Abstract
Background
Testa color is an important trait of peanut (Arachis hypogaea L.) which is closely related with the nutritional and commercial value. Pink and red are main color of peanut testa. However, the genetic mechanism of testa color regulation in peanut is not fully understood. To elucidate a clear picture of peanut testa regulatory model, samples of pink cultivar (Y9102), red cultivar (ZH12), and two RNA pools (bulk red and bulk pink) constructed from F4 lines of Y9102 x ZH12 were compared through a bulk RNA-seq approach.
Results
A total of 2992 differential expressed genes (DEGs) were identified among which 317 and 1334 were up-regulated and 225 and 1116 were down-regulated in the bulk red-vs-bulk pink RNA pools and Y9102-vs-ZH12, respectively. KEGG analysis indicates that these genes were divided into significantly enriched metabolic pathways including phenylpropanoid, flavonoid/anthocyanin, isoflavonoid and lignin biosynthetic pathways. Notably, the expression of the anthocyanin upstream regulatory genes PAL, CHS, and CHI was upregulated in pink and red testa peanuts, indicating that their regulation may occur before to the advent of testa pigmentation. However, the differential expression of down-stream regulatory genes including F3H, DFR, and ANS revealed that deepening of testa color not only depends on their gene expression bias, but also linked with FLS inhibition. In addition, the down-regulation of HCT, IFS, HID, 7-IOMT, and I2’H genes provided an alternative mechanism for promoting anthocyanin accumulation via perturbation of lignin and isoflavone pathways. Furthermore, the co-expression module of MYB, bHLH, and WRKY transcription factors also suggested a fascinating transcriptional activation complex, where MYB-bHLH could utilize WRKY as a co-option during the testa color regulation by augmenting anthocyanin biosynthesis in peanut.
Conclusions
These findings reveal candidate functional genes and potential strategies for the manipulation of anthocyanin biosynthesis to improve peanut varieties with desirable testa color.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-023-04041-0.
Keywords: Peanut; Testa color; Anthocyanin biosynthesis, flavonoids; Bulk RNA-seq (BR-seq); Transcription factors
Background
Anthocyanin is an important color pigment, which is widely distributed across the plant kingdom. The accumulation of anthocyanin pigments including pelargonidin, cyanidin, dalphinidin, petudinin, and malvidin largely contribute to the pink, red, blue, and purple colors found in plant tissues such as flowers, leaves, fruits, and roots [1]. In addition to their important roles as pollinator and insect attractant, anthocyanins can also assist plants against abiotic stresses such as cold, drought stress, and ultraviolet irradiation as well as pathogenic microbes [2, 3]. Anthocyanins and its derivatives are usually believed to possess health-promoting properties through high antioxidant activities, allowing plants to combat ROS-induced risks [4]. Regular consumption of anthocyanins has been shown to lessen the chance of developing atherosclerosis and cardiovascular diseases by suppressing the oxidation of low-density lipids. Previously, many studies have demonstrated anthocyanin biosynthesis in several plant species. In a prior study, the accumulation of anthocyanin and their associated gene expression were shown to be greatly increased in red orange fruit under low temperature storage condition [5]. In ripening grape berry skins, anthocyanin synthesis corresponds to a coordinated increased expression level of a number of anthocyanin biosynthetic genes [6]. It has also been revealed that the TTG1/bHLH/Myb transcriptional complex regulates the anthocyanin biosynthesis pathway in Arabidopsis seedlings [7]. A recent study combining transcriptome and metabolome analysis has shed light on pigment accumulation in peanut testa [8]. However, the underlying molecular mechanism between the peanut testa color and genes that regulate its related trait is still not clearly elucidated.
The biosynthesis of anthocyanin and its derivatives are catalyzed by a number of enzymes through the flavonoid pathway [9]. Initially, the phenylalanine amino acid is converted into 4-coumaryl-CoA with the help of phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), and 4-coumaryl-CoA ligase (4CL). In the following step, 4-coumaryl-CoA and malonyl-CoA are sequentially catalyzed by different enzymes including chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavanone 3-hydroxylase (F3′H), and flavonoid 3′5′-hydroxylase (F3′5 ′H) to produce dihydrokaempferol. Then, colored anthocyanins pigments are synthesized from the colorless anthocyanins with the subsequent reactions of dihydroflavonol 4-reductase (DFR) and anthocyanidin synthase (ANS), and then converted into red, blue or purple glycosides by UDP-glucoside: flavonoid glucosyltransferase (UFGT) [10, 11]. Generally, genes-encoding key enzymes of anthocyanin pathway are highly expressed in fruits [12]. During the ripening process of a dark-colored fig fruit, the expression level of FcANS1 gene was shown to be considerably increased [13]. Similarly, anthocyanin biosynthesis in grapes and strawberries relies on the upregulated expression of UFGT, which is identified as a key enzyme in anthocyanin pathway [14, 15]. Anthocyanins pathway genes are known to be activated at the transcription level by a ternary complex of MYB-bHLH-WD repeat (MBW complex) in many plant species [7, 16].
Peanut (Arachis hypogaea L.), is one of the important cash crops widely known for its high-quality edible oil and rich nutritional value around the world. Peanut testa is rich in amino acids, isoflavones, vitamins (B1, B3, B9, and E), anthocyanin, and procyanidins, which likely affect its economical and nutritional importance [17–19]. Peanut testa appear in a range of colors, the most frequent of which are pink and red. Other hues include purple, black, white, and multicolored testa. Different testa colored peanuts are considered to be linked with varied anthocyanin content and composition. For instance, the red testa is believed to be controlled by a single dominant gene present on chromosome 3, which causes the activation of anthocyanin biosynthesis [20]. In our previous findings, we also identified several up-regulated R2R3-MYB transcription factors associated with anthocyanin biosynthesis in black and pink testa peanut [19, 21]. In addition, our recent study also unleashed the control mechanism of red testa via a recessive gene, AhRt2 located on chromosome 12 through BSA-seq and genetic mapping [22]. Previously, the high expression of genes such as PAL, C4H, CHS, and CHI as well as AtMYB111 homologs may lead to the regulation of pink testa peanut [23]. The metabolome investigation of red and pink testa peanuts suggested that the content of petunidin and cyanidin in red testa was greater compared to the pink testa [8].
In recent years, high-throughput sequencing techniques have been used to investigate the development of pigmentation in a wide range of plant species by combining a number of omics-based methodologies. In fruit and flowers, metabolomic and transcriptome network have revealed a variety of secondary metabolites with altered content and their associated differentially expressed genes, expanding our understanding of how plants regulate their color [24, 25]. Using comparative transcriptome analysis, the significance of flavonoid biosynthesis and their regulatory genes have been investigated in black peanut testa [26]. Similarly, the gene expression profiling of important genes and transcription factors were studied during enhanced anthocyanin biosynthesis in red and yellow fruits of sweet cherries [27]. Another study used comparative transcriptome methods to compare the molecular mechanisms of changes in leaf and peel color found in red and green walnut [28]. Previously, the integrative analysis of multi-omics and miRNA profiling of pink and purple testa peanut uncovered important insights into the regulation of anthocyanin biosynthesis [29]. However, most of these studies were comparative analysis based on different peanut varieties with different testa colors. Besides testa color variation, there are many other trait differences among these varieties which can increase the number of DEGs. For this purpose, we used Bulk RNA-sequencing (BR-seq) analysis of red testa (ZH12) male parent and pink testa (Y9102) female parent with bulk F4 lines of red and pink testa peanuts. The findings of this study will broaden our knowledge regarding the genetic control mechanisms that regulate red and pink peanut testa color, and provide a valuable reference resource for studying gene-to-function relationship that governs pigmentation in peanut testa.
Results
Phenotypic variations and determination of anthocyanin content in red and pink testa peanut
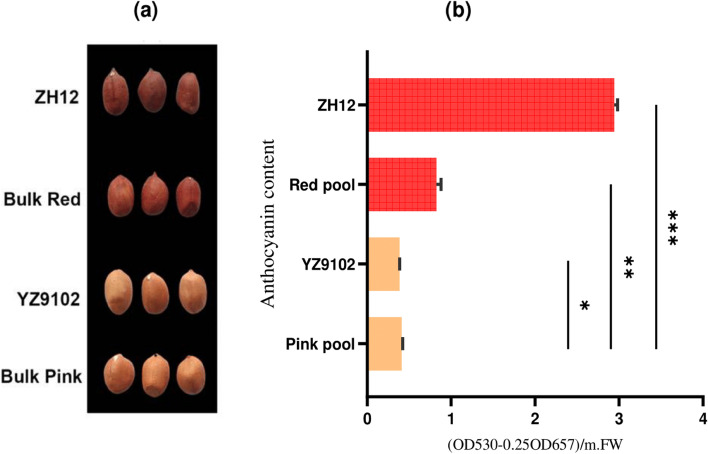
The testa color of ZH12 is red and the testa of Y9012 is traditionally pink. The phenotypic investigation of red testa (ZH12) and pink testa (Y9012) cultivars with the bulk F4 lines of pink and red peanuts showed significant differences. The bulk F4 lines with red testa showed similar color with their counterpart male red parent (ZH12), whereas the bulk F4 lines with pink testa showed similar pink phenotype with their female parent (Y9012) counterpart (Fig. 1a). In addition, the measurement of total anthocyanin content of the fully matured seeds revealed approximately 2–7 times higher content in red testa peanut than the pink testa (Fig. 1b).
Fig. 1.
Phenotypic variations and determination of anthocyanin content (a) The peanut seeds demonstrating red testa parental line (ZH12) and pink testa parental lines (Y9102), with their counterpart bulk F4 lines. (b) relative anthocyanins content in parental lines and bulk F4 lines. The data were presented as SE (n = 3), and the asterisks * denotes P < 0.05, ** denotes P < 0.01 and *** denotes P < 0.001
Data filtration and reference genome mapping
A total of four cDNA libraries were constructed from ZH12, Y9102, bulk red and bulk pink peanuts, each one with three replicates, respectively. The BGISEQ-500 platform was used to sequence the cDNA libraries using the paired-end method. The raw reads were filtered by removing low quality, N-containing and adaptor-contaminated reads and approximately 21.04–22.47 million clean reads on average were acquired from the three replicates of each sample (Table 1). Further analysis showed that the average value of the clean reads Q20 (the proportion of filtered bases with a mass larger than 20 to the total number of bases) was 97.98%, while the average value of clean reads Q30 (the ratio of filtered bases with a mass value higher than 30 to the total number of bases) was 94.07%. The clean reads were mapped to the reference genome using HISAT software package and the results showed that the mapping ratio of each library showed 94.23–96.96% with a perfect average mapping ratio of 95.83%. The analysis of transcript readings using a random number generator revealed that the reads were scattered randomly across different regions of the transcript (Fig. S1). Similarly, it was found that the average proportion of each gene that was spanned by reads was more than 60% (Fig. S2). Hence, the RNA-seq data acquired from this platform might provide a precise reflection of high-quality data and could be applied to study expression analysis and follow-up analysis.
Table 1.
