Abstract
Aeromonas caviae is increasingly being recognized as a cause of gastroenteritis, especially among the young. The adherence of aeromonads to human epithelial cells in vitro has been correlated with enteropathogenicity, but the mechanism is far from well understood. Initial investigations demonstrated that adherence of A. caviae to HEp-2 cells was significantly reduced by either pretreating bacterial cells with an antipolar flagellin antibody or by pretreating HEp-2 cells with partially purified flagella. To precisely define the role of the polar flagellum in aeromonad adherence, we isolated the A. caviae polar flagellin locus and identified five polar flagellar genes, in the order flaA, flaB, flaG, flaH, and flaJ. Each gene was inactivated using a kanamycin resistance cartridge that ensures the transcription of downstream genes, and the resulting mutants were tested for motility, flagellin expression, and adherence to HEp-2 cells. N-terminal amino acid sequencing, mutant analysis, and Western blotting demonstrated that A. caviae has a complex flagellum filament composed of two flagellin subunits encoded by flaA and flaB. The predicted molecular mass of both flagellins was ∼31,700 Da; however, their molecular mass estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis was ∼35,500 Da. This aberrant migration was thought to be due to their glycosylation, since the proteins were reactive in glycosyl group detection assays. Single mutations in either flaA or flaB did not result in loss of flagella but did result in decreased motility and adherence by approximately 50%. Mutation of flaH, flaJ, or both flagellin genes resulted in the complete loss of motility, flagellin expression, and adherence. However, mutation of flaG did not affect motility but did significantly reduce the level of adherence. Centrifugation of the flagellate mutants (flaA, flaB, and flaG) onto the cell monolayers did not increase adherence, whereas centrifugation of the aflagellate mutants (flaH, flaJ, and flaA flaB) increased adherence slightly. We conclude that maximum adherence of A. caviae to human epithelial cells in vitro requires motility and optimal flagellar function.
Aeromonads are ubiquitous waterborne bacteria that cause disease in poikilothermic animals, such as amphibians, fish, and reptiles. They cause furunculosis in salmonid fish and motile aeromonad septicemia in other freshwater species; both diseases are a serious problem for aquaculture (5). In humans, mesophilic aeromonads are associated with gastrointestinal disease, although extraintestinal diseases such as septicemia and wound infections have been reported (49). The three main pathogenic mesophilic species, which account for 85% of all clinical specimens, are Aeromonas hydrophila belonging to hybridization groups 1 and 3 (HG1 and HG3), A. veronii biovar sobria (HG8 and -10), and A. caviae (HG4). A. caviae, in particular, has been reported as the most prevalent paediatric enteropathogenic species of the genus (42, 54).
Aeromonads are efficient colonizers of surfaces and are an important constituent of bacterial biofilms in both water distribution systems and food processing environments. A number of investigators have linked the summer peak of Aeromonas-associated gastroenteritis with increased numbers of Aeromonas in water supplies over the warmer months (7, 25). Indeed, an Australian study correlated the increased incidence of gastroenteritis in households where the water supply system had significant Aeromonas biofilm buildup (25).
Long and/or wavy fimbriae have been implicated as important colonization factors of the main human pathogenic species of the genus (8, 27, 28). However, many clinical isolates have been found to be poorly piliated or nonpiliated (26). Alternative factors which have been suggested to aid in vitro aeromonad adherence to human and fish cell lines are outer membrane proteins, the lipopolysaccharide O-antigen (LPS O-Ag; 1, 12, 37, 38), motility, and the polar flagellum (39).
Flagella are complex bacterial organelles required for motility and are composed of the flagellar filament, hook, and basal body. Flagellar filaments can be simple homopolymers of a single flagellin subunit, as is the case for Escherichia coli, or they can be complex heteropolymers of multiple flagellins as is the case for Caulobacter (53), Campylobacter (16), Rhizobium (44) spp., and Vibrio parahaemolyticus (33). The flagellar filament is linked to the hook via hook-associated proteins (HAPs), which in turn is coupled to the motor located in the basal body (35). Biosynthesis of the complete structure and rotation of the filament that is essential for propelling the bacterium require over 40 genes (35).
Mesophilic aeromonads are motile by the action of a single polar unsheathed flagellum. Several investigators have isolated the flagella from A. hydrophila and A. veronii biovar sobria and have reported diverse flagellin molecular masses of 36 kDa (18) and 44 to 45 kDa (29, 36), but very little is known about this structure and even less is known about its potential role in virulence. Paradoxically, the only two flagellin genes reported have been for A. salmonicida, which is taxonomically defined as nonmotile (53). However, it should be noted that only 1% of the cell population were shown to express any flagella. To date, no flagellar genes have been cloned and sequenced from the mesophilic aeromonads.
Flagella are important in host colonization and biofilm formation in a number of bacterial genera, including Campylobacter (55) and Pseudomonas (3) spp. and within the family Vibrionaceae (13, 34). Recently, we have shown that mutations in the flm locus of A. caviae affect motility, flagella, LPS O-Ag, and adherence (15). Both flagella and LPS O-Ag have been linked to aeromonad adherence. Here we describe five polar flagellum genes of A. caviae, including the two flagellins. Through the creation of defined mutants, we investigate the role of flagella and motility in aeromonad adherence to human epithelial cells in vitro.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids are listed in Table 1. A. caviae strain Sch3 was previously isolated at Sheffield Children's Hospital (54). Bacteria were grown aerobically overnight (16 to 20 h), either statically or with shaking, at 37°C. They were grown either in brain heart infusion broth (BHIB; Oxoid), or on Luria-Bertani agar (LBA), supplemented with the appropriate antibiotics when required. Working stocks of the strains were kept on LBA plates at 4°C for a maximum of 2 weeks. Nalidixic acid (NAL), ampicillin, streptomycin (STR), and kanamycin (KAN) were used at final concentrations of 50 μg/ml, whereas chloramphenicol (CHL) and tetracycline were used at 25 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype and/or phenotypea | Source or reference |
|---|---|---|
| Strains | ||
| A. caviae | ||
| Sch3 | Clinical isolate | 54 |
| Sch3N | Sch3, spontaneous Nalr | This study |
| AAR269 | flaA::Kmr | This study |
| AAR27 | flaB::Kmr | This study |
| AAR31 | flaA::Cmr, flaB::Kmr | This study |
| AAR150 | flaG::Kmr | This study |
| AAR59 | flaH::Kmr | This study |
| AAR8 | flaJ::Kmr | This study |
| E. coli | ||
| CC118λpir | Δ(ara leu)7697 araD139 ΔlacZX74 glaE glaK phoA20 thi-1 rspE rpoB(Rfr) argE(Am) recA1αpir+ | 17 |
| S17-1λpir | hsdR, pro, recA, RP4-2 in chromosome Km::Tn7 (Tc::Mu) λpir, Tpr Smr | 41 |
| XL1-Blue | endA1 recA1 hsdR17 supE44 thi-1 gyrA96 relA1 lac [F′ proAB laclqZΔM15 Tn10 (Tcr)] | Stratagene |
| Plasmids | ||
| pUC19 | High-copy-number cloning vector, MCS, Ampr | Gibco-BRL |
