Highlights
-
•
KIAA1199 promotes oxaliplatin resistance of colorectal cancer.
-
•
KIAA1199 could reduce endoplasmic reticulum stress (ERS) and.
-
•
KIAA1199 enhances the expression of PARP1 via p-ERK/CHOP/Caspase 3.
-
•
Pyrvinium or olaparib could reverse oxaliplatin resistance of colorectal cancer.
-
•
KIAA1199 promotes metastasis of colorectal cancer via EMT mediated by SNAI1.
Keywords: KIAA1199, O-GlcNAcylation, Endoplasmic reticulum stress, PARP1, SNAI1, EMT
Abstract
Oxaliplatin is a commonly used platinum drug for colorectal cancer (CRC). However, the treatment of CRC by oxaliplatin usually fails because of drug resistance, which results in a huge challenge in the therapy of CRC. Elucidation of molecular mechanisms may help to overcome oxaliplatin resistance of CRC. In our study, we revealed that KIAA1199 can promote oxaliplatin resistance of CRC. Mechanistically, KIAA1199 prevents oxaliplatin mediated apoptosis via up-regulated PARP1 derived from reduced endoplasmic reticulum stress induced by protein O-GlcNAcylation. In the meantime, KIAA1199 can also trigger epithelial mesenchymal transition by stabilizing SNAI1 protein via O-GlcNAcylation. Therefore, KIAA1199 has great potential to be a novel biomarker, therapeutic target for oxaliplatin resistance and metastasis of CRC.
Introduction
Colorectal cancer (CRC) is a common malignant tumor with second leading occurrence and deprives approximate one million lives per year globally [1]. Previous studies showed that the mortality of CRC is tightly associated with chemoresistance and metastasis [2,3]. Oxaliplatin is a vital chemotherapeutic drug for CRC. It can trigger apoptosis of cancer cells by formation of platinum-DNA adducts [4,5]. When oxaliplatin is administered with fluorouracil and leucovorin (FOLFOX) in previously untreated CRC, the objective response rate (OR) and median overall survival (OS) can reach 50% and 18–24 months, respectively [4]. Unfortunately, drug resistance will aggravate CRC and remains a challenge in CRC treatment [6,7]. Previous studies suggested the reasons of oxaliplatin resistance including DNA damage repair, apoptosis, NF-κB signaling, and cellular transportation etc. [8]. However, the molecular mechanisms of oxaliplatin resistance of CRC still need further studies.
Nowadays, a novel oncogene, KIAA1199, has been found with key functions in many diseases [9]. KIAA1199 (alias CEMIP, TMEM2L) is located on chromosome 15q25.1 and encodes a 150-kDa protein. KIAA1199 was firstly identified as an inner ear-specific protein and correlated with deafness [10]. Later on, researchers found that KIAA1199 can be expressed in various human tissues and dysregulated expression of KIAA1199 can be detected in numerous cancerous tissues [11]. The basic function of KIAA1199 remains unknown, nevertheless several studies indicate its role in metabolism of hyaluronan [12]. Recently, a growing amount of studies found that KIAA1199 can promote growth and metastasis of various cancers including CRC, breast cancer, pancreatic cancer etc. [2]. Moreover, KIAA1199 has strong relationships with signaling pathways including wnt/β-catenin, MAPK and PI3K/AKT etc. [11].
O-GlcNAcylation is a special type of post-translational modification of protein, which is established by a bond of single N-acetylglucosamine and serine or threonine residue of protein. Protein O-GlcNAcylation responds to changes in the cellular conditions caused by stress, hormones, or nutrition variations by dynamical attachment or removal of sugars on proteins. So far, O-GlcNAcylation modification has been found in more than thousands of nuclear and cytoplasmic proteins widely involved in transcription, translation, signal transduction, cell cycle and other physiological events [13,14]. It is believed that O-GlcNAcylation plays an important role in diabetes, neurodegenerative diseases, heart diseases, tumors and other diseases, and has great potential for diagnosis and treatment of these diseases [15,16]. Amazingly, our previous study revealed that KIAA1199 could promote O-GlcNAcylation of protein via bridging O-GlcNAc transferase (OGT) and substrate protein [17].
In our study, we deciphered that KIAA1199 was up-regulated in oxaliplatin resistant CRC cells compared with parental cells. KIAA1199 can prevent apoptosis of CRC cells by alleviating endoplasmic reticulum stress (ERS) via protein O-GlcNAcylation. Furthermore, KIAA1199 has ability to maintain epithelial mesenchymal transition (EMT) and promotes metastasis of CRC via O-GlcNAcylation modification of SNAI1.
Materials and methods
Cells culture
Human CRC cells (SW480 and HCT116) used in our research were purchased from Chinese Academy of Sciences (CBTCCCAS, China). SW480 and HCT116 were cultured in RPMI 1640 (GIBCO, USA) or DMEM (GIBCO, USA), respectively. Supplementation with 10% fetal bovine serum (BI, Israel), 100 U/mL penicillin and 100 μg/mL streptomycin was used. All cell lines were authenticated by STR profiling.
Plasmid transduction and stable cell lines
Lipofectamine 2000 (Invitrogen, USA) was used to perform transient transduction of cells. Firstly, plasmids and Lipofectamine 2000 were diluted in opti-MEM (GIBCO, USA) respectively. After 5 min, plasmids and Lipofectamine 2000 dilutions were mixed gently and reacted at room temperature for 20 min. Later, the mixture of plasmids and Lipofectamine 2000 was transferred to serum-free growth medium for cells culture. Lastly, the serum-free growth medium was replaced by fresh medium with 10% FBS after 4 h and cells were cultured for 24–48 h. Stable cell lines were made by lentivirus infection and puromycin (AbMole, USA) resistance selection. Detailed transduction procedure was illustrated in our previous study [17]. For silence experiment of gene, short hair RNA (shRNA) was used in our study. The target sequences of shRNAs were listed in supplementary Table 1.
Proteins extraction and western blotting
Whole cell lysates were produced by RIPA lysis buffer (Beyotime, China). Prepared cell lysates were quantified by BCA methods and electrophoresed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Thereafter, the separated proteins were transferred to PVDF membrane (Millipore, USA). After that, the proteins coated PVDF membrane was incubated with primary antibodies at 4 °C overnight. Next day, the membrane was incubated with HRP-conjugated second antibodies at room temperature for 1 h. At last, the target bands were visualized by chemiluminescence system (Bio-Rad, USA).
Several primary antibodies including mouse anti-β-actin (RRID:AB_2,687,938; Proteintech, #66,009–1-Ig, 1:1000), rabbit anti-KIAA1199 (RRID:AB_10,695,771; Proteintech, #21,129–1-AP, 1:1000), HRP-conjugated Affinipure Goat Anti-Rabbit IgG(H + L) (RRID:AB_2,722,564; Proteintech, #SA00001–2, 1:3000), HRP-conjugated Affinipure Goat Anti-Mouse IgG (H + L) (RRID:AB_2,722,565; Proteintech, #SA00001–1, 1:3000), rabbit anti-O-GlcNAc (CST, #82332S, 1:1000), rabbit anti-E-cadherin (CST, #3195S, 1:1000), rabbit anti-SNAI1 (RRID:AB_2,191,756; Proteintech, #13,099–1-AP, 1:1000), mouse anti-PARP1 (RRID:AB_2,881,883; Proteintech, #66,520–1-Ig, 1:1000), mouse anti-caspase 3 (RRID:AB_2,876,892; Proteintech, #66,470–2-Ig, 1:1000), mouse anti-CHOP (RRID:AB_2,882,089; Proteintech, #66,741–1-Ig, 1:1000), mouse anti-ATF4 (RRID:AB_2,058,598; Proteintech, #60,035–1-Ig, 1:1000), rabbit anti-p-eIF2α (CST, #3398S, 1:1000) and rabbit anti-p-PERK (CST, #3179S, 1:1000), were used in western blotting experiments.
