Abstract
The fungal genus Phyllosticta has been reported from all around the world and accommodates numerous pathogenic and endophytic species isolated from a wide range of plant hosts. Based on multilocus phylogenies from a combined dataset of genes encoding internal transcribed spacer (ITS), large subunit of ribosomal RNA (LSU rDNA), translation elongation factor 1 alpha (TEF1α), actin (ACT) and glycerol-3-phosphate dehydrogenase (GPDH), in conjunction with morphological characteristics, we describe two new species P.oblongifoliaesp. nov. and P.pterospermisp. nov., as well as a new Chinese record P.capitalensis. Their similarity and dissimilarity to morphologically-allied and phylogenetically-related species are also annotated and discussed.
Keywords: multigene phylogeny, new species, taxonomy
Introduction
Phyllosticta Pers. was introduced by Persoon (1818) and P.convallariae Pers. was designated as the type species (Donk 1968). Since Phyllosticta is distinct from other genera in that family, Seaver (1922) treated it in the family Phyllostictaceae Fr. of the order Phyllostictites. Nevertheless, Phyllosticta was accommodated in the family Botryosphaeriaceae Theiss. & Syd. (in Botryosphaeriales C.L. Schoch et al.) in several major studies (e.g. Crous et al. 2006; Schoch et al. 2006; Liu et al. 2012). However, the phylogenetic analyses by Wikee et al. (2013a) allocated Phyllosticta in a clade sister to Botryosphaeriaceae. As a result, the genus is currently accepted in the family Phyllostictaceae, in the order Botryosphaeriales.
A total of 3,213 names are documented for Phyllosticta in the Index Fungorum (accessed on 31 March 2022) (Hongsanan et al. 2020; Wijayawardene et al. 2020). However, many of these names have been synonymised (van der Aa and Vanev 2002). Currently, 1499 species are accepted in the genus (Bánki et al. 2022). The majority of the Phyllosticta species are known to infect a broad range of hosts and cause plant diseases, such as leaf and fruit spots (Wikee et al. 2013a; Zhou et al. 2015; Lin et al. 2017). Van der Aa (1973) revised this genus and established his own morphological criteria, i.e. aseptate pycnidia and hyaline conidia that are usually covered by a mucoid layer and bear a single apical appendage. According to these criteria, van der Aa and Vanev (2002) re-classified Phyllosticta and accepted 190 species. Other species were recombined into Asteromella Pass. & Thüm., Diaporthe Fuckel, Guignardia Viala & Ravaz, Leptodothiorella Höhn. and Phoma Sacc. A rare tropical species from the Brazilian Cerrado, P.xylopiae-sericeae Furlan. & Dianese, although morphologically well documented (Furlanetto and Dianese 1998), remains to be molecularly characterised. Recently, DNA sequencing of orthologous genes has greatly improved our knowledge of fungal phylogeny. Since van der Aa and Vanev (2002), several studies have shown that phylogenetic analyses can help delineate species in Phyllosticta (Baayen et al. 2002; Wulandari et al. 2009; Glienke et al. 2011; Wikee et al. 2011). More recently, new species of Phyllosticta have been increasingly described, based on a combination of molecular data and morphological features (Su and Cai 2012; Wang et al. 2012, 2013; Wong et al. 2012; Zhang et al. 2012, 2013; Wikee et al. 2013a; Wulandari et al. 2013; Crous et al. 2014, 2015, 2016, 2017, 2018, 2019, 2021; Zhou et al. 2015; Guarnaccia et al. 2017; Lin et al. 2017; Hattori et al. 2020; Norphanphoun et al. 2020). Norphanphoun et al. (2020) assembled all species denoted as Phyllosticta in GenBank, analysing a comprehensive dataset of five loci and consequently proposing six species complexes, viz. P.capitalensis species complex, P.concentrica species complex, P.cruenta species complex, P.owaniana species complex, P.rhodorae species complex and P.vaccinii species complex.
Hainan Province (18°10'–20°10'N, 108°37'–111°05'E) is an island in southern China, with an annual mean temperature of 22–27 °C and an annual precipitation of 1000–2600 mm. Bawangling National Forest Park is located in the southwest of Hainan, with a typical tropical rainforest climate. Fungi associated with leaf spots were collected from Rhapisexcelsa, Garciniaoblongifolia and Pterospermumheterophyllum. Using sequences of five gene loci, which include the internal transcribed spacer of ribosomal RNA (ITS rDNA), large subunit of ribosomal RNA (LSU rDNA), translation elongation factor 1 alpha (TEF1α), actin (ACT) and glycerol-3-phosphate dehydrogenase (GPDH). We also incorporated their morphology and then identified these fungi as three species of the P.capitalensis species complex, including two new species, as well as a species new to China, based on morphology and phylogenetic analyses.
Materials and methods
Isolation and morphological studies
Leaves of Rhapisexcelsa, Garciniaoblongifolia and Pterospermumheterophyllum showing necrotic spots were collected at the Bawangling National Forest Park, Hainan Province, China. Isolates were obtained using a tissue isolation method (Jiang et al. 2021). Fragments (5 × 5 mm) were taken from the margin of leaf lesions, surface-sterilised by immersing consecutively in 75% ethanol solution for 1 min, 5% sodium hypochlorite solution for 30 s and then rinsed in sterile distilled water for 1 min (Jiang et al. 2021). The sterilised fragments were dried with sterilised paper towels and placed on potato dextrose agar (PDA: 200 g potato, 20 g dextrose, 20 g agar, 1000 ml distilled water, pH 7.0) and incubated at 25 °C for 2–4 days. Subsequently, portions of agar with fungal mycelium from the periphery of the colonies were transferred into new PDA plates and photographed on the 7th and 15th days by a digital camera (Canon Powershot G7X). An inoculum of the purified colonies was placed on 2% malt extract agar (MEA:20 g malt extract, 20 g soy peptone, 15 g agar, 1000 ml distilled water, pH 5.6) and incubated under continuous near-UV light at room temperature to promote sporulation (Braun et al. 2018). Micromorphological characters were observed using an Olympus SZX10 stereomicroscope and Olympus BX53 microscope, all fitted with an Olympus DP80 high-definition colour digital camera to photo-document fungal structures. All fungal strains were stored in 10% sterilised glycerine at 4 °C for further studies. Structural measurements were taken using the Digimizer software (https://www.digimizer.com/), with thirty measurements taken for each character. The holotype specimens were deposited in the Herbarium of Plant Pathology, Shandong Agricultural University (HSAUP). Ex-holotype living cultures were deposited in the Shandong Agricultural University Culture Collection (SAUCC). Taxonomic information of the new taxa was submitted to MycoBank (http://www.mycobank.org).
DNA extraction and sequencing
Genomic DNA was extracted from fungal mycelia grown on PDA, using a modified cetyltrimethylammonium bromide (CTAB) protocol as described in Guo et al. (2000). The internal transcribed spacer region (ITS) with intervening 5.8S rRNA gene, large subunit of rRNA gene (LSU), translation elongation factor 1-alpha gene (tef1), actin gene (ACT) and glyceraldehyde-3-phosphate dehydrogenase gene (GPDH) were amplified and sequenced by using the primer pairs ITS5/ITS4 (White et al. 1990), LROR/LR5 (White et al. 1990), EF1-728F/EF2 (O’Donnell et al. 1998; Carbone and Kohn 1999), ACT-512F/ACT-783R (Carbone and Kohn 1999) and Gpd1-LM/Gpd2-LM (Myllys et al. 2002), respectively.
PCR was performed using an Eppendorf Master Thermocycler (Hamburg, Germany). Amplification reactions were carried out in a 25 μl reaction volume, which contained 12.5 μl 2×Green Taq Mix (Vazyme, Nanjing, China), 1 μl of each forward and reverse primer (10 μM stock; Biosune, Shanghai, China), 1 μl template genomic DNA (approximately 10 ng/μl) and 9.5 μl distilled deionised water. PCR parameters were as follows: 94 °C for 5 min; 35 cycles of denaturation at 94 °C for 30 s, annealing at a suitable temperature for 50 s and extension at 72 °C for 1 min; and a final elongation step at 72 °C for 10 min. The suitable annealing temperatures for the genes were 55 °C for ITS, 51 °C for LSU, 52 °C for ACT, 48 °C for tef1 and 52 °C for GPDH, respectively. PCR products were checked through a 1% agarose gel electrophoresis, stained with GelRed and visualised by a UV light. Sequencing was performed bi-directionally by Biosune Company Limited (Shanghai, China). Consensus sequences were obtained using MEGA v. 7.0 (Kumar et al. 2016). All sequences generated in this study were deposited in GenBank (Table 1).
Table 1.
