Abstract
In the course of characterizing a locus involved in heme utilization, we identified a Legionella pneumophila gene predicted to encode a protein with homology to the product of the Salmonella enterica serovar Typhimurium pagP gene. In Salmonella, pagP increases resistance to the bactericidal effects of cationic antimicrobial peptides (CAMPs). Mutants with insertions in the L. pneumophila pagP-like gene were generated and showed decreased resistance to different structural classes of CAMPs compared to the wild type; hence, this gene was designated rcp for resistance to cationic antimicrobial peptides. Furthermore, Legionella CAMP resistance was induced by growth in low-magnesium medium. To determine whether rcp had any role in intracellular survival, mutants were tested in the two most relevant host cells for Legionnaires' disease, i.e., amoebae and macrophages. These mutants exhibited a 1,000-fold-decreased recovery during a Hartmannella vermiformis coculture. Complementation of the infectivity defect could be achieved by introduction of a plasmid containing the intact rcp gene. Mutations in rcp consistently reduced both the numbers of bacteria recovered during intracellular infection and their cytopathic capacity for U937 macrophages. The rcp mutant was also more defective for lung colonization of A/J mice. Growth of rcp mutants in buffered yeast extract broth was identical to that of the wild type, indicating that the observed differences in numbers of bacteria recovered from host cells were not due to a generalized growth defect. However, in low-Mg2+ medium, the rcp mutant was impaired in stationary-phase survival. This is the first demonstration of a pagP-like gene, involved in resistance to CAMPs, being required for intracellular infection and virulence.
Legionella pneumophila is a facultative intracellular parasite of human monocytes, macrophages, and protozoa (48, 63, 69). In the natural environment, Legionella species occupy the niches of soil and water, either free-living, in biofilms, or, more commonly, residing within amoebal hosts (29, 69). Problems occur when legionellae enter and replicate in human-made water systems, from which inhalation of bacterium-ladened aerosols leads to the establishment of a pneumonia referred to as Legionnaires' disease (28).
Pathogens, such as L. pneumophila, that adopt an intracellular lifestyle must possess multiple strategies to overcome or evade the defense system of the host cell. For example, mammalian macrophages and amoebae are well equipped to bring about the demise of engulfed bacteria via the action of both oxygen-independent and -dependent mechanisms (31, 40, 53). Oxygen-independent mechanisms include cationic antimicrobial peptides (CAMPs), a group of structurally diverse polypeptides that are thought to kill by membrane damage due to pore formation (41, 53). L. pneumophila overcomes much of the killing mechanisms of the host cell by initially residing in a phagosome with limited ability to fuse with lysosomes, the source of the majority of antibacterial substances within the phagocyte (13, 47, 57, 78, 79). However, several lines of evidence suggest that L. pneumophila is exposed to lysosomal factors and likely possesses resistance factors against at least some of them. For example, some L. pneumophila mutants, which are readily delivered to a macrophage phagolysosome, do survive, albeit with little or no replication (9, 46, 56). Also, recent evidence suggests that phagolysosomal fusion does occur late in the intracellular infection cycle (75). Furthermore, extracellular L. pneumophila is inherently resistant to polymyxin B (PmB), a CAMP that is actually used for selection of Legionella spp. from clinical and environmental samples (23).
Although catalase/peroxidase and superoxide dismutase enzymes are known to promote L. pneumophila resistance to oxygen-dependent killing (2, 4, 74), the Legionella factors involved in CAMP resistance are completely unknown. Indeed, knowledge of bacterial CAMP resistance determinants is only beginning to emerge (7, 26, 67). The majority of the literature on CAMP resistance is from Salmonella enterica serovar Typhimurium, where several determinants have been identified, such as pagP (37–39, 81). This gene is transcriptionally activated by PhoPQ (hence the pagP gene designation as PhoP-activated gene P), a two-component regulator of genes involved in both enterocyte invasion and survival within macrophages (5, 39, 62). However, the importance of PagP in intracellular infections is not known (6).
Previous efforts by our group to isolate determinants important for iron acquisition and utilization in L. pneumophila led to the identification of a gene responsible for hemin binding, hbp (65). Here, we demonstrate that in the hbp region of the L. pneumophila chromosome there is an open reading frame whose product has homology with PagP. The aims of our further investigations were to identify whether the pagP-like gene in L. pneumophila has a role in CAMP resistance and, more importantly, whether it has any relevance for intracellular infection and virulence.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains of L. pneumophila and other Legionella spp. used during these studies are listed in Table 1. L. pneumophila strain 130b (Wadsworth) was used for mutagenesis of the Legionella pagP-like gene and subsequently served as the wild-type control. A pBR322 plasmid library of 3- to 6-kb Sau3AI-restricted 130b genomic DNA was maintained in Escherichia coli K-12 strain HB101 (44). In the CAMP susceptibility assay, Pseudomonas aeruginosa strain PAK was used as a sensitive control (26). L. pneumophila strains were routinely grown on standard buffered charcoal-yeast extract (BCYE) agar for 3 days at 37°C (22). For selection of allelic exchange mutations, BCYE agar was supplemented with 5% (wt/vol) sucrose (54). For growth curves, L. pneumophila was grown either in buffered yeast extract broth or in chemically defined liquid medium (CDM) at 37°C with shaking at 250 rpm (Lab-Line Instruments model 3525) (54). The components of CDM have been described elsewhere (68); briefly, the base consisted of 3-(N-morpholino)propanesulfonic acid buffer, NaCl and KH2PO4, which was supplemented with reduced glutathione and a variety of amino acids and trace metals, including 0.7 mM MgSO4. For CDM cultures, the starting inocula were from buffered yeast extract (BYE) cultures in the logarithmic phase of growth, i.e., with an optical density at 660 nm (OD660) of 0.9 to 1.2. Bacteria were washed three times in CDM base before resuspension to an OD660 of 0.15 in CDM containing various amounts of Mg2+. Luria-Bertani broth or agar was used for growth of E. coli and P. aeruginosa (70). Where appropriate, the medium was supplemented with the following antibiotics at final concentrations suitable for L. pneumophila (or E. coli): kanamycin (KAN), 25 μg/ml (50 μg/ml); and chloramphenicol (CHL), 3 or 6 μg/ml (30 μg/ml).
TABLE 1.
Legionella strains
| Species | Strain | Serogroup | Source (reference) |
|---|---|---|---|
| L. pneumophila | ATCCa BAA-74 (130b) | 1 | Clinical (25) |
| ATCC 33154 | 2 | Clinical (59) | |
| ATCC 33155 | 3 | Clinical (59) | |
| ATCC 33156 | 4 | Clinical (59) | |
| ATCC 33216 | 5 | Clinical (35) | |
| ATCC 33215 | 6 | Clinical (60) | |
| ATCC 33823 | 7 | Clinical (10) | |
| ATCC 35096 | 8 | Clinical (12) | |
| ATCC 43736 | 13 | Clinical (55) | |
| ATCC 43703 | 14 | Clinical (8) | |
| L. birminghamensis | ATCC 43702 | 1 | Clinical (80) |
| L. erythra | ATCC 35303 | 1 | Environmental (14) |
| L. feeleii | ATCC 35072 | 2 | Clinical (77) |
| L. longbeachae | ATCC 33462 | 1 | Clinical (58) |
| L. micdadei | Stanford-R | 1 | Clinical (64) |
| L. parisiensis | ATCC 35299 | 1 | Clinical (14) |
ATCC, American Type Culture Collection.
DNA sequence analysis.
Double-stranded sequence was obtained from pEH12, a member of the pBR322 library containing hbp and surrounding genes, by the dideoxy chain termination method with 35S-dATP and Sequenase (Amersham Life Sciences, Arlington Heights, Ill.) (65). Sequencing reactions were performed according to the manufacturer's recommendations using pUC-based subclones of pEH12, with vector-based primers. Nucleotide sequences were analyzed with PCGENE (IntelliGenetics), and homology searches were conducted through GenBank at the National Center for Biotechnology Information.
Mutation and complementation of an L. pneumophila pagP-like gene.