| pUTmini-Tn5Cm | bla, ori R6K, mob RP4, tnp gene of Tn5-IS50r that lacks NotI site, MCS of M13tg131; 8.7 kb, Apr Cmr | 9 |
| pUC4-KIXX | Source of Tn5-derived nptII gene (Kmr) | Pharmacia |
| pBBR1MCS | Broad-host-range vector; IncP, W, and -Q; Col E1; and p15A compatible, containing pBluescript II (KS)- lacZα-polylinker, Cmr | 32 |
| pBBR1MCS-3 | Broad-host-range vector; IncP, -W, and -Q; Col E1; and p15A compatible, carrying Tcr | 31 |
| pKNG101 | ori R6K, mob RK2, strAB sacBR, 6.8 kb; Smr | 21 |
| pARP1 | pUC19 with a 900-bp fla PCR insert | This study |
| pARP2 | pBBR1MCS with a 3.2-kb SacI insert of A. caviae Sch3N DNA | This study |
| pARP126 | pBBR1MCS with a 3.6-kb SacI insert of A. caviae Sch3N DNA | This study |
| pARA100 | 1.5-kb XbaI-SmaI fragment containing flaA from pARP2, inserted at the pKNG101 | This study |
| pARA110 | 1.4-kb Kmr cassette from pUC4-KIXX inserted at the PvuII site of flaA in pARA100 | This study |
| pARA120 | 3.7-kb end-filled HindIII fragment from pUTmini-Tn5Cm, inserted at the PvuII site in pARA100 | This study |
| pARB200 | 1.2-kb SacI-HincII fragment containing flaB from pARP126 in pUC19 | This study |
| pARB210 | 1.8-kb KmrBamHI-SmaI fragment from pUC4-KIXX inserted at the BglII-EcoRV sites of flaB in pARB200 | This study |
| pARB220 | 3.7-kb CmrEcoRI fragment of pUTmini-Tn5Cm inserted at the EcoRI site of pARB210 | This study |
| pARB230 | 2.2-kb BamHI fragment from pARB220 inserted at the BamHI site of pKNG101 | This study |
| pARP400 | 3.6-kb SacI fragment from pARP126 inserted into pUC19 | This study |
| pARG410 | 1.4-kb KmrSmaI fragment from pUC4KIXX inserted at the StuI site of flaG in pARP400 | This study |
| pARG420 | 4.2-kb SalI fragment from pARG410 inserted at the SalI site of pKNG101 | This study |
| pARH500 | 2.2-kb SacI-StuI fragment from pARP400 inserted at SacI-SmaI site on pUC19. | This study |
| pARH510 | 1.4-kb KmrSmaI fragment from pUC4KIXX inserted at the SmaI site of flaH pARH500 | This study |
| pARH520 | 3.6-kb SacI-BamHI fragment from pARH510 inserted at SacI-BamHI sites of pKNG101 | This study |
| pARH530 | 3.7-kb CmrBamHI fragment from pUTmini-Tn5Cm inserted at the BamHI site of pARH520 | This study |
| pARJ600 | 1.4-kb KmrSmaI fragment from pUC4KIXX inserted at BalI site of flaJ in pARH500 | This study |
| pARJ610 | 3.6-kb SacI-BamHI fragment from pARJ600 inserted at the SacI-BamHI sites of pKNG101 | This study |
| pARJ620 | 3.7-kb CmrBamHI fragment from pUTmini-Tn5Cm inserted at the BamHI site of pARJ610 | This study |
| pARP1264 | 1.8-kb NaeI-SacI fragment from pARP2 inserted at the SmaI-SacI site of pBBR1MCS-3 | This study |
| pARP1265 | 3.6-kb SacI insert from pARP126 inserted in the SacI site of pBBR1MCS-3 | This study |
| pARP1266 | 3.6-kb SacI insert from pARP126 inserted in the SacI site of pARP1264 | This study |
Phenotype abbreviations are as follows: resistance (r); Amp (ampicillin); Cm (CHL); Km (KAN); Nal (NAL); Sm (STR); and Tc (tetracycline). MCS, multiple cloning site.
Flagellin purification.
Bacteria from 1 liter of BHIB grown statically (37°C, 16 h) were harvested and resuspended in 50 ml of 20 mM Tris-Cl buffer (pH 8.0). Flagella were removed from the bacterial suspension by homogenization on ice (speed 5, 5 min) with a “whirring type” blender (Kinematica, Lucerne, Switzerland). Bacteria were pelleted by centrifugation (10,000 × g, 4°C, 30 min), and the supernatant was filtered through a 0.2-μm (pore-size) membrane. Partially purified flagellin protein was recovered by the addition of solid ammonium sulfate to a concentration of 20%. This solution was gently agitated overnight at 4°C, and the precipitate was recovered by centrifugation (20,000 × g, 4°C, 30 min). The pellet was resuspended in 1 ml of 20 mM Tris-Cl buffer (pH 8.0) and dialyzed against the same buffer. Flagellin samples were separated on sodium dodecyl sulfate (SDS)-polyacrylamide gels (12%) and transferred onto Hybond-C (Amersham) nitrocellulose membrane. The flagellin protein was visualized on the membrane by (0.2% [wt/vol]) amido black staining, and the band corresponding to the flagellin was excised. Ice-cold acetone was added to the excised membrane to dissolve the nitrocellulose and precipitate the protein. The protein was pelleted by centrifugation (13,000 × g, 4°C, 10 min) and washed three times with ice-cold acetone. Residual acetone was removed under vacuum, and the purified flagellin was resuspended in 500 μl of 20 mM Tris-Cl (pH 8.0).
N-terminal sequence analysis.
After SDS-polyacrylamide gel electrophoresis (PAGE), the purified flagellin band was transferred onto a polyvinylidene difluoride membrane and sequenced using the automated Edman degradation procedure on a Applied Biosystems 470A gas-phase sequenator with a 120A online phenylthiohydantoin analyzer.
Glycosyl group detection.
This procedure was carried out as described by Doig et al. (10). Briefly, after SDS-PAGE, proteins were transferred onto nitrocellulose membranes and the glycosyl groups were oxidized for 30 min with 10 mM sodium periodate in sodium acetate buffer (50 mM; pH 5.5). Following oxidation, 5 mM biotin-hydrazide in sodium acetate buffer (50 mM; pH 5.5) was added (1 h, room temperature). The membrane was washed three times in TBS (10 mM Tris-Cl, pH 7.5; 0.85% NaCl) and blocked by shaking in 1% bovine serum albumin in TBS for 60 min. The membrane was washed again in TBS three times, and avidin-D peroxidase (Vector Labs) was added for 60 min at room temperature. The membrane was then washed in TBS three times and developed by the addition of 2 ml of 0.5% 4-chloro-1-naphthol prepared in methanol and diluted in 8 ml of phosphate-buffered saline (PBS) containing 0.15% (vol/vol) H2O2. Alternatively, the method of Gerard using Schiff reagent was used for direct gel staining to detect the presence of glycosyl groups (14).
Preparation of antibody.
Approximately 200 μg of purified flagellin was emulsified with 1 ml of Freund complete adjuvant and inoculated subcutaneously into dwarf lop-eared rabbits. Booster injections of the flagellin protein were administered 4 and 6 weeks later. Antibodies were obtained by bleeding 10 days after the booster injection.
Whole-cell protein preparation and immunoblotting.
Whole-cell proteins were obtained from A. caviae strains grown statically overnight in BHIB at 37°C. Equivalent numbers of cells were harvested by centrifugation, the cell pellet was resuspended in 50 to 200 μl of SDS-PAGE loading buffer (45) and boiled for 5 min. Following SDS-PAGE and transfer to Hybond-C nitrocellulose membranes, the membranes were blocked with 5% skim milk and probed with a polyclonal rabbit anti-polar flagellin antibody (1:1,000). The unbound antibody was removed by five washes in PBS, and a goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (1:1,000) was added. The unbound secondary antibody was removed by five washes in PBS. The bound conjugate was then detected by the addition of 2 ml of 0.5% (wt/vol) 4-chloro-1-naphthol (Sigma) prepared in methanol and diluted in 8 ml of PBS containing 50 μl of 30% H2O2.
Motility assay.
Freshly grown bacterial colonies were transferred with a sterile toothpick into the center of the motility agar (1% tryptone, 0.5% NaCl, 0.25% agar). The plates were incubated face up at 37°C for 16 to 24 h, and motility was assessed by examining the migration of bacteria through the agar from the center toward the periphery of the plate.
Transmission electron microscopy.
Bacterial suspensions were placed on Formvar-coated copper grids and negatively stained with 2% solution of potassium phosphotungstate. Bacteria were then viewed on a Phillips EM 400 transmission electron microscope.
Adherence assay.
