Abstract
Species of Arthriniums. l. are usually known as endophytes, pathogens or saprobes occurring on various hosts and substrates and are characterised by globose to subglobose, sometimes irregular, dark brown and smooth-walled or finely verruculose conidia, always with a truncate basal scar. Currently, Arthriniums. l. contains two phylogenetically distinct clades, namely, Apiospora and Arthriniums. s. However, Arthriniumtrachycarpi and Ar.urticae have still not been properly classified. With new isolates from diseased leaves of Lithocarpusglaber collected in China, we propose the new Arthrinium-like genus Neoarthrinium in Amphisphaeriales. Based on the morphology and phylogeny of multiple loci, the new genus is established with the type species, N.lithocarpicola and three new combinations, N.moseri (syn. Wardomycesmoseri), N.trachycarpi (syn. Ar.trachycarpi) and N.urticae (syn. Ar.urticae) are added to this genus.
Keywords: Apiospora , Arthrinium , Neoarthrinium , phylogeny, taxonomy
Introduction
Apiosporaceae, including Arthrinium-like taxa, was proposed to accommodate genera with apiosporous hyaline ascospores and a basauxic, Arthrinium-like conidiogenesis (Hyde et al. 1998). In a recent outline of Sordariomycetes, Hyde et al. (2020) accepted five genera viz.Appendicospora, Arthrinium, Dictyoarthrinium, Endocalyx and Nigrospora in family Apiosporaceae. Soon thereafter, Dictyoarthrinium was transferred to Didymosphaeriaceae, based on a multigene phylogeny (Samarakoon et al. 2020). Subsequently, Pintos and Alvarado (2021) separated Apiospora from Arthrinium, based on the study of the type species of both genera and on multigene phylogeny. Recently, Konta et al. (2021) transferred Endocalyx toCainiaceae, based on morphological and phylogenetic evidence and Samarakoon et al. (2022) described the new family Appendicosporaceae forAppendicospora. Therefore, Apiosporaceae currently contains three genera, viz. Apiospora, Arthrinium and Nigrospora.
Until the study of Pintos and Alvarado (2021), the genera Apiospora and Arthrinium were considered synonymous, the first being used for the sexual morph and the second for the asexual morph in dual nomenclature (Réblová et al. 2016). Following the abandonment of dual nomenclature, the older name Arthrinium was recommended for use in unitary nomenclature (Réblová et al. 2016). The genus Arthrinium was proposed by Kunze and Schmidt (1817) and validated by Fries (1832) with Ar.caricicola as the generic type. Apiospora, the type genus of Apiosporaceae, was typified with Ap.montagnei, a new name for Sphaeriaapiospora (Saccardo 1875). However, the phylogenetic identity of Ap.montagnei has been confused because multiple names in Arthrinium have similar sexual morphs that have been referred to as Apiosporamontagnei (Hudson et al. 1976; Pintos et al. 2019; Pintos and Alvarado 2021). New collections from the original region and hosts (Arundo, Piptatherum) of Ap.montagnei have been isolated in pure culture and sequenced (Crous and Groenewald 2013; Pintos et al. 2019). Five species, previously placed in Arthrinium, are classified in Apiospora. Two of these phylogenetically distinct species, Ap.marii and Ap.phragmitis, are morphologically similar to Ap.montagnei (Pintos and Alvarado 2021), but due to a lack of sequence data from the type, it cannot be determined which of these two species should become a synonym of Ap.montagnei. Irrespective of these taxonomic uncertainties in species concept, recent multigene phylogenies revealed that Arthrinium and Apiospora represent two well-supported, distinct lineages close to Nigrospora in Apiosporaceae (Pintos and Alvarado 2021; Samarakoon et al. 2022). However, two Arthrinium species resembling Apiospora in conidial morphology, viz. Ar.trachycarpi and Ar.urticae, were not considered in these studies.
Arthrinium-like species are globally distributed, inhabiting various substrates, mainly associated with plant tissues as endophytes, pathogens and saprobes (Cooke 1954; Minter 1985; Larrondo and Calvo 1992; Senanayake et al. 2020; Feng et al. 2021; Jiang and Tian 2021; Tian et al. 2021). Some species are important plant pathogens; for example, Ap.arundinis causes bamboo brown culm streak, chestnut leaf spot and barley kernel blight (Martínez-Cano et al. 1992; Chen et al. 2014; Jiang et al. 2021), while Ap.sacchari causes damping-off of durum wheat (Mavragani et al. 2007). Another species, Ar.phaeospermum, can cause dermatomycosis in humans (Zhao et al. 1990).
In the present study, new Arthrinium-like isolates were collected and morphologically examined and their phylogenetic affiliation was determined by analyses of a combined matrix of ITS, LSU, tef1 and tub2 sequences. The aim of this study was to determine the phylogenetic placement of Ar.trachycarpi, Ar.urticae and our new isolates within Amphisphaeriales, which resulted in the identification of a new phylogenetic lineage with isolates belonging to neither Arthrinium nor Apiospora. As a result, a new genus is established for these isolates.
Materials and methods
Isolation and morphology
Diseased leaves of Lithocarpusglaber were observed and collected in Guangdong Province of China (39 m elevation; 23°8'52"N, 113°27'18"E), packed in paper bags and transferred to the laboratory for pure culture isolation. The samples were first surface-sterilised for 1 min in 75% ethanol, 3 min in 1.25% sodium hypochlorite and 1 min in 75% ethanol, rinsed for 2 min in distilled water and blotted on dry sterile filter paper. Then, the diseased areas of the leaves were cut into 0.5 × 0.5 cm pieces using an aseptic razor blade, transferred on to the surface of potato dextrose agar plates (PDA; 200 g potatoes, 20 g dextrose, 20 g agar per litre) and incubated at 25 °C to obtain pure cultures. The cultures were deposited in the China Forestry Culture Collection Center (CFCC; http://cfcc.caf.ac.cn/) and the specimen was deposited in the Herbarium of the Chinese Academy of Forestry (CAF; http://museum.caf.ac.cn/).
The morphology of the isolates was studied, based on sporulating axenic cultures grown on PDA in the dark at 25 °C. The conidiomata were observed and photographed under a dissecting microscope (M205 C, Leica, Wetzlar, Germany). The conidiogenous cells and conidia were immersed in tap water and then the microscopic photographs were captured with an Axio Imager 2 microscope (Zeiss, Oberkochen, Germany), equipped with an Axiocam 506 colour camera using differential interference contrast (DIC) illumination. For measurements, 50 conidiogenous cells and conidia were randomly selected. Culture characteristics were recorded from PDA after 10 d of incubation at 25 °C in the dark.
DNA extraction, PCR amplification and phylogenetic analyses
Genomic DNA was extracted from colonies grown on cellophane-covered PDA using a cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle 1990). DNA was checked by electrophoresis in a 1% agarose gel and the quality and quantity were measured using a NanoDrop 2000 (Thermo Scientific, Waltham, MA, USA). The following primer pairs were used for amplification of the gene regions sequenced in the present study: ITS1/ITS4 for the ITS1-5.8S-ITS2 nrDNA region (ITS) (White et al. 1990); LR0R/LR5 for the 28S nrDNA region (LSU) (Vilgalys and Hester 1990); EF1-728F/EF2 for the translation elongation factor 1-α (tef1) gene (O’Donnell and Cigelnik 1997; Carbone and Kohn 1999); Bt2a/Bt2b for the beta-tubulin (tub2) gene (Glass and Donaldson 1995). The PCR conditions were set as follows: an initial denaturation step of 5 min at 94 °C, followed by 35 cycles of 30 s at 94 °C, 50 s at 52 °C (ITS and LSU) or 54 °C (tef1 and tub2) and 1 min at 72 °C and a final elongation step of 7 min at 72 °C. The PCR products were assayed via electrophoresis in 2% agarose gels. DNA sequencing was performed using an ABI PRISM 3730XL DNA Analyser with a BigDye Terminator Kit v.3.1 (Invitrogen, USA) at the Shanghai Invitrogen Biological Technology Company Limited (Beijing, China).