Species and GenBank accession numbers of DNA sequences used in this study.
| Species1 | Voucher2 | Host/Substrate | Country | GenBank accession number | ||||
|---|---|---|---|---|---|---|---|---|
| ITS | LSU | tef1 | ACT | GPDH | ||||
| Phyllostictaacaciigena | CPC 28295 * | Acaciasuaveolens | Australia | KY173433 | KY173523 | ‒ | KY173570 | ‒ |
| P.aloeicola | CPC 21020 * | Aloeferox | South Africa | KF154280 | KF206214 | KF289193 | KF289311 | KF289124 |
| CPC 21021 | Aloeferox | South Africa | KF154281 | KF206213 | KF289194 | KF289312 | KF289125 | |
| P.ardisiicola | NBRC 102261 * | Ardisiacrenata | Japan | AB454274 | AB454274 | ‒ | AB704216 | ‒ |
| P.aristolochiicola | BRIP 53316 * | Aristolochiaacuminata | Australia | JX486129 | ‒ | ‒ | ‒ | ‒ |
| P.azevinhi | MUCC0088 | Ilexpedunculosa | Japan | AB454302 | AB454302 | ‒ | AB704226 | ‒ |
| P.beaumarisii | CBS 535.87 | Muehlenbekiaadpressa | Australia | AY042927 | KF306229 | KF289170 | KF306232 | KF289074 |
| P.brazillianiae | LGMF 330 * | Mangiferaindica | Brazil | JF343572 | KF206217 | JF343593 | JF343656 | JF343758 |
| LGMF 333 | Mangiferaindica | Brazil | JF343574 | KF206216 | JF343595 | JF343658 | JF343760 | |
| P.camelliae | MUCC0059 | Camelliajaponica | Japan | AB454290 | AB454290 | AB704223 | ||
| P.capitalensis | CBS 128856 * | Stanhopeagraveolens | Brazil | JF261465 | KF206255 | JF261507 | KF289289 | JF343776 |
| CBS 226.77 | Baccaurearamiflora | Brazil | FJ538336 | KF206289 | FJ538394 | FJ538452 | JF343718 | |
| CBS 356.52 | Paphiopedilumcallosum | Germany | FJ538342 | KF206300 | FJ538400 | FJ538458 | KF289087 | |
| CBS 100175 | Ilex sp. | Not given | FJ538320 | KF206327 | FJ538378 | FJ538436 | JF343699 | |
| CBS 101228 | Citrus sp. | Brazil | FJ538319 | KF206325 | FJ538377 | FJ538435 | KF289086 | |
| CBS 114751 | Nepheliumlappaceum | Hawaii | EU167584 | EU167584 | FJ538407 | FJ538465 | KF289088 | |
| CBS 115047 | Vaccinium sp. | New Zealand | FJ538323 | KF206318 | FJ538381 | FJ538439 | KF289077 | |
| CBS 115049 | Aspidospermapolyneuron | Brazil | FJ538324 | KF206317 | FJ538382 | FJ538440 | KF289084 | |
| CBS 117118 | Bowdichianitida | Brazil | FJ538339 | JQ743603 | FJ538397 | FJ538455 | KF289090 | |
| CBS 120428 | Musaacuminata | Indonesia | JN692544 | KF206315 | JN692532 | JN692520 | JN692509 | |
| CBS 123373 | Sansevieria sp. | Netherlands | FJ538341 | JQ743604 | FJ538399 | FJ538457 | JF343703 | |
| CPC 13987 | Protearepens | Portugal | KF206183 | KF206281 | KF289176 | KF289263 | KF289083 | |
| CPC 16592 | Citruslimon | Argentina | KF206187 | KF206270 | KF289273 | KF289178 | KF289092 | |
| CPC 17468 | Cymbidium sp. | Brazil | KF206188 | KF206259 | KF289189 | KF289284 | KF289120 | |
| CPC 20256 | Ophiopogonjaponicus | Thailand | KC291337 | KF206247 | KC342557 | KC342534 | KF289089 | |
| CPC 20257 | Ficusbenjamina | Thailand | KC291338 | KF206246 | KC342558 | KC342535 | KF289099 | |
| LGMF219 | Citrussinensis | Brazil | KF206202 | KF206220 | JF261490 | KF289306 | JF343737 | |
| LGMF220 | Citrussinensis | Brazil | KF206203 | KF206219 | JF261488 | KF289307 | JF343735 | |
| LGMF222 | Citrussinensis | Brazil | KF206204 | KF206218 | JF261492 | KF289308 | JF343739 | |
| SAUCC210144 | Rhapisexcelsa | China | OM571175 | OM571179 | OM640045 | OM640047 | OM640049 | |
| SAUCC210148 | Rhapisexcelsa | China | OM571176 | OM571180 | OM640046 | OM640048 | OM640050 | |
| P.carochlae | CGMCC 3.17317 * | Caryotaochlandra | China | KJ847422 | ‒ | KJ847444 | KJ847430 | KJ847438 |
| CGMCC 3.17318 | Caryotaochlandra | China | KJ847423 | ‒ | KJ847445 | KJ847431 | KJ847439 | |
| P.cavendishii | BRIP 554196 * | Musa cv. Formosana | Taiwan | JQ743562 | ‒ | KF009743 | KF014080 | ‒ |
| BRIP 58008 | Banana | Australia | KC988365 | ‒ | KF009742 | KF014071 | ‒ | |
| P.cordylinophila | CPC 20261 * | Cordylinefruticosa | Thailand | KF170287 | KF206242 | KF289172 | KF289295 | KF289076 |
| CPC 20277 | Cordylinefruticosa | Thailand | KF170288 | KF206228 | KF289171 | KF289301 | KF289075 | |
| P.eugeniae | CBS 445.82 | Eugeniaaromatica | Indonesia | AY042926 | KF206288 | KF289208 | KF289246 | KF289139 |
| P.fallopiae | MUCC0113 * | Fallopiajaponica | Japan | AB454307 | AB454307 | ‒ | ‒ | ‒ |
| P.harai | MUCC0043 | Aucubajaponica | Japan | AB454281 | AB454281 | ‒ | AB704219 | ‒ |
| P.hubeiensis | CGMCC 3.14986 * | Viburnumodoratissimim | China | JX025037 | ‒ | JX025042 | JX025032 | JX025027 |
| CGMCC 3.14987 | Viburnumodoratissimim | China | JX025038 | ‒ | JX025043 | JX025033 | JX025028 | |
| P.ilicis-aquifolii | CGMCC 3.14358 * | Ilexaquifolium | China | JN692538 | ‒ | JN692526 | JN692514 | ‒ |
| CGMCC 3.14359 | Ilexaquifolium | China | JN692539 | ‒ | JN692527 | JN692515 | ‒ | |
| P.maculata | CPC 18347 * | Musa cv. Goly-goly pot-pot | Australia | JQ743570 | ‒ | KF009700 | KF014016 | ‒ |
| BRIP 46622 | Musa cv. Goly-goly pot-pot | Australia | JQ743567 | ‒ | KF009692 | KF014013 | ‒ | |
| P.mangiferae | IMI 260.576 * | Mangiferaindica | India | JF261459 | KF206222 | JF261501 | JF343641 | JF343748 |
| CPC 20260 | Arecaceae | Thailand | KF206193 | KF206243 | KF289187 | KF289294 | KF289114 | |
| P.mangifera-indica | MFLUCC 10–0029 * | Mangiferaindica | Thailand | KF170305 | KF206240 | KF289190 | KF289296 | KF289121 |
| P.miurae | MUCC0065 | Linderapraecox | Japan | AB454291 | AB454291 | ‒ | AB704224 | ‒ |
| P.musaechinensis | GZAAS6.1247 | Musa. sp. | China | KF955294 | ‒ | KM816639 | KM816627 | KM816633 |
| GZAAS6.1384 | Musa. sp. | China | KF955295 | ‒ | KM816640 | KM816628 | KM816634 | |
| P.musarum | BRIP57803 | Musa. sp. | Malaysia | JX997138 | ‒ | KF009737 | KF014055 | ‒ |
| BRIP58028 | Musa. sp. | Australia | KC988377 | ‒ | KF009738 | KF014054 | ‒ | |
| P.oblongifolae | SAUCC210055 | Garciniaoblongifolia | China | OM248442 | OM232085 | OM273890 | OM273894 | OM273898 |
| SAUCC210054 | Garciniaoblongifolia | China | OM248443 | OM232086 | OM273891 | OM273895 | OM273899 | |
| SAUCC210053 | Garciniaoblongifolia | China | OM248444 | OM232087 | OM273892 | OM273896 | OM273900 | |
| SAUCC210052 * | Garciniaoblongifolia | China | OM248445 | OM232088 | OM273893 | OM273897 | OM273901 | |
| P.paracapitalensis | CPC 26517 * | Citrusfloridana | Italy | KY855622 | KY855796 | KY855951 | KY855677 | KY855735 |
| CPC 26518 | Citrusfloridana | Italy | KY855623 | KY855797 | KY855952 | KY855678 | KY855736 | |
| CPC 26700 | Citrusfloridana | Italy | KY855624 | KY855798 | KY855953 | KY855679 | KY855737 | |
| CPC 26701 | Citrusfloridana | Italy | KY855625 | KY855799 | KY855954 | KY855680 | KY855738 | |
| CPC 26805 | Citrusfloridana | Italy | KY855626 | KY855800 | KY855955 | KY855681 | KY855739 | |
| CPC 26806 | Citrusfloridana | Italy | KY855627 | KY855801 | KY855956 | KY855682 | KY855740 | |
| CPC 28120 | Citruslimon | Spain | KY855628 | KY855802 | KY855957 | KY855683 | KY855741 | |
| P.paracapitalensis | CPC 28121 | Citruslimon | Spain | KY855629 | KY855803 | KY855958 | KY855684 | KY855742 |
| CPC 28122 | Citruslimon | Spain | KY855630 | KY855804 | KY855959 | KY855685 | KY855743 | |
| CPC 28123 | Citruslimon | Spain | KY855631 | KY855805 | KY855960 | KY855686 | KY855744 | |
| CPC 28127 | Citruslimon | Spain | KY855632 | KY855806 | KY855961 | KY855687 | KY855745 | |
| CPC 28128 | Citruslimon | Spain | KY855633 | KY855807 | KY855962 | KY855688 | KY855746 | |
| CPC 28129 | Citruslimon | Spain | KY855634 | KY855808 | KY855963 | KY855689 | KY855747 | |
| P.parthenocissi | CBS 111645 * | Parthenocissusquinquefolia | USA | EU683672 | ‒ | JN692530 | JN692518 | ‒ |
| P.partricuspidatae | NBRC 9466 * | Parthenocissustricuspidata | Japan | KJ847424 | ‒ | KJ847446 | KJ847432 | KJ847440 |
| NBRC 9757 | Parthenocissustricuspidata | Japan | KJ847425 | ‒ | KJ847447 | KJ847433 | KJ847441 | |
| P.philoprina | CBS 587.69 | Ilexaquifolium | Spain | KF154278 | KF206297 | KF289206 | KF289250 | KF289137 |
| CBS 616.72 | Ilexaquifolium | Germany | KF154279 | KF206296 | KF289205 | KF289251 | KF289136 | |
| P.pterospermi | SAUCC210104 * | Pterospermumheterophyllum | China | OM249954 | OM249956 | OM273902 | OM273904 | OM273906 |
| SAUCC210406 | Pterospermumheterophyllum | China | OM249955 | OM249957 | OM273903 | OM273905 | OM273907 | |
| P.rhizophorae | NCYUCC 19–0352 * | Rhizophorastylosa | Taiwan | MT360030 | MT360039 | ‒ | MT363248 | MT363250 |
| NCYUCC 19–0358 | Rhizophorastylosa | Taiwan | MT360031 | MT360040 | ‒ | MT363249 | MT363251 | |
| P.schimae | CGMCC 3.14354 * | Schimasuperba | China | JN692534 | ‒ | JN692522 | JN692510 | JN692506 |
| P.schimicola | CGMCC 3.17319 * | Schimasuperba | China | KJ847426 | ‒ | KJ847448 | KJ847434 | KJ854895 |
| CGMCC 3.17320 | Schimasuperba | China | KJ847427 | ‒ | KJ847449 | KJ847435 | KJ854896 | |
| P.styracicola | LC1642 * | Styraxgradiflorus | China | JX025040 | ‒ | JX025045 | JX025035 | JX025030 |
| P.vitis-rotundifoliae | CGMCC 3.17321 | Vitisrotundifolia | USA | KJ847429 | ‒ | KJ847451 | KJ847437 | KJ847443 |
| CGMCC 3.17322 * | Vitisrotundifolia | USA | KJ847428 | ‒ | KJ847450 | KJ847436 | KJ847442 | |
1Newly generated sequences in this study are in bold. 2Isolates marked with “*” are ex-type or ex-epitype strains.