Immunohistochemistry (IHC)
Human CRC specimens were collected from Wuhan union hospital. This research was carried out in accordance with the declaration of Helsinki and formally approved by the institutional medical ethics committee in Wuhan union hospital. Informed consents were signed by all included volunteers. The anti-KIAA1199 (1:400), anti-PARP1 (1:500) and anti-SNAI1 (1:500) antibodies were used for the primary reaction. Immunoperoxidase staining was performed later. The semi-quantification of target proteins were calculated by Image J software.
Reverse transcription quantitative PCR (RT-qPCR)
Total RNA Kit (Omega, China) was used to extract the total RNA in CRC cells. One microgram of RNA was used to synthesize the complementary DNA (cDNA) by using UEIris II RT-PCR system for first-strand cDNA synthesis mix (US EVERBRIGHT, China). The mixture of RNA and reagents of first-strand cDNA synthesis mix was reacted for 10 min at 55℃. Then, the reaction ended at 85℃ for 10 s. Later, real-time quantitative PCR experiment was performed in Applied Biosystems 7500 Instrument (Thermo, USA) following the protocol of 2×SYBR Green qPCR Master Mix (US EVERBRIGHT, China). The total volume of reaction mixture was 50 μL. The thermo cycling conditions were: hold at 95℃ for 10 min, (95℃ for 15 s followed by 60℃ for 60 s) × 40 cycles. GAPDH was used as housekeeping gene in our PCR experiments, and the relative quantification of target genes were determined by the ΔΔCq method. The sequences of primers for RT-qPCR were exhibited in Supplementary Table 2.
Cell proliferation and colony assay
CRC cells were seeded and cultured in 96-well plate, and then the cells were treated with indicated drugs. After preset time, cell culture medium with FBS was replaced by serum-free medium. At the same time, CCK-8 solution (Dojindo, Japan) was added and incubated at 37 °C for 2 h. The absorbance values of each well at 450 nm were measured by a microplate reader (Thermo, USA). For colony experiments, CRC cells were seeded and grown in 6-well plates with indicated treatments. Cells were cultured for about2 weeks until obvious colony formation under a microscope. Then, the wells were washed with PBS and the cells were fixed with paraformaldehyde. The fixed cell colony were dyed with 0.5% crystal violet. Colonies containing at least 50 cells were counted manually.
Wound healing and cell migration, invasion assays
For cell wound healing experiments, CRC cells were seeded and grown in 6-well plates. When cells overspread the whole well, several cells were smoothly removed and a wound were created by using a 20 μL pipet. Baseline of the width of wounds were measured by a microscope, and indicated treatments were performed later. Until indicated times for culture in serum-free medium, the width of wounds were measured again and the healed area were calculated by Image J software. For cell migration experiments, CRC cells were suspended in 200 μL serum-free medium and added into the upper chamber of 24-well Transwell inserts (Corning, USA). Meanwhile, 700 μL medium with 10% FBS were added into the lower chamber. The Transwell inserts were incubated at 37 °C for 24 h and allowed the cells migrate across the membrane. After that, the migrated cells were fixed by paraformaldehyde, and dyed by 0.5% crystal violet, and counted manually by an optical microscope. Cell invasion assays were performed similarly with membrane coated with 30 μg Matrigel (BD Biosciences, USA). For both wound healing and transwell experiments, the proliferation of each group of cells cultured in serum-free medium were assessed by CCK-8 experiments. The migration and invasion abilities of cells would be corrected when the proliferation rates of each group of cells were different.
Glycosylated protein purification
For glycosylated proteins purification experiments, the whole cell lysates were prepared by repetitive freeze thaw method. Next, centrifugation of cell lysates were conducted and the supernatants were mixed with wheat germ agglutinin (WGA) agarose (Vector, USA) and incubated overnight at 4 °C. Next day, after centrifugation of the mixture, precipitates were collected and washed with PBS for 5 times. Thereafter, glycosylated proteins were adhered on the surface of WGA agarose.
Animal experiments
Animal experiments in our study were approved by the Medical Ethics Committee of Wuhan union hospital. For mouse model with subcutaneous tumor, four to six weeks female BALB/c nude mice were allocated randomly (by random number method) into indicated groups (5 mice in every group) and subcutaneously injected with 5 × 106 indicated HCT116 cells. Volumes of subcutaneous tumors were measured and calculated as (length (mm) × width (mm) × width (mm))/2 every 3 days. Mice were sacrificed until indicated days and subcutaneous tumors were isolated by surgery. The weights of tumors were measured and compared between groups. To establish mice liver metastasis model, four to six weeks female BALB/c nude mice were allocated randomly into indicated groups (5 mice in every group) and received intrasplenic injection with 3 × 106 indicated HCT116 cells after anesthesia by pentobarbital sodium. After indicated treatments and time, mice were sacrificed for count of liver metastasis. Detailed procedure of animal tumor bearing model establishment was illustrated in our previous study [17].
Statistics analysis
All experiments were repeated in triplicate or more. Data were noted as mean value ± standard deviation (SD). Statistical analyses were performed with SPSS16.0. GraphPad Prism 7 was used to plot figures. The statistical differences were calculated by Student's t-test. The correlations between KIAA1199 and PARP1, SNAI11 expression were analyzed by linear regression. Survival of patient were determined by Kaplan−Meier curve and Log-Rank test. Two-sided P value ≤ 0.05 was considered statistically significant.
Results
KIAA1199 mediates CRC oxaliplatin resistance in vitro and in vivo
To establish oxaliplatin resistant CRC cells (SW480R and HCT116R), parental CRC cells (SW480 and HCT116) were treated with increasing concentrations of oxaliplatin for about 6 months as shown in Supplementary Fig. 1A. The resistance of oxaliplatin was confirmed by comparing the proliferation, colony and apoptosis phenotype between parental and resistant CRC cells in vitro. As shown in Fig. 1A-C, SW480R and HCT116R have higher IC50 and colony formation rates than parental cells. Moreover, oxaliplatin resistant cells have dramatically lower apoptotic rates compared with parental cells (Fig. 1D). To further verify the resistance of oxaliplatin in vivo, HCT116 or HCT116R cells were subcutaneously implanted in nude mice. Oxaliplatin (10 mg/kg) was administrated for tumor bearing mice by intraperitoneal injection once a week (Supplementary Fig. 1B). The volumes of subcutaneous tumor were measured and calculated every 3 days until indicated days. After sacrifice of mice, the tumor were isolated and weighed. Interestingly, the tumor derived from HCT116R cells grew faster than HCT116 cells (Fig. 1E). The volumes and weights of tumors derived from HCT116R cells were significantly higher than control tumors (Supplementary Fig. 1C, Fig. 1F, G).
Fig. 1.