Tissue culture was maintained as described by Thornley et al. (48). The adherence assay was conducted as a slight modification of that described by Carrello et al. (8). Bacteria were grown statically in BHIB at 37°C, harvested by gentle centrifugation (1,600 × g, 5 min), and resuspended in PBS (pH 7.2) at approximately 107 CFU/ml (A600 ∼0.07). The monolayer was infected with 1 ml of the bacterial suspension for 90 min at 37°C in 5% CO2. Following infection, the nonadherent bacteria were removed from the monolayer by three washes with PBS. The remaining adherent bacteria and the monolayers were then fixed in 100% methanol for 5 min. Methanol was removed by washing them with PBS, and the HEp-2 cells with the adherent bacteria were stained for 45 min in 10% (vol/vol) Giemsa stain (BDH) prepared in Giemsa buffer. The coverslips were air dried, mounted, and viewed by oil immersion under a light microscope at ×1,000 magnification. Twenty HEp-2 cells/coverslip were randomly chosen, and the number of bacteria adhering/HEp-2 cell was recorded. Assays were carried out in duplicates or triplicates. For certain experiments, bacteria were deposited onto the monolayer by centrifugation (400 × g, 15 min, room temperature) prior to the 90-min infection period. For flagellin antibody inhibition assays, bacterial suspensions were incubated with antipolar flagellin antibody at a 1:200 final dilution. The suspensions were incubated at 37°C for 15 min before infection of the monolayer. For flagellum competition assays, flagella were partially purified by shearing followed by centrifugation and filtering as described above. Monolayers were incubated with 200 μl of this crude preparation at 37°C for 1 h and washed three times in PBS prior to infection.
Statistical analysis.
The differences in adherence to cell lines between the wild-type and the mutant strains was analyzed by the t test using Microsoft Excel software.
DNA techniques.
Plasmid DNA was isolated by alkaline lysis (45) or by using the Wizard Plus Minipreps DNA purification system (Promega). A. caviae chromosomal DNA isolation was carried out according to standard techniques (45). DNA restriction digestions and T4 ligations were carried out according to the manufacturer's instructions. DNA samples were separated on 0.8% agarose gels; when required, extraction of DNA from gels was carried out using the QIAquick gel extraction kit (Qiagen).
PCR.
Reactions were performed using Pfu DNA polymerase (Stratagene) at 2.5 mM MgCl2 in a Hybaid Omnigene Thermal cycler. Initial DNA denaturation was carried out for 2 min, and amplification reactions were carried out for 30 cycles with denaturation at 95°C for 30 s, primer annealing at 55°C for 1 min, and elongation at 72°C for 2 min.
Southern and dot blot hybridizations.
Southern and dot blot hybridizations were performed by capillary transfer (45). Probe labeling, hybridization, and detection were carried out using the enhanced chemiluminescence labeling and detection system (Amersham) according to the manufacturer's instructions.
Nucleotide sequencing and sequence analysis.
DNA fragments were ligated into pBBR1MCS and sequenced using an ABI PRISM 377 DNA sequencer. The 18-mer forward (5′-TGTAAAACGACGGCCAGT-3′) and the 22-mer reverse (5′-TCACACAGGAAACAGCTATGAC-3′) M13 primers were employed in sequencing the ends of the DNA inserts. Following the first sequencing reaction, custom primers were designed until the insert sequence was complete. The nucleotide sequences obtained were converted into amino acid sequences in all six possible reading frames by using the BLASTX program (2) of the National Center for Biotechnology Information (NCBI), and the homologous proteins on the database were identified. Multiple sequence alignments were carried out using the CLUSTAL W program (47). Putative transcriptional terminator sequences were identified using the Terminator program from the Genetics Computer Group package (Madison, Wis.) in a VAX 4300.
Insertional mutagenesis and complementation.
Mutants were created by the insertion of the Tn5-derived kanamycin resistance cartridge (nptII) from pUC4-KIXX. This cartridge contains an outward reading promoter that drives the transcription of downstream genes when inserted in the correct orientation (6). A flaA mutant was achieved by the insertion of XbaI-SmaI fragment from pARP2 into the XbaI-PvuII sites of the suicide vector pKNG101 to give pARA100. The 1.4-kb SmaI-digested kanamycin resistance cartridge was inserted into the PvuII site and checked for correct orientation to give pARA110; this was transformed into E. coli S17-1λpir. Conjugal transfer of pARA110 from E. coli S17-1λpir to A. caviae Sch3N was performed using a filter mating technique. Bacterial conjugation was allowed to proceed for 6 to 8 h at 37°C on sterile nitrocellulose filters (0.45 μm, pore size) placed onto an LBA plate. Serial dilutions of the mating mix were then plated on LBA supplemented with NAL and KAN, the latter added in order to select for recombination. Colonies that were KAN resistant (Kmr) and STR sensitive (those not likely to have retained the vector) were purified and probed for the KAN cartridge and absence of any plasmid sequences by Southern hybridization, thus demonstrating a double recombination event and allelic exchange. The flaA::Kmr strain was designated AAR269. To mutate flaB, the 1.2-kb SacI-HincII fragment from pARP126 containing flaB was cloned into pUC19 to give pARB200; this was cut with BglII and EcoRV (to delete a 400-bp internal fragment of flaB) and ligated to the 1.8-kb BamHI-SmaI KAN cartridge, resulting in pARB210. The 3.7-kb CHL resistance cassette (Cmr) EcoRI fragment from pUTmini-Tn5Cm was cloned into the EcoRI site to provide two BamHI sites flanking the flaB::Kmr insertion (pARB220). The plasmid pARB220 was digested with BamHI and the 2.2-kb fragment was ligated into pKNG101 to give pARB230. This plasmid was conjugated into Sch3N, and transconjugants were screened as described above; this resulted in the flaB::Kmr mutant AAR27. The flaG gene was mutated by the insertion of the 1.4-kb SmaI-digested Kmr cartridge into the unique StuI site in pARP400 to give pARG410. This plasmid was digested with SalI and the 4.2-kb fragment was inserted into pKNG101, resulting in pARG420; subsequent conjugal transfer and screening was as described above. A flaG::Kmr mutant was identified and designated AAR150. For flaH, a 2.2-kb SacI-StuI fragment from pARP400 was first cloned into pUC19 (pARH500); this was cut with SmaI and ligated to the 1.4-kb SmaI Kmr cassette (pARH510). This vector was digested with SacI-BamHI, and the 3.2-kb fragment inserted at the SacI-BamHI sites of pKNG101 (pARH520). As the SacI site of pKNG101 lies within the STR resistance gene any insertion will truncate it, resulting in STR sensitivity. We therefore inserted a 3.7-kb BamHI Cmr fragment from pUTmini-Tn5Cm into the BamHI site of pARH520, to give pARH530. The Cmr cassette was inserted into the plasmid to help when screening for double homologous recombination events, through the search for CHL sensitivity (Cms) and KAN resistance. The plasmid pARH530 was transferred into Sch3N and transconjugants tested for Kmr and Cms. After screening, the flaH::Kmr mutant was designated AAR59. For the disruption of flaJ, the 1.4-kb SmaI Kmr cartridge was inserted at the BalI site of pARH500 to give pARJ600. The 3.6-kb SacI-BamHI fragment from pARJ600 was ligated into pKNG101, to give pARJ610. As with pARH520, we inserted the Cmr gene at the BamHI site of pARJ610 for ease of selection after conjugation, to give pARJ620. Conjugation and screening was done as described above and gave rise to the flaJ::Kmr mutant AAR8. To create a tandem flaA::Cmr, flaB::Kmr mutant (AAR31), we released the 3.7-kb HindIII Cmr fragment from pUTmini-Tn5Cm, end filled it using Klenow fragment, and ligated it into the PvuII site of pARA100. This was introduced by conjugation into AAR27, resulting in the double flagellin mutant AAR31.
Nucleotide sequence accession number.
The nucleotide sequence reported here has been submitted to GenBank under accession number AF198617.
RESULTS
Adhesion inhibition.
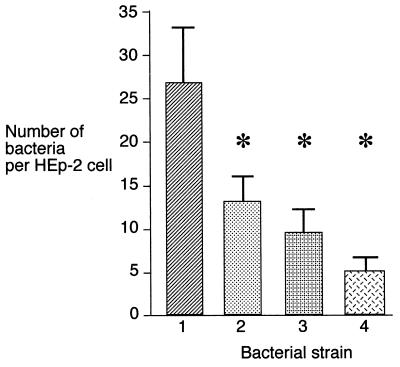
Previous work has indicated that the polar flagella of A. caviae play an essential role in adherence to human epithelial cells. Their removal by mechanical shearing reduces adherence by 75% (48), and nonmotile flm mutants, which lack both of the putative adhesins, polar flagella, and LPS O-antigen, are also nonadherent (15). In this study, a mild shearing force was used to remove the polar flagellum filaments from A. caviae Sch3N cells, and the flagellin subunit was purified to homogeneity as described in Materials and Methods. Polyclonal antibodies were raised against the polar flagellin and used in adhesion inhibition assays. The adhesion of A. caviae Sch3N to HEp-2 cells was reduced by approximately 50% through treatment of the bacteria with the antiflagellin antiserum prior to infection of the cell monolayer (Fig. 1). Adherence could also be reduced by approximately 80% when the cell monolayer was incubated with a concentrated homogenate of sheared flagellum filaments before bacterial infection (Fig. 1). All treatments demonstrated a highly significant reduction in bacterial adherence (P < 0.001).
