Abstract
Purpose: Myelination-related MR signal changes in white matter are helpful for assessing normal development in infants and children. A rule-based myelination evaluation workflow regarding signal changes on T1-weighted images (T1WIs) and T2-weighted images (T2WIs) has been widely used in radiology. This study aimed to simulate a rule-based workflow using a stacked deep learning model and evaluate age estimation accuracy.
Methods: The age estimation system involved two stacked neural networks: a target network-to extract five myelination-related images from the whole brain, and an age estimation network from extracted T1- and T2WIs separately. A dataset was constructed from 119 children aged below 2 years with two MRI systems. A four-fold cross-validation method was adopted. The correlation coefficient (CC), mean absolute error (MAE), and root mean squared error (RMSE) of the corrected chronological age of full-term birth, as well as the mean difference and the upper and lower limits of 95% agreement, were measured. Generalization performance was assessed using datasets acquired from different MR images. Age estimation was performed in Sturge–Weber syndrome (SWS) cases.
Results: There was a strong correlation between estimated age and corrected chronological age (MAE: 0.98 months; RMSE: 1.27 months; and CC: 0.99). The mean difference and standard deviation (SD) were −0.15 and 1.26, respectively, and the upper and lower limits of 95% agreement were 2.33 and −2.63 months. Regarding generalization performance, the performance values on the external dataset were MAE of 1.85 months, RMSE of 2.59 months, and CC of 0.93. Among 13 SWS cases, 7 exceeded the limits of 95% agreement, and a proportional bias of age estimation based on myelination acceleration was exhibited below 12 months of age (P = 0.03).
Conclusion: Stacked deep learning models automated the rule-based workflow in radiology and achieved highly accurate age estimation in infants and children up to 2 years of age.
Keywords: deep learning, infant and child development, magnetic resonance imaging, myelination
Introduction
In humans, the nervous system rapidly develops during infancy and childhood.1 In the brain, the myelination of axons by oligodendrocytes enables high-speed and long-distance transmission of electrical signals.2 Monitoring the progression of myelination is used as an indicator of neurodevelopment in infants and children.3
MRI is a valuable diagnostic tool for visualizing signal changes associated with myelination progression in white matter. The development of myelination is reflected in white matter signal changes in T1-weighted images (T1WIs) and T2-weighted images (T2WIs), and signal changes in each type of image are independent. Brain MRI of neonates with diffuse high signals on T2WIs and low signals on T1WIs develops into low signals on T2WIs and high signals on T1WIs with the progression of myelination.4 These changes are associated with a shortening of relaxation time because of an increase in lipids (cholesterol and galactocerebroside) associated with myelin sheath formation, as well as a decrease in water content.4–10 The signal changes associated with myelination in the brain follow various rules in terms of location, time, and direction. These signal changes progress from inferior to superior, posterior to anterior, and from central to the peripheral in the pediatric brain, where T1WI changes precede T2WI.11–16 Barkovich et al. proposed a timetable of MR signal changes in the pediatric brain with milestones in the pons, midbrain, internal capsule, corpus callosum, and deep and subcortical white matter.11 This timetable is convenient and is still used by radiologists and pediatricians more than 30 years after its proposal. However, although the process is relatively simple, manually matching MR signals to a timetable is time-consuming. In addition, the determination of signal changes can vary among individuals. In 1993, Staudt et al. proposed an objective system for estimating the age of myelination by staging the MR signal pattern in each myelination-related region.17 The age predicted by their system correlated with average growth and revealed delayed myelination in several neurological diseases.17,18 Although their proposed rule-based approach to myelination age estimation exhibited excellent performance, it has not been widely adopted because it is not automated.
In recent years, machine learning and intense convolutional neural networks (CNNs) have assisted manual methods in image recognition and classification. Regarding age estimation of infants and children, two studies have reported a deep learning approach for brain MRI.19,20 One study used sagittal T1WIs of the whole brain, and the other used single T1- and T2WIs at the level of the corpus callosum. The former only uses T1WI features, and the latter only uses one section of the brain. We hypothesized that the age estimation model could be improved by incorporating already established rules and workflow in radiology regarding myelination estimation in MRI into deep learning. The current study involved the simulation of the traditional rule-based myelination evaluation workflow for pediatric brain MRI with a deep learning age estimation model and evaluated its accuracy and generalization performance.
