Abstract
During the past decade, genetic code expansion has been proved to be a powerful tool for protein studies and engineering. As the key part, a series of orthogonal pairs have been developed to site‐specifically incorporate hundreds of noncanonical amino acids (ncAAs) into proteins by using bacteria, yeast, mammalian cells, animals, or plants as hosts. Among them, the pair of tyrosyl‐tRNA synthetase/tRNATyr from Methanococcus jannaschii and the pair of pyrrolysyl‐tRNA synthetase/tRNAPyl from Methanosarcina species are the most popular ones. Recently, other “not‐so‐popular” orthogonal pairs have started to attract attentions, because they can provide more choices of ncAA candidates and are necessary for simultaneous incorporation of multiple ncAAs into a single protein. Here, we summarize the development and applications of those “not‐so‐popular” orthogonal pairs, providing guidance for studying and engineering proteins.
Keywords: aminoacyl‐tRNA synthetases, genetic code expansion, noncanonical amino acids, orthogonal translation systems, tRNA
1. INTRODUCTION
Site‐specific incorporation of noncanonical amino acids (ncAAs) into proteins by genetic code expansion has been widely used for protein studies and engineering (Chen et al., 2018; Chung et al., 2020; de la Torre & Chin, 2021). For this purpose, a series of orthogonal pairs of aminoacyl‐tRNA synthetase (AARS) and tRNA have been engineered to recognize different ncAAs and designated codons, correspondingly. Among them, the pair of tyrosyl‐tRNA synthetase (TyrRS)/tRNATyr from Methanococcus jannaschii and the pair of pyrrolysyl‐tRNA synthetase (PylRS)/tRNAPyl from Methanosarcina species are the most popular ones (Wan et al., 2014; Wang et al., 2001). Although these two pairs can recognize a wide range of ncAAs, their substrates are mostly phenylalanine and lysine analogs. Furthermore, it is beneficial to simultaneously incorporate different ncAAs into a single protein. For example, p‐azido‐phenylalanine and N6‐[(2‐propynyloxy)carbonyl]‐lysine were genetically incorporated into a single protein for double‐labeling and Förster resonance energy transfer (FRET) studies by introducing both MjTyrRS/tRNATyr and PylRS/tRNAPyl pairs (Wang et al., 2014; Wu et al., 2012). Recently, our group utilized the pairs of acetyllysyl‐tRNA synthetase/tRNAPyl and phosphoseryl‐tRNA synthetase/tRNASep to simultaneously incorporate acetyllysine (AcK) and phosphoserine (Sep) into a single protein to study crosstalk between posttranslational modifications (PTMs) (Venkat et al., 2018). Such strategy needs more choices of pairs which are mutually orthogonal to each other. Thus, it is necessary to reapproach those “not‐so‐popular” orthogonal pairs. Here, we summarize the development and applications of them, which include TrpRS/tRNATrp, PheRS/tRNAPhe, LeuRS/tRNALeu, LysRS/tRNALys, and SepRS/tRNASep, as well as GlnRS/tRNAGln, GluRS/tRNAGlu, ProRS/tRNAPro, HisRS/tRNAHis, and SerRS/tRNASer (Table 1).
TABLE 1.
Orthogonal pairs and their origins, hosts, and incorporated ncAAs
| OTSs | Origins | Hosts | Incorporated ncAAs (and corresponding AARS mutants) | References |
|---|---|---|---|---|
|
TrpRS/tRNATrp |
S. cerevisiae | E. coli | NapA (Y105L, E140P, T232V, I252V, F262Y), 1‐MTP (Y105L, E140A, T232C, I252V, F262C), BtA (Y105L, E140A, T232C, I252V, F262C), 6‐MTP (Y105V, E140P, T232C, I252C), 5‐HTP (T107C, P254T, C255A) | 10, 11 |
| E. coli | E. coli (ATM), mammalian cells | 5‐HTP, 5‐MTP, 5‐Brw, 5‐AzW, 5‐PrW, 5‐AmW (V144G, V146C for all above ncAAs) | 12 | |
|
PheRS/tRNAPhe |
S. cerevisiae | E. coli | pFF (WT), pBrF (T415A), 6‐ClW (T415G), 6‐BrW (T415G), BtA (T415G), 2‐NapA (N412G, T415G, S418C, S437F) | 15, 16, 17, 18, 19 |
| Human mitochondria | E. coli, mammalian cells | 4‐AzF (T467G, A507A), 2‐NapA (T467G, A507A), 3‐CNF (T467G, A507A), L‐Dopa (Q356, T467, A507), 6‐CNW (E391D, T467G, A507G), 7CNW (F464V, T467G, A507G) | 21 | |
| LeuRS/tRNALeu | E. coli | S. cerevisiae, mammalian cells, C. elegans | OmeY (M40L, L41E, Y499R, Y527A, H537G), AcA (M40V, L41M, Y499L, Y527L, H537G), nbC (M40W, L41S, Y499I, Y527A, H537G), DanA (M40A, L41N, T252A, Y499I, Y527G, H537T) | 24, 25, 26, 27, 28 |
| M. thermo‐autotrophicum | E. coli | N/A | 29 | |
| LysRS/tRNSLys | P. horikoshii | E. coli | hGln (E41I, Y268S) | 32 |
| SepRS/tRNASep | M. maripaludis | E. coli | Sep (K347E, N352D, E412S, E414I, P495R, I496R, L512I), serine phosphonate (E412P, E414F, P495M, I496W, F529S), pThr (G320A, L321Y) | 36, 37, 38, 42, 43 |
| GlnRS/tRNAGln | S. cerevisiae | E. coli | N/A | 46 |
| E. coli | S. cerevisiae | N/A | 47 | |
| GluRS/tRNAGlu | P. horikoshii | E. coli | N/A | 30 |
| ProRS/tRNAPro | P. horikoshii | E. coli | N/A | 50 |
| HisRS/tRNAHis | C. crescentus | E. coli | N/A | 53 |
| SerRS/tRNASer | M. mazei | E. coli | N/A | 54 |
Abbreviation: N/A, not available.
2. ORTHOGONAL PAIRS BASED ON TRYPTOPHANYL‐TRNA SYNTHETASE (TRPRS)/TRNATRP
Tryptophan has the largest side chain among the set of 20 canonical amino acids, and the capacious tryptophan binding pocket of TrpRS provides potentials for incorporation of ncAAs with bulky side chains. Ellington and coworkers developed an orthogonal pair of TrpRS/tRNATrp from Saccharomyces cerevisiae in Escherichia coli cells (Hughes & Ellington, 2010). They found that the amber suppressor SctRNATrp CUA could be mischarged by E. coli LysRS, as SctRNATrp CUA has a high sequence identity (73%) and shares several identify determinants with EctRNALys. To eliminate the misacylation by EcLysRS, the amber suppressor SctRNATrp CUA was rationally designed by increasing G:C content of the anticodon stem to reduce its structural flexibility. The best tRNA mutant (SctRNATrp CUA‐AS3.5) could reduce misacylation by nearly 50‐fold.
