Abstract
The morphologically variable genus Archidendron is the second largest mimosoid legume genus from the Indomalayan-Australasian region, yet it has not been well represented in phylogenetic studies. Phylogenies that have included multiple representatives of Archidendron suggest it may not be monophyletic, and the same applies to Archidendropsis, another understudied genus of the Archidendron clade. The most comprehensive phylogeny of Archidendron and Archidendropsis to date is presented, based on four nuclear markers (ITS, ETS, SHMT and RBPCO). Exemplars from all genera of the wider Archidendron clade are sampled, including representatives of all series within Archidendron and the two subgenera of Archidendropsis. Our results confirm that Archidendron and Archidendropsis are not monophyletic. Within Archidendron, only one series (ser. Ptenopae) is resolved as monophyletic and species of Archidendron are divided into two primarily geographic lineages. One clade is distributed in western Malesia and mainland Asia and includes most representatives of series Clypeariae, while the other is mostly restricted to eastern Malesia and Australia and includes representatives of the seven other series plus two samples of series Clypeariae. No taxonomic changes are made for Archidendron due to the high level of topological uncertainty and the lack of discrete macromorphological characters separating these two lineages. Each of the two subgenera of Archidendropsis is monophyletic but they are not closely related. A new genus endemic to Queensland (Australia), Heliodendron Gill.K. Br. & Bayly, gen. nov., is described for the former Archidendropsissubg.Basaltica, and combinations for its three species are proposed: Heliodendronbasalticum (F. Muell.) Gill.K. Br. & Bayly, comb. nov., Heliodendronthozetianum (F. Muell.) Gill.K. Br. & Bayly, comb. nov., and Heliodendronxanthoxylon (C.T. White & W.D. Francis) Gill.K. Br. & Bayly, comb. nov.
Keywords: Fabaceae, ingoid clade, legumes, low copy nuclear gene, Malesia, phylogeny, targeted amplicon sequencing
Introduction
The classification of mimosoid legumes has been significantly transformed in the past 20 years since the first comprehensive molecular phylogeny of the then subfamily Mimosoideae (Luckow et al. 2003). Understanding of relationships within the mimosoid legumes has improved through studies at generic, regional, alliance, subfamilial and familial levels (see references in Legume Phylogeny Working Group 2017; Koenen et al. 2020; Ringelberg et al. 2022). In the comprehensive phylogeny and revision of the legume family (Leguminosae or Fabaceae), the mimosoid legumes formed a clade nested within the re-circumscribed subfamily Caesalpinioideae (Legume Phylogeny Working Group 2017). Recent phylogenomic data have sufficiently enhanced resolution to enable recognition of several clades within subfamily Caesalpinioideae, including the mimosoid, core mimosoid and ingoid clades (Koenen et al. 2020; Ringelberg et al. 2022). However, within these clades some large genera, such as Archidendron F. Muell. and allies have remained under-studied relative to Acacia Mill. s.l. and many Neotropical ingoid genera and groups (e.g. Murphy et al. 2010; de Souza et al. 2013; Iganci et al. 2016; Miller et al. 2017; Ferm et al. 2019; Comben et al. 2020).
The two largest mimosoid genera from the Indomalayan-Australasian region are Acacia and Archidendron. These are placed in the Archidendron clade (sensu Koenen et al. 2020), along with Archidendropsis I.C. Nielsen, Falcataria (I.C. Nielsen) Barneby & J.W. Grimes, Pararchidendron I.C. Nielsen, Paraserianthes I.C. Nielsen, Serianthes Benth. and Wallaceodendron Koord. The Archidendron clade is biogeographically distinct within the mimosoid legumes, being primarily restricted to the Indomalayan and Australasian regions, and has been given several names over the years to reflect this: the Australian & SE Asian Ingeae clade (Brown et al. 2008) and the Australo-Malesian mimosoids (Brown et al. 2011). Within the Archidendron clade, Pararchidendron, Paraserianthes and Wallaceodendron are monotypic, and three of the other five genera (Acacia s.s., Falcataria, and Serianthes) are well documented as monophyletic based on morphological and genetic data (Chappill and Maslin 1995; Miller and Bayer 2001; Luckow et al. 2003; Brown et al. 2008, 2011; Murphy et al. 2010; Demeulenaere et al. 2022; Ringelberg et al. 2022). However, Archidendron has been suggested to be paraphyletic (Brown et al. 2008, 2011; Iganci et al. 2016; Demeulenaere et al. 2022; Ringelberg et al. 2022), as has Archidendropsis (Demeulenaere et al. 2022; Ringelberg et al. 2022).
Archidendron is the second largest genus in this clade after Acacia, with 99 described species and an additional 20 putative species that are poorly known due to limited collections or destroyed types (Nielsen et al. 1984b; Cowan 1998; Wu and Nielsen 2010; Dash and Sanjappa 2011). They are small to medium-sized trees found in lowland and montane tropical and subtropical rainforests of the Australo-Malesian and Pacific regions, distributed from Kerala (southern India) and Sri Lanka in the west, to the Solomon Islands in the east; and from Taiwan and the Ryukyu Islands in the north, to Australia in the south (Fig. 1; Nielsen et al. 1984b, 1984a). In the 1970s and 1980s, an extensive revision of the Australo-Malesian and Pacific Ingeae was undertaken (Nielsen 1979, 1981, 1982; Nielsen et al. 1983b, 1983a, 1984b) and Archidendron was expanded based on evidence from wood, pollen, seed and inflorescence characteristics to include species previously referred by Kostermans (1954) to the genera Abarema Pittier, Cylindrokelupha Kosterm., Morolobium Kosterm., Paralbizzia Kosterm., Zygia P. Browne, and by Bentham (1875) to Pithecellobiumsect.Clypearia sensu Benth. (Baretta-Kuipers 1981; Nielsen et al. 1984b; Nielsen 1992). Archidendron now includes unarmed trees or shrubs with bipinnate leaves, mostly opposite leaflets, extrafloral nectaries, and wood anatomy of strictly uniseriate rays and abundant parenchyma with a banded distribution (Nielsen et al. 1984b).
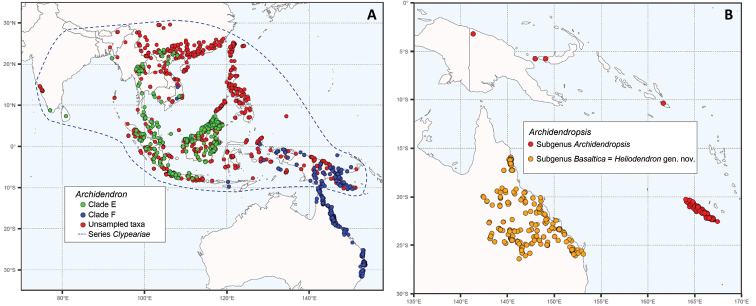
Figure 1.
Distribution maps of the genera Archidendron and Archidendropsis. The maps are based on quality-controlled species-level digitised herbarium specimens from GBIF (www.gbif.org) (Ringelberg et al. 2022). Maps were created using R packages ggplot2 (Wickham 2016), sf (Pebesma 2018), and rnaturalearth (South 2017) AArchidendron. Species distributions are coloured according to the ncDNA phylogeny clades (Fig. 2) except for A.clypearia: Clade E (Clypeariae clade) = green dots; clade F (Archidendron s.s. clade) = blue dots; species not sampled for the phylogeny = red dots. Archidendronclypearia is widespread and falls in both clades E and F, so for this species locations of samples in the ncDNA phylogeny are coloured according to their clade and all other records of this species are coloured red. The overall distribution of series Clypeariae is shown by a blue dashed line BArchidendropsis. All species that belong to subg. Archidendropsis are coloured red and those in subg. Basaltica (= Heliodendron gen. nov.) are coloured orange.
Archidendron is morphologically variable especially in leaf, inflorescence, flower, and pod characteristics, and has been divided into eight series (Nielsen et al. 1984b): Clypeariae (Benth.) I.C. Nielsen, Archidendron, Calycinae I.C. Nielsen, Bellae I.C. Nielsen, Ptenopae I.C. Nielsen, Pendulosae (Mohlenbr.) I.C. Nielsen, Stipulatae (Mohlenbr.) I.C. Nielsen and Morolobiae (Kosterm.) I.C. Nielsen. The largest series, Clypeariae (ca. 51 species) is distributed in mainland southeast Asia, western Malesia, and the Philippines, with only a few species found further east (Fig. 1A). This series is well defined by the absence of stipules and flowers that generally have one carpel per ovary that is often stipitate (Nielsen et al. 1984b). The second largest series, Archidendron (ca. 15 species), is found in eastern Malesia and Australia and is defined by the presence of stipules and stipular glands. Four of the series are largely confined to the island of New Guinea (Nielsen et al. 1984b): series Calycinae (3 species) with strongly ribbed inflated calyces, cauliflorous racemes and sessile ovaries; series Bellae (4 species) with large woody pods without overgrown seeds and cauliflorous paniculate inflorescences; series Ptenopae (2 species), which is defined by the presence of two-winged rachises and pinnae; series Pendulosae (3 species) have inflorescences with lax racemes (Nielsen et al. 1984a). Series Stipulatae (ca. 14 species) are found in New Guinea, the Moluccas, and Queensland (Australia) and have floral bracts with extra floral nectaries, stipular glands and cauliflorous branched racemes (Nielsen et al. 1984b). The three species of series Morolobiae have unifoliolate pinnae, and racemose inflorescences with flowers with single, sessile ovaries, and are disjunctly distributed: A.monopterum (Kosterm.) I.C. Nielsen in Halmahera (North Maluku Islands, Indonesia), A.whitei I.C. Nielsen in northern Queensland (Australia) and A.muellerianum (Maiden & R.T. Baker) I.C. Nielsen in northern New South Wales (Australia) (Nielsen et al. 1984b).
Prior to resolution of the Archidendron clade, the genus Archidendron was suggested to be related to taxa of the Inga-alliance (Barneby and Grimes 1996; Lewis and Rico Arce 2005) or to other Old World genera, such as Archidendropsis, Falcataria, Pararchidendron, Paraserianthes and Serianthes (Baretta-Kuipers 1981; Nielsen et al. 1984a; Nielsen 1992). Archidendron has not been well represented in molecular phylogenies to date with only ten of the 99 species and four of the eight series (Archidendron, Clypeariae, Morolobiae and Ptenopae) included in any one study. In all studies, samples of series Clypeariae are placed distantly from the other series (Brown et al. 2008, 2011; Iganci et al. 2016; Koenen et al. 2020; Demeulenaere et al. 2022; Ringelberg et al. 2022).
The genus Archidendropsis includes 14 species from New Caledonia, the Solomon Islands, New Britain, Papua New Guinea and Australia (Fig. 1B), with all species endemic to their respective region (Nielsen et al. 1983a). Species of Archidendropsis have winged, thin-walled seeds lacking a pleurogram (a mark or depression on both sides of the seed coat; Rodrigues-Junior et al. 2021) and are placed in two subgenera based on pollen and inflorescence characteristics. Species of subgenus Basaltica I.C. Nielsen are restricted to Australia, have smaller polyads (55–60 μm) and globular inflorescences, while species of subgenus Archidendropsis are not found in Australia, have larger polyads (80–120 μm) and flowers arranged in spicate racemes. Like Archidendron, Archidendropsis has been poorly represented in molecular phylogenies with only one or two of the 14 species included in any one study (Brown et al. 2008, 2011; Ferm et al. 2019; Koenen et al. 2020; Demeulenaere et al. 2022; Ringelberg et al. 2022). Only two studies have included representatives of each of the subgenera and in both, Archidendropsis is not resolved as monophyletic (Demeulenaere et al. 2022; Ringelberg et al. 2022).
This study aims to test the monophyly of the genera Archidendron and Archidendropsis and investigate phylogenetic relationships within the large genus Archidendron to test the monophyly of its infrageneric series.
Materials and methods
Taxon sampling and DNA isolation
A total of 87 accessions were sampled, representing 43 species of Archidendron (68 accessions), five species of Archidendropsis (six accessions) and nine species (11 accessions) of the other genera in the Archidendron clade; two species of Old World Albizia Durazz. were included as outgroups (Table 1). In total 43% of the species of Archidendron were sampled including representatives of all eight series. Both subgenera of Archidendropsis were sampled covering 36% of species in the genus. Samples were collected in the field and from herbarium specimens sourced from AAU, BISH, BRI, CANB, CNS, KEP, KUN, L, NY, MEL and MELU (herbarium codes as per Thiers, updated continuously).
Table 1.