The quality of the chromatograms obtained was checked and the nucleotide sequences were assembled using SeqMan v.7.1.0, the DNASTAR lasergene core suite software (DNASTAR Inc, Madison, WI, USA). Reference sequences were retrieved from the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov), based on related publications (Crous and Groenewald 2013; Wang et al. 2018; Liu et al. 2019; Pintos and Alvarado 2021; Samarakoon et al. 2022). Sequences were aligned using MAFFT v. 6 (Katoh and Toh 2010) and corrected manually using MEGA 7.0.21 (Kumar et al. 2016).
The phylogenetic analyses of the combined loci were performed using Maximum Likelihood (ML) and Bayesian Inference (BI) methods. The ML was implemented on the CIPRES Science Gateway portal (https://www.phylo.org) using RAxML-HPC BlackBox 8.2.10 (Stamatakis 2014), employing a GTRGAMMA substitution model with 1000 bootstrap replicates. The Bayesian posterior probabilities (BPP) were determined by Markov Chain Monte Carlo (MCMC) sampling in MrBayes v. 3.2.6 (Ronquist et al. 2012). The six simultaneous Markov chains were run for 1 M generations, starting from random trees and sampling trees every 100th generation and 25% of aging samples were discarded, running until the average standard deviation of the split frequencies dropped below 0.01. The phylogram was visualised in FigTree v.1.3.1 (http://tree.bio.ed.ac.uk/software) and edited in Adobe Illustrator CS5 (Adobe Systems Inc., USA). The newly-generated nucleotide sequences were deposited in GenBank (Table 1).
Table 1.
Isolates and GenBank accession numbers used in the phylogenetic analyses.
| Species | Strain | Host | Origin | GenBank accession numbers | |||
|---|---|---|---|---|---|---|---|
| ITS | LSU | tub2 | tef1 | ||||
| Allelochaetaacuta | CPC 16629 | Eucalyptusdives | Australia | MH554086 | MH554297 | MH554758 | MH554519 |
| Allelochaetaneoacuta | CBS 115131 | Eucalyptussmithii | South Africa | JN871200 | JN871209 | MH704627 | MH704602 |
| Amphisphaeriamicheliae | MFLUCC 20-0121 | Micheliaalba | China | MT756626 | MT756620 | MT774371 | NA |
| Apiosporaacutiapica | KUMCC 20-0209 | Bambusabambos | China | MT946342 | MT946338 | MT947365 | MT947359 |
| Apiosporaacutiapica | KUMCC 20-0210 | Bambusabambos | China | MT946343 | MT946339 | MT947366 | MT947360 |
| Apiosporaarundinis | CBS 114316 | Hordeumvulgare | Iran | KF144884 | KF144928 | KF144974 | KF145016 |
| Apiosporaaurea | CBS 244.83 | Air | Spain | AB220251 | KF144935 | KF144981 | KF145023 |
| Apiosporabalearica | CBS 145129 | Poaceae | Spain | MK014869 | MK014836 | MK017975 | NA |
| Apiosporabiserialis | CGMCC 3.20135 | Bamboo | China | MW481708 | MW478885 | MW522955 | MW522938 |
| Apiosporacamelliae-sinensis | LC8181 | Brassicacampestris | China | KY494761 | KY494837 | KY705229 | NA |
| Apiosporacamelliae-sinensis | CGMCC 3.18333 | Camelliasinensis | China | KY494704 | KY494780 | KY705173 | KY705103 |
| Apiosporacyclobalanopsidis | CGMCC 3.20136 | Cyclobalanopsisglauca | China | MW481713 | MW478892 | MW522962 | MW522945 |
| Apiosporadescalsii | CBS 145130 | Ampelodesmosmauritanicus | Spain | MK014870 | MK014837 | MK017976 | NA |
| Apiosporadichotomanthi | LC8175 | Dichotomanthestristaniiaecarpa | China | KY494755 | KY494831 | KY705223 | KY705151 |
| Apiosporadichotomanthi | CGMCC 3.18332 | Dichotomanthestristaniiaecarpa | China | KY494697 | KY494773 | KY705167 | KY705096 |
| Apiosporaesporlensis | CBS 145136 | Phyllostachysaurea | Spain | MK014878 | MK014845 | MK017983 | NA |
| Apiosporagelatinosa | GZAAS 20-0107 | Bamboo | China | MW481707 | MW478889 | MW522959 | MW522942 |
| Apiosporaguizhouensis | LC5318 | Air | China | KY494708 | KY494784 | KY705177 | KY705107 |
| Apiosporaguizhouensis | CGMCC 3.18334 | Air | China | KY494709 | KY494785 | KY705178 | KY705108 |
| Apiosporahydei | CBS 114990 | Bamboo | China | KF144890 | KF144936 | KF144982 | KF145024 |
| Apiosporaiberica | CBS 145137 | Arundodonax | Portugal | MK014879 | MK014846 | MK017984 | NA |
| Apiosporaintestini | CBS 135835 | Gut of a grasshopper | India | KR011352 | MH877577 | KR011350 | NA |
| Apiosporaitalica | CBS 145138 | Arundodonax | Italy | MK014880 | MK014847 | MK017985 | NA |
| Apiosporajiangxiensis | CGMCC 3.18381 | Maesa sp. | China | KY494693 | KY494769 | KY705163 | KY705092 |
| Apiosporakogelbergensis | CBS 113332 | Cannomoisvirgata | South Africa | KF144891 | KF144937 | KF144983 | KF145025 |
| Apiosporakogelbergensis | CBS 113333 | Restionaceae | South Africa | KF144892 | KF144938 | KF144984 | KF145026 |
| Apiosporamalaysiana | CBS 102053 | Macarangahullettii | Malaysia | KF144896 | KF144942 | KF144988 | KF145030 |
| Apiosporamarii | CBS 497.90 | Air | Spain | AB220252 | KF144947 | KF144993 | KF145035 |
| Apiosporaneobambusae | CGMCC 3.18335 | Bamboo | China | KY494718 | KY494794 | KY705186 | KY806204 |
| Apiosporaneobambusae | LC7107 | Bamboo | China | KY494719 | KY494795 | KY705187 | KY705117 |
| Apiosporaobovata | CGMCC 3.18331 | Lithocarpus sp. | China | KY494696 | KY494772 | KY705166 | KY705095 |
| Apiosporaobovata | LC8177 | Lithocarpus sp. | China | KY494757 | KY494833 | KY705225 | KY705153 |
| Apiosporaovata | CBS 115042 | Arundinariahindsii | China | KF144903 | KF144950 | KF144995 | KF145037 |
| Apiosporaphragmitis | CBS 135458 | Phragmitesaustralis | Italy | KF144909 | KF144956 | KF145001 | KF145043 |
| Apiosporaphyllostachydis | MFLUCC 18-1101 | Phyllostachysheteroclada | China | MK351842 | MH368077 | MK291949 | MK340918 |
