ABSTRACT
Plasmid-encoded virulence factors are important in the pathogenesis of diseases caused by spore-forming bacteria. Unlike many other bacteria, the most common virulence factors encoded by plasmids in Clostridium and Bacillus species are protein toxins. Clostridium perfringens causes several histotoxic and enterotoxin diseases in both humans and animals and produces a broad range of toxins, including many pore-forming toxins such as C. perfringens enterotoxin, epsilon-toxin, beta-toxin, and NetB. Genetic studies have led to the determination of the role of these toxins in disease pathogenesis. The genes for these toxins are generally carried on large conjugative plasmids that have common core replication, maintenance, and conjugation regions. There is considerable functional information available about the unique tcp conjugation locus carried by these plasmids, but less is known about plasmid maintenance. The latter is intriguing because many C. perfringens isolates stably maintain up to four different, but closely related, toxin plasmids. Toxin genes may also be plasmid-encoded in the neurotoxic clostridia. The tetanus toxin gene is located on a plasmid in Clostridium tetani, but the botulinum toxin genes may be chromosomal, plasmid-determined, or located on bacteriophages in Clostridium botulinum. In Bacillus anthracis it is well established that virulence is plasmid determined, with anthrax toxin genes located on pXO1 and capsule genes on a separate plasmid, pXO2. Orthologs of these plasmids are also found in other members of the Bacillus cereus group such as B. cereus and Bacillus thuringiensis. In B. thuringiensis these plasmids may carry genes encoding one or more insecticidal toxins.
INTRODUCTION
Spore-forming bacteria cause some of the most significant diseases of both humans and animals, including tetanus, botulism, gas gangrene, anthrax, and many different enteric or gastroenteritis syndromes. The pathogenesis of most of these diseases involves the production of potent protein toxins, including tetanus and botulinum toxins, anthrax toxin, and alpha-toxin, epsilon-toxin (ETX), and enterotoxin (CPE) from Clostridium perfringens. The genes for many of these toxins, as well as other virulence factors such as the capsule biosynthesis genes of Bacillus anthracis, are located on plasmids, with examples including the tetanus toxin plasmid, the conjugative toxin plasmids of C. perfringens, and the pXO1 and pXO2 virulence plasmids from B. anthracis (1). In this chapter we will review our knowledge of these plasmids, the virulence factors that they encode, and their role in disease.
TOXIN PLASMIDS OF C. PERFRINGENS
C. perfringens causes many histotoxic and gastrointestinal diseases in both humans and domestic livestock, and strains are divided into five toxinotypes (A to E) based on their ability to produce one or more of four typing toxins: alpha-toxin, beta-toxin (CPB), ETX, and iota-toxin (ITX) (2). This classification system is based functionally on the presence or absence of toxin plasmids, specifically plasmids encoding CPB (cpb gene), ETX (etx), or ITX (iap and ibp) (3). Most of these plasmids are very closely related, sharing core plasmid replication, maintenance, and conjugation regions (Fig. 1) (4). These regions are also shared by a group of almost identical conjugative tetracycline resistance plasmids typified by pCW3 (5). Accordingly, the nature of these common regions will be described before the various toxin plasmids are discussed.
FIGURE 1.
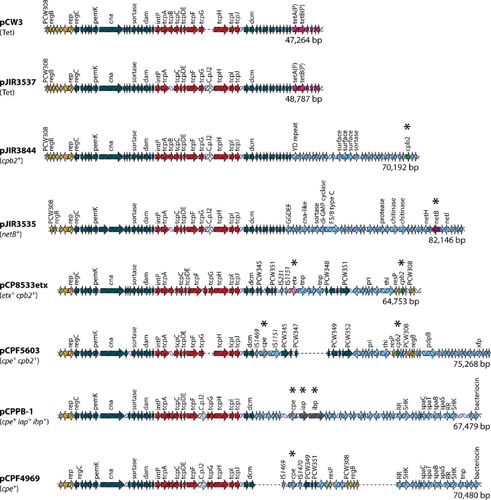
Comparative alignment of C. perfringens plasmids. Open reading frames (ORFs) are indicated by arrows as follows: red, the tcp locus; dark blue, other shared ORFs; light purple, tetracycline resistance genes; green, the cpb2 toxin gene; purple, the netB toxin gene; pink, the etx gene; gray, the cpe gene; dark gray, the iota-toxin gene; yellow, plasmid replication region; light blue, regions unique to each plasmid. Asterisks denote a toxin gene. Reproduced with permission from reference 4.
Replication and Maintenance of Toxin Plasmids of C. perfringens
Determination of the first complete sequences of C. perfringens toxin plasmids, those of the CPE (cpe gene) encoding plasmids pCPF5603 and pCPF4969, did not reveal any obvious plasmid replication genes (6). However, functional studies of the sequenced conjugative tetracycline resistance plasmid, pCW3, led to the identification of the plasmid replication protein, Rep (5). The Rep protein is not related to any other plasmid replication proteins and, although disruption of the rep gene abolishes plasmid replication, the precise mode of action of Rep remains unclear (5). The rep gene subsequently has been shown to be conserved (at least 97% Rep amino acid sequence identity) in all plasmids related to pCW3, including pCPF5603 and pCPF4969, that have been sequenced to date (Fig. 1). Therefore, it is postulated that all members of this plasmid family replicate by the same Rep-dependent mechanism.
In all of these plasmids the gene immediately upstream of the rep gene has similarity to parM genes. The parM gene encodes an actin-like protein that, in conjunction with a downstream parR gene and an upstream parC centromere-like site, constitutes the plasmid partitioning system of several large, low-copy-number plasmids from Gram-negative bacteria (7). The ParMR system relies on the ability of the ParM protein to form long ATP-dependent filaments, the ends of which bind to ParR adaptor proteins that are in turn bound to the centromere-like parC region located upstream of the parM gene (8). Filament extension subsequently results in the separation of similar plasmid molecules with the same parCMR locus to opposite ends of a ParM filament, and therefore to opposite ends of the cell, prior to cell division, resulting in stable plasmid inheritance (8). Potential parR genes have been annotated on several of the sequenced toxin plasmids, and on the other plasmids an unannotated parR gene can be identified downstream of parM. These data suggest that a ParMRC-like partitioning system is utilized by pCW3, the toxin plasmids, and other members of this plasmid family (5, 9, 10).
The other factor relevant to plasmid maintenance is the ability of many spore-forming bacteria to carry multiple plasmids within the same cell. In C. perfringens, many of these multiplasmid strains contain several plasmids that are closely related and encode the same replication and conjugation functions (4). For these very similar plasmids to be stably inherited, distinct plasmid maintenance systems need to be employed. The detailed study of one such multiplasmid strain, EHE-NE18, has demonstrated that three very closely related plasmids are able to coexist in this strain and that they are capable of independent transfer (9). It was postulated that allele differences that can be identified between parC and the ParM and ParR proteins encoded by these different toxin and resistance plasmids determine plasmid incompatibility. Therefore, plasmids with different parCMR alleles can be stably maintained in the same cell; plasmids with the same alleles are predicted to be incompatible (9, 10).
Subsequent analysis of published sequence data has led to the assignment of ParM sequences into one of four distinct groups (10), which have now been designated as the ParMRC clades A to D (J. Rood, V. Adams, and J. Prescott, unpublished). A dendrogram of the ParM proteins from all available C. perfringens plasmid sequences clearly indicates the distinct ParMA, ParMB, ParMC, and ParMD clades and leads to the identification of a fifth C. perfringens-derived group, designated here as the ParME clade (Fig. 2). Recently, another clostridial toxin plasmid has been shown to utilize a ParMR-like system for plasmid maintenance, the pE88 plasmid from Clostridium tetani, which encodes the tetanus toxin structural gene (11). This ParM protein is only distantly related to the C. perfringens ParM proteins and serves to anchor the phylogenetic tree (Fig. 2). There is also a ParM homologue present in Clostridium botulinum Eklund strain 17B, which contains a plasmid-encoded neurotoxin locus (Fig. 3). This putative ParM protein has a much closer relationship to the ParMB family of C. perfringens ParM proteins (Fig. 2). Overall, the data suggest that ParMR systems represent a highly conserved mechanism of plasmid maintenance, not just in the clostridia, but in many other bacterial genera (12).
FIGURE 2.
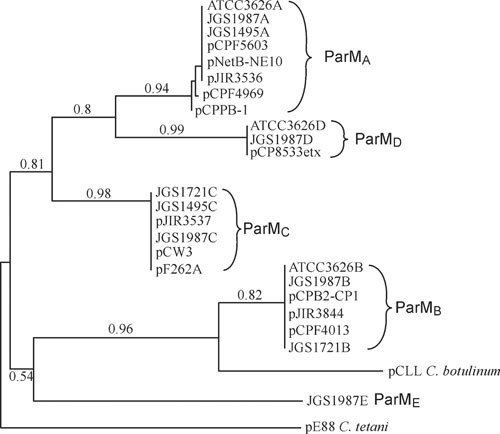
Phylogenetic analysis of ParM variants. The phylogenetic tree was constructed using the amino acid sequences of ParM proteins identified using BlastP searches of the nonredundant NCBI protein database. The phylogenetic tree was constructed using the phylogeny analysis software: http://www.phylogeny.fr/version2_cgi/index.cgi (171, 172). The JGS1495, JGS1987, and ATCC3626 sequences are from genome sequencing projects and yielded multiple ParM homologues from putative plasmid sequences; each ParM homologue was named according to its ParM group.
FIGURE 3.
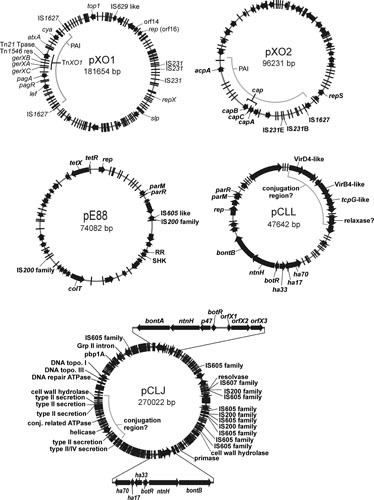
Plasmid maps of toxin plasmids from spore-forming bacteria. The circular maps of the B. anthracis toxin plasmids, pXO1 (AF065404) and pXO2 (AF188935), the C. tetani neurotoxin plasmid pE88 (AF528077), and the conjugative group I (pCLJ, CP001081) and group II (pCLL, CP001057) C. botulinum neurotoxin plasmids are shown. Predicted ORFs are depicted as black arrows or bars along the circular maps. Regions of interest are indicated inside the plasmid circles, such as the pathogenicity islands present on pXO1 and pXO2. Genes of interest are indicated on the outside of the plasmid maps. Gene names are italicized, while ORFs with similarity to known proteins (such as IS elements) are not italicized. The two neurotoxin loci encoded on the pCLJ plasmid are enlarged showing an example of an orfX neurotoxin locus (bontA region, top) and a ha neurotoxin locus (bontB region, bottom); see text.
Mechanism of Conjugative Transfer of Toxin Plasmids of C. perfringens
Conjugative transfer in C. perfringens was recognized first in experiments that involved the transfer of closely related tetracycline resistance plasmids (13, 14). Subsequently, the first toxin plasmid shown to transfer independently was a marked clinically derived CPE plasmid, pMRS4969 (15). Transfer of pMRS4969 resulted in transconjugants that were able to act as plasmid donors, highly suggestive that transfer was mediated by a conjugation-like mechanism encoded on the plasmid. Conjugative transfer has now been demonstrated for several toxin plasmids, including plasmids encoding ETX, NetB toxin, and CPB2, as well as a lincomycin resistance plasmid (9, 16, 17).
Early analysis of the conjugative plasmids from C. perfringens suggested that they shared large regions of similarity (15, 18, 19). Ten toxin and resistance plasmids from C. perfringens have been completely sequenced, including known conjugative plasmids; the data confirmed that these plasmids share a highly conserved 35-kb region that includes the transfer of the clostridial plasmid (tcp) conjugation locus (Fig. 1) (4–6, 9, 10, 20, 21).
The tcp locus has been shown to mediate conjugative transfer of the 47-kb tetracycline resistance plasmid pCW3 (5). The locus originally was identified based on low levels of amino acid identity between several of the encoded Tcp proteins and conjugation proteins from the conjugative transposon Tn916. Significant genetic rearrangements between the conjugation regions from Tn916 and pCW3, as well as limited primary amino acid sequence similarity to Gram-negative systems, suggested that the mechanism of conjugation in C. perfringens was unique. Since this locus is conserved in most of the toxin plasmids, is present in all known conjugative C. perfringens plasmids, and appears to be responsible for the conjugative transfer of toxin genes, it is important to review what is currently known about the function of this conjugation system.
The tcp locus comprises an 11-gene operon (intP, tcpA-tcpJ), and mutagenesis studies have shown that several of the Tcp proteins encoded by this locus are involved in pCW3 transfer (5, 22–24). The genes in the tcp locus are conserved in the toxin plasmids, except that in some plasmids the nonessential tcpB gene is missing and/or there may be insertions of small hypothetical open reading frames (ORFs) and group II introns (5, 6, 9, 16, 17). A model for the C. perfringens conjugation apparatus has been proposed based on functional studies of the Tcp proteins involved in the conjugative transfer of pCW3 (4, 25). Analysis of these Tcp proteins has identified some functional similarity to protein families that form the Gram-negative inner membrane secretion channel (26).
Coupling proteins are an essential component of conjugation systems and are involved in substrate recognition and translocation (27). Two proteins encoded on the tcp locus, TcpA and TcpB, were identified as possessing FtsK-like domains belonging to the FtsK superfamily of DNA translocases (23). TcpA was shown to be essential for transfer, while TcpB was not required, a result consistent with the absence of the tcpB gene in some of the toxin plasmids. Bioinformatic analysis of the TcpA protein identified two N-terminal transmembrane domains (TMDs) and three ATP-binding and hydrolysis motifs in the C-terminal cytoplasmic domain. Supporting the hypothesis that TcpA is an ATPase-dependent DNA translocase, the Walker A, Walker B, and RAAG ATP-binding motifs were shown to be essential for TcpA function (23). Similar to other coupling proteins, the TMDs of TcpA were shown to be required for wild-type TcpA function and to be involved in TcpA homo-oligomerization as well as protein-protein interactions between TcpA and TcpC, and TcpH and TcpG (25). Deletion of 61 amino acids from the C-terminus of TcpA reduced TcpA-TcpC interactions and transfer efficiency, providing evidence that this interaction was important for the transfer process (25). Although TcpA was not identified as a classical coupling protein, these studies provide convincing evidence that TcpA is the DNA translocase of the C. perfringens conjugation system.
