Abstract
A newly discovered natural hybrid, Iris×ampliflora Y.E. Xiao, F.Y. Yu & X.F. Chen (Iridaceae: subgenus LimnirissectionLophiris) from Chongqing, China, is described and illustrated. This hybrid is morphologically similar to I.japonica Thunb. and I.wattii Baker, but can be distinguished by its giant leaves and large purple flowers. Phylogenetic trees based on cpDNA data support the separation of I.×ampliflora from other closely related species in the section Lophiris. According to its morphological features, molecular systematic evidence and chromosome data, we speculate that I.×ampliflora [31 chromosomes] likely is a new hybrid between I.japonica [2n = 32] and I.wattii [2n = 30].
Keywords: chloroplast DNA, Chongqing, Iris × ampliflora, Section Lophiris
Introduction
Iris L. is the largest genus in family Iridaceae with up to 280 species that are mainly distributed in temperate regions of the Northern Hemisphere (Goldblatt and Manning 2008). The characteristics of iris flowers are petaloid-style branches, two obvious perianth whorls, and floral tubes with nectaries (Goldblatt and Manning 2008). Irises produce showy flowers and are well-known and popular ornamental plants worldwide. To gain more cultivars for gardens, many crosses between Iris species have been attempted and achieved success by artificial hybridization (Hu and Xiao 2012; Yu et al. 2017). Natural hybrids between sibling species do occur in the genus Iris, for example in some species within subgen. Limniris in North America (Burke et al. 1998). However, there is no natural hybrid of Iris reported in East Asia.
The outer sepals of irises are equipped with raised beards, crests, signal patches, and midveins, which probably have significance for pollination (Sapir et al. 2005; Guan et al. 2009; Xiao et al. 2019). The sepal crest is an important character in the taxonomic delineation of higher ranks of Iris. Dykes (1913) first placed six rhizomatous species in sect. Evansia. Lawrence (1953) changed Evansia to the subsection level. Rodionenko (1987) elevated subsect. Evansia to subgen. Crossiris. Mathew (1981) elevated subsect. Evansia of Lawrence to section level, namely sect. Lophiris, placing the section within subgen. Limniris. sect. Lophiris contains 11 species, most of which are distributed in Eastern Asia, apart from three species in North America (I.cristata Solander, I.lacustris Nuttall, and I.tenuis S. Watson) (British Iris Society Species Group 1997; Zhao et al. 2000). Mathew’s classification was used in this study.
During field work in Chongqing, we found an interesting specimen originally from the Qingyang Town in Fuling District, Chongqing City, China. Our observations show that it is morphologically similar to I.japonica Thunb. and I.wattii Baker with a yellow and irregularly toothed crest on the outer segments. However, its morphological features differ markedly from those of all known species in sect. Lophiris described by Mathew (1981). We have observed the situation concerning seed set since 2014 when the species was initially introduced to the conservation nursery of Shanghai Botanical Garden. It is sterile with no seed production, which indicates its hybrid origin.
This study was undertaken to assess the status and parentage of the new hybrid by morphological surveys, phylogenetic and chromosome data. The Qingyang collection is a large evergreen plant with large attractive flowers, and it can adapt to warm, wet, and full-sun or partly shady environmental conditions. It has potential uses in breeding and landscaping. We formally publish its description here with the aim of better understanding and utilization of the remarkable morphological divergence of Iris species in subgenus Limnirissect.Lophiris.
Materials and methods
Morphological surveys
Living specimens and vouchers of the new hybrid were examined and compared with four species of sect. Lophiris (I.confusa Sealy, I.japonica Thunb., I.tectorum Maxim. and I.wattii Baker) in Southern China using measurements and descriptions of the main characteristics. Species from the conservation nursery of Shanghai Botanical Garden from mid-March and late-April in 2019 were compared. Eight or ten randomly chosen individuals of each taxon were used for the morphometric surveys (Table 1). Meanwhile, herbarium sheets (CDBI, CSH, IMC, IBSC, LBG, KUN, PE) and type descriptions of I.confusa, I.japonica, I.milesii Baker ex Foster, I.tectorum and I.wattii were compared with the new hybrid.
