Abstract
Recent global health concern motivated the exploration of natural medicinal plant resources as an alternative target for treating COVID-19 infection and associated inflammation. In the current study, a phytochemical, 6-shogaol [1-(4-hydroxy-3-methoxyphenyl)dec-4-en-3-one; 6-SHO] was investigated as a potential anti-inflammatory and anti-COVID-19 agent. In virus release assay, 6-SHO efficiently (94.5%) inhibited SARS-CoV2 replication. When tested in the inflammasome activation model, 6-SHO displayed mechanistic action by regulating the expression of the inflammasome pathway molecules. In comparison to the existing drugs, remdesivir and hydroxy-chloroquine, 6-SHO was not only found to be as effective as the standard anti-viral drugs but also much superior and safe in terms of predicted physicochemical properties and clinical toxicity. Comparative molecular dynamics simulation demonstrated a stable interaction of 6-SHO with NLRP3 (the key inflammasome regulator) in the explicit water environment. Overall, this study provides important cues for further development of 6-SHO as potential anti-inflammatory and anti-viral therapeutic agents.
1. Introduction
The world experienced outbreak of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV2) infection in 2019, which has spread to almost every part of world. The first case of this infection was detected in the city of Wuhan, Hubei province, China, in December 2019.1 Due to the rapid increase in the number of people infected with SARS-CoV2, efforts toward developing effective preventive and therapeutic treatments such as vaccines and antiviral drugs were being made all over the world. Traditional medicines that influence the innate immune system and produce a memory-like response (termed “trained immunity”) help a quicker inflammatory response and have not been explored in greater depth.2 Considering the safety, low cost, and easy availability of these traditional medicines, it is worth considering them as the possible alternative preventive strategy not only in COVID-19 but also for other viral infections.
Pattern recognition receptors (PRRs) expressed in innate immune cells, including long-lived macrophages and their precursors, are involved in the stimulation of trained immunity.3 Trained immunity is crucial in shaping the innate immunity for preventing re-infection with bacteria, fungi, or viruses and controlling inflammation.4 However, in response to viral infection, some PRRs form the inflammasome—a multimeric platform of proteins that initiate inflammation and cell death.5 Among all the inflammasomes discovered, the NLR family pyrin domain containing 3 (NLRP3) inflammasome plays a predominant role in both inflammation regulation and antiviral responses.6 NLRP3 inflammasome has emerged as a crucial investigation point owing to its role in the body’s antiviral defence.6 Therefore, based on this, we focused our study on identifying the phytochemicals that have potential to regulate the NLRP3-associated proteins.
Virus entry into cells is an essential step of cross-species transmission, especially for the beta coronaviruses.7 SARS-CoV2 uses its surface glycoprotein, spike, to recognize and bind to the surface receptor, angiotensin-converting enzyme 2 (ACE2) on the target cell.8 The virus entry requires spike protein cleavage into S1 and S2 subunits by transmembrane protease serine 2 (TMPRSS2) and cathepsin.9 The spike protein S2 subunit mediates the fusion of viral and cellular membranes, ensuring viral entry through endocytosis. The affinity between the virus’s surface proteins and its receptors is a critical step for viral entry.10 SARS-CoV2 primarily infects ciliated bronchial epithelial cells and type-II pneumocytes because these cells express ACE2 and TMPRSS2, required for the spike protein cleavage.11,12 The RNA-dependent RNA polymerase (RdRP) domain of Nsp12 is a key component of the replication-transcription machinery. Hence, RdRP is considered to be one of the primary targets for antiviral drug development, against a wide variety of viruses including SARS-CoV2.13 Recently, remdesivir was shown to inhibit SARS CoV2 viral replication in cell culture, revealing the potential of this nucleoside analogue prodrug to be used for the broad-spectrum treatment of SARS-CoV2 infections.14
1.2. Ginger as One of the Indian Traditional Ayurvedic Medicines
Indian traditional ayurvedic medicines (ITAMs) have been used to treat various infectious diseases since thousands of years (Box S1). An active constituent of ginger (Shunthi), 6-shogaol [(1-(4-hydroxy-3-methoxyphenyl)dec-4-en-3-one; 6-SHO (a monomethoxybenzene derivative of phenol and enone)], is reported to possess anti-inflammatory and anticancer activities.15 Owing to the broad spectral advantages, the aim of this article is to evaluate the importance of ginger and its key constituent phytochemical 6-SHO for their ability to interfere with the host–pathogen interaction and regulating the NLRP3 inflammasome. Our results showed that 6-SHO has the ability to inhibit the replication of SARS-CoV2 and reduce the virus-induced inflammation through inactivation of NLRP3 inflammasome. These results were very well supported by molecular docking study, showing efficient interaction of 6-SHO in the inhibitor binding site of the NRLP3-NACHT. The clinical treatment protocols based on 6-SHO also showed effectiveness in controlling COVID-19 infection. Overall, this study showed that 6-SHO can help in reducing the burden of viral infection by inhibiting its replication and decreasing the virus-induced inflammation. This work highlights the importance of active phytochemical 6-SHO for developing the potential therapeutic intervention.
2. MATERIALS and METHODS
2.1. Extract Preparation from Ginger (Zingiber officinale)
Fine powder of dried rhizomes of ginger was used for extract preparation. The extraction was carried out by refluxing the sample with methanol on a water bath. The sample solvent ratio was 1:20, and extraction time was 8 h. The supernatant of each extraction was combined and filtered through Whatman filter paper. The combined supernatant was concentrated using a rotary evaporator (Heizbad Hei-VAP, Heidolph, Schwabach, Germany temperature = 40 ± 5 °C).
2.2. Identification of 6-SHO from Ginger Extracts
Chromatographic separation was achieved using a high-performance liquid chromatography (HPLC) (Waters, Milford, MA, USA) system consisting of quaternary pumps, an in-line vacuum degasser, and a photodiode array detector (PDA) model: Waters 2996, Waters, Milford, MA, USA. The instrumentation was controlled using Empower 3.0 software (Waters). Separation was achieved on a C-18 reversed phase column (250 × 4.6 mm, 5.0 μm, Waters). The peaks obtained in the HPLC-PDA chromatograms of extracts were monitored in the wavelength range of 210–500 nm using a PDA detector.