Establishment and validation of oxaliplatin resistant CRC cells. A, B Inhibition rate and IC50 of parental and resistant cells treated with oxaliplatin (3 independent replicates). ****P < 0.0001. C Colony formation analysis of parental and resistant cells treated with oxaliplatin (3 independent replicates). ****P < 0.0001. D Apoptosis analysis of parental and resistant cells treated with oxaliplatin (3 independent replicates). ****P < 0.0001. E-G Tumor growth of HCT 116/HCT116R cells bearing mice (n = 5 mice in each group). Comparison of growth rate (E), tumors volume (F) and weight (G) between two groups. **P < 0.01, ****P < 0.0001.
To elucidate the molecular mechanisms of oxaliplatin resistance of CRC, we performed RNA-sequence analysis for both HCT116 parental and resistant cells. As shown in Fig. 2A, thousands differentially expressed genes were identified between HCT116 and HCT116R cells. Surprisingly, one of the differentially expressed genes, KIAA1199, had been previously studied by the authors. Therefore, we focused on this novel gene in the present study. In CRC, the expression level of KIAA1199 was significantly up-regulated compared with normal tissues according to the data in The Cancer Genome Atlas (TCGA) datasets (Fig. 2B). The median disease free survival of patients with higher KIAA1199 is obviously shorter than that with lower KIAA1199 (HR=1.8, log-rank P = 0.026) (Fig. 2C). In addition, there is evident difference of KIAA1199 expression between early and advanced CRC (Supplementary Table 3). The differences between CRC with microsatellite instability-high (MSI-H) and MSI-Low/MSI-stable feature is not statistically significant (Supplementary Table 3). Detailed characteristics of included volunteers from TCGA datasets were showed in Supplementary Table 3. In our previous study, we found that KIAA1199 could promote glutamine metabolic reprogramming and metastasis of CRC via reciprocal regulation of β-catenin. However, the relationships between KIAA1199 and oxaliplatin resistance of CRC remain unclear.
Fig. 2.
KIAA1199 promotes oxaliplatin resistance of CRC. A Volcano plot of identified genes in HCT116 and HCT116R cells. B Comparison of KIAA1199 mRNA expression of tumor and normal tissues included in TCGA datasets. ****P < 0.0001. C Patients with higher level of KIAA1199 have shorter median disease free survival compared with patients with lower KIAA1199 (HR=1.8, log-rank P = 0.026). D Upper panel: Relative KIAA1199 mRNA levels in parental cells compared with resistant cells. Lower panel: western blotting analysis of KIAA1199 protein expression in parental cells compared with resistant cells (3 independent replicates). ***P < 0.001, ****P < 0.0001. E, F Inhibition rate and IC50 of KIAA1199 over-expressed cells and control cells treated with oxaliplatin (3 independent replicates). ****P < 0.0001. G Tumor growth rate of HCT 116/HCT116R cells bearing mice between two groups (n = 5 mice in each group). **P < 0.01.
To confirm the expression level of KIAA1199 in parent and oxaliplatin resistant CRC cells, we performed q-PCR and western blotting experiments, and the outcomes confirmed that KIAA1199 was over-expressed in oxaliplatin resistant CRC cells (Fig. 2D). To further investigate the role of KIAA1199 in oxaliplatin resistance of CRC cells, we evaluated the IC50 of both KIAA1199 over-expressed CRC cells and control cells. As we can see in Fig. 2E and F, the IC50 of CRC cells with over-expressed KIAA1199 were significantly higher than control cells. To confirm the effect of KIAA1199 on growth of CRC in vivo, HCT116 cells with stably over-expressed KIAA1199 and control cells were subcutaneously implanted in nude mice. The volumes of tumors were measured and calculated every 3 days until indicated days. The results demonstrated that KIAA1199 over-expressed tumor grew obviously faster than control tumor under oxaliplatin treatment (Fig. 2G). After sacrifice of mice, the tumor were isolated and weighed. The volumes and weights of tumors derived from HCT116 cells with stably over-expressed KIAA1199 were significantly higher than control tumors (Supplementary Fig. 1D-F).
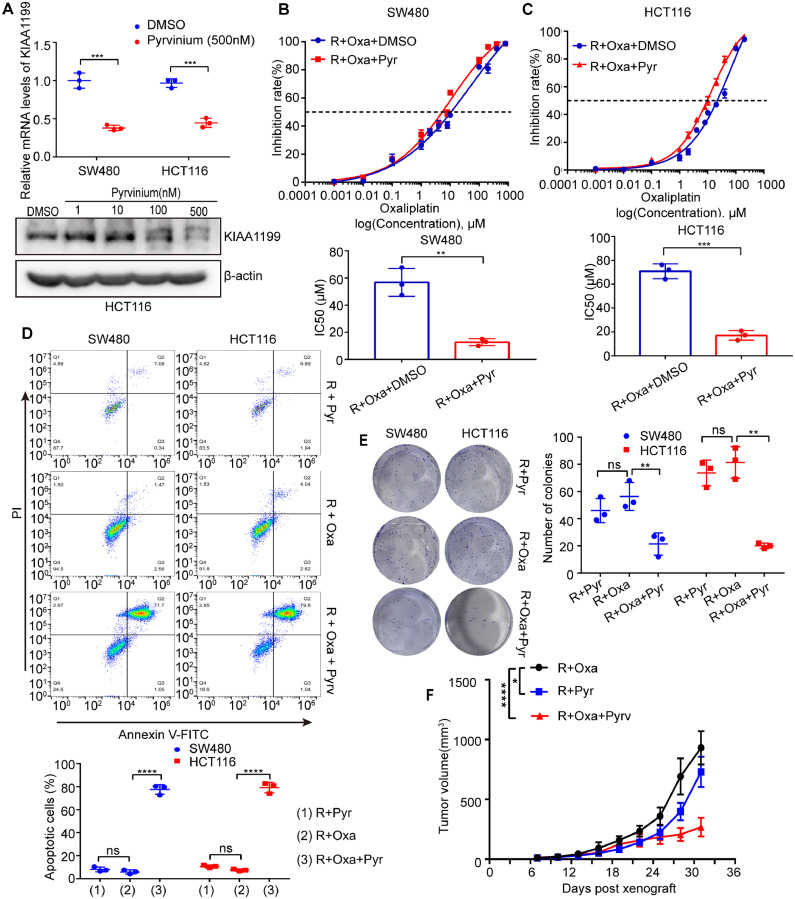
Inhibition of KIAA1199 by pyrvinium reverses oxaliplatin resistance of CRC
It has been demonstrated that KIAA1199 could promote oxaliplatin resistance of CRC cells by our experiments. Thereafter, we tried to address this issue by a convenient strategy. It has been verified that the expression of KIAA1199 was regulated by wnt/β-catenin signaling in our previous study [17]. Pyrvinium, a FDA approved anti-worm drug, can inhibit wnt/β-catenin signaling in CRC cells [18]. Based on these information, we hypothesized that pyrvinium could reverse oxaliplatin resistance of CRC cells. The results of our q-PCR and western blotting experiments indicated that pyrvinium could significantly reduce the mRNA and protein level of KIAA1199 in CRC cells (Fig. 3A). To discover the effect of pyrvinium upon oxaliplatin resistance in vitro, we performed proliferation, colony and apoptosis experiments. Consistent with our hypothesis, pyrvinium can dramatically reduce proliferation rate of resistant CRC cells with oxaliplatin treatment in vitro (Fig. 3B, C). Apoptosis and colony experiments confirmed that pyrvinium could significantly reverse the oxaliplatin resistance of CRC cells (Fig. 3D, E). To verify the effect of pyrvinium on oxaliplatin resistance of CRC in vivo, HCT116R cells were subcutaneously implanted in nude mice. Tumor bearing mice were treated with oxaliplatin (10 mg/kg i.p. every week), pyrvinium (25 mg/kg p.o. every two days), or combination of oxaliplatin and pyrvinium. The volumes of tumors were measured and calculated every 3 days until indicated days. After sacrifice of mice, tumors were isolated and weighed.The results indicated that pyrvinium had potential to reverse oxaliplatin resistance of CRC (Supplementary Fig. 1G, Fig. 3F). To conclude, pyrvinium can reduce the expression of KIAA1199 and reverse oxalipatin resistence of CRC both in vitro and in vivo.
