Abstract
Emerging and existing viruses from various human and animal samples have been studied and analyzed using viral metagenomics, which has proven to be an effective technique. Foxes, as a kind of significant economic animal, are widely raised in China. Viruses carried by foxes may potentially infect humans or other animals. There are currently very few studies of faecal virome in farmed foxes. Using viral metagenomics, we evaluated the faecal virome of twenty-four foxes collected from the same farm in Jilin Province, China. Some sequences more closely related to the families Parvoviridae, Picornaviridae, Smacoviridae, Anelloviridae, and Herpesviridae were detected in the faecal sample. The main animal viruses that infect farmed red foxes were parvovirus and picornavirus. Five smacovirus strains were found and provided evidence for genetic diversity in the genus Smacoviridae. In addition, some viruses infecting avian species or rats were detected in this study. The study helped us better understand faecal virome in farmed red foxes and assisted in the surveillance and prevention of viral diseases in these animals.
Keywords: Red fox, Viral metagenomics, Faecal virome, Viral diversity, Phylogenetic analysis
1. Introduction
Red fox (scientific name: Vulpes) is a member of the Canidae family that shows high ecological adaptability [1], and its fur has high economic value. Foxes have been raised as important economic animals in recent years. However, intensive farming poses an inescapable risk of infectious disease transmission. Viral diseases are an important factor affecting the fox breeding industry. Many viruses can cause diseases in foxes, such as parvovirus, astrovirus, canine distemper virus (CDV), canine adenovirus, and porcine pseudorabies virus (PRV) [[2], [3], [4], [5]]. Besides them, some viruses found in foxes have been clustered into the zoonotic group, such as influenza virus, rabies virus (RV), neurotropic arbovirus (TBEV) and Getah virus [[6], [7], [8]]. Due to frequent contact between foxes and farmers, these zoonotic viruses could easily spread to humans and cause disease [[9], [10], [11]]. There has been no systematic study of viruses that infect farmed foxes.
With the rapid development of next-generation sequencing technologies (NGS), metagenomics has become a powerful tool for analyzing biodiversity, population structure, evolutionary relationships, and potential biological implications [12]. Viral metagenomics has been used to investigate the virus in wild foxes [[13], [14], [15]], but limited virome data is available for farmed foxes. Following the domestication of some wild fox species, the likelihood of spillover effects in contact with humans and domestic animals will increase, so ongoing research into the viral composition of valuable economic animals is warranted. In the present study, viral metagenomics was used to study the faecal virome of farmed red foxes on a farm in Jilin Province, China. Our results showed different viral compositions, including the families of Anelloviridae, Parvoviridae, Smacoviridae, Circoviridae, Genomoviridae, Herpesviridae, Picornaviridae, Picobirnaviridae, Flaviviridae, Iridoviridae, and Poxviridae. The study will advance our understanding of the faecal virome of farmed red foxes and contribute to further surveillance and prevention of viral disease.
2. Materials and methods
2.1. Sample collection and preparation
The purpose of this research was to investigate the virome of farmed foxes. From a single red fox farm in China's Jilin province, 24 faecal samples from adult foxes were randomly taken. Using disposable sterile materials, specialists collected all samples and transported them on dry ice. Resuspended samples were vigorously vortexed in a 2 ml Dulbecco's Phosphate-buffered Saline (DPBS) for 10 min before freeze-thawing three times. After centrifugation (10 min at 15,000×g, 4 °C), supernatants from each sample were then collected in a new 1.5 ml centrifuge tube and stored at −80 °C.
2.2. Viral nucleic acid extraction
The 24 supernatants were randomly and evenly grouped to form two sample pools.
Eukaryotic and some bacterial cell-sized particles were removed from each sample pool using a 0.45 μm filter (Millipore) (5 min at 8,000×g, 4 °C), and DNase (Turbo DNase from Thermo Fisher; Baseline-ZERO from Epicentre; benzonase from Novagen) and RNase (Fermentas) kits were used to digest unprotected nucleic acid in filtrates enriched with viral particles [16,17]. QIAamp viral RNA Minikit (Qiagen) was used to extract the remaining total viral RNA and DNA according to the manufacturer's protocol.
2.3. Library construction and bioinformatics analysis
Reverse transcription reactions with enriched viral RNA from the respective pools were performed using reverse transcriptase (Super-Script IV, Invitrogen) and Random Hexamer Primers, and then double-strand DNA synthesis using Klenow fragment polymerase (New England Biolabs). Using the Nextera XT DNA Sample Preparation Kit (Illumina), two libraries were constructed and then sequenced on the Miseq Illumina platform with 250 bases paired ends.
The 250 bp paired-end reads were debarcoded using Illumina's vendor software. Data was processed using an in-house analysis pipeline running on a 32-node Linux cluster, and reads were treated as duplicates if bases 5 to 55 were identical, with only one random copy of duplicates kept. The Phred quality score of 10 as the threshold was used to trim tails with low sequencing quality, and adaptors were clipped using VecScreen's default parameters, which are NCBI BLASTn with specific adjustment parameters. Using Bowtie2 (v2.2.4), bacterial nucleotide sequences were mapped from the BLAST NT database and subtracted, and cleaned reads were de-novo assembled by SOAPdenovo2 using Kmer size 63 with default settings [18]. With an E-value cutoff of 10−5, the assembled contigs, along with singlets, were aligned to an in-house viral proteome database using BLASTx (v.2.2.7). The virus BLASTx database was compiled using NCBI virus reference proteome (https://ftp.ncbi.nih.gov/refseq/release/viral/) (accessed on December 20, 2021)to which viral protein sequences were added from NCBI nr FASTA file (based on annotation taxonomy in the Virus Kingdom) [17]. Non-viral protein sequences from the NCBI nr FASTA file (based on annotation taxonomy excluding Virus Kingdom) were used to remove false positive viral hits with an E-value cutoff of 10−5. Viral contigs without significant BLASTx similarity were searched for viral protein families in the vFam database [19] using HMMER3 to detect distant viral protein similarities [[20], [21], [22]]. Additionally, MEGAN (v 6.22.2), a metagenomic annotation tool, was used to assign each sequence present in metagenomic data to different taxa using the NCBI taxonomic database.
