Abstract
Biocatalysis is a key tool in both green chemistry and biorefinery fields. NOV1 is a dioxygenase that catalyzes the one-step, coenzyme-free oxidation of isoeugenol into vanillin and holds enormous biotechnological potential for the complete valorization of lignin as a sustainable starting material for biobased chemicals, polymers, and materials. This study integrates computational, kinetic, structural, and biophysical approaches to characterize a new NOV1 variant featuring improved activity and stability compared to those of the wild type. The S283F replacement results in a 2-fold increased turnover rate (kcat) for isoeugenol and a 4-fold higher catalytic efficiency (kcat/Km) for molecular oxygen compared to those of the wild type. Furthermore, the variant exhibits a half-life that is 20-fold higher than that of the wild type, which most likely relates to the enhanced stabilization of the iron cofactor in the active site. Molecular dynamics supports this view, revealing that the S283F replacement decreases the optimal pKa and favors conformations of the iron-coordinating histidines compatible with an increased level of binding to iron. Importantly, whole cells containing the S283F variant catalyze the conversion of ≤100 mM isoeugenol to vanillin, yielding >99% molar conversion yields within 24 h. This integrative strategy provided a new enzyme for biotechnological applications and mechanistic insights that will facilitate the future design of robust and efficient biocatalysts.
The development of biocatalysts and bioprocesses that selectively use lignin as a feedstock for aromatic chemicals is essential to support the economic sustainability of the biorefineries of the 21st century. Lignin is, after cellulose, the second most important source of carbon and the largest reserve of aromatics on Earth and is, therefore, a critical renewable source of chemicals. The pulp and paper industry produces ∼50 million tons of lignin in a year. Most of this is burned for power; only 1 million tons reach the chemical market.1 The successful implementation of strategies for lignin depolymerization enabled the extraction of a range of well-defined compounds in acceptable quantities, bringing its valorization one step closer to reality.1−3 The enzymatic conversion of lignin-derived compounds into drop-in chemicals, polymers, or emerging functional materials has the potential to replace the current petroleum-based synthesis and simultaneously valorize lignin biowaste.4−7
Vanillin exemplifies well the case of a compound that can derive from lignin biowaste and contribute to its valorization. The global demand for this aromatic flavor is enormous. It is a fragrance widely used in food, cleaning products, and perfumes. Its production has been satisfied mainly by chemical synthesis based on fossil feedstocks and chemical catalysts. Therefore, new biotechnological processes for synthesizing this valuable compound are urgently needed.8−12 However, finding a suitable biocatalyst for any fulfilling bioprocess criteria such as eco-friendliness, cost-effectiveness, and long-term sustainability is still a challenging task. NOV1 from Novosphingobium aromaticivorans DSM 12444 is a non-heme iron-dependent dioxygenase that catalyzes the cleavage of double bonds of stilbene compounds such as resveratrol, oxyresveratrol, and piceatannol, yielding small modified benzaldehydes.13,14 NOV1 has activity as compounds comprising a single aromatic ring, such as isoeugenol, which is oxidatively cleaved at the C=C bond of the propenyl functional group to produce vanillin, albeit at low efficiencies (Scheme 1).14 Coenzymes are not required in this reaction, which reduces the complexity and cost of the bioprocess.13,15 The sequence of NOV1 is close to 40% identical to those of isoeugenol oxygenases, IE27 from Pseudomonas putida,16,17 and IEM from Pseudomonas nitroreducens.18−20 Isoeugenol can be obtained from the oxidation of 4-n-propylguaiacol, an intermediate of lignin depolymerization from wood biorefineries,21 and thus, NOV1 and similar enzymes offer the possibility of developing sustainable and environmentally friendly routes for the production of vanillin through an enzymatic bioprocess that employs lignin as a renewable feedstock and notably broadening the biobased product portfolio.22
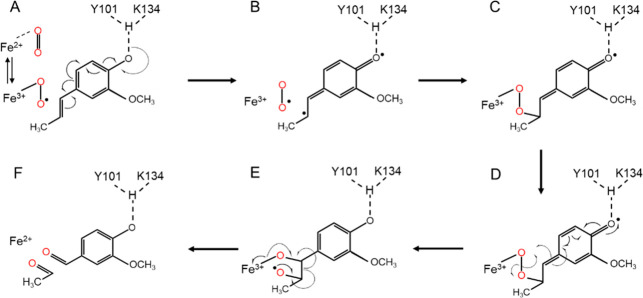
Scheme 1. Proposed NOV1 Reaction Mechanism for Isoeugenol Adapted from ref (14).
(A) Formation of the ternary complex of Fe(III)-superoxo, and the isoeugenol substrate and contributions of deprotonation of 4′-OH by Y101 and K134 in activation of the substrate. (B) Intermediate formed before the formation of the first C–O bond. (C) Intermediate formed after the formation of the first C–O bond. (D) Cleavage of the O–O bond and formation of the second C–O bond. (E) Cleavage of the C–C bond and restoration of the Fe(II) enzyme. (F) Vanillin and acetaldehyde are the reaction products.
In this work, 35 variants were designed on the basis of the structural analysis of the NOV1 enzyme, in silico dockings, comparative structural alignments, and Rosetta computation-based design, constructed, and examined for activity toward the isoeugenol substrate. The S283F variant emerged as the most active and stable variant and a promising biocatalyst for vanillin production. Furthermore, a combination of kinetics, stability measurements, X-ray diffraction, and molecular dynamics allowed identification of the molecular basis behind the improved properties of the S283F variant.
Materials and Methods
Mutant Design with Rosetta Coupled Moves
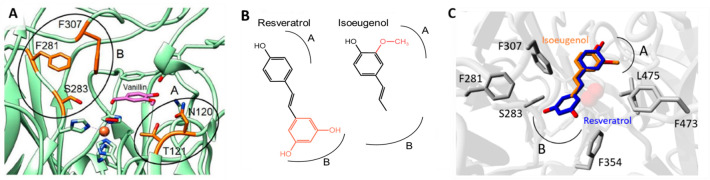
Isoeugenol was included in the active site of wild-type NOV1 using Autodock VINA.23 The vanillin present in the X-ray structure [Protein Data Bank (PDB) entry 5J55] was used to define the center of the simulation box, comprising 3 Å from the ligand, and then it was manually removed. The best scoring binding mode of isoeugenol at the active site was used as the starting point for Rosetta Coupled Moves experiments.24 Three groups of noncatalytic active site residues close to the isoeugenol substrate were selected for mutagenesis (Figure 1C): (i) N120 and T121, (ii) F281, S283, and F307, and (iii) F354, F473, and L475. For each group, a Monte Carlo algorithm explored changes over one, two, or three positions simultaneously, resulting in a library of single, double, and triple variants. Repacking was allowed for all of the residues in contact with any modified position. A Boltzmann constant of 0.6 and a ligand weight of 1.0 were used. Every individual experiment consisted of 100 Coupled Moves runs of 1000 trials. All variants were filtered by protein–ligand docking, retaining the variants that attained precatalytic distances [the distances between the two carbons forming the double bond of isoeugenol and the oxygen molecule bound to Fe (d1) and the hydroxyl group of the substrates and the catalytic residues Y101 and K134 (d2) are <4 Å]. The final list of variants was determined by the TotalScore, LigandScore (from Rosetta), and binding energy (from docking) values (Figure S1).
Figure 1.