One of the first proteins shown to be essential for conjugation was TcpH, with transfer not detected in tcpH mutants (5). Predicted to be an integral membrane protein, this 832-amino acid protein contains 8 TMDs and a large cytoplasmic domain. TcpH has a VirB6 domain between TMD5-TMD8 and was therefore proposed to have a similar role as the VirB6 protein, which forms part of the central channel in the inner membrane of the Gram-negative conjugation apparatus (28). In agreement with a proposed role for TcpH as the major component of the conjugation apparatus in the donor cell wall, TcpH was shown to localize to the cell membrane fraction at the poles of C. perfringens cells (29). The first 581 amino acids of TcpH, which include the TMDs and VirB6 domain, comprised the minimal TcpH derivative required to complement a tcpH mutant. This region was also essential for TcpH homo-oligomerization and interactions with TcpA and TcpC (25, 29). TcpH interactions were not dependent on the VirB6 domain or the conserved 242VQQPW246 motif, also identified as being essential for TcpH function (29). The unique C-terminal cytoplasmic domain was required for wild-type TcpH function but was not involved in TcpH protein-protein interactions. This cytoplasmic domain is not present in other VirB6-like conjugation proteins, demonstrating the functional complexity of TcpH and the unique nature of the C. perfringens conjugation system.
TcpC was originally identified as a bitopic membrane protein with 24% amino acid identity to ORF13 from Tn916 (5). A very significant reduction in transfer efficiency was observed in a tcpC mutant, indicating that TcpC plays a key role in conjugative transfer (24). Determination of the crystal structure of the soluble TcpC99-359 derivative to 1.8 Å identified two domains that each unexpectedly shared a similar fold to members of the nuclear transport factor-2 superfamily, which includes VirB8 proteins that form part of the transfer apparatus at the inner membrane in Gram-negative conjugation systems. VirB8-like proteins act as scaffolding proteins promoting transfer complex assembly and stabilization; TcpC is therefore proposed to have a similar role in C. perfringens. Consistent with this role, TcpC was shown to localize independently to the cell wall of C. perfringens cells as well as interacting with itself, TcpA, TcpH, and TcpG, with all of these interactions requiring the presence of the essential N-terminal TMD. TcpC forms a trimeric structure with the internal central domain shown to be involved in TcpC self-interaction (24). The essential C-terminal domain forms the major exterior surface of the trimer and is required for TcpC interactions with TcpA, TcpH, and TcpG. It is through these protein-protein interactions that TcpC is postulated to direct the formation of the transfer apparatus.
TcpF was identified as a putative ATPase with distant similarity to the VirB4 ATPase of Gram-negative conjugation proteins (29). Like ATPases from other conjugation systems, TcpF was shown to be essential for conjugative transfer (5). Although TcpF was demonstrated to colocalize with TcpH at the poles of C. perfringens donor cells, it remains unclear how TcpF interacts with the conjugation apparatus, with no protein-protein interactions identified between TcpF and the other Tcp proteins (29).
Peptidoglycan hydrolases are another family of proteins commonly present in conjugation systems, playing a role in the assembly of the conjugation apparatus in the cell wall (22). Two genes encoding potential peptidoglycan hydrolases were identified in the tcp locus, tcpG and tcpI (5). Genetic studies showed that the tcpI gene product was not required for conjugative transfer (22). Mutagenesis of the tcpG gene resulted in a reduced transfer frequency, consistent with results for peptidoglycan hydrolases from other conjugation systems. The role of TcpG as a peptidoglycan hydrolase was confirmed by the demonstration that purified TcpG had hydrolase-like activity on cognate C. perfringens peptidoglycan (22).
In the model of the C. perfringens conjugation apparatus (4, 25) the integral membrane protein TcpH, with the atypical VirB8-like protein TcpC acting as a scaffold, forms the core channel of the conjugation apparatus. As well as stabilizing the complex, TcpC is proposed to be involved in recruiting TcpA, TcpH, and TcpG (24). The putative motor ATPases TcpA and TcpF are predicted to energize the system, although it is currently unknown how TcpF interacts with the other components (25, 29). Similarly, the early steps in plasmid transfer are not clear since neither a classical relaxase protein gene nor an origin of transfer (oriT) have been identified on these plasmids (5). The relaxase protein and oriT site are usually essential components that are involved in plasmid recognition and nicking prior to transfer (30). The first gene in the tcp locus, intP, which is conserved in the toxin plasmids, encodes a putative tyrosine recombinase that is postulated to be a potentially novel relaxase (5). Genetic and functional studies are required to determine whether IntP, as well as the remaining highly conserved proteins TcpD, TcpE, and TcpJ, are involved in C. perfringens conjugative transfer. These studies will further define our understanding of the unique conjugation process in C. perfringens, a process that is central to the transfer of both antibiotic resistance and toxin genes.
Enterotoxin (CPE) Plasmids of C. perfringens
C. perfringens type A strains that produce C. perfringens CPE are causative agents of human food poisoning and non-food-borne diarrhea (31). CPE is encoded by the cpe gene and is only produced during sporulation, where its synthesis is controlled by Spo0A and the alternative sigma factors SigF, SigE, and SigK (32–34). The toxin then accumulates intracellularly until it is released when the mother cell lyses to free the mature endospore at the completion of sporulation. CPE then binds to claudin receptors present in the gastrointestinal tract, which induces toxin oligomerization and pore formation in enterocytes. The resultant Ca2+ influx causes cytotoxicity that leads to villus blunting, necrosis, and epithelial desquamation in all sections of the small intestines, along with luminal fluid accumulation (31).
CPE, a 35-kDa polypeptide, has a highly conserved amino acid sequence in all CPE-producing C. perfringens strains, with the exception of type E strains. Structurally, this toxin belongs to the aerolysin family of pore-forming toxins, which also includes ETX (35, 36). The CPE protein consists of a C-terminal domain that confers binding to claudin receptors and an N-terminal domain that mediates both the oligomerization of toxin monomers and membrane insertion during pore formation (37–39).
While some C. perfringens type A, C, D, and E isolates produce CPE, no CPE-producing type B strains have been identified (20, 21, 40–44). In the 1 to 5% of global type A strains that are cpe-positive (45), the cpe gene is located either on the chromosome or on large plasmids that range in size from ∼50 to ∼75 kb (6). Virulence studies testing molecular Koch's postulates have confirmed the importance of CPE expression for the gastrointestinal virulence of both chromosomal and plasmid cpe type A strains (46).
Type A strains with a chromosomal cpe gene are estimated to cause ∼75 to 80% of all C. perfringens type A food poisoning cases (31). This illness is now considered the second most common bacterial food-borne disease in the United States, where it involves one million cases per year (31, 47). The remaining cases of C. perfringens type A food poisoning are caused by strains carrying a cpe plasmid. Type A strains with a cpe plasmid are also responsible for 5 to 10% of all cases of human non-food-borne gastrointestinal diseases, which include both antibiotic-associated and sporadic diarrheas (31, 48). These diseases occur more frequently in the elderly and are more severe and long-lasting than typical type A food poisoning (49). Type A strains carrying a cpe plasmid also reportedly cause enteric disease in domestic animals, particularly dogs (50).
Based upon results from sequencing, pulse-field Southern blot analyses, and overlapping PCR assays, it has been shown that there are two major families of cpe plasmids in type A strains (4). The pCPF5603 cpe plasmid family includes plasmids that are usually ∼75 kb in size and carry genes encoding both CPE and CPB2 (encoded by the cpb2 gene) (6) (Fig. 1). In contrast, the ∼70-kb pCPF4969 cpe plasmid family has the cpe gene, but not the cpb2 gene. Both cpe plasmid families share an ∼35-kb conserved region (6) that contains the tcp region, which mediates the conjugative transfer of several C. perfringens plasmids (5). The presence of this tcp region explains the conjugative transfer of a tagged F4969 plasmid derivative (15). The variable region of pCPF5603-like plasmids contains a cluster of metabolic genes in addition to a functional cpb2 gene. In contrast, the pCPF4969 variable region includes two putative bacteriocin genes and genes encoding a putative two-component regulator with similarity to VirS and VirR from C. perfringens (6).
Whether chromosomal or plasmid-borne, the cpe gene is closely associated with insertion sequences (IS). However, there is considerable variation among type A strains regarding the specific IS elements present in the cpe locus, as well as their arrangement relative to the cpe gene (Fig. 4). In the pCPF5603-like plasmids, the cpe gene is bounded by an upstream IS1469 sequence and a downstream IS1151 sequence, while in pCPF4969-like plasmids; the cpe gene is upstream of an IS1470-like sequence rather than IS1151 (6). By comparison, the chromosomal cpe locus has an IS1469 immediately upstream of the cpe gene (51). This chromosomal IS1469-cpe region is then flanked by IS1470 sequences, suggesting it could be part of an integrated transposon (51).
FIGURE 4.
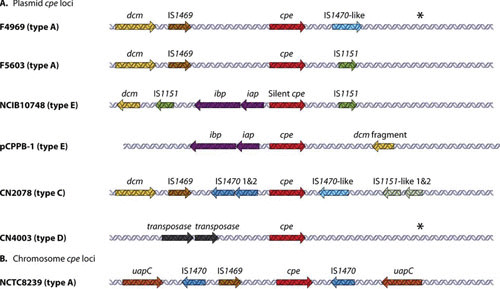
Genetic organization of the cpe gene regions. Organization of plasmid-borne (A) and (B) chromosomal cpe loci from C. perfringens. cpe genes are indicated by red arrows, iap and ibp genes by purple arrows, and dcm genes by yellow arrows. Related IS elements are indicated by identical colors. Reproduced with permission from reference 4.
C. perfringens type C Darmbrand strains produce CPE, as well as CPB, and are genetically related to type A food poisoning strains carrying a chromosomal cpe gene (52). However, these type C strains carry a plasmid-borne cpe gene, as well as a plasmid-borne cpb gene located on ∼75- or ∼85-kb plasmids. In other Darmbrand strains the cpe gene is present on an ∼110-kb plasmid that does not carry the cpb gene. The cpe plasmids in all studied CPE-positive type C strains have the tcp region, suggesting that they are conjugative. Interestingly, the plasmid cpe locus in type C strains is similar to the chromosomal cpe locus of type A isolates, except that an IS1469 sequence is located upstream of IS1470, while an IS1151-like sequence is located downstream of the cpe gene (52) (Fig. 4). Experiments have indicated that IS mobile genetic elements can excise the cpe gene as circular DNA molecules that could be transposition intermediates (52). Those results suggest, but do not yet prove, that the chromosomal cpe locus in type A strains may be derived from insertion of a cpe-carrying transposon originating from a cpe plasmid similar to the cpe plasmid found in type C Darmbrand strains.
Many type D isolates also carry a cpe gene on large plasmids that range in size from 75 to 110 kb (40). In some of these strains, the cpe gene is located on the same plasmid as the etx gene, but other CPE-positive type D strains carry separate cpe and etx plasmids (40). Like cpe plasmids in type A and C strains, most cpe plasmids of type D isolates have a tcp region, suggesting they are conjugative (53). However, in cpe-positive type D strains, the cpe locus has a unique genetic organization differing from the cpe locus of cpe-positive type A or type C strains (53). While the region downstream of the cpe gene in type D strains is similar to the sequences downstream of the pCPF4969 cpe gene, except for the absence of an IS1470-like sequence, two copies of an ORF sharing 67% identity with a Tn1456-like transposase gene are located upstream of the cpe gene in the type D strains that have been studied (Fig. 4). All type E isolates studied to date were shown to carry cpe sequences on large plasmids that also encode ITX (21, 54), which will be discussed in the section describing the ITX plasmids.
Beta Toxin (CPB) Plasmids of C. perfringens
CPB is encoded on large plasmids in both toxinotype B and C isolates (41, 52, 55). The CPB-encoding toxinotypes are responsible for a range of highly significant and often fatal animal diseases. In addition, type C isolates are the only non-type A strains known to cause disease in humans.
Type C-mediated human disease affects undernourished individuals existing on a protein-deficient diet and is often accentuated by the consumption of foods rich in trypsin inhibitors (56). It is known as Darmbrand in Germany (52) and pig bel in Papua New Guinea (56). Symptoms include severe upper abdominal pain, bloody diarrhea, and vomiting (57). As the disease progresses, diarrhea ceases and the bowel becomes obstructed due to damage in the small intestine (56, 57). Enterotoxemia and necrotizing enteritis of livestock are also caused by CPB-producing strains of C. perfringens (58). Type C disease occurs mostly in neonates of several animal species. This age predisposition is believed to be related to the presence of colostrum, which inhibits trypsin activity and in turn allows CPB to remain active in the intestinal lumen. Animal diseases attributed to type B isolates resemble both type C- and/or type D-mediated enteric and/or neurologic syndromes (58). Like type C diseases of livestock, type B infections can involve necrotic enteritis or sudden death, but animals with type B infections may display neurological signs similar to type D disease (59).
Mutational studies combined with the use of a rabbit small intestinal loop model have shown that CPB is essential for the intestinal pathology observed in type C-mediated disease (60). In addition, these type C mutants were used to identify a key role for CPB in lethal enterotoxemia in a mouse model (61), which was confirmed in a large animal model (goat) more recently (62). When cultures of a wild-type strain, a cpb null mutant, and a genetically reversed mutant, were introduced into the goat small intestine, disease progression for the wild type and the complemented mutant occurred in a manner indistinguishable from the natural disease. By contrast, goats inoculated with the cpb mutant showed no sign of disease (62). These studies (60, 62) provide compelling evidence that plasmid-encoded CPB is the most important toxin in type C-mediated disease.
The pathology of type B infection is complicated, as type B isolates typically express the largest number of different lethal toxins: alpha-toxin, CPB, and ETX (41, 58, 59). The only virulence study of type B isolates to date (59) correlated toxin levels in culture supernatants with mouse intravenous lethality. The results indicated that (i) CPB is a major contributor to lethality, (ii) ETX is also implicated, and (iii) other toxins may also be involved. The complexity of disease progression when caused by type B isolates is likely dependent on a number of factors including the protease status of the host and the toxin expression profile of the infecting isolate (59). Such studies are complicated by the fact that while CPB is highly sensitive to trypsin, ETX requires trypsin for activation.
CPB is a 35-kDa pore-forming toxin with about 22 to 28% amino acid sequence similarity to several pore-forming toxins of Staphylococcus aureus (63), 38% amino acid sequence identity to C. perfringens NetB toxin (64), and 43% amino acid sequence identity with delta-toxin of C. perfringens (65). CPB forms an oligomer after binding to an unidentified receptor on sensitive cells, which results in pore formation that produces an ∼12-Å channel that is selective for monovalent cations (66). This toxin is very sensitive to degradation by trypsin or chymotrypsin (67), and intestinal trypsin is therefore the main natural defense against type C infections.
