Abstract
The species of the genus Juniperus L. play an important role in Kazakhstan forest ecosystems and one of them is Juniperus seravschanica Kom. which has been listed as a rare species in the Red Book of Kazakhstan. The distribution area of J. seravschanica extends from Central Asia (Kazakhstan, Uzbekistan, Kyrgyzstan, Tajikistan, and Turkmenistan) to northern and eastern Afghanistan, northern Pakistan, Kashmir, southeastern Iran, and Oman. J. seravschanica occurred in the southern part of Kazakhstan along with the ranges Karatau, Talas Alatau, Kyrgyz Alatau, Chu-Ili, Karzhantau, and Ugam. The distribution area of J. seravschanica is constantly decreasing due to intensive logging, forest fires, and excessive cattle grazing. The species has ecological importance in the stabilization of mountain slopes against erosion, for hydrobiological regulation, and as a significant medicinal herb. The species J. excelsa M. Bieb., J. polycarpos K.Koch (var. polycarpos and var. turcomanica R.P.Adams), and J. seravschanica are morphologically very similar with some difficulties in species identification. For a better understanding of the evolutionary relationship of these species in the Juniperus genus, it is important to obtain genetic information on the highly conserved chloroplast (cp) genome. Due to the conserved genomic structure, the cp genome nucleotide sequences are widely used in species distinguishing and reconstructing phylogenetic relationships. Unfortunately, there are no publicly available nucleotide sequences of cp genomes data for J. polycarpos (var. polycarpos and var. turcomanica), J. excelsa and J. seravschanica. We report the de novo assembly of the J. seravschanica chloroplast genome by applying next-generation sequencing technology based on Illumina NovaSeq 6000. The assembled cp genome of J. seravschanica is 127,609 bp in length and contained 118 genes, including 82 protein-coding genes, 32 transfer RNA genes, and 4 ribosomal RNA genes. In total 152 simple sequence repeats were identified in the chloroplast genome sequence of J. seravschanica. The Bioproject (PRJNA883033), Sequence Read Archive (SRR21673293), and GenBank (OL684343) data were deposited at National Center for Biotechnology Information.
Keywords: Cupressaceae, Juniperus seravschanica, Rare species, Illumina sequencing, Chloroplast genome, De novo assembly, Chloroplast SSRs
Specifications Table
| Subject | Omics: Genomics |
| Specific subject area | Genomics, Forest ecosystem, Environmental science |
| Type of data | Tables, Figure |
| How the data were acquired | The data were acquired using the Illumina NovaSeq 6000 (San Diego, USA) sequencer and assembled with SPAdes v. 3.13.0 |
| Data format | Raw data (fastq) and analyzed data (fasta) |
| Description of data collection | The fresh leaves of J. seravschanica were collected from the Turkistan region of Southern Kazakhstan and desiccated in silica gel. Total DNA was isolated from the leaves using the CTAB protocol [1]. The concentration and quality of the extracted DNA were checked by gel electrophoresis in agarose and Nanodrop 2000 spectrophotometry (Thermo Fisher Scientific, Wilmington, DE, USA). Paired-end sequencing was performed using NovaSeq 6000 platform (Illumina, San Diego, CA, USA). |
| Data source location |
|
| Data accessibility | Repository name: National Center for Biotechnology Information Raw data are available in the Sequence Read Archive (SRA) under BioProject PRJNA883033 with SRA number SRR21673293. The complete chloroplast genome is available under accession number OL684343 Direct URL to data: https://www.ncbi.nlm.nih.gov/sra/PRJNA883033 (SRA) https://www.ncbi.nlm.nih.gov/nuccore/OL684343 (Nucleotide) |
Value of the Data
-
•
The newly sequenced chloroplast genome data of J. seravschanica can be useful in plant molecular identification and evaluating phylogenetic relationships at the Juniperus genus level.
-
•
Researchers in molecular botany, genomics, and bioinformatics will benefit from these data.
-
•
The detected simple sequence repeats can be used in the development of potentially useful molecular markers and evaluation of genetic diversity in J. seravschanica populations and closely related species.
