.289.1.6 [DOI] [Google Scholar]
- Guerra E, Andrade BO, Morim MP, Vieira Iganci JR. (2019) Taxonomic delimitation of species complexes: A challenge for conservation; First steps with the Abaremacochliacarpos complex. Systematic Botany 44(4): 818–825. 10.1600/036364419X15710776741422 [DOI] [Google Scholar]
- Guinet Ph. (1981) Mimosoideae: the characters of their pollen grains. In: Polhill RM, Raven PH (Eds) Advances in Legume Systematics Part 1, Royal Botanic Gardens, Kew.
- Hart ML, Forrest LL, Nicholls JA, Kidner CA. (2016) Retrieval of hundreds of nuclear loci from herbarium specimens. Taxon 65(5): 1081–1092. 10.12705/655.9 [DOI] [Google Scholar]
- Hernández HM. (1986) Zapoteca: A new genus of Neotropical Mimosoideae. Annals of the Missouri Botanical Garden 73(4): 755–763. 10.2307/2399204 [DOI] [Google Scholar]
- Hou D. (1996) Sympetalandra. In: Hou D, Larsen K, Larsen SS, Laferriere JE, Duyfjes BEE. (Eds) Fl.Malesiana Series 1. Rijksherbarium / Hortus Botanicus, Leiden University, Leiden, the Netherlands, 709–714.
- Hoyle AC. (1932) Chidlowia, a new tree genus of Caesalpiniaceae from West Tropical Africa. Kew Bulletin of Miscellaneous Information: 101–103. 10.2307/4113372 [DOI]
- Hu JM, Lavin M, Wojciechowski MF, Sanderson MJ. (2000) Phylogenetic systematics of the tribe Millettieae (Leguminosae) based on chloroplast trnK/matK sequences and its implications for evolutionary patterns in Papilionoideae. American Journal of Botany 87(3): 418–430. 10.2307/2656638 [DOI] [PubMed] [Google Scholar]
- Hughes CE. (1997) Variation in anther and pollen morphology in Leucaena Benth. (Leguminosae-Mimosoideae). Botanical Journal of the Linnean Society 123(3): 177–196. 10.1111/j.1095-8339.1997.tb01412.x [DOI] [Google Scholar]
- Hughes CE, Bailey CD, Krosnick S, Luckow MA. (2003) Relationships among genera of the informal Dichrostachys and Leucaena groups (Mimosoideae) inferred from nuclear ribosomal ITS sequences. In: Klitgaard B, Bruneau A (Eds) Advances in Legume Systematics Part 10, Royal Botanic Gardens, Kew, United Kingdom, 221–238.
- Hughes CE, Ringelberg JJ, Lewis GP, Catalano SA. (2022a) Disintegration of the genus Prosopis L. (Leguminosae, Caesalpinioideae, mimosoid clade). In: Hughes CE, de Queiroz LP, Lewis GP. (Eds) Advances in Legume Systematics 14. Classification of Caesalpinioideae Part 1: New generic delimitations.PhytoKeys 205: 147–190. 10.3897/phytokeys.205.75379 [DOI] [PMC free article] [PubMed]
- Hughes CE, Ringelberg JJ, Luckow M, Jiménez JLC. (2022b) Mezcala – a new segregate genus of mimosoid legume (Leguminosae, Caesalpinioideae, mimosoid clade) narrowly endemic to the Balsas Depression in Mexico. In: Hughes CE, de Queiroz LP, Lewis GP. (Eds) Advances in Legume Systematics 14. Classification of Caesalpinioideae Part 1: New generic delimitations.PhytoKeys 205: 191–202. 10.3897/phytokeys.205.78297 [DOI] [PMC free article] [PubMed]
- Humphreys AM, Linder HP. (2009) Concept versus data in delimitation of plant genera. Taxon 58(4): 1054–1074. 10.1002/tax.584002 [DOI] [Google Scholar]
- Iganci J, Soares M, Guerra E, Morim M. (2016) A Preliminary molecular phylogeny of the Abarema alliance (Leguminosae) and implications for taxonomic rearrangement. International Journal of Plant Sciences 177(1): 34–43. 10.1086/684078 [DOI] [Google Scholar]
- Jiang X, Edwards SV, Liu L. (2020) The multispecies coalescent model outperforms concatenation across diverse phylogenomic data sets. Systematic Biology 69(4): 795–812. 10.1093/sysbio/syaa008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobson RW, Luckow M. (2007) Phylogenetic study of the genus Piptadenia (Mimosoideae: Leguminosae) using plastid trnL-F and trnK/matK sequence data. Systematic Botany 32(3): 569–575. 10.1600/036364407782250544 [DOI] [Google Scholar]
- Johnson MG, Gardner EM, Liu Y, Medina R, Goffinet B, Shaw AJ, Zerega NJC, Wickett NJ. (2016) HybPiper: Extracting coding sequence and introns for phylogenetics from high-throughput sequencing reads using target enrichment. Applications in Plant Sciences 4(7): e1600016. 10.3732/apps.1600016 [DOI] [PMC free article] [PubMed]
