Abstract
A taxonomic revision of Conidiobolus s.l. (Ancylistaceae, Entomophthorales) delimited all members that form capilliconidia into the genus Capillidium. In this study, we report two new species of Capillidium that were isolated in China. Capillidiummacrocapilliconidiumsp. nov. is characterised by large capilliconidia. Capillidiumjiangsuensesp. nov. is differentiated by large capilliconidia and long, slender secondary conidiophores. Phylogenetic analyses were performed using sequences from the nuclear large subunit of rDNA (nucLSU), the mitochondrial small subunit of rDNA (mtSSU) and elongation-factor-like (EFL). The analyses revealed sister relationships between Ca.macrocapilliconidiumsp. nov. and Ca.globuliferus / Ca.pumilum and between Ca.jiangsuensesp. nov. and Ca.denaeosporum. Additionally, a new combination of Ca.rugosum (Drechsler) B. Huang & Y. Nie comb. nov. is proposed herein. An identification key is provided for the ten accepted Capillidium species.
Keywords: Ancylistaceae, Capilliconidia, morphology, new taxa, phylogeny
Introduction
The taxonomic name Capillidium was first introduced as a subgenus within the genus Conidiobolus (Ancylistaceae, Entomophthorales) (Ben-Ze’ev and Kenneth 1982). All its members were clustered into a monophyletic group in the family Ancylistacaceae, based on four molecular loci [i.e. small subunit of nuclear rDNA (nucSSU), large subunit of nuclear rDNA (nucLSU), small subunit of mitochondrial rDNA (mtSSU) and elongation-factor-like (EFL)] (Nie et al. 2020). Species in this genus are typically characterised by capilliconidia protruding from elongated, slender conidiophores (Nie et al. 2020). Based on this synapomorphy and a re-examination of the protologue for Conidiobolus s.l. species (Drechsler 1953a, b, 1954, 1955a, 1957; Srinivasan and Thirumalachar 1967, 1968; Callaghan et al. 2000), seven species so far have been recombined into the monophyletic genus Capillidium, including: Ca.adiaeretum (Drechsler) B. Huang & Y. Nie, Ca.bangalorense (Sriniv. & Thirum.) B. Huang & Y. Nie, Ca.denaeosporum (Drechsler) B. Huang & Y. Nie, Ca.heterosporum (Drechsler) B. Huang & Y. Nie, Ca.lobatum (Sriniv. & Thirum.) B. Huang & Y. Nie, Ca.pumilum (Drechsler) B. Huang & Y. Nie and Ca.rhysosporum (Drechsler) B. Huang & Y. Nie (Nie et al. 2020).
Although Capillidium is a small genus with only seven accepted species, it possesses high morphological diversity. For instance, primary conidia range from 18 μm (Ca.pumilum) to 46 μm (Ca.adiaeretum) in size (Drechsler 1953a, 1955a); resting spores are present in Ca.adiaretum, Ca.bangalorense and Ca.rhysosporum, but not in Ca.denaeosporum, Ca.heterosporum, Ca.lobatum and Ca.pumilum (Drechsler 1953b, 1955a, 1957); Ca.heterosporum has slender conidiophores that are branched at the base and end with 2–6 terminal capilliconidia each (Drechsler 1953b), whereas other members are unbranched and end with one capilliconidia (Nie et al. 2020); although nearly all Capillidium species only produce capilliconidia, Ca.adiaeretum also produces microconidia (Callaghan et al. 2000). These important diagnostic characteristics can help mycologists form a comprehensive understanding of this fungal group.
Two species Ca.adiaeretum and Ca.heterosporum have been identified in China (Wang et al. 2010; Nie et al. 2020). Continuing investigations into Chinese Conidiobolus s.l. led to the discovery of two new species in the genus Capillidium. We describe them herein, suggest a new combination for this genus and provide an updated identification key for the species of Capillidium.
Materials and methods
Isolates and morphology
Plant debris was collected from Wanfo Mountain, Shucheng County, Anhui Province, China and Laoshan National Forest Park and Tianwang Town, Jiangsu Province, China. Pre-sterilised plastic bags were used to pack these plant debris samples. Isolation procedures were the same as described by Drechsler (1952) and King (1976a). Plant debris samples were incubated on inverted Petri dishes containing PDA medium (potato 200 g, dextrose 20 g, agar 20 g, H2O, 1 litre) at 21 °C for 4 days. The incubated dishes were examined daily under a stereomicroscope (SMZ1500, Nikon Corporation, Japan). When an entomophthoroid fungus appeared, it was transferred to a clean PDA plate for purification and then sub-cultivated for morphological studies. Microscopic structure was observed under a light microscope (BX51, Olympus Corporation, Tokyo, Japan) and imaged using a microscope-camera system (DP25, Olympus Corporation, Tokyo, Japan). The size and shape of the primary conidia, primary conidiophores, secondary conidiophores, capilliconidia etc. were measured and described using the method by King (1976a) and the type of replicative conidia were observed on 2% water agar (agar 2 g, H2O, 1 litre). All isolates were deposited at the Research Center for Entomogenous Fungi at Anhui Agricultural University, Anhui Province, China (RCEF) and duplicated at the China General Microbiological Culture Collection Center, Beijing, China (CGMCC). A total of 13 ex-types of Conidiobolus s.l. were acquired from the American Type Culture Collection, Manassas, VA, USA (ATCC).