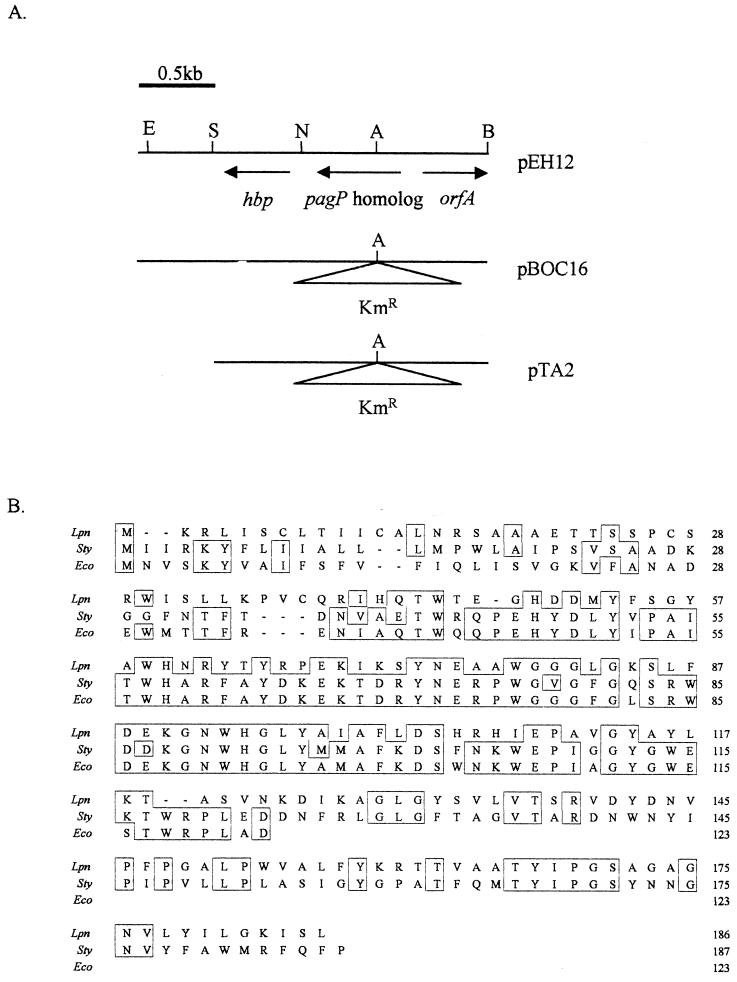
A mutation in the cloned pagP-like gene of strain 130b was generated by insertion of a 1.1-kb KAN resistance cassette at the AflII site of pEH12 (Fig. 1A), resulting in plasmid pBOC16. A 2.9-kb BamHI-SacI fragment from pBOC16 was cloned into pBOC20, resulting in pTA2 (Fig. 1A). Plasmid pBOC20 is based on a ColE1 replicon and facilitates allelic exchange in L. pneumophila by virtue of its sacB gene (54). Production of competent 130b cells and electroporation of pTA2 into L. pneumophila were achieved as previously described (54). Potential mutants were selected based on CHL sensitivity and KAN and sucrose resistance, which are indicative of the introduction of the mutated gene into the 130b chromosome by homologous recombination. Verification of mutant genotypes was carried out by PCR and Southern hybridization (3, 54, 65, 70). A DNA fragment specific to the pagP-like gene encoded within 0.9 kb was generated by PCR using primers PAGP-F (5′-TGA TTC ATT GTC TGG CGA CC-3′) and PAGP-R (5′-GCC AAG ATT ACA GCA CCG AT-3′). Primers were generated by the Northwestern University Biotechnology Center using an Applied Biosystems DNA synthesizer. Genomic DNA was isolated from L. pneumophila as described previously (24). For complementation analysis, the PCR product amplified using PAGP-F and PAGP-R was cloned into pGEMTEasy (Promega, Madison, Wis.) and then subcloned on a 0.9-kb NotI fragment into the low-copy-number CHL resistant pSU2719 (18), resulting in pS14. Another complementing plasmid, pS25, was constructed by cloning the 1.8-kb BamHI-SacI fragment containing the pagP-like gene from pEH12 into pSU2719.
FIG. 1.
Sequence analysis of the L. pneumophila pagP-like gene and PagP-like protein. (A) Sequence organization and restriction analysis of pEH12, pBOC16, and pTA2. For all plasmids, only the L. pneumophila strain 130b DNA fragment is depicted. Restriction sites are as follows: A, AflII; B, BamHI; E, EcoRI; N, NdeI; and S, SacI. (B) Alignment (boxed regions) of PagP-like proteins in L. pneumophila (Lpn), Salmonella serovar Typhimurium (Sty) (GenBank accession number AF057021), and E. coli (Eco) (GenBank accession number AE000167). The Clustal method was used to generate alignment data (45).
CAMP microdilution susceptibility assay.
C18G, a synthetic α-helical peptide based on the carboxy terminus of human platelet factor IV (21), and PmB (7,770 U/mg; U.S. Biochemicals; Swampscott, Mass.) were used to check the resistance of 130b and its mutants to CAMPs. To assess this resistance, a standard microdilution susceptibility assay, which uses minimal amounts of each CAMP, was performed (21, 36, 53, 73, 81). Bacteria grown in BYE, CDM, or CDM containing various amounts of Mg2+ were diluted to 5 × 104 CFU/ml in 2× BYE. In triplicate wells of a 96-well microtiter tray, 50 μl of bacterial suspension was added to an equal volume of various concentrations of C18G or PmB (diluted in sterile distilled water from a fresh stock solution). Thus, the incubation medium throughout the CAMP resistance assays was BYE to prevent any observed differences in MIC being attributable to differences in growth potential. Following incubation at 37°C for 16 h (for P. aeruginosa PAK) or for 30 h (for L. pneumophila strains), the lowest concentration of C18G or PmB resulting in no visible growth was deemed to be the MIC (36). From wells where no growth was evident, four 10-μl samples were spotted onto BCYE agar (for L. pneumophila) or Luria-Bertani agar (for PAK) to assess the minimum bactericidal concentration (MBC) (36). To facilitate maximum availability of the peptides, all stages of the assay were performed using polypropylene plasticware, as the negatively charged surface of polystyrene plasticware has been shown to bind CAMPs (36).
Intracellular infection of macrophages and amoebae by L. pneumophila.
Intracellular infection by L. pneumophila strains was assessed in human U937 cells (1, 54, 65, 66, 76) differentiated into macrophage-like cells by treatment with phorbol myristate acetate and in Hartmannella vermiformis amoebae (17, 19, 29, 54, 71). Assessment of the intracellular growth kinetics of L. pneumophila in U937 macrophages was performed as previously reported (54). Briefly, U937 cells were routinely cultivated in RPMI 1640 medium with l-glutamine (Gibco Life Technologies, Rockville, Md.) supplemented with 10% heat-inactivated fetal calf serum (HyClone, Logan, Utah) and 2.5 ml of amphotericin B (Fungizone; Gibco Life Technologies) in a 5% CO2 incubator at 37°C. Using a multiplicity of infection (MOI) of 1, 106 adherent U937 macrophages were infected with bacteria grown for 3 days on BCYE agar, as standardly performed (1, 3, 21, 56). After a 2-h incubation period to allow bacterial internalization, extracellular bacteria were removed by repeated washing, and then infected monolayers were incubated at 37°C in a 5% CO2 incubator. At various times postinoculation, intracellular bacteria from triplicate wells per strain were released by lysis of the monolayer with 10 μl of 10% saponin (Sigma Chemical Co., St Louis, Mo.). For estimation of viable counts, serial 10-fold dilutions from triplicate wells for each strain were plated on BYCE agar. Initially, cocultures with H. vermiformis and L. pneumophila were performed as described previously (19). Briefly, in 24-well tissue culture trays, 105 H. vermiformis trophozoites were infected with 103 bacteria grown for 3 days on BCYE agar. At 24-h intervals, supernatant samples were taken from triplicate wells and viable counts of extracellular bacteria were estimated. Total numbers of bacteria were also determined in infected H. vermiformis monolayers by lysing amoebae with 10 μl of 10% saponin followed by vigorous pipetting at various time points and assessing viability on BCYE agar. L. pneumophila strains are unable to grow in the medium in which infections of macrophages or H. vermiformis were conducted; thus, increases in bacterial numbers reflect growth within the host cells (19, 66).
Macrophage cytopathicity assay.
To determine whether the numbers of bacteria recovered from U937 cells were truly reflective of a diminished infection process, the viability of the macrophages was assessed by the ability of monolayers to reduce the dye alamarBlue (Biosource International, Camarillo, Calif.). Cytopathicity assays were performed as described previously (1, 3, 32, 50). Briefly, 105 U937 macrophages were infected, as described above, at various MOIs. After 2 h, the infected monolayers were washed twice with RPMI to remove extracellular bacteria and then incubated at 37°C. At various time points, alamarBlue was added (1/11 volume in RPMI 1640 medium) to infected monolayers that had been previously washed twice, the monolayers were incubated at 37°C for 3 h, and fluorescence (at an excitation wavelength of 540 nm and an emission wavelength of 584) was assessed.
Pulmonary infection of A/J mice by L. pneumophila.
The A/J mouse model mimics the acute L. pneumophila-induced pneumonia in humans (1, 15, 16, 20). The role of rcp in vivo was assessed using standard competition assays (30, 52, 61, 82). Six- to 8-week-old female mice (Jackson Laboratories, Bar Harbor, Maine) were anesthetized and then inoculated by intratracheal injection with a 25-μl bacterial suspension in phosphate-buffered saline. Inocula consisted of 106 CFU of a ca. 1:1 ratio of 130b and NU260, which had been previously grown for 3 days on BCYE agar. At various time points, infected mice were sacrificed. Lungs were collected and disrupted by extrusion of the tissue through a fine-mesh grid into 10 ml of phosphate-buffered saline. Cells were lysed by incubation with 100 μl of 10% saponin for 15 min at 37°C followed by vigorous pipetting. The numbers of viable bacteria and the ratio of 130b to NU260 were estimated by plating 10-fold serial dilutions on both plain and KAN-supplemented BCYE agar.
