Abstract
Natural systems use weak interactions and avidity effects to give biological systems high specificity and signal-to-noise ratios. Here we describe design principles for engineering fusion proteins that target therapeutic fusion proteins to membrane-bound signaling receptors by first binding to designer-chosen co-receptors on the same cell surface. The key design elements are separate protein modules, one that has no signaling activity and binds to a cell surface receptor with high affinity and a second that binds to a receptor with low or moderate affinity and carries out a desired signaling or inhibitory activity. These principles are inspired by natural cytokines such as CNTF, IL-2, and IL-4 that bind strongly to nonsignaling receptors and then signal through low-affinity receptors. Such designs take advantage of the fact that when a protein is anchored to a cell membrane, its local concentration is extremely high with respect to those of other membrane proteins, so a second-step, low-affinity binding event is favored. Protein engineers have used these principles to design treatments for cancer, anemia, hypoxia, and HIV infection.
Recombinant DNA technology and synthetic biology have created the potential for multicomponent therapeutics that mimic the complexity of natural systems. This potential has gone unrealized, in part due to the lack of conceptual tools for developing such products. Most of the protein drugs that are currently on the market are either naturally occurring proteins or monoclonal antibodies. This Perspective will discuss engineered fusion protein therapeutics with two or more binding domains that are not naturally attached to each other, specifically focusing on proteins in which each of the two domains binds to different receptors on the same cell surface at the same time (Figure 1).
Figure 1.
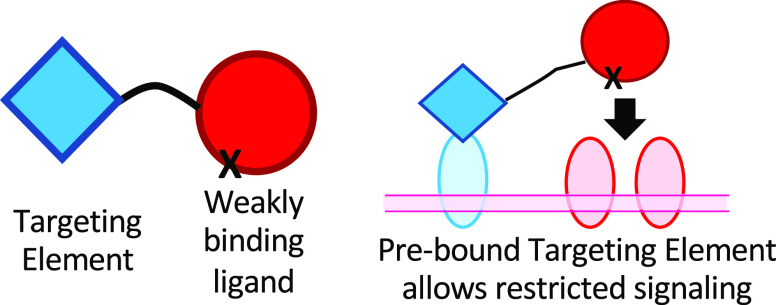
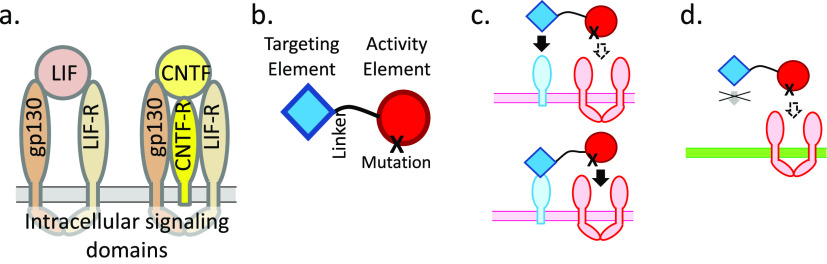
Quantitative aspects of two-component signaling protein design. (a) Signaling by the natural cytokines LIF and CNTF. LIF binds to a heterodimer of the LIF receptor and gp130, both of which have intracellular signaling domains. CNTF has a very low affinity for LIF-R/gp130 but a high affinity for the nonsignaling CNTF receptor, which is expressed on a subset of LIF-R/gp130-expressing cells. When CNTF binds to CNTF-R, the CNTF is positioned to interact with LIF-R and gp130 because of its high concentration on the cell surface.24 (b) A “Chimeric activator”, an engineered protein consisting of a targeting element (typically an scFv or a nanobody) and an activity element (typically a hormone or cytokine) connected by a linker. The activity element has a mutation that decreases its affinity for its receptor. Relative to the natural hormone/cytokine on which it is based, a chimeric activator is analogous to CNTF relative to LIF; i.e., a chimeric activator is a modified form of a signaling protein with weakened affinity for its receptor but can bind to a nonsignaling transmembrane protein present on a subset of cells, so that signaling occurs on those cells. (c and d) Cell-specific signaling of a chimeric activator. (c) On a target cell, the targeting element first binds to a cell-specific surface protein (light blue). Because it is now present at a high local concentration on the cell surface, the activity element then binds to its receptor in spite of its low affinity. (d) On a nontarget cell, there is no receptor for the targeting element, and the activity element does not have a sufficiently high affinity to bind and signal.
When researchers have tried to develop more complex artificial biological systems involving two or more components, their design has posed novel challenges. Consider the repressilator, a transcriptional system in which three repressors turn each other off such that the system should oscillate among three states. In the initially designed repressilator,1 the actual system behaved quite randomly. Only when Pottvin-Trottier et al.2 performed a thorough analysis of the quantitative parameters in the system was it possible to construct a repressilator that oscillates in a reproducible manner.
The same challenges exist with engineered proteins. For example, immunocytokines are engineered proteins in which an immunostimulatory cytokine is fused to an antibody element that typically binds to a tumor-specific antigen.3,4 The concept is that the antibody element will bind to the tumor cell surface and concentrate the cytokine in the tumor. In practice, the slow entry of large proteins into solid tumors means that the cytokine can first bind to its receptor on immune cells in the periphery. Tzeng et al.5 studied antitumor antibodies fused to IL-2 and found that in a mouse tumor model, the biodistribution of an antibody–IL-2 fusion protein was governed by the IL-2 element rather than the antibody V regions. Ribba et al.6 found that in cancer patients, a similar antitumor antibody fused to IL-2 induced expansion of peripheral T cells, which created a sink for the drug in subsequent doses. These examples illustrate how an engineered protein with an appealing qualitative design may fail due to quantitative problems.
Protein and cell engineers have generated a variety of fusion proteins for treatment of cancer and other diseases. Examples of approved therapeutics include bispecific T-cell engagers (BiTEs) and CAR-T cells. The initially approved versions of these drugs use naturally occurring domains as components of their fusion proteins, without regard to whether the quantitative properties of each domain are optimal in the engineered context.
Blinatumomab (Blincyto) consists of an scFv that binds to a tumor antigen, attached to a another scFv that binds to T cells.7 Specifically, the first element of this bispecific T-cell engager binds to CD19, an abundant protein on the surface of B cells, including B-cell lymphomas. The second antibody element binds to the ε subunit of the T-cell receptor.8
This mechanism works well when the target cell is a B-cell-derived lymphoma and the BiTE protein has easy access to tumor cells from the blood. However, application of BiTE technology to solid tumors is challenging because of poor penetration of protein drugs into such tumors.9 In this situation, binding to T cells may occur first, resulting in nonspecific activation, cytokine production, and side effects.
In this case, the problem could be corrected by mutating the anti-TCRε antibody element so that it has an even weaker affinity. Several research groups have pursued this approach in developing next-generation BiTEs for the treatment of solid tumors.10,11 This example illustrates how naturally occurring domains may have quantitative properties that are not optimal in the context of an engineered fusion protein.
