Abstract
In efforts to increase rigor and reproducibility, the USA National Primate Research Centers (NPRCs) have focused on qualification of reagents, cross-lab validations and proficiency testing for methods to detect infectious agents and accompanying immune responses in nonhuman primates. The Pathogen Detection Working Group, comprised of laboratory scientists, colony managers and leaders from the NPRCs, has championed the effort to produce testing that is reliable and consistent across laboratories. Through multi-year efforts with shared proficiency samples, testing percent agreement has increased from as low as 67.1% for SRV testing in 2010 to 92.1% in 2019. The 2019 average agreement for the four basic SPF agents improved to >96% (86.5% BV, 98.9 SIV, 92.1 SRV, 97.0 STLV). As new pathogens such as SARS coronavirus type 2 emerge, these steps can now be quickly replicated to develop and implement new assays that ensure rigor, reproducibly and quality for NHP pathogen detection.
Introduction
Specific Pathogen Free Macaques:
The use of Specific Pathogen Free (SPF) macaques has now become a standard of practice for biomedical research studies in which the presence of viral infections could be a confounding variable that potentially compromises the findings. Figure 1 summarizes the important evolution of proficiency panels used to provide rigor and reproducibility for the testing implemented to develop and maintain SPF colonies. Before the widespread use of SPF animals, numerous studies in the 1980’s that spanned a range of facilities and study types and used macaques as animal models were adversely affected by intercurrent viral infections influencing the outcomes. Table 1 documents a representative subset of these studies (1).
Figure 1.
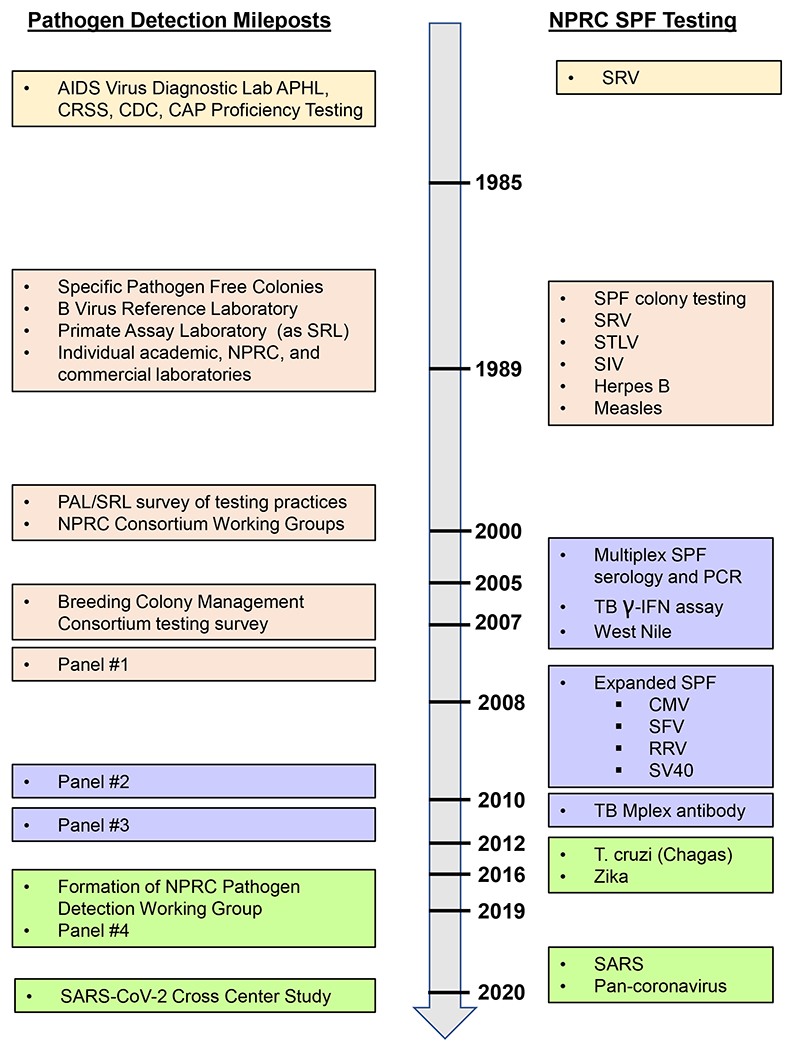
Timeline of events, testing, and shared samples to support SPF and other NHP colonies.
Table 1.
Studies compromised by Simian Betaretrovirus (SRV) infection.
| Facility | Study | Species | Number Animals |
|---|---|---|---|
| Pharmaceutical | Drug safety | M. fasicularis | 48 |
| Pharmaceutical | Drug safety | M. fasicularis | 40 |
| Contract | Gene therapy | M. mulatta | 20 |
| Contract | Teratology | M. fasicularis | 86 |
| University | Addiction | M. fasicularis | 12 |
| Contract | Drug safety | M. fasicularis | 52 |
| University | HIV | M. mulatta | 50 |
| University | Malaria | M. mulatta | 50 |
Adapted from: Lerche NW, Yee JL, Jennings MB. Establishing specific retrovirus-free breeding colonies of macaques: an approach to prelimary screening and surveillance. Anim Sci. 1994 Jun 44(3): 217-21.
The initial development and use of SPF colonies at the NIH sponsored National Primate Research Centers (NPRCs) and other NHP facilities was confounded by a lack of confidence that testing results from different laboratories tests were comparable. Definitions about SPF and the interpretation of results, especially indeterminates and false positives, became the subject of frequent SPF discussion groups. For example, the life cycle of SRV can cause variation in PCR test and antibody results over the course of infection. Interpreting these results, including indeterminate or partial reactivity was a constant challenge for many SPF colonies. Samples from animals determined to be SPF by one facility might be sent to another laboratory where they were found to be positive for an excluded agent by a different test algorithm. This not only had the potential to adversely affect individual research projects; but more importantly, it had the potential to cause an outbreak in an established SPF colony at another facility causing the loss of years of colony development and thousands of dollars spent on testing and breeding programs. This possibility of break through infections led some facilities to institute six month quarantine procedures for new animals entering their SPF breeding colonies. As the biology of the target agents was better understood and the different assays were compared using shared samples, standard definitions and terminology were developed to meet the challenges in SPF colony management with rigor and reproducibility.
To address the need for standardization, in 1989 the National Institutes of Health’s National Center for Research Resources (NCRR), which was reorganized in 2011 to form the Office of Research Infrastructure (ORIP) within NIH’s Office of the Director, initiated the first experimental research contracts to establish and maintain SPF NHP breeding colonies to produce offspring from which selected infectious agents had been eliminated. (2). The original targets for elimination included at least four viruses: i) Macacine herpesvirus 1, formerly known as Cercopithecine herpesvirus 1 and also referred to as B virus or Herpes B virus (BV), ii) Simian immunodeficiency virus (SIV), iii) Simian betaretrovirus formerly known as simian retrovirus type D (SRV), and iv) Simian T-cell lymphotropic virus (STLV-1). (1, 2, 3). Recently, with the emergence of the global COVID 19 pandemic, there is a similar need to establish testing to monitor for SARS coronavirus type 2 (SARS-CoV-2) (4), In addition to these viruses, SPF and other colonies have maintained longstanding practices to surveil and eliminate any Mycobacterium tuberculosis group pathogens (2).