To use the pair of ScTrpRS/tRNATrp to incorporate ncAAs into proteins, Schultz and coworkers further engineered both SctRNATrp CUA and ScTrpRS (Chatterjee et al., 2013). First, the acceptor stem of SctRNATrp CUA‐AS3.5 was randomized and the tRNA mutant library was subjected to three rounds of alternating positive selections by suppressing a permissive amber codon in the gene of chloramphenicol acetyltransferase (CAT) and negative selections by suppressing two permissive amber codons in the gene of barnase which is toxic to E. coli cells. The best tRNA mutant (SctRNATrp CUA‐H14) had a significantly enhanced suppression activity than the initial SctRNATrp CUA‐AS3.5. Then, the tryptophan binding pocket of ScTrpRS (Tyr105, Glu140, Thr232, Ile252, Cys254, and Phe262) was randomized, and variant libraries were screened for an ncAA, 3‐(1‐naphthyl)‐alanine (NapA) by the similar double‐sieve selection scheme. Several NapA‐specific ScTrpRS variants were obtained (Table 1). They further tested the specificities of those variants against other tryptophan analogs. Interestingly, those ScTrpRS variants could recognize 1‐methyl‐tryptophan (1‐MTP), benzothienyl‐alanine (BtA), and 6‐methyl‐tryptophan (6‐MTP), correspondingly (Figure 1). Then, ScTrpRS variants were used to incorporate different tryptophan analogs into the site of a key tryptophan residue in the chromophore of enhanced cyan fluorescent protein (eCFP). Replacement of the tryptophan residue with different tryptophan analogs showed altered spectral features, demonstrating a potential use of the ScTrpRS/tRNATrp pair to monitor tryptophan structures and functions in proteins.
FIGURE 1.
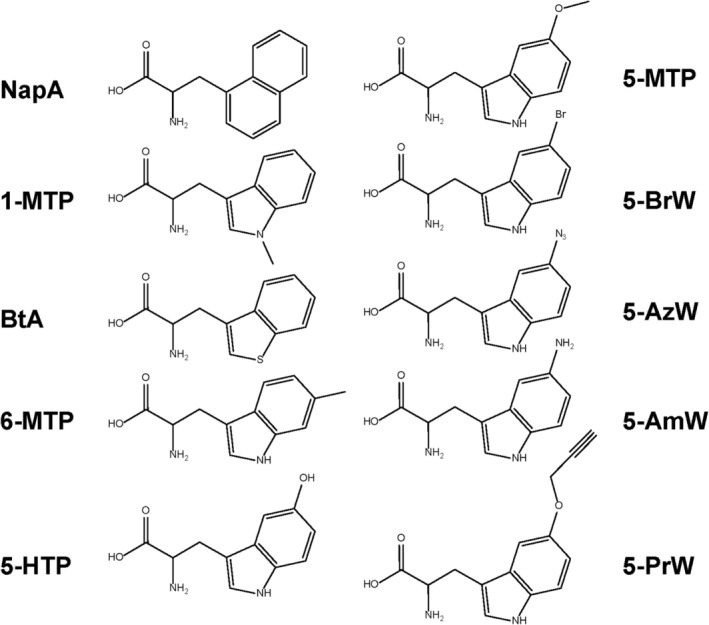
NcAAs which have been incorporated by orthogonal pairs derived from TrpRS/tRNATrp
Later, Ellington and coworkers engineered the orthogonal pair of ScTrpRS/tRNATrp for genetic incorporation of 5‐hydroxy‐tryptophan (5HTP) by directed evolution with compartmentalized partnered replication (CPR), in which synthetic circuits that most efficiently drive Taq DNA polymerase production in cells can be preferentially amplified during the subsequent compartmentalized polymerase chain reaction (PCR) step (Ellefson et al., 2014). First, a library of ScTrpRS variants with randomized three residues around the tryptophan binding pocket (T107, P254, and C255) was screened for 5‐HTP by suppressing an amber codon in the gene of Taq DNA polymerase. After three rounds of selection, the best ScTrpRS variant (T107C, P254T, and C255A) was obtained, showing 1500‐fold improvement in 5‐HTP incorporation than WT ScTrpRS. Then, three libraries of SctRNATrp CUA with randomized mutations at the anticodon stem, the acceptor stem, or loop sequences were subjected to the similar CPR selection, respectively. The best SctRNATrp CUA mutant (U16G, G43U, and U58G) was obtained, showing 3‐fold enhancement than the parental SctRNATrp CUA‐AS3.4.
The orthogonal pairs of ScTrpRS/tRNATrp mentioned above can use E. coli cells at the host but not eukaryotic cells, because S. cerevisiae belongs to the eukaryote, and its AARS/tRNA pairs usually cross‐react with the counterparts in the organism from the same domain. To expand the genetic code in eukaryotic cells, bacterial AARS/tRNA pairs are usually utilized due to their orthogonality to eukaryotic counterparts. However, bacterial AARS/tRNA pairs commonly cannot be evolved by using the facile E. coli‐based selection system for the same reason. To overcome this limit, Chatterjee and coworkers generated an altered translational machinery (ATM) E. coli strain in which evolved orthogonal pairs can be used in both bacteria and eukaryotes (Italia et al., 2017). First, they removed the genes of EcTrpRS and EctRNATrp from the E. coli genome and introduced the pair TrpRS/tRNATrp from S. cerevisiae to decode normal tryptophan codons in the E. coli genome. Then, the pair of EcTrpRS and EctRNATrp UCA opal suppressor was put back into the ATM E. coli strain for further engineering, as it was known that the amber suppressor EctRNATrp CUA could be mischarged by E. coli GlnRS (Jahn et al., 1991). Next, the tryptophan binding pocket of EcTrpRS (Phe7, Ser8, Val144, Pro145, and Val146) was randomized, and the EcTrpRS variant library was screened for 5‐HTP by the double‐sieve selection scheme mentioned above but with opal codons instead of amber codons in selection genes. The best EcTrpRS variant (V144G and V146C) was obtained. They further tested the specificities of this EcTrpRS variant for other 5‐substituted tryptophan analogs and found that it could also genetically incorporate 5‐methoxy‐tryptophan (5‐MTP), 5‐bromo‐tryptophan (5‐BrW), 5‐azido‐tryptophan (5‐AzW), 5‐propargyloxy‐tryptophan (5‐PrW), and 5‐amino‐tryptophan (5‐AmW) in both E. coli and mammalian cells (Figure 1). Among those tryptophan analogs, 5‐PrW and 5‐AzW introduce azido and alkyne groups respectively, which can be used to label proteins by click chemistry.
3. ORTHOGONAL PAIRS BASED ON PHENYLALANYL‐TRNA SYNTHETASE (PHERS)/TRNAPHE
It has been known for a long time that certain AARSs can incorporate analogs of their cognate amino acids into proteins at corresponding codons uniformly through the proteome, such as the encoding of p‐fluoro‐phenylalanine (pFF) at Phe codons by PheRS in Phe‐auxotrophic strains (Richmond, 1962). Furter utilized this fact to site‐specifically incorporate pFF into proteins at introduced amber codons in E. coli cells (Furter, 1998). Site‐specifically fluorinated proteins are ideal for (Kwon & Lim, 2015) F‐nuclear magnetic resonance (NMR) spectroscopy analyses to monitor protein conformational changes and detect protein‐protein interactions. The incorporation of pFF can also serve as a vehicle to deliver isotopically labeled phenylalanine analogues to a position of choice for Fourier‐transform infrared (FTIR) spectroscopy experiments. To avoid the incorporation of pFF at Phe codons by the native E. coli PheRS, a pFF‐resistant E. coli strain was used. It harbors a PheRS(Ser294) variant which cannot recognize pFF (Fangman & Neidhardt, 1964). Then a plasmid with the pair of yeast PheRS and tRNAPhe CUA (orthogonal to E. coli counterparts) was transformed into this pFF‐resistant E. coli strain. A large excess of pFF was supplemented in the growth medium to compete with Phe for the yeast PheRS. Although the system successfully incorporated pFF at the introduced amber codon, the fidelity was not very high with pFF (~65%), Phe (~20%), and Lys (~15%).