2.4. Phylogenetic analysis
The amino acid (aa) sequences of reference strains belonging to different virus groups were downloaded from the NCBI GenBank database to infer phylogenetic relationships. Sequence alignment was performed using MUSCLE with the default settings in MEGA software (version 10.1.8) [23]. The alignment of sites with gaps greater than 50% was temporarily removed, and MrBayes (version 3.2.7) was used to construct Bayesian inference trees [24]. During MrBayes analysis, “lset nst = 6 rates = invgamma” were used for phylogenetic analysis based on nucleotide sequences, which set the evolutionary model to the GTR substitution model with gamma-distributed rate variation across sites and a proportion of invariable sites ("GTR + I+Г"), while we set “prset aamodelpr = mixed” for the phylogenetic analysis using amino acid sequences, which allows the program to utilize the 10 built-in amino acid models, and the number of generations was increased to one million until the standard deviation of split frequencies is below 0.01, sampled every 50 generations, and the first 25% of Markov chain Monte Carlo (MCMC) samples were discarded as burn-in. All Bayesian inference trees were further validated using the maximum likelihood trees constructed by the MEGA software (version 10.1.8).
2.5. Nucleotide sequence accession number
The resulting virus genome and fragments were deposited in GenBank with accession numbers: ON872775 – ON872784. The raw sequence reads were stored in the Sequence Read Archive (SRA) of the GenBank database under accession numbers: SRR19759933 and SRR1759659 (Table. S1).
3. Result
3.1. Overview of viral metagenomic
A total of 236,783 raw reads from the two pools were obtained and classified using Megan.6. With the Evalue cutoff of <10−5, a total of 181,221 sequences (76.5%) demonstrated significant sequence identities for known viruses, the majority of which could be attributed to bacteriophage sequences (98.2%) with a considerable fraction of the family Microviridae, Siphoviridae, and Podoviridae (Table.S2). Sequences from the following families of viruses that could infect eukaryotes were detected: Parvoviridae, Picornaviridae, Smacoviridae, Herpesviridae, Anelloviridae, Genomoviridae, Iridoviridae, Picobirnaviridae, Poxviridae, Flaviviridae, and Circoviridae. Members of these families (including Poxviridae, Herpesviridae, Iridoviridae, Genomoviridae, and Flaviviridae) were disregarded due to insufficient reads. De novo assembly within the families Parvoviridae and Picornaviridae produced the 92 and 109 viral contigs (Table .1), respectively, which account for the major part of viruses observed infecting eukaryotes in this study.
Table 1.
Distribution of metagenomic reads and contigs of viral sequence from farmed foxes faecal.
| Library 11 |
Library 12 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Potential virus family | Number of sequence reads | Number of contig (s) | Mean contig length (nt) | Minimum contig length (nt) | Maximum contig length (nt) | Number of sequence reads | Number of contig (s) | Mean contig length (nt) | Minimum contig length (nt) | Maximum contig length (nt) |
| Parvoviridae | 387 | 33 | 632 | 178 | 3177 | 714 | 59 | 515.7 | 172 | 2456 |
| Picornaviridae | 654 | 63 | 572.9 | 249 | 2119 | 273 | 46 | 465.2 | 196 | 1339 |
| Smacoviridae | 124 | 10 | 644.2 | 256 | 1356 | 230 | 19 | 502.2 | 259 | 1864 |
| Herpesviridae | 51 | 11 | 353.6 | 251 | 457 | 182 | 27 | 375.5 | 230 | 648 |
| Anelloviridae | 100 | 10 | 770.2 | 314 | 1607 | 137 | 8 | 820.1 | 254 | 3521 |
| Genomoviridae | 27 | 4 | 458 | 250 | 713 | 74 | 10 | 548.6 | 254 | 1149 |
| Iridoviridae | 17 | 2 | 255 | 251 | 259 | 38 | 4 | 476.5 | 337 | 601 |
| Picobirnaviridae | 35 | 5 | 391.4 | 278 | 471 | 33 | 3 | 421 | 331 | 478 |
| Poxviridae | 11 | 1 | 26 | 7 | 328.3 | 300 | 362 | |||
| Flaviviridae | 16 | 2 | 419 | 372 | 466 | |||||
| Circoviridae | 65 | 6 | 461.2 | 249 | 786 | |||||
3.2. Viruses belonging to the anelloviridae
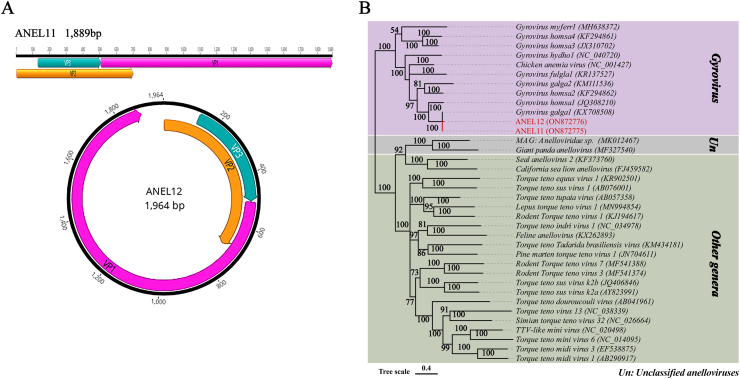
Anellovirus sequences were discovered in both libraries (100 reads in library # 11, 137 reads in library # 12), where a nearly complete genome and a complete genome of anelloviruses were obtained and respectively named ANE11 (mean coverage: 7.5) and ANEL12 (mean coverage: 12.3). The nearly or complete genome of ANEL11 and ANEL12 was 1,889 nt and 1,964 nt, respectively. The genome organization of both viruses presented the typical feature of gyroviruses, with three partially overlapping ORFs in the same direction. For ANEL11, the three ORFs encoded three proteins: VP1 (507–1889 nt, 461 aa), VP2 (4–696 nt, 232 aa) and VP3 (131–505 nt, 125 aa), while the length of VP1, VP2, and VP3 of ANEL12 were 460 aa, 233 aa, and 125 aa, respectively (Fig. 1A). The VP1, VP2 and VP3 of these two viruses shared the highest nt sequence identity of 99.6%, 99.0% and 98.7%, respectively. Chicken anemia virus (CAV, GenBank no. M55918) was the first isolated single-stranded circular DNA virus of the genus Gyrovirus in which the genome has been thoroughly analyzed [25]. At the complete genome level, ANEL11 shared 51% nt sequence identity with CAV strain Cux-1 and ANEL12 shared 50.3% nt identity.