Target Pathogens:
BV is an alphaherpes virus. All species of macaques can be naturally infected; and genotypes vary by species and location (5). Despite its zoonotic potential to cause disseminated infection and severe encephalomyelitis in humans, BV infection is generally asymptomatic in macaques. The primary routes of transmission in colonies are through oral or sexual contact. Grooming, biting, and indirect contact through fomites have also been implicated in transmission. Poor hygiene and overcrowded conditions can also exacerbate transmission (6). The initially low incidence of BV infection in young macaques increases rapidly upon sexual maturity. Rates of 80-90% have been reported in non-SPF colonies (6, 7, 8). Along with proper use of personal protective equipment and safe handling practices, the development of SPF colonies has played an important role in reducing occupational exposure to BV.
SIV is a lentivirus naturally found in African monkeys and apes. In those populations prevalence may exceed 50% (1, 9). Most natural SIV infections in African species in the wild are non-pathogenic, a likely result of their long-shared history of co-evolution (3, 10). Macaques are not naturally infected with SIV in the wild; however, infection can be induced experimentally or by exposure in captivity to African NHPs or their blood or body fluids sexually or through biting and scratching behaviors (1, 3). Transmission between group-housed infected and uninfected animals has been observed (3). Since SIV can cause immunodeficiency disease in macaques, it has become widely used as an animal model to study the biology of human immunodeficiency virus (HIV) and associated AIDS prevention and cure strategies (10). Although SIV is not endemic to macaque populations, the possibility of transmission from experimentally infected or other naturally infected host species in a facility make it an important SPF agent to guard against.
Macaques are natural hosts for exogenous SRV. These type D betaretroviruses are a group of closely related RNA viruses. Serotypes 1-8 have been described (3, 11, 12, 13). Prevalence is highly variable ranging from 0 to 50% and is influenced by geographic origins, and management and husbandry practices in captive macaque populations (3,11). Transmission usually occurs horizontally, through direct contact (parenteral, sexual, perinatal) between infected and susceptible animals, or indirectly through contact with contaminated fomites (instruments or equipment); transplacental transmission has also been documented. Virus shed in saliva, during mutual grooming or aggressive interactions involving biting and scratching, is a major vehicle for transmission (3, 11, 13, 14, 15). SRV infection can lead to immunosuppression and wasting; it was responsible for compromising multiple biomedical research studies in the 1980s and 90s (1). Its elimination was a major driver to establish SPF colonies.
STLV is a member of the Primate T Cell Lymphotropic Virus (PTLV) family. It is a Deltaretrovirus and a C-type member of the oncornavirus subgroup of retroviruses. Macaques and other Asian and African monkeys and apes are natural hosts (1). The seroprevalence in wild populations has been reported to vary considerably from 0-80% and from 3-12% in captive groups (1, 13, 14). STLV is highly cell-associated, and transmission occurs primarily through the transfer of infected cells in blood, breast milk, semen, cervical secretions, and other body fluids; and similarly, to other SPF viruses, transmission increases with increasing age as the animals reach sexual maturity (3, 16). STLV-1-induced overt clinical disease is extremely rare or nonexistent in macaques but has been linked with lymphoma and lymphoproliferative disease in African NHP species (3). Even in the absence of clinical signs, STLV infection has been shown to perturb the immune system and possibly confound research studies (17).
In addition to the four SPF target viruses described above, NHP’s can be infected with several other agents that can compromise the health of breeding colonies and/or represent an important risk of zoonotic infections. With the increased demand for pathogen-free animals for research areas including transplantation studies, congenital infections, disease pathogenesis, and vaccine development among others, as well as the improvement of diagnostic methods for NHPs, several research facilities have expanded their SPF programs incorporating screening methods for other pathogens including Simian foamy virus (SFV), Simian varicella Virus (SVV), Cytomegalovirus (CMV), Lymphocryptovirus virus (LCV), Rhesus rhadinovirus (RRV), Simian vacuolating virus 40 (SV-40), and SARS coronavirus type 2 (SARS-CoV-2).
SFV belongs the subfamily Spumaretrovirinae within the Retroviridae family. They are ancient complex retroviruses that have co-evolved with their NHP hosts for at least 30 million years (18). SFV is not known to cause disease in the natural host. However, it has been shown that it can produce dramatic vacuolizing (foam-like) cytopathic effects in tissue culture and it can exacerbate the pathogenesis of other viruses. SFV replicates primarily in tissues of the oral mucosa. Transmission occurs likely through transfer of infected saliva between animals, and it has been shown that infection can be also achieved through blood transfusion from an infected to an uninfected animal (19). Infection is common in NHPs, and it can be transmitted to humans but there is no evidence of human-to-human transmission.
SVV belongs to the subfamily of Alphaherpesvirinae within the Herpesviridae family. SVV induces disease in NHPs and mortality rate can be as high as 75%. The pathological, virological, and immunological features of the infection with this agent in NHPs are similar to those seen with Varicella Zoster Virus infection in humans. It becomes latent in gangliotic neurons and reactivates after environmental stress or immune suppression (20,21). Natural transmission of SVV occurs via inhalation of aerosols containing the virus or by direct contact with infected skin lesions.
CMV belongs to the subfamily Betaherpesvirinae within the Herpesviridae family. Worldwide, this is the most common pathogen transmitted congenitally in humans and typically it establishes a persistent infection that is generally asymptomatic in immunocompetent individuals; but it may cause severe diseases in patients with congenital or acquired immunodeficiencies such as in organ transplants or AIDS. Rhesus CMV (RhCMV) is the most widely studied NHP CMV due to the high similarity with the human CMV genome, as well as its pathogenesis, prevalence, and transmission. Thus, Rhesus macaques have been used to develop a model of congenital infection of CMV. RhCMV is excreted in saliva, urine, and breast milk of seropositive animals (22, 23). Transmission of the virus occurs horizontally through direct contact between infected and uninfected animals. Unless preventative measures are taken, by 7 months of age the majority of animals in breeding colonies are seropositive and by 1 year of age almost all animals have seroconverted for RhCMV (24).
LCV belongs to the subfamily Gammaherpesvirinae within the Herpesviridae family. The most widely studied LCV is rhesus LCV (rhLCV). Several studies have demonstrated highly genetic and functional homology between LCV and Epstein-Barr virus (EBV) (25), as well as strong similarities in terms of pathogenesis and immunological responses between rhLCV infection in resus macaques and EBV infection in humans (26, 27). LCV infection is very common in rhesus macaques kept in captivity with almost all adult animals testing seropositive.
RRV belongs to the Gammaherpesvirinae subfamily within the Herpesviridae family. Rhesus RRV, classified as a γ2 rhadinovirus of the RV2 lineage, has been described as the simian homologue of Kaposi’s sarcoma-associated herpesvirus (28). Natural infection of macaques with RRV is reported as subclinical in immunocompetent animals. However, disease development has been observed in animals coinfected with RRV and SIV (29, 30). Serologic evidence indicates that RRV infection is highly endemic in socially housed rhesus macaques, with viral DNA detected in blood and saliva samples. As compared to adults, younger animals present with higher viral loads (31, 32).
