Abstract
In malignant primary brain tumors, cancer cells infiltrate into the peritumoral brain structures which results in inevitable recurrence. Quantitative assessment of infiltrative heterogeneity in the peritumoral region, the area where biopsy or resection can be hazardous, is important for clinical decision making. Here, we derive a novel set of Artificial intelligence (AI)-based markers capturing the heterogeneity of tumor infiltration, by characterizing free water movement restriction in the peritumoral region using Diffusion Tensor Imaging (DTI)-based free water volume fraction maps. We leverage the differences in the peritumoral region of metastasis and glioblastomas, the former consisting of vasogenic versus the latter containing infiltrative edema, to extract a voxel-wise deep learning-based peritumoral microenvironment index (PMI). Descriptive characteristics of locoregional hubs of uniformly high PMI values are then extracted as AI-based markers to capture distinct aspects of infiltrative heterogeneity. The proposed markers are utilized to stratify patients’ survival and IDH1 mutation status on a population of 275 adult-type diffuse gliomas (CNS WHO grade 4). Our results show significant differences in the proposed markers between patients with different overall survival and IDH1 mutation status (t test, Wilcoxon rank sum test, linear regression; p < 0.01). Clustering of patients using the proposed markers reveals distinct survival groups (logrank; p < 10−5, Cox hazard ratio = 1.82; p < 0.005). Our findings provide a panel of markers as surrogates of infiltration that might capture novel insight about underlying biology of peritumoral microstructural heterogeneity, providing potential biomarkers of prognosis pertaining to survival and molecular stratification, with applicability in clinical decision making.
Subject terms: Computational models, Machine learning, Biomedical engineering
Introduction
The tumor burden of diffuse gliomas extends beyond the radiographically visible border of tumor margin1–3. However, current clinical practice considers the tumor core (T1 contrast enhancing boundary) as the primary target of treatment4,5; therefore, infiltrative cancer in the peritumoral region (peritumoral T2 hyperintense tissue) remains un-resected which may result in tumor recurrence. Obtaining tissue biopsies can be hazardous in the peritumoral region due to the possibility of functional deficit4,5. As a result, characterization of the infiltrative heterogeneity in the peritumoral region is a critical need to inform clinical decision making. Previous attempts to characterize tumor infiltration applied various imaging modalities using manually delineated or heuristic-based labeling of infiltrative tissue6–12, but approaches using Diffusion Tensor Imaging (DTI) have been limited to clinically used measures7–10.
DTI is the Magnetic Resonance Imaging (MRI) modality that provides insight into tissue microstructure by measuring water diffusivity. In particular, the free water volume fraction, a measure of the amount of extracellular water13, is able to capture differences between infiltrative and vasogenic peritumoral regions by exploiting the variation in water movement restriction14–16. The overarching goal of this paper is to leverage this unique information of DTI and derive artificial intelligence (AI)-based markers to capture infiltrative heterogeneity using the extracellular water-based voxel-wise characterization of tissue in the peritumoral region.
By leveraging the differences in water diffusivity properties in the peritumoral region of brain metastases and glioblastomas (Central Nervous System (CNS) World Health Organization (WHO) grade 4, Isocitrate-Dehydrogenase 1 (IDH1)-wildtype); consisting of purely vasogenic versus infiltrative edema respectively, we train a deep learning model to derive a novel voxel-wise peritumoral microenvironment index (PMI) without using any manual delineation of infiltrative regions. The PMI exploits characterization of water movement restriction in the voxels with and without infiltration and hence captures the infiltrative heterogeneity in the peritumoral region. We later apply connected component analysis to extract locoregional hubs of uniformly high PMI values, hypothesizing them as regions with high infiltration. Descriptive characteristics of locoregional hubs are calculated as AI-based markers of infiltrative heterogeneity, including number and size of hubs and the differences in their shape, direction, and spatial location.
The proposed AI-based markers are utilized for two clinical applications on a population of adult-type diffuse gliomas (CNS WHO grade 4) to demonstrate their potential in capturing distinct locoregional aspects of infiltrative heterogeneity beyond standard diffusion measures: (1) analysis of the duration of survival among IDH1-wildtype glioblastomas, to investigate whether higher infiltration of cancer cells, as measured by the proposed markers, is associated with shorter survival; and (2) differences across patients with varying IDH1 mutation status (astrocytoma IDH1-mutant versus glioblastoma IDH1-wildtype), notably, the IDH1-wildtype tumors, when compared with IDH1-mutants, have poorer prognosis, and higher peritumoral infiltration17,18.
Materials and methods
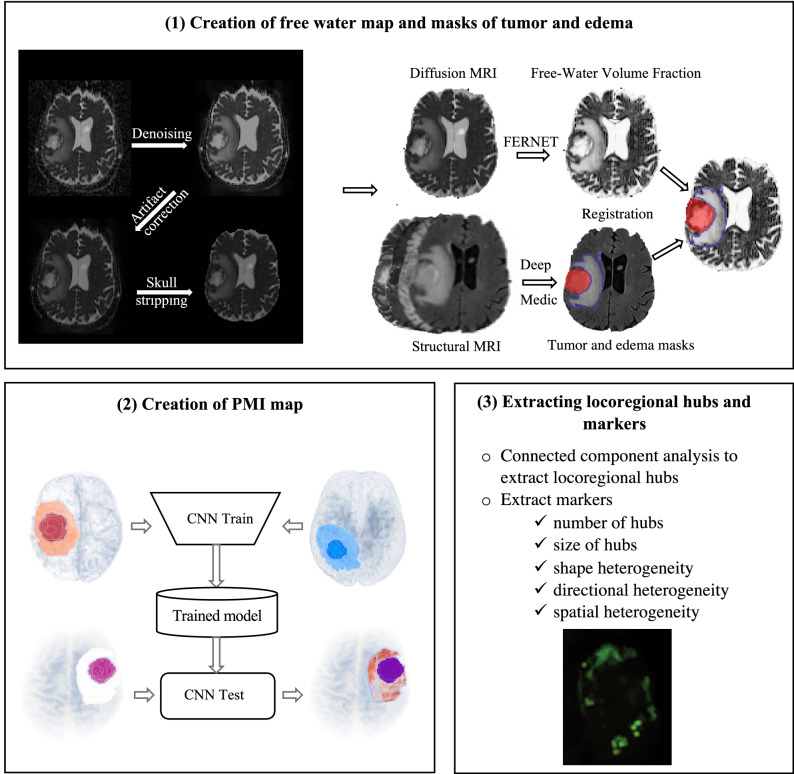
In this section, we describe the details of the datasets and steps of the pipeline for extracting the proposed AI-based markers. The pipeline consists of three steps. (i) creation of free water map and masks of tumor and edema, (ii) creation of the PMI map, (iii) extracting locoregional hubs and AI-based markers. Figure 1 demonstrates the overview of the pipeline. The proposed markers are evaluated on two clinical applications to investigate whether the differences in infiltrative heterogeneity are able to capture differences in overall survival and IDH1 mutation status in adult-type diffuse glioma patients.