FIG. 1.
Inhibition of adherence of A. caviae to HEp-2 cells following treatment of the monolayer or bacteria prior to the adherence assay. Columns: 1, mean number of adherent A. caviae Sch3N bacteria per HEp-2 cell; 2, before addition to the adherence assay Sch3N bacteria were incubated with rabbit polyclonal antibodies raised against the polar flagellin (15 min, 37°C); 3, effect of a mild shearing force for 5 min on the bacteria; 4, monolayers were pretreated with a concentrated homogenate of flagella and washed in PBS prior to bacterial infection. The numbers represent the means of duplicates of at least three experiments. The error bars represent one standard deviation (∗, P < 0.001)
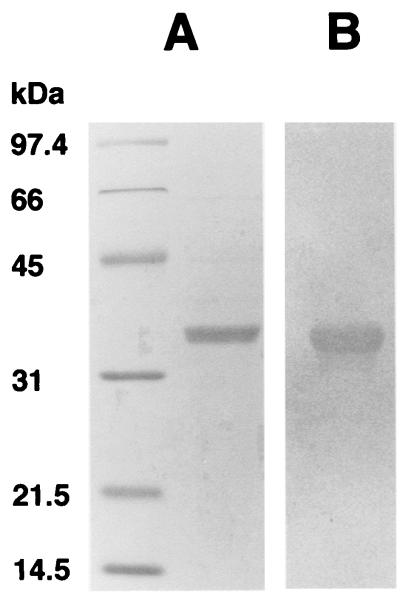
N-terminal sequence of the polar flagellin protein from A. caviae Sch3N.
The molecular mass of the purified flagellin was estimated by SDS-PAGE to be ca. 35,500 Da (Fig. 2A). The purified flagellum subunits were used to determine the N-terminal amino acid sequence of the polar flagellin. The first 21 amino acids were determined, with the exception of the first amino acid, which could not be identified, to be: ALYINTNTSSLNAQRNLMNT (see Fig. 4, boldface type, underlined). The sequence showed homology to a number of flagellins, especially to the recently described flagellins FlaA and FlaB of A. salmonicida (52) and FliC of P. aeruginosa (51) (see Fig. 4).
FIG. 2.
SDS-PAGE and glycosyl group detection of the purified polar flagellin. (A) Purified flagellin subunit on a 12% polyacrylamide Coomassie brilliant blue-stained gel. (B) Demonstration of glycosyl groups on flagellin by periodate oxidation and hydrazine biotinylation.
FIG. 4.
Alignment of the deduced amino acid sequences of the A. caviae (Ac) FlaA and FlaB proteins with their homologues FlaA and FlaB in A. salmonicida (As) and P. aeruginosa (Pa) FliC. The alignment was performed using CLUSTAL W (47). Identical amino acid residues are indicated by an asterisk, and conservatively substituted residues are indicated by one or two dots. The amino acids from the derived A. caviae polar flagellin N-terminal sequence are in boldface and underlined.
Demonstration of glycosylation of the Aeromonas flagellins.
Posttranslational modification of flagella has been reported for a number of bacterial species. To determine whether the A. caviae polar flagellum was glycosylated, the purified Sch3N polar flagellin subunit was treated with periodate to oxidize any glycosyl groups present to produce reactive aldehydes. These were then allowed to react with biotin hydrazide (10). The polar flagellin was biotinylated, indicating that it was glycosylated (Fig. 2B). This result was subsequently confirmed by the use of direct gel staining with Schiff reagent (data not shown).
Cloning of the polar flagellin genes.
The polar flagellin genes were initially isolated by designing two oligonucleotide primers. The first primer (5′-AACGCCCAGCGTAACCTGATG-3′) corresponded to the amino acids NAQRNLM found in the N-terminal sequences of the A. caviae polar flagellin and FlaA and FlaB of A. salmonicida. The second primer (5′-GGVCGYTGGTTKGCYTG-3′) was a degenerate oligonucleotide that corresponded to the conserved amino acids QANQRP found in the C-terminal regions in a number of bacterial flagellins. The primers were used to amplify a single DNA fragment of about 900 bp from A. caviae chromosomal DNA by PCR. This fragment was cloned into pUC19 (pARP1) and then nucleotide sequenced using the universal forward and reverse primers. The sequence was subjected to homology searches using the BLASTX programme at the NCBI and was found to be most similar to the A. salmonicida polar flagellins FlaA and FlaB (52). To locate the flagellin locus on the chromosome, the PCR fragment was labeled and used as a probe in Southern blots of A. caviae Sch3N chromosomal DNA digested with a series of different restriction enzymes. Restriction digestion with SacI resulted in two hybridizing fragments of approximately 3.2 and 3.6 kb. Based on this, chromosomal DNA was digested with SacI and size fractionated by agarose gel electrophoresis, and fragments of 3.0 to 4.0 kb were extracted and ligated into SacI-digested pBBR1MCS. A plasmid minilibrary of 1,200 recombinant clones was generated and screened for flagellin sequences by dot blot hybridization. Two reactive plasmids carrying SacI inserts of 3.6 and 3.2 kb were isolated and designated pARP126 and pARP2, respectively.
Sequencing of the A. caviae flagellin genes.
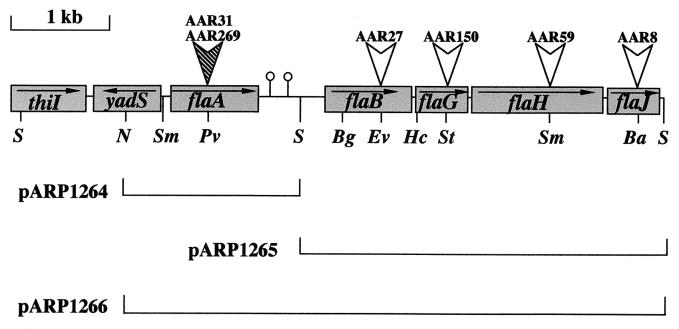
The inserts of pARP2 and pARP126 were nucleotide sequenced from both ends with forward and reverse universal and custom-designed primers. The sequence of pARP2 revealed three open reading frames (ORFs): ORF1 had homology to ThiI a protein involved in thiamine biosynthesis, the ORF2 product was similar to YadS a hypothetical protein of E. coli, whereas ORF3 encoded a protein that was homologous to FlaA of A. salmonicida (52). Sequence analysis of pARP126 revealed four additional ORFs which appeared to have flagellar function, were transcribed in the same direction, and were thought to form an operon. The predicted products of these genes were homologous to FlaB and FlaG of A. salmonicida and to FlaH and FlaJ of V. parahaemolyticus (46). Using PCR, the two SacI fragments were shown to be contiguous, forming a flagellin locus in the order flaA, flaB, flaG, flaH, and flaJ (Fig. 3). This genetic organization was the same as that described for the closely related species A. salmonicida (52) and V. parahaemolyticus (33, 46).
FIG. 3.
Genetic organization of the A. caviae Sch3N polar flagellin locus. Flagellar genes and ORFs are indicated by shaded boxes and named after their homologs in other bacterial species. Horizontal arrows indicate the direction of transcription. Open vertical arrows indicate the site of insertion of the KAN resistance cassette in the single mutants. The closed vertical arrow indicates the site of insertion of the KAN resistance cassette in the single mutant AA269 and the site of insertion of the CHL resistance cassette in the tandem mutant AAR31. Complementation plasmids based on the broad-host-range vector pBBR1MCS-3 are shown (see Table 1). The BalI (Ba), BglII (Bg), EcoRV (Ev), HincII (Hc), NaeI (N), PvuII (Pv), SacI (S), SmaI (Sm), and StuI (St) restriction sites are shown. Lollipop structures depict the approximate position of the putative transcriptional terminators.
