Abstract
Four antigens of Mycobacterium tuberculosis that are expressed in vivo after aerosol infection but prior to the development of clinical tuberculosis (TB) in rabbits were identified by immunoscreening of an expression library of M. tuberculosis genomic DNA with sera obtained 5 weeks postinfection. Three of the proteins identified, PirG (Rv3810), polymorphic GC-repetitive sequence (PE-PGRS; Rv3367), and proline-threonine repetitive protein (PTRP) (Rv0538), have multiple tandem repeats of unique amino acid sequences and have characteristics of surface or secreted proteins. The fourth protein, MtrA (Rv3246c), is a response regulator of a putative two-component signal transduction system, mtrA-mtrB, of M. tuberculosis. All four antigens were recognized by pooled sera from TB patients and not from healthy controls, confirming their in vivo expression during active infection in humans. Three of the antigens (PE-PGRS, PTRP, and MtrA) were also recognized by retrospective preclinical TB sera obtained, prior to the clinical manifestation of TB, from human immunodeficiency virus-TB patients, suggesting that they are potential candidates for devising diagnostic tests for active, preclinical TB.
The vast majority of Mycobacterium tuberculosis-infected individuals develop immune responses that arrest the progression of infection to clinical tuberculosis (TB) and also prevent latent bacilli from reactivating to cause clinical disease, whereas about 10 to 15% of infected individuals progress to developing primary or reactivation TB. An understanding of the host-pathogen interactions that occur after infection but prior to the development of clinical TB (preclinical TB) is required both for the design of effective vaccines and for the development of diagnostic tests for early disease.
Several studies have shown that M. tuberculosis adapts to different environments in broth media (17, 22, 37) and during intracellular residence by altering its gene expression (8, 22, 34). Earlier studies from our laboratory with cavitary and noncavitary TB patients have also shown that the in vivo environment in which the bacilli replicate affects the profile of the antigenic proteins expressed by M. tuberculosis (31). The goal of this study was to identify the antigens expressed by inhaled M. tuberculosis during the preclinical stages of TB. There are no markers to identify nondiseased humans with an active infection with M. tuberculosis, but the rabbit model of TB closely resembles TB in immunocompetent humans in that both species are outbred, both are relatively resistant to M. tuberculosis, and in both the caseous lesions may liquefy and form cavities (10). Studies have shown that on being inhaled, the bacilli are phagocytosed by (nonspecifically) activated alveolar macrophages, which either destroy or allow them to multiply. If the bacilli multiply, the alveolar macrophages die and the released bacilli are phagocytosed by nonactivated monocytes or macrophages that emigrate from the bloodstream. Intracellular replication and host cell death continue for 3 to 5 weeks, when both cellular and humoral immune responses are elicited (11, 24, 25). Lymphocytes and macrophages enter the foci of infection, and if they become activated, bacillary replication is controlled; if not, the infection progresses to clinical disease. During these initial stages of bacillary replication and immune stimulation, there are no outward signs of disease except for the conversion of cutaneous reactivity to purified protein derivative (PPD). The antigens of M. tuberculosis that are expressed and their interaction with the immune system during these preclinical stages of TB have not been delineated.
In view of the paucity of human material available to study the immunological events occurring after the inhalation of virulent bacilli but prior to the development of clinical TB, our studies are based on aerosol-infected rabbits. We reasoned that by 3 to 5 weeks postinfection, the sera from infected rabbits would contain antibodies to the antigens being expressed by the in vivo bacteria. Six pathogen-free rabbits (Covance Research Products, Inc., Denver, Pa.) were infected with aerosols of M. tuberculosis H37Rv, and another six were infected with aerosols of M. tuberculosis CDC1551 (10). The infected rabbits were bled 5 weeks postinfection, at which time all animals were tuberculin positive (average induration diameters, 260 and 130 mm2 in H37Rv- and CDC1551-infected rabbits, respectively); 11 had pulmonary tubercles (averages of 8 tubercles per rabbit in H37Rv infection and 5.2 tubercles per rabbit in CDC1551 infection), with average numbers of CFU of 3,460 in H37Rv- and 51 in CDC1551-infected rabbits (6). Sera from three normal (uninfected) rabbits were obtained as controls.
To obtain the antigenic proteins recognized by antibodies in the sera from the infected rabbits at this early stage postinfection, their serum pool was used to immunoscreen ∼1.2 × 105 PFU from the λgt11 expression library of M. tuberculosis H37Rv genomic DNA obtained from the World Health Organization (38). The library contains random sheared fragments of M. tuberculosis H37Rv genomic DNA cloned into λgt11 phage that express the foreign insert DNA as Escherichia coli β-galactosidase (β-Gal) fusion proteins. Several clones which showed reactivity with the infected rabbit serum pool significantly stronger than background were plaque purified. This article describes the results obtained with five of these clones (designated AD1, AD2, AD9, AD10, and AD16).
