Abstract
The siRNA-loaded lipid nanoparticles have attracted much attention due to its significant gene silencing effect and successful marketization. However, the in vivo distribution and release of siRNA still cannot be effectively monitored. In this study, based on the fluorescence resonance energy transfer (FRET) principle, a fluorescence dye Cy5-modified survivin siRNA was conjugated to nanogolds (Au-DR-siRNA), which were then wrapped with lipid nanoparticles (LNPs) for monitoring the release behaviour of siRNA in vivo. The results showed that once Au-DR-siRNA was released from the LNPs and cleaved by the Dicer enzyme to produce free siRNA in cells, the fluorescence of Cy5 would change from quenched state to activated state, showing the location and time of siRNA release. Besides, the LNPs showed a significant antitumor effect by silencing the survivin gene and a CT imaging function superior to iohexol by nanogolds. Therefore, this work provided not only an effective method for monitoring the pharmacokinetic behaviour of LNP-based siRNA, but also a siRNA delivery system for treating and diagnosing tumors.
Keywords: Survivin siRNA, Lipid nanoparticles, In vivo release, Nanogolds, Fluorescence resonance energy transfer
Graphical abstract
1. Introduction
Onpattro (patisiran) was approved by the Food and Drug Administration (FDA) as the first siRNA drug for the treatment of adult patients with polyneuropathy caused by hereditary transthyretin amyloidosis (hATTR) [1]. RNA interference (RNAi) technology can specifically shut down the expression of specific genes, and has attracted much attention due to its good effect on gene silence and its universality in eukaryotes [2,3]. The launch of siRNA drug is a milestone in the clinical translation of Nobel Prize achievement, which has set off a revolution in the pharmaceutical industry [4]. Targeted delivery and timely release of siRNA drugs in diseased tissues are key challenges affecting their therapeutic effect [5]. Generally, the in vivo efficacy of siRNA drugs is verified by detecting the expression level of the target protein in living tissue [6]. However, the pharmacokinetic properties of siRNA drugs, including the in vivo distribution and release, have not been well-studied yet. The direct cause of this problem is that there is currently no effective means to monitor the dynamic behaviour of siRNA in vivo in real-time.
The key to the successful clinical translation of RNAi technology is its delivery system, lipid nanoparticles (LNPs), which has received extensive attention, especially after the widespread use of mRNA COVID-19 vaccine [7]. The components of LNPs include phospholipid distearate phosphatidylcholine (DSPC), cholesterol, polyethylene glycol dimethacrylate (PEG-C-DMA), and ionizable cationic lipid (DLin-MC3-DMA for Onpattro) [8]. Amongst them, DLin-MC3-DMA is a key component, enabling LNPs to effectively encapsulate negatively charged siRNA under low pH conditions and exhibit a relatively uncharged surface at pH 7.4 [9]. LNPs adsorb endogenous ApoE in the blood circulation and then specifically bind to apolipoprotein (ApoE) receptors on the surface of hepatocytes to trigger the cellular uptake [10]. The success of Onpattro has prompted researchers to expect LNP-based siRNA to be used for more disease treatment [11]. Thus, the effective monitoring of the pharmacokinetic properties of siRNA drugs is particularly important and urgently needed.
The siRNA is a short double-stranded RNA about 21 nucleotides in length, usually with two nucleotides protruding from the 3′-end of each strand [12]. The mechanism of siRNA acting is rough as follows. The siRNA is cleaved into small fragments of approximately 21 bp by the intracellular Dicer enzyme, and then the single strand is combined with protease (endonuclease, exonuclease, helicase, etc.) to form an RNA-induced silencing complex (RISC) [13,14]. RISC bases pair with mRNA and cleaves mRNA at the binding site to exert an RNAi effect [15]. Since the cleavage of siRNA by Dicer is a necessary process for mRNA silencing, the detection of this event may be an effective way to monitor the tissue distribution and release behaviour of siRNA in vivo [16].
Fluorescence resonance energy transfer (FRET), as a highly efficient optical “molecular ruler,” has become a powerful tool for detecting nanoscale distance and nanoscale distance changes of biological macromolecules in living organisms [17]. It has a wide range of applications in biological macromolecular interaction analysis, cell physiology research, and immunoassay [18]. Non-radioactive energy transfer occurs when the emission spectrum of the fluorescent donor molecule overlaps with the absorption spectrum of the acceptor fluorescent molecule and the distance between the two molecules is within 10 nm [19]. The fluorescence intensity of the donor is greatly reduced, while the fluorescence emitted by the acceptor is greatly enhanced [20]. Based on the FRET principle, nanogold (Au) is often used as fluorescent quencher of most fluorophores because of its wide quenching range, high quenching efficiency, good stability, sensitivity, low toxicity, and good biocompatibility [21,22]. In addition, Au can also been used as CT contrast agents due to its high X-ray absorption capacity and good pharmacokinetic properties [23]. Professor Mirkin's group modified the sense and antisense strands of double-stranded mRNA with Cy5 and thiol, respectively, and then connected them to the surface of Au to construct a FRET-based biodetector for monitoring mRNA content in liquid samples or cells [24]. Besides, siRNA has also been attached to the surface of Au to detect the content of Dicer and ribozyme in liquid or cell [25]. However, the FRET principle has not been used for the in vivo monitoring of tissue distribution and release behaviour of siRNA.
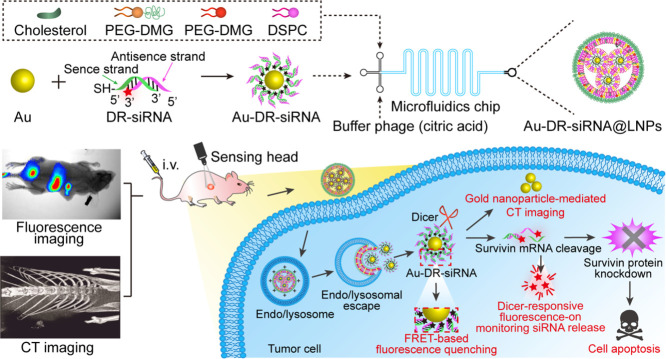
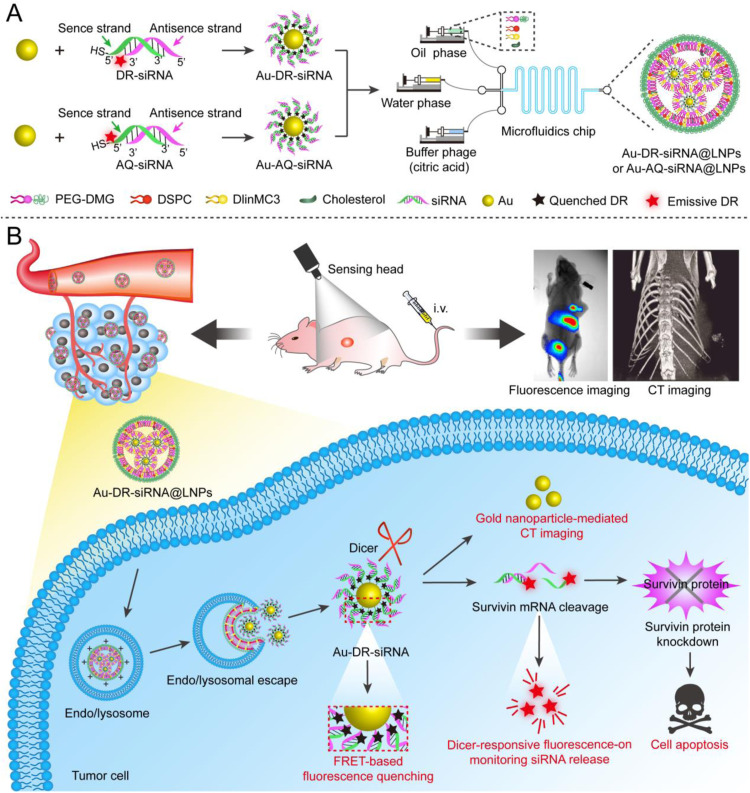
In this work, survivin siRNA was used as the model siRNA to study the monitoring of the in vivo behaviour of siRNA drugs based on FRET principle. Specifically, the siRNA was conjugated with Cy5 at the 3′-end of the antisense strand, then with Au by the strong coordination between Au and the thiol group (-SH) at the 5′ end of the sense chain, and wrapped into LNPs by microfluidic technology (Fig. 1A). Since the Cy5 molecule is close to Au, its fluorescence is in a quenched state. When siRNA was released from LNPs and cleaved by the intracellular Dicer enzyme, siRNA along with Cy5 falls away from the surface of Au to restore the fluorescence of Cy5, thereby monitoring the in vivo distribution and release of siRNA. Besides, the survivin siRNA LNPs could be used for the treatment of tumour by silencing the survivin gene and for CT imaging by Au. In this study, the LNPs were called Dicer-Responsive fluorescence-recovered Au-siRNA LNPs (Au-DR-siRNA@LNPs). In addition, the thiol group and Cy5 were modified to the 5′-end of the siRNA sense chain to form a control LNPs whose fluorescence was still quenched by Au after Dicer enzyme cleavage (Au-AQ-siRNA@LNPs). This study mainly explores the feasibility of monitoring the siRNA release from LNPs by the FRET principle, which provides a powerful tool for studying the pharmacokinetic behaviour of siRNA drugs and promotes their further development (Fig. 1B).
Fig. 1.