Optimization in a Multidimensional Design Space
We see the central conceptual challenge as being how to explore a multidimensional parameter space. In traditional drug development, once a target has been identified, the typical strategy is to create a small molecule or antibody that will bind very tightly to the target; the parameter space is one-dimensional (setting aside bioavailability issues). The goal is always to increase affinity. In the case of immunocytokines, BiTEs, and chimeric activators, there is a two-dimensional parameter space (Figure 2). In the case of the repressilator, there are numerous quantitative parameters: the affinity of each repressor for its operator, the strength of cooperative effects if there are multiple operators, the strength of each promoter and ribosome binding site, the stability of each mRNA, etc. Most of these can be rationally engineered. Promoters and ribosome binding sites are well understood, and the affinity of a protein–protein interaction can be systematically weakened; increasing affinity is usually more difficult but possible with established technology.
Figure 2.
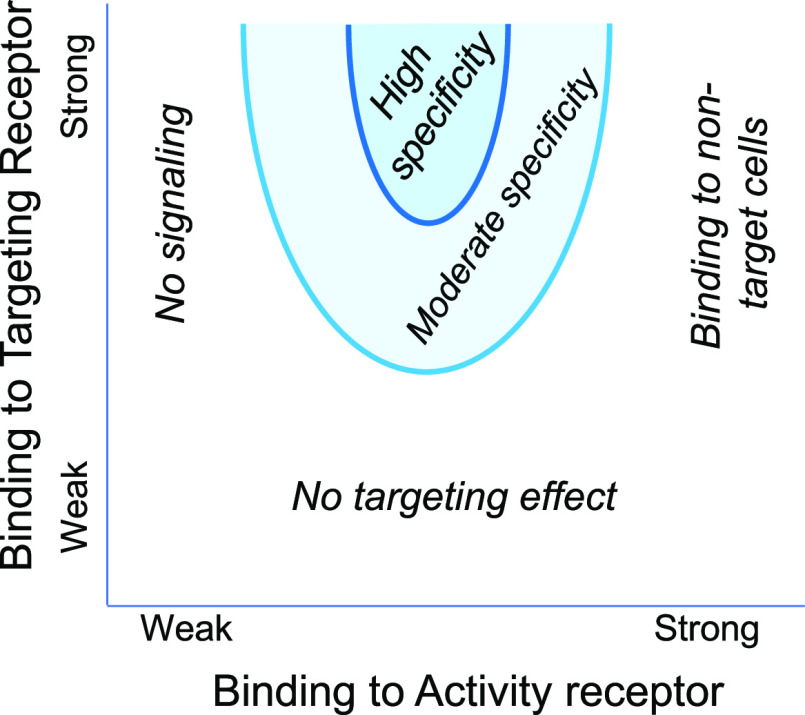
Parameter space for a chimeric activator. To be effective, the level of binding of the activity element to its receptor should have an intermediate value. If the affinity is too high, signaling will occur on nontarget cells. If the affinity is too low, no signaling occurs. The overall level of binding of the targeting element should be greater than for the mutated activity element. This may be achieved if the on rate or affinity constant of the targeting element for its receptor is greater than that of the activity element for its receptor, or if the number of receptors for the targeting element on the cell is much higher than the number of receptors for the activity element, or both.
Two examples from the literature on protein drug development illustrate optimization of proteins with two or more binding elements that act in cis on a cell surface. “Chimeric activators” direct signal transduction to cell types that mediate therapeutic effects and away from cells that cause side effects.12−14 “Adnectins” are antibody-like binders that can be oligomerized to create protein drugs with multiple binding specificities.15 Wensel and colleagues have created a multispecific inhibitor of HIV infection with four elements: two adnectins, an α-helical HIV-cell fusion inhibitor, and albumin to improve plasma half-life.16−18 This molecule is similar to chimeric activators in that it acts in cis, being first localized to a cell surface to concentrate binding elements near a second target. In the development of each type of molecule, researchers needed to optimize the quantitative and spatial characteristics of their molecules.
Chimeric Activators
Natural hormones and cytokines often act on a wide variety of cells. In a therapeutic context, signaling on some cells may be beneficial while action on other cells may lead to side effects. For example, interferon α has been used to treat certain cancers and viral infections but has serious side effects, including flu-like symptoms and suicidal and homicidal ideation.19 Similarly, erythropoietin (EPO) binds to receptors on red blood cell precursors to stimulate production of mature red blood cells (useful for treating anemia) but also stimulates production of activated platelets and pro-thrombotic effects generally.20 Clinically, high erythropoietin use is associated with increased incidence of heart attacks and stroke,21,22 so reduction of EPO-induced thrombosis could have great clinical benefit.
Several groups have independently developed the idea of targeting the activity of a hormone or cytokine to a subset of cells to limit side effects.3,4,12−14,23 In the chimeric activator configuration, a “targeting element” (such as an scFv) is fused via a peptide linker to an “activity element” (a hormone/cytokine) that has been mutated to reduce but not eliminate receptor binding (Figure 1b). The linker length is adjusted so that the scFv and the hormone/cytokine can simultaneously bind to their respective receptors on the same cell surface. Cells that mediate a therapeutic effect should express the antigen bound by the targeting element, while cells that mediate side effects should not.
Several natural cytokines use this principle (Figure 1a).24 For example, the LIF protein (leukemia inhibitor factor) binds strongly to a heterodimeric receptor and signals through both subunits. The structurally homologous CNTF protein (ciliary neurotrophic factor) signals through the same receptor subunits but has a very low affinity for this heterodimer. Signaling occurs only on cells that also express CNTF-Rα, which has a high affinity for CNTF but has no cytoplasmic domain and does not contribute to cytoplasmic signaling events; the only purpose of CNTF-Rα is to localize CNTF to the cell surface, where it will be present in such a high effective concentration that it will bind to LIF receptor subunits in spite of its low affinity for the LIF receptor.24 Other cytokines, including IL-2, IL-4, IL-6, and IL-15, have analogous high-affinity receptor subunits, without cytoplasmic signaling elements, whose sole purpose appears to be to localize the cytokine to a subset of cells. Such nonsignaling receptor subunits are analogous to the receptor for the targeting element in the chimeric activator proteins described here (Figure 1c).
The design space for a chimeric activator includes parameters such as the binding, the on rates and off rates of the targeting element and the activity element for their receptors, and the length of the linker separating these elements. A key design feature of a chimeric activator is the use of a mutation in the activity element that weakens but does not abolish binding to its receptor. If the activity element binds too efficiently to its receptor, this binding will not depend on the targeting element and cell specificity will be lost, but if it binds too poorly, there will be no activity at all (Figure 2). Fortunately, within the overall parameter space, the subspace of working designs is large.