2.3. Evaluation of Anti-viral Efficacy of 6-SHO
2.3.1. Cell Line and Virus
The Vero (African green monkey kidney epithelial cells) cell line was procured from NCCS, Pune. Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, PAN Biotech, Aiden Bach, Germany), supplemented with 10% fetal bovine serum (FBS) (PAN Biotech, Aiden Bach, Germany), gentamicin, and penicillin–streptomycin. ILS-03 (Accession no. EPI_ISL_1196305) strain of SARS-CoV2 was successfully adapted and isolated in Vero cells at ILS, Bhubaneswar, and this strain was used for this study.16
2.3.2. Cytocompatibility of 6-SHO to Vero Cells
MTT assay was performed to determine the cytocompatibility of 6-SHO using the EZcount MTT cell assay kit (HiMedia, Mumbai, India) in Vero E6 cells according to the manufacturer’s protocol. In brief, Vero E6 cells were seeded in a 96-well plate at a density of approximately 10 000 cells per well. After reaching 70% confluency, cells were treated with different concentrations (1, 2.5, 5, 10, 15, and 20 μg/mL) of 6-SHO in the triplicate form in the respective wells along with dimethyl sulfoxide as a reagent control. After 22 h, the plate was washed with 1× phosphate-buffered saline (PBS), and subsequently, 10 μL of the MTT reagent (5 mg/mL) was added to the cells and incubated for 2–3 h at 37 °C. Next, the media was removed, and 100 μL of solubilization buffer was added followed by incubation at 37 °C for 15 min to dissolve the formazan crystals. Finally, the absorbance was measured at 570 nm using a multimode plate reader, and the metabolically active cell percentage was compared with the control cells to determine the cellular cytotoxicity.17
2.3.3. Viral Infection
At 80% confluency, Vero E6 cells were infected with SARS-CoV2 at a multiplicity of infection of 0.1 followed by 90 min incubation with shaking at every 10 to 15 min interval as mentioned before.18 After infection, the cells were washed with 1× sterile PBS followed by treatment with 10 μg/mL 6-SHO diluted in complete DMEM. The cytopathic effect (CPE) was observed under the microscope with 10× magnification at 22 h post infection (hpi), and subsequently, supernatants were collected for RNA extraction.
2.3.4. RNA Extraction and qRT-PCR
To determine the viral copy number and anti-viral effect, quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed according to the method described before.16 For this, viral RNA was isolated from the supernatants using the TAN Bead Maelstrom 4800 automated RNA extraction platform as per manufacturer’s instructions, and cDNA was synthesized using random hexamers by the Prime Script First strand cDNA synthesis kit (Takara, Kusatsu, Japan). The synthesized cDNA was used to amplify the nucleocapsid (NC) gene using specific primers [nucleocapsid forward primer: GTAACACAAGCTTTCGGCAG and nucleocapsid reverse primer: GTGTGACTTCCATGCCAATG)] by qRT-PCR (Mesagreen SYBR Green-No ROX, Eurogentec, Belgium). The viral copy number was determined for the abovementioned samples by generating the standard curve of the SARS-CoV2 NC gene. The percentage of copy number/mL was calculated from the corresponding Ct values of all the samples.
2.3.5. EC50 Estimation of 6-SHO
For EC50 estimation, the Vero cells were infected with SARS-CoV2 as per the protocol mentioned in the website. After infection, the cells were treated with different concentrations of 6-SHO (1, 2.5, 5, and 10 μg/mL) followed by collection of the respective supernatants at 22 hpi.
2.4. Effect of 6-SHO on Inflammasome Pathway Molecules by Immunofluorescence
In brief, A549 cells were dispensed at 5000 cells per well in glass-bottom 35 mm Petri dishes (ThermoFisher Scientific) and allowed for 4 h adherence at 37 °C in a humidified atmosphere of 5% CO2 in air. Cells were exposed to LPS at a concentration of 500 ng/mL for 12 h. This was followed by the known stimulator of the inflammasome pathway Nigericin at a concentration of 5 μM for 45 min. For inhibition experiments, cells were pretreated for 1 h with either 6-SHO at 5 μM or MCC950 at 5 μM before LPS treatment. After assay completion, cells were fixed using 4% paraformaldehyde for 15 min at room temperature and further incubated with primary antibody anti-human NLRP3 (ab263899, Abcam), and it was further labeled with secondary antibody goat anti-rabbit tagged to Alexa 568 (Invitrogen A11011). Cells were also labeled for dual-pathway marker anti-human Caspase-1 (SC 56036) conjugated to secondary antibody goat anti-mouse Alexa-488 (Invitrogen A11001). For mitochondrial activation experiment, cells were labeled first with mitotracker dye (Abcam ab112143) and then fixed with 1% PFA and proceeded with anti-human NLRP3/AIM2 conjugated with Alexa-488. Images were acquired on a laser confocal microscope (LSM780, Carl Zeiss, Germany) and analyzed using LSM Image Browser.
2.5. Physicochemical, Absorption, Distribution, Metabolism, and Excretion/Toxicology, and Toxicity Prediction of Ligands
Physicochemical and absorption, distribution, metabolism, and excretion/toxicology (ADME/T) properties were calculated using the QikProp module of Schrödinger suite 2019-1 (Schrödinger, LLC, New York, NY).19 It predicts both physicochemically significant descriptors and pharmacokinetically important properties of the molecules. QikProp presents ranges to compare the molecular properties of the molecules with those of known drugs. For a quick prediction of the physicochemical and ADME/T properties of all the ligand molecules including 6-SHO, remdesivir, and hydroxy-chloroquine (HCQ), key parameters such as bioavailability, solubility, drug-likeness, gut absorption capacity, cardiotoxicity, and renal toxicity were included in this study. For the toxicity prediction, the TOPKAT (TOxicity Prediction by Komputer Assisted Technology) tool from Discovery studio was used. TOPKAT employs robust and cross-validated quantitative structure toxicity relationship models to assess various measures of toxicity and utilizes the patented Optimal Predictive Space validation method to interpret the results. Few key toxicity predictions of ligand molecules have been included in the study.
2.6. Docking and MD Simulation of 6-SHO with the NLRP3-NACHT Domain
6-SHO is reported to have an antagonistic effect in regulating the NLRP3 inflammasome activation.20 The antagonism of 6-SHO was studied in high-glucose-activated NLRP3 inflammasomes and consequent calcification of human artery smooth muscle cells.21 In order to understand the possible interaction of 6-SHO with NLRP3, molecular docking and MD simulations were carried out. The NLRP3 crystal structure was obtained from PDB (PDBID: 7ALV(22)), and 3D coordinates 6-SHO (CID 5281794) was retrieved from the PubChem database. Prior to docking simulation, both protein and ligand coordinates were prepared by adding polar hydrogen bonds and assigning Kollman charges (to NLRP3 domain) and Gasteiger partial charges (to 6-SHO) using AutoDock Tools 1.5.23 The NP3-146 binding pocket was considered as the target site for molecular docking, where grid boxes were generated. A total of 10 individual docking simulations were carried out using AutoDock Vina.24 To understand the dynamic stability of 6-SHO in the NP3-146 binding pocket, molecular dynamics (MD) simulations were carried out in GROMACS25 using the CHARMM36 force field.26 The ligand topologies for 6-SHO and NP3-146 were obtained from the SwissParam server.27 Both the protein–ligand complexes (NLRP3—NP3-146 and NLRP3—6-SHO) were immersed into individual cubic boxes with TIP3P water models and 0.15 M NaCl. Later, energy minimizations of solvated systems were carried out using the steepest descent algorithm until a tolerance of 1000 kJ/mol to avoid high energy interactions and steric conflicts between atoms. The energy-minimized systems were then equilibrated for 1.1 ns in two different steps, NVT (100 ps) and NPT (1 ns). After system equilibration, the NPT ensemble final production runs were carried out for 60 ns time scale by adopting the detailed simulation procedure from our previous study.28 The trajectory analysis was performed using built-in modules of GROMACS. PyMOL (www.pymol.org) was used for molecular visualization. 2D graphs from trajectory analysis were plotted using the Grace 5.1.23 program (http://plasma-gate.weizmann.ac.il/Grace/).