Table 1.
Differences between Iris×ampliflora, I.confusa, I.tectorum, I.wattii and I.japonica in the conservation nursery of Shanghai Botanical Garden.
| Species | I.tectorum | I.confusa | I.wattii | I.×ampliflora | I.japonica | |
|---|---|---|---|---|---|---|
| (n = 10) | (n = 10) | (n = 8) | (n = 10) | (n = 10) | ||
| Aerial stem Length | No | 67.2 ± 19.9a | 30.3 ± 9.3b | 27.0 ± 2.7b | 12.3 ± 2.7c | |
| Leaf | Waxy | No | Yes | No | No | Yes |
| Longitudinal veins | Clearly | Clearly | Clearly | Clearly | Clearly | |
| Texture | Wrinkled | Smooth | Wrinkled | Wrinkled | Smooth | |
| Length | 49.5 ± 3.5c | 54.8 ± 3.8b | 70.1 ± 9.2a | 74.2 ± 6.4a | 48.2 ± 2.9c | |
| Width | 2.9 ± 0.2d | 5.3 ± 0.6b | 4.3 ± 1.0c | 6.6 ± 0.8a | 3.8 ± 0.7c | |
| Flower | Flowering-stem | 1–2 branches | 2–4 branches | 5–7 branches | 7–10 branches | 5–12 branches |
| Color | Bluish violet | Pale reddish purple | Bluish violet | Violet | Violet / Bluish violet | |
| Size (in diam.) | 9.2 ± 1.1b | 5.1 ± 0.4d | 7.3 ± 0.4 c | 12.5 ± 0.5a | 4.8 ± 0.4d | |
| Crest | White | Yellow | Yellow | Yellow | Yellow | |
| Anthers | White | White | Yellow | White | White | |
| Chromosome number | 28 | 30 | 30 | 31 | 28, 30, 31, 32, 33, 34, 35, 54 and 55 | |
| Distribution | Subtropical and temperate zone of China | Chongqing, Sichuan, Xizang, Yunnan [NW India] | Chongqing, Sichuan, Xizang, Yunnan [NE India, Myanmar]. | Chongqing | Subtropical and temperature zone of China [Japan] | |
Note: n, sample number.
Phylogenetic analyses
We collected samples of five species / hybrid (I.confusa, I.japonica, I.tectorum, I.wattii, and Qingyang collection) of sect. Lophiris, and three species (I.anguifuga Y.T. Zhao, I.henryi Baker and I.proantha Diels) of section Limniris and Irisdomestica (L.) Goldblatt & Mabb. to construct phylogenetic trees based on cpDNA data. The samples of the new hybrid were collected from the type locality. Other iris samples were collected from the conservation nursery of the Shanghai Botanical Garden.
Total genomic DNA was extracted from each sample (30 mg dried leaves) using a DNA Plant Kit (Tiangen, Shanghai, China) according to manufacturer’s instructions. The extracted DNAs were dissolved in 100 μl TE buffer for storage. We amplified and sequenced part of the matK gene (Wilson 2004) and ndhF gene (Shaw et al. 2007) (Table 2). Each PCR mixture (50 μl) contained ddH2O, 1 × buffer (Mg2+ free), 2.5 mM MgCl2, 2.5 mM each dNTP, 0.5 μM each primer, 2 U Taq polymerase (Sangon, Shanghai, China), and 20 ng DNA. The PCR reactions were conducted on a Mastercycler pro Thermal Cycler (Eppendorf, Hamburg, Germany). The procedure was performed with initial denaturing for 5 min at 94 °C followed by 35 cycles of 30 sec at 94 °C (denaturing), 30 sec at 50 °C (annealing), and 45 sec at 72 °C (elongation), and final extension for 10 min at 72 °C. After checking products by electrophoresis on a 1.2% agarose gel, the purified products were bi-directionally sequenced by standard methods on the ABI 3731 automated sequencer (Applied Biosystems, Foster City, CA, USA). The sequences of these irises have been deposited in GenBank (see accession numbers in Appendix 1: Table A1. For the cpDNA datasets, the CLUSTALW program combined with manual adjustment was used for multiple alignments of all sequences (Thompson et al. 1994). We used three phylogenetic methods (i.e., Bayesian inference, maximum parsimony, and maximum likelihood) to analyze the alignments. We conducted the Bayesian inference analysis with MrBayes v.3.2.6 (Ronquist and Huelsenbeck 2003), and the maximum parsimony and maximum likelihood analyses using PAUP v.4.0b10 (Swofford 2002).
