Abstract
This study developed a new single-tube multiplex real-time PCR method for detecting toxigenic C. difficile directly from fecal samples using tcdA, tcdB, cdtB, and internal gene tpi as targets, which could be performed on kinds of polymerase chain reaction device including point-of-care testing (POCT), with improved detection efficiency. The specificity, sensitivity, and repeatability of each gene was evaluated using 69 C. difficile isolates and 74 fecal samples. Results were compared with established PCR, qPCR, and ELISA methods. Interspecies specificity was 100% based on six common intestinal pathogens (Escherichia coli, Enterococcus Faecium, Enterococcus faecalis, Clostridium perfringens, Bacteroides fragilis, Clostridium botulinum). The lower detection limit (LDL) for tcdA, tcdB, and cdtB with pure C. difficile DNA was 101,100, and 100 copies/μL, respectively, the coefficients of variation among different experimental batches and within each experimental batch were both less than 3%, which shows that this method has strong repeatability. And the LDL of fecal DNA was 5 × 100, 5 × 103, and 5 × 102 colony-forming units (CFU)/g, respectively. In addition, the efficiency for detection of tcdA was compared with established PCR and real-time PCR methods, demonstrating high consistency (98.4%) and similar sensitivity. ELISA was used to confirm inconsistent results, which were identical with our method. The sensitivity and specificity for detecting toxigenic C. difficile in fecal samples were 96.49% and 94.12% compared with the toxigenic culture (TC). This method effectively identified the toxigenic and non-toxigenic strains with high specificity, sensitivity, and repeatability, and could reduce the false positive rate of tcdA, and accurately identify the typical Asian strain RT017, making it potentially contribute to the surveillance of CDI in China.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-022-03434-6.
Keywords: Multiplex real-time PCR, Toxigenic Clostridioides difficile, Toxin a/B, Binary toxin
Background
Clostridioides difficile is an anaerobic Gram-positive bacillus that is widespread in the intestines of humans and other animals (Johnson et al. 2021; Czepiel et al. 2019). Risk factors such as antibiotic exposure, older age, and weakened immune system, are known to be closely related to C. difficile infection (CDI) (Leffler and Lamont 2015), which is one of the most common hospital-acquired infections with symptoms of diarrhea, pseudomembranous colitis, toxic megacolon, and even death (McDonald et al. 2018). A multistate point-prevalence survey of healthcare-associated infections in the United States found C. difficile was the most commonly reported pathogen (causing 12.1% of healthcare-associated infections) (Magill et al. 2014), and previous publications which demonstrated the high economic burden of CDI for healthcare settings and health insurance systems (Mollard et al. 2019). In summary, CDI poses a significant burden on both life health and social resources, which should be of great concern.
Toxins produced by C. difficile play an important role in the pathogenic process and two major toxins are produced: toxin A (enterotoxin) and toxin B (cytotoxin), which can be produced alone or simultaneously. Moreover, a third toxin-binary toxin is found in some C. difficile isolates, such as hypervirulent RT027 / NAP1 / BI (Nagy 2018) and RT078. Although the exact role of binary toxin in pathogenesis is unclear, but it is thought to be associated with high incidence, recurrence, and mortality of CDI (Bacci et al. 2011; Stewart et al. 2013). However, typical Asian strain RT017( toxin A-negative toxin B-positive) causing several outbreaks worldwide, has also been identified in recent years (Drudy et al. 2007).
There are three commonly used methods for the identification of toxigenic C. difficile: (i) TC (toxigenic culture) is a two-step method that combined with C. difficile culture and cell cytotoxicity assay (CCNA) (Nagy 2018); (ii) enzyme immunoassay (EIA) for toxin A/B and glutamate dehydrogenase(GDH) (Bagdasarian et al. 2015); (iii) nucleic acid amplification tests (NAATs) targeting toxin-encoding genes, which include PCR, quantitative PCR (QPCR), loop-mediated isothermal amplification (LAMP), and helicase-dependent isothermal DNA amplification (HAD). TC operation is complex and time-consuming, which is mainly used for epidemiological research and evaluation of new methods (Bagdasarian et al. 2015). The sensitivity and specificity of immunological testing can vary, and it must be combined with a high sensitivity-specific approach to make up for its shortcomings. Therefore, NAAT was recommended for diagnosing CDI worldwide in both two-steps and three-steps diagnosis procedures (Lee et al. 2021).
In the present study, our novel method can simultaneously detect tcdA, tcdB, and cdtB genes, which improves the accurate identification of toxigenic C. difficile isolates directly from fecal samples. First, it is noteworthy that the typical Asian strain RT017, which has an A-B + cdtA/B- profile, plays an important role in CDI epidemiology in China. A deletion of 1821 bp was found in the repeat region of tcdA. Although none toxin A was produced due to the presence of a stop codon in these A-B + isolates, but there still are DNA fragments remaining in tcdA-negative isolates (Rupnik et al. 2003), primers that circumvent this deleted region may lead to false-positive results for toxinA in this type strain. Second, other multiple qPCR methods only included tcdA and tcdB as target genes, but binary toxin genes were not included (Luna et al. 2011; Kubota et al. 2014; Bélanger et al. 2003). Additionally, since binary toxin genes cdtA and cdtB may be fused, choosing cdtA as the target gene may lead to false-negative results for binary toxin (Gerding et al. 2014). In other previously published multiple qPCR methods, cdtA rather than cdtB was selected as the target gene for binary toxin detection, which may lead to inaccurate detection of binary toxin (Kilic et al. 2015; Hoegh et al. 2012). Therefore, cdtB was selected as the target gene for binary toxin detection.