3. Results
3.1. ADME and Toxicity Predictions for 6-SHO
The ADME properties of the ligands were predicted using QikProp and TOPKAT. 6-SHO and HCQ conformed to Lipinski’s rule of five, whereas remdesivir violated it (Table 1). All the molecules exhibited appropriate polar surface area (PSA), polarizability, and human serum albumin binding values for biological efficacy. All the molecules have medium risk of cardiotoxicity (HERG K+ inhibition) and central nervous system (CNS) toxicity except HCQ. 6-SHO exhibited the highest gut absorption (Caco-2 cell permeability). Other associated factors, such as blood brain permeability, skin permeability, and percentage of human oral absorption, were also in the acceptable range for all the molecules. The rat oral median lethal dose (LD50) and chronic toxicity properties give the best score to 6-SHO as compared to remdesivir. Thus, 6-SHO has good physicochemical and ADME/T properties followed by HCQ and remdesivir, the last two of which violated some of the physicochemical properties. As evidenced (Table 2), the in silico analysis predicted that 6-SHO is environmentally safe. It is biodegradable and can be easily degraded by microorganisms in the presence of oxygen. However, remdesivir and HCQ have low biodegradable potential. The mutagenicity test (Ames) predicted that 6-SHO and remdesivir are not mutagenic as compared to HCQ. All the three compounds did not exhibit cancer-causing ability in rodents; however, a low possibility of causing cancer was predicted for remdesivir. On testing the irritancy of these compounds, it was found that 6-SHO and remdesivir did not show ocular irritancy but HCQ did. We tested for skin irritancy and found that HCQ was predicted to be a non-irritant, whereas 6-SHO exhibited mild-irritancy potential. The analysis predicted remdesivir to have a moderate to severe skin irritancy potential. Finally, while testing for hepatotoxicity, 6-SHO was predicted to be safe as compared to remdesivir and HCQ, which have the potential to cause hepatotoxicity. Based on the six toxicity predictions, 6-SHO is predicted to be the safest compound, whereas HCQ is predicted to be more toxic followed by remdesivir.
Table 1. Physicochemical Properties and ADME/T Profile of 6-SHO, Remdesivir, and HCQ.
| ADME propertiesa |
||||
|---|---|---|---|---|
| properties | 6-SHO | remdesivir | HCQ | drug likeness |
| rule of five (drug-likeness) | no violation (0) | violated (2) | no violation (0) | 6-SHO > HCQ > Rm |
| PSA (drug-likeness) | very good (57.321) | poor (196.44) | very good (48.802) | 6-SHO > HCQ > Rm |
| binding with proteins (biological activity) | good (0.44) | average (–0.659) | good (0.211) | 6-SHO > HCQ > Rm |
| solubility (hydrophilicity and membrane permeability) | high (4.043) | average (0.897) | high (3.578) | 6-SHO > HCQ > Rm |
| renal absorption (MDCK permeability) | very high (604.025) | poor (13.5) | very high (709.185) | HCQ > 6-SHO > Rm |
| cardiotoxicity (HERG K+) | non-toxic (–5.487) | non-toxic (–5.031) | toxic (–6.494) | Rm > 6-SHO > HCQ |
| gut absorption (Caco-2 permeability) | very high (1202.866) | low (35.568) | good (551.822) | 6-SHO > HCQ > Rm |
| predicted CNS toxicity | safe (–2) | safe (–2) | low risk (1) | 6-SHO > Rm > HCQ |
| predicted brain/blood partition coefficient | very good (–1.068) | average (–2.816) | good (–0.187) | 6-SHO > HCQ > Rm |
| predicted skin permeability | excellent (–1.914) | good (–4.023) | good (–3.195) | 6-SHO > HCQ > Rm |
| percent human oral absorption | high (3) | low (1) | high (3) | 6-SHO > HCQ > Rm |
| rat oral LD50 g/kg | 5.93 | 0.27 | 1.94 | 6-SHO > HCQ > Rm |
| maximum tolerated dose (oral gavage) g/kg | 0.11 | 0.0005 | 0.55 | HCQ > 6-SHO > Rm |
| rat inhalation LC50 mg/m3/h | 13080.40 | 69.14 | 9720.78 | 6-SHO > HCQ > Rm |
| chronic toxicity g/kg | 0.36 | 0.00 | 0.07 | 6-SHO > HCQ > Rm |
Physicochemical properties were evaluated using QikProp Schrodinger version 2019-1 and TOPKAT module of Discovery Studio version 3.5. Normal ranges: PSA (7–200); MDCK (<25 poor, >500 great); HERG K+ channels (concern below −5); Caco-2 cell permeability (<25 poor, >500 great); CNS activity (−2 inactive, +2 active); and brain/blood barrier (−3.0–1.2).
Table 2. Toxicity Predictions of 6-SHO, Remdesivir, and HCQ Based on Similarity with Known Compounds by Discovery Studio.
| toxicity
properties |
||||
|---|---|---|---|---|
| compounda | 6-SHO | remdesivir | HCQ | non-toxicity |
| environment safe (aerobic biodegradability) | yes (3.96) | no (–5.41) | no (–16.3) | 6-SHO > Rm > HCQ |
| mutagenicity (AMES test) | non-mutagen (–1.63) | non-mutagen (–20.9) | mutagen (4.2) | Rm > 6-SHO > HCQ |
| carcinogenic ability (rat/mouse/rodent) | non-carcinogen | non-carcinogen/carcinogen | non-carcinogen | 6-SHO > Rm > HCQ |
| ocular irritancy | non-irritant (–1.21) | non-irritant (–1.34) | irritant (0.562) | Rm > 6-SHO > HCQ |
| skin irritant | mild irritant | mild irritant | non-irritant | HCQ > 6-SHO > Rm |
| hepatotoxicity | no (–10.0442) | yes (–0.643255) | yes (0.437765) | 6-SHO > Rm > HCQ |
Physicochemical properties were evaluated using QikProp Schrodinger version 2019-1 and the TOPKAT module of Discovery Studio version 3.5.