Figure 1.
Different steps of the processing pipeline. The pipeline consists of three steps. (1) creation of free water map and masks of tumor and edema, (2) creation of the PMI map, (3) extracting locoregional hubs and AI-based markers. PMI: Peritumoral microenvironment index, CNN: Convolutional neural networks, FERNET: Freewater estimatoR using Interpolated Initialization, MRI: Magnetic resonance imaging.
Datasets
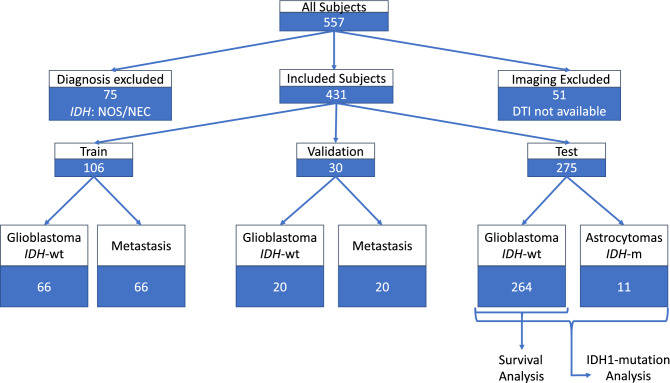
This study was approved by the institutional review board of the University of Pennsylvania. Informed consent was obtained from all participants or their legally authorized representative. All methods were carried out in accordance with relevant guidelines and regulations. The population was identified based on retrospective review of the electronic medical record of patients diagnosed with adult-type diffuse glioma (CNS WHO grade 4). Study inclusion criteria were (i) histopathological tissue diagnosis of adult-type diffuse glioma (CNS WHO grade 4) or brain metastasis and preoperative; (ii) availability of structural sequences (pre (T1) and post-contrast T1 weighted (T1CE), T2 weighted (T2), and T2 weighted fluid attenuated inversion recovery (T2-FLAIR)) and diffusion MRI at time of diagnosis. For the IDH1 mutation study, sufficient tumor tissue collected at time of surgery was required. We identified 381 patients with adult-type diffuse gliomas (CNS WHO grade 4) and 50 patients with brain metastases, and they were randomly divided into three datasets. Figure 2 demonstrates an overall view of the datasets. The training dataset included 106 patients with brain tumors, 66 IDH1-wildtype glioblastomas, aged 23–83 years (mean: 60.5, standard deviation (SD): 11.8), and in a male: female proportion of 37:29; and 40 metastases, aged 29–87 years (mean: 62.12, SD: 12.6), and in a male: female proportion of 18:22. The validation dataset was used to make locoregional hubs. This dataset was independent from the training dataset and consisted of 30 patients with brain tumors, 20 IDH1-wildtype glioblastomas and 10 brain metastases, aged 42–84 years (mean: 64.3, SD: 10.5), and in a male: female proportion of 16:14. The test dataset was used for two different applications, comprising survival among IDH1-wildtype glioblastomas and differences among IDH1 mutation status. The overall survival dataset included 264 IDH1-wildtype glioblastomas in the survival range of 0.43 to 76.9 month, aged 21–88 years (mean: 63.6, SD: 11.4), and in a male: female proportion of 160:104. The mutation dataset consisted of 275 CNS WHO grade 4 adult-type diffuse gliomas, 264 patients with IDH1-wildtype glioblastoma as in the survival data set and 11 IDH1-mutant astrocytomas, aged 21–88 years (mean: 62.5, SD: 12.5), and in a male: female proportion of 165:110. The acquisition parameters for all datasets were based on single-shell diffusion data on two Siemens 3 T scanners, Verio or TrioTim, with TR/TE in range of 4200–7400 ms/83–88 ms, with 1 unweighted volume and either 30 or 12-direction diffusion-weighted volumes, at a b value of 1000 s/mm2. The acquisition was repeated between 1 and 6 times for improved signal to noise ratio (SNR). The spatial resolution was 1.7 × 1.7 × 3 mm.
Figure 2.
Datasets: train, validation, test (survival and IDH1-mutation analysis). IDH1: Isocitrate-Dehydrogenase. NOS/NEC: Not elsewhere classified/Not otherwise specified.
Creation of free water map and masks of tumor and edema
All diffusion data was pre-processed using local Principal Component Analysis (PCA) denoising19, eddy current and motion correction using FSL EDDY20, and skull-stripping with Brain Extraction Tool (BET)21. Fractional anisotropy (FA) and mean diffusivity (MD) maps were calculated after DTI fitting with DIPY using weighted least squares22.
