Abstract
In recent decades, commercial Eucalyptus plantations have expanded toward the warm and humid regions of northern and northeastern Brazil, where Calonectria leaf blight (CLB) has become the primary fungal leaf disease of this crop. CLB can be caused by different Calonectria species, and previous studies have indicated that Calonectria might have high species diversity in Brazil. During a disease survey conducted in three commercial plantations of Eucalyptus in northeastern Brazil, diseased leaves from Eucalyptus trees with typical symptoms of CLB were collected, and Calonectria fungi were isolated. Based on phylogenetic analyses of six gene regions (act, cmdA, his3, rpb2, tef1, and tub2) and morphological characteristics, two new species of Calonectria were identified. Five isolates were named as C.paragominensissp. nov. and four were named as C.imperatasp. nov. The pathogenicity to Eucalyptus of both species was confirmed by fulfilling the Koch’s postulates.
Keywords: Cylindrocladium , GCPSR, phylogenetic network analysis, phylogeny
Introduction
Calonectria species are widely distributed around the world and cause diseases in more than 335 plant species, distributed among nearly 100 plant families, including forestry, agricultural and horticultural crops (Crous 2002; Lombard et al. 2010c; Vitale et al. 2013; Lombard et al. 2016; Li et al. 2021). Most reports of Calonectria from Brazil are focused on forestry crops, such as Acacia, Eucalyptus, and Pinus trees (Alfenas et al. 2015), and mainly evaluate the epidemiology and disease control of Calonectria-associated diseases such as Calonectria leaf blight (CLB), damping-off, cutting rot and root rot in commercial plantations and nurseries of Eucalyptus (Soares et al. 2018).
Currently, 130 Calonectria species have been identified based on DNA phylogenetic analyses and morphological comparisons (Crous et al. 2018, 2019, 2021a, 2021b; Wang et al. 2019; Liu et al. 2020; Mohali and Stewart 2021; Pham et al. 2022). These species are accommodated in eleven species complexes, which are divided into two main phylogenetic groups based on their morphological features: the Prolate Group (C.brassicae, C.candelabrum, C.colhounii, C.cylindrospora, C.gracilipes, C.mexicana, C.pteridis, C.reteaudii and C.spathiphylli species complexes), and the Sphaero-Naviculate Group (C.kyotensis and C.naviculata species complexes) (Lombard et al. 2016; Liu et al. 2020).
In Brazil, a total of 35 species have been described: eleven species isolated from diseased tissues of Eucalyptus, ten species isolated from soil samples of Eucalyptus plantations, seven species isolated from different plant species, six species isolated from soil samples of tropical rainforests, and one mycoparasite species (Crous et al. 2018, 2019; Liu et al. 2020); they belong in the species complexes of C.candelabrum, C.brassicae, C.cylindrospora, C.pteridis, C.gracilipes, and C.naviculata (Crous et al. 2018, 2019; Liu et al. 2020). The results from a previous study indicated high species diversity of Calonectria in Brazil (Alfenas et al. 2015).
Brazil is one of the main producers of pulp, paper, and wood panels in the world, mainly due to the genus Eucalyptus; its hybrids are the most grown trees in the country for these purposes (IBÁ, 2021). In 2020, the total area of Eucalyptus plantations was 7.47 million hectares, with an average productivity of 36.8 m3/ha per year (IBÁ, 2021). However, in recent decades, commercial Eucalyptus plantations have expanded toward the warm and humid regions of northern and northeastern Brazil, where CLB has become the primary fungal leaf disease of this crop (Alfenas et al. 2015). CLB can be caused by different Calonectria species, is widely distributed throughout the country, and affects Eucalyptus plants most severely from six months to 2–3 years after planting (Graça et al. 2009). This disease starts from spores or microsclerotia present in soil or diseased plant debris on the ground and disseminates to lower branches of the tree canopy; lesions start at the base, apex or margins of leaves and can reach a large area of the leaf blade, resulting in leaf drop and, in some cases, severe defoliation in the basal, middle, and apical thirds of the canopy (Alfenas et al. 2009). The defoliation may decrease timber volume as a result of the reduced photosynthetic area and facilitates weed growth due to the increased entrance of light through the subcanopy, leading to competition for nutrients between Eucalyptus and understory plants (Graça et al. 2009; Alfenas et al. 2015).
CLB can be controlled by integrated cultivation and chemical methods as well as by the selection and cultivation of resistant genotypes, which is a much more effective approach (Soares et al. 2018). The demand for new strategies to control this disease requires proper identification of the pathogen species. Additionally, this information may be useful for breeding programs, leading to the development of Eucalyptus genotypes resistant to CLB. Recently, during a disease survey conducted in three commercial plantations of Eucalyptus in northeastern Brazil, diseased leaves from Eucalyptus trees with typical symptoms of CLB were collected, and Calonectria fungi were isolated. Thus, the aims of this study were to identify these isolates based on phylogenetic analyses and morphological characteristics and to confirm their pathogenicity to Eucalyptus.
Materials and methods
Sample collection and fungal isolation
In February 2020, during a disease survey conducted in three commercial plantations of Eucalyptus on six-month-old to one-year-old trees, diseased leaves with typical symptoms of CLB (small, circular or elongated pale grey to pale brown to dark brown spots, that extend throughout the leaf blade), were observed and collected for fungal isolation and species characterization. On average, 50 diseased leaves were sampled from each Eucalyptus genotype, one leaf per tree, depending on the planted areas. The sampled Eucalyptus genotypes corresponded to E.urophylla, localized in the municipalities of Cidelândia (5°09'24"S, 47°46'26"W) and Itinga do Maranhão (4°34'43"S, 47°29'48"W), in the state of Maranhão, and to the E.grandis × E.brassiana hybrid genotype, in the microregion of Paragominas (3°10'51"S, 47°18'49"W), in the state of Pará, Brazil.
Samples were stored in paper bags and transported to the Laboratory of Forest Pathology at the Universidade Federal de Lavras. From each leaf, small segments of 1 cm2 from the transition section between healthy and diseased tissue were cut and the surface was disinfected by washing with 1% sodium hypochlorite for 1 min, with 70% ethanol for 30 s and with sterilized water three times before culture on 2% malt extract agar (MEA; malt extract 20 g·L-1, agar 20 g·L-1, yeast extract 2 g·L-1, sucrose 5 g·L-1) plates at 25 °C. After 48 h of incubation, Calonectria-like mycelial plugs, 5 mm in diameter, were transferred to a fresh MEA plate and incubated at 25 °C until the fungus covered the plate completely. Induction of sporulation on MEA plates and single spore cultures was obtained following the procedures described by Alfenas et al. (2013). Each single spore culture was stored and maintained in a metabolically inactive state in dry culture and sterile water following Castellani’s method (Castellani 1939). Holotypes were deposited as herbaria in the Coleção Micológica do Herbário da Universidade de Brasília (UB). Ex-types were deposited as pure cultures in the Coleção de Culturas de Microrganismos do Departamento de Ciência dos Alimentos/UFLA (CCDCA) at Universidade Federal de Lavras (UFLA), Minas Gerais, Brazil. Ex-paratypes were deposited as pure cultures in the Laboratory of Forest Pathology (PFC) at UFLA.
DNA extraction, PCR amplification, and sequencing
Total genomic DNA was extracted from fresh mycelia of single spore cultures grown on malt extract broth (MEB; malt extract 20 g·L-1, yeast extract 2 g·L-1, sucrose 5 g·L-1) for ten days at 25 °C in the dark. The protocol described by Lee and Taylor (1990) was followed with slight modifications; by adding 1.5 M NaCl and 2% polyvinylpyrrolidone (MW: 40000) to the lysis buffer; the DNA was precipitated directly with isopropanol without the use of 3 M NaOAc, and the DNA pellet was dried at room temperature overnight. A NanoDrop 1,000 spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) was used to quantify its concentration.
Based on a previous study (Liu et al. 2020), actine (act), calmodulin (cmdA), histone H3 (his3), RNA polymerase II (rpb2), translation elongation factor 1-alpha (tef1), and β-tubulin (tub2) genes were used as DNA barcodes due to provide a stable and reliable resolution to distinguish all Calonectria species. The primers ACT-512F and ACT-783R (Carbone and Kohn 1999) were used to amplify the act gene region; CAL-228F and CAL-2Rd (Carbone and Kohn 1999; Quaedvlieg et al. 2011) for the cmdA gene region; CYLH3F and CYLH3R (Crous et al. 2004) for the his3 gene region; fRpb2-5F and fRpb2-7cR (Liu et al. 1999; Reeb et al. 2004) for the rpb2 gene region; EF1-728F (Carbone and Kohn 1999) and EF2 (O’Donnell et al. 1998) for the tef1 gene region and the primer pairs T1 (O’Donnell and Cigelnik 1997) and CYLTUB1R (Crous et al. 2004) for the tub2 gene region.
The PCRs were carried out in a 25 μL final volume containing molecular biology-grade water (Sigma–Aldrich, St. Louis, MO, USA) 1X PCR buffer (Promega, Madison, WI, USA), 2.5 mM MgCl2, 0.2 mM deoxyribonucleotide triphosphate (dNTP) mix (Promega, Madison, WI, USA), 1 U GoTaq Flexi DNA Polymerase (Promega, Madison, WI, USA), 0.2 mM each primer, and 30 ng DNA template. DNA amplifications were conducted in a thermal cycler (5 PRIME G gradient Thermal Cycler, Techne, Staffordshire, UK). The PCR conditions for the act, cmdA, his3, tef1, and tub2 gene regions were as follows: an initial denaturation step at 95 °C for 5 min; then 35 amplification cycles at [94 °C for 30 s; 52 °C for 1 min; 72 °C for 2 min], and a final extension step at 72 °C for 5 min. For the rpb2 gene region, a touchdown PCR protocol was used: an initial denaturation step at 95 °C for 5 min, then (95 °C for 30 s, 57 °C for 30 s, 72 °C for 90 s) × 10 cycles, (95 °C for 30 s, 57 °C for 45 s, 72 °C for 90 s + 5 s/cycle increase) × 30 cycles, and a final extension step at 72 °C for 10 min.
PCR products were separated by electrophoresis at 120 V for 1 h in a 1.2% agarose gel, stained with Diamond Nucleic Acid Dye (Promega, Madison, WI, USA), and visualized using an ultraviolet light transilluminator. Successful PCR products were purified and sequenced in both directions using the same primer pairs used for amplification by Macrogen Inc. (Macrogen, Seoul, Korea). Raw sequences from each gene region were edited, consensus sequences were generated using SeqAssem software ver. 07/2008 (Hepperle 2004), and the sequences generated in this study were deposited in the NCBI/GenBank database (http://www.ncbi.nlm.nih.gov).
Phylogenetic analyses
The generated sequences were aligned with other sequences of closely related Calonectria spp. obtained from GenBank (Table 1), using the online interface of MAFFT v. 7.0 (Katoh et al. 2019, http://mafft.cbrc.jp/alignment/server) with the alignment strategy FFT-NS-i (Slow; interactive refinement method). Alignments were manually corrected using MEGA7 (Kumar et al. 2016).
Table 1.