All type B and C isolates produce CPB toxin during late log-phase growth. Beta toxin production by type C strains is controlled by the chromosomal two-component VirS/VirR regulatory system (68). CPB production is also regulated by a chromosomal Agr-like quorum sensing (QS) system in both the type C strain CN3685 and the type B strains CN1793 and CN1795 (69, 70). CPB production increases when the bacteria closely contact enterocyte-like Caco-2 cells, and this effect requires both the VirS/VirR and Agr-like QS systems (68, 69). Furthermore, both the VirS/VirR and Agr-like QS systems are essential for CN3685 to cause either necrotic enteritis or lethal enterotoxemia in animal models because these regulatory systems are required for in vivo CPB production (68, 69).
In all type B and C strains studied to date, the cpb genes localized on large plasmids (41, 55). Southern hybridization of pulsed-field gels showed that the cpb gene in most type B strains was located on ∼90-kb plasmids, although a few type B strains carried an ∼65-kb cpb plasmid (41). These studies also showed that the cpb plasmid in these type B strains is distinct from their etx plasmid, although a few type B strains possess cpb and etx plasmids of the same (65 kb) size, just to make the picture more confusing (41). While type B strains carry only 65-kb or 90-kb cpb plasmids, the cpb plasmids in type C strains display more diversity, ranging in size from ∼65 to ∼110 kb. The ∼65-kb and 90-kb cpb plasmids present in these type C strains are very similar to the plasmids of matching size found in type B isolates (55). Sequencing and overlapping PCR analysis revealed that in these 65-kb and 90-kb cpb plasmids, a tpeL gene is located ∼3 kb downstream from their cpb gene. This gene encodes a large clostridial glycosylating toxin, TpeL, that is related to the large glucosylating toxins from Clostridium difficile (71). This cpb- and tpeL-containing locus is associated with IS1151 sequences that can excise to form circular molecules that may be transposition intermediates (41, 55).
Type C strains can carry other potential virulence plasmids (55). Type C strains carry either plasmid-borne cpe or tpeL genes, but never both genes. Some type C strains have ∼65- to ∼90-kb plasmids that carry cpb2 genes (72), but these cpb2 plasmids never carry the cpb gene (55).
Almost without exception, all CPB-encoding plasmids appear to encode the tcp conjugation locus (41, 55). Since there have been no reports of a tcp locus-containing plasmid that is nonconjugative, it is highly likely that all cpb plasmids are conjugative. It is postulated that type B strains most likely are derived from a conjugative transfer event from a type C to a type D strain (or vice versa) (55), but more detailed genomic studies are required to validate this hypothesis.
Epsilon-Toxin (ETX) Plasmids of C. perfringens
ETX is produced by C. perfringens type B and type D strains and is expressed as an inactive prototoxin that is activated by cleavage with proteases such as trypsin (73). Livestock diseases caused by C. perfringens type B isolates include both enterotoxemias with neurological clinical signs and lesions and necrotic enteritis (59). C. perfringens type D strains are the most common cause of enterotoxemia in sheep and goats (74), and genetic studies recently have shown that this syndrome is mediated by ETX (75). The disease has been reported, albeit rarely, in cattle. Type D disease in sheep, goats, and cattle is characterized by neurological and respiratory clinical signs and lesions. Enterocolitis with severe diarrhea also occurs in goats.
ETX is a neurotoxin that is able to be absorbed from the gastrointestinal tract, where it is produced, to spread systemically through the circulatory system, where the toxin mostly affects the heart, lungs, kidneys, and brain (74). It is able to bind to vascular endothelial cells and cause cell lysis, leading to increased vascular permeability that causes brain and pulmonary edema. The latter eventually leads to degeneration and necrosis of surrounding tissues. In addition, ETX is also able to cross the blood-brain barrier and target neuronal cells directly. The neurological signs of type B and D disease are believed to be a consequence of the brain damage produced by ETX acting on the vasculature and on neurons (74, 75).
Regulation of ETX production in a type D isolate is controlled by an agr-like QS system (76) and the global virulence regulator CodY (77). How these two regulatory systems interact is not known, but CodY is also responsible for the upregulation of adherence to Caco2 cells in vitro as well as regulating spore production under nutrient-rich conditions. By contrast, regulation studies in two type B isolates have indicated that ETX is not regulated by the agr-like QS system in these strains, unlike other toxins (70). The role of CodY in type B strains is not known.
In all strains examined to date the etx gene is located on a large plasmid (4, 40, 41). In type B strains the cpb and etx plasmids show limited plasmid variation, and no strains have been identified to date that encode both toxins on the same plasmid (20, 41). The cpb gene, as discussed earlier, is encoded on either a 65-kb plasmid or, more commonly, on a 90-kb plasmid in these strains (41). There is no discernable variation in the 65-kb etx plasmids of type B isolates; these plasmids also carry the cpb2 gene (20).
The only complete sequence available for an etx plasmid is from the type B isolate NCTC8533. This plasmid, pCP8533etx (64,753 bp), is closely related at the sequence level to the pCPF5603 family of CPE-encoding plasmids, although it lacks the cpe locus and another large region that encodes putative metabolic genes that are more commonly found on the chromosome (20). Instead of the cpe locus, pCP8533etx carries an etx locus that includes a Mu-like element downstream of the etx gene and IS1151- and IS231-like elements directly upstream of the etx gene (Fig. 5). This etx locus is largely conserved in all type B etx plasmids and some type D etx plasmids (20). The etx gene is located approximately 9 kb downstream of the dcm gene in these isolates, and there appears to have been an IS element-mediated duplication of several genes located downstream of the cpe locus in pCPF5603. These genes now flank the etx locus in pCP8533etx (Fig. 5). Another variant of the etx locus, represented by a plasmid in strain CN1675, has been identified in type D isolates that do not carry the cpe or cpb2 genes. These isolates share the same downstream sequences as the pCP8533etx-like loci, but the upstream sequence is significantly different, encoding an IS1151 sequence and a Tn3 family transposase upstream of the etx gene. In addition, the etx locus is located closer to the dcm gene in these strains (20) (Fig 5).
FIGURE 5.
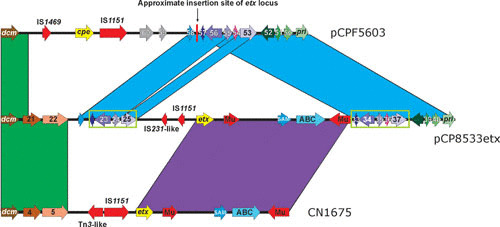
Comparative alignment of etx loci. The two etx (yellow arrows) loci from plasmid pCP8533etx and strain CN1675 are compared to the location of the cpe (yellow arrow) gene and surrounding sequences in plasmid pCPF5603. The aligned region begins with the dcm gene (left side) found downstream of the tcp locus in all sequenced plasmids. Genes (or DNA sequences) with greater than 90% nucleotide identity are colored alike (except for the cpe and etx genes; both are in yellow but are not related). The genes inside the green boxes appear to have been duplicated and flank the etx locus in plasmid pCP8533etx. Numbers are the CDS designations from the respective sequences: pCPF5603 (accession number: NC_007773), pCP8533etx (accession number: NC_011412), and CN1675 (accession number: EU852100). Arrows without numbers represent pseudogenes.
Type D etx plasmids show a marked degree of plasmid diversity, in direct contrast to the situation in type B isolates (20, 40). It is clear that a minority of surveyed type D strains contain a type B-like 65-kb etx plasmid that also carries the cpb2 gene (20). Type D isolates that lack the cpe and cpb2 genes mostly contain ∼48-kb plasmids (although a few contain 75-kb plasmids) that are likely to be conjugative since they appear to carry a tcp locus (40). Most of these strains contain a CN1675-like etx locus (20). Type D isolates that encode the etx, cpe, and/or cpb2 gene(s) are larger, ranging in size from ∼65 to 110 kb. In one type D isolate, carrying all three of these toxin genes, each toxin gene is located on a separate plasmid, while other isolates encode all three genes on the one plasmid (40).
Conjugative transfer of etx plasmids has been demonstrated in two type D isolates, CN1020 and CN3718 (16); these strains do not encode cpe or cpb2 genes and contain CN1675-like etx loci (20, 40). Transfer was initially demonstrated using marked plasmids, where the etx gene was deleted and replaced with a chloramphenicol resistance gene (16). Transfer frequencies from both primary matings (a mating between two unrelated strains) and secondary matings (a mating between two genetically distinct but isogenic strains) were very high, ranging from 1-5 x 10−1 transconjugants/donor cell. Due to the very high frequency of transfer, a wild-type etx plasmid was subsequently transferred to a C. perfringens type A laboratory strain without any genetic selection (16). Plasmid transfer was detected at a frequency of 0.8% and effectively converted a toxinotype A strain to a toxinotype D strain. These results demonstrated for the first time that one of the toxins that is used to determine the toxinotype of an isolate was mobile and therefore that strains that do not contain the etx gene may be able to acquire it laterally (and at high frequency) from a coresident type D isolate perhaps encountered within the milieu of the gastrointestinal tract.
Iota-Toxin (ITX) Plasmids of C. perfringens
C. perfringens type E isolates produce ITX and have been associated with enteric disease in rabbits, sheep, and cattle, although conclusive evidence of the role of type E strains and ITX in the pathogenesis of animal disease is lacking. Type E strains are the only C. perfringens isolates that produce ITX, a member of the clostridial binary toxin family (4, 78–80). This toxin family also includes C. botulinum C2 toxin, as well as the Clostridium spiroforme and C. difficile binary toxins. Consistent with its designation as a binary toxin, ITX is comprised of two polypeptides, IA and IB, that are encoded by the iap and ibp genes, respectively. IA and IB are secreted from C. perfringens as proproteins and must be proteolytically activated by removal of their N-terminal regions by either host proteases or C. perfringens lambda-toxin (78–80).
IB, the receptor-binding component of ITX (80), interacts with the host lipolysis-stimulated lipoprotein (81). Lipolysis-stimulated lipoprotein is also a receptor for C. difficile and C. spiroforme binary toxins, although it is not a C2 toxin receptor (82). The IA component interacts with cell-bound IB, promoting the endocytosis of ITX. IA is transported through the endocytic vacuole, presumably via a heptameric pore formed by IB, and once in the cytoplasm it ADP-ribosylates actin at Arg177, which depolymerizes actin and collapses the cytoskeleton of host cells (78–80). The multifunctional mammalian surface protein CD44 also increases ITX binding, perhaps by acting as a second receptor or a coreceptor (83).
The iap and ibp genes comprise an operon that in “classical” type E strains is located on the largest (∼100 to 135 kb) known toxin plasmids found in C. perfringens (54, 84). These ITX plasmids generally have a pCPF5603-like backbone, plus additional genes that encode several other potential virulence factors, including lambda-toxin and urease. In these classical type E strains, IS1151 sequences are closely associated with the iap/ibp genes, where they may be involved in their movement by a transposition process. These toxin genes can be detected on small circular DNA molecules that could represent transposition intermediates (84). On this basis, it has been suggested that the ITX plasmids in classical type E strains originated from the integration of an IS1151-based, iap/ibp-carrying mobile genetic element into an F5603-like cpe plasmid in a C. perfringens type A strain (54). This putative integration event apparently occurred near the promoter region of a cpe gene (Fig. 4) (84) and thereby inactivated cpe expression, such that these strains produce ITX but not CPE. Subsequent to this silencing of the cpe gene by promoter inactivation, the silent cpe genes have accumulated additional frame-shift and nonsense mutations, converting them to pseudogenes (54).
C. perfringens type E strains have traditionally been viewed as the least important of the C. perfringens types with respect to causing human or animal disease. However, the disease potential of type E strains may require some reevaluation given a recent study reporting that the standard C. perfringens multiplex PCR toxinotyping assay incorrectly classifies some “nonclassical” type E strains as type A due to variations in their iap/ibp gene sequences (21). Since those variant ITX genes are expressed, some strains classified by PCR as type A may actually be type E strains.
The ITX-encoding pCPPB-1 plasmid from one nonclassical type E strain has been sequenced (21). The analysis of this sequence revealed four fundamental differences between pCPPB-1 and the classical ITX plasmids: (i) pCPPB-1 is smaller (∼65 kb), (ii) the backbone of pCPPB-1 is similar to that of pCPF4969, in contrast to the pCPF5603-like backbone of classical ITX plasmids, (iii) pCPPB-1 does not encode lambda-toxin or urease, and (iv) an IS1151 element carrying the iap/ibp genes has apparently inserted near the most upstream of three cpe promoters in pCPPB-1; however, this cpe gene is still expressed, presumably from the other two more downstream cpe promoters. Interestingly, the CPE encoded by pCPPB-1 is a variant compared to the CPE produced by other CPE-positive strains. A tcp locus is present on both the pCPPB-1-like ITX plasmid and the classical ITX plasmids, strongly suggesting that all of these plasmids are conjugative (21, 84). However, conjugative transfer has not yet been experimentally demonstrated.
NetB Plasmids of C. perfringens
NetB, a plasmid-encoded 33-kDa β-barrel pore-forming toxin, is a major virulence factor in C. perfringens-mediated necrotic enteritis of poultry (85). This disease is manifested in two forms: as a frank clinical disease characterized by a sudden increase in flock mortality and as a milder subclinical form characterized by chronic damage to the intestinal mucosa, leading to poor digestion and nutrient absorption and resulting in a loss of bird performance. The subclinical form is of greatest consequence to the industry because of its wider prevalence and the difficulty of detection and amelioration before economic damage occurs. Treatment and control measures are estimated to cost the global poultry industry $2 billion per annum (86).
Structural studies of the secreted soluble monomeric form (87) and the heptameric pore form (88) of NetB have revealed that the conformation of the membrane-binding domain of NetB is significantly different from other beta-barrel pore-forming toxins such as the alpha-hemolysin of S. aureus, indicating that there are likely to be differences in the way these toxins interact with cell membranes. There are clear differences in the toxic activity of NetB toward different cell types; among a number of cultured cell lines, only chicken LMH cells were susceptible (85), and avian red blood cells were much more susceptible than mammalian red blood cells (87). NetB interacts directly with cholesterol (88), and the pores demonstrate a high single-channel conductance and have a preference for cations (87). Mutagenesis studies have shown that key residues in structural elements, such as those required for oligomerization and membrane binding, reduce toxic activity (87, 88). Several recent studies have shown that NetB can be effectively used as an antigen to induce a protective immune response in vaccinated birds (89–92).
Strain surveys have shown that netB is restricted to poultry-derived isolates of C. perfringens (93), apart from a single example of a netB-carrying strain isolated from cattle (94). All C. perfringens isolates that have been rigorously proven to cause necrotic enteritis in experimental infections carry the netB gene (85, 93, 95). Some C. perfringens strains isolated from clinical cases of necrotic enteritis lack the netB gene; however, none of these strains can reproducibly induce disease in experimental infection models (93, 95). We postulate that these disease-derived strains lacking the netB gene may have lost the plasmid carrying the gene during the initial strain isolation from clinical samples. In all isolates for which information is available the netB gene is carried on a large plasmid.