In a word, a multiplex real-time PCR method was developed to detect toxigenic C. difficile directly from fecal samples, involving tcdA, tcdB, cdtB, and internal gene tpi as targets, which could be performed on kinds of polymerase chain reaction devices including POCT for rapid detection and identification of clinical CDI cases. Furthermore, this method could reduce the false positive rate of tcdA, accurately identify the typical Asian strain RT017 with improved detection efficiency, making it potentially contribute to the surveillance of CDI in China.
Methods
Design of primers and probes
Complete sequences of tcdA (NZ_FUUL01000004.1, NC_017178.1, NZ_CDND01000001.1, NC_017174.1, NC_013316.1), tcdB (NZ_FUUL01000004.1, NC_017178.1, NZ_CDND01000001.1, NC_017174.1, NC_013316.1), and cdtB (AF271719.1, HQ639679.1, FN538970.1, NC_017178.1, NZ_CDND01000001.1, NC_017174.1)were download from NCBI GenBank entries and aligned to determine the conserved regions. According to the consensus sequences, primers and probes were designed by Primer Express software v.3.0 (Applied Biosystems, Foster City, CA), and then were BLAST in GenBank to test the specificity. There was no crossover with other pathogens using BLAST searches in the NCBI database. All primers and probes were included in patent application CN202010825309. X. RT017 is a typical Asian strain characterized by a partial deletion in the repeat region of the tcdA gene. Due to the presence of a termination codon, it does not produce toxin A, but most of the tcdA negative isolates still possess remaining DNA fragments. Therefore, it is necessary to design primers targeting the absent region of the tcdA gene for the accurate identification of toxin A.
Reaction system and parameters
The optimized qPCR reaction system (20 μl) is composed of 10 μl Premix (TaKaRa RR390A), 0.4 μl of 10 mM forward and reverse primers (single-tube multiplex), 0.8 μl of 10 mM probe, 0.2 μl of Rox Reference Dye II, 2 μl of template DNA, and 6.2 μl of deionized water. The two-step method was employed by heating at 95 °C for 20 s, followed by 40 cycles at 95 °C for 3 s and 58 °C for 30 s. Reactions were performed on a 7500 Fast Real-Time PCR System (Applied Biosystems), LightCycler 96 (Roche), QuentGene 9600(Bioer), and iPonatic(Sansure).
Construction of standard plasmids and standard curves for the three genes
The C. difficile isolate ATCC-BBA1803 (RT027) was used to amplify the three target toxin genes. PCR products were purified using an EasyPure Quick Gel Extraction Kit (Trans, China), and then ligated into the PMD18-T vector (TaKaRa, Japan), which were subsequently transformed into JM109 competent cells (TaKaRa). Positive plasmids were identified by Blue-White Screening of positive colonies and sequencing of PCR products from plasmids. The standard plasmid concentration was obtained according to the copy number conversion formula (6.02 × 1023copies/M × (concentration)/(MWg/mol) = copies/ml), and ten-fold dilutions of the standard into ten gradients were used as templates to prepare the standard curve.
Evaluation of the specificity, sensitivity, and repeatability of qPCR for pure bacterial DNA
Standard strains of another six common intestinal pathogens (Escherichia coli, Enterococcus Faecium, Enterococcus faecalis, Clostridium perfringens, Bacteroides fragilis, Clostridium botulinum) were used to test interspecies specificity (Table S1). A total of 69 C. difficile isolates with different toxigenic types, including eight ATCC standard strains and 61 clinical isolates, were used to verify intraspecies specificity (Table S1). Through standard curve analysis, the pure bacterial lower detection limit (LDL), also known as the minimum concentration of detected plasmid, was determined. To assess the repeatability of this method, different concentrations (high, medium, and low copies) of three standard plasmids harboring tcdA, tcdB, and cdtB genes were tested in triplicate every other day. Then three parallel DNA samples were tested to obtain the coefficient of variation between batches and the coefficient of variation within groups (Table S2).
Preparation and detection of C. difficile-simulated fecal samples
A pure clone of the C. difficile ATCC1803 toxigenic strain was picked and mixed with 1 ml BHI (Brain Heart Infusion Broth) to achieve an McF 0.5 suspension of ~ 105 colony-forming units (CFU)/ml by the standard plate counting method. Bacterial suspensions at 100 to 105 CFU/ml were obtained by serial dilution, and 0.1 ml of each suspension was transferred into 0.2 mg C. difficile-negative fecal samples from healthy individuals at a final concentration of 5.0 × 10− 1 to 5.0 × 104 CFU/g. Total DNA was extracted from these simulated fecal samples for qPCR evaluation of LDLs for the three target genes (tcdA, tcdB, and cdtB; Table 1).
Table 1.