Calonectria species and GenBank accession numbers of DNA sequences used in this study.
| Species complex | Species | Isolate representing the species‡,§ | Other isolate numbers | Host/ Substrate | Country | Genbank accession numbers | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| act | | cmdA | his3 | rpb2 | tef1 | tub2 | ||||||
| Calonectriaspathiphylli species complex | C.densa | CMW 31182 | Soil | Ecuador | GQ280525 | GQ267444 | GQ267281 | N/A | GQ267352 | GQ267232 | |
| CMW 31184 | Soil | Ecuador | GQ280523 | GQ267442 | GQ267279 | N/A | GQ267350 | GQ267230 | |||
| CMW 31185 | Soil | Ecuador | GQ280524 | GQ267443 | GQ267280 | N/A | GQ267351 | GQ267231 | |||
| C.humicola | CMW 31183 | Soil | Ecuador | GQ280526 | GQ267445 | GQ267282 | N/A | GQ267353 | GQ267233 | ||
| CMW 31186 | Soil | Ecuador | GQ280527 | GQ267446 | GQ267283 | N/A | GQ267354 | GQ267234 | |||
| CMW 31187 | Soil | Ecuador | GQ280528 | GQ267447 | GQ267284 | N/A | GQ267355 | GQ267235 | |||
| C.paragominensis sp. nov. † | CCDCA 11648 | E.grandis × E.brassiana | Brazil | ON009346 | OM974325 | OM974334 | OM974343 | OM974352 | OM974361 | ||
| PFC2 | E.grandis × E.brassiana | Brazil | ON009347 | OM974326 | OM974335 | OM974344 | OM974353 | OM974362 | |||
| PFC3 | E.grandis × E.brassiana | Brazil | ON009348 | OM974327 | OM974336 | OM974345 | OM974354 | OM974363 | |||
| PFC4 | E.grandis × E.brassiana | Brazil | ON009349 | OM974328 | OM974337 | OM974346 | OM974355 | OM974364 | |||
| PFC5 | E.grandis × E.brassiana | Brazil | ON009350 | OM974329 | OM974338 | OM974347 | OM974356 | OM974365 | |||
| C.pseudospathiphylli | CBS 109165 | CPC 1623 | Soil | Ecuador | GQ280493 | GQ267412 | AF348241 | KY653435 | FJ918562 | AF348225 | |
| CPC 1641 | Soil | Ecuador | N/A | N/A | AF348233 | N/A | N/A | AF348217 | |||
| C.spathiphylli | CBS 114540 | ATCC44730, CSF11330 | Spathiphyllum sp. | USA | GQ280505 | GQ267424 | AF348230 | MT412666 | GQ267330 | AF348214 | |
| CBS 116168 | CSF 11401 | Spathiphyllum sp. | Switzerland | GQ280506 | GQ267425 | FJ918530 | MT412667 | FJ918561 | FJ918512 | ||
| Calonectriacandelabrum species complex | C.brasiliana | CBS 111484 | CSF 11249 | Soil | Brazil | MT334968 | MT335198 | MT335438 | MT412502 | MT412729 | MT412951 |
| CBS 111485 | CSF 11250 | Soil | Brazil | MT334969 | MT335199 | MT335439 | MT412503 | MT412730 | MT412952 | ||
| C.brassiana | CBS 134855 | Soil | Brazil | N/A | KM396056 | KM396139 | N/A | KM395882 | KM395969 | ||
| CBS 134856 | Soil | Brazil | N/A | KM396057 | KM396140 | N/A | KM395883 | KM395970 | |||
| C.brevistipitata | CBS 115671 | CSF 11288 | Soil | Mexico | MT334973 | MT335203 | MT335443 | MT412507 | MT412734 | MT412956 | |
| CBS 110928 | CSF 11235 | Soil | Mexico | MT334974 | MT335204 | MT335444 | MT412508 | MT412735 | MT412957 | ||
| C.candelabrum | CMW 31000 | CSF 11404 | Eucalyptus sp. | Brazil | MT334977 | MT335207 | MT335447 | MT412511 | MT412738 | MT412959 | |
| CMW 31001 | CSF 11405 | Eucalyptus sp. | Brazil | MT334978 | MT335208 | MT335448 | MT412512 | MT412739 | MT412960 | ||
| C.colombiana | CBS 115127 | Soil | Colombia | GQ280538 | GQ267455 | FJ972442 | N/A | FJ972492 | FJ972423 | ||
| CBS 115638 | Soil | Colombia | GQ280539 | GQ267456 | FJ972441 | N/A | FJ972491 | FJ972422 | |||
| C.eucalypticola | CBS 134847 | Eucalyptus sp. | Brazil | N/A | KM396051 | KM396134 | N/A | KM395877 | KM395964 | ||
| CBS 134846 | Eucalyptus sp. | Brazil | N/A | KM396050 | KM396133 | N/A | KM395876 | KM395963 | |||
| C.fragariae | CBS 133607 | Fragaria×ananassa | Brazil | N/A | KM998966 | KM998964 | N/A | KM998963 | KM998965 | ||
| LPF141.1 | Fragaria×ananassa | Brazil | N/A | KX500191 | KX500194 | N/A | KX500197 | KX500195 | |||
| C.glaebicola | CBS 134852 | Soil | Brazil | N/A | KM396053 | KM396136 | N/A | KM395879 | KM395966 | ||
| CBS 134853 | Eucalyptus sp. | Brazil | N/A | KM396054 | KM396137 | N/A | KM395880 | KM395967 | |||
| C.hemileiae | COAD 2544 | Hemileiavastatrix | Brazil | N/A | MK037392 | MK006026 | N/A | MK006027 | MK037391 | ||
| Calonectriacandelabrum species complex | C.imperata sp. nov. † | CCDCA 11649 | E.urophylla | Brazil | ON009351 | OM974330 | OM974339 | OM974348 | OM974357 | OM974366 | |
| PFC7 | E.urophylla | Brazil | ON009352 | OM974331 | OM974340 | OM974349 | OM974358 | OM974367 | |||
| PFC8 | E.urophylla | Brazil | ON009353 | OM974332 | OM974341 | OM974350 | OM974359 | OM974368 | |||
| PFC9 | E.urophylla | Brazil | ON009354 | OM974333 | OM974342 | OM974351 | OM974360 | OM974369 | |||
| C.matogrossensis | GFP 006 | E.urophylla | Brazil | N/A | MH837653 | MH837648 | N/A | MH837659 | MH837664 | ||
| GFP 018 | E.urophylla | Brazil | N/A | MH837657 | MH837652 | N/A | MH837663 | MH837668 | |||
| C.metrosideri | CBS 133603 | Metrosiderospolymorpha | Brazil | N/A | KC294304 | KC294307 | N/A | KC294310 | KC294313 | ||
| CBS 133604 | CSF 11309 | Metrosiderospolymorpha | Brazil | MT335056 | MT335288 | MT335528 | MT412585 | MT412819 | MT413033 | ||
| C.nemoricola | CBS 134837 | Soil | Brazil | N/A | KM396066 | KM396149 | N/A | KM395892 | KM395979 | ||
| CBS 134838 | Soil | Brazil | N/A | KM396067 | KM396150 | N/A | KM395893 | KM395980 | |||
| C.pauciramosa | CBS 138824 | CSF 16461 | Soil | South Africa | MT335093 | MT335325 | MT335565 | MT412618 | MT412856 | MT413068 | |
| CMW 31474 | CSF 11422 | E.urophylla × E.grandis | China | MT335104 | MT335336 | MT335576 | MT412629 | MT412867 | MT413079 | ||
| C.piauiensis | CBS 134850 | Soil | Brazil | N/A | KM396060 | KM396143 | N/A | KM395886 | KM395973 | ||
| CBS 134851 | Soil | Brazil | N/A | KM396061 | KM396144 | N/A | KM395887 | KM395974 | |||
| C.pseudometrosideri | CBS 134845 | Soil | Brazil | N/A | KM395995 | KM396083 | N/A | KM395821 | KM395909 | ||
| CBS 134843 | Metrosiderospolymorpha | Brazil | N/A | KM395993 | KM396081 | N/A | KM395819 | KM395907 | |||
| C.pseudospathulata | CBS 134841 | Soil | Brazil | N/A | KM396070 | KM396153 | N/A | KM395896 | KM395983 | ||
| CBS 134840 | Soil | Brazil | N/A | KM396069 | KM396152 | N/A | KM395895 | KM395982 | |||
| C.putriramosa | CBS 111449 | CSF 11246 | Eucalyptus cutting | Brazil | MT335129 | MT335364 | MT335604 | MT412657 | MT412895 | MT413105 | |
| CBS 111470 | CSF 11247 | Soil | Brazil | MT335130 | MT335365 | MT335605 | MT412658 | MT412896 | MT413106 | ||
| C.silvicola | CBS 135237 | LPF081 | Soil | Brazil | N/A | KM396065 | KM396148 | N/A | KM395891 | KM395978 | |
| CBS 134836 | Soil | Brazil | N/A | KM396062 | KM396145 | N/A | KM395888 | KM395975 | |||
| C.spathulata | CMW 16744 | CSF 11331 | E.viminalis | Brazil | MT335139 | MT335376 | MT335616 | MT412668 | MT412907 | MT413117 | |
| CBS 112513 | CSF 11259 | Eucalyptus sp. | Colombia | MT335140 | MT335377 | MT335617 | MT412669 | MT412908 | MT413118 | ||
| C.venezuelana | CBS 111052 | CSF 11238 | Soil | Venezuela | MT335155 | MT335394 | MT335634 | MT412685 | MT412925 | MT413132 | |
| Calonectriagracilipes species complex | C.gracilipes | CBS 115674 | CSF 11289 | Soil | Colombia | MT335022 | MT335252 | MT335492 | MT412554 | MT412783 | MT413001 |
| CBS 111141 | CSF11239 | Soil | Colombia | MT335023 | MT335253 | MT335493 | MT412555 | MT412784 | MT413002 | ||
† New Calonectria species reported in the present study. ‡ Ex-type isolates of the Calonectria species are marked in bold. § ATCC: American Type Culture Collection, Virginia, USA; CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; CCDCA: Coleção de Culturas de Microrganismos do Departamento de Ciência dos Alimentos/UFLA, Lavras, Brazil; CMW: Culture collection of the Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, Pretoria, South Africa; COAD: Coleção Octávio de Almeida Drumond, Universidade Federal de Viçosa, Viçosa, Brazil; CPC: Pedro Crous working collection housed at Westerdijk Fungal Biodiversity Institute; CSF: Culture Collection located at China Eucalypt Research Centre (CERC), Chinese Academy of Forestry, ZhanJiang, GuangDong Province, China; GFP: Universidade Federal de Brasilia, Brasilia, Brazil; LPF: Laboratorio de Patologia Florestal, Universidade Federal de Viçosa, Viçosa, Brazil; PFC: Laboratorio de Patologia Florestal, Universidade Federal de Lavras, Lavras, Brazil. | act: actin; cmdA: calmodulin; his3: histone H3; rpb2: the second largest subunit of RNA polymerase; tef1: translation elongation factor 1-alpha; tub2: β-tubulin. GenBank accession number obtained in this study are marked in bold.
The partition homogeneity test (PHT) described by Farris et al. (1995) was conducted to determine if data for six genes could be combined using PAUP 4.0b10 (Swofford 2003). To determine the phylogenetic relationships among species, phylogenetic analyses based on maximum parsimony (MP), maximum likelihood (ML), and bayesian inference (BI) were conducted on the individual gene regions and their concatenated dataset, depending on the sequence availability.
Maximum parsimony analysis was performed using PAUP 4.0b10 (Swofford 2003), with phylogenetic relationships estimated by heuristic searches, random stepwise addition sequences, and tree bisection and reconnection (TBR) branch swapping. Gaps were treated as missing data, and all characters were unordered and weighted equally. The measures calculated for parsimony included the tree length (TL), consistency index (CI), homoplasy index (HI), retention index (RI), and rescaled consistency index (RC). Statistical support for branch nodes was assessed with 1,000 bootstrap replicates.
The best evolutionary model of nucleotide substitution for each gene region was selected according to the Akaike Information Criterion (AIC) using MODELTEST v. 3.4 (Posada and Crandall 1998) for ML analyses and MRMODELTEST v. 2 (Nylander 2004) for BI analyses.
ML analyses for individual gene regions were performed using PAUP 4.0b10 (Swofford 2003). The ML models used were K80 + G (act), TVM + G (cmdA), TrN + G (his3), SYM + G (rpb2), TVM + I + G (tef1) and HKY + I (tub2). Statistical support for branch nodes was assessed with 1,000 bootstrap replicates. A partitioned ML analysis was performed using IQ-TREE (Nguyen et al. 2015) as implemented in the IQ-TREE web server (http://iqtree.cibiv.univie.ac.at, Trifinopoulos et al. 2016) by using partition models (Chernomor et al. 2016). Branch support values were evaluated based on 10,000 replicates for ultrafast bootstrapping (UFBoot2) (Hoang et al. 2018).
Individual and partitioned BI analyses were performed using MRBAYES v.3.2.7a (Ronquist et al. 2012) on XSEDE at the CIPRES Science Gateway v.3.3 (http://www.phylo.org/). The BI models used were K80 + G (act), GTR + G (cmdA and his3), SYM + G (rpb2), GTR + I + G (tef1) and HKY + I (tub2). A Markov Chain Monte Carlo (MCMC) algorithm was employed, and two independent runs of four MCMC chains (three hot and one cold) were run in parallel simultaneously starting from random trees for 107 generations (individual gene regions) and 307 generations (concatenated dataset), sampling trees every 1,000 generations. The distribution of log-likelihood scores was examined with TRACER v.1.5 (Rambaut and Drummond 2007) to determine the whether the stationary phase of each search was reached and whether chains had achieved convergence. The convergence of the chains was also assessed by the convergent diagnostics of the effective sampling site (ESS), the potential scale reduction factor (PSRF), and the average standard deviation of split frequencies (ASDSF) (Ronquist et al. 2019). The first 25% of saved trees were discarded as the “burn-in” phase, and posterior probabilities (PP) were computed using the remaining trees. Trees were visualized in FIGTREE v. 1.4.4 (Rambaut 2009) and edited in INKSCAPE v. 1.0 (https://inkscape.org).