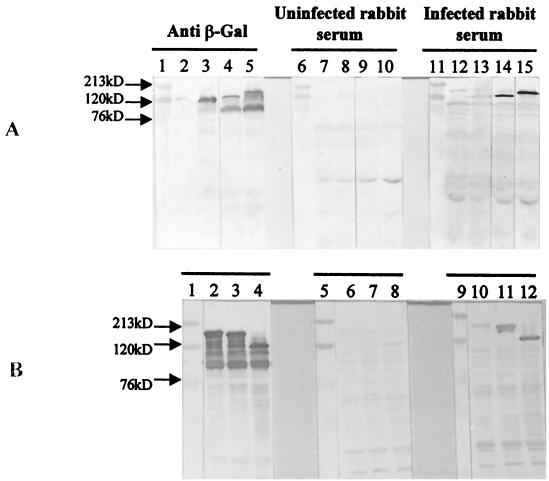
Lysates (5 μg) prepared from cultures of single colonies of lysogens of all five AD clones were fractionated on sodium dodecyl sulfate (SDS)–10% polyacrylamide (PA) gels, and Western blots were probed with the rabbit serum pools from infected or uninfected rabbits or with a mouse anti-β-Gal monoclonal antibody (32). All five recombinant clones produced β-Gal fusion proteins which ranged in size from 125 to 170 kDa (AD1, 130 kDa; AD2, 147 kDa; AD9, 163 kDa; AD10, 157 kDa; and AD16, 125 kDa) and which were recognized by both the anti-β-Gal monoclonal antibody (Fig. 1A, lanes 4 and 5, and B, lanes 2, 3, and 4) and the serum pool from the infected rabbits (Fig. 1A, lanes 14 and 15, and B, lanes 10, 11, and 12) but not the serum pool from the uninfected rabbits (Fig. 1A, lanes 9 and 10, and B, lanes 6, 7, and 8). Neither of the animal serum pools showed reactivity with the β-Gal protein in the control lysates (Fig. 1A, lanes 8 and 13).
FIG. 1.
Reactivity of β-Gal fusion proteins of AD clones with anti β-Gal antibody and sera from uninfected and M. tuberculosis-infected rabbits. Lysates (5 μg) of each AD lysogen and λgt11 vector lysogen were fractionated on SDS–10% PA gels, and Western blots were probed with anti-β-Gal antibody (lanes 2 to 5 in panel A and lanes 2 to 4 in panel B), a serum pool from uninfected rabbits (lanes 7 to 10 in panel A and lanes 6 to 8 in panel B), and a serum pool from infected rabbits (lanes 12 to 15 in panel A and lanes 10 to 12 in panel B). (A) Lanes 1, 6, and 11, molecular mass markers; lanes 2, 7, and 12, uninduced λgt11 lysogens; lanes 3, 8, and 13, induced λgt11 lysogens; lanes 4, 9, and 14, clone AD1; lanes 5, 10, and 15, clone AD2. (B) Lanes 1, 5, and 9, molecular mass markers; lanes 2, 6, and 10, clone AD9; lanes 3, 7, and 11, clone AD10; lanes 4, 8, and 12, clone AD16.
Restriction digestion of the DNA from the five clones with EcoRI yielded single inserts ranging from 3.7 to 5.6 kb (data not shown). The EcoRI inserts were subcloned into plasmid pGEMEX-1 (Promega, Madison, Wis.), and 450- to 700-bp nucleotides from each end were sequenced using SP6 and T3 commercial primers. DNA sequence similarities were identified by using BLAST from the National Center for Biotechnology Information website (2). The orientation of each insert in the AD clones was determined by restriction analysis (data not shown).