3.2. 6-SHO Inhibits SARS-CoV2 Replication
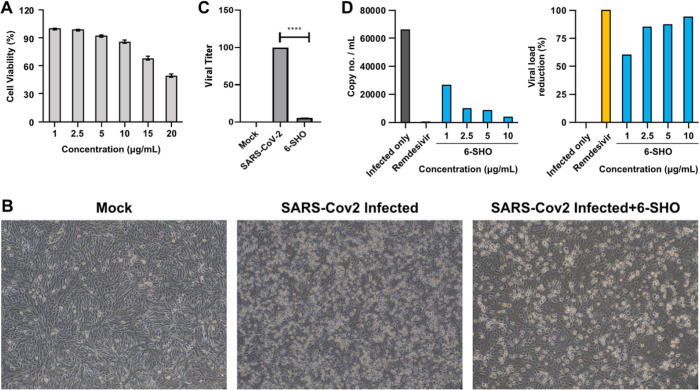
To assess the cytocompatibility of 6-SHO to Vero cells, MTT assay was performed, and 10 μg/mL was found to be the maximum non-toxic concentration (85% cell viability, Figure 1A). Further anti-viral efficacy of 6-SHO on replication of SARS-CoV2 in Vero cells in CPE assay. The anti-SARS-CoV2 potential of 6-SHO was evident as it was showing significant reduction in CPE with comparison to the infection control at 22 hpi (Figure 1B). In addition, the qRT-PCR data also indicated 94.5% reduction in the viral copy number as compared to the infection (Figure 1C). The effective concentration 50 (EC50) dose was found to be 0.95μg/ml (Figure 1D). Altogether, these data indicate that 6-SHO can inhibit SARS-CoV2 infection significantly.
Figure 1.
Effect of 6-SHO on replication of SARS-CoV2 virus in Vero cells. (A) Cytotoxicity of 6-SHO on Vero cells by MTT assay. (B) Bright-field microscopy images of Vero cells in regulating SARS-CoV2 infection in the presence of 6-SHO. (C) Bar graph depicts the real-time PCR, revealing significantly reduced viral titer. (D) Bar graphs display viral load reduction based on the reduction in the CPE of various concentrations of 6-SHO to evaluate the ED50 dose of 6-SHO. Remdesivir was used as the positive control. This experiment was repeated two times.
3.3. Molecular Interaction of 6-SHO with NLRP3
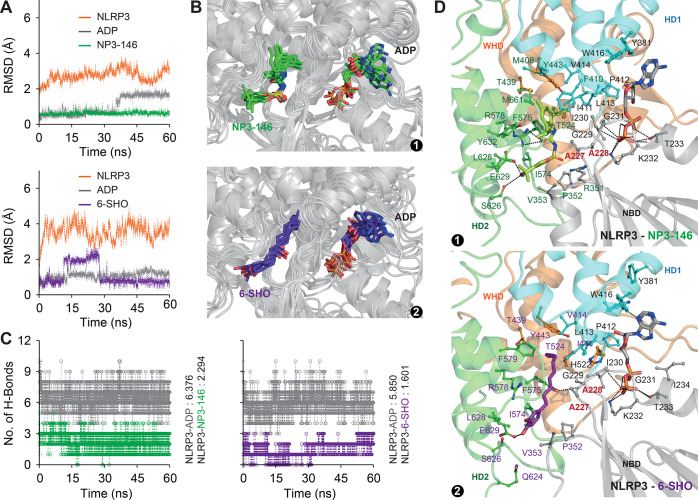
Earlier studies suggested that MCC950 inhibits NLRP3 activation by interaction with its ATP/ADP-binding cavity.29 A recent study revealed that NP3-146, an analogue of MCC950, was found to get accommodated in an additional cavity near the ADP binding cavity.22 As our focus was to understand whether 6-SHO acts through regulating the NLRP3 pathway and prefers the binding mode similar to MCC950/NP3-146, we considered the newly identified ligand-binding cavity for computational study to decipher the binding mode of 6-SHO. Initially, we docked 6-SHO in the NP3-146 binding cavity of NLRP3-NACHT. As shown in Figure S1A,B, we observed that the bound conformation of 6-SHO successfully accommodated in the NP3-146 binding pocket. To check dynamic stability of ligands in the NP3-146-binding cavity, MD simulations lasting up to 60 ns were carried out for both the complexes.
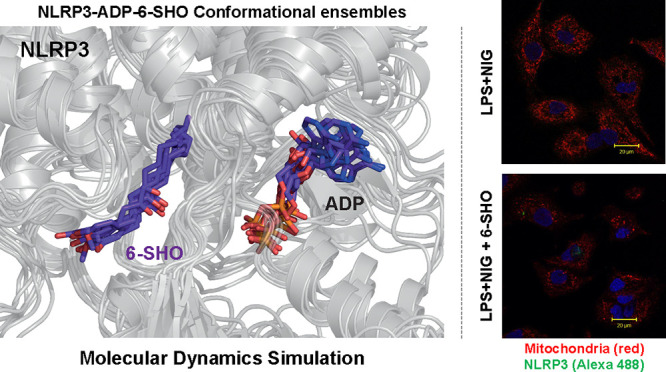
As shown in Figure 2A, we observed stable backbone root mean square deviation (rmsd) of NLRP3-NACHT in both the complexes with minimal drift. The rmsd of ADP, NP3-146, and 6-SHO was found to be highly stable (rmsd: 1–2 Å) during the course of simulation in both complexes (Figure 2A) with minimum deviation. Moreover, the conformational ensemble of NLRP3-ADP-NP3-146/-6-SHO snapshots (Figure 2B) together with intermolecular H-bonds (Figure 2C) suggested a stable association of ligands as a function of simulation time. The in-detail interaction analysis of best-clustered coordinates suggested that 6-SHO is accommodated in the inhibitor-binding cavity with few numbers of H-bonds (highly dominated by hydrophobic forces) as compared to NP3-146 (Figure 2D). Furthermore, the interaction analysis indicated an array of common amino acids that are interacting with ADP, NP3-146, and 6-SHO. Key residues such as A227 and A228, suggested to be critical,22 were noticed to be coordinating the NP3-146-ADP and 6-SHO-ADP (Figure 2D). Overall, the present results suggest that 6-SHO accommodates the same binding space as that of NP3-146.
Figure 2.
MD simulation of NLRP3-ADP-NP3-146 and NLRP3-ADP-6-SHO complexes. (A) Comparative overview of RMSD profiles of NLRP3, ADP, NP3-146, and 6-SHO. (B) Conformational ensembles of (1) NLRP3-ADP-NP3-146 and (2) NLRP3-ADP-6-SHO coordinates. The coordinates were obtained from the last 50 ns trajectory at 10 ns intervals. (C) Total numbers of H-bonds formed between NLRP3 and the ligand across the trajectory. (D) Visualization of key ligand-binding residues in (1) NLRP3-ADP-NP3-146 and (2) NLRP3-ADP-6-SHO complexes. The key ADP-binding residues are highlighted in black font, ligand (NP3-146/6-SHO)-binding residues are highlighted in green/violet, and the residues that interact with both ADP and NP3-146/6-SHO are highlighted in bold red font.