Structural basis for the rational design of NOV1. (A) NOV1 active site in complex with vanillin (PDB entry 5J55) with surrounding residues highlighted. (B) Chemical structures of resveratrol and isoeugenol. The regions at which the different chemical groups of the ligands bind the active site are indicated. (C) Comparison between the binding of isoeugenol and resveratrol (PDB entry 5J54) to the NOV1 binding site. Regions A and B outline the amino acids involved in binding the isoeugenol hydroxyl and propenyl groups.
Construction of Variants
Single-, double-, or triple-amino acid substitutions in the nov1 gene were constructed using the Quick-Change site-directed mutagenesis protocol (Stratagene). Plasmid pET-28a (+) containing the nov1 gene was used as a template using the appropriate primers (Table S1), except triple variants F281M/S283I/F307H and F281M/S283T/F307H, constructed using the DNA template of variants F281M/S283I and F281M/S283T, respectively. Polymerization chain reactions (PCRs) were performed in 50 μL reaction volumes containing 3 ng of DNA template, 2 μM primers, 200 μM dNTPs, NZYProof polymerase buffer, and 1.25 units of NZYProof polymerase (NZYTech). After an initial denaturation period of 5 min at 94 °C, the following steps were repeated for 20 cycles in a thermal cycler (MyCycler thermocycler, Bio-Rad): 1 min at 94 °C, 1 min at 55–58 °C, and 10 min at 72 °C, followed by a final 10 min period at 72 °C. The amplified products were purified using GFX PCR DNA and the gel band purification kit (GE Healthcare). The final PCR products were digested with DpnI to eliminate the wild-type template and were used to transform electrocompetent Escherichia coli strain DH5α (Novagen) cells. The presence of the desired mutation in the resulting plasmid was confirmed by DNA sequencing.
Activity Screenings in 96-Well Plates
The genes encoding the wild-type and variant enzymes were introduced into the host strain E. coli BL21 star (DE3, Novagen) by electroporation. In E. coli BL21 star, the genes are controlled by the T7 promoter and induced by isopropyl β-d-1-thiogalactopyranoside (IPTG). Single colonies were picked from a fresh agar plate and placed in 96-well microplates filled with 200 μL of Luria-Bertani medium (LB) supplemented with 50 μg mL–1 kanamycin. Cultures were cultivated at 37 °C and 750 rpm for 24 h. The following day, 20 μL of these cultures was inoculated with 180 μL of LB medium in 96-well microplates and cultivated for 4 h at 37 °C. At this time, gene expression was induced with 0.1 mM IPTG; the medium was supplemented with 0.5 mM FeSO4 and cultivated at room temperature at 750 rpm. After 24 h, cells were harvested by centrifugation, resuspended in 20 mM Tris-HCl (pH 7.6), disrupted by three cycles of freezing in liquid nitrogen and thawing at room temperature for 5 min, and resuspended in lysozyme (0.5 mg mL–1). After cell disruption, plates were centrifuged at 4000 rpm for 30 min at 4 °C, and supernatants (cell crude extracts) were used for enzymatic activity measurements. The activity was assessed after mixing 20 μL of crude extracts with 160 μL of 100 mM Tris-HCl (pH 9.0) containing 1 mM isoeugenol. Reactions were monitored by following vanillin production at 340 nm. The vanillin molar absorptivity at 340 nm (ε340= 15970 M–1 cm–1) was obtained using a calibration curve (Figure S2).
Enzyme Production and Purification
The recombinant E. coli strains containing plasmids encoding wild-type and S283F were grown in 1 L of LB medium supplemented with kanamycin in a 5 L Erlenmeyer flask with 120 rpm shaking (Innova 44, New Brunswick Scientific). The cells were grown at 37 °C until an optical density of 0.6 at 600 nm was reached, after which 0.1 mM IPTG and 0.5 mM FeSO4 were added to the culture medium, and the temperature was decreased to 25 °C. The incubation was continued for a further 16 h, after which the cells were harvested by centrifugation. The cell pellets were suspended in 20 mM sodium phosphate buffer, 0.5 M NaCl, and 20 mM imidazole (pH 7.6) (buffer A) containing DNase I (2 μg mL–1), MgCl2 (5 mM), and a mixture of protease inhibitors (CompleteTM mini-EDTA-free protease inhibitor mixture tablets; Roche, Basel, Switzerland). Cells were disrupted in a French press cell (at 900 psi), followed by centrifugation (18000g, 1 h, 4 °C). The resulting soluble extracts were loaded onto an affinity chromatography HisTrap column (GE Healthcare) equilibrated with buffer A. Elution was carried out with a linear imidazole gradient (from 0 to 0.5 M in 20 min) in the same buffer. All purification steps were performed at room temperature with an Åkta purifier (GEHealthcare). The purity of eluted fractions was analyzed by SDS–PAGE. The active fractions were pooled and digested by SUMO-protease (1 unit for 5 μg of protein), with removal of the SUMO and His tag overnight at 4 °C, concentrated by ultrafiltration (cutoff of 30 kDa), and equilibrated to 20 mM Tris-HCl and 0.2 M NaCl (pH 7.6).
Apparent Steady-State Kinetic Analysis
The effect of pH on activity was studied in 100 mM Britton-Robinson buffer (pH 3–11), and the optimal temperature was determined in the temperature range of 10–40 °C. Reactions were performed with isoeugenol (0.01–4 mM) in 100 mM Tris-HCl (pH 9) and monitored at 340 nm (see above). Reactions with resveratrol (0.01–0.32 mM) were monitored following the decrease in absorbance at 306 nm (ε = 31800 M–1 cm–1) in 100 mM Tris-HCl (pH 9). Enzyme activity was also measured by monitoring the oxygen consumption with an oxygen electrode (Oxygraph, Hansatech) at 25 °C. Reactions were started by adding the enzyme to a nitrogen-purged mixture containing 4 mM isoeugenol in 100 mM Tris-HCl (pH 9). Kinetic data were fitted directly to the Michaelis–Menten equation using Origin. All enzymatic assays were performed at least in triplicate.
Bioconversions of Isoeugenol to Vanillin
Conversion of isoeugenol to vanillin was studied in a 50 mL Erlenmeyer flask with a reaction volume of 10 mL containing 10, 25, 50, and 100 mM isoeugenol in 0.1 M glycine-NaOH (pH 9) at room temperature and 150 rpm, after the addition of 1 unit mL–1 purified wild-type and S283F variant (defined as the amount of enzyme that produced 1 μmol of vanillin per minute) and whole cells of E. coli overproducing the variant S283F at a final OD600 of 2, which corresponds to 1.3 mg of cell dry weight mL–1. Reaction mixtures contained ≤3.5% ethanol used to prepare the substrate’s stock solution (1–2 M isoeugenol in 70% ethanol). The progress of reactions was monitored at 340 nm by quantifying vanillin production by absorbance (see above) and high-performance liquid chromatography (HPLC). A Waters Alliance 2695 HPLC system with a Purospher STAR RP-18c column [125 mm × 4 mm, 5 μm particle size (Merck KGaA)] was used for HPLC analysis. The column was maintained at 40 °C; the flow rate was set to 1.0 mL min–1, and the volume injected was 25 μL. A linear gradient was set starting from 70% 0.5% acetic acid (solvent A) and 30% methanol with 0.5% acetic acid (solvent B), going from 0% to 80% solvent B in 25 min and maintaining for 10 min. Then, the initial conditions were resumed in 2 min and maintained for 8 min. The absorption was monitored between 200 and 500 nm by a Waters model 2996 photodiode array detector operated by Empower Pro, version 5 (Waters Chromatography).