Structural scans (T1CE, T2, and T2-FLAIR) were registered to the pre-contrast T1 with rigid registration in Advanced Normalization Tools (ANTs)23,24, and then registered to the FA image with a nonlinear registration to account for Echo Planar Imaging (EPI) distortions in the diffusion data in ANTs. Masks of the tumor and edema for each patient were created from the registered structural images using Deep-Medic25. We used Freewater EstimatoR using Interpolated Initialization (FERNET)13, a free water elimination paradigm designed for single-shell diffusion MRI data using a novel interpolated initialization approach, to estimate the free water compartment in single-shell diffusion MRI data. FERNET provides the user with a free water volume fraction map that were resampled to 2×2×2 mm spatial resolution.
Creation of the PMI map
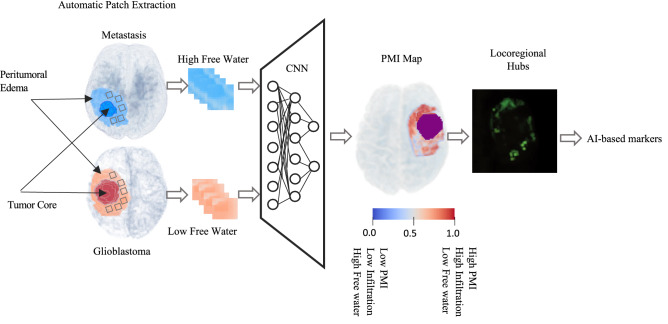
We used the training dataset to train a convolutional neural network (CNN)26 model using the differences in water diffusivity properties of metastasis and glioblastomas. Figure 3 demonstrates a schematic view for the generation of the PMI map. We automatically extracted a set of (16 × 16) voxel patches from the peritumoral edema of metastases and IDH1-wildtype glioblastomas placed at random locations and random directions (within axial, sagittal and coronal planes) using random seed generators. This was the largest patch that could be fit into edema without overlapping into the main tumor mass. Patches in the peritumoral edema of metastases and glioblastomas were labeled as high free water and low free water, respectively. The convolutional neural network consisted of 6 convolutional layers followed by a max-pooling and a fully connected layer. A softmax layer at the end produced a probability value for every input patch that indicated its membership to each class, either high free water or low free water27. Data augmentation was done on the patches by shifting them in random directions, letting a maximum of 20% overlap with the healthy brain. The hyper-parameters of CNN were weight decay 5 × 10−5, momentum 0.9, initial learning rate 10−4. The details of the CNN architecture can be found in supplementary material S.1.
Figure 3.
Generation of the PMI map and locoregional hubs. The inputs to the CNN are patches (boxes) extracted from the free water volume fraction map in the peritumoral region from both glioblastomas (red) and metastases (blue) labeled as low free water and high free water which are used to train the CNN. Locoregional hubs are extracted from PMI. Descriptive characteristics of the locoregional hubs are extracted as AI-based markers. PMI: Peritumoral microenvironment index, CNN: Convolutional neural networks.
To obtain the PMI map for new unseen patients, we placed patches centered at each voxel within the sagittal, axial, and coronal directions. The PMI value for each voxel was then calculated by averaging among the result of the CNN for the patches.
Extracting locoregional hubs and AI-based markers
For extracting locoregional hubs, we used the validation dataset. Connected components (CCs) of high PMI values28 were extracted and defined as locoregional hubs. Descriptive characteristics of locoregional hubs were extracted as AI-based markers for each patient. These characteristics comprised the number and size of the hubs, as well as the differences in their (i) shape (quantified by their individual anisotropic property), (ii) directionality, and (iii) spatial location (see Fig. 4 for a schematic view).
Figure 4.
A schematic view of locoregional hubs descriptive characteristics: number of hubs, size of hubs, shape heterogeneity, directional heterogeneity, and spatial heterogeneity. The purple circle displays the tumor core and blue hubs are located in the peritumoral edema. Arrows are directed toward increasing the values of the descriptive characteristics. Figure in the top-left is the reference figure for the hubs. (i) Moving from left to right, to the top-middle: the spatial heterogeneity of hubs increases; to the top-right: the shape heterogeneity increases. (ii) Moving from top to the bottom, left: the number of hubs increases; center: size of hubs increases; right: directional heterogeneity increases.
CCs were created for patients in different thresholds in range of [0 1] by steps of 0.1. Connected components with diameters less than 2 voxels were not taken into consideration. The threshold which provided the most significant differences in the descriptive characteristics between patients of the validation data set (30 samples of metastases and glioblastomas) was chosen (threshold = 0.9) and used for the mutation and the survival analysis.
For obtaining descriptive characteristics as AI-based markers, the number and size of the locoregional hubs were extracted and normalized based on the number of voxels in edema. The size was averaged among the hubs of each patient. Shape heterogeneity and directional heterogeneity were computed as follows: For each locoregional hub, coordinates for all its voxels along three (axial, sagittal, coronal) directions were extracted. Standard deviation of all voxels coordinate values along those three directions were calculated to make a three directional feature vector called sdvec. Shape heterogeneity was defined as the normalized difference between the maximum and second largest sdvec, which was later averaged among hubs of each patient. For directional heterogeneity, Hausdorff distance29 was applied on the pair-wise cosine distance among sdvec of different hubs. For computing spatial heterogeneity, the center of gravity for each hub and the average Euclidean distance among them was calculated. This distance was divided by the diameter, or maximum possible distance between any two points to make a normalized value between 0 and 1. A prototype code for extracting AI-based markers is provided in supplementary material S.2.
Analysis of survival and IDH1 mutation
The PMI maps were generated for the patients in the survival and mutation datasets. 8 patients were removed from both datasets as they had small edema size (less than two voxels around tumor). Next, the locoregional hubs of high PMI values were generated. 9 patients from the survival dataset and 10 patients from the mutation dataset were removed as they had less than 2 hubs which was the minimum number needed to compute directional and shape heterogeneity.
