Abstract
The cytogenetic relationships in the species of Cucurbitaceae are becoming immensely important to answer questions pertaining to genome evolution. Here, a simplified and updated data resource on cytogenetics of Cucurbitaceae is presented on the basis of foundational parameters (basic, zygotic and gametic chromosome numbers, ploidy, genome size, karyotype) and molecular cytogenetics. We have revised and collated our own findings on seven agriculturally important Indian cucurbit species in a comparative account with the globally published reports. Chromosome count (of around 19% species) shows nearly three-fold differences while genome size (of nearly 5% species) shows 5.84-fold differences across the species. There is no significant correlation between chromosome numbers and nuclear genome sizes. The possible trend of evolution is discussed here based on molecular cytogenetics data, especially the types and distribution of nucleolus organizer regions (NORs). The review supersedes the scopes of general chromosome databases and invites scopes for continuous updates. The offline resource serves as an exclusive toolkit for research and breeding communities across the globe and also opens scope for future establishment of web-database on Cucurbitaceae cytogenetics.
Keywords: chromosome, genome size, karyotype, NORs, ploidy
Introduction
The family Cucurbitaceae contains an extensive range of diversity consisting of about 1000 species spread over 96 genera (Renner and Schaefer 2017). The diversity of plant families is associated with variation in genome sizes and chromosome numbers as a result of enormous adaptive radiation (Soltis et al. 2004; Lysák and Schubert 2013). The viewpoint of evolution has been changed with the understanding of whole genome duplication (WGD) (Soltis et al. 2014) followed by core-eudicot hexaploidy (Wang et al. 2018). A cytogenetic database is essential to gain insights into evolution by supplementing phylogeny trees with chromosome number information (Mota et al. 2016) to upgrade knowledge on plant systematics (Soltis et al. 2014; Viruel et al. 2021). Cucurbitaceae, being the fourth most important and one of the earliest consumed vegetables yielding family, has coped with extreme climates, extensive human intervention and a huge domestication syndrome (Chomicki et al. 2020). Considerable advances have been made in molecular phylogeny (Renner and Schaefer 2016; Bellot et al. 2020; Chomicki et al. 2020; Guo et al. 2020) and genomics (CuGenDB, http://cucurbitgenomics.org) (Zheng et al. 2019). We had previously discussed about the gaps in cytogenetic studies (Bhowmick and Jha 2015b) which has been surmounted with the advent of molecular cytogenetics.
Currently, we have collated the cytogenetic reports of Cucurbitaceae globally and integrated our own findings for a collective interpretation. The review attempts to address i) the trend of chromosome evolution in specific tribes and species based on available information, ii) correlation between chromosome numbers and ploidy or genome size in the studied taxa and iii) the requirement of an exclusive cytogenetic catalogue for genome researchers, taxonomists and breeders working on Cucurbitaceae.
Methodological approaches
Data compilation
The data have been collated as per Schaefer and Renner (2011) after consultation of books, Chromosome atlases, research articles and public resources like Chromosome Counts Database (CCDB; http://ccdb.tau.ac.il/) (Rice et al. 2015), The Index to Plant Chromosome Numbers (IPCN, http://legacy.tropicos.org/Project/IPCN) (Goldblatt and Lowry 2011) and The Plant DNA C-values database (Pellicer and Leitch 2020) (https://cvalues.science.kew.org/).
Chromosome analysis in the Cucurbit species ocurring in India
Presently an enzymatic maceration and air drying (EMA) method followed by flurochrome banding has been employed as per our previous protocols (Bhowmick et al. 2012, 2016; Bhowmick and Jha 2015a, 2019, 2021) to represent fresh karyotypes of seven agriculturally important cucurbit species (Table 1) belonging to Benincaseae and Sicyoeae. Fresh and healthy roots were used from different sources (like germinating seeds, seedlings and underground root stocks). Roots were pretreated with 0.002 M hydroxyquinoline and fixed in 1:3 aceto-methanol solution. The standardization of EMA- fluorescence banding was conducted for the different species. In brief, fixed roots were digested in enzyme mixture [1% Cellulase (Onozuka RS), 0.75% Macerozyme (R-10), 0.15% Pectolyase (Y-23), 1 mM EDTA] for 40–45 min at 37 °C, macerated on slides, air-dried, stained with 2% Giemsa solution (Merck, Germany) and plates selected for karyotyping. After de-staining, slides were kept in McIlvaine buffer, stained with 0.1 µg mL-1 DAPI for 15–20 min in darkness. For CMA staining, slides were incubated in 0.1 mg mL-1 CMA for 15–25 min in darkness. For meiotic chromosomes, fixed anthers were digested in enzyme mixture for 5–8 min, macerated on slides and DAPI staining protocol was followed with minor modifications. All slides were mounted in non-fluorescent glycerol and chromosome plates were observed under a Zeiss Axioscop 2 fluorescence microscope (using UV and BV filter cassettes for DAPI and CMA stains, respectively). Images were captured using the attached ProgRes MFscan Jenoptik D07739 camera and ProgRes CapturePro 2.8.8 software.
Table 1.
Chromosome numbers and nature of fluorescent bands in some cucurbit species occurring in India.
| Tribes | Species (common name, status of cultivation/ wild) | Collection site, Latitude/ Longitude | Fruit image | 2n | CMA bands | DAPI bands (Non-nucleolar) | |
|---|---|---|---|---|---|---|---|
| Nucleolar | Non-nucleolar | ||||||
| Sicyoeae | Luffaacutangula Linnaeus, 1753 (ridged gourd, cultivated) | Bhubaneswar, Odisha, 20.2960°N, 85.8245°E |

|
26 | 11th , 12th, 13th | 12th (centromeric) | 1st to 13th (distal) |
| Luffacylindricaaegyptiaca Miller, 1768 (sponge gourd, cultivated) | Imphal, Manipur, 24.6637°N, 93.906°E |

|
26 | 12th , 13th | 1st , 2nd (distal) | 0 | |
| Luffaechinata Roxburgh, 1814 (wild) | Pantnagar, Uttarakhand, 30.0667°N, 79.019°E |

|
26 | 11th , 12th , 13th | 0 | 1st to 13th (distal) | |
| Trichosanthescucumerina Linnaeus, 1753 (wild) | NBPGR, Thrissur, Kerala, 10.5276°N, 76.2144°E |

|
22 | 10th , 11th | 0 | 1st to 11th (distal) | |
| Trichosanthescucumerinassp.cucumerina Anguina (snake gourd, cultivated) | Bengaluru, 12.9716°N, 77.5946°E |

|
22 | 10th , 11th | 2nd (distal) | 0 | |
| Trichosanthesdioica Roxburgh, 1832 (pointed gourd, cultivated) | Bhagalpur, Bihar, 25.2414°N, 86.9924°E |

|
22 (female) | 0 | 7th , 8th , 10th (distal) | 1st to 11th | |
| 22 (male) | 0 | 0 | 1st to 11th | ||||
| Benincaseae | Benincasahispida Thunberg, 1784 (ash gourd, cultivated) | Imphal, Manipur 24.6637°N, 93.906°E |

|
24 | 12th | 9th (distal) | 0 |
| Cocciniagrandis Linnaeus, 1767 (ivy gourd, restricted cultivation) | Nagpur, Maharashtra, 21.1458°N, 79.0881°E |

|
24 (female) | 8th, 12th * | 1st to 5th, 8th to 12th (centromeric) | 0 | |
| 24 (male) | 8th, 12th * | 1st to 5th, 8th, 10th to 12th (centromeric) | 0 | ||||
Statistical analyses
Statistical analysis involving foundational cytogenetic parameters have been demonstrated to imply significant knowledge on chromosomal evolution within a group (Winterfeld et al. 2020). Considering the lack of hypotheses, we have tested for correlation between the dependent variables (2C genome size, MCL and HCL) and predictor variables [chromosome number (2n) and ploidy level (pl)] and also calculated linear models for regression analysis using IBM SPSS (v23, free).
The modern cytogenetic catalogue of cucurbitaceae
Along with the global review, fresh EMA based somatic plates and idiograms (Figs 1–3) of Indian species are presented here. We retain the previous designation of 10 tribes as ‘understudied’ (Bhowmick and Jha 2015b), excluding Indofevilleeae, having no cytological reports.
Figure 1.
Somatic metaphase chromosomes and idiograms of Luffa species (2n = 26) stained with Giemsa (A, D, G), DAPI (B, E, H) and CMA3 (C, F, I) A–CL.acutangulaD–FL.aegyptiacacylindricaG–IL.echinata. Arrows indicate satellited chromosomes in Giemsa plates and CMA+ve signals in C, F, I. Corresponding somatic idiograms (haploid set) of: JL.acutangulaKL.aegyptiacaLL.echinata, showing DAPI+ve (blue) and CMA+ve (golden yellow) bands. Scale Bars: 5 µm
Figure 3.
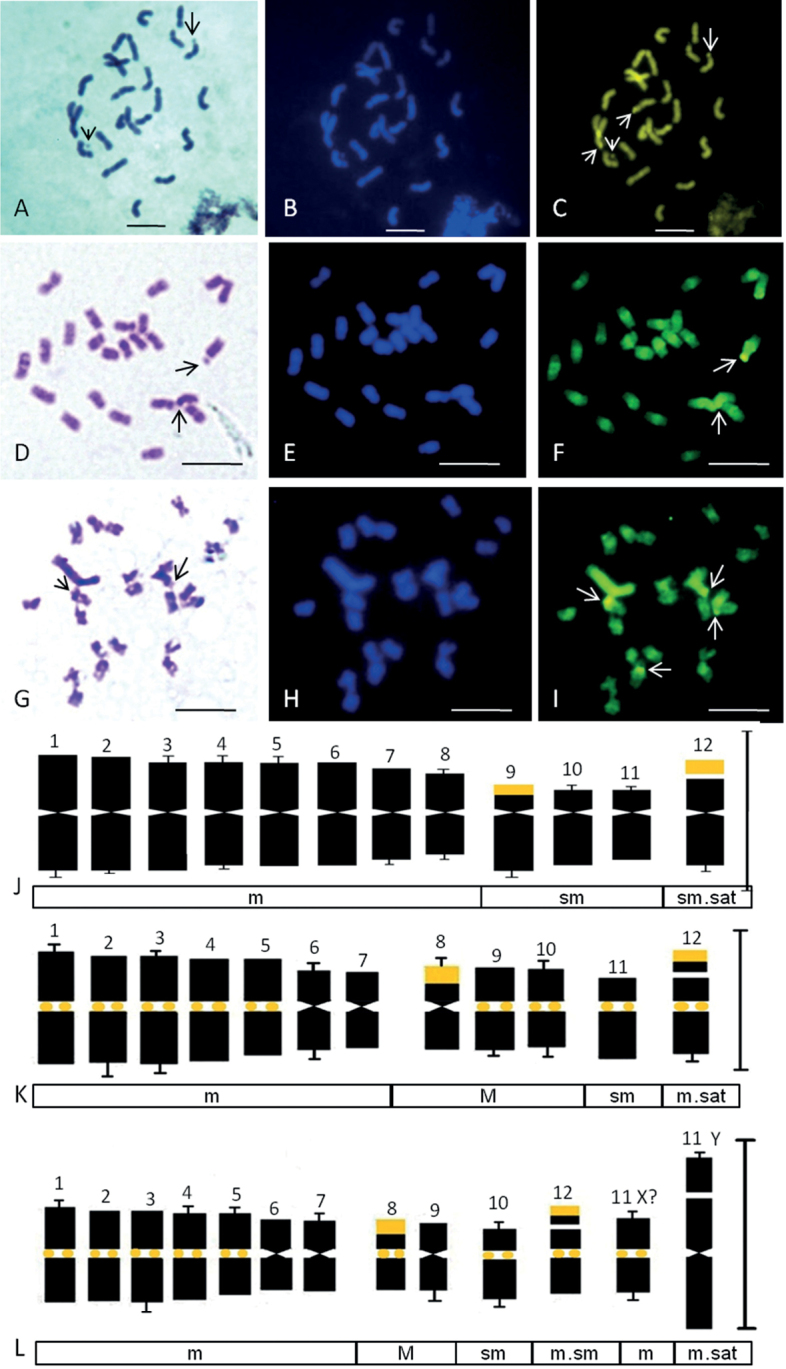
Somatic metaphase chromosomes and idiograms of two Benincaseae species (2n = 24) stained with Giemsa (A, D, G), DAPI (B, E, H) and CMA3 (C, F, I) A–CBenincasahispidaD–FCocciniagrandis (female plant) G–ICocciniagrandis (male plant). Arrows indicate satellited chromosomes in Giemsa plates and distal CMA+ve signals in C, F, I. Note the longest Y chromosome without any CMA band in G–I and centromeric CMA+ve signals in F, I. Corresponding somatic idiograms (haploid set) of: JBenincasahispidaKCocciniagrandis (female plant) LCocciniagrandis (male plant) with CMA+ve (golden yellow) bands. Note the X chromosome remaining indistinguishable in L. Scale Bars: 5 µm
Chromosome numbers
Currently, chromosome counts are available for 188 species (~19%) belonging to about 44 genera (~46%) of the 15 tribes, including the less attended ‘understudied tribes’. Within the ‘understudied tribes’, chromosome counts are available for only 42 species (out of almost 310) belonging to 17 genera (out of nearly 44). The basal number ranges from x/n = 5 (Thladiantha Bunge, 1833) to x/n = 15 (Zanonia Linnaeus, 1753) in these tribes (Table 2). Polyploidy has been abundantly reported in Gomphogyneae. Momordiceae have almost 60 species (Schaefer and Renner 2011) of which reports are known in nearly 11 species. The dibasic condition is noticed in Momordica Linnaeus, 1753 (x = 11 and 14) (Table 3) while polyploidy is detected in M.charantia Linnaeus, 1753 and M.dioica Willdenow, 1805 (2n = 56). M.cymbalaria Hooker, 1871, has the lowest count (2n = 18). In Bryonieae the X-Y sex determination system has been analysed in Bryonia Linnaeus, 1753 as the model along with Ecballium Richard, 1824 (Bhowmick and Jha 2015a). Chromosome counts are reported so far in 10 species of Bryonia (x = 10) and its sister genus Ecballium (x = 12 or x = 9, Table 4). Polyploidy is frequent in Bryonia. Sicyoeae is largest in terms of species (~264–266 species) (Schaefer and Renner 2011) of which cytological reports are known in around 14% species belonging to 9 genera (Table 5). Sicyoeae species range from x = 8 to x = 14 (Table 5). Trichosanthes Linnaeus, 1753 and Luffa Miller, 1754 have x = 11 and x = 13, respectively (Table 1). The less prevalent numbers include x = 12, x = 8 and x = 9 (Table 5). The possibility of multiple base number is noted in Frantzia Pittier, 1910 (x = 12/14) and Sicyos Linnaeus, 1753 (x = 12/13/14). Natural tetraploids are known in two species of Trichosanthes while the majority are diploids. Benincaseae is the second largest tribe comprising of 204–214 species in 24 genera (Schaefer and Renner 2011). Cytological reports are known in around 35% species (76 species of which 41 belong to Cucumis Linnaeus, 1753) of 12 genera (Tables 6, 7). x = 12 is the prevalent condition in Benincaseae (Tables 1, 6, 7). Dual base numbers are noted in the widely studied Cucumis (x = 7, 12). Coccinia Wight et Arnott, 1834 (x = 12) may also possess dual base numbers (x = 10 in C.trilobata Cogniaux, 1895). Molecular cytogenetics of Cucumissativus Linnaeus, 1753 has demonstrated the evolution of x = 7 from x = 12 in Benincaseae. x = 11 has been confirmed in Citrullus Schrader, 1836 and Lagenaria. The base number of Melothria Linnaeus, 1753, Solena Loureiro, 1790 and Zehneria Endlicher, 1833 can be x = 11 or x = 12 or both (Table 6). Cases of natural polyploidy are noted only in four species of Cucumis (Table 7). Cytogenetic information is available for 17 species in three genera of Cucurbiteae with x = 10 and many polyploids (Table 8). The zygotic chromosome numbers of Luffa, Trichosanthes, Benincasa Savi, 1818 and Coccinia, corroborate the previous reports (Figs 1–3, Table 1).