Lower limit of detection of three target genes in simulated feces
| Target genes | CFU/g | 5.0 × 104 | 5.0 × 103 | 5.0 × 102 | 5.0 × 101 | 5.0 × 100 | 5.0 × 10− 1 |
|---|---|---|---|---|---|---|---|
| tcdA | Sample | 26.48415 | 29.63562 | 33.04662 | 36.79008 | 36.71255 | Undetermined |
| Replication1 | 26.55865 | 29.64664 | 32.28347 | 37.06828 | 36.01574 | Undetermined | |
| Replication2 | 26.47834 | 29.70147 | 32.97718 | 36.2962 | 36.13764 | 36.60099 | |
| SD | 0.044782 | 0.035268 | 0.42199 | 0.391029 | 0.37214 | ||
| tcdB | Sample | 31.4739 | 34.99224 | Undetermined | Undetermined | Undetermined | Undetermined |
| Replication1 | 31.56133 | 35.34561 | 38.45827 | Undetermined | Undetermined | Undetermined | |
| Replication2 | 31.53615 | 34.95603 | 37.68927 | Undetermined | Undetermined | Undetermined | |
| SD | 0.045007 | 0.215232 | |||||
| cdtB | Sample | 28.53067 | 31.93304 | 35.61412 | 39.17686 | Undetermined | Undetermined |
| Replication1 | 28.38683 | 31.92636 | 35.62996 | Undetermined | Undetermined | Undetermined | |
| Replication2 | 28.45992 | 31.87342 | 34.94851 | 38.94106 | Undetermined | Undetermined | |
| SD | 0.071922 | 0.032662 | 0.388941 |
Detection of human fecal samples using multiplex real-time PCR
A total of 74 frozen fecal samples in our laboratory were used to evaluate our qPCR detection method. A Fecal DNA Extraction Kit (TIANGEN) was used to extract fecal DNA according to the manufacturer’s instructions. The results were compared with those of the gold standard (TC) method using tpi gene as internal control to evaluate the performance (Table 2).
Table 2.
Fecal samples included in this study and test results
| Number | Samples | Toxigenic | Detection result | |||
|---|---|---|---|---|---|---|
| tcdA | tcdB | cdtB | tpi | |||
| 1 | 10,005 | N | − | − | − | + |
| 2 | 11,032 | N | − | − | − | + |
| 3 | 24,078 | Y | + | + | − | + |
| 4 | 25,049 | Y | + | + | − | + |
| 5 | 0204–001 | N | − | − | − | + |
| 6 | 0204–005 | Y | + | + | − | + |
| 7 | 0205–005 | N | − | − | − | + |
| 8 | 0205–007 | N | − | − | − | + |
| 9 | 0205–009 | N | − | − | − | + |
| 10 | 01,047 | Y | + | + | − | + |
| 11 | 12,038 | Y | + | + | + | + |
| 12 | 25,047 | Y | + | + | − | + |
| 13 | 25,049 | Y | + | + | − | + |
| 14 | 20,051 | Y | + | + | − | + |
| 15 | 25,053 | Y | + | + | + | + |
| 16 | 25,058 | Y | + | + | + | + |
| 17 | 0201–016 | Y | + | + | − | + |
| 18 | 0201–021 | Y | + | − | − | + |
| 19 | 0201–033 | Y | + | + | − | + |
| 20 | 0201–045 | Y | + | + | − | + |
| 21 | 0201–059 | Y | + | + | − | + |
| 22 | 0201–069 | Y | + | + | − | + |
| 23 | 0201–074 | Y | + | + | − | + |
| 24 | 0201–077 | Y | + | + | − | + |
| 25 | 0204–004 | Y | − | − | − | + |
| 26 | 0205–002 | Y | − | + | − | + |
| 27 | 0205–006 | Y | + | + | − | + |
| 28 | 0206–005 | Y | − | + | − | + |
| 29 | 0207–002 | Y | + | + | − | + |
| 30 | 10,122 | N | − | − | − | + |
| 31 | 10,010 | Y | + | − | − | + |
| 32 | 11,034 | Y | + | + | − | + |
| 33 | 20,086 | N | + | − | − | + |
| 34 | 24,078 | Y | + | + | − | + |
| 35 | 0201–018 | Y | − | + | − | + |
| 36 | 0201–029 | Y | + | + | − | + |
| 37 | 0201–041 | Y | + | + | − | + |
| 38 | 0201–080 | Y | + | − | − | + |
| 39 | 0203–004 | Y | + | + | − | + |
| 40 | 0203–006 | Y | − | + | − | + |
| 41 | 0205–001 | Y | + | − | − | + |
| 42 | 0205–008 | Y | − | + | − | + |
| 43 | 0206–003 | Y | + | + | − | + |
| 44 | 0207–003 | Y | − | − | − | + |
| 45 | 0207–006 | Y | + | + | − | + |
| 46 | 0208–002 | N | − | − | − | + |
| 47 | 0208–003 | Y | + | − | − | + |
| 48 | 10,007 | N | − | − | − | + |
| 49 | 10,122–2 | N | − | − | − | + |
| 50 | 21,076 | N | − | − | − | + |