Survival analysis was done in two parts. First, we divided the population into short- and long-survival groups at the median and analyzed their statistical differences. This was done by doing: (i) two sample two tailed t test for each of the markers, followed by Bonferroni correction for multiple comparison; and (ii) linear regression on each of the markers with sex and age as covariates. Second, clusters of patients were generated downstream of the integration of all markers using K-means. We tried different number of clusters in the range of 1 to 9 and the one that maximized the Calinski-Harabasz index30 was chosen. Two clusters were derived, namely, low-PMI and high-PMI patients. Kaplan-Meier (KM)6 curves were fit for low-PMI and high-PMI clusters using nonparametric Turnbull estimator31. Logrank test and Cox hazard ratio were applied to compare KM-curves. The correlation between different markers were also calculated to explore their similarities.
IDH1 mutation analysis was done by statistical group difference in two different ways: (i) to consider differences in the number of samples in each group, Wilcoxon rank sum32 test was done for each of the markers, followed by Bonferroni correction for multiple comparison; (ii) linear regression was performed on each of the markers with sex and age as covariates.
Results
The proposed AI-based markers were evaluated on two clinical applications to investigate whether the differences in infiltrative heterogeneity, as captured by the proposed descriptive characteristics, were able to capture differences overall survival and IDH1 mutation status of adult-type diffuse glioma patients.
Survival analysis in IDH1-wildtype grade 4 gliomas
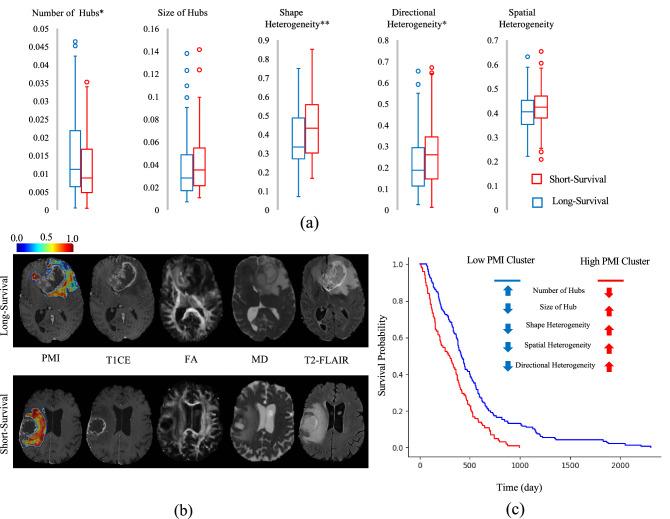
The survival analysis was done on a population of 264 IDH1-wildtype grade 4 glioma patients in the survival range of 0.43 to 76.9 months. The goal was to determine whether the proposed markers contain information pertaining to patient survival. Statistical analysis between short and long-survival groups demonstrated significant differences in the proposed markers, using t test and Bonferroni correction for multiple comparison. The short-survival group had significantly lower number of locoregional hubs (tnumber = 2.54, pnumber = 0.01). Shape and directional heterogeneity were also significantly higher in the short-survival group (tshape = 3.89, pshape < 0.001, tdirectional = 2.79, pdirectional = 0.005). Linear regression analysis of the markers using sex and age as covariates revealed significant differences between short and long-survival groups (Fig. 5a). The short-survival group had significantly lower number of locoregional hubs (tnumber = 2.65, pnumber = 0.008). Shape and directional heterogeneity were significantly higher in the short-survival group (tshape = 3.74, pshape < 0.001, tdirectional = 2.44, pdirectional = 0.015). Size and spatial heterogeneity were not found to be significantly different. Examples of PMI maps (overlaid with structural MRI), along with T1CE, FA, MD, and T2-FLAIR images, for a short and a long-survival patient are presented in Fig. 5b. KM estimates6 of the low-and high-PMI groups of patients (generated using K-means clustering on integration of all markers) are provided in Fig. 5c. A logrank test showed a significant difference between KM curves of low- and high-PMI clusters (t = 19.9, p < 10−5), and the Cox hazard ratio was 1.82 (95% confidence interval: 1.39, 2.37; p < 0.005). Group differences among high- and low-PMI clusters, were similar to short and long-survival groups, respectively (Fig. 5c for details and further information in S.3). The correlations between the proposed markers showed the highest correlations between shape heterogeneity and directional heterogeneity. However, they still provide complementary information as coefficient of determination (R2) showed that only 43% of information in directional heterogeneity was covered by shape heterogeneity (details in S.4).
Figure 5.
The PMI map and AI-based markers for IDH1-wildtype grade 4 glioma patients with different duration of survival. (a) AI-based markers (Descriptive characteristics of PMI locoregional hubs) for long and short survival groups, p value < 0.05 (*), p value < 0.005 (**), Linear regression was used with age and sex as covariates. (b) representative samples of the PMI map with T1CE, FA, MD, and T2-FLAIR images for long and short survival patients. (c) Kaplan–Meier estimates of the two clusters generated by K-means clustering. PMI: Peritumoral Microenvironment Index, IDH1: Isocitrate-Dehydrogenase 1, T1CE: T1 post-contrast, FA: Fractional anisotropy, MD: Mean diffusivity, T2-FLAIR: T2 weighted fluid attenuated inversion recovery.