Table 2.
Cytogenetic reports in the understudied tribes of Cucurbitaceae #.
| Tribe and Genera | Species studied | Chromosome no. | Ploidy, Genome size, Chromosome features | References | ||
|---|---|---|---|---|---|---|
| x | 2n | n | ||||
| GomphogyneaeGomphogyne Griffith, 1845 | G.cissiformis Griffith, 1837 | 32a | 16b | Tetraploidc, autopolyploidd; 10 secondary constrictions, one pair satellitede; II, III, IV in meiosisf | CCDBb; Kumar and Subramaniam (1987)a, Singh (1990)a,d, Roy et al. (1991)a,c,e,f | |
| Hemsleya F.B. Forbes et Hemsley, 1888 | H.amabilis Diels, 1912, H.carnosiflora Wu et Chen, 1985, H.chinensis Forbes et Hemsley, 1888, H.emeiensis Shen et Chang, 1983, H.graciliflora Cogniaux, 1916, H.heterosperma Wallich, 1831, H.macrocarpa Cogniaux, 1916, H.panacis-scandens Wu et Chen, 1985, H.sphaerocarpa Kuang et Lu, 1982 | 7a | 28b, 22c, 24d, 26e, 32f, 40g, 42h | 14h | Tetraploidi, aneuploidsj | Samuel et al. (1995)a-j, Anmin et al. (2011)b, h |
| Gynostemma Blume, 1825 | G.cardiospermum Oliver, 1892 | 11a | 66b | Hexaploidc | IPCN a-c | |
| G.guangxiense Chen et Qin, 1988 | 22 a | Diploid b | IPCN a,b | |||
| G.laxiflorum Wu et Chen, 1983 | 22 a | Diploid b | IPCN a,b | |||
| G.longipes Wu et Chen, 1983 | 22a, 44b | Polyploidc | IPCN a-c | |||
| G.microspermum Wu et Chen, 1983 | 22a | Diploid b | IPCN a,b | |||
| G.pedatum Blume, 1825 | 12a | 24b | Diploidc | Roy et al. (1991) a,b,c | ||
| G.pentagynum Wang, 1989 | 22a | Diploid b | IPCN a,b | |||
| G.pentaphyllum Thunberg, 1784 | 22a, 24b, 64c, 66d | Diploide, triploidf, hexaploidg; 2C (flow cytometry): 3.62pgh; 17M+14sm+2sti; CSR: 2.16–4.09 µmj 5S (8), 45S (10) rDNA and telomeric signalsk | IPCNa,b,c,e,f; Zhang et al. (2013)h, Pellerin et al. (2018)d,g,i,j,k | |||
| G.pentaphyllumvar.dasycarpum Wu, 1983 | 22a, 33b, 44c | Polyploidd | IPCN a-d | |||
| G.pentaphyllumvar.pentaphyllum Thunberg, 1784 | 22a, 44b, 66c, 88d | Polyploide | IPCN a-e | |||
| G.yixingense Wang et Xie, 1981 | 88a | Polyploidb | IPCN a,b | |||
| Triceratieae | 8a | - | Roy et al. (1991) a | |||
| Fevillea Linnaeus, 1753 | ||||||
| ZanonieaeZanonia Linnaeus, 1753 | Z.indica Linnaeus, 1759 | 15a | 30b | 15c | Autoploidd; Metacentric chromosomese; CSR: 1.10-1.98 μmf | Lekhak et al. (2018) a-f |
| Actinostemmateae | A.lobatum (Maxim.) Maxim. ex Franch. & Sav. | 16a | - | IPCN a | ||
| Actinostemma Griffith, 1841 | A.tenerum Griffith, 1837 | 16a | Diploidb; 7M +1smc; CSR: 2.88–4.02 µmd; 45S (1) rDNA and 45S+5S (1) rDNA adjacent signale; telomeric repeat signalsf | Pellerin et al. (2018) a-f | ||
| Thladiantheae | T.calcarata Clarke, 1876, T.cordifolia Blume, 1826 T.davidii Franchet, 1886, T.dentata Cogniaux, 1916, T.lijiangensis Lu et Zhang, 1981, T.nudiflora Hemsley, 1887, T.pustulata Léveillé, 1916 | |||||
| Thladiantha Bunge, 1833 | 3a, 5b, 9c | 18d | 5e, 9f | Diploidg | Darlington and Janaki Ammal (1945)c; Roy et al. (1991)a,b,d,e,g, IPCNd,f | |
| T.dubia Bunge, 1833 | 18a, 22b | Diploidc; 7M+1sm+1std; CSR: 2.60–4.10 µme; 45S (4) and co-localized 45S+5S (1) rDNA signalsf; telomeric repeat signalsg | Samuel et al. (1995)b, Pellerin et al. (2018)a,c,d,e,f,g | |||
| Baijiania Lu et Li, 1993 | B.yunnanensis Lu et Zhang, 1984 | 32a | - | IPCN a | ||
| SiraitieaeSiraitia Merrill, 1934 | S.grosvenorii Swingle, 1941 | 28a | 45S (6) and 5S (2) rDNA signalsb | IPCNa, Li et al. (2007)b | ||
| JoliffieaeTelfairia Hooker, 1827 | T.occidentalis Hooker, 1871 | 22a, 33b, 44c | Diploidd, aneuploide, triploidf, Tetraploidg; 1 Bh | Uguru and Onovo (2011) a-h | ||
| T.pedata Sims, 1826 | 22a | - | Bhowmick and Jha (2015)a | |||
| SchizopeponeaeHerpetospermum Hooker, 1867 | H.pedunculosum Seringe, 1828 | 11a | 45S (14), 5S (2) rDNA signalsb | Xie et al. (2019a) a,b | ||
| Schizopepon Maximowicz, 1859 | S.bryoniifolius Maximowicz, 1859 | 10a | 20b | - | Roy et al. (1991)a, IPCNb | |
| ConiandreaeApodanthera Arnott, 1841 | A.undulata Gray, 1853 | 14a | - | IPCN a | ||
| Corallocarpus Bentham et Hooker, 1867 | C.epigaeus Rottler, 1803 | 26a | 13b | - | Beevy and Kuriachan (1996) a,b | |
| C.welwitschii Naudin, 1863 | 72a | - | Singh (1990)a | |||
| Ibervillea Greene, 1895 | 11a, 12b | - | Darlington and Janaki Ammal (1945) a,b | |||
| Kedrostis Medikus, 1791 | K. africana Linnaeus, 1753 | 40a | 2C (feulgen densitometry): 0.8 pgb; 2C (flow cytometry): 1674 Mbpc | Bennet et al. (1982)a,b, Plant C DNA Values Databasec | ||
| K.foetidissima Jacquin, 1788 | 26a | 13b | Beevy and Kuriachan (1996) a,b | |||
| K.rostrata Rottler, 1803 | 13a | 26b | 13c | - | IPCN a-c | |
| Seyrigia Keraudren, 1960 | 13a | - | IPCN a | |||
# x: base number; 2n: zygotic number; n: gametic number; CSR: chromosome size range; B: B chromosome; II: bivalents, III: trivalent, IV: tetravalent; superscripts correspond to references.
Table 3.
Cytogenetic information in Momordica (Momordiceae)#.
| Species | Chromosome no. | Ploidy, Genome size, Chromosome features | References | |
|---|---|---|---|---|
| 2n | n | |||
| M.balsamina Linnaeus, 1753 | 22a | Diploidb; two chromosomes with double constrictionsc; CSR:0.65–1.98µmd; MCL:1.30µme; TCL: 28.61µmf | Bharathi et al. (2011) a-f | |
| M.charantia | 22a | 11b | Diploide, 2C (Feulgen densitometry): 4.10pg f, 2C (flow cytometry): 1.43pgg; chromosomes mostly metacentric, few submetacentric and subtelocentrich; 2 chromosomes with satellitesi; CSR: 1.26-1.81µmj; 45S (4) and 5S (2) rDNA signalsk | Plant DNA C-Values Databasef; Bharathi et al. (2011)a,i ; Barow and Meister (2003)g; Lombello and Pinto-Maglio (2007)a,b,h; Bharathi et al. (2011); Waminal and Kim (2012)a,e,h,j,k; Kausar et al. (2015)a; Kido et al. (2016)a,i |
| M.charantiavar.charantia | 22a | 11b | Diploidc; 2C (flow cytometry): 0.72pgd; NORs: 4e; nucleolar and centromeric CMA+ bandsf ; CSR: 1.27-3.07µmg; MCL: 1.97 µmh; HCL: 21.77µmi | Ghosh et al. (2018)a-f; Ghosh et al. (2021)a,c,d,g,h,i |
| M.charantiavar.muricata Chakravarty, 1982 | 22a | 11b | Diploidc; 2C (flow cytometry): 1.16pgd; NORs: 6e; nucleolar and centromeric CMA+ bandsf; CSR: 1.64-3.13µmg; MCL: 2.19µmh; HCL: 24.19µmi | Ghosh et al. (2018)a-f; Ghosh et al. (2021)a,c,d,g,h,i |
| M.cochinchinensis Loureiro, 1790 | 28a | 14b | Diploidc, 2C (flow cytometry): 2.64pgd, 6e chromosomes with secondary constrictions; CSR:1.16–2.03µmf /1.71-3.17μmg ; MCL: 2.27µmh; HCL: 31.86μmi; 45S (8) and 5S (2) rDNA signalsj | IPCNb; Xie et al. (2019a)a,j; Bharathi et al. (2011)a,e,f; Ghosh et al. (2021)a,c,d,g,h,i |
| M.cymbalaria | 18a | 8b, 9c, 11d | Diploide, 2C (flow cytometry): 3.74 pgf; 2g–4h chromosomes with secondary constrictions; CSR: 2.71-4.57μmi; MCL:3.75μmj; HCL: 33.79μmk | IPCNb; CCDBb,d; Bharathi et al. (2011)a,c,e,g; Ghosh et al. (2021)a,e,f,h,i,j,k |
| M.denudata Clarke, 1879 | 14a | - | IPCN a | |
| M.dioica Willdenow, 1805 | 28a, 56b | Diploidc; 2C (flow cytometry): 3.36 pgd, 2e-12f chromosomes with secondary constrictions; CSR: 2.04-3.58μmg; MCL: 2.75µmh; HCL: 77.10μmi; 45S (4) and 5S (2) rDNA signalsj | Bharathi et al. (2011)a,c,e; Xie et al. (2019a)a,j; Ghosh et al. (2021)b,d,f,g,h,i | |
| M.foetida Schumacher, 1827 | 44a | - | Behera et al. (2011)a | |
| M.rostrata Zimmermann, 1922 | 22a | - | Behera et al. (2011)a | |
| M.sahyadrica Kattukunnel et Antony, 2007 | 28a | 2 chromosomes with secondary constrictionsb; CSR: 0.73–1.83µmc; TCL: 37.53µmd, MCL: 1.34µme | Behera et al. (2011)a-e | |
| M.subangulata Blume, 1826 | 56a | 2C (flow cytometry): 3.06pgb; 8 chromosomes with secondary constrictionsc; CSR: 1.52-3.11μmd; HCL: 60.30μme | Ghosh et al. (2021) a-e | |
| M.subangulatasubsp.renigera Don, 1834 | 56a | 4 chromosomes with secondary constrictionsb; CSR: 0.52-1.26µmc; MCL: 0.93µmd; TCL 51.88µme | Bharathi et al. (2011) a-e | |
| M.tuberosa Miquel, 1855 | 22a | 11b | - | IPCNa,b; CCDBa,b |
# 2n: Zygotic chromosome number; n: gametic chromosome number; I: univalent; II: bivalent; III: trivalent; CSR: chromosome size range; MCL: mean chromosome length; HCL: total length of haploid set of chromosomes; TCL: total length of diploid set of chromosomes; NOR: nucleolar organizing region, superscripts correspond to references.
Table 4.
Chromosome number and genome size in Bryonieae#.
| Genera | Species studied | Chromosome no. | Genome size | References | ||
|---|---|---|---|---|---|---|
| x | 2n | n | ||||
| Bryonia | 10a | Darlington and Janaki Ammal (1945) a | ||||
| B.alba Linnaeus, 1753 | 10a | 20b | 10c | 2C (flow cytometry): 5827Mbpd | CCDBd , Volz and Renner (2008)a,b,c | |
| B.aspera Ledebour, 1843 | 10a | 40b, 60c | 20d, 10e | - | Kumar and Subramaniam (1987)c, Volz and Renner (2008)a,b,d,e | |
| B . cretica Linnaeus, 1753 | 10a | 60b | 30c | - | Volz and Renner (2008) a,b,c | |
| B.dioica Jacquin, 1774 | 10a | 20b | 10c | 2C (microdensitometry): 4.01pgd; 2C (flow cytometry): 5522Mbpe | CCDBd,e, Volz and Renner (2008)a,b,c | |
| B.macrostylis Heilbronn et Bilge, 1954 | 10a | - | IPCN a | |||
| B . marmorata Petit, 1889 | 40a | 20b | - | Volz and Renner (2008) a,b | ||
| B . monoica Aitchison et Hemsley, 1886 | 10a | 20b | - | Volz and Renner (2008) a,b | ||
| B.multiflora Boissier et Heldreich, 1849 | 10a | - | Volz and Renner (2008) a | |||
| B.syriaca Boissier, 1856 | 10a | 20b | - | Volz and Renner (2008) a,b | ||
| B.verrucosa Aiton, 1789 | 10a | 20b | 10c | 2C (flow cytometry): 2.09pgd; 4504Mbpe | CCDBd,e, Volz and Renner (2008)a–c | |
| Ecballium | 12a | Darlington and Janaki Ammal (1945) a | ||||
| E.elaterium Linnaeus, 1753 | 18a | 12b | 2C (flow cytometry): 2442Mbpc | Veselý (2012)c , Volz and Renner (2008)b | ||
| E.elateriumsubsp.dioicum Battandier, 1989 | 18a, 24b | 9c, 12d | - | Volz and Renner (2008) a-d | ||
| E.elateriumLinnaeus, 1753subsp.elaterium | 18a | 9b | - | Volz and Renner (2008) a,b | ||
# 2n: Zygotic chromosome number; n: gametic chromosome number.
Table 5.