| Apiosporapseudoparenchymatica | CGMCC 3.18336 | Bamboo | China | KY494743 | KY494819 | KY705211 | KY705139 |
| Apiosporapseudospegazzinii | CBS 102052 | Macarangahullettii | Malaysia | KF144911 | KF144958 | KF145002 | KF145045 |
| Apiosporapterosperma | CBS 134000 | Machaerinasinclairii | Australia | KF144913 | KF144960 | KF145004 | KF145046 |
| Apiosporasaccharicola | CBS 191.73 | Air | Netherlands | KF144920 | KF144966 | KF145009 | KF145051 |
| Apiosporaseptata | CGMCC 3.20134 | Bamboo | China | MW481711 | MW478890 | MW522960 | MW522943 |
| Apiosporaserenensis | IMI 326869 | NA | Spain | AB220250 | AB220344 | AB220297 | NA |
| Apiosporasubrosea | LC7291 | Bamboo | China | KY494751 | KY494827 | KY705219 | KY705147 |
| Apiosporasubrosea | CGMCC 3.18337 | Bamboo | China | KY494752 | KY494828 | KY705220 | KY705148 |
| Apiosporaxenocordella | CBS 595.66 | Soil | Austria | KF144926 | KF144971 | KF145013 | KF145055 |
| Arthriniumcaricicola | CBS 145127 | Carexericetorum | Germany | MK014871 | MK014838 | MK017977 | NA |
| Arthriniumcrenatum | CBS 146353 | Grass | France | MW208931 | MW208861 | MW221923 | MW221917 |
| Arthriniumcurvatum | CBS 145131 | Carex sp. | Germany | MK014872 | MK014839 | MK017978 | NA |
| Arthriniumjaponicum | IFO 30500 | Carexdespalata | Japan | AB220262 | AB220356 | AB220309 | NA |
| Arthriniumjaponicum | IFO 31098 | Carexdespalata | Japan | AB220264 | AB220358 | AB220311 | NA |
| Arthriniumluzulae | AP7619-3 | Luzulasylvatica | Spain | MW208937 | MW208863 | MW221925 | MW221919 |
| Arthriniummorthieri | GZU 345043 | Carexdigitata | Austria | MW208938 | MW208864 | MW221926 | MW221920 |
| Arthriniumpuccinioides | CBS 549.86 | Lepidospermagladiatum | Germany | AB220253 | AB220347 | AB220300 | NA |
| Arthriniumsphaerospermum | CBS 146355 | Poaceae | Norway | MW208943 | MW208865 | NA | NA |
| Arthriniumsporophleum | CBS 145154 | Juncus sp. | Spain | MK014898 | MK014865 | MK018001 | NA |
| Bartaliniabella | CBS 125525 | Maytenusabbottii | South Africa | GU291796 | MH554214 | MH554663 | MH554421 |
| Bartaliniapini | CBS 143891 | Pinuspatula | Uganda | MH554125 | MH554330 | MH554797 | MH554559 |
| Beltraniapseudorhombica | CBS 138003 | Pinustabulaeformis | China | MH554124 | KJ869215 | NA | MH554558 |
| Beltraniarhombica | CBS 123.58 | Sand near mangrove swamp | Mozambique | MH553990 | MH554209 | MH704631 | MH704606 |
| Beltraniopsisneolitseae | CPC 22168 | Neolitseaaustraliensis | Australia | KJ869126 | KJ869183 | NA | NA |
| Broomellavitalbae | HPC 1154 | NA | NA | MH554173 | MH554367 | MH554846 | MH554608 |
| Castanediellacagnizarii | CBS 542.96 | Leaf litter | Cuba | MH862597 | MH874222 | NA | NA |
| Ciliochorellaphanericola | MFLUCC 12-0310 | Dead leaves | Thailand | KF827444 | KF827445 | KF827478 | KF827477 |
| Clypeophysalosporalatitans | CBS 141463 | Eucalyptus sp. | Portugal | NR_153929 | NG_058958 | NA | NA |
| Clypeosphaeriamamillana | CBS 140735 | Cornusalba | France | KT949897 | MH554225 | MH704637 | MH704610 |
| Cylindriumelongatum | CBS 115974 | Fagus sp. | The Netherlands | KM231853 | KM231733 | KM232123 | KM231989 |
| Diplocerashypericinum | CBS 109058 | Hypericum sp. | New Zealand | MH553955 | MH554178 | MH554614 | MH554373 |
| Disaetaarbuti | CBS 143903 | Acaciapycnantha | Australia | MH554148 | MH554346 | MH554821 | MH554583 |
| Discosiaartocreas | CBS 124848 | Fagussylvatica | Germany | MH553994 | MH554213 | MH554662 | MH554420 |
| Discosiabrasiliensis | MFLUCC 12-0429 | Dead leaf | Thailand | KF827432 | KF827436 | KF827469 | KF827465 |
| Distononappendiculatabanksiae | CBS 131308 | Banksiamarginata | Australia | JQ044422 | JQ044442 | MH554670 | MH554428 |
| Distononappendiculatacasuarinae | CBS 143884 | Casuarina sp. | Australia | MH554093 | MH554303 | MH554766 | MH554527 |
| Diversimediisporahumicola | CBS 302.86 | Soil | USA | MH554028 | MH554247 | MH554705 | MH554463 |
| Heterotruncatellaacacigena | CBS 143880 | Acaciapedina | Australia | MH554084 | MH554295 | MH554756 | MH554517 |
| Heterotruncatellaaspera | CBS 144140 | Acaciaglaucoptera | Australia | MH554156 | MH554352 | MH554829 | MH554591 |
| Hyalotiellaspartii | MFLUCC 13-0397 | Spartiumjunceum | Italy | KP757756 | KP757752 | NA | NA |
| Hyalotiellatransvalensis | CBS 303.65 | Leaf litter and topsoil of Acaciakarroo community | South Africa | MH554029 | MH554248 | MH554706 | MH554464 |
| Hymenopleellaaustroafricana | CBS 143886 | Gleditsiatriacanthos | South Africa | MH554115 | MH554320 | MH554788 | MH554549 |
| Hymenopleella hippophaëicola | CBS 113687 | Hippophaë rhamnoides | Sweden | MH553969 | MH554188 | MH554628 | MH554387 |
| Immersidiscosiaeucalypti | NBRC 104195 | Quercusmyrsinifolia | Japan | AB594790 | AB593722 | NA | NA |
| Lepteutypafuckelii | CBS 140409 | Tiliacordata | Belgium | NR_154123 | KT949902 | MH554677 | MH554435 |
| Lepteutypasambuci | CBS 131707 | Sambucusnigra | UK | NR_154124 | MH554219 | MH704632 | MH704612 |
| Monochaetiamonochaeta | CBS 115004 | Quercusrobur | Netherlands | AY853243 | MH554198 | MH554639 | MH554398 |
| Monochaetiaquercus | CBS 144034 | Quercuseduardi | Mexico | MH554171 | MH554365 | MH554844 | MH554606 |
| Moriniaacaciae | CBS 137994 | Acaciamelanoxylon | France | MH554002 | MH554221 | MH554673 | MH554431 |
| Moriniacrini | CBS 143888 | Crinumbulbispermum | South Africa | MH554118 | MH554323 | MH554791 | MH554552 |
| Neoarthriniumlithocarpicola | CFCC 54456 | Lithocarpusglaber | China | ON427580 | ON427582 | ON456914 | NA |