SV-40 belongs to the Polyomaviridae family. It is an oncogenic DNA virus discovered in 1960 as a contaminant of polio vaccines. It has been shown that SV-40 can induce tumors in rodents and transforms many types of cells in culture, including human cells (33, 34). Natural infection of rhesus macaques with SV-40 has been reported as subclinical, but disease has been observed in animals coinfected with SV-40 and SIV. SV-40 nucleic acid has been detected in renal tissue in primary infections and brain tissue after viral reactivation (35). In captive macaques. infection with SV-40 is very common with transmission most likely occurring by contact with virus shed in urine.
SARS-CoV-2 can infect NHPs (including Macaque, Cercopithecus, and Papio species) making them both an effective animal model for research and a population at risk for infection (4, 36). Reports in the research literature suggest that this virus follows a typical pattern: Viral RNA is detectable by reverse transcriptase PCR in samples collected by nasopharyngeal or oral swabs 3-21 days after the initial infection; infection elicits host immune response as indicated by IgM antibody as early as day 7 and IgG antibody as early as day 10 with peak antibody responses have been reported at days 21-28 (36, 37). The duration of antibody response has not been well documented; but other coronaviruses are variable at 1 year post initial infection (38). The potential for SARS-CoV-2 to cause clinical disease in NHPs has been variable under experimental conditions (39). Given the susceptibility and presumed lack of pre-existing immunity in NHPs, colony management practices, including surveillance testing, to reduce the risk of virus introduction and transmission became a priority with the emergence of the COVID-19 pandemic (4).
Establishing testing laboratories:
In addition to supporting development of SPF colonies, in 1989 NIH NCRR (presently ORIP) also supported the establishment of dedicated reference laboratories for BV and retrovirus testing. The BV laboratory, now known as the National B Virus Resource Center, directed by Dr. Julia Hilliard is located in the Viral Immunology Center of Georgia State University’s Department of Biology. It serves as a global resource originally funded by NCRR to assist in the identification of zoonotic disease transmissions and develop enhanced strategies to detect virus in macaques. The laboratory maintains a particular focus on the transmission of BV from Asian monkeys to humans who have contact with them (40). Concurrently, NIH established the Simian Retrovirus Laboratory (SRL) at the California National Primate Research Center at the University of California, Davis, directed by Dr. Nicholas Lerche to develop and provide testing for the other targeted viruses to develop SPF colonies and improve overall NHP colony health. SRL expanded to become the Pathogen Detection Laboratory and then the Primate Assay Laboratory (PAL) (41). A similar SPF testing approach was developed collaboratively at the Primate Pathogen Detection Services Laboratory (PDSL) at Washington National Primate Research Center. Multiplex assays for testing multiple antibodies and pathogens were successfully implemented at PAL and PDSL for the basic SPF pathogens BV, SIV, SRV, STLV and measles (42, 43, 44, 45). Over the years, other individual laboratories began testing and these assays have been widely applied and resulted in more accurate and efficient diagnostic tests leading to improved SPF status and better characterized NHP colonies (3). These tests and their methods have also been transferred internationally. Precise testing is especially important in source countries where animals are more likely to be exposed to the natural sources of infection including local wildlife. Examples of local SRV, STLV and herpesvirus infections in Asian macaques at international locations are important examples of the importance of quality testing programs throughout the world (46, 47, 48). Therefore, efforts by PAL, PDSL, and others to train laboratories in China, Indonesia, Vietnam, Cambodia, Thailand, and India have resulted in well characterized SPF status for macaques coming from source countries and leading to safer and healthier domestic SPF colonies.
Initially only a few academic and commercial reference labs provided the majority of SPF testing. The reference laboratories provided important services but frequently there was a need for more rapid turnaround of critical assays, particularly for SRV or BV. The high volume of testing requested sometimes led to delays in receiving test results, particularly if reactivity needed to be repeated for confirmation. These delays posed management challenges because formation of breeding groups and animal moves into new housing often awaited those test results. In ensuing years, as methods were published, reagents were shared, and reference labs provided training, most of the NIH sponsored NPRCs set up on site laboratories. This transition was further facilitated by the rise in commercial availability of reagents and testing services. With the increasing options and variables for testing, the need for standardized definitions of infection and interpretation of laboratory results became apparent.
Rigor and reproducibility:
In the late 1980’s PAL was a sub-section of the University of California Davis AIDS Virus Diagnostic Laboratory (AVL). In addition to participating in regular proficiency testing for HIV sponsored by the Centers for Disease Control Model Performance Evaluation Program and the College of American Pathologists (49), the AVL was one of the founding members of the Consortium for Retrovirus Serology Standardization (50). The purpose of this cooperative group of public health, blood bank, academic, government and commercial laboratories was to address discrepancies and formulate standard interpretative laboratory diagnostic criteria for the western immunoblot and other laboratory assays used to detect HIV infection. The AVL was a leader in sharing protocols, reagents, and control samples; and in distributing and analyzing data from a sample exchange program. AVL leadership also participated in HIV testing consensus conferences sponsored by the Association for State and Territorial Public Health Laboratory Directors (51). The lessons learned from these experiences equipped the PAL to set up a similar program for SPF testing in NHPs.
In the early 2000’s, PAL leadership participated in discussions with other NPRCs and their testing laboratories to collect and distribute information about the availability of various reagents, controls, standards, assay methods and protocols, and equipment platforms. That information exchange was followed up with a sample exchange and proficiency testing as important tools to ensure the rigor and reproducibility in many participating laboratories using different assay platforms for diagnosing animals with a predetermined list of infectious agents.
Improvements in rigor and reproducibility have been achieved by applying some of the guidance used for GLP/GMP/GCP sample analysis. Important considerations for laboratories include: 1) Written SOPs; 2) Personnel training/qualification; 3) Method validations; 4) Instrument qualification and calibration; 5) Qualification of key materials and controls; 6) Data security. All of the assays run for pathogen detection are aimed at high sensitivity with specificity for macaque-specific pathogens. Assays determine the presence or absence of a pathogen by PCR or the host antibody response to the pathogens. Determining if a test result is positive or negative is critical for accurate interpretation. Standard analytical tools such as GLP guidelines for ligand-binding assays for anti-drug antibodies can be useful to determine such cutpoints for virus antibodies in serum (52, 53). Testing a statistically significant number of known negative samples (50 or more) with the calculation (Average + [3 x std. dev]) can give an acceptable and reproducible cut-off point which can be normalized on each run using a small number of known negative and positive samples. GLP guidelines recommend validations prior to sample analysis to ensure the reproducibility of results within a laboratory and the sensitivity and specificity for the target pathogens. Validation parameters of reproducibility (inter- and intra-lab precision), sensitivity (with lower limits of detection characterized) and specificity (to understand and reduce the detection of existing non-specific reactivity) are all addressed and reported among the NHP testing laboratories. The proficiency testing included as part of the collaborations among the NHP testing facilities, helps to ensure the inter- and intra-lab reproducibility and robustness and is useful to compare limits of detection and specificity among different labs with similar or identical methods. These periodic proficiency tests and subsequent face-to-face meetings to report on the results can serve as a model for all laboratories seeking to harmonize assays and results. Such standard practices in different laboratories have led to a high level of agreement of final results.