Cytogenetic information in Sicyoeae#.
| Genera studied | Species studied | Chromosome no. | Ploidy, Genome size, Chromosome features | References | ||
|---|---|---|---|---|---|---|
| x | 2n | n | ||||
| Cyclanthera Lilja, 1870 | 8a | Darlington and Janaki Ammal (1945) a | ||||
| C.pedata (L.) Schrader, 1831 | 16a, 32b | 8c | Diploide | Roy et al. (1991)a,c,e, Samuel et al. (1995)b | ||
| Echinocystis Torrey et Gray, 1840 | 8a | 16b | Bhowmick and Jha (2015b)b, Darlington and Janaki Ammal (1945)a | |||
| E.lobata Michaux, 1803 | 16a, 32b | Tetraploidc; 2C (flow cytometry):1.49pgd | IPCNa,b, Plant DNA C-Values Database c,d | |||
| E.macrocarpa Greene, 1885 | 32a | - | Whitaker (1950) a | |||
| Echinopepon Naudin, 1866 | E.wrightii Gray, 1853 | 12a | - | IPCNa, CCDBa | ||
| Frantzia | 12a, 14b | - | Schaefer and Renner (2011)a,b | |||
| Hodgsonia Persson, 1953 | H.macrocarpavar.capniocarpa Ridley, 1920 | 18a | - | IPCNa, CCDBa | ||
| Luffa | 13a | Darlington and Janaki Ammal (1945) a | ||||
| L.acutangula | 13a | 26b | 13c | Diploidd; CSR:1.39–3.20μme; 18m+2sm+6m.stf; NORs:6g; distal DAPI and nucleolar CMA signalsh | Kumar and Subramaniam (1987)a,b, IPCNc, Bhowmick and Jha (2021)b,d-h | |
| L.acutangulavar.acutangula | 13a | - | Beevy and Kuriachan (1996) a | |||
| L.acutangulavar.amara Clarke, 1879 | 13a | - | Beevy and Kuriachan (1996) a | |||
| L.aegyptiaca (syn L.cylindrica Roemer, 1846) | 13a | 26b | 13c | Diploidd, 2C (flow cytometry): 1.56 pge; 2C (Feulgen densitometry): 1.7pgf; CSR: 1.60–2.06μmg; 24M+1smh; 22m+ 4m.sti; NORs:2j; nucleolar and distal CMA signalsk; 45S (10) and 5S (2) rDNA signalsl | Bennet et al. (1982)b,d,f, Kumar and Subramaniam (1987)a,b, Waminal and Kim (2012)b,d,g,h,l, Bhowmick and Jha (2015a)b,c,d,e,i,j,k | |
| L.echinata | 26a, 39b, 52c | 13d | Diploide; CSR 2.44–3.96 μmf; 16m+4sm+6m.stg; NORs: 6h; Distal and intercalary DAPI and nucleolar CMA signalsi | Kumar and Subramaniam (1987)a-e, Bhowmick and Jha (2021)a,e-i | ||
| L.graveolens Roxburgh, 1832 | 13a | - | Kumar and Subramaniam (1987) a | |||
| L.hermaphrodita Singh et Bhandari, 1963 | 13a | - | IPCN a | |||
| L.operculata Linnaeus, 1759 | 13a | 26b | 13c | - | Kumar and Subramaniam (1987)a,b, IPCNc | |
| Sicyos (75, includes Sechium, Microsechium) | 12a | 24b | - | Darlington and Janaki Ammal (1945) a,b | ||
| S.angulatus | 12a | 24b | Diploidc; CSR: 1.9-4.6µmd; 4 adjacent 45S+5S rDNA signalse | Waminal and Kim (2015)a-e; IPCNb | ||
| S.australis Endlicher, 1833 | 24a, 26b | 12IIc, 13IId | IPCNa-d, CCDBa-d | |||
| S.edulis Jacquin, 1760 (syn of Sechiumedule) | 13a | 26b, 28c | 12d, 13e | Diploidf; metacentric and submetaccentric chromosomesg; CSR: 2.69–5.38µmh; 45S (6), 5S (2) rDNA and telomeric repeat signals (28)i | Beevy and Kuriachan (1996)a,b,e, Pellerin et al. (2018)c,f,g,h,i, Ting et al. (2019)c, IPCNd, CCDBd | |
| S.nihoaensis St. John, 1970 | 12a | - | IPCNa, CCDBa | |||
| Sechiumcompositum Smith, 1903 (syn. Microsechiumcompositum) | 14a | - | IPCNa, CCDBa | |||
| S.hintonii Wilson, 1958 (syn Microsechiumhintonii) | 14a | - | IPCNa, CCDBa | |||
| Trichosanthes (100) | 11a | Darlington and Janaki Ammal (1945) a | ||||
| T.anaimalaiensis Beddome, 1864 | 22a | 11b | - | Beevy and Kuriachan (1996) a,b | ||
| T.boninensis Nakai et Tuyama, 1928 | 22a | - | IPCN a | |||
| T.bracteata Lamarck, 1797 | 11a | 22b, 44c, 66d | - | Kumar and Subramaniam (1987)a,b, Roy et al. (1991)c,d | ||
| T.bracteatavar.bracteata | 11a, 22b | - | Beevy and Kuriachan (1996) a,b | |||
| T.chingiana Handel-Mazzetti, 1936 | 22a | - | IPCN a | |||
| T.costata Blume, 1826 (syn Gymnopetalumchinense Loureiro, 1790) | 22a | Diploidb; 45S (6) and 5S (4) rDNA signalsc | Kumar and Subramaniam (1987)a, Xie et al. (2019a)b,c | |||
| T.cucumerina | 22a | 11b | Diploidc; 12m+4M+2sm+4sm.std; CSR: 2.26–4.99µme; 6 chromosomes with double constrictionsf; NORs: 4g; nucleolar CMA and distal DAPI bandsh | Bhowmick and Jha (2019) a-h | ||
| T.cucumerinassp.cucumerina Anguina | 11a | 22b | 11c, 22d, 32e, 33f | Diploide; 2C (Feulgen densitometry): 2.2pgf; CSR: 2.77–5.01μmg; 12m+4M+2sm+4sm.sth; 6 chromosomes with double constrictionsi; NORs: 4j; nucleolar and distal CMA bandsk; 45S (6) and 5S (2) rDNA signalsl | Kumar and Subramaniam (1987)a, Bhowmick and Jha (2019)b,c,e,g,h,i,j,k, Xie et al. (2019a)b,l, IPCNc-f | |
| T.dioica | 11a | 22b | 11c | Diploidd; 2C (flow cytometry): male-2.27pg, female- 2.32 pge; 12m+6Sm +2St+2Sm.tf; distal DAPI bandsg; distal CMA bands in femalesh; 1 rod bivalent in meiosisi | Kumar and Subramaniam (1987)a, Guha et al. (2004)b,d,f,h, Bhowmick and Jha (2015a)b,c,d,e,f,g,h,i | |
| T.dunniana Léveillé, 1911 | 22a | Diploidb; 45S (6) and 5S (2) rDNA signalsc | Xie et al. (2019a) a-c | |||
| T.himalensis Clarke, 1879 | 11a | - | Roy et al. (1991) a | |||
| T.hupehensis Cheng et Yueh, 1974 | 22a | - | IPCNa, CCDBa | |||
| T.kirilowii Maximowicz, 1859 | 60a, 66b, 88c, 110d | Hexa-, octa-, decaploide;CSR: 2.3-3.5μmf; 45S (4), 5S (4) and 45S +5S (6) adjacent rDNA signalsg | IPCNa, CCDBa, Waminal and Kim (2015)b,c,d,e-g | |||
| T.kirilowiivar.japonica | 11a | - | Roy and Saran (1990)a | |||
| T.lepiniana Naudin, 1868 | 44a | 11b | 1 Bc | Roy et al. (1991)b,c, IPCNa, CCDBa | ||
| T.lobata Roxburgh, 1832 | 11a | 11b | - | Kumar and Subramaniam (1987)a, Beevy and Kuriachan (1996)b | ||
| T.mianyangensis Yueh et. Liao, 1992 | 88a | - | IPCNa, CCDBa | |||
| T.nervifolia Linnaeus, 1753 | 11a | - | Beevy and Kuriachan (1996) a | |||
| T.ovigera Blume, 1826 | 22a | Diploidb; 45S (10) and 5S (2) rDNA signalsc | Xie et al. (2019a) a-c | |||
| T.palmata Linnaeus, 1759 | 22a, 44b, 66c | 11d | IPCN a-d | |||
| T.pedata Merril et Chun, 1934 | 22a | - | IPCNa, CCDBa | |||
| T.truncata Clarke, 1879 | 22a | - | IPCNa, CCDBa | |||
| T.wallichiana Wight, 1840 | 11a | 22b | - | Kumar and Subramaniam (1987) a,b | ||
#x: base number; 2n: zygotic number; n: gametic number; NOR: nucleolar organizing region; B: B chromosome; II: bivalents; superscripts correspond to references.
Table 6.
Cytogenetic information on Benincaseae#.
| Genera studied | Species studied | Chromosome no. | Ploidy, Genome size, Chromosome features | References | ||
|---|---|---|---|---|---|---|
| x | 2n | n | ||||
| Benincasa | B.fistulosa | 24a | Diploidb; 45S (4) and 5S (4) signalsc | Li et al. (2016) a,b,c | ||
| B.hispida | 12a | 24b | 12c | Diploidd; 2C (flow cytometry):1.97pge, 2C (feulgen densitometry):2.1pgf; CSR 2.54-4.59µmg; 16m+6Sm+2Sm.th; NORs:2i; distal CMA signalsj; 45S (2) and 45S+5S (2) adjacent rDNA signalsk | Plant DNA C-Values Databasef, Waminal et al. (2011)b,d,g,k, Bhowmick and Jha (2015a)b,c,d,e,h,i,j | |
| Citrullus | 11a | Darlington and Janaki Ammal (1945) a | ||||
| C.amarus (syn. C.lanatusvar.citroides) | 11a | 22b | Diploidc; CSR: 3.1–4.7μmd; 45S (2) and 5S (4) rDNA signalse | Reddy et al. (2013)b-d, Waminal and Kim (2015)a-e, Renner et al. (2017)b | ||
| C.colocynthis | 22a | 11b | Diploidc; 45S (2) and 45S+5S (2) adjacent rDNA signalsd | Beevy and Kuriachan (1996)b, Reddy et al. (2013)a,c,d, Li et al. (2016)a,c,d | ||
| C.ecirrhosus | 22a | 11b | Diploidc; 2 satellites detected in meiosisd; 45S (2) and 5S (4) rDNA signalse; regular meiosisf | Li et al. (2016)a,c,e, Renner et al. (2017)a,b,c,d,f | ||
| C.lanatus | 22a | 11b | Diploidc; CSR: 1.09μm-1.72μmd; 14m+8sme; 45S (2) and 45S+5S (2) adjacent rDNA signalsf; linkage groups hybridized to chromosomesg | Beevy and Kuriachan (1996)b, Waminal et al. (2011)a,c,d,e,f, Ren et al. (2012)a,c,g | ||
| C.lanatussubsp.lanatus | 22a | Diploidb; 45S (2) and 5S (4) rDNA signalsc | Li et al. (2016) a-c | |||
| C.lanatussubsp.mucosospermus Fursa, 1972 | 22a | Diploidb; 45S (2) and 45S+5S (2) adjacent rDNA signalsc | Li et al. (2016) a-c | |||
| C.lanatussubsp.vulgaris Schrader, 1836 | 22a | Diploidb; 45S (2) and 45S+5S (2) adjacent rDNA signalsc | Li et al. (2016) a-c | |||
| C.lanatusvar.lanatus | 22a | Diploidb; 45S (2) and 45S+5S (2) adjacent rDNA signalsc | Reddy et al. (2013) a-c | |||
| C.naudinianus (syn Acanthosicyosnaudinianus) | 24a | Diploidb; 45S (2) and co-localized 45S+5S (2) rDNA signalsc | Li et al. (2016) a,b,c | |||
| C.rehmii | 22a | Diploidb; 45S (2) and 5S (2) rDNA signalsc | Reddy et al. (2013)a-c, Li et al. (2016)a-c | |||
| C.vulgaris Schrader, 1836 | 22a, 44b | 11c | Diploidd; 2C: 0.88/0.90pge | IPCNa,c,d, Arumuganathan and Earle (1991)e | ||
| Coccinia (30) | 12a | Darlington and Janaki Ammal (1945) a | ||||
| C.abyssinica Lamarck, 1753 | 12a | 24b | - | Kumar and Subramaniam (1987)a, Roy et al. (1991)b | ||
| C.grandis | 12a | 24b | 12c | Diploidd, 2C (Flow cytometry): male- 0.943e/0.92f pg and female- 0.849g/ 0.73h pg; CSR: 1.33-4.71μm (male) and 1.35-2.26µm (female)i; 15m+4M+2sm+2m:sm+1m:st (Y) in male and 14m+6M+2sm+2m:st in femalej; NORs-2k; chromosomal C bandsl; centromeric, nucleolar CMA bandsm; 45S (4)n rDNA signals, 2 signals adjacent to 5So; GISH performedp; repetitive, organellar DNA hybridizedq; centromere immunofluorescencer; heteromorphic sex chromosomes (largest Y)s; X-Y bivalent (meiosis)t | Bhowmick et al. (2012)b,c,d,j,k,m,s,t, (2016) b,d,f,h,j,k,n,s, Sousa et al. (2013)b,d,e,g,i,k,l,n,o,r,s,t, Sousa et al. (2017)b,d,k,n,o,p,q,r,s, Xie et al. (2019a)b,n,o | |
| C.hirtella Cogniaux, 1896 | 24a | Diploidb; 2C (flow cytometry): male-0.988pgc; 45S (4) and 45S+5S (2) adjacent rDNA signalsd, repetitive and organellar DNA hybridizede; centromere immunofluorescence performedf | Sousa et al. (2017) a-f | |||
| C.sessilifolia Sonder, 1881 | 24a | Diploidb; 2C (Flow cytometry): male- 0.984pg, female- 0.998pgc; 45S (4) and 45S+5S (2) adjacent rDNA signalsd; repetitive and organellar DNAe; centromere immunofluorescence performedf | Li et al. (2016)a,b,d, Sousa et al. (2017)a–f | |||
| C.trilobata | 20a | Diploidb; 2C (flow cytometry): male- 1.263pg c; 45S (2) and 45S+5S (2) adjacent rDNA signalsd, repetitive, organellar DNA sequence hybridizede | Sousa et al. (2017) a-e | |||
| Ctenolepis Hooker, 1867 | C.garcinii Burman, 1768 | 24a | 12b | - | Kumar and Subramaniam (1987)a, Beevy and Kuriachan (1996)b | |
| Diplocyclos Endlicher, 1833 | D.palmatus | 24a | Diploidb; 45S (4) and 45S+5S (2) adjacent rDNA signalsc | Li et al. (2016) a-c | ||
| Lagenaria Seringe, 1825 | L.leucantha Rusby, 1896 | 22a | 11b | - | IPCNa,b, CCDBa,b | |
| L.leucanthavar.clavata Makino, 1940 | 22a | - | CCDB a | |||
| L.siceraria | 11a | 22b | 11c | Diploidd, 2C (flow cytometry): 0.734pge; 2C (Feulgen densitometry):1.4pgf; CSR: 0.56–1.06μmg; metacentric and few sub-metacentric chromosomesh; 45S (2) and 45S+5S (2) adjacent rDNA signalsi | Darlington and Janaki Ammal (1945)a, Plant DNA C-Values Databasef, Beevy and Kuriachan (1996)c, Achigan-Dako et al. (2008)d,e, Waminal and Kim (2012)b,d,g,h,i, Li et al. (2016)b,d,i, Xie et al. (2019a)b,i | |
| L.sicerariavar.macrocarpa | 22a | - | CCDB a | |||
| L.vulgaris Seringe, 1825 | 22a | 11b | Diploidc; 2C (Feulgen densitometry): 1.40pgd | Bennet et al. (1982)a,b,c | ||
| Melothria | 11a, 12b | Darlington and Janaki Ammal (1945) a,b | ||||
| M.pendula Linnaeus, 1753 | 24a | Diploidb; 45S (2) and 45S+5S (2) adjacent rDNA signalsc | Li et al. (2016) a-c | |||
| M.perpusilla Blume, 1826 | 48a | - | Kumar and Subramaniam (1987) a | |||
| M.scabra Naudin, 1866 | 24a | - | CCDB a | |||
| Peponium Engler, 1897 | P.betsiliense Keraudren, 1960 | 24a | - | CCDB a | ||
| Solena | S.amplexicaulis Lamarck, 1785 (syn. S.heterophylla, Melothriaheterophylla, Zehneriaumbellata) | 22a, 24b, 26c, 36d, 48e | 11f, 12g, 24h | 2-4 Bi | Kumar and Subramaniam (1987)a,b,c,e, Roy et al. (1991)d,i, Beevy and Kuriachan (1996)b,g,h, IPCNb,d,e,f,g,h | |
| Zehneria | Z.capillacea Jeffrey, 1962 (syn. Melothriacapillacea) | 22a | - | CCDB a | ||
| Z.indica Loureiro, 1790 (syn. Melothriajaponica) | 11a | 22b | 24c | Diploidd; 45S (2) and 45S+5S (2) adjacent rDNA signalse | Waminal and Kim (2015) a,b,d,e | |
| Z.marlothii Cogniaux, 1962 | 24a | Diploidb; 45S (2) and 45S+5S (2) adjacent rDNA signalsc | Li et al. (2016) a,b,c | |||
| Z.maysorensis Wight et Arnott, 1834 | 48a | 24b | 45S (2) and 5S (2) signalsc | Beevy and Kuriachan (1996)a,b, Xie et al. (2019a)a,c | ||
| Z.mucronata Blume, 1856 (syn. Melothriamucronata) | 22a | 12b | - | Darlington et al. (1956)a, CCDBb | ||
| Z.scabra Sonder, 1862 (syn. Melothriapunctata) | 24a, 48b | - | CCDBa, Kumar and Subramaniam (1987)b | |||
| Z.thwaitesii Schweinfurth, 1868 | 44a | - | CCDB a | |||
# genera included other than Cucumis; x: base number; 2n: zygotic number; n: gametic number; NOR: nucleolar organizing region; B: B chromosome; II: bivalents; IPCN: Index to Plant Chromosome Number Reports; CCDB: Chromosome Counts Database; superscripts correspond to references.