Monitoring the in vivo behaviour of siRNA by Au-DR-siRNA@LNPs based on FRET principle. (A) The siRNA was conjugated with Cy5 at the 3′-end of the antisense strand, then with Au by the strong coordination between Au and the thiol group (-SH) at the 5′ end of the sense chain, and wrapped into LNPs (Au-DR-siRNA@LNPs) by microfluidic technology. Au-AQ-siRNA@LNPs were prepared as the control by modifying both thiol group and Cy5 to the 5′-end of the siRNA sense chain, whose fluorescence is always quenched by Au after Dicer enzyme cleavage. (B) When siRNA was released from LNPs and cleaved by the intracellular Dicer enzyme, siRNA along with Cy5 falls away from the surface of Au to restore the fluorescence of Cy5, thereby monitoring the in vivo pharmacokinetic properties of siRNA. Besides, the survivin siRNA LNPs could be used for the treatment of tumour by silencing the survivin gene and for CT imaging by Au.
2. Materials and methods
2.1. Materials
1,2-distearoyl-sn‑glycero-3-phosphocholine (DSPC) and cholesterol were purchased from AVT (Shanghai) Pharmaceutical Tech Co., Ltd. 1640 medium and foetal bovine serum (FBS) were purchased from Gibco Co, USA. LipofectamineTM 2000 kit and Trizol were purchased from Invitrogen, USA. Dicer enzyme and adenosine triphosphate (ATP) were purchased from Aladdin's reagent. CCK8 kit, Annexin V-FITC/PI apoptosis kit, H&E staining reagent, terminal deoxynucleotidyl transferase dUTP nick end labelling (Tunel) staining reagent were purchased from Shanghai Zhongtao Biotechnology Co., Ltd. Rabbit anti-mouse Survivin antibody and DAB chromogenic reagent were purchased from Jackson ImmunoResearch (PA).
Polyethylene glycol-diethylene glycol glyceride (PEG-DMG) and DLin-MC3-DMA were synthsized and provided by Professor Tang Jie of East China Normal University. Three survivin siRNAs, DR-siRNA, and AQ-siRNA were designed and synthesized by Sangon Biotech (Shanghai) Co., Ltd. The three siRNA sequences are shown in Table S1.
DR-siRNA: The Cy5 and sulfhydryl group (-SH) were chemically modified at the 3′-end of the siRNA antisense strand and the 5′ end of the sense strand, respectively.
AQ-siRNA: Both Cy5 and sulfhydryl are modified at the 5′-end of the siRNA sense strand.
The 4T1 cells were purchased from Shanghai Shengbo Biotechnology Co., Ltd. Male Babl/c nude mice were purchased from Shanghai Slack Animal center. All animal studies were approved by the Animal Ethical and Welfare Committee of East China Normal University (m20210228) and conducted according to the institutional guidelines.
2.2. Cell culture and siRNAs transfection assay
The 4T1 cells were cultured in RPMI1640 medium supplemented with 10% (w/v) FBS, 1 mM sodium pyruvate, and 4 mM l-glutamine at 37 °C in 5% CO2. According to the manufacturer's protocol, 4T1 cells were transfected by negative control siRNA (NC-siRNA), siRNA-1, siRNA-2, and siRNA-3 (500 nM siRNA) using lipo2000 (Invitrogen) for 6 h, followed by washing thrice with ice-cold PBS, and incubated for 48 h. After 48 h, tumour cells were collected and the total RNA was extracted according to the method of Trizol kit (Gibco/Life Technologies). Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was used to detect the expression of survivin gene. Quantitative analysis was performed using β-actin as an internal reference. The primers for survivin and β-actin are shown in Table S2. The cycling program was 95 °C for 15 s, 95 °C for 5 s, 60 °C for 30 s (40 cycles). The relative level of RNA was computed using the 2−ΔΔCt analysis method.
2.3. Western blot (WB) analysis
Tumour cells were transfected with Au-DR-siRNA@LNPs prepared by sina1. The 4T1 cells were treated with NC-siRNA, siRNA-1, siRNA-2, and siRNA-3 (500 nM siRNA) using lipo2000 (Invitrogen) for 6 h, followed by washing thrice with ice-cold PBS, and incubated for 48 h. After 48 h, 4T1 cells were incubated with 300 µl RIPA buffer supplemented with protease inhibitor cocktail and phenylmethylsulfonyl fluoride on the ice for 15 min, and whole proteins were extracted and quantified using the bicinchoninic acid (BCA) protein assay kits. Equal amounts of proteins (20 µg) were separated by 10% Sodium Dodecyl Sulphate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride (PVDF) membranes, which were blocked with 5% skimmed milk for 2 h, and incubated with primary antibodies (Survivin, 1:1000) at 4 °C overnight. After washing with Tris-buffered saline with 0.1% Tween 20 detergent (TBST), the membranes were incubated with the peroxidase-conjugated goat-anti-mouse antibody at 1:5000 dilutions for an additional 2 h at room temperature, followed by detecting with the Tanon-5200 chemiluminescent imaging system.
2.4. Preparation of Au-DR-siRNA and Au-AQ-siRNA
First, for the preparation of Au, 1% chloroauric acid solution (500 µl) was added to 40 ml water, and magnetically stirred for 5 min. Then 3.5 ml 1% sodium citrate solution was immediately added to the above solution and continue stirred for 5 min. Sodium citrate solution(3.5 ml, 1% )was added to the above solution immediately, stirred for 5 min, and 4 ml newly prepared 0.05% sodium borohydride was slowly added drop by drop at 4℃ until the solution gradually turned wine red to obtain Au. The pure DEPC solution of diethylpyrocarbonate (1%, v/v) was added to the Au solution, stirred overnight, and autoclaved (120 °C, 15 min) to remove the ribozyme that can degrade siRNA. The amount of gold element in the solution was quantified by inductively coupled plasma mass spectrometry (ICP-MS).
Second, we prepared the Au-DR-siRNA which is responsive to the dicer enzyme. Sulfhydryl-modified siRNA sense strand at 5′-end (1 OD) was dissolved by DEPC water (250 µl) and TCEP (2 µl, 100 nM) and activated for 1 h, and then Cy5-modified siRNA antisense strand at 3′-end (1 OD) dissolved by DEPC water (250 µl) was added and kept in 95 °C water bath for 10 min. After the solution was cooled to room temperature, Au and 10 µl saturated NaCl solution were added and sonicated for 4 h, and then the saturated NaCl solution addition and sonication were repeated 3 times for a total of 12 h of sonication. Then, the solution was added with 500 µl isopropanol, centrifuged at 14 000 rpm/min for 20 min to obtain the precipitate of Au-DR-siRNA, which was washed with DEPC water thrice.
Third, we prepared the Au-AQ-siRNA which is nonresponsive to the dicer enzyme and in a state of fluorescence quenching all the time. Cy5 and Sulfhydryl-modified siRNA sense strand at 5′-end (1 OD) was dissolved by DEPC water (250 µl) and TCEP (2 µl, 100 nM) and activated for 1 h, and then siRNA antisense strand (1 OD) dissolved by DEPC water (250 µl) was added and kept in 95 °C water bath for 10 min. The subsequent steps were the same as those described above.
2.5. Preparation of Au-DR-siRNA@LNPs and Au-AQ-siRNA@LNPs
The Au-DR-siRNA or Au-AQ-siRNA containing 1OD siRNA was dissolved in 2 mL of deionized water as an aqueous phase. The sodium citrate solution (2 ml, pH 5) is used as buffer phase. Dlin-MC3-MDA, PEG-DMG, DSPC, and Cholesterol (40:5:10:45, n:n) were dissolved in 2 ml ethanol as an oil phase. The three solution phases were preheated at 40 °C for 20 min, and loaded into syringes at a volume ratio of 1:1:1, which were connected to the three inlets of the microfluidic chip, respectively. The three solution phases were simultaneously passed through the microfluidic channel at a flow rate of 0.5 ml/min, and the sample is collected at the outlet with a 100k ultrafiltration centrifuge tube. The sample was centrifuged at 4000 rpm/min for 20 min, and the LNPs were resuspended with 1×PBS, passed through 0.22 µm microporous membrane, and then stored at 4 °C.
2.6. Characterization of Au-DR-siRNA@LNPs
The particle size, polydispersity index (PDI), and zeta potential (ζ) of Au, Au-DR-siRNA, and Au-DR-siRNA@LNPs were determined by Dynamic Light Scattering (DLS) method using Zetasizer Nano ZS (Malvern, UK). The morphology of these samples was determined by transmission electron microscopy (TEM) with a JEM2100 (Japan electronics co., ltd) microscope. The ultraviolet-visible (UV–Vis) spectra of Au and Au-DR-siRNA were detected by an UV–Vis absorber Lambda 1050 (PerkinElmer, UK).
2.7. Long-term storage stability and serum stability of Au-DR-siRNA@LNPs
For long-term storage stability, Au (1 ml, 1 mg/ml), Au-DR-siRNA (1 ml, 1 mg/ml), and Au-DR-siRNA@LNPs (1 ml, 1 mg/ml) were incubated in 1×PBS at 4 ℃, respectively, and their sizes were detected at a series of time points (0, 2, 4, 6 and 8 weeks) using DLS.