Pairwise homoplasy index (PHI) test and phylogenetic network analysis
Phylogenetically closely related species were analyzed using the Genealogical Concordance Phylogenetic Species Recognition (GCPSR) model (as described by Taylor et al. 2000) by performing a pairwise homoplasy index (Φw) test (PHI) (Bruen et al. 2006). The PHI test was performed in SPLIT TREE4 v.4.16.1. (https://uni-tuebingen.de) (Huson and Bryant 2006) to determine the recombination level within phylogenetically closely related species. Only the gene regions that were available for all compared individuals were used. Gaps’ sites were excluded. Significant recombination was considered at a PHI index below 0.05 (Φw < 0.05). The relationships between closely related taxa were visualized by constructing a phylogenetic network from the concatenated datasets using the LogDet transformation and the NeighborNet method; the resultant networks were displayed with the EqualAngle algorithm (Dress and Huson 2004). Bootstrap analysis was then conducted with 1,000 replicates.
Mating type and sexual compatibility test
The mating-type idiomorph of each Calonectria species isolate was determined through PCR by using the primer pairs Cal_MAT111_F/Cal_MAT111_R and Cal_MAT121_F/Cal_MAT121_R, which amplify the MAT1-1-1 and MAT1-2-1 genes using the protocol described by Li et al. (2020). Additionally, sexual compatibility tests were performed for all the single-spore isolates of both species on minimal salt agar (MSA; Guerber and Correll 2001) by crossing them in all possible combinations, following the procedure described by Lombard et al. (2010a). The plates were stacked in plastic bags and incubated at 25 °C for 12 weeks.
Morphology
Morphological characterization of representative isolates of each Calonectria species identified by phylogenetic analyses was performed as described by Liu and Chen (2017). Optimal growth temperatures were determined by incubating the representative isolate at temperatures ranging from 5 °C to 30 °C at 5 °C intervals in the dark on MEA plates (three replicates per isolate were used). Colonial characteristics (diameter, color, and texture of colonies) were determined by inoculating the isolates on MEA plates at 25 °C in the dark after seven days of incubation.
Pathogenicity tests
One representative isolate of each Calonectria species was selected for inoculation. Healthy leaves of three short cut branches from an approximately eleven-month-old Eucalyptus plants were inoculated with suspensions of 1 × 104 conidia·mL-1 obtained from single spore cultures. The conidia suspensions for each isolate were prepared using the method described by Graça et al. (2009). Calonectriaparagominensis was inoculated on E.grandis × E.brassiana hybrid genotype and C.imperata on E.urophylla genotype. The inoculation consisted of spraying the conidia suspension until the suspension run off the leaves. Sterile water was sprayed onto healthy leaves as the negative control. The branches with inoculated leaves were covered with plastic bags to maintain high humidity and kept at 25 °C under a photoperiod of 12 h for 72 h. After that time, the plastic bags were removed, and necrotic symptoms were observed.
Results
Fungal isolates
A total of 34 isolates with the typical morphology of Calonectria species were obtained from infected leaves of the Eucalyptus genotypes sampled. Based on preliminary phylogenetic analyses of the tef1 and tub2 gene regions (data not shown), nine isolates were selected for further studies (Table 1).
Phylogenetic analyses
Sequences from 50 isolates corresponding to 25 Calonectria species closely related to the isolates obtained in this study were downloaded from GenBank (Table 1). For the nine isolates selected in this study, five resided in the Calonectriaspathiphylli species complex (CSSC), and four resided in the Calonectriacandelabrum species complex (CCSC). Both Calonectria complexes belong to the Prolate Group, whose species are characterized by their clavate to pyriform to ellipsoidal vesicles (Liu et al. 2020). Therefore, both complexes were combined into a single sequence dataset for phylogenetic analyses, including two strains of Calonectriagracilipes as the outgroup taxa.
Alignments for each gene region and the concatenated dataset were as follows: act (36 isolates, 267 characters), cmdA (58 isolates, 485 characters), his3 (59 isolates, 439 characters), rpb2 (28 isolates, 863 characters), tef1 (58 isolates, 496 characters), tub2 (59 isolates, 511 characters) and concatenated (59 isolates, 3061 characters). The PHT generated a p value of 0.01 for the concatenated dataset, suggesting some incongruence in the datasets for the six regions and the accuracy of the combined data could have suffered relative to the individual partitions (Cunningham 1997). Although the p value was low, the different gene regions were combined because the significance threshold of 0.05 may be too conservative and it has been shown that combining incongruent datasets improves phylogenetic accuracy (Sullivan 1996; Cunningham 1997); moreover, this approach was followed by several previous studies (Lombard et al. 2016; Pham et al. 2019; Liu et al. 2020, 2021).
Tree topologies derived from the MP, ML, and BI analyses of the individual gene regions were similar overall, but the relative positions of some Calonectria species slightly differed. Moreover, the concatenated dataset formed well-supported lineages in the MP, ML, and BI analyses. Only the ML trees are presented in this study (Fig. 1, Suppl. material 1: Figs S1–S6). The concatenated dataset had 466 parsimony-informative characters, 67 parsimony-uninformative characters, and 2,528 constant characters. Analysis of the 466 parsimony-informative characters yielded 2 equally parsimonious trees, with TL = 862, CI = 0.7042, HI = 0.2958, RI = 0.9192, RC = 0.6472. For the partitioned BI analysis, the convergence of the chains was confirmed by an ESS > 200, a PSRF approaching 1, and an ASDSF equal to 0.000793. The aligned sequences were deposited in TreeBASE (http://treebase.org; No. 29573).
Figure 1.
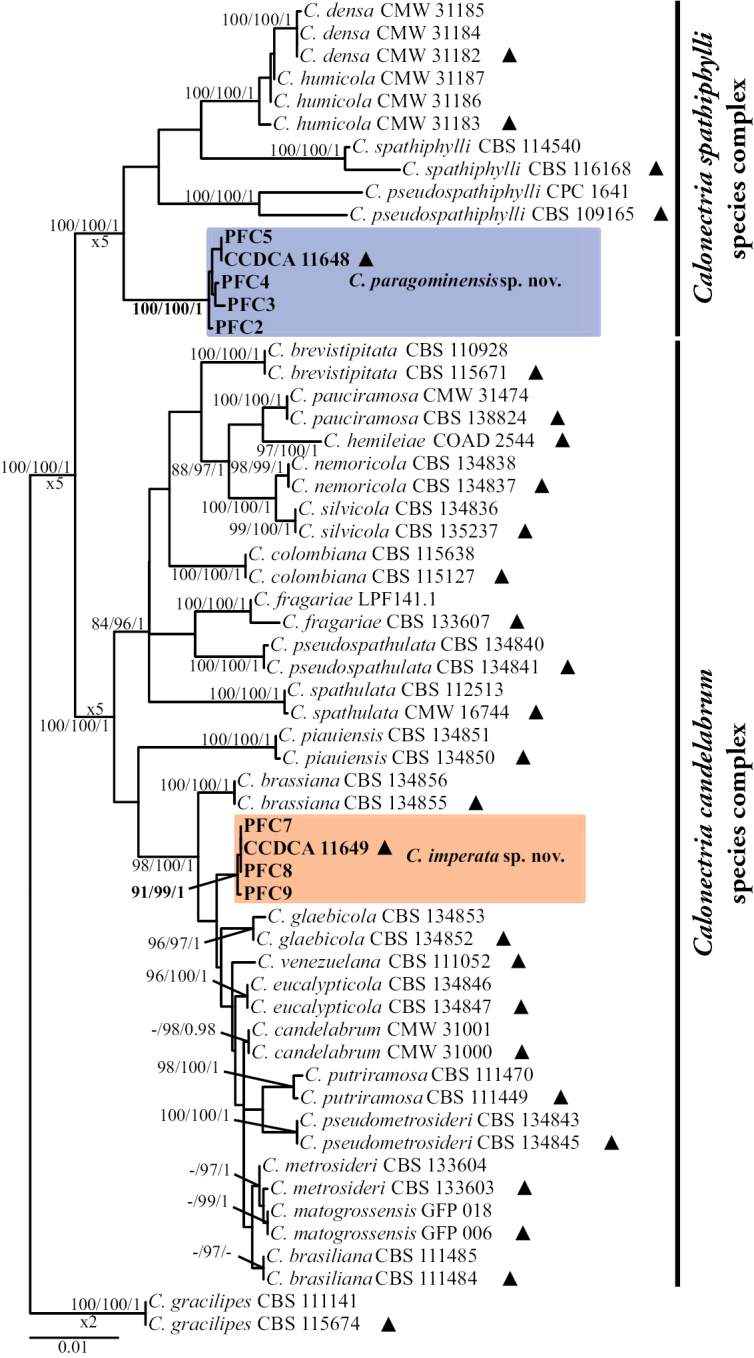
Phylogenetic tree based on maximum likelihood analysis of concatenated act, cmdA, his3, rpb2, tef1 and tub2 gene regions. Bootstrap support values ≥ 80% for maximum parsimony (MP), Ultrafast bootstrap support values ≥ 95% for maximum likelihood (ML), and posterior probability (PP) values ≥ 0.95 from BI analyses are presented at the nodes (MP/ML/PP). Bootstrap values below 80% (MP), 95% (ML) and posterior probabilities below 0.80 are marked with “-”. Ex-type isolates are indicated by “▲”, isolates highlighted in bold were sequenced in this study, and novel species are in blue and orange. C.gracilipes was used as outgroup. The scale bar indicates the number of nucleotide substitutions per site.
Phylogenetic analyses of the six individual gene regions showed that the five isolates from the CSSC were clustered in an independent clade (Suppl. material 1: Figs S1–S6). Based on the concatenated dataset of the six genes, the five isolates formed a new, strongly defined phylogenetic clade that was distinct from the other Calonectria species of the CSSC and was supported by high bootstrap values (MP = 100%, ML = 100%) and high values of posterior probability (1.0) (Fig. 1). A total of 41 fixed unique single nucleotide polymorphisms (SNPs) were identified in the new phylogenetic clade of the five isolates in comparison with their phylogenetically closely related Calonectria species in the six-gene concatenated dataset (Table 2). The results of these phylogenetic and SNP analyses indicate that the five isolates in the CSSC represent a distinct, undescribed species, which we named C.paragominesis.
Table 2.
Single nucleotide polymorphisms unique to C.paragominensis in comparison with their phylogenetically closely related species in the six gene regions.
| Species | act † | cmdA | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 188‡ | 189 | 190 | 191 | 192 | 193 | 194 | 195 | 196 | 197 | 142 | 144 | 170 | 185 | 217 | 270 | 437 | 444 | 455 | 483 | ||
| C.paragominensisCCDCA 11648 | a | g | a | a | a | a | a | g | a | a | t | a | c | c | t | c | a | a | g | a | |
| C.densaCMW 31182 | t | - | - | - | - | - | - | - | - | - | a | c | t | t | c | t | g | g | a | c | |
| C.humicolaCMW 31183 | t | - | - | - | - | - | - | - | - | - | a | c | t | t | c | t | g | g | a | c | |
| C.pseudospathiphylliCBS 109165 | t | - | - | - | - | - | - | - | - | - | a | c | t | t | c | t | g | g | a | c | |
| C.spathiphylliCBS 114540 | t | - | - | - | - | - | - | - | - | - | a | c | t | t | c | t | g | g | a | c | |
| Species | his3 | rpb2 | tef1 | tub2 | |||||||||||||||||
| 6 | 43 | 47 | 52 | 53 | 54 | 247 | 259 | 274 | 81 | 141 | 315 | 474 | 630 | 735 | 32 | 123 | 208 | 434 | 459 | 15 | |
| C.paragominensisCCDCA 11648 | c | t | t | - | - | - | a | g | a | t | c | t | t | a | g | c | t | g | g | c | a |
| C.densaCMW 31182 | t | a | c | c | t | c | t | a | g | - | - | a | t | t | t | ||||||
| C.humicolaCMW 31183 | t | a | c | c | t | c | t | a | g | - | - | a | t | t | t | ||||||
| C.pseudospathiphylliCBS 109165 | - | a | c | c | a | c | c | a | g | c | t | c | c | g | a | - | - | a | a | t | t |
| C.spathiphylliCBS 114540 | - | a | c | c | c | c | c | a | g | c | t | c | c | g | a | - | - | a | t | t | t |
† Only polymorphic nucleotides occurring in all the isolates are shown, not alleles that partially occur in individuals per phylogenetic group. ‡ Numerical positions of the nucleotides in the DNA sequence alignments.