- Johnson MG, Pokorny L, Dodsworth S, Botigué LR, Cowan RS, Devault A, Eiserhardt WL, Epitawalage N, Forest F, Kim JT, Leebens-Mack JH, Leitch IJ, Maurin O, Soltis DE, Soltis PS, Wong GKS, Baker WJ, Wickett NJ. (2019) A universal probe set for targeted sequencing of 353 nuclear genes from any flowering plant designed using k-medoids clustering. Systematic Biology 68(4): 594–606. 10.1093/sysbio/syy086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumah A. (1991) Studies on the morphology of pollen grains of the leguminosae - The mimosoideae. Ghana Journal of Science 31–36: 29–35. 10.4314/gjs.v36i1.47978 [DOI]
- Karimi N, Grover CE, Gallagher JP, Wendel JF, Ané C, Baum DA. (2019) Reticulate evolution helps explain apparent homoplasy in floral biology and pollination in Baobabs (Adansonia; Bombacoideae; Malvaceae). Systematic Biology 69(3): 462–478. 10.1093/sysbio/syz073 [DOI] [PubMed] [Google Scholar]
- Kirk PM, Cannon PF, Minter DW, Stalpers JA. (2008) Dictionary of the Fungi (10th edn.). CABI, Wallingford, United Kingdom, 551 pp. [Google Scholar]
- Koenen EJM. (2022a) Osodendron gen. nov. (Leguminosae, Caesalpinioideae), a new genus of mimosoid legumes of tropical Africa. In: Hughes CE, de Queiroz LP, Lewis GP. (Eds) Advances in Legume Systematics 14. Classification of Caesalpinioideae Part 1: New generic delimitations.PhytoKeys 205: 453–470. 10.3897/phytokeys.205.82821 [DOI] [PMC free article] [PubMed]
- Koenen EJM. (2022b) On the taxonomic affinity of Albiziacarbonaria Britton (Leguminosae, Caesalpinioideae-mimosoid clade). In: Hughes CE, de Queiroz LP, Lewis GP. (Eds) Advances in Legume Systematics 14. Classification of Caesalpinioideae Part 1: New generic delimitations.PhytoKeys 205: 363–370. 10.3897/phytokeys.205.82288 [DOI] [PMC free article] [PubMed]
- Koenen EJM, Ojeda DI, Steeves R, Migliore J, Bakker FT, Wieringa JJ, Kidner C, Hardy OJ, Pennington RT, Bruneau A, Hughes CE. (2020a) Large‐scale genomic sequence data resolve the deepest divergences in the legume phylogeny and support a near‐simultaneous evolutionary origin of all six subfamilies. New Phytologist 225(3): 1355–1369. 10.1111/nph.16290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenen EJM, Kidner CA, de Souza ÉR, Simon MF, Iganci JRV, Nicholls JA, Brown GK, de Queiroz LP, Luckow MA, Lewis GP, Pennington RT, Hughes CE. (2020b) Hybrid capture of 964 nuclear genes resolves evolutionary relationships in the mimosoid legumes and reveals the polytomous origins of a large pantropical radiation. American Journal of Botany 107(12): 1710–1735. 10.1002/ajb2.1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyalangalilwa B, Boatwright JS, Daru BH, Maurin O, van der Bank M. (2013) Phylogenetic position and revised classification of Acacia s.l. (Fabaceae: Mimosoideae) in Africa, including new combinations in Vachellia and Senegalia. Botanical Journal of the Linnean Society 172(4): 500–523. 10.1111/boj.12047 [DOI] [Google Scholar]
- Lartillot N, Rodrigue N, Stubbs D, Richer J. (2013) PhyloBayes MPI: Phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Systematic Biology 62(4): 11–615. 10.1093/sysbio/syt022 [DOI] [PubMed] [Google Scholar]
- Lavin M, Pennington RT, Klitgaard BB, Sprent JI, de Lima HC, Gasson PE. (2001) The dalbergioid legumes (Fabaceae): Delimitation of a pantropical monophyletic clade. American Journal of Botany 88(3): 503–533. 10.2307/2657116 [DOI] [PubMed] [Google Scholar]
- Lavin M, Schrire BP, Lewis GP, Pennington RT, Delgado-Salinas A, Thulin M, Hughes CE, Matos AB, Wojciechowski MF. (2004) Metacommunity process rather than continental tectonic history better explains geographically structured phylogenies in legumes. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 359(1450): 1509–1522. 10.1098/rstb.2004.1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Yaw JA, Grassa CJ, Joly S, Andrew RL, Rieseberg LH. (2019) An evaluation of alternative explanations for widespread cytonuclear discordance in annual sunflowers (Helianthus). New Phytologist 221(1): 515–526. 10.1111/nph.15386 [DOI] [PubMed] [Google Scholar]