For serum stability, Au (1 ml, 1 mg/ml), Au-DR-siRNA (1 ml, 1 mg/ml), and Au-DR-siRNA@LNPs (1 ml, 1 mg/ml) were incubated in 50% foetal bovine serum (FBS) at 37 °C, respectively, and their sizes were detected at a series of time points (0, 2, 4, 6, 8, 12 and 24 h) using DLS.
2.8. In vitro release behaviors of drug in Au-DR-siRNA@LNPs
The solution of Au-DR-siRNA@LNPs (1 ml, 20 mg/ml) with/without Dicer enzyme (10 µl, 20 IU/l) and dNTPs (10 mM) solution were added into dialysis bags (MWCO 5000 Da), which were immersed in 20 ml PBS (pH 7.5, 6.4, 5.5) with orbital shaking at 200 rpm at 37 °C, followed by dialyzing in different buffer solution for 0, 2, 4, 6, 8, 12, 24, 36, 48, 72 h. At each time point, dialysis medium (1.0 ml) was collected, and fresh dialysis medium (1.0 ml) was added to the system. The absorbence of Cy5 was detected at a wavelength of 650/670 nm (Ex/Em) using a microplate reader.
2.9. Investigation of fluorescence quenching-recovery efficiency of nanoparticles
DR-siRNA, Au-DR-siRNA, Au-AQ-siRNA, and Au-DR-siRNA@LNPs were dissolved in 1 ml DEPC water, and their fluorescence values were measured with a fluorescence spectrophotometer F4500 (Hitachi, Ltd, Japan). Dicer enzyme (10 µl, 20 IU/l) and dNTPs (10 mM) solution were added to the above solution, and the fluorescence values were measured again after 5 min.
The 96-well plate was added with 100 µl DR-siRNA, Au-DR-siRNA, Au-AQ-siRNA, Au-DR-siRNA@LNPs with different siRNA content (0.1OD, 0.2OD, 0.5OD, 1OD, 2OD), respectively, and then added with 50 µl fresh blood (containing ribozymes and other ingredients). The plate was imaged with a living imager lumina III (PerkinElmer, USA) at different time points (0 min, 5 min, 12 h, 24 h).
2.10. Cellular uptake of Au-DR-siRNA@LNPs
The 4T1 cells (1 × 105 cells per well) were seeded in 6-well plates in RPMI-1640 complete medium at 37 °C overnight. Au-DR-siRNA@LNPs and Au-AQ-siRNA@LNPs (500 nM siRNA) were incubated with 4T1 cells for 4 h, respectively, and fluorescence intensity of Cy5 was measured by flow cytometry (Ex/ Em = 650/670 nm). For CLSM analysis, 4T1 cells (5 × 104 cells per well) were seeded in confocal dishes in RPMI-1640 complete medium at 37 °C overnight. The 4T1 cells were treated with Au-DR-siRNA@LNPs and Au-AQ-siRNA@LNPs (500 nM siRNA) for 4 h, respectively, washed with PBS, fixed with 4% paraformaldehyde for 15 min, and stained with Hoechst 33,342 for 15 min. At last, the red fluorescence of Cy5 (Ex/Em = 650/670 nm) and blue fluorescence of Hoechst 33,342 (Ex/Em = 350/460 nm) were imaged by confocal laser scanning microscopy (CLSM).
2.11. Lysosomal escape of Au-DR-siRNA@LNPs in 4T1 cells
The 4T1 cells were seeded in confocal dishes (5 × 104 cells per dish), cultured at 37 °C for 24 h, incubated with Au-DR-siRNA or Au-DR-siRNA@LNPs (500 nM siRNA) for 4 h, washed thrice with ice-cold PBS, and stained with Hoechst 33,342 (1 µl, 1 mM) and Lyso-Tracker green (1 µl, 1 mM) for 15 min, respectively. The green fluorescence of Lyso-Tracker green (Ex/Em = 504/511 nm), red fluorescence of Cy5 (Ex/Em = 650/670 nm), and blue fluorescence of Hoechst 33,342 (Ex/Em =350/460 nm) were imaged by CLSM.
2.12. Immunofluorescence of survivin gene expression in 4T1 cells
The 4T1 cells were cultured in confocal dishes and treated with Au, Au-NC-siRNA@LNPs, Au-DR-siRNA, and Au-DR-siRNA@LNPs (500 nM siRNA) for 6 h at 37 °C, respectively. The cells were washed thrice with PBS, cultured for another 48 h, fixed with 4% paraformaldehyde for 15 min at room temperature, permeabilized in PBS containing 0.25% Triton for 15 min, and blocked with 3% BSA for 1 h at room temperature. Subsequently, the cells were stained with Anti-survivin/FITC (1:400; Cell signaling) and Hoechst 33,342 (1 µl, 1 mM), respectively. At last, the green fluorescence of FITC (Ex/Em = 494/520 nm) and blue fluorescence of Hoechst 33,342 (Ex/Em =350/460 nm) were imaged by CLSM.
2.13. Cell proliferation and apoptosis analysis
Cell Counting Kit-8 (CCK8) was used to examine cell proliferation. The 4T1 cells (2 × 103 cells per well) were seeded in 96-well plates in a complete medium overnight, followed by incubating with Au-NC-siRNA@LNPs, Au-DR-siRNA, and Au-DR-siRNA@LNPs (500 nM siRNA) for 6 h, respectively. The cells were washed thrice with PBS and cultured for 2, 3, 4, and 5 d, respectively. CCK8 solution (20 µl) was added to per well and incubated for 4 h, and the absorbence of each well was measured at 490 nm using a Microplate Reader. The cell growth rates (average absorbence of each transfected group/non-transfected group) were calculated.
The AnnexinV-FITC kit was used to detect the cell apoptosis. The 4T1 cells (5 × 104 cells per well) were seeded in 6-well plates in a complete medium overnight, followed by incubating with Au, Au-NC-siRNA@LNPs, Au-DR-siRNA, and Au-DR-siRNA@LNPs (1000 nM siRNA) for 6 h, respectively. The cells were washed thrice with PBS, cultured for another 48 h, and stained with Annexin V-FITC (1 µl, 1 mg/ml) and propidium iodide (PI) (1 µl, 1 mg/ml) for 15 min at 37 °C. The red fluorescence of PI (Ex/Em = 550/605 nm) and green fluorescence of Annexin V-FITC (Ex/Em = 490/520 nm) were quantified by flow cytometry and analysed by FLOWJO programs.
For live/dead staining assay, 4T1 cells (5 × 104 cells per well) and L02 cells (5 × 104 cells per well) were seeded in 6-well plates in RPMI-1640 complete medium overnight, followed by treating with Au, Au-NC-siRNA@LNPs, Au-DR-siRNA, and Au-DR-siRNA@LNPs (1000 nM siRNA) for 6 h, respectively. The cells were washed thrice with PBS, cultured for another 48 h, stained with Calcein-AM (1 µl, 1 mg/ml) and Propidium Iodide (PI) (1 µl, 5 mg/ml) for 10 min at 37 °C, and imaged with CLSM. The red fluorescence of PI (Ex/Em = 550/605 nm) and green fluorescence of calcein (Ex/Em = 490/515 nm) were detected.
2.14. Detection of siRNA release by the in vivo fluorescence imaging
The 4T1 tumour-bearing mice were intravenously injected with Au-DR-siRNA, Au-AQ-siRNA@LNPs, and Au-DR-siRNA@LNPs at a dose of siRNA 1OD/25 g body weight. Mice were anesthetized and imaged in the whole body using an in vivo imaging system (Bruker, Germany) with Ex 610/Em 660 at predetermined time points (0 min, 30 min, 1 h, 3 h, 5 h, 7 h, and 9 h). Mice were sacrificed, and the tumors and major organs were harvested and imaged ex vivo. Furthermore, the distributions of Au-DR-siRNA, Au-AQ-siRNA@LNPs, and Au-DR-siRNA@LNPs in tumour tissues were investigated. The tumour tissues were fixed with 4% paraformaldehyde, sliced, stained with FITC anti-CD31 primary antibody for blood vessel and DAPI for nuclei, and imaged with CLSM.
2.15. Au-DR-siRNA@LNPs enhances CT imaging of tumour sites
The 4T1 tumour-bearing mice were randomly divided to 2 groups, and injected with Au-DR-siRNA@LNPs (200 µl, 10 mM) and pure iodine solution (200 µl, 10 mM), respectively. The mice were anesthetized with isoflurane gas, placed in a small animal living CT imager, and then irradiated with 100 keV intensity X-rays. The tumour regions were imaged with a Micro-CT imaging system (PerkinElmer Co., Ltd.) at different time points (15 min, 30 min, 1 h, and 3 h).
2.16. Therapeutic effects of Au-DR-siRNA@LNPs on 4T1 tumour-bearing mice
Male BABL/c mice of 5-weeks age were purchased from Hayes Lake Laboratory Animal Co., Ltd. (Shanghai, China) and housed at 25 °C at 40%−60% humidity, with a 12-h light/dark cycle. Mice were subcutaneously injected with 4T1 cells (100 ul, 106/ml) into the right axilla, and the length (a) and width (b) of the tumour was measured with a vernier calliper. The tumour volume (V) is: V = 0.5 × a × b2. When the tumour grew to 50 mm3, mice were intravenously injected with Au-DR-siRNA@LNPs (0.15 mg/kg) every2 d for 6 times, and their tumour volume and body weight were recorded every other day for 17 d At last, mice were sacrificed, and tumour tissues were collected, weighed, and imaged. In addition, the survival rates of mice treated by PBS, Au-NC-siRNA, and Au-DR-siRNA@LNPs with the above treatment protocol were observed for 60 d, respectively (n = 6).