Fig. 3.
Inhibition of KIAA1199 by pyrvinium reverses oxaliplatin resistance of CRC. A Upper panel: Relative KIAA1199 mRNA levels in CRC cells treated with pyrvinium compared with control cells. Lower panel: western blotting analysis of KIAA1199 protein expression in CRC cells treated with pyrvinium compared with control cells (3 independent replicates). ***P < 0.001. B, C Inhibition rate and IC50 of resistant cells treated with pyrvinium compared with control cells (3 independent replicates). **P < 0.01, ***P < 0.001. D Apoptosis analysis of resistant cells treated with pyrvinium compared with control cells (3 independent replicates). ****P < 0.0001. ns, not significant. E Colony formation analysis of resistant cells treated with pyrvinium compared with control cells (3 independent replicates). **P < 0.01. ns, not significant. F Tumor growth rate of HCT 116/HCT116R cells bearing mice treated with pyrvinium compared with control mice (n = 5 mice in each group). *P < 0.05, ****P < 0.0001.Pyr, pyrvinium; Oxa, oxaliplatin; R, resistant.
KIAA1199 promotes CRC oxaliplatin resistance via upregulated PARP1 mediated by reducing ERS deriving from protein O-GlcNAcylation
It has been demonstrated that KIAA1199 could promote oxaliplatin resistance of CRC and inhibition of KIAA1199 can reverse the resistance. However, the mechanism of KIAA1199 mediated oxaliplatin resistance was unknown. In vitro, KIAA1199 has no effect upon proliferation, colony and apoptosis of CRC cells without oxaliplatin treatment (Supplementary Fig. 1H-K). Previous studies indicated that oxaliplatin resistance had tight correlation with PARP1 (Poly (ADP-ribose) polymerase 1) [2]. Our bioinformatics analyses showed that the expression level of KIAA1199 had positive correlation with PARP1 (Fig. 4A). Our western blotting and IHC experiments suggested that KIAA1199 could promote the expression of PARP1 (Fig. 4B, C). Furthermore, the increased PARP1 was mediated by p-ERK/CHOP/Caspase 3 signaling axis (Fig. 4B). It's well known that p-ERK/CHOP/Caspase 3 signaling plays a key role in ERS and apoptosis [19]. Based on these results, it can be concluded that overexpression of KIAA1199 could relieve ERS and vice versa.
Fig. 4.
KIAA1199 promotes CRC oxaliplatin resistance via upregulated PARP1 mediated by reducing endoplasmic reticulum stress (ERS). A Correlation analysis of the mRNA level of KIAA1199 and PARP1. B Western blotting analysis of KIAA1199, PARP1, and p-ERK/CHOP/Caspase 3 signaling in CRC cells transducted with indicated plasmid (3 independent replicates). C Representive figure of IHC and correlation analysis of the protein level of KIAA1199 and PARP1 in human CRC tissues. Scale bar, 500μS. D Inhibition rate and IC50 of SW480R cells treated with olaparib compared with control cells (3 independent replicates). ****P < 0.0001. Oxa, oxaliplatin; Ola, olaparib.
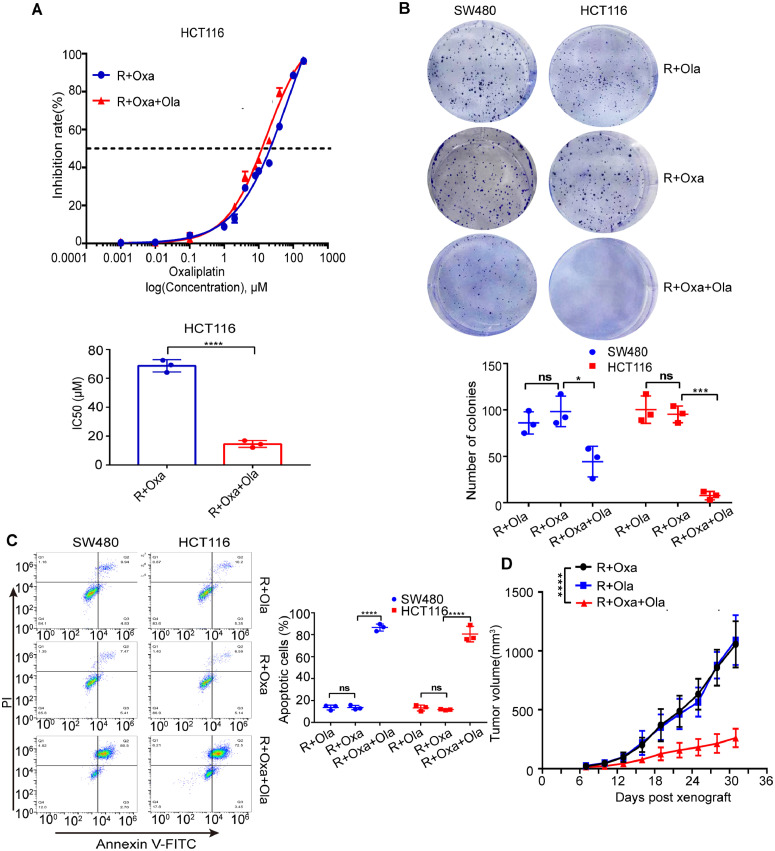
Olaparib is a novel PARP inhibitor which can induce synthetic lethality in BRCA 1/2 deficient cancer cells. It can be used as maintenance treatment in patients with advanced ovarian cancer, fallopian tube cancer, or primary peritoneal cancer with a certain type of abnormal BRCA 1/2 gene [20], [21], [22]. Amazingly, our proliferation, colony and apoptosis experiments showed that olaparib could reverse oxaliplatin resistance of CRC in vitro (Figs. 4D,E and 5A-C). To confirm the effect of olaparib on oxaliplatin resistance of CRC in vivo, HCT116R cells were subcutaneously implanted in nude mice. Tumor bearing mice were treated with oxaliplatin (10 mg/kg i.p. every week), olaparib (50 mg/kg i.p. every day), or combination of oxaliplatin and olaparib. The volumes of tumors were measured and calculated every 3 days until indicated days. After sacrifice of mice, tumors were isolated and weighed. As we can see in Supplementary Fig. 2A and Fig. 5D, olaparib can reverse oxaliplatin resistance of CRC in vivo.
Fig. 5.
Inhibition of PARP1 by olaparib reverses oxaliplatin resistance of CRC. A Inhibition rate and IC50 of HCT116R cells treated with olaparib compared with control cells (3 independent replicates). ****P < 0.0001. B Colony formation analysis of oxaliplatin resistant cells treated with olaparib compared with control cells (3 independent replicates). *P < 0.05, ***P < 0.001. ns, not significant. C Apoptosis analysis of oxaliplatin resistant cells treated with olaparib compared with control cells (3 independent replicates). ****P < 0.0001. ns, not significant. D Tumor growth rate of HCT 116/HCT116R cells bearing mice treated with olaparib compared with control mice (n = 5 mice in each group). ****P < 0.0001. Ola, olaparib; Oxa, oxaliplatin; R, resistant.