Concentration Effect from Prebinding to a Cell Surface
Prebinding of a chimeric activator protein to a cell surface results in a profound increase in the effective concentration of the signaling element. Consider a fusion protein with an antibody and a signaling element, connected by a peptide linker of five amino acids (Figure 3). When the antibody element binds to its antigen on a cell surface, the signaling element will be constrained to lie in a volume defined by the surface of the cell and the distance from the cell surface within which the peptide linker allows movement; the diffusion of the prebound complex is in effect limited to two dimensions.
Figure 3.
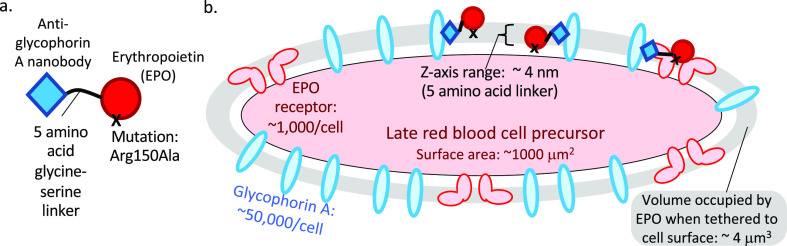
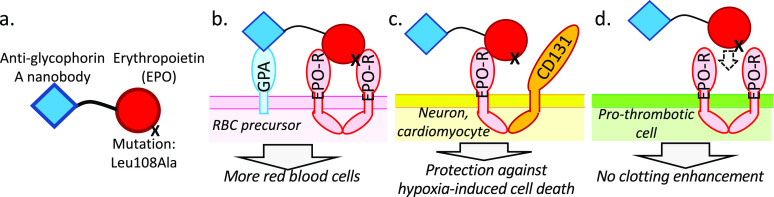
High local concentration of an activity element achieved through anchoring by a targeting element. “Targeted EPO” example. (a) Design of a targeted EPO.26 The targeting element is a nanobody that binds to glycophorin A, which is abundant on late red blood cell precursors; the KD for this interaction is 30 nM.46 The linker is five amino acids in length, such that when fully extended into a β-strand, it is ∼2 Å long. The activity element is erythropoietin (EPO) with a mutation that weakens binding to the EPO receptor by ∼13-fold, so that for this interaction is ∼80 nM.36 (b) Estimation of the local concentration of the EPO-based activity element when prebound to the cell surface by the anti-glycophorin A targeting element. When the anti-glycophorin nanobody binds to its epitope, the EPO element is constrained to lie in a volume defined by the surface area of the cell and the distance from the membrane in which the linker allows the EPO element to diffuse. In this case, the volume is ∼4 μm3, which means that a single EPO activity element will have an effective concentration of ∼400 pM in the neighborhood of its receptor. Schmick and Bastiaens47 perform a similar calculation for intracellular proteins that associate with the cytoplasmic face of the plasma membrane.
The typical surface area of a mammalian cell is in the range of 1000 μm2, and the length of a fully extended five-amino acid linker is ∼2 nm. Thus, the signaling element of this fusion protein sits in a volume of 4 μm3, or 4 fL, and the concentration of only one signaling molecule in this volume is ∼400 pM. If using a short linker is possible, this increases the effective concentration of the signaling molecule in the neighborhood of its receptor on the cell surface. For this calculation to be relevant, either the targeting receptor, the activity receptor, or both must be able to move by two-dimensional diffusion through the cell membrane to be in the proximity (see below).
The choice of a mutation in the hormone/cytokine moiety of a chimeric activator is governed by the following considerations. We want cell binding to be driven by binding to the nonsignaling targeting receptor. In practice, a minimum requirement is that either NTkon,T > NAkon,A or NTKeq,T > NAKeq,A (or preferably both), where NT is the number of targeting receptors per cell, NA is the number of activity receptors per cell, kon,T is the on rate for the targeting element to bind to its receptor, kon,A is the on rate for the activity element to bind to its receptor, Keq,T is the equilibrium binding constant for the targeting element to bind to its receptor, and Keq,A is the equilibrium binding constant for the activity element to bind to its receptor. In some situations, the equilibrium constants may be dominated by a very slow off rate, such that dissociation of a ligand–receptor complex is slower than termination of a binding reaction by receptor-mediated endocytosis. Mutations that change only the off rate may have no effect on signaling, and mutational reduction of kon such that NTkon,T > NAkon,A is more relevant.
For example, the chimeric activators of Cironi et al.12 consist of EGF as a targeting element and IFNα as an activity element and are designed to bind to cells expressing the EGF receptor and to activate type I IFN signaling specifically on those cells. The kon values for EGF and IFNα and their receptors are 106 and 3.7 × 106 M–1 s–1, respectively (i.e., approximately the same), and the Keq values for their receptors are both ∼3 × 108 M. Cironi et al. used Daudi cells engineered to express EGFR, in which there were ∼5600 EGFRs/cell and 3600 IFNAR2s/cell. In this situation, NTkon,T = NAkon,A and NTKeq,T = NAKeq,A (roughly) so no enhancement of IFNα signaling is expected as a result of fusion to EGF, and none was observed. However, when weakening mutations were introduced into IFNα, EGFR-dependent signaling was observed.
The rate of internalization of the targeting receptor should be as low as possible. EGF/EGFR signaling complexes are internalized within a few minutes as a consequence of signaling itself, while nonsignaling EGFR is passively internalized as part of bulk membrane turnover, which occurs on an ∼10-fold slower time scale.25 Thus, it is generally advisable to use an inhibitory antibody as a targeting element, and not a signaling molecule. Moreover, EGF generally promotes tumor progression and so would never be used as part of an anticancer therapeutic.
A striking fact is that once a chimeric activator binds to its targeting receptor, its local concentration is very high within a small volume around the cell surface (Figure 3). It is important to note that in metabolically active cells, there will generally be two-dimensional diffusion of either the targeting receptor, the activity receptor, or both. Such a calculation of the effective concentration of a partially bound fusion protein assumes that two-dimensional diffusion is rapid compared to the time scale of the biological event being measured. These conditions are met in assays in which a fusion protein stimulates cell proliferation in vitro or in vivo on a time scale of several days, because diffusion of a membrane receptor across a significant portion of a cell surface occurs in ≤1 h.