2.17. Immunohistochemical analysis of tumors and major organs
Tumour tissue sections were stained with hematoxylin and eosin (H&E) and terminal deoxynucleotidyl transferase dUTP nick end labelling (Tunel) for detecting apoptosis and necrosis, respectively. In addition, immunohistochemistry was used to detect survivin expression in tumour tissues. Briefly, tumour tissue sections were incubated in a methanol solution containing 3% H2O2 at 37 °C for 10 min to quench the activity of endogenous peroxidase. After the sections were blocked at room temperature for 20 min, they were incubated with survivin antibody (R&D Systems, Germany) at 4 °C overnight and then incubated with horseradish peroxidase coupled secondary antibody (Dako, Kyoto, Japan) at 37 °C for 30 min. Finally, the signals were detected by Diaminobenzidine Substrate Kit (Vector Laboratories, Burlingame, CA, United States), and the cytoplasm or nucleus brown staining showed positive results. The stained sections were imaged with a bright field microscope (Eclipse E800, Nikon, Japan). Furthermore, the heart, liver, spleen, lung, and kidney were harvested, sectioned, and analysed by H&E staining.
2.18. Statistical analysis
All experiments were repeated at least 3 times with 6–12 biological replicates. Data are represented as mean ± SD. Error bars represent standard deviation for independent samples assayed within the experiments. Statistical analysis was performed with GraphPad Prism 8 software. Two groups of data were analysed by using student's t-test (two-tailed). Over three groups of data were analysed with analysis of variance (ANOVA), followed by the multiple comparison of Tukey. * P < 0.05, ** P < 0.01, and *** P < 0.001.
3. Results and discussion
3.1. Characterization of Au-DR-siRNA@LNPs nanoparticles
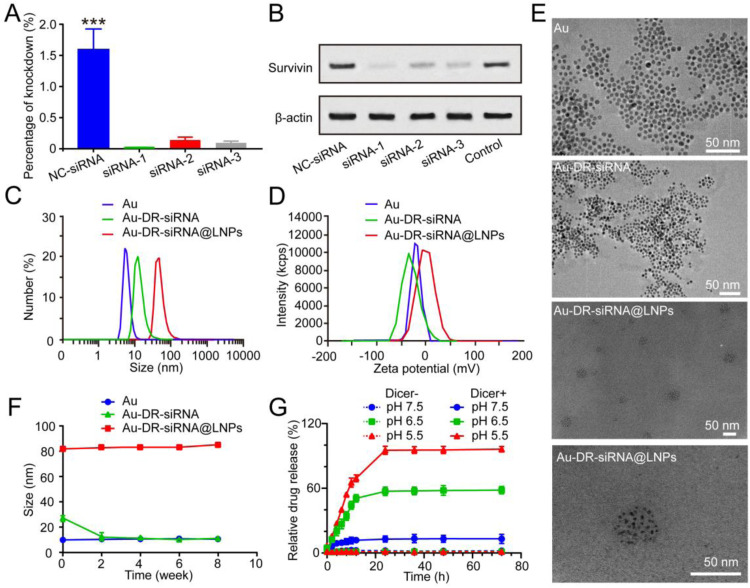
Survivin gene, a member of the inhibitor of apoptosis (IAP) protein family that inhibits caspases and prevents cell death, is highly expressed in most cancers and is associated with poor clinical outcomes [26,27]. It could also affect tumour aggressiveness and cancer patient survival. Compared with normal tissues, the high expression of survivin in cancer cells and its role as a nodal protein in many cellular pathways make it an ideal target for many therapeutic approaches [28]. In this study, we designed and synthesized three siRNA sequences targeting the survivin gene (Tables S1 and S2), transfected them into 4T1 cells with Lipo2000 for 24 h, then extracted the total RNA of the cells for polymerase chain reaction (PCR), and lysed the cells for western blot (WB). PCR and WB results showed that all three siRNAs could significantly inhibit the expression of survivin gene, and the first siRNA had the best gene silencing effect (Fig. 2A and B), which was selected for the subsequent experiments.
Fig. 2.
Preparation and characterization of nanoparticles. (A) Polymerase chain reaction (PCR) map after transfection of 4T1 cells with NC-siRNA, siRNA-1, siRNA-2 and siRNA-3. (B) Western blot (WB) analysis of the changes in survivin expression induced by NC-siRNA, siRNA-1, siRNA-2 and siRNA-3. Particle size distribution (C), zeta potential (D), and TEM images (E) of nanogolds (Au), Au-DR-siRNA, and Au-DR-siRNA@LNPs. (F) The long-term stability of Au, Au-DR-siRNA, Au-DR-siRNA@LNPs at 4℃ for 8 weeks. The results were plotted as mean ± SD (n = 6). (G) Cy5 release curves from Au-DR-siRNA@LNPs in PBS with/without Dicer enzyme at pH 5.5, 6.5, and 7.5 during a 72 h incubation, respectively. The results were plotted as mean ± SD (n = 6). ***P < 0.001. (One-way ANOVA plus Tukey's multiple comparison).
First, we prepared the ultra-small Au according to citrate reduction method. Second, in DR-siRNA, the Cy5 and sulfhydryl group (-SH) were chemically modified at the 3′ end of the antisense strand of the siRNA and the 5′ end of the sense strand, respectively. In AQ-siRNA, both Cy5 and -SH were modified at the 5′-end of the siRNA sense strand (Fig. S1). Third, DR-siRNA was connected to Au to form Au-DR-siRNA, which was further wrapped by LNPs (Au-DR-siRNA@LNPs). Au-DR-siRNA and Au-DR-siRNA@LNPs were also prepared as the control. Then, we investigated the particle size and zeta potential of Au, Au-DR-siRNA and Au-DR-siRNA@LNPs. The results of DLS showed that Au has a particle size of about 5 nm and a zeta potential of about −12 mv. After Au is attached to the DR-siRNA, the particle size increases to about 25 nm, and the zeta potential reaches about −30 mV. The particle size of Au-DR-siRNA@LNPs is about 80 nm with PDI < 0.1, and the zeta potential is nearly neutral (Fig. 2C and D). TEM was used to verify the morphology of Au, Au-DR-siRNA and Au-DR-siRNA@LNPs, which all showed uniform particle sizes and good dispersibility. Importantly, the TEM image of Au-DR-siRNA@LNPs showed that many Au aggregate into clusters, indicating the successful preparation of Au-DR-siRNA@LNPs (Fig. 2E). The UV spectrum of Au showed the characteristic peak at around 520 nm. In addition to this peak, the UV spectrum of Au-DR-siRNA also showed the characteristic peak of siRNA at 260 nm, indicating the successful attachment of siRNA to Au (Fig. S2).
We also investigated the stability of these nanoparticles. The results show that the particle size of Au is very stable at 4 °C, while that of Au-DR-siRNA significantly decreases in one week with an increased PDI. The instability of Au-DR-siRNA may be caused by the siRNA falling off the surface of Au. In contrast, the particle size of Au-DR-siRNA@LNPs kept constant for 6 weeks, indicating their good stability (Fig. 2F). In addition, we evaluated the serum stability of Au, Au-DR-siRNA and Au-DR-siRNA@LNPs. As shown in Fig. S3, the particle sizes of Au and Au-DR-siRNA increase with time, possibly because their negatively charged surfaces would adsorb plasma protein. However, the particle size of Au-DR-siRNA@LNPs did not change during 24 h-incubation in 50% foetal bovine serum at 37℃, possibly because their nearly neutral charge after LNP encapsulation made it have good biostability. DLin-MC3-DMA is a key component in LNP and has a unique pH-dependant charge-variable property: it is positively charged under acidic conditions and promotes cargo release [29]. We next studied the pH responsiveness of Au-DR-siRNA@LNPs to confirm its acid-triggered cargo release capacity. Fig. 2G shows that about 10% of Cy5 is released from Au-DR-siRNA@LNPs after incubation in PBS containing Dicer enzyme at pH 7.5 for 72 h, whereas the drug release rate increases to 56% at pH 6.5, and nearly to 100% at pH 5.5. By contrast, in the absence of Dicer enzyme, the release rate of Cy5 from Au-DR-siRNA@LNPs did not exceed 10% at any pH level, and there was no significant difference. This is because Au-DR-siRNA released from Au-DR-siRNA@LNPs could not cleave cy5 from nanogolds without Dicer enzyme, and the cy5 fluorescence signal was difficult to detect in drug release experiments. However, in the presence of Dicer enzyme, the DR-siRNA moved away from the nanogolds and resulted in the recovery of cy5 fluorescence. Therefore, in the presence of Dicer enzyme, the release of siRNA is pH-dependant. These results indicated the dual acid responsive and Dicer enzyme responsive drug release function of Au-DR-siRNA@LNPs.