| 51 | 22,012 | N | − | − | − | + |
| 52 | 29,033 | N | − | − | − | + |
| 53 | 0204–001 | N | − | − | − | + |
| 54 | 21,074 | Y | + | + | − | + |
| 55 | 12,038–2 | Y | + | + | + | + |
| 56 | 01,047–2 | Y | + | + | − | + |
| 57 | 11,068 | Y | + | + | − | + |
| 58 | 10,115 | Y | + | + | − | + |
| 59 | 20,051–2 | Y | + | + | − | + |
| 60 | 20,054 | Y | + | + | − | + |
| 61 | 21,078 | Y | + | + | − | + |
| 62 | 25,047 | Y | + | + | + | + |
| 63 | 25,058–2 | Y | + | + | + | + |
| 64 | 09,066 | Y | + | + | + | + |
| 65 | 12,038–2 | Y | + | + | + | + |
| 66 | 25,053–2 | Y | + | + | + | + |
| 67 | 0201–006 | Y | + | + | − | + |
| 68 | 0201–014 | N | − | − | − | + |
| 69 | 0201–038 | Y | − | + | − | + |
| 70 | 0201–040 | Y | + | + | − | + |
| 71 | 0201–063 | N | − | − | − | + |
| 72 | 0201–071 | Y | + | + | − | + |
| 73 | 0207–009 | Y | + | + | − | + |
| 74 | 0208–001 | Y | + | + | − | + |
Letter Y refers to positive results and N means negative results. The red letters refer to inconsistent results
Comparison with previously reported detection methods for tcdA and confirmation by ELISA for TcdA
Two pairs of classic PCR primers for gene tcdA (Kato et al. 1999; Lemee et al. 2004) were compared with our method using 33 C. difficile RT017 isolates and 30 other types of clinical isolates preserved in our laboratory (Table S3). For inconsistent results, ELISA was employed for clarification. Briefly, negative control (ATCC43593), positive control (ATCC-BBA1803), and tested C. difficile (N20) isolates (Table S3) were anaerobically cultured in BHI liquid medium at 37 °C for 24 h. After centrifugation, the supernatant was taken and filtered through a 0.45 μm filter, then incubated overnight at 4 °C in a 96-well plate. The plate was washed with PBST(Phosphate Buffered Saline + Tween) and blocked with 5% bovine serum albumin at 37 °C for 2 h. Polyclonal antibody (List Biological Laboratories, Toxin A IgY, 1:1000 dilution) was used for the first hybridization, and horseradish peroxidase (HRP-conjugated goat anti-chicken IgY (Invitrogen, 1:5000 dilution) served as the secondary antibody. Finally, the results were determined using an EL-TMB Chromogenic Reagent Kit (SANGON, China) at an absorbance of 450 nm. Standard curves were prepared using Toxin A (List Biological Laboratories) as a positive control at an initial concentration of 1600 ng/ml that was diluted twice to 0.05 ng/ml. The absorbance ratio (P/N) of the tested sample (N20) and negative control (ATCC43593) was calculated, and results were considered negative at P/N < 1.5, suspicious at P/N ≥ 1.5 and < 2.1, and positive at P/N ≥ 2.1.
In addition, our results were also compared with two previously reported qPCR methods (Luna et al. 2011; Kubota et al. 2014), in which our primers for gene tcdA were located in the missing region to avoid potential false positives (Fig. 1a). The qPCR conditions and mixture were previously described. The lower limits of these three pairs of primers for tcdA were tested and compared by testing the C. difficile-simulated fecal samples (Table 3) prepared as described above.
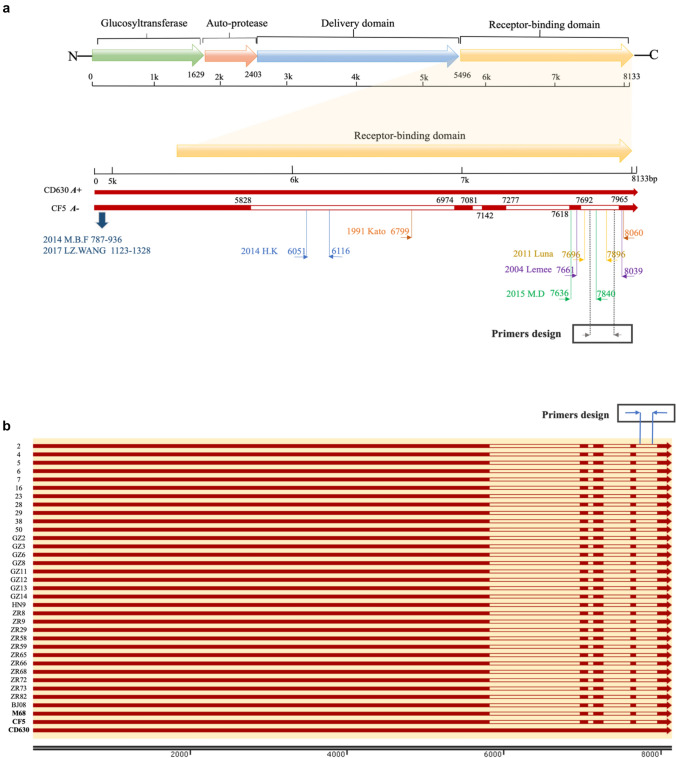
Fig. 1.
Schematic structure of the tcdA gene and the locations of primers in both the schematic diagram and tcdA sequences of 33 previously reported RT017 isolates. a Schematic structure of the tcdA gene and the locations of primers synthesized and tested in this study. b Complete sequences of tcdA extracted from whole genomes of 33 RT017 isolates and the locations of primers were synthesized and tested in this study. M68 (NC_017175.1), CF5 (NC_017173.1) and CD630 (NC_009089.1) were used as reference strains
Table 3.
