Abstract
Two new species of Phylloporia, P.crystallina and P.sumacoensis, are described based on 28S ribosomal RNA phylogeny, morphology, host, and geographic distribution. Phylloporiacrystallina is characterized by pileate, perennial basidiomata with a duplex context, small pores 9–10 per mm, a monomitic hyphal system, absence of cystidia and cystidioles, presence of large rhomboid crystals in tube trama, broadly ellipsoid to subglobose basidiospores measuring 2.8–3 × 2–2.3 μm, and growth on angiosperm stump. Phylloporiasumacoensis is characterized by pileate, perennial basidiomata with a duplex context, very small pores 10–12 per mm, a monomitic hyphal system, hyphae at dissepiment edges bearing fine crystals, presence of cystidioles, broadly ellipsoid to subglobose basidiospores measuring 3–3.7 × 2.1–2.8 μm, and growth on living liana.
Keywords: Hymenochaetaceae, n28S, phylogeny, taxonomy
Introduction
Phylloporia Murrill (Hymenochaetaceae, Hymenochaetales) was established with P.parasitica Murrill as the type (Murrill 1904). The genus is characterized by annual or perennial, effused-reflexed, pileate or stipitate, soft corky to hard corky basidiomata, tomentose to velutinate pileal surface, a context mostly duplex with a black line between upper tomentum and lower contextual layer, a monomitic hyphal system in most species, generative hyphae with simple septa, absence of setal elements (with the exception of Phylloporiamori Wu et al.), and subglobose, ellipsoid or cylindric, hyaline to yellowish, fairly thick-walled basidiospores which are usually collapsed when mature and < 6 µm in the greatest dimension. Phylloporia species mostly grow parasitically on living angiosperm trees, causing a white rot. Phylogenetically, Phylloporia is related to Flaviporellus Murrill and Fulvifomes Murrill, but Flaviporellus and Fulvifomes have mostly homogeneous contexts (Wu et al. 2022).
Seventy-one species are currently recognized in Phylloporia, among them 17 and 37 species from the Neotropics and China, respectively (Wu et al. 2022). Because more tree species occur in Neotropics than in China (Anonymous 1997) and species diversity of Phylloporia is related to tree species diversity (Wu et al. 2019), it seems probable that many unknown species of Phylloporia exist in the Neotropics. During investigations of the neotropical polypores, specimens morphologically corresponding to Phylloporia were collected from Ecuador. Based on morphological, ecological, and phylogenetic evidence, we hereby propose two new species of Phylloporia.
Materials and methods
Studied specimens are deposited in herbaria of the Institute of Microbiology, Beijing Forestry University (BJFC) and the National Museum Prague of Czech Republic (PRM). The sections were prepared in 5% potassium hydroxide (KOH), Melzer’s reagent (IKI), and Cotton Blue (CB). The following abbreviations are used: KOH = 5% potassium hydroxide, IKI = Melzer’s reagent, IKI– = neither amyloid nor dextrinoid, CB = Cotton Blue, CB– = acyanophilous, CB (+) = cyanophilous after 12 hours stained with Cotton Blue, L = mean spore length (arithmetic average of spores), W = mean spore width (arithmetic average of spores), Q = variation in the ratios of L/W between specimens studied, and n = number of spores measured from given number of specimens. The microscopic procedure follows Dai (2010), and the special color terms follow Petersen (1996) and Anonymous (1969). Sections were studied at magnifications up to 1000× using a Nikon Eclipse 80i microscope with phase contrast illumination. Drawings were made with the aid of a drawing tube. Microscopic features, measurements, and illustrations were made from the slide preparations stained with Cotton Blue. Microscopic measurements were made from slide preparations stained with Cotton Blue.