| Neoarthriniumlithocarpicola | CFCC 55883 | Lithocarpusglaber | China | ON427581 | ON427583 | ON456915 | NA |
| Noarthriniummoseri | CBS 164.80 | Dead petiole | Colombia | LN850995 | LN851049 | LN851154 | NA |
| Neoarthriniumtrachycarpi | CFCC 53038 | Trachycarpusfortunei | China | MK301098 | NA | MK303394 | MK303396 |
| Neoarthriniumtrachycarpi | CFCC 53039 | Trachycarpusfortunei | China | MK301099 | NA | MK303395 | MK303397 |
| Neoarthriniumurticae | IMI 326344 | Leaf litter | India | AB220245 | AB220339 | NA | NA |
| Neopestalotiopsiscubana | CBS 600.96 | Leaf litter | Cuba | KM199347 | KM116253 | KM199438 | KM199521 |
| Neophysalosporaeucalypti | CBS 138864 | Corymbiahenryi | Mozambique | KP004462 | MH878627 | NA | NA |
| Nigrosporaaurantiaca | CGMCC 3.18130 | Nelumbo sp. | China | KX986064 | KX986098 | KY019465 | KY019295 |
| Nigrosporacamelliae-sinensis | CGMCC 3.18125 | Camelliasinensis | China | KX985986 | KX986103 | KY019460 | KY019293 |
| Nigrosporachinensis | CGMCC 3.18127 | Machilusbreviflora | China | KX986023 | KX986107 | KY019462 | KY019422 |
| Nigrosporagorlenkoana | CBS 480.73 | Vitisvinifera | Kazakhstan | KX986048 | KX986109 | KY019456 | KY019420 |
| Nigrosporaguilinensis | CGMCC 3.18124 | Camelliasinensis | China | KX985983 | KX986113 | KY019459 | KY019292 |
| Nigrosporahainanensis | CGMCC 3.18129 | Musaparadisiaca | China | KX986091 | KX986112 | KY019464 | KY019415 |
| Nigrosporalacticolonia | CGMCC 3.18123 | Camelliasinensis | China | KX985978 | KX986105 | KY019458 | KY019291 |
| Nigrosporamusae | CBS 319.34 | Musa sp. | Australia | MH855545 | KX986110 | KY019455 | KY019419 |
| Nigrosporaoryzae | LC2693 | Neolitsea sp. | China | KX985944 | KX986101 | KY019471 | KY019299 |
| Nigrosporaosmanthi | CGMCC 3.18126 | Osmanthus sp. | China | KX986010 | KX986106 | KY019461 | KY019421 |
| Nigrosporapyriformis | CGMCC 3.18122 | Citrussinensis | China | KX985940 | KX986100 | KY019457 | KY019290 |
| Nigrosporarubi | LC2698 | Rubus sp. | China | KX985948 | KX986102 | KY019475 | KY019302 |
| Nigrosporasphaerica | LC7298 | Nelumbo sp. | China | KX985937 | KX986097 | KY019606 | KY019401 |
| Nigrosporavesicularis | CGMCC 3.18128 | Musaparadisiaca | China | KX986088 | KX986099 | KY019463 | KY019294 |
| Nonappendiculataquercina | CBS 116061 | Quercussuber | Italy | MH553982 | MH554199 | MH554641 | MH554400 |
| Parabartalinialateralis | CBS 399.71 | Acaciakarroo | South Africa | MH554043 | MH554256 | MH554719 | MH554478 |
| Parapleurotheciopsisinaequiseptata | MUCL 41089 | Rotten leaf | Brazil | EU040235 | EU040235 | NA | NA |
| Parapleurotheciopsiscaespitosa | CBS 519.93 | Syzygiumcordatum | South Africa | MH862437 | NG_066263 | NA | NA |
| Pestalotiopsisadusta | CBS 263.33 | Rhododendronponticum | Netherlands | KM199316 | KM116198 | KM199414 | KM199489 |
| Pestalotiopsisaustralasiae | CBS 114126 | Knightia sp. | New Zealand | KM199297 | KM116218 | KM199409 | KM199499 |
| Phlogicylindriumeucalypti | CBS 120080 | Eucalyptusglobulus | Australia | NR_132813 | DQ923534 | MH704633 | MH704607 |
| Phlogicylindriumeucalyptorum | CBS 120221 | Eucalyptusglobus | Australia | EU040223 | MH554204 | MH704635 | MH704608 |
| Pseudopestalotiopsisampullacea | LC6618 | Camelliasinensis | China | KX895025 | KX895039 | KX895358 | KX895244 |
| Pseudopestalotiopsiscamelliae-sinensis | LC3009 | Camelliasinensis | China | KX894935 | KX895050 | KX895267 | KX895152 |
| Pseudosarcostromaosyridicola | CBS 103.76 | Osyrisalba | France | MH553954 | MH554177 | MH554613 | MH554372 |
| Pseudosporidesmiumknawiae | CBS 123529 | NA | NA | MH863299 | MH874823 | NA | NA |
| Robillardaafricana | CBS 122.75 | NA | South Africa | KR873253 | KR873281 | MH554656 | MH554414 |
| Robillardaterrae | CBS 587.71 | Soil | India | KJ710484 | KJ710459 | MH554734 | MH554493 |
| Sarcostromaafricanum | CBS 143879 | Pelargoniumcucullatum | South Africa | MH554078 | MH554289 | MH554752 | MH554513 |
| Sarcostromaaustraliense | CBS 144160 | Daviesialatifolia | Australia | MH554138 | MH554340 | MH554811 | MH554573 |
| Seimatosporiumgermanicum | CBS 437.87 | NA | Germany | MH554047 | MH554259 | MH554723 | MH554482 |
| Seimatosporiumluteosporum | CBS 142599 | Vitisvinifera | USA | KY706284 | KY706309 | KY706259 | KY706334 |
| Seiridiumcancrinum | CBS 226.55 | Cupressusmacrocarpa | Kenya | LT853089 | MH554241 | LT853236 | LT853186 |
| Seiridiumcupressi | CBS 224.55 | Cupressusmacrocarpa | Kenya | LT853083 | MH554240 | LT853230 | LT853180 |
| Sporocadusbiseptatus | CBS 110324 | NA | NA | MH553956 | MH554179 | MH554615 | MH554374 |
| Sporocaduscornicola | CBS 143889 | Cornussanguinea | Germany | MH554121 | MH554326 | MH554794 | MH554555 |
| Sporocadustrimorphus | CBS 114203 | Rosacanina | Sweden | MH553977 | MH554196 | MH554636 | MH554395 |
| Strickeriakochii | CBS 140411 | Robiniapseudoacacia | Austria | NR_154423 | KT949918 | MH554679 | MH554437 |
| Subramaniomycesfusisaprophyticus | CBS 418.95 | Leaf litter | Cuba | EU040241 | EU040241 | NA | NA |
| Synnemapestaloidesjuniperi | CBS 477.77 | Juniperusphoenicea | France | MH554053 | MH554266 | MH554729 | MH554488 |
| Truncatellaangustata | CBS 113.11 | Piceaabies | Germany | MH553966 | MH554185 | MH554625 | MH554384 |
| Xenoseimatosporiumquercinum | CBS 129171 | Rhododendron sp. | Latvia | MH553997 | MH554216 | MH554666 | MH554424 |
| Xyladictyochaetalusitanica | CBS 143502 | Eucalyptus sp. | Australia | MH107926 | MH107972 | MH108053 | MH108033 |
Note: NA, not applicable. Strains in this study are marked in bold.