Linked data table of specimens sampled for phylogeny. Specimen accession number linking herbarium specimen to sample ID, taxon name with authorities, locality information and geocode (where available) as provided on the specimen/database. GenBank numbers are provided for each marker and where multiple alleles were identified for a specimen, the two GenBank numbers are separated by a semi colon. If the marker was not successfully sequenced for a particular specimen, then the GenBank field is left blank.
| Preserved specimen | Associated sequences | Taxon name/MOTU | Sample ID | Location | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Specimen code (InstCode and/or CollCode + Catalogue #) | SHMT | RBPCO | ITS | ETS | trnK | trnV | psbD | Geolocation name / locality | GPS Coordinates | ||
| MEL 2294706A | OM286906 | OM286992 | ON013654 | Acaciabaueri Benth. | Z176 | Great Sandy National Park, Fraser Island, Woralie track to Moon Point. Queensland, Australia | 153°11'55"E, 25°11'38"S | ||||
| MELU GB309b | OM984488 | OM390190; OM390191 | OM286907 | OM286993 | ON013655 | ON101510 | OM984574 | Acaciamyrtifolia (Sm.) Willd. | JA150 | 0.7km north of Playford Highway on Snug Bay Rd, Kangaroo Island, South Australia | 136°52'51.8"E, 35°46'30.2"S |
| CANB 864530.1 | OM984489 | OM286908 | OM286994 | ON013656 | ON101511 | OM984575 | Albizialebbeck (L.) Benth. | JA137 | Alva, NE of Ayr, Queensland, Australia | 147°28'52"E, 19°27'11"S | |
| MEL 2391890A | OM984490 | OM286909 | OM286995 | ON013657 | ON101512 | OM984576 | Albiziaretusa Benth. | Z106 | Atherton Arboretum. Tag #96. Queensland, Australia | 145°29'8.6"E, 17°15'31.4"S | |
| KUN0599506 | OM984491 | OM286910 | OM286996 | ON013658 | ON101513 | OM984577 | Archidendronalternifoliolatum (T.L.Wu) I.C.Nielsen | JA25 | China | 100.85°E, 24.5667°N | |
| BRI AQ0380081 | OM984492; OM984493 | OM286911 | OM286997 | ON013659 | ON101514 | Archidendronarborescens (Kosterm.) I.C.Nielsen | JA36 | Papua New Guinea, Western Fly; Kwinja Lakes area of the Middle Fly River | 141°41'33.987"E, 7°45'24.772S | ||
| KUN0599551 | OM984494; OM984495 | OM286912 | OM286998 | ON013660 | ON101515 | Archidendronbalansae (Oliv.) I.C.Nielsen | JA26 | China | |||
| AAU D.McKey92-9 | OM984496; OM984497 | OM390192 | OM286913 | OM286999 | ON013661 | ON101516 | OM984578 | Archidendronbigeminum (L.) I.C.Nielsen | JA14 | Sinharaja Forest, SW Sri Lanka | 80°35'23"E, 6°21'17"N |
| AAU Balgooy6063 | OM984498; OM984499 | OM390193 | OM286914 | OM287000 | ON013662 | ON101517 | OM984579 | Archidendronborneense (Benth.) I.C.Nielsen | JA70 | Tanah Merah, Kalimantan Timur | 117°‘E, 1°‘S |
| KEP FRI53789 | OM984500; OM984501 | OM286915 | OM287001 | ON013663 | ON101518 | OM984580 | Archidendronbubalinum (Jack) I.C.Nielsen | JA22 | Pahang, Temerloh, Tasik Bera, Kg. Patihir, Malaysia | 102.4167°E, 3.8167°N | |
| CANB 730419.1 | OM390194; OM390195 | OM286916 | OM287002 | ON013664 | ON101519 | Archidendroncalliandrum de Wit | JA109 | Ambunti District, Waskut Hills, spur ridge NW of Musapien bivouac. East Sepik, PNG | 142°43'55"E, 4°10'36"S | ||
| CANB 211609.1 | OM984502 | OM286917 | OM287003 | ON013665 | ON101520 | OM984581 | Archidendroncalycinum Pulle | JA129 | Saw Mountains, near junction of Tauri and Kapau Rivers. Gulf Province, PNG | 146°8'E, 7°47'S | |
| AAU L.Averyanov4481 | OM984503 | OM286918 | OM287004 | ON013666 | ON101521 | OM984582 | Archidendronchevalieri (Kosterm.) I.C.Nielsen | JA71 | Bi Dup ridge, Vietnam | 108°39'E, 12°6'N | |
| AAU I.Nielsen26 | OM984504 | OM286919 | OM287005 | ON013667 | ON101522 | OM984583 | Archidendronclypearia (Jack) I.C.Nielsen | JA16 | Gunung Mulu National Park, Sarawak | 114°55'E, 4°05'N | |
| AAU H.M.Christensen38 | OM984505 | OM286920 | OM287006 | ON013668 | ON101523 | OM984584 | Archidendronclypearia (Jack) I.C.Nielsen | JA05 | Pa Dalih area, Sarawak | 115°50'E, 3°40'N | |
| AAU L.AveryanovVH3188 | OM984506 | OM286921 | OM287007 | ON013669 | ON101524 | OM984585 | Archidendronclypearia (Jack) I.C.Nielsen | JA15 | Bi Dup mountain system, Vietnam | 108°39'E, 12°8'N | |
| CANB 525617.1 | OM984507 | OM390196; OM390197 | OM286922 | OM287008 | ON013670 | ON101525 | OM984586 | Archidendronclypearia (Jack) I.C.Nielsen | JA95 | East branch of the Avi Avi River. Gulf Province, PNG | 146°30'E, 7°44'S |
| AAU AmbriW838 | OM286923 | OM287009 | ON013671 | ON101526 | OM984587 | Archidendroncockburnii I.C.Nielsen | JA17 | Wanariset research area, Kalimantan Timur | 117°‘E, 1°‘S | ||
| NY03986843 | OM390198 | OM286924 | OM287010 | ON013672 | ON101527 | OM984588 | Archidendroncontortum (Mart.) I.C.Nielsen | T97 | Near Kuala Lumpur, Malaysia | ||
| BRI AQ0380332 | OM286925 | OM287011 | Archidendronforbesii Baker f. | JA38 | Papua New Guinea, Central; Subitana, Sogeri sub-dist., Central, Papua | 147°31'E, 9°25'S | |||||
| BISH752370 | OM984509 | OM390199 | OM286926 | OM287012 | ON013673 | ON101528 | OM984589 | Archidendronglabrum (K.Schum.) Lauterb. & K.Schum. | JA04 | Siboma, Sayama, track along the ridgeline S from Camp 1. PNG | 147.298°E, 7.52857°S |
| BISH763497 | OM984510 | OM390200 | OM286927 | OM287013 | ON013674 | ON101529 | OM984590 | Archidendronglabrum (K.Schum.) Lauterb. & K.Schum. | JA115 | Morobe Province; Oomsis, behind PNG Forestry station. | 146.821°E, 6.71325°S |
| BRI AQ0380375 | OM286928 | OM287014 | ON013675 | ON101530 | OM984591 | Archidendronglandulosum Mohlenbr. ex Verdc. | JA39 | Brown River F.R. Central Province, PNG | 147°10'33.78"E, 9°30'24.60"S | ||
| AAU J.F.Maxwell82-141 | OM984511 | OM286929 | OM287015 | ON013676 | ON101531 | OM984592 | Archidendronglobosum (Blume) I.C.Nielsen | JA20 | Near Bukit Kallang, Singapore | ||
| AAU Bjornland445 | OM984512 | OM286930 | OM287016 | ON101532 | OM984593 | Archidendronglomeriflorum (Kurz) I.C.Nielsen | JA10 | Chiang Mai: Amphoe Muang, Mae Rim, Thailand | |||
| CANB 544379.1 | OM984516 | OM286933 | OM287018 | ON013679 | ON101535 | OM984596 | Archidendrongrandiflorum (Sol. ex Benth.) I.C.Nielsen | JA100 | Gabba Island, Torres Strait. Queensland, Australia | 142°38'22"E, 9°46'8"S | |
| BRI AQ0814833 | OM984513; OM984514 | OM286931 | OM287017 | ON013677 | ON101533 | OM984594 | Archidendrongrandiflorum (Sol. ex Benth.) I.C.Nielsen | JA42 | Curramore Sanctuary Nature Reserve, 14km NW of Maleny. Queensland, Australia | 152°4'05"E, 26°41'43"S | |
| CNS 131336.1 | OM984515 | OM286932 | ON013678 | ON101534 | OM984595 | Archidendrongrandiflorum (Sol. ex Benth.) I.C.Nielsen | JA43 | Mt Lewis, Carbine Tableland. Queensland, Australia | 145°16'E, 16°31'S | ||
| MEL 2391892A | OM984517; OM984518 | OM390201 | OM286934 | OM287019 | ON013680 | ON101536 | OM984597 | Archidendrongrandiflorum (Sol. ex Benth.) I.C.Nielsen | Z109 | Atherton Arboretum. Tag #846. Queensland, Australia | 145°29'8.6"E, 17°15'31.4"S |
| AAU Kostermans22121 | OM286935 | OM287020 | ON013681 | ON101537 | Archidendronharmsii Malm | JA74 | Mbengen, West Flores | ||||
| AAU H.M.Christensen279 | OM984519 | OM390202; OM390203 | OM286936 | OM287021 | ON013682 | ON101538 | OM984598 | Archidendronhavilandii (Ridl.) I.C.Nielsen | JA75 | Pa Dalih area, Sarawak | 115°50'E, 3°40'N |
| CANB 596487.1 | OM984521 | OM390206 | OM286939 | OM287024 | ON013685 | ON101541 | OM984600 | Archidendronhendersonii (F.Muell.) I.C.Nielsen | JA103 | Greenfield Road, Lennox Head. New South Wales, Australia | 153°36'E, 28°49'’S |
| MEL 2293327A | OM984520 | OM390204; OM390205 | OM286937 | OM287022 | ON013683 | ON101539 | OM984599 | Archidendronhendersonii (F.Muell.) I.C.Nielsen | JA44 | Brandy Creek Road, 9 km S of Airlie Beach. Queensland, Australia | 148°43'15"E, 20°21'2"S |
| QRS 18805.2 | OM286938 | OM287023 | ON013684 | ON101540 | Archidendronhendersonii (F.Muell.) I.C.Nielsen | JA45 | Between Starcke homestead and Starcke River. Queensland, Australia | 145°5'E, 14°55'S | |||
| MEL 2391969A | OM984522 | OM390207; OM390208 | OM286940 | OM287025 | ON013686 | ON101542 | OM984601 | Archidendronhendersonii (F.Muell.) I.C.Nielsen | Z114 | Cairns, cultivated in garden. Queensland, Australia | 145°46'15"E, 16°55'13"S |
| QRS 117169.1 | OM984523 | OM390209 | OM286941 | OM287026 | ON013687 | ON101543 | OM984602 | Archidendronhirsutum I.C.Nielsen | JA46 | Claudie River. Queensland, Australia | 143°15'E, 12°44'S |
| CNS 142441.1 | OM984524 | OM390210 | OM286942 | OM287027 | ON013688 | ON101544 | OM984603 | Archidendronhirsutum I.C.Nielsen | JA86 | Umagico, Cape York. Queensland, Australia | 142°21'E, 10°53'19"S |
| MEL 2391887A | OM984525 | OM390211 | OM286943 | OM287028 | ON013689 | ON101545 | OM984604 | Archidendronhirsutum I.C.Nielsen | Z113 | Atherton Arboretum. Tag #482. Queensland, Australia | 145°29'8.6"E, 17°15'31.4"S |
| BISH760310 | OM984526 | OM390212 | OM286944 | OM287029 | ON013690 | ON101546 | OM984605 | Archidendronhispidum (Mohlenbr.) Verdc. | JA02 | Northern Province; Sibium Mountains; W of Akupe Camp, along Afase River. PNG | 148.269°E, 9.28974°S |
| AAU R.Geesink7254 | OM984527 | OM286945 | OM287030 | ON013691 | ON101547 | OM984606 | Archidendronjiringa (Jack) I.C.Nielsen | JA12 | Kao Chong Botanical Garden, Thailand | 99°45'E, 7°40'N | |
| BRI AQ0738090 | OM984528; OM984529 | OM390213 | OM286946 | OM287031 | ON013692 | ON101548 | OM984607 | Archidendronkanisii R.S.Cowan | JA47 | Oliver Creek. Queensland, Australia | 145°26'E, 16°8'S |
| MELUD113392a | OM984530 | OM286947 | OM287032 | ON013693 | ON101549 | OM984608 | Archidendronkanisii R.S.Cowan | JA65 | Shore of creek, end of Stonewood Road, Queensland, Australia | 145.40497°E, 16.16685°S | |
| MELUD113385a | OM984531 | OM390214 | OM286948 | OM287033 | ON013694 | ON101550 | OM984609 | Archidendronkanisii R.S.Cowan | JA66 | Shore of creek, end of Stonewood Road, Queensland, Australia | 145.40497°E, 16.16685°S |
| BRI AQ0733240 | OM984532 | Archidendronkanisii R.S.Cowan | Z49 | NPR133, Daintree, Oliver Creek, Queensland, Australia | 145°26'29.997"E, 16°8'11.708"S | ||||||
| AAU I.Cowley110 | OM286949 | OM287034 | ON013695 | ON101551 | OM984610 | Archidendronkinabaluense (Kosterm.) I.C.Nielsen | JA76 | Melilas. Ulu Belait, Brunei | |||
| AAU H.M.Christensen1719 | OM286950 | OM287035 | ON013696 | ON101552 | OM984611 | Archidendronkunstleri (Prain) I.C.Nielsen | JA07 | near Nanga Sumpa, Sarawak | 112°10'E, 1°20'N | ||
| KUN 0599659 | OM984533; OM984534 | OM390215 | OM286951 | OM287036 | ON013697 | ON101553 | OM984612 | Archidendronlaoticum (Gagnep.) I.C.Nielsen | JA77 | ||
| BRI AQ0835639 | OM984535 | OM286952 | OM287037 | ON013698 | ON101554 | Archidendronlovelliae (F.M.Bailey) I.C.Nielsen | JA48 | Great Sandy National Park; Cooloola Section, Freshwater Road. Queensland, Australia. | 153°6'52"E, 25°57'01S | ||
| BRI AQ0636343 | OM390216; OM390217 | OM286953 | OM287038 | ON013699 | ON101555 | OM984613 | Archidendronlovelliae (F.M.Bailey) I.C.Nielsen | Z112 | Harry’s Hut Road, Cooloola National Park.Queensland, Australia | 153°03'E, 25°26'S | |
| MEL 2034578A | OM984536 | OM390218; OM390219 | OM286954 | OM287039 | ON013700 | ON101556 | OM984614 | Archidendronlucyi F.Muell. | JA49 | Indooroopilly, cultivated. Queensland, Australia | |
| MELUD113387a | OM984537 | OM286955 | OM287040 | ON013701 | ON101557 | OM984615 | Archidendronlucyi F.Muell. | JA62 | Lake Road near Cairns, Queensland, Australia | 145.6693°E, 16.875165°S | |
| MELUD113393a | OM984538 | OM286956 | OM287041 | ON013702 | ON101558 | OM984616 | Archidendronlucyi F.Muell. | JA63 | Lake Road near Cairns, Queensland, Australia | 145.6693°E, 16.875165°S | |
| MELUD113391a | OM984539 | OM286957 | OM287042 | ON013703 | ON101559 | OM984617 | Archidendronlucyi F.Muell. | JA68 | Cape Tribulation Road, adjacent to Coconut Beach resort, Queensland, Australia | 145.45726°E, 16.11345°S | |
| MEL 2391968A | OM984540 | OM390220 | OM286958 | OM287043 | ON013704 | ON101560 | OM984618 | Archidendronlucyi F.Muell. | Z108 | Cairns, cultivated in garden. Queensland, Australia | 145°46'15"E, 16°55'13"S |
| BISH760584 | OM984541 | OM286959 | OM287044 | ON013705 | ON101561 | OM984619 | Archidendronmegaphyllum Merr. & L.M.Perry | JA03 | Central Province; Mt Gerebu, trail towards summit ridge. PNG | 147.646°E, 9.46595°S | |
| AAU H.M.Christensen1282 | OM286960 | OM287045 | ON101562 | OM984620 | Archidendronmicrocarpum (Benth.) I.C.Nielsen | JA06 | Near Sumpa, Sarawak. | 112°10'E, 1°20'N | |||
| BRI AQ0499073 | OM984544 | OM390221; OM390222 | OM286962 | OM287047 | ON013707 | ON101564 | OM984622 | Archidendronmuellerianum (Maiden & R.T.Baker) I.C.Nielsen | JA112 | Big Scrub Flora Reserve, NNE of Lismore, New South Wales, Australia | 153°19'44.880"E, 28°38'18.228"S |
| BRI AQ0763292 | OM984542; OM984543 | OM286961 | OM287046 | ON013706 | ON101563 | OM984621 | Archidendronmuellerianum (Maiden & R.T.Baker) I.C.Nielsen | JA50 | Tallebudgera Creek Road, reveg site. Queensland, Australia | 153°21'57"E, 28°10'37"S | |