Summary of the total read numbers obtained from BulkRed, BulkPink, Y9102 and ZH12 testa peanuts
| Sample Name | Total Raw Reads (M) | Average Reads/sample (M) | Total Clean Reads (M) | Clean Reads (Q20, %) | Clean Reads (Q30, %) | Total Mapping (%) | Uniquely Mapping (%) | |
|---|---|---|---|---|---|---|---|---|
| Pink | Pink-1 | 21.75 | 22.47 | 21.63 | 98.2 | 94.61 | 96.14 | 34.64 |
| Pink-2 | 21.75 | 21.63 | 97.8 | 93.56 | 96.33 | 35.17 | ||
| Pink-3 | 23.92 | 23.75 | 98 | 94.24 | 94.49 | 36.46 | ||
| Red | Red-1 | 21.75 | 21.44 | 21.63 | 97.93 | 93.9 | 95.91 | 34.5 |
| Red-2 | 21.75 | 21.64 | 98.16 | 94.54 | 96.87 | 35.53 | ||
| Red-3 | 20.84 | 20.72 | 98.13 | 94.4 | 96.96 | 35.64 | ||
| Y9102 | Y9102–1 | 21.75 | 21.04 | 21.62 | 98.24 | 94.74 | 94.23 | 33.51 |
| Y9102–2 | 21.75 | 21.61 | 97.72 | 93.34 | 96.07 | 34.93 | ||
| Y9102–3 | 19.63 | 19.52 | 97.84 | 93.7 | 96.33 | 33.9 | ||
| ZH12 | ZH12–1 | 21.75 | 21.75 | 21.64 | 98.08 | 94.28 | 96.3 | 34.53 |
| ZH12–2 | 21.75 | 21.62 | 97.82 | 93.64 | 94.41 | 33.48 | ||
| ZH12–3 | 21.75 | 21.63 | 97.93 | 93.91 | 95.98 | 34.73 | ||
Differential expressed genes (DEGs) identification between red and pink testa peanuts
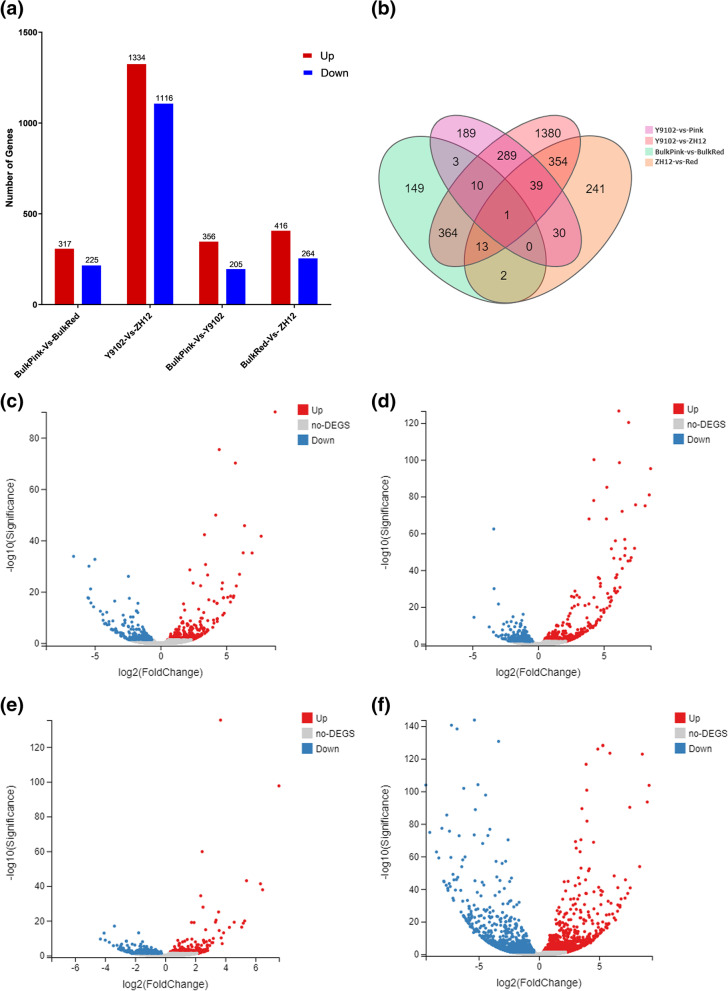
In order to investigate the molecular mechanism governing the variation in red and pink testa color, we screened out DEGs using the following threshold log2 (FC) ≥ 1 and Q-value < 0.001 in bulk red, bulk pink, Y9102 and ZH12 testa peanuts. A total of 4233 DEGs were identified, among which 317 and 1334 in bulk red-vs-bulk pink were up-regulated and 225 and 1116 were down-regulated in Y9102-vs- ZH12 (Table S1 and S2). Similarly, 356 DEGs were up-regulated and 205 DEGs were down-regulated in bulk pink-vs-Y9102 group whereas 414 up-regulated and 264 down-regulated DEGs were detected in bulk red-vs-ZH12 group (Table S3 and S4). Noticeably, the number of up-regulated DEGs were slightly greater than the down-regulated genes between different experimental groups (Fig. 2a). Additionally, the analysis of Venn diagram indicated that the number of commonly expressed DEGs identified in different experimental groups. A total of 1089 DEGs in common were detected in bulk pink-vs-Y1902, bulk red-vs-ZH12, bulk red-vs-bulk pink and ZH12-vs-Y9102 groups, respectively (Fig. 2b). The illustration of Venn diagram depicts how these common or intersected DEGs could be relate to each other during the onset of testa color in peanut. It also elaborates to statistically compare different peanut groups (red and pink) within the same biological process by illustrating the common DEGs from each peanut group (overlap circles) and the individual DEGs that are exclusive to each peanut group (outer circles). Similarly, the volcano plot also enables the visual representation of biologically significant DEGs with large fold changes between each dataset following the negative binomial distribution principle (Fig. 2c-f).
Fig. 2.
Significant changes of differentially expressed genes in in bulk red and bulk pink type peanuts compared to ZH12 (red parent) and Y9102 (pink parent) (a) Statistics of up-regulated and down-regulated differentially expressed genes (DEGs) in each experimental group (b) Venn diagram analysis of pink-vs-Y1902, red-vs-ZH12, ZH12-vs-Y9102 and bulk red-vs-bulk pink dataset. The volcano plot of the both up-regulated and down-regulated DEGs in (c) bulk pink-vs-Y1902 (d) bulk red-vs-ZH12 (e) bulk red -vs-bulk pink type peanuts (f) ZH12-vs-Y9102 type peanuts. The X-axis represents log2-transformed fold-difference values, and the Y-axis represents -log10-transformed significance values. Red dots represent up-regulated DEGs, blue dots represent down-regulated DEGs, and gray represent non-DEGs
Functional enrichment analysis reveals crucial biological pathways during red and pink testa development
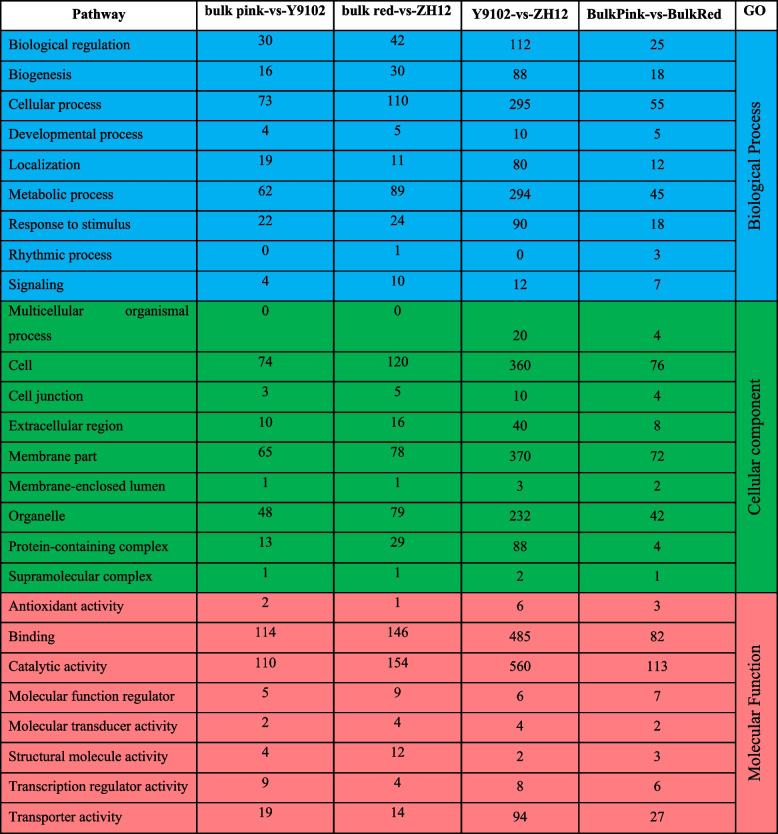
To gain deeper insights into the functions of discovered DEGs between different group of pink and red testa peanuts, the classification of gene ontology (GO) term(s) was carried out, which divided the identified DEGs into three functional terms: biological process, cell component, and molecular function (Table 2). The top 20 GO terms with the smallest Q-value or the selected GO Term (sorted by Q-value, up to 60) were sorted in all four group (Fig. S3). The GO enrichment results of differential genes in Y9102-vs-ZH12 group included hydrolase activity (GO:0016787), carbohydrate metabolic process (GO:0005975), cell periphery (GO: 0071944), and external encapsulating structure (GO:0016787) (Fig. S3a). In case of bulk pink-vs-bulk red group, the most significant enriched GO terms include regulation of gene expression (GO: 0010468), regulation of transcription, DNA-templated (GO: 0006355), secondary active transmembrane transporter activity (GO: 0015291), response to auxin (GO: 0009733), phototropism (GO: 0009638), and auxin-activated signaling pathway (GO: 0009734) (Fig. S3b). Similarly, the GO enriched term in bulk pink-vs-Y9102 classified into nucleobase-containing small molecule biosynthetic (GO: 0034404), signaling receptor binding (GO: 0005102), and glycerol ether metabolic process (GO: 0006662) (Fig. S3c). In the same way, the top GO enriched term in bulk red-vs-ZH12 include cell wall organization or biogenesis (GO: 0071554), external encapsulating structure organization (GO: 0045229), and external encapsulating structure (GO: 0030312) (Fig. S3d). Based on these observations, we speculated that DEGs enriched into different GO categories including response to auxin (GO: 0009733), phototropism (GO: 0009638), and auxin-activated signaling pathway could be crucial in understanding the underlying principles of peanut testa color regulation (Table S3).
Table 2.