The results and comparison of three pairs primers for tcdA detecting the C. difficile-simulated fecal samples
| Target (annealing temperature) | CFU/g | 5.0 × 104 | 5.0 × 103 | 5.0 × 102 | 5.0 × 101 | 5.0 × 100 | 5.0 × 10− 1 |
|---|---|---|---|---|---|---|---|
| Our tcdA (58℃) | Sample | 26.48415 | 29.63562 | 33.04662 | 36.79008 | 36.71255 | Undetermined |
| Replication1 | 26.55865 | 29.64664 | 32.28347 | 37.06828 | 36.01574 | Undetermined | |
| Replication2 | 26.47834 | 29.70147 | 32.97718 | 36.2962 | 36.13764 | 36.60099 | |
| SD | 0.044782 | 0.035268 | 0.42199 | 0.391029 | 0.37214 | ||
| Luna 2011 (57℃) | Sample | 26.76287 | 29.99077 | 33.65609 | 37.25234 | 39.6127 | 38.66362 |
| Replication1 | 26.67333 | 30.12401 | 33.5074 | 36.2081 | Undetermined | Undetermined | |
| Replication2 | 26.61492 | 30.10385 | 33.64701 | 37.33664 | 38.74393 | Undetermined | |
| SD | 0.07452 | 0.071819 | 0.083344 | 0.628642 | |||
| HK 2014 (56℃) | Sample | 26.14689 | 29.84816 | 33.71031 | 37.32001 | 38.19745 | 39.58826 |
| Replication1 | 26.43128 | 29.70106 | 33.79443 | 37.72804 | 39.00477 | 39.58448 | |
| Replication2 | 26.21063 | 29.72093 | 33.90454 | 37.76967 | 38.14799 | 39.7678 | |
| SD | 0.149239 | 0.079814 | 0.097404 | 0.248465 | 0.48102 | 0.104765 |
Flow chart
A flow chart for better understanding of the manuscript (Fig. 2).
Fig. 2.
A flow chart of the manuscript
Results
Primers and probes designed in this study
The sequences of the tcdA gene in the NCBI database for isolates CD630 (A +) and CF5 (A-) were compared, and four deleted and repeated regions of tcdA spanning 1821 bp were identified (Fig. 1a): 5828–6974 (1146 bp), 7081–7142 (61 bp), 7277–618 (341 bp), and 7692–7965 (273 bp). Our primers and probes are located in the deleted region spanning 7692–7965 bp (Fig. 1a). Meanwhile, by blasting against the whole genome sequences of 33 clinical C. difficile isolates (RT017) reported in our previous work (Wu et al. 2019), primers and probes for tcdA in the present study are all within the deleted regions of these 33 strains (Fig. 1b), which further confirmed their accuracy theoretically.
The real-time PCR method is specific, sensitive, and repeatable
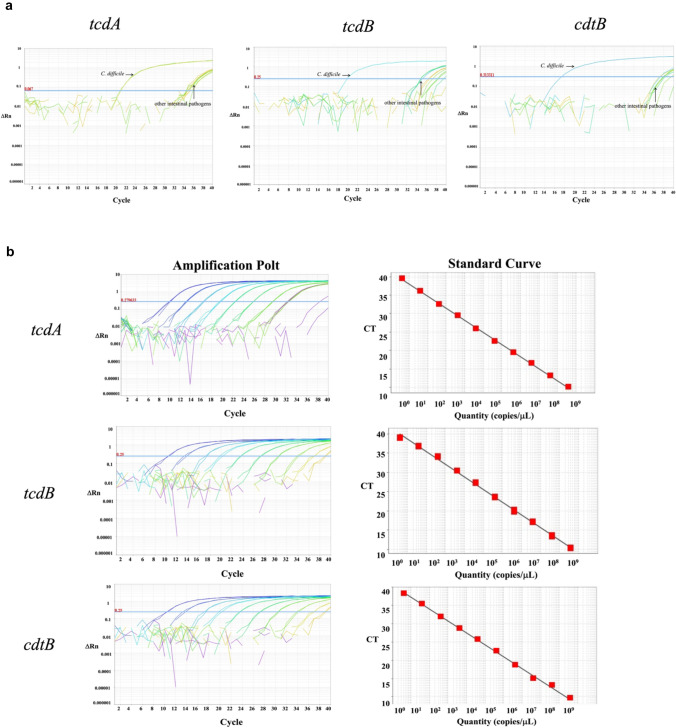
Standard strains of the other six common intestinal pathogens were tested using our primers and probes, and no specific products were amplified (Table S1). ATCC Standard strains of different toxigenic types of C. difficile were used to assess intraspecies specificity, and the results were consistent with the defined toxin profiles (Table S1). According to the above results, the specificity of this method was 100%, confirming its ability to detect toxigenic C. difficile strains accurately and selectively (Fig. 3a).
Fig. 3.
Specificity and sensitivity of the developed qPCR method for detecting tcdA, tcdB, and cdtB. a The specificity for detecting the three target genes was determined by comparing them with another six intestinal bacteria. b Standard curves and amplification curves for the three target genes
Standard curves of the three target genes (tcdA, tcdB, and cdtB) were constructed using plasmids diluted tenfold as templates for qPCR detection (Fig. 3b). The three plasmids were linearly correlated with their corresponding CT (Cycle threshold) values at 100 − 109 copies/μl. Using the lowest concentration of each plasmid that could be detected as the LDL, we verified that the detection limits of DNA from C. difficile isolates for tcdA, tcdB, and cdtB were 101, 100, and 100 copies/μl, respectively. PCR amplification efficiency for each gene was 102%, 103%, and 102%, respectively, and correlation coefficients reached 0.998 (Fig. 3b).