| BISH752405 | OM984545; OM984546 | OM390223 | OM286963 | OM287048 | ON013708 | ON101565 | OM984623 | Archidendronparviflorum Pulle | JA01 | Morobe Province; Siboma, Sayama, above Sayama Creek, to E Camp 1. PNG | 147.302°E, 7.52557°S |
| MEL 2074350A | OM286964 | OM287049 | ON013709 | Archidendronpellitum (Gagnep.) I.C.Nielsen | JA34 | N. de Dalat, prov. Ht. Donnai. Indochina: Annam. Vietnam | 108°27'E, 11°57'N | ||||
| Bell Museum 913425 (WP-3A0575) | OM984547; OM984548 | OM390224 | OM286965 | OM287050 | ON013710 | ON101566 | OM984624 | Archidendronptenopum Verdc. | JA116 | Wanang village, Madang, PNG | 145°10.631'E, 5°14.238'S |
| AAU C.Charoenphol5025 | OM984549; OM984550 | OM286966 | OM287051 | ON013711 | ON101567 | OM984625 | Archidendronquocense (Pierre) I.C.Nielsen | JA13 | Ko Rang Yai, Thailand | 102°23'E, 11°48'N | |
| MEL 2391884A | OM984557 | OM390228 | OM286969 | OM287053 | ON013717 | ON101573 | OM984630 | Archidendronramiflorum (F.Muell) Kosterm. | Z111 | Atherton Arboretum. Tag #1652. Queensland, Australia | 145°29'8.6"E, 17°15'31.4"S |
| MELUD113388a | OM984551 | OM286967 | OM287052 | ON013712 | ON101568 | OM984626 | Archidendronramiflorum (F.Muell) Kosterm. | JA67 | Regeneration plot, Daintree Rainforest Observatory, Queensland, Australia | 145.45004°E, 16.10268°S | |
| BRI AQ0485087 | OM390225 | OM286968 | Archidendronramiflorum (F.Muell) Kosterm. | Z110 | Shiptons Flat. Queensland, Australia | 145°14'E, 15°47'S | |||||
| AAU Balgooy6769 | OM984552 | OM390226 | OM286970 | OM287054 | ON013713 | ON101569 | Archidendron sp. nov. in obs. | JA85 | Pulan Baun, Aru Island Indonesia | 134°35'E, 6°30'S | |
| BRI AQ0052837 | OM984553; OM984554 | OM286971 | OM287055 | ON013714 | ON101570 | OM984627 | Archidendronsyringifolium (Kosterm.) I.C.Nielsen | JA41 | Agu River branch of the middle Fly River, PNG | 141.166667°E, 6.966667°S | |
| MEL 2041191A | OM984555 | OM390227 | OM286972 | OM287056 | ON013715 | ON101571 | OM984628 | Archidendronvaillantii (F.Muell) F.Muell. | JA51 | Cape Tribulation, Queensland, Australia | 145°27'E, 16°6'15"S |
| BRI AQ0558405 | OM984556 | OM286973 | OM287057 | ON013716 | ON101572 | OM984629 | Archidendronvaillantii (F.Muell) F.Muell. | JA52 | Along Paluma Dam Road, Ethel Creek, Queensland, Australia | 146°10'40.222"E, 19°0'7.863"S | |
| MEL 2196304A | OM984558 | OM286974 | OM287058 | ON013718 | ON101574 | OM984631 | Archidendronwhitei I.C.Nielsen | JA53 | State Forest 310 Gadgarra. Queensland, Australia | 145°43'26"E, 17°18'13"S | |
| BRI AQ0824396 | OM390229 | OM286975 | OM287059 | ON013719 | ON101575 | OM984632 | Archidendronwhitei I.C.Nielsen | JA54 | 7km W of Babinda, Queensland, Australia. | 145°54'30"E, 17°20'30"S | |
| KUN 0599686 | OM984559; OM984560 | OM286976 | OM287060 | ON013720 | ON101576 | OM984633 | Archidendronxichouense (C.Chen & H.Sun) X.Y.Zhu | JA84 | China | ||
| BRI AQ0611431 | OM286978 | OM287062 | ON013723 | Archidendropsisbasaltica (F.Muell.) I.C.Nielsen | Z218 | On Isaac River and Hill Creek. 25km S of Glenden, Queensland, Australia | 148°7'E, 21°33'01"S | ||||
| MEL 0290000A | OM286977 | OM287061 | Archidendropsisbasaltica (F.Muell.) I.C.Nielsen | Z44 | Bladensburg National Park, S of Winton, Poison Paddock. Queensland, Australia | 143°2'23"E, 22°41'9"S | |||||
| MEL 2333247A | OM984561; OM984562 | OM286979 | OM287063 | ON013721 | ON101577 | OM984634 | Archidendropsisgranulosa (Labill.) I.C.Nielsen | Z362 | Prov. Sud, near Yate, north side of Yate River. New Caledonia | 166°56'0"E, 22°9'29"S | |
| BRI AQ0430532 | OM286980 | OM287064 | ON013724 | Archidendropsislentiscifolia (Benth.) I.C.Nielsen | Z122 | c. 5km north of Kone, south of Kafeate. New Caledonia. | 164.78333°E, 21.05°S | ||||
| MEL 2095888A | OM286981 | OM287065 | ON013725 | ON101578 | OM984635 | Archidendropsisthozetiana (F.Muell.) I.C.Nielsen | JA144 | Palmgrove National Park, 5km W of Daydream Hill, Queensland, Australia | 149°13'29"E, 24°59'3"S | ||
| BRI AQ0771148 | OM286982 | OM287066 | ON013722 | Archidendropsisxanthoxylon (C.T.White & W.D.Francis) I.C.Nielsen | Z121 | Daintree, narrow ridge above Cassowary Creek, off Stewart Creek road, site 69. Queensland, Australia | 145°17'46"E, 16°17'56"S | ||||
| L.1958248 | OM984563 | OM390230 | OM286983 | OM287067 | ON013726 | ON101579 | OM984636 | Falcatariamoluccana (Miq.) Barneby & J.W.Grimes | JA134 | KPC area, Sebongkok Utara, East Kalimantan, Indonesia. | 117°31'59"E, 0°48'0"N |
| CANB 367091.1 | OM984564 | OM390231 | OM286984 | OM287068 | ON013727 | ON101580 | OM984637 | Falcatariatoona (F.M.Bailey) Gill.K.Br., D.J.Murphy & Ladiges | JA149 | Near Earlando, 27 km N of Proserpine. Queensland, Australia | 148°33'E, 20°10'S |
| MEL 1615244A | OM984567 | OM390234; OM390235 | OM286987 | OM287071 | ON013730 | OM984640 | Pararchidendronpruinosum (Benth.) I.C.Nielsen | Z50 | Palm Tree Creek, W of Mt Whitestone township, Queensland, Australia | 152°4'E, 27°39'S | |
| CNS 134531.1 | OM984565 | OM390232 | OM286985 | OM287069 | ON013728 | ON101581 | OM984638 | Pararchidendronpruinosum (Benth.) I.C.Nielsen | JA55 | CSIRO Arboretum, Queensland, Australia | 145°29'6"E, 17°15'28"S |
| QRS 121813.1 | OM984566 | OM390233 | OM286986 | OM287070 | ON013729 | ON101582 | OM984639 | Pararchidendronpruinosum (Benth.) I.C.Nielsen | JA56 | Clarke Range, Queensland, Australia | 148°31'E, 21°16'S, |
| MEL 2183015A | OM984568; OM984569 | OM390236 | OM286988 | OM287072 | ON013731 | ON101583 | OM984641 | Paraseriantheslophantha (Willd.) I.C.Nielsen | Z43 | Merrimu Reservoir, Victoria, Australia | 144°29'23"E, 37°38'3"S |
| BRI AQ0408829 | OM984570; OM984571 | OM390237 | OM286989 | OM287073 | ON013732 | ON101584 | OM984642 | Serianthesnelsonii Merr. | JA143 | Atop Sailigai Hulo, Rota. Northern Mariana Islands. | 145°12'53"E, 14°09'03"N |
| MEL 2333248A | OM984572 | OM390238 | OM286990 | OM287074 | ON013733 | ON101585 | OM984643 | Serianthespetitiana Guillaumin | Z361 | Prov. Sud, near Prony, New Caledonia | 166°49'52"E, 22°19'4"S |
| MELU SRA051 | OM984573 | OM390239 | OM286991 | OM287075 | ON013734 | ON101586 | OM984644 | Wallaceodendroncellebicum Koord. | Z48 | Bogor Botanic Gardens collection Accession: B19610136 | |
Total genomic DNA (gDNA) was extracted following the CTAB method of Doyle and Doyle (1987) with modifications as per Shepherd and McLay (2011). Isolated gDNA was quantified with a NanoDrop 2000 (ThermoScientific) spectrophotometer and cleaned with a 2.4 M sodium acetate wash. Recalcitrant herbarium material that failed using the CTAB method was extracted using the AccuPrep Stool genomic DNA extraction kit (Bioneer) using the manufacturer’s protocol with some modifications suggested by Schuster (pers. comm.). Only 30 mg of leaf material was used instead of the recommended 100–200 mg. A total of 600 µl of stool lysis buffer (SL) was added to the extraction tube instead of 400 µl, the incubation step was increased to one hour in total, centrifugation was done for 10 minutes at step five, and to maintain equal volumes, 600 µl of binding buffer was added. Two consecutive washes were performed using buffer 1 (W1). The final elution was done by adding 160 µl total elution buffer in two steps (first 60 µl, and then 100 µl) instead of a single elution with 200 µl.
Marker selection, primer design and library preparation
Eight nuclear markers (low copy genes: AIGP, CYB6, Eif3E, SHMT, RBPCO, UDPG; nrDNA: ITS, ETS) and four chloroplast DNA intergenic spacer regions (trnK-matK, trnV-ndhC, psbD-trnT, trnL-rpl32) were assessed for variability between nine individuals spanning the series of Archidendron using Sanger sequencing.
PCR reagents, primers and cycling conditions are described in Suppl. material 1 (Johnson and Soltis 1994; Sun et al. 1994; Käss and Wink 1997; Baldwin and Markos 1998; Miller and Bayer 2001; Ariati et al. 2006; Choi et al. 2006; Shaw et al. 2007; Li et al. 2008). PCR products were visualised on a 1.5% agarose gel with Easy ladder I (Bioline) and cleaned with ExoSAP-IT (USB) as per the manufacturer’s protocol. The purified amplicons were sequenced on an AB3730xl sequencer (Thermo Scientific) at the Australian Genome Research Facility, Melbourne. Sequences were aligned in Geneious v.8.1.4 (Biomatters Ltd.) and assessed for variability between the samples. The most variable loci were then used in a targeted amplicon sequencing (TAS) approach (McLay et al. 2021), sequencing pooled amplicons on an Illumina MiSeq. For this, additional internal primers were designed for the five loci that had a total amplicon length greater than 500 bp, in order to produce shorter amplicons that could be fully sequenced using a 500-cycle sequencing kit. These primers were designed using Primer 3 v.2.3.4 (Rozen and Skaletsky 2000) implemented in Geneious v.8.1.4 (Biomatters Ltd.), selecting priming sites in conserved regions across the nine sequenced individuals.
Library preparation followed the two-step PCR process outlined in McLay et al. (2021). The first step used the region-specific primers to amplify each locus individually for each sample. Initial PCR reactions included 1 × MyTaq Buffer (Bioline), 1.2 µl of MgCl2 2.5 M (Bioline, 100 mg mL), 1.2 µl of dimethyl sulfoxide (DMSO, 99.5%; Sigma-Aldrich), 3 µl of each “tailed” primer (10 µM), 0.375 U of MyTaq (Bioline), 100 ng of gDNA, and ultra-pure water to make up for 16 µl volume. Variations in these reactions are noted in Suppl. material 1 for specific loci. Conditions for PCR were based on those of Choi et al. (2006), Shaw et al. (2007), and Ariati et al. (2006) with modifications as required to obtain successful amplifications (Suppl. material 1). To estimate amplicon concentration to decide the volume of PCR product for amplicon pooling, 2.2 µl of PCR product and 2.5 µl of molecular ladder (Easyladder I, Bioline) were run on 1.5% agarose. A total of 120 ng of each nuclear DNA (ncDNA) region PCR product and 20 ng of each chloroplast region PCR product were pooled in the same well of a 96-well plate. The ncDNA were pooled in a higher concentration to account for the possible presence of different alleles. Pooled samples were cleaned with 1.5 × Serapure beads (Rohland and Reich 2012).
The second step used qPCR to add unique Illumina indexing barcodes to each sample for the pooled amplicons. Indexing PCR reactions consisted of 5 µM of each of index primer (McLay et al. 2021), 3 µl of pooled amplicons, 1 × Kapa HiFi ReadyMix (Biosystems) and ultra-pure water to make up a total of 25 µl reaction. Conditions for PCR were 95 °C for 1 min, followed by 13 cycles of 98 °C for 50 sec, 67 °C for 50 sec, and 72 °C for 20 sec, and a final extension at 72 °C for 30 sec. Each sample was then cleaned with 1.4 × Serapure beads and concentrations were quantified using fluorescence in a EnSpire multimode plate reader. In total, 10 ng of each indexed and cleaned sample was pooled together. The final pooled library was cleaned with 1.5 × Serapure bead-to-sample ratio and the library was submitted to the Australian Genome Research Facility, Melbourne for sequencing on an Illumina MiSeq using a 500 cycle MiSeq v2 Nano Kit.
Data analysis
Sequences obtained by Sanger sequencing were aligned by individual locus in Geneious v.8.1.4 (Biomatters Ltd.) and a consensus sequence was generated and used as the reference for the reads obtained by TAS. The demultiplexed TAS Illumina MiSeq files were imported into Geneious v.8.1.4. Reads were trimmed to remove adapters and low-quality sequence. The map-to-reference option was selected to map reads for each sample to the different reference loci using High Sensitivity/Medium settings and a minimum mapping quality of 20. A consensus sequence for each locus was generated for each individual with Generate Consensus Sequence (Threshold = 65%, with Ns called if coverage was less than 10). The forward and reverse reads of the low-copy nuclear genes (LCNG) overlapped so it was possible to phase these loci into separate alleles, but this was not possible for the nuclear ribosomal DNA loci (ETS and ITS) as the reads were not overlapping due to unexpected length variation in both of these loci. Alignments of individual consensus sequences for each locus were generated using MUSCLE (Edgar 2004) in Geneious v.8.1.4 and adjusted manually. For each LCNG, samples with multiple alleles were assessed for topological concordance between the different copies using neighbour-joining trees (using the Geneious tree-builder, HKY model) and NeighbourNet networks (SplitsTree4, default settings, Huson and Bryant 2006), to ensure that a conflicting signal was not introduced from distantly related allelic variants (see Suppl. material 2: SHMT network and tree and Suppl. material 3: RBPCO network and tree). Allelic variants within samples were largely concordant with one-another permitting consensus sequences for those samples to be used for subsequent phylogenetic analyses.