Significantly enriched GO terms and number of differentially expressed genes between all comparison group with pink and red testa peanuts
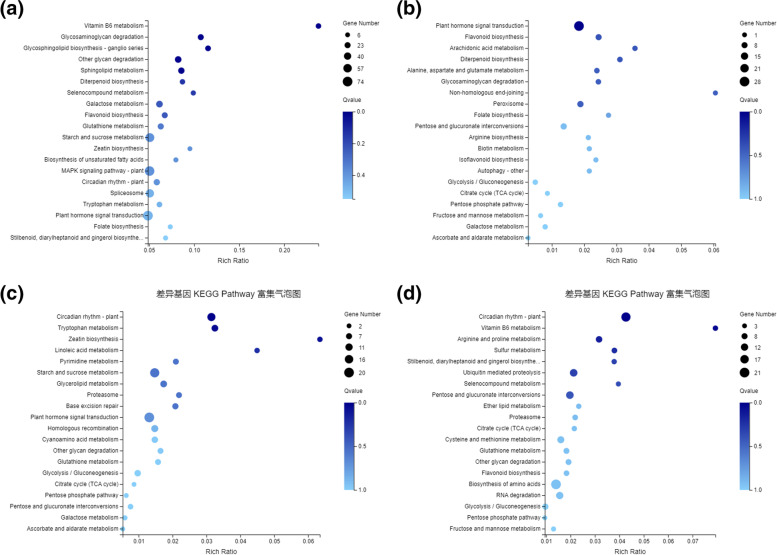
Furthermore, the KEGG pathway classification was carried out to obtain significantly enriched metabolic pathway between Y9102-vs-ZH12, bulk pink-vs-bulk red, bulk pink-vs-Y1902 and bulk red-vs-ZH12 testa peanuts. At first, the KEGG pathways sharing the identified DEGs between each group were categorized into five main branches including cellular processes, environmental information processing, genetic information processing, metabolism, and organic systems (Fig. S4). Further dissection of the enriched metabolic pathway in Y9102-vs-ZH12 indicated that a total of 1124 DEGs were enriched into approximately 129 functional pathways, whereas a total of 239 DEGs in bulk pink-vs-bulk red were significantly enriched into 102 KEGG pathways (Fig. 3a and b Table S4). Similarly, a total 255 DEGs into 95 KEGG pathways were detected in bulk pink-vs-Y9102 group whereas 303 DEGs were categorized into 102 KEGG pathways between bulk red-vs-ZH12 group (Fig. 3c and d, Table S5 and S6). Further analysis demonstrated that the functional pathways related to flavonoid biosynthesis (ko00941), isoflavonoid biosynthesis (ko00943), phenylpropanoid biosynthesis (ko00940), biosynthesis of other secondary metabolites (ko01110), plant hormone signal transduction (ko04075), diterpenoid biosynthesis (ko00904), carotenoid biosynthesis (ko00906) could be associated with the molecular regulation of testa color and these pathways were all significantly enriched in bulk pink-vs-bulk red compared to Y9102-vs-ZH12 group.
Fig. 3.
Elucidation of significantly enriched KEGG pathways involved during testa color in peanut. KEGG Pathway enrichment analysis based on the calculated P-value, and then the P-value was corrected for FDR, usually the function with Q-value <= 0.05 was regarded as a significant enrichment (a) Y9102-vs-ZH12 and (b) bulk pink-vs-bulk red testa peanuts (c) bulk pink-vs-Y1902 (d) bulk red-vs-ZH12. Source: www.kegg.jp/kegg/kegg1.html [30]
DEGs involved in phenylpropanoid and flavonoid/anthocyanin biosynthetic pathways
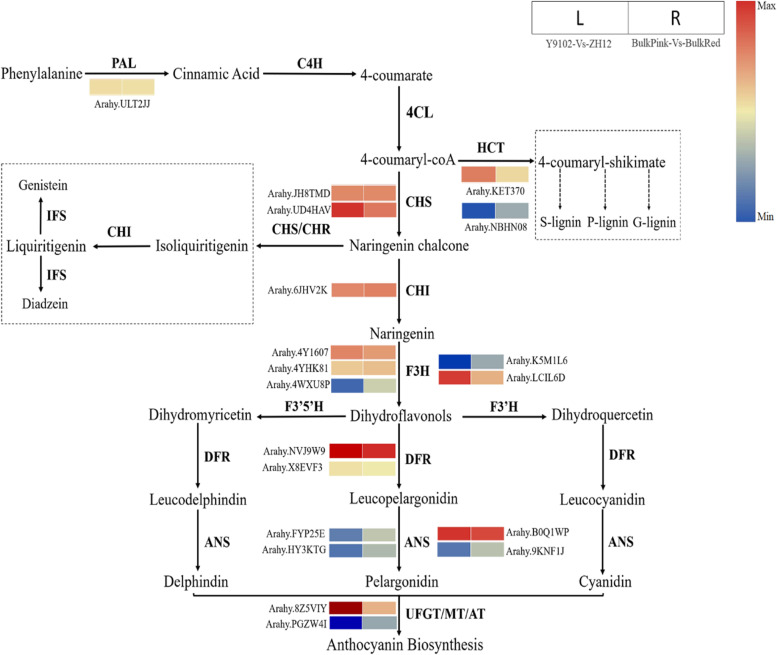
A total of 25 DEGs encoding core structural genes of the phenylpropanoid and flavonoid/anthocyanin biosynthetic pathway were identified between bulk pink-vs-bulk red testa group based on the KEGG pathway assignment (Fig. 4; Table S7). In total, the bulk pink-vs-bulk red testa group demonstrated 15 up-regulated and 10 down-regulated genes, whereas 16 up-regulated and 9 down-regulated genes were identified in Y9102-vs-ZH12 group. Mostly genes encoding PAL, CHS, CHI, F3H, DFR and ANS were preferentially expressed between the two testa peanuts. Interestingly, the expression level of one PAL encoding gene (Arahy.X8EVF3), two CHS encoding genes (Arahy.JH8TMD and Arahy.UD4HAV) and one CHI encoding gene (Arahy.6JHV2K) showed a similar up-regulation pattern of expression in both bulk pink-vs-bulk red testa and Y9102-vs-ZH12 testa group.
Fig. 4.
Reprogrammed expression of genes involved in the phenylpropanoid and flavonoid/anthocyanin biosynthetic pathways regulating red and pink testa development in peanut. The level of transcript abundance in the heatmaps was scored using log2-transformed fold-change values for each experimental group. PAL, phenylalanine ammonia lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate: CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; CHR, chalcone reductase; F3H, flavanone 3-hydroxylase; IFS, 2-hydroxyisoflavanone synthase; F3’H, flavonoid 3′-hydroxylase: flavonoid 3′5′-hydroxylase; DFR, dihydroflavonol 4-reductase; ANS, anthocyanidin synthase; UFGT, UDP glucose-flavonoid 3-O-glcosyl-transferase; MT, methyltransferase
In addition, five F3H encoding genes, which catalyze the conversion of naringenin to different precursor molecules of dihydroflavonol derivatives in anthocyanin biosynthetic pathway were identified as differentially expressed genes. Two F3H genes (Arahy.4WXU8P and Arahy.K5M1L6) showed down-regulation in Y9102-vs-ZH12 testa and bulk pink-vs-bulk red testa groups, although their expression level was differently regulated by 3–4-fold change. Noticeably, the expression level of another F3H gene (Arahy.LCIL6D) showed 3-fold up-regulation in Y9102-vs-ZH12 testa groups compared to bulk pink-vs-bulk red testa group. The remaining two F3H genes (Arahy.4Y1607 and Arahy.4YHK81) showed up-regulation in both testa groups with no significant difference in their expression level (Fig. 4, Table S7). DFR plays an important role in anthocyanin biosynthesis which mainly catalyzes different substrates to produce leucopelargonidin, leucocyanidin and leucodelphinidin. Our results identified two DFR encoding genes of which, one gene (Arahy.NVJ9W9) showed up-regulation in both testa groups, whereas one gene (Arahy.X8EVF3) was significantly down-regulated in the bulk pink-vs-bulk red testa compared to Y9102vsZH12 testa group. Similarly, four ANS encoding genes were identified between the two testa peanuts of which, two genes (Arahy.B0Q1WP and Arahy.33FH8C) demonstrated up-regulation while two genes (Arahy.9KNF1J and Arahy.FYP25E) showed down regulation in bulk pink-vs-bulk red testa and Y9102-vs-ZH12 testa peanut, respectively.
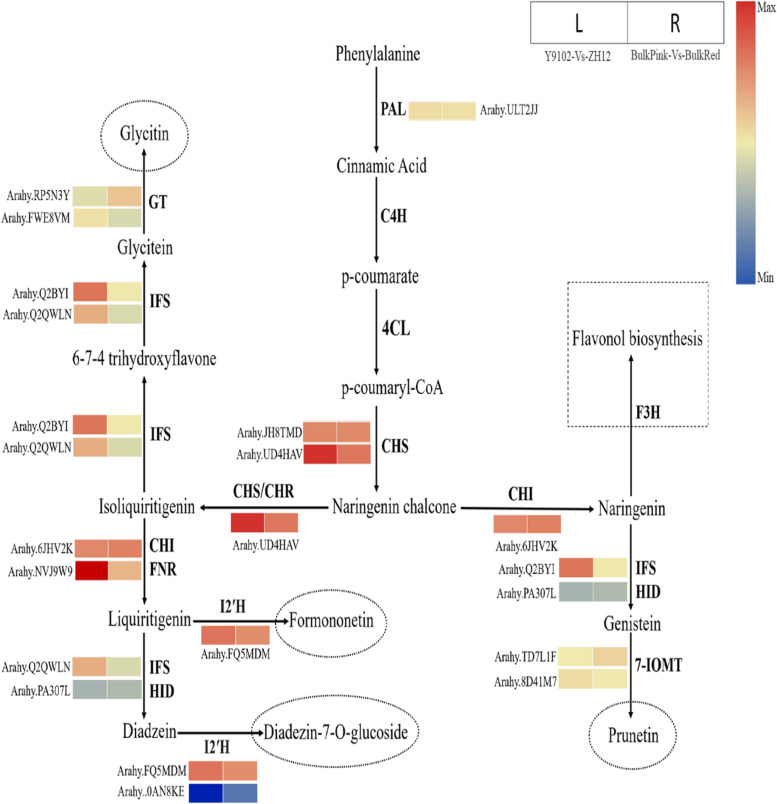
DEGs involved in isoflavonoid biosynthetic pathway
During isoflavonoid biosynthetic pathway, chalcone synthase (CHS) co-acts with chalcone reductase (CHR) to produce isoliquiritigenin chalcone. Then, chalcone isomerase (CHS) or chalcone reductase (CHR) catalyze the conversion of isoliquiritigenin into liquiritigenin, thereby channeling the metabolic flux into isoflavone biosynthesis with the help of isoflavonone synthase (IFS) and 2-hydroxyisoflavanone dehydratase (HID). In this study, two genes encoding IFS were identified with differential expression in bulk pink-vs-bulk red testa and Y9102-vs-ZH12 testa peanuts. One gene encoding IFS (Arahy.Q2QWLN) showed up-regulation in Y9102-vs-ZH12, whereas the expression of Arahy.Q2QWLN was down-regulated in bulk pink-vs-bulk red. Another IFS encoding gene (Arahy.Q2BYIT) showed increased expression level up to (5-fold) in Y9102-vs-ZH12 compare to bulk pink-vs-bulk red peanuts (Fig. 5; Table S8). During isoflavonoid pathway, the HID enzyme generally catalyzes the conversion of liquiritigenin into and into daidzein and naringenin into genistein isoflavones. During our study, we also detected a HID-encoding gene (Arahy.PA307L), which showed down-regulation in both testa peanuts groups.
Fig. 5.
Reprogrammed expression of genes involved in the isoflavonoid biosynthetic pathway regulating red and pink testa development in peanut. The level of transcript abundance in the heatmaps was scored using log2-transformed fold-change values for each experimental group
In the later steps of isoflavonoid pathway, isoflavone 2′-hydroxylase (I2’H) could convert liquiritigenin intermediate into formononetin and diadzein into diadzein-7-0 glucoside. Meanwhile, we found two genes encoding I2’H, of which one gene (Arahy.FQ5MDM) showed up-regulation, whereas another I2’H gene (Arahy.0AN8KE) showed down-regulation in Y9102-vs-ZH12 peanuts (Fig. 5; Table S8). Similarly, isoflavone-7-O-methyltransferase (7-IOMT) catalyze the conversion of genistein into prunetin during the down-stream regulation of isoflavone biosynthesis. Two genes encoding 7-IOMT were also identified with differential expression level between the two peanuts with different testa color (Fig. 5; Table S8).