Materials and Methods
Participants
This retrospective, noninvasive, and non-interventional study was approved by the Juntendo University Hospital Institutional Review Board, with an opt-out alternative to obtaining informed consent from participants. Candidate participants were infants and children up to 2 years of age and below were examined at two MRI units from January 2014 to December 2015. A total of 908 infants and children up to 2 years of age were potentially eligible for inclusion in the study. The candidate selection and exclusion process from 908 participants are shown in Fig. 1. Based on preliminary clinical information in the MR study, we excluded 51 very low birth weight infants (< 1500 g), 12 infants with severe birth hypoxia, 32 infants with intractable epilepsy, and 32 infants with delayed motor development. Diagnostic reports from one of the two board-based neuroradiologists (MS and MH, each with at least 15 years of experience) excluded 320 tumors, 59 vascular diseases, 12 encephalitis or meningitis, 82 chromosomal disorders or neurocutaneous syndromes, and 96 malformations, as well as 12 unexplained abnormal white matter signals and 14 unacceptable artifacts from the description. Among 174 eligible candidates, 55 cases with suspected neurological diseases were excluded by tracking medical records until 2 years after MRI examination. Of the 119 subjects, 40 were boys and 79 were girls; 44% (52/119) were preterm infants with less than 37 weeks of gestation, with a mean birth weight of 2040 g (1674–2406 g). The gestational age at birth was 36.1–40.8 weeks, with a mean gestational age of 38.5 weeks. In all participants, chronological age was corrected by converting gestational age to a full 40 weeks, and the corrected ages ranged from −1.71 to 23.54 months (Fig. 2). Fourteen samples (aged 14 days to 24 months of age) acquired with four different MR systems at our institution from 2018 to 2019 were adopted to evaluate the general performance. Thirteen Sturge–Weber syndrome (SWS) patients aged 15 days–18 months, examined using the same MRI parameters during the current study period, were selected to evaluate the response of our model in comparison to non-normal participants. SWS is a neurocutaneous disorder reported to involve accelerated myelination up to 1 year of age, with a slowdown of myelination occurring beyond that age.21–23
Fig. 1.
Study flowchart.
Fig. 2.
A histogram of participants’ age at the time of MR examination. Participants’ ages are corrected for a full-term birth.
MRI protocol
For the training data, whole-brain 2D T1WI and T2WI were obtained from 95 participants using a 3T MR unit (Achieva; Philips Healthcare, Best, the Netherlands) and from 24 participants using a 1.5T MR unit (MAGNETOM Avanto; Siemens Healthineers, Erlangen, Germany). In addition, the external test data were obtained from four MRI systems (1.5 T Ingenia [Philips Healthcare], 3T MAGNETOM Skyra [Siemens Healthineers], Discovery 750w [GE Healthcare, Chicago, IL, USA], and Vantage Centurian [Canon Medical Systems, Tochigi, Japan]). Details of the sequence parameters and the number of participants for each MR unit are shown in Table 1.
Table 1.
The sequence parameters and the number of participants for each MR unit.
| Manu-facturer | Magnetic field strength | Slice thickness (mm) | Reconstruction matrix | T1WI | T2WI | Number of participants | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequence | TR(msec) | TE(msec) | TI(msec) | FA(degrees) | Echo train length | Sequence | TR(msec) | TE(msec) | FA(degree) | Echo train length | ||||||
| Internal data | ||||||||||||||||
| Achieva | Philips | 3 | 5 | 256×256 | IR | 2155 | 10 | 1000 | 90 | 4 | turbo SE | 4000 | 80 | 90 | 2 | 95 |
| Avanto | Siemens | 1.5 | 5.5 | 256×256 | SE | 530 | 11 | – | 80 | 1 | turbo SE | 4100 | 94 | 90 | 9 | 24 |
| External data | ||||||||||||||||
| Ingenia | Philips | 1.5 | 5 | 256×256 | SE | 620 | 12 | – | 75 | 1 | turbo SE | 4082 | 90 | 90 | 13 | 2 |
| Skyra | Siemens | 3 | 5 | 256×256 | IR | 2540 | 10 | 1045 | 130 | 6 | turbo SE | 7300 | 92 | 127 | 12 | 10 |
| Discovery 750w | GE | 3 | 5 | 256×256 | IR | 2736 | 17 | 832 | 111 | 8 | Fast SE | 4500 | 120 | 90 | 24 | 1 |
| Centurian | Canon | 3 | 5 | 256×256 | IR | 2050 | 15 | 950 | 90 | 7 | Fast SE | 4136 | 84 | 90 | 13 | 1 |
FA, flip angle; IR, inversion recovery; SE, spin echo; TI, inversion time; T1WI, T1-weighted image; T2WI, T2-weighted image.