Materials and Methods- Shared sample panels:
As part of the collaborative work in NHP testing laboratories, an emphasis has been placed on standardization of reagents that are often very specialized for macaques. Standardization does not mean that all laboratories use the same reagents, methods, and instrument platforms; but rather that the final analysis and interpretation of the assay data generated following each participating laboratories’ algorithms agree in correctly detecting all defined infected animals. For example, a viral lysate may have lower specificity than a recombinant protein antigen; but if properly followed up with a more specific confirmatory assay such as a western immunoblot, the final interpretation of the combined screening and confirmatory algorithm would yield the correct result. Similarly, a single recombinant protein antigen might be less sensitive than a viral lysate, but a combination of multiple recombinants could be more sensitive. Once standards are established each laboratory needs to validate that the testing algorithm they are using is appropriately applied and rigorously adhered to in order to accurately detect infection.
The initial panels in 2008 and 2010 consisted of plasma or serum collected at the CNPRC. The 2012, 2017 2019, and 2020 panels included plasma, serum, or DNA collected at several participating facilities. All samples from participating facilities were forwarded to PAL to be de-identified, aliquoted, and compiled as panels. The earliest panel included only well-characterized samples; but historically challenging samples, defined as samples that did not perform equally and could not be clearly interpreted as infected or uninfected using various testing methods, were added to later panels. Selecting panel samples that were appropriate for use in the varying testing methods employed at the different facilities was challenging. Many of the assays were optimized for different, sometimes conflicting, sample types and volumes. A number of potential obstacles had to be addressed: i) Complete clinical and exposure histories to facilitate true infection status of the animal was not always available. ii) Acquiring adequate volume, especially when duplicate or confirmatory testing was part of the algorithm: for some samples the donor size and body condition scores were prohibitively limiting; and for others which were found opportunistically, there was only residual material left. iii) Time, transport, and storage conditions were also variable. In general, fresh samples were not an option for the panels. iv) Laboratory biosafety requirements at some institutions required that all samples received be inactivated. Although not always ideal, most serological assays could accommodate serum or plasma. However, various anticoagulants and inactivation methods are known to cause high background in some assays. Identifying and providing adequate samples for virus detection were more problematic. Both the input sample type and volume required by various DNA extraction methods and the quantity and quality of material needed for the PCR assay had to be addressed. Panels have included pre-extracted DNA and frozen EDTA or heparinized whole blood, neither of which was the ideal sample for all the assays. Some PCR assays were optimized for a specific extraction method. Sample availability and volume continue to be challenging but many issues with sample type and selection and panel preparation and distribution have improved with experience. In this work we present the results of four multi-site proficiency panels for SPF testing and one cross center study for SARS-CoV-2 antibody testing.
Results- Comparison and standardization of tests:
2008
In 2008 PAL selected and aliquoted a set of 8 samples that were previously tested and known to be clearly positive or negative for BV, SIV, SRV, and STLV. These panel samples are described in Supplemental Table 1a. Reference laboratories providing NHP SPF testing were invited to receive and perform antibody testing on the panel samples and report raw data, interpretations, reagents, and methods in as much detail as they choose. PAL agreed not to link the submitted data to the specific laboratory that generated it. All submitted data was summarized and shared back to the participants. Since most SPF colonies did not have their own on-site testing laboratories, these laboratories were providing the majority of SPF testing. Six Laboratories participated using test methods including multiplex microbead immunoassays (Luminex), enzyme immunoassays, western immunoblot, and indirect immunofluorescence. Viral lysate was used as the target antigen for all four SPF viruses. Some SRV assay antigens included serotypes 1,2, and 5 and also recombinant transmembrane glycoprotein. Surrogate viruses (HPV2, HSV) and recombinant proteins were also used in some BV assays. Data was analyzed and the percent agreement (ratio of tests in agreement with the consensus majority result to all tests performed) was calculated. With the exception of one BV result, specificity was very good using these well-characterized negative samples; but there were some discrepancies for BV, SRV, and STLV positive results as summarized in Supplemental Table 1b.
2010
To facilitate increased access and promote sharing of valuable resources among the scientific community, in the early 2000’s ORIP established NPRC Consortiums which include working groups among the eight Centers located throughout the United States. The Consortiums’ overall mission is to improve global health through biomedical research with NHPs and to communicate this mission through outreach. One group, the Breeding Colony - Management Consortium (BCMC) asked PAL to coordinate a survey of their members to determine SPF breeding colony diagnostic testing needs and available resources. The initial survey and follow-up questions were addressed from late 2007 into early 2010. Information collected included current testing practices, available resources, and unmet needs related to viruses, number of samples, methods, protocols, reagents, antigens, controls, standards, and databases. Specific initiatives to explore group purchasing and proficiency testing were also explored. PAL shared its viral testing protocols and provided the remaining sample aliquots from the previously described panel as validation standards and controls to individual NPRC laboratories; and established a working relationship with industry to ensure ongoing availability and professional support and training for reagents for all NPRCs.
Using information from these surveys and discussions, another proficiency panel exchange was coordinated by PAL at the end of 2010. This panel included serum or plasma for antibody testing and DNA. Three of the participating laboratories provided 16 samples for the panel; there were eight parallel plasma and DNA samples and eight serum only samples. Since this panel included samples submitted to various laboratories for routine testing, the descriptive information available was variable. To address reproducibility, there was a pair of samples collected on different dates from the same animal. The panel samples are described in Supplemental Table 2a. After de-identifying, aliquoting and distributing the panels, PAL collated and analyzed the results and then shared them with all participants. Participants were permitted to submit their results without identifying themselves. Eleven sets of results were submitted from six NPRC laboratories, one academic reference laboratory, and four commercial reference laboratories. The antibody results are summarized in Supplemental Table 2b. Technical issues with the DNA quantity, stability, and quality for various methods precluded some laboratories from completing the testing. The PCR results were not analyzed. Unlike the 2008 panel which only included clearly positive or negative samples which addressed laboratory proficiency, this panel included more difficult to interpret samples to better understand and characterize needed testing improvements. More laboratories (including some that recently initiated testing and had only minimal experience) performed testing and more samples were tested. Despite these additional challenges, except for SRV positive samples, the level of agreement between results was at least as favorable as with the earlier, more limited panel. The SRV serological false negative samples would have been correctly identified if the laboratory’s parallel PCR results had been added to the analysis. There were three noteworthy observations from the data: i) The specific target antigen used is more critical than the assay method, platform, or instrument. ii) Some discrepancies are not due to the actual test result but due to how the result is interpreted; highlighting the need for standardized definitions. iii) In truly infected animals, there were negative PCR / positive antibody results and positive PCR / negative antibody results which would still be correctly identified as infection if both tests were run in parallel; thus, supporting the need for SRV testing algorithms to include both antibody and agent detection (3).