Our previous study found that KIAA1199 could enhance O-GlcNAcylation modification level of proteins (Fig. 6A) [17]. Previous articles revealed that unfold proteins derived from non-sufficient glycosylation could trigger ERS and apoptosis [23]. We believed that increased O-GlcNAcylation level of proteins mediated by KIAA1199 could prevent ERS and apoptosis. In agreement with our expectation, decreased O-GlcNAcylation of proteins derived from silence of OGT, could reverse KIAA1199 mediated inhibition of ERS and apoptosis (Supplementary Fig. 2B). Mounting studies discovered that enhanced cellular O-GlcNAcylation could promote tumor growth and progression [24], [25], [26], [27]. Our experiments showed that increased O-GlcNAcylation mediated by thiamet G (an inhibitor of O-GlcNAcase) could promote oxaliplatin resistance of CRC cells. Conversely, decreased O-GlcNAcylation mediated by silence of OGT significantly reverse oxaliplatin resistance of CRC cells (Fig. 6B-K). Collectively, KIAA1199 promotes oxaliplatin resistance of CRC via upregulated PARP1 mediated by reducing ERS deriving from protein O-GlcNAcylation.
Fig. 6.
Protein O-GlcNAcylation promotes oxaliplatin resistance of CRC. A Western blotting analysis of KIAA1199 and protein O-GlcNAcylation in CRC cells transducted with indicated plasmid (3 independent replicates). B Inhibition rate and IC50 of SW480 cells treated with oxaliplatin and thiamet G compared with control cells (3 independent replicates). **P < 0.01. C Inhibition rate and IC50 of HCT116 cells with reduced OGT compared with control cells (3 independent replicates). ***P < 0.001. D, E Colony formation analysis of SW480 cells treated with oxaliplatin and thiamet G compared with control cells (3 independent replicates). ****P < 0.0001. F, G Colony formation analysis of OGT reduced HCT116R cells treated with oxaliplatin compared with control cells (3 independent replicates). ***P < 0.001. H, I Apoptosis analysis of SW480 cells treated with oxaliplatin and thiamet G compared with control cells (3 independent replicates). ***P < 0.001. J, K Apoptosis analysis of OGT reduced HCT116 cells treated with oxaliplatin compared with control cells (3 independent replicates). ***P < 0.001.
KIAA1199 promotes EMT and metastasis of CRC through O-GlcNAcylation of SNAI1
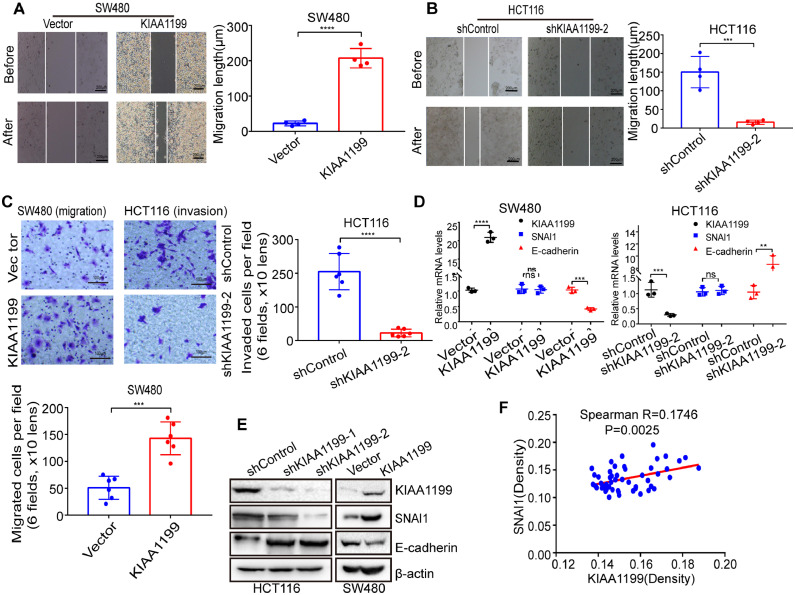
It is well known that KIAA1199 can promote metastasis of different types of cancer [11]. Our wound healing and transwell experiments confirmed that KIAA1199 could enhance migration and invasion of CRC cells in vitro (Fig. 7A-C). More and more studies showed that EMT played a vital role in CRC metastasis and SNAI1 was an important transcriptor involved in EMT [28]. Our experiments indicated that KIAA1199 could enhance the protein expression of SNAI1 but not mRNA. Conversely, both protein and mRNA of E-cadherin were reduced in KIAA1199 over-expressed CRC cells (Fig. 7D, E). IHC of human CRC tissues confirmed the positive association between KIAA1199 and SNAI1 (Supplementary Fig. 2C, Fig. 7F). Knock down and rescue of SNAI1 experiments indicated that SNAI1 was indispensable for KIAA1199 mediated migration and invasion of CRC cells (Fig. 8A, B). Given that KIAA1199 can increase the expression of protein but not mRNA of SNAI1, we hypothesized that KIAA1199 could regulate the stability of SNAI1 protein. Previous study indicated that O-GlcNAcylation modification of SNAI1 protein could prevent its phosphorylation and subsequent proteasome degradation [2]. Our co-immunoprecipitation and western blotting experiments confirmed that SNAI1 interacted with OGT and KIAA1199 could promote O-GlcNAcylation of SNAI1 (Fig. 8C, D). Wound healing and transwell experiments showed that enhanced O-GlcNAcylation mediated by thiamet G could promote migration and invasion of CRC cells. Conversely, reduced O-GlcNAcylation mediated by silence of OGT significantly decrease migration and invasion of CRC cells (Supplementary Fig. 3A-D). To verify the association of KIAA1199 and SNAI1, CRC liver metastasis model were established in nude mice by intrasplenic injection with 3 × 106 indicated HCT116 cells after anesthesia by pentobarbital sodium. After indicated time, mice were sacrificed for count of liver metastasis. As shown in Fig. 8E, KIAA1199 has great potential to promote metastasis of CRC through SNAI1 (Fig. 9).
Fig. 7.
KIAA1199 promotes EMT and metastasis of CRC. A, B Wound healing analysis of CRC cells transducted with indicated plasmid (4 independent replicates). ***P < 0.001, ****P < 0.0001. Scale bar, 200μc. C Transwell analysis of CRC cells transducted with indicated plasmid (6 independent replicates). ***P < 0.001, ****P < 0.0001. Scale bar, 100 μm. D Relative KIAA1199, SNAI1 and E-cadherin mRNA levels in CRC cells transducted with indicated plasmid (3 independent replicates). **P < 0.01, ***P < 0.001, ****P < 0.0001. ns, not significant. E Western blotting analysis of KIAA1199, SNAI1 and E-cadherin proteins expression in CRC cells transducted with indicated plasmid (3 independent replicates). F Correlation analysis of the protein expression of KIAA1199 and SNAI1 in human CRC tissues.
Fig. 8.
KIAA1199 promotes CRC metastasis via SNAI1 mediated EMT. A Wound healing analysis of CRC cells transducted with indicated plasmid (4 independent replicates). ***P < 0.001, ****P < 0.0001. Scale bar, 200 μm. B Transwell analysis of CRC cells transducted with indicated plasmid (6 independent replicates). ***P < 0.001, ****P < 0.0001. Scale bar, 100 μm. C Co-immunoprecipitation experiments of OGT and SNAI1 (3 independent replicates). D Western blotting analysis of SNAI1 and O-GlcNAcylated SNAI1 proteins expression in CRC cells transducted with indicated plasmid (3 independent replicates). E Liver metastasis of mice implanted HCT116 cells stably transducted with indicated plasmid compared with control mice. (n = 5 mice in each group). ***P < 0.001, ****P < 0.0001.