In order to increase the fidelity, Tirrell and coworkers engineered both the yeast PheRS and tRNAPhe CUA for site‐specific incorporation of p‐bromo‐phenylalanine (pBrF) into proteins in E. coli cells (Kwon et al., 2006). Introduction of aryl halides such as pBrF into proteins allows site‐specific modifications via palladium‐catalyzed cross‐coupling reactions with terminal alkene or alkyne reaction partners. First, a UG base pair 30U::40G was introduced to the yeast tRNAPhe CUA amber suppressor to reduce the mischarging by EcLysRS. Then, a point mutation (T415A) was introduced to the yeast PheRS to exclude Trp misincorporation. Besides Phe analogs, Tirrell and coworkers utilized an engineered yeast PheRS(T415G)/tRNAPhe CUA pair to site‐specifically incorporate several Trp analogs, including 6‐chlorotryptophan (6‐ClW), 6‐bromotryptophan (6‐BrW), and BtA (Figure 2) (Kwon & Tirrell, 2007). However, the pair of PheRS(T415A)/tRNAPhe still has Phe misincorporation, while the pair of PheRS(T415G)/tRNAPhe keeps Trp misincorporation. To further increase the specificity of ncAA incorporation against canonical amino acids, Kown and coworkers created a library of yeast PheRS variants with a randomized Phe binding pocket (N412, S418, T415, and S437). Then a high‐throughput screening method based on GFP fluorescence reduction was performed for 2‐naphthylalanine (2‐NapA). Selected variants allow incorporation of either 2‐NapA or canonical amino acids at multiple nonpermissive sites of GFP, where amino acids other than Phe lead to reduction in the fluorescence of cells. The best PheRS variant (N412G, T415G, S418C, and S437F) realized high‐fidelity (98%) incorporation of 2‐NapA into a protein at an introduced amber codon (Kwon & Lim, 2015). As 2‐NapA has a larger hydrophobic side chain than any canonical amino acids, it has been used to tune the active site of proteins to modulate the binding affinity to inhibitors (Zheng & Kwon, 2013).
FIGURE 2.
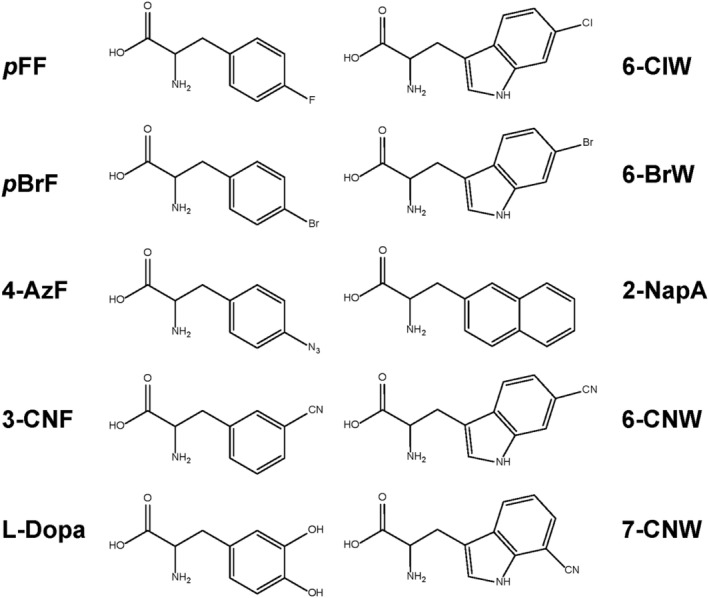
NcAAs which have been incorporated by orthogonal pairs derived from PheRS/tRNAPhe
Recently, Lin and coworkers designed a chimeric PheRS/PylRS system which has broad orthogonality in both bacteria and eukaryotes (Ding et al., 2020). First, they transplanted the key orthogonal components from the PylRS system to human mitochondrial PheRS/tRNAPhe. Briefly, the tRNA binding domain of PylRS was added to the N‐terminal of the PheRS catalytic domain, while the PylRS binding part of tRNAPyl was introduced to tRNAPhe. This chimeric PheRS/tRNAPhe CUA pair showed good orthogonality in both E. coli and mammalian cells (no amber suppression was detected in the presence of chPheT or chPheRS alone). Then, the chimeric PheRS (chPheRS) was evolved for different ncAAs, including both Phe analogs and Trp analogs. For Phe analogs, the chPheRS‐1 variant (T467G and A507A) could incorporate 4‐Azido‐Phe (4‐AzF), 2‐NapA and 3‐Cyano‐Phe (3‐CNF), while the chPheRS‐2 variant (Q356, T467, and A507) incorporated L‐3,4‐Dihydroxy‐Phe (L‐Dopa). For Trp analogs, the chPheRS‐3 variant (E391D, T467G, and A507G) preferred the N1 and C6 substituted Trp analogues such as 6‐Cyano‐Trp (6‐CNW), while the chPheRS‐4 variant (F464V, T467G, and A507G) exhibited higher activity toward the C7 substitute Trp analogues such as 7‐Cyano‐Trp (7CNW) (Figure 2). All these systems were able to genetically incorporate those Trp analogs into proteins by using both E. coli and mammalian cells as hosts. In the same study, such chimeric approach was also applied to HisRS and SerRS systems and showed good orthogonality toward both E. coli and mammalian cells (no amber suppression was detected in the presence of chHisRS or chSerRS alone), demonstrating great potentials of developing other E. coli–mammalian shuttle systems for genetic code expansion.
4. ORTHOGONAL PAIRS BASED ON LEUCYL‐TRNA SYNTHETASE (LEURS)/TRNALEU
Besides Trp, Phe, and Tyr, Leu is also a bulky hydrophobic amino acid. The binding pocket of LeuRS usually has a large cavity formed by side chains without backbone elements, (Cusack et al., 2000; Fukunaga & Yokoyama, 2005) so mutating the side chains around the binding pocket is possible to generate different shapes of cavities for accommodate a wide diversity of ncAAs. To realize this idea, Schultz and coworkers started with E. coli LeuRS/tRNALeu CUA (Wu et al., 2004). This pair was first shown to be orthogonal in S. cerevisiae. Then the amino acids around the Leu binding pocket (M40, L41, Y499, Y527, and H537) were randomized and selected for three ncAAs [O‐methyl tyrosine (OMeY), α‐aminocaprylic acid (AcA), and o‐nitrobenzyl cysteine (nbC)] with distinct electronic and steric properties, separately. The selection was based on the suppression of amber codons at positions 44 and 110 in the gal4 gene which drives the expression of genomic HIS3 and URA3 reporter genes. Finally, three distinct LeuRS variants were obtained, and each one worked with a different ncAA, respectively. The one for OmeY has mutations of M40L, L41E, Y499R, Y527A, and H537G, the one for AcA harbors mutations of M40V, L41M, Y499L, Y527L, and H537G, and the one for nbC brings mutations of M40W, L41S, Y499I, Y527A, and H537G. Among these three ncAAs, nbC is a photocaged amino acid which allows photo‐regulation of proteins with key cysteine residues such as the human proapoptotic protein caspase 3 used in this work (Wu et al., 2004).