3.4. Mechanistic Role of 6-SHO in Regulating the Innate Immune Inflammasome Pathway
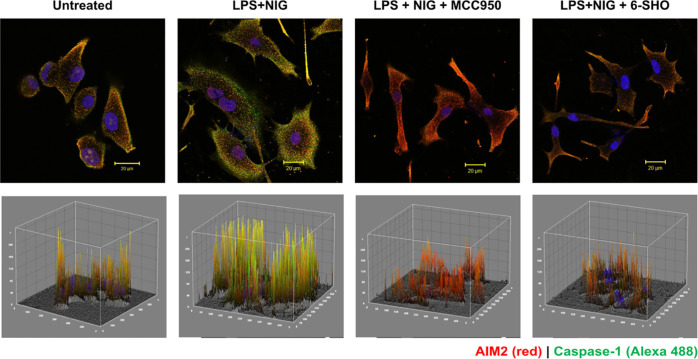
To evaluate if 6-SHO acts by controlling the AIM2/NLRP3 inflammasome pathway, we initially activated the inflammasome pathway in the A549 cell line using damage-inducing agent LPS and stimulator NIG. Then, the expression of AIM2/NLRP3 in the presence of these stimulants was checked by immunofluorescence. Staining of A549 cells without the primary antibody did not show expression of AIM2/Caspase-1 (data not shown). A549 cells activated for the inflammasome pathway and stained with the primary antibody against AIM2 or Caspase-1 showed strong colocalization expression of AIM2 and Caspase-1 (Figure 3), which was significantly inhibited by treatment with 6-SHO and MCC950 at 5 μM.
Figure 3.
Effect of 6-SHO on inflammasome pathway activation and expression of AIM2 and caspase-1 in the A549 lung cancer cell line. The upper panel shows AIM2 (red) and Caspase-1 (green) expression in the A549 lung cancer cell line, cells before and after the activation of the inflammasome pathway. The lower panel shows the corresponding 3D interactive plots for red/ green and dual (yellow) fluorescence intensity. Cells were activated with LPS and nigericin (inflammasome activator). Cells exhibited puncta formation made of multimeric proteins AIM2 and caspase-1. Specific inflammasome pathway inhibitors MCC950 and 6-SHO at 5 μM significantly lowered pathway molecule expression. The scale bar is shown at the right bottom of each image (20 μm).
3.5. Effect of 6-SHO on Mitochondrial Activation and Co-localization with NLRP3
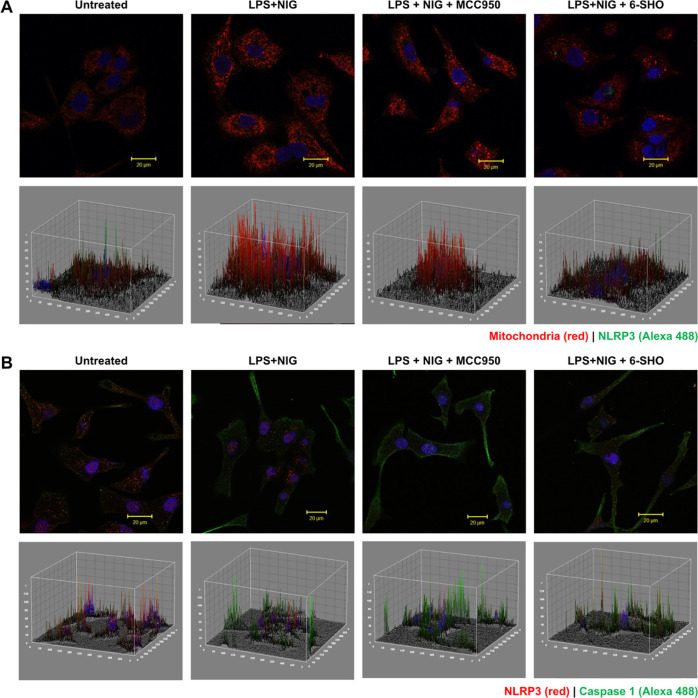
To further investigate the effect of 6-SHO on mitochondrial ROS expression in inflammasome-activated A549 cells, we carried out live imaging. As seen in Figure 4A, A549 exhibited mitochondria with a distinct architecture in live imaging under confocal microscopy. LPS + NIG activation led to an increase in mitochondrial ROS expression. 6-SHO-treatment of inflammasome-activated cells exhibited reduction in mitochondrial ROS expression (Figure 4A). Since mitochondrial expression enhancement was associated with inflammasome activation, we investigated if NLRP3 has any association with mitochondrial protein.
Figure 4.
Effect of 6-SHO on inflammasome pathway activation and association with mitochondrial activation in A549. (A) Upper panel shows mitochondrial (mitotracker red) and NLRP3 co-expression in A549 lung cancer cell lines before and after activation of the inflammasome pathway and inhibition using 6-SHO or MCC950. The lower panel shows the corresponding 3D interactive plots for red/ green and dual (orange) fluorescence intensity. Cells were activated with LPS and nigericin (inflammasome activator). Cells exhibited puncta formation made of multimeric protein mitochondrial ROS and NLRP3. Specific inflammasome pathway inhibitor MCC950 (NLRP3 inhibitor) and 6-SHO at 5 μM brought down pathway molecule expression significantly. The scale bar is shown at the right bottom of each image (20 μm). (B) Effect of 6-SHO on inflammasome pathway activation and expression of NLRP3 and caspase-1 in A549. The upper panel shows inflammasome pathway marker NLRP3 and casapase-1 co-expression in the A549 lung cancer cell line before and after activation of the inflammasome pathway and inhibition using 6-SHO or MCC950. The lower panel shows the corresponding 3D interactive plots for red/ green and dual (orange) fluorescence intensity. The scale bar is shown at the right bottom of each image (20 μm).
When A549 cells were labeled for dual mitochondrial and NLRP3 expression, it was interesting to note that both were found to be colocalized in inflammasome-activated A549 lung cancer cells, while both were found to be significantly reduced in 6-SHO-treated or MCC950-treated A549 cells. Similarly, NLRP3 and caspase-1 expression was affected in a similar way in A549 cells treated for inflammasome activation with 6-SHO (Figure 4B).
3.6. 6-SHO as an Alternative Medicine to Combat COVID-19: Data from Clinical Trials
The U.S. Food and Drug Administration issued an emergency use authorization for the Moderna COVID-19 vaccine and Pfizer-BioNTech COVID-19 vaccine.30 Vaccines stimulate an adaptive immune response that could protect against SARS-CoV2. Traditional medicine may enhance the general innate immune response.31 Efforts have also focused on developing an effective protocol to treat patients infected by COVID-19. Various protocols have been proposed and initiated, which include a combination of antibiotics, various antivirals, steroids, and plasma from recovered patients. Currently, no appropriate drug option is available for this fatal disease. Various compounds derived from natural resources that have previously shown anti-microbial and anti-viral activity are being investigated in clinical trials to treat COVID-19.