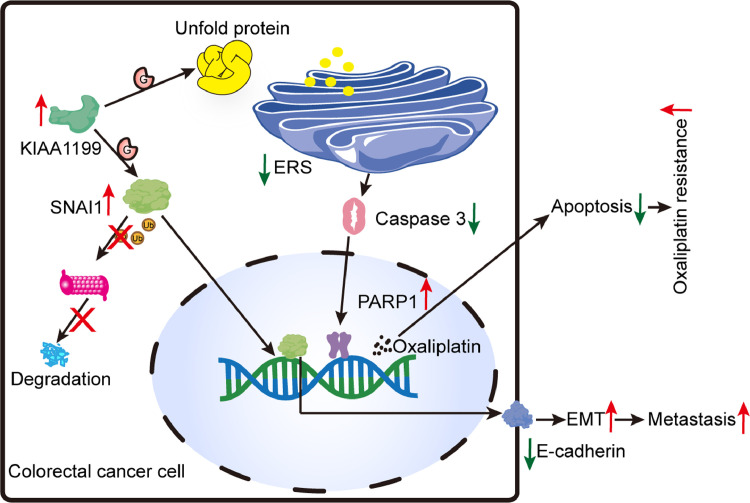
Fig. 9.
Graphical abstract of the present study. KIAA1199 promotes both of oxaliplatin resistance and EMT of CRC cells. Mechanistically, KIAA1199 can enhance protein O-GlcNAcylation that has ability to reduce unfolded protein and prevent ERS and apoptosis in oxaliplatin resistant CRC cells. Specifically, oxaliplatin mediated cellular DNA damage can be reversed by increased PARP1 regulated by p-ERK/CHOP/Caspase 3 signaling axis. In addition,KIAA1199 can promote stability of SNAI1 via protein O-GlcNAcylation, which has a vital role in EMT and metastasis of CRC.
Discussion
Oxaliplatin was considered as standard first-line chemotherapy drug for CRC, which significantly improved survival and decreased recurrence rate of CRC patients. Nevertheless, oxaliplatin chemotherapy often fails because of drug resistance. Previous studies indicated that oxaliplatin resistance was correlated with cellular transport, detoxification, DNA repair, cell death, and epigenetic alternation etc. [8]. In the present study, we found that KIAA1199 can promote oxaliplatin resistance of CRC. As a novel protein that has tight correlation with drug resistance, KIAA1199 has no effect upon proliferation, colony of CRC cells. KIAA1199 maintains oxaliplatin resistance of CRC cells through prevention of apoptosis of CRC cells with exposure to oxaliplatin. Mechanistically, KIAA1199 can prevent apoptosis of CRC cells via relief of ERS induced by oxaliplatin. Other studies also showed the tight association between KIAA1199 and drug resistance. Xu et al. demonstrated that KIAA1199 was over-expressed in sorafenib resistant hepatocellular carcinoma (HCC) and played an important role in metastasis of sorafenib-resistant HCC [29]. Duong et al.'s study indicated that CRC acquired resistance to MEK1 inhibitor by inducing expression of KIAA1199 [30]. In conclusion, KIAA1199 has great potential to be a novel promising therapeutic target to overcome oxaliplatin resistance.
Endoplasmic reticulum (ER) is the major organelle for protein post-translational folding, modification and trafficking. When intrinsic stresses (e.g., oncogene activation) or hostile environmental conditions (e.g., oxidative stress and nutrition deficiency) occur, normal protein folding was disrupted [23]. The accumulation of unfolded or misfolded proteins tend to trigger unfolded protein response (UPR) and ERS, which intersects with a wide variety of physiological and pathological conditions [31]. ERS had been demonstrated with a vital role in the oncogenesis and progression of a wide range of cancers including tumors of stomach, colon, breast, lung and liver etc. [32,33]. In our study, we found that ERS in oxaliplatin resistant CRC cells was relieved by over-expressed KIAA1199. According to published studies, we believe that O-GlcNAcylation of proteins mediated by KIAA1199 is a necessary guarantee for correct folding and normal physiological functions of proteins. In agreement with our results, Ngoh et al. found that enhanced O-GlcNAcylation could attenuate ERS and prevent cardiomyocyte death [34].
A previous study performed by Chen et al. revealed that KIAA1199 was a target gene of β-catenin/TCF4 [35]. Duong et al. found that TCF4 had binding sites in promoter region of KIAA1199 [36]. Moreover, inhibition of wnt/β-catenin signaling significantly reduce protein expression of KIAA1199. Pyrvinium has been demonstrated with promising therapeutic potential for CRC both in vitro and in vivo [2]. Pyrvinium can bind to and activate CK1α, an important component of destruction complex of β-catenin, leading to reduced β-catenin and repression of wnt/β-catenin signaling. In addition, pyrvinium stabilizes CK1α protein via inhibition of cereblon (a component of E3-ubiquitin ligase of CK1α) [37]. In our study, we revealed that pyrvinium could re-sensitize resistant CRC to oxaliplatin through inhibition of KIAA1199. Arena et al.'s study indicated that several CRC cell lines were highly sensitive to olaparib. Moreover, response to olaparib was positively associated with sensitivity to oxaliplatin in CRC cells [38]. In our study, we found that olaparib can reverse the oxaliplatin resistance of CRC, which indicated that combination of olaparib and oxaliplatin may behave better than single drug.The most special characteristic of malignant tumor cells is their ability of metastasis [39]. To establish effective prevention and treatment strategies, molecular mechanisms of tumor metastasis should be uncovered firstly. Tumor metastasis is a complex process involving a series of biological steps [40]. It is believed that EMT plays a central role in tumor metastasis [41]. Mounting studies demonstrated that SNAI1 was an important transcriptor involved in EMT [42]. Mechanistically, SNAI1 attenuates the expression of E-cadherin, a key membrane protein involved in cell adhesion. SNAI1 has been identified with O-GlcNAcylation sites correlated with its protein stability. Specifically, O-GlcNAcylation of SNAI1 prevents its phosphorylation and subsequent proteasome degradation [43]. Herein, we verified that KIAA1199 could increase O-GlcNAcylation level of SNAI1, which resulted in EMT and metastasis of CRC. KIAA1199 may be a potential therapeutic target for CRC metastasis.
Conclusion
In conclusion, KIAA1199 mediates both oxaliplatin resistance and EMT of CRC cells. Mechanistically, KIAA1199 prevents ERS induced apoptosis in oxaliplatin resistant CRC cells and promote stability of SNAI1 via protein O-GlcNAcylation. Inhibition of KIAA1199 can reverse oxaliplatin resistance and reduce metastasis of CRC.
Funding
This work was supported by the scientific research project of Anhui provincial department of education (No. 2022AH051132). This work was supported by the training program foundation of youth scholars by the first affiliated hospital of Anhui medical university (No. 2020kj24).
Ethics approval and consent to participate
Human CRC specimens were collected from Wuhan union hospital. This research was carried out in accordance with the declaration of Helsinki and formally approved by the institutional medical ethics committee in Wuhan union hospital. Informed consents were signed by all included volunteers. Animal experiments in our study were approved by the Medical Ethics Committee of Wuhan union hospital.