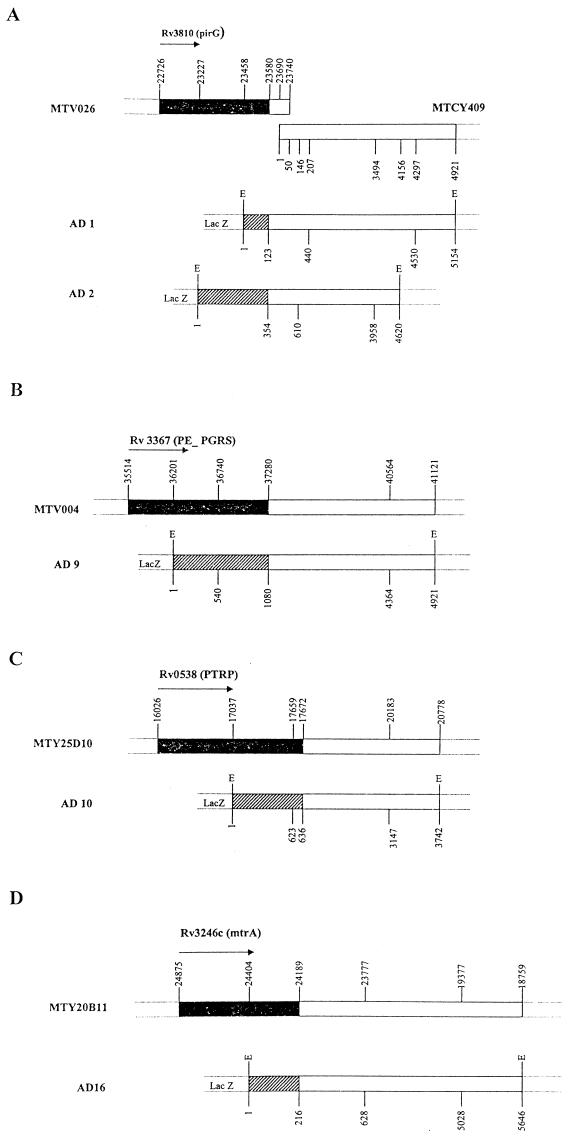
DNA sequence analyses of both ends of the EcoRI inserts of clones AD1 (5.1 kb) and AD2 (4.6 kb) revealed 98% identities with different regions of two overlapping cosmids, MTV026 and MTCY409 (Fig. 2A). Restriction analyses showed that the end of the inserts from both clones which showed homology with cosmid MTV026 was fused with β-Gal. The peptides expressed in clones AD1 (nucleotides 1 to 123) and AD2 (nucleotides 1 to 354) represent amino acids 245 to 284 and 168 to 284, respectively, in the C-terminal region of the Rv3810 (pirG) gene product. The PirG protein is a 284-amino-acid protein with a theoretical molecular mass and pI of 27.6 kDa and 4.34, respectively, and with 12 tandem repeats of five amino acids, PGLTS (Pro-Gly-Leu-Thr-Ser), in the central region. The isolation and partial characterization of this protein have been described earlier (3, 4).
FIG. 2.
Schematic maps showing positions of AD clones on cosmids of M. tuberculosis H37Rv. (A) Clones AD1 and AD2 on cosmids MTV026 and MTCY409. (B) Clone AD9 on cosmid MTV004. (C) Clone AD10 on cosmid MTY25D10. (D) Clone AD16 on cosmid MTY20B11. Black regions represent the gene on the cosmid. Hatched regions indicate regions expressed as β-Gal fusion proteins in AD clones. Arrows indicate the direction of translation. E, EcoRI site.
DNA sequence analyses of both ends of the EcoRI insert of clone AD9 (4.9 kb) revealed 94% identity with different regions of cosmid MTV004 (Fig. 2B). Restriction analysis showed that the end of the insert which was fused with β-Gal started within gene Rv3367 (polymorphic GC-repetitive protein [PE-PGRS]). The peptide expressed in clone AD9 (nucleotides 1 to 1080) represents amino acids 230 to 588 in the C-terminal region of the Rv3367 (PE-PGRS) gene product. This PE-PGRS protein has 588 amino acids and has 39 tandem copies of the motif Gly-Gly-Ala/Asn and 43 tandem copies of the motif Gly-Gly-X (a total of 82 repeats) spanning the entire protein except for the N-terminal region. The deduced amino acid sequence for clone AD9 contains 61 repeats of the motifs. The theoretical molecular mass and pI of the protein are 49.7 kDa and 4.05, respectively.
DNA sequence analyses of both ends of the EcoRI insert of clone AD10 (3.7 kb) revealed 94% identity with different regions of cosmid MTY25D10 (Fig. 2C). Restriction analysis showed that the end of the insert which was fused with β-Gal started within gene Rv0538. The peptide expressed in clone AD10 (nucleotides 1 to 636) represents amino acids 338 to 548 in the C-terminal region of the Rv0538 gene product. The Rv0538 gene product is a 548-amino-acid hypothetical protein with a repetitive proline- and threonine-rich region at the C terminus (proline-threonine repetitive protein [PTRP]). Amino acid sequence analysis of PTRP showed the presence of 23 tandem repeats of motif Pro-Pro-Thr-Thr in the C-terminal region from positions 415 to 516, with positions 2, 3, and 4 being better conserved than position 1. The theoretical molecular mass and pI of the protein are 55 kDa and 4.44, respectively.