Alignments of all nuclear loci (ncDNA; with consensus sequences for LCNG alignments) were analysed individually to explore gene tree topologies in IQ-TREE v.1.6.12 on the web server (http://iqtree.cibiv.univie.ac.at/, Trifinopoulos et al. 2016) with support estimated with 1,000 ultra-fast bootstrap replicates (UFBS) (Minh et al. 2013). After comparing topologies, four ncDNA loci (ETS, ITS, RBPCO, SHMT) were concatenated into a single matrix as no major incongruencies were observed. The combined ncDNA dataset was partitioned into six partitions corresponding to each locus with the ITS region further divided in ITS1, 5.8S and ITS2 for subsequent analyses. IQ-TREE was used to perform maximum likelihood (ML) analyses on the concatenated ncDNA alignment. The analysis was run with the alignment partitioned and allowing ModelFinder (Kalyaanamoorthy et al. 2017) to identify the optimal substitution models for each partition (Table 2). Node support was estimated using 1,000 UFBS. Bayesian Inference (BI) was performed, with the alignment partitioned by locus. The best model of substitution for each partition was estimated with IQ-TREE model selection using the options: selection criteria of Bayesian (BIC), candidate models JC, F81, K80, HKY, SYM, GTR, heterogeneity types I, G, I+G, and the genomic source of nuclear (Table 2). MrBayes v.3.2.7a (Ronquist et al. 2012) was run using the CIPRES Science Gateway (Miller et al. 2010). Two parallel runs each with eight Monte Carlo Markov Chains were run for five million generations, sampling a tree every 1,000 generations and a burn-in of 25%.
Table 2.
ncDNA data partitions and best fit substitution models. Models estimated by IQ-TREE model selection and applied for BI.
| Partition | Model |
|---|---|
| ETS | HKY+F+G4 |
| ITS1 | GTR+F+G4 |
| 5.8S | SYM+I+G4 |
| ITS2 | HKY+F+G4 |
| SHMT | HKY+F+G4 |
| RBPCO | K2P+I |
A consensus network of the combined ncDNA dataset was constructed in SplitsTree4 (Huson and Bryant 2006) using the last 101 sampled BI trees (edge weights = mean, threshold = 0.05). This method allows for the visualisation of conflict in a set of trees and provides an alternative method of interpretation to a single fixed topology of a consensus tree.
All chloroplast (cpDNA) loci were concatenated into a single matrix for phylogenetic analyses. IQ-TREE was used to perform ML analyses on the cpDNA matrix, with the alignment partitioned by locus, using ModelFinder to identify the optimal substitution model for each locus, and support was estimated using 1,000 UFBS replicates. The resulting topology was very poorly supported (though similar groups to the ncDNA phylogeny were discovered within the genus Archidendron). To further investigate cpDNA relationships within Archidendron, the outgroups were removed, and the IQ-TREE analysis was performed on the reduced dataset. The UFBS replicates were then used to create a consensus network in SplitsTree4 (edge-weights = mean, threshold = 0.20).
Pollen morphology of Archidendropsissubg.Basaltica
Pollen size and surface texture are key morphological features differentiating the subgenera of Archidendropsis but one of the three species of subg. Basaltica (A.xanthoxylon (C.T. White & W.D. Francis) I.C. Nielsen) was not examined by Nielsen et al. (1983b). To fill this gap and ensure consistency of results with published data, pollen from A.xanthoxylon (BRI AQ0199126, BRI AQ0874091, BRI AQ0199129 and BRI AQ0648303) and A.basaltica (F. Muell.) I.C. Nielsen (BRI AQ1003764, BRI AQ0199029, BRI AQ0625292 and BRI AQ0648454) of subg. Basaltica was examined. Pollen grains were obtained from flowers of herbarium specimens under a Zeiss dissecting microscope at the Queensland Herbarium (BRI) using clean forceps and a fine brush. Samples were mounted on aluminium stubs using double-sided carbon tabs and coated with gold using an Agar Scientific Automatic Sputter Coater. Pollen grains were observed and photographed using a Phenom G2 5keV (kiloelectron-volt) desktop scanning electron microscope (PhenomWorld). Pollen diameter for 10 grains of A.basaltica and eight grains of A.xanthoxylon was measured using ToupView (TOUPTEK PHOTONICS) software; overall fewer grains were available on specimens of A.xanthoxylon for microscopy.
Results
Targeted amplicon sequencing loci
Of the eight nuclear loci only four were included in the final phylogenetic analyses: SHMT, RBPCO, ITS and ETS. ETS and ITS amplified well, were variable, and are commonly used phylogenetic markers in Caesalpinioideae phylogenetic studies. Of the LCNGs, SHMT was the most informative, followed by RBPCO; allelic variation was found in some individuals for all LCNGs. Exploring allelic variation in the SHMT (36 samples with alleles) and RBPCO (24 samples with alleles) showed that for samples with more than one allele, the copies were closely related to each other (Suppl. material 2: SHMT network and tree and Suppl. material 3: RBPCO network and tree). Two LCNGs were excluded because few individuals of the target genera were successfully sequenced; only 12 sequences of Archidendron and two sequences of Archidendropsis were obtained for AlGP, and only 16 sequences of Archidendron and one Archidendropsis were obtained for Eif3E. The remaining two LCNG loci (CYB6 and UDPG) are not included in the analyses due to their short lengths, 240 bp and 202 bp respectively, and lack of variation.
Of the four chloroplast loci, trnK-matK was the most informative, followed by psbD-trnT and then trnV-ndhC. However, only one of the three blocks of trnV-ndhC was successfully sequenced. The internal primers designed allowed 100% coverage for the trnK-matK, 81% coverage for the psbD-trnT, and less than 30% coverage for the trnV-ndhC. It was not possible to obtain sequences for all samples for all blocks in which the three cpDNA regions were divided; as a result the cpDNA dataset was patchy. The trnL-rpl32 intergenic spacer did not amplify well, with 10 samples partially sequenced, and it was not included in final analyses.
Phylogenetic analyses
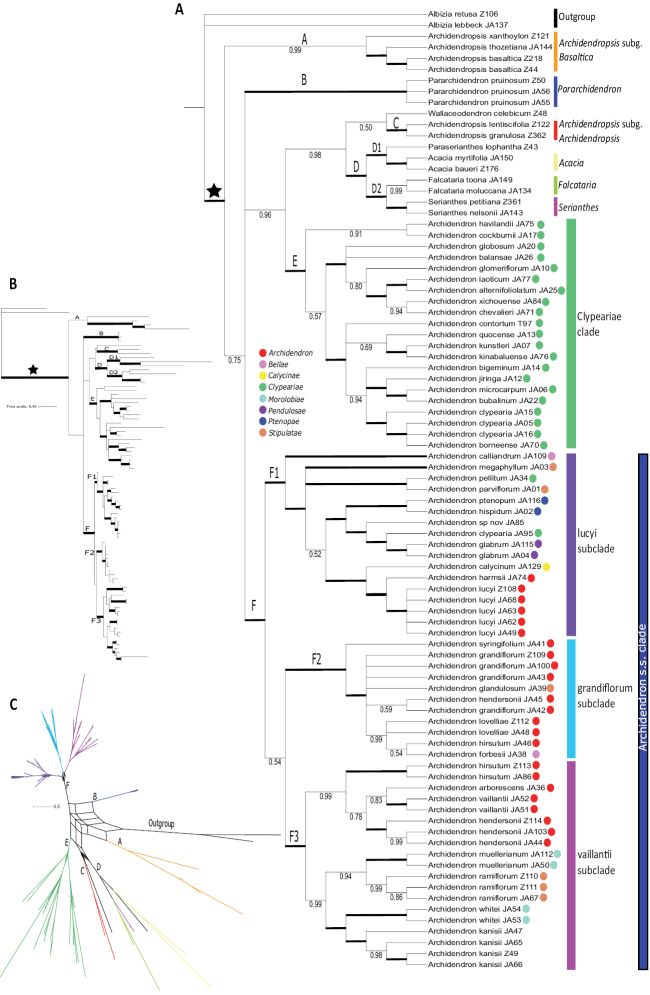
The topologies of the combined ncDNA Bayesian and IQ-TREE analyses were congruent (nodes supported with UFBS ≥ 95; PP ≥ 0.90) and the Bayesian tree is presented (Fig. 2A,B). The Archidendron clade was recovered as monophyletic (PP 1.0) with six well supported clades (A–F) resolved within it. However, the relationships between clades A–F were not well resolved or supported with a polytomy in the backbone of the phylogeny. Clade A (PP 0.99) includes all three species of Archidendropsissubg.Basaltica, clade B (PP 1.0) includes the three samples of Pararchidendronpruinosum (Benth.) I.C. Nielsen, and clade C (PP 1.0) includes the two sampled representatives of Archidendropsissubg.Archidendropsis. Four monophyletic genera are grouped together in clade D (PP 1.0), with Acacia sister to Paraserianthes in clade D1 (PP 1.0) and Falcataria sister to Serianthes (PP 1.0) in clade D2 (Fig. 2A). Clade E (PP 1.0) comprises all but two sampled representatives of Archidendronser.Clypeariae, and all other samples of Archidendron are placed in clade F (PP 1.0). Clades C, D and Wallaceodendron are related (PP 0.98) and together are sister to Clade E (PP 0.96; Fig. 2A).
Figure 2.
Combined ncDNA phylogeny of the Archidendron clade. The Bayesian Inference (BI) cladogram, phylogram, and consensus network for the combined ncDNA dataset are presented A Cladogram: the star indicates the Archidendron clade sensu Koenen et al. (2020). Nodes with PP = 1.0 are shown in bold while other nodes with PP ≥ 0.50 are noted under the node. Clades are labelled with letters above the node. Coloured bars to the right of clades are names discussed in the text. Nielsen’s series of Archidendron are shown as coloured circles next to the sample name; key to colour and series in legend B Phylogram: clades are labelled as per A and nodes with a PP = 1.0 are shown in bold C Consensus network: branches are colour coded and labelled as per the clades of A.
Within Archidendron, only one of Nielsen’s eight series is resolved as monophyletic (ser. Ptenopae) within subclade F1 (Fig. 2A). Clade E, the Clypeariae clade had two main lineages and several smaller supported subclades within them. Clade F, the Archidendron s.s. clade is segregated into three well supported subclades: the lucyi subclade (F1, PP 1.0) that includes three fully supported lineages; the grandiflorum subclade (F2, PP 1.0) that is poorly resolved; and the vaillantii subclade (F3, PP 1.0) that comprises two well supported lineages (PP 0.99; Fig. 2A–C).
Of the 12 species of Archidendron that included more than one accession, seven are monophyletic (A.glabrum (K. Schum.) K. Schum. & Lauterb., A.kanisii R.S. Cowan, A.lucyi F. Muell., A.muellerianum, A.ramiflorum (F. Muell.) Kosterm., A.vaillantii (F. Muell.) F. Muell. and A.whitei), one is unresolved (A.lovelliae (F.M. Bailey) I.C. Nielsen), and four are not monophyletic (A.clypearia (Jack) I.C. Nielsen, A.grandiflorum (Sol. ex. Benth.) I.C. Nielsen, A.hendersonii (F. Muell.) I.C. Nielsen and A.hirsutum I.C. Nielsen). Three of the four samples of A.clypearia form a clade (within clade E, Fig. 2; PP 1.0) with A.borneense (Benth.) I.C. Nielsen nested among them. One sample of A.hendersonii (JA45) is related to A.grandiflorum within clade F2; all other samples of A.hendersonii (Z114, JA103, JA44) form a clade within F3 (PP 1.0; Fig. 2A). Another species falling in both subclades F2 and F3 is A.hirsutum, with one sample (JA46) related to A.forbesii Baker f. and A.lovelliae in subclade F2 (PP 0.99), and the other two (Z113 and JA86) forming a sister pair in subclade F3 (PP 1.0; Fig. 2A).
The consensus network of the final 101-sampled BI trees shows the degree of topological uncertainty between the genera in the Archidendron clade (Fig. 2C). While each respective genus is well-supported as monophyletic (except Archidendropsis and Archidendron as described above) the relationships between the genera are highly uncertain, reflecting the lack of support in the consensus phylogenies. However, the network reinforces the distinction between the two clades of Archidendropsis, and the distinction of the Clyperiae clade from the rest of Archidendron.
The phylogeny of the three cpDNA loci combined lacks support for nearly all nodes (Suppl. material 4: cpDNA tree). Of the supported nodes there are two that are incongruent with the ncDNA tree (Fig. 2): Paraserianthes is sister to Falcataria (UFBS 100), and A.harmsii Malm is supported in the grandiflorum subclade (UFBS 95) sister to A.grandiflorum JA100 (UFBS 97; Suppl. material 4: cpDNA tree). The consensus network of the UFBS replicates (with splits present in at least 20% of trees) reflects the patterns in the ncDNA phylogeny, with four distinct groupings within Archidendron (Fig. 3). Within these groupings, several individuals are placed in different clades to the ncDNA tree: A.hendersonii JA45 is placed in the vaillantii subclade rather than the grandiflorum subclade, and A.harmsii JA74 is in the grandiflorum subclade rather than the lucyi subclade (Fig. 3).
Figure 3.
Combined cpDNA consensus network of clades within the genus Archidendron. The branches are labelled, and colour coded according to clades in Fig. 2A. Samples that have changed position relative to the ncDNA tree (as discussed in the text) are labelled with their name on the network.