Fig. 1.
The genomic organization and phylogenetic analysis of anelloviruses detected in domestic red foxes. (A) The genomic organization of ANEL11 and ANEL12. The viral encoding proteins of ANEL11 and ANEL12 were separately marked with different colors. The arrow represented the direction of gene coding. (B) The phylogenetic analysis is based on the amino acid sequences of VP1 of ANEL11, ANEL12, and different reference strains. ANEL11 and ANEL12, identified in this study, were highlighted using the red font. Un, Unclassified anelloviruses.
ANEL11 and ANEL12 were examined for genetic relationships with other anelloviruses using the VP1 aa sequence phylogenetic tree. The results showed that 10 species of the genus Gyrovirus were clearly delineated in the VP1 phylogenetic tree. ANEL11 and ANEL12 clustered with one avian gyrovirus 2 strain NX1506-1 (GenBank no. KX708508) and formed a clade (Fig. 1B). Sequence analysis showed that ORF1 of ANEL11 and ANEL12 shared the highest nucleotide sequence identity (98.2% of ANEL11, 95.2% of ANEL12) with avian gyrovirus 2 isolate NX1506-1 (GenBank no. KX708508). According to the species classification criteria in the family Anelloviridae, the virus belongs to that specific species if it shares >69% pairwise identification of the complete ORF1 coding region nucleotide sequences with that of any member assigned to a currently classified species [26], so ANEL11 and ANEL12 are classified as the species Gyrovirus galga1.
3.3. Viruses belonging to the parvoviridae
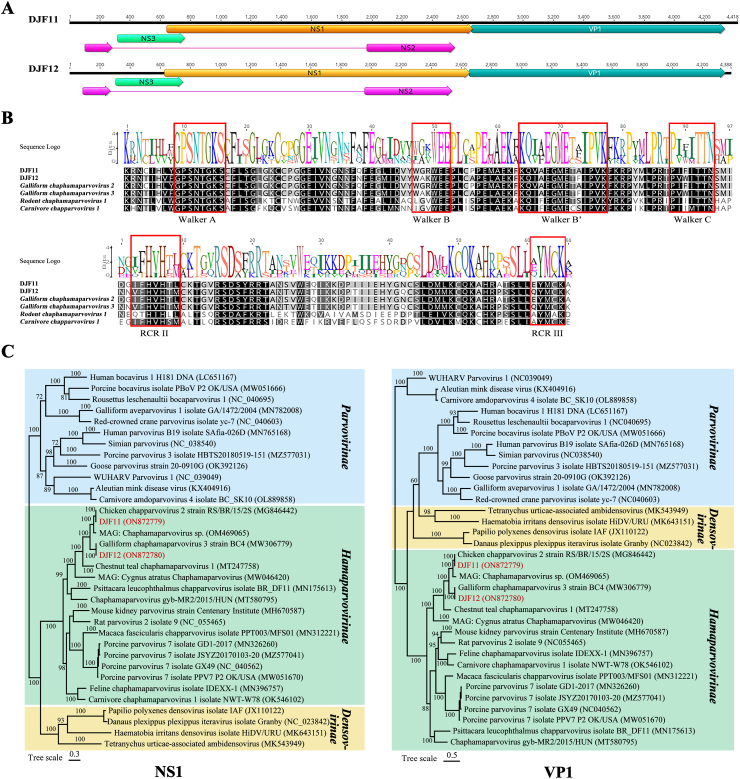
Sequence reads corresponding to the family Parvoviridae were identified from all libraries (387 reads in library #11, 714 reads in library #12). Two nearly complete genomes of parvovirus were obtained from the two libraries and named DJF11 (mean coverage: 6.8) (library #11) and DJF12 (mean coverage: 10.3) (library #12), respectively. The nearly complete genomes of DJF11 and DJF12 were 4,418 nt, and 4,380 nt in length, respectively, and both contain two complete ORFs (ORF1 and ORF2). Both of DJF11 and DJF12 ORF1 were 2,022 nt in length, but their ORF2 were 1,674 and 1,701 nt, respectively. The ORF1 and ORF2 encoded a non-structural protein (NS1) and a structural protein (VP1), respectively. Besides the two ORFs, two additional ORFs overlapping the NS1 ORF were detected and encoded two nonstructural proteins (Fig. 2A). The conserved Walker and rolling circle replication (RCR) motifs typical of parvoviral helicases were present in the NS1 protein, which were similar with the reference strains (GenBank no. MG846442, MW306779, NC_040,843, and MH893826) (Fig. 2B).
Fig. 2.