The crucial roles of transcription factors in the peanut testa color
In this study, a total of 13 MYB TF encoding genes, 9 bHLH TF differential expressed encoding genes were identified between Y9102-vs-ZH12 and bulk pink-vs-bulk red testa peanuts. Among them, 9 up-regulated and 4 down-regulated MYB TF encoding genes, 7 up-regulated and 2 down-regulated bHLH TF encoding genes), and 3 up-regulated and 4 down-regulated WRKY TF encoding genes were identified. Importantly, there is no WD-40 TF encoding genes were detected, instead we detected 7 WRKY TF encoding genes in these two testa peanuts (Table S9). Further dissection of the four R2R3-MYBs (Arahy.S8NMKG, Arahy.12J3VU, Arahy.WVAI17 and Arahy.9UC92R) showed that the expression changes between Y9102-vs-ZH12 and bulk pink-vs-bulk red peanuts. The expression level of Arahy.S8NMKG encoding a MYB6-like TF was significantly up-regulated in Y9102-vs-ZH12, whereas Arahy.9UC92R encoding MYB35 like TF showed up to 11-fold down-regulation in ZH12 (Fig. 6; Table 3). Further comparison of their protein sequences revealed that Arahy.S8NMKG is highly homologous to AtMYB5, while Arahy.9UC92R shared homology with TTG1 and TT2 in Arabidopsis, which are known to be involved in anthocyanin biosynthesis and regulation of outer testa differentiation [31].
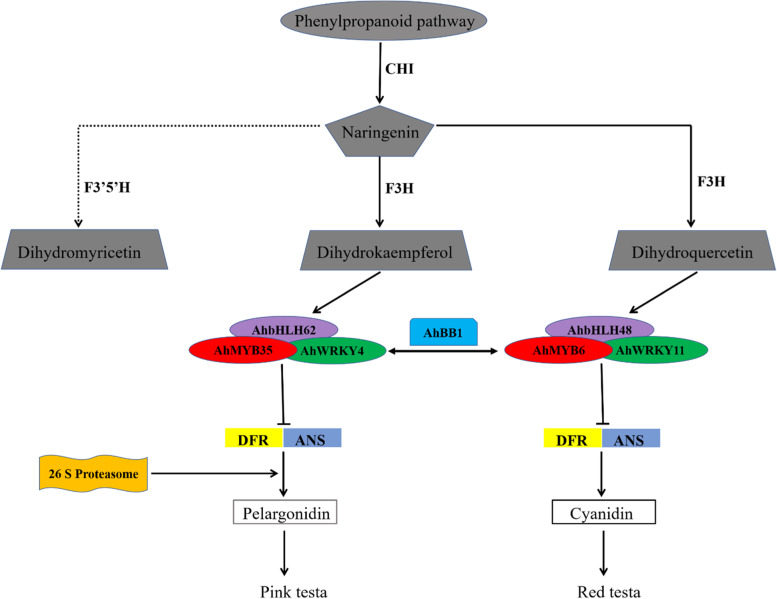
Fig. 6.
The proposed model of transcription factors-induced regulatory mechanism of peanut testa color development via anthocyanin biosynthetic pathway. The different colored oval shapes represent different types of transcription factors i.e. red; MYB, purple; bHLH and green; WRKY
Table 3.
Differentially expressed transcription factors between bulk red-vs-bulk pink and Y9102-vs-ZH12 peanuts
| TF family | Gene name | Y9102-vs-ZH12 | P-value | (bulk red-vs-bulk pink) | P-value | Nr Annotation |
|---|---|---|---|---|---|---|
| MYB | Arahy.S8NMKG | 0.82 | 3.93E-05 | −0.01 | 0.9758 | XP_016205675.1|6.8e-231|transcription repressor MYB6 [Arachis ipaensis] |
| Arahy.12J3VU | 2.30 | 8.02E-12 | 0.60 | 0.2108 | XP_025689501.1|1.4e-173|transcription repressor MYB6-like [Arachis hypogaea] | |
| Arahy.WVAI17 | −3.70 | 1.68E-10 | −0.39 | 0.5161 | XP_025649931.1|2.1e-203|myb-related protein 306-like [Arachis hypogaea] | |
| Arahy.9UC92R | −3.14 | 4.30E-06 | −0.29 | 0.6208 | XP_016194299.1|1.8e-276|transcription factor MYB35-like [Arachis ipaensis] | |
| WRKY | Arahy.V3AFID | −6.73 | 5.33E-51 | −0.59 | 0.3347 | XP_016194312.1|0.0e+ 00|probable WRKY transcription factor 4 [Arachis ipaensis] |
| Arahy.FP3Q5V | 0.54 | 0.0004 | −0.13 | 0.6387 | XP_025688450.1|7.6e-187|probable WRKY transcription factor 11 [Arachis hypogaea] | |
| bHLH | Arahy.GL41VM | 1.04 | 1.38E-06 | 0.22 | 0.4806 | XP_016190307.1|2.1e-202|transcription factor bHLH61 [Arachis ipaensis] |
| Arahy.R1MRXV | −2.78 | 1.27E-06 | 1.76 | 0.005 | XP_025689410.1|5.5e-239|transcription factor bHLH62-like [Arachis hypogaea] | |
| Arahy.26781 N | 2.31 | 6.90E-14 | 1.61 | 1.62E-06 | XP_025634325.1|0.0e+ 00|basic helix-loop-helix protein A-like isoform X1 [Arachis hypogaea] | |
| Arahy.SVL5FL | 2.19 | 4.42E-05 | −1.79 | 5.44E-05 | XP_025637498.1|3.2e-242|transcription factor bHLH48-like [Arachis hypogaea] |
Furthermore, four bHLHs (Arahy.GL41VM, Arahy.R1MRXV, Arahy.26781 N, and Arahy.SVL5FL) also demonstrated preferential expression pattern between the two testa peanuts. The expression level of Arahy.R1MRXV encoding a bHLH62 like TF was down-regulated in Y9102-vs-ZH12, whereas Arahy.26781N encoding a bHLH48 like TF showed down-regulation in bulk pink-vs-bulk red (Fig. 6; Table 3). The alignment of their protein sequences showed close proximity with the Arabidopsis AtTT8 encoding gene, which allows the regulation of cell-specific flavonoids accumulation [32]. In addition, two genes encoding WRKY transcription factors were also identified as differentially expressed genes between the two testa groups. One gene (Arahy.FP3Q5V) showed up-regulation in red peanuts and one gene (Arahy.V3AFID) showed down-regulation in red testa groups however. These findings provide important insights about the crucial role of transcription factors dependent regulation of peanut testa pigmentation. Together, these findings highlighted that the onset of testa color in peanut might involve the regulation of the core structural genes of anthocyanin biosynthetic pathway such as DFR and ANS by tipping off the MYB/bHLH/WRKY transcription factors.
Transcriptomic data validated by quantitative real-time PCR assay
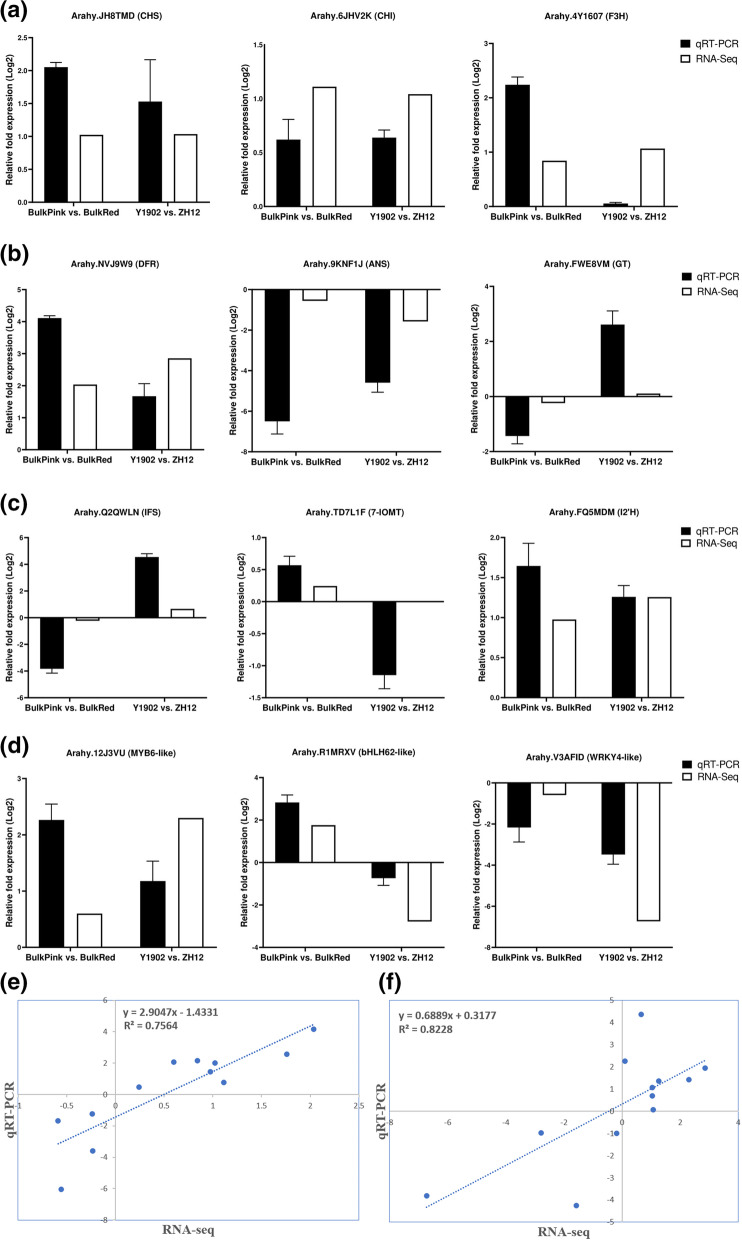
To further confirm the integrity and reliability of RNA-seq data, a total of twelve genes including (9 phenylpropanoid and flavonoid/anthocyanin biosynthetic pathways genes, and 3 transcription factors) were selected and then investigated their transcript abundance in bulk pink-vs-bulk red and Y9102-vs-ZH12 testa peanuts using qRT-PCR analysis. As described in Fig. 7, the relative expression level of these functional genes showed a very similar pattern in both down and up-regulated gene expression with the transcriptome data of the two testa peanuts. For instance, the relative gene expression level of core structural genes of the flavonoid/anthocyanin biosynthetic genes such as CHS, F3H, CHI, DFR and ANS showed a consistent pattern of up and down-regulation in the two testa peanuts compared with the gene expression level of bulk RNA-seq results (Fig. 7). Similarly, the expression level of the functional genes involved in the isoflavonoid biosynthetic pathway including IFS, 7-IOMT, I2’H and GT also mirrored the expression pattern with the RNA-seq results of both testa peanuts. Importantly, three transcription factors (bHLH, MYB and WRKY) likely to be involved in the regulation of key anthocyanin biosynthetic genes, showed a consistent pattern of expression level with their transcriptomic results (Fig. 7). Based on these findings, we assume that quality, accuracy and reliability of the bulk RNA-seq data is sufficient for designing rational molecular studies in the future.
Fig. 7.