Image dataset construction
All MR images were converted from Digital Imaging and Communications in Medicine format into 8-bit grayscale Portable Network Graphics format and resampled to 128 × 128 pixels. Two whole-brain datasets were constructed from 30 pairs of T1WIs and T2WIs. In cases with fewer than 30 images, the missing data were compensated for by including blank images. The 119 datasets were divided into four groups with an equal distribution of corrected ages to apply the four-fold cross-validation method. For the construction of the training dataset for the target image extraction model, five images containing myelination-related changes (pons, midbrain, basal ganglia, centrum semiovale, and subcortical white matter) were selected by a neuroradiologist with 25 years of clinical experience (AW). As the dataset for the second age estimation model, 10 MR images consisting of 5 pairs of T1-and T2WIs output by the first pre-trained image extraction model were adopted. In each training event, image data augmentation was performed randomly with left–right flipping, scaling (0.95–1.05), and rotation (−0.15–0.15 radians), using the Image Data Augmentation layer provided with the neural network software.
Machine learning models
Our deep learning models were designed using the open-source Neural Network Console ver. 1.5 deep learning library, which was developed by Sony Network Communications (Tokyo, Japan, https://dl.sony.com/) and is based on the Python programming language (version 3.6.2; Python Software Foundation, Wilmington, DE, USA), running on a computer (Windows 10 operating system) with an Intel Core i7 2.2 GHz processor, 32 GB RAM, and an NVIDIA GeForce GTX 1070 graphics processing unit. Our deep learning age estimation model was constructed by the stacking of two neural networks that were trained separately (Fig. 3). The first neural network was constructed using a Long Short-Term Memory network (LSTM) with an attention module (Figs. 4 and 5).24–26 This network took in 30 whole-brain 2D images as continuous data and extracted five characteristic images of myelination. The second age estimation neural network was constructed using the Pyramid network (PyramidNet). PyramidNet is an improved version of the residual neural network (ResNet) and fully connected layers (Fig. 6).27 The five pairs of T1- and T2WIs produced from the output of the LSTM are input to two PyramidNets, and the features of T1- and T2WIs were individually extracted and transferred to the fully connected layers, which then performed regression analysis to predict the age. As the loss function of training of image extraction and age estimation networks, the absolute error was employed. The learning parameters were a maximum epoch of 100 in the image extraction network and a maximum epoch of 1000 in the age estimation network, and the optimizer selected was Adam (initial learning rate = 0.001, α: 0.001, and β: 0.9).
Fig. 3.
Deep learning model simulating the rule-based workflow to the pediatric brain. The LSTM network extracts five regions from the whole brain (left). The PyramidNet and fully connected layers extract features of T1- and T2WIs and estimate the age (right). LSTM, Long Short-Term Memory network; PyramidNet, Pyramid network; T1WI, T1-weighted image; T2WI, T2-weighted image.
Fig. 4.
The internal structure of the LSTM unit. Our LSTM unit contained three gates: forget, input, and output (a). The forget and input gates coordinated the long-term memory represented as “C” in the figure, referring to the current input “xt” and transferred short-term memory “ht-1”. The output gate produced “ht” and moved to the following network. Each gate was constructed with a three-layer neural network with convolution, batch normalization, and activate functions (b). LSTM, Long Short-Term Memory network; ReLU, rectified liner unit.
Fig. 6.
