Abstract
The invasion-associated locus A and B genes (ialAB) of Bartonella bacilliformis were previously shown to confer an erythrocyte-invasive phenotype upon Escherichia coli, indirectly implicating their role in virulence. We report the first direct demonstration of a role for ialB as a virulence factor in B. bacilliformis. The presence of a secretory signal sequence and amino acid sequence similarity to two known outer membrane proteins involved in virulence suggested that IalB was an outer membrane protein. To develop an antiserum for protein localization, the ialB gene was cloned in frame into an expression vector with a six-histidine tag and under control of the lacZ promoter. The IalB fusion protein was purified by nickel affinity chromatography and used to raise polyclonal antibodies. IalB was initially localized to the bacterial membrane fraction. To further localize IalB, B. bacilliformis inner and outer membranes were fractionated by sucrose density gradient centrifugation and identified by appearance, buoyant density (ρ), and cytochrome b content. Inner and outer membrane proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and IalB was positively identified by Western blot. Contrary to expectations, IalB was localized to the inner membrane of the pathogen. To directly demonstrate a role for IalB in erythrocyte parasitism, the B. bacilliformis ialB gene was disrupted by insertional mutagenesis. The resulting ialB mutant strain was complemented in trans with a replicative plasmid encoding the full-length ialB gene. PCR and high-stringency DNA hybridization confirmed mutagenesis and transcomplementation events. Abrogation and restoration of ialB expression was verified by SDS-PAGE and immunoblotting. In vitro virulence assays showed that mutagenesis of ialB decreased bacterial association and invasion of human erythrocytes by 47 to 53% relative to controls. Transcomplementation of ialB restored erythrocyte association and invasion rates to levels observed in the parental strain. These data provide direct evidence for IalB's role in erythrocyte parasitism and represent the first demonstration of molecular Koch's postulates for a Bartonella species.
Bartonella bacilliformis is the only bacterium known to invade human erythrocytes. The pathogen is the causative agent of the human disease, Oroya fever, a biphasic illness whose primary-phase symptoms include a severe hemolytic anemia, where up to 100% of the circulating erythrocytes can be parasitized and 80% lysed (1, 15, 31). If untreated, this phase of the disease has a 40% fatality rate (44). Treatment with penicillin, tetracycline, or aminoglycosides is effective (43), but diagnosis can be difficult due to the slow growth and fastidious nature of the bacterium. The secondary phase of Oroya fever occurs 4 to 8 weeks following the primary hemolytic phase and is characterized by hemangiomas, nicknamed verruga peruana, on the patient's head, neck, and extremities. During the secondary phase, bacterial colonization and invasion shifts from erythrocytes to vascular endothelial cells (13, 14, 21) and results in neovascularization (13). This phase of the disease is rarely fatal but can last up to several months (43) and may cause permanent disfigurement. B. bacilliformis is transmitted by the phlebotamine sandfly, Lutzomyia verrucarum. Historically, Oroya fever has been limited to the mountainous regions of South America, presumably due to geographical restriction of its vector (19). However, recent reports of Oroya fever in coastal areas of South America suggest that the range of this pathogen is expanding (1).
Although other bacteria are known to parasitize mammalian erythrocytes (e.g., Anaplasma and Haemobartonella species), B. bacilliformis is unsurpassed among bacteria in its efficiency as an erythrocyte parasite. B. bacilliformis is able to invade nearly all circulating erythrocytes during the acute phase of infection. Erythrocytes lack the actin cytoskeleton necessary for bacterial uptake by induced endocytosis, although endocytosis can be induced under experimental conditions (35, 40). Treatment of erythrocytes with glycolysis and proton-motive-force inhibitors has no effect on Bartonella adhesion, suggesting that these host cells play a passive role in invasion (42). In contrast, B. bacilliformis plays an active role during erythrocyte invasion requiring both respiration and proton motive force (42). Taken together, these data indicate that B. bacilliformis is the only active participant in erythrocyte adherence and invasion. In contrast, B. bacilliformis entry into endothelial and epithelial cells differs significantly from its invasion of erythrocytes. Bacterium-induced rearrangement of the endothelial and epithelial cell cytoskeleton during endocytosis enhances bacterial uptake, while cytochalasin D treatment, inhibiting actin filament formation, reduces internalization by ∼30% (21).
The B. bacilliformis invasion-associated locus A and B genes (ialAB) were indirectly shown to be involved in erythrocyte invasion by conferring an erythrocyte-invasive phenotype upon minimally invasive Escherichia coli strains (27). IalA has since been characterized as a (di)nucleoside polyphosphate hydrolase thought to be involved in reducing levels of stress-induced dinucleotides during invasion, thus aiding bacterial survival (9, 11). IalB was shown to contain a putative 22-amino-acid secretory signal sequence and to have approximately 60% amino acid similarity to the virulence determinants Ail of Yersinia enterocolitica and Rck of Salmonella enterica serovar Typhimurium. The presence of a potential secretory sequence and similarity of IalB to two outer membrane virulence determinants led to our hypothesis that IalB is exported to the bacterial surface, where it functions as an invasion factor. This study was undertaken to localize the IalB protein and directly determine its role in human erythrocyte association by B. bacilliformis.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
B. bacilliformis strains (Table 1) were cultured on heart infusion agar blood (HIAB) plates (heart infusion agar supplemented with 4% sheep erythrocytes and 2% sheep serum) in a water-saturated incubator at 30°C. When required, strains were cultured in the presence of kanamycin (25 μg/ml) and/or chloramphenicol (5 μg/ml). E. coli strains (Table 1) were cultured in Luria-Bertani (LB) broth at 37°C in the presence of antibiotics as needed.
TABLE 1.