Table 7.
Cytogenetic features of Cucumis (Benincaseae)#.
| Species with subspecies/ varieties | Chromosome no. | Ploidy, Genome size, Chromosome features | References | |||||
|---|---|---|---|---|---|---|---|---|
| x | 2n | n | ||||||
| C.aculeatus Cogniaux, 1895 | 48a | Allotetraploidb; 24IIc | IPCNa-c, CCDBa-c | |||||
| C.africanus Linnaeus, 1782 | 12a | 24b, 48c | 12d | Diploide; 2C (Feulgen microdensitometry): 1.782pgf; 4 satellited chromosomesg; 45S (4h/6i) rDNA signals, 2 co-localized 45S+5S signalsj | IPCNb,c, Yadava et al. (1984)b,d, Ramachandran and Narayan (1985)b,e,f, Yagi et al. (2015)a,b,g,i,j, Zhang et al. (2016)b,e,h,j | |||
| C.angolensis Cogniaux, 1881. | 24a | IPCN a | ||||||
| C.anguria Linnaeus, 1753 | 24a | Diploidb; majorly submetacentric and few nearly metacentric chromosomesc; 1 pair satellitedd; 45S (2) and co-localized 45S+5S (2) rDNA signalse, ScgCP enables chromosome identificationf; GISH reveals cross species relationshipsg | Singh and Roy (1974)a-d, Zhang et al. (2015)a,g, (2016)b,e, Li et al. (2018)a,f | |||||
| C.anguriavar.anguria | 12a | 24b | 12c | Diploidd; 4 satellited chromosomese; 45S (2) and co-localized 45S+5S (2) rDNA signalsf | Yadava et al. (1984)b,c, Yagi et al. (2015)a,d,e,f | |||
| C.anguriavar.longipes | 24a | 12b | Diploidc; 2C (Feulgen microdensitometry): 1.587pgd | Yadava et al. (1984)a,b, Ramachandran and Narayan (1985)a,c,d | ||||
| C.anguriavar.longaculeatus | 12a | 12.5 | Diploidc; 4 satellited chromosomesd; 45S (2) and co-localized 45S+5S (2) rDNA signalse | Yagi et al. (2015) a-e | ||||
| C.asper Cogniaux, 1901 | 24a | Diploidb; 45S (4) and 5S (2) signals detectedc | IPCNa, Zhang et al. (2016)a,b,c | |||||
| C.callosus Rottler, 1803 | 14a, 24b | 12c | Diploidd; 2C (Feulgen microdensitometry):1.590pge; 11m+1sm (haploid)f | Ramachandran and Narayan (1985)a,d,e, Rajkumari et al. (2013)b,c, (2015)b,c,f | ||||
| C.cinereusCogniaux, 1901 (syn. Cucumellacinerea) | 2C (Feulgen microdensitometry): 0.5pga | Bennet et al. (1982)a | ||||||
| C.diniae Raamsdonk et Visser, 1992 | 48a | - | IPCN a | |||||
| C.dinteri Cogniaux, 1901 | 24a | Diploidb; 2C (Feulgen microdensitometry): 2.167pgc | IPCNa, Ramachandran and Narayan (1985)a-c | |||||
| C.dipsaceus Spach, 1838 | 24a | 12b | Diploidc; 2C (Feulgen microdensitometry): 2.448pgd; 2m+8sm+2st (haploid)e; 45S (2) and co-localized 45S+5S (2) rDNA signalsf | Yadava et al. (1984)a,b, Ramachandran and Narayan (1985)a,c,d, Rajkumari et al. (2015)a,c,e, Zhang et al. (2016)a,c,f | ||||
| C.ficifoliusRichard, 1847 | 24a, 48b | 12c | Diploidd; 2C (Feulgen microdensitometry):1.373pge; 45S (2) and co-localized 45S+5S (2) rDNA signalsf | Yadava et al. (1984)a,c, Ramachandran and Narayan (1985)a,d,e, Zhang et al. (2016)b,f | ||||
| C.figarei Naudin, 1859 | 48a, 72b | Autoallopolyploidc; 2C (Feulgen microdensitometry): 3.886pge; 36IIf | IPCNa-f, Ramachandran and Narayan (1985)a,c,e | |||||
| C.heptadactylis Naudin, 1859 | 48a | 23b, 24c, 52d | Autotetraploide; 2C (Feulgen microdensitometry): 2.225pgf; 8 satellited chromosomesg; 45S (8) rDNA signalsh of which 4 co-localized to 5S signalsi or separate 5S (4) rDNA signalsj; 10IV+4IIk; irregular meiosisl | IPCNa,e,k, Yadava et al. (1984)a,b,d,e,l, Ramachandran and Narayan (1985)a,e,f, Yagi et al. (2015)a,e,g,h,i, Zhang et al. (2016)a,e,h,j | ||||
| C.hookeri Naudin, 1870 | 24a | 12b | Diploidc | Yadava et al. (1984) a,b,c | ||||
| C.humifructus Stent, 1927 | 24a | Diploidb; 2C (Feulgen microdensitometry): 2.455 pgc | Ramachandran and Narayan (1985) a,b,c | |||||
| C.hystrix Chakravarty, 1952 | 12a | 24b | Diploidc; 2m+10sm (haploid)d; 45S (4) and co-localized 45S+5S (2) rDNA signalse; FISH with bulked oligo probe from cucumber chromosome C7f, GISH reveals cross species relationshipsg | Rajkumari et al. (2015)b,c,d, Han et al. (2015)f, Zhang et al. (2015)b,g, (2016)a,b,c,e | ||||
| C.indicus Ghebretinsae et Thulin, 2007 | 20a | Diploidb; 4m+ 6sm (haploid)c | Rajkumari et al. (2015) a-c | |||||
| C.javanicus Miquel, 1856 (syn. Melothriaassamica) | 12a | 24b, 48c | - | Kumar and Subramaniam (1987)a, CCDBb,c | ||||
| C.leiospermus Wight et Arnott, 1834 (syn. Melothrialeiosperma) | 24a | - | CCDB a | |||||
| C.leptodermis Schweickerdt, 1933 | 24a | 12b | - | Yadava et al. (1984) a,b | ||||
| C.longipes Hooker, 1871 | 24a | - | IPCN a | |||||
| C.meeusei Jeffrey, 1965 | 48b | 22c, 24d | Tetraploide; 2C- 3.203pg (Feulgen microdensitometry)f; 45S (6) and co-localized 45S+5S (2) rDNA signalsg | Yadava et al. (1984)b,c,d, Ramachandran and Narayan (1985)b,e,f, Zhang et al. (2016)b,e,g | ||||
| C.melo Linnaeus, 1753 | 12a | 20b, 22c, 24d | 12e | Diploidf; 2C (Feulgen photometry) : 0.94-1.04pgg, 1.90pgh; 2C (Flow cytometry): 1.05pgi; 14m+10st (2SAT)j; 7m+5sm (haploid)k; 4 satellitesl or 2 satellitesm; CSR1.0-2.1μmn; CMA bands detectedo; 45S (2) and co-localized 45S+5S (2) rDNA signalsp; centromeric, telomeric, nulceolar and SSR probe hybridization reveals chromosomal relationq; ScgCP applied for comparative chromosome rearrangement studywith C.sativusr; FISH with bulked oligo probe from cucumber chromosome C7s; novel centromeric satellite DNA hybridized on chromosomest; GISH reveals cross species relationshipsu; infraspecific positional differences in 45S (terminal and interstitial) -5S (terminal, subterminal and interstitial) rDNA signalsv | CCDBb,c, Plant DNA C-Values Databaseh, Kumar and Subramaniam (1987)a, Arumuganathan and Earle (1991)g, Marie and Brown (1993)i, Zhang (2005)d,f,j,u, (2015)d,t, Song and Kim (2008)d,f,m, Han et al. (2009)d,e,f,q, (2015)s, Liu et al. (2010)d,f,q, Hoshi et al. (2013)d,f,l,n,o,p, Lou et al. (2014)r, Rajkumari et al. (2013)d,e, (2015)d,f,k, Setiawan et al. (2018)d,v, (2020)d,t | |||
| C.melosubsp.melo | 12a | 24b | Diploid; 45S (4) and 5S (2) rDNA signalsc | Zhang et al. (2016) a-c | ||||
| C.melosubsp.agrestis Naudin, 1859 | 12a | 24b | Diploid; 45S (4) and 5S (2) rDNA signalsc | Zhang et al. (2016) a-c | ||||
| C.melovar.agrestis | 12a | 24b | 12c | Diploidd; 2C (Feulgen microdensitometry): 2.483pge; 10m+2sm (haploid)f; 1 pair satellitedg | Singh and Roy (1974)b,d,g, Yadava et al. (1984)a-d, Ramachandran and Narayan (1985)b,d,e, Beevy and Kuriachan (1996)b,c, Rajkumari et al. (2015)b,d,f | |||
| C.melovar.conomon Thunberg, 1780 | 24a | Diploidb; 7m+3sm+2st (haploid)c | Zhang et al. (2005)a, Rajkumari et al. (2015)a,b,c | |||||
| C.melovar.flexuosus Linnaeus, 1763 | 24a | - | IPCN a | |||||
| C.melovar.inodorus Jacquin, 1832 | 24a | Diploidb; 2C (flow cytometry): 0.64pgc | Karimzadeh et al. (2010) a-c | |||||
| C.melovar.melo | 24a | 12b | Diploidc; 4m+8sm (haploid)d | Beevy and Kuriachan (1996)a,b, Rajkumari et al. (2015)b,c,d | ||||
| C.melovar.momordica Roxburgh, 1832 | 24a | 12b | Diploidc; 2C (Feulgen microdensitometry): 2.291pgd; 6m+5sm+1st (haploid)e | Yadava et al. (1984)a,b, Ramachandran and Narayan (1985)a,c,d, Rajkumari et al. (2015)a,c,e | ||||
| C.melovar.muskmelon | 24a | 12b | Yadava et al. (1984) a,b | |||||
| C.melovar.utilissimus Roxburgh, 1832 | 24a | 12b | Diploidc; 2C (Feulgen densitometry): 2.358 pgd | Yadava et al. (1984)a,b; Ramachandran and Narayan (1985)a,c,d | ||||
| C.membranifolius Hooker, 1871 | 48a | 24b | - | Yadava et al. (1984) a,b | ||||
| C.metulifer Naudin, 1859 (syn. C.metuliferus) | 24a | 12b | Diploidc; 2C (Feulgen microdensitometry): 2.391pgd; metacentric, submetacentric, subtelocentric chromosomese; CSR: 0.9–2.0 μmf; 4 satellitesg; nucleolar and centromeric CMA-DAPI bandsh; 45S (2) and co-localized 45S+5S (2) rDNA signalsi, satellite sequencesj and telomeric DNAk hybridized on chromosomes; ScgCP applied for comparative chromosome rearrangement studywith C.sativusl; GISH reveals cross species relationshipsm | Yadava et al. (1984)a,b,c, Ramachandran and Narayan (1985)a,c,g, Ramachandran and Narayan (1990)a,c,i, Hoshi et al. (2013)a,c,e,f,g,h, Lou et al. (2014)l, Yagi et al. (2014)a,c,g,h,i,j,k, Li et al. (2016)a,c,i, Zhang et al. (2015)a,m, (2016)a,c,i | ||||
| C.myriocarpus | 24a | 12b | Diploidc; 45S (2d/4e) and co-localized 45S+5S (2)f rDNA signalsd | CCDBa; Zhang et al. (2016)a,b,c,d,f, Yagi et al. (2015)a-f | ||||
| C.myriocarpussubsp.leptodermis Schweickerdt, 1933 | 12a | 24b | Diploidc; 4 satellited chromosomesd; 45S (3e, 2f) and co-localized 45S+5S (2g) rDNA signals | Yagi et al. (2015) a-g | ||||
| C.myriocarpusvar.myriocarpus | 12a | 48b | Tetraploidc; 8 satellited chromosomesd; 45S (4) and co-localized 45S+5S (4) rDNA signalse | Yagi et al. (2015) a-e | ||||
| C.prophetarum Linnaeus, 1755 | 24a | 12b | Diploidc, 2C (Feulgen Microdensitometry): 1.656 pgd 5m+7sm (haploid)e | Ramachandran and Narayan (1985)a,c,d, Rajkumari et al. (2013)a,b, (2015)a,c,e | ||||
| C.prophetarumsubsp.zeyheri Sonder, 1862 | 48a | - | IPCN a | |||||
| C.pubescens Willdenow, 1805 | 24a | 12b | - | IPCNa; Beevy and Kuriachan (1996)b | ||||
| C.pustulatus Hooker, 1871 | 48a, 72b | 24c | Hexaploidd, 45S (8) and co-localized 45S+5S (2) rDNA signalse; FISH with bulked oligo probe from cucumber chromosome C7f | Yadava et al. (1984)a,c, Han et al. (2015)f, Zhang et al. (2016)b,d,e | ||||
| C.ritchiei Clarke, 1879 | 24a | Diploidb, 8m+4sm (haploid)c | Rajkumari et al. (2015) a,b,c | |||||
| C.sagittatus Peyritsch, 1860 | 24a | 12b | Diploidc, 2C (Feulgen microdensitometry):1.571pgd | Yadava et al. (1984)a,b, Ramachandran and Narayan (1985)a,c,d | ||||
| C.sativus Linnaeus, 1753 | 7a | 14b | 7c | Diploidd, 2C (flow cytometry): 1.03pge /1.77pgf; 12 metacentric and 2 sub-metacentric chromosomesg; CSR: 0.83-1.01μmh, chromosomal C-bandsi; centromeric 45S (10) and distal 5S (2) rDNA signalsj; FISH with centromeric and telomerick and SSR probe reveals chromosome evolutionl; high resolution molecular cytogenetic mapm; ScgCP applied for cross species chromosome rearrangement studyn; FISH with bulked oligo probe from cucumber chromosome C7 in comparison with 5 Cucumis specieso; GISH reveals cross species relationshipsp | Kumar and Subramaniam (1987)a,b, Marie and Brown (1993)f, Beevy and Kuriachan (1996)b,c, Hoshi et al.(2008)b,d,i, Barow and Meister (2003)b,d,e, Han et al (2011)b,d,k,l,m, Liu et al. (2010)b,l, Waminal and Kim (2012)b,d,g,h,j, Rajkumari et al. (2013)b,c,f, Sun et al. (2013)b,m, Lou et al. (2014)b,n, Han et al. (2015)o, Zhang et al. (2015)b,p, Li et al. (2016)b,d,j | |||
| C.sativus var. Hokutosei | 7a | 14b | Diploidc, 12 metacentric, 2 sub-metacentric chromosomesd; centromeric and telomeric signalse | Zhang et al. (2012) a-e | ||||
| C.sativusvar.hardwickii Royle, 1835 | 7a | 14b | Diploidc; 2C (Feulgen Microdensitometry): 1.798pgd; 6m+1sm (haploid)e; centromeric 45S (6) and intercalary 5S (2) rDNA signalsf, centromeric, telomeric and SSR probe hybridizationgh; molecular cytogenetic mapi | Ramachandran and Narayan (1985)b,d, Zhao et al. (2011)b,c,f,g, Yang et al. (2012)b,h, Rajkumari et al. (2015)b,c,e, Zhang et al. (2016)a,b,c,f | ||||
