SUMMARY
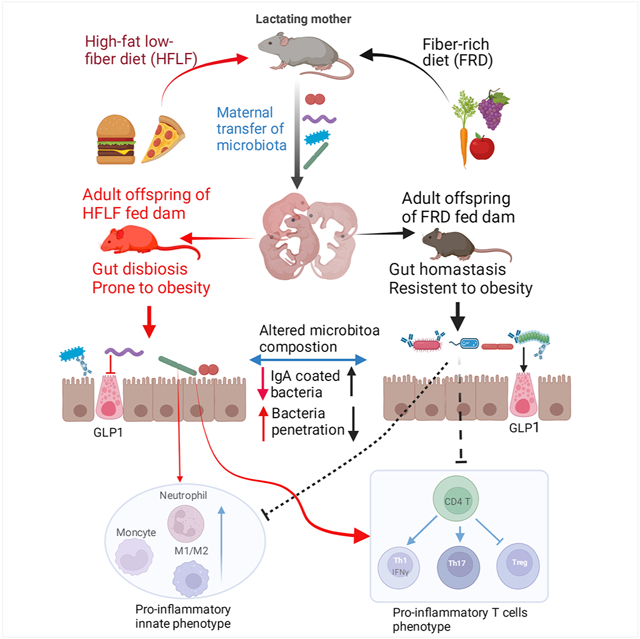
Diet, especially fiber content, plays an important role in sustaining a healthy gut microbiota, which promotes intestinal and metabolic health. Another major determinant of microbiota composition is the specific microbes that are acquired early in life, especially maternally. Consequently, we hypothesized that alterations in maternal diet during lactation might lastingly impact the microbiota composition and health status of offspring. Accordingly, we observed that feeding lactating dams low-fiber diets resulted in offspring with lasting microbiota dysbiosis, including reduced taxonomic diversity and increased abundance of Proteobacteria species, despite the offspring consuming a fiber-rich diet. Such microbiota dysbiosis was associated with increased encroachment of bacteria into inner mucus layers, low-grade gut inflammation and a dramatically exacerbated microbiota-dependent increase in adiposity following exposure to an obesogenic diet. Thus, maternal diet is a critical long-lasting determinant of offspring microbiota composition, impacting gut health and proneness to obesity and its associated disorders.
Keywords: Maternal diets, Inulin, offspring, obesity, gut microbiota
Graphical Abstract
In Brief
Zou et al. show that consumption of low-fiber diets by lactating mice caused offspring to exhibit lasting microbiota dysbiosis and a propensity for obesity despite the offspring consuming fiber-rich diets. These findings indicate that dietary fiber consumption by lactating mothers critically determines the intestinal and metabolic health of offspring.
INTRODUCTION
Humanity is increasingly afflicted with an epidemic of obesity and the inter-related metabolic disorders it promotes, especially type 2 diabetes. 1,2 The obesity epidemic has associated with increased availability of highly processed diets, which are typically high in fat and simple carbohydrate, as low in fiber. Such diets have highly likely been an essential driver of obesity epidemic. 3,4 Yet, the extent to which persons with access to such diets develop obesity is highly variable 5 and, moreover, continues to increase even in regions where such diets have been highly prevalent for decades, thus suggesting additional factors are playing a role in driving the obesity epidemic. Determinants of proneness to obesity are not well understood. 6 Epidemiologic studies indicate that proneness to obesity is heritable but efforts to define specific genetic links to this phenotype have had only limited success thus suggesting roles for other potentially heritable characteristics such as dietary practices and gut microbiota composition, both of which also have potential to influence metabolic phenotype. 7,8 These factors are interrelated in that diet modulates one’s gut microbiota and, consequently, the microbiome that is vertically transmitted to offspring. 9,10 Thus, diet not only influences metabolism of the individual consuming it, rather, may also exert a microbiota-dependent influence over the metabolic phenotype of their offspring. In considering approaches to investigate such impacts, we reasoned that the effect of maternal diet on offspring microbiome might be particularly strong during lactation since this is the window during which microbiota are acquired and shaped. 11-13 Moreover, we reasoned that particularly strong impacts might result from lactating dams consuming low-fiber compositionally defined diets, which we’ve observed result in stark rapid microbiota remodeling. 14,15 Hence, we examined how feeding lactating dams compositionally defined low-fiber diets, with high- or low-fat content, influenced microbiota, intestinal health, and metabolic status of offspring that were, themselves, weaned onto the fiber-rich grain-based chow (GBC) diets typically used to maintain mice in biomedical research. We also examined how maternal diet influenced offspring later in life following exposure to an obesogenic diet. We observed that altering the maternal diet in this manner indeed impacted offspring microbiomes, which associated with low-grade inflammation and mild basal metabolic phenotypes but conferred dramatically elevated proneness to diet-induced obesity.
RESULTS
Feeding lactating dams low-fiber diets impacted offspring microbiome, intestine, and metabolism.
Mice used in biomedical research have classically been maintained on grain-based rodent chow (GBC). GBC is comprised of relatively unrefined ingredients that result in high batch to batch variability that confounds diet-based research thus prompting use “open-source” diets, which are compositionally well-defined and comprised of relatively invariant components. 16,17 Its variability notwithstanding, GBC is naturally rich in fiber (15-25% by weight), including soluble/fermentable fiber, which nourishes gut microbiota thereby promoting intestinal and metabolic health. 14,18 In contrast, open-source control diets classically contain only 5% fiber and are thus considered low-fiber diets. 18 Further, the fiber used is typically cellulose, which is insoluble and highly resistant to fermentation. Consequently, such low-fiber diets fail to support a healthy gut microbiota, resulting in epithelial stagnation, low-grade gut inflammation, and promotion of obesity, especially when such low-fiber diets are also rich in saturated fats. 14 While diet-induced changes in gut microbiota are reversible, 19 we hypothesized that diet-induced microbiota dysbiosis occurring during lactation might lastingly and detrimentally imprint the host-microbiota relationship in their offspring. As schematized in Figure 1A, we tested this hypothesis by switching the diet of lactating dams from GBC to a low-fiber low-fat diet (LF-LFD) or a low-fiber high-fat diet (LF-HFD) shortly following delivery of pups. LF-HFD, which derives 60% of its calories from fat, seeks to model a “fast-food” diet while our use of LF-LFD aimed to elucidate relative roles of high-fat vs. low-fiber. Lactating diets were maintained until pups were weaned at 3 weeks, of age, at which time fecal microbiomes and metabolic phenotype of dams and pups were examined.
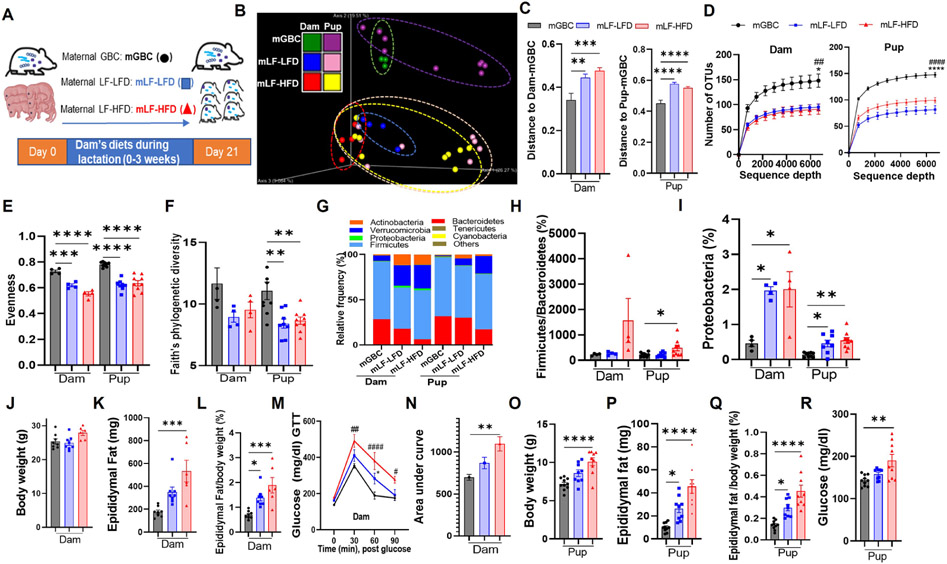
Figure 1. Consumption of low-fiber diets, during lactation, impacts microbiota and metabolic health of offspring.
(A) Scheme; Dams were fed with grain-based chow (GBC), low-fiber low-fat diet (LF-LFD) or low-fiber high-fat diet (LF-HFD) during lactation. Dams and female pups were euthanized at the end of lactation.
(B-D) Fecal microbiota composition (N=4 for dams, N=8-9 for pups) via 16S rRNA gene sequencing, as expressed by unweighted UniFrac PCoA analysis (B), Unifrac distances between microbiota of GBC group and LF-LFD/LF-HFD group (C), and rarefaction curves of the observed-OTU richness (D).
(E&F) Alpha diversity of gut microbiota measured by Pielou's evenness (E) and Faith’s phylogenetic diversity (F).
(G-I) Taxonomic analysis of relative abundance of bacteria at the phylum level (G), ratio of Firmicutes to Bacteroidetes (H). Percent of Proteobacteria (I).
(J-N) Metabolic data of dams (N=6-8) including body weight (J), epididymal fat pad weight (K), fat pad to body weight ratio (L), intraperitoneally glucose tolerance testing (GTT) (M) and area under curve of GTT calculated (N) at the end of lactation.
(O-R) Pup metabolic data (N=9-10) including body weight (O), absolute epididymal fat mass (P), epididymal fat percent (Q), and non-fasting glucose measured (R) at weaning time.
One-way ANOVA: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; #p<0.05, ##p < 0.01, ####p < 0.0001. In D and M, * indicated mLF-LFD vs mGBC; # indicated mLF-HFD VS mGBC. Also, see Figure S1.
Analogous to our published worked in non-lactating mice, 14 analysis of fecal microbiomes by 16S rRNA gene sequencing followed by Uni-Frac/principal coordinate analysis (PCoA) revealed stark diet-based microbiome clustering between dams fed GBC vs. those fed either low-fiber diet (Figure 1B). Moreover, microbiomes of all groups of pups were similar to their respective dams indicating that, as predicted, diet-induced impacts on gut microbiota had indeed been transmitted to offspring. Measure of UniFrac distances confirmed that these differences were statistically significant (Figure 1C). Furthermore, the predominant features of dysbiosis previously observed in non-lactating mice fed low-fiber diets were also observed in fiber-deprived lactating dams and their pups. Specifically, fiber-deprived dams, and their pups, exhibited reduced α-diversity based on number of taxa observed (Figure 1D) and measures of evenness (Figure 1E). A concomitant trend of reduced Faith’s phylogenetic diversity was also observed but this did not reach statistical significance in the dams that consumed fiber deprived diet (Figure 1F). Additionally, fiber deprivation altered gut microbiota composition at the phylum level, including increased abundance of Proteobacteria in dams and, albeit to a lesser extent, their weanlings (Figure 1G-1I). Similar features of dysbiosis were also observed in the small intestinal microbiome (Figure S1A-S1E). Thus, in accord with the notion that the microbiome is vertically transmitted, newly weaned mouse pups exhibited the microbiota dysbiosis that their mothers had acquired via alteration of diet during lactation.