2012
A third shared proficiency panel that included both characterized and unknown or “problem” serum or plasma and DNA was prepared by PAL in 2012. This 24 member panel was comprised of DNA and plasma/serum specimens from 20 animals, serum or plasma only from three animals, and DNA only from one animal. The panel samples, as described in Supplemental Table 3a, were chosen to reflect practical, real world testing situations; thus, availability of donor information was variable. Continued technical difficulties with providing adequate DNA quantity and quality as well as the small number of laboratories performing PCR precluded statistically significant conclusions for virus detection; but antibody results from six NPRC laboratories were successfully generated, collated and compared as shown in Supplemental Table 3b. There were four false negative BV results - generated by two different laboratories using the same commercial reagent; but there were no false positive results. The laboratories reporting the BV false negative results were using the proficiency panel samples to evaluate that particular assay as a supplemental test. The validated assays they were routinely using did detect antibody and did not yield false negative results. Noting the discrepancy, the laboratories worked with the supplier and have not had this issue recur with other reagent lots; however, neither laboratory is now relying on this assay for routine testing. There were several apparent false positive results for SIV or STLV antibody, but no false negative results. Some of the apparent false positive results generated by screening tests in laboratories that did not have confirmatory testing available on-site and would normally have referred the samples to another laboratory with the subsequent testing correctly identifying the samples as uninfected. A false positive screening test is not necessarily problematic or unexpected. By design, SPF testing algorithms use exquisitely sensitive screening assays paired with a more specific confirmatory assay (3). For SRV, four false-negative samples test results were generated in two different laboratories, one using commercial reagents and the other used in laboratory-developed reagents. As previously noted, SRV testing algorithms strongly recommend including both antibody and virus detection since one test or the other may be negative at any given time point during infection. Thus, it is possible that these false negative SRV antibody results would still yield a final correct interpretation of infection if coupled with a positive PCR result. Similarly to the situation with SIV and STLV, there were SRV false positive screening results which were correctly overridden and interpreted as uninfected by subsequent negative confirmatory test results.
2019
In 2019, ORIP sponsored the formation of the Pathogen Detection Working Group (PDWG). In addition to surveying, compiling, and publishing the available resources from 6 NPRC laboratories on a website (54), another round of surveys and proficiency testing was included in the first year’s accomplishments. As in previous iterations, the primary purpose of this panel was to provide proficiency testing for BV, SIV, SRV, and STLV. In addition, the samples were used to develop and validate assays for other expanded SPF or emerging pathogens including measles virus (MV), rhesus CMV, RRV, SFV, LCV, SV40, SVV, flaviviruses (Zika, West Nile), T. Cruzi, and Burkholderia pseudomallei. Five NPRCs submitted the 32 serum, plasma, whole blood, or DNA samples as described in Supplemental Table 4a. A special feature of this panel were DNA standards for SRV serotypes 1-5 provided by the PDSL. Antibody results generated using both in-laboratory prepared and commercial multiplex liquid and solid arrays, singleplex microbead immunoassays, enzyme immunoassays, western immunoblots and immunofluorescent assays. There were no discrepant BV antibody results. Although two labs each reported non-negative SIV screening results in samples that were reported as negative in all other laboratories, those samples were not positive in their subsequent confirmatory test in the same laboratory so therefore were correctly identified as uninfected; thus, there were no discrepant final interpretations. Although different SRV antibody screening assays and reagents yielded different false positives, confirmatory tests brought results for all but four samples into agreement. Two samples yielded antibody false negative results possibly due to the same antigen reagent and there was no consensus agreement for two other samples, highlighting the continuing need to design improved antigens and test methods. The challenges of distinguishing apparent SRV antibody from simian endogenous virus and correlating it with infection have been documented (55, 56). As with SRV antibody, different STLV antibody screening reagents yielded different false positives. This is partially explained by known false positive cross reactivity between SRV and STLV envelope proteins (env) that result in false positive screening tests with one commercial reagent; however, confirmatory testing brought the final determination of infected or uninfected for all but one sample into agreement. The single discrepancy was not due to a difference in actual test results but due to a difference in how the western immunoblot band pattern was interpreted. In addition, although not all laboratories submitted results, reasonably good testing agreement was also observed for SFV, CMV, and T. Cruzi. Assays for some of the other agents of interest need further improvement and that work is in progress. Antibody and real time PCR results are summarized in Supplemental Table 4b.
2020
With the emergence of the COVID-19 pandemic and the potential threat to NHPs with the PDWG laboratories were tasked with providing testing for SARS-CoV-2 surveillance. Although some commercial and research reagents and protocols were available, none were well validated for use in NHP. The laboratories worked together to quickly evaluate various assays for viral RNA and antibody and establish standards for testing. Initial surveillance data along with a number of enhancements to the sample collection and RT-PCR source materials and protocols were posted on the group’s website (54). A cross center study including 10 human or NHP antibody detection immunoassays was performed at 6 sites. These included commercially available and laboratory developed enzyme immunoassays, multiplex microbead assays, and solid phase arrays. A panel of 52 samples from known negative rhesus or pig tailed macaques was shared for testing by all the assays across various sites; in addition, each laboratory tested available samples from experimentally infected or vaccinated rhesus macaques, African green monkeys, or baboons shared by researchers at their facilities (54). Due to logistical issues (biosafety, materials transfer, research protocols), some but not all the positive samples were tested on multiple assays at multiple sites. Some assays included seasonal coronavirus antigens. There were some differences in interpretation based on the specific antigens tested and reactivity pattern in the various assays, all performed well. Fifteen samples reported as reactive to a single antigen in five assays, but none were reported reactive to both spike and nucleocapsid antigens. Seven samples collected from animals at least 14 days post experimental infection were reported negative in 2 different assays. There were no other discrepancies. The assays and results are shown in Tables 2a and 2b.
Table 2a.
SARS-CoV-2 Antibody Assay Target Antigens
| Assay | S-CoV-2 Antigens | Other Antigens |
|---|---|---|
| Xpress Spike ELISA | S1, S2 | |
| Xpress Nucleocapsid ELISA | NC | |
| Intuitive Bioscience Panel | S1, S2, NC | |
| MesoScale Dx Panel (human) | S, NC, RBD, NTD | HKU1 S, OC43 S, NL63S, 229ES, SARS-CoV-1 S |
| Charles River Labs MMIA | S, NP | HKU1 S, OC43 S, NL63S, 229ES |
| Tetracore MMIA (human) | S1, NC, RBD | S-CoV-2 Variants S |
| Xmap MMIA (human) | S, NC, RBD, NTD | |
| CNPRC MMIA | S trimer, NC, RBD, Viral Lysate | |
| Xmap MMIA (human) | S, NC, RBD, NTD | |
| WANPRC MMIA | S, NC | HKU1S1, OC43S, NL63S1, 229ES1 OC43NP, NL63NP, 229ENP |
Table 2b.
Aggregate SARS-CoV-2 antibody testing results. Samples were characterized as either positive or negative based on the sample history and results reported from all participants. The positive samples were collected at least 14 days post experimental inoculation. The number of tests performed per virus and per sample varied for each laboratory and each assay.
| SARS-CoV-2 | |
|---|---|
| Number of positive samples tested | 260 |
| Number of positive test results | 255 |
| Number of total negative samples tested | 450 |
| Number of negative test results | 435 |
| Percent Agreement | 94.2% |
Discussion and Conclusions
Thirty years ago, baseline serosurveys performed at the CNPRC for five facilities establishing SPF colonies demonstrated a prevalence of 1-3% for SRV in three facilities, 1-4% for STLV in four colonies and no SIV as summarized in Table 3(1). After the early years of colony development, only rare potential breakthroughs have been found. Similarly, the cumulative rate of non-negative BV results from six SPF colonies changed from 0.132 to 0.036 after one year of aggressive SPF management, and then declined further to 0.018 in year two and 0.004-0.006 in years three to six yielding a nearly 20-fold reduction in risk in occupational exposure to BV (3, 57).
Table 3.