DNA extraction, PCR amplification and sequencing
Total cellular DNA was extracted using the method by Watanabe et al. (2010). For phylogenetic analyses, three loci were amplified using relevant primer pairs: LR0R (5’-ACC CGC TGA ACT TAA GC-3’) / LR5 (5’-TCC TGA GGG AAA CTT CG-3’) for nucLSU (Vilgalys and Hester 1990), mtSSU1 (5’-GCW GCA GTG RGG AAT NTT GGR CAA T-3’) / mtSSU2R (5’-GTR GAC TAM TSR GGT ATC TAA TC-3’) for mtSSU (Zoller et al. 1999) and EF983 (5’-GCY CCY GGH CAY CGT GAY TTY AT-3’) / EF1aZ-1R (5’-ACA TCW CCG ACA CCC TTG ATC TTG -3’) for EFL (Nie et al. 2012).
PCR amplification was carried out in a 50 µl mixture containing 1 μl dNTPs (200 μM), 1 μl MgCl2 (2.5 mM), 10 µl Phusion HF buffer (5×), 1 μl primers each (0.5 μM), 100 ng genomic DNA and 0.5 μl Taq polymerase (0.04 Unit/l, Super Pfx DNA Polymerase, Cowinbioscience Co. Ltd., Shanghai, China). PCR runs were conducted under the following conditions: an initial denaturation step at 94 °C for 3 min followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 55 / 54 / 57 °C (nucLSU / mtSSU / EFL), extension at 72 °C for 1 min; a final extension step at 72 °C for 7 min. DNA sequences were generated on both strands by performing dideoxy-nucleotide chain termination on an ABI 3700 automated sequencer at the Shanghai Genecore Biotechnologies Company (Shanghai, China). Sequences were processed with Geneious 9.0.2 (http://www.geneious.com, Kearse et al. 2012) and deposited in GenBank under the accession numbers listed in Table 1.
Table 1.
The species used in phylogenetic analyses.
| Species | Strains* | GenBank accession numbers | References | ||
|---|---|---|---|---|---|
| nucLSU | EFL | mtSSU | |||
| Azygosporusmacropapillatus | CGMCC 3.16068 (T) | MZ542006 | MZ555650 | MZ542279 | Cai et al. (2021) |
| A.parvus | ATCC 14634 (T) | KX752051 | KY402207 | MK301192 | Cai et al. (2021) |
| Capillidiumadiaeretum | ARSEF 451 (T) | KC461182 | – | – | GenBank |
| Ca.adiaeretum | CGMCC 3.15888 | MN061284 | MN061481 | MN061287 | Nie et al. (2020) |
| Ca.bangalorense | ARSEF 449 (T) | DQ364204 | – | DQ364225 | Chen and Huang (2018) |
| Ca.denaeosporum | ATCC 12940 (T) | JF816215 | JF816228 | MK301181 | Nie et al. (2012, 2020) |
| Ca.globuliferum | CBS 152.56 (T) | MH869095 | – | – | Vu et al. (2019) |
| Ca.heterosporum | CBS 543.63 | MH869973 | – | – | Vu et al. (2019) |
| Ca.heterosporum | RCEF 4430 | JF816225 | JF816239 | MK301183 | Nie et al. (2012, 2020) |
| Ca.lobatum | ATCC 18153 (T) | JF816218 | JF816233 | MK301187 | Nie et al. (2012, 2020) |
| Ca.pumilum | ARSEF 453 (T) | EF392383 | – | EF392496 | GenBank |
| Ca.rhysosporum | ATCC 12588 (T) | JN131540 | JN131546 | MK301195 | Nie et al. (2018, 2020) |
| Ca.rhysosporum | CBS 141.57 | MH869215 | – | – | Vu et al. (2019) |
| Ca.rugosum | CBS 158.56 (T) | MH869097 | – | – | Vu et al. (2019) |
| Ca.marcocapilliconidium | CGMCC 3.16169 (T) | OL830454 | OL801337 | OL830457 | This article |
| Ca.marcocapilliconidium | RCEF 6332 | OL830455 | OL801338 | OL830458 | This article |
| Ca.jiangsuense | CGMCC 3.16168 (T) | OL830456 | OL801339 | OL830459 | This article |
| Conidioboluscoronatus | NRRL 28638 | AY546691 | DQ275337 | – | Lutzoni et al. (2004) |
| C.humicolus | ATCC 28849 (T) | JF816220 | JF816231 | MK301184 | Nie et al. (2012, 2020) |
| C.khandalensis | ATCC 15162 (T) | KX686994 | KY402204 | MK301185 | Nie et al. (2012, 2020) |
| C.lichenicolus | ATCC 16200 (T) | JF816216 | JF816232 | MK301186 | Nie et al. (2012, 2020) |
| C.polytocus | ATCC 12244 (T) | JF816213 | JF816227 | MK301194 | Nie et al. (2012, 2020) |
| Microconidiobolusnodosus | ATCC 16577 (T) | JF816217 | JF816235 | MK333388 | Nie et al. (2012, 2020) |
| M.paulus | ARSEF 450 (T) | KC788409 | – | – | Gryganskyi et al. (2013) |
| M.terrestris | ATCC 16198 (T) | KX752050 | KY402208 | MK301199 | Nie et al. (2016, 2020) |
| Neoconidioboluscouchii | ATCC 18152 (T) | JN131538 | JN131544 | MK301179 | Nie et al. (2016, 2020) |
| N.mirabilis | CGMCC 3.17763 (T) | MH282852 | MH282853 | MK333389 | Nie et al. (2018, 2020) |
| N.pachyzygosporus | CGMCC 3.17764 (T) | KP218521 | KP218524 | MK333390 | Nie et al. (2018, 2020) |
| N.stromoideus | ATCC 15430 (T) | JF816219 | JF816229 | MK301198 | Nie et al. (2012, 2020) |
| N.thromboides | ATCC 12587 (T) | JF816214 | JF816230 | MK301200 | Nie et al. (2012, 2020) |
*ARSEF, ARS Entomopathogenic Fungus Collection (Ithaca, U.S.A.). ATCC, American Type Culture Collection (Manassas, U.S.A). CBS, Westerdijk Fungal Biodiversity Institute (Utrecht, The Netherlands). CGMCC, China General Microbiological Culture Collection Center (Beijing, China). NRRL, ARS Culture Collection (Peoria, U.S.A). RCEF, Research Center for Entomogenous Fungi (Hefei, China). T = ex-type.