The genomic organization and phylogenetic analysis of the parvoviruses detected in domestic red foxes. (A) The genomic organization of the DJF11 and DJF12 genes was identified in this study. The viral encoding proteins of DJF11 and DJF12 were marked with different colors. (B) The comparison of aa sequences of helicase motifs in members of the genus Chaphamaparvivirus. The conserved rolling circle replication (RCRs) and Walker motifs of SF3 helicases were shown in the red square. (C) The phylogenetic analysis of parvoviruses identified in this study is based on the amino acid sequences of NS1 and VP1. The DJF11 and DJF12 identified in this study were marked with red font.
Phylogenetic analysis was carried out based on NS1 and VP1 aa sequences including reference sequences from subfamilies Densovirinae, Hamaparvovirinae, and Parvovirinae. The results showed that DJF11 and DJF12 clustered with other viruses of the subfamily Hamaparvovirinae in the NS1 and VP1 phylogenetic trees. DJF11 clustered with one chicken chapparvovirus 2 strain RS/BR/15/2S (GenBank no. MG846442) formed a clade, while DJF12 clustered with one galliform chaphamaparvovirus 3 strain BC4 formed a clade (GenBank no. MW306779) (Fig. 2C). Sequence analysis showed that NS1 of DJF11 shared the highest aa sequence identity of 99.11% with strain RS/BR/15/2S, and VP1 of DJF11 had the highest aa identity of 98.20% with the same strain. Alignment with the strain RS/BR/15/2S showed that NS1 and VP1 in DJF11 had only 6 aa and 10 aa mutations (data not shown). NS1 and VP1 of DJF12 shared the highest aa sequence identity (99.0% and 97.7%) with strain BC4, respectively. Viruses within a species usually encode NS1 protein that exhibits >85% aa sequence identity, according to the International Committee on Taxonomy of Viruses (ICTV) classification criteria for species in the family Parvoviridae [27]. Therefore, DJF11 and DJF12 belong to the subfamily Hamaparvovirinae, genus Chaphamaparvovirus, species Galliform Chaphamaparvovirus 2 and Galliform Chaphamaparvovirus 3, respectively. This is the first time these viruses have been detected in farmed fox faecal samples.
3.4. Smacovirus and smaco-like viruses detected here
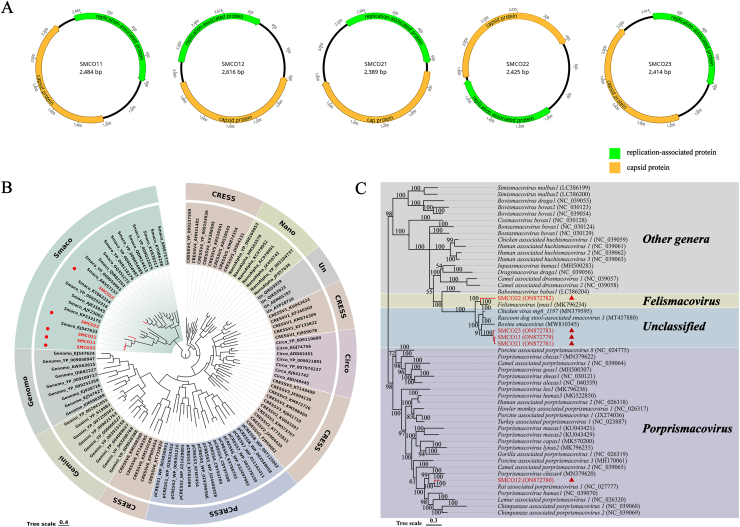
Five complete genomes of smacoviruses were obtained from these two libraries. Two of them from library # 11 were named SMCO11 (mean coverage: 9.4) and SMCO12 (mean coverage: 6.2), while the remaining three from library # 12 were named SMCO21 (mean coverage: 27.1), SMCO22 (mean coverage: 8.1), and SMCO23 (mean coverage: 41.8) respectively. The complete genomes of these five viruses are 2,389 to 2,616 nt in length and have two ORFs encoding the putative Rep and Cap proteins separately. As shown in Fig. 3A, the five virus genomes had two types of gene structures, and except for SMCO12, which contains two bidirectional ORFs, the other four viruses (SMCO11, SMCO21, SMCO22, and SMCO23) contain two ORFs in the same direction. The nucleotide sequence identity among them was 39.7%∼68.4%. Amino acid sequences analysis based on the Rep protein showed that SMCO11 shared the highest aa identity of 95.02% with the syrmaticus ellioti CRESS-DNA virus sp. isolate eph225sma2 (GenBank no. MW182789), SMCO12 shared the highest aa identity of 82.61% with the Rat stool-associated circular ssDNA virus isolate Mu/10/1799 (GenBank no. KP860907), SMCO21 shared the highest aa identity of 96.14% with the CRESS virus sp. isolate 16806 × 66_211 (GenBank no. MH111087), SMCO22 shared the highest aa identity of 90.94% with the Smacoviridae sp. isolate w3chi090cir1 (GenBank no. MT138076), while SMCO23 shared the highest identity of 95.75% to the Smacoviridae sp. isolate wbp226sma2 (GenBank no. MT138085).
Fig. 3.
Schematic genome organization and phylogenetic analysis of five smacoviruses. (A) Genome organizations of SMCO11, SMCO12, and SMCO21-23. The replication-associated protein was marked with green and the capsid protein with yellow. The arrow represented the direction of gene coding. (B) The phylogenetic tree was constructed based on the Rep protein of reference strains of CRESS-DNA virus (Circoviridae, Genomoviridae, Geminiviridae, Nanoviridae, Bacilladnaviridae, and Smacoviridae), unclassified CRESS-DNA virus (CRESSV1-6), Bacterial plasmids (pCRESS1-8), and five viruses identified in this study. The viruses identified in this study were shown in red font. (C) Phylogenetic analysis of Rep from various smacovirus genera (Bovismacovirus, Cosmacovirus, Dragsmacovirus, Drosmacovirus, Huchismacovirus, Porprismacovirus, Inpeasmacovirus, Bostasmacovirus, Bonzesmacovirus, Simismacovirus, Babosmacovirus, Felismacovirus, and Unclassified Smacoviridae). The viruses detected in this study were highlighted using the red font.