In the extensively studied strain, EHE-NE18, the netB plasmid pJIR3535 is one of three large conjugative plasmids (9). Each of these plasmids has been sequenced, as have two plasmids from a Canadian necrotic enteritis isolate (10), and all have very similar conjugation loci and plasmid replication and maintenance regions, as already discussed (Figs. 1 and 2). The plasmids are differentiated by the other genes they encode: pJIR3535 carries netB and a series of other genes that could potentially have a role in virulence, pJIR3844 carries a cpb2 gene encoding an atypical form of CPB2 plus genes that encode putative surface proteins of unknown function, and pJIR3537 carries the tet(P) tetracycline resistance operon (Fig. 1). All of these plasmids can transfer at high frequency. The extensive region of sequence similarity (∼35 kb) made it difficult to assemble whole-genome sequence data and to complete the sequence of each of these plasmids. This problem is common to the analysis of many C. perfringens strains, of all toxin types, where the presence of multiple, closely related large plasmids is very common. Sophisticated bioinformatic approaches, such as the construction of de Bruijn graphs (9), were used to assist in determining whole-plasmid sequences, but the clearest and most readily assembled results can be obtained by transferring each of the plasmids to C. perfringens strains lacking endogenous plasmids and then sequencing the whole genomes of transconjugant clones. Multiple large plasmid carriage has been found in all necrotic enteritis-derived strains investigated, with some reported to have up to five large plasmids (96).
Two netB plasmids (pJIR3535 and pNetB-NE10) have been sequenced and shown to be very similar 82-kb plasmids despite being derived from geographically distinct C. perfringens strains (9, 10). Analysis of the genomes of other strains by microarray hybridization suggests that the netB plasmid gene content is largely conserved across strains carrying the plasmid (96). One smaller netB plasmid has been reported (in strain CP2 [88]), but the alterations in the plasmid have not been characterized. The netB plasmids contain other genes that may have a role in virulence, for example, genes homologous to internalins, chitinases, leukocidins, proteases, and carbohydrate binding proteins. Expression of netB is under the control of the VirS/VirR two-component regulatory system (97), and the adjacent netI gene also has a predicted VirR binding box in the upstream promoter region and thus may also be under VirS/VirR control (96).
The mobility of virulence factors such as NetB is potentially important in the origin and spread of pathogenic strains. It appears that NetB is absolutely required for the pathogenesis of avian necrotic enteritis, but it is not yet clear what other plasmid- or chromosomally encoded genes may also be required to arm C. perfringens with the ability to cause necrotic enteritis. Is it simply enough for a C. perfringens strain capable of colonizing chickens to carry a netB plasmid, or are there wider requirements? Other genes that are associated to varying degrees with strains isolated from diseased birds have been identified (98), and it will be important to determine the contribution that these genes make toward the pathogenic phenotype. Multilocus sequence typing has shown that necrotic enteritis-derived C. perfringens isolates cluster into two major clonal groups that are associated with carriage of netB (99). It will be of interest to determine whether the clonal grouping is related to other properties, besides netB, that pathogenic strains may require (e.g., the ability to colonize the appropriate gastrointestinal niche) or whether it indicates some selective ability of these clonal groups to stably maintain the plasmids carrying netB.
Beta2-Toxin (CPB2) Plasmids of C. perfringens
CPB2 is a putative pore-forming cytolysin, but it is generally regarded as an accessory toxin that is not yet associated with any specific disease. Although there has been much speculation about the role of CPB2 in animal disease, most studies were based on detection of cpb2-positive isolates in the intestinal tract. However, the prevalence of such isolates seems to be similar in diseased and healthy individuals of most animal species (with the possible exception of the pig), which renders this diagnostic criterion unreliable. Most experimental studies have failed find any role for CPB2 in the pathogenesis of animal disease (100–102).
The cpb2 gene has been identified in some strains of all five C. perfringens toxinotypes and is always located on a large plasmid, although there is considerable variation in plasmid size (45 to 90 kb). In cpe/cpb2-positive type A strains the two toxin genes can be carried on separate plasmids that are characterized by cpe genes with downstream IS1470-like sequences. In some of these isolates they are on the same plasmid, which is characterized by IS1151 sequences downstream of the cpe gene (103, 104).
Several cpb2-encoding plasmids have been fully sequenced (6, 9, 10, 105). The cpb2 gene locus is conserved (Fig. 6) between sequenced plasmids from diverse origins such as the human gas gangrene isolate type A strain 13 (105), the lamb dysentery-derived type B strain NCTC8533 (20), the pig necrotizing enterocolitis type C strain CWC245 (72), two type A human gastrointestinal disease isolates (F5603 and F4013 [6]), a type A strain (F262) from a bovine clostridial abomasitis case (106), and two type A chicken necrotic enteritis strains isolated from two different countries (9, 10).
FIGURE 6.
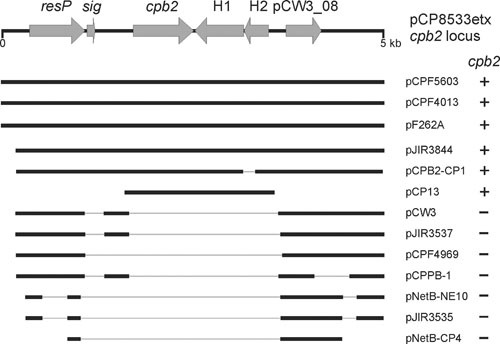
Genetic conservation of cpb2 locus. Labeled arrows indicate the cpb2 locus from pCP8533etx: resP encoding a putative resolvase, sig encoding a putative signal peptidase I, the cpb2 gene, two hypothetical genes (H1 and H2), and a conserved hypothetical (pCW3_08). Below the locus are heavy lines indicating nucleotide sequence homology of 68% (pCP13) or >95% (all others). The gray lines indicate gaps in the sequence alignment where the sequence is absent from that particular plasmid sequence. Whether the cpb2 gene is present in each plasmid is indicated on the right.
The location of the cpb2 gene is also conserved in these plasmids. It is found downstream of a putative resolvase gene, resP, and is flanked by the conserved hypothetical gene, pcw308 (pCW3 nomenclature [5]), that is located downstream of the resP gene in pCW3-like cpb2-negative plasmids (Fig. 6). pCP13 from strain 13 (107) has a silent cpb2 gene and is different from the conjugative toxin and antibiotic resistance plasmids described in detail in this chapter; for example, it does not carry the tcp conjugation locus and has a very different plasmid replication and maintenance region, but it still has some sequence conservation with this plasmid family. Despite the more distant relationship between pCP13 and other cpb2 plasmids, the genes located downstream of the cpb2 gene are still conserved (Fig. 6), suggesting that a cpb2 cassette, consisting of the cpb2 gene and the two downstream hypothetical genes, may have been mobile at some stage. The plasmid encoding the cpb2 gene from the chicken necrotic enteritis strain EHE-NE18 encodes an intact tcp locus, and conjugative transfer at a high frequency has been demonstrated both from the original host as well as from secondary hosts, in the absence of the other conjugative plasmids (9). Finally, expression of the cpb2 gene has been shown to be under the regulatory control of both the VirS/VirR two-component regulatory system (108) and the Agr-like QS system (109).
THE NEUROTOXIN PLASMIDS OF C. BOTULINUM AND C. TETANI
The most potent bacterial toxins are the neurotoxins encoded by C. botulinum and C. tetani. These bacteria are the causative agents of botulism and tetanus, respectively, and the pathogenesis of these often fatal conditions is mediated by similar neurotoxins: botulinum toxin (BoNT) and tetanus toxin (TeNT). Both BoNT and TeNT are related zinc metalloproteases that cleave either vSNARE or tSNARE proteins that are associated with neuronal synaptic vesicles. Cleavage of SNARE proteins stops neural signal transmission; for BoNT, the neuromuscular junctions are affected, leading to flaccid paralysis, while TeNT affects the relaxation pathway in the spinal cord, leading to the characteristic spastic paralysis of tetanus (110).
TeNT is encoded by the tetX gene, which is highly conserved between strains and is located on a 74-kb plasmid in the sequenced strain E88 (111, 112) (Fig. 3). This plasmid, pE88, encodes 61 ORFs that include a collagenase gene, colT, genes encoding a two-component signal transduction system that may be involved in the regulation of TeNT production (113), and several putative alternative sigma factors. Regulation of toxin production is mediated, at least in part, by the tetR gene, which is located immediately upstream of the tetX gene (Fig. 3). TetR is an alternative sigma factor that is closely related to the BotR protein, which in C. botulinum regulates the expression of BoNT and associated proteins. TetR and BotR also are similar to the TcdR and UviA proteins, which positively regulate the expression of toxins A and B in C. difficile and bacteriocin production in C. perfringens, respectively. These alternative sigma factors interact with RNA polymerase to facilitate toxin gene expression (114).
In addition, it was recently shown that several two-component signal transduction systems are involved in both positive and negative regulation of the toxin locus in C. botulinum (115, 116). C. botulinum group I organisms have two agr-like quorum sensing systems, one regulating sporulation and the other BoNT production (117). It is likely that the regulation of TeNT production in C. tetani is also complex (118).
C. botulinum isolates were historically designated to belong to the species C. botulinum based on the single phenotype of BoNT production. However, it now is clear that strains able to produce BoNT are, in some instances, only distantly related at the phylogenetic level (119). As a result, BoNT-producing bacteria are classified into four groups: group I or proteolytic C. botulinum, group II or nonproteolytic C. botulinum, group III encoding the BoNT/C and BoNT/D serotypes, and group IV (often referred to as Clostridium argentinense). In addition, Clostridium baratii strains producing BoNT/F and strains of Clostridium butyricum producing BoNT/E have been identified (120). Until very recently there were seven serologically distinct BoNT types (BoNT/A to BoNT/G), but an eighth serotype, BoNT/H, recently was identified from a case of infant botulism (121, 122). Most of the serotypes have been further divided into subtypes (for example A1-A5). The BoNT serotype and the mechanism of toxin carriage often are common among members of the same group.
The BoNT locus is also variable and consists of two polycistronic operons: the orfX and HA loci. The bont and ntnh genes are common to all strains; the botR gene is also well conserved, but is not present in serotype E strains, and is often located between the two operons (123). Some bivalent strains have more than one BoNT locus, encoding BoNT proteins of different serotypes. In this situation, one BoNT serotype is generally found to be more abundant than the other, and the toxin designation used is, for example, Bf, where serotype B will demonstrate higher expression levels than toxin serotype F (123). Some chimeric Cd and Dc strains have been identified that have undergone recombination within the bont gene (113, 123). The variability of BoNT distribution is most likely due to mobile genetic elements such as plasmids and bacteriophages as well as transposons (120, 124).
BoNT loci can be chromosomally encoded, located on large plasmids, or encoded by lysogenic bacteriophages (123). In group III C. botulinum type C and D strains, the BoNT locus is located on large 186- to 203-kb pseudolysogenic bacteriophages (125, 126). These bacteriophage genomes are not integrated into the chromosome but exist as plasmids, although functional virus particles can be purified and used to infect susceptible strains (126). Group III isolates may also encode the C2 toxin, a binary toxin related to the ITX toxin of C. perfringens (127). The C2 toxin in these strains is also plasmid encoded, on large plasmids in excess of 100 kb. Several type C and D strains have been shown to contain other plasmids that encode putative toxin genes including a homologue of C. septicum alpha-toxin and two orthologues of ETX from C. perfringens (125).
For many years it was thought that, apart from the association of type C and D BoNT loci with bacteriophages, most other serotypes encoded chromosomal neurotoxin loci, an assumption borne out by the release of the first type A genome sequence—strain Hall (128). However, it is now clear that the toxin genes in all of the toxin serotypes studied to date may be either chromosomally encoded or plasmid-determined. This variability was first demonstrated in C. botulinum type A strains such as Loch Maree and 657Ba (pCLJ; Fig. 3), which encode BoNT/A loci on 267- and 270-kb plasmids, respectively (115, 129, 130). Plasmid-borne toxin loci in type B strains have also been demonstrated. In group I BoNT/B isolates, the plasmids range in size from 140 to 245 kb, while group II BoNT/B strains encode the toxin locus on 48- to 55-kb plasmids (Fig. 3) (130, 131). Some BoNT/E strains have been shown to harbor BoNT/E plasmids, although most appear to have chromosomally located neurotoxin genes (130, 132).
Notwithstanding the variation in the BoNT loci and their very different locations (chromosome, plasmid, or bacteriophage) the available sequence data indicate that the BoNT gene regions integrate into only one of three chromosomal locations or one of two plasmid insertion points, indicating that this locus, which is flanked by IS elements in many strains, potentially constitutes a mobile genetic element. Transfer of these loci could easily be facilitated by their carriage on elements such as bacteriophages, and recently it has been demonstrated that three type A and B toxin plasmids, two from group I organisms (pCLJ from strain 657Ba and pBotCDC-A3 from strain CDC-A3) and one from group II (pCLL from strain Eklund 17B), are capable of conjugative transfer (133). Transfer rates varied between the three plasmids, but all transferred at a relatively low level; however, plasmid transfer frequencies increased for the group I strains when a different nontoxigenic C. botulinum recipient was used. The plasmid genes responsible for the conjugative transfer remain to be identified, although pCLL showed some limited homology to the nonconjugative C. perfringens plasmid pCP13 as well as to regions of the conjugative C. perfringens plasmids (133). The similarity of several predicted ORFs to components of type II and IV bacterial secretion systems has suggested potential regions of pCLL and pCLJ that may be involved in conjugative transfer (Fig. 3). Conjugation adds yet another mode of potential transfer for these highly potent neurotoxins, which are clearly not restricted to a single clostridial strain.
TOXIN AND CAPSULE PLASMIDS OF B. ANTHRACIS
B. anthracis belongs to a family of six closely related pathogenic bacilli that includes Bacillus cereus and Bacillus thuringiensis and is known as the B. cereus group; members of this group are very closely related and are probably derived from a common ancestor (134). B. anthracis is the causative agent of anthrax, an important disease of both humans and domestic livestock. In recent years it has had increased attention as a result of its use as a bioterrorism agent. There are two major forms of anthrax: pulmonary anthrax, which is often fatal, and cutaneous anthrax, which is a more localized infection that is often restricted to workers in the wool, hide, or meat industries. The epidemiology of anthrax involves the acquisition of spores or vegetative cells by inhalation or by contact with contaminated animal material or soil (135). Disease pathogenesis is somewhat unusual, as it involves two major toxin subunits that enter the target host cell by forming heterooligomeric complexes with the third, binding and pore-forming component, protective antigen (PA, encoded by the pagA gene). The toxic components are edema factor (EF, cya gene), a calmodulin-dependent adenylyl cyclase, and lethal factor (LF, lef gene), a zinc metalloprotease that cleaves the N-terminal region of MAPKK. Another important B. anthracis virulence factor is its ability under appropriate conditions to produce a poly-d-glutamic acid capsule that protects the invading bacterial cell from phagocytosis (136).