Phylogenetic analyses of the individual gene regions of act, cmdA, his3, rpb2, and tub2 showed that the four isolates that resided in the CCSC were clustered in an independent clade (Suppl. material 1: Figs S1–S4, S6). However, the phylogenetic tree based on tef1 showed that three of those isolates formed an independent clade, while one isolate was closely related to C.metrosideri, C.pseudometrosideri, and C.candelabrum (Suppl. material 1: Fig. S5). Based on the concatenated dataset of the six genes, the four isolates formed a new, strongly defined phylogenetic clade that was distinct from other Calonectria species in the CSSC and was supported by high bootstrap values (MP = 91%, ML = 99%) and high values of posterior probability (1.0) (Fig. 1). The four isolates of the new phylogenetic clade were distinguished from their phylogenetically closely related Calonectria species using SNP analyses for the six-gene concatenated dataset, by presenting eight unique SNPs from a total of 78 SNPs (Table 3). The results of these phylogenetic and SNP analyses indicate that the four isolates in the CCSC represent a distinct, undescribed species, which we named C.imperata.
Table 3.
Single nucleotide polymorphisms found in Calonectriaimperata and its phylogenetically closely related species in the six gene regions.
| Species | act † | cmdA | his3 | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 57‡ | 62 | 71 | 121 | 171 | 187 | 210 | 319 | 376 | 403 | 405 | 418 | 444 | 8 | 44 | 46 | 53 | 56 | 60 | 66 | |
| C.imperataCCDCA 11649 | c | a | c | c | g | g | c | c | t | t | c | t | t | c | c | t | c | c | g | a |
| C.brassianaCBS 134855 | c | g | t | g | g | c | c | c | c | c | a | t | t | c | t | c | c | g | a | |
| C.glaebicolaCBS 134852 | a | c | c | g | g | c | g | t | c | c | a | t | c | t | c | c | c | g | a | |
| C.piauiensisCBS 134850 | a | g | c | c | a | c | c | c | c | c | a | c | c | c | - | a | c | a | c | |
| C.venezuelanaCBS 111052 | t | a | c | c | g | g | a | g | t | c | t | a | t | c | c | t | c | t | g | a |
| Species | his3 | |||||||||||||||||||
| 93 | 99 | 105 | 114 | 156 | 189 | 234 | 235 | 238 | 244 | 245 | 250 | 251 | 252 | 254 | 255 | 257 | 262 | 275 | 276 | |
| C.imperataCCDCA 11649 | c | c | c | a | t | t | t | t | c | c | a | c | c | a | g | c | a | a | t | g |
| C.brassianaCBS 134855 | c | c | c | t | t | t | t | t | c | c | a | c | c | a | g | c | a | a | t | g |
| C.glaebicolaCBS 134852 | c | c | c | a | t | t | t | t | c | a | a | c | c | a | g | c | a | a | t | g |
| C.piauiensisCBS 134850 | t | t | c | a | c | t | c | g | t | g | g | t | a | g | a | t | g | g | c | a |
| C.venezuelanaCBS 111052 | c | c | a | a | t | c | t | t | c | c | a | c | c | a | g | c | a | a | t | g |
| Species | his3 | rpb2 | tef1 | |||||||||||||||||
| 277 | 278 | 333 | 336 | 351 | 405 | 420 | 105 | 603 | 624 | 693 | 840 | 47 | 81 | 110 | 112 | 135 | 220 | 239 | 357 | |
| C.imperataCCDCA 11649 | c | t | t | c | g | c | t | g | a | c | t | a | c | g | a | t | t | c | c | c |
| C.brassianaCBS 134855 | c | t | t | c | g | t | t | c | g | t | t | t | c | c | c | |||||
| C.glaebicolaCBS 134852 | c | t | t | c | a | c | t | c | a | a | t | t | c | c | c | |||||
| C.piauiensisCBS 134850 | t | t | c | t | g | c | g | t | g | a | a | c | a | t | c | |||||
| C.venezuelanaCBS 111052 | c | c | t | c | g | c | t | t | g | t | c | t | c | g | a | t | t | c | c | t |
| Species | tef1a | tub2 | ||||||||||||||||||
| 417 | 421 | 422 | 425 | 453 | 50 | 99 | 120 | 132 | 174 | 175 | 188 | 191 | 220 | 377 | 398 | 408 | 409 | |||
| C.imperataCCDCA 11649 | c | c | c | a | a | c | a | g | t | c | c | g | c | t | t | t | - | - | ||
| C.brassianaCBS 134855 | c | t | t | a | a | c | a | g | t | c | c | g | c | t | t | t | a | c | ||
| C.glaebicolaCBS 134852 | c | t | t | a | a | c | a | g | t | c | c | g | c | t | t | t | a | c | ||
| C.piauiensisCBS 134850 | t | c | c | a | a | g | g | a | c | t | t | a | a | c | c | g | a | c | ||
| C.venezuelanaCBS 111052 | c | c | c | c | g | c | a | g | t | c | c | g | c | t | t | t | a | c | ||
† Only polymorphic nucleotides occurring in all the isolates are shown, not alleles that partially occur in individuals per phylogenetic group. ‡ Numerical positions of the nucleotides in the DNA sequence alignments.
Species delimitation by GCPSR analysis
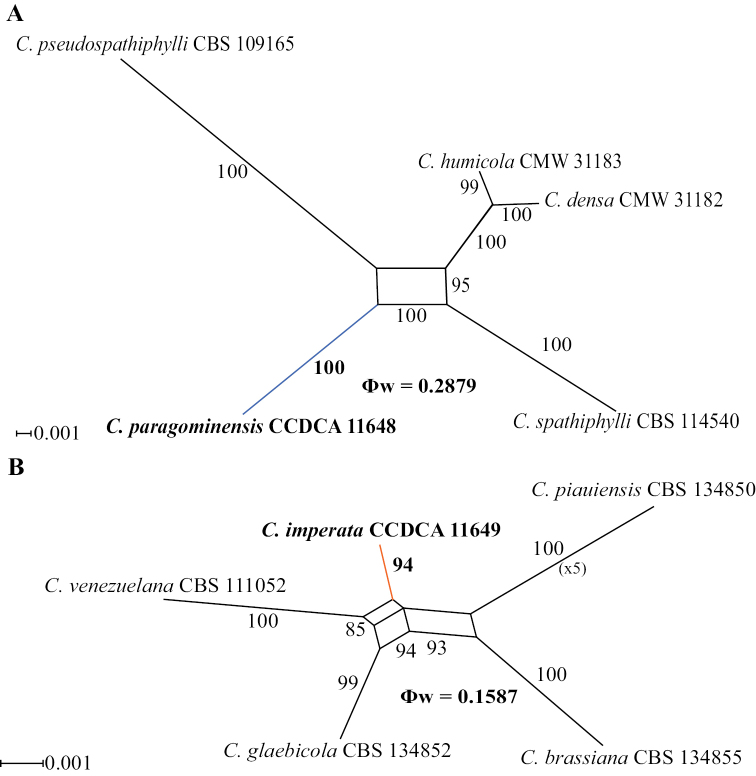
A PHI test using a five-locus concatenated dataset (act, cmdA, his3, tef1, tub2) was performed to determine the recombination level among C.paragominensis and its phylogenetically closely related species, C.densa, C.humicola, C.spathiphylli and C.pseudospathiphylli. A value of Φw = 0.2879 revealed no significant genetic recombination events, and this relationship was supported with a high bootstrap value (100%) in the phylogenetic network analysis, indicating that they are different species (Fig. 2A).
Figure 2.
Results of the pairwise homoplasy index (PHI) test for C.paragominensis and C.imperata. Phylogenetic networks constructed using the LogDet transformation and the NeighborNet method and displayed with the EqualAngle algorithm. Bootstrap support values > 80% are shown. Φw < 0.05 indicate significant recombination. New species described in this study are highlighted in bold, with blue (A) and orange (B) lines.
A PHI test using a four-locus concatenated dataset (cmdA, his3, tef1, tub2) was performed to determine the recombination level among C.imperata and its phylogenetically closely related species, C.brassiana, C.glabeicola, C.piauiensis, and C.venezuelana. A value of Φw = 0.1587 revealed no significant genetic recombination events, and this relationship was supported with a high bootstrap value (94%) in the phylogenetic network analysis, indicating that they are different species (Fig. 2B).
Mating-type and sexual compatibility test
MAT1-1-1 and MAT1-2-1 genes were amplified in all isolates of each identified species, indicating that they are putatively homothallic. However, after a twelve-week mating test on MSA, all isolates failed to yield sexual structures, indicating that they have lost the ability to be self-fertile or have retained the ability to favor outcrossing rather than selfing.
Taxonomy
Based on phylogenetic analyses, GCPSR, and network analyses, the nine isolates presented two strongly defined phylogenetic clades in both the Calonectriaspathiphylli species complex and the Calonectriacandelabrum species complex. Morphological differences, especially in the macroconidia and stipe dimensions, were observed between each phylogenetic clade and its phylogenetically closely related species (Table 4). Thus, the fungi isolated in this study represent two new species of Calonectria and are described as follows:
Table 4.
Morphological characteristics of two new Calonectria species and their phylogenetically closely related species.
| Species complex | Species | Conidiogenus apparatus | Stipe | Macroconidia | Vesicle | Reference | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Size (L × W)†,‡,§ | Branches | Size (L × W)†,‡,§ | Extension (L × W)†,‡,§ | Size (L × W)†,‡,§,| | Average (L × W)†,‡,§ | Septa | Diameter†,§ | Shape | |||
| Calonectriaspathiphylli species complex | C.paragominensis | 40–113 × 45–129 | (–4) | 112–281 × 2–4 | 123–295 × 1.5–3 | (47–)56–66(–71) × (4–)4.8–5.9(–7) | 61 × 5 | 1(–3) | 8–12 | globoid to sphaeropedunculate | This study |
| C.densa | 49–78 × 63–123 | (–4) | 54–90 × 6–10 | 149–192 × 5–6 | (47–)50–58(–62) × (5–)6 | 54 × 6 | 1 | 10–12 | ovoid to ellipsoid to sphaeropedunculate | Lombard et al. 2010b | |
| C.humicola | 43–71 × 42–49 | 3 | 44–90 × 6–8 | 126–157 × 4–5 | (45–)48–54(–56) × (4–)5 | 51 × 5 | 1 | 10–12 | globoid to ovoid to sphaeropedunculate | Lombard et al. 2010b | |
| C.pseudospathiphylli | 70–100 × 25–70 | 4 | 100–350 × 5–6 | 100–250 × 2.5–3.5 | (40–)47–55(–60) × 4–5 | 52 × 4 | 1(–3) | 8–12 | sphaeropedunculate to ellipsoid | Kang et al. 2001; Crous 2002 | |
| C.spathiphylli | 60–150 × 40–90 | 4 | 120–150 × 6–8 | 170–260 × 3–4 | (45–)65–80(–120) × (5–)6(–7)¶ | 70 × 6 | 1(–3) | 8–15 | globoid or ellipsoid to obpyriform | Crous 2002 | |
| Calonectriacandelabrum species complex | C.imperata | 50–127 × 41–110 | (–3) | 135–227 × 2–4 | 151–254 × 1.5–3 | (38–)43–49(–52) × (2–)2.7–3.2(–4) | 46 × 3 | (–1) | 3–6 | ellipsoid to narrowly obpyriform | This study |
| C.piauiensis | 20–60 × 35–80 | 2 | 50–110 × 4–6 | 95–130 × 2–3 | (38–)47–52(–60) × 3–5 | 49 × 4.5 | 1 | 3–7 | ellipsoid to narrowly obpyriform | Alfenas et al. 2015 | |
| C.brassiana | 50–80 × 50–135 | 3 | 55–155 × 5–8 | 90–172 × 2–3 | (35–)50–56(–65) × 3–5 | 53 × 4 | 1 | 3–7 | ellipsoid to narrowly obpyriform | Alfenas et al. 2015 | |
| C.glaebicola | 27–45 × 25–40 | 2 | 50–130 × 5–7 | 100–165 × 2–4 | (45–)50–52(–55) × 3–5 | 50 × 4 | 1 | 3–5 | ellipsoid to narrowly obpyriform | Alfenas et al. 2015 | |
| C.venezuelana | 25–65 × 25–60 | 3 | 35–100 × 4–8 | 85–190 × 3–6 | (48–)54–62(–65) × (4–)4.5–5.5(–7) | 58 × 5 | 1 | 5–9 | fusiform to ovoid to ellipsoid | Lombard et al. 2016 | |
† All measurements are in μm. ‡ L × W = length × width. § Minimum–maximum. | Measurements are presented in the format [(minimum–) (average – standard deviation) – (average + standard deviation) (–maximum)]. ¶ Measurements are presented in the format [(minimum–) (average) (–maximum)].
. Calonectria paragominensis
E.I.Sanchez, T.P.F.Soares & M.A.Ferreira sp. nov.
F9C48BD4-5C29-5A44-9515-0AD7F88CEAA8
843460
Figure 3.