Phylogenetic analyses
The generated consensus sequences were subjected to BLAST searches to identify closely-related sequences in the NCBI’s GenBank nucleotide database (Zhang et al. 2000). For phylogenetic inferences, based on ITS-LSU-tef1-ACT-GPDH sequences, a subset of sequences from the alignments of Norphanphoun et al. (2020) was used as the backbone. Newly-generated sequences in this study were aligned with related sequences retrieved from GenBank (Table 1) using MAFFT 7 online tool with the Auto strategy (Katoh et al. 2019; http://mafft.cbrc.jp/alignment/server/). To establish the identity of the isolates at species level, phylogenetic analyses were first performed for each locus individually and then all loci were concatenated together for a unified analysis (ITS-LSU-tef1-ACT-GPDH).
Phylogenetic analyses were carried out with Maximum Likelihood (ML) and Bayesian Inference (BI) algorithms. The best evolutionary model for each partition was determined using MrModelTest v. 2.3 (Nylander 2004) and incorporated into the BI analyses. ML and BI run on the CIPRES Science Gateway portal (https://www.phylo.org/; Miller et al. 2012) using RAxML-HPC2 on XSEDE v. 8.2.12 (Stamatakis 2014) and MrBayes on XSEDE v. 3.2.7a (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003; Ronquist et al. 2012), respectively. Default parameters were used for the ML analyses and the rapid bootstrapping with the automatic halt option was set for the BI analyses. Bayesian Inference included four parallel runs of 10,000,000 generations, with the stop rule option and a sampling frequency of 1,000 generations. Burn-in fraction was set to 0.25 and posterior probabilities (PP) were determined from the remaining trees. All resultant trees were plotted using FigTree v. 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree) and the layout of the trees was edited in Adobe Illustrator CC 2019.
Results
Phylogenetic analyses
A total of 86 isolates representing the Phyllosticta species were phylogenetically analysed, of which 84 isolates in the P.capitalensis species complex were considered as ingroup and two strains of Phyllostictahubeiensis (CGMCC 3.14986, CGMCC 3.14987) in the P.cruenta species complex were used as outgroup. The final alignment contained 2665 concatenated characters, viz. 1–733 (ITS), 734–1499 (LSU), 1500–1790 (tef1), 1791–2042 (ACT), 2043–2665 (GPDH). Of these characters, 1964 were constant, 126 were variable and parsimony-uninformative and 575 were parsimony-informative. MrModelTest recommended that the Bayesian Inference should use Dirichlet base frequencies for the ITS, LSU, tef1, ACT and GPDH data partitions. The GTR+I+G model was proposed for ITS, LSU and GPDH, while HKY+G for tef1 and ACT. The MCMC analysis of the five concatenated genes was run for 1,520,000 generations, resulting in 30,402 trees. The initial 7,600 trees generated in the burn-in phase were discarded, while the remaining trees were used to calculate posterior probabilities in the majority rule consensus trees. The alignment contained a total of 876 unique site patterns (ITS: 358, LSU: 69, tef1: 170, ACT: 137, GPDH: 142). The topology of the ML tree confirmed the tree topology obtained from the Bayesian Inference and, therefore, only the ML tree is presented (Fig. 1). The 86 strains were assigned to 34 species, based on the five-gene phylogeny (Fig. 1). The present study revealed three species, viz. Phyllostictaoblongifolae sp. nov., P.pterospermi sp. nov. and P.capitalensis. The P.oblongifolae sp. nov. was a sister group to P.eugeniae (0.98/81) and the P.pterospermi sp. nov. was closely related to P.mangiferae (0.99/92).
Figure 1.
Phylogram of the Phyllostictacapitalensis species complex, based on a concatenated ITS, LSU, tef1, ACT and GPDH sequence alignment, with Phyllostictahubeiensis (CGMCC 3.14986, CGMCC 3.14987) of the P.cruenta species complex serving as outgroup. Bayesian Inference posterior probabilities and Maximum Likelihood bootstrap support values above 0.70 and 70% are shown at the first and second position, respectively. Ex-type cultures are indicated in bold face. Strains obtained in the current study are in red. Some branches are shortened for layout purposes – these are indicated by two diagonal lines with the number of times. The bar at the left-bottom represents substitutions per site.
Taxonomy
The taxa described belong in family Phyllostictaceae.
. Phyllosticta oblongifoliae
Z.X. Zhang, X.Y. Liu, Z. Meng & X.G. Zhang sp. nov.
CAC8EE11-40B5-55FF-9260-B396E6E58C74
843232
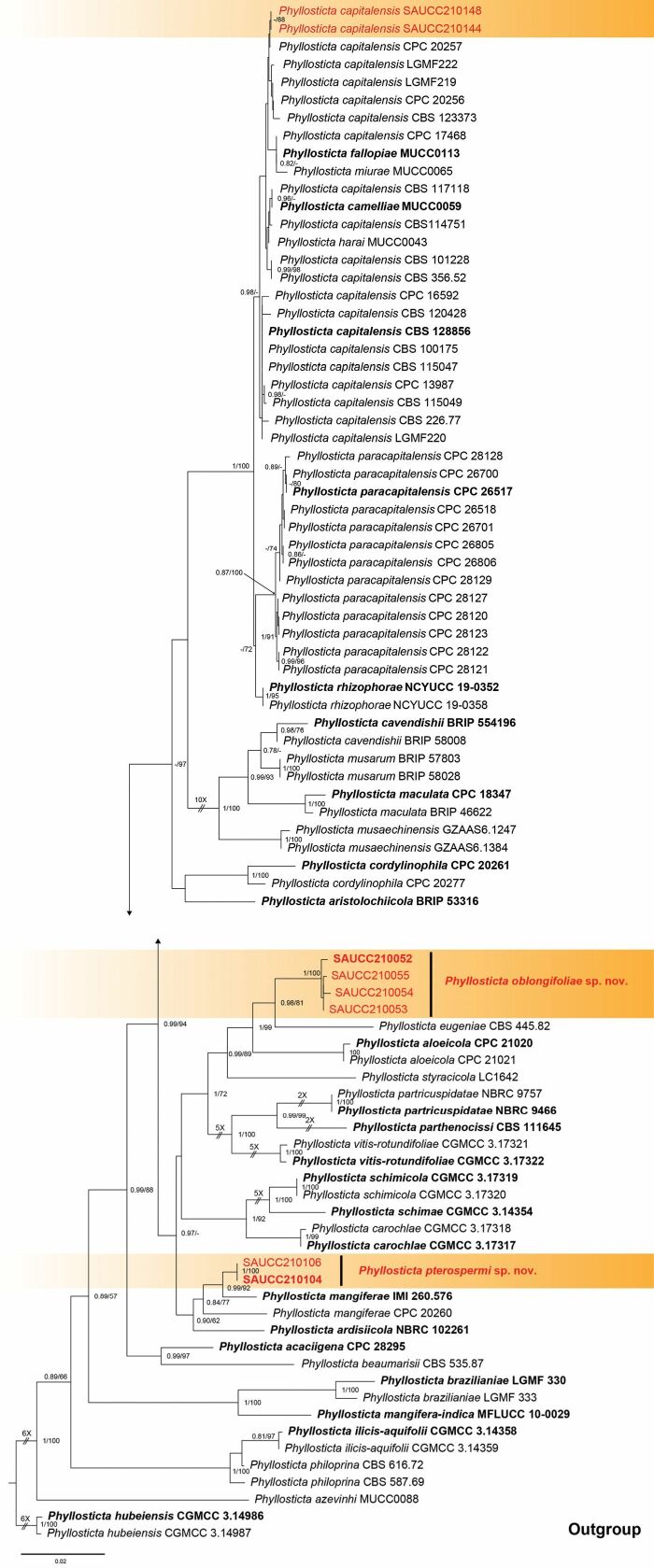
Figure 2.
Phyllostictaoblongifoliae (SAUCC210052) a diseased leaf of Garciniaoblongifoliab, c colonies (left-above, right-reverse) after 15 days on PDA (b) and MEA (c) d conidiomata e–h conidiogenous cells with conidia i–j conidia. Scale bars: 10 μm (e–j).
Etymology.
The specific epithet “oblongifoliae” refers to the host plant Garciniaoblongifolia.
Type.
China, Hainan Province: Bawangling National Forest Park, on diseased leaves of Garciniaoblongifolia, 19 May 2021, Z.X. Zhang (holotype, HSAUP210052; ex-type SAUCC210052).