Clinical trials for COVID-19 using traditional medicine or alternative medicine have been compiled (Table S1; adapted from NIH clinical trial registry). One trial # ChiCTR2000032400 is testing the benefit of vitamin C via the intravenous route for COVID-19 pneumonia treatment. Siddha treatment has also been initiated in Trial # ISRCTN13311119. The NCT04323345 trial is testing the use of honey which is FDA-approved and has previously proven broad-spectrum clinical benefits as an anti-microbial, anti-viral, anti-mycobacterial, and anti-fungal nutraceutical. Yang et al. has reviewed the use of traditional Chinese medicine for identification of the therapeutic agent.32
A series of ITAM-based clinical trials were registered at ‘The Clinical Trials Registry India (CTRI) [CTRI/2020/05/025276, registered on: 20/05/2020; CTRI/2020/05/025335, registered on: 24/05/2020] and have been initiated by Ch. Brahm Prakash Ayurved Charak Sansthan (CBPACS), an autonomous institute of Ayurveda under the Government of NCT Delhi at Khera Dabar, New Delhi, India 110073. These clinical trial studies were approved by ethics committees of participating institutes and were in compliance with the regulations of the Ministry of AYUSH, Government of India. As per the guidelines, RT-PCR tests were conducted on 2540 patients. A Task Force clinical team chaired and led by Prof. Dr. Vidula Gujjarwar, Director and Principal at CBPACS, had enrolled in the trial, and 2208 active positive COVID cases having mild to moderate severe symptoms (Figure S3) were found. Ayurveda medicines were prescribed to the patients under trial as per protocol guidelines set by the Ministry of AYUSH, Government of India. No other drugs were administered. Around 1970 patients were placed under home isolation after hospitalization with the same medication. These patients showed clearance of viral load after 7–10 days and were discharged after being clinically cured. A total of 237 patients were referred to a different center for further management. One of the components used in this successful trial was Nagaradi Kwath (ginger-based formulation). The importance of the current study could be appreciated by the fact that administration of the formulation with 6-SHO as an active component proved to be beneficial for the patients. In another study at Sukhayu Ayurveda Panchakarma Research Institute, Kolhapur, Dr. Sunil Inamdar administered the herbal power mixture to more than 80 patients with COVID-19 between 10 to 70 years of age. The herbal powder showed clinically promising results, and the major content of herbal mixture was Z. officinale (Shunthi). Taken together, these findings showed that natural compounds such as 6-SHO could help in the management of infection and inflammation in patients with COVID-19, and to the best of our knowledge, this is the first study reporting the anti-viral and anti-inflammatory properties and underlying mechanism of 6-SHO.
3.7. Extraction of 6-SHO from Ginger
It is necessary to understand the host–pathogen interaction in patients with COVID-19 and to identify the potential compounds that block the process of viral entry and replication. The identity of 6-SHO in the ginger extract was confirmed by matching the spectra in the HPLC-PDA chromatogram (Figure S2).
4. Discussion
Natural compounds have been used for treating diseases since thousands of years. Therefore, natural compounds should be regarded as a complementary therapy for patients with COVID-19. Several clinical trials on natural compounds have reported their antiviral and anti-inflammatory properties. Patients with severe symptoms of COVID-19 have shown the presence of cytokine storm33 that involves the elevation of the levels of several pro-inflammatory cytokines including IL-6, IP-10, macrophage inflammatory protein 1α (MIP1α), MIP1β, MCP1, and IL-1β,34 which in turn are associated with activation of the inflammasome pathway.33 Upon activation, the NLRP3 inflammasome induces a programmed cell death pathway assisted by IL-1β and IL-18.35 These inflammatory cytokines recruit neutrophils to the inflammatory sites for protection against the virus through interactions with the adaptive immune system.36 These proteins attract monocytes, macrophages, and T cells to the site of infection, and the addition of IFNγ produced by T cells could reach an over-whelming pro-inflammatory feedback loop.33 A defective immune response could damage the lung infrastructure significantly. Thus, without a certain level of control on the persistent activation of NLRP3 inflammasomes, the inflammatory immune response and pyroptosis-mediated cell damage could lead to organ injury.12,33 Ginger enhances the immune response and is known to be few of the best drugs among traditional medicines. An active constituent of ginger (Shunthi), 6-SHO, is reported to possess anti-inflammatory and anticancer activities.31−33 Thus, this phytochemical could be employed for treating patients with COVID-19 to decrease the severity of inflammation.
The predicted toxicity of HCQ that we observed could be the reason why clinical trials with HCQ have been retracted as they reported decreased survival, and the treatment was associated with an increase in serious adverse events such as ventricular arrhythmias and liver toxicity. In another study, it was reported that treatment of patients with COVID-19 with remdesivir did not provide significant clinical benefits to patients.37 In fact, in 12% of the cases, the treatment was stopped due to serious adverse events observed. It was also reported that remdesivir alleviates immunopathological condition through the cytokine storm which is responsible for aggravating COVID-19 disease. Furthermore, they commented that the anti-viral efficacy of remdesivir needs to be enhanced using either a higher dose (which could aggravate toxicity further) or in combination with other antiviral drugs or neutralizing antibodies for improved clinical benefit.
In summary, we can say that ITAMs are optimal in binding affinities to important druggable targets and blocking vital functions of SARS-CoV2 compared to allopathy drugs such as remdesivir and HCQ. In practice, herbal compounds or extracts are used in combination or as a concoction. An interesting aspect is that they are eco-friendly and are safe for humans. Since they are well tolerated in the body and have good oral absorption, they can be consumed daily or regularly. This study opens up new avenues for potential sources of 6-SHO from plants (spices, medicinal plants, etc.) so that these could be incorporated at an appropriate concentration level in herbal formulations for combating/managing viral infections. Phytoformulations comprising sources of these two molecules could also be developed to have better safety and efficacy. Currently, in one such ongoing clinical trial of Dr Vidula Gujjarwar with a large cohort, one of the components of the treatment includes ginger, and it has indicated a promising clinical outcome in fighting COVID-19. Furthermore, in another ongoing clinical study with both dry ginger and yashtimadhu, nanoemulsions showed a positive clinical outcome. Our study using key phytochemicals has provided a systematic approach by which components of ITAMs can be evaluated for their scientific knowledge (Figure S3). Comparative MD analysis of 6-SHO explains the conformational stability and natural dynamics of the interaction under physiological environmental conditions. ADME/T prediction on the ligands enabled us to analyze their physicochemical and pharmacokinetic profiles. The obtained results will aid in the launch of new research protocols for discovery of potential drugs for the treatment of COVID-19. A similar approach can be followed to increase awareness and wider acceptance of ITAMs as a treatment option for various other viral diseases at a global level. This provides a basis for initiating systematic pharmacokinetic studies using herbal formulations or natural products. The results of this research are intriguing, and they open up new possibilities for treating patients with COVID-19 by exploring alternative treatment approaches. However, additional studies are required to determine the efficacy and mechanism of action of these phytochemicals using systematic in vitro and in vivo preclinical approaches in healthy lung epithelial cells or an animal model of lung fibrosis, respectively. Furthermore, phase I to IV clinical trials on optimized leads from natural products or herbal formulations are warranted to evaluate anti COVID-19 efficacy and safety in humans for developing potential anti-viral therapeutics.