The 918-bp ORF encompassing the flaA gene encoded a 306-amino-acid protein with a predicted molecular mass of 31,776 Da. Approximately 600 bp downstream of flaA is flaB (915 bp) encoding a protein of 305 amino acids with a predicted molecular mass of 31,731 Da. Both FlaA and FlaB were most similar to the polar flagellins of A. salmonicida and P. aeruginosa, with the highest homology occurring at the N and C termini (Fig. 4). FlaA and FlaB shared 92% identity at the amino acid level and 84% identity at the nucleotide level. As reported for other flagellins, cysteine, histidine, and tryptophan were absent from both FlaA and FlaB. The first 21 amino acids of the predicted sequences of both FlaA and FlaB did not exactly correspond to that derived by N-terminal sequencing. In fact, the N-terminal amino acid sequence obtained from the purified flagellin appeared to be a mixture of the predicted sequences of the flaA and flaB products. This suggests that the polar flagellar filament of A. caviae Sch3N consists of both flagellins (Fig. 4).
A number of possible ς28 promoter sequences were found upstream of the flaA, and the putative Shine-Dalgarno sequence is 7 bp above the start codon. Upstream of flaB there are several potential ς28 and ς54 promoter sequences, and the putative Shine-Dalgarno sequence is 9 bp above the start codon. The two genes are transcribed in the same direction but seem to be part of different transcriptional units, since downstream of the flaA TAA stop codon there are two potential stem-loop structures. The first sequence, AGGGCGGTTGTTCAGGCCGCCCT, is 25 bp downstream of the stop codon and has a ΔG0 value of −19.4 kcal (50). The second sequence, GGCCGACATTGTGTCGGCCTTTTTT, is 193 bp downstream of the stop codon and is a possible rho-independent sequence with a ΔG° value of −17.8 kcal.
The next ORF was located 41 bp downstream of flaB and encoded a 154-amino-acid protein with a predicted molecular mass of 16,739 Da. The deduced amino acid sequence shared similarity with a series of FlaG proteins which have little amino acid identity and whose role in flagellum biogenesis remains unknown (33, 34), although FlaG has been suggested to have a role in the regulation of flagellin export or flagellin gene expression (34). Starting 34 bp downstream of flaG was another ORF of 1,287 bp (flaH) that encoded a protein of 429 amino acids with a predicted molecular mass of 45,094 Da. FlaH shared homology to a number of HAPs (e.g., HAP-2), such as FliD of P. aeruginosa, E. coli, and Salmonella enterica serovar Typhimurium. HAP-2 proteins cap the distal tip of the flagellar filament and prevent loss of unpolymerized flagellin. The final ORF (flaJ) began 127 bp downstream of flaH. The deduced amino acid sequence encoded a protein of 15,812 Da that shared homology to FlaJ of V. parahaemolyticus and FliS proteins of a number of bacteria. FliS is thought to be a chaperone protein that facilitates flagellin export (56).
Mutant analysis.
To determine the role of the identified genes in motility, polar flagellin biosynthesis, and subsequently adherence, mutations were created in each gene using a Kmr cartridge, as described in Materials and Methods. The antibiotic resistance cartridge was inserted in the same transcriptional orientation with respect to the target gene; the presence of an outward reading promoter on the cartridge ensures the expression of downstream genes (6).
The correct construction of all mutants was verified by Southern hybridization of chromosomal DNA with the Kmr cartridge and vector probes (data not shown). The positions of the cartridge insertions are presented in Fig. 3. The motility of the A. caviae mutant strains was then assessed in semisolid motility plates and by light microscopy. The motility phenotypes of the mutant strains are shown in Table 2. Mutation of either flaA (AAR269) or flaB (AAR27) did not abolish motility but did reduce the swarm size in soft agar plates. Mutation of flaH (AAR59) and flaJ (AAR8) completely abolished motility, whereas mutation of flaG (AAR150) did not appear to have any effect. Because single insertions in flaA or flaB did not abolish motility, a double-mutant strain (AAR31), in which both genes were mutated in tandem, was constructed. The double mutant was constructed in the flaB mutant background (AAR27) by inserting a Cmr cassette in flaA. As assessed by swarm size, motility was completely abolished in this double-flagellin mutant.
TABLE 2.
Relative swimming motility phenotypes of defined and complemented insertion mutants
| Strain | Genotype | Motility phenotypea
|
|||
|---|---|---|---|---|---|
| Defined (none) | Complementation plasmid
|
||||
| pARP1264 | pARP1265 | pARP1266 | |||
| Sch3N | Wild type | ++++ | ND | ND | ND |
| AAR269 | flaA | ++ | ++++ | ND | ++++ |
| AAR27 | flaB | ++ | ND | +++ | +++ |
| AAR31 | flaA flaB | − | ++ | ++ | ++ |
| AAR150 | flaG | ++++ | ND | ND | ND |
| AAR59 | flaH | − | ND | ++ | ++ |
| AAR8 | flaJ | − | ND | ++ | ++ |
Motility phenotypes were examined in semisolid motility plates (see Materials and Methods). Plus signs indicate spreading. The diameters of the spread zones are as follows: ++, 2.0 to 3.0 cm; +++, 4.0 to 5.0 cm; ++++, >6.0 cm; −, nonmotile. ND, not determined.
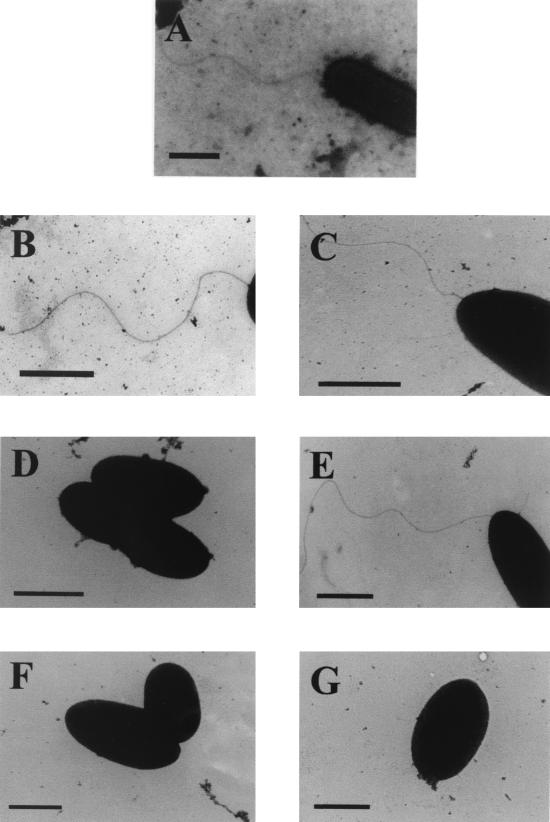
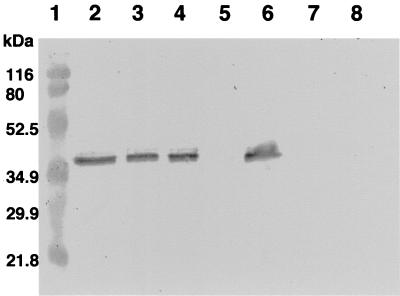
To determine if the flagellar mutants still possessed a polar flagellum, which could have possibly explained the alterations in motility, the flagellum filament of each mutant was examined by electron microscopy (Fig. 5). Both the flaA (AAR269) and flaB (AAR27) single mutants had flagella of the wild-type length, while the mutation in AAR150 (flaG) resulted in cells with a longer flagellar filament. No flagella were observed for the mutants AAR59 (flaH), AAR8 (flaJ), and AAR31 (flaA flaB). To assess flagellin protein expression in the mutants, flagellin immunoblots of whole-cell protein were carried out (Fig. 6). The flagellin protein was detected in the strains carrying single mutations in either flaA (AAR269), flaB (AAR27), or flaG (AAR150). This suggests that either of the flagellin proteins is able to substitute for each other to a certain extent, only causing a slight loss of motility. Flagellins were not detected in the flaH (AAR59) and flaJ (AAR8) single mutants and the flaA flaB (AAR31) tandem mutant.
FIG. 5.