IDH1 mutation analysis of grade 4 gliomas
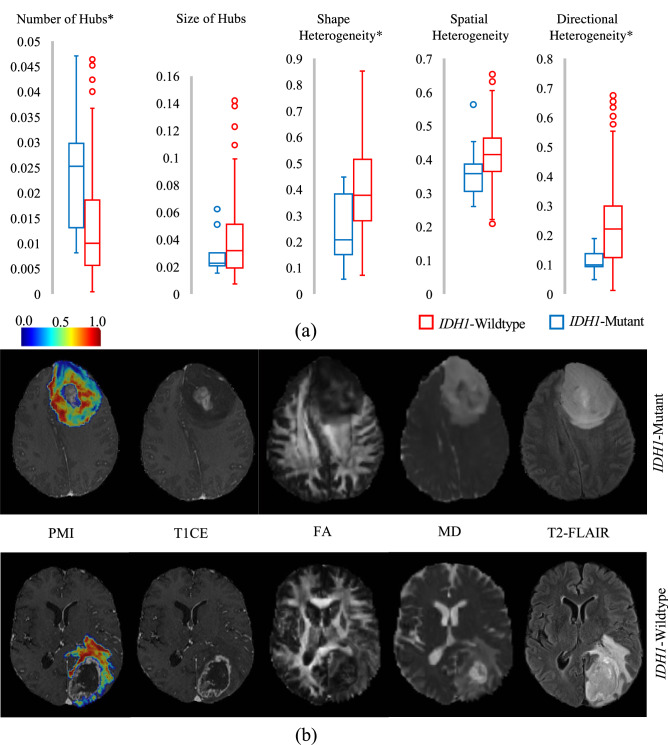
IDH1 mutation analysis was done on a population of 275 CNS WHO grade 4 adult-type diffuse gliomas with different IDH1 mutation status (i.e., 264 IDH1-wildtype glioblastomas vs. 11 IDH1-mutant astrocytomas) to investigate whether the proposed markers can characterize mutation status. Statistical analysis across the population, using Wilcoxon rank sum test and Bonferroni correction for multiple comparison, demonstrated significant differences in the proposed markers between IDH1-mutants and IDH1-wildtypes. The IDH1-wildtype group had significantly lower number of locoregional hubs (znumber = 4.11, pnumber < 10−4). Shape heterogeneity and directional heterogeneity were also significantly higher in IDH1-wildtypes (zshape = 3.15, pshape = 0.001, zdirectional = 3.5, pdirectional < 10−3). Linear regression analysis of the proposed markers using sex and age as covariates revealed significant differences between the two groups (Fig. 6a). IDH1-wildtypes had significantly lower number of locoregional hubs, when compared with IDH1-mutants (tnumber = 2.861, pnumber = 0.005). Shape heterogeneity and directional heterogeneity were significantly higher in IDH1-wildtypes comparing to IDH1-mutants (tshape = 2.407, pshape = 0.01, tdirectional = 2.380, pdirectional = 0.01). Size and spatial heterogeneity were not found to be significantly different. Examples of PMI maps (overlaid with structural MRI), along with the corresponding T1CE, FA, MD, and T2-FLAIR images for two patients (IDH1-mutant astrocytoma and IDH1-wildtype glioblastoma) are presented in Fig. 6(b). AI-based markers of the complete population of IDH1-mutants versus IDH1-wildtype long-survival group versus IDH1-wildtype short-survival group are provided in S.5.
Figure 6.
The PMI map and AI-based markers for grade 4 glioma patients with different IDH1 mutation status. (a) AI-based markers (Descriptive characteristics of PMI locoregional hubs) for IDH1-mutant and IDH1-wildtype groups, p value < 0.05 (*), p value < 0.005 (**), Linear regression was used with age and sex as covariates. (b) representative samples of the PMI map with T1CE, FA, MD, and T2-FLAIR images for IDH1-mutant and IDH1-wildtype patients. IDH1: Isocitrate-Dehydrogenase 1, T1CE: T1- and post-contrast, FA: Fractional anisotropy, MD: Mean diffusivity, T2-FLAIR: T2 weighted fluid attenuated inversion recovery.
Discussion
We introduced fully automated, novel AI-based markers of the peritumoral microenvironment (PME) of adult-type diffuse gliomas using unexplored information of water restriction extracted from DTI. The markers were based on descriptive characteristics derived from locoregional hubs of the PMI map that captured a unique aspect of the infiltrative heterogeneity from the extracellular water properties of the peritumoral regions of metastases and glioblastomas. Our findings identified a panel of markers as surrogates of infiltration, that could be considered as potential biomarkers of prognosis based on survival and IDH1 mutation status.
Previous methods on characterizing the infiltrative heterogeneity utilized various imaging modalities using manual or heuristic-based labeling of infiltrative tissue6–12. DTI, the modality to characterize water movement, has been used for defining infiltrative heterogeneity in the peritumoral region33. However, the extracellular free water movement differences have not been explored for peritumoral region. Therefore, the proposed AI-based markers presented here quantified the heterogeneity in the PME which could complement and further strengthen other diffusion-based measures and MRI parameters to aid in personalized treatment planning toward optimizing patient outcome.
Peritumoral edema, happens due to excess accumulation of fluid in the brain parenchyma which results from infiltrating tumor cells as well as biological responses to the permeability of the spatially adjacent tumor cells6–8. The spatial distribution and level of water dynamics pattern in edema has not been well defined yet6–8. Using a CNN model trained by characteristics of water diffusivity in glioblastomas versus brain tumor metastases, we were able to capture peritumoral heterogeneity in adult-type diffuse gliomas, without using any manual labeling, to capture surrogates of infiltration and learn how brain parenchyma and immunes system responded to different malignancies.
Brain peritumoral tissue is a heterogeneous microenvironment and accurately quantifying this heterogeneity can be crucial for understanding tumor progression. The proposed markers described regionally distinct water movement centers that contained voxels sharing the same brain tissue microstructures and we demonstrated their utility in two clinical applications.