Validation of gene expression level of the key functional genes likely to be involved during the regulation of pink and red testa in peanut using qRT-PCR assay (a) DEGs selected from the up-stream regulatory anthocyanin pathway (b) DEGs selected from the down-stream regulatory anthocyanin pathway (c) DEGs selected from the iso-flavonoid regulatory pathway (d) DEGs encoding important transcription factors involved in anthocyanin pathway in peanut. Pearson’s correlation analysis of the RNA-seq and qRT-PCR in (e) bulk pink-vs-bulk red peanuts and (f) Y9102-vs-ZH12 peanuts. The black bars in the y-axis indicate the relative expression level of genes quantified by qRT-PCR, whereas the white bars indicate the expression results from the transcriptome data. The x-axis demonstrates the expression results obtained from bulk pink-vs-bulk red and Y9102-vs-ZH12 testa peanuts. The data were presented as means of three independent biological replicates, and error bars denote ± SE (n = 3)
Discussion
Anthocyanin is widely known as the water-soluble bioactive flavonoids abundantly found in deep-colored peanuts [17]. The accumulation level of anthocyanins was found to have a strong correlation with peanut pigmentation and nutrient quality. Several classes of flavonoids, such as anthocyanins and flavonols were identified in different peanut cultivars [17, 33]. Similarly, the testa of the two black peanut cultivars were analyzed for the identification of two anthocyanins classes and four types of flavonols [26]. The important role of flavonoid biosynthetic genes have been implicated during the regulation of black testa in peanut by utilizing comparative transcriptome profiling [26]. A previous study demonstrated that pink testa in peanut is regulated by the upregulation of genes encoding important structural genes of anthocyanin biosynthesis such as PAL, C4H, CHS, and CHI as well as transcription factor genes including MYB, bHLH, and WRKY [8]. Recent studies have shown that peanut seed contains several essential nutrients, and the color of the testa has a significant impact on the nutritional and economic value of peanuts [21, 34, 35]. Presently, red and pink testa containing peanuts are the most widely consumed types in China. Several efforts have been carried out to uncover the regulatory mechanism of testa colors in peanut [8, 21, 23, 36]. However, the existing knowledge is still limited to genome-wide and comparative transcriptomic and metabolome analysis. In this study, as an effort to elucidate a clear picture of the differential regulation of peanut testa color via flavonoid/anthocyanin biosynthesis, we established a bulk RNA-seq approach by comparing the bulk F4 generation of red and pink testa and their correspondent parents. Our findings provided in depth insights into the regulatory network that governs testa pigmentation and as a result, will aid in the development of improved peanut varieties through selective breeding.
The orchestrated link between peanut testa color and anthocyanin metabolic flux
Plants produce anthocyanins, isoflavonoids, and flavonols, owing to a branch point of the phenylpropanoid system known as the flavonoid biosynthetic route. Among flavonoids, anthocyanins are the most important class which accumulate in different tissues (seeds, flowers, leaves and fruits) and contribute to their pigmentation, thus aiding seed and pollen dispersal [37]. In our study, we first analyzed the accumulation level of anthocyanin in red and pink testa peanuts along with their bulked F4 generation. The results suggested that the total anthocyanin content of the fully matured seeds of red testa peanut was higher than the pink testa peanut (Fig. 1B), indicating that the pigmentation of the peanut testa is strongly correlated with the accumulation level of anthocyanins. Previous studies confirmed that different plant species contained different level and types of anthocyanins resulting in multi-color hues. For instance, the integrated analysis of metabolomic and transcriptomic studies in green and purple fig cultivars showed significant differences in flavonoids metabolites in fruit peels [38]. It was further demonstrated that the cyanidin o-malonylhexoside was 3992-fold higher while different cyanidin glucosides were 100-fold higher in the mature purple peel, suggesting that anthocyanins are the key regulators of purple color in fig peels. Similarly, the transcriptome and metabolome analysis of jujube fruit peel during ripening periods also presented the underlying mechanism of red color formation. The reddening of jujube peel was shown to be strongly linked with the increase of malvidin and delphinidin content during ripening. It was also suggested that genes involved in the initial stages of the flavonoid biosynthetic pathway as well UFGT genes could also play important roles in anthocyanins accumulation when the fruit began the ripening process [39]. In addition, the acyl-modified type of anthocyanins are the most widespread class of anthocyanin found in Arabidopsis [40], whereas the only pigment detected in red leaves of cultivated lettuce is cyanidin 3-O-(malonyl)-glucoside [41]. A comparison of several asparagus varieties revealed that the major pigments in different varieties include, cyanidin, and peonidin type of anthocyanins [42]. Based on these informations, we further illustrate a deeper overview of the red and pink testa development in peanut with differently accumulated level of anthocyanins and thoroughly investigating the major alterations in gene-expression networks involved in anthocyanin biosynthetic pathway.
The crucial role of the upstream flavonoid pathway during testa color development
The biosynthetic pathway of anthocyanins is generally regulated by a multi-enzyme complex in plants [9]. At first, the phenylpropanoid pathway is activated through the conversion of phenylalanine amino acid into 4-coumaryl-CoA by PAL, C4H and 4CL enzymes. Then, 4-coumaryl-CoA and malonyl-CoA are sequentially converted to naringenin and dihydrokaempferol through the action of CHS, CHI, F3H, F3′H, and flavonoid F3′5 ′H enzymes. At this stage, when naringin is converted into dihydrokaempferol, the anthocyanin biosynthesis pathway is activated. The colored anthocyanin pigments are then synthesized from their colorless precursor molecules using DFR, ANS, and UFGT enzymes into red, blue or purple glycosides [10, 11]. Noticeably, our results demonstrated the large-scale transcription expression changes of major flavonoid biosynthetic pathway genes in red and pink testa peanuts, supporting the existing paradigm of flavonoid/anthocyanin biosynthesis. Recently, studies have shown that upstream regulatory genes of the anthocyanin and flavonoid biosynthesis in blueberries and pears were found to be upregulated throughout the primary stages of fruit development [43, 44]. CHS has been discovered as key regulator of the red petal color in crabapple cultivars [45], whereas its expression was significantly inhibited in the colorless (white) petals in mustard crop, and the transcription of many other genes associated with anthocyanin biosynthesis pathway was comparable to that detected in the colored petals [11]. In peaches, the increased anthocyanin content of peaches was found to be associated with higher CHS expression [46]. In our study, we identified two CHS (Arahy.JH8TMD and Arahy.UD4HAV) with up-regulated expression in red testa peanuts, implying that the fundamental transcriptional regulation of the up-stream regulatory genes could occur far earlier than the development of the testa phenotype.
Multi-channel regulation of core structural genes of anthocyanin pathway during peanut testa color formation
Transcriptional activation or inhibition of the F3H as well as the competitive expression of FLS and DFR genes particularly crucial in determining the production of anthocyanin component during anthocyanin pathway. Several studies have demonstrated that the synthesis of anthocyanins could be directed by the up-regulation of F3′H genes in Rosa hybrida and Vitis vinifera [47, 48]. The DFR derived from various plants have been shown to exhibit distinct substrate preferences for dihydrmyricetin, dihydroquercetin, and dihydrokaempferol [49, 50]. A previous study suggested that petunia flowers turn red when DFR enzyme catalyze the synthesis of pelargonin [51]. Recently, the positive role of DFR enzyme has been shown during the effective production of leucoanthocyanidin biosynthesis in safflower [52]. Pelargonin is accumulated in the petals of transgenic plants when maize DFR gene is introduced into white petunias, resulting in red petals [53]. Similarly, by overexpressing a number of DFR genes, the anthocyanin content in the flower tissues of tobacco was increased, which corresponded to a rise in red pigment [54]. Similar to other studies, our results also demonstrated that two F3H genes (Arahy.4WXU8P and Arahy.K5M1L6) were down-regulated in both pink testa and red testa peanuts, however, their expression level was differentially regulated by 3–4-fold change. Noticeably, the expression level of another F3H gene (Arahy.LCIL6D) showed 3-fold up-regulation in pink testa peanut (Fig. 4; Table S7). We found that one DFR (Arahy.NVJ9W9) showed up-regulation in both red testa groups. Furthermore, we found one ANS gene (Arahy.33FH8C) with up-regulated expression in red testa. While, one ANS gene (Arahy.BOQ1WP) showed up-regulation in both testa peanuts. These findings implied that the deepening of peanut testa color not only depends on the transcriptional activation of the key genes encoding F3H, DFR, and ANS, but also linked with the inhibition of FLS expression and bypassing flavonol biosynthesis.
Regulation of peanut testa color via perturbation of lignin and isoflavone pathway
In plants, the biosynthesis of isoflavones and lignin both share a same upstream pathway with the common pathway of flavonoid. Hence, the regulatory pathway of anthocyanin is also depending on the inhibition of lignin and isoflavone synthesis. During the lignin biosynthetic pathway, a key enzyme Hydroxycinnamoyl: CoA transferase (HCT) competes with 4CL and catalyze the conversion of 4-coumaryl-CoA into 4-coumaryl-shikimate and then bifurcates the production of S-P-G type of lignin through the action of downstream regulatory enzymes [55]. The suppression of HCT gene in transgenic alfalfa showed a decreased content of lignin, resulting a clear shift in the composition of lignin [56]. In this study, we identified two HCT genes, of which one gene (Arahy.NBHN08) showed reduced expression level in ZH12 compared with Y9102. Isoflavanones, on the other hand, are produced by isoflavone synthase (IFS) and 2-hydroxyisoflavanone dehydratase (HID) enzyme using naringenin and/or liquiritigenin and then undergoes several tailoring steps such as glycosylation, methylation, and hydroxylation [57]. In a previous work, the agrobacterium rhizogenes transformation method was shown to confirm isoflavones accumulation in soybeans through the enhanced expression of CHS and IFS genes [58]. We also detected the reduced expression and/or down-regulation of IFS, HID, 7-IOMT and I2’H in both testa peanuts, suggesting an obvious switch from isoflavone biosynthetic pathway towards anthocyanin biosynthesis.
MYB–bHLH with a co-option of WRKY transcription factors regulatory model in peanut testa color
In combination with known functional genes, flavonoid/anthocyanin biosynthesis is transcriptionally regulated by MBW complex [16]. Similarly, anthocyanin and flavonoid biosynthetic genes are regulated by MYB and bHLH transcription factors in numerous plant species [59–61]. One of the most important roles of MYB TFs is to regulate anthocyanin pathway genes, owing to pigmentation in fruits and flowers [45, 62]. Similar findings were found for the pear PpMYB10 and PpMYB114, which are responsible for regulating anthocyanin biosynthesis [63]. In Arabidopsis, MYB TFs have been reported, of which PAP1 and PAP2 shared important role as the common transcription factors to regulate the transcription of main anthocyanin structural genes [7]. Recently, the expression of the two R2R3-MYB genes, namely AhMYB1 and AhMYB2 was significantly increased in black peanuts [26]. Previously, we reported the identification of candidate gene (AhTc1) encoding a R2R3-MYB TF regulates the testa color in purple peanuts [19]. In this study, the expression level of MYB6-like TF (Arahy.S8NMKG) demonstrated high homology with the Arabidopsis (AtMYB5) and was significantly up-regulated in Y9102-vs-ZH12 testa peanut.