| C.sativus var. Long green | 7a | 14b | Diploidc, 12 metacentric, 2 sub-metacentric chromosomesd; centromeric and telomeric sequence signalse | Zhang et al. (2012) a-e | ||||
| C.sativusvar.sativus (CSS) | 7a | 14b | Diploidc, centromeric 45S (10) and intercalary 5S (2) rDNA signalsd; centromeric and distal repetitive sequence probese; molecular cytogenetic mapf | Zhao et al. (2011)b-e, Yang et al. (2012)b,f, Zhang et al. (2016)a-d | ||||
| C.sativus cv. Winter Long | 14a | 7b | Diploidc, C- bandingd, DAPI bandinge, 45S (6) and 5 S (2) rDNA signalsf, repetitive sequence based molecular karyotype in somatic and pachytene chromosomesg | Koo et al. (2002)a-f, (2005)a,b,g | ||||
| C.sativusvar.xishuangbannesis Qi et Yuan Zhenzhen, 1983 | 7a | 14b | Diploidc, centromeric 45S (10) and intercalary 5S (2) rDNA signalsd; centromeric and telomeric signalse | Zhao et al. (2011)b,c,e, Zhang et al. (2016)a-d | ||||
| C.setosus Cogniaux, 1881 | 24a | 12b | Diploidc; 4m+5sm+3st (haploid)d | Rajkumari et al. (2013)a-c, (2015)a,c,d | ||||
| C.silentvalleyii Manilal et Sabu et Mathew, 1985 | 24a | 12b | - | Rajkumari et al. (2013) a,b | ||||
| C.trigonus Roxb. | 24a | 12b | - | Rajkumari et al. (2013) a,b | ||||
| C.zambianus Widrl., J.H.Kirkbr., Ghebret. and K.R.Reitsma | 12a | 24b | Diploidc; 45S (2) and co-localized 45S+5S (2) signalsd | Zhang et al. (2016) a-d | ||||
| C.zeyheri Sond. | 24a, 48b | Diploidc, Allotetraploidd; 2C (Feulgen densitometry): 1.682e/2.846 pgf; 4 satellitesg; 45S (2) and co-localized 45S+5S (2) rDNA signalsh; FISH with bulked oligo probe from cucumber schromosome C7i; 24IIj , 12IIk, 11II+2Il | IPCNa,b,d,j,k,l, Ramachandran and Narayan (1985)a-f, Han et al. (2015)i, Yagi et al. (2015)a,c,g,h | |||||
| Cucumellacinerea (Cogn.) C.Jeffrey | 2C (Feulgen Microdensitometry): 0.50pga | Bennet et al. (1982)a | ||||||
| Mukiamaderaspatana (L.) M.Roem. (syn. Cucumismaderaspatanas and Melothriamaderaspatana) | 12a | 24b | 11c, 12d | - | CCDBb,c, Rajkumari et al. (2015)b,d | |||
| Oreosyceafricana Hook.f. (syn. Cucumissubsericeus) | 12a | 48b | Tetraploidc; co-localized 45S and 5S rDNA signals (2)d; FISH with bulked oligo probe from cucumber chromosome C7e | Han et al. (2015)e, Zhang et al. (2016)a-d | ||||
#x: base number; 2n: zygotic number; n: gametic number; NOR: nucleolar organizing region; SAT: satellite chromosome; ScgCP: Single-copy gene-based chromosome painting (Lou et al. 2014); I: univalent, II: bivalent, IV: tetravalent; CCDB: Chromosome Counts Database; superscripts correspond to reference.
Table 8.
Cytogenetic information in Cucurbiteae #.
| Genera studied | Species studied | Chromosome no. | Ploidy, Genome size, Chromosome features | References | ||
|---|---|---|---|---|---|---|
| x | 2n | n | ||||
| Cayaponia Silva Manso, 1836 | C.laciniosa Linnaeus, 1753 | 24a | - | Kumar and Subramaniam (1987) a | ||
| Cucurbita | 10a, 12b | - | Darlington and Janaki Ammal (1945) a,b | |||
| C.andreana Naudin, 1896 | 40a | CCDB a | ||||
| C.argyrosperma Huber, 1867 (syn. C.mixta Pangalo, 1930) | 40a | 2C (flow cytometry): 0.748 pgb | Sisko et al. (2003)a,b | |||
| C.cylindrata Bailey, 1943 | 40a | 20b | - | CCDB a,b | ||
| C.digitata Gray, 1853 | 10a, 12b | 40c | 20d | - | Darlington and Janaki Ammal (1945)a,b, CCDBc,d | |
| C.ecuadorensis Cutler et Whitaker, 1969 | 2C: 0.72pga | Plant DNA C Value databasea | ||||
| C.ficifolia Bouché, 1837 (syn. C.melanosperma Gasparrini, 1847) | 40a | 2C (flow cytometry): 0.933pgb | Plant DNA C- Values Databasea,b | |||
| C.foetidissima Kunth, 1817 | 10a, 12b | 40c, 42d | 2C (flow cytometry): 0.686pge | Darlington and Janaki Ammal (1945)a,b, Plant DNA C- Values Databasec,e, CCDBc,d | ||
| C.indica (unresolved) | 40a | - | IPCN a | |||
| C.lundelliana Bailey, 1943 | 20a | 2C (flow cytometry): 0.72pgb | CCDBa, Plant DNA C Value databaseb | |||
| C.maxima Duchesne, 1786 | 20a | 24b, 40c, 44d, 48e | 20f | Kumar and Subramaniam (1987)a,c,d,e, Beevy and Kuriachan (1996)f, CCDBc,f | ||
| C.moschata Duchesne, 1786 | 10a, 12b | 24c, 40d, 44e,48f | Diploidg; 2C (Feulgen microdensitometry): 0.90pgh; 2C (flow cytometry): 0.708i/ 0.97jpg; 36 metacentric and 4 sub-metacentric chromosomesk; CSR: 1.05-1.78μml, 45S (10) and 5S (4) rDNA signalsm | CCDBf, Plant DNA C- Values Databaseh,i, Kumar and Subramaniam (1987)a-f, Barrow and Meister (2003)j, Xu et al. (2007)d,m, Waminal et al. (2011)g,d,k,l,m | ||
| C.okeechobeensisssp.martinezii Bailey, 1943 | 40a | 2C (flow cytometry): 0.74pgb | Plant DNA C- Values Databasea,b | |||
| C.palmata Watson, 1876 | 10a, 12b | 40c, 42d | 20e | - | Kumar and Subramaniam (1987)a,b, CCDBc,d,e | |
| C.pedatifolia Bailey, 1943 | 40a | - | CCDB a | |||
| C.pepo Linnaeus, 1753 | 10a, 12b | 22c, 24d, 28e, 40f, 42g, 44h, 46i, 80j | 20k | 2C (flow cytometry): 0.74pgl; 0.864m; 1.109 pg-1.064 pgn; 1.18pgo; 45S (10) and 5S (4) rDNA signalsp | Kumar and Subramaniam (1987)a-j, CCDBf,k, Marie and Brown (1993)l, Barow and Meister (2003)o, Rayburn (2008)n, Plant DNA C- Values Databasem, Xie et al. (2019b)f, p | |
| Sicana Naudin, 1862 | S.odorifera Vellozo, 1831 | 40a | 20b | - | IPCN a,b | |
# x: base number; 2n: zygotic number; n: gametic number; CCDB: Chromosome Counts Database; superscripts correspond to references.
Nuclear genome contents
Nuclear genome sizes are reported in 49 species (~5% of total species) belonging to 15 genera (~16% of total genera) of Cucurbitaceae. Among the understudied tribes, 2C genome content is known for one species each from Gomphogyneae and Coniandreae (Table 2). Within the Momordiceae species of India, significant interspecific genome size differences have been reported (Ghosh et al. 2021). The species differed 5.19-fold in their genome sizes (2C = 0.72-3.74 pg) (Table 3) (Ghosh et al. 2021). Interestingly, the species with lowest chromosome number (M.cymbalaria, 2n = 18) contained highest nuclear DNA content among the four Momordica species (Table 3). In Bryonieae, flow cytometric genome size of Bryonia shows a 2.2-fold increase than Ecballium (Table 4). In case of Sicyoeae, flow cytometric 2C DNA content ranges from 1.49–2.32 pg/2C, indicating 1.55-fold differences in genome size. Echinocystislobata Michaux, 1803, in spite of tetraploid condition, shows lowest genome size (Table 5). There is no significant difference in genome size between the genders of Trichosanthesdioica Roxburgh, 1832 (Table 5). Genome size estimates are known from 24 Benincaseae species of which 17 species belong to Cucumis (Tables 6, 7). Highest 2C nuclear genome is known in Benincasahispida Thunberg, 1784 (1.97 pg) (Bhowmick and Jha 2015a) while the lowest is known in Cucumismelovar.inodorus Harz, 1885 (0.64 pg) (Karimzadeh et al. 2010). In case of Cucumis, there is yet no consensus on whether the taxa with different base numbers (x = 7, 12) have correspondingly dissimilar genome sizes since the researchers depended on diverse methods of genome size estimation. Lower 2C genome size was reported in C. Cocciniagrandis Linnaeus, 1767 (2n = 24) while C.trilobata (2n = 20) had higher 2C DNA content (Table 6). The divergence in genome size between genders was found to be highest in dioecious C.grandis (Table 6), a sharp contrast to dioecious Trichosanthesdioica (Table 5). Benincaseae shows a 3.07-fold overall difference in genome size. Genome sizes are known in eight species of Cucurbita Jussieu, 1789. Flow cytometric genome size ranges from 0.686–0.933 pg/2C, indicating a 1.36-fold variation (Table 8). Despite polypoidy, the nuclear DNA content of Cucurbita species is comparable to many diploids.
Karyotypes, chromosome banding and molecular cytogenetics
Among the understudied tribes, information on chromosome morphology, size and karyotype are reported in very few taxa (Table 2). In Gynostemmapentaphyllum Thunberg, 1784, the number of rDNA loci was suggested to reduce during polyploidization (Pellerin et al. 2018). The Actinostemmatenerum Griffith, 1837, genome contained interstitial telomeric repeats which were suggested to be the result of chromosome fusion from ancestral genome. The co-localization of 45S and 5S rDNA loci in A.tenerum and Thladianthadubia Bunge, 1833, have been thought to imply regional synteny and shared ancestral traits (Xie et al. 2019b). In the tribe Cucurbiteae, detailed karyotype analysis is known only in Cucurbitamoschata Duchesne, 1786 and C.pepo Linnaeus, 1753, showing conserved 45S and 5S rDNA signals (non-co-localized) in independent analyses (Table 8).
Karyotypes and chromosome sizes are reported in ten species of Momordiceae (Table 3). Interspecific differences have been observed and found to correlate with phylogenetic relationship within Momordica (Ghosh et al. 2021). Infraspecific delimitation of Indian M.charantia varieties was based on fluorochrome banding pattern and genome size divergence (Table 3), corresponding to infraspecific distinction reported in the Japanese bitter gourd cultivars (Kido et al. 2016). FISH in three Momordica species revealed 45S and 5S rDNA sites to be localised on different chromosomes (Table 3). In context of the genome sequence of bitter gourds (Matsumura et al. 2020), further scopes for cytogenetic and genomic investigation remain open.
Karyotype and chromosome size is reported in eight 8 species of Sicyoeae (Table 5). Fluorochrome banding pattern has facilitated comparative analysis in Luffa species occurring in India (Tables 1, 5) (Bhowmick and Jha 2015a, 2021). The cultivated ridged gourd (L.acutangula Linnaeus, 1753) showed three CMA+ satellite bearing pairs (Fig. 1A–C, J) as in the wild L.echinata Roxburgh, 1814 (Fig. 1G–I, L), while the sponge gourd (L.aegyptiaca Miller, 1768 has two satellited pairs (Fig. 1D–F, K). Luffaacutangula and L.echinata also showed up distal DAPI bands (Fig. 1J, L), absent in L.aegyptiaca (Fig. 1K). Trichosanthes species (2n = 22) have inter-specific differences (Fig. 2) as well as infraspecific distinction (T.cucumerina Linnaeus, 1753) in fluorochrome banding pattern (Tables 1, 5, Fig. 2A–H). The male and female plants of T.dioica show similar chromosome number, morphology and genome size but show differences in fluorochrome banding pattern (Fig. 2I–P, Table 5). The 11th, 12th and 13th pairs (CMA+) are marker chromosomes in Luffa (Fig. 1, Table 1) while the 10th and 11th pairs are conserved CMA+ satellited pairs in Trichosanthes (Fig. 2, Table 1). Eight species of Sicyoeae have been subjected to FISH (Table 5). The polyploid and diploid species have differences in the number of rDNA loci, showing separate localization of the 45S and 5S rDNA signals except Sicyosangulatus Linnaeus, 1753 and Trichosantheskirilowii Maximowicz, 1859 (Table 5).
Figure 2.