In the absence of such diffusion and if the number of receptors is low, most targeting receptors may be too far from an activity receptor for meaningful stimulation to occur. TF-1 erythroleukemia cells, which are used to test chimeric activators based on erythropoietin and anti-glycophorin A, have ∼3900 glycophorin A molecules per cell.13 Assuming a cell surface area of 150 μm2, there will be a glycophorin A approximately every 200 nm, so each EPO receptor will be on average approximately 50–100 nm from a glycophorin. The linker in the optimized anti-GPA/EPO chimeric activator of Lee et al.26 was only five amino acids in length (with an extended length of ∼20 nm), so the initial binding to glycophorin A would generally not be productive in the absence of lateral diffusion. Moreover, systematically changing the length of the flexible glycine-serine from five to 35 amino acids had no discernible effect on the potency of this type of chimeric activator.26
Two-dimensional diffusion is rapid compared to the off rate of a typical antibody–antigen interaction that might be used in a chimeric activator. Glycophorin A is a relatively slowly diffusing protein; most of this protein has a two-dimensional diffusion constant of 5 × 10–3 μm2/s.27 Some EPO-containing chimeric activators used the anti-glycophorin antibody 10F7, which has a monovalent KD of 10–7 M and an estimated off rate of 1/100 s (i.e., this is a weakly binding antibody28). Using the two-dimensional diffusion equation ⟨r2⟩ = 4Dt, where r is a typical distance traveled in time t, we find that in 100 s, glycophorin A with its bound chimeric activator would travel, on average, ∼1.4 μm within a cell membrane. In this time, the chimeric activator would have ample opportunity to be in the proximity of an EPO receptor so that a second-step binding could occur. The glycophorin A/10F7 combination is a worst-case scenario. For example, unliganded EGFR has a two-dimensional diffusion constant of 0.11 μm2/s29 and the koff of a typical anti-EGFR antibody, cetuximab, is ∼1.1 × 10–3 s–1,30 together giving an end-to-end travel distance of ∼20 μm in the time frame of an EGFR–cetuximab complex.
(This is in contrast to a situation in which avidity effects of antibodies are measured with immobilized antigens on plates or on cells maintained at 4 °C. In these cases, two immobilized antigens are close enough to allow divalent binding or are not. In such cases, description of divalent binding requires knowing ⟨r⟩, the average distance between two ligands on a surface, which has been modeled by Crothers and Metzger31 and Kaufman and Jain.32)
It is important to consider the spatial configuration of the active signaling complex when designing a targeted fusion protein. One key parameter is the length of the linker between the targeting element and the activity element, which must be long enough that both elements can simultaneously bind to their targets. When the targeting element is first bound to its receptor, the activity element will undergo constrained Brownian motion, with the linker forming a random coil. Simple entropic considerations indicate that the linker will almost never be fully extended.33 Coarse-grained molecular dynamics simulations and theoretical considerations indicate that the average distance from one end of the linker to the other will increase roughly with a less-than-linear function of the linker length, characteristic of a volume-excluded random walk in three dimensions.33,34 However, course-grained molecular dynamics also showed that in many simulations of a chimeric activator with a bound targeting element and an unbound activity element, the linker tended to spend much time wrapped around the activity element, such that its binding to the activity receptor could be sterically blocked; this effect may occur because rotational Brownian motion is significant relative to translational Brownian motion of globular proteins, so that the linker is pulled around a rotating protein.34 In any case, it is best to have a linker that is somewhat longer than is required to simply bridge the distance between the receptor-bound states of the targeting and activity elements.
Specific Examples of Targeted Cytokines and Hormones
The group of Silver and the group of Tavernier and Uze have developed targeted forms of interferon α, erythropoietin (EPO), and leptin in which a mutant, reduced-function signaling protein is fused to a targeting scFv or nanobody.12−14 (In a coincidental convergence, both groups independently developed fusion proteins using the interferon α mutation Arg149Ala, which reduces receptor binding activity by ∼200-fold.35)
Choosing a mutation in the activity element is a straightforward matter. It is not necessary to construct large libraries of fusion proteins, and typically, it is sufficient to test a handful of mutations. Taking advantage of an extensive site-directed mutagenesis study of IFNα,35 the Silver group showed that when EGF was used as a targeting element and IFNα was used as an activity element, IFNα mutations Lys133Ala, Arg144Ala, and Arg149Ala all showed enhanced signaling on cells expressing the EGF receptor.12 These mutations reduce the affinity of IFNα for its receptor by 9-, 40-, and 200-fold, respectively.35 Similarly, Garcin et al. used an anti-leptin receptor nanobody to target IFNα to cells expressing the leptin receptor.14 They found that the IFNα mutations Met148Ala, Arg149Ala, and Leu153Ala all led to enhanced IFNα signaling of 100–1000-fold, while a corresponding fusion with wild-type IFNα showed an ∼10-fold targeting effect. Taylor et al.13 constructed a set of fusions between mutant erythropoietin proteins and an scFv that targets glycophorin A and found that the three EPO mutations tested all showed enhanced glycophorin-dependent signaling relative to a fusion with wild-type EPO. Xu et al.23 constructed a form of IL-15 targeted to PD1-expressing T cells by fusion of an anti-PD1 antibody to a low-affinity form of IL-15. These groups used mutations that replace wild-type side chains with smaller or equal-sized variants; thus, there is no steric hindrance in binding and only a loss of a contact.
Animal studies confirm that chimeric activators exhibit enhanced cell-type specificity in vivo. Burrill et al.36 performed an in vivo structure–function analysis and showed that an anti-glycophorin A (scFv)–EPO(Arg150Ala) chimeric activator (“targeted EPO”) specifically stimulated production of red blood cells but not platelets in mice. The experiments were performed using a transgenic mouse expressing human glycophorin A on its late red blood cell precursors and mature red blood cells. The late red blood cell precursors thus co-express the EPO receptor and human glycophorin A, allowing for simultaneous binding of the chimeric activator to these two surface proteins and activation of maturation into red blood cells, in spite of the weakening Arg150Ala mutation. Mice treated with an anti-glycophorin A–EPO(wild type) showed a stimulation of both red blood cell and platelet production, while nontransgenic mice (whose endogenous glycophorin A is not recognized by the antibody element) did not produce extra red blood cells in response to anti-glycophorin A–EPO(Arg150Ala). A loss of function mutation in the anti-glycophorin scFv portion of this fusion protein also blocked stimulation of red blood cell production. Thus, the anti-glycophorin A–EPO(Arg150Ala) chimeric activator showed in vivo activity and specificity exactly as predicted: cell-type specificity depended on the weakening mutation in EPO, and erythropoietic activity depended on binding of the scFv to its receptor.
Xu et al.23 constructed a chimeric activator that used anti-PD1 to target T cells in the tumor microenvironment. IL-15 binds to the same signaling receptors as IL-2 but has a different nonsignaling receptor α subunit. IL-15 binds with a moderate affinity to the IL-2Rβ/γ signaling receptor, so there is a risk that an IL-15-based cancer treatment would bind to peripheral immune cells and be drained into this pharmacokinetic sink before being distributed into a solid tumor. Xu et al. used the fact that PD1 is upregulated on tumor-infiltrating lymphocytes. IL-15 was mutated to eliminate binding to the IL-15α receptor subunit and to weaken binding to IL-2Rβ/γ, so that signaling activity depends on prior binding to PD1 via the attached antibody element. In mouse experiments, PD1-targeted IL-15(mutant) localized to solid tumors, unlike the IL-2 immunocytokines discussed above,5,6 and induced less cytokine release than a comparable nontargeted form of IL-15. PD1-targeted IL-15 also showed a superior therapeutic index for tumor inhibition versus weight loss when compared to the nontargeted IL-15 control.