In another study, Schultz and coworkers evolved the pair of E. coli LeuRS/tRNALeu CUA to genetically encode a fluorescent amino acid, 2‐amino‐3‐(5‐(dimethylamino)naphthalene‐1‐sulfonamide) propanoic acid (DanA) in S. cerevisiae (Summerer et al., 2006). DanA has a small size, relatively large Stokes shift, and high sensitivity to be an ideal fluorophore to label proteins without affecting protein structures, which is usually a concern for using fluorescent proteins (commonly >20 kDa). DanA also has a distinct electronic and steric property from those three ncAAs (OmeY, AcA, and nbC) incorporated by LeuRS variants mentioned above (Figure 3a), supporting the idea that mutating the Leu binding pocket of LeuRS is expected to accommodate various ncAAs. The LeuRS evolved for DanA has mutations of M40A, L41N, Y499I, Y527G, and H537T. To further increase the fidelity of DanA incorporation, a T252A mutation was introduced to the LeuRS variant. This mutations site is at the edit domain of LeuRS, and the T252A mutation results in efficient hydrolysis of aminoacylated tRNALeu (Mursinna & Martinis, 2002). Then, genetically encoded DanA was used as a probe to monitor protein unfolding. Besides S. cerevisiae, the pair of EcLeuRS/tRNALeu CUA for DanA has been transplanted to mammalian cells, neurons, and Caenorhabditis elegans, providing practical applications in cell biology and neurobiology studies (Parrish et al., 2012; Wang et al., 2007).
FIGURE 3.
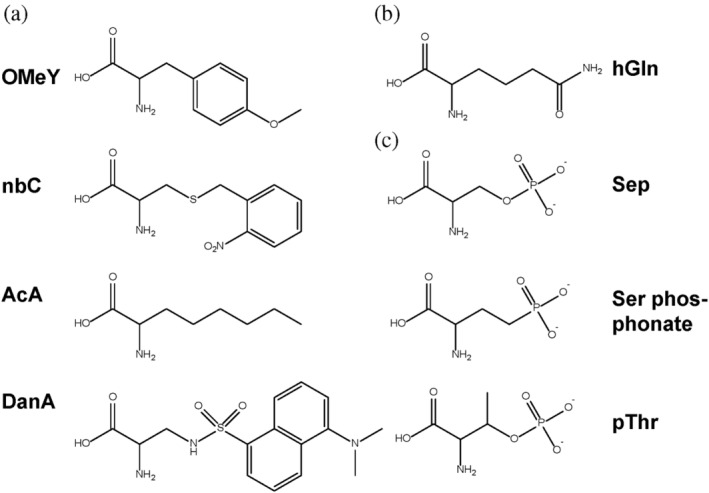
NcAAs which have been incorporated by orthogonal pairs derived from (a) LeuRS/tRNALeu; (b) LysRS/tRNALys; and (c) SepRS/tRNASep
On the other end, a pair of archaeal LeuRS and tRNALeu CUA was developed to be orthogonal in E. coli cells (Anderson & Schultz, 2003). First, the anticodons of leucyl tRNAs from different archaeal strains (Archaeoglobus fulgidus, Halobacterium sp. NRC‐1, M. jannaschii, Pyrococcus furiosus, and Pyrococcus horikoshii) were mutated to CUA for amber suppression. Then, different combinations of tRNALeu CUA amber suppressors and LeuRSs from different archaeal strains were tested for their efficiencies of amber suppression in E. coli cells. The best one was the pair of Methanobacterium thermoautotrophicum LeuRS and Halobacterium sp. NRC‐1 tRNALeu CUA. Then a G37A mutation was introduced to tRNALeu CUA for better amber suppression, and the acceptor stem of tRNALeu CUA was optimized (a discriminator base A73 and all Watson‐Crick base pairs in the stem) for better orthogonality in E. coli cells. To expand the list of codons used for genetic code expansion, Halobacterium sp. NRC‐1 tRNALeu CUA were further mutated and optimized for opal (UGA) and four‐based codon (AGGA) suppression. However, no ncAAs were incorporated into proteins by using this pair, possibly because of the relatively low aminoacylation activity (Santoro et al., 2003).
5. ORTHOGONAL PAIRS BASED ON LYSYL‐TRNA SYNTHETASE (LYSRS)/TRNALYS
Besides three‐base suppressor tRNAs, those with four‐base anticodons have been proposed to provide more codon choices for genetic code expansion. Schultz and coworkers selected LysRS from P. horikoshii as a candidate for a new orthogonal pair toward quadruplet codons because PhLysRS is flexible for substitutions in the anticodon loop of its cognate tRNALys (Anderson et al., 2004). To solve the problem that PhLysRS is toxic to E. coli cells, serial culture of the E. coli mutator strain XL1‐red with a plasmid containing PhLysRS was performed. The resulting nontoxic variant PhLysRS∆ lost its residues after S357, which contains the anticodon recognition site (Terada et al., 2002). Then they designed a cognate suppressor tRNA for PhLysRS∆. The conserved recognition sites among all the available archaeal tRNALys for the archaeal LysRS were retained, while nonessential mismatched base pairs deleterious to suppression efficiency were removed. The four‐base anticodon UCCU was added to such tRNALys for decoding the AGGA quadruplet codon, but it is not orthogonal in E. coli. To obtain orthogonal tRNALys UCCU mutants, a library was constructed in which the last 4 bp of the acceptor stem were randomized, as this region is essential for the recognition of most AARSs and cognate tRNAs. The library was screened based on ampicillin resistance by suppression the AGGA codon for the position A184 of β‐lactamase. The best tRNALys UCCU mutant contains a mutated acceptor stem (G1U::C72A, C71U, and C4U::G69A) and a A37C substitution in the anticodon loop. Next, PhLysRS∆ was evolved for homoglutamine (hGln) (Figure 3b), which is structurally close to lysine. Two residues (E41 and Y268) recognizing the ε‐amino group of lysine were randomized, and the variant library was screened by a GFP‐based reporting system (Santoro et al., 2002). The PhLysRS∆ variant specific for hGln contains two mutations of E41I and Y268S. To prove the concept of simultaneous incorporation of two ncAAs into one protein, this evolved orthogonal pair of PhLysRS∆/tRNALys UCCU was used together with another orthogonal pair of M. jannaschii TyrRS/tRNATyr CUA to decode an AGGA four‐base codon and a TAG three‐base codon as hGln and OmeY in myoglobin, respectively. This work also opens an avenue of using other quadruplet codons such as CCCU and CUAG as signals for ncAA incorporation in E. coli cells.
6. ORTHOGONAL PAIRS BASED ON PHOSPHOSERYL‐TRNA SYNTHETASE (SEPRS)/TRNASEP
Phosphorylation is one of the most abundant PTMs in nature. Because of its importance, many groups have developed orthogonal translation systems (OTSs) for genetic incorporation of phosphoamino acids. Site‐specific incorporation of phosphotyrosine utilizes orthogonal pairs derived from MjTyrRS/tRNATyr CUA, (Fan et al., 2016; Luo et al., 2017) while Sep and phosphothreonine (pThr) has been installed into proteins by orthogonal pairs derived from the pair M. maripaludis SepRS/MjtRNACys CUA (Park et al., 2011; Rogerson et al., 2015; Zhang et al., 2017).