The extraction of total genomic DNA from frozen specimens followed Góes-Neto et al. (2005) using the protocol of CTAB 2%. The CTAB rapid plant genome extraction kit-DN14 (Aidlab Biotechnologies Co., Ltd, Beijing) was used to obtain PCR products from dried specimens, following the manufacturer’s instructions with some modifications (Chen et al. 2015, 2016). The primer pairs LR0R and LR7 (Vilgalys and Hester 1990) and LR0R and LR5 (White et al. 1990) were used for PCR amplification. The PCR procedure for 28S was as follows: initial denaturation at 94 °C for 1 min, followed by 35 cycles at 94 °C for 30 s, 50 °C for 1 min and 72 °C for 1.5 min, and a final extension of 72 °C for 10 min. The PCR products were purified and directly sequenced at Beijing Genomics Institute. All newly generated sequences were deposited at GenBank (Sayers et al. 2022) and are shown in the tree (Fig. 1).
Figure 1.
Phylogeny of Phylloporia inferred from the 28S dataset. The topology is from the ML analysis, and the support values from ML (former) and BI (latter) analyses simultaneously greater than or equal to 50% and 0.90 are indicated at the nodes, respectively. The new species are in boldface.
In addition to the newly generated sequences, 28S sequences (Fig. 1) from Zhou (2016), Ren and Wu (2017), Qin et al. (2018), and Wu et al. (2019, 2022) were downloaded from GenBank and included in the dataset for phylogenetic analysis. Inonotushispidus (Bull.) P. Karst. was selected as outgroup following Zhou (2016). The dataset was aligned using MAFFT v. 7.0 with the Q-INS-i strategy with default parameters (Katoh et al. 2019), and then edited as necessary in BioEdit v. 7.0.5.3 (Hall 1999). Sequence alignments were deposited at TreeBase (submission ID: 29564). Maximum Likelihood (ML) and Bayesian Inference (BI) methods were used for the phylogenetic analysis. The GTR+I+ G model was estimated as the best-fit evolutionary model by MrModeltest v. 2.3 (Nylander 2004) using the Akaike information criterion (AIC). The ML analysis was carried out with raxmlGUI v. 1.2 (Stamatakis 2006; Silvestro and Michalak 2012), and the BI tree reconstruction was carried out with MrBayes v. 3.2.5 (Ronquist et al. 2012). Four Markov chains were run for two runs from random starting trees for 10 million generations, and trees were sampled every 100 generations. BI analysis stopped after effective sample sizes (ESSs) reached more than 200 and the potential scale reduction factors (PSRFs) were close to 1.000 for all parameters. Branches that received bootstrap support for ML (BS) and Bayesian Posterior Probability (BPP) methods greater than or equal to 75% (BS) and 0.95 (BPP) were considered as significantly supported, respectively.
Results
Phylogenetic results
Two 28S sequences were generated in this study and were deposited in GenBank. Their accession numbers are specified in the phylogenetic tree (Fig. 1). The final 28S dataset included 135 sequences and resulted in an alignment of 993 characters. The ML and BI analyses resulted in nearly identical topologies, and thus only the ML tree is presented with the BS and BPP when they were greater than or equal to 50% and 0.90, respectively (Fig. 1).
The phylogeny inferred from the 28S dataset (Fig. 1) shows that the specimen JV 2106/102 together with one specimen (MUCL 45062 from Cuba) form a distinct lineage and that the specimen JV2109/73 forms another independent Phylloporia lineage.
Taxonomy
. Phylloporia crystallina
Y.C. Dai, F. Wu, Meng Zhou & Vlasák sp. nov.
2EAC5E3B-F249-57ED-84EC-9C68089ABD5A
843482
Figure 2.
Basidiomata of Phylloporiacrystallina (holotype, JV2106/102). Scale bar: 1 cm.
Figure 3.