Detection of pagP-like sequences in other Legionella spp. and SG.
Southern hybridization was carried out using BamHI-restricted genomic DNAs from strains representing several L. pneumophila serogroups (SG) and a variety of Legionella spp. The digoxigenin nonradioactive labeling and detection system (Roche Molecular Biochemicals, Indianapolis, Ind.) was used. The probe was produced by PCR incorporation according to the manufacturer's recommendations using primers PAGP-F and PAGP-R and 130b genomic DNA as a template. High-stringency washes (0 to 10% base pair mismatch) were employed for hybridization of the probe to L. pneumophila DNAs (two washes with 2×SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]–0.1% sodium dodecyl sulfate [SDS] at room temperature for 15 min and two washes with 0.1× SSC–0.1% SDS at 68°C for 15 min). Low-stringency washes (∼30% base pair mismatch) were employed for hybridization of the probe to genomic DNAs from other Legionella spp. (two washes with 5× SSC–0.1% SDS at 50°C for 15 min).
Nucleotide sequence accession number.
The GenBank accession number for the L. pneumophila pagP-like gene is AF348322.
RESULTS
Identification and mutation of an L. pneumophila pagP-like gene.
Sequence analysis of the region surrounding hbp encoded on the 130b library clone pEH12 led to the discovery of a gene whose product has 42% identity and 57% similarity to PagP in Salmonella serovar Typhimurium (Fig. 1). The products of the Salmonella pagP gene and the homologous gene in L. pneumophila are both predicted to consist of 186 amino acid residues, providing substantiating evidence that these genes are related (Fig. 1B). The L. pneumophila pagP-like gene product also shows homology, i.e., 43% identity and 57% similarity, to the crcA (camphor resistance A) gene in E. coli K-12 (Fig. 1B). Initially, crcA was characterized as conferring camphor resistance when encoded on a high-copy-number plasmid in combination with cspE and crcB (49). However, recent evidence suggests that the product of crcA is functionally homologous to the Salmonella PagP, prompting an alternative designation from CrcA to the E. coli PagP (11). PagP and CrcA are the only proteins that show strong homology to the product of the pagP-like gene in L. pneumophila (data not shown). Upstream from the 559-bp pagP-like gene on pEH12 is an incomplete open reading frame, orfA, which is transcribed divergently from the pagP-like gene (Fig. 1B). Homology searches suggest that orfA encodes an amino acid permease (data not shown). The pagP-like gene and the downstream hbp are transcribed in the same orientation. Since Northern blot analysis indicates that transcription of hbp is monocistronic (65), the pagP-like gene and hbp are not thought to constitute an operon.
To be able to determine whether the L. pneumophila pagP-like gene has a role similar to that of its Salmonella counterpart, a mutation was introduced into the gene in virulent strain 130b. Mutants were isolated by allelic exchange, whereby the chromosomal copy of the pagP-like gene was replaced by a mutant form containing a KAN insertion cassette at its AflII restriction site, 100 bp from its putative translational start site (Fig. 1A). Two independent mutants whose CHL sensitive and KAN- and sucrose-resistant phenotypes were indicative of homologous recombination of the mutant gene into the chromosome were isolated from separate electroporation events. The mutants, designated NU260 and NU261, were verified by PCR and Southern hybridization to contain the 1.1-kb insertion in the pagP-like gene, using a probe specific to the pagP-like gene (data not shown). All further experiments described here were performed with both NU260 and NU261 with similar results, although, for clarity, only data from NU260 are presented in this paper. Thus, the phenotypes observed result directly or proximately from the mutation in the pagP-like gene and not from spontaneous second-site mutations. However, this does not rule out the possibility of the additional involvement of unlinked genes.
Extracellular growth of the L. pneumophila pagP-like mutant.
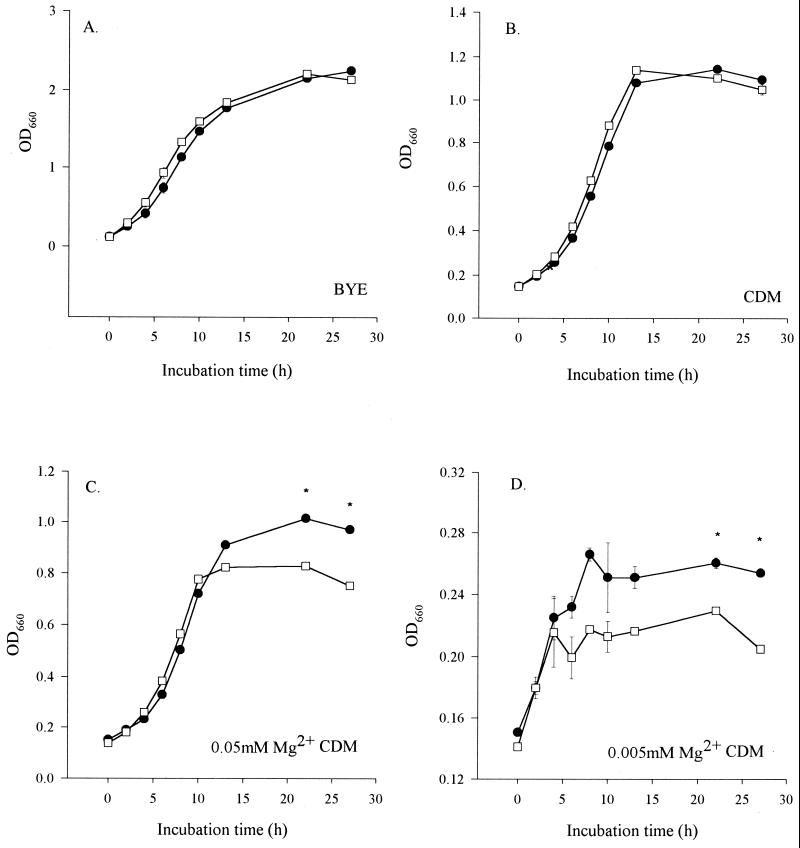
The behavior of NU260 in liquid media was assessed to determine if loss of the pagP-like gene resulted in gross changes in extracellular growth. No differences were observed in the growth kinetics of the mutant compared to 130b in standard BYE and CDM broths (Fig. 2A and B). In Salmonella, the PhoPQ regulon senses Mg2+ cations, and bacterial growth in low-Mg2+ minimal medium increases the CAMP resistance generated by pagP (39, 72). Thus, we hypothesized that the L. pneumophila mutants might have reduced survival in low-Mg2+ broth. The growth maxima of wild type 130b were considerably reduced in standard CDM compared to BYE and decreased as the concentration of Mg2+ in CDM was lowered from 0.7 to 0.005 mM (Fig. 2), which would be predicted from the known metal requirements of L. pneumophila (68). No growth was observed in CDM lacking added Mg2+ (data not shown). More importantly, as the Mg2+ concentration of CDM was decreased, NU260 entered into stationary phase earlier than 130b, with an overall reduction in the growth maxima of the mutant, although the growth rate was unaffected (Fig. 2C and D and data not shown). Furthermore, bacterial viability, as judged by CFU/per milliliter normalized to OD660, was decreased 20-fold in stationary-phase NU260 grown in 0.005 mM Mg2+–CDM medium compared to 130b (data not shown). Thus, the L. pneumophila pagP-like gene appears to be involved in growth and/or survival in low-Mg2+ medium.
FIG. 2.
Extracellular growth kinetics of L. pneumophila strains. Wild-type strain 130b (●) and mutant NU260 (□) were compared for growth in BYE (A), CDM (0.7 mM Mg2+) (B), and CDM with 0.05 mM (C), or 0.005 mM (D) Mg2+ cations. Growth was assessed by measuring the OD660 of triplicate cultures at various times of incubation. Asterisks indicate significant differences (P < 0.05; Student's t test) between the 130b and NU260 cultures. Standard deviations are shown, but the error bars are too small to be observed in panels A to C. This experiment was performed in triplicate with comparable results, and representative data are presented here.
Resistance of L. pneumophila strains to CAMPs.