- Lewis GP. (1998) Caesalpinia: a revision of the Poincianella-Erythrostemon group. Royal Botanic Gardens, Kew, United Kingdom, 233 pp. [Google Scholar]
- Lewis GP, Guinet P. (1986) Notes on Gagnebina (Leguminosae: Mimosoideae) in Madagascar and neighbouring islands. Kew Bulletin 41(2): 463–470. 10.2307/4102962 [DOI] [Google Scholar]
- Lewis GP, Schrire BD. (2003) Thailentadopsis Kostermans (Leguminosae: Mimosoideae: Ingeae) resurrected. Kew Bulletin 58(2): 491–494. 10.2307/4120634 [DOI] [Google Scholar]
- Lewis GP, Schrire B, Mackinder B, Lock M. (2005) Legumes of the World. Royal Botanic Gardens, Kew, United Kingdom, 592 pp. [Google Scholar]
- Lima A, de Paula-Souza J, Ringelberg JJ, Simon MF, de Queiroz L, Borges L, Mansano V, Souza V, Scalon V. (2022) New Segregates from the Neotropical genus Stryphnodendron (Leguminosae, Caesalpinioideae, mimosoid clade). In: Hughes CE, de Queiroz LP, Lewis GP. (Eds) Advances in Legume Systematics 14. Classification of Caesalpinioideae Part 1: New generic delimitations.PhytoKeys 205: 203–238. 10.3897/phytokeys.205.82220 [DOI] [PMC free article] [PubMed]
- von Linnaeus C. (1753) Species Plantarum 1.
- LPWG (2013) Legume phylogeny and classification in the 21st century: Progress, prospects and lessons for other species-rich clades. Taxon 62(2): 217–248. 10.12705/622.8 [DOI] [Google Scholar]
- LPWG (2017) A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny – The Legume Phylogeny Working Group (LPWG). Taxon 66(1): 44–77. 10.12705/661.3 [DOI] [Google Scholar]
- Luckow MA. (1993) Monograph of Desmanthus (Leguminosae-Mimosoideae). Systematic Botany Monographs 38: 1–166. 10.2307/25027822 [DOI] [Google Scholar]
- Luckow MA, Grimes J. (1997) A survey of anther glands in the mimosoid legume tribes Parkieae and Mimoseae. American Journal of Botany 84(3): 285–297. 10.2307/2446002 [DOI] [PubMed] [Google Scholar]
- Luckow MA, White PJ, Bruneau A. (2000) Relationships among basal genera of mimosoid legumes. In: Herendeen PS, Bruneau A. (Eds) Advances in Legume Systematics, part 9.Royal Botanic Gardens, Kew, United Kingdom, 165–180.
- Luckow MA, Miller JT, Murphy DJ, Livshultz T. (2003) A phylogenetic analysis of the Mimosoideae (Leguminosae) based on chloroplast DNA sequence data. In: Klitgaard BB, Bruneau A. (Eds) Advances in Legume Systematics, part 10, Higher Level Systematics.Royal Botanic Gardens, Kew, United Kingdom, 197–220.
- Luckow MA, Fortunato RH, Sede S, Livshultz T. (2005) The phylogenetic affinities of two mysterious monotypic mimosoids from southern South America. Systematic Botany 30(3): 585–602. 10.1600/0363644054782206 [DOI] [Google Scholar]
- Manzanilla V, Bruneau A. (2012) Phylogeny reconstruction in the Caesalpinieae grade (Leguminosae) based on duplicated copies of the sucrose synthase gene and plastid markers. Molecular Phylogenetics and Evolution 65(1): 149–162. 10.1016/j.ympev.2012.05.035 [DOI] [PubMed] [Google Scholar]
- Marazzi B, Sanderson MJ. (2010) Large-scale patterns of diversification in the widespread legume genus Senna and the evolutionary role of extrafloral nectaries. Evolution 64(12): 3570–3592. 10.1111/j.1558-5646.2010.01086.x [DOI] [PubMed] [Google Scholar]
- Marazzi B, Gonzalez AM, Delgado-Salinas A, Luckow MA, Ringelberg JJ, Hughes CE. (2019) Extrafloral nectaries in Leguminosae: Phylogenetic distribution, morphological diversity and evolution. Australian Systematic Botany 32: 409–458. 10.1071/SB19012 [DOI] [Google Scholar]
- Miller JT, Bayer RJ. (2000) Molecular systematics of the tribe Acaciae (Leguminosae: Mimosoideae). In: Herendeen PS, Bruneau A. (Eds) Advances in Legume Systematics 9.Royal Botanic Gardens, Kew, United Kingdom, 181–200.