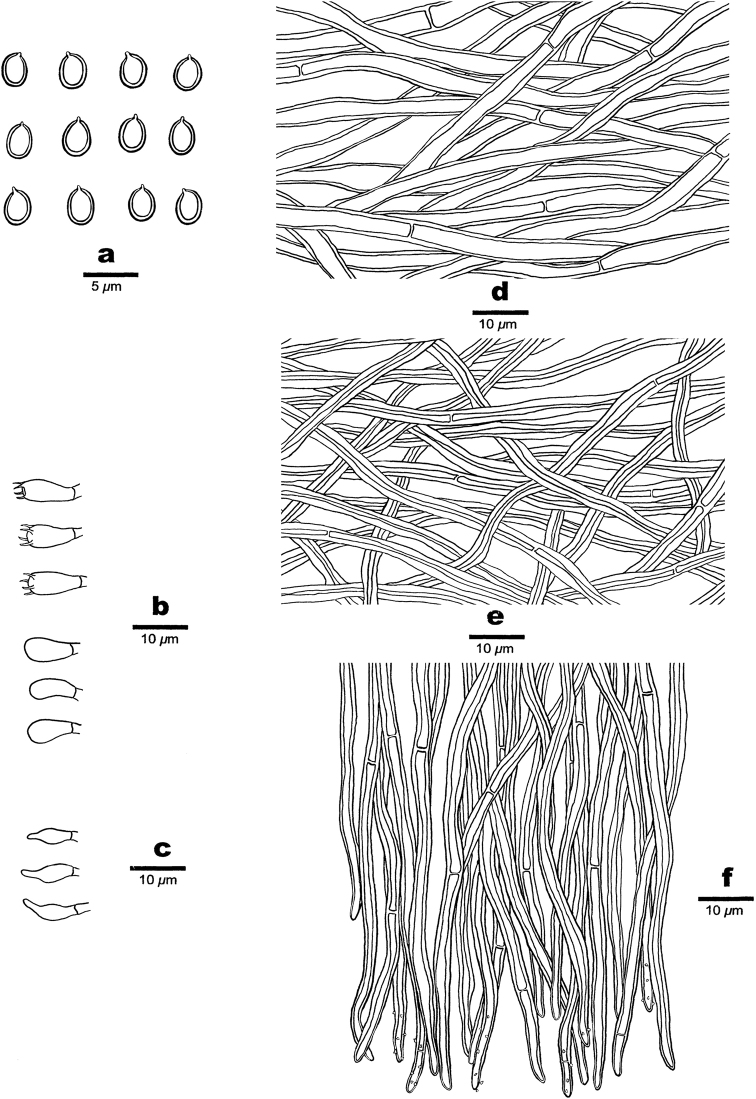
Microscopic structures of Phylloporiacrystallina (drawn from the holotype, JV2106/102). a basidiospores b basidia and basidioles c hyphae from upper tomentum d hyphae from lower compacted context e hyphae from dissepiment edge.
Type.
Ecuador, Mindo Valley, San Carlos, Cascadas; alt. 1400m; 0°4'S, 78°45'W; 20 Jun. 2021; Vlasák leg.; on angiosperm freshly dead stump in tropical cloud forest; JV2106/102 (holotype BJFC038563, isotype PRM957106). GenBank: ON129551 (ITS); ON006467 (LSU)
Etymology.
— Crystallina (Lat.): refer to the species having abundant large rhomboid crystals in tube trama.
Diagnosis.
Phylloporiacrystallina is characterized by pileate, perennial basidiomata with a thin layer of context between individual tube layers, a duplex context with a black line separating the upper tomentum and a lower compacted layer, small pores 9–10 per mm, a monomitic hyphal system, generative hyphae thin- to distinctly thick-walled with simple septa, the absence of cystidia and cystidioles, the presence of large rhomboid crystals in tube trama, broadly ellipsoid basidiospores measuring 2.8–3 × 2–2.3 μm, and growth on angiosperm stump in the Neotropics.
Basidiomata.