Table 2.
Primers used in this study.
| Gene name | Primer | Sequence (5′ to 3′) |
|---|---|---|
| matK | matK-3914F | ATCTGGGTTGCTAACTCAATGG |
| (Wilson 2004) | matK-1235R | GGAGTGGGGTATTAGTATA |
| matK-1176F | CTATTCATTCCATTTTTCCT | |
| matK-trnK2R | AACTAGTCGGATGGAGTAG | |
| ndhF | ndhF-pair1 | ATGGAACA(GT)ACATAT(CG)AATATGC |
| (Shaw et al. 2007) | ndhF-1201ir | GGAATACCACAAAGAGAAAGTGTACCT |
| ndhF-972i | GTCTCAATTGGGTTATATTATG | |
| ndhF-2210R | CCCCCTA(CT)ATATTTGATACCTTCTCC |
Chromosome number analyses
To determine the chromosome number of the new hybrid from somatic cells. Root tips (1–1.5 cm in length) were collected and washed with distilled water and immersed in 0.002 mol/L 8-hydroxyquinoline with dark pretreatment for 2–2.5 h. These roots then were fixed in Carnoy solution (volume ratio: 95% ethanol: acetic acid = 3:1) at 4 °C for 2–3 h. The fixed roots were dissociated in 5 mol/L hydrochloric acid for 8–10 min and washed with distilled water, then stained with carbol fuchsin and squashed on glass slides. Finally, the samples were observed and photographed using a Motic BA400 optical microscope. More than 20 cells were observed to determine the number of chromosomes for each specimen examined.
Results
Morphological comparisons
The Qingyang collection is morphologically similar to the species of section Lophiris, I.japonica which has 5–10 branches of the flowering stem, a yellow crest on the outer sepals, and is the most common species of Iris in Southern China (Zhao et al. 2000). However, the Qingyang collection has several characteristics that distinguish it from I.japonica, I.confusa, I.milesii, I.wattii and I.tectorum, including plant larger flowers, larger leaves, and other features as described in Table 1. The flowering stem of this specimen has 7–10 branches and an aerial stem (mean length = 27.0 ± 2.7 cm, n = 10) (Fig. 1). Compared with other species in sect. Lophiris, the Qingyang collection has larger leaves (mean length = 74.2 ± 6.4 cm, n = 10; mean width = 6.6 ± 0.8 cm, n = 10) and larger purple flowers (mean diameter = 12.5 ± 0.5 cm, n = 10) (Fig. 2).
Figure 1.
Line drawings of Iris×ampliflora based on photos and the type specimens (Drawn by Tian-Yi Yu).
Figure 2.
AIris×ampliflora in flower B flower anatomy of Iris×ampliflora.
Phylogenetics
Among eight individuals of different species / hybrid, there were 318 variable sites, 187 singleton variable sites, and 131 parsimony informative sites across 4779 bp aligned positions of two cpDNA fragments. There were 22 and 39 mutations between the sequences of I.japonica / I.wattii and the new hybrid, respectively.