DNA sequence analyses of both ends of the EcoRI insert of clone AD16 (5.6 kb) revealed 98% identity with different regions of cosmid MTY20B11 (Fig. 2D). Restriction analysis showed that the end of the insert which was fused with β-Gal started within gene Rv3246c (mtrA). The peptide expressed in clone AD16 (nucleotides 1 to 216) represents amino acids 157 to 228 in the C-terminal region of the Rv3246c (mtrA) gene product. The Rv3246c gene product is a 228-amino-acid MtrA response regulator protein, a putative transcriptional activator, which is identical (100% identity in a 225-amino-acid overlap) to the previously described response regulator protein MtrA of a putative two-component system, mtrA-mtrB, of M. tuberculosis H37Rv (36). The theoretical molecular mass and pI of MtrA are 25.2 kDa and 5.34, respectively.
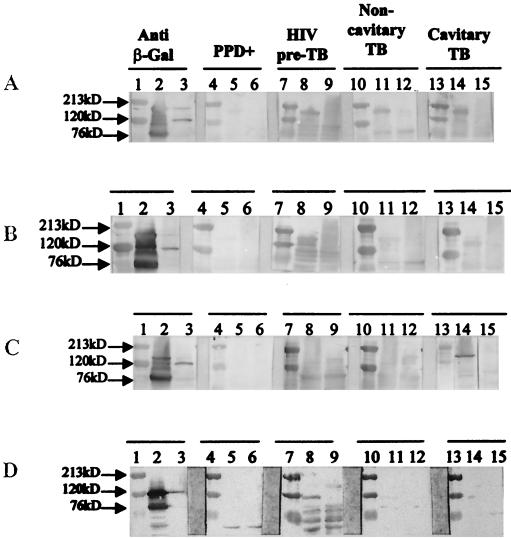
To determine the relevance of these proteins during human infection with M. tuberculosis, the reactivity of the four fusion proteins with sera from TB patients at different stages of disease progression was evaluated by Western blotting. A serum pool from five PPD-positive healthy controls showed poor or no reactivity with any of the fusion proteins (Fig. 3). In contrast, the fusion proteins of PE-PGRS (Fig. 3A), PTRP (Fig. 3B), and MtrA (Fig. 3D) were strongly reactive with pooled preclinical TB sera from five human immunodeficiency virus (HIV)-TB patients. These sera have been described in detail earlier (21). Briefly, these are sera from HIV-infected individuals who were routinely being monitored for their CD4+ T-cell numbers, and when these individuals developed TB, it was possible to obtain retrospective preclinical TB sera that had been saved (and frozen) from their previous hospital visits. These sera, obtained during the preclinical hospital visits from confirmed TB patients, represent the earliest stage of active M. tuberculosis infection that can be recognized in humans. Although multiple samples from each individual were available, to maintain the consistency of the preclinical TB time point only sera obtained 3 to 6 months prior to the diagnosis of clinical TB were included. The PE-PGRS fusion protein was also well recognized by the serum pools from two noncavitary, acid-fast bacillus sputum smear-negative, culture-positive TB patients and five cavitary, acid-fast bacillus-positive TB patients (Fig. 3A), but the PTRP and MtrA fusion proteins showed poorer reactivity with these serum pools (Fig. 3B and D). In contrast, the PirG fusion protein reacted only with the serum pool from the cavitary TB patients (Fig. 3C). In view of the observed reactivity of the preclinical TB serum pools with fusion proteins of three antigens, reactivity with individual preclinical TB serum samples from 10 patients and 3 PPD-positive controls was also assessed. All 10 preclinical TB serum samples recognized the PE-PGRS and PTRP fusion proteins, and 6 of the 10 patient serum samples had antibodies to the MtrA fusion protein (data not shown).
FIG. 3.
Reactivity of β-Gal fusion proteins with human sera. Lysates (5 μg) of each AD lysogen and λgt11 vector lysogen were fractionated on SDS–10% PA gels, and Western blots were probed with human sera. (A) Clone AD9 (PE-PGRS). (B) Clone AD10 (PTRP). (C) Clone AD2 (PirG). (D) Clone AD16 (MtrA). Lanes 1, 4, 7, 10, and 13, molecular mass markers; lanes 2, 5, 8, 11, and 14, respective AD clones; lanes 3, 6, 9, 12, and 15, λgt11 vector lysogens. Lanes 2 and 3 were probed with anti-β-Gal antibody, lanes 5 and 6 were probed with pooled sera from PPD-positive healthy individuals, lanes 8 and 9 were probed with pooled preclinical TB sera from HIV-TB patients, lanes 11 and 12 were probed with pooled sera from noncavitary TB patients, and lanes 14 and 15 were probed with pooled sera from cavitary TB patients.