Pollen morphology of Archidendropsissubg.Basaltica
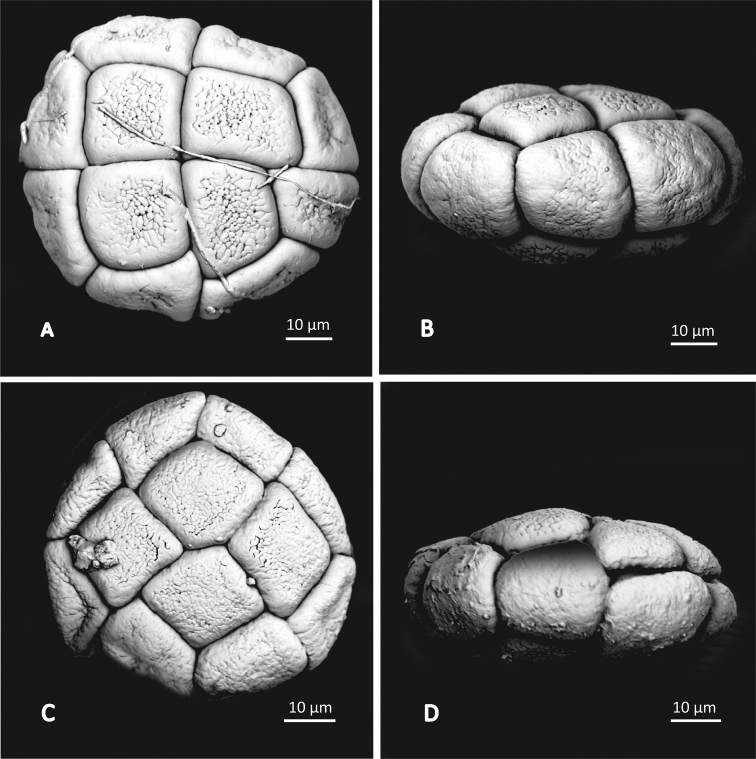
The pollen measurement results are consistent with Nielsen et al. (1983a, 1983b). The pollen of the two species examined (A.basaltica and A.xanthoxylon) are aggregated into symmetrical 16-celled polyads with a diameter of 55–62 μm for A.basaltica and 62–68 μm for A.xanthoxylon (Fig. 4). Fossules were present on the surface of all grains of both species, but they were fainter on the peripheral cells compared to the central ones and overall fainter on A.basaltica compared to A.xanthoxylon (Fig. 4).
Figure 4.
Scanning electron micrographs of Archidendropsissubg.Basaltica pollen. Archidendropsisxanthoxylon (A BRI AQ0199126 and B BRI AQ0874091) and Archidendropsisbasaltica (C BRI AQ0199029 and D BRI AQ01003764).
Discussion
Phylogeny of the Archidendron clade
Our study presents the most taxon-rich sampling of the Archidendron clade of any phylogenetic analyses to date. We confirm that the Archidendron clade sensu Koenen et al. (2020) of Indomalayan-Australasian genera (Acacia, Archidendron, Archidendropsis, Falcataria, Serianthes, Pararchidendron, Paraserianthes and Wallaceodendron) is robustly supported, yet the relationships between the constituent clades are poorly resolved and lack support. This result is not unexpected given we used only four ncDNA loci and that phylogenomic studies based on hundreds of loci also yield short branches with low support across the backbone of the Archidendron clade (Koenen et al. 2020; Demeulenaere et al. 2022; Ringelberg et al. 2022). It has been suggested that this lack of resolution may be the result of extremely rapid speciation and that the backbone of this clade could be best regarded as a polytomy within the Ingoid legumes (Koenen et al. 2020). The differences in published topologies of the Archidendron clade are illustrated in Demeulenaere et al. (2022) but it is clear that further work based on increased sampling of phylogenomic data is required to uncover the evolutionary history of the clade.
Despite the poorly resolved backbone of the Archidendron clade, many clades within it are robustly supported and corroborate published phylogenies, as well as shedding new light on the genera Archidendron and Archidendropsis (Fig. 2). Four genera of the Archidendron clade are confirmed to be monophyletic – Acacia (Miller and Bayer 2001; Luckow et al. 2003; Miller et al. 2003; Brown et al. 2008), Falcataria (Brown et al. 2011), Pararchidendron and Serianthes (Demeulenaere et al. 2022) – and the previously suggested non-monophyly of Archidendron and Archidendropsis (Brown et al. 2008, 2011; Iganci et al. 2016; Demeulenaere et al. 2022; Ringelberg et al. 2022) is confirmed and clarified by increased sampling within these genera.
Phylogenetic relationships within Archidendron
The genus Archidendron is not monophyletic, and the eight series, while useful for identification purposes, do not coincide with evolutionary lineages (Fig. 2). The only series confirmed to be monophyletic was series Ptenopae from the island of New Guinea, the smallest series comprising just two species with two-winged leaf rachises and pinnae: A.ptenopum Verdc. and A.hispidum (Mohlenbr.) Verdc. (Nielsen et al. 1984b). The monophyly of series Calycinae and Pendulosae was not tested, as only one species of each was sampled, however, all other series (Archidendron, Bellae, Clypeariae, Morolobiae, and Stipulatae) are not monophyletic. Archidendron is instead resolved into two well supported lineages, one of which is primarily distributed in western Malesia and mainland Asia (the Clypeariae clade; clade E, Figs 1–3) and the other (the Archidendron s.s. clade; clade F, Figs 1–3) mostly restricted to eastern Malesia and Australia. These two lineages have been identified in previous phylogenetic studies but the sampling for each was extremely limited, with at most seven species of one lineage included (Brown et al. 2008, 2011; Iganci et al. 2016; Demeulenaere et al. 2022; Ringelberg et al. 2022). The further segregation of the Archidendron s.s. clade into three well supported lineages, the lucyi (F1), the grandiflorum (F2), and the vaillantii subclades (F3; Figs 2–3), is novel.
These three subclades of the Archidendron s.s. clade reflect geographic distributions to some extent, but no macromorphological characters have been identified to clearly delineate them. The grandiflorum and vaillantii subclades are predominantly Australian with some southern New Guinean species included, while the lucyi subclade is geographically more broadly distributed in the Lesser Sunda Islands, the Moluccas, through New Guinea to the Solomon Islands with only one species, A.lucyi, extending into northern Australia. Morphologically, the lucyi subclade includes all the sampled species lacking stipules that are not from ser. Clypeariae (i.e. A.calliandrum de Wit, A.harmsii, and A.glabrum), although stipules are reported for other species in this clade, three with stipular glands (A.lucyi, A.megaphyllum Merr. & L.M. Perry, Archidendron sp. nov. JA85), two with stipules only (A.ptenopum and A.hispidum) and A.parviflorum Pulle having both stipular glands and stipules (AAU Balgooy 6769; Nielsen et al. 1984b). All sampled species in the grandiflorum and vaillantii subclades have stipules, except A.arborescens (Kosterm.) I.C. Nielsen and A.forbesii, which have stipular glands (BM000946689; BRI AQ0380081; BRI AQ052589; Nielsen et al. 1984b) The placement of an undescribed species (Archidendron sp. nov. JA85) from the Aru Islands (Moluccas) in the lucyi subclade fits the geographic range. Ivan Nielsen noted this as a putative new species in October 1998 (AAU Balgooy 6769) but it does not align with any of the 20 imperfectly known species he outlined (Nielsen et al. 1984b), highlighting that further taxonomic work is required.
Three species in the Archidendron s.s. clade were not resolved as monophyletic (Fig. 2A), although it is unlikely these are issues with species delimitation. The paraphyly of A.grandiflorum (Fig. 2), a morphologically consistent species across a large geographic range (Brown pers. obs.), could be the result of potentially rapid and recent divergence or may be due to insufficient phylogenetically informative characters in this study. The latter could also apply to the polyphyletic species (A.hendersonii and A hirsutum), as A.hendersonii JA45, which is placed separately from the other conspecific samples is missing data for two of the four ncDNA loci (Table 1). However, this was not the case for A.hirsutum JA46. Re-examination of the vouchers of all accessions of A.hendersonii and A.hirsutum confirmed their identifications, suggesting that incomplete lineage sorting or paralogy problems associated with one or more nuclear loci could explain these non-monophyletic species; further data are required to investigate this.
The Clypeariae clade (clade E, Figs 2–3) includes all sampled species of ser. Clypeariae (19/51), except one accession of A.clypearia (JA95) from Papua New Guinea and A.pellitum (Gagnep.) I.C. Nielsen from Vietnam. Series Clypeariae was previously recognised in Pithecellobium as section Clypearia until Nielsen et al. (1984b) expanded Archidendron based on evidence from shared wood anatomy, inflorescence and pod morphology (Nielsen et al. 1984b). Characters of the pods are also useful to differentiate series Clypeariae from the rest of Archidendron. Nielsen et al. (1984b) described six pod types and most species of ser. Clypeariae have pod type 2 (long funicle, opens ventral suture first) or 6 (straight pods with overgrown seeds), while the other series primarily have pod type 1 (opens dorsal suture first, short funicles). Seeds of ser. Clypeariae are usually flattened and are not embedded in the pericarp, which is possibly linked to characteristics of the pod, such as dryness (de Wit 1942; Nielsen 1981, 1992; Nielsen et al. 1984b). Additionally, the combination of lack of stipules and solitary, stipitate ovaries delineates ser. Clypeariae (Nielsen et al. 1984b). Individually though, these characters are not diagnostic, as some species with sessile ovaries are placed in ser. Clypeariae (e.g. A.occultatum (Gagnep.) I.C. Nielsen and A.turgidum (Merr.) I.C. Nielsen), other species lacking stipules are placed in series Archidendron (e.g. A.harmsii and A.tjendana (Kosterm.) I.C. Nielsen), and two Philippine species of ser. Clypeariae (A.apoense (Elmer) I.C. Nielsen and A.merrillii (J.F. Macbr.) I.C. Nielsen) have more than one ovary but both are stipitate (Nielsen et al. 1984b). Given these morphological differences of ser. Clypeariae from the rest of Archidendron, together with the non-monophyly of the genus, there are grounds for segregating Clypeariae as a distinct genus; however, we are not proposing such a taxonomic change here for several reasons. First, there are many shared morphological characters between species of Archidendron s.l.; second, the shallow backbone of the ncDNA tree remains poorly supported with topological uncertainty between lineages; third, the placement of two species of ser. Clypeariae within the Archidendron s.s. clade (clade F; A.clypeariavar.velutinum (Merr. & L.M. Perry) I.C. Nielsen and A.pellitum) raises further doubts; and fourth, phylogenetic sampling of species remains incomplete. All these issues suggest that denser taxon sampling and larger phylogenomic datasets are required before re-classifying Archidendron as two genera.
Archidendronclypearia is the most widespread species of Archidendron, found from India through to Papua New Guinea. The morphological variation within A.clypearia has been used to recognise four infraspecific taxa (Legume Phylogeny Working Group 2021): subsp. clypearia, subsp.subcoriaceum (Thwaites) M.G. Gangop & Chakrab., var.sessiliflorum (Merr.) I.C. Nielsen, and var.velutinum. The one accession of A.clypearia placed outside the Clypeariae clade (JA95) (Fig. 2A) has been identified as var.velutinum (Brown, pers. obs. of CANB525617; previously only identified to species level by the collector), the only infraspecific taxon found in eastern Malesia (Sulawesi, Moluccas and PNG). The three other samples of A.clypearia included in the phylogeny have not been assigned to infraspecific taxa but they are not likely var.velutinum, as they are from Malaysia and Vietnam and lack the woolly to velutinous hairs on the lower surface of the leaflets (Brown per. obs.). Taxonomic revision and denser phylogenetic sampling of A.clypearia from across its morphological and geographic range is required to verify this placement, delineate the taxa and investigate if var.velutinum should be raised to species level (Merrill and Perry 1942) or if there are intermediate forms as suggested by Kostermans (1966). The only other species of series Clypeariae that extends into eastern Malesia, A.palauense (Kaneh.) I.C. Nielsen, from the Moluccas through to the Solomon Islands (Nielsen et al. 1984b), was not sampled here. There are no obvious morphological characters that support placement of A.pellitum outside the Clypeariae clade, as it has the full combination of diagnostic characters of ser. Clypeariae: compressed pods with a long (3–5 mm) funicle, stipitate single ovary and no visible stipules (US 2515891; P01818442; Nielsen 1981). In addition, no evidence of paralogy in the nuclear loci of A.pellitum and A.clypeariavar.velutinum (JA95) was noted in this study; all sequences suggest they fall in the A.lucyi subclade.
The last revision of the genus Archidendron (Nielsen et al. 1984b) significantly advanced our understanding of the genus but more detailed taxonomic study is still required, focusing especially on the large number of species known from incomplete material and widespread morphologically variable species, such as A.clypearia. To resolve the backbone of the Archidendron clade and inform decisions about generic delimitation to deal with the non-monophyly of Archidendron, we recommend further sampling of ser. Clypearia, particularly from the Wallacean region of Malesia (i.e. Moluccas, Sulawesi, Philippines), together with further genomic sampling.
Phylogenetic relationships within Archidendropsis
While Archidendropsis is not monophyletic, its two subgenera (Archidendropsis and Basaltica) are (Fig. 2). The species within each subgenus have long been recognised as closely related (Bentham 1875; Nielsen 1981) but the two subgenera themselves have not always been associated with each other. For example, Bentham (1875) placed the species of each subgenus in different sections of Albiziabased on inflorescence shape. Species ofsubgenusArchidendropsis that have flowers arranged in cylindrical spikes were placed by Bentham (1875) in AlbiziasectionLophantha Benth. (an illegitimate name later corrected to AlbiziasectionPachysperma (Benth.) Fosberg by Fosberg (1965)). Within this section they were separated from the other taxa, which are now recognised as Paraserianthes, into series Platyspermae Benth. because they have flattened, broadly orbiculate seeds (Bentham 1875). The two species of subgenus Basaltica known at that time (A.basaltica and A.thozetiana (F. Muell.) I.C. Nielsen) were placed by Bentham in his large section Eualbizzia distinguished by flowers in globular heads and flattened orbicular seeds (Bentham 1875). Within that section, these taxa were placed into series Obtusifolia, which corresponds to the Australian species with 1–2 jugate leaves, ovate, oblong or obtuse leaflets, short petioles, pedunculate heads in the axils, and small sessile flowers.
It was only recently that the species of the two subgenera were united within Archidendropsis by Nielsen (1983) based on characters of the fruit and seed: pods dehiscent along both sutures, and seeds that are winged, thin-walled and lack a pleurogram. However, Nielsen himself questioned whether the subgenera should be congeneric, noting that if they were not, “the evolution of the winged thin walled seeds without pleurogram should have happened twice” (Nielsen et al. 1983a: p. 337). The results presented here (Fig. 2) alongside two recent phylogenomic analyses (Demeulenaere et al. 2022; Ringelberg et al. 2022) show that the two subgenera of Archidendropsis do not form a monophyletic group, suggesting these seed characteristics are indeed the result of convergent evolution.