- Miller JT, Bayer RJ. (2001) Molecular phylogenetics of Acacia (Fabaceae: Mimosoideae) based on the chloroplast matK coding sequence and flanking trnK intron spacer regions. Australian Journal of Botany 88(4): 697–705. 10.2307/2657071 [DOI] [PubMed] [Google Scholar]
- Miller JT, Bayer RJ. (2003) Molecular phylogenetics of Acacia subgenera Acacia and Aculeiferum (Fabaceae: Mimosoideae), based on the chloroplast matK coding sequence and flanking trnK intron spacer regions. Australian Systematic Botany 16(1): 27–33. 10.1071/SB01035 [DOI] [PubMed] [Google Scholar]
- Miller JT, Seigler D. (2012) Evolutionary and taxonomic relationships of Acacia s.l. (Leguminosae: Mimosoideae). Australian Systematic Botany 25(3): 217–224. 10.1071/SB11042 [DOI] [Google Scholar]
- Miller JT, Andrew R, Bayer RJ. (2003) Molecular phylogenetics of the Australian acacias of subg. Phyllodineae (Fabaceae: Mimosoideae) based on the trnK intron. Australian Journal of Botany 51(2): 167–177. 10.1071/BT01099 [DOI] [Google Scholar]
- Miller JT, Murphy DJ, Ho SYW, Cantrill DJ, Seigler D. (2013) Comparative dating of Acacia: Combining fossils and multiple phylogenies to infer ages of clades with poor fossil records. Australian Journal of Botany 61(6): 436–445. 10.1071/BT13149 [DOI] [Google Scholar]
- Miller JT, Terra V, Riggins C, Ebinger JE, Seigler DS. (2017) Molecular phylogenetics of Parasenegalia and Pseudosenegalia (Fabaceae: Mimosoideae). Systematic Botany 42(3): 465–469. 10.1600/036364417X696140 [DOI] [Google Scholar]
- Mishler BD, Knerr N, González-Orozco CE, Thornhill AD, Laffan SW, Miller JT. (2014) Phylogenetic measures of biodiversity and neo- and paleo-endemism in Australian Acacia. Nature Communications 5(1): e4473. 10.1038/ncomms5473 [DOI] [PubMed]
- Moore AJ, De Vos JM, Hancock LP, Goolsby E, Edwards EJ. (2018) Targeted enrichment of large gene families for phylogenetic inference: Phylogeny and molecular evolution of photosynthesis genes in the Portullugo clade (Caryophyllales). Systematic Biology 67(3): 367–383. 10.1093/sysbio/syx078 [DOI] [PubMed] [Google Scholar]
- Muñoz-Rodríguez P, Carruthers T, Wood JRI, Williams BRM, Weitemier K, Kronmiller B, Goodwin Z, Sumadijaya A, Anglin NL, Filer D, Harris D, Rausher MD, Kelly S, Liston A, Scotland RW. (2019) A taxonomic monograph of Ipomoea integrated across phylogenetic scales. Nature Plants 5(11): 1136–1144. 10.1038/s41477-019-0535-4 [DOI] [PubMed] [Google Scholar]
- Murphy DJ, Miller JT, Bayer RJ, Ladiges PY. (2003) Molecular phylogeny of AcaciasubgenusPhyllodineae (Mimosoideae: Leguminosae) based on DNA sequences of the internal transcribed spacer region. Australian Systematic Botany 16(1): 19–26. 10.1071/SB01042 [DOI] [Google Scholar]
- Nicholls JA, Pennington RT, Koenen EJM, Hughes CE, Hearn J, Bunnefeld L, Dexter KG, Stone GN, Kidner CA. (2015) Using targeted enrichment of nuclear genes to increase phylogenetic resolution in the neotropical rain forest genus Inga (Leguminosae: Mimosoideae). Frontiers in Plant Science 6. 10.3389/fpls.2015.00710 [DOI] [PMC free article] [PubMed]
- Nielsen I. (1981) Tribe 5. Ingeae. In: Polhill RM, Raven PH. (Eds) Advances in Legume Systematics Part 1.Royal Botanic Gardens, Kew, United Kingdom, 173–190.