It is interesting that three of the four proteins identified in this study have multiple repeats of different amino acid motifs, and all four proteins are either known to be or to have signatures of surface or secreted proteins of M. tuberculosis. Thus, the PirG gene product is identical (99.3% identity in a 284-amino-acid overlap) to the previously described cell surface protein ERP (exported repetitive protein) of M. tuberculosis (4) and to secreted antigen P36/P34 of Mycobacterium bovis (5) and has been shown to be a cell surface-exposed protein expressed by the bacilli during residence in the phagosomes of in vitro-maintained macrophages (3). Sequence analysis with SignalP version 2.0 software using neural networks and hidden Markov models trained on gram-positive bacteria (Center for Biological Sequence Analyses, Lyngby, Denmark) showed that the PE-PGRS protein possesses an N-terminal signal peptide with a putative signal peptidase cleavage site between amino acids 44 and 45; TMpred analysis predicted five transmembrane helices at amino acid positions 24 to 43, 166 to 186, 194 to 218, 351 to 368, and 431 to 451. PTRP was also predicted to contain four transmembrane domains at amino acid positions 97 to 114, 198 to 218, 278 to 299, and 379 to 398. The cellular location of M. tuberculosis MtrA is not known, although the homolog of MtrA was isolated from cell walls of Mycobacterium leprae (26).
Proteins with tandem repetitive amino acid sequences are found in several eukaryotic (1, 18, 20, 23, 30) and prokaryotic (13, 15, 16) organisms. In fact, the vast majority of gram-positive cell wall-associated proteins have tandem repeats of amino acid sequences which are associated with domains that bind to host cell ligands (7, 19). In many instances, the ability to alter the number of repetitive domains contributes to antigenic variations and to adaptation to environmental changes (19). Although the role of the repetitive proteins of M. tuberculosis obtained in this study is not yet known, PTRP (Rv0538) is structurally similar to other recently described mycobacterial proteins in that it has repeat motifs clustered in the C-terminal region (27, 33). A 21-kDa surface protein of M. leprae which has been shown to bind laminin-2 of peripheral nerves, thus facilitating the entry of the bacilli into Schwann cells, has 11 repeats of the XKKX motif at the C terminus (33). Also, a heparin-binding hemagglutinin of M. tuberculosis has been shown to bind to epithelial cells via the Pro-Lys repeats in the C-terminal region (27, 28). The structural similarities suggest that PTRP may have a similar function.
The PE-PGRS (Rv3367) protein belongs to the PE protein family, which is one of the two large clustered multigene families of glycine-rich acidic proteins discovered when the genome sequence of M. tuberculosis was determined (9). Some information is now available regarding the expression, subcellular location, and function of a few PE-PGRS proteins (14, 29). PE-PGRS proteins of Mycobacterium marinum which are homologous to M. tuberculosis PE-PGRS proteins (Rv3812 and Rv1651c) have been shown to be induced in cultured macrophages as well as in frog granulomas (29). Also, another member of the M. tuberculosis PE-PGRS family (Rv1759c) has been reported to be expressed in vivo but absent from antigen preparations made from bacteria grown in bacteriological media (14). Recent studies showed that M. tuberculosis aerosol-infected mice possess antibodies to the repetitive region of a PE-PGRS protein (Rv1818) but not to the PE region (12). Also, the C terminus (475 amino acids) of a PE-PGRS protein (Rv1759c) which has a large number of Gly-Gly-X repeats was shown to bind fibronectin (14). Preliminary experiments in which the above peptide (obtained from Clara Espitia) and the β-Gal fusion protein containing the C-terminal repetitive region of PE-PGRS (Rv3367) were evaluated showed that only the former and not the latter bound fibronectin (data not shown). Thus, it appears that not all PE-PGRS proteins bind fibronectin.
Although a recent analysis of ∼4,000 open reading frames from the genome sequence carried out to predict their subcellular location showed that, in contrast to Bacillus subtilis, M. tuberculosis has four times as many proteins with basic pIs (35), all four proteins identified in this study have acidic pIs, ranging between 4 and 6. Since ERP and MtrA are known to be expressed during intracellular residence (3, 36, 39), it is possible that the PTRP and PE-PGRS proteins are also expressed (or upregulated) under similar conditions. Interestingly, retrospective preclinical TB sera from HIV-TB patients contained antibodies to both of these proteins, and since none of these patients had recognizable cavitary lesions even at the time of the clinical manifestation of TB, bacterial replication would be intracellular during the preclinical TB stages. However, the presence of antibodies to these proteins in sera from cavitary TB patients suggests that they are also expressed during the later stages of the disease.