Transmission electron microscopy of the A. caviae strains Sch3N (wild type) (A), AAR269 (flaA) (B), AAR27 (flaB) (C), AAR31 (flaA flaB) (D), AAR150 (flaG) (E), AAR59 (flaH) (F), and AAR8 (flaJ) (G) grown at 37°C in BHIB. Bacteria were gently placed onto Formvar-coated copper grids and negatively stained using 2% potassium phosphotungstate. Bar, 0.6 μm.
FIG. 6.
Western blot analysis of the A. caviae wild-type and mutant strain whole-cell protein preparations using polyclonal antipolar flagellin antibodies (1:1,000). Lane 1, molecular weight markers; lane 2, Sch3N (wild type); lane 3, AAR269 (flaA); lane 4, AAR27 (flaB); lane 5, AAR31 (flaA flaB); lane 6, AAR150 (flaG); lane 7, AAR59 (flaH); lane 8, AAR8 (flaJ). Whole-cell proteins were obtained from bacteria grown at 37°C in BHIB.
Complementation studies were undertaken to determine if wild-type motility could be restored to the mutants by providing the fla genes in trans. The plasmids, pARP1264 (flaA+), pARP1265 (flaB+ flaG+ flaH+ flaJ+), and pARP1266 (flaA+ flaB+ flaG+ flaH+ flaJ+) were constructed using the broad-host-range mobilizable vector pBBR1MCS-3. Plasmids were introduced into each mutant strain by conjugation, and the resulting phenotypes shown in Table 2. The motility of strain AAR269 was fully restored to the wild-type level by providing pARP1264 or pARP1266 in trans, as judged by swarm size on soft agar. The motility of the flaB mutant AAR27 was also increased by providing either pARP1265 or pARP1266 in trans; however, this was not to wild-type levels. Motility was restored in the nonmotile double-mutant AAR31 (flaA flaB) by providing either or both of the flagellin genes in trans, but again the swarm size was not restored to the wild-type level. Complementation analysis of the nonmotile strains AAR59 (flaH) and AAR8 (flaJ) demonstrated that motility could be rescued by providing either pARP1265 or pARP1266 in trans, but swarm size in soft agar again could not be restored to wild-type levels. Partial complementation could be due to altering the levels of expression of the flagellar genes by providing them on multicopy plasmids. Although the cartridge insertions ensure transcription of downstream genes, the regulation and expression levels of these genes could be altered, thus resulting in partial complementation. However, the genetic information required to complement motility (not to wild-type levels in certain cases) is present on these plasmids.
Adherence.
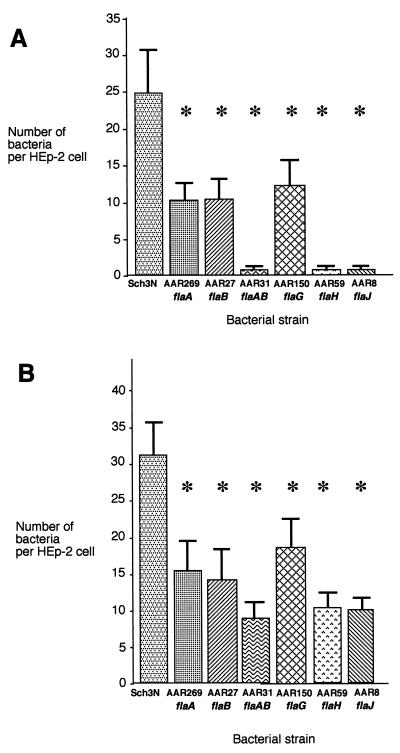
In vitro adherence of the mutants was tested by HEp-2 adherence assay. The overall pattern of adherence observed for the mutants was similar to that seen for motility (Fig. 7A). Single mutations in either flaA (AAR269) or flaB (AAR27) resulted in a reduction of adhesion to 41 and 42% of the wild-type level, respectively. Mutations that caused the loss of flagella and therefore impaired motility, such as those in flaH (AAR59), flaJ (AAR8), and flaA flaB (AAR31), completely abolished adherence. Mutation of flaG had the least effect on adherence, reducing the level to 50% of the wild-type level. However, all mutants demonstrated a highly significant reduction in adherence compared to the wild type as determined by the t test (P < 0.0001). To determine whether motility or the possession of flagella per se was required for adherence to HEp-2 cells, the motility defect was overcome through centrifugation of the bacteria onto the monolayer (Fig. 7B). Centrifugation increased the average number of the wild-type bacteria per HEp-2 cell from approximately 25 to 32. Again, the single mutations in flaA, flaB, and flaG resembled the adhesion pattern observed in the uncentrifuged adherence assay, with the levels of adherence being 52, 47, and 58% of the wild-type level, respectively. However, the largest increases in adhesion were seen for the tandem mutant AAR31 and the flaH and flaJ mutants, whose adherence levels increased to 30, 34, and 33% of the wild-type level, respectively. However, the reduction in adherence of the mutant strains was still highly significant compared to the wild-type level (P < 0.0001). Thus, the nonadherent phenotype could only be partially rescued by centrifugation, suggesting that motility and optimal polar flagellar function are required for full aeromonad adherence to HEp-2 cells.
FIG. 7.
Adherence of A. caviae strains to HEp-2 cells without (A) or with (B) centrifugation of the bacteria onto the monolayer. Bacteria were grown statically in BHIB at 37°C, and adherence assays carried out (see Materials and Methods). For assays that were preceded by centrifugation of the bacteria onto the monolayer, bacteria were centrifuged at 400 × g for 15 min at room temperature prior to the 90-min infection period. Assays were carried out in duplicate on three separate occasions, and the mean numbers of adherent bacteria per HEp-2 cell were recorded. The error bars represent one standard deviation (∗, P < 0.0001).
DISCUSSION
The ability of aeromonads to adhere to the HEp-2 human epithelial cell model has been correlated with the enteropathogenicity of the genus (8). Recent studies investigating the adherence of A. caviae to HEp-2 cells suggested the involvement of the polar flagellum in this process (48). This was further supported by the loss of adherence recorded by the flm mutants of A. caviae Sch3N which lost motility, flagella, and LPS O-Ag (15). Both of these extracellular structures have been implicated in aeromonad adherence (37, 38, 39), and we were unable to differentiate if one or both were essential for adhesion. We therefore initiated this study to characterize and mutate the polar flagellin locus of A. caviae which would allow us to investigate its role in adherence.
In preliminary experiments, antiflagellin antibodies and purified flagella significantly (P < 0.001) reduced A. caviae adherence to the HEp-2 model, suggesting a role for the flagellum in the process. The subsequent use of defined flagellar mutants confirmed this hypothesis and indeed showed that motility and optimal flagellar function are required. Mutants with single insertions in the flagellin genes flaA or flaB had both reduced motility and adherence, while nonmotile mutants which lacked the flagellar filament (flaA flaB, flaH, and flaJ) were completely nonadherent. Interestingly, cells of the flaG mutant were fully motile but had a significant reduction in adherence (P < 0.0001). The motility defect of the aflagellate mutants (AAR31, AAR59, and AAR8) could only be partially compensated for by centrifugation of the bacteria onto the monolayer. This process had no effect on the adherence phenotypes of the slightly motile A. caviae strains (AAR269, AAR27, and AAR150) and only recovered adherence to approximately 30% of the wild-type level for the nonmotile aflagellate mutants (AAR31, AAR59, and AAR8). Centrifugation may not be as efficient as bacterial motility at bringing the bacterium into contact with the HEp-2 cells. These findings suggest that motility is important for A. caviae adherence to HEp-2 cells. Mutation of flaG results in wild-type motility but significantly reduces bacterial adhesion, indicating that some component of the flagellum is involved in the adherence process or that the mutation does alter the flagellum somehow. Although we were unable to detect this using the crude motility assay, we conclude that the flagellum needs to function in an optimal wild-type manner for adherence.
Studies with nonflagellated isolates of C. jejuni (43), H. felis (19), and P. aeruginosa (11) have demonstrated the requirement of both flagellum and motility for virulence and colonization. In P. aeruginosa the flagellum is thought to act in the initial interactions and tether the bacterium to the host epithelium (11). In nonpiliated strains of P. aeruginosa, FliD (FlaH homologue) is required for full motility and is thought to be essential for adherence to respiratory mucin in the airways of cystic fibrosis patients (4). Motility per se has been shown to be essential for the adherence to HEp-2 cells by V. cholerae (13) and for in vivo virulence of the fish pathogen V. anguillarum (34, 40).