Thermodynamic Stability
Protein unfolding was monitored using a Cary Eclipse spectrofluorimeter with an excitation wavelength of 296 nm and by recording the fluorescence emission of tryptophyl residues at 340 nm. Guanidine hydrochloride (GdnHCl) concentrations in the range of 0–3 M in 20 mM Tris-HCl and 0.2 M NaCl (pH 7.6) were used to induce protein unfolding after incubation at room temperature for 5 min. The fraction of unfolded protein (fU) was plotted versus GdnHCl concentration and fitted to the equation fU = exp(−ΔG°/RT)/[1 + exp(−ΔG°/RT)]. The chemical-induced unfolding was analyzed using the equations previously described on the basis of a two-state process.25 Thermal unfolding was performed for the iron-depleted enzyme preparations (10 μM) with EDTA for 12 h at room temperature. Before measurement, EDTA was removed by ultrafiltration using Vivaspin. Enzyme preparations in 20 mM Tris-HCl and 0.2 M NaCl (pH 7.6) were placed onto a thermostatically thermal block and heated at a rate of 1 °C/min to 100 °C. Thermal unfolding was analyzed according to an N ⇌ U two-state model.26 Static light scattering using excitation and emission wavelengths of 500 nm in the spectrofluorimeter was used to follow enzyme aggregation.
Kinetic Stability
Thermal inactivation was studied by incubating enzyme preparations in 20 mM Tris-HCl (pH 7.6) at 25 °C. FeSO4 was added to the solution at a concentration that was 100-fold higher than the enzyme molarity when required. At appropriate time points, aliquots were withdrawn and examined for activity. Inactivation constants (kin) were obtained by linear regression of logarithm activity versus time. Half-life t1/2 was calculated using the equation t1/2 = ln 2/kin.
Protein Crystallization, Data Collection, and X-ray Structure Determination and Refinement
The S283F enzyme was crystallized according to the crystallization conditions reported in the literature for wild-type NOV1 [sitting drop vapor diffusion, 0.16 M MgCl2, 0.08 M Tris-HCl (pH 8.5), 18% (w/v) PEG 4000, and 20% (v/v) glycerol at 20 °C]. However, the shape of the crystals did not correspond to the cubic geometry described,14 but clusters of large transparent crystal plates were obtained instead. For data collection, crystals were flash-cooled in a stream of gaseous nitrogen at 100 K. X-ray diffraction data were measured at beamlines of the Swiss Light Source (SLS, Villigen, Switzerland). Data processing and scaling were performed using XDS27 and the CCP4 package28 (Table S2). The coordinates of the NOV1 bound to vanillin (PDB entry 5J55) deprived of all ligands and waters were used as the initial model for structure determination. Coot29 was used for electron density inspection and model building, whereas crystallographic refinement was performed with REFMAC5.30
Molecular Dynamics Simulations
The 800 ns trajectories (200 ns per four replicas) were run for four different systems: (1) wild-type NOV1 in its Fe(II)-O2 substrate-free form, (2) S283F variant in its Fe(II)-O2 substrate-free form, (3) wild-type NOV1 in its apo form (no metal and no ligand), and 4) S283F variant in its apo form. The NOV1 structures and the MD simulations of apoproteins were prepared using Yasara (details in the Supporting Information).31 The protein and the water solvent were described on the basis of the AMBER14 force field32 and the TIP3P model,33 respectively. The resulting conformations of the holoproteins were used for structural analysis (free volume analysis and inter-residue interactions) and ensemble docking. Those of the apoproteins were used for pKa studies on the histidines composing the catalytic center to assess the effect of the S283F mutation on their flexibility and orientation.
Ensemble Protein–Ligand Docking
The isoeugenol and resveratrol substrates were systematically docked every 40 ps along with the MD simulations of the ligand-free systems (20000 structures for each system) using a simulation box of 8 Å from the oxygen molecule. For each MD snapshot, 96 docking runs and a clustering cutoff of 2 Å of the RMSD were established. The protein was kept rigid while the rotors of the substrate were allowed to move. The distances between the two carbons forming the double bond of the ligands and the oxygen molecule bound to Fe (d1) and the hydroxyl group of the substrates and catalytic residues Y101 and K134 (d2) were measured. The dockings were performed with AutoDock VINA23 using the YAMBER force field,34 and the structural analysis was done with the Yasara Python module. For all of the binding modes identified as precatalytic structures (the distances d1 and d2 were <4 Å), the ΔGbinding (it estimates the quality of ligand–protein interactions) and ΔEdihedral (it measures how strong the distortion of the ligand is in comparison to that of its protein-free form) energies were assessed.
Inter-residue Interactions
The Protein Interactions Calculator35 Web server was used to identify the hydrophobic patches on the PDB structures of both wild-type NOV1 and S283F.
Other Methods
The concentration of purified protein preparations was estimated using the molar absorption coefficient of NOV1 (ε280 = 81930 M–1 cm–1) calculated from the protein sequence using the ExPASy Bioinformatics Resource Portal (http://web.expasy.org). The iron content of purified protein samples was determined by a colorimetric assay resorting to the chelator TPTZ [2,4,6-tris(2-pyridyl)-s-triazine]36 and by atomic emission spectroscopy at Laboratório Central de Análises, Universidade de Aveiro (Aveiro, Portugal).
Results and Discussion
Active Site Design: Construction and Characterization of Variants
The analysis of the binding of vanillin and resveratrol to NOV1 (PDB entries 5J55 and 5J54, respectively) suggested that isoeugenol can be docked into the NOV1 active site with the hydroxyl group pointed toward Y101 and K134 residues and the methoxy substituent pointed toward the pocket created by N120, T121, and L475 (Figure 1A–C, region A). In this position, the isoeugenol aromatic ring plane sits parallel to the side chain of F59. Moreover, the reactive double bond of isoeugenol is located right above the iron center. In contrast, the terminal methyl group is hosted in a large and hydrated niche in the rear of the cavity close to F281, S283, F307, and F354 (Figure 1A–C, region B). The alignment of NOV1 with P. nitroreducens isoeugenol monooxygenase shows a phenylalanine residue at position S283 in this enzyme.16 Furthermore, analysis of the X-ray crystal structure supported the reasoning that the S283F replacement could improve isoeugenol catalysis by filling the free space left in the active site upon binding the single-ring isoeugenol substrate.
Mutations N120L, F281W, S283Q, F307H, F307W, and L475S were suggested by sequence comparisons and biochemical information from the literature.14,16,18,19 Positions 120, 121, 281, 283, 307, 354, 473, and 475 were submitted to mutagenesis using Rosetta and filtered by protein–ligand docking (see the computational methods section for further details). Thirty-five variants based on the rational and computational approaches were constructed using site-directed mutagenesis and characterized (Tables S1 and S3). The enzymatic activity of isoeugenol was tested in crude cell extracts in 96-well microplates. The results showed that mutations at positions 120, 121, 473, and 475 impaired isoeugenol activity. Three double variants, F281M/S283T, F281M/S283V, and F281C/S283I, showed specific activities comparable to that of the wild type, and single variant S283F stood out with almost 2-fold higher enzymatic activity compared to that of the wild type.