Prevalence of antibodies to Simian retroviruses in the 1990’s.
| Colony | # Animals | SIV | SRV | STLV |
|---|---|---|---|---|
| A | 663 | 0% | 0% | 2.9% |
| B | 51 | 0% | 0% | 0% |
| C | 332 | 0% | 2.9% | 0.9% |
| D | 640 | 0% | 1.0% | 2.1% |
| E | 351 | 0% | 3.1% | 3.9% |
Adapted from: Lerche NW, Yee JL, Jennings MB. Establishing specific retrovirus-free breeding colonies of macaques: an approach to preimary screening and surveillance. Anim Sci. 1994 Jun 44(3): 217-21.
The establishment of SPF colonies has led to improved individual animal and colony health and reproduction; more well characterized animal models free of potentially confounding infections for biomedical research; and reduced potential exposure to zoonotic pathogens in occupational settings. The original SPF program targeted elimination of four viruses: BV, SIV, SRV, and STLV in closed groups of Indian origin Macaca mulatta or Indonesian origin Macaca nemistrina. Success has been achieved using a two-part test and removal strategy: extensive laboratory testing to detect the agents and / or the host antibody responses and barrier management to prevent direct or indirect contact with potentially infected or untested / unknown animals (1,2,3). As a result of the successful test and removal strategy, nearly all NPRC facilities maintain at least some or even all of their colony as SPF for the original four agents. In addition, some colonies have added subsets of animals free of combinations of additional agents. Some of the additional agents targeted for elimination include RhCMV, SFV, LCV, RRV, SV40, and others. These expanded SPF animals are required to meet the challenge of emerging agents and the increasing need for new research models. The numbers, species, and origin of animals have also expanded. For example, an SPF baboon resource supported by ORIP is free of at least 18 recommended pathogens, including herpesviruses, retroviruses, polyomavirus, paramyxovirus, arterivirus, and monkeypox virus (59). In order to encompass the different agents, combinations, and animals at the various NPRCs and other colonies, this manuscript uses the term SPF generically; But it is important to note that until nomenclature conventions are established, the term SPF alone is not meaningful when referring to a specific situation unless it is defined to include a description of both the agents and the animals.
The origin of the Primate Assay Laboratory out of the University of California Davis AIDS Virus Diagnostic Laboratory (AVL) in the 1980’s provided a foundation in rigor and reproducibility evolving from critical human HIV testing. This culture to address discrepancies and formulate standard interpretative laboratory diagnostic criteria for the western immunoblot and other laboratory assays used to detect HIV infection has provided a useful example to apply to SPF testing in NHP. The comparison of test algorithms began as an informal collaboration by participating laboratories; and the establishment by ORIP and the NPRC leadership of the PDWG has formalized this process and greatly facilitated reproducibility in the field. Over the course of five panel exchanges from 2010-2020 we have shown increasing percent agreement for SPF and other viruses, even with more difficult, challenge samples; thus validating our hypothesized outcome that proficiency testing improves testing quality and standardization. The most recent results have shown improved percent agreement: BV 86.5%, SIV 98.9%, SRV 92.1%, STLV 97%, CMV 94.4%, RRV 94.9%, SFV 96.9%, T. cruzi 92.4%, SARS-CoV-2 94.2%, as partially summarized in Figure 2.
Figure 2.
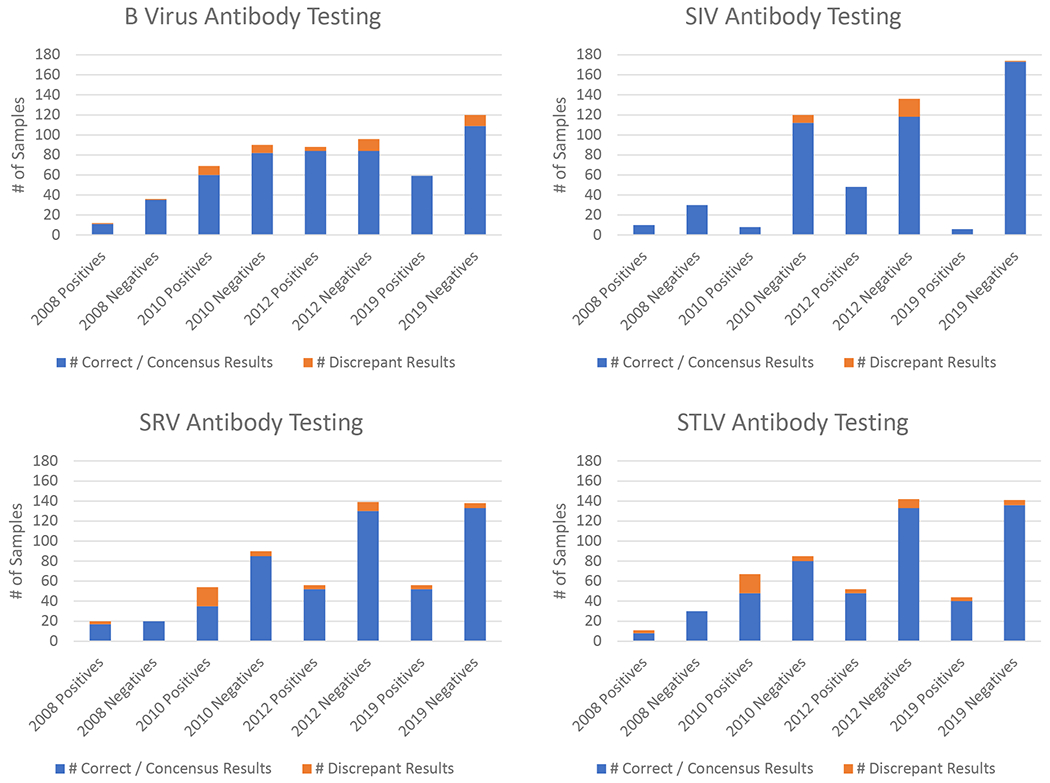
Summary of increasing percent agreement of antibody testing results for the four initial SPF target viruses from exchange panels from 2008 to 2019. Note that the 2008 panel included only prototypical positive and negative samples while subsequent panels included more difficult to interpret challenge samples.
Moving forward, the PDWG’s ongoing exchange of information and focus on regularly scheduled proficiency testing will lead to continued quality improvement as demonstrated by an increased number of results in agreement with the correct or consensus antibody results from all laboratories. The current shortage of NHPs to meet critical needs in biomedical research (58, 59) highlights the need to derive and expand SPF colonies. This effort will require pathogen testing that meets the highest quality standards of rigor and reproducibility. As summarized in this report, testing characterized samples has and will continue to provide a means to assess assay sensitivity, specificity, reproducibility, and accuracy in multiple laboratories. This will be critical as reagent availability changes, technology advances, and new scientific discoveries are made. Analysis of data generated using the current and future test reagents and protocols will be important to determine and interpret exactly what is being measured in order to minimize variation and provide standardized definitions. The compiled data will also provide guidance in selecting the most appropriate test for specific questions or situations. Shared reagents, controls, standards, assay methods, protocols and proficiency testing ensure the integrity of testing results generated for the NIH/OAR SPF Macaque Breeding Colony Program and informs the pathway to continue to improve quality and address the testing needs for new and emerging pathogens as they are added to expanded SPF colonies.