The presence of a pleurogram is common in mimosoid genera (Gunn 1984), and is considered to have evolved multiple times (Maumont 1993). Within the Archidendron clade, Archidendron and Archidendropsis are the only two genera whose seeds lack a pleurogram (Nielsen 1992). The absence of a pleurogram has been associated with short-lived ‘recalcitrant’ seeds (i.e. seeds which lack dormancy and can be viviparous; Nielsen 1992) and has been thought to be an adaptive response to humid environments (Corner 1951 in Nielsen 1992; Maumont 1993). Like the absence of a pleurogram, winged seeds are also rare in mimosoids occurring in only eight genera, including Archidendropsis (Gunn 1984). The possession of a winged seed has been suggested to be an adaptation for wind-dispersal but there have been no published observations of this in Archidendropsis (Gunn 1984; Nielsen 1992). The short viability of Archidendropsis seeds has been linked to the restricted geographic ranges of individual species (Nielsen 1983). However, humidity may be a more important determinant of these distributions, as the ranges of the two Australian species occurring in drier, non-rainforest habitats are more than 10 times larger than the rainforest species (e.g. A.basaltica ≥ 750,000 km2 compared to A.xanthoxylon c. 8,750 km2 (AVH 2021)). The habitats of A.basaltica and A.thozetiana are also more open than for A.xanthoxylon, but these two species generally have narrower wings on their seeds than the rainforest species A.xanthoxylon (Cowan 1998), suggesting that the wing is unlikely to have an impact on wind dispersal. Morphological features that have been used to unite the two subgenera in Archidendropsis are thus homoplasious and not useful for generic delimitation.
The non-monophyly and clear morphological distinctions between them means that the two subgenera can no longer be treated as congeneric and need to be placed in separate genera. As the type of Archidendropsis (A.fulgens (Labill.) I.C. Nielsen) is from subg. Archidendropsis, it is subg. Basaltica that requires a new name. No name exists at the generic level for these taxa, as they have previously been placed in Acacia, Albizia and Archidendropsis (Mueller 1859; Bentham 1875; Fosberg 1965; Nielsen 1983), names which are all typified by other taxa.
In addition to the aforementioned morphological differences between the two subgenera, species of subg. Basaltica are endemic to Australia, whereas those of subg. Archidendropsis are found in New Caledonia, New Britain, the Solomon Islands and on the island of New Guinea (Fig. 1B). Furthermore, there are several pollen characters separating the two subgenera (Nielsen et al. 1983a). Pollen of subg. Basaltica has isometric channels in the tectum and is aggregated into smaller polyads (55–68 μm), cf (80–120 μm) for subg. Archidendropsis where the tectum has non-isometric channels (Fig. 4; Nielsen et al. 1983a). The pollen surface of subg. Basaltica has fossules on the central cells, with either faint fossules or smooth peripheral cells, while in subg. Archidendropsis the surface of all pollen cells has small rounded areoles or deep fossules (Fig. 4; Nielsen et al. 1983a). Species of subg. Basaltica have sessile flowers arranged in globular pedunculate heads, rather than in spikes or racemes. Although one species of subg. Archidendropsis, A.fournieri (Vieill.) I.C. Nielsen, also has flowers arranged in globular pedunculate heads, it does not share the other diagnostic characters of subg. Basaltica, it is endemic to New Caledonia, its seeds are not winged, and the diameter of the pollen polyads is larger, fitting within the size range for subg. Archidendropsis (Nielsen 1983). Another character noted by Nielsen et al. (1983a) to differentiate the two subgenera, was the shape of the stipules, with those of subg. Basaltica being small and often developed into stipular spines (to 1.2 mm long; Brown pers. obs.; Fig. 5F) that are early caducous. However, the stipules of A.xanthoxylon were not recorded by Nielsen et al. (1983a) and are not like other Australian species being 1.2–3 mm long, ovate to triangular, dark gland-like and persistent (Brown, pers. obs., BRI AQ022813, BRI AQ0234095, BRI AQ0771148, BRI AQ199127, BRI AQ0199128; Fig. 5G). These stipules do differ, however, from those of the species of subg. Archidendropsis which, if present, are usually small (c. 1 mm), ovate or filiform and often caducous (Nielsen 1983).
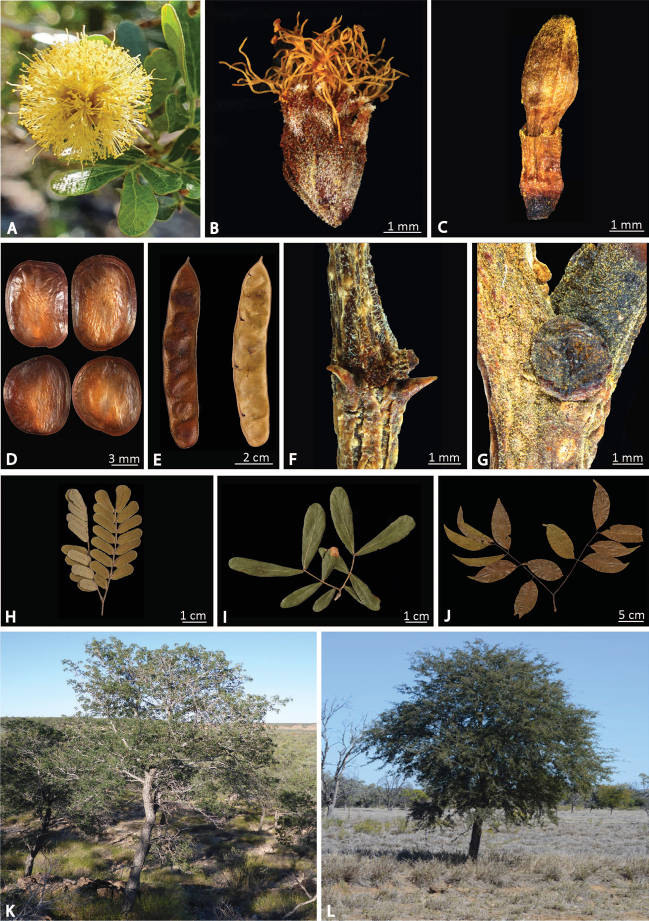
Figure 5.
Morphology of Heliodendron. Plate showing diagnostic features of the new genus HeliodendronA inflorescence of H.thozetianum, Hazelwood Gorge, west of Mackay, Queensland (photo, Stuart Worboys, Australian Tropical Herbarium) B single flower of H.basalticum (BRI AQ0648454) showing hairs on calyx and corolla C mature bud of H.xanthoxylon (BRI AQ0874091) showing hairs on the lobes of the calyx and corolla D seeds of H.basalticum (BRI AQ0746724) E overall pod shape of H.xanthoxylon (BRI AQ0234095) F small rigid stipules of H.basalticum (BRI AQ0673898) G glandular stipule of H.xanthoxylon (BRI AQ0771148). Whole leaf showing overall leaflet size and shape of HH.basalticum (BRI AQ0648454) IH.thozetianum (BRI AQ0611464), and JH.xanthoxylon (BRI AQ0874091). Habit of H.basalticum from K Bladensberg National Park, Queensland (photo, Dale Richter, Queensland Herbarium) L 65 km west south-west of Blackall, Queensland (photo, Murray Fagg, Australian Plant Image Index, Australian National Botanic Gardens).
Flowers arranged in globular heads, seeds lacking a pleurogram with a narrow peripheral membranous wing and flat, narrowly oblong, brown pods opening along both sutures distinguish this new genus from other Australian mimosoid legumes, and the keys in Flora of Australia (Cowan 1998) and available on KeyBase (Bean 2021; KeyBase 2021) still remain suitable.
Taxonomic treatment
. Heliodendron
Gill.K. Br. & Bayly gen. nov.
2B4ED609-159C-5CE2-8571-9E5607762628
urn:lsid:ipni.org:names:77303797-1
Diagnosis.
A genus of mimosoid legumes similar to Archidendropsis but differing in the following combination of features: inflorescences of glomerules, calyx and corolla with hairs (restricted to the lobes in H.xanthoxylon); stipules either small (to 1.2 mm) rigid and caducous or glandular (1.2–3 mm long) and persistent; pollen arranged in polyads diameter of 55–68 μm; pollen tectum with isometric channels. In contrast, Archidendropsis has inflorescences of spikes, spiciform racemes, racemes or in one species glomerules, but when in glomerules the calyx and corolla are glabrous; stipules (if present) either small (c. 1mm) ovate or filiform and often caducous, or large auriculate, orbicular, or cordate and persistent; pollen polyad diameter of 80–120 μm, pollen tectum with non-isometric channels.
Description.
Trees or shrubs, with terete branchlets. Stipules either resembling small thorns to 1.2 mm long that are early caducous, or persistent circular-ovate glands 1–3 mm in diameter. Leaves bipinnate, pinnae 1–2 pairs with 1.5–11 leaflet pairs per pinna; glands at the junction of pinnae circular or triangular to rhombic, +/- circular glands at the junction of leaflet petiolules. Leaflets opposite, subsessile (0.2–0.7 mm) or long (3.5–7 mm) petiolulate; elliptic to elliptic-lanceolate or oblong, 2–38 mm × 1.5–15 mm, glabrous to puberulous. Inflorescence of globular heads 0.5–1.7 mm in diameter, either simple or arranged into a panicle up to 35 cm long. Flowers: homomorphic, yellow to cream, sessile. Calyx 1.5–3 mm long, tubular to subcampanulate; corolla 2.5–7 mm long, tubular to narrowly campanulate. Ovary 0.8–2 mm long, solitary and shortly stipitate; stamens numerous 5–9 mm long, united basally into a tube that equals or slightly exceeds the corolla tube. Pollen 16-celled polyads with a diameter of 55–68 μm, tectum with isometric channels. Pod brown, valves chartaceous, 6–22 cm × 0.5–2.5 mm, oblong, flat and dehiscing along both sutures. Seeds lacking a pleurogram, flat, circular to ovate or obliquely ovate, 5–13 mm, with a narrow 0.2–1 mm peripheral, membranous wing. Fig. 5.
Type.
Heliodendronbasalticum (F. Muell.) Gill.K. Br. & Bayly ≡ Acaciabasaltica F. Muell., Journal of the Proceedings of the Linnean Society, Botany 3: 146 (1859)
Etymology.
From the Greek helios (sun) and dendron (tree) alluding to the endemic distribution of the genus in the Australian state of Queensland, widely known as the “sunshine state”, the globular, sun-like inflorescences of yellow flowers, and the tree habit (Fig. 5A, K, L) and also in reference to the genera Archidendropsis (in which the species were previous placed) and Archidendron (which they resemble).
Homotypic synonym.
Archidendropsissubg.Basaltica I.C. Nielsen, Bulletin du Muséum National d’Histoire Naturelle. Section B, Adansonia: Botanique Phytochimie 5(3): 325 (1983).
Notes.
We have chosen to create a new name for this genus rather than making a new combination based on the name Archidendropsissubg.Basaltica. This is because using the name “Basaltica” at generic rank would require a change of epithet for the most widespread species in the genus under Art. 23.4 of the International Code of Nomenclature for algae, fungi, and plants (Turland et al. 2018). To minimise taxonomic change, and to avoid potential confusion, we would rather that the species retains its well-known epithet, which has been in continuous use since 1859.
The genus includes the following three species, all endemic to Queensland, Australia (Fig. 1B).
. Heliodendron basalticum
(F. Muell.) Gill.K. Br. & Bayly comb. nov.
2ABA19F9-2B7A-5BF4-A0B8-465C8D36B716
urn:lsid:ipni.org:names:77303798-1
Basionym.
Acaciabasaltica F. Muell., Journal of the Proceedings of the Linnean Society, Botany 3: 146 (1859). ≡ Albiziabasaltica (F. Muell.) Benth., Flora Australiensis 2: 422 (1864); Archidendropsisbasaltica (F. Muell.) I.C. Nielsen, Bulletin du Muséum National d’Histoire Naturelle. Section B, Adansonia: Botanique Phytochimie 5(3): 326 (1983).
Type.
Peak Downs, F. Mueller 42 (holotype: MEL 594732A image!; isotype K000822321 image!).
. Heliodendron thozetianum
(F. Muell.) Gill.K. Br. & Bayly comb. nov.
DBDDAF57-645B-5E72-A76E-BFC43E16744E
urn:lsid:ipni.org:names:77303799-1
Basionym.
Acaciathozetiana F. Muell., Fragmenta Phytographiae Australiae 4(24): 9 (1863). ≡ Albiziathozetiana (F. Muell.) F. Muell. ex Benth., Flora Australiensis 2: 422 (1864); Archidendropsisthozetiana (F. Muell.) I.C. Nielsen, Bulletin du Muséum National d’Histoire Naturelle. Section B, Adansonia: Botanique Phytochimie 5(3): 326 (1983).
Type.
Fort Cooper, [A. Thozet?] no. 29. (Lectotype, designated by R.S. Cowan, Nuytsia 11: 13 (1996)): MEL 595338A image!; residual syntypes: MEL 595339A, MEL 595340A, MEL 595342A, MEL 595377A].
. Heliodendron xanthoxylon
(C.T. White & W.D. Francis) Gill.K. Br. & Bayly comb. nov.
645F8512-B16D-5E54-AA5A-0A8389874408
urn:lsid:ipni.org:names:77303800-1
Basionym.
Albiziaxanthoxylon C.T. White & W.D. Francis, Proceedings of the Royal Society of Queensland 41: 141, t. X (1929). Archidendropsisxanthoxylon (C.T. White & W.D. Francis) I.C. Nielsen, Bulletin du Muséum National d’Histoire Naturelle. Section B, Adansonia: Botanique Phytochimie 5(3): 326 (1983).
Type.
Atherton District, North Queensland, Overseer brothers s.n. (Provisional Forestry Board), end of October, 1927 (Lectotype, designated by I.C. Nielsen as “Type”, Bulletin du Muséum National d’Histoire Naturelle. Section B, Adansonia: Botanique Phytochimie 5(3): 341 (1983): BRI AQ022813! [2 sheets]; isolectotypes: DNA D0053218 image!, K000822329 image!, MEL 1562403A image!).
Notes.
The protologue of Albiziaxanthoxylon (White and Francis 1929) gave a location, collector name and month of the collection but did not indicate the herbarium in which the type was held, thus meaning that all specimens of this gathering could be considered syntypes. However, it appears that Nielsen inadvertently typified this taxon, according to Art. 7.11 of the ICN (Turland et al. 2018), when providing the description for the new combination of Archidendropsisxanthoxylon with the text “Type: Overseer Brothers, Australia, N. Queensland, Atherton District, Oct 1927, fl. fr. (holo-,BRI; iso-K)” (Nielsen et al. 1983a: p. 341). We believe this satisfies the requirements of Art. 7.11 to effectively lectotypify the name, which means that the BRI specimen is the lectotype and the K specimen is the isolectotype. Interestingly, the material illustrated in the protologue is clearly the isolectotype at K, as it is the only type specimen of Heliodendronxanthoxylon with pods, and the structure of the inflorescence and leaves is almost identical (K000822329; White and Francis 1929).