3.2. Dicer enzyme responsive fluorescence recovery
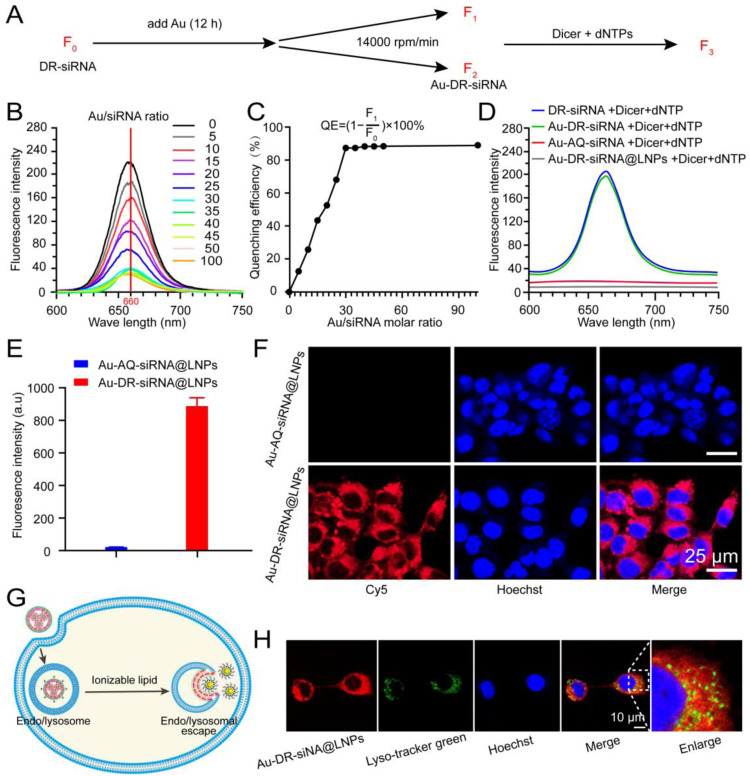
We investigated the fluorescence quenching and recovery of Au-DR-siRNA@LNPs based on the FRET principle. DR-siRNA was reacted with Au for 12 h, and centrifuged at 14,000 rpm/min for 30 min to obtain Au-DR-siRNA, which was incubated with Dicer enzyme and dNTPs to get the final product. By detecting the fluorescence of DR-siRNA (F0), supernatant (F1), Au-DR-siRNA (F2), and the final product (F3), we calculated the Dicer-responsive fluorescence recovery ability of Au-DR-siRNA@LNPs (Fig. 3A).
Fig. 3.
The screen of the feed ratio and the fluorescence recovery function of LNPs under Dicer enzyme. (A) Schematic for calculating the fluorescence quenching efficiency of Au-DR-siRNA. (B) Fluorescence spectrum of the supernatant obtained from Au and DR-siRNA at different feed ratios. (C) Fluorescence quenching efficiency of Au and DR-siRNA at different feed ratios. (D) Fluorescence spectrum of the supernatant after the incubation of Dicer with DR-siRNA, Au-DR-siRNA, Au-AQ-siRNA, and Au-DR-siRNA@LNPs for 5 min, respectively. (E) Fluorescence quantification of 4T1 cells treated with Au-AQ-siRNA@LNPs and Au-DR-siRNA@LNPs. The results were plotted as mean ± SD (n = 6). (F) CLSM images of 4T1 cells treated Au-AQ-siRNA@LNPs and Au-DR-siRNA@LNPs. Scale bar: 25 µm. (G) Schematic of lysosomal escape of Au-DR-siRNA@LNPs in tumour cells. (H) CLSM images of 4T1 cells indicating the lysosomal escape of Au-DR-siRNA@LNPs. Scale bar: 10 µm.
Firstly, different amounts of Au were reacted with 0.2 OD DR-siRNA at a molar ratio of 0, 5, 10, 15, 20, 25, 30, 35, 40, 45, 50 and 100, respectively. Fig. 3B and C show that when the molar ratio is 30, the fluorescence value of the supernatant is the smallest and reaches the plateau phase, and the fluorescence quenching efficiency is the highest (about 90%). However, the fluorescence value of Au-DR-siRNA was almost zero. This indicated that the molar ratio of 30 was the best choice, which we adopted as the feeding ratio of subsequent experiments. The ribozymes in the blood are similar to Dicer enzymes in the cytoplasm, being able to recognize the tail of siRNA and cut it. Hence, the wrapping of Au-DR-siRNA with LNP could also protect Au-DR-siRNA from being cut in the blood. We added Dicer enzyme and dNTPs to Au-DR-siRNA, Au-AQ-siRNA, and Au-DR-siRNA@LNPs, and measured the fluorescence value after 5 min. Fig. 3D shows that the fluorescence of Au-AQ-siRNA was not restored, while that of Au-DR-siRNA was significantly restored to be equivalent to DR-siRNA, demonstrating the function of Dicer responsive fluorescence recovery. Interestingly, the fluorescence of Au-DR-siRNA@LNPs did not recover, indicating that the LNP wrapping would hinder the Dicer enzyme from recognizing and shearing Au-DR-siRNA outside the cells.
To investigate whether Au-DR-siRNA@LNPs retains the Dicer-responsive fluorescence recovery function in tumour cells, we co-cultured 4T1 cells with Au-DR-siRNA@LNPs for 4 h and characterized them by flow cytometry and CLSM. The results showed that Au-AQ-siRNA@LNPs showed no fluorescence in 4T1 cells with high Dicer expression, while Au-DR-siRNA@LNPs displayed significant fluorescence in cells, indicating the function of Dicer-responsive fluorescence recovery (Fig. 3E and F). In addition, efficient lysosomal escape is a crucial ability for the delivery system of siRNA drugs. Many studies have shown that the ionizable lipid DLin-MC3-DMA plays a key role in the lysosomal escape of LNPs. DLin-MC3-DMA is composed of two unsaturated acyl chains and an amino function exhibiting a pKa between 6.2 and 6.4 and, therefore, is positively charged inside the endosome (pH ∼5) [30,31]. These characteristics result in association of DLin-MC3-DMA from the LNP with anionic lipids from the endosomal membrane, leading to endosomal membrane disruption (from bilayer structure to hexagonal structure) and endosomal siRNA release into cytoplasm (Fig. 3G) [32]. Therefore, we studied the lysosomal escape behaviour of Au-DR-siRNA and Au-DR-siRNA@LNPs in tumour cells. Fig. S4 shows that Au-DR-siRNA was accumulated in lysosomes, and a high overlap (yellow fluorescence) of Cy5 red fluorescence with Lyso-Tracker green fluorescence was observed after 4 h of incubation. By contrast, the CLSM images of Au-DR-siRNA@LNPs showed that the Cy5 fluorescence was distributed in the whole cytoplasm, indicating that the siRNA had escaped from lysosomes and been cleaved by Dicer enzyme (Fig. 3H).
3.3. In vitro antitumor effects of Au-DR-siRNA@LNPs
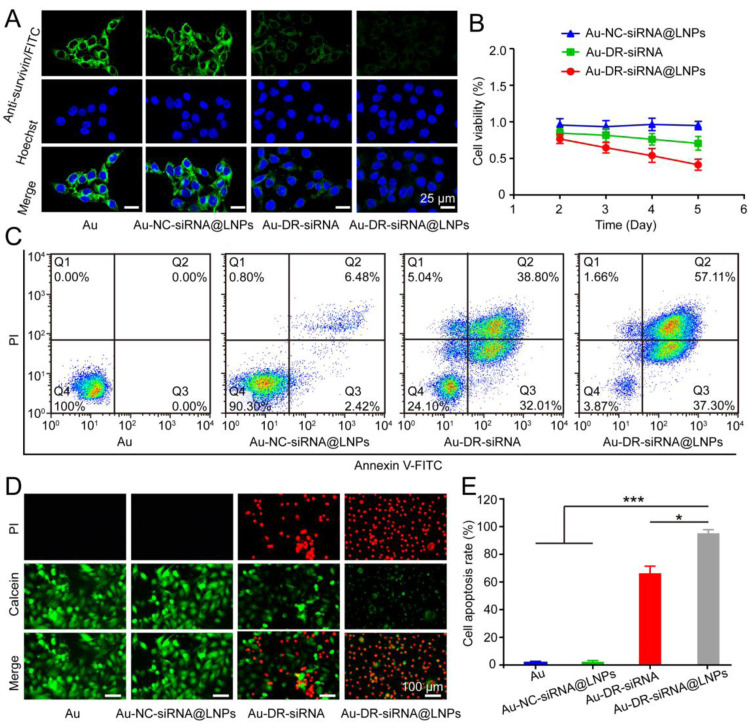
We continued to investigate the in vitro antitumor effect of Au, Au-NC-siRNA, Au-DR-siRNA and Au-DR-siRNA@LNPs. First, we used Anti-survivin/FTIC to examine whether Au-DR-siRNA@LNPs could deliver siRNA into 4T1 cells and silence survivin gene. Fig. 4A shows that Au-DR-siRNA@LNPs-treated 4T1 cells have the weakest green fluorescence compared with the other three control groups, indicating the successful survivin gene silencing by Au-DR-siRNA@LNPs. In addition, to investigate the cytotoxicity of LNPs on 4T1 cells, we incubated Au-DR-siRNA@LNPs (500 nM siRNA) with 4T1 cells for 6 h on the first day followed by the replacement of a new medium, and measured the cell viability after 2, 3, 4, 5 d using CCK8 kit. Fig. 4B shows that Au-NC-siRNA@LNPs have almost no effect on the 4T1 cell viability, whereas Au-DR-siRNA@LNPs can significantly decreased it, which is better than Au-DR-siRNA. The results indicate that Au-DR-siRNA@LNPs can effectively enter tumour cells, release siRNA drugs, and exert anti-tumour effects. To further study apoptosis, 4T1 cells were incubated with Au-DR-siRNA@LNPs for 6 h followed by the replacement of a new medium, cultured for 48 h, double-stained with Annexin V-FITC/PI, and measured of apoptosis by flow cytometry. The apoptosis rate of Au-DR-siRNA@LNPs treated 4T1 cells was 96.07%, which was significantly higher than Au-DR-siRNA (75.85%) and Au-NC-siRNA@LNPs (9.70%) (Fig. 4C). Meanwhile, a similar trend was observed by the staining of live/dead cells by calcein and PI, demonstrating the enhanced inhibitory effect of Au-DR-siRNA@LNPs on tumour cells than Au-DR-siRNA and Au-NC-siRNA@LNPs (Fig. 4D and E). Interestingly, Au-DR-siRNA showed good effect of cellular uptake, apoptosis, and tumour cell inhibition, indicating that the conjugation of siRNA to Au could increase the cellular uptake of siRNA and produce effective gene silencing effect.