Perennial, effused reflexed, imbricate, broadly attached to the substrate, hard corky when fresh, woody hard when dry. Pilei applanate to semi-circular, projecting up to 2 cm and 3 cm wide. Pileal surface curry yellow to cinnamon buff when fresh, become purplish chestnut when dry, concentrically sulcate with narrow zones, densely tomentum when juvenile, become velutinate to matted with age, the tomentum up to 1 mm thick, wearing off, leaving a dense trichoderm, sometime covered by mosses; margin sharp, entire. Pore surface pinkish buff to buff yellow and glancing when fresh, become honey yellow when dry; pores round, 9–10 per mm; dissepiments thin, entire. Context umber, up to 3 mm thick, duplex, with a black line separating the upper tomentum and a lower compacted layer, the upper tomentum soft corky, the lower layer hard corky. Tubes fulvous, paler than context, up to 5 mm long, distinctly stratified, usually filled a thin context among tube layers.
Hyphal structure.
Hyphal system monomitic; generative hyphae simple septate; tissue darkening but otherwise unchanged in the shape of the hyphae in KOH.
Context. Hyphae in the lower context golden yellow, fairly thick-walled with a wide lumen, unbranched, frequently simple-septate, loosely interwoven, slightly CB+, 3–5 μm diam.; hyphae in the upper tomentum yellow, fairly thick-walled with a wide lumen, unbranched, frequently simple septate, straight, regularly arranged, 5–7 μm diam.
Tubes. Tramal hyphae hyaline to yellow, thin- to thick-walled with a narrow to medium lumen, rarely branched, frequently to occasionally simple septate, flexuous, loosely interwoven, slightly CB+, 2–3.5 μm diam.; hyphae at dissepiment edges smooth; large rhomboid crystals abundant among tube trama.
Hymenium. Cystidia and cystidioles absent; basidia barrel-shaped with four sterigmata and a simple septum at the base, 5–7 × 3.5–4 μm. Basidioles similar to basidia in shape, but slightly smaller. Basidiospores broadly ellipsoid to subglobose, yellowish, thick-walled, smooth, not collapsed, IKI–, CB–, (2.7–) 2.8–3 (–3.1) × 2–2.3 (–2.4) μm, L = 2.9 μm, W = 2.1 μm, Q = 1.38 (n = 30/1).
Notes.
Phylogenetically (Fig. 1), Phylloporiacrystallina is related to P.montana Oliveira-Filho & Gibertoni (Wu et al. 2019). However, P.montana has wider pores (3–5 per mm vs. 9–10 per mm) and larger and cylindrical basidiospores (4–5 × 2–3 μm vs. 2.8–3 × 2–2.3 μm) (Wu et al. 2019). Morphologically, P.crystallina resembles P.crataegi L.W. Zhou & Y.C. Dai by sharing perennial and pileate basidiomata with duplex context, a monomitic hyphal system, interwoven tramal hyphae, the absence of cystidia and cystidioles, and broadly ellipsoid to subglobose basidiospores (Zhou and Dai 2012). However, the latter species differs from P.crystallina by the absence of rhomboid crystals, distinctly longer basidia (8–11 μm vs. 5–7 μm), and growth on living Crataegus in temperate China (Zhou and Dai 2012). In addition, P.crystallina and P.crataegi are phylogenetically distantly related (Fig. 1). P.chrysites (Berk.) Ryvarden is a Neotropical species. It has similar basidiospores as P.crystallina, but the former is readily distinguished from the latter by its annual habit and larger pores (9–10 per mm vs. pores 6–8 per mm, Wu et al. 2022).
Trameteslilliputiana Speg. and Pyropolyporussubpectinatus Murrill were originally described from Brazil and Cuba, respectively (Spegazzini 1889; Murrill 1908), and they were treated as synonyms of Phylloporiapectinata (Klotzsch) Ryvarden (Bresadola 1912; Ryvarden 1985; Rajchenberg and Wright 1987). However, these two taxa may be different from Phylloporiapectinata because its type locality is in India (Wu et al. 2022). The type of T.lilliputiana is sterile, but its pilei are confluent and thin, and its upper surface is smooth according to its original description (Spegazzini 1889). P.subpectinatus has globose basidiospores (Murrill 1908). So, these two taxa are closer or identical to P.pectinata which has a dimitic hyphal structure and globose basidiospores (Ryvarden 2004); while P.crystallina has a monomitic hyphal system and broadly ellipsoid basidiospores.