In Flora of Australia, Cowan (1998) cited BRI as holding an isotype as well as the holotype of this taxon; however, the two sheets have the same collection details, are labelled as sheet 1 of 2 and sheet 2 of 2, and share a single accession number (BRI AQ022813). Therefore, it is herein determined that these are the one collection, and both represent the holotype (now lectotype; BRI AQ022813).
Conclusion
We present the most densely sampled phylogeny of the genera Archidendron and Archidendropsis to date and confirm that both genera are not monophyletic. The well supported clades within the Archidendron clade based on four nuclear markers agree with more data-rich phylogenomic data sets now being generated. A new genus, Heliodendron, endemic to Queensland (Australia), is described for the Australian members of the former Archidendropsissubg.Basaltica. Further sampling of species from subg. Archidendropsis would be beneficial, particularly to ascertain the relationships of the globular flowered A.fournieri and the non-New Caledonian representatives of Archidendropsis s.s. While Archidendron is also not monophyletic, no nomenclatural changes are made, because low phylogenetic support and high topological uncertainty between genera of the Archidendron clade mean that the relationships between the two clades of Archidendron remain uncertain. In addition, discrete macromorphological characters need to be identified to distinguish the two lineages of Archidendron as the basis for generic re-delimitation. A taxonomic revision of the widespread polymorphic A.clypearia would aid this, as our results indicate var.velutinum from eastern Malesia may represent a distinct species. Phylogenomic data and additional sampling of this species would be beneficial before taxonomic changes are made.
Supplementary Material
Acknowledgements
This research was funded by an Australia and Pacific Science Foundation grant (APSF14-4: Systematics and evolution of Archidendron, the largest group of tropical legumes in the Austral region) and an Australia Awards Scholarship to Javier Aju to undertake his Masters. We thank the following herbaria for providing access to specimens, including destructive sampling: AAU, BISH, BRI, CANB, CNS, KEP, KUN, L, NY, MEL and MELU. We thank Shelly James (then BISH now PERTH), Stuart Worboys (CNS) and George Weiblen (Bell Museum) for providing material, and Jo Palmer and Kirsten Cowley (CANB) who sampled and imaged specimens for us. Field work for this project was conducted in Queensland under the permit WITK15692815, issued by the Department of Environment and Heritage Protection, Queensland Government. Darren Crayn is thanked for his support and guidance in the fieldwork to collect samples of Archidendron in north Queensland. Melodina Fabillo is appreciatively recognised for her work on the pollen morphology of Heliodendron, generating Figure 5, and providing feedback on the draft manuscript. Jens Ringelberg is thanked for generating the maps of Archidendron and Archidendropsis s.l. in Figure 1. We also thank Tony Bean, David Halford and Peter Bostock for nomenclatural discussions. Thank you to Else Demeulenaere and Erik Koenen for providing helpful comments and suggestions during the review process to improve the manuscript. Lastly, we thank Colin Hughes for inviting us to be involved in this special issue of Advances in Legume Systematics.
Citation
Brown GK, Aju J, Bayly MJ, Murphy DJ, McLay TGB (2022) Phylogeny and classification of the Australasian and Indomalayan mimosoid legumes Archidendron and Archidendropsis (Leguminosae, subfamily Caesalpinioideae, mimosoid clade). In: Hughes CE, de Queiroz LP, Lewis GP (Eds) Advances in Legume Systematics 14. Classification of Caesalpinioideae Part 1: New generic delimitations. PhytoKeys 205: 299–333. https://doi.org/10.3897/phytokeys.205.79381
Supplementary materials
Primer sequences and PCR variations
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Gillian K. Brown, Javier Aju, Michael J. Bayly, Daniel J. Murphy, Todd G.B. McLay
Data type
Pdf file.
Explanation note
The reference for the primer and their PCR conditions are provided, along with the variations for PCR reagents and cycling conditions for the initial PCR in the two-step PCR process. * only used for sanger sequencing so no variations to note. Standard PCR reagents prior to variation consisted of 2X QIAGEN PCR buffer (QIAGEN), 5 mM of each dNTP (Bioline), 1 µl of each primer (10 µM), 1.25 µl of dimethyl sulfoxide (DMSO, 99.5%; Sigma-Aldrich), 1 U of Taq DNA polymerase, 100 ng of template and made up to 25 µl with ultra pure water per reaction. Reagent variations, A: not varied; B: 200 ng DNA, 1.2 µl BSA instead of DMSO; C: 200 ng DNA; D: 200ng DNA, 6 μM each primer, 1.5 µl MgCl2, 0.9 µl DMSO and 0.1 µl Taq; E: 6 µM each primer. Cycle variations: Z: 94 °C for 15 mins; 30 cycles of 94 °C for 20 sec, 61 °C for 20 sec, 72 °C for 2 mins; 72 °C for 5 mins; Y: 94 °C for 15 mins; 35 cycles of 94 °C for 20 sec, 61 °C for 20 sec, 72 °C for 2 mins; 72 °C for 5 mins; X: 94 °C for 15 mins; 35 cycles of 94 °C for 20 sec, 55 °C for 30 sec, 72 °C for 2 mins; 72 °C for 7 mins; W: 94 °C for 15 mins; 40 cycles of 94 °C for 20 sec, 50 °C for 1 min, 72 °C for 3 mins; 72 °C for 7 mins; V: 80 °C for 5 mins; 40 cycles of 95 °C for 1 min, 50 °C for 1 min with 0.3 °C/sec ramp, 65 °C for 4 mins; 65 °C for 5 mins; U: 94 °C for 5 mins; 30 cycles of 94 °C for 30 sec, 53 °C for 30 sec, 72 °C for 1 min; 72 °C for 7 mins; T: 80 °C for 5 mins; 30 cycles of 95 °C for 1 min, 50 °C for wwith 0.3 °C/sec ramp, 65 °C for 4 mins; 65 °C for 5 mins.
SHMT network and tree
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Gillian K. Brown, Javier Aju, Michael J. Bayly, Daniel J. Murphy, Todd G.B. McLay
Data type
Pdf file.
Explanation note
Neighbour-joining tree and NeighbourNet network are presented with individual samples with more than one allele coloured to highlight their positions. The samples are coloured the same in both the tree and network. The clades that are congruent with Fig. 2 (B, C, D, D1, D2, F1, F2) are labelled. The sequences from species of Albizia (Z106, JA137) were removed as they occur on a very long branch relative to the rest of the samples in the network.
RBPCO network and tree
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Gillian K. Brown, Javier Aju, Michael J. Bayly, Daniel J. Murphy, Todd G.B. McLay
Data type
Pdf file.
Explanation note
Neighbour-joining tree and NeighbourNet network are presented with individual samples with more than one allele coloured to highlight their positions. The samples are coloured are the same in both the tree and network. Clade B, which is congruent with Fig. 2 is labelled.
cpDNA tree
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Gillian K. Brown, Javier Aju, Michael J. Bayly, Daniel J. Murphy, Todd G.B. McLay
Data type
Pdf file.
Explanation note
IQ-Tree of combined cpDNA loci, with all UFBS values shown. The two clades that are congruent with of Fig. 2 are labelled (A and F). Arrows indicate the placement of the two supported incongruences mentioned in the results text.
References
- Ariati SR, Murphy DJ, Udovicic F, Ladiges PY. (2006) Molecular phylogeny of three groups of acacias (AcaciasubgenusPhyllodineae) in arid Australia based on the internal and external transcribed spacer regions of nrDNA. Systematics and Biodiversity 4(4): 417–426. 10.1017/S1477200006001952 [DOI] [Google Scholar]
- AVH (2021) Archidendropsis occurrence data. 10.26197/ala.c6b96911-99fa-4dea-a032-f20b59be877d [DOI]
- Baldwin BG, Markos S. (1998) Phylogenetic utility of the external transcribed spacer (ETS) of 18S-26S rDNA: Congruence of ETS and ITS trees of Calycadenia (Compositae). Molecular Phylogenetics and Evolution 10(3): 449–463. 10.1006/mpev.1998.0545 [DOI] [PubMed] [Google Scholar]
- Baretta-Kuipers T. (1981) Wood anatomy of Leguminosae: its relevance to taxonomy. In: Polhill R, Raven P. (Eds) Advances in Legume Systematics, Part 2.Royal Botanic Gardens, Kew, 677–705.
- Barneby RC, Grimes JW. (1996) Silk tree, guanacaste, monkey’s earring : A generic system for the synandrous Mimosaceae of the Americas. Memoirs of the New York Botanical Garden, 74 pp. http://mertzdigital.nybg.org/digital/collection/p9016coll16/id/5691
- Bean AR. (2021) Species of Archidendropsis, in: KeyBase: Flowering plants of Queensland. https://keybase.rbg.vic.gov.au/keys/show/11753 [December 3, 2021]
- Bentham G. (1875) Revision of the suborder Mimosaeae. Transactions of the Linnean Society 30: 335–664. 10.1111/j.1096-3642.1875.tb00005.x [DOI] [Google Scholar]
- Brown GK, Murphy DJ, Miller JT, Ladiges PY. (2008) Acacias.s. and its relationship among tropical legumes, tribe Ingeae (Leguminosae: Mimosoideae). Systematic Biology 33: 739–751. 10.1600/036364408786500136 [DOI] [Google Scholar]
- Brown GK, Murphy DJ, Ladiges PY. (2011) Relationships of the Australo-Malesian genus Paraserianthes (Mimosoideae: Leguminosae) identifies the sister group of Acaciasensu stricto and two biogeographical tracks. Cladistics 27(4): 380–390. 10.1111/j.1096-0031.2011.00349.x [DOI] [PubMed] [Google Scholar]
- Chappill J, Maslin BR. (1995) A phylogenetic assessment of tribe Acacieae. In: Crisp M, Doyle JJ. (Eds) Advances in Legume Systematics, Part 7, Phylogeny.Royal Botanic Gardens, Kew, 77–99.
- Choi HK, Luckow MA, Doyle JJ, Cook DR. (2006) Development of nuclear gene-derived molecular markers linked to legume genetic maps. Molecular Genetics and Genomics 276(1): 56–70. 10.1007/s00438-006-0118-8 [DOI] [PubMed] [Google Scholar]
- Comben DF, McCulloch GA, Brown GK, Walter GH. (2020) Phylogenetic placement and the timing of diversification in Australia’s endemic Vachellia (Caesalpinioideae, Mimosoid Clade, Fabaceae) species. Australian Systematic Botany 33(1): 103–109. 10.1071/SB19013 [DOI] [Google Scholar]
- Cowan RS. (1996) Notes on miscellaneous mimosoid legumes (Leguminosae: Mimosoideae), mostly Australian. Nuytsia 11: 11–19. https://www.biodiversitylibrary.org/page/53381804#page/16/mode/1up [Google Scholar]
- Cowan RS. (1998) Mimosaceae (excl. Acacia), Flora of Australia 12. CSIRO Australia, Melbourne, 49 pp. [Google Scholar]
- Dash SS, Sanjappa M. (2011) Two new species and a new distributional record of Archidendron F. Muell. (Leguminosae: Mimosoideae) from India. Nelumbo 53: 7–16. [Google Scholar]
- de Souza É, Lewis G, Forest F, Schnadelbach AS, van den Berg C, de Queiroz LP. (2013) Phylogeny of Calliandra (Leguminosae: Mimosoideae) based on nuclear and plastid molecular markers. Taxon 62(6): 1200–1219. 10.12705/626.2 [DOI] [Google Scholar]
- de Wit HCD. (1942) Conspectus of the genus Archidendron F. von Mueller (Legum). Bulletin of the Botanic Gardens of Buitenzorg, Series III 17: 256–272. [Google Scholar]
- Demeulenaere E, Schils T, Burleigh JG, Ringelberg JJ, Koenen EJM, Ickert-Bond SM. (2022) Phylogenomic assessment prompts recognition of the Serianthes clade and confirms the monophyly of Serianthes and its relationship with Falcataria and Wallaceodendron in the wider ingoid clade (Leguminosae, Caesalpinioideae). In: Hughes CE, de Queiroz LP, Lewis GP. (Eds) Advances in Legume Systematics 14. Classification of Caesalpinioideae Part 1: New generic delimitations.PhytoKeys 205: 335–362. 10.3897/phytokeys.205.79144 [DOI] [PMC free article] [PubMed]
- Doyle JJ, Doyle JL. (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19: 11–15. [Google Scholar]
- Edgar RC. (2004) MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32(5): 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferm J, Korall P, Lewis GP, Ståhl B. (2019) Phylogeny of the Neotropical legume genera Zygia and Marmaroxylon and close relatives. Taxon 68(4): 661–672. 10.1002/tax.12117 [DOI] [Google Scholar]
- Fosberg FR. (1965) AlbiziaSect.Pachysperma (Leguminosae-Mimosoideae). Reinwardtia 7: 71–90. [Google Scholar]
- Gunn CR. (1984) Fruits and seeds of genera in the subfamily Mimosoideae (Fabaceae). U.S. Department of Agriculture, Technical Bulletin 1681: 1–194. https://naldc.nal.usda.gov/download/CAT85842079/PDF [Google Scholar]
- Huson DH, Bryant D. (2006) Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution 23(2): 254–267. 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- Iganci JRV, Soares MV, Guerra E, Morim MP. (2016) A preliminary molecular phylogeny of the Abarema alliance (Leguminosae) and implications for taxonomic rearrangement. International Journal of Plant Sciences 177(1): 34–43. 10.1086/684078 [DOI] [Google Scholar]
- Johnson LA, Soltis DE. (1994) matK DNA sequences and phylogenetic reconstruction in Saxifragaceae s. str. Systematic Botany 19(1): 143–156. 10.2307/2419718 [DOI] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. (2017) ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods 14(6): 587–589. 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käss E, Wink M. (1997) Molecular phylogeny and phylogeography of Lupinus (Leguminosae) inferred from nucleotide sequences of the rbcL gene and ITS 1+2 regions of rDNA. Plant Systematics and Evolution 208: 139–167. 10.1007/BF00985439 [DOI] [Google Scholar]
- KeyBase (2021) Flora of Australia: vascular plants: Species of Archidendropsis. https://keybase.rbg.vic.gov.au/keys/show/518
- Koenen EJM, Kidner C, de Souza ÉR, Simon MF, Iganci JR, Nicholls JA, Brown GK, de Queiroz LP, Luckow M, Lewis GP, Pennington RT, Hughes CE. (2020) Hybrid capture of 964 nuclear genes resolves evolutionary relationships in the mimosoid legumes and reveals the polytomous origins of a large pantropical radiation. American Journal of Botany 107(12): 1710–1735. 10.1002/ajb2.1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostermans AJGH. (1954) A monograph of the Asiatic, Malaysian, Australian and Pacific species of Mimosaceae, formerly included in Pithecellobium Mart. The Bulletin of the Organization for Scientific Research in Indonesia 20: 1–122. [Google Scholar]
- Kostermans AJGH. (1966) Notes on some Asian mimosaceous genera. Adansonia 6: 351–373. [Google Scholar]
- Legume Phylogeny Working Group (2017) A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny. Taxon 66(1): 44–77. 10.12705/661.3 [DOI] [Google Scholar]
- Legume Phylogeny Working Group (2021) The World Checklist of Vascular Plants (WCVP): Fabaceae. https://hp-legume.gbif-staging.org/taxonomy/search?TAXON_ID=x4&facet=rank&facet=issue&facet=status&facet=nomStatus&facet=nameType&facet=field&facet=authorship&facet=extinct&facet=environment&limit=50&offset=0&q=Archidendron%20clypearia
- Lewis GP, Rico Arce ML. (2005) Tribe Ingeae. Legumes of the World. Royal Botanic Gardens Kew, Richmond, Surry UK, 193–213.