Phylogenetic analyses
The data for the three target loci (nucLSU, mtSSU and EFL) were produced during this study and during our previous study (Nie et al. 2020). Sequences were retrieved from GenBank and concatenated using SequenceMatrix 1.7.8 (Vaidya et al. 2011). For this analysis, fifteen species in four closely-related genera (Azygosporus, Conidiobolus s.s., Neoconidiobolus and Microconidiobolus) served as outgroups (Table 1). Local alignment was conducted with MUSCLE 3.8.31 (Edgar 2004) and manually refined with BioEdit v. 7.2.6 (Hall 1999). The aligned sequence matrix was deposited in TreeBase (https://treebase.org) under the submission ID S29102.
Phylogenetic analyses were performed using three different methods: Maximum Likelihood (ML), Maximum Parsimony (MP) and Bayesian Inference (BI). For ML and BI analyses, best-fit substitution models for each locus were estimated in Modeltest 3.7 using the Akaike Information Criterion (AIC) value (Posada and Crandall 1998). The ML phylogenetic analysis was statistically tested in RAxML 8.1.17 with 1000 bootstrap replicates (Stamatakis 2014). The BI analysis was carried out in MrBayes v.3.1.2 using Markov Chain Monte Carlo (MCMC) methods (Ronquist and Huelsenbeck 2003). Beginning with random starting trees, four MCMC chains ran simultaneously for 1 million generations. The trees were sampled once every 100 generations. These chains stopped when all convergences met and the standard deviation fell below 0.01. MP analyses were conducted using a heuristic search in PAUP* 4.0b10 (Swofford 2002). Bootstrap analyses were conducted with 1000 bootstrap replicates to determine the confidence levels of the nodes within the inferred tree topologies (Felsenstein 1985). Tree bisection-reconnection (TBR) was selected for branch swapping. Phylogenetic trees were checked with FigTree 1.4 (Rambaut 2012) and further modified with iTOL (https://itol.embl.de/).
Results
Phylogenetic analyses
The concatenated alignment included 30 strains, 15 of which were outgroups from Azygosporus, Conidiobolus s.s., Microconidiobolus and Neoconidiobolus (Table 1). The aligned three-locus datasets contained 1861 characters. Amongst these, 852 characters were constant, 159 were parsimony-uninformative and 850 were parsimony informative. The most parsimonious tree had a tree length (TL) consisting of 3445 steps, a consistency index (CI) of 0.5068, a homoplasy index (HI) of 0.4932, a retention index (RI) of 0.7145 and a rescaled consistency index (RC) of 0.3621. The ML and BI analyses were performed using the best models for nucLSU (TrNef+G), EFL (TIMef) and mtSSU (K81) partitioning. The final average standard deviation of the split frequencies was 0.0059 and the final likelihood value was -17189. The tree topology from ML analysis was identical to those obtained from MP and BI analyses. The final ML tree was generated with bootstrap support values from MP/ML analyses, as well as posterior probability values from BI analysis at each branch.
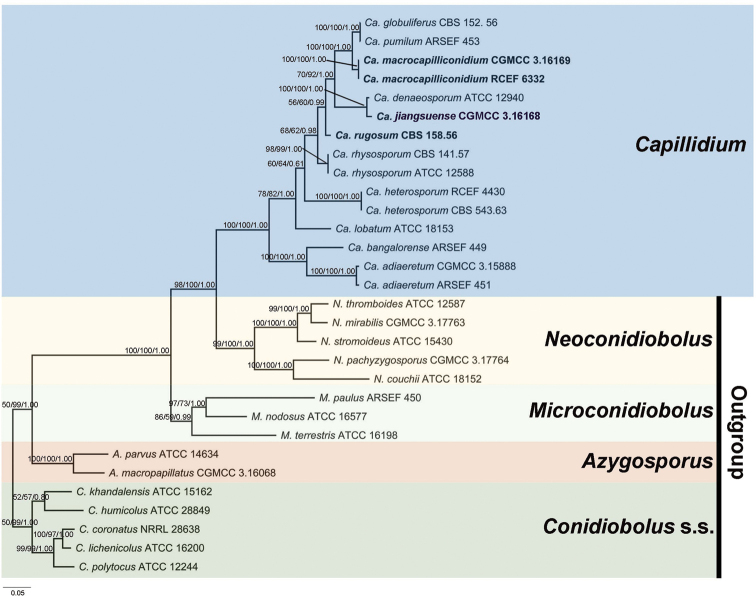
The phylogeny revealed that three strains belong to the genus Capillidium. The strains CGMCC 3.16169 / RCEF 6332 and CGMCC 3016168 were grouped closely with Ca.pumilum / Ca.globuliferus (100/100/1.00) and Ca.denaeosporum (100/100/1.00), respectively.