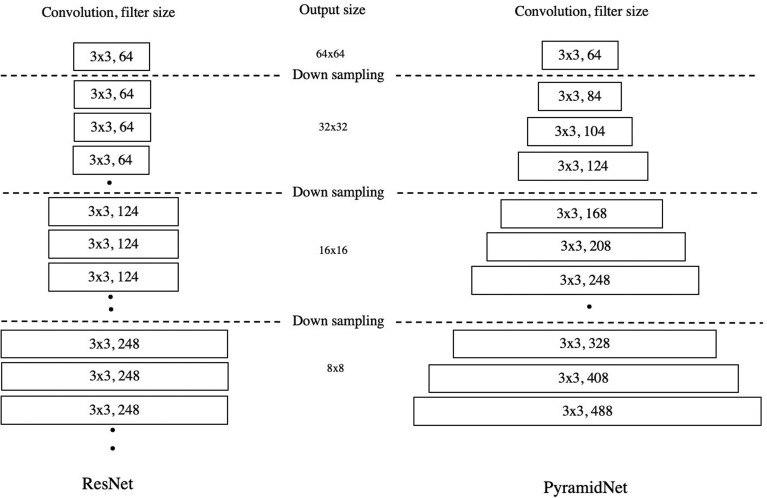
The architecture of ResNet and PyramidNet. The structure of ResNet (left) is a stack of multilayers convolutional units, and the number of filters increases after downsampling. In PyramidNet (right), the number of filters in the convolutional layer rises gradually. PyramidNet, Pyramid network; ResNet, residual neural network.
Performance evaluation of the machine learning model
In evaluating age estimation accuracy, the mean absolute error (MAE), root mean square error (RMSE), and correlation coefficient (CC) between the estimated age and the corrected chronological age were assessed using four-fold cross-validation. Because the age of the 119 participants was not found to be normally distributed in the Shapiro–Wilk normality test (P = 0.747 × 10-11), a nonparametric method was adopted for statistical analysis. Spearman’s rank-order correlation test was used to evaluate the correlation, and the statistical significance was determined with a significance threshold of P < 0.05. The mean difference, standard deviation (SD), and the limits of 95% agreement between the estimated age and the corrected chronological age were evaluated using Bland–Altman analysis. Statistical analyses were performed with SPSS (IBM SPSS Statistics 37; IBM, Armonk, NY, USA).
Results
Our stacked model of LSTM and PyramidNets showed high accuracy with CC of 0.99 (P < 0.01), MAE of 0.98 months, and RMSE of 1.27 months for internal validation data extracted from the same population as training data (Fig. 7). The mean difference between the estimated age and the corrected chronological age was −0.15 months, with an SD of 1.26 months, and the upper and lower limits of 95% agreement (mean ± 1.96 SD) were 2.33 and −2.63 months, respectively.
Fig. 7.
Correlation between estimated age and corrected chronological age. The CC was 0.99, MAE was 0.98 months, and RMSE was 1.85 months. CC, correlation coefficient; MAE, mean absolute error; RMSE, root mean square error.
The generalization performance evaluation with external data from different magnetic field strengths, models, and vendors MRI showed a decrease in CC to 0.93, an increase in MAE to 1.85 months, and an increase in RMSE to 2.59 months (Fig. 8). The Bland–Altman plot of SWS cases showed that 7 of the 13 patients exceeded the 95% limits of agreement (LOA). In particular, the difference in patients between 6 and 12 months of age was higher than the mean + 1.96 SD (Fig. 9). A paired samples t-test showed a positive proportional bias up to 12 months of age (P = 0.03).
Fig. 8.
Correlation between estimated age and corrected chronological age in external MR data. The CC was 0.93, MAE was 1.27 months, and RMSE was 2.59 months. CC, correlation coefficient; MAE, mean absolute error; RMSE, root mean square error.
Fig. 9.
Scatter diagram of estimated age and corrected chronological age in SWS patients and Bland–Altman plot of 119 participants (●) and 13 SWS patients (×). In SWS patients, the estimated age was higher than the corrected age until 1 year (a). Seven of the 13 SWS patients (×) were beyond the limits of 95% agreement from −2.63 to 2.33 months (mean difference ± 1.96 SD) (b). SD, standard deviation; SWS, Sturge–Weber syndrome.