Supplementary Material
Acknowledgements:
We thank Thomas Vanderford, David Lee, Mary Barnes, Kathrine Falkenstein, Elizabeth Didier, Heidi Palmer, Danny De Los Reyes, Kamm Prongay, Luis Giavedoni, Laura Parodi, LaRene Kuller, Peter Nham, Bryson Halley, and other members of the NIH P51 supported National Primate Research Centers Pathogen Detection Working and their laboratory staff for their testing and analysis expertise that made this study possible. The members of the PDWG provided valuable scientific discussion.We thank Sheri Hild from NIH for encouraging, reading, and providing valuable insights for this manuscript.
This work was supported in part by the Office of Research Infrastructure Programs/OD grant Nos. P51 OD011107 and 5U42OD010990 (California NPRC), P51 OD011092 and U42 OD010426 (Oregon NPRC), P51OD010425 and U42OD011123 (Washington NPRC), and P51OD011104, U42 OD024282, and U42OD010568 (Tulane NPRC).
Footnotes
The authors confirm that the ethical policies of the journal, as noted on the journal’s author guidelines page, have been adhered to. Ethical approval was not required because no animals were used for research in this study. Samples used were from surplus material remaining from standard of care medical treatment or research studies approved by appropriate institutional review committees.
References
- 1.Lerche NW, Yee JL, Jennings MB Establishing specific retrovirus-free breeding colonies of macaques: an approach to primary screening and surveillance. Lab Anim Sci. 44(3):217–21 (1994). [PubMed] [Google Scholar]
- 2.Morton WR, et al. Specific pathogen-free macaques: definition, history, and current production. ILAR J. 49(2): p. 137–44 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Yee JL, Vanderford TH, Didier ES, et al. Specific pathogen free macaque colonies: a review of principles and recent advances for viral testing and colony management. J Med Primatol. 45(2):55–78 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yee JL, Van Rompay KKA, Carpenter AB, et al. SARS-CoV-2 surveillance for a non-human primate breeding research facility. J Med Primatol. 49: 322–331 (2020). 10.1111/jmp.12483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weigler BJ, Hird DW, Hilliard JK, et al. Epidemiology of cercopithecine herpesvirus 1 (B virus) infection and shedding in a large breeding cohort of rhesus macaques. J Infect Dis. 167:257–263 (1993). [DOI] [PubMed] [Google Scholar]
- 6.Andrade MR, Yee J, Barry P, et al. Prevalence of antibodies to selected viruses in a long-term closed breeding colony of rhesus macaques (Macaca mulatta) in Brazil. Am J Primatol. 59:123–128 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Sariol CA, Gonzalez-Martinez J, Arana T, et al. Differential distribution of antibodies to different viruses in young animals in the free-ranging rhesus macaques of Cayo Santiago. J Med Primatol. 35:369–375 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Lowenstine LJ, Pedersen NC, Higgins J, et al. Seroepidemiologic survey of captive Old-World primates for antibodies to human and simian retroviruses, and isolation of a lentivirus from sooty mangabeys (Cercocebus atys). Int J Cancer. 38:563–574 (1986). [DOI] [PubMed] [Google Scholar]
- 9.Hahn BH, Shaw GM, De Cock KM, et al. AIDS as a zoonosis: scientific and public health implications. Science 287:607–614 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Gardner MB, Luciw PA. Animal models of AIDS. FASEB J. 3(14):2593–606 (1989). [DOI] [PubMed] [Google Scholar]
- 11.Montiel NA An updated review of simian betaretrovirus (SRV) in macaque hosts. J Med Primatol. 39:303–314 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Zao CL, Armstrong K, Tomanek L, et al. The complete genome and genetic characteristics of SRV-4 isolated from cynomolgus monkeys (Macaca fascicularis). Virology. 405:390–396 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lerche NW: Simian retroviruses: infection and disease-implications for immunotoxicology research in primates. Journal of immunotoxicology. 7:93–101 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Lerche NW,, Osbornm KG Simian retrovirus infections: potential confounding variables in primate toxicology studies. Toxicol Pathol. 31 Suppl:103–110 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Daniel MD, Letvin NL, Sehgal PK, et al. Prevalence of antibodies to 3 retroviruses in a captive colony of macaque monkeys. Int J Cancer. 41:601–608 (1988). [DOI] [PubMed] [Google Scholar]
- 16.Parrish SW, Brown AE, Chanbancherd P, et al. Transmission of STLV in a closed colony of macaques. Am J Primatol. 63:103–109 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Yee JL, Montiel NA, Ardeshr A, et al. Constitutive release of IFNgamma and IL2 from peripheral blood mononuclear cells of rhesus macaques (Macaca mulatta) infected with simian T-lymphotropic virus type 1. Comp Med. 63:508–514 (2013). [PMC free article] [PubMed] [Google Scholar]
- 18.Switzer WM, Salemi M, Shanmugam V, et al. Ancient co-speciation of simian foamy viruses and primates. Nature.434(7031):376–80 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Khan AS, Kumar D, Simian foamy virus infection by whole-blood transfer in rhesus macaques: potential for transfusion transmission in humans. Transfusion. 46(8):1352–9 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Messaoudi I, Barron A, Wellish M, et al. Simian varicella virus infection of rhesus macaques recapitulates essential features of varicella zoster virus infection in humans. PLoS Pathog 5(11): e1000657 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Traina-Dorge V, Doyle-Meyers LA, Sanford R et al. Simian Varicella Virus Is Present in Macrophages, Dendritic Cells, and T Cells in Lymph Nodes of Rhesus Macaques after Experimental Reactivation. J Virol. 89(19):9817–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asher DM, Gibbs CJ Jr, Lang DJ, et al. Persistent shedding of cytomegalovirus in the urine of healthy rhesus monkeys. Proc Soc Exp Biol Med. 145:794–801 (1974). [DOI] [PubMed] [Google Scholar]
- 23.Huff JL, Eberle R, Capitanio J, et al. Differential detection of B virus and rhesus cytomegalovirus in rhesus macaques. J Gen Virol. 84(Pt 1):83–92 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Vogel P, Weigler BJ, Kerr H, et al. Seroepidemiologic studies of cytomegalovirus infection in a breeding population of rhesus macaques. Lab Anim Sci. 44(1):25–30 (1994). [PubMed] [Google Scholar]
- 25.Rivailler P, Jiang H, Cho YG, et al. Complete nucleotide sequence of the rhesus lymphocryptovirus: genetic validation for an Epstein-Barr virus animal model. J Virol. 76(1):421–6 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moghaddam A, Rosenzweig M, Lee-Parritz D, et al. An animal model for acute and persistent Epstein-Barr virus infection. Science. 276(5321):2030–3 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Rivailler P, Carville A, Kaur A, Rao P, et al. Experimental rhesus lymphocryptovirus infection in immunosuppressed macaques: an animal model for Epstein-Barr virus pathogenesis in the immunosuppressed host. Blood 104(5):1482–9 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Desrosiers RC, Sasseville VG, Czajak SC, et al. A herpesvirus of rhesus monkeys related to the human Kaposi’s sarcoma-associated herpesvirus. J Virol. 71(12):9764–9 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong SW, Bergquam EP, Swanson RM, et al. Induction of B cell hyperplasia in simian immunodeficiency virus-infected rhesus macaques with the simian homologue of Kaposi’s sarcoma-associated herpesvirus. J Exp Med. 190(6):827–40 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orzechowska BU, Powers MF, Sprague J et al. Rhesus macaque rhadinovirus-associated non-Hodgkin lymphoma: animal model for KSHV-associated malignancies. Blood.112(10):4227–34 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White JA, Todd PA, Yee JL et al. Prevalence of viremia and oral shedding of rhesus rhadinovirus and retroperitoneal fibromatosis herpesvirus in large age-structured breeding groups of rhesus macaques (Macaca mulatta). Comp Med. 59(4):383–90 (2009). [PMC free article] [PubMed] [Google Scholar]