Taxonomy
. Capillidium macrocapilliconidium
B. Huang & Y. Nie sp. nov.
9433C5CA-C055-5CE6-A061-0EF291C0CDC4
842227
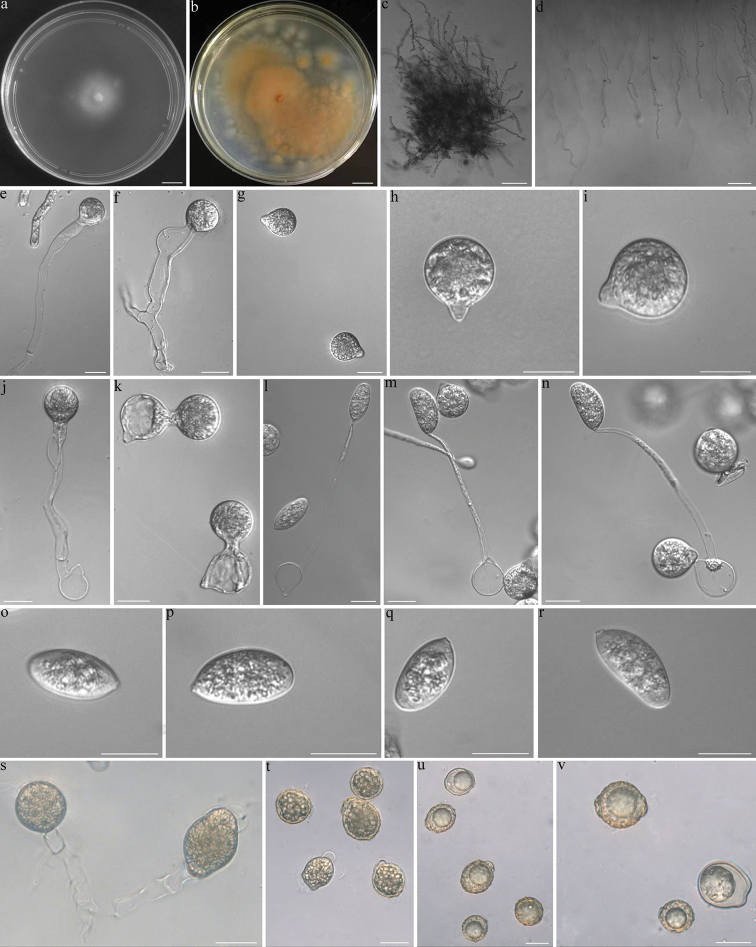
Figure 2.
Capillidiummacrocapilliconidiuma colony on PDA after 3 d at 21 °C b colony on PDA after 10 d at 21 °C c Mycelia d Mycelia unbranched at the edge of the colony e, f primary conidiophores bearing primary conidia g, h, i primary conidia j, k primary conidia bearing a single secondary conidium i, m, n a primary conidium bearing a single capilliconidium o, p, q, r Capilliconidia s zygospores that were formed on adjacent segments of the same hypha t immature zygospores u, v mature zygospores. Scale bars: 10 mm (a–b); 100 μm (c–d); 20 μm (e–v).
Etymology.
macrocapilliconidium (Lat.), referring to the large size of its capilliconidia.
Known distribution.
Jiangsu Province, China.
Typification.
China, Jiangsu Province, Nanjing City, Laoshan National Forest Park, 32°5'52"N, 118°35'37"E, from plant debris, 1 Dec 2018, Y. Nie and Y. Gao, culture ex-holotype CGMCC 3.16169 (=RCEF 6553).
Additional specimens examined.
China, Anhui Province, Shucheng County, Wanfo Mountain, 31°9'51"N, 116°57'86"E, from plant debris, 13 Mar 2016, X.X. Tang, culture RCEF 6332. GenBank: nrLSU = OL830455; EFL = OL801338; mtSSU = OL830458.
Description.
Colonies on PDA at 21 °C after 3 d white, reaching ca. 28 mm in diameter, yellowish after 10 d. Mycelia hyaline, 5.5–10 μm wide, often branched. Primary conidiophores arising from hyphal segments, hyaline, 70–250 × 5–13 μm, unbranched and producing a single globose primary conidium, widening upwards near the tip. Primary conidia forcibly discharged, globose to subglobose, 25–34 × 20–28 μm, papillate or conical, 7–10 μm wide, 3–8 μm long. Secondary conidiophores short or long, arising from primary conidia, bearing a single replicative conidium similar to, but smaller than those primary ones and forcibly discharged, producing another kind of replicative conidia called capillidiconidia from slender secondary conidiophores on the 2% water agar. Capillidiconidia colourless, elongate ellipsoidal, 25–37 μm long, 14–17 μm wide. Slender secondary conidiophores unbranched, 85–130 μm long, 4–6 μm wide at the base, tapering gradually to a width of 1–2 μm at the tip. Zygospores usually formed between adjacent segments of the same hypha after 10 d, yellowish, mostly boldly wrinkled, sometimes smooth, globose, elongate ellipsoidal or irregular, 18–35 μm long, 17–28 μm wide, with a wall 1–2 μm thick.
Notes.
Capillidiummacrocapilliconidium is characterised by having larger capilliconidia compared to other Capillidium species. It produces yellowish and wrinkled zygospores like Ca.rhysosporum (Drechsler 1954). However, Ca.macrocapilliconidium has larger capilliconidia than Ca.rhysosporum (25–37 × 14–17 μm in Ca.macrocapilliconidium vs. 12–32 × 6.5–16 μm in Ca.rhysosporum). Ca.macrocapilliconidium is phylogenetically distant from Ca.rhysosporum (Fig. 1) and most closely related to Ca.pumilum. It is distinguished from Ca.pumilum by larger primary conidia (25–34 × 20–28 μm in Ca.macrocapilliconidium vs. 9–18 × 7.3–14 μm in Ca.pumilum) and capilliconidia (25–37 × 14–17 μm in Ca.macrocapilliconidium vs. 8.8–12 × 5–7.5 μm in Ca.pumilum) (Drechsler 1955b).