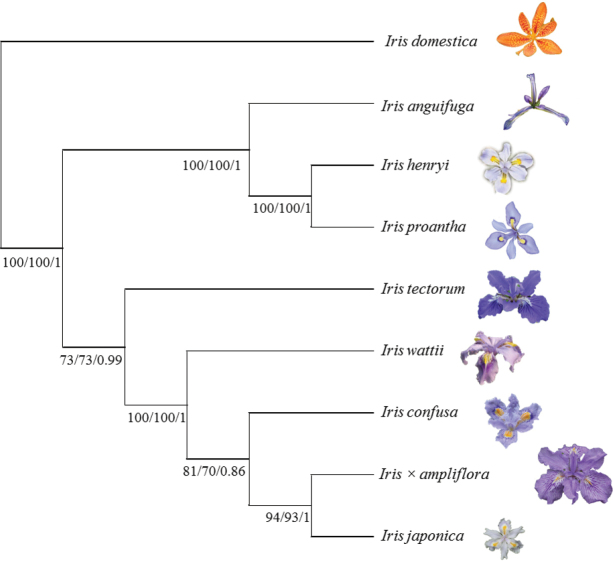
In the molecular tree based on cpDNA data, the sampled new hybrid was resolved as sister to the sample of I.japonica (Fig. 3). In Bayesian, maximum parsimony, and maximum likelihood trees based on cpDNA data, this specimen clustered into a clade with I.japonica (Fig. 3). The inter-clade sequence difference (I.japonica vs. I.wattii, 0.11) was 22 times greater than the intra-clade sequence differences (the new hybrid vs. I.japonica, 0.05).
Figure 3.
Phylogenetic relationships of Iris×ampliflora based on two plastid DNA fragments. Numbers around nodes are Bayesian posterior probabilities and bootstrap percentages (PP /BSMP/BSML). (Pictures of flowers are relatively proportional sizes among different Iris species).
Chromosome number
Cytological study indicated that the Qingyang collection has 31 chromosomes (Fig. 4). No evidence of chromosome pairing could be examined and it is unknown if it functions as a diploid.
Figure 4.
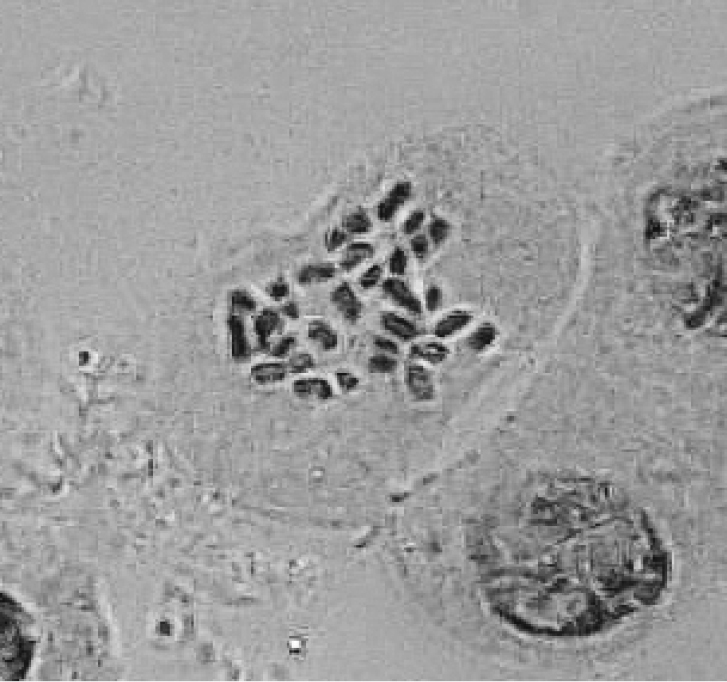
Mitotic metaphase chromosomes of Iris×ampliflora [number of chromosomes, 31].
Taxonomy
Iris × ampliflora
Y.E. Xiao, F.Y. Yu & X.F. Chen nothosp. nov.