Results
Phylogenetic analyses
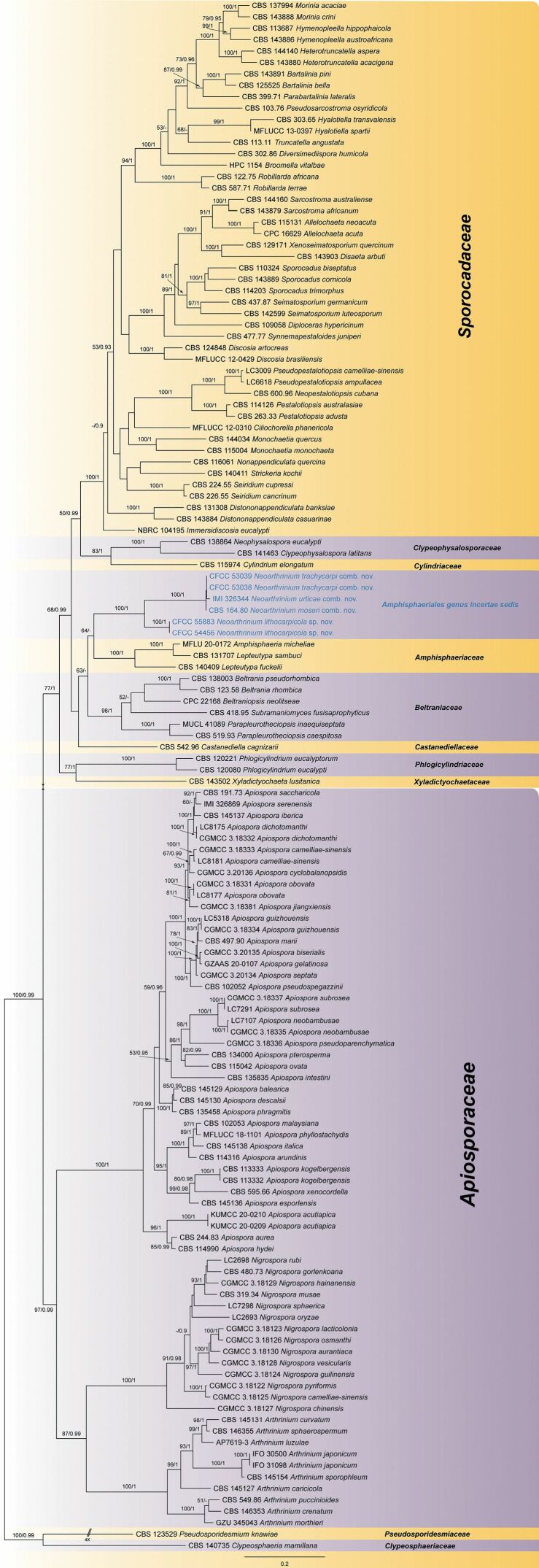
The combined sequence dataset (ITS, LSU, tef1 and tub2) was analysed to infer the phylogenetic placement of our new isolates within Amphisphaeriales. The dataset consisted of 136 sequences, including two outgroup taxa, Clypeosphaeriamamillana (CBS 140735) and Pseudosporidesmiumknawiae (CBS 123529). A total of 3526 characters, including gaps (793 for ITS, 859 for LSU, 762 for tef1 and 1112 for tub2), were included in the phylogenetic analysis. Of these characters, 1543 were constant, 284 were variable, but parsimony-uninformative and 1699 were parsimony-informative. The best ML tree (lnL = - 72640.48) revealed by RAxML is shown in Fig. 1. The topologies resulting from ML and BI analyses of the concatenated dataset were congruent (Fig. 1). Isolates CFCC 54456 and CFCC 55883 from the present study, together with CFCC 53038, CFCC 53039, CBS 164.80 and IMI 326344, formed a clade distinct from Apiosporaceae and the other families in Amphisphaeriales. Hence, a new genus named Neoarthrinium is proposed herein for this clade. Arthriniumtrachycarpi, Ar.urticae and Wardomycesmoseri are transferred to Neoarthrinium. In addition, the two new isolates (CFCC 54456 and CFCC 55883) that form a sister clade to N.moseri, N.trachycarpi and N.urticae are described here as the new species N.lithocarpicola.
Figure 1.
Phylogram of Amphisphaeriales resulting from a Maximum Likelihood analysis, based on a combined matrix of ITS, LSU, tef1 and tub2. Numbers above the branches indicate ML bootstraps (left, ML BS ≥ 50%) and Bayesian Posterior Probabilities (right, BPP ≥ 0.90). The tree is rooted with Clypeosphaeriamamillana (CBS 140735) and Pseudosporidesmiumknawiae (CBS 123529). New species and combinations proposed in the present study are marked in blue.
Taxonomy
. Neoarthrinium
Ning Jiang gen. nov.
0F22D7F7-D920-516B-90DB-F1A07FFDFA3C
843845
Etymology.
Named after its morphological similarity to Arthrinium.
Type species.
Neoarthriniumlithocarpicola Ning Jiang
Description.
Hyphae formed on PDA hyaline, branched, septate. Asexual morph: Conidiophores cylindrical, septate, verrucose, flexuous, sometimes reduced to conidiogenous cells. Conidiogenous cells erect, blastic, aggregated in clusters on hyphae, hyaline to pale brown, smooth, doliiform, subglobose to lageniform, branched. Conidia brown to dark brown, smooth to finely roughened, subglobose, ellipsoid to lenticular, with a longitudinal germ slit, occasionally elongated to ellipsoidal. Sexual morph: Undetermined.
. Neoarthrinium lithocarpicola
Ning Jiang sp. nov.
080D8973-BDAF-5A96-AB80-82D4041D2664
843846
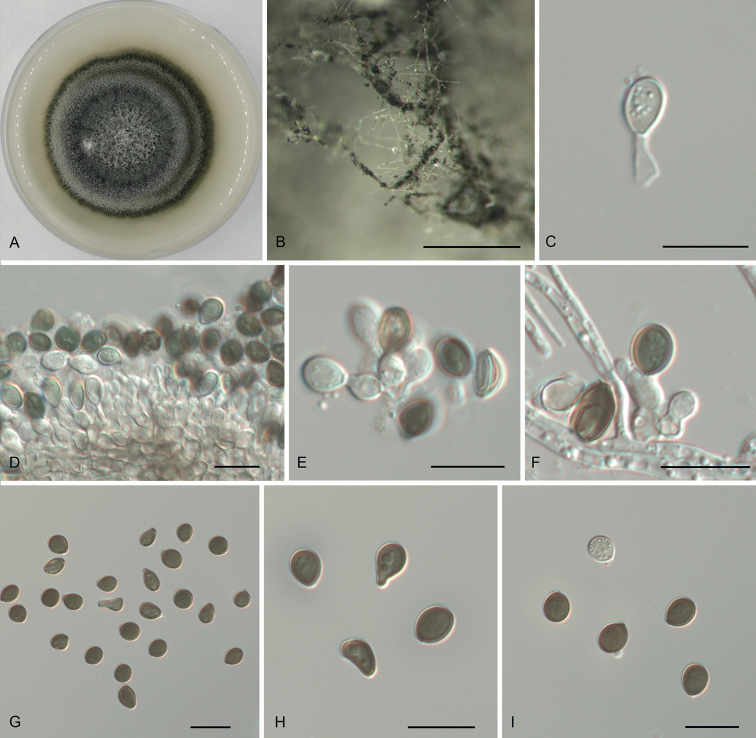
Figure 2.
NeoarthriniumlithocarpicolaA colony on PDAB conidiomata formed in culture C–F conidiogenous cells giving rise to conidia G–I conidia. Scale bars: 500 μm (B), 10 μm (C–I).
Etymology.
Named for its host genus “Lithocarpus” and “-cola” = inhabiting.
Description.
Hyphae 1.5–4.5 μm diam., hyaline, branched, septate. Asexual morph: Conidiophores cylindrical, septate, verrucose, flexuous, sometimes reduced to conidiogenous cells. Conidiogenous cells erect, blastic, aggregated in clusters on hyphae, hyaline to pale brown, smooth, globose to subglobose, branched, (4–)5.5–8 × 2.5–3.5(–4) μm, mean ± SD = 6.6 ± 1.3 × 3.1 ± 0.4 μm, n = 50. Conidia brown to dark brown, smooth to finely roughened, subglobose to lenticular, with a longitudinal germ slit, occasionally elongated to ellipsoidal, (5–)6–8(–8.5) × (4.5–)5–5.5(–6) μm, mean ± SD = 7 ± 0.8 × 5.3 ± 0.5 μm, L/W = 1.1–1.8, n = 50. Sexual morph: Undetermined.
Culture characters.