The presence in sera from TB patients of antibodies to all four proteins and their absence in sera from PPD-positive healthy individuals show that these proteins are expressed in vivo during active infection with M. tuberculosis in humans and that the native molecules (without β-Gal) are immunogenic. Sera from PPD-negative and HIV-positive asymptomatic individuals also failed to show any reactivity with these proteins (data not shown). It was shown previously that an 88-kDa culture filtrate protein (now identified as the 81-kDa M. tuberculosis malate synthase, GlcB) was recognized by antibodies in preclinical TB sera from about 75% of HIV-TB patients (21). Since the β-Gal fusion proteins of PE-PGRS, PTRP, and MtrA were also recognized by preclinical TB sera, these proteins may be useful for developing surrogate markers for identifying early, preclinical TB stages in HIV- and M. tuberculosis-coinfected individuals. Such markers have the potential to make a significant contribution to TB control in countries with a high incidence of coinfection.
Antibodies to M. bovis protein P36/P34, which is a homolog of PirG (ERP), are present in M. bovis-infected cattle and in leprosy patients (5). Our results show that cavitary TB patients have antibodies to the PirG fusion protein, but sera from noncavitary TB patients and preclinical TB sera did not show reactivity even when individual patients were tested (data not shown). It is possible that in the human tissue environment, this protein is not well expressed and therefore is immunogenic only when the bacterial load is high.
In summary, we have identified four antigenic proteins of M. tuberculosis that are immunodominant during relatively early, preclinical TB stages of an active M. tuberculosis infection. Three of the four antigens are proteins containing repetitive amino acid sequences and having characteristics of surface or secreted molecules. The involvement of these proteins in bacillary adhesion and/or invasion is currently under investigation. Three of the four antigens are potential candidates for devising immunodiagnostic tests for the identification of individuals with active preclinical TB.
Acknowledgments
We are indebted to Arthur M. Dannenberg, Jr., for providing the rabbit sera and for critical reading of the manuscript. We also thank Josephine E. Clark-Curtiss for critical reading of the manuscript.
This work was supported by the Research Center for AIDS and HIV Infection, Veterans Affairs Medical Center, Department of Veterans Affairs, and by NIH grant AI36984.
REFERENCES
- 1.Allred D R, Mcguire T C, Palmer G H, Leib S R, Harkins T M, McElwain T F, Barbet A F. Molecular basis for surface antigen size polymorphisms and conservation of a neutralization-sensitive epitope in Anaplasma marginale. Proc Natl Acad Sci USA. 1990;87:3220–3224. doi: 10.1073/pnas.87.8.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berthet F-X, Lagranderie M, Gounon P, Laurent-Winter C, Ensergueix D, Chavarot P, Thouron F, Maranghi E, Pelicic V, Portnoi D, Marchal G, Gicquel B. Attenuation of virulence by disruption of the Mycobacterium tuberculosis erp gene. Science. 1998;282:759–762. doi: 10.1126/science.282.5389.759. [DOI] [PubMed] [Google Scholar]
- 4.Berthet F-X, Rauzier J, Lim E M, Philipp W, Gicquel B, Portnoi D. Characterization of the Mycobacterium tuberculosis erp gene encoding a potential cell surface protein with repetitive structures. Microbiology. 1995;141:2123–2130. doi: 10.1099/13500872-141-9-2123. [DOI] [PubMed] [Google Scholar]
- 5.Bigi F, Alito A, Fisanotti J C, Romano M I, Cataldi A. Characterization of a novel Mycobacterium bovis secreted antigen containing PGLTS repeats. Infect Immun. 1995;63:2581–2586. doi: 10.1128/iai.63.7.2581-2586.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bishai W R, Dannenberg A M, Jr, Parrish N, Ruiz R, Chen P, Zook B C, Johnson W, Boles J W, Pitt M L M. Virulence of Mycobacterium tuberculosis CDC1551 and H37Rv in rabbits evaluated by Lurie's pulmonary tubercle count method. Infect Immun. 1999;67:4931–4934. doi: 10.1128/iai.67.9.4931-4934.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chhatwal G S, Preissner K T. Extracellular matrix interactions with gram-positive pathogens. In: Fischetti V A, Novick R P, Ferretti J J, Portnoy D A, Rood J I, editors. Gram-positive pathogens. Washington, D.C.: American Society for Microbiology; 2000. pp. 78–86. [Google Scholar]