B2DF36F7-DC0C-5D58-B02A-5ACCE9B11ECB
Figures 1 , 2 , Appendix 2 : Figure A1
Diagnosis.
Morphologically similar to I.japonica, the new hybrid differs in having 7–10 branches and an aerial stem (25–29 cm compared to 10–15 cm), larger leaves (length and width, 68–80 cm and 6–7 cm compared to 45–51 cm and 3–4.5 cm) and larger purple flowers (diam., 11–13 cm compared to 4–5 cm).
Type.
Shanghai Botanical Garden, grown from collection from the Qingyang Town of Fuling District, Chongqing, 14 June 2014, Y.E. Xiao XYE20140614 (holotype: CSH-0180673!; isotypes: CSH!). A photo of the holotype is shown in Appendix 2: Figure A1.
Description.
Rhizomes creeping, thick, ca. 1.5–2 cm in diam. Overall plant up to 85.4–125.5 cm. Stem ascending upright, 24–31 cm. Leaves mainly in basal fans, green, broadly sword-shaped, curved, midrib evident, 69.0–82.9 × 5.5–7.5 cm, the basal leaves fibrous. Flowering stems with 7–10 branched that arise in the axillary leaves, 1- or 2 leaved subtend the flower on the branch, ca. 50.0 × 4.5 cm. Spathes 2 or 3, green, lanceolate, ca. 2 cm, 3- or 4- flowered, seldom 5, apex acuminate. Flowers blueish violet, 11.5–12.8 cm in diam.; pedicel 1.5–3.0 cm, perianth tube slender, ca. 1.5 cm; outer segments mottled darker around conspicuous, yellow, irregularly toothed crest, broadly ovate, 6.8–7.3 × 5.2–5.5 cm; inner segments spreading horizontally at anthesis, elliptical, 5.5–6.2 × 3.9–4.2 cm. Stamens ca. 2 cm; anthers bright white without pollen. Style branches pale bluish violet, 4–5 cm, feathery apex, terminal lobes fimbriate. Ovary cylindrical, ca. 2 cm. Flowering season, March–April. Sterile.
Etymology.
The new hybrid is named for the large flower.
Distribution and habitat.
Iris×ampliflora was collected from the Qingyang Town, Fuling District, Chongqing, China (29°31'40.8"N, 107°12'54"E). With complicated mountainous topography, the Qingyang Town is located in the range of the Dalou Mountains with an average altitude of 750 m. There were about 10 clones each with 6–10 individuals in the population of I.×ampliflora, covering an area of 200 m2. Plants of I.×ampliflora grow well on roadsides of subtropical mixed evergreen deciduous broad-leaved forest in full-sun and partly-shaded environments at an altitude of about 650 m. The lowest and highest temperature of the original site are about -5 °C and 38 °C, respectively.
Phenology and reproductive characteristics.
Iris×ampliflora blooms in March to April in Chongqing, and it blooms in April in Shanghai. It is evergreen. No fruits have been observed, but it can reproduce vegetatively.
Preliminary conservation status.
Only one population of I.×ampliflora was found by our investigation in Qingyang Region and there are risks of disturbance by human activities. I.×ampliflora is currently cultivated in the conservation nurseries of Shanghai Botanical Garden and the Flower Fragrance Horticulture Limited Company in Chongqing.
Other Iris species examined.
I.japonica Chongqing Municipality, WUK0495843; PE01012482; PE01012483; PE01012489; PE01012492; IMC0013795. I.confusa Chongqing Municipality, CDBI0169691, IMC0013989, IMC0013998; IMC0014000; IMC0014010. I.milesii Yunnan Province, LBG 00106670; LBG00106671; KUN0360444. I.wattii Chongqing Municipality, KUN0360622. Mount Emei, Sichuan Province, CSH0086611. Liangshan Prefecture, Sichuan Province, PE01013840. I.tectorum, Chongqing Municipality, IBSC0629040; IBSC0629027; CDBI0169658.