Colonies on PDA flat, spreading, with flocculent aerial mycelium forming concentric rings, edge entire, mouse grey to greyish-green, reaching 60 mm diam. after 10 d at 25 °C, forming abundant conidiomata.
Specimens examined.
China. Guangdong Province, Guangzhou City, on leaf spots of Lithocarpusglaber (Thunb.) Nakai, Shang Sun (holotype CAF800050 = JNH0046; ex-type living culture: CFCC 54456; other living culture: CFCC 55883).
Notes.
Two isolates of Neoarthriniumlithocarpicola from Lithocarpusglaber (Thunb.) Nakai formed a well-supported monophyletic clade, distinct from N.moseri, N.trachycarpi and N.urticae (Fig. 1). Morphologically, N.lithocarpicola is distinguished from N.moseri in smaller conidia (5–8.5 × 4.5–6 µm in N.lithocarpicola vs. 10–14 × 3–4.5 µm in N.moseri; Gams 1995). Neoarthriniumlithocarpicola is different from N.urticae by lacking thick blackish septa in conidiophores (Ellis 1965). Neoarthriniumlithocarpicola is similar to N.trachycarpi in the size of its conidiogenous cells and conidia, but it can be distinguished by its globose to subglobose conidiogenous cells (Yan et al. 2019).
. Neoarthrinium moseri
(W. Gams) Voglmayr comb. nov.
F199C77A-6306-51CB-A79A-84705D6FABDA
844772
Basionym.
Wardomycesmoseri W. Gams, Beih. Sydowia 10: 67 (1995)
Notes.
Based on a placement within Xylariales in phylogenetic analyses, Sandoval-Denis et al. (2016) excluded this species from the genus (Microascales); however, they did not suggest an alternative generic classification. The blastic hyaline, smooth, lageniform conidiogenous cells aggregated in clusters and the subglobose to ellipsoid dark brown conidia with a longitudinal germ slit (Gams 1995) fully matched the genus Neoarthrinum. The ITS, LSU and tub2 sequences of the ex-holotype strain of N.moseri (CBS 164.80) are almost identical to those of N.trachycarpi, indicating that they may be synonymous. Both species were isolated from petioles of palms: N.moseri from Mauritiaminor Burret in Colombia and N.trachycarpi from Trachycarpusfortunei (Hook.) H.Wendl. in China. However, the two species were reported to differ in conidial size (10–14 × 3–4.5 µm in N.moseri vs. 6.1–8.5 × 4.2–5.8 μm in N.trachycarpi; Gams 1995; Yan et al. 2019) and for the time being, we therefore kept them separate.
. Neoarthrinium trachycarpi
(C.M. Tian & H. Yan) Ning Jiang comb. nov.
FDA9EAE0-58E6-5223-B458-E47E1D6B83C2
843847
Basionym.
Arthriniumtrachycarpi C.M. Tian & H. Yan [as ‘trachycarpum’], Phytotaxa 400(3): 208 (2019)
. Neoarthrinium urticae
(M.B. Ellis) Ning Jiang comb. nov.
AC635989-1929-5909-8FE9-29C2E571041F
843848
Basionym.
Arthriniumurticae M.B. Ellis, Mycol. Pap. 103: 16 (1965)
Notes.
The possibility that Apiosporellaurticae (Rehm) Höhn. is the sexual morph of Arthriniumurticae is raised by the fact that both share the same host (Urtica) and are classified as members of the Apiosporaceae (Index Fungorum, accessed 4 July 2022). This evidence would have far reaching nomenclatural consequences not only for species, but also for generic classification, as Apiosporella (Höhnel 1909) may then qualify for an older genus name to be used for Neoarthrinium. However, according to L. Holm, the holotype specimen of its basionym, Apiosporaurticae (S-F12119), represents a very different fungus, Didymellaeupyrena (Didymellaceae, Pleosporales, Dothideomycetes; https://herbarium.nrm.se/specimens/F12119, accessed 4 July 2022). The status of the genus Apiosporella is still unclear because Höhnel (1909) did not choose a type from the six different species included in the genus. However, none of the original species is a close relative of Apiosporaceae or Neoarthrinium; therefore, Apiosporella should be excluded from Apiosporaceae.
No sequence data are available for isolates from the type host Urticadioica L. (Urticaceae). The single culture sequenced (IMI 326344) was isolated from unidentified leaf litter collected in India. Additional molecular studies on verified isolates from Urtica collected in Europe are necessary to reveal whether IMI 326344 represents true N.urticae. However, N.urticae appears to be very rare and we are unaware of any additional collections with the exception of the type.
Discussion
Arthrinium and related genera are important fungal taxa whose concepts and classification have undergone many changes and additions (e.g. Cooke 1954; Samuels et al. 1981; Larrondo and Calvo 1990; Hyde et al. 1998; Jaklitsch and Voglmayr 2012; Crous and Groenewald 2013; Singh et al. 2013; Sharma et al. 2014; Dai et al. 2016, 2017; Hyde et al. 2016; Jiang et al. 2018, 2020; Wang et al. 2018; Pintos et al. 2019; Pintos and Alvarado 2021). In recent years, substantial changes in classification were implemented in the course of unitary nomenclature. A large number of newly-discovered species have been described as a result of extensive sampling of new isolates, based on multigene phylogenies (e.g. Crous and Groenewald 2013; Wang et al. 2018; Pintos and Alvarado 2021). Currently, Arthrinium-like asexual morphs are shared by three distinct lineages within Amphisphaeriales, viz. Apiospora, Arthriniums. s. and Neoarthrinium as shown in Fig. 1. Arthriniums. s. is the sister genus to Nigrospora, which morphologically differs from Apiospora, Arthrinium and Neoarthrinium in conidial ontogeny (Wang et al. 2017). The phylogram shown in Fig. 1 is consistent with that shown in Tian et al. (2021) in placing Apiospora, Arthrinium and Nigrospora within a clade that is distinct from the new genus Neoarthrinium, although Apiospora and Arthrinium share conidial morphology similar to that of Neoarthrinium.
Morphologically, Apiospora, Arthrinium and Neoarthrinium are similar in having basauxic conidiogenesis. Conidia of Apiospora and Neoarthrinium are generally more or less rounded in face view and lenticular in side view, while those of Arthrinium are variously shaped, viz. globose, angular, polygonal, curved, fusiform or navicular (Yan et al. 2019; Pintos and Alvarado 2021). However, the conidiophores of several Arthrinium and Neoarthrinium species have thick blackish septa, which are rarely observed in Apiospora (Ellis 1965; Wang et al. 2018; Pintos and Alvarado 2021). Hence, these three genera are difficult to distinguish by only asexual morphology.
Regarding their hosts, there are some tendencies in host preferences, while Arthrinium species are predominantly found in Cyperaceae and Juncaceae (Pintos and Alvarado 2021) and species of Apiospora primarily occur on Poaceae (but also on many other hosts; Wang et al. 2018). Four Neoarthrinium species were discovered on four hosts from three distantly-related host families (i.e. N.lithocarpicola from Lithocarpusglaber (Thunb.) Nakai, Fagaceae; N.moseri from Mauritiaminor Burret, Arecaceae; N.trachycarpi from Trachycarpusfortune (Hook.) H.Wendl., Arecaceae; and N.urticae from Urticadioica L., Urticaceae; Ellis 1965; Yan et al. 2019). Hence, host association is not a fully reliable feature to distinguish Apiospora, Arthrinium and Neoarthrinium.