Somatic metaphase chromosomes and idiograms of Trichosanthes species stained with Giemsa (A, D, I, L), DAPI (C, E, J, M) and CMA3 (B, F, K, N) A–CT.cucumerinassp.cucumerina (2n = 22), D–FTrichosanthescucumerinassp.cucumerina ‘Anguina’ (2n = 22) I–KT.dioica (male, 2n = 22) L–NT.dioica (female, 2n = 22). Arrows indicate satellited chromosomes in Giemsa plates and CMA+ve signals in B, F, K, N. Corresponding somatic idiograms (haploid set) of: GT.cucumerinassp.cucumerinaHTrichosanthescucumerinassp.cucumerina ‘Anguina’ OT.dioica male plant PT.dioica female plant. Blue and golden yellow bands in idiograms indicate DAPI+ve and CMA+ve signals, respectively. Scale Bars: 5 µm
Benincaseae generally reveal two distal 45S rDNA loci of which at least one locus is either adjacent to 5S rDNA locus (Table 6) or co-localized in the same chromosome as in most of the Cucumis species (Table 7). Exceptionally, a wild species of Benincasa (B.fistulosa Stocks, 1851) has non-adjacent 45S and 5S signals (Li et al. 2016). GC rich satellites were observed in the 12th pair of chromosomes showing CMA+ bands in cultivated Indian ashgourd (B.hispida) (Fig. 3 A–C, J, Tables 1, 6). Lagenariasiceraria Molina, 1782 and Cucumismelo Linnaeus, 1753 are the other two genera having similarity in rDNA hybridization profile, agreeing with phylogenetic affinity (Li et al. 2016).
Citrulluscolocynthis Linnaeus, 1753 and C.lanatus Thunberg, 1794 may share a common ancestor both having two 45S rDNA loci and one 5S locus. Loss of one 45S rDNA locus has given way to C.rehmii De Winter, 1990 while gain of one 5S rDNA locus has been proposed to lead to C.ecirrhosus Cogniaux, 1888 and C.lanatusvar.citroides Bailey, 1930 (presently C.amarus Schrader, 1836) (Reddy et al. 2013; Li et al. 2016). GISH using C.lanatusvar.citroides genome has revealed divergence from C.lanatusvar.lanatus (Reddy et al. 2013).
The genus Cucumis is the largest in Benincaseae with 65 species of which 39 have been studied (Table 7). Among the Cucumis species with x = 12, co-localization rDNA loci (45S and 5S rDNA) have been documented in 14 species, including C.melo (Table 7). However, the number of 45S sites is generally four, which may be six or eight in some cases (Table 7). rDNA hybridization data strongly corroborated with the ‘fusion’ theory for derivation of x = 7 (C.sativus) from x = 12 (C.melo) (Waminal and Kim 2012) which is substantiated by genomic studies (Li et al. 2011). There are ten pericentromeric/ centromeric 45S and two distal 5S rDNA sites in C.sativus while six 45S rDNA sites were reported in C.sativusvar.hardwickii Royle, 1835 (Koo et al. 2005; Zhang et al. 2012). Comparative chromosome painting (Lou et al. 2014) and GISH (Zhang et al. 2015) proved high colinearity between cucumber and melon. Based on chloroplast and nuclear DNA (ITS) phylogeny, C.melo (melon) has been found to be sister to a clade comprising C.sativus and related genera (Dicaelospermum Clarke, 1879 and Mukia Arnott, 1840) (Renner et al. 2007). rDNA site co-localization was found to coincide with geographical origin of 12 Cucumis species (Zhang et al. 2016). The chromosomal affinity between C.metuliferus Schrader, 1838, C.anguira Linnaeus, 1753, C.zeyheri Sonder, 1862, C.myriocarpus Naudin, 1859 and polyploid C.heptadactylis Naudin, 1859 (dioecious) (Yagi et al. 2015) can be substantiated by their phylogenetic proximity based on chloroplast and nuclear DNA (ITS) sequences (Renner et al. 2007). rDNA distribution of C.metuliferus was also the reason to consider proximity with Citrullusnaudinianus Sonder, 1862, (previously Acanthosicyosnaudinianus Sonder, 1862) (Reddy et al. 2013). Infraspecific differences were documented in Cucumismelo on the basis of 45S- 5S rDNA signals (linked or separated) which also possessed unique centromeric satellites (Setiawan et al. 2018, 2020). Moreover, chromosome painting method elucidated chromosomal rearrangement in some Cucumis species (Lou et al. 2014; Li et al. 2018).
The dramatic evolution of Y chromosome was validated in karyotypes (Fig. 3 D–I, K–L) of Cocciniagrandis (Table 6). The 45S rDNA sites enabled confirmation of NORs in the 8th and 12th pair containing distal GC rich CMA+ signals in C.grandis (Fig. 3 D–I, K–L, Tables 1, 6). 45S and 5S rDNA hybridization pattern was similar in three other Coccinia species and Diplocyclospalmatus Linnaeus, 1753 (Table 6). The three closely related dioecious species of Coccinia accumulated Y chromosome repeats and displayed sex chromosome turnover (Sousa et al. 2017). Strong centromeric CMA bands (Fig. 3 D–I, K–L, Table 1) were observed in C.grandis except Y chromosome (Fig. 3 I, L), presenting a possibility that CgCent (CL1) is a feature of centromeres of dioecious Coccinia species (Sousa et al. 2017). In addition, non-nucleolar CMA+ heterochromatin might be associated with sexual differentiation of autosomes in dioecious C.grandis (Fig. 3) which is also a marker in Trichosanthesdioica (Fig. 2, Table 1), opening good scope for further study.
Distinct 45S rDNA sites are higher in number than 5S rDNA sites in Cucurbitaceae (Fig. 4) (Waminal and Kim 2012). The distal 45S rDNA loci are conserved genomic landmarks (Fig. 4) while 5S rDNA loci are relatively diverse (Fig. 4). Based on the literature reports, some NORs (Type I) included chromosomes showing non-colocalized 45S and 5S rDNA sites in seven species of Benincaseae, one species each from Cucurbiteae and Momordiceae and two species of Sicyoeae. The rearrangement of 45S rDNA site in Cucumissativus, probes for chromosome number reduction which may be a consequence of diploidization. The second type (Type II) shows colocalised 45S and 5S rDNA loci, either adjacent or distant, but always on the same chromosome and found in one species each of Benincaseae, Sicyoeae and Actinostemmateae. The third type (Type III) was characterized by chromosomes with non-colocalized and colocalised 45S and 5S rDNA loci, as in 14 species of Benincaseae and one species each of Sicyoeae and Thladiantheae. The rDNA sites of majority of Cucumis species were of non-adjacent type. Hence, type III NORs in majority of Benincaseae genera advocates conservation of the marker chromosomes having distal NOR (45S rDNA). Gynostemmapentaphyllum and some polyploid Cucumis reveal rDNA loci reduction after polyploidization (Zhang et al. 2016; Pellerin et al. 2018).
Figure 4.
Types of chromosomes bearing the NORs as per available reports of rDNA hybridization in Cucurbitaceae. Type I: Chromosomes with only non-colocalised 45S and 5S rDNA sites, Type II: Chromosomes with colocalised 45S and 5S rDNA sites, Type III: Chromosomes with both non- colocalised and colocalised 45S and 5S rDNA sites. See text for explanation.
Correlation between parameters
Chromsome numbers in Cucurbitaceae range from x = 5 to x = 16. The most prevalent number x = 12 (Fig. 5) is considered ancestral (Xie et al. 2019b), followed by x = 11, 13, 14 and 10 (Fig. 5). The present regression analyses for 41 taxa (including 16 Indian taxa) (Table 9) revealed significant linear correlation between 2n and HCL, between ploidy and genome size and between ploidy and HCL (Fig. 6). Therefore, an increase in ploidy/ 2n number is linked with increase in HCL. There was no significant correlation between 2C genome size and chromosome numbers. Cytogenetic parameters may not reflect residual evidence of CCT in Cucurbitraceae at present, as reasoned by Alix et al. (2017).
Figure 5.
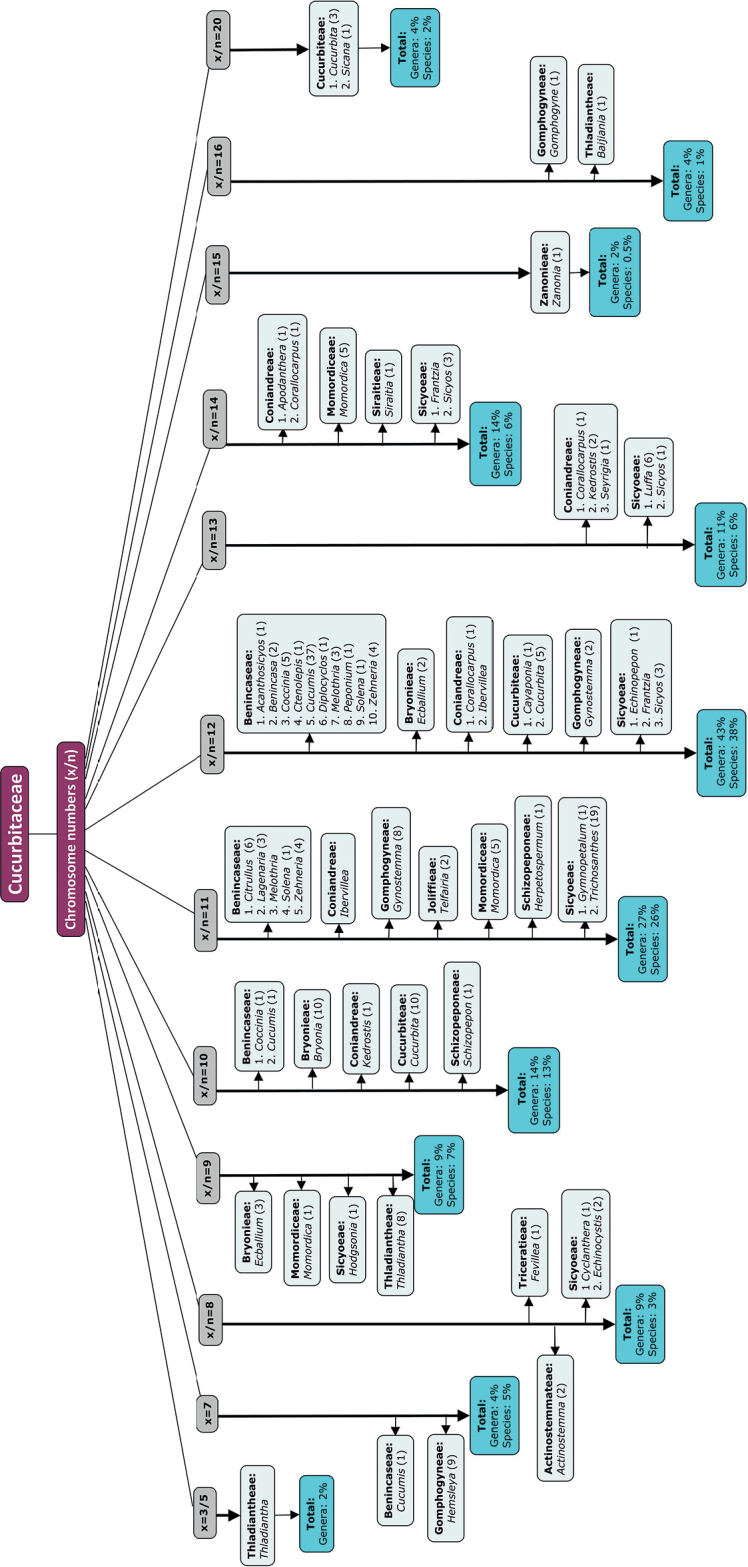
The types of different base numbers (x, based on published reports) or possible base numbers (x/n, based on reported haploid counts) in Cucurbitaceae. The numbers in brackets beside names of genera signify the number of species whose chromosome counts are reported. The % of genera and species with a particular chromosome number, is indicated at the end arrow (out of a total of 44 genera and 188 species with chromosome counts).
Table 9.
Data on fundamental cytogenetic parameters utilized for statistical analysis.
| Species | 2n Chromosome no. | Ploidy | 2C genome size (pg) | MCL (μm) | HCL (μm) | References |
|---|---|---|---|---|---|---|
| Gynostemmapentaphyllum | 66 | 6 | 3.62 | Zhang et al. (2013), Pellerin et al. (2018) | ||
| Zanoniaindica | 30 | 2 | 1.47 | 22.12 | Lekhak et al. (2018) | |
| Momordicabalsamina | 22 | 2 | 1.30 | 14.3# | Bharathi et al. (2011) | |
| Momordicacharantiavar.charantia | 22 | 2 | 0.72 | 1.97 | 21.77 | Ghosh et al. (2018) |
| Momordicacharantiavar.muricata | 22 | 2 | 1.16 | 2.19 | 24.19 | Ghosh et al. (2018) |
| Momordicacochinchinensis | 28 | 2 | 2.64 | 2.27 | 31.86 | Ghosh et al. (2021) |
| Momordicacymbalaria | 18 | 2 | 3.74 | 3.75 | 33.79 | Ghosh et al. (2021) |
| Momordicadioica | 56 | 4 | 3.36 | 2.75 | 77.1 | Ghosh et al. (2021) |
| Momordicasahyadrica | 28 | 2 | 1.34 | 18.76 | Bharathi et al. (2011) | |
| Momordicasubangulata | 56 | 4 | 3.06 | 2.15 | 60.3 | Ghosh et al. (2021) |
| Luffaacutangula | 26 | 2 | 2.20 | 28.63 | this study | |
| Luffacylindrica | 26 | 2 | 1.56 | 2.98 | 38.77 | Bhowmick and Jha (2015a), this study |
| Luffaechinata | 26 | 2 | 3.17 | 41.26 | this study | |
| Trichosanthescucumerina | 22 | 2 | 3.47 | 37.855 | Bhowmick and Jha (2019), this study | |
| Trichosanthescucumerinasubsp.cucumerina Anguina | 22 | 2 | 3.43 | 37.74 | Bhowmick and Jha (2019), this study | |
| Trichosanthesdioica Male | 22 | 2 | 2.27 | 3.71 | 40.82 | Bhowmick and Jha (2015a), this study |
| Trichosanthesdioica Female | 22 | 2 | 2.32 | 3.71 | 40.82 | Bhowmick and Jha (2015a), this study |
| Benincasahispida | 24 | 2 | 1.97 | 3.17 | 38.08 | Bhowmick and Jha (2015a), this study |
| Citrulluslanatus | 22 | 2 | 1.33# | 14.67 | Waminal et al. (2011) | |
| Cocciniagrandis male | 24 | 2 | 0.92 | 1.80 | 20.32 | Bhowmick et al. (2012, 2016), this study |
| Cocciniagrandis female | 24 | 2 | 0.73 | 1.86 | 19.85 | Bhowmick et al. (2012, 2016), this study |
| Cocciniahirtella | 24 | 2 | 0.988 | Sousa et al. (2017) | ||
| Cocciniasessilifolia Male | 24 | 2 | 0.984 | Sousa et al. (2017) | ||
| Cocciniasessilifolia Female | 24 | 2 | 0.998 | Sousa et al. (2017) | ||
| Cocciniatrilobata | 20 | 2 | 1.263 | Sousa et al. (2017) | ||
| Lagenariasiceraria | 22 | 2 | 0.734 | 1.79 | 20.06 | Achigan-Dako et al. (2008) |
| Cucumisafricanus | 24 | 2 | 2.08 | 25.045 | Yagi et al. (2015) | |
| Cucumisanguriavar.anguria | 24 | 2 | 2.13 | 25.6 | Yagi et al. (2015) | |
| Cucumisanguriavar.longaculeatus | 24 | 2 | 2.10 | 25.195 | Yagi et al. (2015) | |
| Cucumisheptadactylus | 48 | 4 | 2.09 | 50.225 | Yagi et al. (2015) | |
| Cucumismelo | 24 | 2 | 1.05 | 1.50 | 17.8# | Marie and Brown (1993), Hoshi et al. (2013) |
| Cucumismelovar.inodorus | 24 | 2 | 0.64 | Karimzadeh et al. (2010) | ||
| Cucumismyriocarpusvar.leptodermis | 24 | 2 | 1.93 | 23.19 | Yagi et al. (2015) | |
| Cucumismyriocarpusvar.myriocarpus | 48 | 4 | 2.25 | 53.985 | Yagi et al. (2015) | |
| Cucumiszeyheri | 24 | 2 | 2.30 | 27.56 | Yagi et al. (2015) | |
| Cucumissativus | 14 | 2 | 1.03, 1.77## | 2.07# | 14.50 | Barow and Meister (2003), Marie and Brown (1993), Waminal and Kim (2012) |
| Cucurbitaargyrosperma | 40 | 0.748 | Roy et al. (1991), Sisko et al. (2003) | |||
| Cucurbitaecuadorensis | 40 | 0.933 | Sisko et al. (2003) | |||
| Cucurbitafoetidissima | 40 | 0.686 | Sisko et al. (2003) | |||
| Cucurbitamoschata | 40 | 2 | 0.708, 0.97## | 1.26# | 25.19 | Sisko et al. (2003), Barrow and Meister (2003), Waminal et al. (2011) |
| Cucurbitaokeechobeensisssp.martinezii | 40 | 0.74 | Sisko et al. (2003) |
# calculated from chromosome measurements reported in publications, ## different entries for same taxa were taken from different reports
Figure 6.