1. Objective
Chloroplast genome data can be used in species distinguishing and reconstructing plant evolutionary relationships due to the highly conserved genome structure. There are some difficulties in species identification for morphologically very similar Juniperus species J. excelsa, J. polycarpos (var. polycarpos and var. turcomanica), and J. seravschanica. Unfortunately, presently there are no publicly available nucleotide sequences of cp genomes data for these listed species. In the present study, we report de novo assembled data of the J. seravschanica cp genome by applying next-generation sequencing technology based on Illumina NovaSeq 6000. The genome assembly details and annotation for the J. seravschanica cp genome were described. The obtained data will provide valuable resources for plant molecular identification and evaluation of phylogenetic relationships at the genus level.
2. Data Description
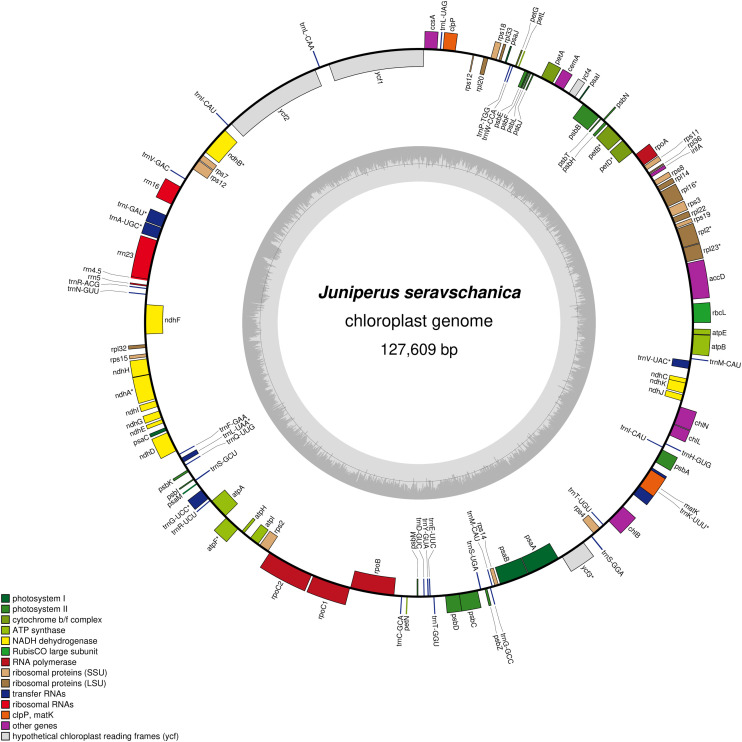
Complete chloroplast genome sequencing using Illumina NovaSeq 6000 of J. seravschanica generated about 4 GB of raw data which consisting 24,772,052 paired-end reads with GC content of 34,45% and phred score of 94,39% (Q30) and 98,85% (Q20). The assembled chloroplast genome size of the J. seravschanica was 127,609 bp. The structure of the chloroplast genome is circular with a small single-copy region (SSC) and a large single-copy region (LSC). Fig. 1 presented a circular gene map of J. seravschanica chloroplast genome.
Fig. 1.
Gene map of the J. seravschanica chloroplast genome.
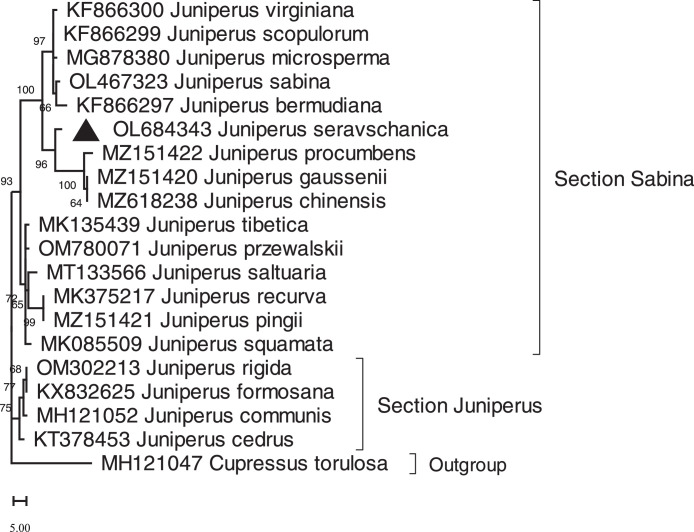
Maximum parsimony concatenated phylogenetic tree based on matK and rbcL nucleotide sequences is given in Fig. 2. The phylogenetic tree separated Juniperus species into two clades which corresponding to the sections Juniperus and Sabina.