Toxic activity is initiated by the binding of the 83-kDa PA protein to one of two receptors, TEM8 or CMG2, on the host cell surface (137, 138). The PA protein then is proteolytically cleaved by host furins, and the resultant activated PA63 derivative then oligomerizes to form a heptameric prepore. This PA63 complex can bind up to three EF or LF monomers, in any combination. After subsequent endocytosis, acidification of the endosome leads to the formation of a heptameric PA63 pore in the membrane; conformational changes then lead to the active translocation of EF and LF into the cytoplasm, where they have their lethal biological effects (139, 140).
The genes for the three components of anthrax toxin and an operon that encodes capsule production (capBCADE), together with genes involved in the regulation of their production, are all carried on two virulence plasmids, pXO1 (181.6 kb) and pXO2 (96.2 kb) (141, 142) (Fig. 3). Both plasmids are required for virulence; strains without pXO1 are avirulent because they do not produce the anthrax toxins, whereas strains lacking pXO2, such as the Sterne vaccine strain, are greatly attenuated because they do not have the capsule.
pXO1 has 203 ORFs including the structural genes for all three components of anthrax toxin (142). These toxin genes are located on a 44.8-kb pathogenicity island that is flanked by inverted copies of IS1627 and includes an 8.7-kb Tn3-like transposon, TnXO1, that carries the gerX operon, which is involved in spore germination (143). Toxin and capsule production, and therefore virulence, is regulated by AtxA, which is encoded by the pathogenicity island located on pXO1, and AcpA, which is encoded by pXO2. There is regulatory crosstalk between genes that are encoded on the plasmids and the chromosome; for example, AtxA, which has been designated as a global regulator, also regulates chromosomal genes, including S-layer genes (144, 145). The pXO1 pathogenicity island also encodes an S-layer adhesin, BslA, which is essential for the adherence of vegetative B. anthracis cells to human cells (146).
pXO2 has 110 ORFS, including the cap capsule operon and the acpAB regulatory genes (141, 147, 148). Capsule production is tightly regulated and is only produced in the presence of bicarbonate ions.
Virulence Plasmids of B. cereus and B. thuringiensis
The carriage of different virulence plasmids traditionally was considered a major contributor to the phenotypic properties that were critical for the speciation of B. anthracis, B. cereus, and B. thuringiensis. For example, pXO1 and pXO2 were regarded as major defining characteristics of B. anthracis. However, recent findings suggest a need to reconsider traditional species assignments that were based largely upon plasmid-mediated pathogenic phenotypes. B. thuringiensis strains have been identified that produce the pXO2-encoded polyglutamate capsule that was historically associated with B. anthracis (149). Additionally, several B. cereus strains have been isolated from patients in Louisiana and Texas who were suffering from severe pneumonias that clinically presented like inhalational anthrax (150). Analysis of G9241, one of those B. cereus strains, reveals that it carries a pXO1 plasmid that is 99.6% identical to the B. anthracis pXO1 plasmid and expresses all three toxin components (151). Interestingly, G9241 is encapsulated but lacks pXO2; instead, it carries another large plasmid, pBC210, that is predicted to encode a polysaccharide (not polypeptide) capsule. Animal studies indicate that both pBC210 and pXO1 are required for the full virulence of G9241 (151). Finally, B. cereus strains carrying both pXO1 and pXO2 have been isolated in Africa from great apes suffering from anthrax (152, 153). Collectively, these studies indicate that possession of pXO1 and pXO2 no longer defines B. anthracis.
Although pXO1 and pXO2 do not appear to be conjugative, they carry genes encoding components of a type IV secretion system similar to those of a classical conjugation apparatus (154) and are capable of being mobilized by conjugative plasmids (155, 156). While pXO2 does not encode its own transfer, a plasmid from B. thuringiensis, pAW63, which is closely related to pXO2, is conjugative (148). Comparative analysis has shown that 50 of the 76 ORFs found on pXO2 have significant sequence similarity to genes on pAW63; the genetic organization of these genes is also very similar and is closely related to that of pBT9727 from B. thuringiensis (Fig. 7). By contrast, pXO2 carries a 37-kb pathogenicity island that is absent from the B. thuringiensis plasmids (148). pAW63 carries a 42-kb conjugation region, and most of these genes are also present on pXO2, but the precise reason why pXO2 is not conjugative is not known. These data provide clear evidence that although pXO1 and pXO2 are not conjugative, there are conjugative plasmids that are closely related to these plasmids and that conjugation and plasmid mobilization have played a major role in the wide distribution of these plasmids throughout the B. cereus group.
FIGURE 7.
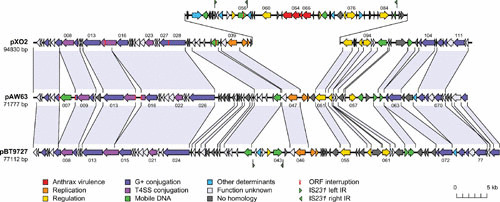
Comparative genetic organization of pXO2, pAW63, and pBT9727. Shared regions are indicated by shaded segments. The pathogenicity island on pXO2 is raised above the map. Reproduced from reference 148 with permission of the authors.
B. cereus is commonly responsible for two different types of human food poisoning, an emetic form and a diarrheal form (157). The diarrheal form is caused by several enterotoxins that are encoded by genes with a chromosomal location in B. cereus, although genes encoding one of these enterotoxins (Nhe) have been identified on a large plasmid in Bacillus weihenstephanensis (157). The emetic form of B. cereus food poisoning is caused by cereulide, a cyclic dodecapeptide with a molecular mass of 1.2 kDa (157). The ces genes responsible for cereulide synthesis are present on pCER270 (originally named pBCE4810). This large (∼270 kb), low-copy-number plasmid is related to pXO1 but lacks the pathogenicity island encoding EF and LF (pCER270 is apparently restricted to only a few clonal clusters of B. cereus). B. cereus also causes opportunistic human infections, but the involvement of plasmids in those diseases remains to be determined (157).
B. thuringiensis is an important insect and nematode pathogen but only very rarely a mammalian pathogen. The B. thuringiensis Cry (crystal) or Cyt (cytolytic) toxins are produced during stationary phase and then localize in paracrystalline bodies (158–160). Sequestration of these toxins in crystalline inclusions offers protection from bacterial cytoplasmic proteases; these inclusions are eventually solubilized by the alkaline pH of the insect midgut, and the solubilized toxins are then proteolytically activated by insect intestinal proteases (159). Different members of the Cry and Cyt toxin family exhibit toxicity for specific insects or nematodes, and some of these toxins act synergistically. Receptors for these toxins vary and can include insect plasma membrane proteins such as cadherin-like proteins, aminopeptidase N, and alkaline phosphatase (161). The Cry and Cyt toxins are considered beta pore-forming toxins that create channels in epithelial cells to damage the insect midgut (162). A single B. thuringiensis strain can carry up to seven different genes encoding crystal-associated toxins (163). Expression of these toxin genes occurs during stationary phase, and these toxins can represent up to 20 to 30% of the protein in sporulating cells (159), due to a combination of transcriptional, posttranscriptional, and posttranslational regulatory effects (159, 160, 164).
B. thuringiensis strains carry from 1 to 17 plasmids, with sizes ranging from 3 to ∼120 kb (164, 165). Most cry and cyt genes are encoded on large (>70 kb) plasmids that are often transmissible, due to either their own conjugative ability or to mobilization by helper plasmids (164, 166–168). A single plasmid can encode up to six different Cry and Cyt toxins (169). Recent studies showed that pBMB0228, which is an unusual 18-kb plasmid encoding two nematicidal Cry toxins, is a co-integrate of two plasmids, each with a cry gene (168). pBMB0228 is mobilizable and can resolve into its two separate plasmids after conjugative transfer; those plasmids can later fuse back together. In addition to often being located on conjugative plasmids, the dispersion of cry genes is probably assisted by their association with various IS elements, including IS231, IS232, IS240, ISBt1, and ISBt2, and transposons, including Tn4430 and Tn5401 (159, 160, 170).
CONCLUSIONS
We now know that toxin plasmids play an important role in the virulence of the spore-forming bacteria, but there is still a lot to learn about the biology of these plasmids and their molecular epidemiology. These plasmids represent fertile ground for future research. We need to elucidate the precise mechanism by which the conjugation process occurs in C. perfringens and how C. perfringens cells can maintain so many closely related but distinct plasmids. The role of several C. perfringens toxins in disease still remains to be determined, as do any synergistic effects on disease. There is little functional data available about the mechanism of replication and conjugative transfer of the toxin plasmids of the neurotoxic clostridia or the bacilli. Finally, there is a need for a systematic plasmid sequencing approach to determine the molecular epidemiology and evolution of the virulence plasmids of the B. cereus group.
ACKNOWLEDGMENTS
The authors gratefully acknowledge research support provided by U.S. Public Health Service grants R37AI19844-30 (B.A.M.) and R01AI056177-09 (B.A.M., J.I.R., and F.A.U.), by the Australian Research Council (ARC) through funding of the ARC Centre of Excellence in Structural and Functional Microbial Genomics (J.I.R.,), by the Australian National Health and Medical Research Council (J.I.R.), and by the Australian Poultry CRC (R.J.M. and J.I.R.). J.A.W. was supported by the provision of an Australian Postgraduate Award.
Conflict of interest: We disclose no conflicts.
Note Added in Proof
Recently other workers have identified a novel binary toxin, BEC, that is produced by C. perfringens isolates from two outbreaks of human food-borne gastroenteritis in Japan. The BECa and BECb components are related to the equivalent ITX proteins and the becAB genes are encoded on 54.5-kb plasmids that are related to pCP13 (173).
REFERENCES
- 1.Rood JI. 2004. Virulence plasmids of spore-forming bacteria, p 413–422. In Funnell BE, Phillips GJ (ed), The Biology of Plasmids. ASM Press, Washington, DC. 10.1128/9781555817732.ch19 [DOI] [Google Scholar]
- 2.Rood JI. 1998. Virulence genes of Clostridium perfringens. Annu Rev Microbiol 52:333–360. [PubMed] 10.1146/annurev.micro.52.1.333 [DOI] [PubMed] [Google Scholar]
- 3.Petit L, Gibert M, Popoff MR. 1999. Clostridium perfringens: toxinotype and genotype. Trends Microbiol 7:104–110. [PubMed] 10.1016/S0966-842X(98)01430-9 [DOI] [PubMed] [Google Scholar]
- 4.Li J, Adams V, Bannam TL, Miyamoto K, Garcia JP, Uzal FA, Rood JI, McClane BA. 2013. Toxin plasmids of Clostridium perfringens. Microbiol Mol Biol Rev 77:208–233. [PubMed] 10.1128/MMBR.00062-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bannam TL, Teng WL, Bulach D, Lyras D, Rood JI. 2006. Functional identification of conjugation and replication regions of the tetracycline resistance plasmid pCW3 from Clostridium perfringens. J Bacteriol 188:4942–4951. [PubMed] 10.1128/JB.00298-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyamoto K, Fisher DJ, Li J, Sayeed S, Akimoto S, McClane BA. 2006. Complete sequencing and diversity analysis of the enterotoxin-encoding plasmids in Clostridium perfringens type A non-food-borne human gastrointestinal disease isolates. J Bacteriol 188:1585–1598. [PubMed] 10.1128/JB.188.4.1585-1598.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerdes K, Howard M, Szardenings F. 2010. Pushing and pulling in prokaryotic DNA segregation. Cell 141:927–942. [PubMed] 10.1016/j.cell.2010.05.033 [DOI] [PubMed] [Google Scholar]
- 8.Salje J, Gayathri P, Lowe J. 2010. The ParMRC system: molecular mechanisms of plasmid segregation by actin-like filaments. Nat Rev Microbiol 8:683–692. [PubMed] 10.1038/nrmicro2425 [DOI] [PubMed] [Google Scholar]
- 9.Bannam TL, Yan XX, Harrison PF, Seemann T, Keyburn AL, Stubenrauch C, Weeramantri LH, Cheung JK, McClane BA, Boyce JD, Moore RJ, Rood JI. 2011. Necrotic enteritis-derived Clostridium perfringens strain with three closely related independently conjugative toxin and antibiotic resistance plasmids. mBio 2:e00190–00111. [PubMed] 10.1128/mBio.00190-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parreira VR, Costa M, Eikmeyer F, Blom J, Prescott JF. 2012. Sequence of two plasmids from Clostridium perfringens chicken necrotic enteritis isolates and comparison with C. perfringens conjugative plasmids. PLoS One 7:e49753. [PubMed] 10.1371/journal.pone.0049753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Popp D, Narita A, Lee LJ, Ghoshdastider U, Xue B, Srinivasan R, Balasubramanian MK, Tanaka T, Robinson RC. 2012. Novel actin-like filament structure from Clostridium tetani. J Biol Chem 287:21121–21129. [PubMed] 10.1074/jbc.M112.341016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derman AI, Becker EC, Truong BD, Fujioka A, Tucey TM, Erb ML, Patterson PC, Pogliano J. 2009. Phylogenetic analysis identifies many uncharacterized actin-like proteins (Alps) in bacteria: regulated polymerization, dynamic instability and treadmilling in Alp7A. Mol Microbiol 73:534–552. [PubMed] 10.1111/j.1365-2958.2009.06771.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brefort G, Magot M, Ionesco H, Sebald M. 1977. Characterization and transferability of Clostridium perfringens plasmids. Plasmid 1:52–66. [PubMed] 10.1016/0147-619X(77)90008-7 [DOI] [PubMed] [Google Scholar]