It was known that certain methanogenic archaea used an unusual AARS, SepRS to catalyzes the formation of Sep‐tRNACys for cysteine biosynthesis (Sauerwald et al., 2005). Söll and coworkers utilized this pathway to develop the first OTS of Sep incorporation in E. coli (Park et al., 2011). First, MjtRNACys was mutated to an amber suppressor tRNACys CUA (named tRNASep). An additional C20U mutation was introduced to tRNASep to improve aminoacylation by SepRS (Hohn et al., 2006). The endogenous Sep phosphatase gene serB was deleted from the E. coli genome to stabilize Sep in cells. However, Sep‐tRNASep cannot be delivered to ribosome efficiently by the elongation factor‐Tu (EF‐Tu), because EF‐Tu binds poorly to tRNAs carrying negatively charged amino acids (Sanderson & Uhlenbeck, 2007). Thus, the amino acid binding pocket of EF‐Tu (H67, D216, E217, F219, T229, and N274) was randomized, and the library of EF‐Tu variants was selected for Sep‐tRNASep recognition based on suppression of an amber codon in the CAT gene. The resulting EF‐Tu variant (EF‐Sep) harbors mutations of H67R, D216N, E217G, F219Y, T229S, and N274W. Then, the OTS containing SepRS, tRNASep, and EF‐Sep successfully incorporated Sep into a human protein MEK1, demonstrating a biological application in kinase signaling studies. Later, Park and coworkers further optimized this Sep‐OTS (Lee et al., 2013). SepRS binds to the anticodon region of tRNACys, so the introduced CUA anticodon for amber codon suppression decreased the aminoacylation activity. To improve aminoacylation, the anticodon‐binding region of SepRS (E412, E414, P495, and I496) was randomized and the library of SepRS variant was selected based on suppression of an amber codon in the CAT gene. Additional DNA shuffling was conducted. The resulting SepRS variant (SepRS9) has four mutations in the anticodon‐binding region (E412S, E414I, P495R, and I496R) and three additional mutations in other regions (K347E, N352D, and L512I). Then, they identified key residues of EF‐Sep for Sep‐tRNASep binding by alanine scanning and randomized N216, of which the substitution with Ala increased the binding with Sep‐tRNASep. The best EF‐Sep variant (EF‐Sep21) has the mutation of N216V. Together with SepRS9 and tRNASep, the optimized Sep‐OTS (SepRS9, tRNASep, and EF‐Sep21) increased Sep‐incorporation nearly 10‐fold of the original Sep‐OTS. Then, Sep was genetically incorporated into H3 protein to investigate histone acetyltransferase activity of Gcn5p and the SAGA complex. Later, Rinehart and coworkers optimized Sep‐OTSs in recoded E. coli strains by increasing the copy number of tRNASep from one to five in the plasmid, and the purity of Sep‐incorporation approached ~90% (Pirman et al., 2015).
Phosphorylated residues usually undergo hydrolysis by specific or non‐specific phosphatases inside cells, so it is useful to incorporate nonhydrolyzable analogs of phosphorylated amino acids into proteins to isolate and study bound phosphatases. For this purpose, Chin and coworkers genetically incorporate a nonhydrolyzable analog of Sep, serine phosphonate (replacing the O‐P bond in Sep with a C‐P bond) (Rogerson et al., 2015). First, they further engineered tRNAsep with randomized sequences around the anticodon (G29‐U33 and G37‐C41) and SepRS with randomized anticodon‐binding region (E412, E414, K471, P495, I496, and F529). The best pair of SepRS(2)/tRNASep(B4) had an additional 20‐fold than pervious Sep‐OTSs. Then, the E. coli strain was genetically modified for the incorporation of the nonhydrolyzable analog serine phosphonate. Serine phosphonate has a very similar structure to Sep, and SepRS cannot discriminate it from Sep, so the intracellular concentration of Sep needs to be decreased. To do so, SerB (the phosphatase to hydrolyze Sep) was overexpressed and SerC (the aminotransferase to produce Sep) was inactivated.
As Sep and pThr have very similar structures (Figure 3C), it is feasible to start with Sep‐OTS for pThr incorporation. However, Sep is naturally existing in cells for serine biosynthesis, while pThr is not involved in any metabolic pathways in E. coli cells. Furthermore, negatively charged compounds like pThr cannot go through the cell membrane easily, so it could not be sufficient for pThr incorporation by simply supplementing media with pThr. In Salmonella, a threonine kinase (PduX) phosphorylates free threonine to form pThr for coenzyme B12 biosynthesis (Fan & Bobik, 2008). Chin and coworkers introduced PduX into E. coli cells to produce a high level of intracellular pThr (Zhang et al., 2017). Another challenge for pThr incorporation is also derived from the structural similarity between pThr and Sep. It is difficult to evolve SepRS to recognize pThr but not Sep. To solve this problem, Chin and coworkers randomized a group of amino acids (M317, N318, L319, G320, and L321) in the Sep binding pocket of SepRS which were modeled to interact with the additional methyl‐group of pThr. The SepRS variant library was screened by parallel positive selections in cells with PduX or without PduX combined with deep sequencing and statistical analysis to that compare sequencing read counts for each synthetase clone in the presence and absence of pThr. The resulting pThrRS variant has the mutations G320A and L321Y. Together with an optimized tRNASep and EF‐Sep, this pThr‐OTS successfully incorporated pThr into ubiquitin and a protein kinase Cdk2 for phosphoprotein structure determination and synthetic protein kinase activation.
7. OTHER ORTHOGONAL PAIRS
Beside the five orthogonal pairs discussed above which have successfully incorporated ncAAs into proteins, several other orthogonal pairs have also been developed but without ncAA incorporation so far, providing bases for further engineering for ncAAs with distinct structures.
7.1. GlnRS/tRNAGln
Motivated by the fact that E. coli GlnRS does not charge S. cerevisiae tRNAGln, (Whelihan & Schimmel, 1997) Schultz and coworkers developed an orthogonal pair based on ScGlnRS/tRNAGln in E. coli cells (Liu & Schultz, 1999). First, the anticodon of SctRNAGln was mutated from CUG to CUA for amber suppression. An additional mutation U38A was introduced to SctRNAGln CUA to increase suppression efficiency. Both suppressor tRNAs cannot be acylated by any E. coli AARSs. In addition, ScGlnRS could aminoacylate SctRNAGln CUA and SctRNAGln CUA (A38) but none of E. coli tRNAs, making them orthogonal pairs in E. coli cells. From the other end, RajBhandary and coworkers developed an orthogonal pair of E. coli GlnRS and a mutant human initiator tRNAi for S. cerevisiae (Kowal et al., 2001). The mutant tRNAi has U35A36 mutations for amber suppression and the U50G51:C63A64 mutations in the TψC stem to allows it to participate in elongation. Such mutant tRNAi cannot be charged by any yeast AARSs but only by E. coli GlnRS, showing good orthogonality in S. cerevisiae cells.
7.2. GluRS/tRNAGlu
Archaeal GluRS recognizes both tRNAGlu and tRNAGln and charges them with Glu to form Glu‐tRNAGlu and Glu‐tRNAGln, respectively. Then Glu‐tRNAGln is converted to Gln‐tRNAGln by transamidation (Tumbula et al., 2000). GluRS is able to recognize four tRNA anticodons (UUC, CUC, UUG, CUG) for both tRNAGlu and tRNAGln, so it is expected to tolerate mutations in tRNA anticodon for amber suppression. In the meantime, archaeal tRNAGlu cannot be aminoacylated by E. coli AARSs (Kwok & Wong, 1980). Based on these facts, Schultz and coworkers developed an orthogonal pair of GluRS/tRNAGlu in E. coli cells (Santoro et al., 2003). All available sequences of archaeal tRNAGlu were aligned, and the consensus sequences were retained. The anticodon was mutated to CUA for amber codon suppression, and the nucleotide to the 3′ side of the anticodon was replaced by A to improve suppression efficiency in E. coli. The resulting tRNAGlu CUA was shown to be orthogonal to E. coli AARSs. However, none of the GluRSs from selected archaea species could aminoacylate such tRNAGlu CUA efficiently. The consensus sequence of archaeal tRNAGlu has a non‐canonical G10‐U28 base which could lower the aminoacylation activity, so a library of tRNAGlu CUA variants in positions 10 and 28 was created and screened. All the active clones have the canonical G10‐C28 base pair, which has a 5‐fold increase in amber suppression. Then GluRSs from A. fulgidus, Methanosarcina mazei, M. thermoautotrophicum, and P. horikoshii were evaluated for the orthogonality with tRNAGlu CUA (GC) in E. coli, individually. The best GluRS was from P. horikoshii. The pair of PhGluRS and the consensus tRNAGlu CUA (GC) forms an orthogonal pair for E. coli cells.