- Nielsen I. (1992) Flora Malesiana - series I: Spermatophyta. Volume 11, part 1: Mimosaceae (Leguminosae - Mimosoideae). Rijksherbarium / Hortus Botanicus, Leiden University, Leiden, the Netherlands.
- O’Donnell S, Ringelberg JJ, Lewis GP. (2022) Re-circumscription of the mimosoid genus Entada including new combinations for all species of the phylogenetically nested Elephantorrhiza (Leguminosae, Caesalpinioideae, mimosoid clade). In: Hughes CE, de Queiroz LP, Lewis GP. (Eds) Advances in Legume Systematics 14. Classification of Caesalpinioideae Part 1: New generic delimitations.PhytoKeys 205: 99–146. 10.3897/phytokeys.205.76790 [DOI] [PMC free article] [PubMed]
- Orthia LA, Cook LG, Crisp MD. (2005) Generic delimitation and phylogenetic uncertainty: An example from a group that has undergone an explosive radiation. Australian Systematic Botany 18(1): 41–47. 10.1071/SB04016 [DOI] [Google Scholar]
- Paul SR. (1979) The genus Calliandra (Mimosaceae) in the Indian subcontinent. Feddes Repertorium 90(3): 155–164. 10.1002/fedr.19790900303 [DOI] [Google Scholar]
- Pease JB, Brown JW, Walker JF, Hinchliff CE, Smith SA. (2018) Quartet sampling distinguishes lack of support from conflicting support in the green plant tree of life. American Journal of Botany 105(3): 385–403. 10.1002/ajb2.1016 [DOI] [PubMed] [Google Scholar]
- Pfeil BE, Crisp MD. (2005) What to do with Hibiscus? A proposed nomenclatural resolution for a large and well known genus of Malvaceae and comments on paraphyly. Australian Systematic Botany 18(1): 49–60. 10.1071/SB04024 [DOI] [Google Scholar]
- Phillipson PB, Lewis GP, Andriambololonera S, Rakotonirina N, Thulin M, Wilding N. (2022) Leguminosae (Fabaceae). In: Goodman SM (Ed.) The New Natural History of Madagascar. Princeton University Press, Princeton, New Jersey, United States of America.
- Polhill RM. (1994) Classification of the Leguminosae and complete synopsis of legume genera. In: Bisby FA, Buckingham J, Harborne JB (Eds) Phytochemical Dictionary of the Leguminosae. Chapman and Hall, Cambridge, United Kingdom, xxxv–lvii.
- Polhill RM, Vidal JE. (1981) Caesalpinieae. In: Polhill RM, Raven PH. (Eds) Advances in Legume Systematics Part 1.Royal Botanic Gardens, Kew, United Kingdom, 81–95.
- R Core Team (2022) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/
- Revell LJ. (2012) phytools: An R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution 3(2): 217–223. 10.1111/j.2041-210X.2011.00169.x [DOI] [Google Scholar]
- Ribeiro PG, Luckow M, Lewis GP, Simon MF, Cardoso D, de Souza ÉR, Silva APC, Jesus MC, dos Santos FAR, Azevedo V, de Queiroz LP. (2018) Lachesiodendron, a new monospecific genus segregated from Piptadenia (Leguminosae: Caesalpinioideae: Mimosoid clade): Evidence from morphology and molecules. Taxon 67(1): 37–54. 10.12705/671.3 [DOI] [Google Scholar]
- Ringelberg JJ, Zimmermann NE, Weeks A, Lavin M, Hughes CE. (2020) Biomes as evolutionary arenas: Convergence and conservatism in the trans-continental succulent biome. Global Ecology and Biogeography 29(7): 1100–1113. 10.1111/geb.13089 [DOI] [Google Scholar]
- Ringelberg JJ, Koenen EJM, Sauter B, Aebli A, Rando JG, Iganci JR, de Queiroz LP, Murphy DJ, Gaudeul M, Bruneau A, Luckow M, Lewis GP, Miller JT, Simon MF, Jordão LSB, Morales M, Bailey CD, Nageswara-Rao M, Loiseau O, Pennington RT, Dexter KG, Zimmermann NE, Hughes CE. (2022) Precipitation is the main axis of tropical phylogenetic turnover across space and time. https://www.biorxiv.org/content/10.1101/2022.05.27.493777 [DOI] [PMC free article] [PubMed]
- Robinson J, Harris SA. (2000) A plastid DNA phylogeny of the genus Acacia Miller (Acacieae, Leguminoseae). Botanical Journal of the Linnean Society 132(3): 195–222. 10.1111/j.1095-8339.2000.tb01527.x [DOI] [Google Scholar]