Bacteria and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| B. bacilliformis | ||
| JB584 | Transformation-competent strain of B. bacilliformis | 5 |
| SC1 | JB584 with ialB interrupted by pSAC100 (Kmr, ialB mutant) | This study |
| SC2 | SC1 complemented in trans with pSAC200 (Kmr Cmr, ialB+) | This study |
| E. coli | ||
| DH5α | Host strain for cloning and plasmid propagation | Gibco-BRL |
| M15 | Host strain for fusion protein expression | 41 |
| Plasmids | ||
| pIAL1 | pUC19 containing ialA and ialB of B. bacilliformis | 27 |
| pIALB | pUC19 containing ialB of B. bacilliformis | 27 |
| pQE-31 | Expression vector | Qiagen Inc. |
| pREP4 | Plasmid encoding lacI | Qiagen Inc. |
| pQIALB | pQE-31 with 574-bp PvuII-PstI fragment encoding ialB minus its secretory signal sequence plus 15 nucleotides | This study |
| pUB1 | B. bacilliformis suicide plasmid; Kmr | 5 |
| pSAC100 | pUB1 with an internal 430-bp PvuII-MfeI fragment of ialB; Kmr | This study |
| pBBR1MCS | B. bacilliformis shuttle vector; Cmr | 18 |
| pSAC200 | Complementation plasmid; pBBR1MCS with 756-bp SwaI-BamHI fragment containing intact ialB; Cmr | This study |
Preparation and manipulation of DNA.
Plasmids used or generated in this study are given in Table 1. Plasmids were propagated in E. coli DH5α and isolated by the methods of Birnboim and Doly (7), a Perfectprep kit (Eppendorf Scientific, Westbury, N.Y.), or a Qiagen Midiprep kit (Qiagen, Inc., Valencia, Calif.). Restriction digests and agarose gel electrophoresis were done using standard protocols (2). DNA fragments from restriction digests were purified from ethidium bromide-stained agarose gels with a GeneClean II kit (Bio 101, La Jolla, Calif.). Ligations were performed by standard protocol (2), and transformations were done by the method of Chung et al. (10). Genomic DNA was isolated using cetyltrimethylammonium bromide (CTAB) (2). Electroporation of B. bacilliformis was done as previously described (5).
PCR and oligonucleotide primers.
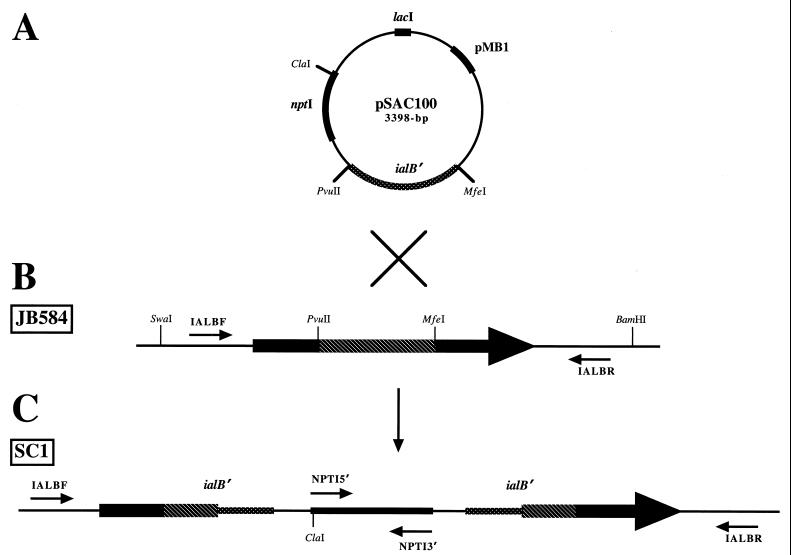
PCR amplification was done in a GeneAmp 2400 thermocycler (Perkin-Elmer, Norwalk, Conn.) as previously described (5). DNA was denatured at 94°C for 5 min, amplified for 30 cycles (1 min at each of the following temperatures: 94, 59 or 65, and 72°C), and extended for 10 min at 72°C. Single-strand oligonucleotide primers for the ialB gene, IALBF (5′-GTATTATGAATTACTATCGAGAATAA-3′) and IALBR (5′-ATCCGACCATAATACTTATCTTCT-3′), and for the neomycin phosphotransferase I gene (nptI), NPT15′ (5′-AGCCACGTTGTGTCTCAAAATCTC-3′) and NPTI3′ (5′-CGTCCCGTCAAGTCAGCGTAATGC-3′), were used. A “junction” primer set consisting of IALBR and NPTI5′ was designed to amplify the site of homologous recombination between the chromosomal ialB gene and the suicide plasmid, pSAC100. Annealing sites for all primers are depicted in Fig. 2.
FIG. 2.
Schematic representation depicting site-directed mutagenesis of the B. bacilliformis ialB gene. (A) The suicide plasmid, pSAC100, was created by cloning a 426-bp PvuII-MfeI internal fragment of the B. bacilliformis ialB gene (ialB′) into pUB1. (B) The JB584 strain of B. bacilliformis encodes the wild-type ialB gene. (C) Electroporation of JB584 with pSAC100, followed by recombination between the suicide plasmid, pSAC100, and the chromosomal ialB gene, results in insertional disruption and generation of the B. bacilliformis ialB mutant strain, SC1. Primer sites for ialB (IALBF and IALBR) and nptI (NPTI5′ and NPTI3′) are indicated by small arrows. (The figure is not drawn to scale.)
DNA hybridization analysis.
Genomic DNA from B. bacilliformis and plasmid DNA were digested to completion with ClaI and separated on a 1.2% (wt/vol) agarose gel stained with ethidium bromide. DNA was transferred to a supported nitrocellulose membrane (pore size, 0.45 μm; Schleicher & Schuell, Keene, N.H.) by the method of Southern (37) and then baked for 1 h at 80°C. DNA probes were made by random primer extension (2) with [α-32P]dCTP (New England Nuclear, Boston, Mass.). High-stringency hybridization, washes, and visualization were done as previously described (6).
SDS-PAGE.
Protein concentrations were determined using a bicinchoninic acid protein kit per the manufacturer's instructions (Sigma Chemical Co., St. Louis, Mo.). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was done following the general procedures of Laemmli (20) with either 12.5, 15, or 15 to 20% gradient polyacrylamide (wt/vol) gels. Either 20 or 100 μg of protein was loaded per lane for gels that were Coomassie blue stained (33) and 2.5 μg was loaded per lane on gels that were silver stained (45).
Preparation of polyclonal antibodies and immunoblotting.
To prepare antibodies against IalB, E. coli M15 (pQIALB, pREP4) was grown overnight with vigorous shaking in LB broth containing ampicillin and kanamycin. The overnight culture was used to inoculate LB broth plus antibiotics and grown to an optical density at 600 nm (OD600) of 0.7 to 0.9, and ialB expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG; 2 mM final concentration). Cultures were induced for 3 h, and the bacterial pellet was harvested by centrifugation at 4,000 × g for 20 min at 4°C. Bacterial pellets were solubilized in Laemmli sample buffer and proteins separated by SDS-PAGE. The IalB protein was excised from unfixed Coomassie blue-stained gels, minced, suspended in 1 ml of phosphate-buffered saline (PBS; pH 7.4), and used to generate antibody in a female New Zealand White rabbit as previously described (34).