To evaluate the repeatability of this method, coefficients of variation among different experimental batches were calculated. Three replicate samples for each gene within each experiment batch were also tested. The coefficients of variation among different experimental batches and within each experimental batch were both less than 3%, which shows that this method has strong repeatability (Table S2).
Detection of toxigenic C. difficile in simulated fecal samples
When 0.2 g of feces was mixed with 100 CFU bacteria, the method could stably detect tcdA, and the bacteria concentration in feces was 5.0 × 100 CFU/g. Gene tcdB could be stably detected when 0.2 g of feces was mixed with 103 CFU bacteria, and the bacterium content in feces was 5.0 × 103 CFU/g.
And when 0.2 g of feces were mixed with 102 CFU bacteria, the method could stably detect cdtB, and the bacteria content in feces was 5.0 × 102 CFU/g (Table 1). Therefore, the LDL of simulated fecal samples for the three genes was 5 × 100, 5 × 103, and 5 × 102 colony-forming units (CFU)/g, respectively.
The real-time PCR method detects toxigenic C. difficile in human fecal samples
A total of 74 fecal samples, including 57 samples with toxigenic C. difficile and 17 samples with non-toxigenic C. difficile, all confirmed by TC, were randomly selected to evaluate our method (Table 2). Using our method, we detected 56 toxin-positive samples, including one false-positive, and 18 toxin-negative samples, including two false-negative (Table 2). Therefore, the true-positive and true-negative values were 96.49% (55/57) and 94.12% (16/17), respectively.
Evaluation of the detection of the tcdA gene
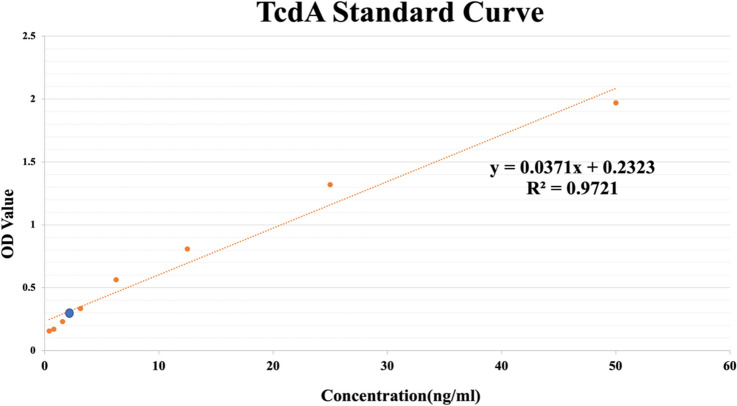
The developed method demonstrated high consistency (98.4%) with two sets of standard PCR primers reported previously for detecting tcdA in 63 C. difficile isolates, and only one strain (N20) yielded different results (Table S3). Therefore, the gold standard ELISA method for detecting toxin A in isolate N20 was used for further confirmation, from which the standard curve shows that the concentration of TcdA was linear at eight concentrations (0.4 ng/ml, 0.8 ng/ml, 1.6 ng/ml, 3.2 ng/ml, 6.4 ng/ml, 12.8 ng/ml, 25 ng/ml, and 50 ng/ml). The ratio of the absorbance between N20 and the negative control, P(0.363)/N (0.15), was 2.42, confirming that isolate N20 is a TcdA-positive strain, with a corresponding toxin A concentration was 3.53 ng/ml (Fig. 4). The ELISA results support this study’s detection results for the tcdA gene.
Fig. 4.
The toxin A in N20 is determined by ELISA. The concentration of toxin A in isolate N20 is indicated by a blue circle
Discussion
Toxigenic culture and CCNA (cell cytotoxicity assay) are considered the gold standards for CDI detection (Crobach et al. 2009). However, the CCNA procedure is complicated, time-consuming, and limited by cell types and equipment (Buss et al. 2015). Nowadays, immunological techniques are used as the preliminary screening method for CDI, but the sensitivity and specificity can be unstable and the results must be interpreted together with those of PCR approaches (Guery et al. 2019). In recent years, NAAT has been recommended for auxiliary diagnosis of CDI cases due to its high sensitivity, specificity, and time-saving. To date, at least 15 kinds of C. difficile nucleic acid detection methods have been approved by the United States Food and Drug Administration (FDA), of which 12 methods rely on specific instruments, 6 methods only detect the tcdB gene, and no methods detect tcdA, tcdB, and cdtB simultaneously (https://www.fda.gov/medical-devices/vitro-diagnostics/nucleic-acid-based-tests).
Our novel method can simultaneously detect tcdA, tcdB, and cdtB genes, which improves the accurate identification of toxigenic C. difficile isolates directly from fecal samples. In four previously reported qPCR methods, primers for tcdA were not located in the deletion region (Fig. 1a) (Kilic et al. 2015; Hoegh et al. 2012; Avbersek et al. 2011), which might cause the inaccurate identification of tcdA-negative isolates including RT017/ST37. In the present study, primers for tcdA were situated in the deletion region (7692–7965; Fig. 1a), and results with these primers were highly consistent (98.4%) with those of standard PCR primers reported previously for detecting tcdA in 63 C. difficile isolates, except for only one strain (N20) showing different results (Lemee et al. 2004; Kato et al. 1999) (Table S3). ELISA of the inconsistent N20 strain confirmed the results for our method (Fig. 4).