Declaration of Competing Interest
The authors declare that they have no conflicts of interest.
Data availability statement
All the data from the current study are included in the article or uploaded as supplementary information.
CRediT authorship contribution statement
Qingling Hua: Conceptualization, Supervision, Investigation, Methodology, Funding acquisition, Project administration, Writing – review & editing. Yuanyuan Lu: Investigation, Methodology, Formal analysis. Dingxiang Wang: Investigation, Methodology, Formal analysis. Jie Da: Resources, Supervision. Wanren Peng: Resources, Supervision, Conceptualization, Supervision, Investigation, Methodology, Project administration, Writing – review & editing. Guoping Sun: Resources, Supervision. Kangsheng Gu: . Hua Wang: Conceptualization, Supervision, Investigation, Methodology, Project administration, Writing – review & editing. Yanzhe Zhu: Conceptualization, Supervision, Investigation, Methodology, Project administration, Writing – review & editing.
Acknowledgment
Not applicable.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2023.101617.
Appendix. Supplementary materials
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Lambert A.W., Pattabiraman D.R., Weinberg R.A. Emerging biological principles of metastasis. Cell. 2017;168:670–691. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D. Kanojia, M. Garg, S. Gupta, A. Gupta, and A. Suri, Sperm-associated antigen 9 is a novel biomarker for colorectal cancer and is involved in tumor growth and tumorigenicity. 178 (2011) 630–690. [DOI] [PMC free article] [PubMed]
- 4.Rottenberg S., Disler C., Perego P. The rediscovery of platinum-based cancer therapy. Nat. Rev. Cancer. 2021;21:37–50. doi: 10.1038/s41568-020-00308-y. [DOI] [PubMed] [Google Scholar]
- 5.De Mattia E., Cecchin E., Toffoli G. Pharmacogenomics of intrinsic and acquired pharmacoresistance in colorectal cancer: toward targeted personalized therapy. Drug Resist. Updat. 2015;20:39–70. doi: 10.1016/j.drup.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Crea F., Nobili S., Paolicchi E., Perrone G., Napoli C., Landini I., Danesi R., Mini E. Epigenetics and chemoresistance in colorectal cancer: an opportunity for treatment tailoring and novel therapeutic strategies. Drug Resist. Updat. 2011;14:280–296. doi: 10.1016/j.drup.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 7.V.U. Warrier, A.I. Makandar, M. Garg, G. Sethi, R. Kant, J.K. Pal, E. Yuba, and R.K. Gupta, Engineering anti-cancer nanovaccine based on antigen cross-presentation. 39 (2019) BSR20193220. [DOI] [PMC free article] [PubMed]
- 8.Nussinov R., Tsai C.J., Jang H. Anticancer drug resistance: an update and perspective. Drug Resist. Updat. 2021;59 doi: 10.1016/j.drup.2021.100796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deroyer C., Charlier E., Neuville S., Malaise O., Gillet P., Kurth W., Chariot A., Malaise M., de Seny D. CEMIP (KIAA1199) induces a fibrosis-like process in osteoarthritic chondrocytes. Cell Death Dis. 2019;10:103. doi: 10.1038/s41419-019-1377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abe S., Usami S.I., Nakamura Y. Mutations in the gene encoding KIAA1199 protein, an inner-ear protein expressed in Deiters' cells and the fibrocytes, as the cause of nonsyndromic hearing loss. J. Hum. Genet. 2003;48:564–570. doi: 10.1007/s10038-003-0079-2. [DOI] [PubMed] [Google Scholar]
- 11.J. Liu, W. Yan, P. Han, and D. Tian, The emerging role of KIAA1199 in cancer development and therapy. 138 (2021) 111507. [DOI] [PubMed]
- 12.Yoshida H., Nagaoka A., Kusaka-Kikushima A., Tobiishi M., Kawabata K., Sayo T., Sakai S., Sugiyama Y., Enomoto H., Okada Y., Inoue S. KIAA1199, a deafness gene of unknown function, is a new hyaluronan binding protein involved in hyaluronan depolymerization. Proc. Natl. Acad. Sci. U.S.A. 2013;110:5612–5617. doi: 10.1073/pnas.1215432110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng C., Zhu Y., Zhang W., Liao Q., Chen Y., Zhao X., Guo Q., Shen P., Zhen B., Qian X., Yang D., Zhang J.S., Xiao D., Qin W., Pei H. Regulation of the Hippo-YAP Pathway by Glucose Sensor O-GlcNAcylation. Mol. Cell. 2017;68 doi: 10.1016/j.molcel.2017.10.010. 591-604.e5. [DOI] [PubMed] [Google Scholar]
- 14.Wu D., Cai Y., Jin J. Potential coordination role between O-GlcNAcylation and epigenetics. Protein Cell. 2017;8:713–723. doi: 10.1007/s13238-017-0416-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quik M., Hokke C.H., Everts B. The role of O-GlcNAcylation in immunity against infections. Immunology. 2020;161:175–185. doi: 10.1111/imm.13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.B. Gupta, D. Sadaria, V.U. Warrier, A. Kirtonia, R. Kant, A. Awasthi, P. Baligar, J.K. Pal, E. Yuba, G. Sethi, M. Garg, and R.K. Gupta, Plant lectins and their usage in preparing targeted nanovaccines for cancer immunotherapy. 80 (2022) 87–106. [DOI] [PubMed]
- 17.Hua Q., Zhang B., Xu G., Wang L., Wang H., Lin Z., Yu D., Ren J., Zhang D., Zhao L., Zhang T. CEMIP, a novel adaptor protein of OGT, promotes colorectal cancer metastasis through glutamine metabolic reprogramming via reciprocal regulation of β-catenin. Oncogene. 2021;40:6443–6455. doi: 10.1038/s41388-021-02023-w. [DOI] [PubMed] [Google Scholar]
- 18.Senkowski W., Zhang X., Olofsson M.H., Isacson R., Höglund U., Gustafsson M., Nygren P., Linder S., Larsson R., Fryknäs M. Three-dimensional cell culture-based screening identifies the anthelmintic drug nitazoxanide as a candidate for treatment of colorectal cancer. Mol. Cancer Ther. 2015;14:1504–1516. doi: 10.1158/1535-7163.MCT-14-0792. [DOI] [PubMed] [Google Scholar]
- 19.Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Bio. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 20.Arora S., Balasubramaniam S., Zhang H., Berman T., Narayan P., Suzman D., Bloomquist E., Tang S., Gong Y., Sridhara R., Turcu F.R., Chatterjee D., Saritas-Yildirim B., Ghosh S., Philip R., Pathak A., Gao J.J., Amiri-Kordestani L., Pazdur R., Beaver J.A. FDA approval summary: olaparib monotherapy or in combination with bevacizumab for the maintenance treatment of patients with advanced ovarian cancer. Oncologist. 2021;26:e164–e172. doi: 10.1002/onco.13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chase D.M., Patel S., Shields K. Profile of olaparib in the treatment of advanced ovarian cancer. Int. J. Womens Health. 2016;8:125–129. doi: 10.2147/IJWH.S55906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakamoto I., Hirotsu Y., Nakagomi H., Ikegami A., Teramoto K., Omata M. Durable response by olaparib for a Japanese patient with primary peritoneal cancer with multiple brain metastases: a case report. J. Obstet. Gynaecol. Res. 2019;45:743–747. doi: 10.1111/jog.13851. [DOI] [PubMed] [Google Scholar]