- Rokas A, Carroll SB. (2006) Bushes in the tree of life. PLoS Biology 4(11): 1899–1904. 10.1371/journal.pbio.0040352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JP, Toledo CAP, Lemmon EM, Lemmon AR, Sytsma KJ. (2021) Out of sight, out of mind: widespread nuclear and plastid-nuclear discordance in the flowering plant genus Polemonium (Polemoniaceae) suggests widespread historical gene flow despite limited nuclear signal. Systematic Biology 70(1): 162–180. 10.1093/sysbio/syaa049 [DOI] [PubMed] [Google Scholar]
- Salichos L, Rokas A. (2013) Inferring ancient divergences requires genes with strong phylogenetic signals. Nature 497(7449): 327–331. 10.1038/nature12130 [DOI] [PubMed] [Google Scholar]
- Sandwith NY. (1932) Contributions to the flora of tropical America. XIV. Bulletin of Miscellaneous Information (Royal Botanic Gardens, Kew) 8: 395–406. 10.2307/4113483 [DOI] [Google Scholar]
- Sauter B. (2019) Evolutionary diversification of Mimosa and the Piptadenia Group – An example of phylogenetic biome conservatism. Unpublished MSc thesis, University of Zürich.
- Sayyari E, Mirarab S. (2018) Testing for polytomies in phylogenetic species trees using quartet frequencies. Genes 9(3): e132. 10.3390/genes9030132 [DOI] [PMC free article] [PubMed]
- Scalon VR. (2007) Revisão Taxonômica do gênero Stryphnodendron Mart. (Leguminosae-Mimosoideae). PhD thesis, University of São Paulo.
- Schrire BD, Lavin M, Lewis GP. (2005) Global distribution patterns of the Leguminosae: Insights from recent phylogenies. Biologiske Skrifter 55: 375–422. [Google Scholar]
- Scotland RW, Olmstead RG, Bennett JR. (2003) Phylogeny reconstruction: The role of morphology. Systematic Biology 52(4): 539–548. 10.1080/10635150309309 [DOI] [PubMed] [Google Scholar]
- Seigler DS, Ebinger JE, Miller JT. (2006a) Mariosousa, a new segregate genus from Acacia s.l. (Fabaceae, Mimosoideae) from Central and North America. Novon 16: 413–420. 10.3417/1055-3177(2006)16[413:mansgf]2.0.co;2 [DOI]
- Seigler DS, Ebinger JE, Miller JT. (2006b) The genus Senegalia (Fabaceae: Mimosoideae) from the New World. Phytologia 88: 38–93. 10.5962/bhl.part.17845 [DOI] [Google Scholar]
- Seigler DS, Ebinger JE, Riggins CW, Terra V, Miller JT. (2017) Parasenegalia and Pseudosenegalia (Fabaceae): New genera of the Mimosoideae. Novon 25: 180–205. 10.3417/2015050 [DOI] [Google Scholar]
- Shen X-X, Hittinger CT, Rokas A. (2017) Contentious relationships in phylogenomic studies can be driven by a handful of genes. Nature Ecology & Evolution 1: e0126. 10.1038/s41559-017-0126 [DOI] [PMC free article] [PubMed]
- Simon MF, Grether R, de Queiroz LP, Särkinen TE, Dutra VF, Hughes CE. (2011) The evolutionary history of Mimosa (Leguminosae): Toward a phylogeny of the sensitive plants. American Journal of Botany 98(7): 1201–1221. 10.3732/ajb.1000520 [DOI] [PubMed] [Google Scholar]
- Simon MF, Pastore JFB, Souza AF, Borges LM, Scalon VR, Ribeiro PG, Santos-Silva J, Souza VC, Queiroz LP. (2016) Molecular phylogeny of Stryphnodendron (Mimosoideae, Leguminosae) and generic delimitations in the Piptadenia group. International Journal of Plant Sciences 177(1): 44–59. 10.1086/684077 [DOI] [Google Scholar]
- Sinou C, Cardinal-McTeague W, Bruneau A. (2020) Testing generic limits in Cercidoideae (Leguminosae): Insights from plastid and duplicated nuclear gene sequences. Taxon 69(1): 67–86. 10.1002/tax.12207 [DOI] [Google Scholar]
- Smith SA, Moore MJ, Brown JW, Yang Y. (2015) Analysis of phylogenomic datasets reveals conflict, concordance, and gene duplications with examples from animals and plants. BMC Evolutionary Biology 15(1): e150. 10.1186/s12862-015-0423-0 [DOI] [PMC free article] [PubMed]
- Soares MVB, Guerra E, Morim MP, Iganci JRV. (2021) Reinstatement and recircumscription of Jupunba and Punjuba (Fabaceae) based on phylogenetic evidence. Botanical Journal of the Linnean Society 196(4): 456–479. 10.1093/botlinnean/boab007 [DOI] [Google Scholar]