The CRESS-DNA viruses associated with eukaryotic hosts have been classified into six families by ICTV, namely Circoviridae, Genomoviridae, Geminiviridae, Nanoviridae, Bacilladnaviridae, and Smacoviridae [28]. To investigate the genetic relationship of these five viruses with other CRESS-DNA viruses, phylogenetic trees were constructed based on the Rep protein. As shown in Fig. 3B, the five viruses in this study clustered with other smacoviruses. We further explored the evolutionary relationships of these five viruses by constructing phylogenetic trees with other smacoviruses of different genera (Bovismacovirus, Cosmacovirus, Dragsmacovirus, Drosmacovirus, Huchismacovirus, Porprismacovirus, Inpeasmacovirus, Bostasmacovirus, Bonzesmacovirus, Simismacovirus, Babosmacovirus, and Felismacovirus) (Fig. 3C). Phylogenetic analysis showed that SMCO12 and SMCO22 were delineated in the genus Porprismacovirus and Felismacovirus, respectively. SMCO12 formed a clade with Rat associated porprismacovirus 1 (GenBank no. NC_027777) and shared 81.5% aa identity of Rep protein, while SMCO22 formed a clade with Felismacovirus lynas1 (GenBank no. MK796234) and shared 65.4% aa identity of Rep protein. According to the ICTV criteria for smacovirues, the 40% aa sequence identity of Rep with strong phylogenetic support was proposed as a genus-level demarcation threshold [28]. The SMCO12 and SMCO22 belonged to the genus Porprismacovirus and Felismacovirus, respectively. Otherwise, SMCO11, SMCO21, and SMCO23, together with the Bovine smacovirus strain 68-Smacoviridae-2 (GenBank no. MW810345) formed a clade and shared 60.4%, 62.2%, and 61.5% aa sequence identity of Rep protein with it, respectively, so all of them belonged to the unclassified smacovirus.
3.5. An enterovirus D-like strain detected in this study
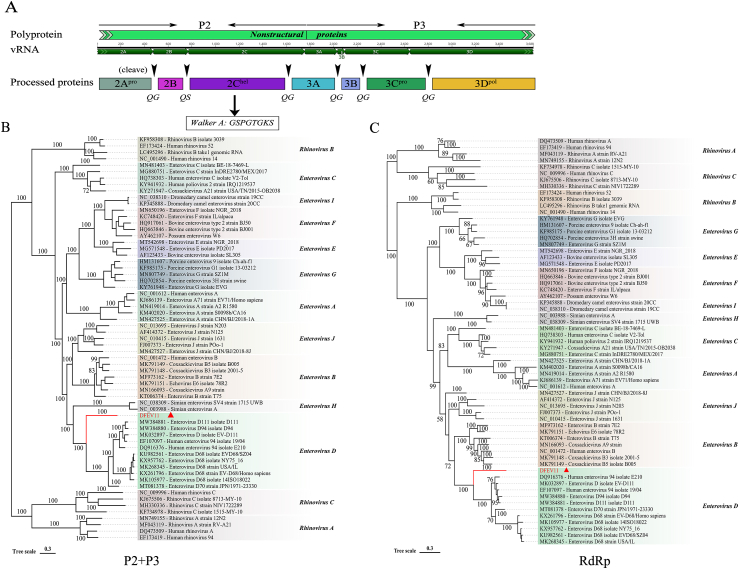
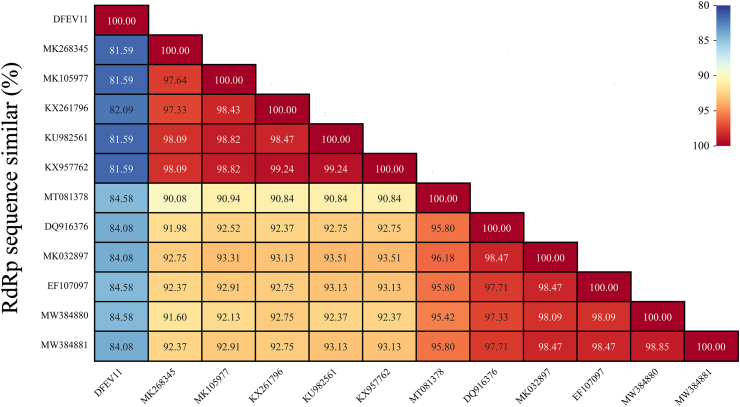
An incomplete enterovirus genome was identified and named DFEV11 (mean coverage: 12.2) in Library #11. The incomplete enterovirus genome is 3,688 nt in length, including partial P2 and P3 regions. The aa sequence of P2+P3 has 72.62% identity with enterovirus D94 (GenBank no. DQ916376). The P2+P3 sequence was cut into several nonstructural proteins, including partial 2A, 2B (99 aa), and 2C (328 aa), 3A (92 aa), 3B (22 aa), 3C (183 aa), and partial 3D, respectively. 2A and 3C proteins were proteases involved in the cleavage of polyprotein [29]. The putative cleavage sites were QG (2A/2B), QS (2B/2C), QG (2C/3A), QG (3A/3B), QG (3B/3C), and QG (3C/3D), respectively. The 2C protein contained a conserved nucleotide phosphate-binding motif, also known as the Walker A motif GSPGTGKS (GXXGXGK [S/T]), which belonged to the P-loop NTPase domain superfamily (Fig. 4A). 3A protein was a critical component of the enterovirus replication complex [30,31]. Orthologous proteins 2C, 3C, and 3D were conserved in all picornaviruses. The incomplete 3D protein in this study also contained a conserved RdRp domain. Based on RdRp aa sequence alignment, DFEV11 showed 81.59%∼84.58% aa sequence identity with other enterovirus D strains (Fig.S1).