Previous studies showed that higher infiltration of tumor cells is associated with poorer prognosis6,7,34,35. The PMI maps also demonstrated higher PMI values in the voxels of the short-survival patients. Previous literature reporting differences in the levels of FA and MD in altered infiltrated tissues which might be related to higher cellularity and lower water content7,8,36,37. Our result provides complementary information in regard to extracellular water movement restriction differences. Analysis of the proposed markers between poor prognosis (short-survivors and high-PMI cluster) and good prognosis (long-survivors and low-PMI cluster), showed a significantly lower number of locoregional hubs in the poor prognosis group which implies larger hubs corresponding to a more infiltrative and aggressive type of tumor. A significantly higher value of shape heterogeneity in the poor prognosis group could reflect the fact that in adult-type diffuse gliomas, cancer cells mainly infiltrate along the white matter38–40. Likewise, higher value of directional heterogeneity for poor prognosis patients could reflect the fact that cancer spreads in different directions comparing to good prognosis group. This is consistent with previous literature showing that glioma cells infiltrate brain tissue by at least two topographic paths, including perivascular invasion along the vascular system or infiltration along the extracellular matrix, nerve, and astrocytic tracts41.
IDH1-wildtype glioblastoma has a poorer prognosis than the IDH1-mutant astrocytoma17,18,34 which shows higher PMI values. The statistical differences of the proposed markers between IDH1-mutants and IDH1-wildtypes suggests them as powerful markers to discriminate mutation status, even prior to biopsy. IDH1-wildtypes had a lower number of locoregional hubs, which could reflect bigger size of adjacent tumoral cells compared to IDH1-mutants. This might represent extracellular vesicles which are involved in the rich network of intercellular connections and develop pathologic cascade leading to neurological diseases42. Likewise, higher shape and directional heterogeneity in IDH1-wildtypes, could demonstrate that in IDH1-wildtypes, cancer spreads more throughout the surrounding brain tissue, reflecting poorer prognosis which might result in higher tumor cellularity and infiltration, causing FA and MD changes43,44.
The locoregional hubs-based markers introduced here capture the heterogeneity in masses of adjacent voxels with high infiltration which could correspond to a connected set of glioma cells in the peritumoral region. Multicellular networks with filamentous microtubes are found to connect tumoral cells to a network in the peritumoral space in animal models41,45. These cellular networks allow rapid progression and are likely related to treatment resistance41,45. Therefore, biologically applicable markers can be identified in highly connected tumor cells that provide insights into the tumor microenvironment and offer translation to the clinic41,45.
The proposed markers could be interpreted as signatures of connected tumoral cells. Size and number of hubs quantitated modularity which is a measure of the structure of networks. Lower number of hubs in the poor prognosis group demonstrated their higher modularity. Higher values of shape and directional heterogeneity in poor prognosis group suggest connected tumoral cells with higher shape and directional heterogeneity. This might provide potentially important biomarkers that could be further explored as key targets for treatment planning and patient selection for clinical trials, including novel immunotherapy and anti-invasion therapy41,45,46.
The data in this study was collected from a single institution. This limitation was addressed by utilizing different acquisition settings and use of an independent cohort for testing, which provides strong sign of a generalizability of our results. The number of IDH1 mutant subjects were limited in our study since only approximately 5.6% of primary glioblastomas are IDH1 mutant47. The statistical significance of our findings can be improved by higher number of subjects. Our markers can be extracted fully automatic and do not require any manual intervention or reference labels and are easily translatable to the clinic. Although the proposed markers without a corresponding resected specimen cannot pathologically prove the infiltrative heterogeneity of the peritumoral region, they can be used as surrogate markers for prognosis.
In the future, the proposed markers could be integrated with other imaging modalities to provide biologically relevant characterization of the PME to quantitate invasion and microenvironmental heterogeneity for effective cancer therapy48–50. Further studies using the markers described here could be useful to elucidate biological processes linked to the IDH1 mutation status and better understand multiple sources of heterogeneity in adult-type diffuse gliomas which has major clinical implications51–53.
Supplementary Information
Acknowledgements
Research reported in this publication was supported by the National Institutes of Health (NIH) Grant R01NS096606 (PI: Ragini Verma), National Cancer Institute (NCI) of the National Institutes of Health (NIH) under award numbers: NCI: U01CA242871, NCI: U24CA189523, NINDS: R01NS042645. The content of this publication is solely the responsibility of the authors and does not represent the official views of the NIH.
Author contributions
Conceptualization: Z.R.S., R.V.; Methodology: Z.R.S.; Experiments: Z.R.S.; Data processing: D.P.; Analysis: Z.R.S., D.P., H.A.; Interpretation: Z.R.S., D.P., H.A., R.L.W., S.B., Sp.B., R.V.; Writing and editing: Z.R.S., D.P., H.A., R.L.W., S.B., Sp.B., R.V.; Supervision: S.B., Sp.B., R.V.
Data availability
The datasets generated during the current study are not publicly available due to the IRB requirements of the Hospital of University of Pennsylvania. The datasets can be made available on request to the corresponding author, after required data transfer and IRB paperwork is completed. The code is available here: https://github.com/zrsamani/AI-Based-Locoregional-Markers.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-26448-9.