Since PagP of Salmonella serovar Typhimurium and E. coli are involved in resistance to CAMPs, we sought to assess the relative resistance of NU260 to two CAMPs, the cyclical PmB and the α-helical C18G (Table 2). Initially, experiments determined that CAMP resistance was similar in both logarithmic and stationary phase (data not shown). Subsequent investigations were performed with logarithmic-phase bacteria to ensure maximum viability of the culture. After growth of 130b in BYE, the MIC and MBC of PmB were 12.5 and 25 to 50 μg/ml, respectively (Table 2). The MIC of PmB for P. aeruginosa PAK was ≤3.1 μg/ml, a fourfold difference compared to that for 130b, in accordance with Legionella spp. being especially PmB resistant. The MIC and MBC of PmB for NU260 were 6.2 and 25 μg/ml, respectively (Table 2), suggesting a role for the pagP-like gene in L. pneumophila resistance to CAMPs. The MIC for C18G of PAK was 32 to 64 μg/ml following growth in BYE. However, L. pneumophila strain 130b appeared to be much more sensitive to the effects of this CAMP, with the C18G MIC and MBC being 16 and 32 μg/ml, respectively. The MIC and MBC of C18G for NU260 were 8 and 16 to 32 μg/ml, respectively. The modest difference in both PmB and C18G resistance between 130b and NU260 was highly reproducible in independent experiments (Table 2). Taken together with the significant degree of sequence homology with PagP proteins in both Salmonella and E. coli, these data indicate that the L. pneumophila counterpart of PagP plays a role in CAMP resistance. Hence, we designated the L. pneumophila pagP-like gene rcp, for resistance to cationic antimicrobial peptides. We avoided the pag (PhoP-activated gene) designation because PhoP has not yet been identified in L. pneumophila and the genetic regulation of rcp is wholly uncharacterized.
TABLE 2.
CAMP susceptibilities of L. pneumophila strainsa
| CAMP | Growth medium | Strain | MIC (μg/ml) | MBC (μg/ml) |
|---|---|---|---|---|
| PmB | BYE | 130b | 12.5 | 25–50 |
| NU260 | 6.2 | 25 | ||
| CDM | 130b | 3.1 | 25 | |
| NU260 | 3.1 | 12.5–25 | ||
| 0.05 mM Mg2+–CDM | 130b | 6.2–12.5 | 25 | |
| NU260 | 6.2 | 12.5–25 | ||
| 0.005 mM Mg2+–CDM | 130b | 12.5 | 25 | |
| NU260 | 6.2 | 12.5–25 | ||
| C18G | BYE | 130b | 16 | 32 |
| NU260 | 8 | 16–32 | ||
| CDM | 130b | 16 | 32 | |
| NU260 | 4–8 | 8–16 | ||
| 0.05 mM Mg2+–CDM | 130b | 16 | 32 | |
| NU260 | 4 | 8–16 | ||
| 0.005, mM Mg2+–CDM | 130b | 64 | 128 | |
| NU260 | 16 | 16–32 |
The bacteria were grown in triplicate to mid-exponential phase in the indicated media and then, after dilution to 5 × 104 CFU/ml, were exposed to PmB or C18G. The incubation medium during all CAMP resistance assays was BYE. Similar MICs and MBCs were obtained in a replicate experiment (data not shown). A range of concentrations indicates the range of MICs of MBCs obtained from triplicate samples.
Given the reduced ability of the L. pneumophila rcp mutant to grow in low-Mg2+ medium (Fig. 2), we tested its relative resistance to C18G and PmB after growth in CDM containing various amounts of Mg2+. Growth in low-Mg2+ CDM increased the resistance of 130b toward both CAMPs (Table 2). The MIC of PmB increased from 3.1 μg/ml in standard CDM to 6.2 to 12.5 μg/ml as the Mg2+ content of CDM was lowered to 0.05 mM and finally to 12.5 μg/ml in 0.005 mM Mg2+–CDM. Using 0.005 mM Mg2+–CDM yielded the greatest CAMP resistance difference between 130b and NU260 but was also associated with poor growth, raising the possibility that complex physiological changes, other than Mg2+ depletion, could be responsible for the phenotype. However, reasonable growth was achieved using 0.05 mM Mg2+–CDM, which also resulted in increased PmB resistance in both 130b and NU260 compared to CDM (Table 2; Fig. 2C). Taken together, these results suggest that low Mg2+ does increase the PmB resistance of L. pneumophila. The MIC of C18G increased from 16 μg/ml in standard CDM to 64 μg/ml with 0.005 mM Mg2+ (Table 2). For NU260, the MIC of PmB increased from 3.1 μg/ml in standard CDM to 6.2 μg/ml in both 0.05 mM– and 0.005 mM Mg2+–CDM, and the MIC for C18G increased from between 4 and 8 μg/ml in standard CDM to 16 μg/ml in 0.005 mM–Mg2+ CDM (Table 2). Although CAMP resistance in both 130b and NU260 was modestly increased as the Mg2+ concentration in the CDM was lowered, there was a greater increase in the wild type, suggesting that rcp is partly responsible for the Mg2+ -dependent CAMP resistance. In summary, these experiments represent the first identification of an L. pneumophila gene involved in CAMP resistance and the first demonstration of increased Legionella CAMP resistance due to low-Mg2+ conditions.
Intracellular infections by L. pneumophila strains.
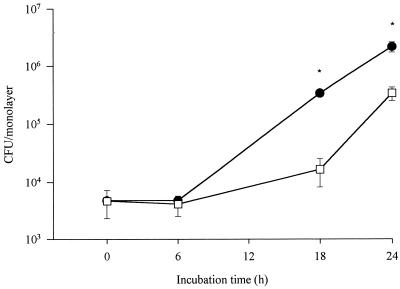
To assess the role of rcp in an intracellular environment, a phenotype not previously investigated for any pagP-like gene, U937 macrophages were infected with either 130b or NU260 and then numbers of bacteria were monitored over time. At 0 h, equivalent numbers of bacteria 130b and NU260 were recovered (Fig. 3). Thus, there was no obvious defect in the initial stages of infection, i.e., attachment and invasion. However, the mutant exhibited reduced recovery compared to the wild-type parent, by 1 log unit at 18 h postinoculation (p.i.) (Fig. 3). The difference in numbers of recovered bacteria diminished at later time points, until by 72 h similar numbers of mutant and wild-type bacteria were recovered (Fig. 3 and data not shown). No difference was noted in the abilities of NU260 and 130b to survive in the medium used to culture the macrophages (data not shown). Taken together, these data suggest that rcp promotes the ability of L. pneumophila to replicate and/or survive in macrophages. Since the PmB resistances of wild-type and mutant bacteria grown for 3 days on BCYE are comparable (data not shown), we suspect that the difference in numbers of recovered bacteria is due to intracellular induction of rcp-dependent CAMP resistance.
FIG. 3.
Intracellular infection of human macrophages with L. pneumophila strains. U937 cell monolayers (n = 3) were infected at an MOI of 1 with strain 130b (●) and NU260 (□). Asterisks indicate significant differences (P < 0.01; Student's t test) in numbers of recovered bacteria between 130b and NU260. Comparable results were obtained in triplicate experiments, and the data presented here are from one representative experiment. Error bars indicate standard deviations.
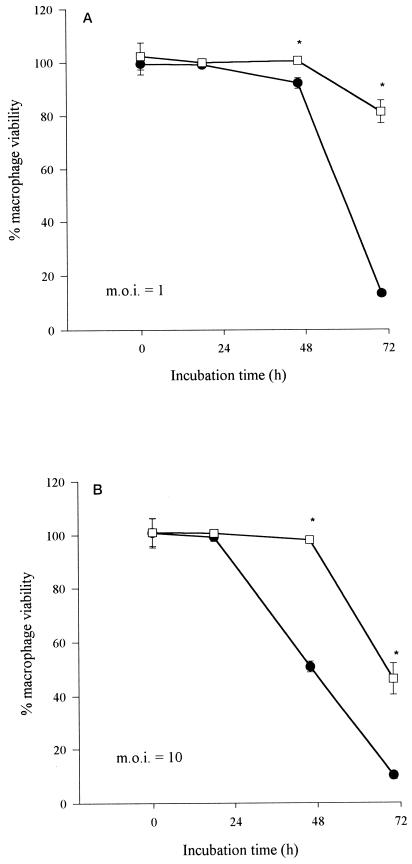
Since a low MOI with L. pneumophila ultimately leads to host cell death (1, 66), corroborating evidence of a role for rcp in macrophage infection was sought using a standard cytopathicity assay. Using the same MOI as during kinetic studies, the cytopathic effect of the wild type became apparent at 48 h p.i. and then proceeded to increase until by 72 h survival was only ∼10% compared with uninfected cells (Fig. 4A). In contrast, the viability of macrophages infected with NU260 did not decrease until 72 h p.i., when survival was still ∼80% compared to uninfected cells (Fig. 4A). A defect in cytopathicity was also observed using an MOI of 10; i.e., whereas 130b elicited 50% macrophage survival at 48 h, NU260 did so only after 72 h (Fig. 4B). Thus, rcp appears to be important for the ability of L. pneumophila to kill macrophages.
FIG. 4.
Cytopathicity assay to assess the viability of L. pneumophila-infected human macrophages. U937 cell monolayers (n = 6) were infected at MOIs of 1 (A) and 10 (B) with wild-type strain 130b (●) and mutant NU260 (□). Macrophage viability is expressed as the percentage of alamarBlue dye reduction in infected compared to uninfected monolayers. Asterisks indicate significant differences in cytopathicity (P < 0.0005; Student's t test) between 130b and NU260. Comparable results were obtained from an additional experiment (data not shown). Error bars indicate standard deviations.