Figure 1.
The phylogenetic tree of Capillidium constructed using Maximum Likelihood analyses on nucLSU, EFL and mtSSU sequences. Conidiobolus s.l. species were used as outgroups. New taxa are indicated by bold text. Maximum Parsimony bootstrap values (≥ 50%) / Maximum Likelihood bootstrap values (≥ 50%) / Bayesian posterior probabilities (≥ 0.50) of clades are provided alongside the branches. The scale bar at the lower left indicates substitutions per site.
. Capillidium jiangsuense
B. Huang & Y. Nie sp. nov.
FEEECB5A-1DCC-5FA1-9DFF-C083E5561569
842228
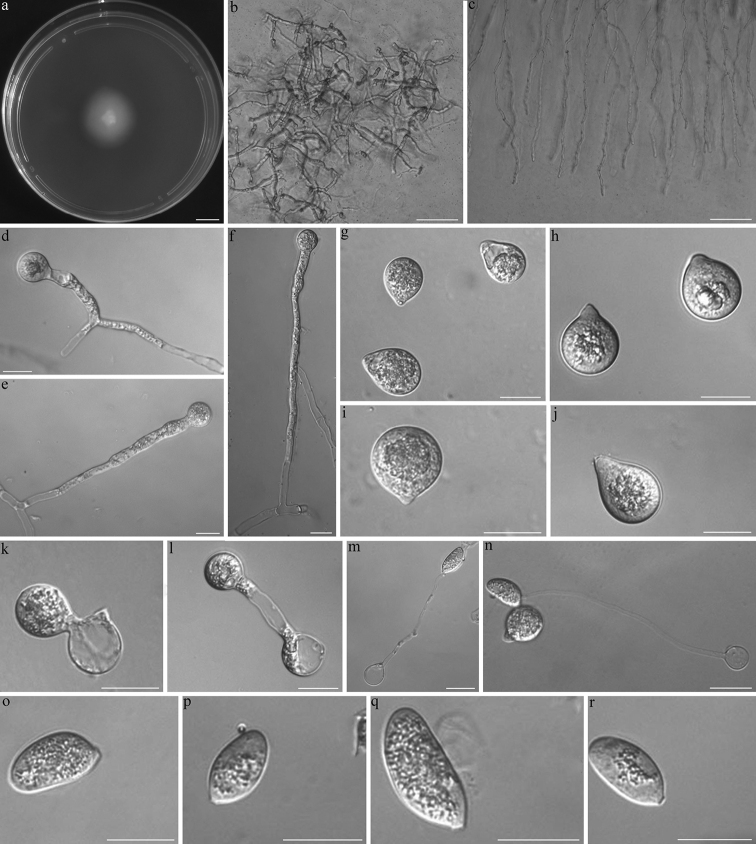
Figure 3.
Capillidiumjiangsuensea colony on PDA after 3 d at 21 °C b Mycelia c Mycelia unbranched at the edge of the colony d, e, f primary conidiophores arising from mycelia segments g, h, i, j primary conidia k, i secondary conidia arising from primary conidia m, n primary conidia bearing a single capilliconidium o, p, q, r Capilliconidia. Scale bars: 10 mm (a); 100 μm (b, c); 20 μm (d–r).
Etymology.
jiangsuense (Lat.), referring to the region where the fungus was isolated.
Known distribution.
Jiangsu Province, China.
Typification.
China, Jiangsu Province, Jurong City, Tianwang Town, 31°6'94"N, 119°26'91"E, from plant debris, 25 Mar 2018, Y. Nie, culture ex-holotype CGMCC 3.16168 (=RCEF 6545).
Description.
Colonies on PDA at 21 °C after 3 d white, reaching ca. 21 mm in diameter. Mycelia haline, often unbranched, vegetative hyphae filamentous, 5–10 μm wide. Primary conidiophores unbranched, producing a single primary conidium, widening upwards near the tip, 50–240 × 6–10 μm. Primary conidia forcibly discharged, subglobose to turbinate, 21–31 × 12–29 μm. Papilla 4–10 μm wide, 2–4 μm long. Replicative conidia two kinds on 2% water agar, arising from primary conidia, one similar and smaller to the primary conidia, the other elongate and passively detached, 17–32 × 10–15 μm. Slender secondary conidiophores unbranched, 65–120 μm long, 2.5–3 μm wide at the base, tapering gradually to a width of 1 μm at the tip. Resting spore not observed.
Notes.
Morphologically, the present isolate resembles Ca.denaeosporum because of the size of its primary conidia (13–32 × 6–21 μm in Ca.denaeosporum vs. 21–31 × 12–29 μm in Ca.jiangsuense) (Drechsler 1957). However, Ca.denaeosporum has larger capilliconidia (10–18 × 6–10 μm in Ca.denaeosporum vs. 17–32 × 10–15 μm in Ca.jiangsuense) and longer, more slender secondary conidiophores (35–65 μm in Ca.denaeosporum vs. 65–120 μm in Ca.jiangsuense) (Drechsler 1957). Although they grouped together with relatively little divergence on the phylogram, DNA similarity levels between the two species are only around 97.9% (nucLSU) (Nie et al. 2012). This evidence supports the present isolate being a new species, which we have named Capillidiumjiangsuense sp. nov.