Consumption of low-fiber diets by lactating dams resulted in a metabolic phenotype reminiscent of what has been previously observed in non-lactating mice. Specifically, 3 weeks consumption of LF-HFD resulted in a non-significant increase in body weight while both low-fiber diets resulted in increased adiposity and reduced glucose tolerance relative to GBC-fed dams (Figure 1J-1N). The low-fiber diets also resulted in reduced colon weight and length indicating that previously described intestinal phenotypes had also manifested in lactating mice (Figure S1F and S1G). Analogous metabolic phenotypes were also evident in newly weaned offspring. Specifically, relative to pups whose mothers consumed GBC, dam consumption of LF-LFD and, to a greater extent, LF-HFD resulted in 3-week-old pups exhibiting increased weight/adiposity and elevated non-fasting glucose levels (Figure 1O-1R). These metabolic phenotypes were not accompanied by the overt changes in pups’ intestinal morphology (Figure S1H and S1I), that were observed in the dams that directly consumed these diets, but profiling of intestinal innate immune cells by flow cytometry found pups had modest evidence for the low-grade intestinal inflammation exhibited by their dams, including an increased ratio of M1/M2 macrophages and elevated levels of neutrophils and monocytes in small intestine and colon. These changes were associated with modest variable changes in CD4 and CD8 T cells, and a reduction in B-cell frequency, which likely reflected the increased inflammatory cells (Figure S1J-S1Q).
Impacts of maternal fiber deprivation on offspring microbiome are long-lasting
We next examined the extent to which the impacts of maternal lactation diet on microbiome and phenotype that were observed in newly weaned offspring would persist once such mice were maintained on a fiber-rich diet, namely GBC (Figure 2A). Analysis of fecal microbiomes in 12-week-old offspring (i.e. after 9 weeks of GBC consumption) found that the reduction in taxonomic α-diversity that resulted from maternal fiber deprivation was not remediated by GBC consumption (Figure 2B-2D). Specifically, PCoA analysis revealed stark differences in microbiota composition between mice whose mothers had consumed GBC vs. low-fiber diets during lactation (Figure 2E and 2F). This difference remained evident at the phylum level (Figure 2G) with significantly elevated abundance of Proteobacteria in offspring of both LF-LFD and LF-HFD fed dams (Figure 2H). Total levels of bacteria per mg feces, which changes rapidly and proportionally to fermentable fiber content, 14 did not differ in offspring indicating that, bacterial density was not imprinted by maternal diet (Figure S1R).
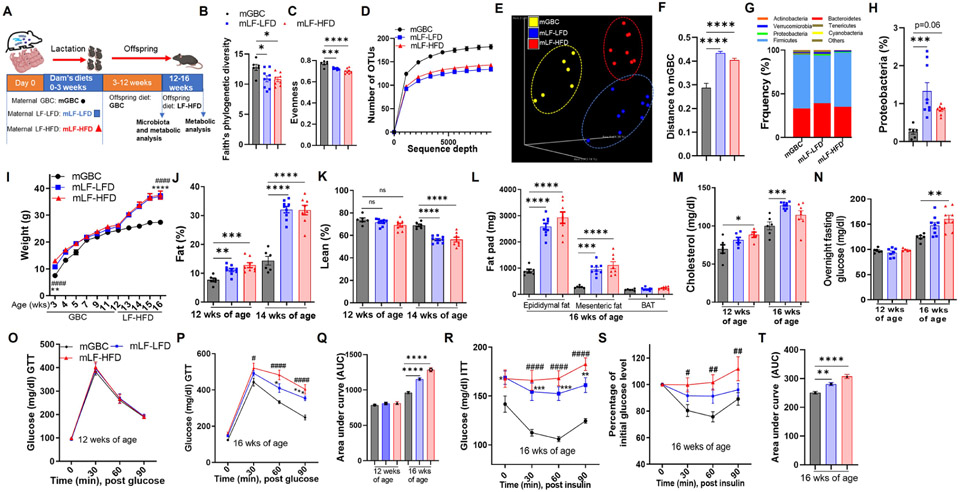
Figure 2. Maternal fiber deprivation lastingly impacted offspring microbiota and exacerbated diet-induced obesity in offspring.
(A) Scheme; Lactating dams were fed with indicated diets upon giving birth. Male offspring (N=6-9) mice were weaned at 3 weeks of age and administered GBC until 12 weeks of age at which time feces were collected for 16S rRNA gene sequencing, then administered LF-HFD for 4 weeks.
(B-D) Alpha diversity in gut microbiota of offspring at 12 weeks of age, measured faith’s phylogenetic diversity (B), Pielou’s evenness (C) and Rarefaction curves of the observed-OTU richness (D).
(E-F) Beta diversity of intestinal microbiota in 12-weeks old offspring was measured by PCoA via unweighted UniFrac (E) and plot of Unifrac distances (F).
(G&H) Taxonomic analysis including relative phylum abundance (G) and percentage of Proteobacteria (H).
(I-L) Metabolic data of offspring including body weight recorded (I), whole-body fat (J) and lean (K) measured by using MRI, Epididymal, mesenteric fat and brown adipose tissue (BAT) weight (L) measured at the end of experiment,
(M-Q) Cholesterol (M) and fasting glucose (N) in blood measured, glucose tolerance test (GTT) by intraperitoneally injecting with glucose (O&P), AUC of GTT calculated (Q).
(R-T) Blood glucose during insulin tolerance test as showed by absolute concentration (R) or as relative changes compared with initial value (S). AUC from S calculated (T).
One-way ANOVA: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; #p<0.05, ##p < 0.01, ####p < 0.0001. ns, not significant. For I, P, R and S, * indicated mLF-LFD VS mGBC; # indicated mLF-HFD vs mGBC. Also, see Figure S1.
Maternal fiber deprivation dramatically enhanced offspring proneness to diet-induced obesity
We next examined impacts of maternal fiber deprivation on offspring metabolic phenotype, while mice were maintained on GBC, and in response to LF-HFD, which has long been widely used to model diet-induced obesity (DIO) (Figure 2A). We found that offspring of both LF-LFD and LF-HFD dams displayed elevations in body weight upon weaning that gradually dissipated over the subsequent weeks that these mice were maintained on GBC (Figure 2I and S1S). Nonetheless, modest but statistically significant differences in adiposity remained at 12 and 16 weeks of age when mice were maintained on GBC (Figure 2J, S1V and S1W). However, far more striking was the impact of maternal diet on proneness to DIO. Specifically, maternal feeding of either low-fiber diet resulted in severe DIO phenotypes as indicated by rapid gain in body weight, increased fat mass, decreased lean mass by MRI, and increased mass of major fat depots following 4 weeks exposure to LF-HFD (i.e. 16-week-old mice) (Figure 2I-2L). Such severe DIO was paralleled by other indices of metabolic syndrome, namely hypercholesterolemia, dysglycemia, and insulin resistance (Figure 2M-2T). Thus, lasting metabolic impacts of maternal fiber deprivation are modest while offspring are maintained on a fiber-rich diet but confer high proneness to DIO and its consequences.
Maternal fiber deprivation promotes DIO via impacts on microbiota
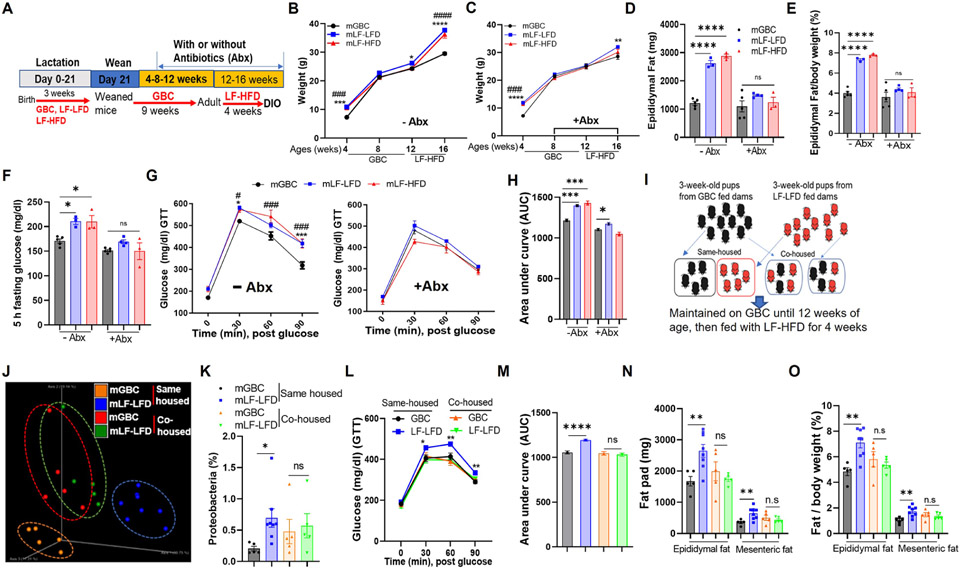
In mice, culling of litter size increases caloric consumption of suckling mice, resulting in lastingly potentiated proneness to experimentally induced colitis. 20 Obesity can also be viewed as an inflammatory disease, prompting us to speculate that the highly refined nature of the low-fiber diets may have resulted in dams ingesting more calories, which could then be passed along during lactation to offspring thereby priming them to develop DIO. However, culling of litters did not alter proneness of 12-week-old mice to DIO (Figure S2A) arguing against this notion. Thus, we next hypothesized that the exacerbated DIO in offspring of fiber-deprived dams was driven by their persisting microbiota dysbiosis. To test this notion, we compared offspring of GBC-fed or fiber-deprived dams under conditions wherein offspring were, or were not, subjected to microbiota ablation via broad-spectrum antibiotics, starting at 8 weeks of age and maintained throughout the DIO challenge (Figure 3A). This antibiotic regimen did not, by itself, ameliorate DIO in offspring of GBC-fed dams but largely eliminated the exacerbated DIO that was otherwise exhibited by offspring of fiber-deprived dams (Figure 3B-3E). In accord with previous studies, 21 antibiotics reduced DIO-induced dysglycemia (Figure 3F-3H). Moreover, maternal fiber deprivation did not exacerbate dysglycemia under conditions in which all mice had undergone antibiotic-mediated microbiota ablation (Figure 3F-3H). A slightly modified antibiotic regimen, namely discontinuing their use shortly before the 4-week exposure to LF-HFD also ablated the impact of maternal fiber deprivation on DIO (Figure S2B-S2D). This result suggested that proneness of offspring from fiber-deprived dams to DIO was dictated by microbiota present upon exposure to the obesogenic diet rather than reflect that severe DIO required microbiota be present during DIO per se. We also investigated the role of microbiota in maternal fiber deprivation-induced exacerbation of DIO via co-housing. Specifically, offspring of dams fed GBC or LF-LFD were, upon weaning, placed in a single cage to enable natural exchange of gut microbiota (Figure 3I) resulting in the cohoused mice having similar microbiomes as reflected by unweighted Unifrac distance and relative proteobacteria abundance (Figure 3J and 3K). Such normalization of their microbiomes associated with similar immune cell profiles (Figure S2E-S2K) and, moreover, similar proneness to DIO (Figure 3L-3O) thus further supporting the notion that the impact of maternal fiber deprivation on microbiota composition mediates exacerbation of DIO. Furthermore, that the extent of DIO in the cohoused mice was similar to that of non-cohoused offspring of GBC-fed dams suggests a protective role for microbes that such dams pass along to their offspring.
Figure 3. Maternal fiber deprivation’s exacerbation of DIO was eliminated by antibiotics or co-housing.
(A) Scheme, Male offspring (N=3-5) of dams fed indicated lactating diet were subjected to microbiota ablation via administration of drinking water containing broad-spectrum antibiotics starting 8 weeks of age and maintained throughout the LF-HFD exposure.
(B-C) Body weight of offspring without (B) or with (C) antibiotics treatment recorded.
(D-E) Epididymal adipose mass (D) and its percentage in relative to body weight (E) upon euthanasia.
(F-H) Mice were fasted for 5h and basal glucose level was measured (F), then subjected to intraperitoneally glucose tolerance testing (G). AUC from G calculated (H).