Pogue et al.37 constructed an anti-CD38 antibody fused to attenuating mutants of IFNα. CD38 is a marker of multiple myeloma. Previously, IFNα alone had been clinically tested for treatment of multiple myeloma, but its side effects were severe. Similarly to other targeted IFNα fusions,12,14 significant targeting effects were seen in vitro. In a mouse xenograft model, good control of tumor growth was observed, but because human IFNα does not interact well with mouse IFN receptors, no inference could be made about a therapeutic index. To address this, the authors treated cynomolgus monkeys with wild-type IFNα and mutant fusion proteins; not surprisingly, the mutant fusions induced lower levels of IFNα-inducible biomarkers. Together, these results indicate that genuine improvements in specificity in vivo can be achieved with chimeric activators.
A spatial consideration in the design of chimeric activators is that the targeting and activity elements in the fusion protein may mediate adhesion between different cells. A chimeric activator is designed to bind to receptors on the same cell surface, but it is sometimes possible for fusion protein consisting of two ligands connected by a flexible linker to bridge two cells. The bispecific T-cell engagers deliberately take advantage of this principle; these proteins are a single polypeptide chain with a flexible linker connecting two scFvs: one scFv binds to T cells, and the other binds to tumor cells, so that the two cells become cross-linked.
Lee et al.26 found that undesired cell–cell interaction appeared to be a potential problem with the “targeted EPO” chimeric activator described above.13,36 The goal for this molecule is to treat anemia, but without the prothrombotic side effects of erythropoietin. The scFv–EPO fusion protein was designed to bind to glycophorin A on late red blood cell precursors and activate EPO receptors on those cells. A bonus feature of targeted EPO is that it also binds to mature red blood cells, which have approximately 800 000 glycophorin A proteins on their surface. There is no EPO receptor on mature red blood cells and thus no signaling, but these cells act as a pharmacokinetic reservoir: most of the fusion protein binds noncovalently to the RBCs and is slowly released. As a result, the plasma half-life of targeted EPO is profoundly improved.
The first-generation targeted EPO stimulated production of red blood cells but not platelets, unlike EPO itself, which stimulates production of both cell types.36 Platelets were taken as a surrogate marker for blood clotting. Even though targeted EPO did not stimulate platelet production in vivo, treatment of mice with the first-generation protein led to an enhancement of blood clotting (as did EPO itself). Lee et al.26 hypothesized that the binding of the fusion protein to glycophorin A on mature red blood cells could cause adherence to cells with EPO receptors, such as macrophages or vascular endothelial cells. Such adhesion was demonstrated in vitro: red blood cells adhered to tumor cells expressing the EPO receptor in the presence of the first-generation targeted EPO.
To correct the problem of protein-mediated cell–cell adhesion, Lee et al. took advantage of the fact that glycophorin A forms a dense canopy on red blood cells; at 800 000 copies per cell, the average distance between between glycophorin A molecules is <200 Å, and it extends approximately 90–130 Å from the red cell surface. The glycophorin binding element in targeted EPO was changed from an scFv that binds ∼35 amino acids from the membrane-distal tip of glycophorin A to a nanobody that binds 55 amino acids from the tip. In addition, the linker was shortened from 35 to only five amino acids. The targeted EPO with the short linker and membrane-proximal binding site no longer mediated adhesion between red blood cells and EPO receptor-bearing tumor cells, presumably because when this protein is bound to RBCs, the EPO element is effectively hidden within the forest of glycophorin A.
A Targeted Fusion Protein That Retains Two Activities While Eliminating a Third
In addition to inducing the production of red blood cells, erythropoietin also promotes survival of neurons, cardiac cells, and other cells in response to hypoxia and other insults. This protective response is thought to be mediated by a heterodimeric receptor consisting of EPO-R and CD131.38 In the complex of EPO with its homodimer receptor, one face of EPO strongly interacts with the receptor, with a KD of ∼1 nM, and the other face of EPO interacts weakly, with a KD of ∼1 μM.39 When EPO binds to the EPO-R/CD131 heterodimer, by comparison to the EPO/(EPO-R)2 complex, the CD131 receptor replaces the weakly interacting EPO receptor. On the basis of its design, the targeted EPO described above is not expected to have the tissue-protective activity, because the activity-reducing mutation in EPO lies in the interface that strongly interacts with EPO receptor, and this mutation would also weaken formation of the EPO-R/CD131 signaling complex.
To treat hypoxia, it would be ideal to have an engineered protein that would retain both the red blood cell-producing and tissue-protective activities of erythropoietin, while eliminating the blood-clotting activity. To achieve this, Lee et al.39 took advantage of the fact that mutations on the “weak face” of EPO that weaken activation of (EPO-R)2 often have no effect on signaling through EPO-R/CD131; i.e., such mutations disrupt interaction with the second EPO-R but not with CD131. These authors constructed a chimeric activator consisting of the anti-glycophorin nanobody and an EPO element with a mutation on the weak face (Figure 4). According to this design, on red blood cell precursors the poor activation of (EPO-R)2 receptors will be rescued by prior binding to glycophorin A (Figure 4B), signaling will still occur through EPO-R/CD131 heterodimers (Figure 4C), but signaling will not occur via (EPO-R)2 receptors on cells mediating thrombosis (Figure 4D). The resulting fusion protein protected neuroblastoma cells against a hypoxia-mimetic challenge in vitro and induced production of red blood cells but not platelets in vivo, indicating that the protein behaves as designed.29
Figure 4.
Targeted fusion protein for treatment of hypoxia. This engineered protein takes advantage of the fact that red blood cell production and thrombosis mediated by EPO are mediated by EPO receptor homodimers, while protection of cells against hypoxia-induced death is mediated by a heterodimer of the EPO receptor and CD131. (a) Molecular design of a fusion protein that stimulates red blood cell production and protection of cells against hypoxia-induced death, but without inducing thrombosis.39 This protein consists of an anti-glycophorin A nanobody,46 a short linker, and erythropoietin containing the Leu108Ala mutation. This mutation is on the face of EPO opposite from Arg150 and weakens its binding to the more weakly interacting EPO receptor. (b) On red blood cell precursors, the protein is anchored to the cell surface by binding of the nanobody to glycophorin A, so that it will bind to EPO-R homodimers in spite of the mutation that weakens this interaction. (c) On cells such as neurons and heart cells, the EPO element binds to an EPO-R/CD131 heterodimer that signals cells to survive in spite of hypoxia. The Leu108Ala mutation does not affect signaling through EPO-R/CD131 heterodimers. (d) The fusion protein does not bind to EPO-R homodimers on cells that mediate thrombosis, due to the Leu108Ala mutation.