- 8.Clark-Curtiss J E, Graham J E. Proceedings of the Thirty-Fourth Tuberculosis-Leprosy Research Conference. Bethesda, Md: National Institute of Allergy and Infectious Diseases, National Institutes of Health; 1999. Unraveling the secrets of mycobacterial pathogenesis; pp. 206–210. [Google Scholar]
- 9.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M A, Rogers J, Rutter S, Seegar K, Skelton J, Squares R, Squares S, Sulston J E, Taylor K, Whitehead S, Barell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 10.Converse P J, Dannenberg A M, Jr, Estep J E, Sugisaki K, Abe Y, Schofield B H, Pitt M L M. Cavitary tuberculosis produced in rabbits by aerosolized virulent tubercle bacilli. Infect Immun. 1996;64:4776–4787. doi: 10.1128/iai.64.11.4776-4787.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dannenberg A M., Jr Delayed-type hypersensitivity and cell mediated immunity in the pathogenesis of tuberculosis. Immunol Today. 1991;12:228–233. doi: 10.1016/0167-5699(91)90035-R. [DOI] [PubMed] [Google Scholar]
- 12.Delogu G, Chen Y, Brennan M J. ASM Conference on Tuberculosis: Past, Present, and Future. Washington, D.C.: American Society for Microbiology; 2000. Immunological characterization of a Mycobacterium tuberculosis PE-PGRS protein; p. 20. [Google Scholar]
- 13.Dramsi S, Dehoux P, Cossart P. Common features of gram-positive bacterial proteins involved in cell recognition. Mol Microbiol. 1993;9:1119–1122. doi: 10.1111/j.1365-2958.1993.tb01241.x. [DOI] [PubMed] [Google Scholar]
- 14.Espitia C, Laclette J P, Mondragon-Palomino M, Amador A, Campuzano J, Martens A, Singh M, Cicero R, Zhang Y, Moreno C. The PE-PGRS glycine-rich proteins of Mycobacterium tuberculosis: a new family of fibronectin-binding proteins. Microbiology. 1999;145:3487–3495. doi: 10.1099/00221287-145-12-3487. [DOI] [PubMed] [Google Scholar]
- 15.Fischetti V A, Jarymowycz M, Jones K F, Scott J R. Streptococcal M protein size mutants occur at high frequency within a single strain. J Exp Med. 1986;164:971–980. doi: 10.1084/jem.164.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaillard J-L, Berche P, Frehel C, Gouin E, Cossart P. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell. 1991;65:1127–1141. doi: 10.1016/0092-8674(91)90009-n. [DOI] [PubMed] [Google Scholar]
- 17.Garbe T R, Hibler N S, Deretic V. Response to reactive nitrogen intermediates in Mycobacterium tuberculosis: induction of the 16-kilodalton alpha-crystallin homolog by exposure to nitric oxide donors. Infect Immun. 1999;67:460–465. doi: 10.1128/iai.67.1.460-465.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ibanez C F, Affranchino J L, Macina R A, Reyes M B, Leguizamon S, Camargo M E, Aslund L, Pettersson U, Frasch A C C. Multiple Trypanosoma cruzi antigens containing tandemly repeated amino acid sequence motifs. Mol Biol Parasitol. 1988;30:27–34. doi: 10.1016/0166-6851(88)90129-6. [DOI] [PubMed] [Google Scholar]
- 19.Kehoe M A. Cell-wall-associated proteins in gram-positive bacteria. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science B. V.; 1994. pp. 217–261. [Google Scholar]
- 20.Kemp D J, Coppel R L, Anders R F. Repetitive proteins and genes of malaria. Annu Rev Microbiol. 1987;41:181–208. doi: 10.1146/annurev.mi.41.100187.001145. [DOI] [PubMed] [Google Scholar]
- 21.Laal S, Samanich K M, Sonnenberg M G, Belisle J T, O'Leary J, Simberkoff M S, Zolla-Pazner S. Surrogate marker of preclinical tuberculosis in human immunodeficiency virus infection: antibodies to an 88-kDa secreted antigen of Mycobacterium tuberculosis. J Infect Dis. 1997;176:133–143. doi: 10.1086/514015. [DOI] [PubMed] [Google Scholar]
- 22.Lee B-Y, Horwitz M A. Identification of macrophage and stress-induced proteins of Mycobacterium tuberculosis. J Clin Investig. 1995;96:245–249. doi: 10.1172/JCI118028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Longacre S, Hibner U, Raibaud A, Eisen H, Baltz T, Giroud C, Baltz D. DNA rearrangements and antigenic variation in Trypanosoma equiperdum: multiple expression-linked sites in independent isolates of trypanosomes expressing the same antigen. Mol Cell Biol. 1983;3:399–409. doi: 10.1128/mcb.3.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lurie M B. Host-parasite relations in natively resistant and susceptible rabbits on quantitive inhalation of human and bovine tubercle bacilli, and nature of genetic resistance to tuberculosis. In: Lurie M B, editor. Resistance to tuberculosis: experimental studies in native and acquired defensive mechanisms. Cambridge, Mass: Harvard University Press; 1964. pp. 192–222. [Google Scholar]