(I) Scheme. 3-week-old pups (N=5-8) were weaned from dams fed indicated diet were co-housed as indicated while administered GBC until 12 weeks of age and then fed LF-HFD.
(J&K) Microbiota of same-housed and co-housed mice was analyzed by 16S rRNA gene sequencing, global composition as expressed by PCoA via unweighted UniFrac analysis (J), percentage of proteobacteria (K) calculated.
(L&M) Intraperitoneally glucose tolerance test (GTT) measured for co-housed and same housed mice (L) and AUC of GTT (M).
(N&O) Epididymal fat and mesenteric fat mass including absolute value (N) and in relation to body weight (O) measured.
One-way ANOVA (B, C, D, E, F, G, H) or Student’s t test (K, L, M, N, O). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; #p<0.05, ##p<0.01, ####p < 0.0001, ns, not significant. In B, C and G, * indicated mLF-LFD VS mGBC; # indicated mLF-HFD vs mGBC. In L, * indicated mLF-LFD vs mGBC in same housed mice. Also, See Figure S2
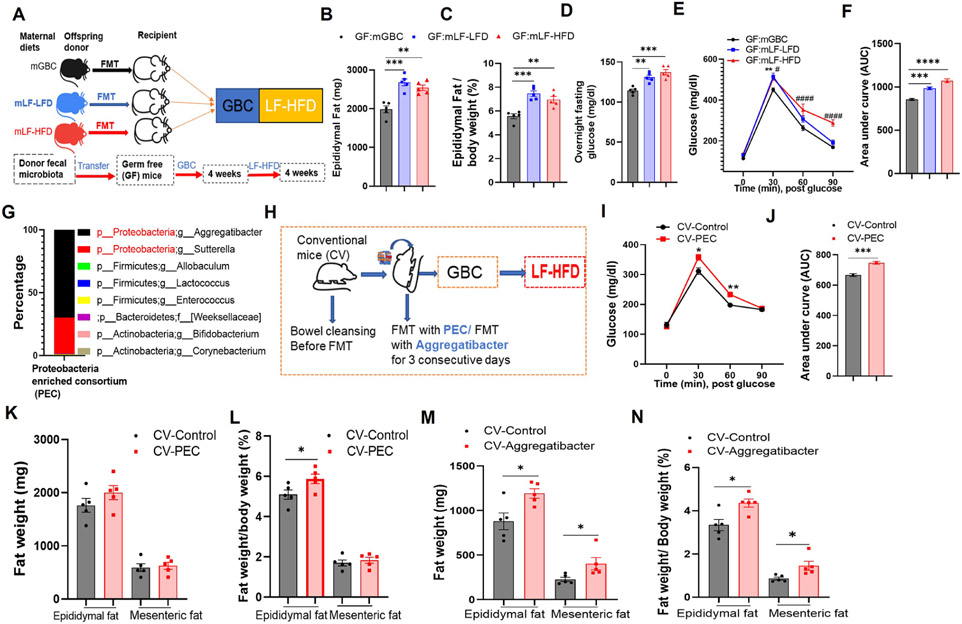
The extent to which alterations in microbiome that resulted from maternal fiber deprivation might be sufficient to exacerbate DIO was investigated via fecal microbiota transplants. First, we administered 8-week-old germfree mice suspensions of feces from 12-week-old GBC-fed offspring of dams that had consumed GBC, LF-LFD or LF-HFD during lactation. 4 weeks later, such mice were exposed to LF-HFD for 4 weeks (Figure 4A). Moderately greater adiposity and dysglycemia was observed in recipients of feces from mice whose dams had been fiber deprived supporting the notion that microbiota composition contributed to the maternal fiber deprivation DIO phenotype (Figure 4B-4F). Next, we probed the extent to which enriching microbiota with Proteobacteria, which was enriched in offspring of dams fed low-fiber diets, might impact DIO. Mice were subjected to microbiota cleanout and then administered GBC feces, supplemented, or not, with a proteobacteria enriched consortium (PEC), which 16S rRNA gene sequencing indicated was comprised of 98% of proteobacteria, predominantly of the Aggregatibacter and Sutterella genera (Fig 4G). Such mice were maintained on GBC for 4 weeks, then subjected to 4 weeks of LF-HFD administration (Figure 4H). Addition of PEC to the fecal transplant did not significantly change body weight, but yet resulted in moderately increased glucose intolerance and altered the ratio of epididymal fat mass to body wight (Figure 4I-4L). A similar pattern of enhanced DIO (Figure 4M and 4N) was attained by supplementing fecal suspensions with only a single Proteobacteria isolate, namely an Aggregatibacter strain that had been previously isolated from dysbiotic mice in our vivaria. 22 Such enhanced DIO associated with an increased colonic M1/M2 ratio and mild neutrophil influx 4 weeks post-FMT (Figure S2L and S2M). These results further support the hypothesis that alterations in gut microbiota, particularly those that result in elevated levels of Proteobacteria, contribute to the proneness to obesity that results from maternal fiber deprivation.
Figure 4. Microbiota from offspring of fiber-deprived dams exacerbated diet-induced obesity in receipt mice after transplantation.
(A) Scheme. Germ free mice (N=5) were administered a fecal microbiota transplant (FMT) from 12-week-old offspring of GBC, LF-LFD, or LF-HFD fed dams. 4 weeks post-FMT, mice were administered a 4-week course of LF-LFD feeding and then euthanized.
(B&C) Epididymal adipose tissue of the recipient mice (B) and calculated as percentage of body weight (C).
(D-F) 28d post-LF-HFD treatment, mice were fasted overnight and basal glucose (D), intraperitoneal glucose tolerance (E) assayed. AUC of E is calculated (F).
(G) Genus level of composition of the fecal proteobacteria enriched consortium (PEC) utilized in H-L.
(H) Experimental scheme for I-L and M-N. 3-week-old conventional mice (CV) were “bowel cleansed” (via PEG) and then administered GBC-fed mouse fecal suspension +/− PEC (I-L) or an Aggregatibacter isolate (M-N). Mice (N=5) were maintained on GBC followed a 4-week (I-L) or 2-week (M-N) course of LF-HFD feeding.
(I&J) Glucose tolerance (I) and resulting AUC (J).
(K-N) Adiposity. One-way ANOVA (B, C, D, E, F) or Student’s t test (I, J, K, L, M, N). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; #p<0.05, #### p < 0.0001. In E, * indicated GF: mLF-LFD vs GF: mGBC; # indicated GF: mLF-HFD vs GF: mGBC. Also, see Figure S2.
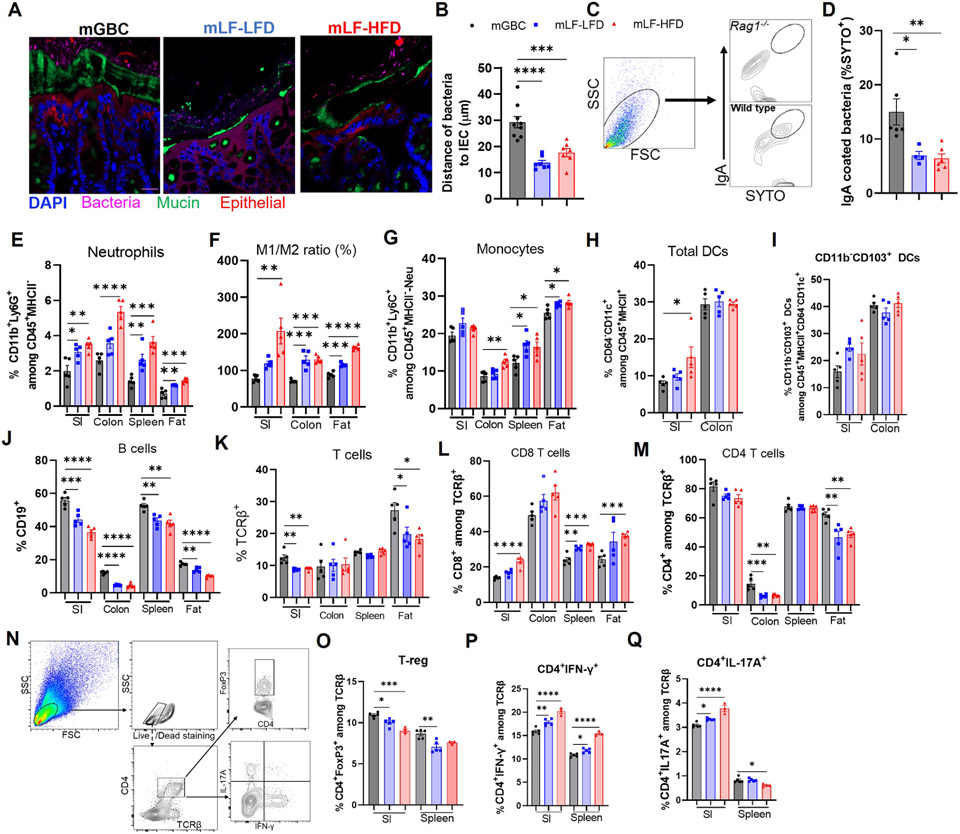
Maternal fiber deprivation results in microbiota encroachment and low-grade inflammation
We next sought to investigate how lack of maternal fiber impacted the mucosa in offspring at 12-weeks of age, at which time they had consumed GBC for 9 weeks and thus had only modest adiposity but were very prone to DIO. Assay of microbiota localization by confocal microscopy using non-dehydrating tissue fixation, which is known to preserve mucoid structure, 23 found a more generally frequent encroachment of bacteria into the inner mucus of colon (Figure 5A). Measuring the distances of the closest bacteria to the epithelium across multiple view fields and specimens indicated that, indeed, maternal fiber deprivation resulted in a significant degree of microbiota encroachment in 12-week-old GBC-fed offspring (Figure 5B). This phenotype associated with a reduction in the percentage of fecal bacteria coated with IgA (Figure 5C and 5D). In these offspring mice, we did not observe alterations in expression of tight junction protein claudin-1 or mucus thickness (Figure S3A and S3B), which are reduced in mice that directly consume low fiber diet. 18,24 Maternal fiber deprivation-induced microbiota encroachment associated with increased fecal levels of the antibacterial protein/inflammation marker lipocalin-2 (Figure S3C) and higher levels of serum anti-flagellin and anti-LPS antibodies, whereas total serum concentration of IgG1 and IgG2C did not change (Figure S3D-S3G). Consistent with these indices of low-grade inflammation, flow cytometric analysis, found that, in both ileum and colon, maternal fiber deprivation resulted in offspring having significantly increased neutrophils, monocytes, M1 macrophage as well as an elevated M1/M2 macrophage ratio, and a trend of elevated ileal DC in small intestine, especially CD11b− CD103+, which have potential to promote inflammatory T cells and cross-prime CD8 T cells (Figure 5E-5I, S3H-S3K). Analysis of lymphocytes revealed reduced B- and T-cell frequencies and number, especially in small intestine (Figure 5J and 5K, S3L and S3M). Analysis of T-cells subsets showed a modest increase in CD8, and decrease in CD4, cells in intestines of offspring of fiber-deprived dams (Figure 5L and 5M, S3N-S3O). A similar pattern was observed in spleen and adipose tissue, which also showed an increase in neutrophils, monocytes and the M1/M2 ratio (Figure 5E-5G, S3P-S3S). We also observed modest reduced frequencies of B and T lymphocytes. A portion of this difference associated with a reduction in the absolute number of these cell types but also likely reflected total increase in splenocytes in that, for example, the absolute number of B cells in spleen did not change significantly (Figure 5J-5M, S3T-S3W). Further CD4 T-cell analysis revealed that maternal fiber deprivation led to modest reductions in regulatory T cells and a modest increase in IL-17+ and IFNγ+ cells (Figure 5N-5Q, S3X-S3Z). Collectively, these findings support the notion of lasting low-grade inflammation in offspring of fiber-deprived dams.