- 32.White JA, Yang X, Todd PA, et al. Longitudinal patterns of viremia and oral shedding of rhesus rhadinovirus and retroperitoneal fibromatosis herpesviruses in age-structured captive breeding populations of rhesus Macaques (Macaca mulatta). Comp Med. 61(1):60–70 (2011). [PMC free article] [PubMed] [Google Scholar]
- 33.Carbone M, Stach R, Di Resta I, et al. Simian virus 40 oncogenesis in hamsters. Dev Biol Stand. 94:273–9 (1998). [PubMed] [Google Scholar]
- 34.Bocchetta M, Di Resta I, Powers A, et al. Human mesothelial cells are unusually susceptible to simian virus 40-mediated transformation and asbestos cocarcinogenicity. Proc Natl Acad Sci USA.97(18):10214–9 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horvath CJ, Simon MA, Bergsagel DJ, et al. Simian virus 40-induced disease in rhesus monkeys with simian acquired immunodeficiency syndrome. Am J Pathol. 140(6):1431–40 (1992). [PMC free article] [PubMed] [Google Scholar]
- 36.Shaan Lakshmanappa Y, Elizaldi SR, Roh JW et al. SARS-CoV-2 induces robust germinal center CD4 T follicular helper cell responses in rhesus macaques. Nat Commun 12, 541 (2021). 10.1038/s41467-020-20642-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang A, Garcia-Carreras B, Hitchings M, et al. A systematic review of antibody mediated immunity to coronaviruses: antibody kinetics, correlates of protection, and association of antibody responses with severity of disease. medRxiv. 2020. 10.1101/2020.04.14.20065771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chandrashekar A, Liu J, Martinot AJ, et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 369(6505): 812–81 (2020). doi: 10.1126/science.abc4776. Epub 2020 May 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rockx B, Kuiken T, Herfst S, et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368(6494):1012–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. http://biotech.gsu.edu/virology/index.html .
- 41. https://cnprc.ucdavis.edu/primate-assay-laboratory-core/
- 42.Kuller L, Watanabe R, Anderson D, et al. Development of a whole-virus multiplex flow cytometric assay for antibody screening of a specific pathogen-free primate colony. Diagn Microbiol Infect Dis. 53(3):185–93 (2005). doi: 10.1016/j.diagmicrobio.2005.05.012. Epub 2005 Oct 21. [DOI] [PubMed] [Google Scholar]
- 43.Liao Q, Guo H, Tang M, et al. Simultaneous detection of antibodies to five simian viruses in nonhuman primates using recombinant viral protein based multiplex microbead immunoassays. J Virol Methods. 178(1-2):143–52 (2011). doi: 10.1016/j.jviromet.2011.09.004. Epub 2011 Sep 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khan IH, Mendoza S, Yee J, et al. Simultaneous detection of antibodies to six nonhuman-primate viruses by multiplex microbead immunoassay. Clin Vaccine Immunol. 13(1):45–52 (2006). doi: 10.1128/CVI.13.1.45-52.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White JA, Todd PA, Rosenthal AN, et al. Development of a generic real-time PCR assay for simultaneous detection of proviral DNA of simian Betaretrovirus serotypes 1, 2, 3, 4 and 5 and secondary uniplex assays for specific serotype identification. J Virol Methods.162(1–2):148–54 (2009). doi: 10.1016/j.jviromet.2009.07.030. Epub 2009 Aug 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iskandriati D, Saepuloh U, Mariya S, et al. Isolation and Characterization of Simian Retrovirus Type D from Macaca fascicularis and M. nemestrina in Indonesia. Microbiology Indonesia. 4(3):132–136 (2010). [Google Scholar]
- 47.Richards AL, Giri A, Iskandriati D, et al. Simian T-lymphotropic virus type I infection among wild-caught Indonesian pig-tailed macaques (Macaca nemestrina). J Acquir Immune Defic Syndr Hum Retrovirol. 19(5):542–5 (1998). doi: 10.1097/00042560-199812150-00015. [DOI] [PubMed] [Google Scholar]
- 48.Jones-Engel L, Engel GA, Heidrich J, et al. Temple monkeys and health implications of commensalism, Kathmandu, Nepal. Emerg Infect Dis. 12(6):900–6 (2006). doi: 10.3201/eid1206.060030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hearn TL Standards of Laboratory Practice for HIV Testing. In: Schochetman G, George JR (eds) AIDS Testing. Springer, New York, NY: (1994). [Google Scholar]
- 50.Serological Diagnosis of Human Immunodeficiency Virus Infection by Western Blot Testing. JAMA. 260(5):674–679 (1988). doi: 10.1001/jama.1988.03410050094037 [DOI] [PubMed] [Google Scholar]
- 51. https://www.cdc.gov/mmwr/preview/mmwrhtml/00001431.htm .
- 52.Ronald RB ViswanathDevanarayan. Are Lessons Learned in Setting Cut Points for Detection of Anti-Drug Antibodies Also Useful in Serology Assays for Robust Detection of SARS-CoV-2 Reactive Antibodies? The AAPS Journal 22:127 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen M Statistical Evaluation of Several Methods for Cut-Point Determination of Immunogenicity Screening Assay. Journal of biopharmaceutical statistics. 25(2): 269–279 (2015). [DOI] [PubMed] [Google Scholar]
- 54. https://nprcresearch.org/primate/pathogen-detection/pathogen-detection-working-group.php .
- 55.Yee JL, Grant R, Van Rompay KK, et al. Emerging diagnostic challenges and characteristics of simian betaretrovirus infections in captive macaque colonies. J Med Primatol.46(4):149–153 (2017). doi: 10.1111/jmp.12295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grant R, Keele B, Kuller L, Watanabe R, Perret A, Smedley J. Identification of novel simian endogenous retroviruses that are indistinguishable from simian retrovirus (SRV) on current SRV diagnostic assays. J Med Primatol. 2017;46(4):158–161. doi: 10.1111/jmp.12297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hilliard JK, Ward JA: B-virus specific-pathogen-free breeding colonies of macaques (Macaca mulatta): retrospective study of seven years of testing. Lab Anim Sci. 49:144–148 (1999). [PubMed] [Google Scholar]
- 58.Subbaraman N The US is boosting funding for research monkeys in the wake of COVID. Nature. 595, 633–634 (2021). [DOI] [PubMed] [Google Scholar]
- 59.Contreras MA, Arnegard ME, Chang MC et al. Nonhuman primate models for SARS-CoV-2 Research: Managing demand for specific-pathogen-free (SPF) animals. Lab Anim 50, 200–201 (2021). 10.1038/s41684-021-00810-2 [DOI] [PubMed] [Google Scholar]
- 60.Hild SA, Chang MC, Murphy SJ et al. Nonhuman primate models for SARS-CoV-2 research: Infrastructure needs for pandemic preparedness. Lab Anim 50, 140–141 (2021). 10.1038/s41684-021-00760-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