A Multicomponent Inhibitor of HIV Fusion to Target Cells
HIV treatment is a challenge because of the high rate of mutation of the virus. Combination therapy is the norm. Typically, approximately three different drugs are combined, so that in a person with a high viral load, the probability of any virus simultaneously mutating to resist all three drugs is low. The initial small molecule-based combination therapies involved a complex regimen of pills taken with different schedules over a day, because each drug had different pharmacokinetic properties. The development of once-a-day pills such as Biktarvy (Bictegravir/Emtricitabine/Tenofovir Alafenamide) has taken many years and represents a triumph of formulation, allowing slow and coordinated release of three different drugs.40 The same challenges around resistance mutations and pharmacokinetics apply to protein drugs.
However, engineered fusion proteins allow for the possibility of combining multiple activities in a single protein. Thus, simultaneous mutation to resistance to different elements can be greatly reduced, and the pharmacokinetics of the different elements will be uniform because they are all attached to each other.
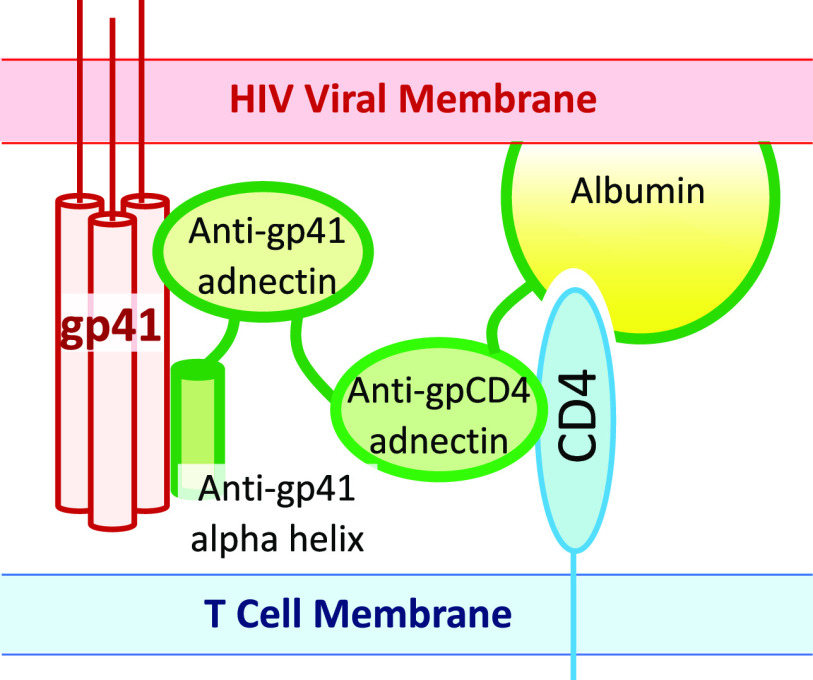
Wensel et al.16−18 developed a remarkable four-component fusion protein with an element that binds to CD4, two elements that bind to gp41 of the HIV envelope, and an albumin moiety to extend the plasma half-life (Figure 5). This group developed binding elements (“adnectins”) derived from the fibronectin repeat FnIII10,15 a small, tightly folded β-strand protein domain. It is possible to construct display libraries of FnIII10 in which the loops at one end are varied and then select variants that bind to an arbitrarily chosen target. Wensel et al.16 first identified an adnectin that bound to a region of CD4 that was not needed for the immunological function of this protein but still inhibits binding of the gp120 subunit of the HIV envelope (IC50 = 6 nM).
Figure 5.
Inhibition of HIV infection with a cell-targeted, multidomain engineered protein. GSK3732394 consists of an adnectin that binds to CD4, a second adnectin and an α helix that bind to adjacent segments of gp41, and serum albumin to extend the plasma half-life.16−18 During HIV infection, after gp120 has engaged with CD4, it is stripped off and the underlying α helices of gp41 are transiently exposed. The rearrangement of these α helices into a fusogenic conformation can be inhibited with proteins that bind to and stabilize the nonfusogenic conformation. In a treated patient, before the virus binds to a target T cell, GSK3732394 will bind to CD4 on the T-cell surface. When a virus binds, the gp41 binding elements of the drug are in a high local concentration and rapidly bind to gp41 to inhibit virus–cell fusion.
Wensel et al. then identified adnectins that bind to gp41.17 Just to recall, the HIV envelope proteins gp120 and gp41 are derived from the large homotrimeric precursor gp160 by a cleavage event. On the free virus, before it binds to a target cell, gp120 sits on top of gp41 and holds it in a metastable state.41 When gp120 interacts with CD4 and a chemokine receptor, gp120 comes off of the virus and gp41 rearranges to a fusion-promoting conformation. This process takes some time, and drugs that bind to and stabilize non-rearranged gp41 are used clinically (e.g., enfuvirtide42). These anti-gp41 adnectins efficiently inhibit HIV infection in vitro (IC50 = 5 nM), but the virus can mutate to resistance in a single step. However, when the anti-CD4 and anti-gp41 adnectins are fused, the resulting inhibitor is extremely potent, with an IC50 of 1 pM. The synergistic effect is not seen when the two inhibitors are mixed; they must be attached to each other. The authors hypothesize that the anti-CD4/anti-gp41 fusion protein binds rapidly to CD4 and becomes concentrated on the surface of the target cell, so that when the virus is undergoing the infection process, the anti-gp41 adnectin is at a high local concentration and binds rapidly to gp41 before it rearranges.
The anti-CD4/anti-gp41 double adnectin was further improved by addition of an α-helical region that binds to gp41. Here the design was based on rational and/or structural considerations. Others had characterized how various α-helical peptides could bind to gp41 and stabilize the non-rearranged state. Wensel and colleagues knew where their anti-gp41 adnectin bound, because this was part of the selection process. On the basis of the gp41 structure, they fused the anti-gp41 adnectin to an α helix predicted to bind to an adjacent segment of gp41, to achieve synergistic binding. The resulting protein binds tightly to gp41, and its binding is relatively resistant to mutations. Finally, addition of serum albumin to the fusion protein increases the plasma half-life of the drug; this is a standard part of the protein engineering tool kit.43
Conclusion
In 1968, Adam and Delbruck44 proposed that biological processes could be accelerated by “reduction of dimensionality”. An initial rapid binding to a large structure would be followed by constrained diffusion on that structure to find a target. One example is the binding of proteins such as the Lac repressor to its operator; the protein first binds nonspecifically to DNA (driven by phosphate contacts) and then undergoes one-dimensional diffusion until it finds the specific sequence to which it binds tightly.45 In natural signaling systems, it is common for cytokines to bind to receptors that have no apparent purpose except to position the cytokine on the cell surface, at the proper distance from the membrane and orientation to interact with low-affinity receptors that actually signal. It is possible for biological engineers to mimic these processes in constructing new systems, and the good news is that while a great deal of thought and design may be required, the amount of lab work needed for implementation is often modest.