- 14.Rood JI, Maher EA, Somers EB, Campos E, Duncan CL. 1978. Isolation and characterization of multiple antibiotic-resistant Clostridium perfringens strains from porcine feces. Antimicrob Agents Chemother 13:871–880. [PubMed] 10.1128/AAC.13.5.871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brynestad S, Sarker MR, McClane BA, Granum PE, Rood JI. 2001. Enterotoxin plasmid from Clostridium perfringens is conjugative. Infect Immun 69:3483–3487. [PubMed] 10.1128/IAI.69.5.3483-3487.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes ML, Poon R, Adams V, Sayeed S, Saputo S, Uzal FA, McClane BA, Rood JI. 2007. Epsilon toxin plasmids of Clostridium perfringens type D are conjugative. J Bacteriol 189:7531–7538. [PubMed] 10.1128/JB.00767-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyras D, Adams V, Ballard SA, Teng WL, Howarth PM, Crellin PK, Bannam TL, Songer JG, Rood JI. 2009. tISCpe8, an IS1595-family lincomycin resistance element located on a conjugative plasmid in Clostridium perfringens. J Bacteriol 191:6345–6351. [PubMed] 10.1128/JB.00668-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abraham LJ, Rood JI. 1985. Molecular analysis of transferable tetracycline resistance plasmids from Clostridium perfringens. J Bacteriol 161:636–640. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abraham LJ, Wales AJ, Rood JI. 1985. Worldwide distribution of the conjugative Clostridium perfringens tetracycline resistance plasmid, pCW3. Plasmid 14:37–46. [PubMed] 10.1016/0147-619X(85)90030-7 [DOI] [PubMed] [Google Scholar]
- 20.Miyamoto K, Li J, Sayeed S, Akimoto S, McClane BA. 2008. Sequencing and diversity analyses reveal extensive similarities between some epsilon-toxin-encoding plasmids and the pCPF5603 Clostridium perfringens enterotoxin plasmid. J Bacteriol 190:7178–7188. [PubMed] 10.1128/JB.00939-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyamoto K, Yumine N, Mimura K, Nagahama M, Li J, McClane BA, Akimoto S. 2011. Identification of novel Clostridium perfringens type E strains that carry an iota toxin plasmid with a functional enterotoxin gene. PLoS One 6:e20376. [PubMed] 10.1371/journal.pone.0020376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bantwal R, Bannam TL, Porter CJ, Quinsey NS, Lyras D, Adams V, Rood JI. 2012. The peptidoglycan hydrolase TcpG is required for efficient conjugative transfer of pCW3 in Clostridium perfringens. Plasmid 67:139–147. [PubMed] 10.1016/j.plasmid.2011.12.016 [DOI] [PubMed] [Google Scholar]
- 23.Parsons JA, Bannam TL, Devenish RJ, Rood JI. 2007. TcpA, an FtsK/SpoIIIE homolog, is essential for transfer of the conjugative plasmid pCW3 from Clostridium perfringens. J Bacteriol 189:7782–7790. [PubMed] 10.1128/JB.00783-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porter CJ, Bantwal R, Bannam TL, Rosado CJ, Pearce MC, Adams V, Lyras D, Whisstock JC, Rood JI. 2012. The conjugation protein TcpC from Clostridium perfringens is structurally related to the type IV secretion system protein VirB8 from Gram-negative bacteria. Mol Microbiol 83:275–288. [PubMed] 10.1111/j.1365-2958.2011.07930.x [DOI] [PubMed] [Google Scholar]
- 25.Steen JA, Bannam TL, Teng WL, Devenish RJ, Rood JI. 2009. The putative coupling protein TcpA interacts with other pCW3-encoded proteins to form an essential part of the conjugation complex. J Bacteriol 191:2926–2933. [PubMed] 10.1128/JB.00032-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goessweiner-Mohr N, Arends K, Keller W, Grohmann E. 2013. Conjugative type IV secretion systems in Gram-positive bacteria. Plasmid 70:289–302. [PubMed] 10.1016/j.plasmid.2013.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gomis-Rüth FX, Coll M. 2001. Structure of TrwB, a gatekeeper in bacterial conjugation. Int J Biochem Cell Biol 33:839–843. [PubMed] 10.1016/S1357-2725(01)00060-7 [DOI] [PubMed] [Google Scholar]
- 28.Fronzes R, Christie PJ, Waksman G. 2009. The structural biology of type IV secretion systems. Nat Rev Microbiol 7:703–714. [PubMed] 10.1038/nrmicro2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teng WL, Bannam TL, Parsons JA, Rood JI. 2008. Functional characterization and localization of the TcpH conjugation protein from Clostridium perfringens. J Bacteriol 190:5075–5086. [PubMed] 10.1128/JB.00386-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de la Cruz F, Frost LS, Meyer RJ, Zechner EL. 2010. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol Rev 34:18–40. [PubMed] 10.1111/j.1574-6976.2009.00195.x [DOI] [PubMed] [Google Scholar]
- 31.McClane BA, Robertson SL, Li J. 2013. Clostridium perfringens, p 465–490. In Doyle MP, Buchanan RA (ed), Food Microbiology: Fundamentals and Frontiers. ASM Press, Washington, DC. [Google Scholar]
- 32.Huang IH, Waters M, Grau RR, Sarker MR. 2004. Disruption of the gene (spo0A) encoding sporulation transcription factor blocks endospore formation and enterotoxin production in enterotoxigenic Clostridium perfringens type A. FEMS Microbiol Lett 233:233–240. [PubMed] 10.1111/j.1574-6968.2004.tb09487.x [DOI] [PubMed] [Google Scholar]
- 33.Harry KH, Zhou R, Kroos L, Melville SB. 2009. Sporulation and enterotoxin (CPE) synthesis are controlled by the sporulation-specific sigma factors SigE and SigK in Clostridium perfringens. J Bacteriol 191:2728–2742. [PubMed] 10.1128/JB.01839-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, McClane BA. 2010. Evaluating the involvement of alternative sigma factors SigF and SigG in Clostridium perfringens sporulation and enterotoxin synthesis. Infect Immun 78:4286–4293. [PubMed] 10.1128/IAI.00528-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Briggs DC, Naylor CE, Smedley JG, 3rd, Lukoyanova N, Robertson S, Moss DS, McClane BA, Basak AK. 2011. Structure of the food-poisoning Clostridium perfringens enterotoxin reveals similarity to the aerolysin-like pore-forming toxins. J Mol Biol 413:138–149. [PubMed] 10.1016/j.jmb.2011.07.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitadokoro K, Nishimura K, Kamitani S, Fukui-Miyazaki A, Toshima H, Abe H, Kamata Y, Sugita-Konishi Y, Yamamoto S, Karatani H, Horiguchi Y. 2011. Crystal structure of Clostridium perfringens enterotoxin displays features of beta-pore-forming toxins. J Biol Chem 286:19549–19555. [PubMed] 10.1074/jbc.M111.228478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katahira J, Sugiyama H, Inoue N, Horiguchi Y, Matsuda M, Sugimoto N. 1997. Clostridium perfringens enterotoxin utilizes two structurally related membrane proteins as functional receptors in vivo. J Biol Chem 272:26652–26658. [PubMed] 10.1074/jbc.272.42.26652 [DOI] [PubMed] [Google Scholar]
- 38.Hanna PC, Mietzner TA, Schoolnick GK, McClane BA. 1991. Localization of the receptor-binding region of Clostridium perfringens enterotoxin utilizing cloned toxin fragments and synthetic peptides. The 30 C-terminal amino acids define a functional binding region. J Biol Chem 266:11037–11043. [PubMed] [PubMed] [Google Scholar]
- 39.Kokai-Kun JF, Benton K, Wieckowski EU, McClane BA. 1999. Identification of a Clostridium perfringens enterotoxin region required for large complex formation and cytotoxicity by random mutagenesis. Infect Immun 67:5634–5641. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sayeed S, Li J, McClane BA. 2007. Virulence plasmid diversity in Clostridium perfringens type D isolates. Infect Immun 75:2391–2398. [PubMed] 10.1128/IAI.02014-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sayeed S, Li J, McClane BA. 2010. Characterization of virulence plasmid diversity among Clostridium perfringens type B isolates. Infect Immun 78:495–504. [PubMed] 10.1128/IAI.00838-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fisher DJ, Fernandez-Miyakawa ME, Sayeed S, Poon R, Adams V, Rood JI, Uzal FA, McClane BA. 2006. Dissecting the contributions of Clostridium perfringens type C toxins to lethality in the mouse intravenous injection model. Infect Immun 74:5200–5210. [PubMed] 10.1128/IAI.00534-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collie R, McClane B. 1998. Phenotypic characterization of enterotoxigenic Clostridium perfringens isolates associated with nonfoodborne human gastrointestinal diseases. Anaerobe 4:69–79. [PubMed] 10.1006/anae.1998.0152 [DOI] [PubMed] [Google Scholar]
- 44.Fernandez-Miyakawa ME, Sayeed S, Fisher DJ, Poon R, Adams V, Rood JI, McClane BA, Saputo J, Uzal FA. 2007. Development and application of an oral challenge mouse model for studying Clostridium perfringens type D infection. Infect Immun 75:4282–4288. [PubMed] 10.1128/IAI.00562-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kokai-Kun JF, Songer JG, Czeczulin JR, Chen F, McClane BA. 1994. Comparison of Western immunoblots and gene detection assays for identification of potentially enterotoxigenic isolates of Clostridium perfringens. J Clin Microbiol 32:2533–2539. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarker MR, Carman RJ, McClane BA. 1999. Inactivation of the gene (cpe) encoding Clostridium perfringens enterotoxin eliminates the ability of two cpe-positive C. perfringens type A human gastrointestinal disease isolates to affect rabbit ileal loops. Mol Microbiol 33:946–958. [PubMed] 10.1046/j.1365-2958.1999.01534.x [DOI] [PubMed] [Google Scholar]
- 47.Scallan E, Griffin PM, Angulo FJ, Tauxe RV, Hoekstra RM. 2011. Foodborne illness acquired in the United States: unspecified agents. Emerg Infect Dis 17:16–22. [PubMed] 10.3201/eid1701.P21101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carman RJ. 1997. Clostridium perfringens in spontaneous and antibiotic-associated diarrhoea of man and other animals. Rev Med Microbiol 8(Suppl. 1):S43–S45. 10.1097/00013542-199712001-00024 [DOI] [Google Scholar]
- 49.Bos J, Smithee L, McClane B, Distefano R, Uzal F, Songer JG, Mallonee S, Crutcher JM. 2005. Fatal necrotizing enteritis following a foodborne outbreak of enterotoxigenic Clostridium perfringens type A infection. Clin Infect Dis 15:e78–e83. [PubMed] 10.1086/429829 [DOI] [PubMed] [Google Scholar]
- 50.Marks SL, Kather EJ, Kass PH, Melli AC. 2002. Genotypic and phenotypic characterization of Clostridium perfringens and Clostridium difficile in diarrheic and healthy dogs. J Vet Intern Med 16:533–540. [PubMed] 10.1111/j.1939-1676.2002.tb02383.x [DOI] [PubMed] [Google Scholar]
- 51.Brynestad S, Synstad B, Granum PE. 1997. The Clostridium perfringens enterotoxin gene is on a transposable genetic element in type A human food poisoning strains. Microbiology 143:2109–2115. [PubMed] 10.1099/00221287-143-7-2109 [DOI] [PubMed] [Google Scholar]
- 52.Ma M, Li J, McClane BA. 2012. Genotypic and phenotypic characterization of Clostridium perfringens isolates from darmbrand cases in post-World War II Germany. Infect Immun 80:4354–4363. [PubMed] 10.1128/IAI.00818-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J, Miyamoto K, Sayeed S, McClane BA. 2010. Organization of the cpe locus in CPE-positive Clostridium perfringens type C and D isolates. PLoS One 5:e10932. [PubMed] 10.1371/journal.pone.0010932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Billington SJ, Wieckowski EU, Sarkar MR, Bueschel D, Songer JG, McClane BA. 1998. Clostridium perfringens type E animal enteritis isolates with highly conserved, silent enterotoxin gene sequences. Infect Immun 66:4531–4536. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Gurjar A, Li J, McClane BA. 2010. Characterization of toxin plasmids in Clostridium perfringens type C isolates. Infect Immun 78:4860–4869. [PubMed] 10.1128/IAI.00715-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lawrence G. 2005. The pathogenesis of pig-bel in Papua New Guinea. P N G Med J 48:39–49. [PubMed] [PubMed] [Google Scholar]
- 57.Murrell TG, Walker PD. 1991. The pigbel story of Papua New Guinea. Trans R Soc Trop Med Hyg 85:119–122. [PubMed] 10.1016/0035-9203(91)90183-Y [DOI] [PubMed] [Google Scholar]
- 58.McClane BA, Uzal FA, Fernandez Miyakawa ME, Lyerly D, Wilkins TD. 2006. The enterotoxic clostridia, p 698–752. In Dworkin M, Falkow S, Rosenberg E, Schleifer K, Stackebrandt E (ed), The Prokaryotes. Springer, New York, NY. 10.1007/0-387-30744-3_22 [DOI] [Google Scholar]
- 59.Fernandez-Miyakawa ME, Fisher DJ, Poon R, Sayeed S, Adams V, Rood JI, McClane BA, Uzal FA. 2007. Both epsilon-toxin and beta-toxin are important for the lethal properties of Clostridium perfringens type B isolates in the mouse intravenous injection model. Infect Immun 75:1443–1452. [PubMed] 10.1128/IAI.01672-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sayeed S, Uzal FA, Fisher DJ, Saputo J, Vidal JE, Chen Y, Gupta P, Rood JI, McClane BA. 2008. Beta toxin is essential for the intestinal virulence of Clostridium perfringens type C disease isolate CN3685 in a rabbit ileal loop model. Mol Microbiol 67:15–30. [PubMed] 10.1111/j.1365-2958.2007.06007.x [DOI] [PubMed] [Google Scholar]