. Phylloporia sumacoensis
Y.C. Dai, F. Wu, Meng Zhou & Vlasák sp. nov.
C58E74F9-09D6-5885-A372-835B90CFB8D1
843484
Figure 4.
A basidiomata of Phylloporiasumacoensis (holotype, JV2109/73). Scale bar: 1 cm.
Figure 5.
Microscopic structures of Phylloporiasumacoensis (drawn from the holotype, JV2109/73). a basidiospores b basidia and basidioles c cystidioles d hyphae from upper tomentum e hyphae from lower compacted context f hyphae from dissepiment edge.
Type.
Ecuador, Guamani, Wild Sumaco Lodge; alt. 1200m; 0°40'S, 77°36'W; 30. Sep. 2021; Vlasák leg.; on living liana in tropical cloud forest; JV2109/73 (holotype BJFC038576, isotype PRM957107). GenBank: ON129552 (ITS); ON006468 (LSU).
Etymology.
— Sumacoensis (Lat.): refer to the species being found close to Sumaco Vulcan, Ecuador.
Diagnosis.
Phylloporiasumacoensis is characterized by pileate, perennial basidiomata with a thin layer of context between individual tube layers, a duplex context with a black line separating the upper tomentum and a lower compacted layer, very small pores 10–12 per mm, a monomitic hyphal system, generative hyphae thin- to distinctly thick-walled with simple septa, the hyphae at dissepiment edges bearing fine crystals, presence of cystidioles, broadly ellipsoid to subglobose basidiospores as 3–3.7 × 2.1–2.8 μm, and growth on living liana at medium elevation in the Neotropical cloud forest.
Basidiomata.
Perennial, pileate, solitary, broadly attached to the substrate, corky when fresh, hard corky when dry. Pilei applanate to semi-circular, projecting up to 4 cm, 5 cm wide and 15 mm thick at base. Pileal surface fuscous to vinaceous gray when fresh, become fulvous to date brown when dry, concentrically zonate and sulcate, densely tomentose, the tomentum up to 4 mm thick; margin obtuse, entire. Pore surface brownish gray to yellowish gray and glancing when fresh, become snuff brown when dry; pores round, 10–12 per mm; dissepiments thick, entire. Context fulvous, up to 8 mm thick, duplex, with a black line separating an upper soft corky tomentum, up to 4 mm thick and the lower compacted layer, hard corky, up to 4 mm thick. Tubes fawn, darker than context, up to 7 mm long, distinctly stratified, usually with a thin layer of context between individual tube layers.
Hyphal structure.
Hyphal system monomitic; generative hyphae simple septate; tissue darkening but otherwise unchanged in the shape of the hyphae in KOH.
Context. Hyphae in the lower context golden yellow, thick-walled with a narrow to medium lumen, unbranched, occasionally simple septate, interwoven, 3–5 μm diam.; hyphae in the tomentum brownish yellow, fairly thick-walled with a wide lumen, unbranched, frequently simple septate, some collapsed, loosely interwoven, 5–7 μm diam.
Tubes. Tramal hyphae hyaline to golden yellow, thin- to thick-walled with a narrow to medium lumen, rarely branched, frequently to occasionally simple septate, parallel or subparallel along the tubes, 2–4 μm diam.; hyphae at dissepiment edges bearing fine crystals.
Hymenium. Cystidia absent, fusoid cystidioles rarely present; basidia barrel-shaped with four sterigmata and a simple septum at the base, 10–12 × 4.5–5 μm. Basidioles similar to basidia in shape, but slightly smaller. Basidiospores broadly ellipsoid to subglobose, yellowish, thick-walled, smooth, some collapsed, IKI–, CB–, (2.9–)3–3.7(–3.9) × 2.1–2.8 μm, L = 3.18 μm, W = 2.48 μm, Q = 1.28 (n = 30/1).
Notes.