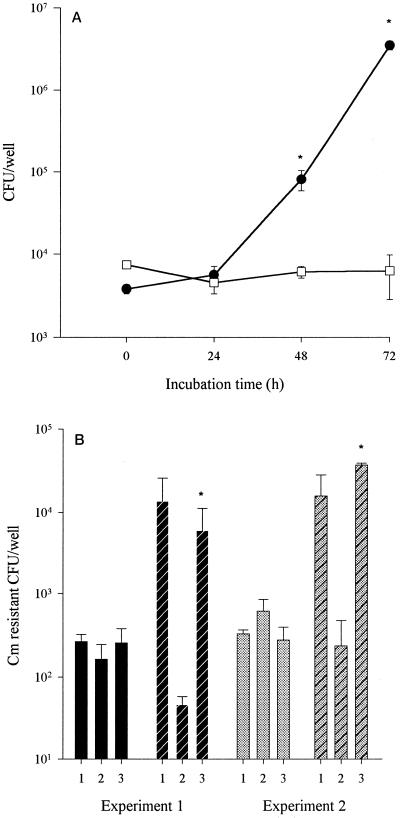
As amoebae are the natural host for L. pneumophila in the environment, intracellular growth of the mutant was tested in a coculture assay with H. vermiformis. This species of amoeba is known to support the growth of L. pneumophila and was isolated from a clinical case of legionellosis (29). Both the rcp mutant and 130b were recovered in similar numbers from the supernatants overlying the infected monolayers until 24 h p.i. (Fig. 5A). However, by 72 h p.i., the mutant exhibited a nearly 1,000-fold reduction in numbers compared with the wild type (Fig. 5A). By visual inspection, it was apparent that more cellular debris, indicative of cell lysis or necrosis, occurred in monolayers infected with 130b compared to NU260. When infected amoebal monolayers were lysed, the total numbers of bacteria were identical to that observed during coculture (data not shown and Fig. 5A). No difference in the abilities of the rcp mutant and 130b to survive in the assay media were noted (data not shown). Similar to the case with macrophages, the growth defect of the mutant during coculture with H. vermiformis could be due to a decreased ability to replicate and/or survive intracellularly.
FIG. 5.
Coculture of L. pneumophila strains with H. vermiformis. (A) Monolayers of amoebae (n = 3) were infected with wild-type strain 130b (●) and mutant NU260 (□) at an MOI of 0.01. Asterisks indicate significant differences (P < 0.05 at 48 h or P < 0.005 at 72 h; Student's t test) in numbers of recovered bacteria between 130b and NU260. The experiment was performed five times with comparable results, and representative data are presented here. (B) Genetic complementation of the rcp mutant's infectivity defect. Monolayers of amoebae were cocultured with either 130b containing the CHL resistance vector pSU2719 (bars 1), NU260 containing the vector (bars 2), or NU260 containing rcp cloned into pSU2719 (bars 3), and the numbers of recovered bacteria were determined at 0 h (solid bars) and 72h (hatched bars) p.i. In experiment 1 (black bars), the complementing plasmid was pS14. In experiment 2 (grey bars), which is representative of a replicate experiment, the complementing plasmid was pS25. Asterisks indicate significant differences (P < 0.005; Student's t test) in numbers of recovered bacteria between NU260 containing the vector and NU260 containing rcp. Error bars indicate standard deviations.
Convincing evidence exists that the mutant's infectivity defects are associated with loss of rcp function. First, the infection phenotypes of two independent rcp mutants (NU260 and NU261) were identical (Fig. 3, 4, and 5 and data not shown). Second, rcp is not part of an operon, as the upstream gene is in the opposite orientation and the downstream hbp gene is monocistronic (Fig. 1A) (65). Third, a mutant bearing an insertion in hbp is not defective for intracellular infection of macrophages or amoebae (65). Finally, trans complementation of the amoebal infection defect was achieved with plasmids containing rcp (Fig. 5B), which rules out the possibility that these phenotypes are due to the effect of unlinked genes.
In vivo virulence studies in A/J mice.
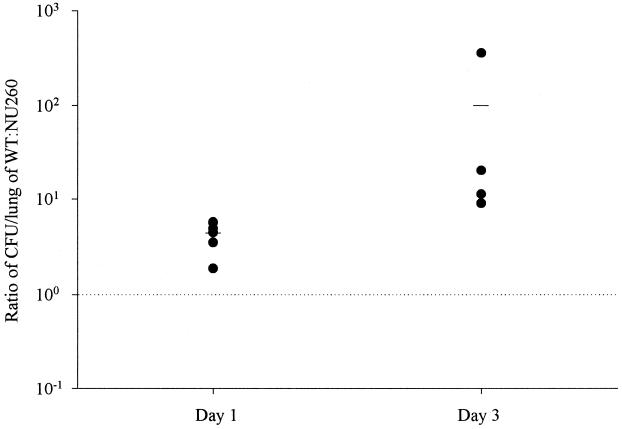
Due to the intracellular infectivity defect observed in the rcp mutant, in vivo virulence studies in A/J mice were undertaken (1, 15, 16, 20). Competition assays in which a mixture of 130b and NU260 in a ca. 1:1 ratio was inoculated by intratracheal injection into A/J mice were performed to assess the role of rcp in vivo. In competition assays, increases in the ratio of wild type to mutant over time reflect the ability of the wild type to outcompete the mutant. Following 24 h of incubation, the ratio of the wild type to the mutant recovered from infected lungs increased to 4 (Fig. 6). By 72 h, the ratio of wild type to mutant had further increased to 100 (Fig. 6). NU260 recovered from infected animals retained KAN resistance. Thus, we suspect that rcp promotes L. pneumophila virulence.
FIG. 6.
In vivo competition assays with L. pneumophila 130b and NU260. The wild type (WT) and mutant were inoculated intratracheally into A/J mice (n = 4 to 6). The ratio of wild type to mutant in infected lungs was assessed by dividing the CFU per lung for the wild type by the CFU per lung for NU260. The values expressed are normalized to the deviation observed in the ratio of 130b to NU260 in the initial inoculum (i.e., 0.61).
Detection of pagP-homologous sequences in L. pneumophila SG and other Legionella spp.
Probes designed against the rcp gene of 130b were used to check for the presence of pagP-like sequences in other L. pneumophila SG and Legionella spp. Sequences homologous to the probe were observed in all SG tested, i.e., SG 2 to 8, 13, and 14 (Table 1). Under low-stringency conditions (30% base pair mismatch), sequences homologous to the probe were observed in L. birminghamenesis, L. erythra, L. feeleii, L. longbeachae, L. micdadei, and L. parisienesis.
DISCUSSION
In this study, we have identified a new L. pneumophila infectivity determinant, Rcp, a protein with homology to PagP of Salmonella serovar Typhimurium and E. coli. Sequences homologous to the rcp gene were detected in nine SG of L. pneumophila and in six Legionella spp., suggesting distribution of rcp throughout the genus. The loss of Rcp resulted in modestly decreased Legionella resistance to C18G, a CAMP based on human platelet-derived factor IV, and PmB, a bacterially derived CAMP. This result is entirely in keeping with the established link between PagP and CAMP resistance (39). Growth in low-Mg2+ medium, a condition known to induce Salmonella CAMP resistance, promoted L. pneumophila resistance to C18G and PmB (39). Although induction of CAMP resistance by low Mg2+ concentrations was evident in both the wild type and the rcp mutant, increased induction was demonstrated for 130b, especially against PmB. These results provide substantiating evidence that Rcp is, in part, responsible for resistance to CAMPs in L. pneumophila. Although rcp in L. pneumophila and pagP in Salmonella are genetically and functionally similar, differences between them are apparent. For example, in Legionella the rcp gene is involved in resistance to both C18G and PmB, whereas pagP mediates C18G resistance but not PmB resistance (39). The highly complex composition of CAMPs encountered by the organism in vivo is undefined, and therefore, it is possible that a larger difference could be obtained using a CAMP mixture more similar to that encountered in the intracellular niche occupied by legionellae. In Salmonella and E. coli, PagP functions as a palmitoyl transferase, able to modify the lipid A component of lipopolysaccharide by addition of the fatty acid palmitate (11, 39). The increased acylation is believed to promote resistance to CAMPs by decreasing membrane fluidity and preventing insertion of the peptide (39). Lipid A modifications which increase bacterial CAMP resistance are an emerging theme in gram-negative bacteria (26, 39). For example, in P. aeruginosa, the addition of palmitate to lipid A also increases CAMP resistance in a PhoP-dependent fashion in response to low Mg2+ concentrations (26). Based upon the strong sequence and functional homology with PagP of S. enterica, we strongly suspect that L. pneumophila rcp promotes similar lipid A modifications. Preliminary data suggest that NU260 contains lipopolysaccharide molecules with lower molecular masses (approximately 250 to 300 kDa) than 130b, in accordance with the expected loss of one fatty acid, as judged by electrophoretic separation (U. Zähringer, M. Robey, and N. P. Cianciotto, unpublished results). Detailed analysis to formally prove this hypothesis is currently being pursued. Since the rcp mutant was only partly defective in CAMP resistance, we suspect that additional factors are involved in Legionella resistance to antimicrobial peptides. In Salmonella, CAMP resistance is clearly promoted by several determinants, including the PhoPQ-regulated pagP gene. For example, PmrAB is a two-component regulator of PmB resistance which promotes aminoarabinose additions to lipid A, PgtE is an outer membrane CAMP protease, and waaP is involved in lipopolysaccharide core modifications (37, 38, 81). Thus, in L. pneumophila, as in Salmonella, CAMP resistance appears to be multifactorial.