Discussion
With the characters of maternal inheritance in cpDNA genes, I.japonica was most likely postulated as maternal parent of the hybrid I.×ampliflora because these two cluster as sister taxa in a clade in the phylogenetic tree. It is difficult to find the actual maternal parent of I.×ampliflora because the chromosome number of I.japonica is variable, 2n = 28, 30, 31, 32, 33, 34, 35, 54 and 55 (British Iris Society Species Group 1997). However, the chromosome number of the paternal parent of the hybrid can be speculated, 2n = 34, 31, 30, 29, 28, 27, since I.×ampliflora has 31 chromosomes. Thus, the possibilities about the parentage of I.×ampliflora are: I.japonica (2n = 34) × I.tectorum (2n = 28) or I.japonica (2n = 32) × I.wattii / I.confusa (2n = 30).
Furthermore, the parents of I.×ampliflora can be deduced according to morphological features. The hybrid has aerial stems (mean length = 27.0 ± 2.7 cm, n = 10). Without an aerial stem, I.tectorum cannot be the paternal parent. Iriswattii and I.confusa both have aerial stems (I.wattii, mean length = 67.2 ± 19.9 cm, n = 10; I.confusa, mean length = 30.3 ± 9.3 cm, n = 10). However, the leaf surface of I.×ampliflora has no waxy coat; it is dull and ruffled similar to that of I.wattii. The leaf surfaces of I.confusa and I.japonica have a waxy coat, glossy and smooth. Thus, compared with I.confusa, I.wattii is possibly more likely to be the paternal parent of I.×ampliflora.
Though I.japonica and I.×ampliflora are clustered into one clade, there are 22 mutations between the sequences of these two species. Compared with leaves (length 45–51 cm and width 3–4.5 cm) and flowers (diam. 4–5 cm) of I.japonica, I.×ampliflora has larger leaves (length 68–80 cm and width 6–7 cm) and larger purple flowers (diam. 11–13 cm). It cannot be determined which population of I.japonica it is derived from for the maternal parent since intraspecific chromosome numbers are variable. Thus, the sequence divergence could not reflect the real evolutionary distance between I.japonica and I.×ampliflora. The evolution of I.×ampliflora is in need of further study.
Supplementary Material
Acknowledgements
We thank Tian-Yi Yu, Shucheng Feng and Kai Jiang. Tianyi Yu kindly made the line drawing of I.×ampliflora. Kai Jiang conducted the phylogenetic analysis of irises presented here. Shu-Cheng Feng helped to collect the materials.
This work was supported by the Science and Technology Commission of Shanghai Municipality Project (Grant No 19DZ1204004) in Shanghai, P. R. China.
Appendix 1
Table A1.
Genbank accession of two chloroplast fragments for nine Iris species (Mathew 1981; Goldblatt and Manning 2008) in the study.
| Species | Collect location | Collector | Genbank accession | |
|---|---|---|---|---|
| matK | ndhF | |||
| I.×ampliflora | Shanghai Botanical Garden | Yue-E Xiao | MW203044 | MW203053 |
| I.anguifuga | Yue-E Xiao | MW203045 | MW203054 | |
| I.confusa | Yue-E Xiao | MW203046 | MW203055 | |
| I.domestica | Yue-E Xiao | MW203043 | MW203052 | |
| I.henryi | Yue-E Xiao | MW203047 | MW203056 | |
| I.japonica | Yue-E Xiao | MW203048 | MW203057 | |
| I.proantha | Yue-E Xiao | MW203049 | MW203058 | |
| I.tectorum | Yue-E Xiao | MW203050 | MW203059 | |
| I.wattii | Yue-E Xiao | MW20305 | MW203060 | |
Appendix 2
Figure A1.
The photo of holotype of Iris×ampliflora.