Fig. 4.
The genome organization and phylogenetic analysis of enterovirus (A) The partial genome structure of P2 and P3 region and putative cleavage sites of enterovirus D-like virus were shown. (B) The phylogenetic tree based on the nucleotide sequences of the partial P2 and P3 of DFEV11, the reference strains of 11 Enterovirus species and 3 Rhinovirus species. (C) The phylogenetic tree based on the RdRp nucleotide sequences of DFEV11. The virus detected in this study was marked with red triangle.
The VP1 sequence variant is used to classify enterovirus serotypes. It has been proposed that enteroviruses should be categorized as belonging to the same serotype if their VP1-coding sequence shares at least 75% nucleotide similarity (>85% aa sequence similarity) [32]. Because the whole gene sequence was not available, we investigated the genetic relationship based on the P2+P3 nucleotide sequences of DFEV11. The results showed that DFEV11, as a separate branch, was clustered with 11 enterovirus D strains (Fig. 4B). Furthermore, RNA-dependent RNA-polymerase (RdRp) was used to examine evolutionary relationships among picornaviruses [33]. Similarly, phylogenetic analysis using RdRp nucleotide sequences showed that DFEV11 remained a separate branch, clustering with 11 enterovirus D strains (Fig. 4C). Although DFEV11 fell outside the enterovirus D clade, RdRp sequence analysis showed that DFEV11 shared a high level of nucleotide and amino acid identity with other enterovirus species in this genus, implying that DFEV11 was closely related to the enterovirus D strain.
4. Discussion
Viral metagenomics is an attractive tool for discovering broad-based pathogens and highly mutated viruses. Since the 20th century, identifying new pathogens has significantly impacted infectious diseases, microbiology, and human health [[34], [35], [36]]. Zoonotic diseases are diseases transmitted from animals to humans through direct contact, food, water, and the environment. It was estimated that rough 61% of emerging infectious diseases are zoonoses, caused mainly by viruses [37,38]. Diversification of market demands, such as the fur industry, petting zoos, and specialty food products, accelerates the spread of numerous undiscovered pathogens. Domestic animals may act as amplifiers for wild pathogens; therefore, changes in viral community diversity in livestock farms should be monitored more closely.
In this study, faecal samples from a red fox farm in Jilin Province, China were analyzed using high-throughput sequencing to predict the viral community composition of farmed red foxes. Canidae, such as wolves, foxes, raccoon dogs and dogs, are the natural hosts of the rabies virus (RV), canine distemper virus (CDV), and canine parvovirus (CPV) [13,39,40]. However, RV and CDV were not detected in this study, possibly because of standardized vaccination procedures and the change in living habits which may lead to differences in the viral composition in domestic and wild foxes. In previous studies, a great proportion of viral reads were mapped to circovirus in juveniles and picobirnavirus in wild adult foxes [15]. Similarly, astroviruses were also identified from faecal samples of wild red foxes [41]. However, the majority virus communities in this study were parvoviruses and picornaviruses, which also reflected the unique distribution of red fox virus communities in different areas under rearing methods compared with wild foxes.
Following rigorous assembly parameter setting and Polymerase Chain Reaction (PCR) validation, several virus strains were identified. Parvovirus is small, non-enveloped virus with linear, single-stranded DNA genomes of about 4–6 kb in size. Some members of the family Parvoviridae can cause diseases that range from subclinical to lethal alone, while some require co-infection with helper viruses from other families [27,42,43]. Two chaphamaparvoviruses discovered in this study were clustered with other galliform chaphamaparvoviruses. In addition, two anelloviruses were firstly detected in this study and clustered together with avian-associated gyrovirus strains. Since chicken flesh was a staple of the farmed fox’s diet, this might be the explanation for where these viruses came from. Meanwhile, animal research is needed to confirm whether amino acid deletions or mutations of the DJF11 and DJF12 capsid proteins impact host range and pathogenicity.
Members of the family Smacoviridae have small, circular, single-stranded DNA genomes encoding replication-associated proteins (Rep) and capsid proteins (Cap), with a genome length of approximately 2.3–2.9 kb. Although these viruses are thought to infect eukaryotes, their actual host remains unconfirmed [44]. In this study, five smacoviruses were detected. Phylogenetic analysis showed that SMCO12 and SMCO22 were clustered with rat and lynx-associated virus strains, respectively, and were assigned to two distinct recognized genera (GenBank no. NC_027777, no. MK796234). SMCO11, SMCO21, and SMCO23 were clustered with unclassified smacoviruses and belonged to samco-like viruses. Unlike members of the family Smacoviridae, the Rep and Cap encoding in their genome were in the same orientation rather than in the bidirectional orientation, suggesting that these smaco-like viruses may represent a new genus. In addition, the smacovirus from lynx was thought to be a common felid gut microbiome associated virus in previous study [45]. Two smacoviruses belonging to the genus Porprismacovirus and Felismacovirus were first detected in faecal samples from healthy farmed red foxes. In this study, we suspected that smacoviruses may be the common viruses associated with the fox gut microbiome. We cannot also rule out the possibility that these five smacoviruses, particularly SMCO12, arose from foodborne consumption due to environmental complexity.
Enterovirus, a member of the family Picornaviridae, has a genome of about 7.3 kb and is a non-enveloped, single-stranded, positive-sense RNA virus. The P1 region encodes the structural polypeptide and further cleaves it into four structural proteins (VP1∼VP4). The P2 and P3 regions encode replication-associated nonstructural proteins and cleave into three nonstructural proteins (2A∼2C) and four nonstructural proteins (3A∼3D), respectively. Based on sequence diversity, enteroviruses (EVs) are currently divided into 15 species, including Rhinovirus A-C and Enterovirus A-L. An enterovirus D-like strain (known as DFEV11) was detected in red fox faeces samples in this study. Enteroviruses have been shown to be responsible for varying degrees of disease and may infect multiple mammals, including humans and non-human primates [46]. However, foxes did not show clinical symptoms during sample collection, indicating that DFEV11 may not be pathogenic for red foxes.