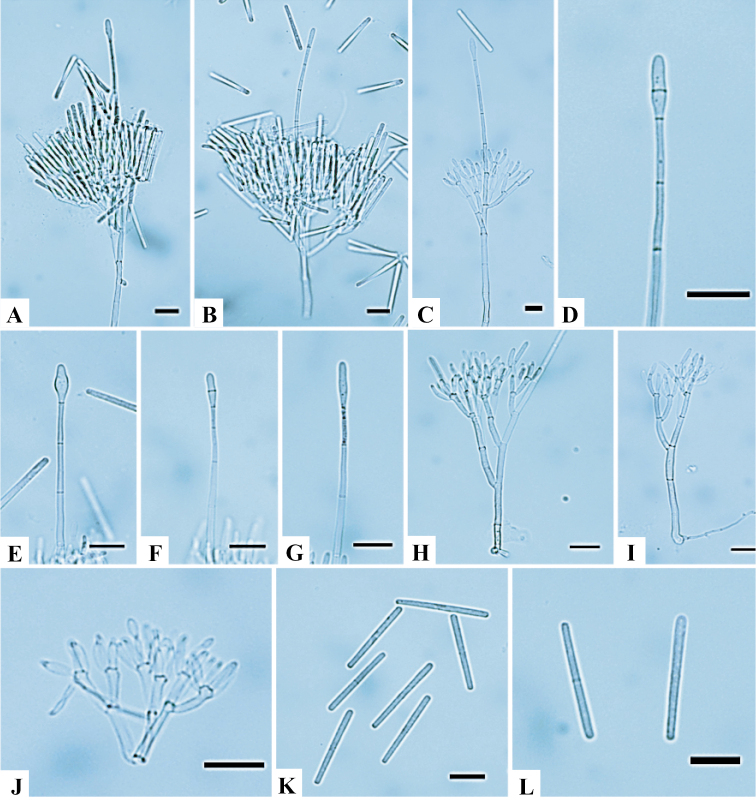
CalonectriaparagominensisA, B macroconidiophore C lateral stipe extensions D, E conidiogenous apparatus with conidiophore branches and doliiform to reniform phialides F, G globose to sphaeropedunculate vesicles H, I one, two, and three-septate macroconidia. Scale bars: 20 μm.
Etymology.
The term “paragominensis” refers to the microregion of Paragominas, Brazil, which is the place where the fungus was collected.
Diagnosis.
Calonectriaparagominensis differs from the phylogenetically closely related species C.densa, C.humicola, C.spathiphylli and C.pseudospathiphylli with respect to its macroconidia dimensions.
Type.
Brazil,• Pará state, Paragominas microregion; 3°10'51"S, 47°18'49"W; From infected leaves of E.grandis × E.brassiana; 20 Feb. 2020; M.A. Ferreira; holotype: UB24349, ex-type: CCDCA 11648 = PFC1. GenBank: act = ON009346; cmdA = OM974325; his3 = OM974334; rpb2 = OM974343; tef1 = OM974352; tub2 = OM974361.
Description.
Sexual morph unknown. Macroconidiophores consisted of a stipe, a suite of penicillate arrangements of fertile branches, a stipe extension, and a terminal vesicle; stipe septate, hyaline, smooth, (112–)135–207(–281) × (2–)2.6–3.5(–4) μm; stipe extension septate, straight to flexuous, (123–)147–220(–295) μm long, (1.5–)1.9–2.4(–3) μm wide at the apical septum, terminating in a globose to sphaeropedunculate vesicle, (8–)8.5–10.5(–12) μm diam; lateral stipe extensions (90° to the axis) also present. Conidiogenous apparatus was (40–)56–88(–113) μm long, (45–)67–107(–129) μm wide; primary branches aseptate or 1-septate, (15.7–)18.4–25.9(–30.6) × (3.3–)4–6(–6.5) μm; secondary branches aseptate, (12.7–)14.3–19.6(–22.1) × (3–)3.5–5(–6) μm; tertiary branches aseptate, (9.9–)11.6–15.3(–17.9) × (2.8–)3.6–5.3(–6.4) μm; additional branches (–4), aseptate, (10.3–)11–13.2(–14) × (3–)3.2–4.4(–5) μm; each terminal branch produced 2–4 phialides; phialides doliiform to reniform, hyaline, aseptate, (8–)9.1–11.8(–14) × (2–)2.7–4.1(–6) μm, apex with minute periclinal thickening and inconspicuous collarette. Macroconidia were cylindrical, rounded at both ends, straight, (47–)56–66(–71) × (4–)4.8–5.9(–7) μm (av. = 61 × 5 μm), (1–3) septate, lacking a visible abscission scar, held in parallel cylindrical clusters by colorless slime. Megaconidia and microconidia were not observed.
Culture characteristics.
Colonies formed abundant white aerial mycelium on MEA at 25 °C after seven days, with irregular margins and moderate sporulation. The surface had white to buff outer margins, and sienna to amber in reverse with abundant chlamydospores throughout the medium, forming microsclerotia. The optimal growth temperature was 23.8 °C, with no growth at 5 °C; after seven days, colonies at 10 °C, 15 °C, 20 °C, 25 °C, and 30 °C reached 7 mm, 23 mm, 38.3 mm, 36.1 mm, and 31.8 mm, respectively.
Substratum.
Leaves of E.grandis × E.brassiana.
Distribution.
Northeast Brazil.
Other specimens examined.
Brazil,• Pará state, Paragominas microregion; From infected leaves of E.grandis × E.brassiana; 20 Feb. 2020; M.A. Ferreira; cultures PFC2, PFC3, PFC4, PFC5.
Notes.
C.paragominensis is a new species in the C.spathiphylli species complex (Liu et al., 2020). Morphologically, C.paragominensis is very similar to C.densa, since both form lateral stipe extensions, which have not been reported for the other three species in the complex. However, the macroconidia of C.paragominensis (av. 61 × 5 μm) are longer than those of C.densa (av. 54 × 6 μm), C.humicola (av. 51 × 5 μm) and C.pseudospathiphylli (av. 52 × 4 μm) but smaller than those of C.spathiphylli (av. 70 × 6 μm).
. Calonectria imperata
E.I.Sanchez, T.P.F.Soares & M.A.Ferreira sp. nov.
4843A76F-0B34-56D4-B28F-1E0EEFAC60E7
843461
Figure 4.
CalonectriaimperataA–C macroconidiophore D–G ellipsoidal to narrowly obpyriform vesicles H–J conidiogenous apparatus with conidiophore branches and doliiform to reniform phialides K, L macroconidia. Scale bars: 20 μm.
Etymology.
The term “imperata” is in honor of the city of Imperatriz, Brazil, which was close to the place where the fungus was collected.
Diagnosis.
Calonectriaimperata differs from the phylogenetically closely related species C.brassiana, C.glaebicola, C.piauiensis and C.venezuelana with respect to the number of unique alleles and stipe dimensions.
Type.
Brazil,• Maranhão state, Cidelândia municipality; 5°09'24"S, 47°46'26"W; From infected leaves of E.urophylla; 20 Feb. 2020; M.A. Ferreira; holotype: UB24350, ex-type: CCDCA 11649 = PFC6. GenBank: act = ON009351; cmdA = OM974330; his3 = OM974339; rpb2 = OM974348; tef1 = OM974357; tub2 = OM974366.
Description.
Sexual morph unknown. Macroconidiophores consisted of a stipe, a suite of penicillate arrangements of fertile branches, a stipe extension, and a terminal vesicle; stipe septate, hyaline, smooth, (135–)151–198(–227) × (2–)2.6–3.4(–4) μm; stipe extension septate, straight to flexuous, (151–)169–220(–254) μm long, (1.5–)1.9–2.7(–3) μm wide at the apical septum, terminating in an ellipsoidal to narrowly obpyriform vesicle (3–)3.1–4.6(–6) μm diam. Conidiogenous apparatus was (50–)66–100(–127) μm long, (41–)62–89(–110) μm wide; primary branches aseptate, (14.6–)19–24.8(–28.5) × (2.5–)3.2–4(–4.5) μm; secondary branches aseptate, (12.1–)13.5–18.2(–24.2) × (2.3–)2.8–3.7(–4) μm; tertiary branches aseptate, (10.1–)11–15(–18.1) × (1.9–)2.3–3.2(–4.1) μm; each terminal branch producing 2–4 phialides; phialides doliiform to reniform, hyaline, aseptate, (8–)9.1–13(–15) × (2–)2.7–3.3(–4) μm, apex with minute periclinal thickening and inconspicuous collarette. Macroconidia were cylindrical, rounded at both ends, straight, (38–)43–49(–52) × (2–)2.7–3.2(–4) μm (av. = 46 × 3 μm), (–1) septate, lacking a visible abscission scar, held in parallel cylindrical clusters by colorless slime. Megaconidia and microconidia were not observed.
Culture characteristics.
Colonies formed moderate aerial mycelium on MEA at 25 °C after seven days, with moderate sporulation. The surface had white to buff outer margins, and sepia to umber in reverse with abundant chlamydospores throughout the medium, forming microsclerotia. The optimal growth temperature was 25 °C, with no growth at 5 °C; after seven days, colonies at 10 °C, 15 °C, 20 °C, 25 °C, and 30 °C reached 10.1 mm, 25.5 mm, 29.1 mm, 44.5 mm, and 40.6 mm, respectively.
Substratum.
Leaves of E.urophylla.
Distribution.
Northeast Brazil.
Other specimens examined.
Brazil,• Maranhão state, Cidelândia municipality; 5°09'24"S, 47°46'26"W; From infected leaves of E.urophylla; 20 Feb. 2020; M.A. Ferreira; cultures PFC7, PFC8, PFC9. Brazil• Maranhão state, Itinga do Maranhão; 4°34'43"S, 47°29'48"W; from infected leaves of E.urophylla; 20 Feb. 2020; M.A. Ferreira; culture PFC9.
Notes.
C.imperata is a new species in the C.candelabrum species complex (Liu et al., 2020). Morphologically, C.imperata is very similar to its closest relatives, from which it can be distinguished based on stipe dimensions and phylogenetic inference. Stipe of C.imperata (135–227 × 2–4 μm) is larger than those of C.piauiensis (50–110 × 4–6 μm), C.glaebicola (50–130 × 5–7 μm), and C.venezuelana (35–100 × 4–8 μm) but narrower than those of C.brassiana (55–155 × 5–8 μm). Additionally, C.imperata lacks lateral stipe extensions, which are present in C.piauiensis.
Pathogenicity tests
The conidia suspensions of the representative isolates of C.paragominensis and C.imperata produced lesion symptoms on leaves (Fig. 5E, F, I, J), but no lesions were observed on the negative control inoculations (Fig. 5G, H, K). The pathogens were reisolated from inoculated leaves but not from the negative controls and identified by the same morphological characteristics as the originally inoculated species, thus, fulfilling the requirements of Koch’s postulates.
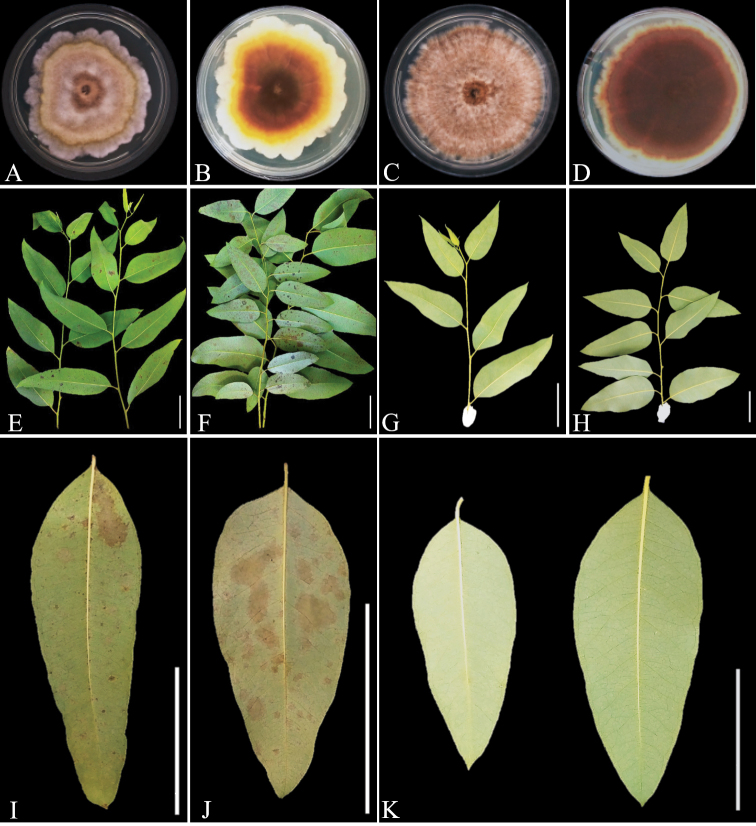
Figure 5.