Figure 5. Maternal diet impacts microbiota localization and pro-inflammatory potential in offspring.
(A-B) Microbiota-mucus-epithelial localization in colon of 12-week-old offspring via FISH. Representative confocal images (A) and average bacterial-epithelial distances per HPF (B).
(C&D) Bacterial IgA coating. Gating scheme in (C), using Rag−/− feces as neg control, was used to discern the percentage of fecal bacteria coated with IgA (D).
(E-M) Flow cytometric analysis of intestinal immune cells in 12-week-old offspring (N=5) as gated in Figure S1, Neutrophils (Neu), monocytes, M1/M2 macrophage, Dendritic cells (DC), B and T lymphocytes were quantitated.
(N-Q) Functional T-cell subset analysis (N=3-5). Small intestinal lymphocytes and splenocytes were stimulated with PMA ex vivo and analyzed by FACS gating in (N). Percentage of regulatory T cells (O), IFN-γ+ (P), IL-17+ T cells(Q) were calculated. One-way ANOVA: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. See Figure S3&S4
Maternal fiber deprivation associated with increased energy intake in offspring
Low-grade inflammation and perturbations of microbiota homeostasis can associate with increased epithelial proliferation. 25 Accordingly, maternal fiber deprivation resulted in increased Ki67 staining and increased migration of administered BrDu, indicating increased epithelial proliferation and migration, respectively (Figure S4A-S4D). Such alterations associated with longer villi (Figure S4E), which we hypothesized might facilitate uptake of lipids that are abundant in the LF-HFD diet. In accord with this notion, we observed elevated serum TG following an administered lipid challenge (Figure S4F). We envisioned that another possible consequence of increased proliferation/migration might be reductions in populations of more differentiated enterocytes such as L-cells that produce metabolic regulators, including GLP-1. Accordingly, we observed fewer GLP-1+ L-cells and reduced GLP1 mRNA levels in offspring of fiber-deprived dams (Figure S4G-S4I). GLP-1’s broad impacts include promoting satiety thus potentially explaining why maternal fiber deprivation led to a 15% increase in GBC consumption in 12-week-old offspring (Figure S4J). The crumbly consistency of LF-HFD makes measuring its consumption technically challenging prompting us to examine the role of hyperphagia in exacerbation of DIO by administering LF-HFD ad libitum or via pair-feeding. Specifically, offspring of GBC, LF-LFD, or LF-HFD fed dams were administered a precise amount of LF-HFD each day equal to about 90% of the amount that age-matched control mice ate when fed ad libitum. Such food restriction largely, albeit not completely normalized adiposity and blood glucose levels (Figure S4K-S4N) indicating hyperphagia plays a major role in but may not entirely explain, the exacerbated DIO that results from maternal fiber deprivation. More generally, these results indicate that offspring of fiber-deprived dams are primed for high energy intake and storage, particularly when given access to energy-rich diets.
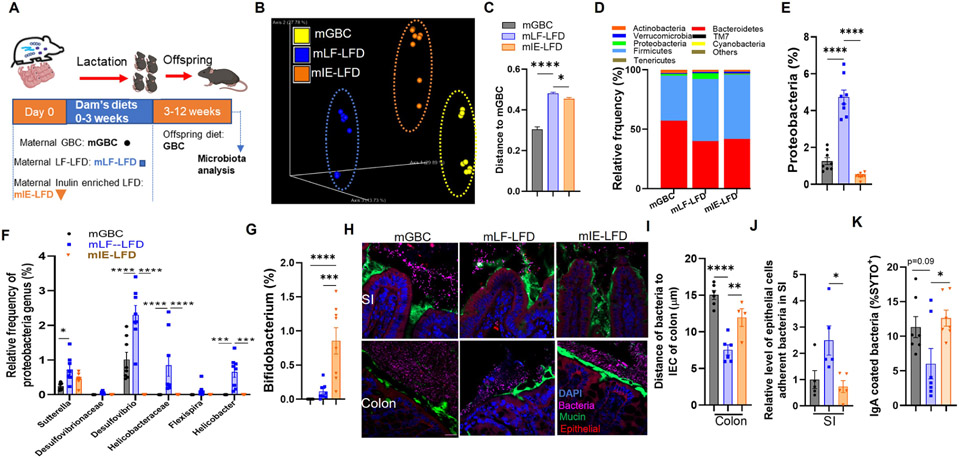
Enrichment of maternal diet with inulin ameliorates maternal fiber deprivation induced dysbiosis and its associated consequences
Many of the detrimental impacts of low-fiber diets on intestinal and metabolic phenotypes in non-lactating mice can be prevented by enriching such diets with the fermentable fiber inulin. 14,26 We thus investigated whether enriching dams’ low-fiber diets with inulin might protect their offspring from severe DIO. We compared offspring of dams administered lactating diets of GBC, LF-LFD, or inulin-enriched low-fat diet (IE-LFD) (Figure 6A). First, we examined impacts on microbiota composition, which, in non-lactating mice, is altered by inulin but not in a manner that restores microbiomes to those of GBC-fed mice. 14 We herein observed an analogous result in offspring of mice consuming these diets. Specifically, 12-week-old offspring, which had consumed GBC for 9 weeks, displayed fecal microbiomes that clustered based on dam diet but measure of Uni-Frac distances showed that the difference of maternal GBC vs. IE-LFD feeding was only slightly less than that of GB vs. LF-LFD (Figure 6B and 6C). Nonetheless, offspring of mice fed IE-LFD lacked a cardinal feature of dysbiosis, namely the expansion of Proteobacteria (Figure 6D and 6E). Analysis of this phyla at the family level showed that the reduction in Protobacteria was seen across multiple families, including Desulfovibrio, which was been associated with severe DIO (Figure 6F).27 An additional known and presumed beneficial impact of inulin consumption is a marked increase in Bifidobacterium to levels well beyond that observed in GBC-fed mice. 28 Such elevated Bifidobacterium manifested in offspring of dams consuming IE-LFD compared to those LF-LFD fed dams (Figure 6G), while in vitro studies found that Bifidobacterium was capable of impeding growth of a proteobacteria isolate, namely an Aggregatibacter strain (Figure S5A). Enriching maternal LF-LFD with inulin also alleviated aberrant microbiota localization. Specifically, assaying of microbiota localization by confocal microscopy found that offspring of mice fed LF-LFD, but not GBC or IE-LFD, revealed bacteria very close to or beyond the epithelium (Figure 6H). Measure of microbiota-epithelial distance across multiple fields confirmed that such microbiota encroachment was present in offspring of dams fed LF-LFD but not IE-LFD fed mice (Figure 6I) while qPCR indicated higher levels of bacteria adherent to the ileal epithelium in offspring of dams fed LF-LFD (Figure 6J). Prevention of microbiota encroachment in offspring also associated with increased IgA coated bacteria in the feces (Figure 6K). Enriching maternal LF-LFD with inulin also corrected the above-described impacts on the intestinal mucosa of offspring. Specifically, maternal feeding of LF-LFD but not IE-LFD altered intestinal levels of immune cells (Figure S5B-S5F) and GLP-1-positive cells in colon (Figure S5G-S5I).
Figure 6. Enrichment of maternal diet with inulin ameliorates maternal fiber deprivation-induced dysbiosis in offspring.
(A) Scheme. Male offspring mice of dams fed indicated diets during lactation were weaned onto GBC at 3 weeks of age and feces collected 12 weeks of age.
(B-C) Fecal microbiota composition via 16S rRNA gene sequencing (N=8) as expressed by unweighted UniFrac PCoA plots (B) and Unifrac distances (C).
(D-G) Relative phylum abundance (D), including proteobacteria phylum (E), proteobacteria (F) and bifidobacterial genera (G).
(H-J) Microbiota-mucus-epithelial localization in colon of 12-week-old offspring via FISH. Representative confocal images (H), average bacterial-epithelial distances per HPF (I).
(J) Bacteria adherent to epithelial cells in small intestine was measured by qPCR.
(K) Bacterial IgA coating. One-way ANOVA: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. See Figure S5
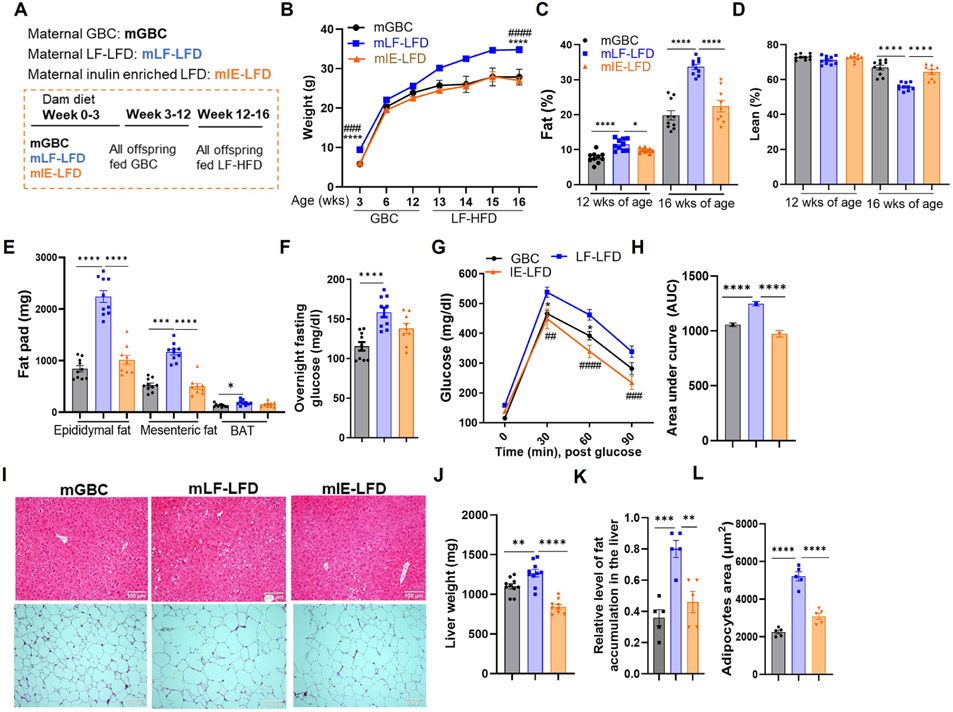
Enriching maternal LF-LFD with inulin also prevented their offspring from being highly prone to development of obesity (Figure 7). Specifically, both the modest basal increase in basal weight and adiposity and the striking degree of DIO was normalized by enriching the low-fiber maternal diet with inulin (Figure 7A-7H). Analysis of livers and adipose tissue yielded parallel results indicating that the inulin enrichment protected against other manifestation of diet-induced obesity (Figure 7I-7L). Moreover, a very similar pattern of results was observed in female offspring of dams fed GBC, LF-LFD, and IE-LFD, particularly in response to the LF-HFD (Figure S6A-S6E). Such alleviation of exacerbated DIO by enrichment of the maternal diet with inulin associated with reduced indices of inflammation as reflected by levels of proinflammatory cytokines, neutrophils, and the M1/M2 macrophage ratio in liver, adipose tissue, and intestine (Figure S6F-S6O). Collectively, these results indicate that consumption of fermentable fiber by lactating mothers can have lasting impacts on their offspring’s host-microbiota relationship and, consequently, metabolic phenotypes.
Figure 7. Enrichment of maternal diet with fermentable fiber inulin reduced offspring DIO.
(A) Scheme. Pups (N=9-10) from dams fed indicated lactating diet were weaned onto a GBC diet until 12 weeks of age, then fed LF-HFD.
(B-D) Body weight (B) and whole-body fat (C) and lean (D) mass content via MRI.