5. Conclusions
In this study, we demonstrated the crucial role of phytochemical 6-SHO in exhibiting a growth inhibition effect on SARS-CoV2. 6-SHO has the potential to regulate lung pathology by ameliorating the effects of the inflammasome pathway, which was further complemented with computational findings. Moreover, the safe and drug-like properties of 6-SHO during ADME predictions strongly support 6-SHO phytochemical medications as a combined multi-target therapy. However, additional in vivo studies are necessary to develop 6-SHO into SARS-CoV2 treatment.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c07138.
Brief information on ITAMs; molecular docking of 6-SHO in the NP3-146 binding site of NLRP3; HPLC-PDA chromatogram of 6-SHO (from the ginger extract); and clinical trial data on ITAMs for COVID-19 treatment (PDF)
Author Present Address
△ Institute of Biological Chemistry, Academia Sinica, Taipei 11529, Taiwan
Author Contributions
∇∇ J.K., J.M., and A.A.D. contributed equally. J.K. conceived the idea and designed and supervised the study; J.M. and D.P. carried out the NLRP3-ADP-NP-146/-6-SHO simulation study; N.G. and D.S. performed toxicity evaluation study; T.P. performed innate immune inflammasome assays and immunofluorescence staining. VK was involved in imaging and data analysis; S.K. contributed to extract preparation and HPLC-PDA analysis; S.G., S.S.K., P.M., and S.C. contributed in checking the drug cytotoxicity in Vero cells and estimated the efficacy of 6-SHO against SARS CoV-2 in vitro; V.G. and S.I. contributed to clinical benefits from Ayurveda practitioners and provided clinical observations from their ongoing clinical study on COVID-19 patients; J.K., J.M., A.A.D., and S.M. analyzed the data and critically examined the results; A.A.D., J.M., S.M., and J.K. drafted the manuscript; all coauthors revised and approved the final version of the manuscript.
No specific grant was received to carry out this research. Institutional funds were utilized for generating biological and computational data. ACTREC received grant from Department of Atomic Energy, Government of India as support to ACTREC with grant sanction order no. 1/3(7)/2020/TMC/R&D-II/8823 Dt.30.07.2021 (for basic and translational research in cancer), sanction order no. 1/3(6)/2020/TMC/R&D-II/3805 Dt.18.03.2021(for animal imaging at ACTREC), and sanction order no. 1/3(4)/2021/TMC/R&D-II/15063 Dt.15.12.2021 (for capacity building and development of novel and cutting-edge research activities).
The authors declare no competing financial interest.
Notes
COVID-19 patients who were treated at two different centers had provided written consent for getting treatment on the designated protocol. The clinical trial studies were approved by ethics committees of participating institutes and were in compliance with the regulations of the Ministry of AYUSH, Government of India.
Supplementary Material
References
- Wu F.; Zhao S.; Yu B.; Chen Y. M.; Wang W.; Song Z. G.; Hu Y.; Tao Z. W.; Tian J. H.; Pei Y. Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covián C.; Fernández-Fierro A.; Retamal-Díaz A.; Díaz F. E.; Vasquez A. E.; Lay M. K.; Riedel C. A.; González P. A.; Bueno S. M.; Kalergis A. M. BCG-Induced Cross-Protection and Development of Trained Immunity: Implication for Vaccine Design. Front Immunol. 2019, 10, 2806. 10.3389/fimmu.2019.02806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea M. G.; Giamarellos-Bourboulis E. J.; Domínguez-Andrés J.; Curtis N.; van Crevel R.; van de Veerdonk F. L.; Bonten M. Trained Immunity: a Tool for Reducing Susceptibility to and the Severity of SARS-CoV-2 Infection. Cell 2020, 181, 969–977. 10.1016/j.cell.2020.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea M. G.; Joosten L. A.; Latz E.; Mills K. H.; Natoli G.; Stunnenberg H. G.; O’Neill L. A.; Xavier R. J. Trained immunity: A program of innate immune memory in health and disease. Science 2016, 352, aaf1098. 10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I. Y.; Ichinohe T. Response of host inflammasomes to viral infection. Trends Microbiol. 2015, 23, 55–63. 10.1016/j.tim.2014.09.007. [DOI] [PubMed] [Google Scholar]
- Allen I. C.; Scull M. A.; Moore C. B.; Holl E. K.; McElvania-TeKippe E.; Taxman D. J.; Guthrie E. H.; Pickles R. J.; Ting J. P. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity 2009, 30, 556–565. 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Receptor recognition and cross-species infections of SARS coronavirus. Antiviral Res. 2013, 100, 246–254. 10.1016/j.antiviral.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Ou X.; Liu Y.; Lei X.; Li P.; Mi D.; Ren L.; Guo L.; Guo R.; Chen T.; Hu J.; et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Shang J.; Wan Y.; Luo C.; Ye G.; Geng Q.; Auerbach A.; Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 2020, 117, 11727–11734. 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S.; Chu V. C.; Whittaker G. R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 5871–5876. 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Hoffmann M.; Kleine-Weber H.; Schroeder S.; Krüger N.; Herrler T.; Erichsen S.; Schiergens T. S.; Herrler G.; Wu N. H.; Nitsche A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Li W.; Moore M. J.; Vasilieva N.; Sui J.; Wong S. K.; Berne M. A.; Somasundaran M.; Sullivan J. L.; Luzuriaga K.; Greenough T. C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X.; Chen K.; Zou J.; Han P.; Hao J.; Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. 2020, 14, 185–192. 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W.; Huang N.; Huang C.; Bécavin M.; Berg R.; Queen M.; Litvinukova C.; Talavera-López H.; Maatz D.; Reichart F.; et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020, 26, 681–687. 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon A.; Le N. T.; Selisko B.; Eydoux C.; Alvarez K.; Guillemot J. C.; Decroly E.; Peersen O.; Ferron F.; Canard B. Remdesivir and SARS-CoV-2: Structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antiviral Res. 2020, 178, 104793. 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.; Xu Y.; Gao R.; Lu R.; Han K.; Wu G.; Tan W.; Agostini M. L.; Andres E. L.; Sims A. C.; Graham R. L.; Sheahan T. P.; Lu X.; Smith E. C.; Case J. B.; Feng J. Y.; Jordan R.; et al. Detection of SARS-CoV-2 in Different Types of Clinical SpecimensCoronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. JAMAmBio 20202018, 9, 1843–1844. 10.1001/jama.2020.378610.1128/mBio.00221-18. [DOI] [Google Scholar]