7.3. ProRS/tRNAPro
Proline is unique among the set of 20 canonical amino acids, because it is the only one with a secondary amine. N‐modified amino acids could endow proteins with unique properties, thus OTSs based on ProRS will be significant for protein engineering. For this purpose, Schultz and coworkers tested different combinations of ProRS and tRNAPro CUA from several archaeal strains (A. fulgidus, P. horikoshii, M. mazei, Sulfolobus solfataricus, M. jannaschii, Halobacterium sp. (NRC‐1), and Thermus Thermophilus) for their amber suppression efficiencies (Chatterjee et al., 2012). The best pair appeared to be the pair of P. horikoshii ProRS and A. fulgidus tRNAPro CUA. Since the anticodon is a major identify element for the tRNAPro recognition, the anticodon‐binding region of PhProRS was evolved. Six residues around the anticodon‐binding pocket (Y335, E338, R345, E347, D352, and K368) were randomized, and the PhProRS variant library was selected based on suppression of amber codons in both genes of GFP and CAT. The best one has mutations of Y335N, E338C, R345I, E347T, D352E, and K368S. Then, the anticodon of AftRNAPro was mutated to AGGG and CUAG, respectively, and the same PhProRS variant library was selected toward CCCT and CTAG suppression, correspondingly. The best ProRS variant for tRNAPro AGGG only has two mutations of D352E and K368S, because tRNAAGGG Pro is similar to the native tRNAPro NGG. The best ProRS variant for tRNAPro CUAG harbors mutations of Y335W, E338D, R345T, E347W, D352S, and K368S. Importantly, due to the strong recognition of ProRS with the anticodon of tRNAPro, those three pairs of ProRS variants and tRNAPro with different anticodons are mutually orthogonal to each other, providing possibility of simultaneous incorporation of different N‐modified ncAAs.
7.4. HisRS/tRNAHis
Different from other tRNAs, tRNAHis has an additional guanylate (G−1) at its 5′ terminus, which is a major recognition element for HisRS (Francklyn et al., 1992; Himeno et al., 1989). There are few exceptions. Söll and coworkers showed that tRNAHis from Caulobacter crescentus lacks such essential G−1, and C. crescentus HisRS aminoacylates tRNAHis without G−1 (Yuan et al., 2011). Instead, C. crescentus HisRS recognize the anticodon and the acceptor stem of ∆G−1 tRNAHis. Since the conventional HisRS does not recognize the anticodon of tRNAHis, the pair of ∆G−1 type HisRS and tRNAHis could be a potential orthogonal pair to the conventional pair of HisRS and tRNAHis. In this case, the anticodon of conventional tRNAHis can be mutated to CUA for amber suppression, while the conventional HisRS could be engineered to recognize His analogs and incorporate them into proteins at amber codons. On the other side, the host HisRS/tRNAHis can be replaced by ∆G−1 type HisRS and tRNAHis to decode normal histidine codons in the genome.
7.5. SerRS/tRNASer
Most of the ncAAs which have been genetically incorporated into proteins have relatively large side chains. To develop an orthogonal pair for ncAAs with small side chains, Schultz and coworkers recently started with SerRS which does not rely on anticodon recognition (Zambaldo et al., 2020). First, combinations of five different archaeal tRNASer CUA (A. fulgidus [CGA and GCU], M. jannaschii, M. mazei, and P. horikoshii) and six different archaeal SerRSs (A. fulgidus, P. horikoshii, M. mazei, S. solfataricus, M. jannaschii, Halobacterium sp. [NRC‐1], and P. horikoshii) were tested for suppression efficiency of an amber codon in the CAT gene. The best combination is MmSerRS and AftRNASer CUA. Then the AftRNASer CUA were further engineered for higher suppression efficiency by randomizing 11 nucleotides in the acceptor stem. The best variant has a substantially changed acceptor stem. An A to G mutation next the 3′ side of anticodon showed additional increase in amber suppression. The crystal structure of MmSerRS has also been solved, providing a solid base for incorporation of ncAAs with small side chains (Zambaldo et al., 2020).
8. PERSPECTIVES
Not all the existing orthogonal pairs have been used for ncAA incorporation, mostly because those AARSs cannot sufficiently aminoacylate corresponding suppressor tRNAs (Santoro et al., 2003). The amount of ncAA‐acylated suppressor tRNAs cannot fully compete with near cognate suppression from host endogenous tRNAs for designated codons. As most of AARSs recognize the anticodons of tRNAs, mutating original anticodons will affect aminoacylation significantly. Thus, further engineering the anticodon binding regions of AARSs is essential for ncAA incorporation. Another possible reason is that the expression, folding, or stability of introduced AARSs may not be optimal in host cells since they are usually from different domains. In this case, approaches to evaluate and increase their expression, folding, or stability in hosts will help for ncAA incorporation.
It is significant to simultaneously incorporate different ncAAs into a single protein to expand the applications of genetic code expansion. Although we have popular TyrRS and PylRS systems as well as those “not‐so‐popular” ones, we still need to pay attention to several issues. First, the poly‐specificity of evolved AARSs. Because most of the evolved AARSs were only screened against canonical amino acids in negative selections but not for other ncAAs, so they usually cannot distinguish ncAAs with similar or even distinct structures (Figure 4a). For example, the PylRS variant originally selected for AcK is 30‐fold more efficient with Phe derivatives such as 3‐iodo‐Phe than it is with AcK (Guo et al., 2014). Thus, selected orthogonal pairs should be first tested for their specificities for target ncAAs which will be simultaneously incorporated into a single protein, individually. If they can cross‐react with ncAAs for other pairs, a mixture of ncAAs will be incorporated at designated codons. To overcome this issue, engineered AARSs should be further evolved for high specificity toward the target ncAA but not for other ncAAs. To realize this aim, other ncAAs which will be simultaneously incorporated into the protein should be added into the medium in the negative selection to exclude AARS variants recognizing those undesirable ncAAs. Second, the orthogonality between selected pairs. Selected pairs should be mutually orthogonal to each other. Otherwise, we cannot obtain proteins with ncAAs at corresponding positions correspondingly (Figure 4b). All the combinations of evolved AARSs and suppressor tRNAs from selected orthogonal pairs should be tested for their mutual orthogonality. To date, several mutually orthogonal pairs have already been used to incorporate multiple ncAAs into the same protein, including SepRS/PylRS, mjTyrRS/PylRS, SepRS/mjTyrRS, PhLysRS/mjTyrRS, PylRS/EcLeuRS (Vargas‐Rodriguez et al., 2018). Third, the codon selection for individual ncAA. Most AARSs use anticodon recognition to distinguish noncognate tRNAs, thus mutating anticodons may affect aminoacylation significantly. Since most of orthogonal pairs were developed for amber suppression, their aminoacylation activity for other anticodons need to be measured before use. Fortunately, several AARSs mentioned above are tolerant to anticodon changes such as PylRS, PhLysRS, archaea GluRS, HisRS, and SerRS. If they are chosen for multiple ncAA incorporation, the anticodons of their cognate suppressor tRNAs could be flexible.
FIGURE 4.