Scatter plots of 2n chromosome number and ploidy level (predictor variables) versus 2C genome size, MCL (mean chromosome length) and HCL (total length of haploid chromosome set) in Cucurbitaceae taxa. Symbols below plots depict regression analysis parameters; square: adjusted R square, circle: standard error of the estimate, triangle: Pearson Correlation, star: 2-tailed significance of Pearson Correlation. Regular lines indicate significant linear regression and dotted lines indicate not significant linear regress
Future directions
Chromosome number and genome size information in the basal clades (understudied tribes) should be given attention to infer ancient base numbers. The parameters of fundamental and molecular cytogenetics are inevitable for genomic interpretation (Weiss-Schneeweiss and Schneeweiss 2013; Deakin et al. 2019) and hence relevant to spot genetic resources and relationships with wild relatives. The current review is not exhaustive but supersedes the scopes of general web resources and brings an offline resource exclusive for Cucurbitaceae.
Acknowledgements
SJ is thankful to the National Academy of Sciences (NASI, Allahabad, India) for the NASI Senior Scientist Fellowship award. BKB gratefully acknowledges Principal, Scottish Church College, India for continuous support and encouragement in research activities.
Citation
Bhowmick BK, Jha S (2022) A critical review on cytogenetics of Cucurbitaceae with updates on Indian taxa. Comparative Cytogenetics 16(2): 93–125. https://doi.org/10.3897/compcytogen.v16.i2.79033
ORCID
Biplab Kumar Bhowmick https://orcid.org/0000-0001-6029-1098
Sumita Jha https://orcid.org/0000-0002-1375-2768
References
- Achigan-Dako EG, Fuchs J, Ahanchede A, Blattner FR. (2008) Flow cytometric analysis in Lagenariasiceraria (Cucurbitaceae) indicates correlation of genome size with usage types and growing elevation. Plant Systematics and Evolution 276: e9. 10.1007/s00606-008-0075-2 [DOI]
- Alix K, Gérard PR, Schwarzacher T, Heslop-Harrison JS. (2017) Polyploidy and interspecific hybridization: partners for adaptation, speciation and evolution in plants. Annals of Botany 120: 183–194. 10.1093/aob/mcx079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anmin L, Luqi H, Shukun C, Jeffrey C. (2011) Flora of China 19. http://www.efloras.org/flora_page.aspx?flora_id=2
- Arumuganathan K, Earle ED. (1991) Nuclear DNA content of some important plant species. Plant Molecular Biology Reporter 9: 208–218. 10.1007/BF02672069 [DOI] [Google Scholar]
- Barow M, Meister A. (2003) Endopolyploidy in seed plants is differently correlated to systematics, organ, life strategy and genome size. Plant, Cell & Environment 26: 571–584. 10.1046/j.1365-3040.2003.00988.x [DOI] [Google Scholar]
- Beevy SS, Kuriachan PH. (1996) Chromosome numbers of south Indian Cucurbitaceae and a note on the cytological evolution in the family. Journal of Cytology and Genetics 31: 65–71. [Google Scholar]
- Bellot S, Mitchell TC, Schaefer H. (2020) Phylogenetic informativeness analyses to clarify past diversification processes in Cucurbitaceae. Scientific Reports 10: 1–13. 10.1038/s41598-019-57249-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Smith JB, Heslop-Harrison JS. (1982) Nuclear DNA amounts in angiosperms. Proceedings of the Royal Society of London – Series B, Biological Sciences 216: 179–199. 10.1098/rspb.1982.0069 [DOI] [PubMed] [Google Scholar]
- Bharathi LK, Munshi AD, Vinod SC, Behera TK, Das AB, John KJ. (2011) Cytotaxonomical analysis of Momordica L. (Cucurbitaceae) species of Indian occurrence. Journal of Genetics 90: 21–30. 10.1007/s12041-011-0026-5 [DOI] [PubMed] [Google Scholar]
- Bhowmick BK, Jha S. (2015a) Differential heterochromatin distribution, flow cytometric genome size and meiotic behavior of chromosomes in three Cucurbitaceae species. Scientia Horticulturae 193: 322–329. 10.1016/j.scienta.2015.07.006 [DOI] [Google Scholar]
- Bhowmick BK, Jha S. (2015b) Dynamics of sex expression and chromosome diversity in Cucurbitaceae: A story in the making. Journal of Genetics 94: 793–808. 10.1007/s12041-015-0562-5 [DOI] [PubMed] [Google Scholar]
- Bhowmick BK, Jha S. (2019) Differences in karyotype and fluorochrome banding patterns among variations of Trichosanthescucumerina with different fruit size. Cytologia 84: 237–245. 10.1508/cytologia.84.237 [DOI] [Google Scholar]
- Bhowmick BK, Jha S. (2021) A comparative account of fluorescent banding pattern in the karyotypes of two Indian Luffa species. Cytologia 86: 35–39. 10.1508/cytologia.86.35 [DOI] [Google Scholar]
- Bhowmick BK, Jha TB, Jha S. (2012) Chromosome analysis in the dioecious cucurbit Cocciniagrandis (L.) Voigt. Chromosome Science 15: 9–15. [Google Scholar]
- Bhowmick BK, Yamamoto M, Jha S. (2016) Chromosomal localization of 45S rDNA, sex specific C values and heterochromatin distribution in Cocciniagrandis (L.) Voigt. Protoplasma 253: 201–209. 10.1007/s00709-015-0797-2 [DOI] [PubMed] [Google Scholar]
- Chomicki G, Schaefer H, Renner SS. (2020) Origin and domestication of Cucurbitaceae crops: insights from phylogenies, genomics and archaeology. New Phytologist 226: 1240–1255. 10.1111/nph.16015 [DOI] [PubMed] [Google Scholar]
- Darlington CD, Janaki Ammal EK. (1945) Chromosome atlas of cultivated plants. George Allen and Unwin LTD, London.
- De Donato M, Cequea H. (1994) A cytogenetic study of six cultivars of the chayote, Sechiumedule Sw. (Cucurbitaceae). Journal of Heredity 85: 238–241. 10.1093/oxfordjournals.jhered.a111444 [DOI] [Google Scholar]
- Deakin JE, Potter S, O’Neill R, Ruiz-Herrera A, Cioffi MB, Eldridge MD, Fukui K, Marshall Graves JA, Griffin D, Grutzner F, Kratochvíl L. (2019) Chromosomics: bridging the gap between genomes and chromosomes. Genes 10: e627. 10.3390/genes10080627 [DOI] [PMC free article] [PubMed]
- Ghosh I, Bhowmick BK, Jha S. (2018) Cytogenetics of two Indian varieties of Momordicacharantia L. (bittergourd). Scientia Horticulturae 240: 333–343. 10.1016/j.scienta.2018.06.027 [DOI] [Google Scholar]
- Ghosh I, Saha PS, Bhowmick BK, Jha S. (2021) A phylogenetic analysis of Momordica (Cucurbitaceae) in India based on karyo-morphology, nuclear DNA content and rDNA ITS1–5.8S–ITS2 sequences. Protoplasma 258: 347–60. 10.1007/s00709-020-01576-z [DOI] [PubMed] [Google Scholar]
- Goldblatt P, Lowry PP. (2011) The Index to Plant Chromosome Numbers (IPCN): three decades of publication by the Missouri Botanical Garden come to an end. Annals of the Missouri Botanical Garden 98: 226–227. 10.3417/2011027 [DOI] [Google Scholar]
- Guo J, Xu W, Hu Y, Huang J, Zhao Y, Zhang L, Huang CH, Ma H. (2020) Phylotranscriptomics in cucurbitaceae reveal multiple whole-genome duplications and key morphological and molecular innovations. Molecular Plant 13: 1117–1133. 10.1016/j.molp.2020.05.011 [DOI] [PubMed] [Google Scholar]
- Han Y, Zhang T, Thammapichai P, Weng Y, Jiang J. (2015) Chromosome-specific painting in Cucumis species using bulked oligonucleotides. Genetics: 200(3): 771–779. 10.1534/genetics.115.177642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Zhang Z, Huang S, Jin W. (2011) An integrated molecular cytogenetic map of Cucumissativus L. chromosome 2. BMC Genetics 12: e18. 10.1186/1471-2156-12-18 [DOI] [PMC free article] [PubMed]
- Han Y, Zhang Z, Liu C, Liu J, Huang S, Jiang J, Jin W. (2009) Centromere repositioning in cucurbit species: implication of the genomic impact from centromere activation and inactivation. Proceedings of the National Academy of Sciences 106: 14937–14941. 10.1073/pnas.0904833106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi Y, Kido M, Yagi K, Tagashira N, Morikawa A, Nagano K. (2013) Somatic chromosome differentiation in Cucumismelo L. and C.metuliferus E. Mey. ex Naudin. Chromosome Botany 8: 7–12. 10.3199/iscb.8.7 [DOI] [Google Scholar]
- Hoshi Y, Mori M, Matoba H, Tagashira N, Murata T, Plader W, Malepszy S. (2008) Chromosomal polymorphism of two pickling cucumbers (Cucumissativus L.) revealed by fluorescent staining with CMA and DAPI. Cytologia 73: 41–48. 10.1508/cytologia.73.41 [DOI] [Google Scholar]
- Jha (2021) Karyotype diversity in cultivated and wild Indian rice through EMA-based chromosome analysis. Journal of Genetics 100: 1–15. 10.1007/s12041-021-01332-z [DOI] [PubMed] [Google Scholar]
- Karimzadeh G, Mousavi SH, Jafarkhani-Kermani M, Jalali-Javaran M. (2010) Karyological and nuclear DNA variation in Iranian endemic muskmelon (Cucumismelovar.inodorus). Cytologia 75: 451–461. 10.1017/S147926211400077X [DOI] [Google Scholar]
- Kausar N, Yousaf Z, Younas A, Ahmed HS, Rasheed M, Arif A, Rehman HA. (2015) Karyological analysis of bitter gourd (Momordicacharantia L., Cucurbitaceae) from Southeast Asian countries. Plant Genetic Resources 13: 180–182. 10.1017/S147926211400077X [DOI] [Google Scholar]
- Kido M, Morikawa A, Saetiew K, Hoshi Y. (2016) A cytogenetic study of three Japanese cultivars of Momordicacharantia L. Cytologia 81: 7–12. 10.1508/cytologia.81.7 [DOI] [Google Scholar]
- Koo DH, Hur Y, Jin DC, Bang JW. (2002) Karyotype analysis of a Korean cucumber cultivar (Cucumissativus L. cv. Winter Long) using C-banding and bicolor fluorescence in situ hybridization. Molecules and Cells 13: 413–418. [PubMed] [Google Scholar]
- Koo DH, Choi HW, Cho J, Hur Y, Bang JW. (2005) A high-resolution karyotype of cucumber (Cucumissativus L. ‘Winter Long’) revealed by C-banding, pachytene analysis, and RAPD-aided fluorescence in situ hybridization. Genome 48: 534–540. 10.1139/g04-128 [DOI] [PubMed] [Google Scholar]
- Kumar V, Subramaniam B. (1987) Chromosome atlas of flowering plants of the Indian subcontinent. Vol 1 and 2. Botanical Survey of India.
- Lekhak MM, Yadav PB, Attar UA, Rajput KS, Ghane SG. (2018) Cytopalynological studies in Zanoniaindica (Cucurbitaceae), a monotypic genus. The Nucleus 61: 105–109. 10.1007/s13237-018-0234-y [DOI] [Google Scholar]
- Li D, H Cuevas E, Yang L, Li Y, Garcia-Mas J, Zalapa J, J Staub E, Luan F, Reddy U, He X, Gong Z, Weng Y. (2011) Syntenic relationships between cucumber (Cucumissativus L.) and melon (C.melo L.) chromosomes as revealed by comparative genetic mapping. BMC Genomics 12: e396. 10.1186/1471-2164-12-396 [DOI] [PMC free article] [PubMed]
- Li K, Wang H, Wang J, Sun J, Li Z, Han Y. (2016) Divergence between C.melo and african Cucumis species identified by chromosome painting and rDNA distribution pattern. Cytogenetic and Genome Research 150: 150–155. 10.1159/000453520 [DOI] [PubMed] [Google Scholar]
- Li Q, Ma L, Huang J, Li LJ. (2007) Chromosomal localization of ribosomal DNA Sites and karyotype analysis in three species of Cucurbitaceae. Journal-Wuhan University Natural Sciences Edition 53: e449.