The growth of both the wild type and NU260 was similar in BYE or CDM, suggesting that mutation of rcp did not cause a generalized growth defect. However, inoculation into low-Mg2+ CDM resulted in lower total growth and decreased bacterial viability of the rcp mutant compared to wild-type 130b. These phenotypes were very similar to the kinetics and viability of Salmonella serovar Typhimurium phoP, mgtA, and mgtCB mutants in low-Mg2+ media (72). The Salmonella mutants are purported to have reduced growth in low-Mg2+ media due to the loss of the Mg2+ transporters (MgtA and MgtCB) or their activator (PhoP) (72). At present, there is no knowledge of Legionella magnesium transport. Thus, clarification of why an rcp mutant has extracellular growth similar to that of Salmonella phoP, mgtA, and mgtCB mutants requires detailed further characterization of rcp.
Here, we demonstrate, for the first time, that a pagP-like gene is important for intracellular growth and/or survival. Indeed, rcp promotes L. pneumophila growth in both H. vermiformis and U937 macrophages, representatives of the two most relevant host cells for Legionella infections. Preliminary data indicate that the rcp mutants are also defective for intracellular infection of peripheral blood monocytes (M. Robey and N. P. Cianciotto, unpublished results). Although other scenarios are possible, it is reasonable to postulate that the decreased infectivity of the rcp mutant is due, at least in part, to decreased CAMP resistance. In U937 macrophages, the difference between the numbers of intracellular bacteria for 130b and the rcp mutant diminished over time. This suggests adaptation of the rcp mutants to their environment, possibly involving the induction of other CAMP resistance mechanisms. Interestingly, a typical cytopathic effect was not elicited by the mutant; i.e., the extent of cytopathicity did not closely correlate with numbers of recovered CFU (3, 32, 42, 50). Recently, mutants have been characterized which are defective for cytopathicity but have no intracellular growth defect and thus remain trapped inside the macrophage (1). However, to our knowledge, NU260 is the first example of an L. pneumophila mutant with an infectivity defect that ultimately achieves a wild-type level of replication in macrophages but lacks a full cytopathic effect. The intracellular infectivity defect of the rcp mutant was more pronounced in H. vermiformis than in U937 macrophages, suggesting greater importance for rcp in amoebal infections. It is possible that (i) amoebae contain more CAMPs than macrophages, (ii) amoebal CAMPs are more accessible to the Legionella intracellular environment, or (iii) amoebal CAMPs are more active against Rcp-induced resistance mechanisms. The notion that the protozoan environment is less permissive has been suggested by the behavior of other Legionella mutants that are more defective in amoebae than in macrophages (17, 27, 71).
Importantly, we have also shown that rcp is important for colonization of the mammalian lung, which is the first demonstration of a pagP-like gene being involved in virulence. The in vivo defect noted in NU260 could be due to changes in the ability of the bacterium to resist CAMPs intracellularly and/or extracellularly. In Salmonella, loss of pagP had no effect on virulence as judged by 50% lethal dose evaluation during intraperitoneal inoculation of BALB/c mice (6). It is possible that the differences in the virulence of pagP in Salmonella and rcp in L. pneumophila reflect inherent differences in assay conditions (i.e., competition assay versus 50% lethal dose or use of BALB/c versus A/J mice). Alternatively, pagP-like genes maybe more important for virulence in legionellosis.
The ability to sense and adapt to changing environmental conditions is highly advantageous to L. pneumophila, an organism that occupies diverse niches, from free-living in water to intracellular in protozoa or macrophages. As the PhoPQ regulon regulates pagP in Salmonella, the question arises whether L. pneumophila rcp is similarly regulated. The unfinished L. pneumophila genome database (www.genome3.cpmc.columbia.edu) contains an open reading frame the product of which has 37% identity and 60% similarity to PhoP in Salmonella serovar Typhimurium. However, the product of this open reading frame also appears to be homologous (44% identity and 61% similarity) to the Salmonella regulatory gene product PmrA, which is involved in PmB resistance. That rcp may be controlled by a PhoPQ-like regulator is suggested by the fact that upstream of the rcp putative transcription start site lie sequences homologous to a motif involved in PhoPQ-dependent Mg2+-responsive induction of transcription in E. coli (51). In Salmonella, Mg2+ is the signal for PhoPQ to transcriptionally activate genes involved in intramacrophage survival, leading to speculation that this cation is limiting within the phagosome (33, 34, 43). That an L. pneumophila rcp mutant shows decreased growth in low-Mg2+ media and decreased infectivity for both amoebae and macrophages suggests that Mg2+ limitation may be an environmental cue for Legionella. Thus, Legionella, like the successful intracellular pathogen Salmonella, may respond to its environment by activating various virulence determinants that facilitate survival and proliferation within the host. Rcp appears to be one such determinant, important for intracellular infection by mediating resistance to CAMPs.
ACKNOWLEDGMENTS
This work was funded in part by grant A134937 from the National Institutes of Health awarded to N.P.C.
We thank Tracey Aber Scheel for construction of pTA2 and Joseph Gawronski-Salerno, Ombeline Rossier, Virginia Aragon, and Antje Flieger for assistance during animal studies. We also thank R. Darveau for the kind gift of defensin C18G and A. Hauser for providing the P. aeruginosa strain used during these studies. For helpful discussions and comments, we thank past and present members of the Cianciotto laboratory.
REFERENCES
- 1.Alli O A, Gao L Y, Pedersen L L, Zink S, Radulic M, Doric M, Abu Kwaik Y. Temporal pore formation-mediated egress from macrophages and alveolar epithelial cells by Legionella pneumophila. Infect Immun. 2000;68:6431–6440. doi: 10.1128/iai.68.11.6431-6440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amemura-Maekawa J, Mishima-Abe S, Kura F, Takahashi T, Watanabe H. Identification of a novel periplasmic catalase-peroxidase KatA of Legionella pneumophila. FEMS Microbiol Lett. 1999;176:339–344. doi: 10.1111/j.1574-6968.1999.tb13681.x. [DOI] [PubMed] [Google Scholar]
- 3.Aragon V, Kurtz S, Flieger A, Neumeister B, Cianciotto N P. Secreted enzymatic activities of wild-type and pilD-deficient Legionella pneumophila. Infect Immun. 2000;68:1855–1863. doi: 10.1128/iai.68.4.1855-1863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bandyopadhyay P, Steinman H M. Legionella pneumophila catalase-peroxidases: cloning of the katB gene and studies of KatB function. J Bacteriol. 1998;180:5369–5374. doi: 10.1128/jb.180.20.5369-5374.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behlau L, Miller S I. A PhoP-repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J Bacteriol. 1993;175:4475–4484. doi: 10.1128/jb.175.14.4475-4484.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belden W J, Miller S I. Further characterization of the PhoP regulon: identification of new PhoP-activated virulence loci. Infect Immun. 1994;62:5095–5101. doi: 10.1128/iai.62.11.5095-5101.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bengoechea J A, Skurnik M. Temperature-regulated efflux pump/potassium antiporter system mediates resistance to cationic antimicrobial peptides in Yersinia. Mol Microbiol. 2000;37:67–80. doi: 10.1046/j.1365-2958.2000.01956.x. [DOI] [PubMed] [Google Scholar]
- 8.Benson R F, Thacker W L, Wilkinson H W, Fallon R J, Brenner D J. Legionella pneumophila serogroup 14 isolated from patients with fatal pneumonia. J Clin Microbiol. 1988;26:382. doi: 10.1128/jcm.26.2.382-.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berger K H, Isberg R R. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 10.Bibb W F, Arnow P M, Dellinger D L, Perryman S R. Isolation and characterization of a seventh serogroup of Legionella pneumophila. J Clin Microbiol. 1983;17:346–348. doi: 10.1128/jcm.17.2.346-348.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bishop R E, Gibbons H S, Guina T, Trent M S, Miller S I, Raetz C R. Transfer of palmitate from phospholipids to lipid A in outer membranes of Gram-negative bacteria. EMBO J. 2000;19:5071–5080. doi: 10.1093/emboj/cdd507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bissett M L, Lee J O, Lindquist D S. New serogroup of Legionella pneumophila, serogroup 8. J Clin Microbiol. 1983;17:887–891. doi: 10.1128/jcm.17.5.887-891.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bozue J A, Johnson W. Interaction of Legionella pneumophila with Acanthamoeba castellanii: uptake by coiling phagocytosis and inhibition of phagosome-lysosome fusion. Infect Immun. 1996;64:668–673. doi: 10.1128/iai.64.2.668-673.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brenner D J, Steigerwalt A G, Gorman G W, Wilkinson H W, Bibb W F, Hackel M, Tyndall R L, Campbell J, Feeley J C, Thacker W L, Skaliy P, Martin W T, Brake B J, Fields B S, McEachern H V, Corcoran L K. Ten new species of Legionella. Int J Syst Bacteriol. 1985;35:50–59. [Google Scholar]