Basic helix-loop-helix (bHLH) TFs, are known to govern control the expression of genes such as DFR, ANS, and ANR in the seedlings and pods of Arabidopsis [64]. The bHLH TF also participates in the assembly of MBW complex, which regulates both anthocyanin and procyanidin synthesis in Arabidopsis, Petunia hybrid, Antirrhinum majus and Zea mays [7, 31, 65–67]. In this study, the expression level of Arahy.R1MRXV encoding a bHLH62 like TF was significantly down-regulated in Y9102-vs-ZH12, whereas Arahy.26781 N encoding a bHLH48 like TF showed up-regulation in both bulk pink-vs-bulk red and Y9102-vs-ZH12 testa peanuts. However, no such genes that code for anthocyanin-related WD-40 containing transcription factors were identified in both peanut testa groups. The findings were found consistent with previous findings, who provides the underlying network in black peanut testa color development [26]. Furthermore, and one gene (Arahy.V3AFID) showed down-regulation in red testa peanut ZH12. Previous, researches have shown that a WRKY TF (TTG2) plays an important role in the development of trichome and testa formation in Arabidopsis [68]. Recent research has shown that the WRKY-based regulation mechanism is present in Petunia but absent in Arabidopsis [69]. Nevertheless, in contrast to tt2 and tt8 mutants, the color morphology of ttg2 mutants is confined to the testa and no deficit of anthocyanins was detected in the plant body of ttg2 mutants. Hence, we proposed that WRKY TF alone could not sufficiently and directly induce anthocyanin biosynthesis in peanut testa, however, the co-expression of MYB and bHLH TFs could stimulate the transcription of WRKY encoding genes. In conclusion, the co-option of WRKY-induced MYB-bHLH regulatory complex could provide a fascinating insight into the regulation of peanut testa color development via anthocyanin biosynthesis.
Conclusions
The present study describes the identification of key functional genes that are most likely involved during the regulation of testa color in red and pink-types peanuts using a combination of BGISEQ-500 platform and bulk RNA-seq technology. Our findings revealed that the expression of PAL, CHS, CHI, F3H, DFR, and ANS encoding genes could be the key players governing the deepening of peanut testa color. In addition, the down-regulation of HCT, IFS, HID, 7-IOMT and I2’H genes suggested an alternative route for anthocyanin accumulation through perturbation of lignin and isoflavone pathway. In addition, the role of MYB, bHLH, and WRKY transcription factors also provides an intriguing transcriptional activation module in deciphering the underlying regulatory mechanism of peanut testa color via anthocyanin biosynthesis.
Materials and methods
Plant materials and phenotypic analysis
Peanut cultivars including red testa Zhonghua12 (ZH12), and pink testa Yuanza9102 (Y9102) were kindly provided by oil crop research institute of Chinese Academy of Agricultural Sciences. The two cultivars Zhonghua12 (ZH12), and Yuanza9102 (Y9102) along with F2:4 lines of population YZZH12 (Y9102 x ZH12) were grown in the experimental field located at Shandong Academy of Agricultural Sciences, Jinan, China under the controlled agricultural conditions. The population of YZZH12 was constructed using single seed descent which has been described in our previous study [22]. The phenotypic homozygous F4 lines were selected for mixing RNA pools as the method describe in our previous study [70]. The seeds of ZH12 and Y9102 were further selected, and allowed to grow until maturity. Similarly, the F4 bulk red and bulk pink lines were obtained, respectively. The testa was carefully excised from the seeds of both pink and red peanuts at fully mature stage and then analyzed for further investigation.
Anthocyanin quantification
Three sets of more than eight peanut seeds per sample were used to quantify the anthocyanin content of each testa peanut, following the previously reported protocol with only small alterations [19]. The anthocyanin content of peanut seeds was measured at a development stage of 50 days after pegging. Briefly, 50 mg of frozen seeds were ground to an extremely fine powder in liquid nitrogen. Then, 700 μl of acidic methanol (99:1) was added to the homogenized testa, and the extraction was performed at 4 °C. Following an overnight incubation time, the homogenates were subjected to centrifugation for 10 minutes at 12000 rpm at room temperature. The filtrates (roughly 600 μl) was separated and thoroughly mixed with 1 mL of trichloromethane and 400 μl of distilled water. The mixture was then centrifuged for 10 minutes at 4 °C and 12,000 rpm. With the help of a spectrophotometer (U-3000, HITACHI, Japan), the absorbance of the supernatant was measured at 530 and 657 nm. The relative anthocyanin content was calculated using the absorbance eq. [A530–0.25 × A657)/FW] unit/mg Fresh Weight. and then adjusted by weight of the sample. Three biological replicates were chosen from each plant material (ZH12, Y9102, bulk red and bulk pink lines) for RNA-seq analysis.
Library construction and RNA sequencing
The total RNA content was isolated from the testa with Trizol Reagent kit (TaKaRa, Inc., Dalian, China) following the manufacturer’s protocol. The purity and amount of the RNA from each sample was determined by Agilent 2100 and NanoDrop. The total RNA was treated with mRNA enrichment method using polyA tail connected magnetic beads with Oligo (dT). The DNA/RNA hybrid strand was selectively digested by RNaseH, and then the DNA probe was digested by DNaseI, and the desired RNA was obtained after purification. The mRNA was treated with fragmentation buffer to cleave into short fragments. Reverse transcription with random N6 primers was carried out, and then double-stranded DNA was synthesized. The double-stranded DNA was linearized and phosphorylate the 5′ end, and produce a sticky end with an “A” protruding from the 3′ end, and finally connected to a bubble-shaped linker with a protruding “T” at the 3′ end. The ligated product was then amplified by PCR using specific primers. The PCR products obtained from the previous step were thermally denatured into single-stranded DNA, and then a single-stranded circular DNA library was obtained by circularizing the single-stranded DNA with the help of a bridge primer. After that, sequencing was carried out using DNBSEQ platform.
Data analysis and reference genome alignment
Raw reads were thoroughly filtered using the in-house filtering software SOAPnuke (v1.5.2) (https://github.com/BGI-flexlab/SOAPnuke) to obtain the clean reads. The removal reads of reads containing adapters (adapter contamination), reads with unknown base N content greater than 10%, and low-quality reads (the bases with a quality value below 15 accounting for more than 50% of the total bases in the read) was conducted. Finally, the filtered clean reads were assembled in a FASTQ file. After obtaining the clean reads, we used HISAT (Hierarchical Indexing for Spliced Alignment of Transcripts) [71] to align the clean reads to the reference genome sequence of Arachis hypogaea cv. Tifrunner (https://www.peanutbase.org/data/public/Arachis_hypogaea/).
Identification of differentially expressed genes (DEGs)
High-quality clean reads were aligned to the reference gene sequences using Bowtie2 software package (http://bowtie-bio.sourceforge.net/Bowtie2/index.shtml) [72]. Then, the expression level of genes and transcripts were calculated from fragments per kb per million reads (FPKM) values using RSEM software [73]. The R package of DEseq2 was used to perform DEG detection based on the negative binomial distribution principle following the instructions described by [74], and the P-values were adjusted according to normal distribution. The (DEGs) were scored based on the thresholds log2 (FC) ≥ 1 and Q-value < 0.001.
Gene ontology (GO) and KEGG enrichment analysis of DEGs
To investigate the functional annotation, the DEGs identified in different groups of red and pink testa peanuts were mapped to the corresponding Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analysis. Then, the classification of DEGs was performed with GO functional terms and biological pathways using the phyper function in R software for enrichment analysis. The p-value was calculated according to the following method:
The p-values were then FDR corrected and typically features with Qvalue <= 0.05 were considered significantly enriched.
Transcription factors and plant disease resistance encoding DEGs
For plant transcription factors, the ORF of Unigene was identified by getorf, and then used hmmsearch (http://hmmer.org/) to align the ORF to the transcription factor protein domain (data from TF). The Unigene’s ability was identified according to the transcription factor family characteristics described in PlantTFDB. Similarly, the DIAMOND software (https://github.com/bbuchfink/diamond) was used to align the genes to the plant disease resistance gene database PRGdb for annotation, and the annotation results were further screened according to conditions such as query coverage and identity, in order to obtain possible disease resistance genes.
Validation of RNA-seq data using qRT-PCR
The expression results of twelve selected unigenes were chosen for confirmation with qRT-PCR assay. The total RNA content was extracted from individual samples and the first strand cDNA templates were prepared using the PrimeScript II reverse transcriptase system (TaKaRa). Primers for each unigene were designed using Primer3 software. The experiment was performed using the qRT PCR system ABI7500 Real Time System (Applied Biosystems) using SYBR Green I (Roche). Each reaction was carried out in a total of 20 μL volume following the reaction conditions: 94 °C 10 min; 94 °C 15 s, 60 °C 10 s and 72 °C 25 s for 40 cycles. Three independent biological replicates were used for each sample and the actin gene was kept as an internal reference gene. The relative expression of genes was calculated by using the 2 − △△Ct method.
Supplementary Information
Additional file 1: Fig. S1. The random reads distribution of bulk pink, bulk red, Y9102 and ZH12 samples. Fig. S2. Reads coverage of bulk pink, bulk red, Y9102 and ZH12 samples. Fig. S3. GO enrichment analysis. The enrichment bubble chart shows the enrichment degree of GO Term from three dimensions. By default, the top 20 GO Term with the smallest Qvalue or the selected GO Term (sorted by Q-value, up to 60) are plotted. The figure below shows the GO enrichment results of differential genes in (a) Y9102-Vs-ZH12 and (b) bulk pink-vs-bulk red peanuts. Fig. S4. KEGG Pathway Classification. The KEGG metabolic pathway is divided into 7 branches: Cellular Processes, Environmental Information Processing, Genetic Information Processing, Metabolism, Organic Systems. (a) Y9102-Vs-ZH12 and (b) bulk pink-vs-bulk red peanuts. Fig. S4. Multiple sequence alignment between Arahy.S8NMKG and AtMYB5. Fig. S5. Multiple sequence alignment between Arahy.9UC92R and TTG1 / TT2.
Additional file 2: Table S1. Statistics of up-regulated and down-regulated differentially expressed genes (DEGs) in each comparison group.
Additional file 3: Table S2. Expression level of DEGS in all comparison groups with red and pink testa peanuts.
Additional file 4: Table S3. Significantly enriched GO terms between different comparison groups with pink and red testa peanuts.
Additional file 5: Table S4. KEGG Pathway enrichment analysis between different comparison groups with pink and red testa peanuts.
Additional file 6: Table S5. DEGS distribution into different KEGG pathways between bulk pink-vs-bulk red testa peanuts.
Additional file 7: Table S6. DEGS distribution into different KEGG pathways between Y9102-vs-ZH12 testa peanuts.
Additional file 8: Table S7. KEGG pathway enrichment of DEGs involved in phenylpropanoid and flavonoid/anthocyanin biosynthetic pathways.
Additional file 9: Table S8. KEGG pathway enrichment of DEGS involved in iso-flavonoid pathway.
Additional file 10: Table S9. Expression level of Transcription factors encoding genes in bulk pink-vs-bulk red and Y9102-vs-ZH12 testa peanuts.
Acknowledgements
Not applicable.
Abbreviations
- DEGs
Differentially expressed genes
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- GO
Gene ontology
- FDR
The False Discovery Rate
- qRT-PCR
Quantitative real-time PCR
- TF
Transcription factor
- μl
Microliter
- mg
Milligram
- rpm
Revolutions per minute
- nm
Nanometer
Authors’ contributions
NA and KZ analyzed and interpreted the data. CZ, AL and XW supervised this work. SZ, MW, and JM performed statistical analysis. MY, DL and LR conducted the expression data analysis. BG, SG and CL revised the final version of this manuscript. JP, CM and RM provided technical support in this study. All authors read and approved the final manuscript.