Fig. 4.
Au-DR-siRNA@LNPs inhibit the proliferation of 4T1 cells. (A) Confocal images of survivin protein expression in 4T1 cells treated with Au, Au-NC-siRNA@LNPs, Au-DR-siRNA, and Au-DR-siRNA@LNPs at 37℃. Scale bar: 25 µm. (B) Cell viability of 4T1 cells treated by Au-NC-siRNA@LNPs, Au-DR-siRNA, and Au-DR-siRNA@LNPs. The results were plotted as mean ± SD (n = 6). (C) Annexin V-FITC/PI flow cytometry analysis of 4T1 cells treated with Au, Au-NC-siRNA@LNPs, Au-DR-siRNA, and Au-DR-siRNA@LNPs. (d) Apoptosis of 4T1 cells treated with Au, Au-NC-siRNA@LNPs, Au-DR-siRNA, and Au-DR-siRNA@LNPs. Scale bar: 100 µm. (E) Quantification of cell apoptosis detected by flow cytometry. The data are presented as mean ± SD (n = 6). *P < 0.05, ***P < 0.001 (One-way ANOVA plus Tukey's multiple comparison).
3.4. In vivo fluorescence imaging and CT imaging in 4T1 tumour-bearing mice
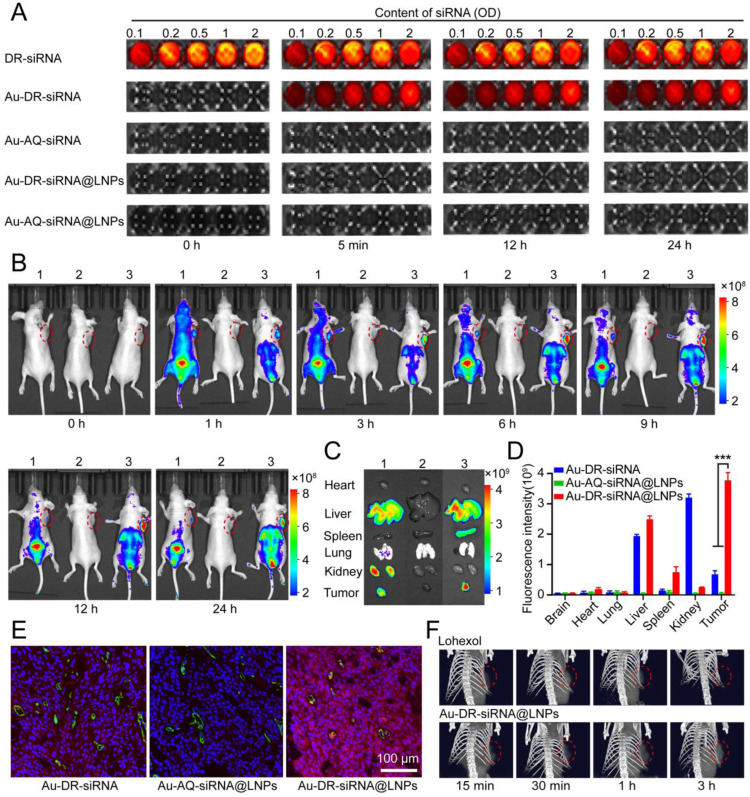
The ribozymes in the blood are similar to Dicer enzymes in the cytoplasm, being able to recognize the tail of siRNA and cut it [33,34]. To investigate whether the LNPs wrapping increases the stability of Au-DR-siRNA in the blood, we added DR-siRNA, Au-DR-siRNA, Au-AQ-siRNA, Au-DR-siRNA@LNPs, and Au-AQ-siRNA@LNPs in a 96-well plate, respectively; 50 µl fresh blood was added to each well; the fluorescence recovery was examined with the in vivo imager at different time points. Fig. 5A shows that the fluorescence of Au-AQ-siRNA and Au-AQ-siRNA@LNPs was always in a quenched state, while the fluorescence of Au-DR-siRNA recovered at 5 min, and that of Au-DR-siRNA@LNPs did not recover until 24 h. This indicates that the naked Au-DR-siRNA is easily cleaved by ribozymes in the blood, and the blood stability of Au-AQ-siRNA@LNPs is greatly enhanced, being suitable for in vivo application.
Fig. 5.
Fluorescence imaging and CT imaging of Au-DR-siRNA@LNPs. (A) Fluorescence images of DR-siRNA, Au-DR-siRNA, Au-AQ-siRNA, Au-DR-siRNA@LNPs, and Au-AQ-siRNA@LNPs containing different concentrations of siRNA incubated with blood containing ribozymes for different times. (B) In vivo fluorescence images of 4T1 tumour-bearing mice at different time points after intravenous injection of Au-DR-siRNA (1), Au-AQ-siRNA@LNPs (2), and Au-DR-siRNA@LNPs (3). Red dotted circle: tumour. (C) Ex vivo fluorescence images of major organs and tumors. (D) Ex vivo fluorescence intensity of major organs and tumors. (E) CLSM images of tumour tissue sections treated with Au-DR-siRNA, Au-AQ-siRNA@LNPs and Au-DR-siRNA@LNPs. Green: CD31, red:Cy5, blue: hoechst 33,342. Scale bar: 100 µm. (F) CT imaging images of 4T1 tumour-bearing mice at different time points after intravenous injection of Au-DR-siRNA@LNPs and iohexol. The data are presented as mean ± SD (n = 6). ***P < 0.001 (One-way ANOVA analysis with Tukey's post-hoc test).
We further investigated the tumour-targeting ability of Au-DR-siRNA@LNPs in vivo and the release site and time of siRNA. The 4T1 tumour-bearing nude mice were intravenously injected with Au-DR-siRNA, Au-AQ-siRNA@LNPs, and Au-DR-siRNA@LNPs, respectively. Fluorescence images of the whole body and the isolated organs were taken with maestro EX fluorescence imaging system instrument at a series of time points (0, 1, 3, 6, 9, 12 and 24 h). Fig. 5B shows that Au-AQ-siRNA@LNPs were non-fluorescent in vivo in the whole process, indicating that the Cy5 fluorescent molecule was in a quenched state. One hour after injection of Au-DR-siRNA nanoparticles, the fluorescence had spread all over the body, indicating that the siRNA had been cleaved by ribozymes in the blood. After that, a large amount of fluorescence was distributed in the kidney and bladder, proving that Cy5 had been eliminated through the kidney. By contrast, after injection of Au-DR-siRNA@LNPs, the fluorescence was concentrated in the tumour, liver and bladder, rather than distributed throughout the body. In addition, the tumors and major organs were harvested and imaged after 24 h injection. Fig. 5C and D show that Au-DR-siRNA nanoparticles displayed significant fluorescence in the liver and kidney, weak fluorescence in the tumour. In contrast, Au-DR-siRNA@LNPs showed significant fluorescence in the tumour, and decreased fluorescence in the kidney. Importantly, the CLSM images of tumour tissue sections showed that the red fluorescence of Au-DR-siRNA@LNPs was distributed inside all cells of tumour tissue, suggesting the siRNA had been cleaved by Dicer enzyme in the cytoplasm (Fig. 5E). These results indicated that the LNP wrapping protected the naked Au-DR-siRNA from being cleaved by ribozymes in the blood, and promoted its delivery into the tumour cells. Importantly, the siRNA release behaviour of Au-DR-siRNA@LNPs could be monitored by the in vivo fluorescence imaging system.
Many studies have shown that Au can enhance the effect and time window of computed tomography (CT) imaging [35,36], hence Au-DR-siRNA@LNPs may have the application potential in CT imaging. To investigate the CT contrast effect in tumors, we intravenously injected iohexol and Au-DR-siRNA@LNPs into 4T1 tumour-bearing mice, respectively, and irradiated the chest with 100 Kev intensity X-rays at different time points (15 min, 30 min, 1 h and 3 h). Fig. 5F shows that Au-DR-siRNA@LNPs could be quickly delivered to the tumour site within 15 min and kept the CT signal for 3 h, while the CT signal of iohexol in tumour was relatively weak and almost disappeared at 3 h, indicating that Au-DR-siRNA@LNPs had better CT imaging function and could be used for tumour diagnosis and monitoring.
In brief, in vivo fluorescence imaging and CT imaging demonstrated that the liposome membrane in Au-DR-siRNA@LNPs can protect siRNA from degradation by nucleases in the blood and effectively deliver to the tumour site. In addition, gold nanoparticles and cy5 fluorescence Molecules can be excreted from the body by metabolism, indicating that Au-DR-siRNA@LNPs has high biological safety and can be used for tumour diagnosis and monitoring.