For immunoblots, 20 to 80 μg of protein were separated by SDS-PAGE (12.5 or 15% [wt/vol] acrylamide), electrophoretically transferred to a supported nitrocellulose membrane (pore size, 0.45 μm; Schleicher & Schuell), and reacted with anti-IalB antiserum (diluted 1:1,000) as previously described (34).
Localization of IalB.
Accessible outer membrane proteins of intact B. bacilliformis were extrinsically radioiodinated as previously described (24) and then analyzed by SDS-PAGE. Whole bacteria were extrinsically treated with various proteases (proteinase K, trypsin, subtilisin, papain, and thermolysin) to cleave any accessible, sensitive surface proteins as previously described (24), and protein profiles were analyzed by gradient SDS-PAGE. Immunofluorescent labeling of intact B. bacilliformis strains using anti-IalB polyclonal antibodies was done according to standard protocols (2). Twenty plates of 3-day-old B. bacilliformis were harvested into 1 ml of ice-cold Dulbecco's PBS, and membranes were isolated and fractionated as previously described for B. quintana (8). Cytochrome assays were performed using inner and outer membrane fractions (final protein concentration, 1 μg/μl) by the methods of Osborn et al. (30).
Human erythrocyte association assay.
Blood was drawn from human volunteers into an acid citrate-dextrose Vacutainer tube and stored overnight at 4°C to separate plasma from the erythrocytes. After removal of the plasma, erythrocytes were washed with 10 ml of sterile saline (0.9%, wt/vol) and centrifuged at 700 × g for 5 min. Erythrocytes were washed a second time, counted, and resuspended in recovery broth (5) to a final concentration of 109 erythrocytes per ml.
Three- to four-day-old B. bacilliformis cultures were harvested into recovery broth and diluted to an OD600 of 1.0 (∼1.6 × 109 CFU/ml). Approximately 5 × 108 bacteria were gently mixed with 108 erythrocytes (multiplicity of infection 5:1) in a total volume of 0.5 ml of recovery broth. Association reactions were incubated for 8 h at 30°C in a water-saturated environment. Erythrocytes and parasitized erythrocytes were separated from free bacteria by Percoll gradient centrifugation. Briefly, 1 ml of 70% Percoll (Sigma) containing 154 mM NaCl was centrifuged at 16,000 × g for 10 min to create a continuous gradient. Then, 0.1 ml of each association reaction was carefully layered onto the preformed Percoll gradient and centrifuged at 1,500 × g for 5 min. The erythrocyte-bacterium band was collected, washed twice with sterile saline, and pelleted by centrifugation at 1,000 × g for 15 s. The pellet was resuspended in 0.5 ml of heart infusion broth, serially diluted, and then plated onto HIAB plates. Plates were incubated at 30°C in a water-saturated incubator for 12 days and then counted for CFU.
Statistical analysis.
Numerical data reported for human erythrocyte association assays are the means of three independent samples ± the standard errors of the mean (SEM). The statistical significance of the data was determined by use of the Student's t test. A P value of <0.05 was considered significant.
RESULTS
Expression and purification of IalB fusion protein.
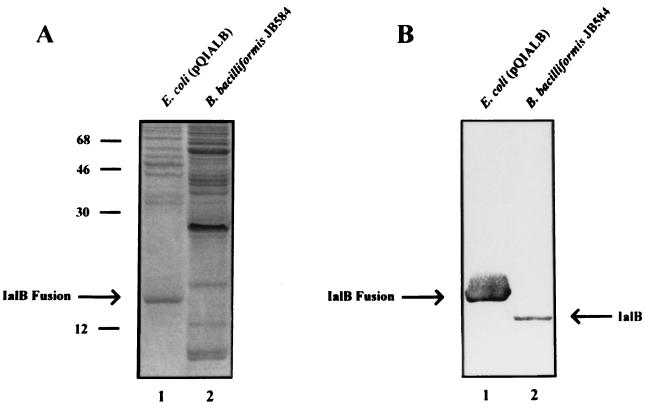
To obtain sufficient amounts of IalB protein to generate antibodies, the ialB gene (excluding the portion encoding its secretory signal sequence plus five N-terminal amino acids) was cloned in frame into the expression vector pQE-31. This vector contains a six-histidine tag and a polylinker under the control of the lacZ promoter. The resulting construct, pQIALB, was transformed into E. coli M15, and ialB expression was induced with IPTG. The IalB fusion protein was synthesized at high levels and localized to the insoluble fraction of E. coli. The insoluble fraction was treated with a strong denaturant (6 M guanidine hydrochloride), and the recombinant IalB was purified using nickel affinity chromatography. IalB was purified to apparent homogeneity when analyzed by using Coomassie blue-stained SDS-PAGE gels (data not shown). Polyclonal anti-IalB antibodies were generated and found to recognize both the IalB fusion protein synthesized in E. coli and wild-type IalB synthesized by B. bacilliformis in Western blots (Fig. 1B). On Western blots, the IalB fusion protein and IalB from B. bacilliformis have estimated masses of 18.6 and 17.1 kDa, respectively. From its DNA sequence, the mature B. bacilliformis IalB protein was predicted to be 17.5 kDa (27), in close agreement with our finding. Presumably, the larger estimated mass of the IalB fusion protein is due to the presence of the charged, six-histidine tag.
FIG. 1.
Recognition of B. bacilliformis IalB and the IalB fusion protein by polyclonal anti-IalB antibodies. Cell lysates of IPTG-induced E. coli M15(pQIALB) (lane 1) and B. bacilliformis (lane 2) analyzed by an SDS-PAGE gel stained with Coomassie blue (A) or immunoblot reacted with polyclonal anti-IalB antibodies (B). Both B. bacilliformis IalB and the IalB fusion protein are specifically detected with the antiserum. Molecular mass standards in kilodaltons are indicated on the left.
Generating an ialB mutant and a transcomplemented strain of B. bacilliformis.