Discussion
In infants and children up to 2 years of age, the myelination-related signal change in brain MRI is a valuable indicator for confirming normal neurological development. In the current study, we successfully automated the age estimation model from myelination-related MR signals by constructing a deep learning model that simulated the radiology workflow and training with brain MRI without abnormalities.
To simulate the workflow of the myelination-related MR signal evaluation in radiology using deep learning, we separated the workflow into two tasks: extraction of regions with a myelination-related signal change from the whole brain, and feature extraction of T1- and T2WI and regression analysis of age estimation. The first task, in which five images, including the pons, midbrain, internal capsule, corpus callosum, and white matter, were extracted from 30 whole-brain images, can be regarded as specific scene extraction from a movie, considering the 30 images as continuous movie data. We selected the LSTM network, a variant of the recurrent neural network architecture, to simulate this task. The recurrent neural network was suitable for handling continuous data, such as text, audio, and movies, using its characteristic “memory transmission” function (Fig. 4).24,25,28 For image extraction of a single region, the LSTM receives continuous input of 30 T1WIs from bottom to top and outputs the similarity to the target region. Based on the similarity, only one image was selected via the attention module with a one-hot and multiplication process (Fig. 5).26 Five independently trained LSTMs built a dataset consisting of five pairs of T1- and T2WIs and transmitted the data to the following age estimation network. The second step was feature extraction of myelination-related findings on T1- and T2WIs using an adapted PyramidNet, which is an advanced type of ResNet.27,28 ResNet was designed by combining a multilayer CNN with a skip connection.29–31 The skip connection is a short-circuit in the convolutional layer. Placing a skip connection in multiple convolution units can provide a potentially large number of feature transfer paths in the network, and its ensemble effect contributes to the high image recognition accuracy of ResNet. However, in ResNet, using the same number of channels in multilayer convolutional layers is not beneficial for feature extraction in image recognition. In PyramidNet, this disadvantage is improved by gradually increasing the number of channels, like the base of a pyramid (Fig. 6).27 In our model, two parallel PyramidNets extracted the features of T1- and T2WIs independently, and the following fully connected layer output the predicted age. Finally, the stacked deep learning model with an individually trained LSTM network and PyramidNets provided a fully automatic age estimation system (Fig. 3).
Fig. 5.
The attention module selected one image from 30 images of the whole brain. The attention module consisted of two 1 × 1 convolution layers and an activation function. The output of the LSTM network to the similarity to the target image was converted, and the one-hot representation was multiplied to select the most suitable image from 30 images. Conv, convolutional layer; LSTM, Long Short-Term Memory network; ReLU, rectified liner unit.
The previous two studies have reported age estimation from brain MRI using deep learning in infants and children up to 2 years of age.19,20 Hong et al. constructed an optimized 10-layer 3D CNN with whole-brain sagittal T1WI and reported MAE of 67.6 days (2.3 months), RMSE of 96.1 days (3.2 months), and CC of 0.9685, and the mean difference and the upper and lower limits of 95% agreement (mean difference ± 1.96 SD) between the estimated age and the corrected chronological age were 4.16 ± 73.5 days (0.14 ± 2.45 months), respectively, in Bland–Altman analysis.19 Moreover, Kawaguchi et al. constructed a multilayer CNN consisting of six convolutional layers and fully connected layers with independent inputs of single T1- and T2WI at the level of the corpus callosum splenium.20 They reported an MAE of 8.2 weeks (1.9 months), RMSE of 12.6 weeks (2.9 months), and CC of 0.94, and Bland–Altman analysis was only adopted to compare the performance of humans and artificial intelligence. The current results revealed an MAE of 0.98 months, RMSE of 1.27 months, CC of 0.99, and a mean difference ± 1.96 SD of −0.15 ± 2.48 months, exhibiting a higher CC and smaller error values compared with the previous studies. We consider that simulating the radiology workflow of focusing on the location and progression of myelination-related MR signals in T1- and T2WIs contributed to these results. However, it is not appropriate to compare regression models trained on different data only using CCs and error values. Anomaly detection trials of machine learning models trained only on normal data can be considered as one performance indicator. In our model, 7 of the 13 cases of SWS were beyond the LOA of the Bland–Altman plot, and myelination acceleration below 12 months of age exhibited a proportional error. Kawaguchi et al.’s model estimated a younger age than the corrected chronological age in six of the seven cases of myelination delay.20 Hong et al. did not validate their model with myelination abnormalities.19 Another proposed method of comparison is to evaluate the general performance with external data. Our model’s MAE, RMSE, and CC values were worsened from 0.98, 1.27, and 0.99 to 1.85, 2.59, and 0.93, respectively, with external data validation in MR systems with different static magnetic field strengths. The previous two studies mentioned above did not examine accuracy with comparable external data.