- 15.Brieland J, Freeman P, Kunkel R, Chrisp C, Hurley M, Fantone J, Engleberg C. Replicative Legionella pneumophila lung infection in intratracheally inoculated A/J mice. A murine model of human Legionnaires' disease. Am J Pathol. 1994;145:1537–1546. [PMC free article] [PubMed] [Google Scholar]
- 16.Brieland J, McClain M, Heath L, Chrisp C, Huffnagle G, LeGendre M, Hurley M, Fantone J, Engleberg C. Coinoculation with Hartmannella vermiformis enhances replicative Legionella pneumophila lung infection in a murine model of Legionnaires' disease. Infect Immun. 1996;64:2449–2456. doi: 10.1128/iai.64.7.2449-2456.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brieland J, McClain M, LeGendre M, Engleberg C. Intrapulmonary Hartmannella vermiformis: a potential niche for Legionella pneumophila replication in a murine model of legionellosis. Infect Immun. 1997;65:4892–4896. doi: 10.1128/iai.65.11.4892-4896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandler M S. New shuttle vectors for Haemophilus influenzae and Escherichia coli: P15A-derived plasmids replicate in H. influenzae Rd. Plasmid. 1991;25:221–224. doi: 10.1016/0147-619x(91)90016-p. [DOI] [PubMed] [Google Scholar]
- 19.Cianciotto N P, Fields B S. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc Natl Acad Sci USA. 1992;89:5188–5191. doi: 10.1073/pnas.89.11.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cirillo J D, Cirillo S L, Yan L, Bermudez L E, Falkow S, Tompkins L S. Intracellular growth in Acanthamoeba castellanii affects monocyte entry mechanisms and enhances virulence of Legionella pneumophila. Infect Immun. 1999;67:4427–4434. doi: 10.1128/iai.67.9.4427-4434.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darveau R P, Blake J, Seachord C L, Cosand W L, Cunningham M D, Cassiano-Clough L, Maloney G. Peptides related to the carboxyl terminus of human platelet factor IV with antibacterial activity. J Clin Invest. 1992;90:447–455. doi: 10.1172/JCI115880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edelstein P H. Improved semiselective medium for isolation of Legionella pneumophila from contaminated clinical and environmental specimens. J Clin Microbiol. 1981;14:298–303. doi: 10.1128/jcm.14.3.298-303.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edelstein P H, Finegold S M. Use of a semiselective medium to culture Legionella pneumophila from contaminated lung specimens. J Clin Microbiol. 1979;10:141–143. doi: 10.1128/jcm.10.2.141-143.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engleberg N C, Carter C, Weber D R, Cianciotto N P, Eisenstein B I. DNA sequence of mip, a Legionella pneumophila gene associated with macrophage infectivity. Infect Immun. 1989;57:1263–1270. doi: 10.1128/iai.57.4.1263-1270.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engleberg N C, Drutz D J, Eisenstein B I. Cloning and expression of Legionella pneumophila antigens in Escherichia coli. Infect Immun. 1984;44:222–227. doi: 10.1128/iai.44.2.222-227.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ernst R K, Yi E C, Guo L, Lim K B, Burns J L, Hackett M, Miller S I. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science. 1999;286:1561–1565. doi: 10.1126/science.286.5444.1561. [DOI] [PubMed] [Google Scholar]
- 27.Fettes P S, Susa M, Hacker J, Marre R. Characterisation of the Legionella pneumophila gene ligA. Int J Med Microbiol. 2000;290:239–250. doi: 10.1016/S1438-4221(00)80121-6. [DOI] [PubMed] [Google Scholar]
- 28.Fields B S. The molecular ecology of legionellae. Trends Microbiol. 1996;4:286–290. doi: 10.1016/0966-842x(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 29.Fields B S, Nerad T A, Sawyer T K, King C H, Barbaree J M, Martin W T, Morrill W E, Sanden G N. Characterization of an axenic strain of Hartmannella vermiformis obtained from an investigation of nosocomial legionellosis. J Protozool. 1990;37:581–583. doi: 10.1111/j.1550-7408.1990.tb01269.x. [DOI] [PubMed] [Google Scholar]
- 30.Francis M S, Thomas C J. Mutants in the CtpA copper transporting P-type ATPase reduce virulence of Listeria monocytogenes. Microb Pathog. 1997;22:67–78. doi: 10.1006/mpat.1996.0092. [DOI] [PubMed] [Google Scholar]
- 31.Ganz T, Lehrer R I. Defensins. Curr Opin Immunol. 1994;6:584–589. doi: 10.1016/0952-7915(94)90145-7. [DOI] [PubMed] [Google Scholar]
- 32.Gao L Y, Harb O S, Abu Kwaik Y. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant host cells, mammalian macrophages and protozoa. Infect Immun. 1997;65:4738–4746. doi: 10.1128/iai.65.11.4738-4746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-del Portillo F, Foster J W, Maguire M E, Finlay B B. Characterisation of the micro-environment of Salmonella typhimurium-containing vacuoles within MDCK epithelial cells. Mol Microbiol. 1992;6:3289–3297. doi: 10.1111/j.1365-2958.1992.tb02197.x. [DOI] [PubMed] [Google Scholar]
- 34.Garcia Vescovi E, Soncini F C, Groisman E A. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 35.Garrity G M, Elder E M, Davis B, Vickers R M, Brown A. Serological and genotypic diversity among serogroup 5- reacting environmental Legionella isolates. J Clin Microbiol. 1982;15:646–653. doi: 10.1128/jcm.15.4.646-653.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giacometti A, Cirioni O, Barchiesi F, Del Prete M S, Fortuna M, Caselli F, Scalise G. In vitro susceptibility tests for cationic peptides: comparison of broth microdilution methods for bacteria that grow aerobically. Antimicrob Agents Chemother. 2000;44:1694–1696. doi: 10.1128/aac.44.6.1694-1696.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guina T, Yi E C, Wang H, Hackett M, Miller S I. A PhoP-regulated outer membrane protease of Salmonella enterica serovar Typhimurium promotes resistance to alpha-helical antimicrobial peptides. J Bacteriol. 2000;182:4077–4086. doi: 10.1128/jb.182.14.4077-4086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gunn J S, Lim K B, Krueger J, Kim K, Guo L, Hackett M, Miller S I. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol. 1998;27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 39.Guo L, Lim K B, Poduje C M, Daniel M, Gunn J S, Hackett M, Miller S I. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell. 1998;95:189–198. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- 40.Halablab M A, Bazin M, Richards L, Pacy J. Ultra-structure and localisation of formazan formed by human neutrophils and amoebae phagocytosing virulent and avirulent Legionella pneumophila. FEMS Microbiol Immunol. 1990;2:295–301. doi: 10.1111/j.1574-6968.1990.tb03532.x. [DOI] [PubMed] [Google Scholar]
- 41.Hancock R E, Scott M G. The role of antimicrobial peptides in animal defenses. Proc Natl Acad Sci USA. 2000;97:8856–8861. doi: 10.1073/pnas.97.16.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harb O S, Abu Kwaik Y. Essential role for the Legionella pneumophila Rep helicase homologue in intracellular infection of mammalian cells. Infect Immun. 2000;68:6970–6978. doi: 10.1128/iai.68.12.6970-6978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heithoff D M, Conner C P, Hentschel U, Govantes F, Hanna P C, Mahan M J. Coordinate intracellular expression of Salmonella genes induced during infection. J Bacteriol. 1999;181:799–807. doi: 10.1128/jb.181.3.799-807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hickey E K, Cianciotto N P. Cloning and sequencing of the Legionella pneumophila fur gene. Gene. 1994;143:117–121. doi: 10.1016/0378-1119(94)90615-7. [DOI] [PubMed] [Google Scholar]