Fig. 2.
Maximum parsimony concatenated phylogenetic tree based on matK and rbcL nucleotide sequences with 1000 bootstraps. ▲ denotes species analyzed in this study.
The Bioproject (PRJNA883033), Sequence Read Archive (SRR21673293), and GenBank (OL684343) data were deposited at National Center for Biotechnology Information.The chloroplast genome of J. seravschanica encoded 118 genes, including 82 protein-coding genes, 32 transfer RNA (tRNA) genes, and 4 ribosomal RNA (rRNA) genes (Table 1).
Table 1.
List of genes identified in the J. seravschanica cp genome.
| Category | Group of genes | Name of genes |
|---|---|---|
| Self-replication | Transfer RNA | trnA-UGC*, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC*, trnH-GUG, trnI-CAU (x2), trnI-GAU*, trnK-UUU*, trnL-CAA, trnL-UAA*, trnL-UAG, trnM-CAU (x2), trnN-GUU, trnP-UGG, trnQ-UUG (x2), trnR-ACG, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC, trnV-UAC*, trnW-CCA, trnY-GUA |
| Ribosomal RNA | rrn16, rrn23, rrn5, rrn4.5 | |
| Small subunit of ribosome | rps2, rps3, rps4, rps7, rps8, rps11, rps12*, rps14, rps15, rps18, rps19 | |
| Large subunit of ribosome | rpl14, rpl16*, rpl2*, rpl20, rpl22, rpl23*, rpl32, rpl33, rpl36 | |
| DNA-dependent RNA polymerase | rpoA, rpoB, rpoC1*, rpoC2 | |
| Translational initiation factor | infA | |
| Genes for photosynthesis | Rubisco | rbcL |
| Photosystem I | psaA, psaB, psaC, psaI, psaJ, psaM | |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| ATP synthase | atpA, atpB, atpE, atpF*, atpH, atpI | |
| Subunits of cytochrome | petA, petB*, petD*, petG, petL, petN | |
| Chlorophyll biosynthesis | chlB, chlL, chlN | |
| NADH dehydrogenase | ndhA*, ndhB*, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Other genes | Maturase | matK |
| Protease | clpP | |
| Envelope membrane protein | cemA | |
| Subunit of acetyl-CoA | accD | |
| C-type cytochrome synthesis gene | ccsA | |
| Genes of unknown function | Conserved open reading frames | ycf1, ycf2, ycf3**, ycf4 |
Note: One or two asterisks indicate one or two intron-containing genes, respectively, (x2) indicates duplicated genes.
Among these 118 genes, three genes (trnI-CAU, trnM-CAU and trnQ-UUG) are duplicated, 16 genes (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC, rps12, rpl16, rpl2, rpl23, rpoC1, atpF, petB, petD, ndhA and ndhB) contain one intron and one gene (ycf3) contain two introns. The overall GC content of the J. seravschanica assembled chloroplast genome was 35.05%.
In total, 152 simple sequence repeats (SSRs) were determined in J. seravschanica plastome by MicroSAtellite (MISA) [2]. Four types of SSRs were detected: 108 mononucleotides, 33 dinucleotides, 5 trinucleotides, and 6 tetranucleotides. Types and amounts of identified SSRs are provided in Table 2.
Table 2.
Types and amounts of simple sequence repeats (SSRs) in the J. seravschanica chloroplast genome.
| SSR type | Repeat Unit | Ammount | Ratio (%) |
|---|---|---|---|
| Mono | A/T | 106 | 98.1 |
| C/G | 2 | 1.9 | |
| Di | AC/GT | 5 | 15.1 |
| AG/CT | 13 | 39.4 | |
| AT/AT | 15 | 45.5 | |
| Tri | AAG/CTT | 3 | 60 |
| AAT/ATT | 2 | 40 | |
| Tetra | AAAC/GTTT | 1 | 16.7 |
| AAAG/CTTT | 2 | 33.2 | |
| AAGT/ACTT | 1 | 16.7 | |
| ACCT/AGGT | 1 | 16.7 | |
| ATCC/ATGG | 1 | 16.7 |
Among these 152 SSR markers (Supplementary Table 1), 77 (50.7%) SSRs were located in intergenic region, 56 (36.8%) in protein-coding genes, 14 (9.2%) in introns, 3 (2%) in rRNA and 2 in tRNA (1.3%) (rrn23 and trnS-UGA, trnS-GCU, respectively). Most of the SSRs identified in J. seravschanica chloroplast genome were located in the intergenic and genic regions (87.5%).