The current study involved several limitations. First, our dataset was biased and was not an ideal sample of healthy participants. Approximately half of the participants were preterm infants with less than 37 weeks’ gestation, and the aim of the MRI examination was to screen at the full-term equivalent. The other participants had a clinical course for which the pediatrician ordered a screening head MRI. Although abnormal participants were excluded wherever possible on the basis of medical records up to 2 years after the examination, the inclusion of unhealthy participants cannot be ruled out. Second, the size of our dataset was relatively small because it is ethically challenging to justify obtaining a large amount of brain MRI data from infants and children, who typically need to be sedated to undergo scanning. However, our sample size was close to the median value of recent machine learning research in medical imaging reported in a recent review of machine learning studies.32
Conclusion
An automated age estimation model from brain MRI was constructed using stacked deep learning simulating the rule-based workflow in radiology and achieved highly accurate age estimation in infants and children up to 2 years of age.
Acknowledgment
We thank Benjamin Knight, MSc., for editing a draft of this manuscript.
Funding Statement
This work was supported in part by JSPS KAKENHI Grant Number 18K07730.
Footnotes
Conflicts of Interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
References
- 1.Roche AF, Garn SM, Reynolds EL, et al. The first seriatim study of human growth and middle aging. Am J Phys Anthropol 1981; 54:23–24. [DOI] [PubMed] [Google Scholar]
- 2.Dobbing J, Sands J. Quantitative growth and development of human brain. Arch Dis Child 1973; 48:757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Squires LA, Krishnamoorthy KS, Natowicz MR. Delayed myelination in infants and young children: radiographic and clinical correlates. J Child Neurol 1995; 10:100–104. [DOI] [PubMed] [Google Scholar]
- 4.Holland BA, Haas DK, Norman D, et al. MRI of normal brain maturation. AJNR Am J Neuroradiol 1986; 7:201–208. [PMC free article] [PubMed] [Google Scholar]
- 5.McArdle CB, Richardson CJ, Nicholas DA, et al. Developmental features of the neonatal brain: MR imaging. Part I. Gray-white matter differentiation and myelination. Radiology 1987; 162:223–229. [DOI] [PubMed] [Google Scholar]
- 6.Martin E, Kikinis R, Zuerrer M, et al. Developmental stages of human brain: an MR study. J Comput Assist Tomogr 1988; 12:917–922. [DOI] [PubMed] [Google Scholar]
- 7.Koenig SH, Brown RD, Spiller M, et al. Relaxometry of brain: why white matter appears bright in MRI. Magn Reson Med 1990; 14:482–495. [DOI] [PubMed] [Google Scholar]
- 8.Kucharczyk W, Macdonald PM, Stanisz GJ, et al. Relaxivity and magnetization transfer of white matter lipids at MR imaging: importance of cerebrosides and pH. Radiology 1994; 192:521–529. [DOI] [PubMed] [Google Scholar]
- 9.Barkovich AJ. Concepts of myelin and myelination in neuroradiology. AJNR Am J Neuroradiol 2000; 21:1099–1109. [PMC free article] [PubMed] [Google Scholar]
- 10.Welker KM, Patton A. Assessment of normal myelination with magnetic resonance imaging. Semin Neurol 2012; 32:15–28. [DOI] [PubMed] [Google Scholar]
- 11.Barkovich AJ, Kjos BO, Jackson DE, et al. Normal maturation of the neonatal and infant brain: MR imaging at 1.5 T. Radiology 1988; 166: 173–180. [DOI] [PubMed] [Google Scholar]