- 23.Hetz C., Zhang K., Kaufman R.J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Bio. 2020;21:421–438. doi: 10.1038/s41580-020-0250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.C.H. Lin, C.C. Liao, S.Y. Wang, C.Y. Peng, Y.C. Yeh, M.Y. Chen, and T.Y. Chou, Comparative O-GlcNAc proteomic analysis reveals a role of O-GlcNAcylated SAM68 in lung cancer aggressiveness. 14 (2022) 243. [DOI] [PMC free article] [PubMed]
- 25.Y. Liu, Y. Cao, X. Pan, M. Shi, Q. Wu, T. Huang, H. Jiang, W. Li, and J. Zhang, O-GlcNAc elevation through activation of the hexosamine biosynthetic pathway enhances cancer cell chemoresistance. 9 (2018) 485. [DOI] [PMC free article] [PubMed]
- 26.H. Nie, H. Ju, J. Fan, X. Shi, Y. Cheng, X. Cang, Z. Zheng, X. Duan, and W. Yi, O-GlcNAcylation of PGK1 coordinates glycolysis and TCA cycle to promote tumor growth. 11 (2020) 36. [DOI] [PMC free article] [PubMed]
- 27.D. Wu, J. Jin, Z. Qiu, D. Liu, and H. Luo, Functional analysis of O-GlcNAcylation in cancer metastasis. 10 (2020) 585288. [DOI] [PMC free article] [PubMed]
- 28.Dongre A., Weinberg R.A. New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Bio. 2019;20:246–248. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 29.Xu Y., Xu H., Li M., Wu H., Guo Y., Chen J., Shan J., Chen X., Shen J., Ma Q., Liu J., Wang M., Zhao W., Hong J., Qi Y., Yao C., Zhang Q., Yang Z., Qian C., Li J. KIAA1199 promotes sorafenib tolerance and the metastasis of hepatocellular carcinoma by activating the EGF/EGFR-dependent epithelial-mesenchymal transition program. Cancer Lett. 2019;454:78–89. doi: 10.1016/j.canlet.2019.03.049. [DOI] [PubMed] [Google Scholar]
- 30.Duong H.Q., Nemazanyy I., Rambow F., Tang S.C., Delaunay S., Tharun L., Florin A., Buttner R., Vandaele D., Close P., Marine J.C., Shostak K., Chariot A. The endosomal protein CEMIP links WNT signaling to MEK1–ERK1/2 activation in selumetinib-resistant intestinal organoids. Cancer Res. 2018;78:4533–4548. doi: 10.1158/0008-5472.CAN-17-3149. [DOI] [PubMed] [Google Scholar]
- 31.Lebeaupin C., Vallée D., Hazari Y., Hetz C., Chevet E., Bailly-Maitre B. Endoplasmic reticulum stress signalling and the pathogenesis of non-alcoholic fatty liver disease. J. Hepatol. 2018;69:927–947. doi: 10.1016/j.jhep.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 32.Cubillos-Ruiz J.R., Bettigole S.E., Glimcher L.H. Tumorigenic and immunosuppressive effects of endoplasmic reticulum stress in cancer. Cell. 2017;168:692–706. doi: 10.1016/j.cell.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oakes S.A. Endoplasmic reticulum stress signaling in cancer cells. Am. J. Pathol. 2020;190:586–597. doi: 10.1016/j.ajpath.2020.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ngoh G.A., Hamid T., Prabhu S.D., Jones S.P. O-GlcNAc signaling attenuates ER stress-induced cardiomyocyte death. Am. J. Physiol. Heart Circ. Physiol. 2009;297:H1711–H1719. doi: 10.1152/ajpheart.00553.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.C. Chen, Y. Lu, J. Liu, L. Li, N. Zhao, and B. Lin, Genome-wide ChIP-seq analysis of TCF4 binding regions in colorectal cancer cells. 7 (2014) 4253–4259. [PMC free article] [PubMed]
- 36.Duong H.Q., Nemazanyy I., Rambow F., Tang S.C., Delaunay S., Tharun L., Florin A., Buttner R., Vandaele D., Close P., Marine J.C., Shostak K., Chariot A. The endosomal protein CEMIP links WNT signaling to MEK1-ERK1/2 activation in selumetinib-resistant intestinal organoids. Cancer Res. 2018;78:4533–4548. doi: 10.1158/0008-5472.CAN-17-3149. [DOI] [PubMed] [Google Scholar]
- 37.C.A. Thorne, A.J. Hanson, J. Schneider, E. Tahinci, D. Orton, C.S. Cselenyi, K.K. Jernigan, K.C. Meyers, B.I. Hang, A.G. Waterson, K. Kim, B. Melancon, V.P. Ghidu, G.A. Sulikowski, B. Lafleur, A. Salic, L.A. Lee, D.M. Miller, and E. Lee, Small-molecule inhibition of Wnt signaling through activation of casein kinase 1α. 6 (2010) 829–836. [DOI] [PMC free article] [PubMed]
- 38.Arena S., Corti G., Durinikova E., Montone M., Reilly N.M., Russo M., Lorenzato A., Arcella P., Lazzari L., Rospo G., Pagani M., Cancelliere C., Negrino C., Isella C., Bartolini A., Cassingena A., Amatu A., Mauri G., Sartore-Bianchi A., Mittica G., Medico E., Marsoni S., Linnebacher M., Abrignani S., Siena S., Nicolantonio F.D., Bardelli A. A subset of colorectal cancers with cross-sensitivity to olaparib and oxaliplatin. Clin. Cancer Res. 2020;26:1372–1384. doi: 10.1158/1078-0432.CCR-19-2409. [DOI] [PubMed] [Google Scholar]
- 39.Kelly H., Goldberg R.M. Systemic therapy for metastatic colorectal cancer: current options, current evidence. J. Clin. Oncol. 2005;23:4553–4560. doi: 10.1200/JCO.2005.17.749. [DOI] [PubMed] [Google Scholar]
- 40.Zhang N., Ng A.S., Cai S., Li Q., Yang L., Kerr D. Novel therapeutic strategies: targeting epithelial–mesenchymal transition in colorectal cancer. Lancet Oncol. 2021;22:e358–e368. doi: 10.1016/S1470-2045(21)00343-0. [DOI] [PubMed] [Google Scholar]
- 41.Francou A., Anderson K.V. The epithelial-to-mesenchymal transition in development and cancer. Ann. Rev. Cancer Biol. 2020;4:1129–1143. doi: 10.1146/annurev-cancerbio-030518-055425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taki M., Abiko K., Baba T., Hamanishi J., Yamaguchi K., Murakami R., Yamanoi K., Horikawa N., Hosoe Y., Nakamura E., Sugiyama A., Mandai M., Konishi I., Matsumura N. Snail promotes ovarian cancer progression by recruiting myeloid-derived suppressor cells via CXCR2 ligand upregulation. Nat. Commun. 2018;9:1685. doi: 10.1038/s41467-018-03966-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park S.Y., Kim H.S., Kim N.H., Ji S., Cha S.Y., Kang J.G., Ota I., Shimada K., Konishi N., Nam H.W., Hong S.W., Yang W.H., Roth J., Yook J.I., Cho J.W. Snail1 is stabilized by O-GlcNAc modification in hyperglycaemic condition. EMBO J. 2010;29:3787–3796. doi: 10.1038/emboj.2010.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data from the current study are included in the article or uploaded as supplementary information.