3. Experimental Design, Materials and Methods
3.1. Plant Material and DNA Extraction
In this study, the fresh leaves of J. seravschanica were collected from the Turkistan region of Southern Kazakhstan (42.331250N, 70.372583E). Fresh leaves from J. seravschanica samples were desiccated in silica gel and stored at room temperature until DNA extraction. Then, the total DNA was isolated from the leaves under highly sterile conditions using the CTAB protocol [1]. The concentration and quality of the extracted DNA were checked by gel electrophoresis in agarose and Nanodrop 2000 spectrophotometry (Thermo Fisher Scientific, Wilmington, DE, USA).
3.2. Library Preparation and Sequencing
Library preparation and cp genome sequencing were conducted by Macrogen Inc. (Seoul, Korea). The library was performed with the TruSeq Nano DNA Kit (Illumina, USA). Paired-end sequencing was performed on Illumina NovaSeq 6000 sequencer based on sequencing by synthesis technology. Generated raw read Fastq format files were used for the genome assembly.
3.3. Genome Assembly and Annotation
For accurate genome assembly raw data were quality filtered. Reads in which 90% of the bases had a phred score of 20 or higher were used for assembly. After quality filtering, poly-G trimming was performed using fastp 0.19.4 with a quality phred option as 10 and an unqualified percent limit as 50. In order to reduce biases in the analysis, low-quality reads were removed using Trimmomatic [3]. After filtering, the library for J. seravschanica included 24,772,052 total reads. Trimmed reads were used for de novo assembly by SPAdes 3.13.0 [4] assembler approach. The complete genome contigs were combined into one contig by joining overlapping DNA segments of each contig. After the draft genome was assembled, the locations of protein genes were predicted and their functions were annotated using Prokka [5]. The circular map (Fig. 1) of the J. seravschanica cp genome was generated using Organellar Genome DRAW (OGDRAW) software [6].
3.4. Detection of SSR Markers in the Chloroplast Genome
Simple sequence repeats (SSRs) were determined by MISA software [2] with the following thresholds: eight for mononucleotide repeats, four for dinucleotide repeats, four for trinucleotide repeats, three for tetranucleotide repeats, three for pentanucleotide repeats, and three for hexanucleotide repeats. A total of 152 putative SSR markers were identified in the chloroplast genome sequence of J. seravschanica (Table 2).
Ethics Statements
The manuscript adheres to Ethical requirements for publication. The work does not involve studies with animals and humans.
CRediT authorship contribution statement
Moldir Yermagambetova: Methodology, Investigation, Data curation. Saule Abugalieva: Investigation, Writing – review & editing. Yerlan Turuspekov: Conceptualization, Investigation, Writing – review & editing. Shyryn Almerekova: Supervision, Conceptualization, Investigation, Software, Writing – original draft.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was funded by the Science Committee of the Ministry of Education and Science of the Republic of Kazakhstan (Grant number AP09259027).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dib.2022.108866.
Appendix. Supplementary materials
Data Availability
Sequence Read Archive (Original data) (National Center for Biotechnology Information).
Complete chloroplast genome of Juniperus seravschanica (Original data) (National Center for Biotechnology Information).
References
- 1.Doyle J.J., Doyle J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- 2.Beier S., Thiel T., Münch T., Scholz U., Mascher M. MISA-web: a web server for microsatellite prediction. Bioinformatics. 2017;33:2583–2585. doi: 10.1093/bioinformatics/btx198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., Pyshkin A.V., Sirotkin A.V., Vyahhi N., Tesler G., Alekseyev M.A., Pevzner P.A. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 6.Lohse M., Drechsel O., Kahlau S., Bock R. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013;41:W575–W581. doi: 10.1093/nar/gkt289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence Read Archive (Original data) (National Center for Biotechnology Information).
Complete chloroplast genome of Juniperus seravschanica (Original data) (National Center for Biotechnology Information).