3.5. Therapeutic effect of Au-DR-siRNA@LNPs on 4T1 tumour-bearing nude mice
The in vivo fluorescence imaging and CT imaging of Au-DR-siRNA@LNPs demonstrated that it could not only monitor the release behaviour of siRNA in vivo but also be used for tumour diagnosis. We further investigated the antitumor effect and mechanism of Au-DR-siRNA@LNPs on 4T1 tumour-bearing nude mice. The mice were randomly divided into 3 groups and intravenously injected with PBS, Au-DR-siRNA@LNPs, and Au-NC-siRNA@LNPs (0.15 mg/kg) every 2 d for 6 times, respectively, and their tumour volumes and body weights were measured for 17 d After that, the mice were sacrificed, and tumour tissues were collected, weighted, imaged, sectioned, and analysed by immunohistochemistry. In addition, major organs were collected, sectioned, and analysed by hematoxylin and eosin (H&E) staining. At last, we investigated the survival rate of mice and monitored for 60 d
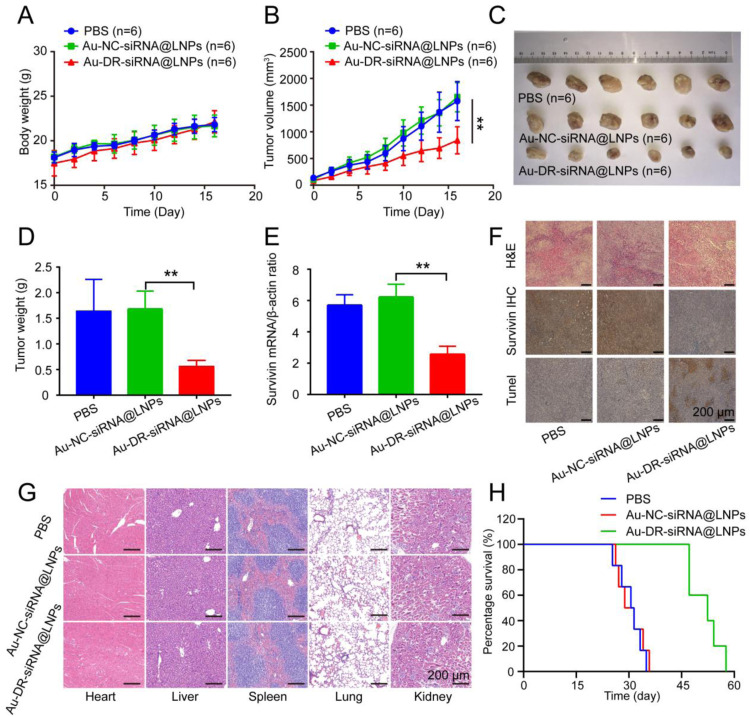
The results showed that the body weights of the mice in all the three groups gradually increased with time (Fig. 6A). The tumour volumes of mice in PBS and Au-NC-siRNA@LNPs groups increased significantly over time with similar growth curves, while Au-DR-siRNA@LNPs had a significantly enhanced inhibitory effect on tumour growth (Fig. 6B). Au-DR-siRNA@LNPs show a tumour suppression rate of 70.59%, compared with the other two groups (Fig. 6C and D), indicating its potent antitumor effect in vivo. The PCR results showed that the survivin gene expression in the tumour tissues of Au-DR-siRNA@LNPs group was significantly lower than that of PBS and Au-NC-siRNA@LNPs groups, indicating that the siRNA in Au-DR-siRNA@LNPs could effectively silence the survivin gene and achieve anti-tumour effects (Fig. 6E). Importantly, to further evaluate the antitumor effects of Au-DR-siRNA@LNPs in vivo, immunohistochemical staining of tumour tissues was performed, including H&E, survivin, TdT-dependant dUTP-biotin gap end labelling (Tunel). H&E staining showed that the tumour tissues treated with PBS and Au-NC-siRNA@LNPs displayed a high nuclear-to-cytoplasmic ratio, accumulated and grew tumour cells, and angiogenesis. However, the tumour tissue treated with Au-DR-siRNA@LNPs had a relatively low nucleus and large areas of necrosis, indicating the better anti-tumour effect. Immunohistochemical staining showed that the brown area of the tumour tissue in PBS and Au-NC-siRNA@LNPs groups was broad, while that in Au-DR-siRNA@LNPs group had almost no brown area, indicating that Au-DR-siRNA@LNPs could inhibit survivin protein expression. Tunel staining showed that Au-DR-siRNA@LNPs treated tumour tissue had significantly larger brown areas than the other two groups, indicating the better effect of tumour apoptosis (Fig. 6F). Importantly, H&E staining showed no tissue damages in the heart, liver, spleen, lung, and kidney, indicating the good biosafety of Au-DR-siRNA@LNPs (Fig. 6G). In addition, the median survival times for PBS, Au-NC-siRNA@LNPs, and Au-DR-siRNA@LNPs groups were 29, 31, and 50 d, respectively, indicating that Au-DR-siRNA@LNPs can significantly prolong the survival of tumour-bearing mice (Fig. 6H). Therefore, Au-DR-siRNA@LNPs is an effective nucleic acid drug delivery system with good biocompatibility, biosafety, and enhanced antitumor efficacy.
Fig. 6.
Antitumor effect of 4T1 tumour-bearing mice after tail vein administration of nanoparticles. (A) Body weights of 4T1 tumour-bearing mice treated with PBS, Au-NC-siRNA@LNPs, and Au-DR-siRNA@LNPs for 17 d (B) tumour volume of 4T1 tumour-bearing mice treated with PBS, Au-NC-siRNA@LNPs, and Au-DR-siRNA@LNPs for 17 d (C) Representative images of tumors after 17-d treatment with PBS, Au-NC-siRNA@LNPs, and Au-DR-siRNA@LNPs. (D) Average weights of tumors after 17-d treatments. (E) RT-PCR quantification of survivin gene expression in tumour tissues after 17-d treatments. (F) H&E, survivin immunohistochemical, and Tunel staining of tumour tissue sections in 4T1 tumour-bearing mice after 17-d treatments. Scale bar: 200 µm. (G) H&E histopathological analysis of major organ tissues in 4T1 tumour-bearing mice after 17-d treatments. Scale bar: 200 µm. (H) Survival curves of 4T1 tumour-bearing mice treated with PBS, Au-NC-siRNA@LNPs, and Au-DR-siRNA@LNPs. The data are presented as mean ± SD (n = 6). **P < 0.01 (One-way ANOVA analysis with Tukey's post-hoc test).
4. Conclusion
The ribozyme in the blood and the dicer enzyme in the cytoplasm can cleave the siRNA and make it fall off the surface of Au, and both can use the FRET principle to excite fluorescence. In order to protect siRNA from being degraded by ribozymes in blood, Cy5 has been in a fluorescence quenching state in blood circulation. We prepared stable nucleic acid liposomes to encapsulate Au-DR-siRNA to obtain Au-DR-siRNA@LNPs for fluorescence/CT dual-mode imaging of tumors. In vitro experiments, the nanoparticles can not only detect siRNA in living cells, but also effectively silence the expression of survivin protein in tumour cells and inhibit the growth of tumour cells. Currently, the main target organ of LNPs after intravenous administration is the liver. The results of in vivo tumour-bearing animal models show that LNPs can also selectively enter tumour tissues and be taken up by tumour cells, thereby realizing fluorescence imaging and CT imaging. It can be explained from two perspectives: the PEG on the surface of LNPs does not fall off completely and quickly, and the LNPs can enter the tumour through the EPR effect before falling off [37,38]; or even after falling off, LNPs may adsorb other plasma proteins except ApoE, so that part of it entered the tumour tissue [10]. In addition, Au-DR-siRNA@LNPs have potent antitumor effect and good biocompatibility and biosafety. However, the Au-DR-siRNA@LNPs in this study lacked active targeting to tumour tissue. Next, we will adopt new methods to solve this problem, such as adding targeting peptides, designing and utilizing protein coronas, developing targeted phospholipids, etc.
In summary, this work provided not only an effective method for monitoring the pharmacokinetic behaviour of LNPs-based siRNA, but also a siRNA delivery system for treating and diagnosing tumors, which would promote the further development of siRNA drugs.
Conflicts of interest
The authors have declared that no competing interest exists.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81872812, 82073800), the China Postdoctoral Science Fundation (2021TQ0111, 2021M691040).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ajps.2022.11.003.
Contributor Information
Shulei Zhu, Email: slzhu@chem.ecnu.edu.cn.
Zhiqiang Yan, Email: zqyan@sat.ecnu.edu.cn.