According to previously reported qPCR methods for detecting toxigenic C. difficile isolates, two pairs of tcdA primers from Luna and Kubota are located in the missing region, which were selected and compared with our method. The LDLs for these three qPCR methods were compared using simulated fecal samples. The LDL for tcdA in fecal samples was 2.5 × 102 CFU/ml according to Luna et al., while the LDL reported by Kubota et al. was 103 cells/g (Luna et al. 2011; Kubota et al. 2014). The LDLs for the methods of Kubota and Luna were 5.0 × 100 CFU/g and 5.0 × 101 CFU/g, respectively (Table 3), and our method yielded an LDL comparable with that of the Kubota team (Kubota et al. 2014).
In conclusion, we established and validated a real-time PCR method for the detection of toxigenic C. difficile simultaneously targeting the tcdA, tcdB, and cdtB genes. The method displayed good performance for the specificity, sensitivity, and repeatability which has been submitted for a Chinese invention patent (CN202010825309.X). Despite the great advantages mentioned above there are some limitations: the reaction parameters and system should be further optimized to improve the overall sensitivity of the method and to optimize the amplification efficiency, in addition, the sample size should also be increased to obtain more accurate evaluation data.
It is worth mentioning that the method improved the identification of the A-B + straind, including typical Asian strain RT017. It could also be combined with the method in Chinese invention patent CN202010821402.3 for early warning testing of the highly toxigenic strain BI/NAPI/027, and be applicable to CDI epidemiological surveillance and potential outbreak control. In addition, our method is applicable to a variety of fluorescence quantitative PCR instruments, and can also be combined with POCT for rapid detection and identification of clinical CDI cases.
Conflict of interest
The authors declare that they have no competing interests.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contributions
LU Jin xing, WU Yuan conceived and designed the assays. JIA Xiao xi, WANG Yuan yuan, ZHANG Wen zhu, LI Wen ge, Bai Lulu conducted experimental work. JIA Xiao xi, was in charge of draft the manuscript. WU Yuan and MA Chao feng revised and approved the final version of the manuscript. All authors read and approved the final manuscript.
Funding
This work were supported by the National Key Research and Development Program of China (2021YFC2301000) and the National Sci-Tech Key Project (grant No. 2018ZX10733402).
References
- Avbersek J, Cotman M, Ocepek M. Detection of Clostridium difficile in animals: comparison of real-time PCR assays with the culture method. J Med Microbiol. 2011;60:1119–1125. doi: 10.1099/jmm.0.030304-0. [DOI] [PubMed] [Google Scholar]
- Bacci S, Mølbak K, Kjeldsen M, Olsen K. Binary toxin and death after Clostridium difficile infection. Emerg Infect Dis. 2011;17(6):976–982. doi: 10.3201/eid/1706.101483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdasarian N, Rao K, Malani P. Diagnosis and treatment of Clostridium difficile in adults: a systematic review. JAMA. 2015;313(4):398–408. doi: 10.1001/jama.2014.17103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bélanger S, Boissinot M, Clairoux N, Picard F, Bergeron M. Rapid detection of Clostridium difficile in feces by real-time PCR. J Clin Microbiol. 2003;41(2):730–734. doi: 10.1128/jcm.41.2.730-734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss S, Leber A, Chapin K, Fey P, Bankowski M, Jones M, Rogatcheva M, Kanack K, Bourzac K. Multicenter evaluation of the BioFire FilmArray gastrointestinal panel for etiologic diagnosis of infectious gastroenteritis. J Clin Microbiol. 2015;53(3):915–925. doi: 10.1128/jcm.02674-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crobach M, Dekkers O, Wilcox M, Kuijper E. European Society of Clinical Microbiology and Infectious Diseases (ESCMID): data review and recommendations for diagnosing Clostridium difficile-infection (CDI) Clin Microbiol Infect. 2009;15(12):1053–1066. doi: 10.1111/j.1469-0691.2009.03098.x. [DOI] [PubMed] [Google Scholar]
- Czepiel J, Dróżdż M, Pituch H, Kuijper E, Perucki W, Mielimonka A, Goldman S, Wultańska D, Garlicki A, Biesiada G. Clostridium difficile infection: review. Eur J Clin Microbiol Infect Dis. 2019;38(7):1211–1221. doi: 10.1007/s10096-019-03539-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drudy D, Harnedy N, Fanning S, O'Mahony R, Kyne L. Isolation and characterisation of toxin A-negative, toxin B-positive Clostridium difficile in Dublin, Ireland. Clin Microbiol Infect Dis. 2007;13(3):298–304. doi: 10.1111/j.1469-0691.2006.01634.x. [DOI] [PubMed] [Google Scholar]