References
- 1.Sebök M, et al. Hypermetabolism and impaired cerebrovascular reactivity beyond the standard MRI-identified tumor border indicate diffuse glioma extended tissue infiltration. Neuro Oncol. Adv. 2021;3:1–9. doi: 10.1093/noajnl/vdab048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Claes A, Idema AJ, Wesseling P. Diffuse glioma growth: A guerilla war. Acta Neuropathol. 2007;114:443–458. doi: 10.1007/s00401-007-0293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J, et al. Glioma invasion along white matter tracts: A dilemma for neurosurgeons. Cancer Lett. 2022;526:103–111. doi: 10.1016/j.canlet.2021.11.020. [DOI] [PubMed] [Google Scholar]
- 4.Cuddapah VA, Robel S, Watkins S, Sontheimer H. A neurocentric perspective on glioma invasion. Nat. Rev. Neurosci. 2014;15:455–465. doi: 10.1038/nrn3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelly PJ, Daumas-Duport C, Kispert DB, Kall BA, Scheithauer BW, Illig JJ. Imaging-based stereotaxic serial biopsies in untreated intracranial glial neoplasms. J. Neurosurg. 1987;66:865–874. doi: 10.3171/jns.1987.66.6.0865. [DOI] [PubMed] [Google Scholar]
- 6.Akbari H, et al. Pattern analysis of dynamic susceptibility contrast-enhanced MR imaging demonstrates peritumoral tissue heterogeneity. Radiology. 2014;273:502–510. doi: 10.1148/radiol.14132458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akbari H, et al. Imaging surrogates of infiltration obtained via multiparametric imaging pattern analysis predict subsequent location of recurrence of glioblastoma. Neurosurgery. 2016;78:572–580. doi: 10.1227/NEU.0000000000001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rathore S, et al. Radiomic signature of infiltration in peritumoral edema predicts subsequent recurrence in glioblastoma: Implications for personalized radiotherapy planning. J. Med. Imaging. 2018;5(021219):021211–21210. doi: 10.1117/1.JMI.5.2.021219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan J-L, et al. Multimodal MRI characteristics of the glioblastoma infiltration beyond contrast enhancement. Therap. Adv. Neurol. Disord. 2019;12:1–10. doi: 10.1177/1756286419844664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.d’Este SH, Nielsen MB, Hansen AE. Visualizing glioma infiltration by the combination of multimodality imaging and artificial intelligence, a systematic review of the literature. Diagnostics. 2021;11:592. doi: 10.3390/diagnostics11040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan J-L, Li C, van der Hoorn A, Boonzaier NR, Matys T, Price SJ. A neural network approach to identify the peritumoral invasive areas in glioblastoma patients by using MR Radiomics. Sci. Rep. 2020;10:1–10. doi: 10.1038/s41598-020-66691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dasgupta A, et al. Quantitative mapping of individual voxels in the peritumoral region of IDH-wildtype glioblastoma to distinguish between tumor infiltration and edema. J. Neuro Oncol. 2021;153(2):1–11. doi: 10.1007/s11060-021-03762-2. [DOI] [PubMed] [Google Scholar]
- 13.Parker D, et al. Freewater estimator using interpolated initialization (FERNET): Characterizing peritumoral edema using clinically feasible diffusion MRI data. PLoS ONE. 2020;15:e0233645. doi: 10.1371/journal.pone.0233645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samani ZR, Parker D, Wolf R, Hodges W, Brem S, Verma R. Distinct tumor signatures using deep learning-based characterization of the peritumoral microenvironment in glioblastomas and brain metastases. Sci. Rep. 2021;11:14469. doi: 10.1038/s41598-021-93804-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samani ZR, Parker D, Alappatt JA, Brem S, Verma R. Deep learning-based peritumoral microstructure mapping in glioblastomas using free water volume fraction. Neuro Oncol. 2020;22:157–158. doi: 10.1093/neuonc/noaa215.658. [DOI] [Google Scholar]
- 16.Samani ZR, Parker D, Alappatt JA, Brem S, Verma R. NIMG-21. Differentiating tumor types based on the peritumoral microenvironment using convolutional neural networks. Neuro Oncol. 2020;22:ii151. doi: 10.1093/neuonc/noaa215.634. [DOI] [Google Scholar]
- 17.Aliotta E, et al. Increased intratumoral infiltration in IDH wild-type lower-grade gliomas observed with diffusion tensor imaging. J. Neurooncol. 2019;145:257–263. doi: 10.1007/s11060-019-03291-z. [DOI] [PubMed] [Google Scholar]
- 18.Chang Y-Z, et al. Transcriptional characteristics of IDH-wild type glioma subgroups highlight the biological processes underlying heterogeneity of IDH-wild type WHO grade IV gliomas. Front. Cell Dev. Biol. 2020;8:1132. doi: 10.3389/fcell.2020.580464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manjón JV, Coupé P, Concha L, Buades A, Collins DL, Robles M. Diffusion weighted image denoising using overcomplete local PCA. PLoS ONE. 2013;8:e73021. doi: 10.1371/journal.pone.0073021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersson JL, Sotiropoulos SN. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage. 2016;125:1063–1078. doi: 10.1016/j.neuroimage.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith SM. Fast robust automated brain extraction. Hum. Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garyfallidis E, et al. Dipy, a library for the analysis of diffusion MRI data. Front. Neuroinformatics. 2014;8:1–17. doi: 10.3389/fninf.2014.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54:2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tustison NJ. Explicit B-spline regularization in diffeomorphic image registration. Front. Neuroinform. 2013;7:39. doi: 10.3389/fninf.2013.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamnitsas K, et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 2017;36:61–78. doi: 10.1016/j.media.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 26.Samani ZR, Alappatt JA, Parker D, Ismail AAO, Verma R. QC-Automator: Deep learning-based automated quality control for diffusion mr images. Front. Neurosci. 2020;13:1456. doi: 10.3389/fnins.2019.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012;5:1106–1114. [Google Scholar]
- 28.He L, Ren X, Gao Q, Zhao X, Yao B, Chao Y. The connected-component labeling problem: A review of state-of-the-art algorithms. Pattern Recogn. 2017;70:25–43. doi: 10.1016/j.patcog.2017.04.018. [DOI] [Google Scholar]