A 426-bp, PvuII-MfeI internal fragment of the ialB gene was cloned into pUB1 to create the suicide vector, pSAC100. The pMB1 origin of pSAC100 is not functional in B. bacilliformis (5); therefore, expression of the nptI gene, conferring kanamycin resistance, would only occur following recombination of the suicide plasmid into the chromosome. Cloning an internal fragment of the ialB gene ensured that homologous recombination between pSAC100 and the chromosome would not result in reconstitution of a full-length gene.
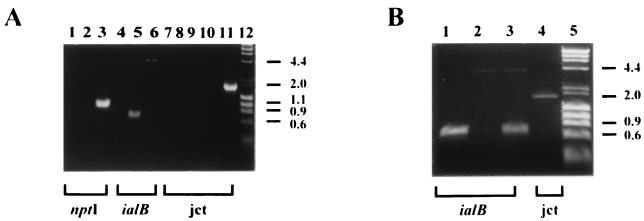
The JB584 strain of B. bacilliformis was electroporated with pSAC100. Kanamycin-resistant colonies were isolated, cultured, and initially characterized by PCR. The ialB gene, the nptI gene, or the junction where pSAC100 recombined with the chromosomal ialB gene were PCR amplified as depicted in Fig. 2. The nptI gene primer set (NPTI5′ and NPTI3′) amplified a 983-bp segment of the nptI gene in the kanamycin-resistant strain, SC1, but not the parental strain, JB584 (Fig. 3A, lanes 3 and 2, respectively), showing that kanamycin resistance in SC1 was due to nptI and not to selection of spontaneous kanamycin-resistant mutants. The ialB gene primer set (IALBF and IALBR) was expected to produce a 4,097-bp product from the site of homologous recombination or a 688-bp product from an intact ialB gene. Upon analysis, an amplicon of ∼4,000-bp was obtained from the kanamycin-resistant strain, SC1, indicating that pSAC100 had recombined with the chromosomal ialB (Fig. 3A, lane 6). No PCR product would be amplified from unintegrated pSAC100 since the ialB primers are complementary to chromosomal sequences flanking the ialB gene and absent in pSAC100. As expected, a 688-bp amplicon was obtained from the intact ialB gene in JB584 (Fig. 3A, lane 5).
FIG. 3.
Electrophoretic analysis of PCR products derived from ialB mutant strain, SC1, and transcomplemented strain, SC2. PCR products were generated by amplification of genomic DNA from parent and recombinant strains using three amplimer sets (nptI [NPTI3′ and NPTI5′], ialB [IALBF and IALBR], and junction [jct] [NPTI5′ and IALBR]). Brackets below the gel indicate the amplimer set used in each reaction. Amplimer sets and template DNA for PCR used in this analysis are as follows. (A) Lane 1, NPTI3′ and NPTI5′, no template; lane 2, NPTI3′ and NPTI5′, JB584; lane 3, NPTI3′ and NPTI5′, SC1; lane 4, IALBF and IALBR, no template; lane 5, IALBF and IALBR, JB584; lane 6, IALBF and IALBR, SC1; lane 7, NPTI5′ and IALBR, no template; lane 8, NPTI5′ and IALBR, JB584; lane 9, NPTI5′ and IALBR, pSAC100; lane 10, NPTI5′ and IALBR, JB584 and pSAC100; lane 11, NPTI5′ and IALBR, SC1; lane 12, lambda DNA/HindIII and φX174 DNA/HaeIII markers. (B) Lane 1, IALBF, and IALBR, JB584; lane 2, IALBF and IALBR, SC1; lane 3, IALBF and IALBR, SC2; lane 4, NPTI5′ and IALBR, SC2; lane 5, lambda DNA/HindIII and φX174 DNA/HaeIII markers. PCR products were analyzed by ethidium bromide-stained agarose (1.2%, wt/vol) gel electrophoresis. Size standards in kilobase pairs are indicated on the right.
The junction primer set (NPTI5′ and IALBR) produced an amplicon of approximately 1,700-bp from SC1 and no product from the parental strain, JB584 (Fig. 3A, lanes 11 and 8, respectively). As expected, no amplicon was obtained when pSAC100 DNA was added to JB584 genomic DNA and then amplified with the junction primer set (Fig. 3A, lane 10). From these data we concluded that homologous recombination had occurred between pSAC100 and the chromosomal ialB gene, creating an ialB mutant strain, SC1.
We then proceeded to create a transcomplemented strain using SC1 as the parental strain. The pIALB plasmid was digested with SwaI and BamHI, the 756-bp fragment containing the intact ialB gene isolated, and cloned into the broad-host-range plasmid, pBBR1MCS to produce the shuttle plasmid, pSAC200. pSAC200 was subsequently electroporated into SC1, and transformants were selected on HIAB plates supplemented with both kanamycin and chloramphenicol. Potential transcomplemented strains were isolated, cultured, and characterized by PCR.
The ialB gene primer set (IALBF and IALBR) was used to screen for potential transcomplemented strains. One strain, SC2, produced amplicons of 4,097 and 688 bp representing the interrupted ialB gene on the bacterial chromosome and the intact ialB gene on pSAC200, respectively (Fig. 3B, lane 3). PCR amplification of SC2 DNA using the junction primer set (NPT15′ and IALBR) resulted in a product of approximately 1,700 bp (Fig. 3B, lane 4), indicating that the original site of integration was intact.
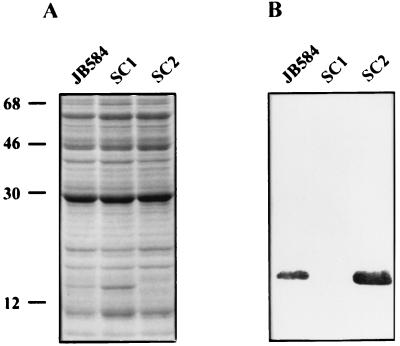
To determine whether expression of the ialB gene had been disrupted in SC1 and transcomplemented in SC2, cell lysates of the bacteria were analyzed by SDS-PAGE and Western blot (Fig. 4). A 17.1-kDa band was present in both JB584 and SC2 lysates but absent in SC1 lysates. This protein was positively identified as IalB by Western blots (Fig. 4B). We consistently observed more IalB in cell lysates of SC2 relative to JB584, by both SDS-PAGE and Western blots. Presumably, increased synthesis in SC2 is due to the multiple copies of pSAC200 encoding ialB (Fig. 5B).
FIG. 4.