Biochemical and Kinetic Characterization
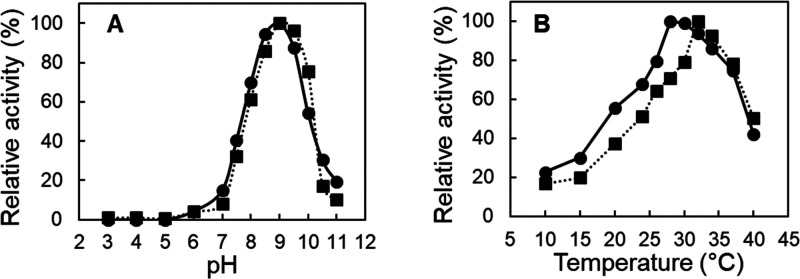
The wild type and variant S283F were produced at an Erlenmeyer scale and purified. Both enzymes showed an optimal pH of 9 and optimal temperatures of ∼28 °C for the wild-type enzyme and 32 °C for the S283F variant (Figure 2A,B). Purified enzyme preparations displayed approximately 0.5 mol of iron/mol of protein and were partially iron-depleted, similar to those observed in isoeugenol oxygenases from P. putida IE2716 and P. nitroreducens IEM.18
Figure 2.
pH and temperature profiles. (A) pH–activity profile of wild-type (●) and S283F (■) NOV1. Reactions were performed using Britton Robinson buffer (in the pH range from 3 to 11) in the presence of 1 mM isoeugenol at room temperature. (B) Temperature dependence of enzymatic activity in reactions performed in 100 mM Tris-HCl buffer (pH 9).
The steady-state kinetic analysis revealed that the S283F variant features a kcat 2-fold higher than that of the wild type, and a slightly higher Km value for isoeugenol, leading to a catalytic efficiency (kcat/Km) comparable to that of the wild type (Table 1). The activity using resveratrol as a substrate was also investigated; the S283F variant showed a 5-fold lower kcat and a 4-fold higher Km, resulting in a sharp 20-fold decrease in kcat/Km, compared to that of the wild type. Notably, the kinetic analysis for O2 in the presence of isoeugenol showed that the S283F variant displayed enhanced binding to molecular oxygen, with an ∼2-fold lower Km and a 4-fold higher catalytic efficiency compared to those of the wild type (Table 1 and Figure S3). This is an important asset for overcoming the usually limiting levels of soluble O2 in large-scale industrial processes; indeed, enhancing O2 binding and catalysis has been a critical challenge in the application of oxygenases.37,38 The results obtained supported the prediction that the introduction of the bulky and hydrophobic phenylalanine at position 283 impairs resveratrol oxidation by shifting the specificity of NOV1 toward smaller substrates such as isoeugenol and molecular oxygen.
Table 1. Apparent Steady-State Kinetic Parameters of Wild-Type and S283F NOV1 for Isoeugenol, Molecular Oxygen (in the presence of isoeugenol), and Resveratrola.
| isoeugenol | O2 (isoeugenol) | resveratrol | ||
|---|---|---|---|---|
| wild type | kcat (s–1) | 7.3 ± 0.2 | 10.9 ± 0.8 | 0.38 ± 0.02 |
| Km (mM) | 0.6 ± 0.1 | (0.7 ± 0.1) × 10–2 | 0.06 ± 0.01 | |
| kcat/Km (M–1 s–1) | (12.2 ± 2) × 103 | (15.6 ± 4) × 105 | (6.3 ± 0.2) × 103 | |
| S283F | kcat (s–1) | 14.5 ± 0.8 | 17.3 ± 0.6 | 0.078 ± 0.004 |
| Km (mM) | 0.8 ± 0.1 | (0.29 ± 0.04) × 10–2 | 0.227 ± 0.002 | |
| kcat/Km (M–1 s–1) | (18.1 ± 4) × 103 | (59.7 ± 10) × 105 | (0.34 ± 0.02) × 103 |
Kinetic assays were performed at room temperature in 0.1 M Tris-HCl (pH 9).
Enzymatic Vanillin Bioproduction
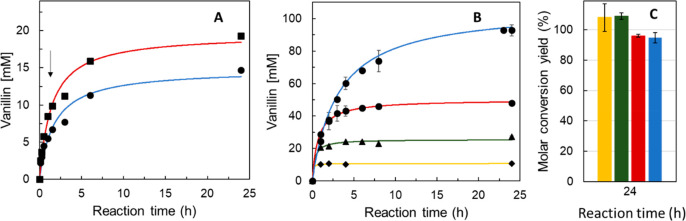
Bioconversion assays confirmed that purified S283F converts isoeugenol to vanillin more efficiently than does the wild type. A >99% conversion of 10 mM isoeugenol after reaction for 1.5 h was achieved, which favorably compares with the 67% conversion obtained using the wild-type enzyme (Figure 3A). The reaction product was confirmed to be vanillin by HPLC (Figure S4) in accordance with the previous identification.14 Further addition of isoeugenol (10 mM) resulted in 96% and 74% conversion yields after 24 h for S283F and the wild type, respectively. The total turnover numbers (TTNs), defined as the total moles of vanillin produced per mole of enzyme over the entire length of the reaction, are 5.1 and 2.2 for S283F and the wild type, respectively. Remarkably, the kcat of the S283F variant for isoeugenol is 2–10 times higher than the values previously reported for isoeugenol oxygenases,16,18,19,39 further endorsing this enzyme as a superior biocatalyst at nonlimiting substrate concentrations, typical of industrial setups. We set up time-course bioconversion assays using whole cells that overproduced the S283F variant to reduce the costs associated with enzyme purification (Figure 3B,C).40,41 Our results revealed that excellent molar conversion yields (>99%) were achievable within 24 h of reaction using ≤100 mM isoeugenol in the presence of small amounts (3.5%) of ethanol, a biosolvent, in the whole cell catalysis mixture.
Figure 3.
Bioconversions of isoeugenol to vanillin. (A) Time course of vanillin production using 1 unit mL–1 wild-type (blue) and S283F NOV1 (red) purified enzymes; reactions started with 10 mM substrate, and additional supplementation with 10 mM isoeugenol occurred after reaction for 1.5 h (arrow). (B) Time course of vanillin production using recombinant E. coli whole cells overproducing the S283F NOV1 variant (final OD600 of 2) in reaction mixtures containing initial concentrations of 10 (yellow), 25 (green), 50 (red), and 100 (blue) mM isoeugenol. (C) Molar conversion yields after 24 h for reaction mixtures containing initial concentrations of 10 (yellow), 25 (green), 50 (red), and 100 (blue) mM isoeugenol. Reactions were performed in glycine-NaOH buffer (pH 9) at room temperature and 150 rpm.