(E) Epididymal, mesenteric and brown adipose tissue (BAT) weight measured upon euthanasia.
(F-H) Fasting glucose level (F), glucose tolerance (G), and resulting AUC (H).
(I) H&E staining of liver and adipose fat (representative images).
(J-L) Liver weight (J), Relative level of fat accumulation in liver (K) and average adipose cells area (L). One-way ANOVA: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ##p<0.01, ###p<0.001, #### p < 0.0001. See Figure S6. In B and G, * indicated mLF-LFD vs mGBC; # indicated mLF-LFD vs mIE-LFD. See Figure S6
DISCUSSION
While a broad range of environmental (i.e. non-genetic) factors has likely contributed to the obesity epidemic, changes in diet, particularly the ready-availability of energy-rich foods, has likely played a central role. Yet, the energy-density of western style diets is not their only obesogenic feature. Rather, they are also deficient in fiber, which, at least in mice, leads to changes in gut microbiota that lead to low-grade inflammation, which promotes many of the detrimental metabolic impacts of such diets. 14 In this sense, the low-fiber diets used in this study, which have 25% of the total fiber of standard mouse chow (GBC), are generally reminiscent of fiber consumption of most people living in developed countries whom typically consume only 15 grams of fiber per day; less than half of USDA recommendations, which themselves may reflect less than ideal levels of fiber consumption.29 Gut microbiotas are acquired from one’s early life environment prompting us to investigate if low-fiber diets not only impact those who consume them but may impact their offspring as well. We herein report that maternal fiber deprivation, during lactation, resulted in gut microbiota dysbiosis, low-grade inflammation, and increased adiposity in dams and, moreover, their adult offspring, despite such offspring, themselves, consuming a fiber-rich diet. Furthermore, such dysbiotic microbiotas rendered offspring of fiber-deprived dams extremely prone to DIO. These results support the plausibility of the notion that societal transgenerational changes in gut microbiota composition have contributed to the obesity epidemic. More specifically, our findings highlight the importance of maternal fiber consumption in promoting healthy offspring and ameliorating their proneness to obesity.
Benefits of fiber fermentation by gut microbiota include those conferred by fermentation products, namely short-chain fatty acids (SCFA), which provide energy to gut epithelial cells and, furthermore, an array of systemic benefits including improving glycemic control and keeping immune responses in check. 30 Additionally, and possibly more importantly, nourishment of microbiota by fermentable fiber promotes a dense diverse microbiota, which in turn provides an array of SCFA-independent benefits to the host including colonization resistance and promoting mucosal dynamism, as characterized by robust enterocyte proliferation and secretion of mucus and antimicrobial peptides. 14,31 Collectively, such benefits of fermentable fiber result in a healthy microbiota-host relationship characterized by low levels of pathobionts, which are frequently Proteobacteria, and absence of bacteria in the inner mucus layer. Conversely, the absence of fiber decimates microbiota density and results in those bacteria that remain penetrating, and digesting, the mucus layer and thus encroaching upon the host. This leads to recruitment of inflammatory cells and alterations in lymphocytes. 18 Such low-grade inflammation promotes metabolic dysfunction in general, insulin resistance in particular, and may potentiate severity of DIO.
Our previous work, particularly that enriching a low-fiber diet with inulin restored microbiota density, mucosal dynamism, and metabolic health, but not microbiota composition, argued that total gut bacterial load is a key parameter while high variance in species composition may be well tolerated. 14 In accord with this notion, we herein observed that mice whose dams had been deprived fiber during lactation, but themselves consumed fiber-rich diets exhibited normal bacterial density and grossly normal gut morphology, i.e. lacked the stark reductions in cecal/colon mass, and crypt depth, observed in mice fed low-fiber diets. However, their alterations in microbiota composition that had resulted from maternal fiber deprivation nonetheless led to microbiota encroachment, low-grade inflammation and conferred proneness to DIO. Enriching dam LF-LFD with inulin prevented these outcomes in offspring but, again, but did not restore the microbiota composition per se. Collectively, these results accord with the general notion that microbiota density may be a determinant of gut health in general, but also underscores the role of microbiota composition in maintaining healthy gut mucosa. In particular, our findings suggest that “inheritance” of microbiotas enriched in Proteobacteria, may be one factor that predisposes mice to obesity and perhaps other diseases promoted by low-grade inflammation. 32 Components of Proteobacteria, including flagellin and especially LPS, are potent activators of inflammatory signaling. Thus, we speculate that elevated Proteobacteria abundance and/or their encroachment may increase signaling through innate immune receptors such as TLR4. Indeed, we observed increased anti-LPS antibodies suggesting increased exposure to this product and note that extended exposure to LPS via subcutaneous minipump can directly promoted obesity and its consequences. 33
That newly weaned offspring of fiber-deprived dams exhibited elevations in Proteobacteria reminiscent, albeit to a lesser degree, than that of their dams was not surprising. However, while we anticipated that several weeks of consuming the fiber-rich diet would alleviate the inherited dysbiosis, in fact, elevations in Proteobacteria were greater following 9 weeks of GBC feeding. Viewed together with our observations that co-housing offspring of fiber-deprived dams with those of GBC-fed dams partially normalized level of Proteobacteria and prevented severe DIO, suggests that the dysbiosis that resulted from maternal fiber deprivation may in-part reflect lack of inheritance of beneficial microbes that, when properly nourished by fiber, can keep pathobionts in-check. Thus, our findings can be viewed as an example of the “missing microbes” hypothesis put forth by Blaser 34 and are also reminiscent of findings from Sonnenburg and colleagues that microbiota diversity is progressively reduced over several generations of consuming low-fiber diets. 35 Our experimental approaches were not designed to identify the specific bacteria that help keep pathobionts in-check but, nonetheless we noted that enrich of maternal diet with inulin resulted in offspring having increased abundance of Bifidobacterium, which inhibited Proteobacteria growth in vitro (Figure S5A). Meanwhile, enriching the maternal diet with inulin also reduced offspring Proteobacteria in vivo, and ameliorated proneness to DIO suggesting that some variety of bacteria, likely working in concert can keep pathobionts in-check when adequately nourished. We speculate that the window for such beneficial microbial consortia to engraft and symbiotically may be optimal during periods of immune system development and thus, therapeutic interventions to prevent future obesity might work best in this time window.
The exacerbated DIO associated with greater caloric intake. Yet, increased food intake was just one aspect of their metabolic phenotype. Rather these mice displayed intestinal physiology seemingly geared for maximum energy intake, including overt morphologic changes in the intestine, namely longer villi, that associated with greater ability to harvest lipids when provided in bolus. Such phenotypes associated with lack of intestinal L-cells and concomitant reductions in some of their products, namely GLP-1. We speculate that reduced GLP-1 levels contributed to their hyperphagia and reduced insulin responsiveness, and that this metabolic phenotype was imprinted early in life and, thereafter, maintained by alterations in microbiota. A variety of factors limit the extent to which our studies in mice are applicable to humans. For example, while young humans have a broad range of potential exposures to microbes, mice in our study were only able to acquire microbes from their mother, siblings, or cagemates with whom we deliberately placed them. This raises the possibility that perhaps the beneficial microbes that might be transmitted by human mothers consuming a fiber-rich diet can, in fact, also be readily acquired from other sources. It is also important to acknowledge that alteration of offspring microbiota may be but one of many mechanisms by which maternal fiber deprivation promotes proneness to obesity. Indeed, transplant of feces from mice subjected to maternal fiber deprivation to germfree mice only partially recapitulated the maternal fiber deprivation phenotype. We speculate that additional factors, including impact of diet on milk composition may also play a role. More generally, mice and humans have different nutritional requirements making translation of mouse dietary studies to humans inherently difficult. Such caveats notwithstanding, we submit our results suggest that poorly understood early life events, including microbiota acquisition, can imprint metabolic phenotype thus influencing proneness to obesity.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Andrew Gewirtz (agewirtz@gsu.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The raw sequencing data have been deposited at GenBank with access number: PRJNA873095, and all other data used for this study will be shared upon reasonable request from the lead contact.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Diet fed mice model
C57BL/6 mice were purchased from Jackson Labs (Bar Harbor, ME) and then bred at Georgia State University under approved animal protocols (IACUC # A20043). Mating cages of 2F and 1M 8-week-old mice per cage were administered a grain-based breeder diet (LabDiet). Upon birth, the male mice were removed, and two dams nursing 15-16 liters per cage were fed grain-based chow (Table S1, LabDiet), or a compositionally-defined diet, namely low fiber low fat diet (LF-LFD), Inulin enriched low fat diet (IE-LFD) and low fiber high fat diet (LF-HFD) (Table S1, Research Diets, Inc). At 3 weeks of age, pups were separated from dams (weaned) and by placed in a new cage and fed GBC until reaching 12-weeks of age, at which time they were administered LF-HFD for 4 weeks. Both male and female offspring mice were used in the study as indicated. The mice were euthanized at pup-weaning time, 12 weeks of age or 16 weeks of age as indicated. Serum, feces, and various organs including small intestine, colon, liver, and adipose tissue, were collected to measure or store for later analysis.
Bacteria strain and culture condition
Bifidobacterium longum subsp. longum Reuter was obtained from the American Type Culture Collection (ATCC, 15707). Aggregatibacter was isolated from immunodeficient mice in our animal core facility as described in our previous publication. 22 Both bacteria strains were cultured in brain heart infusion (BHI) medium at 37 °C in anaerobic condition.
METHODS DETAILS
Antibiotic treatment
Where indicated, 8-week-old offspring were administered antibiotics via drinking water containing ampicillin (1 g/L) and neomycin (0.5 g/L). These mice were maintained on GBC until 12 weeks of age, then fed with LF-HFD until 16 weeks of age. The antibiotic treatment was maintained throughout the LF-HFD exposure or discontinued shortly before the 4-week LF-HFD exposure as indicated.
Co-housing experiment
For co-housing, male pups from dams consumed GBC (Pup-GBC) or LF-LFD (Pup-LF-LFD) during lactation were weaned at three weeks old, Pups-GBC and Pups-LF-LFD were co-housed in single cages with non-co-housed siblings serving as controls. These co-housed and non-co-housed siblings were fed with GBC until 12 weeks of age, then administered LF-HFD for 4 weeks. Feces were collected at 12 weeks of age for microbiota analysis. Mice were euthanized at 16 weeks of age after LF-HFD, adipose tissue was weighted and immune cells in the colon and adipose tissue were analyzed by FACS.
Fecal microbiota/bacteria transplantation
To transplant feces from offspring of fiber-deprived mice, feces were collected from offspring mice at 12 weeks of age and suspended in PBS as 100 mg/ml. Germ-free 8-week-old C57BL/6 male mice were purchased from Taconic Inc, and orally administered with 200 μl of above fecal suspension. Recipient mice were maintained in isocages to prevent other bacterial exposures. Transplants were also conducted using fecal suspensions of unmanipulated mice +/− proteobacteria-enriched consortia (PEC) or an Aggregatibacter isolate. As previously described, 36 PEC was generated from feces was collected from mice 1 day after the cessation of antibiotic cocktail including ampicillin (1 g/liter), vancomycin (250 mg/liter), neomycin (1 g/liter), and metronidazole (1 g/liter), and was resuspended in PBS as 100 mg/ml. PEC and isolated Aggregatibacter, were added to fecal suspension and transplanted as previously describe: 37 3-week old C57BL/6 male mice was treated with a bowel cleansing with 1.2 ml of polyethylene glycol solution (PEG 3350 (77.5 g/L), sodium chloride (1.9 g/L), sodium sulfate (7.4 g/L), potassium chloride (0.98 g/L) and sodium bicarbonate (2.2 g/L), then orally gavage with 100 μl of suspension of feces from control GBC-fed mice supplemented, with 100 μl proteobacteria enriched feces, or 105 CFU of Aggregatibacter for 3 consecutive days. After transplantation, these mice were fed with autoclaved GBC for 4 weeks, then administered with LF-HFD for another 4 weeks, except conventional mice with Aggregatibacter for only 2 weeks of LF-HFD.