Compared to species, generic delimitation is much more subjective. However, there is a broad agreement that genera, along with all taxonomic classification units at all ranks, should be monophyletic. As morphology is frequently insufficient for phylogenetic classification, molecular evidence is regarded as significant data or even an essential characteristic in the classification and identification of fungal taxa. In the present study, Neoarthrinium is proposed as a new genus for a group of species phylogenetically distinct from Apiospora, Arthrinium and Nigrospora to maintain monophyletic Arthrinium-like genera. Using morphological and phylogenetic data, however, we need more samples to improve our understanding of Arthrinium-like taxa and genera in the Amphisphaeriales.
Supplementary Material
Acknowledgements
This research was funded by the National Microbial Resource Center of the Ministry of Science and Technology of the People’s Republic of China (NMRC-2021-7). The three anonymous reviewers are also acknowledged for their useful comments.
Citation
Jiang N, Voglmayr H, Ma C-Y, Xue H, Piao C-G, Li Y (2022) A new Arthrinium-like genus of Amphisphaeriales in China. MycoKeys 92: 27–43. https://doi.org/10.3897/mycokeys.92.86521
Funding Statement
National Microbial Resource Center of the Ministry of Science and Technology of the People’s Republic of China (NMRC-2021-7).
Contributor Information
Ning Jiang, Email: n.jiang@caf.ac.cn.
Yong Li, Email: lylx@caf.ac.cn.
References
- Carbone I, Kohn LM. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 3(3): 553–556. 10.1080/00275514.1999.12061051 [DOI] [Google Scholar]
- Chen K, Wu XQ, Huang MX, Han YY. (2014) First report of brown culm streak of Phyllostachyspraecox caused by Arthriniumarundinis in Nanjing, China. Plant Disease 98(9): 1274. 10.1094/PDIS-02-14-0165-PDN [DOI] [PubMed] [Google Scholar]
- Cooke WB. (1954) The genus Arthrinium. Mycologia 46(6): 815–822. 10.1080/00275514.1954.12024418 [DOI]
- Crous PW, Groenewald JZ. (2013) A phylogenetic re-evaluation of Arthrinium. IMA Fungus 4(1): 133–154. 10.5598/imafungus.2013.04.01.13 [DOI] [PMC free article] [PubMed]
- Dai DQ, Jiang HB, Tang LZ, Bhat DJ. (2016) Two new species of Arthrinium (Apiosporaceae,Xylariales) associated with bamboo from Yunnan, China. Mycosphere: Journal of Fungal Biology 7(9): 1332–1345. 10.5943/mycosphere/7/9/7 [DOI] [Google Scholar]
- Dai DQ, Phookamsak R, Wijayawardene NN, Li WJ, Bhat DJ, Xu JC, Taylor JE, Hyde KD, Chukeatirote E. (2017) Bambusicolous fungi. Fungal Diversity 82(1): 1–105. 10.1007/s13225-016-0367-8 [DOI] [Google Scholar]
- Doyle JJ, Doyle JL. (1990) Isolation of plant DNA from fresh tissue. Focus (San Francisco, Calif. ) 12: 13–15. [Google Scholar]
- Ellis MB. (1965) Dematiaceous Hyphomycetes. VI. Mycological Papers 103: 1–46. [Google Scholar]
- Feng Y, Liu JK, Lin CG, Chen YY, Xiang MM, Liu ZY. (2021) Additions to the genus Arthrinium (Apiosporaceae) from bamboos in China. Frontiers in Microbiology 12: 661281. 10.3389/fmicb.2021.661281 [DOI] [PMC free article] [PubMed]
- Fries EM. (1832) Systema Mycologicum vol. 3(2). Sumptibus Ernesti Mauritii, Greifswald.
- Gams W. (1995) An unusual species of Wardomyces (Hyphomycetes). Beihefte zur Sydowia 10: 67–72. [Google Scholar]
- Glass NL, Donaldson GC. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61(4): 1323–1330. 10.1128/aem.61.4.1323-1330.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhnel FXR. (1909) Fragmente zur Mykologie (VIII. Mitteilung, Nr. 354 bis 406). Sitzungsberichte der Kaiserlichen Akademie der Wissenschaften in Wien Mathematisch-Naturwissenschaftliche Klasse, Abt. 1(118): 1157–1246. [Google Scholar]
- Hudson HJ, McKenzie EHC, Tommerup IC. (1976) Conidial states of Apiospora Sacc. Transactions of the British Mycological Society 66(2): 359–362. 10.1016/S0007-1536(76)80075-7 [DOI] [Google Scholar]
- Hyde KD, Fröhlich J, Taylor JE. (1998) Fungi from palms. XXXVI. Reflections on unitunicate ascomycetes with apiospores. Sydowia 50: 21–80. [Google Scholar]
- Hyde KD, Hongsanan S, Jeewon R, Bhat DJ, McKenzie EHC, Jones EBG, Phookamsak R, Ariyawansa HA, Boonmee S, Zhao Q, Abdel-Aziz FA, Abdel-Wahab MA, Banmai S, Chomnunti P, Cui B-K, Daranagama DA, Das K, Dayarathne MC, de Silva NI, Dissanayake AJ, Doilom M, Ekanayaka AH, Gibertoni TB, Góes-Neto A, Huang S-K, Jayasiri SC, Jayawardena RS, Konta S, Lee HB, Li W-J, Lin C-G, Liu J-K, Lu Y-Z, Luo Z-L, Manawasinghe IS, Manimohan P, Mapook A, Niskanen T, Norphanphoun C, Papizadeh M, Perera RH, Phukhamsakda C, Richter C, de A. Santiago ALCM, Drechsler-Santos ER, Senanayake IC, Tanaka K, Tennakoon TMDS, Thambugala KM, Tian Q, Tibpromma S, Thongbai B, Vizzini A, Wanasinghe DN, Wijayawardene NN, Wu H-X, Yang J, Zeng X-Y, Zhang H, Zhang J-F, Bulgakov TS, Camporesi E, Bahkali AH, Amoozegar MA, Araujo-Neta LS, Ammirati JF, Baghela A, Bhatt RP, Bojantchev D, Buyck B, da Silva GA, de Lima CLF, de Oliveira RJV, de Souza CAF, Dai Y-C, Dima B, Duong TT, Ercole E, Mafalda-Freire F, Ghosh A, Hashimoto A, Kamolhan S, Kang J-C, Karunarathna SC, Kirk PM, Kytövuori I, Lantieri A, Liimatainen K, Liu Z-Y, Liu X-Z, Lücking R, Medardi G, Mortimer PE, Nguyen TTT, Promputtha I, Raj KNA, Reck MA, Lumyong S, Shahzadeh-Fazeli SA, Stadler M, Soudi MR, Su H-Y, Takahashi T, Tangthirasunun N, Uniyal R, Wang Y, Wen T_C, Xu J-C, Zhang Z-K, Zhao Y-C, Zhou J-L, Zhu L. (2016) Fungal diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 80: 1–270. 10.1007/s13225-016-0373-x [DOI] [Google Scholar]
- Hyde KD, Norphanphoun C, Maharachchikumbura SSN. (2020) Refined families of Sordariomycetes. Mycosphere: Journal of Fungal Biology 11(1): 305–1059. 10.5943/mycosphere/11/1/7 [DOI]
- Jaklitsch WM, Voglmayr H. (2012) Phylogenetic relationships of five genera of Xylariales and Rosasphaeria gen. nov. (Hypocreales). Fungal Diversity 52(1): 75–98. 10.1007/s13225-011-0104-2 [DOI] [Google Scholar]
- Jiang N, Tian CM. (2021) The holomorph of Arthriniumsetariae sp. nov. (Apiosporaceae,Xylariales) from China. Phytotaxa 483(2): 149–159. 10.11646/phytotaxa.483.2.7 [DOI] [Google Scholar]
- Jiang N, Li J, Tian CM. (2018) Arthrinium species associated with bamboo and reed plants in China. Fungal Systematics and Evolution 2: 1–9. 10.3114/fuse.2018.02.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Liang YM, Tian CM. (2020) A novel bambusicolous fungus from China, Arthriniumchinense (Xylariales). Sydowia 72: 77–83. 10.12905/0380.sydowia72-2020-0077 [DOI] [Google Scholar]
- Jiang N, Fan XL, Tian CM. (2021) Identification and characterization of leaf-inhabiting fungi from Castanea plantations in China. Journal of Fungi 7(1): 64. 10.3390/jof7010064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. (2010) Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26(15): 1899–1900. 10.1093/bioinformatics/btq224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konta S, Hyde KD, Eungwanichayapant PD, Karunarathna SC, Samarakoon MC, Xu J, Dauner LAP, Aluthwattha ST, Lumyong S, Tibpromma S. (2021) Multigene phylogeny reveals Haploanthostomellaelaeidis gen. et sp. nov. and familial replacement of Endocalyx (Xylariales,Sordariomycetes, Ascomycota). Life 11(6): 486. 10.3390/life11060486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33(7): 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G, Schmidt JC. (1817) Mykologische 1: 1–109.