- 29.Huttenlocher DP, Klanderman GA, Rucklidge WJ. Comparing images using the Hausdorff distance. IEEE Trans. Pattern Anal. Mach. Intell. 1993;15:850–863. doi: 10.1109/34.232073. [DOI] [Google Scholar]
- 30.Caliński T, Harabasz J. A dendrite method for cluster analysis. Commun. Stat. Theory Methods. 1974;3:1–27. doi: 10.1080/03610927408827101. [DOI] [Google Scholar]
- 31.Hudgens MG. On nonparametric maximum likelihood estimation with interval censoring and left truncation. J. Royal Stat. Soc. Ser. B Stat. Methodol. 2005;67:573–587. doi: 10.1111/j.1467-9868.2005.00516.x. [DOI] [Google Scholar]
- 32.Fay MP, Proschan MA. Wilcoxon-Mann-Whitney or t-test? On assumptions for hypothesis tests and multiple interpretations of decision rules. Stat. Surveys. 2010;4:1. doi: 10.1214/09-SS051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoefnagels FW, et al. Differentiation of edema and glioma infiltration: Proposal of a DTI-based probability map. J. Neurooncol. 2014;120:187–198. doi: 10.1007/s11060-014-1544-9. [DOI] [PubMed] [Google Scholar]
- 34.Di Jia SL, Li D, Xue H, Yang D, Liu Y. Mining TCGA database for genes of prognostic value in glioblastoma microenvironment. Aging Albany N. Y. 2018;10:592. doi: 10.18632/aging.101415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brancato V, Nuzzo S, Tramontano L, Condorelli G, Salvatore M, Cavaliere C. Predicting survival in glioblastoma patients using diffusion MR imaging metrics—A systematic review. Cancers. 2020;12:2858. doi: 10.3390/cancers12102858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akbari H, et al. Histopathology-validated machine learning radiographic biomarker for noninvasive discrimination between true progression and pseudo-progression in glioblastoma. Cancer. 2020;126:2625–2636. doi: 10.1002/cncr.32790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Metz M-C, et al. Predicting glioblastoma recurrence from preoperative MR scans using fractional-anisotropy maps with free-water suppression. Cancers. 2020;12:728. doi: 10.3390/cancers12030728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharifi G, Pajavand AM, Nateghinia S, Meybodi TE, Hasooni H. Glioma migration through the corpus callosum and the brainstem detected by diffusion and magnetic resonance imaging: Initial findings. Front. Hum. Neurosci. 2020;13:472. doi: 10.3389/fnhum.2019.00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hara A, et al. Treatment strategies based on histological targets against invasive and resistant glioblastoma. J. Oncol. 2019;2019:1–10. doi: 10.1155/2019/2964783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferrer VP, Moura Neto V, Mentlein R. Glioma infiltration and extracellular matrix: Key players and modulators. Glia. 2018;66:1542–1565. doi: 10.1002/glia.23309. [DOI] [PubMed] [Google Scholar]
- 41.Gritsenko PG, et al. p120-catenin-dependent collective brain infiltration by glioma cell networks. Nat. Cell Biol. 2020;22:97–107. doi: 10.1038/s41556-019-0443-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ciccocioppo F, Lanuti P, Marchisio M, Miscia S. Extracellular vesicles involvement in the modulation of the glioblastoma environment. J. Oncol. 2020;2020:1–8. doi: 10.1155/2020/3961735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rathore S, et al. Radiomic MRI signature reveals three distinct subtypes of glioblastoma with different clinical and molecular characteristics, offering prognostic value beyond IDH1. Sci. Rep. 2018;8:1–12. doi: 10.1038/s41598-018-22739-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee S, et al. Evaluation of the microenvironmental heterogeneity in high-grade gliomas with IDH1/2 gene mutation using histogram analysis of diffusion-weighted imaging and dynamic-susceptibility contrast perfusion imaging. J. Neurooncol. 2015;121:141–150. doi: 10.1007/s11060-014-1614-z. [DOI] [PubMed] [Google Scholar]
- 45.Hai L, et al. A connectivity signature for glioblastoma. bioRxiv. Preprint. 2021;2208:14445. doi: 10.1101/20211107465791. [DOI] [Google Scholar]
- 46.Verma R, et al. Prediction of response to combination of nivolumab and bevacizumab in patients with recurrent glioblastoma via radiomic analysis on clinical MRI scans. Neuro Oncol. Adv. 2021;3:iv6–iv6. doi: 10.1093/noajnl/vdab112.022. [DOI] [Google Scholar]
- 47.Liu A, Hou C, Chen H, Zong X, Zong P. Genetics and epigenetics of glioblastoma: Applications and overall incidence of IDH1 mutation. Front. Oncol. 2016;6:16. doi: 10.3389/fonc.2016.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marusyk A, Janiszewska M, Polyak K. Intratumor heterogeneity: The rosetta stone of therapy resistance. Cancer Cell. 2020;37:471–484. doi: 10.1016/j.ccell.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khalafallah AM, Huq S, Jimenez AE, Serra R, Bettegowda C, Mukherjee D. “Zooming in” on glioblastoma: Understanding tumor heterogeneity and its clinical implications in the era of single-cell ribonucleic acid sequencing. Neurosurgery. 2021;88:477–486. doi: 10.1093/neuros/nyaa305. [DOI] [PubMed] [Google Scholar]
- 50.Majumder B, et al. Predicting clinical response to anticancer drugs using an ex vivo platform that captures tumour heterogeneity. Nat. Commun. 2015;6:1–14. doi: 10.1038/ncomms7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neftel C, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178:835–849. doi: 10.1016/j.cell.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qazi M, et al. Intratumoral heterogeneity: pathways to treatment resistance and relapse in human glioblastoma. Ann. Oncol. 2017;28:1448–1456. doi: 10.1093/annonc/mdx169. [DOI] [PubMed] [Google Scholar]
- 53.Gill BJ, et al. MRI-localized biopsies reveal subtype-specific differences in molecular and cellular composition at the margins of glioblastoma. Proc. Natl. Acad. Sci. 2014;111:12550–12555. doi: 10.1073/pnas.1405839111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are not publicly available due to the IRB requirements of the Hospital of University of Pennsylvania. The datasets can be made available on request to the corresponding author, after required data transfer and IRB paperwork is completed. The code is available here: https://github.com/zrsamani/AI-Based-Locoregional-Markers.