. Capillidium rugosum
(Drechsler) B. Huang & Y. Nie comb. nov.
3D839536-F9ED-520C-8BB6-F183B93B704D
842229
Basionym.
Conidiobolusrugosus Drechsler, Am. J. Bot. 42: 437 (1955).
Description.
Refer to Drechsler (1955a).
Notes.
The ex-type living culture is ATCC 12586 (United States, New Jersey, Moorestown, 25 February 1954, Drechsler). Historically, Conidiobolusrugosus was synonymised with Co.heterosporus (King 1976b). However, we have re-established its taxonomic status at the species level, based on the phylogeny herein and the morphological traits of the capilliconidia.
Discussion
From the 1950s-1970s, a total of eight Conidiobolus species have been reported to produce capilliconidia, including Conidiobolusdenaeosporus, Co.globuliferus, Co.heterosporus, Co.inordinatus, Co.lobatus, Co.pumilus, Co.rhysosporus and Co.rugosus (Drechsler 1953a, b, 1954, 1955a, b, 1956, 1957; Srinivasan and Thirumalachar 1968). Based on the numerical taxonomy of Conidiobolus (King 1976a, b, 1977), four species were rejected. Co.rugosus was considered synonymous with C.heterosporus. On the other hand, Conidiobolusdenaeosporus, Co.globuliferus and Co.inordinatus were considered synonymous with Co.pumilus. Consequently, only four species forming capilliconidia were accepted into this genus. Based on this synapomorphy, the subgenusCapillidium was erected in the latter taxonomic study of Conidiobolus (Ben-Ze’ev and Kenneth 1982; Humber 1989). Interestingly, it appears that Co.adiaeretus and Co.bangalorensis develop both microconidia and capilliconidia (Callaghan et al. 2000). Unfortunately, there was no molecular evidence at the time to support these morphological results. Recently, we summarised molecular data from available Conidiobolus s.l. ex-types and identified a monophyletic lineage of Capillidium producing capilliconidia. Since then, some taxonomic revisions have been conducted. For example, Co.denaeosporus was separated from Co.pumilus and recombined into Capillidium. Co.adiaeretus and Co.bangalorensis were also recombined into Capillidium. In total, Capillidium now has seven accepted species.
Conidiobolusheterosporus (= Capillidiumheterosporum) and Co.rugosus share distinct morphological characteristics. For instance, Co.heterosposus bears no resting spores and has conidiophores that are often branched at the base and bear 2–6 terminal capilliconidia (Drechsler 1953a). The conidiophores of Co.rugosus, though, have yellowish zygospores with wrinkled or smooth surfaces, are unbranched and bear a single capilllicondiuma (Drechsler 1955b).
Based on a phylogenetic analysis of three gene regions (nucLSU, mtSSU and EFL), the ex-type of Co.rugosus (Strain No: CBS 158.56) and Co.heterosposus diverged into two distinct lineages. Consequently, we identified Co.rugosus as an independent species and recombined it into Capillidium as a new combination: Capillidiumrugosum (Drechsler) B. Huang & Y. Nie comb. nov. On a side note, while researchers previously considered Co.denaeosporus (= Ca.denaeosporum), Co.globuliferus and Co.inordinatus to be synonymous with Co.pumilus (= Ca.pumilum) (King 1976b), the present phylogeny confirmed that Co.denaeosporus (= Ca.denaeosporum) is an independent species and Co.globuliferus is synonymous with Co.pumilus (= Ca.pumilum). More molecular evidence is needed to clarify the taxonomic status of Co.inordinatus.
Capillidiumbangalorense may be another Capillidium species that forms microspores, based on its close phylogenetic relationship with Ca.adiaeretum. Besides microspores, these two species possess another morphological characteristic that is distinctive compared with the other members of Capillidium, that being the width between the primary conidiophores and the hyphae (Drechsler 1955a; Srinivasan and Thirumalachar 1967). This could explain why Ca.adiaeretum and Ca.bangalorense are grouped into a single clade in the phylogenetic tree (Fig. 1). However, Ca.bangalorense should be re-examined and more evidence should be supplied to confirm that this clade is in a separate taxon.
With the current description of Azygosporus, most members of Conidiobolus s.l. have now received suitable taxonomic placements. Yet, there are still many other taxonomic challenges to be resolved in the future, such as replacing the missing ex-type Co.utriculosis and assigning Co.coronatus as the epitype of Conidiobolus s.s., isolating lost ex-types to confirm their taxonomic placements etc. (Nie et al. 2018, 2020, 2021; Cai et al. 2021). For the first time, this study used partial sequence data from nucLSU, mtSSU and EFL genes to identify two new species of Capillidium from China, increasing the total number of species in the genus to ten. A key to the species of Capillidium is provided below.