Description.
Leaf endogenic and associated with leaf spots. Asexual morph: Conidiomata pycnidial, mostly aggregated in clusters, black, erumpent. In MEA culture exuding colourless to opaque conidial masses within 10 days or longer. Pycnidial wall multilayered, textura angularis, brown to dark brown, up to 30 μm thick; inner walls hyaline. Conidiophores indistinct, often reduced to conidiogenous cells. Conidiogenous cells terminal, subcylindrical, ampulliform, hyaline, smooth, 9.0–14.0 × 2.5–4.5 μm. Conidia 8.0–13.0 × 6.0–8.0 μm, mean ± SD = 10.0 ± 1.3 × 7.2 ± 0.5 μm, hyaline, aseptate, thin and smooth walled, coarsely guttulate or with a single large central guttule, ovoid, ampulliform, ellipsoidal to subglobose, enclosed in a thin mucoid sheath, 1.0–2.0 μm thick and bearing a hyaline, apical mucoid appendage, 3.0–8.5 × 1.0–1.5 μm, flexible, unbranched, tapering towards an acutely rounded tip.
Culture characteristics
. Colonies on PDA occupying an entire 90 mm Petri dish in 14 days at 25 °C in darkness, with a growth rate of 6.0–6.5 mm/day, greenish-black in obverse and reverse. Colonies on MEA 82–86 mm in diameter after 14 days at 25 °C in darkness, with a growth rate of 5.7–6.2 mm/day, undulate at edge, white to grey white in obverse and reverse, with moderate aerial mycelia on the surface, with black, gregarious conidiomata.
Additional specimens examined.
China, Hainan Province: Bawangling National Forest Park, on diseased leaves of Garciniaoblongifolia, 19 May 2021, Z.X. Zhang, HSAUP210053, living culture SAUCC210053; on diseased leaves of Garciniaoblongifolia, 19 May 2021, Z.X. Zhang, paratype HSAUP210054, ex-paratype living culture SAUCC210054; on diseased leaves of Garciniaoblongifolia, 19 May 2021, Z.X. Zhang, paratype HSAUP210055, ex-paratype living culture SAUCC210055.
Notes.
Phyllostictaoblongifoliae is introduced, based on the multi-locus phylogenetic analysis as the strain clustered into a well-supported clade (Fig. 1; 1.00/100), which is closely related to Phyllostictaugeniae (0.98/81), but distinguished, based on molecular data, ITS, LSU, tef1, ACT and GPDH loci by 57 nucleotide differences in the concatenated alignment. Morphologically, P.oblongifoliae (SAUCC210052) differs from P.ugeniae (CBS 445.82) in its shorter and wider conidia (8.0–13.0 × 6.0–8.0 vs. 9.6–16.8 × 4.8–6.0 μm) (Wikee et al. 2013a). Therefore, we establish this fungus as a novel species (Jeewon and Hyde 2016).
. Phyllosticta pterospermi
Z.X. Zhang, X.Y. Liu, Z. Meng & X.G. Zhang sp. nov.
38604F69-9BB1-52E0-B304-8DE69A1F8755
843233
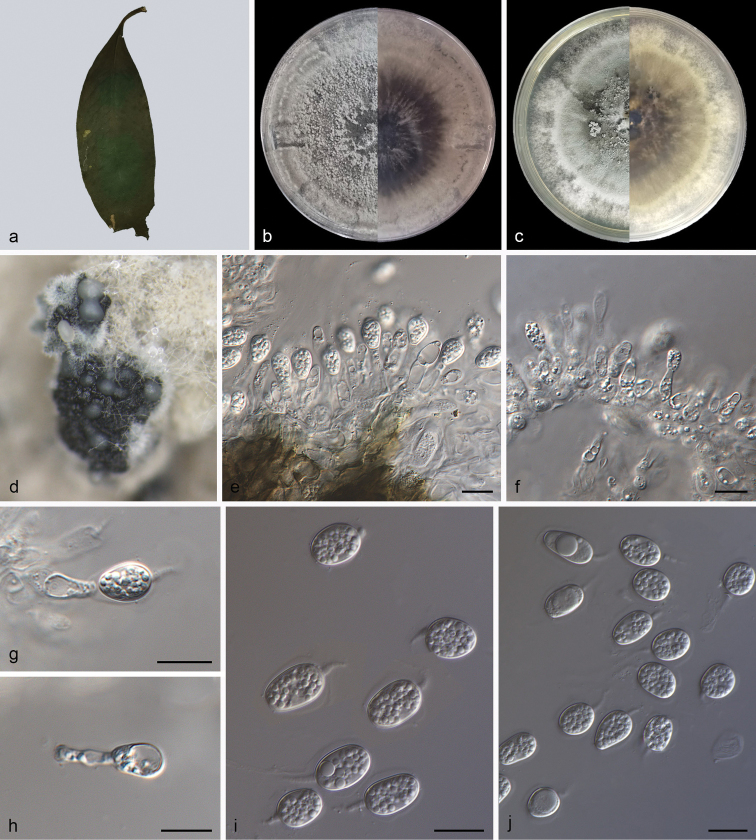
Figure 3.
Phyllostictapterospermi (holotype SAUCC210104) a diseased leaf of Pterospermumheterophyllumb, c colonies (left-above, right-reverse) after 15 days on PDA (b) and MEA (c) d conidiomata e–h conidiogenous cells with conidia i–j conidia. Scale bars: 10 μm (e–j).
Type.
China, Hainan Province: Bawangling National Forest Park, on diseased leaves of Pterospermumheterophyllum, 19 May 2021, Z.X. Zhang (holotype, HSAUP210104; ex-holotype living culture SAUCC210104).
Etymology.
The specific epithet “pterospermi” refers to the genus name of the host plant Pterospermumheterophyllum.
Description.
Leaf endogenic and associated with leaf spots. Asexual morph: Conidiomata pycnidial, mostly aggregated in clusters, black, erumpent. On MEA, pycnidia exudes yellow conidial masses, within 15 days or longer. Pycnidial walls multilayered, textura angularis, brown, up to 30 μm thick; inner walls of hyaline. Conidiophores indistinct, often reduced to conidiogenous cells. Conidiogenous cells, cylindrical, hyaline, smooth, 7.5–11.0 × 2.5–4.5 μm. Conidia 8.0–12.0 × 4.5–8.5 μm, mean ± SD = 9.8 ± 0.9 × 7.3 ± 0.7 μm, hyaline, aseptate, thin and smooth-walled, coarsely guttulate or with a single large central guttule, obovoid, ellipsoidal to subglobose, enclosed in a thin mucoid sheath, 1.0–2.0 μm thick and bearing a hyaline, apical mucoid appendage, 4.0–6.8 × 1.5–3.0 μm, flexible, unbranched, tapering towards an acutely rounded tip.
Culture characteristics.
Colonies on PDA 80–90 mm in diameter after 14 days at 25 °C in darkness, with a growth rate of 5.7–6.5 mm/day, undulate at edge, grey white to greyish-green in obverse and reverse. Colonies on MEA 82–86 mm in diameter after 14 days at 25 °C in darkness, with a growth rate of 5.8–6.2 mm/day, undulate at edge, grey white to yellow in obverse and reverse, with moderate aerial mycelia on the surface, with black, gregarious conidiomata.
Additional specimen examined.
China, Hainan Province: Bawangling National Forest Park, on diseased leaves of Pterospermumheterophyllum. 19 May 2021, Z.X. Zhang, paratype HSAUP210106, ex-paratype living culture SAUCC210106.
Notes.
Two isolates from leaf spots of Pterospermumheterophyllum phylogenetically clustered into a well-supported clade (1.00/100), which is closely related to P.ardisiicola (0.90/62) and P.mangiferae (0.99/91; Fig. 1). However, P.pterospermi differs from P.ardisiicola by 30 nucleotides (13/603 in ITS, 3/553 in LSU and 14/248 ACT) and from P.mangiferae by 29 nucleotides (7/567 in ITS, 2/763 in LSU, 3/215 in tef1, 3/226 in ACT and 14/643 in GPDH). In morphology, they are distinguished by hosts and conidial size (8.0–12.0 × 4.5–8.5 μm in P.pterospermi vs. 7.0–11.0 × 5.0–7.5 μm in P.ardisiicola vs. 10.0–12.0 × 6.0–7.0 μm in P.mangiferae). Furthermore, P.pterospermi differs from P.ardisiicola and P.mangiferae by wider conidiogenous cells (7.5–11.0 × 2.5–4.5 μm vs. 5.0–12.5 × 1.2–2.5 μm) and from P.mangiferae in having longer conidiogenous cells (7.5–11.0 × 2.5–4.5 μm vs. 6.0–10.0 × 3.0–4.0 μm) (Motohashi et al. 2008; Glienke et al. 2011). Therefore, we establish this strain as P.pterospermi sp. nov. (Jeewon and Hyde 2016).
. Phyllosticta capitalensis
Henn., Hedwigia 48: 13. 1908
1AD5AD6B-00B6-58A2-A45B-892AFC4C2BE8
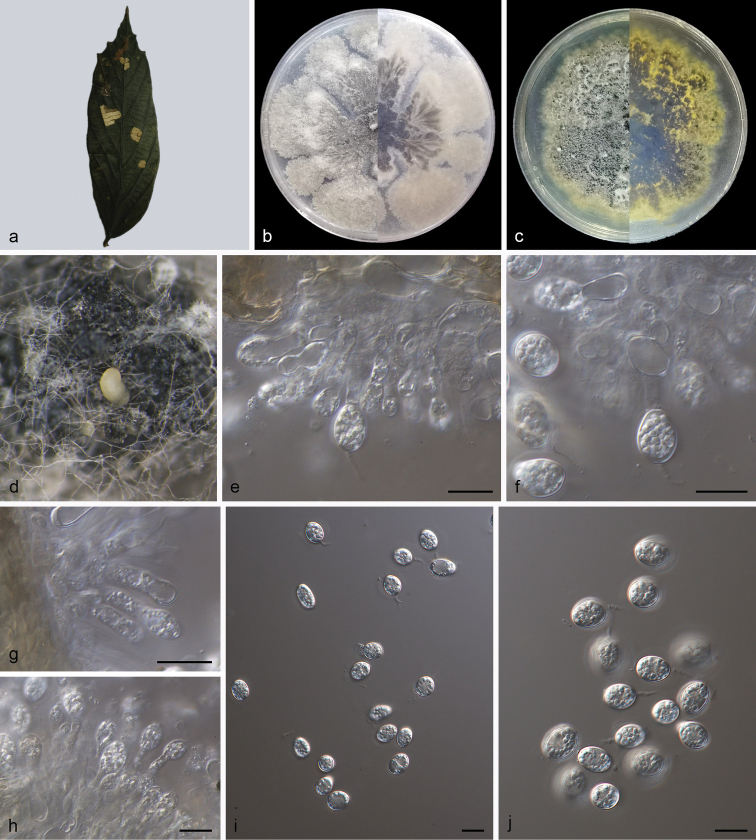
Figure 4.