Citation
Xiao Y-e, Yu F-y, Zhou X-f (2021) A new natural hybrid of Iris (Iridaceae) from Chongqing, China. PhytoKeys 174: 1–12. https://doi.org/10.3897/phytokeys.174.62306
Funding Statement
the Science and Technology Commission of Shanghai Municipality Project (Grant No 19DZ1204004) in Shanghai, P. R. China.
References
- British Iris Societwy Species Group (1997) A guide to species irises: their identification and cultivation, eds. The Species Group of the British Iris Society. Cambridge University Press, Cambridge, 109–121.
- Burke JM, Voss TJ, Arnold ML. (1998) Genetic interactions and natural selection in Louisiana iris hybrids. Evolution; International Journal of Organic Evolution 52(5): 1304–1310. 10.1111/j.1558-5646.1998.tb02012.x [DOI] [PubMed] [Google Scholar]
- Dykes WR. (1913) The genus Iris. Dover Publications, New York, 106 pp. https://www.biodiversitylibrary.org/page/50320633 [Google Scholar]
- Goldblatt P, Manning JC. (2008) The Iris Family: Natural History and Classification. Timber Press, Portland, 200–204.
- Guan WL, Li YF, Chen X, Yang D. (2009) Flower structure and biological characteristics of flowering and pollination in Irisjaponica Thunb. Acta Horticulture Sinica 36(10): 1485–1490. [Google Scholar]
- Hu YH, Xiao YE. (2012) The wetland irises: appreciation, cultivation and application. Science Press, Beijing, 44 pp. [Google Scholar]
- Lawrence GHM. (1953) A reclassification of the genus Iris. Gentes Herbarum 8: 346–371. [Google Scholar]
- Mathew B. (1981) The Iris. B. T. Batsford Ltd., London, 69–78.
- Rodionenko GI. (1987) [1964 in Russian] The genus Iris L. The British Iris Society, London, 19 pp. [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics (Oxford, England) 19(12): 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Sapir Y, Shmida A, Ne’eman G. (2005) Pollination of Oncocyclus irises (Iris: Iridaceae) by night-sheltering male bees. Plant Biology 7(4): 417–424. 10.1055/s-2005-837709 [DOI] [PubMed] [Google Scholar]
- Shaw J, Lickey EB, Schilling EE, Small RL. (2007) Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. American Journal of Botany 94(3): 275–288. 10.3732/ajb.94.3.275 [DOI] [PubMed] [Google Scholar]
- Swofford DL. (2002) PAUP*. Phylogenetic analysis using parsimony (*and other methods), v. 4.0 beta 10. Sinauer, Sunderland. 10.1111/j.0014-3820.2002.tb00191.x [DOI]
- Thompson JD, Higgins DG, Gibson TJ. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22(22): 4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CA. (2004) Phylogeny of Iris based on chloroplast matK gene and trnK intron sequence data. Molecular Phylogenetics and Evolution 33(2): 402–412. 10.1016/j.ympev.2004.06.013 [DOI] [PubMed] [Google Scholar]
- Xiao YE, Jin DM, Jiang K, Hu YH, Tong X, Mazer SJ, Chen XY. (2019) Pollinator limitation causes sexual reproductive failure in ex situ populations of self-compatible Irisensata. Plant Ecology & Diversity 12(1): 21–35. 10.1080/17550874.2019.1569170 [DOI] [Google Scholar]
- Yu FY, Xu WJ, Xiao YE, Luo GJ, Jia QX, Bi XY. (2017) Identifying apomixis in matroclinal progeny from an interspecific crossing between Irisdomestica and three different colors of Irisdichotoma. Euphytica 213(12): e273. 10.1007/s10681-017-2065-3 [DOI]
- Zhao Y, Noltie H, Mathew B. (2000) Iridaceae. In: Wu ZY, Raven PH. (Eds) Flora of China. Science Press, Beijing & Missouri Botanical Garden Press, St.Louis 24: 297–313.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.