Sequence analysis of the polar flagellin locus revealed a genetic organization similar to that reported for A. salmonicida, with two tandemly linked flagellin genes followed by genes encoding FlaG and FlaH homologues (52). A similar organization is also seen in V. parahaemolyticus (24), V. anguillarum (34), and V. cholerae (30). However, these organisms possess between four and six polar flagellin genes, which all share significant homology to each other. The A. caviae polar flagellum consists of two flagellin subunits encoded by flaA and flaB, since both of the predicted amino acid sequences of the flagellins were present in the derived N-terminal amino acid sequence. Additionally, mutation of flaA and flaB in tandem resulted in the complete loss of motility and flagellin production, and no other polar flagellin genes were detected by Southern hybridization (data not shown).
Even though the A. caviae flagellins shared 92% amino acid identity, mutation of either caused a decrease in motility, suggesting that one cannot fully compensate for the loss of the other but that both are required for optimal flagellum function. For complex multisubunit flagella such as those present in V. parahaemolyticus, the flagellins are thought to work best with their contiguous partners (33); this seems to be the case for A. caviae.
Mutation of flaG did not affect motility but resulted in an abnormally long flagellum filament. A comparable phenotype was observed for flaG mutants of V. anguillarum (34). In Vibrio spp. the role of FlaG is not yet clearly defined since it has been implicated in either flagellar export regulating flagellum length or in the regulation of flagellum gene expression (34). FlaH is a homologue of FliD, a HAP-2 distal capping protein. The A. caviae flaH mutant did not express any flagellin and, hence, was completely nonmotile and aflagellate. A similar observation has been made for the FliD mutants of a number of bacteria, including P. aeruginosa, E. coli, and serovar Typhimurium (4). Interestingly, this is not the case for the flaH mutants of V. parahaemolyticus, which were reported to be swimming impaired but were still able to produce a truncated flagellum. This was proposed to be due to V. parahaemolyticus possessing a flagellum sheath, which was able to substitute for FlaH the flagellar capping protein and help with the polymerization of flagellin subunits (33). Helicobacter pylori possesses a sheathed polar flagellum; however, fliD mutants produce truncated flagella which are functionally defective and result in a nonmotile phenotype (23). FlaJ is a homologue of FliS, which is thought to be a chaperone required for the export of the flagellin subunits. Mutation of flaJ in A. caviae resulted in the complete loss of motility and flagellin expression; the same phenotype was reported for V. parahaemolyticus (46). Polar mutations which affect FliS expression in P. aeruginosa were demonstrated to be unimportant for motility (4); however, this organism possesses at least two fliS genes (4).
The predicted molecular masses of the two A. caviae flagellins were almost identical at 31,700 Da and could not be differentiated by one-dimensional PAGE. The flagellins exhibited aberrant migration on SDS-PAGE, with an estimated apparent molecular mass of 35,500 Da, which was significantly higher than the predicted molecular mass. An explanation for this discrepancy could be posttranslational modification. The preliminary data presented here indicated that the A. caviae polar flagellins are indeed glycosylated. Posttranslational modification was previously believed to be rare in eubacteria, but recently a number of bacterial flagellins have been demonstrated to be modified through glycosylation (10) or phosphorylation (22) or to contain N-methyl-lysine (20). Glycosylation of the A. caviae flagellin could be executed by the flm locus recently described for mesophilic Aeromonas spp. (15).
ACKNOWLEDGMENTS
We are grateful to the Saudi Arabian Ministry of Higher Education for the Scholarship to A.A.R. and thank Ann Cooke and Margaret Lee for their help with tissue culture. We thank Maite Polo for technical assistance.
Part of this work was supported by grants from DGICYT and Plan Nacional de I+D (Ministerio de Educación y Cultura, Spain).
REFERENCES
- 1.Aguilar A, Merino S, Rubirés X, Tomás J M. Influence of osmolarity on lipopolysaccharides and virulence of Aeromonas hydrophila serotype O:34 strains grown at 37°C. Infect Immun. 1997;65:1245–1250. doi: 10.1128/iai.65.4.1245-1250.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul F S, Madden T L, Schaffer A A, Zhang J, Zang Z, Miller W, Lipman J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arora S K, Ritchings B W, Almira E C, Lory S, Ramphal R. Cloning and characterization of Pseudomonas aeruginosa fliF, necessary for flagellar assembly and bacterial adherence to mucin. Infect Immun. 1996;64:2130–2136. doi: 10.1128/iai.64.6.2130-2136.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arora S K, Ritchings B W, Almira E C, Lory S, Ramphal R. The Pseudomonas aeruginosa flagellar cap protein, FliD, is responsible for mucin adhesion. Infect Immun. 1998;66:1000–1007. doi: 10.1128/iai.66.3.1000-1007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Austin B, Adams C. Fish pathogens. In: Austin B, et al., editors. The genus Aeromonas. Chichester, West Sussex, United Kingdom: John Wiley and Sons; 1996. pp. 197–243. [Google Scholar]
- 6.Bott M, Meyer M, Dimroth P. Regulation of anaerobic citrate metabolism in Klebsiella pneumoniae. Mol Microbiol. 1995;18:533–546. doi: 10.1111/j.1365-2958.1995.mmi_18030533.x. [DOI] [PubMed] [Google Scholar]
- 7.Burke V, Robinson J, Gracey M, Petersen D, Partridge K. Isolation of Aeromonas hydrophila from a metropolitan water supply: seasonal correlation with clinical isolates. Appl Environ Microbiol. 1984;49:361–366. doi: 10.1128/aem.48.2.361-366.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrello A, Silburn K A, Budden J R, Chang B J. Adhesion of clinical and environmental Aeromonas isolates to HEp-2 cells. J Med Microbiol. 1988;26:19–27. doi: 10.1099/00222615-26-1-19. [DOI] [PubMed] [Google Scholar]
- 9.De Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doig P, Kinsella N, Guerry P, Trust T J. Characterization of a post-translational modification of Campylobacter flagellin: identification of a sero-specific glycosyl moiety. Mol Microbiol. 1996;19:379–387. doi: 10.1046/j.1365-2958.1996.370890.x. [DOI] [PubMed] [Google Scholar]
- 11.Feldman M, Bryan R, Rajan S, Scheffler L, Brunnert S, Tang H, Prince A. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect Immun. 1998;66:43–51. doi: 10.1128/iai.66.1.43-51.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francki K T, Chang B J. Variable expression of O-antigen and the role of lipopolysaccharide as an adhesin in Aeromonas sobria. FEMS Microbiol Lett. 1994;122:97–102. doi: 10.1111/j.1574-6968.1994.tb07150.x. [DOI] [PubMed] [Google Scholar]
- 13.Gardel C L, Mekalanos J J. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect Immun. 1996;64:2246–2255. doi: 10.1128/iai.64.6.2246-2255.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerard C. Purification of glycoproteins. Methods Enzymol. 1990;182:529–539. doi: 10.1016/0076-6879(90)82042-z. [DOI] [PubMed] [Google Scholar]
- 15.Gryllos I, Shaw J G, Gavín R, Merino S, Tomás J M. Role of flm locus in mesophilic Aeromonas adherence. Infect Immun. 2001;69:65–74. doi: 10.1128/IAI.69.1.65-74.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerry P, Alm R A, Power M E, Logan S M, Trust T J. The role of two flagellin genes in Campylobacter motility. J Bacteriol. 1991;173:4757–4764. doi: 10.1128/jb.173.15.4757-4764.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honma Y, Nakasone N. Pili of Aeromonas hydrophila: purification, characterisation and biological role. Microbiol Immunol. 1990;34:83–98. doi: 10.1111/j.1348-0421.1990.tb00995.x. [DOI] [PubMed] [Google Scholar]