Key to the Species of Capillidium
| 1 | Capilliconidia and microconidia produced, the width of primary conidiophores offers a pronounced dimensional contrast with the mycelial filaments | 2 |
| – | Only capilliconidia produced, the width of primary conidiophores offers a similar dimensional contrast with the mycelial | 3 |
| 2 | Primary conidia larger, up to 46 μm, chlamydospores produced | Ca.adiaeretum |
| – | Primary conidia smaller, less than 25 μm, zygospores produced | Ca.bangalorense |
| 3 | Slender conidiophores branched at the base, bearing 2–6 terminal capilliconidia | Ca.heterosporum |
| – | Slender conidiophores unbranched at the base, bearing a single capilliconidia | 4 |
| 4 | Resting spores of zygospores produced, yellowish, mostly wrinkled, sometimes smooth | 5 |
| – | Resting spores not observed | 6 |
| 5 | Primary conidia and zygospores larger, more than 30 μm | 7 |
| – | Primary conidia and zygospores smaller, less than 25 μm | Ca.rugosum comb. nov. |
| 6 | Primary conidia larger, more than 30 μm | 8 |
| – | Primary conidia smaller, less than 26 μm | 9 |
| 7 | Capilliconidia larger, up to 37 μm | Ca.macrocapilliconidium sp. nov. |
| – | Capilliconidia smaller, less than 32 μm | Ca.rhysosporum |
| 8 | Capilliconidia larger, 17–32 × 10–15 μm, primary conidiophores longer, 50–240 μm | Ca.jiangsuense sp. nov. |
| – | Capilliconidia smaller, 10–18 × 6–10 μm, primary conidiophores shorter, 15–50 μm | Ca.denaeosporum |
| 9 | Primary conidia larger, 21–26 × 20–24 μm, capilliconidia larger, 18–25 × 8–10 μm | Ca.lobatum |
| – | Primary conidia smaller, 9–18 × 7.3–14 μm, capilliconidia smaller, 8.8–12 × 5–7.5 μm | Ca.pumilum |
Supplementary Material
Acknowledgements
We thank Dr. Yang Gao (Jiangxi Agricultural University) for helping with sample collection. We also thank Dr. Jeffery Hannahn (Michigan State University) for his assistance with English language and grammatical editing. This study was supported by the National Natural Science Foundation of China (Nos. 31900008, 30770008 31670019 and 31970009).
Citation
Nie Y, Zhao H, Wang ZM, Zhou ZY, Liu XY, Huang B (2022) Two new species in Capillidium (Ancylistaceae, Entomophthorales) from China, with a proposal for a new combination. MycoKeys 89: 139–153. https://doi.org/10.3897/mycokeys.89.79537
Contributor Information
XiaoYong Liu, Email: 896760569@qq.com.
Bo Huang, Email: bhuang@ahau.edu.cn.
References
- Ben-Ze’ev IS, Kenneth RG. (1982) Features-criteria of taxonomic value in the Entomophthorales: I. A revision of the Batkoan classification. Mycotaxon 14: 393–455. [Google Scholar]
- Cai Y, Nie Y, Zhao H, Wang ZM, Zhou ZY, Liu XY, Huang B. (2021) Azygosporus gen. nov., a synapmorphic clade in the family Ancylistaceae. MycoKeys 85: 161–172. 10.3897/mycokeys.85.73405 [DOI] [PMC free article] [PubMed]
- Callaghan AA, Waters SD, Manning RJ. (2000) Alternative repetitional conidia in Conidiobolusadiaeretus: Development and germination. Mycological Research 104(10): 1270–1275. 10.1017/S0953756200003063 [DOI] [Google Scholar]
- Chen MJ, Huang B. (2018) Conidiobolusantarcticus, a synonym of C.osmodes. Mycotaxon 133(4): 635–641. 10.5248/133.635 [DOI]
- Drechsler C. (1952) Widespread distribution of Delacroixiacoronata and other saprophytic Entomophthoraceae in plant detritus. Science 115(2995): 575–576. 10.1126/science.115.2995.575 [DOI] [PubMed] [Google Scholar]
- Drechsler C. (1953a) Three new species of Conidiobolus isolated from leaf mold. Journal of the Washington Academy of Sciences 43(2): 29–34. [Google Scholar]
- Drechsler C. (1953b) Two new species of Conidiobolus occurring in leaf mold. American Journal of Botany 40(3): 104–115. 10.1002/j.1537-2197.1953.tb06458.x [DOI] [Google Scholar]
- Drechsler C. (1954) Two species of Conidiobolus with minutely ridged zygospores. American Journal of Botany 41(7): 567–575. 10.1002/j.1537-2197.1954.tb14380.x [DOI] [Google Scholar]
- Drechsler C. (1955a) A small Conidiobolus with globose and with elongated secondary conidia. Journal of the Washington Academy of Sciences 45(4): 114–117. [Google Scholar]
- Drechsler C. (1955b) Three new species of Conidiobolus isolated from decaying plant detritus. American Journal of Botany 42(5): 437–443. 10.1002/j.1537-2197.1955.tb11144.x [DOI] [Google Scholar]
- Drechsler C. (1956) Two new species of Conidiobolus. American Journal of Botany 43(10): 778–787. 10.1002/j.1537-2197.1956.tb11168.x [DOI] [Google Scholar]
- Drechsler C. (1957) Two medium-sized species of Conidiobolus occurring in Colorado. Journal of the Washington Academy of Sciences 47: 309–315. [Google Scholar]
- Edgar RC. (2004) MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32(5): 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1985) Confidence limits on the bootstrap: An approach using the bootstrap. Evolution; International Journal of Organic Evolution 38(4): 783–791. 10.1111/j.1558-5646.1985.tb00420.x [DOI] [PubMed] [Google Scholar]
- Gryganskyi AP, Humber RA, Smith ME, Hodge K, Huang B, Voigt K, Vilgalys R. (2013) Phylogenetic lineages in Entomophthoromycota. Persoonia 30(1): 94–105. 10.3767/003158513X666330 [DOI] [PMC free article] [PubMed]