Notably, the obtained conversion yields were similar to those obtained with Pseudomonas isoeugenol monooxygenases in the presence of organic solvents or enzyme aggregates17,20 and higher than those obtained with oxygenases from Herbaspirillum seopedicae and Rhodobacteraceae bacterium (Table S4).39 Furthermore, substrate concentrations of ≤100 mM were tolerated by the NOV1 system in contrast to other enzymatic systems inhibited by isoeugenol and force reactions to be performed at significantly lower substrate concentrations.39,42
NOV1 Stability and Iron Incorporation
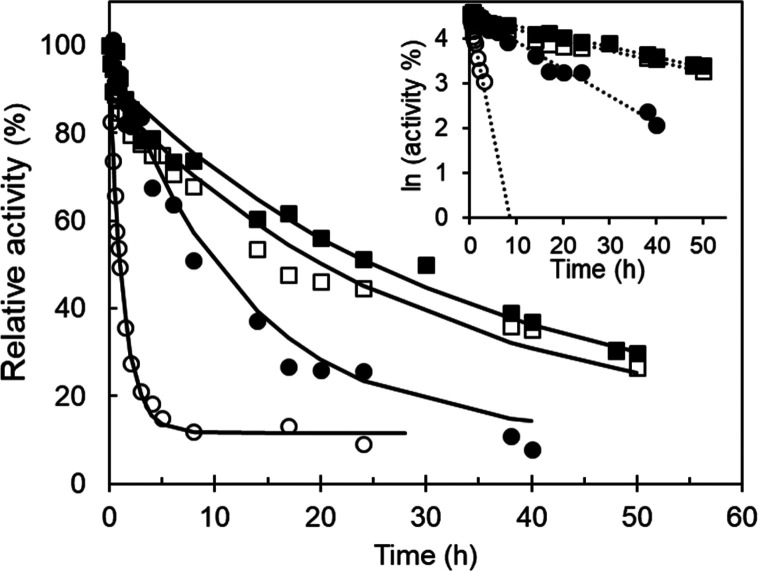
The kinetic or operational stability is relevant for assessing the biocatalyst performance under specific operating conditions, for example, at a given temperature, and for studying pathways that lead to the formation of irreversibly inactivated states of the enzyme.43 The thermal inactivation assays revealed that the S283F amino acid replacement drastically improves the enzyme kinetic stability. While wild-type NOV1 displayed inferior stability with a half-life (t1/2) of ∼1 h at 25 °C, variant S283F exhibited a half-life of ∼29 h (Figure 4). Notably, incubation of the wild type with iron increased the half-life 10-fold but not that of the variant, indicating that (i) binding of the iron cofactor seems to be the critical determinant of thermostability of the NOV1 enzyme and (ii) the wild-type enzyme loses iron more quickly than the S283F variant. The chemical unfolding of the wild type and S283F was assessed by using the fluorescence emission of tryptophan residues.
Figure 4.
Kinetic stability. Stability of wild-type (circles) and S283F (squares) NOV1 at 25 °C in the absence (empty symbols) and presence of 100 equiv of FeSO4 (filled symbols). The inset shows the linear regression of logarithm activity vs time. In the absence of iron, the half-lives at 25 °C were 1.3 ± 0.2 and 29 ± 3.4 h for the wild type and S283F variant, respectively. The addition of iron increased 10-fold the half-life (11.4 ± 0.7 h) of the wild type, whereas the stability of S283F remained similar (30.4 ± 1.2 h).
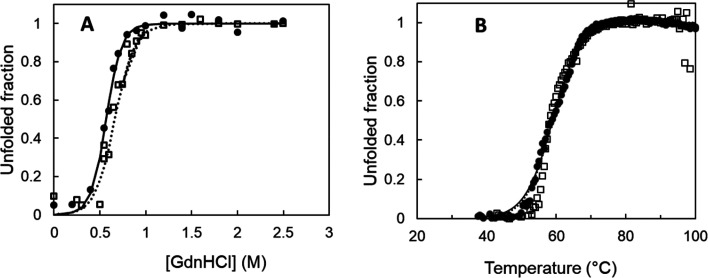
The folded and unfolded states were the only states that accumulated in significant amounts. A two-stage process accurately fit the unfolding process (Figure 5A), and both enzymes displayed similar stability. The guanidinium hydrochloride midpoint concentration is ∼0.6 M (where 50% of molecules are unfolded), and the native-state free energy is 3.5 kcal mol–1. In thermal unfolding experiments, the fluorescence emission from tryptophan residues increased in the range of 30–40 °C (Figure S5), indicating the iron cofactor’s release, a known fluorescence quencher.44 Incubation of the wild type with 2000 molar equivalents of EDTA abolished this effect (Figure S5A), in contrast to S283F, which shows a persisting “iron quenching effect” even after incubation with EDTA, suggesting a significantly higher affinity for iron (Figure S5B), in line with the longer half-lives of kinetic stability. The apparent melting temperatures (Tm) are very similar in both enzymes at 57–59 °C (Figure 5B); static light scattering at 500 nm revealed a strong aggregation tendency with an onset of aggregation (Tagg) at 47 °C before enzyme unfolding (Figure S6). These results showed that both enzymes share a relatively high thermal robustness of the enzymes’ native state. The higher kinetic thermostability observed in the S283F variant compared to that of the wild type most likely results from an enhanced stabilization of the iron cofactor inside the catalytic cavity.
Figure 5.
(A) Fraction of wild-type (●) and S283F (□) NOV1 unfolded by guanidinium chloride as measured by fluorescence emission of tryptophyl residues at 340 nm. Measurements were performed by reading the fluorescence at excitation wavelengths of 296 nm and emission wavelengths of 340 nm. The solid line is the fit according to the equation fU = exp(−ΔG°/RT)/[1 + exp(−ΔG°/RT)], which assumes the N ↔ U equilibrium.25 (B) Thermal unfolding following fluorescence emission of tryptophan residues at 340 nm (Tm = 57–59 °C) for the wild type (●) and S283F variant (□).
Structural Characterization of the S283F Variant
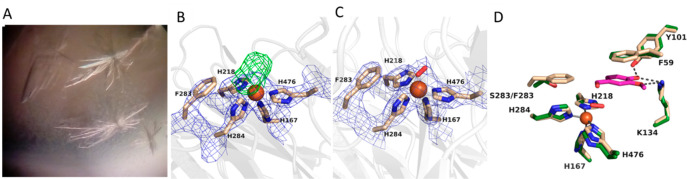
The crystal structure of NOV1-S283F was determined at 2.9 Å using synchrotron radiation (Table S2), revealing the recognizable electron density of a phenylalanine side chain at position 283 (Figure 6A–C). The aromatic ring of F283 was found to occupy the active site, cluttering the catalytic cavity. The Fe(II)-O2 complex, required for activity, is coordinated by four histidine residues: H167, H218, H284, and H476 (Figure 6B,C). F283 is predicted to reach out and interact with isoeugenol through hydrophobic contacts (Figure 1C), whereas K134 and Y101 create the proper hydrogen bonding environment with the 4-hydroxy group of the substrate (Figure 6C,D).
Figure 6.
(A) Crystals of the S283F NOV1 variant. (B) Weighted 2Fo – Fc electron density of the active site. The contour level is 1.2σ. The side chain of F283 is quite recognizable with its aromatic ring close to the His-coordinated iron. The difference Fourier Fo – Fc map [contoured at the 3.0σ level (green)] showed a residual electron density interpreted as bound oxygen. (C) Oxygen-bound active site structure of the final model. (D) S283F NOV1 crystal structure with the reaction product (vanillin, carbons colored purple) modeled in the cavity. The model was generated using the structure of the complex between the wild-type enzyme and vanillin as a reference (carbons colored green, PDB entry 5J55). F283 interacts with the edge of the substrate ring, whereas F59 is involved in π–π stacking with the aromatic ring of the substrate.