Funding
This research is supported by National Natural Science Foundation of China (32072090), National Key Research and Development Program (2022YFD1200402), Key Research and Development Project of Shandong Province (2020LZGC001, 2021LZGC025), Agricultural Scientific and Technological Innovation Project of Shandong Academy of Agricultural Sciences, Famous subjects construction project of Shandong Agricultural and Engineering University, and Taishan Scholar Project of Shandong Province (ts20190964).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the Sequence Read Archive (SRA repository), under the Accession number PRJNA886491 [https://www.ncbi.nlm.nih.gov/sra/PRJNA886491] at NCBI.
Declarations
Ethics approval and consent to participate
The authors confirmed that the experimental research and field studies on plants (either cultivated or wild), including the collection of plant material comply with relevant institutional, national, and international guidelines and legislation.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Naveed Ahmad and Kun Zhang contributed equally to this work.
Contributor Information
Aiqin Li, Email: liaiqin1970@163.com.
Chuanzhi Zhao, Email: chuanzhiz@126.com.
References
- 1.Jaakola L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013;18(9):477–483. doi: 10.1016/j.tplants.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Hu Z, Zhu M, Zhu Z, Wang Z, Tian S, Chen G. Anthocyanin accumulation and molecular analysis of correlated genes in purple kohlrabi (Brassica oleracea var. gongylodes L.) J Agric Food Chem. 2015;63(16):4160–4169. doi: 10.1021/acs.jafc.5b00473. [DOI] [PubMed] [Google Scholar]
- 3.Castellarin SD, Pfeiffer A, Sivilotti P, Degan M, Peterlunger E, Di Gaspero G. Transcriptional regulation of anthocyanin biosynthesis in ripening fruits of grapevine under seasonal water deficit. Plant Cell Environ. 2007;30(11):1381–1399. doi: 10.1111/j.1365-3040.2007.01716.x. [DOI] [PubMed] [Google Scholar]
- 4.Shin W-H, Park S-J, Kim E-J. Protective effect of anthocyanins in middle cerebral artery occlusion and reperfusion model of cerebral ischemia in rats. Life Sci. 2006;79(2):130–137. doi: 10.1016/j.lfs.2005.12.033. [DOI] [PubMed] [Google Scholar]
- 5.Lo Piero AR, Puglisi I, Rapisarda P, Petrone G. Anthocyanins accumulation and related gene expression in red orange fruit induced by low temperature storage. J Agric Food Chem. 2005;53(23):9083–9088. doi: 10.1021/jf051609s. [DOI] [PubMed] [Google Scholar]
- 6.Boss PK, Davies C, Robinson SP. Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv shiraz grape berries and the implications for pathway regulation. Plant Physiol. 1996;111(4):1059–1066. doi: 10.1104/pp.111.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53(5):814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- 8.Xue Q, Zhang X, Yang H, Li H, Lv Y, Zhang K, et al. Transcriptome and metabolome analysis unveil anthocyanin metabolism in pink and red testa of peanut (Arachis hypogaea L.). Int J Genom. 2021;5883901. [DOI] [PMC free article] [PubMed]
- 9.Falcone Ferreyra ML, Rius S, Casati P. Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front Plant Sci. 2012;3:222. doi: 10.3389/fpls.2012.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng Y, Lin-Wang K, Cooney JM, Wang T, Espley RV, Allan AC. Differential regulation of the anthocyanin profile in purple kiwifruit (Actinidia species). Hortic Res. 2019;1:6:3. [DOI] [PMC free article] [PubMed]
- 11.Dick CA, Buenrostro J, Butler T, Carlson ML, Kliebenstein DJ, Whittall JB. Arctic mustard flower color polymorphism controlled by petal-specific downregulation at the threshold of the anthocyanin biosynthetic pathway. PLoS One. 2011;6(4):e18230. doi: 10.1371/journal.pone.0018230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han Y, Vimolmangkang S, Soria-Guerra RE, Korban SS. Introduction of apple ANR genes into tobacco inhibits expression of both CHI and DFR genes in flowers, leading to loss of anthocyanin. J Exp Bot. 2012;63(7):2437–2447. doi: 10.1093/jxb/err415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao L, Xu X, Chen S, Ma H. Cloning and expression analysis of Ficus carica anthocyanidin synthase 1 gene. Sci Hortic. 2016;211:369–375. doi: 10.1016/j.scienta.2016.09.015. [DOI] [Google Scholar]
- 14.Kobayashi S, Goto-Yamamoto N, Hirochika H. Retrotransposon-induced mutations in grape skin color. Science. 2004;304(5673):982. doi: 10.1126/science.1095011. [DOI] [PubMed] [Google Scholar]
- 15.Griesser M, Hoffmann T, Bellido ML, Rosati C, Fink B, Kurtzer R, Aharoni A, Munoz-Blanco J, Schwab W. Redirection of flavonoid biosynthesis through the down-regulation of an anthocyanidin glucosyltransferase in ripening strawberry fruit. Plant Physiol. 2008;146(4):1528–1539. doi: 10.1104/pp.107.114280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao L, Gao L, Wang H, Chen X, Wang Y, Yang H, Wei C, Wan X, Xia T. The R2R3-MYB, bHLH, WD40, and related transcription factors in flavonoid biosynthesis. Funct Integr Genom. 2013;13(1):75–98. doi: 10.1007/s10142-012-0301-4. [DOI] [PubMed] [Google Scholar]
- 17.Kuang Q, Yu Y, Attree R, Xu B. A comparative study on anthocyanin, saponin, and oil profiles of black and red seed coat peanut (Arachis hypogacea) grown in China. Int J Food Prop. 2017;20(sup1):S131–S140. doi: 10.1080/10942912.2017.1291676. [DOI] [Google Scholar]
- 18.Pandey MK, Monyo E, Ozias-Akins P, Liang X, Guimarães P, Nigam SN, Upadhyaya HD, Janila P, Zhang X, Guo B. Advances in Arachis genomics for peanut improvement. Biotechnol Adv. 2012;30(3):639–651. doi: 10.1016/j.biotechadv.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Y, Ma J, Li M, Deng L, Li G, Xia H, Zhao S, Hou L, Li P, Ma C. Whole-genome resequencing-based QTL-seq identified AhTc1 gene encoding a R2R3-MYB transcription factor controlling peanut purple testa colour. Plant Biotechnol J. 2020;18(1):96–105. doi: 10.1111/pbi.13175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhuang W, Chen H, Yang M, Wang J, Pandey MK, Zhang C, Chang W-C, Zhang L, Zhang X, Tang R. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat Genet. 2019;51(5):865–876. doi: 10.1038/s41588-019-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia H, Zhu L, Zhao C, Li K, Shang C, Hou L, Wang M, Shi J, Fan S, Wang X. Comparative transcriptome analysis of anthocyanin synthesis in black and pink peanut. Plant Signal Behav. 2020;15(2):1721044. doi: 10.1080/15592324.2020.1721044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang K, Yuan M, Xia H, He L, Ma J, Wang M, et al. BSA-Seq and genetic mapping reveals AhRt2 as a candidate gene responsible for red Testa of peanut. Theor Appl Genet. 2022;135(5):1529–40. [DOI] [PubMed]
- 23.Wan L, Li B, Lei Y, Yan L, Huai D, Kang Y, Jiang H, Tan J, Liao B. Transcriptomic profiling reveals pigment regulation during peanut testa development. Plant Physiol Biochem. 2018;125:116–125. doi: 10.1016/j.plaphy.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 24.Lou Q, Liu Y, Qi Y, Jiao S, Tian F, Jiang L, Wang Y. Transcriptome sequencing and metabolite analysis reveals the role of delphinidin metabolism in flower colour in grape hyacinth. J Exp Bot. 2014;65(12):3157–3164. doi: 10.1093/jxb/eru168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matus JT. Transcriptomic and metabolomic networks in the grape berry illustrate that it takes more than flavonoids to fight against ultraviolet radiation. Front Plant Sci. 2016;7:1337. doi: 10.3389/fpls.2016.01337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang J, Xing M, Li Y, Cheng F, Gu H, Yue C, Zhang Y. Comparative transcriptome analysis of the skin-specific accumulation of anthocyanins in black peanut (Arachis hypogaea L.) J Agric Food Chem. 2019;67(4):1312–1324. doi: 10.1021/acs.jafc.8b05915. [DOI] [PubMed] [Google Scholar]
- 27.Wei H, Chen X, Zong X, Shu H, Gao D, Liu Q. Comparative transcriptome analysis of genes involved in anthocyanin biosynthesis in the red and yellow fruits of sweet cherry (Prunus avium L.) PLoS One. 2015;10(3):e0121164. doi: 10.1371/journal.pone.0121164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Luo X, Wu C, Cao S, Zhou Y, Jie B, Cao Y, Meng H, Wu G. Comparative transcriptome analysis of genes involved in anthocyanin biosynthesis in red and green walnut (Juglans regia L.) Molecules. 2017;23(1):25. doi: 10.3390/molecules23010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Ma Y, Hu M, Zhao Y, Liu B, Wang C, Zhang M, Zhang L, Yang X, Mu G. Multi-omics and miRNA interaction joint analysis highlight new insights into anthocyanin biosynthesis in peanuts (Arachis hypogaea L.) Front Plant Sci. 2022;13:818345. doi: 10.3389/fpls.2022.818345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalez A, Mendenhall J, Huo Y, Lloyd A. TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev Biol. 2009;325(2):412–421. doi: 10.1016/j.ydbio.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 32.Baudry A, Caboche M, Lepiniec L. TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 2006;46(5):768–779. doi: 10.1111/j.1365-313X.2006.02733.x. [DOI] [PubMed] [Google Scholar]
- 33.Zhao Z, Wu M, Zhan Y, Zhan K, Chang X, Yang H, Li Z. Characterization and purification of anthocyanins from black peanut (Arachis hypogaea L.) skin by combined column chromatography. J Chromatogr A. 2017;1519:74–82. doi: 10.1016/j.chroma.2017.08.078. [DOI] [PubMed] [Google Scholar]
- 34.Zhao X, Chen J, Du F. Potential use of peanut by-products in food processing: a review. J Food Sci Technol. 2012;49(5):521–529. doi: 10.1007/s13197-011-0449-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chukwumah Y, Walker LT, Verghese M. Peanut skin color: a biomarker for total polyphenolic content and antioxidative capacities of peanut cultivars. Int J Mol Sci. 2009;10(11):4941–4952. doi: 10.3390/ijms10114941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wan L, Lei Y, Yan L, Liu Y, Pandey MK, Wan X, Varshney RK, Fang J, Liao B. Transcriptome and metabolome reveal redirection of flavonoids in a white testa peanut mutant. BMC Plant Biol. 2020;20(1):1–19. doi: 10.1186/s12870-020-02383-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001;126(2):485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z, Cui Y, Vainstein A, Chen S, Ma H. Regulation of fig (Ficus carica L.) fruit color: metabolomic and transcriptomic analyses of the flavonoid biosynthetic pathway. Front Plant Sci. 2017;20(8):1990. [DOI] [PMC free article] [PubMed]
- 39.Zhang Q, Wang L, Liu Z, Zhao Z, Zhao J, Wang Z, Zhou G, Liu P, Liu M. Transcriptome and metabolome profiling unveil the mechanisms of Ziziphus jujuba mill. Peel coloration. Food Chem. 2020;312:125903. doi: 10.1016/j.foodchem.2019.125903. [DOI] [PubMed] [Google Scholar]