- a Ali B. H.; Blunden G.; Tanira M. O.; Nemmar A. Some phytochemical, pharmacological and toxicological properties of ginger (Zingiber officinale Roscoe): a review of recent research. Food Chem. Toxicol. 2008, 46, 409–420. 10.1016/j.fct.2007.09.085. [DOI] [PubMed] [Google Scholar]; b Pan M. H.; Hsieh M. C.; Hsu P. C.; Ho S. Y.; Lai C. S.; Wu H.; Sang S.; Ho C. T. 6-Shogaol suppressed lipopolysaccharide-induced up-expression of iNOS and COX-2 in murine macrophages. Mol. Nutr. Food Res. 2008, 52, 1467–1477. 10.1002/mnfr.200700515. [DOI] [PubMed] [Google Scholar]; c Ling H.; Yang H.; Tan S. H.; Chui W. K.; Chew E. H. 6-Shogaol, an active constituent of ginger, inhibits breast cancer cell invasion by reducing matrix metalloproteinase-9 expression via blockade of nuclear factor-κB activation. Br. J. Pharmacol. 2010, 161, 1763–1777. 10.1111/j.1476-5381.2010.00991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath P.; Chauhan N. R.; Jena K. K.; Datey A.; Kumar N. D.; Mehto S.; De S.; Nayak T. K.; Priyadarsini S.; Rout K.; et al. Inhibition of IRGM establishes a robust antiviral immune state to restrict pathogenic viruses. EMBO Rep. 2021, 22, e52948 10.15252/embr.202152948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P.; Kumar A.; Mamidi P.; Kumar S.; Basantray I.; Saswat T.; Das I.; Nayak T. K.; Chattopadhyay S.; Subudhi B. B.; et al. Inhibition of Chikungunya Virus Replication by 1-[(2-Methylbenzimidazol-1-yl) Methyl]-2-Oxo-Indolin-3-ylidene] Amino] Thiourea(MBZM-N-IBT). Sci. Rep. 2016, 6, 20122. 10.1038/srep20122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A.; Mamidi P.; Das I.; Nayak T. K.; Kumar S.; Chhatai J.; Chattopadhyay S.; Suryawanshi A. R.; Chattopadhyay S. A novel 2006 Indian outbreak strain of Chikungunya virus exhibits different pattern of infection as compared to prototype strain. PLoS One 2014, 9, e85714 10.1371/journal.pone.0085714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qikprop Schrödinger suite 2019-1; Schrödinger, LLC: New York, 2019.
- Burley S. K.; Bhikadiya C.; Bi C.; Bittrich S.; Chen L.; Crichlow G. V.; Christie C. H.; Dalenberg K.; Di Costanzo L.; Duarte J. M.; et al. RCSB Protein Data Bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021, 49, D437–D451. 10.1093/nar/gkaa1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X. Q.; Wang X. Y.; Gong F. T.; Feng M.; Bai J. J.; Zhang R. R.; Dang X. Q. Oral treatment with glycyrrhizin inhibits NLRP3 inflammasome activation and promotes microglial M2 polarization after traumatic spinal cord injury. Brain Res. Bull. 2020, 158, 1–8. 10.1016/j.brainresbull.2020.02.009. [DOI] [PubMed] [Google Scholar]
- Dekker C.; Mattes H.; Wright M.; Boettcher A.; Hinniger A.; Hughes N.; Kapps-Fouthier S.; Eder J.; Erbel P.; Stiefl N.; et al. Crystal Structure of NLRP3 NACHT Domain With an Inhibitor Defines Mechanism of Inflammasome Inhibition. J. Mol. Biol. 2021, 433, 1843–1844. 10.1016/j.jmb.2021.167309. [DOI] [PubMed] [Google Scholar]
- Morris G. M.; Huey R.; Lindstrom W.; Sanner M. F.; Belew R. K.; Goodsell D. S.; Olson A. J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O.; Olson A. J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk S.; Páll S.; Schulz R.; Larsson P.; Bjelkmar P.; Apostolov R.; Shirts M. R.; Smith J. C.; Kasson P. M.; van der Spoel D.; et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.; MacKerell A. D. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoete V.; Cuendet M. A.; Grosdidier A.; Michielin O. SwissParam: A Fast Force Field Generation Tool for Small Organic Molecules. J. Comput. Chem. 2011, 32, 2359–2368. 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]
- Maharana J.; Panda D.; De S. Deciphering the ATP-binding mechanism(s) in NLRP-NACHT 3D models using structural bioinformatics approaches. PLoS One 2018, 13, e0209420 10.1371/journal.pone.0209420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia-Abellán A.; Angosto-Bazarra D.; Martínez-Banaclocha H.; de Torre-Minguela C.; Cerón-Carrasco J. P.; Pérez-Sánchez H.; Arostegui J. I.; Pelegrin P. MCC950 closes the active conformation of NLRP3 to an inactive state. Nat. Chem. Biol. 2019, 15, 560–564. 10.1038/s41589-019-0278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Baden L. R.; El Sahly H. M.; Essink B.; Kotloff K.; Frey S.; Novak R.; Diemert D.; Spector S. A.; Rouphael N.; Creech C. B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Polack F. P.; Thomas S. J.; Kitchin N.; Absalon J.; Gurtman A.; Lockhart S.; Perez J. L.; Pérez Marc G.; Moreira E. D.; Zerbini C.; et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.; Conlon M.; Ren W.; Chen B. B.; Bączek T. Natural Products as Targeted Modulators of the Immune System. J. Immunol. Res. 2018, 2018, 1. 10.1155/2018/7862782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Islam M. S.; Wang J.; Li Y.; Chen X. Traditional Chinese Medicine in the Treatment of Patients Infected with 2019-New Coronavirus (SARS-CoV-2): A Review and Perspective. Int. J. Biol. Sci. 2020, 16, 1708–1717. 10.7150/ijbs.45538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay M. Z.; Poh C. M.; Rénia L.; MacAry P. A.; Ng L. F. P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020, 20, 363–374. 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T. C.; Yen C. K.; Lu Y. C.; Shi C. S.; Hsieh R. Z.; Chang S. F.; Chen C. N. The antagonism of 6-shogaol in high-glucose-activated NLRP3 inflammasome and consequent calcification of human artery smooth muscle cells. Cell Biosci. 2020, 10, 5. 10.1186/s13578-019-0372-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.; Cao R.; Zhang L.; Yang X.; Liu J.; Xu M.; Shi Z.; Hu Z.; Zhong W.; Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderdys V.; Allak A.; Guessous F.; Benamar M.; Read P. W.; Jameson M. J.; Abbas T. The Neddylation Inhibitor Pevonedistat (MLN4924) Suppresses and Radiosensitizes Head and Neck Squamous Carcinoma Cells and Tumors. Mol. Cancer Ther. 2018, 17, 368–380. 10.1158/1535-7163.MCT-17-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Zhang D.; Du G.; Du R.; Zhao J.; Jin Y.; Fu S.; Gao L.; Cheng Z.; Lu Q.; et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet 2020, 395, 1569–1578. 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.