The authors declare the following competing financial interest(s): J.C.W., D.R.B., and P.A.S. are shareholders of General Biologics, Inc., which is commercializing certain technologies described in this Perspective.
References
- Elowitz M. B.; Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature 2000, 403, 335–8. 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- Potvin-Trottier L.; Lord N. D.; Vinnicombe G.; Paulsson J. Synchronous long-term oscillations in a synthetic gene circuit. Nature 2016, 538, 514–517. 10.1038/nature19841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisfeld R. A.; Becker J. C.; Gillies S. D. Immunocytokines: a new approach to immunotherapy of melanoma. Melanoma Res. 1997, 7 (Suppl. 2), S99–106. [PubMed] [Google Scholar]
- Hutmacher C.; Neri D. Antibody-cytokine fusion proteins: Biopharmaceuticals with immunomodulatory properties for cancer therapy. Adv. Drug Delivery Rev. 2019, 141, 67–91. 10.1016/j.addr.2018.09.002. [DOI] [PubMed] [Google Scholar]
- Tzeng A.; Kwan B. H.; Opel C. F.; Navaratna T.; Wittrup K. D. Antigen specificity can be irrelevant to immunocytokine efficacy and biodistribution. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 3320–3325. 10.1073/pnas.1416159112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribba B.; Boetsch C.; Nayak T.; Grimm H. P.; Charo J.; et al. Prediction of the Optimal Dosing Regimen Using a Mathematical Model of Tumor Uptake for Immunocytokine-Based Cancer Immunotherapy. Clin. Cancer Res. 2018, 24, 3325–33. 10.1158/1078-0432.CCR-17-2953. [DOI] [PubMed] [Google Scholar]
- Przepiorka D.; Ko C.-W.; Deisseroth A.; Yancey C. L.; Candau-Chacon R.; Chiu H.-J.; et al. FDA Approval: Blinatumomab. Clin. Cancer Res. 2015, 21, 4035–4039. 10.1158/1078-0432.CCR-15-0612. [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen L.; Dunstone M. A.; Kostenko L.; Ely L. K.; Beddoe T.; Mifsud N. A.; Purcell A. W.; Brooks A. G.; McCluskey J.; Rossjohn J. Crystal structure of the human T cell receptor CD3 epsilon gamma heterodimer complexed to the therapeutic mAb OKT3. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 7675–80. 10.1073/pnas.0402295101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber G. M.; Schmidt M. M.; Wittrup K. D. Antibody tumor penetration: transport opposed by systemic and antigen-mediated clearance. Adv. Drug Delivery Rev. 2008, 60, 1421–34. 10.1016/j.addr.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang K.; Castello G.; Clarke S. C.; et al. Attenuating CD3 affinity in a PSMAxCD3 bispecific antibody enables killing of prostate tumor cells with reduced cytokine release. Journal for ImmunoTherapy of Cancer 2021, 9, e002488. 10.1136/jitc-2021-002488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortoletto N.; Scotet E.; Myamoto Y.; D’Oro U.; Lanzavecchia A. Optimizing anti-CD3 affinity for effective T cell targeting against tumor cells. Eur. J. Immunol. 2002, 32, 3102–7. . [DOI] [PubMed] [Google Scholar]
- Cironi P.; Swinburne I. A.; Silver P. A. Enhancement of cell type specificity by quantitative modulation of a chimeric ligand. J. Biol. Chem. 2008, 283, 8469–76. 10.1074/jbc.M708502200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor N. D.; Way J. C.; Silver P. A.; Cironi P. Anti-glycophorin single-chain Fv fusion to low-affinity mutant erythropoietin improves red blood cell-lineage specificity. Protein Eng. Des. Sel. 2010, 23, 251–60. 10.1093/protein/gzp085. [DOI] [PubMed] [Google Scholar]
- Garcin G.; Paul F.; Staufenbiel M.; Bordat Y.; Van der Heyden J.; Wilmes S.; Cartron G.; Apparailly F.; De Koker S.; Piehler J.; Tavernier J.; Uzé G. High efficiency cell-specific targeting of cytokine activity. Nat. Commun. 2014, 5, 3016. 10.1038/ncomms4016. [DOI] [PubMed] [Google Scholar]
- Lipovsek D. Adnectins: engineered target-binding protein therapeutics. Protein Eng. Des. Sel. 2011, 24, 3–9. 10.1093/protein/gzq097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensel D.; Sun Y.; Li Z.; Zhang S.; Picarillo C.; McDonagh T.; Fabrizio D.; Cockett M.; Krystal M.; Davis J. Discovery and characterization of a novel CD4-binding Adnectin with potent anti-HIV activity. Antimicrob. Agents Chemother. 2017, 61, e00508-17. 10.1128/AAC.00508-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensel D.; Sun Y.; Davis J.; Li Z.; Zhang S.; McDonagh T.; Fabrizio D.; Cockett M.; Krystal M. A novel gp41-binding adnectin with potent anti-HIV activity is highly synergistic when linked to a CD4-binding adnectin. J. Virol. 2018, 92, e00421-18. 10.1128/JVI.00421-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensel D.; Sun Y.; Davis J.; Li Z.; Zhang S.; McDonagh T.; Langley D.; Mitchell T.; Tabruyn S.; Nef P.; Cockett M.; Krystal M. GSK3732394: a multi-specific inhibitor of HIV entry. J. Virol. 2019, 93, e00907-19. 10.1128/JVI.00907-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PRODUCT INFORMATION INTRON A. Interferon alfa-2b, recombinant for injection. https://www.accessdata.fda.gov/drugsatfda_docs/label/2014/103132s5190lbl.pdf.