- Soares M, Koenen E, Iganci J, Morim M. (2022) A new generic circumscription of Hydrochorea (Leguminosae, Caesalpinioideae, mimosoid clade) with an amphi-Atlantic distribution. In: Hughes CE, de Queiroz LP, Lewis GP. (Eds) Advances in Legume Systematics 14. Classification of Caesalpinioideae Part 1: New generic delimitations.PhytoKeys 205: 401–438. 10.3897/phytokeys.205.82775 [DOI] [PMC free article] [PubMed]
- Sprent JI, Ardley J, James EK. (2017) Biogeography of nodulated legumes and their nitrogen-fixing symbionts. New Phytologist 215(1): 40–56. 10.1111/nph.14474 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics (Oxford, England) 30(9): 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo-Cen I, Torke BM, López Contreras JE, Carnevali Fernández-Concha G, Ramírez Morillo I, Can Itza LL, Duno de Stefano R. (2022) Revisiting the phylogeny and taxonomy of the Pithecellobium clade (Leguminosae, Caesalpinioideae) with new generic circumscriptions. In: Hughes CE, de Queiroz LP, Lewis GP. (Eds) Advances in Legume Systematics 14. Classification of Caesalpinioideae Part 1: New generic delimitations.PhytoKeys 205: 279–298. 10.3897/phytokeys.205.82728 [DOI] [PMC free article] [PubMed]
- Terra V, Garcia FCP, Queiroz LP de. (2017) Phylogenetic relationships in Senegalia (Leguminosae-Mimosoideae) emphasizing the South American lineages. Systematic Botany 42(3): 458–464. 10.1600/036364417X696122 [DOI] [Google Scholar]
- Terra V, Ringelberg JJ, Maslin B, Koenen EJM, Ebinger J, Seigler D, Hughes CE. (2022) Dilemmas in generic delimitation of Senegalia and allies (Caesalpinioideae, mimosoid clade): how to reconcile phylogenomic evidence with morphology and taxonomy? In: Hughes CE, de Queiroz LP, Lewis GP. (Eds) Advances in Legume Systematics 14. Classification of Caesalpinioideae Part 1: New generic delimitations.PhytoKeys 205: 261–278. 10.3897/phytokeys.205.79378 [DOI] [PMC free article] [PubMed]
- Thielsch A, Knell A, Mohammadyari A, Petrusek A, Schwenk K. (2017) Divergent clades or cryptic species? Mito-nuclear discordance in a Daphnia species complex. BMC Evolutionary Biology 17(1): 1–9. 10.1186/s12862-017-1070-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulasne ML-R. (1844) Legumineuses arborescentes de L’Amerique du Sud - Dimorphandra. Archives du Muséum d’Histoire Naturelle Paris 4: 182–198. [Google Scholar]
- Turland NJ, Wiersema JH, Barrie FR, Greuter W, Hawksworth DL, Herendeen PS, Knapp S, Kusber W-H, Li D-Z, Marhold K, May TW, McNeill J, Monro AM, Price MJ, Smith GF [Eds] (2018) International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code) adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017. Regnum Vegetabile 159. Koeltz Botanical Books, Glashütten, Germany.
- van Steenis CGGJ. (1975) A review of the genus Sympetalandra Stapf and its position in Caesalpinioideae. Blumea 22: 159–167. [Google Scholar]
- Villiers J-F. (1984) Le genre Calpocalyx (Legiminosae, Mimosoideae) en Afrique. Adansonia 3: 297–311. 10.1017/CBO9781107415324.004 [DOI] [Google Scholar]
- Villiers J-F. (1994) Alantsilodendron Villiers, genre nouveau de Leguminosae-Mimosoideae de Madagascar. Bulletin du Muséum National d’Histoire Naturelle Paris, 4e sér., Section B. Adansonia 16: 65–70. [Google Scholar]
- Wang N, Yang Y, Moore MJ, Brockington SF, Walker JF, Brown JW, Liang B, Feng T, Edwards C, Mikenas J, Olivieri J, Hutchison V, Timoneda A, Stoughton T, Puente R, Majure LC, Eggli U, Smith SA. (2019) Evolution of Portulacineae marked by gene tree conflict and gene family expansion associated with adaptation to harsh environments. Molecular Biology and Evolution 36(1): 112–126. 10.1093/molbev/msy200 [DOI] [PubMed] [Google Scholar]
- Wight R, Walker-Arnott GA. (1834) Prodromus Florae Peninsulae Indiae Orientalis, volume 1. Parbury, Allen, & Co, London, United Kingdom.