- Gerding D, Johnson S, Rupnik M, Aktories K. Clostridium difficile binary toxin CDT: mechanism, epidemiology, and potential clinical importance. Gut Microbes. 2014;5(1):15–27. doi: 10.4161/gmic.26854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guery B, Galperine T, Barbut F. Clostridioides difficile: diagnosis and treatments. BMJ (clinical Research Ed) 2019;366:l4609. doi: 10.1136/bmj.l4609. [DOI] [PubMed] [Google Scholar]
- Hoegh A, Nielsen J, Lester A, Friis-Møller A, Schønning K. A multiplex, internally controlled real-time PCR assay for detection of toxigenic Clostridium difficile and identification of hypervirulent strain 027/ST-1. Eur J Clin Microbiol Infect Dis. 2012;31(6):1073–1079. doi: 10.1007/s10096-011-1409-5. [DOI] [PubMed] [Google Scholar]
- Johnson S, Lavergne V, Skinner A, Gonzales-Luna A, Garey K, Kelly C, Wilcox M. Clinical Practice Guideline by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA): 2021 Focused Update Guidelines on Management of Clostridioides difficile Infection in adults. Clin Infect Dis. 2021;73(5):755–757. doi: 10.1093/cid/ciab718. [DOI] [PubMed] [Google Scholar]
- Kato H, Kato N, Katow S, Maegawa T, Nakamura S, Lyerly D. Deletions in the repeating sequences of the toxin A gene of toxin A-negative, toxin B-positive Clostridium difficile strains. FEMS Microbiol Lett. 1999;175(2):197–203. doi: 10.1111/j.1574-6968.1999.tb13620.x. [DOI] [PubMed] [Google Scholar]
- Kilic A, Alam M, Tisdel N, Shah D, Yapar M, Lasco T, Garey K. Multiplex real-time PCR method for simultaneous identification and toxigenic type characterization of clostridium difficile from stool samples. Annal Lab Med. 2015;35(3):306–313. doi: 10.3343/alm.2015.35.3.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H, Sakai T, Gawad A, Makino H, Akiyama T, Ishikawa E, Oishi K. Development of TaqMan-based quantitative PCR for sensitive and selective detection of toxigenic Clostridium difficile in human stools. PLoS ONE. 2014;9(10):e111684. doi: 10.1371/journal.pone.0111684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Plechot K, Gohil S, Le J. Clostridium difficile: diagnosis and the consequence of over diagnosis. Infect Dis Ther. 2021;10(2):687–697. doi: 10.1007/s40121-021-00417-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffler DA, Lamont JT. Clostridium difficile infection. N Engl J Med. 2015;372(16):1539–1548. doi: 10.1056/NEJMra1403772. [DOI] [PubMed] [Google Scholar]
- Lemee L, Dhalluin A, Testelin S, Mattrat M, Maillard K, Lemeland J, Pons J. Multiplex PCR targeting tpi (triose phosphate isomerase), tcdA (Toxin A), and tcdB (Toxin B) genes for toxigenic culture of Clostridium difficile. J Clin Microbiol. 2004;42(12):5710–5714. doi: 10.1128/jcm.42.12.5710-5714.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna R, Boyanton B, Mehta S, Courtney E, Webb C, Revell P, Versalovic J. Rapid stool-based diagnosis of Clostridium difficile infection by real-time PCR in a children's hospital. J Clin Microbiol. 2011;49(3):851–857. doi: 10.1128/jcm.01983-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, Ray SM, Thompson DL, Wilson LE, Fridkin SK, Healthcare-Associated EIP, I, Antimicrobial Use Prevalence Survey T, Multistate point-prevalence survey of health care-associated infections. N Engl J Med. 2014;370(13):1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald L, Gerding D, Johnson S, Bakken J, Carroll K, Coffin S, Dubberke E, Garey K, Gould C, Kelly C, Loo V, Shaklee Sammons J, Sandora T, Wilcox M. Clinical practice guidelines for clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA) Clin Infect Dis. 2018;66(7):e1–e48. doi: 10.1093/cid/cix1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollard S, Lurienne L, Heimann SM, Bandinelli PA. Burden of Clostridium (Clostridioides) difficile infection during inpatient stays in the USA between 2012 and 2016. J Hosp Infect. 2019;102(2):135–140. doi: 10.1016/j.jhin.2019.01.020. [DOI] [PubMed] [Google Scholar]
- Nagy E. What do we know about the diagnostics, treatment and epidemiology of Clostridioides (Clostridium) difficile infection in Europe? J Infect Chemother. 2018;24(3):164–170. doi: 10.1016/j.jiac.2017.12.003. [DOI] [PubMed] [Google Scholar]
- Rupnik M, Kato N, Grabnar M, Kato H. New types of toxin A-negative, toxin B-positive strains among Clostridium difficile isolates from Asia. J Clin Microbiol. 2003;41(3):1118–1125. doi: 10.1128/jcm.41.3.1118-1125.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart D, Berg A, Hegarty J. Predicting recurrence of C. difficile colitis using bacterial virulence factors: binary toxin is the key. J Gastrointest Surg. 2013;17(1):118–124. doi: 10.1007/s11605-012-2056-6. [DOI] [PubMed] [Google Scholar]
- Wu Y, Liu C, Li W, Xu J, Zhang W, Dai Y, Lu J (2019) Independent microevolution mediated by mobile genetic elements of individual clostridium difficile isolates from clade 4 revealed by whole-genome sequencing. mSystems 4(2): e00252–18. doi:10.1128/mSystems.00252-18 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.