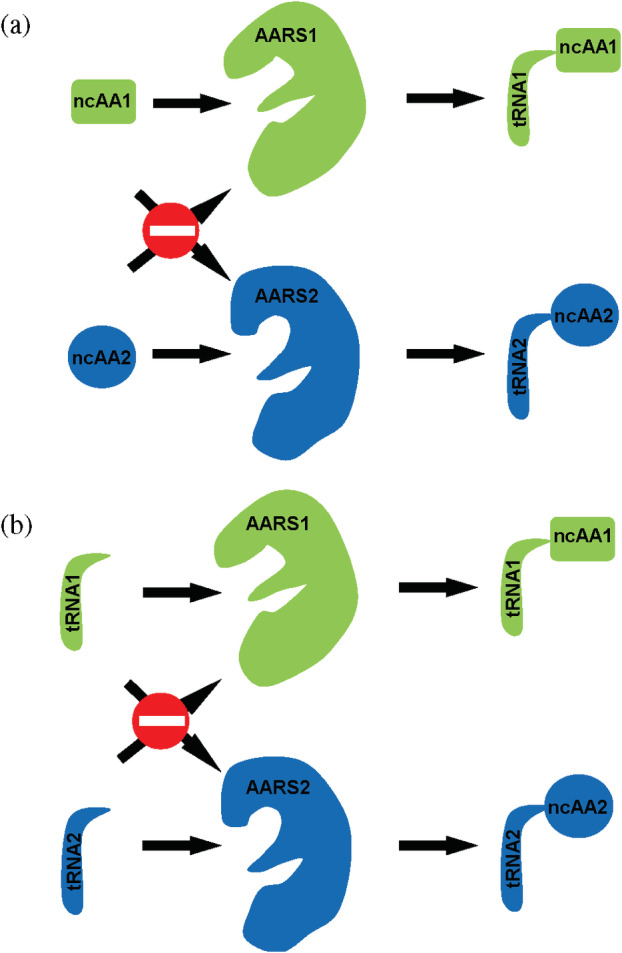
Mutual orthogonality between selected pairs for (a) ncAAs and (b) tRNAs
AUTHOR CONTRIBUTIONS
Joseph Andrews: Writing – original draft (lead). Qinglei Gan: Writing – review and editing (supporting). Chenguang Fan: Writing – review and editing (lead).
CONFLICT OF INTEREST
All authors declare no conflict of interest.
ACKNOWLEDGMENT
This work was funded by the National Institutes of Health R15GM140433 and P20GM139768.
Andrews J, Gan Q, Fan C. “Not‐so‐popular” orthogonal pairs in genetic code expansion. Protein Science. 2023;32(2):e4559. 10.1002/pro.4559
Review Editor: John Kuriyan
Funding information National Institutes of Health, Grant/Award Numbers: P20GM139768, R15GM140433
REFERENCES
- Anderson JC, Schultz PG. Adaptation of an orthogonal archaeal leucyl‐tRNA and synthetase pair for four‐base, amber, and opal suppression. Biochemistry. 2003;42:9598–608. [DOI] [PubMed] [Google Scholar]
- Anderson JC, Wu N, Santoro SW, Lakshman V, King DS, Schultz PG. An expanded genetic code with a functional quadruplet codon. Proc Natl Acad Sci U S A. 2004;101:7566–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Xiao H, Schultz PG. Evolution of multiple, mutually orthogonal prolyl‐tRNA synthetase/tRNA pairs for unnatural amino acid mutagenesis in Escherichia coli. Proc Natl Acad Sci U S A. 2012;109:14841–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Xiao H, Yang PY, Soundararajan G, Schultz PG. A tryptophanyl‐tRNA synthetase/tRNA pair for unnatural amino acid mutagenesis in E. coli. Angew Chem‐Int Edit. 2013;52:5106–9. [DOI] [PubMed] [Google Scholar]
- Chen H, Venkat S, McGuire P, Gan Q, Fan C. Recent development of genetic code expansion for posttranslational modification studies. Molecules. 2018;23:1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CZ, Amikura K, Soll D. Using genetic code expansion for protein biochemical studies. Front Bioeng Biotechnol. 2020;8:598577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack S, Yaremchuk A, Tukalo M. The 2 Å crystal structure of leucyl‐tRNA synthetase and its complex with a leucyl‐adenylate analogue. EMBO J. 2000;19:2351–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre D, Chin JW. Reprogramming the genetic code. Nat Rev Genet. 2021;22:169–84. [DOI] [PubMed] [Google Scholar]
- Ding WL, Zhao HX, Chen YL, Zhang B, Yang Y, Zang J, et al. Chimeric design of pyrrolysyl‐tRNA synthetase/tRNA pairs and canonical synthetase/tRNA pairs for genetic code expansion. Nat Commun. 2020;11:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellefson JW, Meyer AJ, Hughes RA, Cannon JR, Brodbelt JS, Ellington AD. Directed evolution of genetic parts and circuits by compartmentalized partnered replication. Nat Biotechnol. 2014;32:97–101. [DOI] [PubMed] [Google Scholar]
- Fan C, Bobik TA. The PduX enzyme of Salmonella enterica is an L‐threonine kinase used for coenzyme B12 synthesis. J Biol Chem. 2008;283:11322–9. [DOI] [PubMed] [Google Scholar]
- Fan C, Ip K, Soll D. Expanding the genetic code of Escherichia coli with phosphotyrosine. FEBS Lett. 2016;590:3040–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangman WL, Neidhardt FC. Protein and ribonucleic acid synthesis in a mutant of Escherichia coli with an altered aminoacyl ribonucleic acid synthetase. J Biol Chem. 1964;239:1844–7. [PubMed] [Google Scholar]
- Francklyn C, Shi JP, Schimmel P. Overlapping nucleotide determinants for specific aminoacylation of RNA microhelices. Science. 1992;255:1121–5. [DOI] [PubMed] [Google Scholar]
- Fukunaga R, Yokoyama S. Crystal structure of leucyl‐tRNA synthetase from the archaeon Pyrococcus horikoshii reveals a novel editing domain orientation. J Mol Biol. 2005;346:57–71. [DOI] [PubMed] [Google Scholar]
- Furter R. Expansion of the genetic code: site‐directed p‐fluoro‐phenylalanine incorporation in Escherichia coli. Protein Sci. 1998;7:419–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo LT, Wang YS, Nakamura A, Eiler D, Kavran JM, Wong M, et al. Polyspecific pyrrolysyl‐tRNA synthetases from directed evolution. Proc Natl Acad Sci U S A. 2014;111:16724–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeno H, Hasegawa T, Ueda T, Watanabe K, Miura K, Shimizu M. Role of the extra g‐C pair AT the end of the acceptor stem of tRNA (His) in aminoacylation. Nucleic Acids Res. 1989;17:7855–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn MJ, Park HS, O'Donoghue P, Schnitzbauer M, Soll D. Emergence of the universal genetic code imprinted in an RNA record. Proc Natl Acad Sci U S A. 2006;103:18095–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes RA, Ellington AD. Rational design of an orthogonal tryptophanyl nonsense suppressor tRNA. Nucleic Acids Res. 2010;38:6813–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Italia JS, Addy PS, Wrobel CJJ, Crawford LA, Lajoie MJ, Zheng Y, et al. An orthogonalized platform for genetic code expansion in both bacteria and eukaryotes. Nat Chem Biol. 2017;13:446–50. [DOI] [PubMed] [Google Scholar]
- Jahn M, Rogers MJ, Soll D. Anticodon and acceptor stem nucleotides in tRNA(Gln) are major recognition elements for E. coli glutaminyl‐tRNA synthetase. Nature. 1991;352:258–60. [DOI] [PubMed] [Google Scholar]
- Kowal AK, Kohrer C, RajBhandary UL. Twenty‐first aminoacyl‐tRNA synthetase‐suppressor tRNA pairs for possible use in site‐specific incorporation of amino acid analogues into proteins in eukaryotes and in eubacteria. Proc Natl Acad Sci U S A. 2001;98:2268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok Y, Wong JT. Evolutionary relationship between Halobacterium cutirubrum and eukaryotes determined by use of aminoacyl‐tRNA synthetases as phylogenetic probes. Can J Biochem. 1980;58:213–8. [DOI] [PubMed] [Google Scholar]
- Kwon I, Lim SI. Tailoring the substrate specificity of yeast phenylalanyl‐tRNA synthetase toward a phenylalanine analog using multiple‐site‐specific incorporation. ACS Synth Biol. 2015;4:634–43. [DOI] [PubMed] [Google Scholar]
- Kwon I, Tirrell DA. Site‐specific incorporation of tryptophan analogues into recombinant proteins in bacterial cells. J Am Chem Soc. 2007;129:10431–7. [DOI] [PubMed] [Google Scholar]