Pathogenicity tests on leaves of Eucalyptus genotypes A, B surface and reverse of C.paragominensis on MEA plates after 14 days grown at 25 °C C, D surface and reverse of C.imperata on MEA plates after 14 days grown at 25 °C E, I lesions on leaves of E.grandis × E.brassiana induced by C.paragominensis 72 h after inoculation F, J lesions on leaves of E.urophylla induced by C.imperata 72 h after inoculation G, H, K no disease symptoms on leaves inoculated with sterile water (negative controls). Scale bars: 5 cm (E–K).
Discussion
Two new species of Calonectria isolated from diseased Eucalyptus leaves were identified based on phylogenetic analyses of six gene regions and on morphological comparisons. These two species were named C.paragominensis and C.imperata.
Calonectriaparagominensis is a new species in the C.spathiphylli complex. The five species identified and described in C.spathiphylli complex are C.densa, C.humicola, C.spathiphylli, C.pseudospathiphylli, and C.paragominensis, where C.paragominensis can be differentiated morphologically with respect to the macroconidia dimensions (El-Gholl et al. 1992; Kang et al. 2001; Crous 2002; Lombard et al. 2010b). These species are characterized by presenting globoid to ovoid to sphaeropedunculate terminal vesicles (Kang et al. 2001; Crous 2002; Lombard et al. 2010b). Calonectriaspathiphylli is described as heterothallic (El-Gholl et al. 1992), C.densa as putatively heterothallic (Li et al. 2020) and C.pseudospatiphilly as homothallic (Kang et al. 2001). The C.humicola mating type has not been indicated (Lombard et al. 2010b). Here, C.paragominensis is described as putatively homothallic based on PCR amplification of the mating-type genes. Regarding pathogenicity, C.paragominensis is pathogenic to Eucalyptus sp., C.spathiphylli is pathogenic to Sapthiphyllum sp. Heliconia sp. Ludwigia sp. Strelitzia sp. and Eugenia sp. (El-Gholl et al. 1992; Poltronieri et al. 2011). Calonectriadensa, C.humicola, and C.pesudospathiphylli were isolated from soil, and their pathogenicity has not been indicated (Kang et al. 2001; Lombard et al. 2010b). In addition to C.paragominensis, only C.spathiphylli has been indicated to be present in Brazil (Reis et al. 2004; Poltronieri et al. 2011).
Calonectriaimperata is a new species in the C.candelabrum complex. Species in this complex are characterized by presenting ellipsoidal to obpyriform terminal vesicles, in both heterothallic and homothallic species, and occur in Africa, Asia, Europe, North and South America, and Oceania (Liu et al. 2020). Of the 21 species in the C.candelabrum complex (Crous et al. 2018, 2019; Liu et al. 2020), 17 have been found in Brazil (Schoch et al. 1999; Crous et al. 2018, 2019; Liu et al. 2020). Calonectriaimperata is phylogenetically closely related to C.brassiana, C.glaebicola, C.piauiensis and C.venezuelana, which can be differentiated with respect to the number of unique alleles and stipe dimensions (Alfenas et al. 2015; Lombard et al. 2016). Calonectriabrassiana, C.glaebicola, and C.piauiensis were found in Brazil, isolated from soil samples of Eucalyptus plantations, but only C.glaebicola has been confirmed to be pathogenic to Eucalyptus sp. (Alfenas et al. 2015). Calonectriavenezuelana was reported in Venezuela, similarly, isolated from soil samples, but its pathogenicity has not been indicated (Lombard et al. 2016). The mating-type for C.brassiana, C.glaebicola, C.piauiensis and C.venezuelana has not been determined (Liu et al. 2020).
Pathogenicity tests showed that C.paragominensis and C.imperata are pathogenic to E.grandis × E.brassiana hybrid genotype and E.urophylla genotype, respectively. Although the death of Eucalyptus trees due to CLB is not common, it affects Eucalyptus plants most severely from six months to 2–3 years after planting (Graça et al. 2009). Although the economic loss due to defoliation caused by CLB has not been quantified directly, according to artificial pruning studies conducted by Pulrolnik et al. (2005) and Pires (2000), when the loss of branches is equal to or greater than 75% of E.grandis seedlings of one year of age, the volumetric productivity has decreased by 45% by the time they reach seven years old. Therefore, it has been inferred that in susceptible clones, the pathogen can cause economic losses, since under favorable conditions, infection by Calonectria can result in severe defoliation (Soares et al. 2018). Additionally, Miranda et al. (2021) indicated a potential growth loss of 19.8 to 39.6% due to CLB, and by using the estimates of growth reduction from Pires (2000) as a baseline, concluded that a reduction in the volumetric increment to the order of 39.6% may result in an economic loss of R$ 4291.00 per ha, considering a price of Eucalyptus wood as R$ 38.70 per m3 (IEA 2020) and a production of 280 m3·ha-1 in the first 7-year rotation. Therefore, accurate diagnoses of plant diseases and identification of their casual agents are fundamental in promoting the development of effective disease management strategies (Wingfield et al. 2015; Liu and Chen 2017).
In this study, we described two new Calonectria species, both isolated from diseased Eucalyptus leaves from commercial plantations localized in a tropical zone. These results suggest that there are still more Calonectria species to be discovered in Brazil, and that they require careful monitoring, since this knowledge could facilitate the development of resistant Eucalyptus clones.
Supplementary Material
Acknowledgements
We thank Heitor S. Dallapiccola and Francisco J. A. Gomes for their help during sample collection. We thank the laboratories of Molecular Biology, Plant Virology, and Nematology at Universidade Federal de Lavras (UFLA) for the facilities granted during the completion of this study. The first author thanks the “Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES)” for the doctorate scholarship assigned through Brazil’s PAEC OAS-GCUB Scholarships Program.
Citation
Sanchez-Gonzalez EI, Farias Soares TdP, Galafassi Zarpelon T, Valverde Zauza EA, Gonçalves Mafia R, Alves Ferreira M (2022) Two new species of Calonectria (Hypocreales, Nectriaceae) causing Eucalyptus leaf blight in Brazil. MycoKeys 91: 169–197. https://doi.org/10.3897/mycokeys.91.84896
Supplementary materials
Figures S1–S6
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Enrique I. Sanchez-Gonzalez, Thaissa de Paula Farias Soares, Talyta Galafassi Zarpelon, Edival Angelo Valverde Zauza, Reginaldo Gonçalves Mafia, Maria Alves Ferreira
Data type
Phylogenetic trees (pdf file)
Explanation note
Figure S1. Phylogenetic tree based on maximum likelihood analysis of act gene region. Bootstrap support values ≥ 80% for maximum parsimony (MP), Ultrafast bootstrap support values ≥ 95% for maximum likelihood (ML), and posterior probability (PP) values ≥ 0.95 from BI analyses are presented at the nodes (MP/ML/PP). Bootstrap values below 80% (MP), 95% (ML) and posterior probabilities below 0.80 are marked with “-”. Ex-type isolates are indicated by “▲”, isolates highlighted in bold were sequenced in this study, and novel species are in blue and orange. C.gracilipes was used as outgroup. The scale bar indicates the number of nucleotide substitutions per site. Figure S2. Phylogenetic tree based on maximum likelihood analysis of cmdA gene region. Bootstrap support values ≥ 80% for maximum parsimony (MP), Ultrafast bootstrap support values ≥ 95% for maximum likelihood (ML), and posterior probability (PP) values ≥ 0.95 from BI analyses are presented at the nodes (MP/ML/PP). Bootstrap values below 80% (MP), 95% (ML) and posterior probabilities below 0.80 are marked with “-”. Ex-type isolates are indicated by “▲”, isolates highlighted in bold were sequenced in this study, and novel species are in blue and orange. C.gracilipes was used as outgroup. The scale bar indicates the number of nucleotide substitutions per site. Figure S3. Phylogenetic tree based on maximum likelihood analysis of his3 gene region. Bootstrap support values ≥ 80% for maximum parsimony (MP), Ultrafast bootstrap support values ≥ 95% for maximum likelihood (ML), and posterior probability (PP) values ≥ 0.95 from BI analyses are presented at the nodes (MP/ML/PP). Bootstrap values below 80% (MP), 95% (ML) and posterior probabilities below 0.80 are marked with “-”. Ex-type isolates are indicated by “▲”, isolates highlighted in bold were sequenced in this study, and novel species are in blue and orange. C.gracilipes was used as outgroup. The scale bar indicates the number of nucleotide substitutions per site. Figure S4. Phylogenetic tree based on maximum likelihood analysis of rpb2 gene region. Bootstrap support values ≥ 80% for maximum parsimony (MP), Ultrafast bootstrap support values ≥ 95% for maximum likelihood (ML), and posterior probability (PP) values ≥ 0.95 from BI analyses are presented at the nodes (MP/ML/PP). Bootstrap values below 80% (MP), 95% (ML) and posterior probabilities below 0.80 are marked with “-”. Ex-type isolates are indicated by “▲”, isolates highlighted in bold were sequenced in this study, and novel species are in blue and orange. C.gracilipes was used as outgroup. The scale bar indicates the number of nucleotide substitutions per site. Figure S5. Phylogenetic tree based on maximum likelihood analysis of tef1 gene region. Bootstrap support values ≥ 80% for maximum parsimony (MP), Ultrafast bootstrap support values ≥ 95% for maximum likelihood (ML), and posterior probability (PP) values ≥ 0.95 from BI analyses are presented at the nodes (MP/ML/PP). Bootstrap values below 80% (MP), 95% (ML) and posterior probabilities below 0.80 are marked with “-”. Ex-type isolates are indicated by “▲”, isolates highlighted in bold were sequenced in this study, and novel species are in blue and orange. C.gracilipes was used as outgroup. The scale bar indicates the number of nucleotide substitutions per site. Figure S6. Phylogenetic tree based on maximum likelihood analysis of tub2 gene region. Bootstrap support values ≥ 80% for maximum parsimony (MP), Ultrafast bootstrap support values ≥ 95% for maximum likelihood (ML), and posterior probability (PP) values ≥ 0.95 from BI analyses are presented at the nodes (MP/ML/PP). Bootstrap values below 80% (MP), 95% (ML) and posterior probabilities below 0.80 are marked with “-”. Ex-type isolates are indicated by “▲”, isolates highlighted in bold were sequenced in this study, and novel species are in blue and orange. C.gracilipes was used as outgroup. The scale bar indicates the number of nucleotide substitutions per site.