Abrogation and complementation of ialB expression in B. bacilliformis strains. (A) Cell lysate proteins (80 μg/lane) separated by SDS-PAGE (12.5%, wt/vol) and stained with Coomassie blue. Lane 1, JB584; lane 2, SC1; lane 3, SC2. (B) Corresponding immunoblot reacted with polyclonal anti-IalB antibodies showing IalB is present in the parental B. bacilliformis strain, JB584 (lane 1), and the transcomplemented mutant strain, SC2 (lane 3), but is absent in the ialB mutant strain, SC1 (lane 2). Molecular mass standards in kilodaltons are indicated on the left.
FIG. 5.
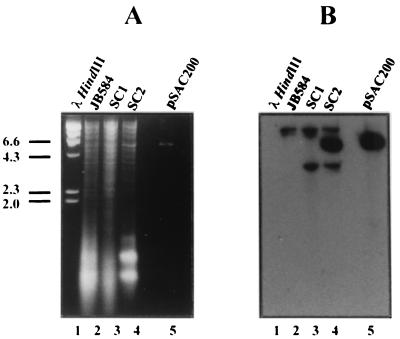
Detection of the wild-type and mutated ialB genes in B. bacilliformis strains using DNA hybridization. (A) Ethidium bromide-stained agarose gel (1.2%, wt/vol) of ClaI-digested genomic DNA from the parental B. bacilliformis strain, JB584 (lane 2), the ialB mutant strain, SC1 (lane 3), and the transcomplemented strain, SC2 (lane 4). The shuttle plasmid used in transcomplementation, pSAC200, digested with ClaI is shown in lane 5, and DNA size standards (Lambda DNA/HindIII markers) are provided in lane 1. (B) Corresponding Southern blot hybridized with the ialB probe. Lane 1, DNA size standards; lane 2, single hybridization band of ∼23 kbp from the parental B. bacilliformis strain, JB584; lane 3, two-band hybridization pattern from the disrupted ialB gene in the ialB mutant strain, SC1; lane 4, two-band hybridization pattern from the disrupted ialB gene, as well as the ∼5.4-kbp hybridization band from pSAC200 in the transcomplemented strain, SC2; lane 5, single hybridization band from pSAC200. Size standards in kilobase pairs are indicated on the left.
Genotypes of the mutant and transcomplemented strains were corroborated using DNA hybridization (Fig. 5). Restriction endonuclease digestion of pIALB with KpnI and HindIII yielded a 744-bp fragment containing ialB that was used to probe Southern blots of ClaI-digested genomic DNA from each strain (Fig. 5A). Hybridization of the probe with JB584 DNA showed a single, distinct band of ∼23 kbp (Fig. 5B, lane 2), while hybridization with the ialB mutant strain, SC1, gave two bands of ∼23 and ∼3.7 kbp (Fig. 5B, lane 3). The two hybridization products in SC1 are due to the presence of a ClaI restriction enzyme site in the integrated suicide plasmid (Fig. 2). Each band contains a portion of the ialB gene. The insertionally mutagenized ialB gene of the transcomplemented strain SC2 gives the expected two-band pattern like SC1, plus an additional hybridization band of ∼5.4 kbp from ialB on pSAC200 (Fig. 5B, lane 4).
No overt phenotypic differences between the parental, ialB mutant, and transcomplemented strains were apparent.
Localization of IalB in the bacterium.
As expected, SDS-PAGE analysis of total membranes showed that IalB was present in the membrane fraction of JB584 and SC2 but not the mutant strain, SC1, and its identity as IalB was verified by Western blot (data not shown). Extrinsic radioiodination of intact JB584 and SC1 showed no difference in protein profiles when analyzed by SDS-PAGE (data not shown). Whole JB584 bacteria extrinsically treated with several proteases showed no alteration in the migration of IalB on gradient SDS-PAGE gels (data not shown). No difference in immunofluorescence was seen when whole JB584 and SC1 bacteria were surface labeled using anti-IalB polyclonal antibodies (data not shown). Radioiodination, proteolysis, and immunofluorescence data suggested that IalB is an inner membrane protein.
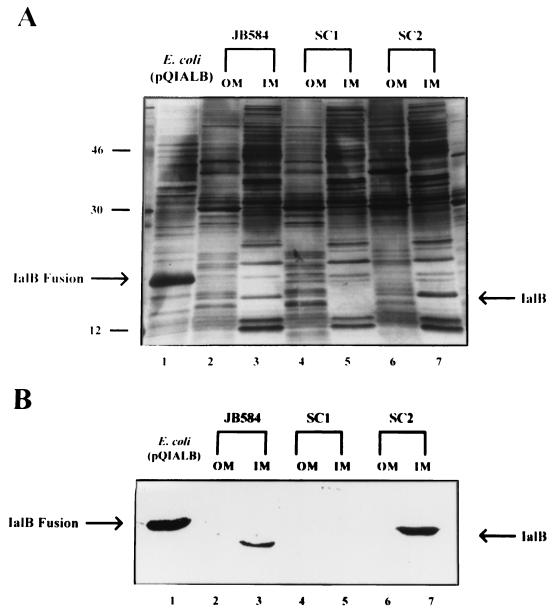
To conclusively localize IalB to the inner membrane, crude lysates were subjected to sucrose density gradient centrifugation as we previously described for B. quintana (8). Inner and outer membrane bands were collected from gradients and identified on the basis of their appearance. Outer membrane fractions typically showed a white flocculent appearance, while inner membrane fractions were typically tea colored (28). The average buoyant densities (ρ) were determined from three membrane preparations and calculated to be 1.08 g/cm3 for the inner membrane and 1.22 g/cm3 for the outer membrane. These values are very similar to the buoyant densities for the outer and inner membranes of E. coli (28) and Salmonella spp. (30) and are nearly identical to those we obtained from B. quintana membrane fractions (8). Outer membrane fractions analyzed by SDS-PAGE on a 15 to 20% gradient gel and stained with silver gave a protein profile similar to that previously reported for B. bacilliformis (24). In addition, the outer, but not the inner, membrane fractions contained the 42-kDa flagellin protein (34) and three bacteriophage proteins with molecular masses of 32, 34, and 36 kDa (4). The identity of the inner membrane fraction was unequivocally established by the presence of cytochrome b. Difference spectra for the inner and outer membrane fractions were obtained between 499 and 600 nm. The inner, but not the outer, membrane fraction had an absorbance peak at 558 nm, which is characteristic of cytochrome b. Once the identity of the inner and outer membrane fractions was established, their respective protein profiles were analyzed using SDS-PAGE. Contrary to our hypothesis that IalB was an outer membrane protein, the protein was found in the inner membrane fractions of both JB584 and SC2 (Fig. 6, lanes 3 and 7). The identity of IalB was confirmed by Western blot (Fig. 6B).