- Vaziri N. D.; Zhou X. J.Potential mechanisms of adverse outcomes in trials of anemia correction with erythropoietin in chronic kidney disease. Nephrol. Dial. Transplant. 2008, 24, 1082–1088. 10.1093/ndt/gfn601. [DOI] [PubMed] [Google Scholar]
- Besarab A.; Bolton W. K.; Browne J. K.; Egrie J. C.; Nissenson A. R.; Okamoto D. M.; Schwab S. J.; Goodkin D. A. The effects of normal as compared with low hematocrit values in patients with cardiac disease who are receiving hemodialysis and epoetin. N. Engl. J. Med. 1998, 339, 584–90. 10.1056/NEJM199808273390903. [DOI] [PubMed] [Google Scholar]
- Pfeffer M. A.; Burdmann E. A.; Chen C. Y.; Cooper M. E.; de Zeeuw D.; et al. A trial of darbepoetin alfa in type 2 diabetes and chronic kidney disease. N. Engl. J. Med. 2009, 361, 2019–32. 10.1056/NEJMoa0907845. [DOI] [PubMed] [Google Scholar]
- Xu Y.; Carrascosa L. C.; Yeung Y. A.; Chu M. L.-H.; et al. An engineered IL15 cytokine mutein fused to an anti-PD1 improves intratumoral T-cell function and antitumor immunity. Cancer Immunol. Res. 2021, 9, 1141–1157. 10.1158/2326-6066.CIR-21-0058. [DOI] [PubMed] [Google Scholar]
- Rose-John S. Interleukin-6 family cytokines. Cold Spring Harb. Perspect. Biol. 2018, 10, a028415. 10.1101/cshperspect.a028415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley H. S. Trafficking of the ErbB receptors and its influence on signaling. Exp. Cell Res. 2003, 284, 78–88. 10.1016/S0014-4827(03)00002-8. [DOI] [PubMed] [Google Scholar]
- Lee J.; Vernet A.; Redfield K.; Lu S.; Ghiran I. C.; Way J. C.; Silver P. A. Rational design of a bifunctional AND-gate ligand To modulate cell-cell interactions. ACS Synth. Biol. 2020, 9, 191–197. 10.1021/acssynbio.9b00273. [DOI] [PubMed] [Google Scholar]
- Giger K.; Habib I.; Ritchie K.; Low P. S. Diffusion of glycophorin A in human erythrocytes. Biochim. Biophys. Acta 2016, 1858, 2839–2845. 10.1016/j.bbamem.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catimel B.; Wilson K. M.; Kemp B. E. Kinetics of the autologous red cell agglutination test. J. Immunol. Methods 1993, 165, 183–192. 10.1016/0022-1759(93)90344-7. [DOI] [PubMed] [Google Scholar]
- Vamosi G.; Friedlander-Brock E.; Ibrahim S. M.; Brock R.; Szollosi J.; Vereb G. EGF receptor stalls upon activation as evidenced by complementary fluorescence correlation spectroscopy and fluorescence recovery after photobleaching measurements. International J. Mol. Sci. 2019, 20, 3370. 10.3390/ijms20133370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D.; Lahiji A.; Patel S.; Franklin M.; Jimenez X.; Hicklin D. J.; Kang X. Monoclonal antibody cetuximab binds to and down-regulates constitutively activated epidermal growth factor receptor vIII on the cell surface. Anticancer Res. 2007, 27, 3355–3366. [PubMed] [Google Scholar]
- Crothers D. M.; Metzger H. The influenze of polyvalency on the binding properties of antibodies. Immunochemistry 1972, 9, 341–357. 10.1016/0019-2791(72)90097-3. [DOI] [PubMed] [Google Scholar]
- Kaufman E. N.; Jain R. K. Effect of bivalent interaction upon apparent antibody affinity: Experimental confirmation of theory using fluorescence photobleaching and implications for antibody binding assays. Cancer Res. 1992, 52, 4157–4167. [PubMed] [Google Scholar]
- Zhou H.-X. Quantitative account of the enhanced affinity of two linked scFvs specific for different epitopes on the same antigen. J. Mol. Biol. 2003, 329, 1–8. 10.1016/S0022-2836(03)00372-3. [DOI] [PubMed] [Google Scholar]
- Robinson-Mosher A.; Shinar T.; Silver P. A.; Way J. Dynamics simulations for engineering macromolecular interactions. Chaos 2013, 23, 025110. 10.1063/1.4810915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piehler J.; Roisman L. C.; Schreiber G. New structural and functional aspects of the type I interferon-receptor interaction revealed by comprehensive mutational analysis of the binding interface. J. Biol. Chem. 2000, 275, 40425–33. 10.1074/jbc.M006854200. [DOI] [PubMed] [Google Scholar]
- Burrill D. R.; Vernet A.; Collins J. J.; Silver P. A.; Way J. C. Targeted erythropoietin selectively stimulates red blood cell expansion in vivo. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 5245–50. 10.1073/pnas.1525388113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue S. L.; Taura T.; Bi M.; Yun Y.; Sho A.; Mikesell G.; et al. Targeting attenuated interferon-α to myeloma cells with a CD38 antibody induces potent tumor regression with reduced off-target activity. PLoS One 2016, 11, e0162472. 10.1371/journal.pone.0162472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brines M.; Cerami A. The receptor that tames the innate immune response. Mol. Med. 2012, 18, 486–96. 10.2119/molmed.2011.00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.; Vernet A.; Gruber N. G.; Kready K. M.; Burrill D. R.; Way J. C.; Silver P. A. Rational engineering of an erythropoietin fusion protein to treat hypoxia. Protein Eng., Des. Sel. 2021, 34, gzab025. 10.1093/protein/gzab025. [DOI] [PubMed] [Google Scholar]
- BIKTARVY (bictegravir, emtricitabine, and tenofovir alafenamide) tablets, for oral use. https://www.gilead.com/~/media/files/pdfs/medicines/hiv/biktarvy/biktarvy_pi.pdf.
- Pancera M.; Majeed S.; Ban Y. E.; Chen L.; Huang C. C.; Kong L.; Kwon Y. D.; Stuckey J.; Zhou T.; Robinson J. E.; Schief W. R.; Sodroski J.; Wyatt R.; Kwong P. D. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 1166–71. 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamjian M. C.; McNicholl I. R. Enfuvirtide: first fusion inhibitor for treatment of HIV infection. Am. J. Health Syst. Pharm. 2004, 61, 1242–7. 10.1093/ajhp/61.12.1242. [DOI] [PubMed] [Google Scholar]
- Sleep D.; Cameron J.; Evans L. R. Albumin as a versatile platform for drug half-life extension. Biochim. Biophys. Acta 2013, 1830, 5526–34. 10.1016/j.bbagen.2013.04.023. [DOI] [PubMed] [Google Scholar]
- Adam G.; Delbrück M.. Reduction of Dimensionality in Biological Diffusion Processes. In Structural Chemistry and Molecular Biology; Rich A., Davidson N., Eds.; W. H. Freeman and Co.: San Francisco, 1968; pp 198–215. [Google Scholar]
- Winter R. B.; von Hippel P. H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 2. The Escherichia coli repressor--operator interaction: equilibrium measurements. Biochemistry. 1981, 20, 6948–60. 10.1021/bi00527a029. [DOI] [PubMed] [Google Scholar]
- Habib I.; Smolarek D.; Hattab C.; Grodecka M.; Hassanzadeh-Ghassabeh G.; et al. V(H)H (nanobody) directed against human glycophorin A: a tool for autologous red cell agglutination assays. Anal. Biochem. 2013, 438, 82–9. 10.1016/j.ab.2013.03.020. [DOI] [PubMed] [Google Scholar]
- Schmick M.; Bastiaens P. I. H. The interdependence of membrane shape and cellular signal processing. Cell 2014, 156, 1132–1138. 10.1016/j.cell.2014.02.007. [DOI] [PubMed] [Google Scholar]