Body fat and lean percentage measurement.
Offspring mice from dams consumed GBC, LF-LFD, LF-HFD or IE-LFD were subjected to body composition analysis at 12, 14 or 16 weeks of age, by using Minispec NMR body composition analyzer (Bruker Biospin Corporation; Billeria, MA). Data were expressed as percentage of fat or lean.
Glucose measurement
Mice were provided with water without food for 5 or 16 h in a clean cage as indicated. Fasting blood glucose level was measured by using a Nova Max Plus Glucose Meter and expressed in mg/dL. Glucose tolerance test was conducted as following: after fasting for 5 h or 16 h, the baseline blood glucose was measured by using a Nova Max plus Glucose meter, then injected with 2 mg of glucose per gram of body weight intraperitoneally. To conduct insulin tolerance test, 5-h-fasted mice were intraperitoneally injected with 0.75 U insulin/kg body weight. After injection with glucose or insulin, the blood glucose levels were measured at 30, 60, 90 min post-dosing.
Measurement of food consumption
Offspring mice from dams fed with maternal diets including GBC, LF-LFD and LF-HFD, were fed with GBC until 12 weeks of age, then were placed in a clean cage with a known amount of GBC food. Twenty-four hours later, the remaining food was measured to calculate the food consumption per day.
Food restriction experiment
Mice were individually placed in a clean cage supplied with 2.8 g LF-HFD every day for 10 days, which we estimated to be 80-90% of what age-matched offspring of GBC-fed mice would eat when administered LF-HFD ad libitum.
Lipid absorption measurement
Mice were fasted for 16 h, then orally gavaged with olive oil (10μ1/ g body weight), after 30 min post tyloxapol i.p injection (250 μg/g of body weight). Blood was collected from these mice via tail or retro-orbital bleed at 0, 2, 4 and 6 h after oil administration. Serum was collected after centrifugation to measure triglyceride concentrations using Infinity triglyceride Reagent (Thermo Scientific, TR22421) according to manufacturer’s instructions.
Measurement of Triglyceride and Cholesterol
Infinity Triglyceride (Thermo Scientific, TR22421) and Cholesterol Reagent (Thermo Scientific, TR13421) were used to measure Triglyceride and cholesterol levels in serum following manufacturer’s instruction. Briefly, 96-well plate was sequentially added with 2 μl serum and 200 μl Triglyceride or Cholesterol Reagent, then incubated for 5 min at 37°C before measuring by using a 96-well plate reader at wavelength of 550 nm.
Immunofluorescence Staining
Intestine, including jejunum and ileum, colon was collected, embedded in OCT, then be sectioned at 4 μm thickness. The tissue sections were then fixed with 4% paraformaldehyde for 30 min at room temperature (RT), and then washed with PBS before being permeabilized in cold methanol for 5 min. The intestinal section was blocked with 5% FBS, then incubated with anti-GLP1 antibody (Abcam, Ab22625) or Anti-Ki67 antibody (Abcam, Ab15580) overnight at 4°C. After washing with PBS for three times, the sections were stained with Alexa Fluor 488 Goat Anti-Rabbit IgG or Alexa Fluor 555 Goat Anti-Rabbit IgG. The tissues were counterstained with mounting medium containing DAPI (SouthemBiotech, 0100-20), then imaged by fluorescent microscope. The density of GLP1 positive L cells and fluorescent intensity of Ki67 were measured by using image J software.
Brdu Staining
Epithelial cells migration was measured as previously described. 38 Briefly, Offspring mice at 12 weeks of age were injected with 5-bromo-2-deoxyuridine (BrdU) with 50 μg per mg of mice body weight, then were euthanized after 24 h post injection. 1 cm of jejunum was collected and imbedded in OCT, then sectioned at 4 μm thickness before fixed with 4% paraformaldehyde for 30 min at RT. After washing with PBS, the section was denatured by incubating in prewarmed 1.5 N HCl for 30 min at 37 °C, then washed with PBS for three time. Sections were blocked with 5% FBS for 1 h at RT, then stained with Alexa Fluor® 488 Anti-BrdU antibody (Abcam, ab74545) for 2 hours at RT, and counterstained with mounting medium containing DAPI. The BrdU-labeled cells were visualized by fluorescence microscopy, and the migration was measured by Image J software.
Hematoxylin and eosin stain
Following euthanasia, livers and epididymal fat was fixed in 10% phosphate-buffered formalin for at least one week, before transfer into 70% ethanol. The tissue was embedded in paraffin and sectioned at 4 μm thickness before hematoxylin and eosin (H&E) staining according to a standard protocol. 39 The relative level of fat accumulation in the liver was assessed after HE staining. The size of adipocytes was measured by using image J software.
Flow cytometry analysis
Mouse splenocytes, immune cells in fat, and intestinal lamina propria cells were prepared as previously described. 40 Briefly, splenocytes were released by using the flat end of plunger to mince the spleen, then passed through 70 μm cell strainer. The cells suspension was centrifuged at 1,600 rpm for 5 minutes. The cell pellet was resuspended in 3 mL of red blood cells lysing buffer, then washed with PBS. To isolate immune cells in epididymal fat, fat was digested in 10 mL of 1 mg/mL of type IV collagenase in HBSS with 5% FBS for 20 minutes at 37°C as previously described. 21 After digestion, EDTA was added to a final concentration of 10 mmol/L and incubated at 37°C for an additional 10 minutes. The suspension was passed through 100 μm and 40 μm strainer sequentially, then centrifuged at 1600 rpm for 10 minutes at 4°C. The pellet was resuspended in 3 mL of red blood cell lysis buffer before washing with PBS. To isolate lamina propria cells in colon and small intestine, the intestines were cut into 1 cm long pieces and placed into 20 mL of HBSS with 5% fetal bovine serum (FBS) and 2 mmol/L EDTA and incubated with shaking for 20 minutes at 37°C for two times, then incubated in 20 mL HBSS with 5% FBS, 1 mg/mL type IV collagenase (Sigma, C5138-5G), and 0.1 mg/mL DNase I (Sigma, 10104159001) with shaking for 10 minutes at 37°C. Finally, cells were passed through 40 μm cell strainer, and centrifuged. After isolation, all cells were washed with 200 ul of cold phosphate-buffered saline (PBS) twice and incubated with anti-mouse CD16/CD32 for Fc-blocking (BioXCel). Cells were then washed twice with fluorescence-activated cell sorting (FACs) buffer (phosphate-buffered saline with 0.5% bovine serum albumin). Cells were stained with fluorescently labeled antibodies purchased from Biolegend, BD Biosciences, eBioscience, or Thermofisher in FACS buffer as listed in the key resources table. Staining cocktails included Anti-CD45 BV605 (1:800), anti-Ly6G AF700 (1:200), anti-Ly6C PerCp-Cy5.5 (1:200), anti-CD11b APC-Cy7 (1:200), anti-CD11c BV785 (1:100), anti-IA/IE BV650 (1:1000), anti-CD64 BV421 (1:400), anti-F4/80 (1:100), anti-CD103 PE (1:200), and anti-CD301 APC (1:200), or a mixture of antibodies containing anti-TCRβ APC (1:200), anti-CD4 PerCP-Cy5.5 (1:200), anti-CD8α pacific blue (1:100) and anti-CD19 FITC (1:200), was added into the cells and incubated on ice, in dark for 30 minutes. For analysis of T cell function, lamina propria cells from intestine and splenocytes isolated from spleen of offspring mice, were administered a stimulation cocktail, with protein transported inhibitor (Thermo Scientific, 00-4975-93), for 3 hours in vitro. Cells were the stained with anti-CD4 APC-AF780 (1:200) before fixing and permeabilizing using Fix/Perm buffer set, then intracellularly stained with antibodies including anti-FoxP3 APC (1:100), anti-IFN-γ BV785 (1:100) and anti-IL-17 PE (1:100) in FoxP3 staining buffer (Thermo Scientific, 00-5523-00). Cells were washed three times with FACs buffer and resuspended in 200 ul of FACs buffer. Multi-parameter analysis was performed on a CytoFlex (Beckman Coulter) and the percentage of different immune cells population analyzed using FlowJo software (Tree Star). For measurement of monocytes, we subtracted neutrophils from CD11b+&Ly6C+ population.