Phylogenetically, Phylloporiasumacoensis is closely related to two other Neotropical species, P.spathulata (Hook.) Ryvarden sensu auctore and P.ulloae R. Valenz. et al. (Fig. 1). However, P.spathulata differs from P.sumacoensis in having stipitate basidiomata, wider pores (7–9 per mm vs. 10–12 per mm), and the absence of cystidioles (Ryvarden 2004). Phylloporiaulloae differs from P.sumacoensis in having wider pores (6–8 per mm vs. 10–12 per mm) and longer basidia (14.5–16 μm vs. 10–12 μm) (Valenzuela et al. 2011). Morphologically, P.sumacoensis is similar to P.fontanesiae L.W. Zhou & Y.C. Dai by sharing same pores size and broadly ellipsoid basidiospore (Zhou and Dai 2012), but the latter species has an annual habit, shorter basidia (6–7 × 3.5–4 µm vs. 10–12 × 4.5–5 μm), shorter basidiospores (2.5–3 μm vs. 3–3.7 μm), and growth on living Fontanesia in Asia (Zhou and Dai 2012). In addition, P.sumacoensis and P.fontanesiae are phylogenetically distantly related (Fig. 1).
Discussion
Most Phylloporia species grow parasitically on living hardwoods, and speciation in the genus seems to be driven by the process of colonizing and adapting to new hosts (Wu et al. 2019). The genus is taxonomically difficult, however, because of the similar morphology among species. Before the era of molecular phylogenetics, the diversity of Phylloporia was grossly underestimated. The genus has received considerable attention through LSU-based phylogenetic studies (Wagner and Ryvarden 2002). So far, 73 species in the genus are accepted, and most of them were described and confirmed by molecular data recently (Wu et al. 2019, 2022). In addition, the majority of the recently described species were from subtropical and tropical areas, pointing to a remarkable species richness of the genus in tropical regions. There can be little doubt that more undescribed taxa of Phylloporia are present in the Neotropics, and the more samples are collected the better our understanding of species diversity of the genus will be.
Unlike other wood-inhabiting fungal genera, very long and complex ITS sequences are present in most Phylloporia species. These are difficult to align confidently. Accordingly, most phylogenies were based on LSU sequences (Decock et al. 2015; Yombiyeni et al. 2015; Ferreira-Lopes et al. 2016; Zhou 2016; Rajchenberg et al. 2019; Wu et al. 2019, 2022). Although Phylloporia is shown to be monophyletic based on LSU sequences, the genus is composed of a non-trivial number of subclades (Fig. 1). In addition, in some cases, phylogenetic estimates of Phylloporia look strikingly different depending on what exact taxa are included in the analyses. Phylogenetic inference based on multiple genetic markers is a better solution, but most described taxa are represented by only a very limited number of sequences and different genetic markers in GenBank. Sequences from multiple genetic markers from samples of Phylloporia are much needed, from fairly conserved genetic markers. Ideally, the next version of MAFFT and other multiple sequence alignment tools will furthermore be able to handle the ITS sequences of the genus Phylloporia in a better way.
Supplementary Material
Acknowledgements
The research was financed by National Natural Science Foundation of China (Project Nos. 32161143013 and 32011540380) and the Second Tibetan Plateau Scientific Expedition and Research Program (STEP, Grant No. 2019QZKK0503), and by the institutional support of the Academy of Sciences of the Czech Republic RVO: 60077344.
Citation
Zhou M, Wu F, Dai Y-C, Vlasák J (2022) Two new species of Phylloporia (Hymenochaetales) from the Neotropics. MycoKeys 90: 71–83. https://doi.org/10.3897/mycokeys.90.84767
Funding Statement
National Natural Science Foundation of China (Project Nos. 32161143013 and 32011540380) and the Academy Sciences of the Czech Republic RVO: 60077344.
Contributor Information
Yu-Cheng Dai, Email: yuchengdai@bjfu.edu.cn.
Josef Vlasák, Email: vlasak@umbr.cas.cz.