Molecular Dynamics Simulations and Substrate Docking
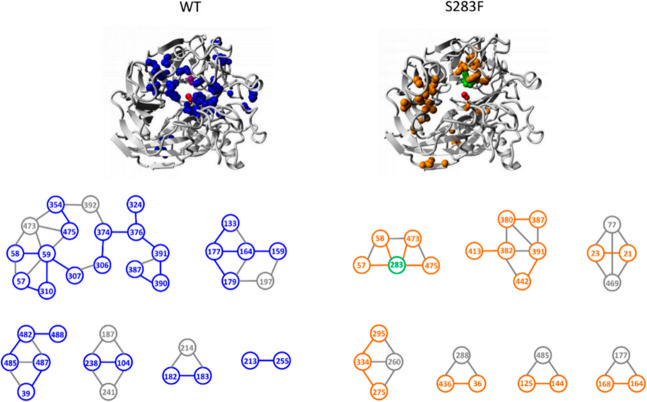
Starting from the experimental wild-type (PDB entry 5J55) and S283F (this work) structures, the computational analysis showed that a comparable number of interactions is present in both wild-type and S283F enzymes and the active site cavity along the MD trajectories (800 ns), which remained in a relatively stable conformation with a constant volume in both structures (Figure S7). However, the S283F replacement resulted in local rearrangement of a sizable hydrophobic core present in the wild type to create small hydrophobic patches near the active site (Figure 7); the average cavity volume is expanded in S283F where, additionally, longer distances were measured between molecular oxygen and F59 (Figure S8A,B). Consequently, a higher number of water molecules was identified inside the active site of the mutant (Figure S8C). The more expanded shape of the S283F active site could make the catalytic center more accessible to the solvent and, by extension, other small molecules, such as molecular oxygen and isoeugenol.
Figure 7.
Inter-residue interaction analysis performed with PIC (Protein Interaction Calculator) using the X-ray structures of wild-type NOV1 (PDB entry 55j5) and the S283F variant (this work). The side chain–side chain hydrophobic interactions that appear in the wild type but not in the variant (left) or the variant but not in the wild type (right) are shown as blue and orange spheres. The S283F mutation is highlighted with green spheres. The gray spheres and sticks comprise interactions shared by both systems.
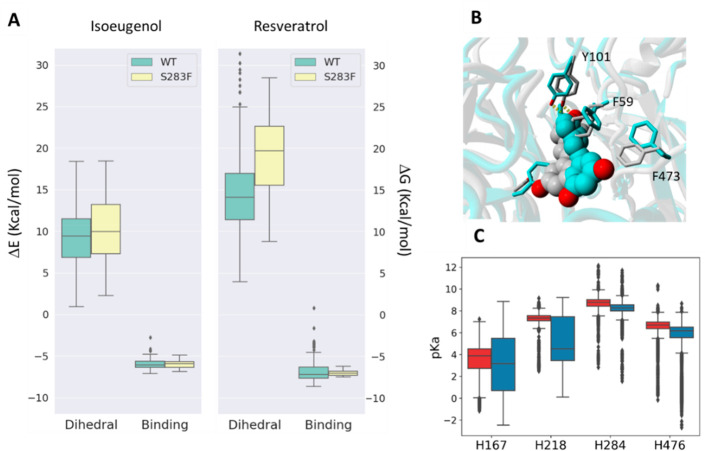
Furthermore, the presence of the bulky phenylalanine at the active site seems to distort the resveratrol conformation significantly when approaching precatalytic states (Figure 8A,B and Table S5). In contrast, the distortion of the isoeugenol structure when approaching precatalytic binding modes appeared only slightly higher in the variant than in the wild type. These data support the experimental observation that resveratrol is a poorer substrate for the variant than is the wild type (4-fold higher Km and 5-fold lower kcat). Furthermore, MD trajectories of the ligand-, oxygen-, and iron-free systems of the wild type showed that the four iron binding histidines are kept in a stable conformation that is not compatible with iron coordination, particularly for H167 and H218 (Figure S9). This was different from the case for S283F, where the presence of phenylalanine in the iron neighborhood increased the fluctuation of the His residues, which visited conformations compatible with iron binding (Figure S9), indicating a lower relative transition energy barrier between iron-free and iron-bound forms in S283F. This increased flexibility of the iron-coordinating histidines could be related to alterations in the H-bond network of the active site, where three conserved carboxylate residues (E135, E353, and E418) are responsible for stabilizing H218, H284, and H476.14,45 In addition, lower pKa values for all histidine side chains, particularly H167 and H218, were found in the S283F variant (Figure 8C), suggesting that their dissociation equilibrium favors the unprotonated state required to bind the iron metal. These electrostatic calculations support a higher affinity of iron for the S283F variant with kinetic stability and thermal unfolding/EDTA chelation, which showed that the wild type loses iron more readily than does S283F (Figure 4 and Figure S5A,B).
Figure 8.
(A) Dihedral (ΔE) and binding energies (ΔG) estimated for ensemble docking of isoeugenol and resveratrol to wild-type NOV1 and the S283F variant. ΔEdihedral measures how strong the ligand distortion is compared to its protein-free form, and ΔGbinding estimates the quality of ligand–protein interactions. (B) Representation of resveratrol binding to wild-type NOV1 (gray) and the S283F variant (blue). The substrate is represented as spheres, and the residues are represented as sticks. (C) pKa values of iron-coordinating histidines 167, 218, 284, and 476 were measured every 40 ps along the 800 ns MD trajectory for the apo form of both the wild type (red) and the S283F mutant (blue).
Concluding Remarks
In this work, 35 variants were rationally designed, constructed, and tested for activity. The S283F variant was selected for further investigations on the basis of increased selectivity and catalytic rates for isoeugenol compared to those of wild-type enzymes. This variant also showed an enhanced kinetic (operational) thermostability due to enhanced stabilization of the iron cofactor inside the catalytic cavity. Incidentally, the primary molecular determinant of NOV1 kinetic stability is suggested to be iron deletion of the active site. MD analyses supported increased iron retention in the active site of S283F and, therefore, enhanced kinetic stability. Biotransformation of the plant-derived phenylpropanoid compound, isoeugenol, using whole cells overproducing the improved variant showed remarkable levels of conversion to vanillin at concentrations of ≤100 mM, at room temperature, and in relatively short periods. The deactivation and stability of the enzymes under harsh conditions are some of the main limitations of the industrial application of biocatalysts. Therefore, the S283F variant, which shows increased activity and stability, is an exciting candidate for industrial bioprocesses targeted in valorizing lignin-related phenolics for biovanillin production in the lignocellulosic biorefinery realm.
Acknowledgments
The authors thank Margarida Ruivo for their help constructing mutants and Tiago N. Cordeiro for valuable discussions.
Glossary
Abbreviations
- EDTA
ethylenediaminetetraacetic acid
- fU
fraction of unfolded protein
- IPTG
isopropyl β-d-1-thiogalactopyranoside
- LB
Luria-Bertani medium
- GdnHCl
guanidine hydrochloride
- MD
molecular dynamics
- RMSD
root-mean-square deviation
- SDS–PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- t1/2
half-life
- Tagg
onset of aggregation temperature
- Tm
melting temperature
- TPTZ
2,4,6-tris(2-pyridyl)-S-triazine
- TTN
total turnover number.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.2c00168.