- Larrondo JV, Calvo MA. (1990) Two new species of Arthrinium from Spain. Mycologia 82(3): 396–398. 10.1080/00275514.1990.12025899 [DOI] [Google Scholar]
- Larrondo JV, Calvo MA. (1992) New contributions to the study of the genus Arthrinium. Mycologia 84(3): 475–478. 10.1080/00275514.1992.12026164 [DOI]
- Liu F, Bonthond G, Groenewald JZ, Cai L, Crous PW. (2019) Sporocadaceae, a family of coelomycetous fungi with appendage-bearing conidia. Studies in Mycology 92(1): 287–415. 10.1016/j.simyco.2018.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Cano C, Grey WE, Sands DC. (1992) First report of Arthriniumarundinis causing kernel blight on barley. Plant Disease 76(10): 1077. 10.1094/PD-76-1077B [DOI] [Google Scholar]
- Mavragani DC, Abdellatif L, McConkey B, Hamel C, Vujanovic V. (2007) First report of damping-off of durum wheat caused by Arthriniumsacchari in the semi-arid Saskatchewan fields. Plant Disease 91(4): 469. 10.1094/PDIS-91-4-0469A [DOI] [PubMed] [Google Scholar]
- Minter DW. (1985) A re-appraisal of the relationships between Arthrinium and other hyphomycetes. Proceedings of the Indiana Academy of Sciences 94(2–3): 281–308. 10.1007/BF03053145 [DOI] [Google Scholar]
- O’Donnell K, Cigelnik E. (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7(1): 103–116. 10.1006/mpev.1996.0376 [DOI] [PubMed] [Google Scholar]
- Pintos A, Alvarado P. (2021) Phylogenetic delimitation of Apiospora and Arthrinium. Fungal Systematics and Evolution 7(1): 197–221. 10.3114/fuse.2021.07.10 [DOI] [PMC free article] [PubMed]
- Pintos A, Alvarado P, Planas J, Jarling R. (2019) Six new species of Arthrinium from Europe and notes about A.caricicola and other species found in Carex spp. hosts. MycoKeys 49: 15–48. 10.3897/mycokeys.49.32115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Réblová M, Miller AN, Rossman AY, Seifert KA, Crous PW, Hawksworth DL, Abdel-Wahab MA, Cannon PF, Daranagama DA, De Beer ZW, Huang S-K, Hyde KD, Jayawardena R, Jaklitsch W, Jones EBG, Ju Y-M, Judith C, Maharachchikumbura SSN, Pang K-L, Petrini LE, Raja HA, Romero AI, Shearer C, Senanayake IC, Voglmayr H, Weir BS, Wijayawarden NN. (2016) Recommendations for competing sexual-asexually typified generic names in Sordariomycetes (except Diaporthales, Hypocreales, and Magnaporthales). IMA Fungus 7(1): 131–153. 10.5598/imafungus.2016.07.01.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61(3): 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccardo PA. (1875) Fungi veneti novi vel critici. Series II. Nuovo Giornale Botanico Italiano 7: 299–329. [Google Scholar]
- Samarakoon BC, Wanasinghe DN, Samarakoon MC, Phookamsak R, McKenzie EHC, Chomnunti P, Hyde KD, Lumyong S, Karunarathna SC. (2020) Multi-gene phylogenetic evidence suggests Dictyoarthrinium belongs in Didymosphaeriaceae (Pleosporales, Dothideomycetes) and Dictyoarthriniummusae sp. nov. on Musa from Thailand. MycoKeys 71: 101–118. 10.3897/mycokeys.71.55493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarakoon MC, Hyde KD, Maharachchikumbura SSN, Stadler M, Gareth Jones EB, Promputtha I, Suwannarach N, Camporesi E, Bulgakov TS, Liu J-K. (2022) Taxonomy, phylogeny, molecular dating and ancestral state reconstruction of Xylariomycetidae (Sordariomycetes. Fungal Diversity 112(1): 1–88. 10.1007/s13225-021-00495-5 [DOI] [Google Scholar]
- Samuels GJ, McKenzie EHC, Buchanan DE. (1981) ascomycetes of New Zealand. 3. Two new species of Apiospora and their Arthrinium anamorphs on bamboo. New Zealand Journal of Botany 19(2): 137–149. 10.1080/0028825X.1981.10425113 [DOI] [Google Scholar]
- Sandoval-Denis M, Guarro J, Cano-Lira JF, Sutton DA, Wiederhold NP, de Hoog GS, Abbott SP, Decock C, Sigler L, Gené J. (2016) Phylogeny and taxonomic revision of Microascaceae with emphasis on synnematous fungi. Studies in Mycology 83(1): 193–233. 10.1016/j.simyco.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake IC, Bhat JD, Cheewangkoon R, Xie N. (2020) Bambusicolous Arthrinium species in Guangdong Province, China. Frontiers in Microbiology 11: 602773. 10.3389/fmicb.2020.602773 [DOI] [PMC free article] [PubMed]
- Sharma R, Kulkarni G, Sonawane MS, Shouche YS. (2014) A new endophytic species of Arthrinium (Apiosporaceae) from Jatrophapodagrica. Mycoscience 55(2): 118–123. 10.1016/j.myc.2013.06.004 [DOI]
- Singh SM, Yadav LS, Singh PN, Hepat R, Sharma R, Singh SK. (2013) Arthriniumrasikravindrii sp. nov. from Svalbard, Norway. Mycotaxon 122(1): 449–460. 10.5248/122.449 [DOI] [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9): 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Karunarathna SC, Mapook A, Promputtha I, Xu J, Bao D, Tibpromma S. (2021) One new species and two new host records of Apiospora from bamboo and maize in northern Thailand with thirteen new combinations. Life 11(10): 1071. 10.3390/life11101071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172(8): 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Liu F, Crous PW, Cai L. (2017) Phylogenetic reassessment of Nigrospora: Ubiquitous endophytes, plant and human pathogens. Persoonia 39(1): 118–142. 10.3767/persoonia.2017.39.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Tan X-M, Liu F, Cai L. (2018) Eight new Arthrinium species from China. MycoKeys 34: 1–24. 10.3897/mycokeys.34.24221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications 18: 315–322. [Google Scholar]
- Yan H, Jiang N, Liang LY, Yang Q, Tian CM. (2019) Arthriniumtrachycarpum sp. nov. from Trachycarpusfortuneii in China. Phytotaxa 400(3): 203–210. 10.11646/phytotaxa.400.3.7 [DOI] [Google Scholar]
- Zhao YM, Deng CR, Chen X. (1990) Arthriniumphaeospermum causing dermatomycosis, a new record of China. Acta Mycologica Sinica 9: 232–235. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.