KEY RESOURCES TABL
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-BrdU antibody | Abcam | Cat# ab74545 RRID: AB_1523224 |
| Rabbit polyclonal to Ki67 | Abcam | Cat# Ab15580 RRID: AB_443209 |
| Anti-Mucin 2 | Santa Cruz Biotechnology | Cat# H-300 |
| Anti-GLP1 | Abcam | Cat# Ab22625 RRID: AB_447206 |
| Alexa Fluor 488 Goat Anti-Rabbit IgG | Invitrogen | Cat# A11008 RRID: AB_143165 |
| Alexa Fluor 555 Goat Anti-Rabbit IgG | Abcam | Cat# Ab150078 RRID: AB_2722519 |
| Anti-mouse CD16/CD32 | BioXCel | Cat# BE0307 |
| Anti-CD45 | Biolegend | Cat# 103155 RRID: AB_2650656 |
| Anti-CD11b | BD Biosciences | Cat# 557657 RRID: AB_396772 |
| Anti-Ly6G | BD Biosciences | Cat# 561236 RRID: AB_10611860 |
| Anti-Ly6C | Thermo fisher | Cat# 45-5932 RRID: AB_2723342 |
| Anti-CD11c | BD Biosciences | Cat# 563786 RRID: AB_2732919 |
| Anti-MHCII | BD Biosciences | Cat# 563415 RRID: AB_2738192 |
| Anti-CD64 | Biolegend | Cat# 139309 RRID: AB_2562694 |
| Anti-CD19 | Biolegend | Cat# 152404 RRID: AB_2629813 |
| Anti-IFN-γ | Biolegend | Cat# 505838 RRID: AB_2629667 |
| Anti-mouse IgA | eBioscience | Cat# 12-4204-82 |
| Anti-CD8α | Biolegend | Cat# 100725 RRID: AB_493425 |
| Anti-CD45 | Biolegend | Cat# 103155 RRID: AB_2650656 |
| Anti-CD4 | Thermo fisher | Cat# 47-0042-82 RRID: AB_1272183 |
| Anti-IL17A | BD Biosciences | Cat# 559502 RRID: AB_397256 |
| Anti-FoxP3 | Biolegend | Cat# 320014 RRID: AB_439750 |
| Anti-TCRβ | Biolegend | Cat# 109212 RRID: _313435 |
| Anti-CD4 | Biolegend | Cat# 100434 RRID: AB_893324 |
| Anti-mouse IgG antibody | Sigma-Aldrich | Cat# M2650-1ML RRID: AB_260486 |
| Anti-mouse IgG1 | SouthernBiotech | Cat# 1070-08 AB_2794413 |
| Anti-mouse IgG2C | SouthernBiotech | Cat# 1079-08 AB_2794467 |
| Normal Rat Serum | Thermo Scientific | Cat# 10710C RRID: AB_2532985 |
| Bacterial and virus strains | ||
| Bifidobacterium longum subsp. longum Reuter | ATCC | Cat# 15707 |
| Chemicals, peptides, and recombinant proteins | ||
| D- (+)-GLUCOSE | Sigma-Aldrich | Cat# G8270-1KG |
| Insulin | Sigma-Aldrich | Cat# 12643-25MG |
| Ampicillin | Sigma-Aldrich | Cat# A9518-100G |
| Neomycin | Sigma-Aldrich | Cat# N1876-100G |
| PEG 3350 | Sigma-Aldrich | Cat# P4338 |
| Tyloxapol | MedChemExpress | Cat # HY-B1068 |
| SytoBC | Invitrogen | Cat# S34855 |
| Lipopolysaccharides | Sigma-Aldrich | Cat# l2887-10MG |
| Phalloidin Tetramethyl rhodamine B isothiocyanate | Sigma-Aldrich | Cat# P1951-.1MG |
| DAPI Fluoromount-G® Mounting Medium | SouthernBiotech | Cat# 0100-20 |
| Stimulation cocktail | Thermo Scientific | Cat# 00-4975-93 |
| Brain heart infusion (BHI) medium | Sigma-Aldrich | Cat# 237500 |
| Paraformaldehyde | Thermo Scientific | Cat# J19943-K2 |
| Type IV collagenase | Sigma-Aldrich | Cat# C5138-5G |
| DNase I | Sigma-Aldrich | Cat# 10104159001 |
| Critical commercial assays | ||
| Triglycerides Reagent | Thermo Scientific | Cat# TR22421 |
| Cholesterol Reagent | Thermo Scientific | Cat# TR13421 |
| One-Step RT–PCR Kit with SYBR Green | Bio-Rad | Cat# 172-5151 |
| QIAamp DNA Stool Mini Kit | Qiagen | Cat # 51504 |
| QuantiFast SYBR Green PCR kit | Bio-Rad | Cat# 204054 |
| Lipocalin-2/NGAL ELISA | R&D Systems | Cat# DY1857 |
| FoxP3 staining buffer | Thermo Scientific | Cat# 00-5523-00 |
| Deposited data | ||
| 16S rRNA sequence | NCBI Sequence Read Archive | PRJNA873095 |
| Experimental models: Organisms/strains | ||
| Mice: C57BL/6 | The Jackson Laboratory | Cat # 000664 |
| Oligonucleotides | ||
| EUB338 probe: 5’-GCTGCCTCCCGTAGGAGT-3’, with a 5' labeling using Alexa 647 | Invitrogen | NA |
| 16S rRNA: 5′-AGAGTTTGATCCTGGCTCAG-3′ | Invitrogen | NA |
| 16S rRNA: 5′CTGCTGCCTCCCGTAGGAGT-3′ | Invitrogen | NA |
| 16S rRNA sequence primers of V4: 515FB 5′TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGTGYCAGCMGCCGCGGTAA-3′ | Invitrogen | NA |
| 16S rRNA sequence primers of V4: 806RB 5′GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGGACTACNVGG | Invitrogen | NA |
| 16S rRNA sequence primers of V3-V4: 5′TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′; | Invitrogen | NA |
| 16S rRNA sequence primers of V3-V4: 5′GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3′. | Invitrogen | NA |
| 18S rRNA: 5′GTAACCCGTTGAACCCCATT3′ | Invitrogen | NA |
| 18S rRNA: 5′CCATCCAATCGGTAGTAGCG3′ | Invitrogen | NA |
| IL-6: 5′GTGGCTAAGGACCAAGACCA3′ | Invitrogen | NA |
| IL-6: 5′GGTTTGCCGAGTAGACCTCA3′ | Invitrogen | NA |
| TNF-α: 5′CGAGTGACAAGCCTGTAGCC3′ | Invitrogen | NA |
| TNF-α: 5′CATGCCGTTGGCCAGGA3′ | Invitrogen | NA |
| MCP-1: 5′GCTGGAGCATCCACGTGTT3′ | Invitrogen | NA |
| MCP-1: 5′TGGGATCATCTTGCTGGTGAA3′ | Invitrogen | NA |
| Claudin-1: 5′ TCCTTGCTGAATCTGAACA3′ | Invitrogen | NA |
| Claudin-1: 5′AGCCATCCACATCTTCTG3′ | Invitrogen | NA |
| 36B4: 5′TCCAGGCTTTGGGCATCA3′ | Invitrogen | NA |
| 36B4:5′CTTTATTCAGCTGCACATCACTCAGA3′ | Invitrogen | NA |
| Software and algorithms | ||
| GraphPad Prism | GraphPad Software | NA |
| Image J | NIH | NA |
IgA-coated bacteria analysis
IgA coated bacteria were analyzed as previously described. 41,42 Briefly, fecal pellets were collected from the offspring mice at 12 weeks of age or Rag1 ko mice (as negative control), resuspended in PBS as 100mg/ml, before filtering through 100 μm and 40 μm nylon meshes. 200 μl passthrough was pelleted by centrifugation at 10,000 rpm for 1 min. The resulting bacterial pellet was washed with PBS for 2 times before blocked with 20% normal rat serum and incubated for 20 min on ice. After blocking, the bacteria were stained with PE-conjugated Anti-mouse IgA (eBioscience, 12-4204-82) with a dilution of 1:50 in staining buffer (1% BSA in PBS) for 30 min on ice. After three-time washes with staining buffer, bacterial pellet was resuspended with staining buffer containing a 1:5000 dilution of SytoBC (Invitrogen, S34855). The samples were washed with staining buffer for two times before analyzed by CytoFlex (Beckman Coulter).
Microbial co-culture in vitro
Bifidobacterium longum subspecies and Aggregatibacter were pre-cultured in BHI media overnight at 37 °C in anaerobic conditions, and bacteria enumerated by serial dilution and colony counting. Aggregatibacter was cultured alone or co-cultured with Bifidobacterium at a ratio 1:1 (6×l05cfu/ml VS 6×105 cfu/ml) in BHI media in anaerobic conditions for 30 h. Samples were harvested after culture and the relative level of Aggregatibacter analyzed by qPCR. Briefly, 0.5 ml medium of bacteria culture was centrifuged, and subsequently the pellet was resuspended in 0.5 ml PBS and boiled for 10 min before centrifuging at 12000 rpm for 10 min to collect the supernatant. 1 μl of the supernatant was used as DNA template for qPCR using Pasteurellaceae specific primers: 5’-CATAAGATGAGCCCAAG-3; 5’ -GTCAGTACATTCCCAAGG-3 with using QuantiFast SYBR Green PCR kit (Bio-Rad, Hercules, CA). Results are expressed as the ratio of 2−CT between non-cocultured and cocultured.
ELISA
Fecal samples were collected from offspring mice at 12 weeks of age, homogenized in PBS at 100 mg/ml, then centrifuged at 12,000 rpm for 10 min and the supernatant were stored at −80°C. A DuoSet Mouse Lipocalin-2/NGAL ELISA (R&D Systems, DY1857) kit was used to measure the level of Lipocalin-2 in the supernatant of the feces according to its provided instructions. Flagellin and LPS specific antibody in serum was measured according to previous described. 43 Briefly, 96 well EIA/RIA corning plate were coated with purified LPS (2 mg/well) or flagellin (100ng/well) before serum with 1:500 dilution was applied to the coated plate. After washing for three times, the wells were incubated with anti-mouse IgG-HRP (1:1000), followed by standard ELISA analysis. To measure total serum IgG1 and IgG2C, 96 well EIA/RIA coming plate was coated with anti-mouse IgG antibody (Sigma, M2650-1M) overnight at RT. The plate was washed with washing buffer (0.05% Tween 20 in PBS) and blocked with 1% milk in PBS for 1 hour at RT. The serum was dilated 1:500 with 0.5% normal goat serum in PBS and added in the plate. After incubation for 1h, the plate was washed and added with 100 ul/well of biotin labeled anti-mouse IgG1 (Southern Biotech, 1070-08) or IgG2C (Southern Biotech, 1079-08) at 1:500 dilution in 1% milk in PBS for 1 hour at RT before incubating with streptavidin-HRP. After three washings, TMB solution was added to develop color.
qRT-PCR
Following euthanasia, colon, liver and epididymal fat were collected. Total RNA was isolated from colon, liver and epididymal fat using TRIzol (Invitrogen, Carlsbad, CA) according to its provided instructions, mRNAs of liver and fat were further purified using the RNeasy mini kit RNA cleanup procedure (Qiagen, Valenica, CA). The expression level of IL-6, TNF-α and MCP-1 in liver and fat, claudin-1 in colon was analyzed by using quantitative RT-PCR (qRT-PCR) according to the Biorad iScript™ One-Step RT-PCR Kit in a CFX96 apparatus (Bio-Rad, Hercules, CA) with the primers listed in the key resources table. Difference in transcript levels were quantified by normalization of each amplicon to housekeeping gene 36B4.
Bacterial quantification in feces and ileum tissue
To measure the total fecal bacterial load, total DNA was isolated from weighted feces using QIAamp DNA Stool Mini Kit (Qiagen, 51504). The DNA was then subjected to quantitative PCR using QuantiFast SYBR Green PCR kit (Biorad, 204054) with universal 16S rRNA primers 8F: 5′-AGAGTTTGATCCTGGCTCAG-3′ and 338R: 5′-CTGCTGCCTCCCGTAGGAGT-3′ to measure total bacteria number. Results are expressed as bacteria number per mg of stool, using a standard curve. To measure bacteria that adherent to the ileum tissue, 2 cm distal ileum was cut, and the content was flushed out, ileum tissue was washed in PBS for three times before subjected to extract DNA by using QIAamp DNA Stool Mini Kit. The extracted DNA was used as template to measure bacteria and host cells by using universal 16S rRNA primers 8F and 338R, 18S rRNA (5′-GTAACCCGTTGAACCCCATT-3′, 5′-CCATCCAATCGGTAGTAGCG-3′) respectively. Difference in the relative abundance of bacteria in ileum tissue were quantified by normalization of each amplicon to host cells housekeeping gene 18S rRNA.
Bacteria localization by FISH staining
Ileum and colon were placed in methanol-Carnoy's fixative solution (60% methanol, 30% chloroform, 10% glacial acetic acid) for a minimum of 3 h at room temperature. The localization of bacteria in ileum and colon was stained by using 16s rRNA FISH as previously described. 14 Briefly, tissues were sequentially washed in methanol 2x 30 min, ethanol 2x 20 min, and xylene 2x 20 min, then embedded in paraffin to section. The tissue sections were dewaxed by preheating at 60°C for 10 min, followed by xylene 60°C for 10 min, xylene for 10 min and 99.5% ethanol for 10 minutes. Deparaffinized sections were incubated at 50°C overnight with EUB338 probe (5’-GCTGCCTCCCGTAGGAGT-3’, with a 5' labeling using Alexa 647) diluted to 10 μg/mL in hybridization buffer (20 mM Tris–HCl, pH 7.4, 0.9 M NaCl, 0.1% SDS, 20% formamide). After incubating with wash buffer (20 mM Tris-HCl, pH 7.4, 0.9 M NaCl) for 10 min and washing 3x in PBS, allowed to dry, the slides were blocked and stained with anti-mucin 2 antibody (Santa Cruz Biotechnology, H-300) before incubated with anti-rabbit Alexa 488 secondary antibody and Phalloidin Tetramethyl rhodamine B isothiocyanate (Sigma, P1951-.1MG), then mounted in DAPI containing mounting medium. The thickness of mucin stained by anti mucin-2 antibody and the distance of bacteria to epithelial cells was measured by using image J software.