- 25.Lurie M B, Dannenberg A M., Jr Macrophage function in infectious disease with inbred rabbits. Bacteriol Rev. 1965;29:466–476. doi: 10.1128/br.29.4.466-476.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marques M A M, Chitale S, Brennan P J, Pessolani M C V. Mapping and identification of the major cell wall-associated components of Mycobacterium leprae. Infect Immun. 1998;66:2625–2631. doi: 10.1128/iai.66.6.2625-2631.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menozzi F D, Bischoff R, Fort E, Brennan M J, Locht C. Molecular characterization of the mycobacterial heparin-binding hemagglutinin, a mycobacterial adhesin. Proc Natl Acad Sci USA. 1998;95:12625–12630. doi: 10.1073/pnas.95.21.12625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pethe K, Aumercier M, Fort E, Gatot C, Locht C, Menozzi F D. Characterization of the heparin-binding site of the mycobacterial heparin-binding hemagglutinin adhesin. J Biol Chem. 2000;275:14273–14280. doi: 10.1074/jbc.275.19.14273. [DOI] [PubMed] [Google Scholar]
- 29.Ramakrishnan L, Federspiel N A, Falkow S. Granuloma-specific expression of mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science. 2000;288:1436–1439. doi: 10.1126/science.288.5470.1436. [DOI] [PubMed] [Google Scholar]
- 30.Richardson J P, Beecroft R P, Tolson D L, Liu M K, Pearson T W. Procyclin: an unusual immunodominant glycoprotein surface antigen from the procyclic stage of African trypanosomes. Mol Biochem Parasitol. 1988;31:203–216. doi: 10.1016/0166-6851(88)90150-8. [DOI] [PubMed] [Google Scholar]
- 31.Samanich K M, Belisle J T, Sonnenberg M G, Keen M A, Zolla-Pazner S, Laal S. Delineation of human antibody responses to culture filtrate antigens of Mycobacterium tuberculosis. J Infect Dis. 1998;178:1534–1538. doi: 10.1086/314438. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Shimoji Y, Ng V, Matsumura K, Fischetti V A, Rambukkana A. A 21-kDa surface protein of Mycobacterium leprae binds peripheral nerve laminin-2 and mediates Schwann cell invasion. Proc Natl Acad Sci USA. 1999;96:9857–9862. doi: 10.1073/pnas.96.17.9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith I, Duserget O, Rodriquez G M, Timm J, Gomez M, Dubnau J, Gold B, Manganelli R. Extra and intracellular expression of Mycobacterium tuberculosis genes. Tuber Lung Dis. 1998;79:91–97. doi: 10.1054/tuld.1998.0010. [DOI] [PubMed] [Google Scholar]
- 35.Tekaia F, Gordon S V, Garnier T, Brosch R, Barrell B G, Cole S T. Analysis of the proteome of Mycobacterium tuberculosis in silico. Tuber Lung Dis. 1999;79:329–342. doi: 10.1054/tuld.1999.0220. [DOI] [PubMed] [Google Scholar]
- 36.Via L E, Curcic R, Mudd M H, Dhandayuthapani S, Ulmer R J, Deretic V. Elements of signal transduction in Mycobacterium tuberculosis: in vitro phosphorylation and in vivo expression of the response regulator MtrA. J Bacteriol. 1996;178:3314–3321. doi: 10.1128/jb.178.11.3314-3321.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong D K, Lee B-Y, Horwitz M A, Gibson B W. Identification of Fur, aconitase, and other proteins expressed by Mycobacterium tuberculosis under conditions of low and high concentrations of iron by combined two-dimensional gel electrophoresis and mass spectrometry. Infect Immun. 1999;67:327–336. doi: 10.1128/iai.67.1.327-336.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young R A, Bloom B R, Grosskinsky C M, Ivannyi J, Thomas D, Davis R W. Dissection of Mycobacterium tuberculosis antigens using recombinant DNA. Proc Natl Acad Sci USA. 1985;82:2583–2587. doi: 10.1073/pnas.82.9.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zahrt T C, Deretic V. An essential two-component signal transduction system in Mycobacterium tuberculosis. J Bacteriol. 2000;182:3832–3838. doi: 10.1128/jb.182.13.3832-3838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]