- Hall TA. (1999) BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Humber RA. (1989) Synopsis of a revised classification for the Entomophthorales (Zygomycotina). Mycotaxon 34: 441–460. 10.1098/rspa.1937.0056 [DOI] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. (2012) Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics (Oxford, England) 28(12): 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DS. (1976a) Systematics of Conidiobolus (Entomophthorales) using numerical taxonomy I. Taxonomic considerations. Canadian Journal of Botany 54: 45–65. 10.1139/b76-008 [DOI] [Google Scholar]
- King DS. (1976b) Systematics of Conidiobolus (Entomophthorales) using numerical taxonomy II. Taxonomic considerations. Canadian Journal of Botany 54(12): 1285–1296. 10.1139/b76-141 [DOI] [Google Scholar]
- King DS. (1977) Systematics of Conidiobolus (Entomophthorales) using numerical taxonomy III. Descriptions of recognized species. Canadian Journal of Botany 55(6): 718–729. 10.1139/b77-086 [DOI] [Google Scholar]
- Lutzoni F, Kauff F, Cox CJ, McLaughlin D, Celio G, Dentinger B, Padamsee M, Hibbett DS, James TY, Baloch E, Grube M, Reeb V, Hofstetter V, Schoch C, Arnold AE, Miadlikowska J, Spatafora J, Johnson D, Hambleton S, Crockett M, Schoemaker R, Sun GH, Lücking R, Lumbsch HT, O’Donnell K, Binder M, Diederich P, Ertz D, Gueidan C, Hall B, Hansen K, Harris RC, Hosaka K, Lim YW, Liu Y, Matheny B, Nishida H, Pfister D, Rogers J, Rossman A, Schmitt I, Sipman H, Stone J, Sugiyama J, Yahr R, Vilgalys R. (2004) Where are we in assembling the Fungal Tree of Life, classifying the fungi and understanding the evolution of their subcellular traits? American Journal of Botany 91(10): 1446–1480. 10.3732/ajb.91.10.1446 [DOI] [PubMed]
- Nie Y, Yu CZ, Liu XY, Huang B. (2012) A new species of Conidiobolus (Ancylistaceae) from Anhui, China. Mycotaxon 120(1): 427–435. 10.5248/120.427 [DOI] [Google Scholar]
- Nie Y, Tang XX, Liu XY, Huang B. (2016) Conidiobolusstilbeus, a new species with mycelial strand and two types of primary conidiophores. Mycosphere. Journal of Fungal Biology 7(6): 801–809. 10.5943/mycosphere/7/6/11 [DOI] [Google Scholar]
- Nie Y, Qin L, Yu DS, Liu XY, Huang B. (2018) Two new species of Conidiobolus occurring in Anhui, China. Mycological Progress 17(10): 1203–1211. 10.1007/s11557-018-1436-z [DOI] [Google Scholar]
- Nie Y, Yu DS, Wang CF, Liu XY, Huang B. (2020) A taxonomic revision of the genus Conidiobolus (Ancylistaceae, Entomophthorales): Four clades including three new genera. MycoKeys 66: 55–81. 10.3897/mycokeys.66.46575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Y, Wang ZM, Liu XY, Huang B. (2021) A morphological and molecular survey of Neoconidiobolus reveals a new species and two new combinations. Mycological Progress 20(10): 1233–1241. 10.1007/s11557-021-01720-w [DOI] [Google Scholar]
- Posada D, Crandall KA. (1998) MODELTEST: Testing the model of DNA substitution. Bioinformatics (Oxford, England) 14(9): 817–818. 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Rambaut A. (2012) FigTree version 1.4.0. http://tree.bio.ed.ac.uk/software/figtree/
- Ronquist F, Huelsenbeck JP. (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics (Oxford, England) 19(12): 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Srinivasan MC, Thirumalachar MJ. (1967) Evaluation of taxonomic characters in the genus Conidiobolus with key to known species. Mycologia 59(4): 698–713. 10.1080/00275514.1967.12018462 [DOI] [Google Scholar]
- Srinivasan MC, Thirumalachar MJ. (1968) Two new species of Conidiobolus from India. Journal of the Mitchell Society 84: 211–212. 10.2307/3757098 [DOI] [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics (Oxford, England) 30(9): 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. (2002) PAUP*: Phylogenetic analysis using parsimony (*and other methods), Version 4.0b10. Sinauer Associates, Sunderland.
- Vaidya G, Lohman DJ, Meier R. (2011) SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27(2): 171–180. 10.1111/j.1096-0031.2010.00329.x [DOI] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172(8): 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu D, Groenewald M, de Vries M, Gehrmann T, Stielow B, Eberhardt U, Al-Hatmi A, Groenewald JZ, Cardinali G, Houbraken J, Boekhout T, Crous PW, Robert V, Verkley GJM. (2019) Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Studies in Mycology 92(1): 135–154. 10.1016/j.simyco.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CF, Li KP, Liu YJ, Li ZZ, Huang B. (2010) Three new Chinese records of Conidiobolus. Junwu Xuebao 29: 595–599.
- Watanabe M, Lee K, Goto K, Kumagai S, Sugita-Konishi Y, Hara-Kudo Y. (2010) Rapid and effective DNA extraction method with bead grinding for a large amount of fungal DNA. Journal of Food Protection 73(6): 1077–1084. 10.4315/0362-028X-73.6.1077 [DOI] [PubMed] [Google Scholar]
- Zoller S, Scheideggera C, Sperisena C. (1999) PCR primers for the amplification of mitochondrial small subunit ribosomal DNA of lichen-forming ascomycetes. Lichenologist (London, England) 31(5): 511–516. 10.1006/lich.1999.0220 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.