Computational methods, primers used for the construction of variants (Table S1), data collection and refinement statistics for S283F (Table S2), mutant design and variants’ enzymatic activity (Table S3), comparison of whole cell reactions in the conversion of isoeugenol in vanillin (Table S4), binding energy and dihedral energy values for isoeugenol and resveratrol at the binding site of WT and S2583F in the ensemble docking experiments (Table S5), Rosetta ligand score, Rosetta total score, and VINA Autodocking binding energy values for the in silico variants (Figure S1), calibration curve for vanillin quantification (Figure S2), steady-state kinetic analysis of isoeugenol and O2 catalyzed by the wild type and S283F (Figure S3), HPLC chromatograms (Figure S4), tryptophan fluorescence emission at 340 nm at increasing temperatures of the wild type and S283F (Figure S5), static light scattering at 500 nm for the wild type and S283F (Figure S6), inter-residue interaction analysis performed with PIC (Figure S7), empty volumes of the active site of the wild type and S283F calculated with MDpocket (Figure S8), and dihedral angles calculated by MD simulation (Figure S9) (PDF)
Author Contributions
M.D.S. performed most kinetic and biochemical experiments, including enzyme characterization, and wrote the first draft of the manuscript. V.B. supervised mutagenesis, cloning, and gene expression. E.P.M. supervised the stability experiments. L.A., J.C., and A.M. crystallized the variant, collected data, and analyzed the structure. L.A.-C., E.M., and M.F.L. designed the variants with Rosetta and performed molecular dynamics and docking experiments. L.O.M. and A.M. supervised and reviewed all results. All authors read and approved the final version of the manuscript.
This project has received funding from the Biobased Industries Joint Undertaking (JU) under Grant Agreement 837890 (SMARTBOX) and from Marie Skłodowska-Curie Research and Innovation Staff Exchange (RISE) Grant 824017 (B-Ligzymes). The JU receives support from the European Union’s Horizon 2020 research and innovation program and the Biobased Industries Consortium (BIC). Fundação para a Ciência e a Tecnologia (FCT) additionally supported this work through R&D Unit MOSTMICRO-ITQB (UIDB/04612/2020 and UIDP/04612/2020), LS4FUTURE Associated Laboratory (LA/P/0087/2020), and Project UID/Multi/04326/2019, from the operational programs CRESC Algarve 2020 and COMPETE for 2020 through Project EMBRC.PT ALG-01-0145-FEDER-022121. M.D.S. acknowledges a Ph.D. grant (2020.08246.BD) from FCT.
The authors declare no competing financial interest.
Supplementary Material
References
- Sun Z.; Fridrich B.; de Santi A.; Elangovan S.; Barta K. Bright side of lignin depolymerization: toward new platform chemicals. Chem. Rev. 2018, 118, 614–678. 10.1021/acs.chemrev.7b00588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Bosch S.; Koelewijn S. F.; Renders T.; Van den Bossche G.; Vangeel T.; Schutyser W.; Sels B. F. Catalytic Strategies Towards Lignin-Derived Chemicals. Top. Curr. Chem. 2018, 376, 36. 10.1007/s41061-018-0214-3. [DOI] [PubMed] [Google Scholar]
- Hamalainen V.; Gronroos T.; Suonpaa A.; Heikkila M. W.; Romein B.; Ihalainen P.; Malandra S.; Birikh K. R. Enzymatic Processes To Unlock The Lignin Value. Front. Bioeng. Biotechnol. 2018, 6, 20. 10.3389/fbioe.2018.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runeberg P. A.; Brusentsev Y.; Rendon S. M. K.; Eklund P. C. Oxidative Transformations of Lignans. Molecules 2019, 24, 300. 10.3390/molecules24020300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llevot A.; Grau E.; Carlotti S.; Grelier S.; Cramail H. From Lignin-derived Aromatic Compounds to Novel Biobased Polymers. Macromol. Rapid Commun. 2016, 37, 9–28. 10.1002/marc.201500474. [DOI] [PubMed] [Google Scholar]
- Natte K.; Narani A.; Goyal V.; Sarki N.; Jagadeesh R. V. Synthesis of Functional Chemicals from Lignin-derived Monomers by Selective Organic Transformations. Adv. Synth Catal 2020, 362, 5143–5169. 10.1002/adsc.202000634. [DOI] [Google Scholar]
- Zakzeski J.; Bruijnincx P. C.; Jongerius A. L.; Weckhuysen B. M. The Catalytic Valorization Of Lignin For The Production Of Renewable Chemicals. Chem. Rev. 2010, 110, 3552–99. 10.1021/cr900354u. [DOI] [PubMed] [Google Scholar]
- Fache M.; Boutevin B.; Caillol S. Vanillin Production from Lignin and Its Use as a Renewable Chemical. ACS Sustain Chem. Eng. 2016, 4, 35–46. 10.1021/acssuschemeng.5b01344. [DOI] [Google Scholar]
- Fache M.; Boutevin B.; Caillol S. Epoxy thermosets from model mixtures of the lignin-to-vanillin process. Green Chem. 2016, 18, 712–725. 10.1039/C5GC01070E. [DOI] [Google Scholar]
- Smolarski N. High-Value Opportunities for Lignin: Unlocking its Potential Lignin potential. The Frost & Sullivan Blog 2012, 1–15. [Google Scholar]
- Havkin-Frenkel D. Vanillin. Kirk-Othmer Encyclopedia of Chemical Technology 2018, 1–12. 10.1002/0471238961.2201140905191615.a01.pub3. [DOI] [Google Scholar]
- Kaur B.; Chakraborty D. Biotechnological and Molecular Approaches for Vanillin Production: a Review. Appl. Biochem. Biotechnol. 2013, 169, 1353–1372. 10.1007/s12010-012-0066-1. [DOI] [PubMed] [Google Scholar]
- Marasco E. K.; Schmidt-Dannert C. Identification Of Bacterial Carotenoid Cleavage Dioxygenase Homologues That Cleave The Interphenyl Alpha, Beta Double Bond Of Stilbene Derivatives Via A Monooxygenase Reaction. Chembiochem 2008, 9, 1450–1461. 10.1002/cbic.200700724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAndrew R. P.; Sathitsuksanoh N.; Mbughuni M. M.; Heins R. A.; Pereira J. H.; George A.; Sale K. L.; Fox B. G.; Simmons B. A.; Adams P. D. Structure and mechanism of NOV1, a resveratrol-cleaving dioxygenase. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 14324–14329. 10.1073/pnas.1608917113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui X. W.; Golczak M.; Zhang J. Y.; Kleinberg K. A.; von Lintig J.; Palczewski K.; Kiser P. D. Utilization of Dioxygen by Carotenoid Cleavage Oxygenases. J. Biol. Chem. 2015, 290, 30212–30223. 10.1074/jbc.M115.696799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M.; Okada Y.; Yoshida T.; Nagasawa T. Purification, Characterization And Gene Cloning Of Isoeugenol-Degrading Enzyme from Pseudomonas putida IE27. Arch. Microbiol. 2007, 187, 511–517. 10.1007/s00203-007-0218-9. [DOI] [PubMed] [Google Scholar]
- Yamada M.; Okada Y.; Yoshida T.; Nagasawa T. Vanillin Production Using Escherichia Coli Cells Over-Expressing Isoeugenol Monooxygenase of Pseudomonas putida. Biotechnol. Lett. 2008, 30, 665–670. 10.1007/s10529-007-9602-4. [DOI] [PubMed] [Google Scholar]