References
- Alfenas AC, Zauza EAV, Mafia RG, de Assis TF. (2009) Clonagem e doenças do eucalipto. 2nd ed. , Editora UFV, Viçosa, MG, Brazil, 500 pp. [Google Scholar]
- Alfenas RF, Pereira OL, Freitas RG, Freitas CS, Dita MA, Alfenas AC. (2013) Mass spore production and inoculation of Calonectriapteridis on Eucalyptus spp. under different environmental conditions. Tropical Plant Pathology 38(5): 406–413. 10.1590/S1982-56762013000500005 [DOI] [Google Scholar]
- Alfenas RF, Lombard L, Pereira OL, Alfenas AC, Crous PW. (2015) Diversity and potential impact of Calonectria species in Eucalyptus plantations in Brazil. Studies in Mycology 80(1): 89–130. 10.1016/j.simyco.2014.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruen TC, Philippe H, Bryant D. (2006) A simple and robust statistical test for detecting the presence of recombination. Genetics 172(4): 2665–2681. 10.1534/genetics.105.048975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone I, Kohn LM. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91(3): 553–556. 10.1080/00275514.1999.12061051 [DOI] [Google Scholar]
- Castellani A. (1939) Viability of some pathogenic fungi in distilled water. The Journal of Tropical Medicine and Hygiene 42: 225–226. [Google Scholar]
- Chernomor O, Von Haeseler A, Minh BQ. (2016) Terrace aware data structure for phylogenomic inference from supermatrices. Systematic Biology 65(6): 997–1008. 10.1093/sysbio/syw037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW. (2002) Taxonomy and pathology of Cylindrocladium (Calonectria) and allied genera. APS Press, St. Paul, Minnesota, USA, 278 pp. [Google Scholar]
- Crous PW, Groenewald JZ, Risède JM, Simoneau P, Hywel-Jones NL. (2004) Calonectria species and their Cylindrocladium anamorphs: Species with sphaeropedunculate vesicles. Studies in Mycology 50: 415–430. 10.3114/sim.55.1.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Luangsa-ard JJ, Wingfield MJ, Carnegie AJ, Hernández-Restrepo M, Lombard L, Roux J, Barreto RW, Baseia IG, Cano-Lira JF, Martín MP, Morozova OV, Stchigel AM, Summerell BA, Brandrud TE, Dima B, García D, Giraldo A, Guarro J, Gusmão LFP, Khamsuntorn P, Noordeloos ME, Nuankaew S, Pinruan U, Rodríguez-Andrade E, Souza-Motta CM, Thangavel R, van Iperen AL, Abreu VP, Accioly T, Alves JL, Andrade JP, Bahram M, Baral H-O, Barbier E, Barnes CW, Bendiksen E, Bernard E, Bezerra JDP, Bezerra JL, Bizio E, Blair JE, Bulyonkova TM, Cabral TS, Caiafa MV, Cantillo T, Colmán AA, Conceição LB, Cruz S, Cunha AOB, Darveaux BA, da Silva AL, da Silva GA, da Silva GM, da Silva RMF, de Oliveira RJV, Oliveira RL, De Souza JT, Dueñas M, Evans HC, Epifani F, Felipe MTC, Fernández-López J, Ferreira BW, Figueiredo CN, Filippova NV, Flores JA, Gené J, Ghorbani G, Gibertoni TB, Glushakova AM, Healy R, Huhndorf SM, Iturrieta-González I, Javan-Nikkhah M, Juciano RF, Jurjević Ž, Kachalkin AV, Keochanpheng K, Krisai-Greilhuber I, Li Y-C, Lima AA, Machado AR, Madrid H, Magalhães OMC, Marbach PAS, Melanda GCS, Miller AN, Mongkolsamrit S, Nascimento RP, Oliveira TGL, Ordoñez ME, Orzes R, Palma MA, Pearce CJ, Pereira OL, Perrone G, Peterson SW, Pham THG, Piontelli E, Pordel A, Quijada L, Raja HA, Rosas de Paz E, Ryvarden L, Saitta A, Salcedo SS, Sandoval-Denis M, Santos TAB, Seifert KA, Silva BDB, Smith ME, Soares AM, Sommai S, Sousa JO, Suetrong S, Susca A, Tedersoo L, Telleria MT, Thanakitpipattana D, Valenzuela-Lopez N, Visagie CM, Zapata M, Groenewald JZ. (2018) Fungal Planet description sheets: 785–867. Persoonia: Molecular Phylogeny and Evolution of Fungi 41(1): 238–417. 10.3767/persoonia.2018.41.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Carnegie AJ, Wingfield MJ, Sharma R, Mughini G, Noordeloos ME, Santini A, Shouche YS, Bezerra JDP, Dima B, Guarnaccia V, Imrefi I, Jurjević Ž, Knapp DG, Kovács GM, Magistà D, Perrone G, Rämä T, Rebriev YA, Shivas RG, Singh SM, Souza-Motta CM, Thangavel R, Adhapure NN, Alexandrova AV, Alfenas AC, Alfenas RF, Alvarado P, Alves AL, Andrade DA, Andrade JP, Barbosa RN, Barili A, Barnes CW, Baseia IG, Bellanger J-M, Berlanas C, Bessette AE, Bessette AR, Biketova AYu, Bomfim FS, Brandrud TE, Bransgrove K, Brito ACQ, Cano-Lira JF, Cantillo T, Cavalcanti AD, Cheewangkoon R, Chikowski RS, Conforto C, Cordeiro TRL, Craine JD, Cruz R, Damm U, de Oliveira RJV, de Souza JT, de Souza HG, Dearnaley JDW, Dimitrov RA, Dovana F, Erhard A, Esteve-Raventós F, Félix CR, Ferisin G, Fernandes RA, Ferreira RJ, Ferro LO, Figueiredo CN, Frank JL, Freire KTLS, García D, Gené J, Gęsiorska A, Gibertoni TB, Gondra RAG, Gouliamova DE, Gramaje D, Guard F, Gusmão LFP, Haitook S, Hirooka Y, Houbraken J, Hubka V, Inamdar A, Iturriaga T, Iturrieta-González I, Jadan M, Jiang N, Justo A, Kachalkin AV, Kapitonov VI, Karadelev M, Karakehian J, Kasuya T, Kautmanová I, Kruse J, Kušan I, Kuznetsova TA, Landell MF, Larsson K-H, Lee HB, Lima DX, Lira CRS, Machado AR, Madrid H, Magalhães OMC, Majerova H, Malysheva EF, Mapperson RR, Marbach PAS, Martín MP, Martín-Sanz A, Matočec N, McTaggart AR, Mello JF, Melo RFR, Mešič A, Michereff SJ, Miller AN, Minoshima A, Molinero-Ruiz L, Morozova OV, Mosoh D, Nabe M, Naik R, Nara K, Nascimento SS, Neves RP, Olariaga I, Oliveira RL, Oliveira TGL, Ono T, Ordoñez ME, de M Ottoni A, Paiva LM, Pancorbo F, Pant B, Pawłowska J, Peterson SW, Raudabaugh DB, Rodríguez-Andrade E, Rubio E, Rusevska K, Santiago ALCMA, Santos ACS, Santos C, Sazanova NA, Shah S, Sharma J, Silva BDB, Siquier JL, Sonawane MS, Stchigel AM, Svetasheva T, Tamakeaw N, Telleria MT, Tiago PV, Tian CM, Tkalčec Z, Tomashevskaya MA, Truong HH, Vecherskii MV, Visagie CM, Vizzini A, Yilmaz N, Zmitrovich IV, Zvyagina EA, Boekhout T, Kehlet T, Læssøe T, Groenewald JZ. (2019) Fungal Planet description sheets: 868–950. Persoonia: Molecular Phylogeny and Evolution of Fungi 42: 291–473. 10.3767/persoonia.2019.42.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Hernández-Restrepo M, Schumacher RK, Cowan DA, Maggs-Kölling G, Marais E, Wingfield MJ, Yilmaz N, Adan OCG, Akulov A, Duarte E, Berraf-Tebbal A, Bulgakov TS, Carnegie AJ, de Beer ZW, Decock C, Dijksterhuis J, Duong TA, Eichmeier A, Hien LT, Houbraken JAMP, Khanh TN, Liem NV, Lombard L, Lutzoni FM, Miadlikowska JM, Nel WJ, Pascoe IG, Roets F, Roux J, Samson RA, Shen M, Spetik M, Thangavel R, Thanh HM, Thao LD, van Nieuwenhuijzen EJ, Zhang JQ, Zhang Y, Zhao LL, Groenewald JZ. (2021a) New and Interesting Fungi 4. Fungal Systematics and Evolution 7(1): 255–343. 10.3114/fuse.2021.07.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Cowan DA, Maggs‐Kölling G, Yilmaz N, Thangavel R, Wingfield MJ, Noordeloos ME, Dima B, Brandrud TE, Jansen GM, Morozova OV, Vila J, Shivas RG, Tan YP, Bishop‐Hurley S, Lacey E, Marney TS, Larsson E, Le Floch G, Lombard L, Nodet P, Hubka V, Alvarado P, Berraf‐Tebbal A, Reyes JD, Delgado G, Eichmeier A, Jordal JB, Kachalkin AV, Kubátová A, Maciá‐Vicente JG, Malysheva EF, Papp V, Rajeshkumar KC, Sharma A, Spetik M, Szabóová D, Tomashevskaya MA, Abad JA, Abad ZG, Alexandrova AV, Anand G, Arenas F, Ashtekar N, Balashov S, Bañares Á, Baroncelli R, Bera I, Biketova AYu, Blomquist CL, Boekhout T, Boertmann D, Bulyonkova TM, Burgess TI, Carnegie AJ, Cobo‐Diaz JF, Corriol G, Cunnington JH, da Cruz MO, Damm U, Davoodian N, de A Santiago ALCM, Dearnaley J, de Freitas LWS, Dhileepan K, Dimitrov R, Di Piazza S, Fatima S, Fuljer F, Galera H, Ghosh A, Giraldo A, Glushakova AM, Gorczak M, Gouliamova DE, Gramaje D, Groenewald M, Gunsch CK, Gutiérrez A, Holdom D, Houbraken J, Ismailov AB, Istel Ł, Iturriaga T, Jeppson M, Jurjević Ž, Kalinina LB, Kapitonov VI, Kautmanova I, Khalid AN, Kiran M, Kiss L, Kovács Á, Kurose D, Kusan I, Lad S, Læssøe T, Lee HB, Luangsa‐ard JJ, Lynch M, Mahamedi AE, Malysheva VF, Mateos A, Matočec N, Mešić A, Miller AN, Mongkolsamrit S, Moreno G, Morte A, Mostowfizadeh‐Ghalamfarsa R, Naseer A, Navarro‐Ródenas A, Nguyen TTT, Noisripoom W, Ntandu JE, Nuytinck J, Ostrý V, Pankratov TA, Pawłowska J, Pecenka J, Pham THG, Polhorský A, Posta A, Raudabaugh DB, Reschke K, Rodríguez A, Romero M, Rooney‐Latham S, Roux J, Sandoval‐Denis M, Smith MTh, Steinrucken TV, Svetasheva TY, Tkalčec Z, van der Linde EJ, vd Vegte M, Vauras J, Verbeken A, Visagie CM, Vitelli JS, Volobuev SV, Weill A, Wrzosek M, Zmitrovich IV, Zvyagina EA, Groenewald JZ. (2021b) Fungal planet description sheets: 1182–1283. Persoonia: Molecular Phylogeny and Evolution of Fungi 46: 313–528. 10.3767/persoonia.2021.46.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CW. (1997) Can three incongruence tests predict when data should be combined? Molecular Biology and Evolution 14(7): 733–740. 10.1093/oxfordjournals.molbev.a025813 [DOI] [PubMed]
- Dress AW, Huson DH. (2004) Constructing splits graphs. IEEE/ACM Transactions on Computational Biology and Bioinformatics 1(3): 109–115. 10.1109/TCBB.2004.27 [DOI] [PubMed] [Google Scholar]
- El-Gholl NE, Uchida JY, Alfenas AC, Schubert TS, Alfieri SA. (1992) Induction and description of perithecia of Calonectriaspathiphylli sp. nov. Mycotaxon 45: 285–300. [Google Scholar]
- Farris JS, Kallersjo M, Kluge AG, Bult C. (1995) Testing significance of incongruence. Cladistics 10(3): 315–319. 10.1111/j.1096-0031.1994.tb00181.x [DOI] [Google Scholar]
- Graça RN, Alfenas AC, Maffia LA, Titon M, Alfenas RF, Lau D, Rocabado JMA. (2009) Factors influencing infection of eucalypts by Cylindrocladiumpteridis. Plant Pathology 58(5): 971–981. 10.1111/j.1365-3059.2009.02094.x [DOI] [Google Scholar]
- Guerber JC, Correll JC. (2001) Characterization of Glomerellaacutata, the teleomorph of Colletotrichumacutatum. Mycologia 93(1): 216–229. 10.1080/00275514.2001.12063151 [DOI] [Google Scholar]
- Hepperle D. (2004) SeqAssem. Win32–Version. A sequence analysis tool contig assembler and trace data visualization tool for molecular sequences. Win32-Version. [Distributed by the author via:] http://www.sequentix.de
- Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, Vinh LS. (2018) UFBoot2: Improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35(2): 518–522. 10.1093/molbev/msx281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D. (2006) Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution 23(2): 254–267. 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- IBÁ (2021) IBÁ–Indústria Brasileira de Árvores. Relatório 2020. https://iba.org/datafiles/publicacoes/relatorios/relatorio-iba-2020.pdf
- IEA (2020) IEA–Instituto de Economia Agrícola. https://www.noticiasagricolas.com.br/cotacoes/silvicultura/preco-eucalipto
- Kang JC, Crous PW, Schoch CL. (2001) Species concepts in the Cylindrocladiumfloridanum and Cy.spathiphylli complexes (Hypocreaceae) based on multi-allelic sequence data, sexual compatibility and morphology. Systematic and Applied Microbiology 24(2): 206–217. 10.1078/0723-2020-00026 [DOI] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. (2019) MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20(4): 1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33(7): 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Taylor JW. (1990) Isolation of DNA from fungal mycelia and single spores. In: Innis MA, Gelfand DV, Sninsky JJ, White TJ. (Eds) PCR protocols: a guide to methods and applications.Academic Press, New York, 282–287. 10.1016/B978-0-12-372180-8.50038-X [DOI]
- Li JQ, Wingfield BD, Wingfield MJ, Barnes I, Fourie A, Crous PW, Chen SF. (2020) Mating genes in Calonectria and evidence for a heterothallic ancestral state. Persoonia. Molecular Phylogeny and Evolution of Fungi 45(1): 163–176. 10.3767/persoonia.2020.45.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JQ, Barnes I, Liu FF, Wingfield MJ, Chen SF. (2021) Global genetic diversity and mating type distribution of Calonectriapauciramosa: An important wide host-range plant pathogen. Plant Disease 105(6): 1648–1656. 10.1094/PDIS-05-20-1050-RE [DOI] [PubMed] [Google Scholar]