FIG. 6.
Localization of IalB to the B. bacilliformis inner membrane. (A) Proteins (2.5 μg/lane) were separated by SDS-PAGE (15 to 20% [wt/vol] gradient), and the gel was silver stained. IalB was found in the inner membrane fractions of JB584 (lane 3) and SC2 (lane 7), the parental and transcomplemented B. bacilliformis strains, respectively. IalB was absent from all outer membrane fractions and the inner membrane fraction of SC1, the ialB mutant strain. (B) Corresponding immunoblot reacted with polyclonal anti-IalB antibodies. IalB localized to the inner membrane fractions of JB584 and SC1 (lanes 3 and 7, respectively). IPTG-induced E. coli M15(pQIALB) cell lysate is provided as a control in lane 1.
Role of IalB in erythrocyte adhesion and invasion.
Following the 8-h association assays, Percoll gradient centrifugation was used to separate erythrocytes from free bacteria. Since both adherent and invaded bacteria were complexed with erythrocytes, CFU counts from these assays include bacteria that are adhering to, or have invaded, erythrocytes.
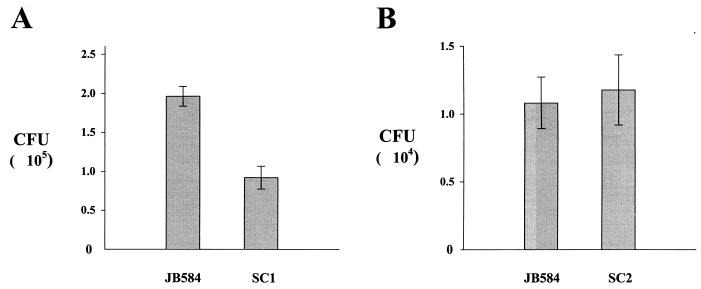
Association assays were carried out at least four times, with each experiment containing two to five independent samples. While the number of CFU varied between experiments, the data trends remained consistent. For the association assays conducted with the ialB mutant strain, SC1, and the parental strain, JB584, SC1 adherence and invasion decreased 47 to 53% compared to JB584. In a representative experiment, SC1 showed a significant decrease (P < 0.05) of 53% in adherence and invasion compared to JB584 (mean CFU of 91,750 ± 14,655 versus 196,300 ± 12,537, respectively) (Fig. 7A). Association assays conducted with JB584 and the complemented strain, SC2, showed statistically insignificant differences in adherence and invasion, although the range of values varied more than that observed in assays with JB584 and SC1. This increased scatter in SC2 values may be due to multiple plasmid copies of the ialB gene in SC2. In a representative experiment, the trans-complemented strain, SC2, showed no significant change (P = 0.7825) in association assays when compared to JB584 (mean CFU of 10,833 ± 1,906 versus 11,775 ± 2,575, respectively) (Fig. 7B).
FIG. 7.
Effect of ialB mutagenesis and transcomplementation on human erythrocyte adherence and invasion on B. bacilliformis strains. (A) The ialB mutant strain, SC1, shows a 53% decrease in erythrocyte adherence and invasion compared to the parental strain, JB584. The n values for JB584 and SC1 are 5 and 4, respectively. (B) Transcomplementation of ialB in SC2 restores erythrocyte adherence and invasion to parental strain levels. The n values for JB584 and SC2 are 3 and 2, respectively. Experimental data presented graphically in panels A and B are the mean ± the SEM from two separate but representative experiments.
DISCUSSION
This study is the first demonstration of molecular Koch's postulates (12) for a Bartonella species. Insertional mutagenesis of ialB, creating the B. bartonella mutant strain, SC1, resulted in a 47 to 53% decrease in human erythrocyte adherence and invasion compared to the parental strain, JB584. Transcomplementation of ialB, creating the SC2 strain, restored erythrocyte adherence and invasion to parental levels. These data clearly establish IalB as a virulence determinant for B. bacilliformis erythrocyte parasitism.
Mitchell and Minnick originally isolated and characterized the two-gene locus, ialAB, reporting that both ialA and ialB were necessary to confer an invasive phenotype upon E. coli (27). However, the results of the present study demonstrate that ialB has a significant effect on B. bacilliformis erythrocyte parasitism. In vivo experiments with the rat pathogen, B. tribicorum, support our findings that ialB is a virulence factor. Specifically, an ialB mutant strain of B. tribicorum failed to develop bacteremia and to invade rat erythrocytes in vivo (C. Gille, C. Lanz, and C. Dehio, Abstr. 1st Int. Conf. Bartonella Emerging Pathogens, abstr. 28, 1999).
ialA and ialB homologues are present in the three most prevalent, human pathogenic species of Bartonella: B. henselae, B. quintana, and B. bacilliformis (26). B. henselae and B. quintana cause cat-scratch disease and trench fever, respectively. All three species share phenotypic similarities: they are transmitted by arthropod vector, are intracellular parasites, and have an absolute growth requirement for hemin. All three species invade or attach to erythrocytes during the course of infection (17, 22, 23) and can cause neovascularization of infected tissue (25). Erythrocyte parasitism and neovascularization may provide the blood and heme required for these pathogenic bacteria. Given the phenotypic similarities of B. bacilliformis, B. quintana, and B. henselae, IalA and IalB may share similar functions contributing to the virulence of all three species.
Homologues of ialA and ialB have been found in other gram-negative pathogenic bacteria. Brucella melitensis is a facultative intracellular pathogen and the causative agent of ovine brucellosis. The ability of B. melitensis to cause disease is tied to its ability to adapt and survive in a range of environments. B. melitensis' adaptive responses to heat, oxidative, and acid stress were recently characterized (39). Protein levels, in response to these stresses, were analyzed by two-dimensional PAGE. In response to heat shock (a temperature shift from 37 to 42°C), an appreciable reduction in synthesis was observed for a protein with homology to the IalB protein of B. bacilliformis. No change in synthesis was seen for the IalB homologue in response to either oxidative or acid stress. Brucella and Bartonella are closely related α-proteobacteria, and their phylogenetic relationship is underscored by the ability of both genera to interact with eukaryotic cells in a parasitic or mutualistic association. In light of these similarities, it is interesting that these two species may share a virulence factor associated with eukaryotic cell invasion. We are currently examining the effect of environmental cues on ialB expression, as the transfer of B. bacilliformis from sandfly to human would be associated with significant changes in temperature, iron availability, pH, and oxidative stress. These environmental cues could serve as signals for expression of virulence factors necessary for human infection.