- Willdenow CL. (1805) Species Plantarum. Editio quarta. Tomus 4, pars 1. G.C. Nauk, Berolini, 629 pp. [Google Scholar]
- Yang Y, Smith SA. (2014) Orthology inference in nonmodel organisms using transcriptomes and low-coverage genomes: improving accuracy and matrix occupancy for phylogenomics. Molecular Biology and Evolution 31(11): 3081–3092. 10.1093/molbev/msu245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Su D, Chang X, Foster CSP, Sun L, Huang C-H, Zhou X, Zeng L, Ma H, Zhong B. (2020) Phylogenomic insights into deep phylogeny of Angiosperms based on broad nuclear gene sampling. Plant Communications 1(2): e100027. 10.1016/j.xplc.2020.100027 [DOI] [PMC free article] [PubMed]
- Zhang C, Rabiee M, Sayyari E, Mirarab S. (2018) ASTRAL-III: Polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics 19(S6): 15–30. 10.1186/s12859-018-2129-y [DOI] [PMC free article] [PubMed]
- Zhang R, Wang YH, Jin JJ, Stull GW, Bruneau A, Cardoso D, De Queiroz LP, Moore MJ, Zhang SD, Chen SY, Wang J, Li DZ, Yi TS. (2020) Exploration of plastid phylogenomic conflict yields new insights into the deep relationships of Leguminosae. Systematic Biology 69(4): 613–622. 10.1093/sysbio/syaa013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zhang R, Jiang K, Qi J, Hu Y, Guo J, Zhu R, Zhang T, Egan AN, Yi T-S, Huang C-H, Ma H. (2021) Nuclear phylotranscriptomics/phylogenomics support numerous polyploidization events and hypotheses for the evolution of rhizobial nitrogen-fixing symbiosis in Fabaceae. Molecular Plant 14(5): 748–773. 10.1016/j.molp.2021.02.006 [DOI] [PubMed] [Google Scholar]
- Zhou X, Lutteropp S, Czech L, Stamatakis A, Von Looz M, Rokas A. (2020) Quartet-based computations of internode certainty provide robust measures of phylogenetic incongruence. Systematic Biology 69(2): 308–324. 10.1093/sysbio/syz058 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Jens J. Ringelberg, Erik J.M. Koenen, João R. Iganci, Luciano P. de Queiroz, Daniel J. Murphy, Myriam Gaudeul, Anne Bruneau, Melissa Luckow, Gwilym P. Lewis, Colin E. Hughes
Data type
excel file.
Explanation note
Samples included in this study.
Table S2
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Jens J. Ringelberg, Erik J.M. Koenen, João R. Iganci, Luciano P. de Queiroz, Daniel J. Murphy, Myriam Gaudeul, Anne Bruneau, Melissa Luckow, Gwilym P. Lewis, Colin E. Hughes
Data type
excel file.
Explanation note
Trait data used for character evolution analyses.
Figure S1
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Jens J. Ringelberg, Erik J.M. Koenen, João R. Iganci, Luciano P. de Queiroz, Daniel J. Murphy, Myriam Gaudeul, Anne Bruneau, Melissa Luckow, Gwilym P. Lewis, Colin E. Hughes
Data type
Pdf file.
Explanation note
Chloroplast phylogeny of Caesalpinioideae. Only bootstrap support values lower than 100% are shown.
Supplementary tree file
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Jens J. Ringelberg, Erik J.M. Koenen, João R. Iganci, Luciano P. de Queiroz, Daniel J. Murphy, Myriam Gaudeul, Anne Bruneau, Melissa Luckow, Gwilym P. Lewis, Colin E. Hughes
Data type
Tree file (Newick format).
Explanation note
Tree file of the ASTRAL phylogeny based on the single-copy genes (depicted in Figs 2–12), in which taxon labels have been updated to reflect taxonomic changes made in all the entries in Advances in Legume Systematics 14 Part 1.