- Liu Q, Chen S. (2017) Two novel species of Calonectria isolated from soil in a natural forest in China. MycoKeys 26: 25–60. 10.3897/mycokeys.26.14688 [DOI] [Google Scholar]
- Liu YJ, Whelen S, Hall BD. (1999) Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16(12): 1799–1808. 10.1093/oxfordjournals.molbev.a026092 [DOI] [PubMed] [Google Scholar]
- Liu QL, Li JQ, Wingfield MJ, Duong TA, Wingfield BD, Crous PW, Chen SF. (2020) Reconsideration of species boundaries and proposed DNA barcodes for Calonectria. Studies in Mycology 97: e100106. [71 pp.] 10.1016/j.simyco.2020.08.001 [DOI] [PMC free article] [PubMed]
- Liu L, Wu W, Chen S. (2021) Species diversity and distribution characteristics of Calonectria in five soil layers in a eucalyptus plantation. Journal of Fungi (Basel, Switzerland) 7(10): e857. 10.3390/jof7100857 [DOI] [PMC free article] [PubMed]
- Lombard L, Crous PW, Wingfield BD, Wingfield MJ. (2010a) Multigene phylogeny and mating tests reveal three cryptic species related to Calonectriapauciramosa. Studies in Mycology 66: 15–30. 10.3114/sim.2010.66.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L, Crous PW, Wingfield BD, Wingfield MJ. (2010b) Phylogeny and systematics of the genus Calonectria. Studies in Mycology 66: 31–69. 10.3114/sim.2010.66.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L, Crous PW, Wingfield BD, Wingfield MJ. (2010c) Species concepts in Calonectria (Cylindrocladium). Studies in Mycology 66: 1–13. 10.3114/sim.2010.66.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L, Wingfield MJ, Alfenas AC, Crous PW. (2016) The forgotten Calonectria collection: Pouring old wine into new bags. Studies in Mycology 85(1): 159–198. 10.1016/j.simyco.2016.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda IDS, Auer CG, dos Santos ÁF, Ferreira MA, Tambarussi EV, da Silva RAF, Rezende EH. (2021) Occurrence of Calonectria leaf blight in Eucalyptusbenthamii progenies and potential for disease resistance. Tropical Plant Pathology 46(3): 254–264. 10.1007/s40858-021-00426-4 [DOI] [Google Scholar]
- Mohali SR, Stewart JE. (2021) Calonectriavigiensis sp. nov. (Hypocreales, Nectriaceae) associated with dieback and sudden-death symptoms of Theobromacacao from Mérida state, Venezuela. Botany 99(11): 683–693. 10.1139/cjb-2021-0050 [DOI] [Google Scholar]
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. (2015) IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32(1): 268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander JAA. (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
- O’Donnell K, Cigelnik E. (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7(1): 103–116. 10.1006/mpev.1996.0376 [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Kistler HC, Cigelnik E, Ploetz RC. (1998) Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences of the United States of America 95(5): 2044–2049. 10.1073/pnas.95.5.2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham NQ, Barnes I, Chen S, Liu F, Dang QN, Pham TQ, Lombard L, Crous PW, Wingfield MJ. (2019) Ten new species of Calonectria from Indonesia and Vietnam. Mycologia 111(1): 78–102. 10.1080/00275514.2018.1522179 [DOI] [PubMed] [Google Scholar]
- Pham NQ, Marincowitz S, Chen S, Yaparudin Y, Wingfield MJ. (2022) Calonectria species, including four novel taxa, associated with Eucalyptus in Malaysia. Mycological Progress 21(1): 181–197. 10.1007/s11557-021-01768-8 [DOI] [Google Scholar]
- Pires BM. (2000) Efeito da desrama artificial no crescimento e qualidade da madeira de Eucalyptusgrandis para serraria. MSc Dissertation, Universidade Federal de Viçosa, Viçosa, Brasil. https://locus.ufv.br//handle/123456789/11170
- Poltronieri LS, Alfenas RF, Verzignassi JR, Alfenas AC, Benchimol RL, Poltronieri TPDS. (2011) Leaf blight and defoliation of Eugenia spp. caused by Cylindrocladiumcandelabrum and C.spathiphylli in Brazil. Summa Phytopathologica 37(2): 147–149. 10.1590/S0100-54052011000200011 [DOI] [Google Scholar]
- Posada D, Crandall KA. (1998) Modeltest: Testing the model of DNA substitution. Bioinformatics (Oxford, England) 14(9): 817–818. 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Pulrolnik K, Reis GGD, Reis MDGF, Monte MA, Fontan IDCI. (2005) Crescimento de plantas de clones de Eucalyptusgrandis [Hill ex Maiden] submetidas a diferentes tratamentos de desrama artificial, na região do cerrado. Revista Árvore 29(4): 495–505. 10.1590/S0100-67622005000400001 [DOI] [Google Scholar]
- Quaedvlieg W, Kema GHJ, Groenewald JZ, Verkley GJM, Seifbarghi S, Razavi M, Gohari AM, Mehrabi R, Crous PW. (2011) Zymoseptoria gen. nov.: A new genus to accommodate Septoria-like species occurring on graminicolous hosts. Persoonia. Molecular Phylogeny and Evolution of Fungi 26(1): 57–69. 10.3767/003158511X571841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. (2009) FigTree, a graphical viewer of phylogenetic trees. http://tree.bio.ed.ac.uk/software/figtree/
- Rambaut A, Drummond AJ. (2007) Tracer v1.5. http://tree.bio.ed.ac.uk/software/tracer/
- Reeb V, Lutzoni F, Roux C. (2004) Contribution of RPB2 to multilocus phylogenetic studies of the euascomycetes (Pezizomycotina, Fungi) with special emphasis on the lichen-forming Acarosporaceae and evolution of polyspory. Molecular Phylogenetics and Evolution 32(3): 1036–1060. 10.1016/j.ympev.2004.04.012 [DOI] [PubMed] [Google Scholar]
- Reis A, Mafia RG, Silva PP, Lopes CA, Alfenas AC. (2004) Cylindrocladiumspathiphylli, causal agent of Spathiphyllum root and collar rot in the Federal District-Brazil. Fitopatologia Brasileira 29(1): 102–102. 10.1590/S0100-41582004000100017 [DOI] [Google Scholar]
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61(3): 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck J, Teslenko M, Nylander JAA. (2019) MrBayes version 3.2 manual: tutorials and model summaries. https://nbisweden.github.io/MrBayes/manual.html
- Schoch CL, Crous PW, Wingfield BD, Wingfield MJ. (1999) The Cylindrocladiumcandelabrum species complex includes four distinct mating populations. Mycologia 91(2): 286–298. 10.1080/00275514.1999.12061019 [DOI] [Google Scholar]
- Soares TP, Pozza EA, Pozza AA, Mafia RG, Ferreira MA. (2018) Calcium and potassium imbalance favours leaf blight and defoliation caused by Calonectriapteridis in Eucalyptus plants. Forests 9(12): e782. 10.3390/f9120782 [DOI]
- Sullivan J. (1996) Combining data with different distributions of among-site rate variation. Systematic Biology 45(3): 375–380. 10.1093/sysbio/45.3.375 [DOI] [Google Scholar]
- Swofford DL. (2003) PAUP*. Phylogenetic analysis using parsimony (*and other methods). V. 4.0b10. Sinauer Associates, Sunderland, Massachusetts, USA.
- Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC. (2000) Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology 31(1): 21–32. 10.1006/fgbi.2000.1228 [DOI] [PubMed] [Google Scholar]
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. (2016) W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research 44(W1): W232–W235. 10.1093/nar/gkw256 [DOI] [PMC free article] [PubMed]
- Vitale A, Crous PW, Lombard L, Polizzi G. (2013) Calonectria diseases on ornamental plants in Europe and the Mediterranean basin: An overview. Journal of Plant Pathology 95(3): 463–476. 10.4454/JPP.V95I3.007 [DOI] [Google Scholar]
- Wang QC, Liu QL, Chen SF. (2019) Novel species of Calonectria isolated from soil near Eucalyptus plantations in southern China. Mycologia 111(6): 1028–1040. 10.1080/00275514.2019.1666597 [DOI] [PubMed] [Google Scholar]
- Wingfield MJ, Brockerhoff EG, Wingfield BD, Slippers B. (2015) Planted forest health: The need for a global strategy. Science 349(6250): 832–836. 10.1126/science.aac6674 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1–S6
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Enrique I. Sanchez-Gonzalez, Thaissa de Paula Farias Soares, Talyta Galafassi Zarpelon, Edival Angelo Valverde Zauza, Reginaldo Gonçalves Mafia, Maria Alves Ferreira
Data type
Phylogenetic trees (pdf file)
Explanation note
Figure S1. Phylogenetic tree based on maximum likelihood analysis of act gene region. Bootstrap support values ≥ 80% for maximum parsimony (MP), Ultrafast bootstrap support values ≥ 95% for maximum likelihood (ML), and posterior probability (PP) values ≥ 0.95 from BI analyses are presented at the nodes (MP/ML/PP). Bootstrap values below 80% (MP), 95% (ML) and posterior probabilities below 0.80 are marked with “-”. Ex-type isolates are indicated by “▲”, isolates highlighted in bold were sequenced in this study, and novel species are in blue and orange. C.gracilipes was used as outgroup. The scale bar indicates the number of nucleotide substitutions per site. Figure S2. Phylogenetic tree based on maximum likelihood analysis of cmdA gene region. Bootstrap support values ≥ 80% for maximum parsimony (MP), Ultrafast bootstrap support values ≥ 95% for maximum likelihood (ML), and posterior probability (PP) values ≥ 0.95 from BI analyses are presented at the nodes (MP/ML/PP). Bootstrap values below 80% (MP), 95% (ML) and posterior probabilities below 0.80 are marked with “-”. Ex-type isolates are indicated by “▲”, isolates highlighted in bold were sequenced in this study, and novel species are in blue and orange. C.gracilipes was used as outgroup. The scale bar indicates the number of nucleotide substitutions per site. Figure S3. Phylogenetic tree based on maximum likelihood analysis of his3 gene region. Bootstrap support values ≥ 80% for maximum parsimony (MP), Ultrafast bootstrap support values ≥ 95% for maximum likelihood (ML), and posterior probability (PP) values ≥ 0.95 from BI analyses are presented at the nodes (MP/ML/PP). Bootstrap values below 80% (MP), 95% (ML) and posterior probabilities below 0.80 are marked with “-”. Ex-type isolates are indicated by “▲”, isolates highlighted in bold were sequenced in this study, and novel species are in blue and orange. C.gracilipes was used as outgroup. The scale bar indicates the number of nucleotide substitutions per site. Figure S4. Phylogenetic tree based on maximum likelihood analysis of rpb2 gene region. Bootstrap support values ≥ 80% for maximum parsimony (MP), Ultrafast bootstrap support values ≥ 95% for maximum likelihood (ML), and posterior probability (PP) values ≥ 0.95 from BI analyses are presented at the nodes (MP/ML/PP). Bootstrap values below 80% (MP), 95% (ML) and posterior probabilities below 0.80 are marked with “-”. Ex-type isolates are indicated by “▲”, isolates highlighted in bold were sequenced in this study, and novel species are in blue and orange. C.gracilipes was used as outgroup. The scale bar indicates the number of nucleotide substitutions per site. Figure S5. Phylogenetic tree based on maximum likelihood analysis of tef1 gene region. Bootstrap support values ≥ 80% for maximum parsimony (MP), Ultrafast bootstrap support values ≥ 95% for maximum likelihood (ML), and posterior probability (PP) values ≥ 0.95 from BI analyses are presented at the nodes (MP/ML/PP). Bootstrap values below 80% (MP), 95% (ML) and posterior probabilities below 0.80 are marked with “-”. Ex-type isolates are indicated by “▲”, isolates highlighted in bold were sequenced in this study, and novel species are in blue and orange. C.gracilipes was used as outgroup. The scale bar indicates the number of nucleotide substitutions per site. Figure S6. Phylogenetic tree based on maximum likelihood analysis of tub2 gene region. Bootstrap support values ≥ 80% for maximum parsimony (MP), Ultrafast bootstrap support values ≥ 95% for maximum likelihood (ML), and posterior probability (PP) values ≥ 0.95 from BI analyses are presented at the nodes (MP/ML/PP). Bootstrap values below 80% (MP), 95% (ML) and posterior probabilities below 0.80 are marked with “-”. Ex-type isolates are indicated by “▲”, isolates highlighted in bold were sequenced in this study, and novel species are in blue and orange. C.gracilipes was used as outgroup. The scale bar indicates the number of nucleotide substitutions per site.