- 40.D’Auria JC, Reichelt M, Luck K, Svatoš A, Gershenzon J. Identification and characterization of the BAHD acyltransferase malonyl CoA: anthocyanidin 5-O-glucoside-6 ″-O-malonyltransferase (At5MAT) in Arabidopsis thaliana. FEBS Lett. 2007;581(5):872–878. doi: 10.1016/j.febslet.2007.01.060. [DOI] [PubMed] [Google Scholar]
- 41.Becker C, Klaering H-P, Kroh LW, Krumbein A. Cool-cultivated red leaf lettuce accumulates cyanidin-3-O-(6 ″-O-malonyl)-glucoside and caffeoylmalic acid. Food Chem. 2014;146:404–411. doi: 10.1016/j.foodchem.2013.09.061. [DOI] [PubMed] [Google Scholar]
- 42.Dong T, Han R, Yu J, Zhu M, Zhang Y, Gong Y, Li Z. Anthocyanins accumulation and molecular analysis of correlated genes by metabolome and transcriptome in green and purple asparaguses (Asparagus officinalis, L.) Food Chem. 2019;271:18–28. doi: 10.1016/j.foodchem.2018.07.120. [DOI] [PubMed] [Google Scholar]
- 43.Lin Y, Wang Y, Li B, Tan H, Li D, Li L, Liu X, Han J, Meng X. Comparative transcriptome analysis of genes involved in anthocyanin synthesis in blueberry. Plant Physiol Biochem. 2018;127:561–572. doi: 10.1016/j.plaphy.2018.04.034. [DOI] [PubMed] [Google Scholar]
- 44.Yang Y-n, Zhao G, Yue W-q, Zhang S-l, Gu C, Wu J. Molecular cloning and gene expression differences of the anthocyanin biosynthesis-related genes in the red/green skin color mutant of pear (Pyrus communis L.) Tree Genet Genom. 2013;9(5):1351–1360. doi: 10.1007/s11295-013-0644-6. [DOI] [Google Scholar]
- 45.Tai D, Tian J, Zhang J, Song T, Yao Y. A Malus crabapple chalcone synthase gene, McCHS, regulates red petal color and flavonoid biosynthesis. PLoS One. 2014;9(10):e110570. doi: 10.1371/journal.pone.0110570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao K, Ding T, Mao D, Zhu G, Fang W, Chen C, Wang X, Wang L. Transcriptome analysis reveals novel genes involved in anthocyanin biosynthesis in the flesh of peach. Plant Physiol Biochem. 2018;123:94–102. doi: 10.1016/j.plaphy.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 47.Katsumoto Y, Fukuchi-Mizutani M, Fukui Y, Brugliera F, Holton TA, Karan M, Nakamura N, Yonekura-Sakakibara K, Togami J, Pigeaire A. Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant Cell Physiol. 2007;48(11):1589–1600. doi: 10.1093/pcp/pcm131. [DOI] [PubMed] [Google Scholar]
- 48.Jeong ST, Goto-Yamamoto N, Hashizume K, Esaka M. Expression of the flavonoid 3′-hydroxylase and flavonoid 3′, 5′-hydroxylase genes and flavonoid composition in grape (Vitis vinifera) Plant Sci. 2006;170(1):61–69. doi: 10.1016/j.plantsci.2005.07.025. [DOI] [Google Scholar]
- 49.Hua C, Linling L, Shuiyuan C, Fuliang C, Feng X, Honghui Y, Conghua W. Molecular cloning and characterization of three genes encoding dihydroflavonol-4-reductase from Ginkgo biloba in anthocyanin biosynthetic pathway. PLoS One. 2013;8(8):e72017. doi: 10.1371/journal.pone.0072017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saito K, Yonekura-Sakakibara K, Nakabayashi R, Higashi Y, Yamazaki M, Tohge T, Fernie AR. The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity. Plant Physiol Biochem. 2013;72:21–34. doi: 10.1016/j.plaphy.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 51.Keting H, Ke H, Silan D. Flower color breeding by molecular design in ornamentals. Mol Plant Breed. 2008;6(1):16–24
- 52.Ahmad N, Li T, Liu Y, Hoang NQV, Ma X, Zhang X, Liu J, Yao N, Liu X, Li H. Molecular and biochemical rhythms in dihydroflavonol 4-reductase-mediated regulation of leucoanthocyanidin biosynthesis in Carthamus tinctorius L. Industrial Crops Products. 2020;156:112838. doi: 10.1016/j.indcrop.2020.112838. [DOI] [Google Scholar]
- 53.Meyer P, Heidmann I, Forkmann G, Saedler H. A new petunia flower colour generated by transformation of a mutant with a maize gene. Nature. 1987;330(6149):677–678. doi: 10.1038/330677a0. [DOI] [PubMed] [Google Scholar]
- 54.Luo P, Ning G, Wang Z, Shen Y, Jin H, Li P, Huang S, Zhao J, Bao M. Disequilibrium of flavonol synthase and dihydroflavonol-4-reductase expression associated tightly to white vs. red color flower formation in plants. Front Plant Sci. 2016;6:1257. doi: 10.3389/fpls.2015.01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Q, Luo L, Zheng L. Lignins: biosynthesis and biological functions in plants. Int J Mol Sci. 2018;19(2):335. doi: 10.3390/ijms19020335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shadle G, Chen F, Reddy MS, Jackson L, Nakashima J, Dixon RA. Down-regulation of hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry. 2007;68(11):1521–1529. doi: 10.1016/j.phytochem.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 57.Wang X, Li S, Li J, Li C, Zhang Y. De novo transcriptome sequencing in Pueraria lobata to identify putative genes involved in isoflavones biosynthesis. Plant Cell Rep. 2015;34(5):733–743. doi: 10.1007/s00299-014-1733-1. [DOI] [PubMed] [Google Scholar]
- 58.Yi J, Xu Z, Wang J, Zhang D, He X, Ali Z, Zhu H, Ma H, Sangeeta D. GmCHS8 and GmIFS2 gene co-determine accumulation of isoflavonoid in soybean. Acta Agron Sin. 2011;37(4):571–578. doi: 10.3724/SP.J.1006.2011.00571. [DOI] [Google Scholar]
- 59.An JP, Wang XF, Zhang XW, Xu HF, Bi SQ, You CX, Hao YJ. An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation. Plant Biotechnol J. 2020;18(2):337–353. doi: 10.1111/pbi.13201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiang L, Liu X, Li H, Yin X, Grierson D, Li F, Chen K. CmMYB# 7, an R3 MYB transcription factor, acts as a negative regulator of anthocyanin biosynthesis in chrysanthemum. J Exp Bot. 2019;70(12):3111–3123. doi: 10.1093/jxb/erz121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deng J, Li J, Su M, Lin Z, Chen L, Yang P. A bHLH gene NnTT8 of Nelumbo nucifera regulates anthocyanin biosynthesis. Plant Physiol Biochem. 2021;158:518–523. doi: 10.1016/j.plaphy.2020.11.038. [DOI] [PubMed] [Google Scholar]
- 62.Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007;49(3):414–427. doi: 10.1111/j.1365-313X.2006.02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ni J, Zhao Y, Tao R, Yin L, Gao L, Strid Å, Qian M, Li J, Li Y, Shen J. Ethylene mediates the branching of the jasmonate-induced flavonoid biosynthesis pathway by suppressing anthocyanin biosynthesis in red Chinese pear fruits. Plant Biotechnol J. 2020;18(5):1223–1240. doi: 10.1111/pbi.13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004;39(3):366–380. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- 65.Spelt C, Quattrocchio F, Mol J, Koes R. ANTHOCYANIN1 of petunia controls pigment synthesis, vacuolar pH, and seed coat development by genetically distinct mechanisms. Plant Cell. 2002;14(9):2121–2135. doi: 10.1105/tpc.003772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carey CC, Strahle JT, Selinger DA, Chandler VL. Mutations in the pale aleurone color1 regulatory gene of the Zea mays anthocyanin pathway have distinct phenotypes relative to the functionally similar TRANSPARENT TESTA GLABRA1 gene in Arabidopsis thaliana. Plant Cell. 2004;16(2):450–464. doi: 10.1105/tpc.018796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lloyd A, Brockman A, Aguirre L, Campbell A, Bean A, Cantero A, Gonzalez A. Advances in the MYB–bHLH–WD repeat (MBW) pigment regulatory model: addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 2017;58(9):1431–1441. doi: 10.1093/pcp/pcx075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johnson CS, Kolevski B, Smyth DR. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell. 2002;14(6):1359–1375. doi: 10.1105/tpc.001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Verweij W, Spelt CE, Bliek M, de Vries M, Wit N, Faraco M, Koes R, Quattrocchio FM. Functionally similar WRKY proteins regulate vacuolar acidification in petunia and hair development in Arabidopsis. Plant Cell. 2016;28(3):786–803. doi: 10.1105/tpc.15.00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ahmad N, Hou L, Ma J, Zhou X, Xia H, Wang M, Leal-Bertioli S, Zhao S, Tian R, Pan J. Bulk RNA-Seq analysis reveals differentially expressed genes associated with lateral branch angle in Peanut. Genes. 2022;13(5):841. doi: 10.3390/genes13050841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12(1):1–16. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. The random reads distribution of bulk pink, bulk red, Y9102 and ZH12 samples. Fig. S2. Reads coverage of bulk pink, bulk red, Y9102 and ZH12 samples. Fig. S3. GO enrichment analysis. The enrichment bubble chart shows the enrichment degree of GO Term from three dimensions. By default, the top 20 GO Term with the smallest Qvalue or the selected GO Term (sorted by Q-value, up to 60) are plotted. The figure below shows the GO enrichment results of differential genes in (a) Y9102-Vs-ZH12 and (b) bulk pink-vs-bulk red peanuts. Fig. S4. KEGG Pathway Classification. The KEGG metabolic pathway is divided into 7 branches: Cellular Processes, Environmental Information Processing, Genetic Information Processing, Metabolism, Organic Systems. (a) Y9102-Vs-ZH12 and (b) bulk pink-vs-bulk red peanuts. Fig. S4. Multiple sequence alignment between Arahy.S8NMKG and AtMYB5. Fig. S5. Multiple sequence alignment between Arahy.9UC92R and TTG1 / TT2.
Additional file 2: Table S1. Statistics of up-regulated and down-regulated differentially expressed genes (DEGs) in each comparison group.
Additional file 3: Table S2. Expression level of DEGS in all comparison groups with red and pink testa peanuts.
Additional file 4: Table S3. Significantly enriched GO terms between different comparison groups with pink and red testa peanuts.
Additional file 5: Table S4. KEGG Pathway enrichment analysis between different comparison groups with pink and red testa peanuts.
Additional file 6: Table S5. DEGS distribution into different KEGG pathways between bulk pink-vs-bulk red testa peanuts.
Additional file 7: Table S6. DEGS distribution into different KEGG pathways between Y9102-vs-ZH12 testa peanuts.
Additional file 8: Table S7. KEGG pathway enrichment of DEGs involved in phenylpropanoid and flavonoid/anthocyanin biosynthetic pathways.
Additional file 9: Table S8. KEGG pathway enrichment of DEGS involved in iso-flavonoid pathway.
Additional file 10: Table S9. Expression level of Transcription factors encoding genes in bulk pink-vs-bulk red and Y9102-vs-ZH12 testa peanuts.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the Sequence Read Archive (SRA repository), under the Accession number PRJNA886491 [https://www.ncbi.nlm.nih.gov/sra/PRJNA886491] at NCBI.