Phyllostictacapitalensis (holotype SAUCC210144) a diseased leaf of Rhapisexcelsab, c colonies (left-above, right-reverse) after 15 days on PDA (b) and MEA (c) d conidiomata e asci and ascospores f asci, ascospores and conidia g conidiogenous cells with conidia h conidia i spermatia. Scale bars: 10 μm (e–i).
Description.
Leaf endogenic and associated with leaf spots. Asexual morph: Conidiomata pycnidial, mostly aggregated in clusters, black, erumpent. In MEA, cultures exuded colourless to opaque conidial masses, appeared on pycnidia after 10 days or longer. Pycnidial walls of multilayered, textura angularis, brown to dark brown, up to 35 μm thick; inner walls hyaline. Conidiophores subcylindrical to ampulliform, frequently reduced to conidiogenous cells or branching from a basal supporting cell, coated in mucoid layer, 8.0–14.0 × 3.0–5.0 μm. Conidiogenous cells terminal, subcylindrical to ampulliform, hyaline, smooth, 8.0–11.0 × 3.0–4.5 μm. Conidia 9.0–12.5 × 5.0–7.0 μm, mean ± SD = 10.6 ± 0.9 × 6.2 ± 0.5 μm, solitary, hyaline, aseptate, thin and smooth walled, coarsely guttulate or with a single large central guttule, ovoid, ampulliform, ellipsoidal to subglobose, enclosed in a thin mucoid sheath, 1.3–2.7 μm thick and bearing a hyaline, apical mucoid appendage, 3.0–8.5 × 1.0–1.5 μm, flexible, unbranched, tapering towards an acutely rounded tip. Spermatia hyaline, smooth, guttulate to granular, bacilliform, 6.0–8.2 × 1.3–2.0 μm, occurring in conidioma with conidia. Sexual morph: Ascomata shape and wall like those of the conidiomata. Asci bitunicate, hyaline, clavate to broadly fusoid-ellipsoid, with visible apical chamber, 2 μm diam., 45–85 × 9–13 μm. Ascospores bi- to multiseriate, hyaline, smooth, granular to guttulate, aseptate, straight, rarely curved, widest in the middle, limoniform with obtuse ends, 15–18 × 6–7 μm.
Culture characteristics.
Colonies on PDA occupying an entire 90 mm Petri dish in 14 days at 25 °C in darkness, with a growth rate of 6.0–6.5 mm/day, greenish-black in obverse and reverse. Colonies on MEA 82–86 mm in diameter after 14 days at 25 °C in darkness, with a growth rate of 5.7–6.2 mm/day, undulate at edge, white to grey white in obverse and reverse, with moderate aerial mycelia on the surface, with black, gregarious conidiomata.
Specimens examined.
China, Hainan Province: Bawangling National Forest Park, on diseased leaves of Rhapisexcelsa (Thunb.) Henry ex Rehd, 19 May 2021, Z.X. Zhang, HSAUP210144, living culture SAUCC210144; on diseased leaves of Rhapisexcelsa. 19 May 2021, Z.X. Zhang, HSAUP210148, living culture SAUCC210148.
Notes.
Based on morphological features, Hennings (1908) described Phyllostictacapitalensis and Glienke et al. (2011) added molecular data. The holotype (CBS 128856) of P.capitalensis was collected from Stanhopeagraveolens (Glienke et al. 2011). In our current study, two isolates (SAUCC210144, SAUCC210148), collected from diseased leaves of Rhapisexcelsa, cluster in the P.capitalensis clade (Fig. 1). Although four other species are also in this clade, we consider these two isolates as P.capitalensis, based on their morphological characters, such as granular to guttulate ascospores (15–18 × 6–7 vs. 15–17 × 5–6 μm), subcylindrical to ampullate conidiogenous cells (8.0–11.0 × 3.0–4.5 vs. 7–10 × 3–5 μm), ellipsoidal to subglobose conidia (9–12.5 × 5–7 vs. 11–12 × 6–7 μm) and hyaline, apical mucoid appendages (3–8.5 × 1–1.5 vs. 6–8 × 1–1.5 μm).
Discussion
Compared to other parts of China, species richness is highly diverse in Hainan Province, especially in Bawangling National Forest Park, which has a typical tropical rainforest climate. The environment favours growth of unusual microbial species. Historically, Phyllosticta species have been identified by morphology and host association. However, overlapping morphology makes it difficult to pinpoint homologous characters and, consequently, traditional identification of Phyllosticta species has long been a complicated endeavour (Norphanphoun et al. 2020). This issue has led to confusion in the taxonomy of Phyllosticta. Molecular phylogenetics has promoted species delimitation and species complex determination (Baayen et al. 2002; Okane et al. 2003; Motohashi et al. 2009; Wulandari et al. 2009; Glienke et al. 2011; Wikee et al. 2012). Norphanphoun et al. (2020) introduced six species complexes in Phyllosticta, based on five gene loci encoding the internal transcribed spacer of ribosomal RNA (ITS rDNA), large subunit of ribosomal RNA (LSU rDNA), translation elongation factor 1 alpha (TEF1α), actin (ACT) and glycerol-3-phosphate dehydrogenase (GPDH). Amongst these, the P.capitalensis species complex consisted of 28 cryptic species, P.acaciigena, P.aloeicola, P.ardisiicola, P.aristolochiicola, P.azevinhi, P.beaumarisii, P.brazilianiae, P.capitalensis, P.carochlae, P.cavendishii, P.cordylinophila, P.eugeniae, P.fallopiae, P.ilicis-aquifolii, P.maculata, P.mangiferae, P.mangifera-indicae, P.musaechinensis, P.musarum, P.paracapitalensis, P.parthenocissi, P.partricuspidatae, P.philoprina, P.rhizophorae, P.schimae, P.schimicola, P.styracicola and P.vitis-rotundifoliae. In this study, we focus our analyses on the P.capitalensis species complex and report two new species and one new Chinese record.
Multilocus phylogeny, as well as morphological characters observed in culture, described and illustrated herein eight isolates of Phyllosticta species from three host genera, which contributed knowledge to the diversity of Phyllosticta species in Hainan, China. Two new species are proposed: P.oblongifoliae sp. nov. and P.pterospermi sp. nov. This is the first time we report Phyllosticta species from Pterospermumheterophyllum (Sterculiaceae). In a recent study, Allophomapterospermicola was reported as pathogenic to Pterospermum (Marin-Felix et al. 2019). In reality, the number of phytopathogenic fungi from the Pterospermum host is inherently small. The known species Phyllostictacapitalensis (synonym Guignardiamangiferae; Baayen et al. 2002) was described multiple times from Stanhopeagraveolens (Orchidaceae) in Brazil (Glienke et al. 2011). In this study, we describe and illustrate Phyllostictacapitalensis again. Each of these species show typical morphological characteristics of Phyllosticta, i.e. conidia with mucilaginous sheaths and an apical appendage (van der Aa 1973).
Phyllostictacapitalensis is a cosmopolitan endophytic species reported in more than 300 host records in Fungal Databases (https://nt.ars-grin.gov/fungaldatabases/index.cfm) (Okane et al. 2001, 2003; Baayen et al. 2002; Glienke et al. 2011; Wikee et al. 2013b; Wu et al. 2014; Zhang et al. 2015; Tran et al. 2019; Hattori et al. 2020). As a weak pathogen, P.capitalensis causes leaf spots on tea (Camelliasinensis), oil palm (Elaeisguineensis), Ricinuscommunis and black spot disease on Psidiumguajava (Cheng et al. 2019; Nasehi et al. 2019; Liao et al. 2020; Tang et al. 2020).
Supplementary Material
Acknowledgements
This work was jointly supported by the National Natural Science Foundation of China (nos. 31900014, U2002203, 31750001)
Citation
Zhang Z, Liu X, Zhang X, Meng Z (2022) Morphological and phylogenetic analyses reveal two new species and a new record of Phyllosticta (Botryosphaeriales, Phyllostictaceae) from Hainan, China. MycoKeys 91: 1–23. https://doi.org/10.3897/mycokeys.91.84803
Funding Statement
This work was jointly supported by the National Natural Science Foundation of China (nos. 31900014, U2002203, 31750001)
Supplementary materials
The combined ITS, LSU, tef1, ACT and GAPDH sequences
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Zhaoxue Zhang, Xiaoyong Liu, Xiuguo Zhang, Zhe Meng
Data type
Phylogenetic.
Explanation note
The combined ITS, LSU, tef1, ACT and GAPDH sequences.