Supplementary materials
The sequence alignments and tree
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Meng Zhou, Fang Wu, Yu-Cheng Dai, Josef Vlasák
Data type
NEX file
Explanation note
The NEX file includes Phylloporia phylogenetic sequence alignments inferred from the 28S dataset and the topology of the ML analysis.
References
- Anonymous (1969) Flora of British fungi. Colour identification chart. Her Majesty’s Stationery Office, London, 3 pp. [Google Scholar]
- Anonymous (1997) The forests in China 1. The Chinese Forest Press, Beijing, 584 pp. [Google Scholar]
- Bresadola G. (1912) Basidiomycetes Philippinenses 2. Hedwigia 53: 46–80. [Google Scholar]
- Chen JJ, Cui BK, Zhou LW, Korhonen K, Dai YC. (2015) Phylogeny, divergence time estimation, and biogeography of the genus Heterobasidion (Basidiomycota, Russulales). Fungal Diversity 71(1): 185–200. 10.1007/s13225-014-0317-2 [DOI] [Google Scholar]
- Chen JJ, Cui BK, Dai YC. (2016) Global diversity and molecular systematics of Wrightoporia s.l. (Russulales, Basidiomycota). Persoonia 37(1): 21–36. 10.3767/003158516X689666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai YC. (2010) Hymenochaetaceae (Basidiomycota) in China. Fungal Diversity 45(1): 131–343. 10.1007/s13225-010-0066-9 [DOI] [Google Scholar]
- Decock C, Yombiyeni P, Memiaghe H. (2015) Hymenochaetaceae from the Guineo-Congolian Rainforest: Phylloporiaflabelliforma sp. nov. and Phylloporiagabonensis sp. nov., two undescribed species from Gabon. Cryptogamie. Mycologie 36(4): 449–467. 10.7872/crym/v36.iss4.2015.449 [DOI] [Google Scholar]
- Ferreira-Lopes V, Robledo GL, Reck MA, Góes-Neto A, Drechsler-Santos ER. (2016) Phylloporiaspathulata sensu stricto and two new South American stipitate species of Phylloporia (Hymenochaetaceae). Phytotaxa 257(2): 133–148. 10.11646/phytotaxa.257.2.3 [DOI] [Google Scholar]
- Góes-Neto A, Loguercio-Leite C, Guerrero RT. (2005) DNA Extraction from frozen field-collected and dehydrated herbarium fungal basidiomata: Performance of SDS and CTAB-based methods. Biotemas 18: 19–32. [Google Scholar]
- Hall TA. (1999) Bioedit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. 10.1021/bk-1999-0734.ch008 [DOI] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. (2019) MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20(4): 1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrill WA. (1904) The Polyporaceae of North America 9. Bulletin of the Torrey Botanical Club 31: 593–610. 10.2307/2478612 [DOI] [Google Scholar]
- Murrill WA. (1908) Polyporaceae 2. North American Flora 9(2): 73–131. [Google Scholar]
- Nylander JAA. (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University, Uppsala.