In another study, differential fluorescence induction was used to identify E. coli K1 genes expressed under environmental conditions favoring bacterial invasion of human brain microvascular endothelial cells (HBMEC) (3). One gene identified in that study was an IalA homologue (38% homology). Site-directed mutagenesis of this E. coli gene reduced HBMEC invasion twofold, and transcomplementation restored the invasive phenotype to wild-type levels. IalA and IalB homologues are being identified in a number of bacterial species, all of which invade eukaryotic cells. Additionally, experimental evidence for the role of these proteins in virulence is accumulating.
We originally hypothesized that IalB is exported to the bacterial surface, where it functions as an invasion factor. Contrary to our hypothesis, IalB was localized to the inner membrane in this study. Our original hypothesis was, in part, based on the reported ∼60% amino acid sequence similarity of IalB to Ail and Rck (27). However, although these proteins have significant amino acid similarity, their amino acid identity is actually quite low (∼11%). The IalB protein also lacks a terminal phenylalanine amino acid residue characteristic of most outer membrane proteins (38), including Ail and Rck.
Localization of IalB to the cytoplasmic membrane necessitated rethinking of its function as a virulence factor. Virulence-related activities for inner membrane proteins include transport of virulence factors, uptake of nutrients, response to environmental stresses, chemotaxis, cell motility, and intracellular survival, to name a few. These various functions fall into one of two general categories: transport or signal transduction. For example, the virB operon of Brucella suis and Brucella abortus was found to be essential for virulence and intracellular survival of these mammalian pathogens. The virB operon encodes homologues to a type IV secretory system including putative inner membrane proteins (29, 36). An intriguing example of a signal-transducing, inner membrane protein is found in Pseudomonas aeruginosa. Normally, the sigma factor responsible for expression of a mucoid phenotype is sequestered at the cytoplasmic membrane by an inner membrane protein. Release of this sigma factor into the cytosol, presumably in response to some signal, results in the expression of mucoidy (32). Phosphorylation is another mechanism by which an inner membrane protein could facilitate signal transduction. The etk gene of E. coli encodes an inner membrane protein capable of autophosphorylation (16). Interestingly, while all E. coli strains possess the etk gene, it is only expressed by a subset of pathogenic strains.
With these examples as precedents for cytoplasmic membrane proteins serving as virulence factors, we are currently investigating whether IalB functions as a transporter or signal transduction protein. To date, database searches for proteins with homology to IalB have not suggested any function. This lack of homology to known proteins may reflect IalB's unique and unusual role in erythrocyte parasitism by B. bacilliformis.
ACKNOWLEDGMENTS
We thank Karen Behan of The University of Montana Curry Student Health Center for providing human blood samples, George Card for technical assistance with cytochrome assays, Scott Samuels for critical review of the manuscript, and Laura Smitherman for technical assistance.
This work was supported by Public Health Service grant AI34050 from the National Institutes of Health (NIAID) (to M.F.M.) and a Predoctoral Honors Fellowship from The University of Montana (to S.A.C.).
REFERENCES
- 1.Alexander B. A review of bartonellosis in Ecuador and Colombia. Am J Trop Med Hyg. 1995;52:354–359. doi: 10.4269/ajtmh.1995.52.354. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 3.Badger J L, Wass C A, Kim K S. Identification of Escherichia coli K1 genes contributing to human brain microvascular endothelial cell invasion by differential fluorescence induction. Mol Microbiol. 2000;36:174–182. doi: 10.1046/j.1365-2958.2000.01840.x. [DOI] [PubMed] [Google Scholar]
- 4.Barbian K D, Minnick M F. A bacteriophage-like particle from Bartonella bacilliformis. Microbiology. 2000;146:599–609. doi: 10.1099/00221287-146-3-599. [DOI] [PubMed] [Google Scholar]
- 5.Battisti J M, Minnick M F. Development of a system for genetic manipulation of Bartonella bacilliformis. Appl Environ Microbiol. 1999;65:3441–3448. doi: 10.1128/aem.65.8.3441-3448.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Battisti J M, Smitherman L S, Samuels D S, Minnick M F. Mutations in Bartonella bacilliformis gyrB confer resistance to coumermycin A1. Antimicrob Agents Chemother. 1998;42:2906–2913. doi: 10.1128/aac.42.11.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroll J A, Coleman S A, Smitherman L S, Minnick M F. Hemin-binding surface protein from Bartonella quintana. Infect Immun. 2000;68:6750–6757. doi: 10.1128/iai.68.12.6750-6757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cartwright J L, Britton P, Minnick M F, McLennan A G. The ialA invasion gene of Bartonella bacilliformis encodes a (di)nucleoside polyphosphate hydrolase of the MutT motif family and has homologs in other invasive bacteria. Biochem Biophys Res Commun. 1999;256:474–479. doi: 10.1006/bbrc.1999.0354. [DOI] [PubMed] [Google Scholar]
- 10.Chung C T, Niemela S L, Miller R H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conyers G B, Bessman M J. The gene, ialA, associated with the invasion of human erythrocytes by Bartonella bacilliformis, designates a nudix hydrolase active on dinucleoside 5′-polyphosphates. J Biol Chem. 1999;274:1203–1206. doi: 10.1074/jbc.274.3.1203. [DOI] [PubMed] [Google Scholar]
- 12.Falkow S. Molecular Koch's postulates applied to microbial pathogenicity. Rev Infect Dis. 1988;10(Suppl.):S274–S276. doi: 10.1093/cid/10.supplement_2.s274. [DOI] [PubMed] [Google Scholar]
- 13.Garcia F U, Wojta J, Broadley K N, Davidson J M, Hoover R L. Bartonella bacilliformis stimulates endothelial cells in vitro and is angiogenic in vivo. Am J Pathol. 1990;136:1125–1135. [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia F U, Wojta J, Hoover R L. Interactions between live Bartonella bacilliformis and endothelial cells. J Infect Dis. 1992;165:1138–1141. doi: 10.1093/infdis/165.6.1138. [DOI] [PubMed] [Google Scholar]