References
- Baayen RP, Bonants P, Verkley G, Carroll GC, van der Aa HA, de Weerdt M, van Brouwershaven IR, Schutte GC, Maccheroni Jr W, Glienke de Blanco C, Azevedo JL. (2002) Nonpathogenic isolates of the citrus black spot fungus, Guignardiacitricarpa, identified as a cosmopolitan endophyte of woody plants, G.mangiferae (Phyllostictacapitalensis). Phytopathology 92(5): 464–477. 10.1094/PHYTO.2002.92.5.464 [DOI] [PubMed] [Google Scholar]
- Bánki O, Roskov Y, Döring M, Ower G, Vandepitte L, Hobern D, Remsen D, Schalk P, DeWalt RE, Keping M, Miller J, Orrell T, Aalbu R, Adlard R, Adriaenssens EM, Aedo C, Aescht E, Akkari N, Alfenas-Zerbini P. (2022) Catalogue of Life Checklist (Version 2022-03-21). Catalogue of Life. 10.48580/dfpd [DOI]
- Braun U, Nakashima C, Crous PW, Groenewald JZ, Moreno-Rico O, Rooney-Latham S, Blomquist CL, Haas J, Marmolejo J. (2018) Phylogeny and taxonomy of the genus Tubakia s. lat. Fungal Systematics and Evolution 1(1): 41–99. 10.3114/fuse.2018.01.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone I, Kohn LM. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91(3): 553–556. 10.1080/00275514.1999.12061051 [DOI] [Google Scholar]
- Cheng LL, Thangaraj K, Deng C, Deng WW, Zhang ZZ. (2019) Phyllostictacapitalensis causes leaf spot on tea plant (Camelliasinensis) in China. Plant Disease 103(11): e2964. 10.1094/PDIS-04-19-0768-PDN [DOI]
- Crous PW, Slippers B, Wingfield MJ, Rheeder J, Marasas WFO, Philips AJL, Alves A, Burgess T, Barber P, Groenewald JZ. (2006) Phylogenetic lineages in the Botryosphaeriaceae. Studies in Mycology 55: 235–253. 10.3114/sim.55.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Schumacher RK, Summerell BA, Giraldo A, Gené J, Guarro J, Wanasinghe DN, Hyde KD, Camporesi E, Garethjones EB, Thambugala KM, Malysheva EF, Malysheva VF, Acharya K, Álvarez J, Alvarado P, Assefa A, Barnes CW, Bartlett JS, Blanchette RA, Burgess TI, Carlavilla JR, Coetzee MPA, Damm U, Decock CA, Denbreeÿen A, Devries B, Dutta AK, Holdom DG, Rooney-Latham S, Manjón JL, Marincowitz S, Mirabolfathy M, Moreno G, Nakashima C, Papizadeh M, Shahzadehfazeli SA, Amoozegar MA, Romberg MK, Shivas RG, Stalpers JA, Stielow B, Stukely MJC, Swart WJ, Tan YP, Vanderbank M, Wood AR, Zhang Y, Groenewald JZ. (2014) Fungal Planet description sheets: 281–319. Persoonia 33(1): 212–289. 10.3767/003158514X685680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Roux JJL, Richardson DM, Strasberg D, Shivas RG, Alvarado P, Edwards J, Moreno G, Sharma R, Sonawane MS, Tan YP, Altés A, Barasubiye T, Barnes CW, Blanchette RA, Boertmann D, Bogo A, Carlavilla JR, Cheewangkoon R, Daniel R, de Beer ZW, Yáñez-Morales MJ, Duong TA, Fernández-Vicente J, Geering ADW, Guest DI, Held BW, Heykoop M, Hubka V, Ismail AM, Kajale SC, Khemmuk W, Kolařík M, Kurli R, Lebeuf R, Lévesque CA, Lombard L, Magista D, Manjón JL, Marincowitz S, Mohedano JM, Nováková A, Oberlies NH, Otto EC, Paguigan ND, Pascoe IG, Pérez-Butrón JL, Perrone G, Rahi P, Raja HA, Rintoul T, Sanhueza RMV, Scarlett K, Shouche YS, Shuttleworth LA, Taylor PWJ, Thorn RG, Vawdrey LL, Solano-Vidal R, Voitk A, Wong PTW, Wood AR, Zamora JC, Groenewald JZ. (2015) Fungal Planet description sheets: 371–399. Persoonia 35(1): 264–327. 10.3767/003158515X690269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI, St J, Hardy GE, Crane C, Barrett S, Cano-Lira JF, Leroux JJ, Thangavel R, Guarro J, Stchigel AM, Martín MP, Alfredo DS, Barber PA, Barreto RW, Baseia IG, Cano-Canals J, Cheewangkoon R, Ferreira RJ, Gené J, Lechat C, Moreno G, Roets F, Shivas RG, Sousa JO, Tan YP, Wiederhold NP, Abell SE, Accioly T, Albizu JL, Alves JL, Antoniolli ZI, Aplin N, Araújo J, Arzanlou M, Bezerra JDP, Bouchara JP, Carlavilla JR, Castillo A, Castroagudín VL, Ceresini PC, Claridge GF, Coelho G, Coimbra VRM, Costa LA, da cunha KC, da silva SS, Daniel R, de beer ZW, Dueñas M, Edwards J, Enwistle P, Fiuza PO, Fournier J, García D, Gibertoni TB, Giraud S, Guevara-Suarez M, Gusmão LFP, Haituk S, Heykoop M, Hirooka Y, Hofmann TA, Houbraken J, Hughes DP, Kautmanová I, Koppel O, Koukol O, Larsson E, Latha KPD, Lee DH, Lisboa DO, Lisboa WS, López-Villalba Á, Maciel JLN, Manimohan P, Manjón JL, Marincowitz S, Marney TS, Meijer M, Miller AN, Olariaga I, Paiva LM, Piepenbring M, Poveda-Molero JC, Raj KNA, Raja HA, Rougeron A, Salcedo I, Samadi R, Santos TAB, Scarlett K, Seifert KA, Shuttleworth LA, Silva GA, Silva M, Siqueira JPZ, Souza-Motta C.M, Stephenson SL. (2016) Fungal Planet description sheets: 469–557. Persoonia 37: 218–403. 10.3767/003158516X694499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI, Carnegie AJ, St J, Hardy GE, Smith D, Summerell BA, Cano-Lira JF, Guarro J, Houbraken J, Lombard L, Martín MP, Sandoval-Denis M, Alexandrova AV, Barnes CW, Baseia IG, Bezerra JDP, Guarnaccia V, May TW, Hernández-Restrepo M, Stchigel AM, Miller AN, Ordoñez ME, Abreu VP, Accioly T, Agnello C, Agustincolmán A, Albuquerque CC, Alfredo DS, Alvarado P, Araújo-Magalhães GR, Arauzo S, Atkinson T, Barili A, Barreto RW, Bezerra JL, Cabral TS, Rodríguez Camello F, Cruz RHSF, Daniëls PP, da silva BDB, de Almeida DAC, de Carvalhojúnior AA, Decock CA, Delgat L, Denman S, Dimitrov RA, Edwards J, Fedosova AG, Ferreira RJ, Firmino AL, Flores JA, García D, Gené J, Giraldo A, Góis JS, Gomes AAM, Gonçalves CM, Gouliamova DE, Groenewald M, Guéorguiev BV, Guevara-Suarez M, Gusmão LFP, Hosaka K, Hubka V, Huhndorf SM, Jadan M, Jurjevi Kraak B, Kuera V, Kumar TKA, Kušan I, Lacerda SR, Lamlertthon S, Lisboa WS, Loizides M, Luangsa-Ard JJ, Lysková P, Maccormack WP, Macedo DM, Machado AR, Malysheva EF, Marinho P, Matoec N, Meijer M, Meši A, Mongkolsamrit S, Moreira KA, Morozova OV, Nair KU, Nakamura N, Noisripoom W, Olariaga I, Oliveira RJV, Paiva LM, Pawar P, Pereira OL, Peterson SW, Prieto M, Rodríguez-Andrade E, Rojodeblas C, Roy M, Santos ES, Sharma R, Silva GA, Souza-Motta CM, Takeuchi-Kaneko Y, Tanaka C, Thakur A, Smith MTH, Tkalec Z, Valenzuela-Lopez N, Vanderkleij P, Verbeken A, Viana MG, Wang XW, Groenewald JZ. (2017) Fungal Planet description sheets: 625–715. Persoonia 39: 270–467. 10.3767/persoonia.2017.39.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Schumacher RK, Wingfield MJ, Akulov A, Denman S, Roux J, Braun U, Burgess TI, Carnegie AJ, Váczy KZ, Guatimosim E, Schwartsburd PB, Barreto RW, Hernández-Restrepo M, Lombard L, Groenewald JZ. (2018) New and interesting fungi. 1. Fungal Systematics and Evolution 1(1): 169–215. 10.3114/fuse.2018.01.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Carnegie AJ, Wingfield MJ, Sharma R, Mughini G, Noordeloos ME, Santini A, Shouche YS, Bezerra JDP, Dima B, Guarnaccia V, Imrefi I, Jurjević Ž, Knapp DG, Kovács GM, Magistà D, Perrone G, Rämä T, Rebriev YA, Shivas RG, Singh SM, Souza-Motta CM, Thangavel R, Adhapure NN, Alexandrova AV, Alfenas AC, Alfenas RF, Alvarado P, Alves AL, Andrade DA, Andrade JP, Barbosa RN, Barili A, Barnes CW, Baseia IG, Bellanger JM, Berlanas C, Bessette AE, Bessette AR, Biketova AYu, Bomfim FS, Brandrud TE, Bransgrove K, Brito ACQ, Cano-Lira JF, Cantillo T, Cavalcanti AD, Cheewangkoon R, Chikowski RS, Conforto C, Cordeiro TRL, Craine JD, Cruz R, Damm U, de Oliveira RJV, de Souza JT, de Souza HG, Dearnaley JDW, Dimitrov RA, Dovana F, Erhard A, Esteve-Raventós F, Félix CR, Ferisin G, Fernandes RA, Ferreira RJ, Ferro LO, Figueiredo CN, Frank JL, Freire KTLS, García D, Gené J, Gęsiorska A, Gibertoni TB, Gondra RAG, Gouliamova DE, Gramaje D, Guard F, Gusmão LFP, Haitook S, Hirooka Y, Houbraken J, Hubka V, Inamdar A, Iturriaga T, Iturrieta-González I, Jadan M, Jiang N, Justo A, Kachalkin AV, Kapitonov VI, Karadelev M, Karakehian J, Kasuya T, Kautmanová I, Kruse J, Kušan I, Kuznetsova TA, Landell MF, Larsson KH, Lee HB, Lima DX, Lira CRS, Machado AR, Madrid H, Magalhães OMC, Majerova H, Malysheva EF, Mapperson RR, Marbach PAS, Martín MP, Martín-Sanz A, Matočec N, McTaggart AR, Mello JF, Melo RFR, Mešič A, Michereff SJ, Miller AN, Minoshima A, Molinero-Ruiz L, Morozova OV, Mosoh D, Nabe M, Naik R, Nara K, Nascimento SS, Neves RP, Olariaga I, Oliveira RL, Oliveira TGL, Ono T, Ordoñez ME, de M Ottoni A, Paiva LM, Pancorbo F, Pant B, Pawłowska J, Peterson SW, Raudabaugh DB, Rodríguez-Andrade E, Rubio E, Rusevska K, Santiago ALCMA, Santos ACS, Santos C, Sazanova NA, Shah S, Sharma J, Silva BDB, Siquier JL, Sonawane MS, Stchigel AM, Svetasheva T, Tamakeaw N, Telleria MT, Tiago PV, Tian CM, Tkalčec Z, Tomashevskaya MA, Truong HH, Vecherskii MV, Visagie CM, Vizzini A, Yilmaz N, Zmitrovich IV, Zvyagina EA, Boekhout T, Kehlet T, Læssøe T, Groenewald JZ. (2019) Fungal Planet description sheets: 868–950. Persoonia 39: 291–473. 10.3767/persoonia.2019.42.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Hernández-Restrepo M, Schumacher RK, Cowan DA, Maggs-Kölling G, Marais E, Wingfield MJ, Yilmaz N, Adan OCG, Akulov A, Duarte EÁ, Berraf-Tebbal A, Bulgakov TS, Carnegie AJ, de Beer ZW, Decock C, Dijksterhuis J, Duong TA, Eichmeier A, Hien LT, Houbraken JAMP, Khanh TN, Liem NV, Lombard L, Lutzoni FM, Miadlikowska JM, Nel WJ, Pascoe IG, Roets F, Roux J, Samson RA, Shen M, Spetik M, Thangavel R, Thanh HM, Thao LD, van Nieuwenhuijzen EJ, Zhang JQ, Zhang Y, Zhao LL, Groenewald JZ. (2021) New and interesting fungi. 4. Fungal Systematics and Evolution 7(1): 255–343. 10.3114/fuse.2021.07.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donk MA. (1968) Report of the committee for fungi and lichen 1964–1968. Taxon 17(5): 578–581. 10.2307/1216075 [DOI] [Google Scholar]