- Petersen JH. (1996) Farvekort. The Danish Mycological Society’s colour-chart. Foreningen til Svampekundskabens Fremme, Greve, 6 pp. [Google Scholar]
- Qin WM, Wang XW, Sawahata T, Zhou LW. (2018) Phylloporialonicerae (Hymenochaetales, Basidiomycota), a new species on Lonicerajaponica from Japan and an identification key to worldwide species of Phylloporia. MycoKeys 30: 17–30. 10.3897/mycokeys.30.23235 [DOI] [PMC free article] [PubMed]
- Rajchenberg M, Wright JE. (1987) Type studies of Corticiaceae and Polyporaceae (Aphyllophorales) described by C. Spegazzini. Mycologia 79(2): 246–464. 10.1080/00275514.1987.12025704 [DOI] [Google Scholar]
- Rajchenberg M, Pildain MB, Cajas Madriaga D, de Errasti A, Riquelme C, Becerra J. (2019) New poroid Hymenochaetaceae (Basidiomycota, Hymenochaetales) from Chile. Mycological Progress 18(6): 865–877. 10.1007/s11557-019-01495-1 [DOI] [Google Scholar]
- Ren GJ, Wu F. (2017) Phylloporialespedezae sp. nov. (Hymenochaetaceae, Basidiomycota) from China. Phytotaxa 299(2): 243–251. 10.11646/phytotaxa.299.2.8 [DOI] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Avres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes3.2: Efficient Bayesian phylogenetic inference and model choice, across a large model space. Systematic Biology 61(3): 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryvarden L. (1985) Type studies in the Polyporaceae 17. Species described by W.A. Murrill. Mycotaxon 23: 169–198. [Google Scholar]
- Ryvarden L. (2004) Neotropical polypores 1. Introduction, Hymenochaetaceae and Ganodermataceae. Synopsis Fungorum 19: 1–227. [Google Scholar]
- Sayers EW, Cavanaugh M, Clark K, Pruitt KD, Schoch CL, Sherry ST, Karsch-Mizrachi I. (2022) GenBank. Nucleic Acids Research 50(D1): 161–164. 10.1093/nar/gkab1135 [DOI] [PMC free article] [PubMed]
- Silvestro D, Michalak I. (2012) raxmlGUI: A graphical front-end for RAxML. Organisms, Diversity & Evolution 12(4): 335–337. 10.1007/s13127-011-0056-0 [DOI] [Google Scholar]
- Spegazzini C. (1889) Fungi Puiggariani. Pugillus 1. Boletín de la Academia Nacional de Ciencias en Córdoba 11: 381–622. 10.5962/bhl.title.3624 [DOI] [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics (Oxford, England) 22(21): 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Valenzuela R, Raymundo T, Cifuentes J, Castillo G, Amalfi M, Decock C. (2011) Two undescribed species of Phylloporia are evidenced in Mexico based on morphological, phylogenetic, and ecological data. Mycological Progress 10: 341–349. 10.1007/s11557-010-0707-0 [DOI] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172(8): 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T, Ryvarden L. (2002) Phylogeny and taxonomy of the genus Phylloporia (Hymenochaetales). Mycological Progress 1(1): 105–106. 10.1007/s11557-006-0009-8 [DOI] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor JW. (1990) Amplification and direct sequencing of ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (Eds) PCR Protocols, a Guide to Methods and Applications.Academic Press, New York, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Wu F, Ren GJ, Wang L, Oliveira-Filho JRC, Gibertoni TB, Dai YC. (2019) An updated phylogeny and diversity of Phylloporia (Hymenochaetales): Eight new species and keys to species of the genus. Mycological Progress 18(5): 615–639. 10.1007/s11557-019-01476-4 [DOI] [Google Scholar]
- Wu F, Zhou LW, Vlasák J, Dai YC. (2022) Global diversity and systematics of Hymenochaetaceae with poroid hymenophore. Fungal Diversity 113(1): 1–192. 10.1007/s13225-021-00496-4 [DOI] [Google Scholar]
- Yombiyeni P, Balezi A, Amalfi M, Decock C. (2015) Hymenochaetaceae from the Guineo-Congolian rainforest: Three new species of Phylloporia based on morphological, DNA sequences and ecological data. Mycologia 107(5): 996–1011. 10.3852/14-298 [DOI] [PubMed] [Google Scholar]
- Zhou LW. (2016) Phylloporiaminutipora and P.radiata spp. nov. (Hymenochaetales, Basidiomycota) from China and a key to worldwide species of Phylloporia. Mycological Progress 15(6): e57. 10.1007/s11557-016-1200-1 [DOI]
- Zhou LW, Dai YC. (2012) Phylogeny and taxonomy of Phylloporia (Hymenochaetales): New species and a worldwide key to the genus. Mycologia 104(1): 211–222. 10.3852/11-093 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The sequence alignments and tree
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Meng Zhou, Fang Wu, Yu-Cheng Dai, Josef Vlasák
Data type
NEX file
Explanation note
The NEX file includes Phylloporia phylogenetic sequence alignments inferred from the 28S dataset and the topology of the ML analysis.