- Ryu J. Y.; Seo J.; Park S.; Ahn J. H.; Chong Y.; Sadowsky M. J.; Hur H. G. Characterization of an Isoeugenol Monooxygenase (Iem) from Pseudomonas nitroreducens Jin1 That Transforms Isoeugenol to Vanillin. Biosci Biotechnol. Biochem. 2013, 77, 289–294. 10.1271/bbb.120715. [DOI] [PubMed] [Google Scholar]
- Zhao L. Q.; Xie Y. M.; Chen L. Y.; Xu X. F.; Zhao C. X.; Cheng F. Efficient Biotransformation Of Isoeugenol To Vanillin In Recombinant Strains Of Escherichia coli By Using Engineered Isoeugenol Monooxygenase And Sol-Gel Chitosan Membrane. Proc. Biochem. 2018, 71, 76–81. 10.1016/j.procbio.2018.05.013. [DOI] [Google Scholar]
- Zhao L. Q.; Jiang Y. Z.; Fang H. Y.; Zhang H. C.; Cheng S.; Rajoka M. S. R.; Wu Y. G. Biotransformation of Isoeugenol into Vanillin Using Immobilized Recombinant Cells Containing Isoeugenol Monooxygenase Active Aggregates. Appl. Biochem. Biotechnol. 2019, 189, 448–458. 10.1007/s12010-019-02996-1. [DOI] [PubMed] [Google Scholar]
- Renders T.; Van den Bosch S.; Koelewijn S. F.; Schutyser W.; Sels B. F. Lignin-First Biomass Fractionation: The Advent Of Active Stabilisation Strategies. Energy Environ. Sci. 2017, 10, 1551. 10.1039/C7EE01298E. [DOI] [Google Scholar]
- Lange L.; Connor K. O.; Arason S.; Bundgard-Jorgensen U.; Canalis A.; Carrez D.; Gallagher J.; Gotke N.; Huyghe C.; Jarry B.; Llorente P.; Marinova M.; Martins L. O.; Mengal P.; Paiano P.; Panoutsou C.; Rodrigues L.; Stengel D. B.; van der Meer Y.; Vieira H. Developing a Sustainable and Circular Bio-Based Economy in EU: By Partnering Across Sectors, Upscaling and Using New Knowledge Faster, and For the Benefit of Climate, Environment & Biodiversity, and People & Business. Front. Bioeng. Biotechnol. 2021, 8, 619066. 10.3389/fbioe.2020.619066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O.; Olson A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–61. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollikainen N.; de Jong R. M.; Kortemme T. Coupling Protein Side-Chain and Backbone Flexibility Improves the Re-design of Protein-Ligand Specificity. PLoS Comput. Biol. 2015, 11, e1004335 10.1371/journal.pcbi.1004335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes A. T.; Martins L. O.; Melo E. P. The Hyperthermophilic Nature Of The Metallo-Oxidase from Aquifex aeolicus. BBA-Proteins Proteom 2009, 1794 (1), 75–83. 10.1016/j.bbapap.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Eftink M. R. The Use of Fluorescence Methods to Monitor Unfolding Transitions in Proteins. Biophys. J. 1998, 63, 276–84. 10.1016/s0006-3495(94)80799-4. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66 (2), 125–32. 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The CCP4 suite: programs for protein crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1994, 50 (5), 760–3. 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. Features and Development of Coot. Acta Crystallogr. D 2010, 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G. N.; Skubak P.; Lebedev A. A.; Pannu N. S.; Steiner R. A.; Nicholls R. A.; Winn M. D.; Long F.; Vagin A. A. REFMAC5 for the Refinement Of Macromolecular Crystal Structures. Acta Crystallogr. D 2011, 67, 355–67. 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger E.; Vriend G. New Ways To Boost Molecular Dynamics Simulations. J. Comput. Chem. 2015, 36, 996–1007. 10.1002/jcc.23899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornak V.; Abel R.; Okur A.; Strockbine B.; Roitberg A.; Simmerling C. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins 2006, 65, 712–725. 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark P.; Nilsson L. Structure and Dynamics of the TIP3P, SPC, and SPC/E Water Models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. 10.1021/jp003020w. [DOI] [Google Scholar]
- Krieger E.; Darden T.; Nabuurs S. B.; Finkelstein A.; Vriend G. Making Optimal Use Of Empirical Energy Functions: Force-Field Parameterization In Crystal Space. Proteins 2004, 57, 678–83. 10.1002/prot.20251. [DOI] [PubMed] [Google Scholar]
- Tina K. G.; Bhadra R.; Srinivasan N. PIC: Protein Interactions Calculator. Nucleic Acids Res. 2007, 35, W473–6. 10.1093/nar/gkm423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer D. S.; Price D. C. A Simple Serum Iron Method Using the New Sensitive Chromogen Tripyridyl-s-Triazine. Clin. Chem. 1964, 10, 21–31. 10.1093/clinchem/10.1.21. [DOI] [PubMed] [Google Scholar]
- Garcia-Ochoa F.; Gomez E.; Santos V. E.; Merchuk J. C. Oxygen uptake rate in microbial processes: An overview. Biochem. Eng. J.l 2010, 49, 289–307. 10.1016/j.bej.2010.01.011. [DOI] [Google Scholar]
- Sigurdardóttir S. B.; Lehmann J.; Ovtar S.; Grivel J.-C.; Negra M. D.; Kaiser A.; Pinelo M. Enzyme Immobilization on Inorganic Surfaces for Membrane Reactor Applications: Mass Transfer Challenges, Enzyme Leakage and Reuse of Materials. Adv. Synth. Catal. 2018, 360, 2578–2607. 10.1002/adsc.201800307. [DOI] [Google Scholar]
- Han Z. C.; Long L. K.; Ding S. J. Expression and Characterization of Carotenoid Cleavage Oxygenases From Herbaspirillum seropedicae and Rhodobacteraceae bacterium Capable of Biotransforming Isoeugenol and 4-Vinylguaiacol to Vanillin. Front. Microbiol. 2019, 10, 01869. 10.3389/fmicb.2019.01869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B. X.; Tao Y. Whole-cell Biocatalysts by Design. Microb. Cell Fact. 2017, 16, 106. 10.1186/s12934-017-0724-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes A.; Pinto B.; Bonardo L.; Royo B.; Robalo M. P.; Martins L. O. Wasteful Azo Dyes as a Source of Biologically Active Building Blocks. Front. Bioeng. Biotechnol. 2021, 9, 672436. 10.3389/fbioe.2021.672436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya T.; Miura M.; Kuroiwa M.; Kino K. High-yield Production Of Vanillin From Ferulic Acid By A Coenzyme-Independent Decarboxylase/Oxygenase Two-Stage Process. New Biotechnol 2015, 32, 335–339. 10.1016/j.nbt.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Sanchez-Ruiz J. M. Protein Kinetic Stability. Biophys Chem. 2010, 148, 1–15. 10.1016/j.bpc.2010.02.004. [DOI] [PubMed] [Google Scholar]
- Dunning Hotopp J. C.; Auchtung T. A.; Hogan D. A.; Hausinger R. P. Intrinsic Tryptophan Fluorescence As A Probe Of Metal And Alpha-Ketoglutarate Binding To Tfda, A Mononuclear Non-Heme Iron Dioxygenase. J. Inorg. Biochem 2003, 93, 66–70. 10.1016/S0162-0134(02)00436-1. [DOI] [PubMed] [Google Scholar]
- Daruwalla A.; Kiser P. D. Structural And Mechanistic Aspects Of Carotenoid Cleavage Dioxygenases (CCDs). BBA Mol. Cell Biol. Lipids 2020, 1865, 158590. 10.1016/j.bbalip.2019.158590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.