In addition, viruses from other families have also been found, such as Genomoviridae, Herpesviridae, Iridoviridae, Picobirnaviridae, and Poxviridae. However, relatively complete genetic information cannot be obtained, which may be caused by the loss of some gene fragments in the process of library enrichment. Despite the inherent limitions of sample size and sequencing method, the research also demonstrates to some extent the relationship between viral community diversity and the environment, which eventually provides valuable information for monitoring the health of domestic animals.
5. Conclusion
Fecal virome in farmed foxes are examined in this study, which dramatically increases our understanding of viral diversity in samples from members of the Canidae family. Information from this study may help prevent viral diseases in farmed foxes and help monitor the health of these animals.
Funding statement
This work was supported by National Key Research and Development Programs of China No. 2022YFC2603801, Jiangsu Provincial Key Research and Development Projects No. BE2017693, National Natural Science Foundation of China No. 81741062 and Independent Project of Chengdu Research Base of Giant Panda Breeding No. 2020CPB-C11.
Ethics statements
Studies involving animal subjects
Regarding the use of animals, the Jiangsu University Ethics Committee reviewed and approved the animal study, and it complied with Chinese ethics regulations and laws. Written informed consent to participate in this study was sought from the owners of their animals.
Studies involving human subjects
No human studies were involved in this research.
Data availability statement
These datasets are available in online repositories. A list of repository names and accession numbers can be found in the supplementary material.
Author contribution statement
Shixing Yang and Wen Zhang conceived and designed the experiments. Dianqi Zhang, Yan Wang, and Xu Chen performed the experiments. Dianqi Zhang, Yumin He, Min Zhao, Xiang Lu, Juan Lu, Wen Zhang, Yan Wang, Likai Ji, and Xiaochun Wang analyzed and interpreted the data. Wen Zhang, Shixing Yang, Likai Ji, and Quan Shen contributed reagents, materials, analysis tools or data. Dianqi Zhang wrote the paper, and all authors substantially reviewed and revised the manuscript.
Declaration of competing interest
The authors state that there are no competing interests.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.heliyon.2023.e12826.
Contributor Information
Shixing Yang, Email: 1000004113@ujs.edu.cn.
Wen Zhang, Email: z0216wen@yahoo.com.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
FigS1.
References
- 1.Edwards C.J., et al. Temporal genetic variation of the red fox, Vulpes vulpes, across western Europe and the British Isles. Quat. Sci. Rev. 2012;57:95–104. doi: 10.1016/j.quascirev.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin H.L., Gao S.M., Liu Y., Zhang S.F., Hu R.L. Pseudorabies in farmed foxes fed pig offal in Shandong province, China. Arch. Virol. 2016;161:445–448. doi: 10.1007/s00705-015-2659-9. [DOI] [PubMed] [Google Scholar]
- 3.Denzin N., Herwig V., van der Grinten E. Occurrence and geographical distribution of Canine Distemper Virus infection in red foxes (Vulpes vulpes) of Saxony-Anhalt, Germany. Vet. Microbiol. 2013;162:214–218. doi: 10.1016/j.vetmic.2012.08.031. [DOI] [PubMed] [Google Scholar]
- 4.Balboni A., et al. Unique genetic features of canine adenovirus type 1 (CAdV-1) infecting red foxes (Vulpes vulpes) in northern Norway and arctic foxes (Vulpes lagopus) in Svalbard. Vet. Res. Commun. 2019;43:67–76. doi: 10.1007/s11259-019-09746-y. [DOI] [PubMed] [Google Scholar]
- 5.Campbell S.J., et al. Red fox viromes in urban and rural landscapes. Virus Evol. 2020;6:veaa065. doi: 10.1093/ve/veaa065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haut M., et al. The red fox (Vulpes vulpes) as sentinel for tick-borne encephalitis virus in endemic and non-endemic areas. Microorganisms. 2020;8 doi: 10.3390/microorganisms8111817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi N., Li L., Lu R., Yan X., Liu H. Highly pathogenic swine Getah virus in blue foxes, eastern China. Emerg. Infect. Dis. 2019;25:1252–1254. doi: 10.3201/eid2506.181983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qian Z., et al. Molecular characteristics of H9N2 influenza viruses isolated from farmed raccoon dogs and arctic foxes in China. Vet Sci. 2021;135:542–546. doi: 10.1016/j.rvsc.2020.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Taubenberger J.K., Morens D.M. Pandemic influenza--including a risk assessment of H5N1. Revue Scientifique et Technique (International Office of Epizootics) 2009;28:187–202. doi: 10.20506/rst.28.1.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Human rabies: 2016 updates and call for data. Releve Epidemiologique Hebdomadaire. 2017;92:77–86. [PubMed] [Google Scholar]
- 11.Lukman N., et al. A review of hantavirus research in Indonesia: prevalence in humans and rodents, and the discovery of serang virus. Viruses. 2019;11 doi: 10.3390/v11080698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiu C.Y. Viral pathogen discovery. Curr. Opin. Microbiol. 2013;16:468–478. doi: 10.1016/j.mib.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber M.N., et al. Virome of crab-eating (Cerdocyon thous) and pampas foxes (Lycalopex gymnocercus) from southern Brazil and Uruguay. Infect. Genet. Evol. :J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2020;85 doi: 10.1016/j.meegid.2020.104421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J., et al. Identification of a novel norovirus species in fox. Infect. Genet. Evol. :J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2022;98 doi: 10.1016/j.meegid.2022.105214. [DOI] [PubMed] [Google Scholar]
- 15.Lojkić I., et al. Faecal virome of red foxes from peri-urban areas. Comp. Immunol. Microbiol. Infect. Dis. 2016;45:10–15. doi: 10.1016/j.cimid.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Virome in the cloaca of wild and breeding birds revealed a diversity of significant viruses. Microbiome. 2022 doi: 10.1186/s40168-022-01246-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gut Virome of the World’s Highest-Elevation Lizard Species (Phrynocephalus Erythrurus and Phrynocephalus Theobaldi) Reveals Versatile Commensal Viruses. [DOI] [PMC free article] [PubMed]