- 45.Higgins D G, Sharp P M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 46.Horwitz M A. Characterisation of avirulent mutant Legionella pneumophila that survive but do not multiply within human monocytes. J Exp Med. 1987;166:1310–1328. doi: 10.1084/jem.166.5.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horwitz M A. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J Exp Med. 1983;158:2108–2126. doi: 10.1084/jem.158.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horwitz M A, Silverstein S C. Legionnaires' disease bacterium (Legionella pneumophila) multiples intracellularly in human monocytes. J Clin Invest. 1980;66:441–450. doi: 10.1172/JCI109874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu K H, Liu E, Dean K, Gingras M, DeGraff W, Trun N J. Overproduction of three genes leads to camphor resistance and chromosome condensation in Escherichia coli. Genetics. 1996;143:1521–1532. doi: 10.1093/genetics/143.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joshi A D, Swanson M S. Comparative analysis of Legionella pneumophila and Legionella micdadei virulence traits. Infect Immun. 1999;67:4134–4142. doi: 10.1128/iai.67.8.4134-4142.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kato A, Tanabe H, Utsumi R. Molecular characterization of the PhoP-PhoQ two-component system in Escherichia coli K-12: identification of extracellular Mg2+-responsive promoters. J Bacteriol. 1999;181:5516–5520. doi: 10.1128/jb.181.17.5516-5520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krogfelt K A, Hjulgaard M, Sorensen K, Cohen P S, Givskov M. rpoS gene function is a disadvantage for Escherichia coli BJ4 during competitive colonization of the mouse large intestine. Infect Immun. 2000;68:2518–2524. doi: 10.1128/iai.68.5.2518-2524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leippe M, Andra J, Nickel R, Tannich E, Muller-Eberhard H J. Amoebapores, a family of membranolytic peptides from cytoplasmic granules of Entamoeba histolytica: isolation, primary structure, and pore formation in bacterial cytoplasmic membranes. Mol Microbiol. 1994;14:895–904. doi: 10.1111/j.1365-2958.1994.tb01325.x. [DOI] [PubMed] [Google Scholar]
- 54.Liles M R, Edelstein P H, Cianciotto N P. The prepilin peptidase is required for protein secretion by and the virulence of the intracellular pathogen Legionella pneumophila. Mol Microbiol. 1999;31:959–970. doi: 10.1046/j.1365-2958.1999.01239.x. [DOI] [PubMed] [Google Scholar]
- 55.Lindquist D S, Nygaard G, Thacker W L, Benson R F, Brenner D J, Wilkinson H W. Thirteenth serogroup of Legionella pneumophila isolated from patients with pneumonia. J Clin Microbiol. 1988;26:586–587. doi: 10.1128/jcm.26.3.586-587.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marra A, Blander S J, Horwitz M A, Shuman H A. Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc Natl Acad Sci USA. 1992;89:9607–9611. doi: 10.1073/pnas.89.20.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matthews M, Roy C R. Identification and subcellular localization of the Legionella pneumophila IcmX protein: a factor essential for establishment of a replicative organelle in eukaryotic host cells. Infect Immun. 2000;68:3971–3982. doi: 10.1128/iai.68.7.3971-3982.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McKinney R M, Porschen R K, Edelstein P H, Bissett M L, Harris P P, Bondell S P, Steigerwalt A G, Weaver R E, Ein M E, Lindquist D S, Kops R S, Brenner D J. Legionella longbeachae species nova, another etiologic agent of human pneumonia. Ann Intern Med. 1981;94:739–743. doi: 10.7326/0003-4819-94-6-739. [DOI] [PubMed] [Google Scholar]
- 59.McKinney R M, Thacker L, Harris P P, Lewallen K R, Hebert G A, Edelstein P H, Thomason B M. Four serogroups of Legionnaires' disease bacteria defined by direct immunofluorescence. Ann Intern Med. 1979;90:621–624. doi: 10.7326/0003-4819-90-4-621. [DOI] [PubMed] [Google Scholar]
- 60.McKinney R M, Wilkinson H W, Sommers H M, Fikes B J, Sasseville K R, Yungbluth M M, Wolf J S. Legionella pneumophila serogroup six: isolation from cases of legionellosis, identification by immunofluorescence staining, and immunological response to infection. J Clin Microbiol. 1980;12:395–401. doi: 10.1128/jcm.12.3.395-401.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Merrell D S, Camilli A. The cadA gene of Vibrio cholerae is induced during infection and plays a role in acid tolerance. Mol Microbiol. 1999;34:836–849. doi: 10.1046/j.1365-2958.1999.01650.x. [DOI] [PubMed] [Google Scholar]
- 62.Miller S I, Mekalanos J J. Constitutive expression of the phoP regulon attenuates Salmonella virulence and survival within macrophages. J Bacteriol. 1990;172:2485–2490. doi: 10.1128/jb.172.5.2485-2490.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nash T W, Libby D M, Horwitz M A. Interaction between the Legionnaires' disease bacterium (Legionella pneumophila) and human alveolar macrophages. Influence of antibody, lymphokines, and hydrocortisone. J Clin Invest. 1984;74:771–782. doi: 10.1172/JCI111493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O'Connell W A, Bangsborg J M, Cianciotto N P. Characterization of a Legionella micdadei mip mutant. Infect Immun. 1995;63:2840–2845. doi: 10.1128/iai.63.8.2840-2845.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O'Connell W A, Hickey E K, Cianciotto N P. A Legionella pneumophila gene that promotes hemin binding. Infect Immun. 1996;64:842–848. doi: 10.1128/iai.64.3.842-848.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pearlman E, Jiwa A H, Engleberg N C, Eisenstein B I. Growth of Legionella pneumophila in a human macrophage-like (U937) cell line. Microb Pathog. 1988;5:87–95. doi: 10.1016/0882-4010(88)90011-3. [DOI] [PubMed] [Google Scholar]
- 67.Peschel A, Otto M, Jack R W, Kalbacher H, Jung G, Gotz F. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J Biol Chem. 1999;274:8405–8410. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 68.Reeves M W, Pine L, Hutner S H, George J R, Harrell W K. Metal requirements of Legionella pneumophila. J Clin Microbiol. 1981;13:688–695. doi: 10.1128/jcm.13.4.688-695.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rowbotham T J. Current views on the relationships between amoebae, legionellae and man. Isr J Med Sci. 1986;22:678–689. [PubMed] [Google Scholar]
- 70.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 71.Segal G, Shuman H A. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect Immun. 1999;67:2117–2124. doi: 10.1128/iai.67.5.2117-2124.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Soncini F C, Garcia Vescovi E, Solomon F, Groisman E A. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol. 1996;178:5092–5099. doi: 10.1128/jb.178.17.5092-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Steinberg D A, Hurst M A, Fujii C A, Kung A H, Ho J F, Cheng F C, Loury D J, Fiddes J C. Protegrin-1: a broad-spectrum, rapidly microbicidal peptide with in vivo activity. Antimicrob Agents Chemother. 1997;41:1738–1742. doi: 10.1128/aac.41.8.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.St. John G, Steinman H M. Periplasmic copper-zinc superoxide dismutase of Legionella pneumophila: role in stationary-phase survival. J Bacteriol. 1996;178:1578–1584. doi: 10.1128/jb.178.6.1578-1584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sturgill-Koszycki S, Swanson M S. Legionella pneumophila replication vacuoles mature into acidic, endocytic organelles. J Exp Med. 2000;192:1261–1272. doi: 10.1084/jem.192.9.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sundstrom C, Nilsson K. Establishment and characterisation of a human histiocytic lymphoma cell line (U-937) Int J Cancer. 1976;17:565–577. doi: 10.1002/ijc.2910170504. [DOI] [PubMed] [Google Scholar]
- 77.Thacker W L, Wilkinson H W, Plikaytis B B, Steigerwalt A G, Mayberry W R, Moss C W, Brenner D J. Second serogroup of Legionella feeleii strains isolated from humans. J Clin Microbiol. 1985;22:1–4. doi: 10.1128/jcm.22.1.1-4.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vogel J P, Andrews H L, Wong S K, Isberg R R. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- 79.Wiater L A, Dunn K, Maxfield F R, Shuman H A. Early events in phagosome establishment are required for intracellular survival of Legionella pneumophila. Infect Immun. 1998;66:4450–4460. doi: 10.1128/iai.66.9.4450-4460.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wilkinson H W, Thacker W L, Benson R F, Polt S S, Brookings E, Mayberry W R, Brenner D J, Gilley R G, Kirklin J K. Legionella birminghamensis sp. nov. isolated from a cardiac transplant recipient. J Clin Microbiol. 1987;25:2120–2122. doi: 10.1128/jcm.25.11.2120-2122.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yethon J A, Gunn J S, Ernst R K, Miller S I, Laroche L, Malo D, Whitfield C. Salmonella enterica serovar Typhimurium waaP mutants show increased susceptibility to polymyxin and loss of virulence in vivo. Infect Immun. 2000;68:4485–4491. doi: 10.1128/iai.68.8.4485-4491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yildiz F H, Schoolnik G K. Role of rpoS in stress survival and virulence of Vibrio cholerae. J Bacteriol. 1998;180:773–784. doi: 10.1128/jb.180.4.773-784.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]