- Li M, Wunder J, Bissoli G, Scarponi E, Gazzani S, Barbaro E, Saedler H, Varotto C. (2008) Development of COS genes as universally amplifiable markers for phylogenetic reconstructions of closely related plant species. Cladistics 24(5): 727–745. 10.1111/j.1096-0031.2008.00207.x [DOI] [Google Scholar]
- Luckow M, Miller J, Murphy DJ, Livshultz T. (2003) A phylogenetic analysis of the Mimosoideae (Leguminosae) based on chloroplast DNA sequence data. In: Klitgaard BB, Bruneau A. (Eds) Advances in Legume Systematics Part 10, Higher-level Systematics.Kew, Royal Boatnic Gardens, 197–220. http://www.worldwidewattle.com/infogallery/publications/lucknow-et-al-2003.pdf
- Maumont S. (1993) Seed-coat anatomy of the non-pleurogrammic seeds in the tribe Ingeae (Leguminosae, Mimosoideae). Brittonia 45(3): 249–259. 10.2307/2807111 [DOI] [Google Scholar]
- McLay TGB, Ladiges PY, Doyle SR, Bayly MJ. (2021) Phylogeographic patterns of the Australian grass trees (XanthorrhoeaAsphodelaceae) shown using targeted amplicon sequencing. Australian Systematic Botany 34(2): 206. 10.1071/SB20013 [DOI] [Google Scholar]
- Merrill ED, Perry LM. (1942) PlantaePapuanae Archbolianae, X. Journal of the Arnold Arboretum 23(4): 383–416. 10.5962/p.185463 [DOI] [Google Scholar]
- Miller JT, Bayer RJ. (2001) Molecular phylogenetics of Acacia (Fabaceae: Mimosoideae) based on the chloroplast matK coding sequence and flanking trnK intro spacer regions. American Journal of Botany 88(4): 697–705. 10.2307/2657071 [DOI] [PubMed] [Google Scholar]
- Miller JT, Grimes JW, Murphy DJ, Bayer RJ, Ladiges PY. (2003) A phylogenetic analysis of the Acacieae and Ingeae (Mimosoideae: Fabaceae) based on trnK, matK, psbA-trnH, and trnL/trnF sequence data. Systematic Botany 28: 558–566. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. 2010 Gateway Computing Environments Workshop, GCE 2010. 10.1109/GCE.2010.5676129 [DOI]
- Miller JT, Terra V, Riggins C, Ebinger JE, Seigler DS. (2017) Molecular Phylogenetics of Parasenegalia and Pseudosenegalia (Fabaceae: Mimosoideae). Systematic Botany 42(3): 465–469. 10.1600/036364417X696140 [DOI] [Google Scholar]
- Minh BQ, Nguyen MAT, von Haeseler A. (2013) Ultrafast approximation for phylogenetic bootstrap. Molecular Biology and Evolution 30(5): 1188–1195. 10.1093/molbev/mst024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller FJH. (1859) Contributiones ad Acaciarum Australiae Cognitionem. Journal of the Proceedings of the Linnean Society. Botany 3: 114–148. 10.1111/j.1095-8339.1859.tb02045.x [DOI] [Google Scholar]
- Murphy DJ, Brown GK, Miller JT, Ladiges PY. (2010) Molecular phylogeny of Acacia Mill. (Mimosoideae: Leguminosae): Evidence for major clades and informal classification. Taxon 59(1): 7–19. 10.1002/tax.591002 [DOI] [Google Scholar]
- Nielsen IC. (1979) Notes on the genera Archidendron F. v. Mueller and Pithecellobium Martius in Mainland S.E. Asia. Adansonia 19: 3–37. https://www.biodiversitylibrary.org/part/297165 [Google Scholar]
- Nielsen IC. (1981) Flore du Cambodge, du Laos et du Vietnam 19 Légumineuses-Mimosoïdées. Aubréville A, Leroy J-F (Eds), 1–164. https://www.biodiversitylibrary.org/bibliography/166374
- Nielsen IC. (1982) The Australian species of Archidendron. Nordic Journal of Botany 2(5): 479–490. 10.1111/j.1756-1051.1982.tb01213.x [DOI] [Google Scholar]
- Nielsen IC. (1983) Mimosoideae. Flore de la Nouvelle-Caledonie et Dependances 12: 3–103. [Google Scholar]
- Nielsen IC. (1992) Mimosaceae (Leguminosae-Mimosoideae). Flora Malesiana Series I. Spermatophyta 11: 1–226. https://www.biodiversitylibrary.org/item/90410#page/233/mode/1up [Google Scholar]
- Nielsen IC, Guinet P, Baretta-Kuipers T. (1983a) Studies in the Malesian, Australian and Pacific Ingeae (Leguminosae-Mimosoideae): the genera Archidendropsis, Wallaceodendron, Paraserianthes, Pararchidendron and Serianthes (part 2). Bulletin du Muséum National d’Histoire Naturelle Section B, Adansonia, botanique, phytochimie 5: 335–360. https://www.biodiversitylibrary.org/part/276288 [Google Scholar]
- Nielsen IC, Guinet P, Baretta-Kuipers T. (1983b) Studies in the Malesian, Australian and Pacific Ingeae (Leguminosae-Mimosoideae): the genera Archidendropsis, Wallaceodendron, Paraserianthes, Pararchidendron and Serianthes (part 1). Bulletin du Muséum National d’Histoire Naturelle Section B, Adansonia, botanique, phytochimie 5: 303–329. https://www.biodiversitylibrary.org/part/276287 [Google Scholar]
- Nielsen IC, Guinet P, Baretta-Kuipers T. (1984a) Studies in the Malesian, Australian and Pacific Ingeae (Leguminosae-Mimosoideae): the genera Archidendropsis, Wallaceodendron, Paraserianthes, Pararchidendron and Serianthes (part 3). Bulletin du Muséum National d’Histoire Naturelle Section B, Adansonia, botanique, phytochimie 6: 79–111. https://www.biodiversitylibrary.org/part/274937 [Google Scholar]
- Nielsen IC, Baretta-Kuipers T, Guinet P. (1984b) The genus Archidendron (Leguminosae – Mimosoideae). Opera Botanica 4: 1–120. 10.1111/j.1756-1051.1984.tb02008.x [DOI] [Google Scholar]
- Pebesma E. (2018) Simple Features for R: Standardized support for spatial vector data. The R Journal 10(1): 439–446. 10.32614/RJ-2018-009 [DOI] [Google Scholar]
- Ringelberg JJ, Koenen EJM, Iganci JR, de Queiroz LP, Murphy DJ, Gaudeul M, Bruneau A, Luckow M, Lewis GP, Hughes CE. (2022) Phylogenomic analysis of 997 nuclear genes reveals the need for extensive generic re-delimitation in Caesalpinioideae (Leguminosae). In: Hughes CE, de Queiroz LP, Lewis GP. (Eds) Advances in Legume Systematics 14. Classification of Caesalpinioideae Part 1: New generic delimitations.PhytoKeys 205: 3–58. 10.3897/phytokeys.205.85866 [DOI] [PMC free article] [PubMed]
- Rodrigues-Junior AG, Baskin CC, Baskin JM, De-Paula OC. (2021) The pleurogram, an under-investigated functional trait in seeds. Annals of Botany 127(2): 167–174. 10.1093/aob/mcaa161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohland N, Reich D. (2012) Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Research 22(5): 939–946. 10.1101/gr.128124.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61(3): 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. (2000) Primer3 on the WWW for general users and for biologist programmers. Methods in Molecular Biology (Clifton, N.J.) 132: 365–386. 10.1385/1-59259-192-2:365 [DOI] [PubMed]
- Shaw J, Lickey EB, Schilling EE, Small RL. (2007) Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. American Journal of Botany 94(3): 275–288. 10.3732/ajb.94.3.275 [DOI] [PubMed] [Google Scholar]
- Shepherd LD, McLay TGB. (2011) Two micro-scale protocols for the isolation of DNA from polysaccharide-rich plant tissue. Journal of Plant Research 124(2): 311–314. 10.1007/s10265-010-0379-5 [DOI] [PubMed] [Google Scholar]
- South A. (2017) rnaturalearth: World Map Data from Natural Earth. R package version 0.1.0. https://CRAN.R-project.org/package=rnaturalearth
- Sun Y, Skinner DZ, Liang GH, Hulbert SH. (1994) Phylogenetic analysis of Sorghum and related taxa using internal transcribed spacers of nuclear ribosomal DNA. Theoretical and Applied Genetics 89(1): 26–32. 10.1007/BF00226978 [DOI] [PubMed] [Google Scholar]
- Thiers BM. (updated continuously). Index Herbariorum. http://sweetgum.nybg.org/science/ih/
- Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ. (2016) W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research 44(W1): W232–W235. 10.1093/nar/gkw256 [DOI] [PMC free article] [PubMed]
- Turland N, Wiersema J, Barrie F, Greuter W, Hawksworth D, Herendeen P, Knapp S, Kusber W-H, Li D-Z, Marhold K, May T, McNeill J, Monro A, Prado J, Price M, Smith G [Eds] (2018) 159 International Code of Nomenclature for Algae, Fungi, and Plants. Koeltz Botanical Books. 10.12705/Code.2018 [DOI]
- White CT, Francis WD. (1929) Contributions to the Queensland Flora, No. 4. Proceedings of the Royal Society of Queensland 1(41): 139–143. 10.1126/science.24.602.58 [DOI] [Google Scholar]
- Wickham H. (2016) ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag, New York, United States. 10.1007/978-3-319-24277-4_9 [DOI]
- Wu D, Nielsen IC. (2010) Tribe Ingeae. Archidendron. In: Wu ZU, Raven PH, Hong DY. (Eds) , Flora of China.Science Press, Beijing, and Missouri Botanical Garden Press, St. Louis, 66–71.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sequences and PCR variations
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Gillian K. Brown, Javier Aju, Michael J. Bayly, Daniel J. Murphy, Todd G.B. McLay
Data type
Pdf file.
Explanation note
The reference for the primer and their PCR conditions are provided, along with the variations for PCR reagents and cycling conditions for the initial PCR in the two-step PCR process. * only used for sanger sequencing so no variations to note. Standard PCR reagents prior to variation consisted of 2X QIAGEN PCR buffer (QIAGEN), 5 mM of each dNTP (Bioline), 1 µl of each primer (10 µM), 1.25 µl of dimethyl sulfoxide (DMSO, 99.5%; Sigma-Aldrich), 1 U of Taq DNA polymerase, 100 ng of template and made up to 25 µl with ultra pure water per reaction. Reagent variations, A: not varied; B: 200 ng DNA, 1.2 µl BSA instead of DMSO; C: 200 ng DNA; D: 200ng DNA, 6 μM each primer, 1.5 µl MgCl2, 0.9 µl DMSO and 0.1 µl Taq; E: 6 µM each primer. Cycle variations: Z: 94 °C for 15 mins; 30 cycles of 94 °C for 20 sec, 61 °C for 20 sec, 72 °C for 2 mins; 72 °C for 5 mins; Y: 94 °C for 15 mins; 35 cycles of 94 °C for 20 sec, 61 °C for 20 sec, 72 °C for 2 mins; 72 °C for 5 mins; X: 94 °C for 15 mins; 35 cycles of 94 °C for 20 sec, 55 °C for 30 sec, 72 °C for 2 mins; 72 °C for 7 mins; W: 94 °C for 15 mins; 40 cycles of 94 °C for 20 sec, 50 °C for 1 min, 72 °C for 3 mins; 72 °C for 7 mins; V: 80 °C for 5 mins; 40 cycles of 95 °C for 1 min, 50 °C for 1 min with 0.3 °C/sec ramp, 65 °C for 4 mins; 65 °C for 5 mins; U: 94 °C for 5 mins; 30 cycles of 94 °C for 30 sec, 53 °C for 30 sec, 72 °C for 1 min; 72 °C for 7 mins; T: 80 °C for 5 mins; 30 cycles of 95 °C for 1 min, 50 °C for wwith 0.3 °C/sec ramp, 65 °C for 4 mins; 65 °C for 5 mins.
SHMT network and tree
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Gillian K. Brown, Javier Aju, Michael J. Bayly, Daniel J. Murphy, Todd G.B. McLay
Data type
Pdf file.
Explanation note
Neighbour-joining tree and NeighbourNet network are presented with individual samples with more than one allele coloured to highlight their positions. The samples are coloured the same in both the tree and network. The clades that are congruent with Fig. 2 (B, C, D, D1, D2, F1, F2) are labelled. The sequences from species of Albizia (Z106, JA137) were removed as they occur on a very long branch relative to the rest of the samples in the network.
RBPCO network and tree
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Gillian K. Brown, Javier Aju, Michael J. Bayly, Daniel J. Murphy, Todd G.B. McLay
Data type
Pdf file.
Explanation note
Neighbour-joining tree and NeighbourNet network are presented with individual samples with more than one allele coloured to highlight their positions. The samples are coloured are the same in both the tree and network. Clade B, which is congruent with Fig. 2 is labelled.
cpDNA tree
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Gillian K. Brown, Javier Aju, Michael J. Bayly, Daniel J. Murphy, Todd G.B. McLay
Data type
Pdf file.
Explanation note
IQ-Tree of combined cpDNA loci, with all UFBS values shown. The two clades that are congruent with of Fig. 2 are labelled (A and F). Arrows indicate the placement of the two supported incongruences mentioned in the results text.