- 18.Luo R., et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:18. doi: 10.1186/2047-217x-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skewes-Cox P., Sharpton T.J., Pollard K.S., DeRisi J.L. Profile hidden Markov models for the detection of viruses within metagenomic sequence data. PLoS One. 2014;9 doi: 10.1371/journal.pone.0105067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finn R.D., Clements J., Eddy S.R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eddy S.R. A new generation of homology search tools based on probabilistic inference. Genome informatics. International Conference on Genome Informatics. 2009;23:205–211. [PubMed] [Google Scholar]
- 22.Johnson L.S., Eddy S.R., Portugaly E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinf. 2010;11:431. doi: 10.1186/1471-2105-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. Mega X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ronquist F., et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noteborn M.H., et al. Characterization of cloned chicken anemia virus DNA that contains all elements for the infectious replication cycle. J. Virol. 1991;65:3131–3139. doi: 10.1128/jvi.65.6.3131-3139.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varsani A., et al. Taxonomic update for mammalian anelloviruses (family Anelloviridae) Arch. Virol. 2021;166:2943–2953. doi: 10.1007/s00705-021-05192-x. [DOI] [PubMed] [Google Scholar]
- 27.Cotmore S.F., et al. The family Parvoviridae. Arch. Virol. 2014;159:1239–1247. doi: 10.1007/s00705-013-1914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smacoviridae: a new family of animal-associated single-stranded DNA viruses. Arch. Virol. 2018 doi: 10.1007/s00705-018-3820-z. [DOI] [PubMed] [Google Scholar]
- 29.Laitinen O.H., et al. Enteroviral proteases: structure, host interactions and pathogenicity. Rev. Med. Virol. 2016;26:251–267. doi: 10.1002/rmv.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu J.Y., et al. Secretory carrier membrane protein 3 interacts with 3A viral protein of enterovirus and participates in viral replication. Microbiol. Spectr. 2021;9 doi: 10.1128/Spectrum.00475-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Filipe I.C., Guedes M.S., Zdobnov E.M., Tapparel C. Enterovirus D: a small but versatile species. Microorganisms. 2021;9 doi: 10.3390/microorganisms9081758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown B.A., et al. Resolving ambiguities in genetic typing of human enterovirus species C clinical isolates and identification of enterovirus 96, 99 and 102. J. Gen. Virol. 2009;90:1713–1723. doi: 10.1099/vir.0.008540-0. [DOI] [PubMed] [Google Scholar]
- 33.Lewis-Rogers N., Crandall K.J. M.p. & evolution. Evolution of Picornaviridae: an examination of phylogenetic relationships and cophylogeny. Mol. Phylogenet. Evol. 2010;54:995–1005. doi: 10.1016/j.ympev.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 34.Rota P.A., et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science (New York, N.Y.) 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 35.Gao R., et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 36.MacLean O.A., et al. Natural selection in the evolution of SARS-CoV-2 in bats created a generalist virus and highly capable human pathogen. PLoS Biol. 2021;19 doi: 10.1371/journal.pbio.3001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor L.H., Latham S.M., Woolhouse M.E. Risk factors for human disease emergence. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryu S., Kim B.I., Lim J.S., Tan C.S., Chun B.C. One health perspectives on emerging public health threats. J. Prevent. Med. Public Health = Yebang Uihakhoe chi. 2017;50:411–414. doi: 10.3961/jpmph.17.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conceição-Neto N., et al. Viral gut metagenomics of sympatric wild and domestic canids, and monitoring of viruses: insights from an endangered wolf population. Ecol. Evol. 2017;7:4135–4146. doi: 10.1002/ece3.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Almeida Curi N.H., et al. Wild canids, domestic dogs and their pathogens in Southeast Brazil: disease threats for canid conservation. Biodivers. Conserv. 2010;19:3513–3524. doi: 10.1007/s10531-010-9911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bodewes R., van der Giessen J., Haagmans B.L., Osterhaus A.D., Smits S.L. Identification of multiple novel viruses, including a parvovirus and a hepevirus, in feces of red foxes. J. Virol. 2013;87:7758–7764. doi: 10.1128/jvi.00568-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pénzes J.J., et al. Reorganizing the family Parvoviridae: a revised taxonomy independent of the canonical approach based on host association. Arch. Virol. 2020;165:2133–2146. doi: 10.1007/s00705-020-04632-4. [DOI] [PubMed] [Google Scholar]
- 43.Cotmore S.F., et al. ICTV virus taxonomy profile: Parvoviridae. J. Gen. Virol. 2019;100:367–368. doi: 10.1099/jgv.0.001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ng T.F., et al. A diverse group of small circular ssDNA viral genomes in human and non-human primate stools. Virus Evol. 2015;1:vev017. doi: 10.1093/ve/vev017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kraberger S., et al. Identification of circular single-stranded DNA viruses in faecal samples of Canada lynx (Lynx canadensis), moose (Alces alces) and snowshoe hare (Lepus americanus) inhabiting the Colorado San Juan Mountains. Infect. Genet. Evol. :J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2018;64:1–8. doi: 10.1016/j.meegid.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 46.Mombo I.M., et al. African non-human primates host diverse enteroviruses. PLoS One. 2017;12 doi: 10.1371/journal.pone.0169067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
These datasets are available in online repositories. A list of repository names and accession numbers can be found in the supplementary material.