- Furlanetto C, Dianese JC. (1998) Some coelomycetes from Central Brazil. Mycological Research 102(1): 19–29. 10.1017/S0953756297004656 [DOI] [Google Scholar]
- Glienke C, Pereira OL, Stringari D, Fabris J, Kava-Cordeiro V, Galli-Terasawa L, Cunnington J, Shivas RG, Groenewald JZ, Crous PW. (2011) Endophytic and pathogenic Phyllosticta species, with reference to those associated with citrus black spot. Persoonia 26(1): 47–56. 10.3767/003158511X569169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnaccia V, Groenewald JZ, Li H, Glienke C, Carstens E, Hattingh V, Fourie PH, Crous PW. (2017) First report of Phyllostictacitricarpa and description of two new species, P.paracapitalensis and P.paracitricarpa, from citrus in Europe. Studies in Mycology 87(1): 161–185. 10.1016/j.simyco.2017.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo LD, Hyde KD, Liew ECY. (2000) Identification of endophytic fungi from Livistonachinensis based on morphology and rDNA sequences. The New Phytologist 147(3): 617–630. 10.1046/j.1469-8137.2000.00716.x [DOI] [PubMed] [Google Scholar]
- Hattori Y, Motohashi K, Tanaka K, Nakashima C. (2020) Taxonomical re-examination of the genus Phyllosticta – Parasitic fungi on Cupressaceae trees in Japan. Forest Pathology 50(5): 1–14. 10.1111/efp.12630 [DOI] [Google Scholar]
- Hennings P. (1908) FungiS. paulenses IV a cl. Puttmans collecti. Hedwigia 48: e13.
- Hongsanan S, Hyde KD, Phookamsak R, Wanasinghe DN, McKenzie EHC, Sarma VV, Boonmee S, Lücking R, Bhat DJ, Liu NG, Tennakoon DS, Pem D, Karunarathna A, Jiang SH, Jones EBG, Phillips AJL, Manawasinghe IS, Tibpromma S, Jayasiri SC, Sandamali DS, Jayawardena RS, Wijayawardene NN, Ekanayaka AH, Jeewon R, Lu YZ, Dissanayake AJ, Zeng XY, Luo ZL, Tian Q, Phukhamsakda C, Thambugala KM, Dai DQ, Chethana KWT, Samarakoon MC, Ertz D, Bao DF, Doilom M, Liu JK, PérezOrtega S, Suija A, Senwanna C, Wijesinghe SN, Konta S, Niranjan M, Zhang SN, Ariyawansa HA, Jiang HB, Zhang JF, Norphanphoun C, de Silva NI, Thiyagaraja V, Zhang H, Bezerra JDP, Miranda-González R, Aptroot A, Kashiwadani H, Harishchandra D, Sérusiaux E, Aluthmuhandiram JVS, Abeywickrama PD, Devadatha B, Wu HX, Moon KH, Gueidan C, Schumm F, Bundhun D, Mapook A, Monkai J, Chomnunti P, Suetrong S, Chaiwan N, Dayarathne MC, Yang J, Rathnayaka AR, Bhunjun CS, Xu JC, Zheng JS, Liu G, Feng Y, Xie N. (2020) Refined families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere 11(1): 1553–2107. 10.5943/mycosphere/11/1/13 [DOI] [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17(17): 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Jeewon R, Hyde KD. (2016) Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere 7(11): 1669–1677. 10.5943/mycosphere/7/11/4 [DOI] [Google Scholar]
- Jiang N, Voglmayr H, Bian DR, Piao CG, Wang SK, Li Y. (2021) Morphology and phylogeny of Gnomoniopsis (Gnomoniaceae, Diaporthales) from Fagaceae leaves in China. Journal of Fungi 7(10): e792. 10.3390/jof7100792 [DOI] [PMC free article] [PubMed]
- Katoh K, Rozewicki J, Yamada KD. (2019) MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20(4): 1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33(7): 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao YM, Wang ZX, Wei MC, Wang C. (2020) First report of Phyllostictacapitalensis causing black spot disease on Psidiumguajava in mainland China. Plant Disease 104(12): e3252. 10.1094/PDIS-02-20-0338-PDN [DOI]
- Lin S, Sun X, He W, Zhang Y. (2017) Two new endophytic species of Phyllosticta (Phyllostictaceae, Botryosphaeriales) from southern China. Mycosphere 8(2): 1273–1288. 10.5943/mycosphere/8/2/11 [DOI] [Google Scholar]
- Liu JK, Phookamsak R, Doilom M, Wikee S, Li YM, Ariyawansha H, Boonmee S, Chomnunti P, Dai DQ, Bhat JD, Romero AI, Zhuang WY, Monkai J, Jones EBG, Chukeatirote E, Zhao YC, Wang Y, Hyde KD. (2012) Toward a natural classification of Botryosphaeriales. Fungal Diversity 57(1): 149–210. 10.1007/s13225-012-0207-4 [DOI] [Google Scholar]
- Marin-Felix Y, Hernández-Restrepo M, Iturrieta-Gonzalez I, García D, Gené J, Groenewald JZ, Cai L, Chen Q, Quaedvlieg W, Schumacher RK, Taylor PWJ, Ambers C, Bonthond G, Edwards J, Krueger-Hadfield SA, Luangsa-ard JJ, Morton L, Moslemi A, Sandoval-Denis M, Tan YP, Thangavel R, Vaghefi N, Cheewangkoon R, Crous PW. (2019) Genera of phytopathogenic fungi: GOPHY 3. Studies in Mycology 94(1): 1–124. 10.1016/j.simyco.2019.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2012) The CIPRES science gateway: enabling high-impact science for phylogenetics researchers with limited resources. In: Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment. Bridging from the extreme to the campus and beyond. Association for Computing Machinery, USA, 1–8. 10.1145/2335755.2335836 [DOI]
- Motohashi K, Anzai K, Nakashima C. (2008) Four new species of Phyllosticta, one new species of Pseudocercospora, and one new combination in Passalora from Japan. Mycoscience 49(2): 138–146. 10.1007/S10267-007-0395-Z [DOI] [Google Scholar]
- Motohashi K, Inaba S, Anzai K, Takamatsu S, Nakashima C. (2009) Phylogenetic analyses of Japanese species of Phyllosticta sensu stricto. Mycoscience 50(4): 291–302. 10.1007/S10267-009-0487-Z [DOI] [Google Scholar]
- Myllys L, Stenroos S, Thell A. (2002) New genes for phylogenetic studies of lichenized fungi, glyceraldehyde-3-phosphate dehydrogenase and beta-tubulin genes. Lichenologist (London, England) 34(3): 237–246. 10.1006/lich.2002.0390 [DOI] [Google Scholar]
- Nasehi A, Sathyapriya H, Wong MY. (2019) First report of leaf spot on oil palm caused by Phyllostictacapitalensis in Malaysia. Plant Disease 103(11): 2964. 10.1094/PDIS-04-19-0768-PDN [DOI] [Google Scholar]
- Norphanphoun C, Hongsanan S, Gentekaki E, Chen YJ, Kuo CH, Hyde KD. (2020) Differentiation of species complexes in Phyllosticta enables better species resolution. Mycosphere 11(1): 2542–2628. 10.5943/mycosphere/11/1/16 [DOI] [Google Scholar]
- Nylander JAA. (2004) MrModelTest v. 2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
- O’Donnell K, Kistler HC, Cigelnik E, Ploetz RC. (1998) Multiple evolutionary origins of the fungus causing panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences of the United States of America 95(5): 2044–2049. 10.1073/pnas.95.5.2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okane I, Nakagiri A, Ito T. (2001) Identity of Guignardia sp. inhabiting ericaceous plants. Canadian Journal of Botany 79(1): 101–109. 10.1139/b00-136 [DOI] [Google Scholar]
- Okane I, Lumyong S, Ito T, Nakagiri A. (2003) Extensive host range of an endophytic fungus, Guignardiaendophyllicola (anamorph, Phyllostictacapitalensis). Mycoscience 44(5): 353–363. 10.1007/S10267-003-0128-X [DOI] [Google Scholar]
- Persoon CH. (1818) Traité sur les champignons comestibles, contenant l’indication des espèces nuisibles; a l’histoire des champignons. Belin-Leprieur, Paris, France. 10.5962/bhl.title.110115 [DOI]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12): 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61(3): 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch CL, Shoemaker RA, Seifert KA, Hambleton S, Spatafora JW, Crous PW. (2006) A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia 98(6): 1041–1052. 10.1080/15572536.2006.11832632 [DOI] [PubMed] [Google Scholar]
- Seaver FJ. (1922) Phyllostictaceae. North American Flora 6: 3–84. [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9): 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YY, Cai L. (2012) Polyphasic characterisation of three new Phyllosticta spp. Persoonia 28(1): 76–84. 10.3767/003158512X645334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JR, Liu YL, Yin XG, Lu JN, Zhou YH. (2020) First report of castor dark leaf spot caused by Phyllostictacapitalensis in Zhanjiang, China. Plant Disease 104(6): 1856. 10.1094/PDIS-11-19-2490-PDN [DOI] [Google Scholar]
- Tran NT, Miles AK, Dietzgen RG, Drenth A. (2019) Phyllostictacapitalensis and P.paracapitalensis are endophytic fungi that show potential to inhibit pathogenic P.citricarpa on citrus. Australasian Plant Pathology 48(3): 281–296. 10.1007/s13313-019-00628-0 [DOI] [Google Scholar]
- van der Aa HA. (1973) Studies in Phyllosticta. Studies in Mycology 5: 1–110. [Google Scholar]
- van der Aa HA, Vanev S. (2002) A revision of the species described in Phyllosticta. Utrecht, The Netherlands: Centraalbureau voor Schimmelcultures (CBS).