- 19.Josenhans C, Ferrero R L, Labigne A, Suerbaum S. Cloning and allelic exchange mutagenesis of two flagellin genes in Helicobacter felis. Mol Microbiol. 1999;33:350–362. doi: 10.1046/j.1365-2958.1999.01478.x. [DOI] [PubMed] [Google Scholar]
- 20.Joys T M, Kim H. Identification of N-methyl-lysine residues in the phase-1 flagellar protein of Salmonella typhimurium. Microbios Lett. 1979;7:65–68. [Google Scholar]
- 21.Kaniga K, Delor I, Cornelis G R. A wide-host-range suicide vector for improving reverse genetics in Gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene. 1991;109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- 22.Kelly-Winterberg K, South S L, Montie T C. Tyrosine phosphate in a- and b-type flagellins of Pseudomonas aeruginosa. J Bacteriol. 1993;175:2458–2461. doi: 10.1128/jb.175.8.2458-2461.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J S, Chang J H, Chung S I, Yum J S. Molecular cloning and characterisation of the Helicobacter pylori fliD gene, an essential factor in flagellar structure and motility. J Bacteriol. 1999;181:6969–6976. doi: 10.1128/jb.181.22.6969-6976.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim Y-K, McCarter L L. Analysis of the polar flagellar gene system of Vibrio parahaemolyticus. J Bacteriol. 2000;182:3693–3704. doi: 10.1128/jb.182.13.3693-3704.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirk M, Dalton C, Cameron S. An exploratory case control study of Aeromonas gastroenteritis—is water the vehicle for transmission. Aust Microbiol. 1997;18:5.3. [Google Scholar]
- 26.Kirov S M. Adhesion and piliation of Aeromonas spp. Med Microbiol Lett. 1993;2:274–280. [Google Scholar]
- 27.Kirov S M, Jacobs I, Hayward L J, Hapin R H. Electron microscopic examination of factors influencing the expression of filamentous surface structures on clinical and environmental isolates of Aeromonas veronii biotype sobria. Microbiol Immunol. 1995;39:329–338. doi: 10.1111/j.1348-0421.1995.tb02209.x. [DOI] [PubMed] [Google Scholar]
- 28.Kirov S M, O'Donovan L, Sanderson K. Functional characterisation of type IV pili expressed on diarrhea-associated isolates of Aeromonas species. Infect Immun. 1999;67:5447–5454. doi: 10.1128/iai.67.10.5447-5454.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirov S M, Sanderson K. Characterisation of a type IV bundle forming pilus (SFP) from a gastroenteritis-associated strain of Aeromonas veronii biovar sobria. Microb Pathog. 1996;21:23–34. doi: 10.1006/mpat.1996.0039. [DOI] [PubMed] [Google Scholar]
- 30.Klose K E, Mekalanos J J. Differential regulation of multiple flagellins in V. cholerae. J Bacteriol. 1998;180:303–316. doi: 10.1128/jb.180.2.303-316.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kovach M E, Elzer P H, Hill D S, Robertson G T, Farris M A, Roop II R M, Peterson K. Four new derivatives of the broad-host-range cloning vector pBBR1MCS carrying different antibiotic resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 32.Kovach M E, Phillips R W, Elzer P H, Roop II R M, Peterson K M. pBBR1MCS: a broad-host-range cloning vector. BioTechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- 33.McCarter L L. Genetic and molecular characterisation of the polar flagellum of Vibrio parahaemolyticus. J Bacteriol. 1995;178:1310–1319. doi: 10.1128/jb.177.6.1595-1609.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGee K, Horstedt P, Milton D L. Identification and characterization of additional flagellin genes from Vibrio anguillarum. J Bacteriol. 1996;178:5188–5198. doi: 10.1128/jb.178.17.5188-5198.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacNab R M. Flagella and motility. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 123–145. [Google Scholar]
- 36.Merino S, Campuri S, Tomás J M. Isolation and characterisation of bacteriophage PM3 from Aeromonas hydrophila the bacteriological receptor for which is the monopolar flagellum. FEMS Microbiol Lett. 1990;69:277–282. doi: 10.1016/0378-1097(90)90080-a. [DOI] [PubMed] [Google Scholar]
- 37.Merino S, Rubirés X, Aguilar A, Guillot J F, Tomás J M. The role of the O-Ag lipopolysaccharide on the colonisation in vivo of the germ free chicken gut by Aeromonas hydrophila serogroup O:34. Microb Pathog. 1996;20:325–333. doi: 10.1006/mpat.1996.0031. [DOI] [PubMed] [Google Scholar]
- 38.Merino S, Rubirés X, Aguilar A, Tomás J M. The O:34-antigen lipopolysaccharide as an adhesin in Aeromonas hydrophila. FEMS Microbiol Lett. 1996;139:97–101. doi: 10.1111/j.1574-6968.1996.tb08186.x. [DOI] [PubMed] [Google Scholar]
- 39.Merino S, Rubirés X, Aguilar A, Tomás J M. The role of flagella and motility in the adherence and invasion to fish cell lines by Aeromonas hydrophila serogroup O:34 strains. FEMS Microbiol Lett. 1997;151:213–217. doi: 10.1111/j.1574-6968.1997.tb12572.x. [DOI] [PubMed] [Google Scholar]
- 40.Milton D L, O'Toole R, Horstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. J Bacteriol. 1996;178:1310–1319. doi: 10.1128/jb.178.5.1310-1319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Namdari H, Bottone E J. Microbiologic and clinical evidence supporting the role of Aeromonas caviae as pediatric enteric pathogen. J Clin Microbiol. 1990;28:837–840. doi: 10.1128/jcm.28.5.837-840.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ottemann K M, Miller J F. Roles for motility in bacterial-host interactions. Mol Microbiol. 1997;24:1109–1117. doi: 10.1046/j.1365-2958.1997.4281787.x. [DOI] [PubMed] [Google Scholar]
- 44.Pleier E, Schmitt R. Identification and sequence of two related flagellin genes in Rhizobium meliloti. J Bacteriol. 1989;171:1467–1475. doi: 10.1128/jb.171.3.1467-1475.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Stewart B J, McCarter L L. Vibrio parahaemolyticus FlaJ, a homologue of FliS, is required for production of flagellin. Mol Microbiol. 1996;20:137–149. doi: 10.1111/j.1365-2958.1996.tb02496.x. [DOI] [PubMed] [Google Scholar]
- 47.Thompson J D, Higgins D G, Gibson T J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thornley J P, Shaw J G, Gryllos I A, Eley A. Adherence of Aeromonas caviae to human cell lines HEp-2 and Caco-2. J Med Microbiol. 1996;45:445–451. doi: 10.1099/00222615-45-6-445. [DOI] [PubMed] [Google Scholar]
- 49.Thornley J P, Shaw J G, Gryllos I, Eley A. Virulence properties of clinically significant Aeromonas species. Rev Med Microbiol. 1997;8:61–72. [Google Scholar]
- 50.Tinoco I, Borer P N, Dengler B, Levire M D, Uhlenbeck O C, Crothers D M, Gralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973;246:40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- 51.Totten P A, Lory S. Characterization of the type a flagellin gene from Pseudomonas aeruginosa PAK. J Bacteriol. 1990;172:7188–7199. doi: 10.1128/jb.172.12.7188-7199.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Umelo E, Trust T J. Identification and molecular characterization of two tandemly located flagellin genes from Aeromonas salmonicida A449. J Bacteriol. 1997;179:5292–5299. doi: 10.1128/jb.179.17.5292-5299.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weissborn A, Steiman H M, Shapiro L. Characterization of the proteins of the Caulobacter crescentus flagellar filament. J Biol Chem. 1982;257:2066–2074. [PubMed] [Google Scholar]
- 54.Wilcox M H, Cook A M, Eley A R, Spencer R C. Aeromonas spp. as a potential cause of diarrhoea in children. J Clin Pathol. 1992;45:959–963. doi: 10.1136/jcp.45.11.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yao R, Burr D H, Doig P, Trust T J, Guerry P. Isolation of motile and non-motile insertional mutants of Campylobacter jejuni: the role of motility in adherence and invasion of eukaryotic cells. Mol Microbiol. 1994;14:883–893. doi: 10.1111/j.1365-2958.1994.tb01324.x. [DOI] [PubMed] [Google Scholar]
- 56.Yokoseki T, Kutsukake K, Ohnishi K, Lino T. Functional analysis of the flagellar genes in the fliD operon of Salmonella typhimurium. Microbiology. 1995;141:1715–1722. doi: 10.1099/13500872-141-7-1715. [DOI] [PubMed] [Google Scholar]