- 12.Bird CR, Hedberg M, Drayer BP, et al. MR assessment of myelination in infants and children: usefulness of marker sites. AJNR Am J Neuroradiol 1989; 10:731–740. [PMC free article] [PubMed] [Google Scholar]
- 13.van der Knaap MS, Valk J. MR imaging of the various stages of normal myelination during the first year of life. Neuroradiology 1990; 31:459–470. [DOI] [PubMed] [Google Scholar]
- 14.Branson HM. Normal myelination: a practical pictorial review. Neuroimaging Clin N Am 2013; 23:183–195. [DOI] [PubMed] [Google Scholar]
- 15.Lee K, Cherel M, Budin F, et al. Early Postnatal Myelin Content Estimate of White Matter via T1w/T2w Ratio. Proc SPIE Int Soc Opt Eng 2015; 9417:94171R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soun JE, Liu MZ, Cauley KA, et al. Evaluation of neonatal brain myelination using the T1- and T2-weighted MRI ratio. J Magn Reson Imaging 2017; 46:690–696. [DOI] [PubMed] [Google Scholar]
- 17.Staudt M, Schropp C, Staudt F, et al. Myelination of the brain in MRI: a staging system. Pediatr Radiol 1993; 23:169–176. [DOI] [PubMed] [Google Scholar]
- 18.Staudt M, Schropp C, Staudt F, et al. MRI assessment of myelination: an age standardization. Pediatr Radiol 1994; 24:122–127. [DOI] [PubMed] [Google Scholar]
- 19.Hong J, Feng Z, Wang SH, et al. Brain Age Prediction of Children Using Routine Brain MR Images via Deep Learning. Front Neurol 2020; 11:584682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawaguchi M, Kidokoro H, Ito R, et al. Age estimates from brain magnetic resonance images of children younger than two years of age using deep learning. Magn Reson Imaging 2021; 79:38–44. [DOI] [PubMed] [Google Scholar]
- 21.Jacoby CG, Yuh WT, Afifi AK, et al. Accelerated myelination in early Sturge-Weber syndrome demonstrated by MR imaging. J Comput Assist Tomogr 1987; 11:226–231. [DOI] [PubMed] [Google Scholar]
- 22.Andica C, Hagiwara A, Hori M, et al. Aberrant myelination in patients with Sturge-Weber syndrome analyzed using synthetic quantitative magnetic resonance imaging. Neuroradiology 2019; 61:1055–1066. [DOI] [PubMed] [Google Scholar]
- 23.Adamsbaum C, Pinton F, Rolland Y, et al. Accelerated myelination in early Sturge-Weber syndrome: MRI-SPECT correlations. Pediatr Radiol 1996; 26:759–762. [DOI] [PubMed] [Google Scholar]
- 24.Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput 1997; 9:1735–1780. [DOI] [PubMed] [Google Scholar]
- 25.Gers FA, Schmidhuber J, Cummins F. Learning to forget: continual prediction with LSTM. Neural Comput 2000; 12:2451–2471. [DOI] [PubMed] [Google Scholar]
- 26.Hu J, Shen L, Albanie S, et al. Squeeze-and-Excitation Networks. IEEE Trans Pattern Anal Mach Intell 2020; 42:2011–2023. [DOI] [PubMed] [Google Scholar]
- 27.Han D, Kim J, Kim J. Deep Pyramidal Residual Networks. 2017 Ieee Conf Comput Vis Pattern Recognit (CVPR), 2017; 6307–6315. [Google Scholar]
- 28.Elman JL. Finding Structure in Time. Cogn Sci 1990; 14:179–211. [Google Scholar]
- 29.He K, Zhang X, Ren S, et al. Deep Residual Learning for Image Recognition. 2016 Ieee Conf Comput Vis Pattern Recognit (CVPR), 2016; 770–778. [Google Scholar]
- 30.Fukushima K. Neocognitron: A new algorithm for pattern recognition tolerant of deformations and shifts in position. Pattern Recogn 1982; 15:455–469. [Google Scholar]
- 31.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015; 521:436–444. [DOI] [PubMed] [Google Scholar]
- 32.Sakai K, Yamada K. Machine learning studies on major brain diseases: 5-year trends of 2014-2018. Jpn J Radiol 2019; 37:34–72. [DOI] [PubMed] [Google Scholar]