- Li Z, Bi Y, Wang X, Wang Y, Yang S, Zhang Z, Chen J, Lou Q. (2018) Chromosome identification in Cucumisanguria revealed by cross-species single-copy gene FISH. Genome 61: 397–404. 10.1139/gen-2017-0235 [DOI] [PubMed] [Google Scholar]
- Liu C, Liu J, Li H, Zhang Z, Han Y, Huang S, Jin W. (2010) Karyotyping in melon (Cucumismelo L.) by cross-species fosmid fluorescence in situ hybridization. Cytogenetic and Genome Research 129: 241–249. 10.1159/000314343 [DOI] [PubMed] [Google Scholar]
- Lombello RA, Pinto-Maglio CAF. (2007) Cytomolecular Studies in Momordicacharantia L. (Cucurbitaceae), a potential medicinal plant. Cytologia 72: 415–418. 10.1508/cytologia.72.415 [DOI] [Google Scholar]
- Lou Q, Zhang Y, He Y, Li J, Jia L, Cheng C, Guan W, Yang S, Chen J. (2014) Single‐copy gene‐based chromosome painting in cucumber and its application for chromosome rearrangement analysis in Cucumis. The Plant Journal 78: 169–179. 10.1111/tpj.12453 [DOI] [PubMed] [Google Scholar]
- Lysák MA, Schubert I. (2013) Mechanisms of chromosome rearrangements. In: Greilhuber J, Dolezel J, Wendel JF. (Eds) Plant genome diversity volume 2: Physical structure, behaviour and evolution of plant genomes.Springer, Vienna, 137–147. 10.1007/978-3-7091-1160-4_9 [DOI]
- Marie D, Brown SC. (1993) A cytometric exercise in plant DNA histograms, with 2C values for 70 species. Biology of the Cell 78: 41–51. 10.1016/0248-4900(93)90113-S [DOI] [PubMed] [Google Scholar]
- Matsumura H, Hsiao MC, Lin YP, Toyoda A, Taniai N, Tarora K, Lee CR. (2020) Long-read bitter gourd (Momordicacharantia) genome and the genomic architecture of nonclassic domestication. Proceedings of the National Academy of Sciences 117: 14543–14551. 10.1073/pnas.1921016117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mota L, Torices R, Loureiro J. (2016) The evolution of haploid chromosome numbers in the sunflower family. Genome Biology and Evolution 8: 3516–3528. 10.1093/gbe/evw251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellerin RJ, Waminal NE, Kim HH. (2018) Triple-color FISH karyotype analysis of four korean wild Cucurbitaceae species. Horticultural Science and Technology 36: 98–107. 10.12972/kjhst.20180011 [DOI] [Google Scholar]
- Pellicer J, Leitch IJ. (2020) The Plant DNA C‐values database (release 7.1): an updated online repository of plant genome size data for comparative studies. New Phytologist 226: 301–305. 10.1111/nph.16261 [DOI] [PubMed] [Google Scholar]
- Rajkumari K, Joseph John K, Yadav SR, Bhat KV, Rao SR. (2013) Comparative male meiotic studies in some Indian representative species of Cucumis L. (Cucurbitaceae) Caryologia 66: 313–320. 10.1080/00087114.2013.854613 [DOI] [Google Scholar]
- Rajkumari K, Joseph John K., Yadav SR, Bhat K V, Shamurailatpam A, Rao SR. (2015) Cytogenetical treatise of Indian representative species of Cucumis. A karyotypic approach. Cytology and Genetics 49: 388–396. 10.3103/S0095452715060079 [DOI] [PubMed] [Google Scholar]
- Rayburn AL, Kushad MM, Wannarat W. (2008) Intraspecific genome size variation in pumpkin (Cucurbitapeposubsp.pepo). Horticultural Science 43: 949–951. 10.21273/HORTSCI.43.3.949 [DOI] [Google Scholar]
- Ramachandran C, Narayan RKJ. (1985) Chromosomal DNA variation in Cucumis. Theoretical and Applied Genetics 69: 497–502. 10.1007/BF00251092 [DOI] [PubMed] [Google Scholar]
- Ramachandran C, Narayan RKJ. (1990) Satellite DNA specific to knob heterochromatin in Cucumismetuliferus (Cucurbitaceae). Genetica 80: 129–138. 10.1007/BF00127133 [DOI] [Google Scholar]
- Reddy UK, Aryal N, Islam-Faridi N, Tomason YR, Levi A, Nimmakayala P. (2013) Cytomolecular characterization of rDNA distribution in various Citrullus species using fluorescent in situ hybridization. Genetic Resources and Crop Evolution 60: 2091–2100. 10.1007/s10722-013-9976-1 [DOI] [Google Scholar]
- Ren Y, Zhao H, Kou Q, Jiang J, Guo S, Zhang H, Hou W, Zou X, Sun H, Gong G, Levi A, Xu Y. (2012) A high resolution genetic map anchoring scaffolds of the sequenced watermelon genome. PloS ONE 7: e1. 10.1371/journal.pone.0029453 [DOI] [PMC free article] [PubMed]
- Renner SS, Schaefer H. (2017) Phylogeny and evolution of the Cucurbitaceae. In: Grumet R, Katzir N, Garcia-Mas J. (Eds) Genetics and genomics of Cucurbitaceae.Springer, Cham, 13–23. 10.1007/7397_2016_14 [DOI]
- Renner SS., Schaefer H, Kocyan A. (2007) Phylogenetics of Cucumis (Cucurbitaceae): Cucumber (C.sativus) belongs in an Asian/Australian clade far from melon (C.melo). BMC Evolutionary Biology 7: 1–11. 10.1186/1471-2148-7-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner SS, Sousa A, Chomicki G. (2017) Chromosome numbers, Sudanese wild forms, and classification of the watermelon genus Citrullus, with 50 names allocated to seven biological species. Taxon 66: 1393–1405. 10.12705/666.7 [DOI] [Google Scholar]
- Rice A, Glick L, Abadi S, Einhorn M, Kopelman NM, Salman-Minkov A, Mayzel J, Chay O, Mayrose I. (2015) The Chromosome Counts Database (CCDB)–a community resource of plant chromosome numbers. New Phytologist 206: 19–26. 10.1111/nph.13191 [DOI] [PubMed] [Google Scholar]
- Roy RP, Saran S, Dutt B. (1991) Cytogenetics of the Cucurbitaceae. In: Tsuchiya T, Gupta PK. (Eds) Chromosome engineering in plants: genetics, breeding, evolution.Part B. Elsevier Science Publishers BV, Amsterdam, The Netherlands, 181–200. 10.1016/B978-0-444-88260-8.50015-X [DOI]
- Samuel R, Balasubramaniam S, Morawetz W. (1995) The karyology of some cultivated Cucurbitaceae of Srilanka. Ceylon Journal of Science (Biological Sciences) 24: 17–22. [Google Scholar]
- Setiawan AB, Teo CH, Kikuchi S, Sassa H, Kato K, Koba T. (2018) Cytogenetic variation among Cucumis accessions revealed by fluorescence in situ hybridization using ribosomal RNA genes as the probes. Chromosome Science 21: 67–73. [Google Scholar]
- Setiawan AB, Teo CH, Kikuchi S, Sassa H, Kato K, Koba T. (2020) Centromeres of Cucumismelo L. comprise Cmcent and two novel repeats, CmSat162 and CmSat189. PloS ONE 15(1): e0227578. 10.1371/journal.pone.0227578 [DOI] [PMC free article] [PubMed]
- Soltis DE, Soltis PS, Tate JA. (2004) Advances in the study of polyploidy since plant speciation. New Phytologist 161: 173–191. 10.1046/j.1469-8137.2003.00948.x [DOI] [Google Scholar]
- Soltis DE, Visger CJ, Soltis PS. (2014) The polyploidy revolution then and now: Stebbins revisited. American Journal of Botany 101: 1057–1078. 10.3732/ajb.1400178 [DOI] [PubMed] [Google Scholar]
- Song K, Kim HH. (2008) Chromosome compositions of four cultivated Cucurbitaceae species. Journal of Life Sciences 18: 1019–1022. 10.5352/JLS.2008.18.7.1019 [DOI] [Google Scholar]
- Sousa A, Fuchs J, Renner SS. (2013) Molecular cytogenetics (FISH, GISH) of Cocciniagrandis: A ca. 3 myr-old species of Cucurbitaceae with the largest Y/Autosome divergence in flowering plants. Cytogenetic and Genome Research 139: 107–118. 10.1159/000345370 [DOI] [PubMed] [Google Scholar]
- Sousa A, Fuchs J, Renner SS. (2017) Cytogenetic comparison of heteromorphic and homomorphic sex chromosomes in Coccinia (Cucurbitaceae) points to sex chromosome turnover. Chromosome Research 25: 191–200 10.1007/s10577-017-9555-y [DOI] [PubMed] [Google Scholar]
- Soza VL, Haworth KL, Di Stilio VS. (2013) Timing and consequences of recurrent polyploidy in meadow-rues (Thalictrum, Ranunculaceae). Molecular Biology and Evolution 30: 1940–1954. 10.1093/molbev/mst101 [DOI] [PubMed] [Google Scholar]
- Ting Y, Ya W, Su-qin Z, Guang-dong GENG. (2019) Chromosomal karyotype analysis of Sechiumedule Swartz. Hubei Agricultural Sciences 58: 215.
- Uguru MI, Onovo JC. (2011) Evidence of polyploidy in fluted pumpkin (Telfairiaoccidentalis Hook F.). African Journal of Plant Science 5: 287–290. [Google Scholar]
- Viruel J, Kantar MB, Gargiulo R, Hesketh-Prichard P, Leong N, Cockel C, Forest F, Gravendeel B, Pérez-Barrales R, Leitch IJ, Wilkin P. (2021) Crop wild phylorelatives (CWPs): Phylogenetic distance, cytogenetic compatibility and breeding system data enable estimation of crop wild relative gene pool classification. Botanical Journal of the Linnean Society 195: 1–33. 10.1093/botlinnean/boaa064 [DOI] [Google Scholar]
- Volz SM, Renner SS. (2008) Hybridization, polyploidy and evolutionary transitions between monoecy and dioecy in Bryonia (Cucurbitaceae). American Journal of Botany 95: 1297–1306. 10.3732/ajb.0800187 [DOI] [PubMed] [Google Scholar]
- Wang J, Sun P, Li Y, Liu Y, Yang N, Yu J, Ma X, Sun S, Xia R, Liu X, Ge D, Luo S, Liu Y, Kong Y, Cui X, Lei T, Wang L, Wang Z, Ge W, Zhang L, Song X, Yuan M, Guo D, Jin D, Chen W, Pan Y, Liu T, Yang G, Xiao Y, Sun J, Zhang C, Li Z, Xu H, Duan X, Shen S, Zhang Z, Huang S, Wang X. (2018) An overlooked paleotetraploidization in Cucurbitaceae. Molecular Biology and Evolution 35: 16–26. 10.1093/molbev/msx242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waminal NE, Kim HH. (2012) Dual-color FISH karyotype and rDNA distribution analyses on four Cucurbitaceae species. Horticulture, Environment, and Biotechnology 53: 49–56 . 10.1007/s13580-012-0105-4 [DOI] [Google Scholar]
- Waminal NE, Kim HH. (2015) FISH karyotype analysis of four wild cucurbitaceae species using 5S and 45S rDNA probes and the emergence of new polyploids in Trichosantheskirilowii Maxim. Korean Journal of Horticultural Science and Technology 33: 869–876. 10.7235/hort.2015.15101 [DOI] [Google Scholar]
- Waminal NE, Kim NS, Kim HH. (2011) Dual‐color FISH karyotype analyses using rDNAs in three Cucurbitaceae species. Genes and Genomics 33: 521–528. 10.1007/s13258-011-0046-9 [DOI] [Google Scholar]
- Winterfeld G, Ley A, Hoffmann MH, Paule J, Röser M. (2020) Dysploidy and polyploidy trigger strong variation of chromosome numbers in the prayer-plant family (Marantaceae). Plant Systematics and Evolution 306: 1–7. 10.1007/s00606-020-01663-x [DOI] [Google Scholar]
- Whitaker TW. (1950) Polyploidy in Echinocystis. Madrono 10: 209–10. [Google Scholar]
- Xie W, Huang J, Ma X. (2019a) Localization of 45S and 5S rDNA sequences on chromosomes of 20 species of Cucurbitaceous plants. Journal of South China Agricultural University 40: 74–81. [Google Scholar]
- Xie D, Xu Y, Wang J, Liu W, Zhou Q, Luo S, Huang W, He X, Li Q, Peng Q, Yang X, Yuan J, Yu J, Wang X, Lucas WJ, Huang S, Jiang B, Zhang Z. (2019b) The wax gourd genomes offer insights into the genetic diversity and ancestral cucurbit karyotype. Nature Communications 10: 1–12. 10.1038/s41467-019-13185-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YH, Yang F, Cheng YL, Ma L, Wang J-B, Li L-J. (2007) Comparative analysis of rDNA distribution in metaphase chromosomes of Cucurbitaceae species. Yi Chuan Hereditas 29(5): 614–620. 10.1360/yc-007-0614 [DOI] [PubMed] [Google Scholar]
- Yadava KS, Singh AK, Arya HC. (1984) Cytogenetic investigation in Cucumis L. I. Meiotic analysis in twenty four Cucumis species. Cytologia 49: 1–9. 10.1508/cytologia.49.1 [DOI] [Google Scholar]
- Yagi K, Siedlecka E, Pawełkowicz M, Wojcieszek M, Przybecki Z, Tagashira N, Hoshi Y, Malepszy S, Pląder W. (2014) Karyotype analysis and chromosomal distribution of repetitive DNA sequences of Cucumismetuliferus using fluorescence in situ hybridization. Cytogenetic and Genome Research 144: 237–242. 10.1159/000369183 [DOI] [PubMed] [Google Scholar]
- Yagi K, Pawełkowicz M, Osipowski P, Siedlecka E, Przybecki Z, Tagashira N, Hoshi Y, Malepszy S, Pląder W. (2015) Molecular cytogenetic analysis of Cucumis wild species distributed in southern Africa: physical mapping of 5S and 45S rDNA with DAPI. Cytogenetic and Genome Research 146: 80–87. 10.1159/000433572 [DOI] [PubMed] [Google Scholar]
- Yang L, Koo D-H, Li Y, Zhang X, Luan F, Havey MJ, Jiang J, Weng Y. (2012) Chromosome rearrangements during domestication of cucumber as revealed by high-density genetic mapping and draft genome assembly. The Plant Journal 71: 895–906. 10.1111/j.1365-313X.2012.05017.x [DOI] [PubMed] [Google Scholar]
- Zhang L, Cao B, Bai C. (2013) New reports of nuclear DNA content for 66 traditional Chinese medicinal plant taxa in China. Caryologia 66: 375–383. 10.1080/00087114.2013.859443 [DOI] [Google Scholar]
- Zhao X, Lu J, Zhang Z, Hu J, Huang S, Jin WW. (2011) Comparison of the distribution of the repetitive DNA sequences in three variants of Cucumissativus reveals their phylogenetic relationships. Journal of Genetics and Genomics 38: 39–45. 10.1016/j.jcg.2010.12.005 [DOI] [PubMed] [Google Scholar]
- Zhang C, Kikuchi S, Koba T. (2012) Karyotype comparison of Indian and Japanese cucumber cultivars by fluorescence in situ hybridization probed with tandem repeat sequences. Chromosome Science 15: 17–21. [Google Scholar]
- Zhang Y, Chen J, Yi H, Feng J, Wu M. (2005) Staining and slide-preparing technique of mitotic chromosomes and its use in karyotype determination of Cucumismelo L. Acta Botanica Boreali-Occidentalia Sinica 25: 1735–1739. [Google Scholar]
- Zhang Y, Cheng C, Li J, Yang S, Wang Y, Li Z, Chen J, Lou Q. (2015) Chromosomal structures and repetitive sequences divergence in Cucumis species revealed by comparative cytogenetic mapping. BMC Genomics 16: 1–3. 10.1186/s12864-015-1877-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Wu S, Bai Y, Sun H, Jiao C, Guo S, Zhao K, Blanca J, Zhang Z, Huang S, Xu Y, Weng Y, Mazourek M, Reddy UK, Ando K, McCreight JD, Schaffer AA, Burger J, Tadmor Y, Katzir N, Tang X, Liu Y, Giovannoni JJ, Kai-Ling S, Wechter WP, Levi A, Garcia-Mas J, Grumet R, Fei Z. (2019) Cucurbit Genomics Database (CuGenDB): a central portal for comparative and functional genomics of cucurbit crops. Nucleic Acids Research 47(D1): D1128–D1136. 10.1093/nar/gky944 [DOI] [PMC free article] [PubMed]
- Zhang Z-T, Yang S-Q, Li Z-A, Zhang Y-X, Wang Y-Z, Cheng C-Y, J Li, Chen J-F, Lou Q-F. (2016) Comparative chromosomal localization of 45S and 5S rDNAs and implications for genome evolution in Cucumis. Genome 59: 449–457. 10.1139/gen-2015-0207 [DOI] [PubMed] [Google Scholar]