Gut microbiota analysis by using 16S rRNA gene sequencing
Fecal DNA extraction, 16S rRNA gene amplification and sequencing analysis were conducted as described. 44 Briefly, fecal DNA was extracted by using the DNeasy 96 PowerSoil Pro QIAcube HT Kit, and the extracted DNA was used to amplify the region V4 of 16S rRNA genes by the following forward and reverse primers at Georgia Institute of Technology Molecular Evolution Core (Atlanta GA): 515FB 5′TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGTGYCAGCMGCCGCGGTAA-3′; 806RB 5′GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGGACTACNVGGGTWTCTAAT-3′; or amplify the region V3-V4 in our lab with the following primers: 5′TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′; 5′GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3′. Ampure XP magnetic purification beads were used to purify PCR products of each sample, of which the size of the amplicon was verified by using the bioanalyzer High Sensitivity Chip or running on DNA gel. A second PCR was performed to attach dual indices and Illumina sequencing adapters using Nextera XT Index kit. Products were then quantified before the DNA pool was generated from the purified products in equimolar ratios. The diluted library was spiked with 10% PhiX control and then sequenced by using iSeq™ 100 Sequencing System in our lab (paired-end reads, 2 × 150 base pairs), or on the Illumina MiSeq sequencer (paired-end reads, 2 × 250 base pairs) at Georgia Institute of Technology Molecular Evolution Core (Atlanta GA). After the sequenced was demultiplexed, the DADA2 plugin in Qiime2 was used to quality filter, denoise, and remove the chimera. Principal Coordinate Analysis (PCoA) plot based on unweighted UniFrac tables was visualized using Emperor in Qiime2 pipeline. Taxonomy was assigned based on Greengenes 16S rRNAgene database.
QUANTIFICATION AND STATISTICAL ANALYSES
Data are expressed as means +/− SEM, unless other indicated. Statistical significances of results were analyzed by unpaired student t test or using the one-way analysis of variance (ANOVA) as indicated in Figure legends. N represents number of mice. Differences between experimental groups were considered significant at *P < 0.05, **P <0.01, ***P < 0.001. ****P < 0.0001; #p<0.05, ##p < 0.01, ##p < 0.001, ####p < 0.0001, ns, not significant. Analysis was performed using GraphPad Prism 9.
Supplementary Material
Highlights.
Maternal fiber deprivation lastingly and detrimentally influenced microbiome of offspring.
Maternal fiber deprivation resulted in low-grade gut inflammation in offspring.
Offspring of fiber-deprived mice were highly prone to develop obesity.
Enriching maternal diet with fiber restored microbiome of offspring and prevented obesity.
ACKNOWLEDGEMENT
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grants DK099071 (ATG) and DK083890 (ATG). J.Z. is supported by the American Diabetes Association (#1-19-JDF-077).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Saklayen MG (2018). The Global Epidemic of the Metabolic Syndrome. Curr Hypertens Rep 20, 12. 10.1007/s11906-018-0812-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pulgaron ER, and Delamater AM (2014). Obesity and type 2 diabetes in children: epidemiology and treatment. Curr Diab Rep 14, 508. 10.1007/s11892-014-0508-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeda Y, Fujihara K, Nedachi R, et al. (2021). Comparing Associations of Dietary Energy Density and Energy Intake, Macronutrients with Obesity in Patients with Type 2 Diabetes (JDDM 63). Nutrients 13. 10.3390/nu13093167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendoza JA, Drewnowski A, and Christakis DA (2007). Dietary energy density is associated with obesity and the metabolic syndrome in U.S. adults. Diabetes Care 30, 974–979. 10.2337/dc06-2188. [DOI] [PubMed] [Google Scholar]
- 5.Lee A, Cardel M, and Donahoo WT (2000). Social and Environmental Factors Influencing Obesity. In Endotext, Feingold KR, Anawalt B, Boyce A, Chrousos G, de Herder WW, Dhatariya K, Dungan K, Hershman JM, Hofland J, Kalra S, et al. , eds. [Google Scholar]
- 6.Romieu I, Dossus L, Barquera S, et al. (2017). Energy balance and obesity: what are the main drivers? Cancer Causes Control 28, 247–258. 10.1007/s10552-017-0869-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breuninger TA, Wawro N, Breuninger J, et al. (2021). Associations between habitual diet, metabolic disease, and the gut microbiota using latent Dirichlet allocation. Microbiome 9, 61. 10.1186/s40168-020-00969-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Velasquez MT (2018). Altered Gut Microbiota: A Link Between Diet and the Metabolic Syndrome. Metab Syndr Relat Disord 16, 321–328. 10.1089/met.2017.0163. [DOI] [PubMed] [Google Scholar]
- 9.Tochitani S (2021). Vertical transmission of gut microbiota: Points of action of environmental factors influencing brain development. Neurosci Res 168, 83–94. 10.1016/j.neures.2020.11.006. [DOI] [PubMed] [Google Scholar]
- 10.Asnicar F, Manara S, Zolfo M, et al. (2017). Studying Vertical Microbiome Transmission from Mothers to Infants by Strain-Level Metagenomic Profiling. mSystems 2. 10.1128/mSystems.00164-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fehr K, Moossavi S, Sbihi H, et al. (2020). Breastmilk Feeding Practices Are Associated with the Co-Occurrence of Bacteria in Mothers' Milk and the Infant Gut: the CHILD Cohort Study. Cell Host Microbe 28, 285–297 e284. 10.1016/j.chom.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 12.Granger CL, Embleton ND, Palmer JM, et al. (2021). Maternal breastmilk, infant gut microbiome and the impact on preterm infant health. Acta Paediatr 110, 450–457. 10.1111/apa.15534. [DOI] [PubMed] [Google Scholar]
- 13.Cox LM, Yamanishi S, Sohn J, et al. (2014). Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158, 705–721. 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou J, Chassaing B, Singh V, et al. (2018). Fiber-Mediated Nourishment of Gut Microbiota Protects against Diet-Induced Obesity by Restoring IL-22-Mediated Colonic Health. Cell Host Microbe 23, 41–53 e44. 10.1016/j.chom.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zou J, Reddivari L, Shi Z, et al. (2021). Inulin Fermentable Fiber Ameliorates Type I Diabetes via IL22 and Short-Chain Fatty Acids in Experimental Models. Cell Mol Gastroenterol Hepatol 12, 983–1000. 10.1016/j.jcmgh.2021.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pellizzon M (2016). Choice of laboratory animal diet influences intestinal health. Lab Anim (NY) 45, 238–239. 10.1038/laban.1014. [DOI] [PubMed] [Google Scholar]
- 17.Tuck CJ, De Palma G, Takami K, et al. (2020). Nutritional profile of rodent diets impacts experimental reproducibility in microbiome preclinical research. Sci Rep 10, 17784. 10.1038/s41598-020-74460-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desai MS, Seekatz AM, Koropatkin NM, et al. (2016). A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 167, 1339–1353 e1321. 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turnbaugh PJ, Backhed F, Fulton L, et al. (2008). Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3, 213–223. 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al Nabhani Z, Dulauroy S, Lecuyer E, et al. (2019). Excess calorie intake early in life increases susceptibility to colitis in adulthood. Nat Metab 1, 1101–1109. 10.1038/s42255-019-0129-5. [DOI] [PubMed] [Google Scholar]
- 21.Tran HQ, Bretin A, Adeshirlarijaney A, et al. (2020). "Western Diet"-Induced Adipose Inflammation Requires a Complex Gut Microbiota. Cell Mol Gastroenterol Hepatol 9, 313–333. 10.1016/j.jcmgh.2019.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zou J, Zhao X, Shi Z, et al. (2021). Critical Role of Innate Immunity to Flagellin in the Absence of Adaptive Immunity. J Infect Dis 223, 1478–1487. 10.1093/infdis/jiaa521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansson ME, Phillipson M, Petersson J, et al. (2008). The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A 105, 15064–15069. 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian B, Zhao J, Zhang M, et al. (2021). Lycium ruthenicum Anthocyanins Attenuate High-Fat Diet-Induced Colonic Barrier Dysfunction and Inflammation in Mice by Modulating the Gut Microbiota. Mol Nutr Food Res 65, e2000745. 10.1002/mnfr.202000745. [DOI] [PubMed] [Google Scholar]
- 25.Curciarello R, Canziani KE, Docena GH, et al. (2019). Contribution of Non-immune Cells to Activation and Modulation of the Intestinal Inflammation. Front Immunol 10, 647. 10.3389/fimmu.2019.00647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guess ND, Dornhorst A, Oliver N, et al. (2015). A randomized controlled trial: the effect of inulin on weight management and ectopic fat in subjects with prediabetes. Nutr Metab (Lond) 12, 36. 10.1186/s12986-015-0033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petersen C, Bell R, Klag KA, et al. (2019). T cell-mediated regulation of the microbiota protects against obesity. Science 365. 10.1126/science.aat9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vandeputte D, Falony G, Vieira-Silva S, et al. (2017). Prebiotic inulin-type fructans induce specific changes in the human gut microbiota. Gut 66, 1968–1974. 10.1136/gutjnl-2016-313271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Keefe SJ (2019). The association between dietary fibre deficiency and high-income lifestyle-associated diseases: Burkitt's hypothesis revisited. Lancet Gastroenterol Hepatol 4, 984–996. 10.1016/S2468-1253(19)30257-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koh A, De Vadder F, Kovatcheva-Datchary P, et al. (2016). From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 165, 1332–1345. 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 31.Chassaing B, Koren O, Goodrich JK, et al. (2016). Corrigendum: Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 536, 238. 10.1038/nature18000. [DOI] [PubMed] [Google Scholar]
- 32.Carvalho FA, Koren O, Goodrich JK, et al. (2012). Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe 12, 139–152. 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cani PD, Amar J, Iglesias MA, et al. (2007). Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56, 1761–1772. 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 34.Blaser MJ (2018). Our missing microbes: Short-term antibiotic courses have long-term consequences. Cleve Clin J Med 85, 928–930. 10.3949/ccjm.85gr.18005. [DOI] [PubMed] [Google Scholar]
- 35.Sonnenburg ED, Smits SA, Tikhonov M, et al. (2016). Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215. 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Ran Z, Tian M, et al. (2019). Commensal Microbes Affect Host Humoral Immunity to Bordetella pertussis Infection. Infect Immun 87. 10.1128/IAI.00421-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le Roy T, Debedat J, Marquet F, et al. (2018). Comparative Evaluation of Microbiota Engraftment Following Fecal Microbiota Transfer in Mice Models: Age, Kinetic and Microbial Status Matter. Front Microbiol 9, 3289. 10.3389/fmicb.2018.03289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z, Zou J, Shi Z, et al. (2020). IL-22-induced cell extrusion and IL-18-induced cell death prevent and cure rotavirus infection. Sci Immunol 5. 10.1126/sciimmunol.abd2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zou J, and Shankar N (2016). Surface protein Esp enhances pro-inflammatory cytokine expression through NF-kappaB activation during enterococcal infection. Innate Immun 22, 31–39. 10.1177/1753425915611237. [DOI] [PubMed] [Google Scholar]
- 40.Zhang B, Oyewole-Said D, Zou J, et al. (2017). TLR5 signaling in murine bone marrow induces hematopoietic progenitor cell proliferation and aids survival from radiation. Blood Adv 1, 1796–1806. 10.1182/bloodadvances.2017006981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palm NW, de Zoete MR, Cullen TW, et al. (2014). Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010. 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kau AL, Planer JD, Liu J, et al. (2015). Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci Transl Med 7, 276ra224. 10.1126/scitranslmed.aaa4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hagan T, Cortese M, Rouphael N, et al. (2019). Antibiotics-Driven Gut Microbiome Perturbation Alters Immunity to Vaccines in Humans. Cell 178, 1313–1328 e1313. 10.1016/j.cell.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.An J, Zhao X, Wang Y, et al. (2021). Western-style diet impedes colonization and clearance of Citrobacter rodentium. PLoS Pathog 17, e1009497. 10.1371/journal.ppat.1009497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data have been deposited at GenBank with access number: PRJNA873095, and all other data used for this study will be shared upon reasonable request from the lead contact.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.