- Wang X, Chen G, Huang F, Zhang J, Hyde KD, Li H. (2012) Phyllosticta species associated with citrus diseases in China. Fungal Diversity 52(1): 209–224. 10.1007/s13225-011-0140-y [DOI] [Google Scholar]
- Wang Y, Jin L, Chen XR, Lin L, Chen HG. (2013) Phyllostictaephedricola sp. nov. on Ephedraintermedia. Mycotaxon 125(1): 165–167. 10.5248/125.165 [DOI] [Google Scholar]
- White T, Burns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of ribosomal RNA genes for phylogenetics. In: Innis MA. (Ed.) PCR Protocols: A Guide to Methods and Applications.Academic Press, New York, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Wijayawardene NN, Hyde KD, Al-Ani LKT, Tedersoo L, Haelewaters D, Rajeshkumar KC, Zhao RL, Aptroot A, Leontyev DV, Saxena RK, Tokarev YS, Dai DQ, Letcher PM, Stephenson SL, Ertz D, Lumbsch HT, Kukwa M, Issi IV, Madrid H, Phillips AJL, Selbmann L, Pfliegler WP, Horváth E, Bensch K, Kirk PM, Kolaříková K, Raja HA, Radek R, Papp V, Dima B, Ma J, Malosso E, Takamatsu S, Rambold G, Gannibal PB, Triebel D, Gautam AK, Avasthi S, Suetrong S, Timdal E, Fryar SC, Delgado G, Réblová M, Doilom M, Dolatabadi S, Pawłowska JZ, Humber RA, Kodsueb R, Sánchez-Castro I, Goto BT, Silva DKA, de Souza FA, Oehl F, da Silva GA, Silva IR, Błaszkowski J, Jobim K, Maia LC, Barbosa FR, Fiuza PO, Divakar PK, Shenoy BD, Castañeda-Ruiz RF, Somrithipol S, Lateef AA, Karunarathna SC, Tibpromma S, Mortimer PE, Wanasinghe DN, Phookamsak R, Xu J, Wang Y75, Tian F, Alvarado P, Li DW, Kušan I, Matočec N, Mešić A, Tkalčec Z, Maharachchikumbura SSN, Papizadeh M, Heredia G, Wartchow F, Bakhshi M, Boehm E, Youssef N, Hustad VP, Lawrey JD87, Santiago ALCMA, Bezerra JDP, Souza-Motta CM, Firmino AL, Tian Q, Houbraken J, Hongsanan S, Tanaka K, Dissanayake AJ, Monteiro JS, Grossart HP, Suija A, Weerakoon G, Etayo J, Tsurykau A, Vázquez V, Mungai P, Damm U, Li QR, Zhang H, Boonmee S, Lu YZ, Becerra AG, Kendrick B, Brearley FQ, Motiejūnaitė J, Sharma B, Khare R, Gaikwad S, Wijesundara DSA, Tang LZ, He MQ, Flakus A, Rodriguez-Flakus P, Zhurbenko MP, McKenzie EHC, Stadler M, Bhat DJ, Liu JK, Raza M, Jeewon R, Nassonova ES, Prieto M, Jayalal RGU, Erdoğdu M, Yurkov A, Schnittler M, Shchepin ON, Novozhilov YK, Silva-Filho AGS, Gentekaki E, Liu P, Cavender JC, Kang Y, Mohammad S, Zhang LF, Xu RF, Li YM, Dayarathne MC, Ekanayaka AH, Wen TC, Deng CY, Pereira OL, Navathe S, Hawksworth DL, Fan XL, Dissanayake LS, Kuhnert E, Grossart HP, Thines M. (2020) Outline of fungi and fungus-like taxa. Mycosphere : Journal of Fungal Biology 11(1): 1060–1456. 10.5943/mycosphere/11/1/8 [DOI] [Google Scholar]
- Wikee S, Udayanga D, Crous PW, Chukeatirote E, McKenzie EHC, Bahkali AH, Dai DQ, Hyde KD. (2011) Phyllosticta – an overview of current status of species recognition. Fungal Diversity 51(1): 43–61. 10.1007/s13225-011-0146-5 [DOI] [Google Scholar]
- Wikee S, Wulandari NF, McKenzie EHC, Hyde KD. (2012) Phyllostictaophiopogonis sp. nov. from Ophiopogonjaponicas (Liliaceae). Saudi Journal of Biological Sciences 19(1): 13–16. 10.1016/j.sjbs.2011.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikee S, Lombard L, Nakashima C, Motohashi K, Chukeatirote E, Cheewangkoon R, McKenzie EHC, Hyde KD, Crous PW. (2013a) A phylogenetic re-evaluation of Phyllosticta (Botryosphaeriales). Studies in Mycology 76: 1–29. 10.3114/sim0019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikee S, Lombard L, Crous PW, Nakashima C, Motohashi K, Chukeatirote E, Alias SA, McKenzie EHC, Hyde KD. (2013b) Phyllostictacapitalensis, a widespread endophyte of plants. Fungal Diversity 60(1): 91–105. 10.1007/s13225-013-0235-8 [DOI] [Google Scholar]
- Wong MH, Crous PW, Henderson J, Groenewald JZ, Drenth A. (2012) Phyllosticta species associated with freckle disease of banana. Fungal Diversity 56(1): 173–187. 10.1007/s13225-012-0182-9 [DOI] [Google Scholar]
- Wu SP, Liu YX, Yuan J, Wang Y, Hyde KD, Liu ZY. (2014) Phyllosticta species from banana (Musa sp.) in Chongqing and Guizhou Provinces, China. Phytotaxa 188(3): 135–144. 10.11646/phytotaxa.188.3.2 [DOI] [Google Scholar]
- Wulandari NF, Hyde KD, Duong LM, De Gruyter J, Meffert JP, Groenewald JZ, Crous PW. (2009) Phyllostictacitriasiana sp. nov., the cause of citrus tan spot of Citrusmaxima in Asia. Fungal Diversity 34: 23–39. [Google Scholar]
- Wulandari NF, Bhat DJ, To-anun C. (2013) A modern account of the genus Phyllosticta. Plant Pathology & Quarantine 3(2): 145–159. 10.5943/ppq/3/2/4 [DOI] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. (2000) A greedy algorithm for aligning DNA sequences. Journal of Computational Biology 7(1–2): 203–214. 10.1089/10665270050081478 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Crous PW, Schoch CL, Hyde KD. (2012) Pleosporales. Fungal Diversity 53(1): 1–221. 10.1007/s13225-011-0117-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Zhang N, Cai L. (2013) Typification and phylogenetic study of Phyllostictaampelicida and P.vaccinii. Mycologia 105(4): 1030–1042. 10.3852/12-392 [DOI] [PubMed] [Google Scholar]
- Zhang K, Shivas RG, Cai L. (2015) Synopsis of Phyllosticta in China. Mycology 6(1): 50–75. 10.1080/21501203.2015.1027507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N, Chen Q, Carroll G, Zhang N, Shivas RG, Cai L. (2015) Polyphasic characterization of four new plant pathogenic Phyllosticta species from China, Japan, and the United States. Fungal Biology 119(5): 433–446. 10.1016/j.funbio.2014.08.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The combined ITS, LSU, tef1, ACT and GAPDH sequences
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Zhaoxue Zhang, Xiaoyong Liu, Xiuguo Zhang, Zhe Meng
Data type
Phylogenetic.
Explanation note
The combined ITS, LSU, tef1, ACT and GAPDH sequences.