- Kwon I, Wang P, Tirrell DA. Design of a bacterial host for site‐specific incorporation of p‐bromophenylalanine into recombinant proteins. J Am Chem Soc. 2006;128:11778–83. [DOI] [PubMed] [Google Scholar]
- Lee S, Oh S, Yang A, Kim J, Soll D, Lee D, et al. A facile strategy for selective incorporation of phosphoserine into histones. Angew Chem Int Ed Engl. 2013;52:5771–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DR, Schultz PG. Progress toward the evolution of an organism with an expanded genetic code. Proc Natl Acad Sci U S A. 1999;96:4780–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Fu G, Wang RE, Zhu X, Zambaldo C, Liu R, et al. Genetically encoding phosphotyrosine and its nonhydrolyzable analog in bacteria. Nat Chem Biol. 2017;13:845–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mursinna RS, Martinis SA. Rational design to block amino acid editing of a tRNA synthetase. J Am Chem Soc. 2002;124:7286–7. [DOI] [PubMed] [Google Scholar]
- Park HS, Hohn MJ, Umehara T, Guo LT, Osborne EM, Benner J, et al. Expanding the genetic code of Escherichia coli with phosphoserine. Science. 2011;333:1151–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish AR, She X, Xiang Z, Coin I, Shen Z, Briggs SP, et al. Expanding the genetic code of Caenorhabditis elegans using bacterial aminoacyl‐tRNA synthetase/tRNA pairs. ACS Chem Biol. 2012;7:1292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirman NL, Barber KW, Aerni HR, Ma NJ, Haimovich AD, Rogulina S, et al. A flexible codon in genomically recoded Escherichia coli permits programmable protein phosphorylation. Nat Commun. 2015;6:8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond MH. The effect of amino acid analogues on growth and protein synthesis in microorganisms. Bacteriol Rev. 1962;26:398–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogerson DT, Sachdeva A, Wang KH, Haq T, Kazlauskaite A, Hancock SM, et al. Efficient genetic encoding of phosphoserine and its nonhydrolyzable analog. Nat Chem Biol. 2015;11:496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson LE, Uhlenbeck OC. Exploring the specificity of bacterial elongation factor Tu for different tRNAs. Biochemistry. 2007;46:6194–200. [DOI] [PubMed] [Google Scholar]
- Santoro SW, Anderson JC, Lakshman V, Schultz PG. An archaebacteria‐derived glutamyl‐tRNA synthetase and tRNA pair for unnatural amino acid mutagenesis of proteins in Escherichia coli. Nucleic Acids Res. 2003;31:6700–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro SW, Wang L, Herberich B, King DS, Schultz PG. An efficient system for the evolution of aminoacyl‐tRNA synthetase specificity. Nat Biotechnol. 2002;20:1044–8. [DOI] [PubMed] [Google Scholar]
- Sauerwald A, Zhu WH, Major TA, Roy H, Palioura S, Jahn D, et al. RNA‐dependent cysteine biosynthesis in archaea. Science. 2005;307:1969–72. [DOI] [PubMed] [Google Scholar]
- Summerer D, Chen S, Wu N, Deiters A, Chin JW, Schultz PG. A genetically encoded fluorescent amino acid. Proc Natl Acad Sci U S A. 2006;103:9785–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada T, Nureki O, Ishitani R, Ambrogelly A, Ibba M, Soll D, et al. Functional convergence of two lysyl‐tRNA synthetases with unrelated topologies. Nat Struct Biol. 2002;9:257–62. [DOI] [PubMed] [Google Scholar]
- Tumbula DL, Becker HD, Chang WZ, Soll D. Domain‐specific recruitment of amide amino acids for protein synthesis. Nature. 2000;407:106–10. [DOI] [PubMed] [Google Scholar]
- Vargas‐Rodriguez O, Sevostyanova A, Soll D, Crnkovic A. Upgrading aminoacyl‐tRNA synthetases for genetic code expansion. Curr Opin Chem Biol. 2018;46:115–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkat S, Sturges J, Stahman A, Gregory C, Gan Q, Fan C. Genetically incorporating two distinct post‐translational modifications into one protein simultaneously. ACS Synth Biol. 2018;7:689–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan W, Tharp JM, Liu WR. Pyrrolysyl‐tRNA synthetase: an ordinary enzyme but an outstanding genetic code expansion tool. Biochim Biophys Acta. 2014;1844:1059–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KH, Sachdeva A, Cox DJ, Wilf NW, Lang K, Wallace S, et al. Optimized orthogonal translation of unnatural amino acids enables spontaneous protein double‐labelling and FRET. Nat Chem. 2014;6:393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. [DOI] [PubMed] [Google Scholar]
- Wang W, Takimoto JK, Louie GV, Baiga TJ, Noel JP, Lee KF, et al. Genetically encoding unnatural amino acids for cellular and neuronal studies. Nat Neurosci. 2007;10:1063–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelihan EF, Schimmel P. Rescuing an essential enzyme‐RNA complex with a non‐essential appended domain. EMBO J. 1997;16:2968–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Wang Z, Huang Y, Liu WR. Catalyst‐free and site‐specific one‐pot dual‐labeling of a protein directed by two genetically incorporated noncanonical amino acids. Chembiochem. 2012;13:1405–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N, Deiters A, Cropp TA, King D, Schultz PG. A genetically encoded photocaged amino acid. J Am Chem Soc. 2004;126:14306–7. [DOI] [PubMed] [Google Scholar]
- Yuan J, Gogakos T, Babina AM, Soll D, Randau L. Change of tRNA identity leads to a divergent orthogonal histidyl‐tRNA synthetase/tRNA(His) pair. Nucleic Acids Res. 2011;39:2286–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambaldo C, Koh M, Nasertorabi F, Han GW, Chatterjee A, Stevens RC, et al. An orthogonal seryl‐tRNA synthetase/tRNA pair for noncanonical amino acid mutagenesis in Escherichia coli. Bioorg Med Chem. 2020;28:115662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MS, Brunner SF, Huguenin‐Dezot N, Liang AD, Schmied WH, Rogerson DT, et al. Biosynthesis and genetic encoding of phosphothreonine through parallel setection and deep sequencing. Nat Methods. 2017;14:729–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Kwon I. Controlling enzyme inhibition using an expanded set of genetically encoded amino acids. Biotechnol Bioeng. 2013;110:2361–70. [DOI] [PubMed] [Google Scholar]