- 15.Hurtado A, Musso J P, Meriono C. La anemia en la enfermedad de Carrion (verruga peruana) Ann Fac Med Lima. 1938;28:154–168. [Google Scholar]
- 16.Ilan O, Bloch Y, Frankel G, Ullrich H, Geider K, Rosenshine I. Protein tyrosine kinases in bacterial pathogens are associated with virulence and production of exopolysaccharide. EMBO J. 1999;18:3241–3248. doi: 10.1093/emboj/18.12.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kordick D L, Breitschwerdt E B. Intraerythrocytic presence of Bartonella henselae. J Clin Microbiol. 1995;33:1655–1656. doi: 10.1128/jcm.33.6.1655-1656.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kovach M E, Phillips R W, Elzer P H, Roop II R M, Peterson K M. pBBR1MCS: a broad-host-range cloning vector. BioTechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- 19.Kreier J P, Ristic M. The biology of hemotrophic bacteria. Annu Rev Microbiol. 1981;35:325–338. doi: 10.1146/annurev.mi.35.100181.001545. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.McGinnis Hill E, Raji A, Valenzuela M S, Garcia F, Hoover R. Adhesion to and invasion of cultured human cells by Bartonella bacilliformis. Infect Immun. 1992;60:4051–4058. doi: 10.1128/iai.60.10.4051-4058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehock J R, Greene C E, Gherardini F C, Hahn T-W, Krause D C. Bartonella henselae invasion of feline erythrocytes in vitro. Infect Immun. 1998;66:3462–3466. doi: 10.1128/iai.66.7.3462-3466.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merrell B R, Weiss E, Dasch G A. Morphological and cell association characteristics of Rochalimaea quintana: comparison of the vole and Fuller strains. J Bacteriol. 1978;135:633–640. doi: 10.1128/jb.135.2.633-640.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minnick M F. Identification of outer membrane proteins of Bartonella bacilliformis. Infect Immun. 1994;62:2644–2648. doi: 10.1128/iai.62.6.2644-2648.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minnick M F, Anderson B E. Bartonella interactions with host cells. In: Oelschlaeger T A, Hacker J, editors. Subcellular biochemistry: bacterial invasion into eukaryotic cells. Vol. 33. New York, N.Y: Kluwer Academic/Plenum Publishers; 2000. pp. 97–123. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell S J, Minnick M F. A carboxy-terminal processing protease gene is located immediately upstream of the invasion-associated locus from Bartonella bacilliformis. Microbiology. 1997;143:1221–1233. doi: 10.1099/00221287-143-4-1221. [DOI] [PubMed] [Google Scholar]
- 27.Mitchell S J, Minnick M F. Characterization of a two-gene locus from Bartonella bacilliformis associated with the ability to invade human erythrocytes. Infect Immun. 1995;63:1552–1562. doi: 10.1128/iai.63.4.1552-1562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikaido H. Outer membrane. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 29–47. [Google Scholar]
- 29.O'Callaghan D, Cazevieille C, Allardet-Servent A, Boschiroli M L, Bourg G, Foulongne V, Frutos P, Kulakov Y, Ramuz M. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essental for intracellular survival of Brucella suis. Mol Microbiol. 1999;33:1210–1220. doi: 10.1046/j.1365-2958.1999.01569.x. [DOI] [PubMed] [Google Scholar]
- 30.Osborn M J, Gander J E, Parisi E, Carson J. Mechanism of assembly of the outer membrane of Salmonella typimurium. J Biol Chem. 1972;247:3962–3972. [PubMed] [Google Scholar]
- 31.Reynafarje C, Ramos J. The hemolytic anemia of human bartonellosis. Blood. 1961;17:562–578. [PubMed] [Google Scholar]
- 32.Rowen D W, Deretic V. Membrane-to-cytosol redistribution of ECF sigma factor AlgU and conversion to mucoidy in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Mol Microbiol. 2000;36:314–327. doi: 10.1046/j.1365-2958.2000.01830.x. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Scherer D C, DeBuron-Connors I, Minnick M F. Characterization of Bartonella bacilliformis flagella and effect of antiflagellin antibodies on invasion of human erythrocytes. Infect Immun. 1993;61:4962–4971. doi: 10.1128/iai.61.12.4962-4971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schrier S L, Bensch K G, Johnson M, Junga I. Energized endocytosis in human erythrocyte ghosts. J Clin Investig. 1975;56:8–22. doi: 10.1172/JCI108083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sieira R, Comerci D J, Sanchez D O, Ugalde R A. A homologue of an operon required for DNA transfer in Agrobacterium is required in Brucella abortus for virulence and intracellular multiplication. J Bacteriol. 2000;182:4849–4855. doi: 10.1128/jb.182.17.4849-4855.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 38.Struyve M, Moons M, Tommassen J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J Mol Biol. 1991;218:141–148. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- 39.Teixeira-Gomes A P, Cloeckaert A, Zygmunt M S. Characterization of heat, oxidative, and acid stress responses in Brucella melitensis. Infect Immun. 2000;68:2954–2961. doi: 10.1128/iai.68.5.2954-2961.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsang H, Mollenhauer H, Ihler G. Entrapment of proteins, viruses, bacteria and DNA in erythrocytes during endocytosis. J Appl Biochem. 1982;4:418–435. [Google Scholar]
- 41.Villarejo M R, Zabin I. Beta-galactosidase from termination and deletion mutant strains. J Bacteriol. 1974;120:466–74. doi: 10.1128/jb.120.1.466-474.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker T S, Winkler H H. Bartonella bacilliformis: colonial types and erythrocyte adherence. Infect Immun. 1981;31:480–486. doi: 10.1128/iai.31.1.480-486.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinman D. The bartonella group. In: Dubos R J, Hirsch J G, editors. Bacterial and mycotic infections of man. 4th ed. Philadelphia, Pa: Lippincott Co.; 1965. pp. 775–785. [Google Scholar]
- 44.Weinman D. Infectious anemias due to Bartonella and related red cell parasites. Trans Am Philos Soc. 1944;33:243–287. [Google Scholar]
- 45.Wray W, Boulikas T, Wray V P, Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem. 1981;118:197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]