Appendix. Supplementary materials
References
- 1.Akinc A., Maier M., Manoharan M., Fitzgerald K., Jayaraman M., Barros S., et al. The onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat Nanotechnol. 2019;14(12):1084–1087. doi: 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- 2.Zhu K.Y., Palli S. Applications, and challenges of insect RNA interference. Annu Rev Entomol. 2020;65(1):293–311. doi: 10.1146/annurev-ento-011019-025224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J., Shen H., Xu J., Liu L., Tan J., Li M., et al. Liver-targeted siRNA lipid nanoparticles treat hepatic cirrhosis by dual antifibrotic and anti-inflammatory activities. ACS Nano. 2020;14(5):6305–6322. doi: 10.1021/acsnano.0c02633. [DOI] [PubMed] [Google Scholar]
- 4.Bernards R. Exploring the uses of RNAi — Gene knockdown and the nobel prize. N Engl J Med. 2006;355(23):2391–2393. doi: 10.1056/NEJMp068242. [DOI] [PubMed] [Google Scholar]
- 5.Zou Y., Sun X., Wang Y., Yan C., Liu Y., Li J., et al. Single sirna nanocapsules for effective sirna brain delivery and glioblastoma treatment. Adv Mater. 2020;32(24) doi: 10.1002/adma.202000416. [DOI] [PubMed] [Google Scholar]
- 6.Overhoff M., Wünsche W., Sczakiel G. Quantitative detection of siRNA and single-stranded oligonucleotides: relationship between uptake and biological activity of siRNA. Nucleic Acids Res. 2004;32(21):e170. doi: 10.1093/nar/gnh168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miao L., Zhang Y., Huang L. mRNA vaccine for cancer immunotherapy. Mol Cancer. 2021;20(1):41. doi: 10.1186/s12943-021-01335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou J.E., Sun L., Liu L., Jia Y., Han Y., Shao J., et al. Hepatic macrophage targeted siRNA lipid nanoparticles treat non-alcoholic steatohepatitis. J Control Release. 2022;343:175–186. doi: 10.1016/j.jconrel.2022.01.038. [DOI] [PubMed] [Google Scholar]
- 9.Miao L., Lin J., Huang Y., Li L., Delcassian D., Ge Y., et al. Synergistic lipid compositions for albumin receptor mediated delivery of mRNA to the liver. Nat Commun. 2020;11(1):2424. doi: 10.1038/s41467-020-16248-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sebastiani F., Yanez Arteta M., Lerche M., Porcar L., Lang C., Bragg R., et al. Apolipoprotein E binding drives structural and compositional rearrangement of mrna-containing lipid nanoparticles. ACS Nano. 2021;15(4):6709–6722. doi: 10.1021/acsnano.0c10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong Y., Siegwart D.J., Anderson ADG. Strategies, design, and chemistry in siRNA delivery systems. Adv Drug Deliv Rev. 2019;144:133–147. doi: 10.1016/j.addr.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X., Yang H.Y., Giachelli C.M. Role of the sodium-dependent phosphate cotransporter, pit-1, in vascular smooth muscle cell calcification. Circ Res. 2006;98(7):905–912. doi: 10.1161/01.RES.0000216409.20863.e7. [DOI] [PubMed] [Google Scholar]
- 13.Sledz C.A., Williams B.R.G. RNA interference and double-stranded-RNA-activated pathways. Biochem Soc Trans. 2004;32(6):952–956. doi: 10.1042/BST0320952. [DOI] [PubMed] [Google Scholar]
- 14.Kawamata T., Tomari Y. Making RISC. Trends Biochem Sci. 2010;35(7):368–376. doi: 10.1016/j.tibs.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 15.Revia R.A., Stephen Z.R., Zhang M. Theranostic nanoparticles for RNA-based cancer treatment. Acc Chem Res. 2019;52(6):1496–1506. doi: 10.1021/acs.accounts.9b00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Z., Wang J., Cheng H., Ke X., Sun L., Zhang Q.C., et al. Cryo-EM structure of human dicer and its complexes with a pre-mirna substrate. Cell. 2018;173(5) doi: 10.1016/j.cell.2018.03.080. 1191-203.e12. [DOI] [PubMed] [Google Scholar]
- 17.Algar W.R., Hildebrandt N., Vogel S.S., Medintz I.L. FRET as a biomolecular research tool — understanding its potential while avoiding pitfalls. Nat Methods. 2019;16(9):815–829. doi: 10.1038/s41592-019-0530-8. [DOI] [PubMed] [Google Scholar]
- 18.Chen T., He B., Tao J., He Y., Deng H., Wang X., et al. Application of förster resonance energy transfer (FRET) technique to elucidate intracellular and in vivo biofate of nanomedicines. Adv Drug Deliv Rev. 2019;143:177–205. doi: 10.1016/j.addr.2019.04.009. [DOI] [PubMed] [Google Scholar]
- 19.Baibakov M., Patra S., Claude J.B., Moreau A., Lumeau J., Wenger J. Extending single-molecule förster resonance energy transfer (FRET) range beyond 10 nanometers in zero-mode waveguides. ACS Nano. 2019;13(7):8469–8480. doi: 10.1021/acsnano.9b04378. [DOI] [PubMed] [Google Scholar]
- 20.Laskaratou D., Fernández G.S., Coucke Q., Fron E., Rocha S., Hofkens J., et al. Quantification of FRET-induced angular displacement by monitoring sensitized acceptor anisotropy using a dim fluorescent donor. Nat Commun. 2021;12(1):2541. doi: 10.1038/s41467-021-22816-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma F., Wang Q., Xu Q., Zhang C. Self-assembly of superquenched gold nanoparticle nanosensors for lighting up BACE-1 in live cells. Anal Chem. 2021;93(45):15124–15132. doi: 10.1021/acs.analchem.1c03430. [DOI] [PubMed] [Google Scholar]
- 22.Gasparri A.M., Sacchi A., Basso V., Cortesi F., Freschi M., Rrapaj E., et al. Boosting interleukin-12 antitumor activity and synergism with immunotherapy by targeted delivery with isodgr-tagged nanogold. Small. 2019;15(45) doi: 10.1002/smll.201903462. [DOI] [PubMed] [Google Scholar]
- 23.Perets N., Betzer O., Shapira R., Brenstein S., Angel A., Sadan T., et al. Golden exosomes selectively target brain pathologies in neurodegenerative and neurodevelopmental disorders. Nano Lett. 2019;19(6):3422–3431. doi: 10.1021/acs.nanolett.8b04148. [DOI] [PubMed] [Google Scholar]
- 24.Sun C., Gradzielski M. Fluorescence sensing of cyanide anions based on Au-modified upconversion nanoassemblies. Analyst. 2021;146(7):2152–2159. doi: 10.1039/d0an01954b. [DOI] [PubMed] [Google Scholar]
- 25.Akashi H., Matsumoto S., Taira K. Gene discovery by ribozyme and siRNA libraries. Nat Rev Mol Cell Biol. 2005;6(5):413–422. doi: 10.1038/nrm1646. [DOI] [PubMed] [Google Scholar]
- 26.Li F., Aljahdali I., Ling X. Cancer therapeutics using survivin BIRC5 as a target: what can we do after over two decades of study? J Exp Clin Cancer Res. 2019;38(1):368. doi: 10.1186/s13046-019-1362-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelly R.J., Lopez-Chavez A., Citrin D., Janik J.E., Morris J.C. Impacting tumor cell-fate by targeting the inhibitor of apoptosis protein survivin. Mol Cancer. 2011;10(1):35. doi: 10.1186/1476-4598-10-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim W., Ryu J., Kim J.E. CCAR2/DBC1 and Hsp60 positively regulate expression of survivin in neuroblastoma cells. Int J Mol Sci. 2019;20(1):131. doi: 10.3390/ijms20010131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jayaraman M., Ansell S.M., Mui B.L., Tam Y.K., Chen J., Du X., et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew Chem Int Ed Engl. 2012;51(34):8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biscans A., Ly S., McHugh N., Cooper D.A., Khvorova A. Engineered ionizable lipid siRNA conjugates enhance endosomal escape but induce toxicity in vivo. J Control Release. 2022;349:831–843. doi: 10.1016/j.jconrel.2022.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karam M., Daoud G. mRNA vaccines: past, present, future. Asian J Pharm Sci. 2022;17(4):491–522. doi: 10.1016/j.ajps.2022.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Semple S.C., Akinc A., Chen J., Sandhu A.P., Mui B.L., Cho C.K., et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28(2):172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 33.Hoehener C., Hug I., Nowacki M. Dicer-like enzymes with sequence cleavage preferences. Cell. 2018;173(1) doi: 10.1016/j.cell.2018.02.029. 234-47.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rouge J.L., Sita T.L., Hao L., Kouri F.M., Briley W.E., Stegh A.H., et al. Ribozyme–spherical nucleic acids. J Am Chem Soc. 2015;137(33):10528–10531. doi: 10.1021/jacs.5b07104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun G., Wang T., Li X., Li D., Peng Y., Wang X., et al. Sub-micrometer Au@PDA-125I particles as theranostic embolism beads for radiosensitization and spect/ct monitoring. Adv Healthc Mater. 2018;7(16) doi: 10.1002/adhm.201800375. [DOI] [PubMed] [Google Scholar]
- 36.Kush P., Kumar P., Singh R., Kaushik A. Aspects of high-performance and bio-acceptable magnetic nanoparticles for biomedical application. Asian J Pharm Sci. 2021;16(6):704–737. doi: 10.1016/j.ajps.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mui B.L., Tam Y.K., Jayaraman M., Ansell S.M., Du X., Tam Y.Y., et al. Influence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of sirna lipid nanoparticles. Mol Ther Nucleic Acids. 2013;2(12):e139. doi: 10.1038/mtna.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki T., Suzuki Y., Hihara T., Kubara K., Kondo K., Hyodo K., et al. PEG shedding-rate-dependent blood clearance of PEGylated lipid nanoparticles in mice: faster PEG shedding attenuates anti-PEG IgM production. Int J Pharm. 2020;588 doi: 10.1016/j.ijpharm.2020.119792. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.