- 61.Vidal JE, Ohtani K, Shimizu T, McClane BA. 2009. Contact with enterocyte-like Caco-2 cells induces rapid upregulation of toxin production by Clostridium perfringens type C isolates. Cell Microbiol 11:1306–1328. [PubMed] 10.1111/j.1462-5822.2009.01332.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garcia JP, Beingesser J, Fisher DJ, Sayeed S, McClane BA, Posthaus H, Uzal FA. 2012. The effect of Clostridium perfringens type C strain CN3685 and its isogenic beta toxin null mutant in goats. Vet Microbiol 157:412–419. [PubMed] 10.1016/j.vetmic.2012.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hunter SEC, Brown JE, Oyston PCF, Sakurai J, Titball RW. 1993. Molecular genetic analysis of beta-toxin of Clostridium perfringens reveals sequence homology with alpha-toxin, gamma-toxin, and leukocidin of Staphylococcus aureus. Infect Immun 61:3958–3965. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Keyburn AL, Bannam TL, Moore RJ, Rood JI. 2010. NetB, a pore-forming toxin from necrotic enteritis strains of Clostridium perfringens. Toxins 2:1913–1927. [PubMed] 10.3390/toxins2071913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Manich M, Knapp O, Gibert M, Maier E, Jolivet-Reynaud C, Geny B, Benz R, Popoff MR. 2008. Clostridium perfringens delta toxin is sequence related to beta toxin, NetB, and Staphylococcus pore-forming toxins, but shows functional differences. PLoS One 3:e3764. [PubMed] 10.1371/journal.pone.0003764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shatursky O, Bayles R, Rogers M, Jost BH, Songer JG, Tweten RK. 2000. Clostridium perfringens beta-toxin forms potential-dependent, cation-selective channels in lipid bilayers. Infect Immun 68:5546–5551. [PubMed] 10.1128/IAI.68.10.5546-5551.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vidal JE, McClane BA, Saputo J, Parker J, Uzal FA. 2008. Effects of Clostridium perfringens beta-toxin on the rabbit small intestine and colon. Infect Immun 76:4396–4404. [PubMed] 10.1128/IAI.00547-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ma M, Vidal J, Saputo J, McClane BA, Uzal F. 2011. The VirS/VirR two-component system regulates the anaerobic cytotoxicity, intestinal pathogenicity, and enterotoxemic lethality of Clostridium perfringens type C isolate CN3685. MBio 2:e00338–00310. [PubMed] 10.1128/mBio.00338-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vidal JE, Ma M, Saputo J, Garcia J, Uzal FA, McClane BA. 2012. Evidence that the Agr-like quorum sensing system regulates the toxin production, cytotoxicity and pathogenicity of Clostridium perfringens type C isolate CN3685. Mol Microbiol 83:179–194. [PubMed] 10.1111/j.1365-2958.2011.07925.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen J, McClane BA. 2012. Role of the Agr-like quorum-sensing system in regulating toxin production by Clostridium perfringens type B strains CN1793 and CN1795. Infect Immun 80:3008–3017. [PubMed] 10.1128/IAI.00438-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Amimoto K, Noro T, Oishi E, Shimizu M. 2007. A novel toxin homologous to large clostridial cytotoxins found in culture supernatant of Clostridium perfringens type C. Microbiology 153:1198–1206. [PubMed] 10.1099/mic.0.2006/002287-0 [DOI] [PubMed] [Google Scholar]
- 72.Gibert M, Jolivet-Renaud C, Popoff MR. 1997. Beta2 toxin, a novel toxin produced by Clostridium perfringens. Gene 203:65–73. [PubMed] 10.1016/S0378-1119(97)00493-9 [DOI] [PubMed] [Google Scholar]
- 73.Popoff MR, Bouvet P. 2009. Clostridial toxins. Future Microbiol 4:1021–1064. [PubMed] 10.2217/fmb.09.72 [DOI] [PubMed] [Google Scholar]
- 74.Uzal FA, Songer JG. 2008. Diagnosis of Clostridium perfringens intestinal infections in sheep and goats. J Vet Diagn Invest 20:253–265. [PubMed] 10.1177/104063870802000301 [DOI] [PubMed] [Google Scholar]
- 75.Garcia JP, Adams V, Beingesser J, Hughes ML, Poon R, Lyras D, Hill A, McClane BA, Rood JI, Uzal FA. 2013. Epsilon toxin is essential for the virulence of Clostridium perfringens type D infection in sheep, goats, and mice. Infect Immun 81:2405–2414. [PubMed] 10.1128/IAI.00238-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen J, Rood JI, McClane BA. 2011. Epsilon toxin production by Clostridium perfringens type D strain CN3718 is dependent on the agr operon but not the VirS/VirR two component regulatory system. MBio 2:e00275–00211. [PubMed] 10.1128/mBio.00275-11 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 77.Li J, Ma M, Sarker MR, McClane BA. 2013. CodY is a global regulator of virulence-associated properties for Clostridium perfringens type D strain CN3718. MBio 4:e00770–00713. [PubMed] 10.1128/mBio.00770-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stiles BG, Wigelsworth DJ, Popoff MR, Barth H. 2011. Clostridial binary toxins: iota and C2 family portraits. Front Cell Infect Microbiol 1:11. [PubMed] 10.3389/fcimb.2011.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Aktories K, Schwan C, Papatheodorou P, Lang AE. 2012. Bidirectional attack on the actin cytoskeleton. Bacterial protein toxins causing polymerization or depolymerization of actin. Toxicon 60:572–581. [PubMed] 10.1016/j.toxicon.2012.04.338 [DOI] [PubMed] [Google Scholar]
- 80.Sakurai J, Nagahama M, Oda M, Tsuge H, Kobayashi K. 2009. Clostridium perfringens iota-toxin: structure and function. Toxins 1:208–228. [PubMed] 10.3390/toxins1020208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Papatheodorou P, Carette JE, Bell GW, Schwan C, Guttenberg G, Brummelkamp TR, Aktories K. 2011. Lipolysis-stimulated lipoprotein receptor (LSR) is the host receptor for the binary toxin Clostridium difficile transferase (CDT). Proc Natl Acad Sci USA 108:16422–16427. [PubMed] 10.1073/pnas.1109772108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Papatheodorou P, Wilczek C, Nolke T, Guttenberg G, Hornuss D, Schwan C, Aktories K. 2012. Identification of the cellular receptor of Clostridium spiroforme toxin. Infect Immun 80:1418–1423. [PubMed] 10.1128/IAI.06378-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wigelsworth DJ, Ruthel G, Schnell L, Herrlich P, Blonder J, Veenstra TD, Carman RJ, Wilkins TD, Van Nhieu GT, Pauillac S, Gibert M, Sauvonnet N, Stiles BG, Popoff MR, Barth H. 2012. CD44 promotes intoxication by the clostridial iota-family toxins. PLoS One 7:e51356. [PubMed] 10.1371/journal.pone.0051356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li J, Miyamoto K, McClane BA. 2007. Comparison of virulence plasmids among Clostridium perfringens type E isolates. Infect Immun 75:1811–1819. [PubMed] 10.1128/IAI.01981-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Keyburn AL, Boyce JD, Vaz P, Bannam TL, Ford ME, Parker D, Di Rubbo A, Rood JI, Moore RJ. 2008. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathog 4:e26. [PubMed] 10.1371/journal.ppat.0040026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van der Sluis W. 2000. Clostridial enteritis is an often underestimated problem. World Poultry 16:42–43. [Google Scholar]
- 87.Yan X, Porter CJ, Hardy SP, Steer D, Smith AI, Quinsey NS, Hughes V, Cheung JK, Keyburn AL, Kaldhusdal M, Moore RJ, Bannam TL, Whisstock JC, Rood JI. 2013. Structural and functional analysis of the pore-forming toxin NetB from Clostridium perfringens. mBio 4:e00019–00013. [PubMed] 10.1128/mBio.00019-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Savva CG, Fernandes da Costa SP, Bokori-Brown M, Naylor CE, Cole AR, Moss DS, Titball RW, Basak AK. 2013. Molecular architecture and functional analysis of NetB, a pore-forming toxin from Clostridium perfringens. J Biol Chem 288:3512–3522. [PubMed] 10.1074/jbc.M112.430223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fernandes da Costa SP, Savva CG, Bokori-Brown M, Naylor CE, Moss DS, Basak AK, Titball RW. 2014. Identification of a key residue for oligomerisation and pore-formation of Clostridium perfringens NetB. Toxins 6:1049–1061. [PubMed] 10.3390/toxins6031049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Keyburn AL, Portela RW, Ford ME, Bannam TL, Yan XX, Rood JI, Moore RJ. 2013. Maternal immunization with vaccines containing recombinant NetB toxin partially protects progeny chickens from necrotic enteritis. Vet Res 44:108. [PubMed] 10.1186/1297-9716-44-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Keyburn AL, Portela RW, Sproat K, Ford ME, Bannam TL, Yan X, Rood JI, Moore RJ. 2013. Vaccination with recombinant NetB toxin partially protects broiler chickens from necrotic enteritis. Vet Res 44:54. [PubMed] 10.1186/1297-9716-44-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jang SI, Lillehoj HS, Lee SH, Lee KW, Lillehoj EP, Hong YH, An DJ, Jeong W, Chun JE, Bertrand F, Dupuis L, Deville S, Arous JB. 2012. Vaccination with Clostridium perfringens recombinant proteins in combination with Montanide ISA 71 VG adjuvant increases protection against experimental necrotic enteritis in commercial broiler chickens. Vaccine 30:5401–5406. [PubMed] 10.1016/j.vaccine.2012.06.007 [DOI] [PubMed] [Google Scholar]
- 93.Keyburn AL, Yan XX, Bannam TL, Van Immerseel F, Rood JI, Moore RJ. 2010. Association between avian necrotic enteritis and Clostridium perfringens strains expressing NetB toxin. Vet Res 41:21. [PubMed] 10.1051/vetres/2009069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Martin TG, Smyth JA. 2009. Prevalence of netB among some clinical isolates of Clostridium perfringens from animals in the United States. Vet Microbiol 136:202–205. [PubMed] 10.1016/j.vetmic.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 95.Smyth JA, Martin TG. 2010. Disease producing capability of netB positive isolates of C. perfringens recovered from normal chickens and a cow, and netB positive and negative isolates from chickens with necrotic enteritis. Vet Microbiol 146:76–84. [PubMed] 10.1016/j.vetmic.2010.04.022 [DOI] [PubMed] [Google Scholar]
- 96.Lepp D, Roxas B, Parreira VR, Marri PR, Rosey EL, Gong J, Songer JG, Vedantam G, Prescott JF. 2010. Identification of novel pathogenicity loci in Clostridium perfringens strains that cause avian necrotic enteritis. PLoS One 5:e10795. [PubMed] 10.1371/journal.pone.0010795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cheung JK, Low L-Y, Hiscox TJ, Rood JI. 2013. Regulation of extracellular toxin production in Clostridium perfringens, p 281-294. In Vasil ML, Darwin AJ (ed), Regulation of Bacterial Virulence. ASM Press, Washington, DC. [Google Scholar]
- 98.Lepp D, Gong J, Songer JG, Boerlin P, Parreira VR, Prescott JF. 2013. Identification of accessory genome regions in poultry Clostridium perfringens isolates carrying the NetB plasmid. J Bacteriol 195:1152–1166. [PubMed] 10.1128/JB.01032-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hibberd MC, Neumann AP, Rehberger TG, Siragusa GR. 2011. Multilocus sequence typing subtypes of poultry Clostridium perfringens isolates demonstrate disease niche partitioning. J Clin Microbiol 49:1556–1567. [PubMed] 10.1128/JCM.01884-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Manteca C, Daube G, Jauniaux T, Linden A, Pirson V, Detilleux J, Ginter A, Coppe P, Kaeckenbeeck A, Mainil JG. 2002. A role for the Clostridium perfringens beta2 toxin in bovine enterotoxaemia? Vet Microbiol 86:191–202. [PubMed] 10.1016/S0378-1135(02)00008-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dray T. 2004. Clostridium perfringens type A and beta2 toxin associated with enterotoxemia in a 5-week-old goat. Can Vet J 45:251–253. [PubMed] [PMC free article] [PubMed] [Google Scholar]
- 102.Uzal FA, Vidal JE, McClane BA, Gurjar AA. 2010. Clostridium perfringens toxins involved in mammalian veterinary diseases. Open Toxinology J 2:24–42. [PubMed] 10.2174/1875414701003020024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fisher DJ, Miyamoto K, Harrison B, Akimoto S, Sarker MR, McClane BA. 2005. Association of beta2 toxin production with Clostridium perfringens type A human gastrointestinal disease isolates carrying a plasmid enterotoxin gene. Mol Microbiol 56:747–762. [PubMed] 10.1111/j.1365-2958.2005.04573.x [DOI] [PubMed] [Google Scholar]
- 104.Harrison B, Raju D, Garmory HS, Brett MM, Titball RW, Sarker MR. 2005. Molecular characterization of Clostridium perfringens isolates from humans with sporadic diarrhea: evidence for transcriptional regulation of the beta2-toxin-encoding gene. Appl Environ Microbiol 71:8362–8370. [PubMed] 10.1128/AEM.71.12.8362-8370.2005 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 105.Shimizu T, Ohtani K, Hirakawa H, Ohshima K, Yamashita A, Shiba T, Ogasawara N, Hattori M, Kuhara S, Hayashi H. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc Natl Acad Sci USA 99:996–1001. [PubMed] 10.1073/pnas.022493799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nowell VJ, Kropinski AM, Songer JG, MacInnes JI, Parreira VR, Prescott JF. 2012. Genome sequencing and analysis of a type A Clostridium perfringens isolate from a case of bovine clostridial abomasitis. PLoS One 7:e32271. [PubMed] 10.1371/journal.pone.0032271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shimizu T, Shima K, Yoshino K, Yonezawa K, Hayashi H. 2002. Proteome and transcriptome analysis of the virulence genes regulated by the VirR/VirS system in Clostridium perfringens. J Bacteriol 184:2587–2594. [PubMed] 10.1128/JB.184.10.2587-2594.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ohtani K, Kawsar HI, Okumura K, Hayashi H, Shimizu T. 2003. The VirR/VirS regulatory cascade affects transcription of plasmid-encoded putative virulence genes in Clostridium perfringens strain 13. FEMS Microbiol Lett 222:137–141. [PubMed] 10.1016/S0378-1097(03)00255-6 [DOI] [PubMed] [Google Scholar]
- 109.Li J, Chen J, Vidal JE, McClane BA. 2011. The Agr-like quorum-sensing system regulates sporulation and production of enterotoxin and beta2 toxin by Clostridium perfringens type A non-food-borne human gastrointestinal disease strain F5603. Infect Immun 79:2451–2459. [PubMed] 10.1128/IAI.00169-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Caleo M, Schiavo G. 2009. Central effects of tetanus and botulinum neurotoxins. Toxicon 54:593–599. [PubMed] 10.1016/j.toxicon.2008.12.026 [DOI] [PubMed] [Google Scholar]
- 111.Finn C, Jr, Silver R, Habig W, Hardegree M, Zon G, Garon C. 1984. The structural gene for tetanus neurotoxin is on a plasmid. Science 224:881–884. [PubMed] 10.1126/science.6326263 [DOI] [PubMed] [Google Scholar]
- 112.Brüggemann H, Bäumer S, Fricke WF, Wiezer A, Liesegang H, Decker I, Herzberg C, Martinez-Arias R, Merkl R, Henne A, Gottschalk G. 2003. The genome sequence of Clostridium tetani, the causative agent of tetanus disease. Proc Natl Acad Sci USA 100:1316–1321. [PubMed] 10.1073/pnas.0335853100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bruggemann H, Bauer R, Raffestin S, Gottschalk G. 2004. Characterization of a heme oxygenase of Clostridium tetani and its possible role in oxygen tolerance. Arch Microbiol 182:259–263. [PubMed] 10.1007/s00203-004-0721-1 [DOI] [PubMed] [Google Scholar]
- 114.Raffestin S, Dupuy B, Marvaud JC, Popoff MR. 2005. BotR/A and TetR are alternative RNA polymerase sigma factors controlling the expression of the neurotoxin and associated protein genes in Clostridium botulinum type A and Clostridium tetani. Mol Microbiol 55:235–249. [PubMed] 10.1111/j.1365-2958.2004.04377.x [DOI] [PubMed] [Google Scholar]
- 115.Zhang Z, Korkeala H, Dahlsten E, Sahala E, Heap JT, Minton NP, Lindstrom M. 2013. Two-component signal transduction system CBO0787/CBO0786 represses transcription from botulinum neurotoxin promoters in Clostridium botulinum ATCC 3502. PLoS Pathog 9:e1003252. [PubMed] 10.1371/journal.ppat.1003252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Connan C, Brueggemann H, Mazuet C, Raffestin S, Cayet N, Popoff MR. 2012. Two-component systems are involved in the regulation of botulinum neurotoxin synthesis in Clostridium botulinum type A strain Hall. PLoS One 7:e41848. [PubMed] 10.1371/journal.pone.0041848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cooksley CM, Davis IJ, Winzer K, Chan WC, Peck MW, Minton NP. 2010. Regulation of neurotoxin production and sporulation by a putative agrBD signaling system in proteolytic Clostridium botulinum. Appl Environ Microbiol 76:4448–4460. [PubMed] 10.1128/AEM.03038-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Connan C, Deneve C, Mazuet C, Popoff MR. 2013. Regulation of toxin synthesis in Clostridium botulinum and Clostridium tetani. Toxicon 75:90–100. [PubMed] 10.1016/j.toxicon.2013.06.001 [DOI] [PubMed] [Google Scholar]
- 119.Hill KK, Smith TJ, Helma CH, Ticknor LO, Foley BT, Svensson RT, Brown JL, Johnson EA, Smith LA, Okinaka RT, Jackson PJ, Marks JD. 2007. Genetic diversity among botulinum neurotoxin-producing clostridial strains. J Bacteriol 189:818–832. [PubMed] 10.1128/JB.01180-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hill KK, Xie G, Foley BT, Smith TJ, Munk AC, Bruce D, Smith LA, Brettin TS, Detter JC. 2009. Recombination and insertion events involving the botulinum neurotoxin complex genes in Clostridium botulinum types A, B, E and F and Clostridium butyricum type E strains. BMC Biol 7:66. [PubMed] 10.1186/1741-7007-7-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Barash JR, Arnon SS. 2014. A novel strain of Clostridium botulinum that produces type B and type H botulinum toxins. J Infect Dis 209:183–191. [PubMed] 10.1093/infdis/jit449 [DOI] [PubMed] [Google Scholar]
- 122.Dover N, Barash JR, Hill KK, Xie G, Arnon SS. 2014. Molecular characterization of a novel botulinum neurotoxin type H gene. J Infect Dis 209:192–202. [PubMed] 10.1093/infdis/jit450 [DOI] [PubMed] [Google Scholar]
- 123.Hill KK, Smith TJ. 2013. Genetic diversity within Clostridium botulinum serotypes, botulinum neurotoxin gene clusters and toxin subtypes. Curr Top Microbiol Immunol 364:1–20. [PubMed] 10.1007/978-3-642-33570-9_1 [DOI] [PubMed] [Google Scholar]
- 124.Popoff MR, Bouvet P. 2013. Genetic characteristics of toxigenic clostridia and toxin gene evolution. Toxicon 75:63–89. [PubMed] 10.1016/j.toxicon.2013.05.003 [DOI] [PubMed] [Google Scholar]
- 125.Skarin H, Segerman B. 2011. Horizontal gene transfer of toxin genes in Clostridium botulinum: involvement of mobile elements and plasmids. Mobile Genetic Elements 1:213–215. [PubMed] 10.4161/mge.1.3.17617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sakaguchi Y, Hayashi T, Kurokawa K, Nakayama K, Oshima K, Fujinaga Y, Ohnishi M, Ohtsubo E, Hattori M, Oguma K. 2005. The genome sequence of Clostridium botulinum type C neurotoxin-converting phage and the molecular mechanisms of unstable lysogeny. Proc Natl Acad Sci USA 102:17472–17477. [PubMed] 10.1073/pnas.0505503102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sakaguchi Y, Hayashi T, Yamamoto Y, Nakayama K, Zhang K, Ma S, Arimitsu H, Oguma K. 2009. Molecular analysis of an extrachromosomal element containing the C2 toxin gene discovered in Clostridium botulinum type C. J Bacteriol 191:3282–3291. [PubMed] 10.1128/JB.01797-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sebaihia M, Peck MW, Minton NP, Thomson NR, Holden MT, Mitchell WJ, Carter AT, Bentley SD, Mason DR, Crossman L, Paul CJ, Ivens A, Wells-Bennik MH, Davis IJ, Cerdeno-Tarraga AM, Churcher C, Quail MA, Chillingworth T, Feltwell T, Fraser A, Goodhead I, Hance Z, Jagels K, Larke N, Maddison M, Moule S, Mungall K, Norbertczak H, Rabbinowitsch E, Sanders M, Simmonds M, White B, Whithead S, Parkhill J. 2007. Genome sequence of a proteolytic (group I) Clostridium botulinum strain Hall A and comparative analysis of the clostridial genomes. Genome Res 17:1082–1092. [PubMed] 10.1101/gr.6282807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Marshall KM, Bradshaw M, Pellett S, Johnson EA. 2007. Plasmid encoded neurotoxin genes in Clostridium botulinum serotype A subtypes. Biochem Biophys Res Commun 361:49–54. [PubMed] 10.1016/j.bbrc.2007.06.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang Z, Hintsa H, Chen Y, Korkeala H, Lindstrom M. 2013. Plasmid-borne type E neurotoxin gene clusters in Clostridium botulinum strains. Appl Environ Microbiol 79:3856–3859. [PubMed] 10.1128/AEM.00080-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Franciosa G, Maugliani A, Scalfaro C, Aureli P. 2009. Evidence that plasmid-borne botulinum neurotoxin type B genes are widespread among Clostridium botulinum serotype B strains. PLoS One 4:e4829. [PubMed] 10.1371/journal.pone.0004829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang X, Maegawa T, Karasawa T, Kozaki S, Tsukamoto K, Gyobu Y, Yamakawa K, Oguma K, Sakaguchi Y, Nakamura S. 2000. Genetic analysis of type E botulinum toxin-producing Clostridium butyricum strains. Appl Environ Microbiol 66:4992–4997. [PubMed] 10.1128/AEM.66.11.4992-4997.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Marshall KM, Bradshaw M, Johnson EA. 2010. Conjugative botulinum neurotoxin-encoding plasmids in Clostridium botulinum. PLoS One 5:e11087. [PubMed] 10.1371/journal.pone.0011087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pilo P, Frey J. 2011. Bacillus anthracis: molecular taxonomy, population genetics, phylogeny and patho-evolution. Infect Genet Evol 11:1218–1224. [PubMed] 10.1016/j.meegid.2011.05.013 [DOI] [PubMed] [Google Scholar]
- 135.Koehler TM. 2009. Bacillus anthracis physiology and genetics. Mol Aspects Med 30:386–396. [PubMed] 10.1016/j.mam.2009.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fouet A. 2009. The surface of Bacillus anthracis. Mol Aspects Med 30:374–385. [PubMed] 10.1016/j.mam.2009.07.001 [DOI] [PubMed] [Google Scholar]
- 137.Bradley KA, Mogridge J, Mourez M, Collier RJ, Young JA. 2001. Identification of the cellular receptor for anthrax toxin. Nature 414:225–229. [PubMed] 10.1038/n35101999 [DOI] [PubMed] [Google Scholar]
- 138.Scobie HM, Rainey GJ, Bradley KA, Young JA. 2003. Human capillary morphogenesis protein 2 functions as an anthrax toxin receptor. Proc Natl Acad Sci USA 100:5170–5174. [PubMed] 10.1073/pnas.0431098100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Young JA, Collier RJ. 2007. Anthrax toxin: receptor binding, internalization, pore formation, and translocation. Annu Rev Biochem 76:243–265. [PubMed] 10.1146/annurev.biochem.75.103004.142728 [DOI] [PubMed] [Google Scholar]
- 140.Collier RJ. 2009. Membrane translocation by anthrax toxin. Mol Aspects Med 30:413–422. [PubMed] 10.1016/j.mam.2009.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Okinaka R, Cloud K, Hampton O, Hoffmaster A, Hill K, Keim P, Koehler T, Lamke G, Kumano S, Manter D, Martinez Y, Ricke D, Svensson R, Jackson P. 1999. Sequence, assembly and analysis of pX01 and pX02. J Appl Bacteriol 87:261–262. [PubMed] 10.1046/j.1365-2672.1999.00883.x [DOI] [PubMed] [Google Scholar]
- 142.Okinaka RT, Cloud K, Hampton O, Hoffmaster AR, Hill KK, Keim P, Koehler TM, Lamke G, Kumano S, Mahillon J, Manter D, Martinez Y, Ricke D, Svensson R, Jackson PJ. 1999. Sequence and organization of pXO1, the large Bacillus anthracis plasmid harboring the anthrax toxin genes. J Bacteriol 181:6509–6515. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Van der Auwera G, Mahillon J. 2005. TnXO1, a germination-associated class II transposon from Bacillus anthracis. Plasmid 53:251–257. [PubMed] 10.1016/j.plasmid.2004.08.004 [DOI] [PubMed] [Google Scholar]
- 144.Bourgogne A, Drysdale M, Hilsenbeck SG, Peterson SN, Koehler TM. 2003. Global effects of virulence gene regulators in a Bacillus anthracis strain with both virulence plasmids. Infect Immun 71:2736–2743. [PubMed] 10.1128/IAI.71.5.2736-2743.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mignot T, Mock M, Fouet A. 2003. A plasmid-encoded regulator couples the synthesis of toxins and surface structures in Bacillus anthracis. Mol Microbiol 47:917–927. [PubMed] 10.1046/j.1365-2958.2003.03345.x [DOI] [PubMed] [Google Scholar]
- 146.Kern JW, Schneewind O. 2008. BslA, a pXO1-encoded adhesin of Bacillus anthracis. Mol Microbiol 68:504–515. [PubMed] 10.1111/j.1365-2958.2008.06169.x [DOI] [PubMed] [Google Scholar]
- 147.Candela T, Mock M, Fouet A. 2005. CapE, a 47-amino-acid peptide, is necessary for Bacillus anthracis polyglutamate capsule synthesis. J Bacteriol 187:7765–7772. [PubMed] 10.1128/JB.187.22.7765-7772.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Van der Auwera GA, Andrup L, Mahillon J. 2005. Conjugative plasmid pAW63 brings new insights into the genesis of the Bacillus anthracis virulence plasmid pXO2 and of the Bacillus thuringiensis plasmid pBT9727. BMC Genomics 6:103. [PubMed] 10.1186/1471-2164-6-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kolstø A, Tourasse NJ, Økstad OA. 2009. What sets Bacillus anthracis apart from other Bacillus species? Annu Rev Microbiol 63:451–476. [PubMed] 10.1146/annurev.micro.091208.073255 [DOI] [PubMed] [Google Scholar]
- 150.Hoffmaster AR, Ravel J, Rasko DA, Chapman GD, Chute MD, Marston CK, De BK, Sacchi CT, Fitzgerald C, Mayer LW, Maiden MC, Priest FG, Barker M, Jiang L, Cer RZ, Rilstone J, Peterson SN, Weyant RS, Galloway DR, Read TD, Popovic T, Fraser CM. 2004. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc Natl Acad Sci USA 101:8449–8454. [PubMed] 10.1073/pnas.0402414101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wilson MK, Vergis JM, Alem F, Palmer JR, Keane-Myers AM, Brahmbhatt TN, Ventura CL, O'Brien AD. 2011. Bacillus cereus G9241 makes anthrax toxin and capsule like highly virulent B. anthracis Ames but behaves like attenuated toxigenic nonencapsulated B. anthracis Sterne in rabbits and mice. Infect Immun 79:3012–3019. [PubMed] 10.1128/IAI.00205-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Klee SR, Brzuszkiewicz EB, Nattermann H, Bruggemann H, Dupke S, Wollherr A, Franz T, Pauli G, Appel B, Liebl W, Couacy-Hymann E, Boesch C, Meyer FD, Leendertz FH, Ellerbrok H, Gottschalk G, Grunow R, Liesegang H. 2010. The genome of a Bacillus isolate causing anthrax in chimpanzees combines chromosomal properties of B. cereus with B. anthracis virulence plasmids. PLoS One 5:e10986. [PubMed] 10.1371/journal.pone.0010986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Leendertz FH, Ellerbrok H, Boesch C, Couacy-Hymann E, Matz-Rensing K, Hakenbeck R, Bergmann C, Abaza P, Junglen S, Moebius Y, Vigilant L, Formenty P, Pauli G. 2004. Anthrax kills wild chimpanzees in a tropical rainforest. Nature 430:451–452. [PubMed] 10.1038/nature02722 [DOI] [PubMed] [Google Scholar]
- 154.Grynberg M, Li Z, Szczurek E, Godzik A. 2007. Putative type IV secretion genes in Bacillus anthracis. Trends Microbiol 15:191–195. [PubMed] 10.1016/j.tim.2007.03.002 [DOI] [PubMed] [Google Scholar]
- 155.Green BD, Battisti L, Koehler TM, Thorne CB, Ivins BE. 1985. Demonstration of a capsule plasmid in Bacillus anthracis. Infect Immun 49:291–297. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Reddy A, Battisti L, Thorne CB. 1987. Identification of self-transmissible plasmids in four Bacillus thuringiensis subspecies. J Bacteriol 169:5263–5270. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Stenfors Arnesen LP, Fagerlund A, Granum PE. 2008. From soil to gut: Bacillus cereus and its food poisoning toxin. FEMS Microbiol Rev 32:579–606. [PubMed] 10.1111/j.1574-6976.2008.00112.x [DOI] [PubMed] [Google Scholar]
- 158.Bravo A, Gill SS, Soberon M. 2007. Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon 49:423–435. [PubMed] 10.1016/j.toxicon.2006.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler DR, Dean DH. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol Rev 62:775–806. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lereclus D, Agaisse H, Grandvalet C, Salamitou S, Gominet M. 2000. Regulation of toxin and virulence gene transcription in Bacillus thuringiensis. Int J Med Microbiol 290:295–299. [PubMed] 10.1016/S1438-4221(00)80024-7 [DOI] [PubMed] [Google Scholar]
- 161.Pardo-Lopez L, Soberon M, Bravo A. 2013. Bacillus thuringiensis insecticidal three-domain Cry toxins: mode of action, insect resistance and consequences for crop protection. FEMS Microbiol Rev 37:3–22. [PubMed] 10.1111/j.1574-6976.2012.00341.x [DOI] [PubMed] [Google Scholar]
- 162.Soberon M, Lopez-Diaz JA, Bravo A. 2013. Cyt toxins produced by Bacillus thuringiensis: a protein fold conserved in several pathogenic microorganisms. Peptides 41:87–93. [PubMed] 10.1016/j.peptides.2012.05.023 [DOI] [PubMed] [Google Scholar]
- 163.Doggett NA, Stubben CJ, Chertkov O, Bruce DC, Detter JC, Johnson SL, Han CS. 2013. Complete genome sequence of Bacillus thuringiensis serovar israelensis strain HD-789. Genome Announc 1:e01023–13. [PubMed] 10.1128/genomeA.01023-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Baum JA, Malvar T. 1995. Regulation of insecticidal crystal protein production in Bacillus thuringiensis. Mol Microbiol 18:1–12. [PubMed] 10.1111/j.1365-2958.1995.mmi_18010001.x [DOI] [PubMed] [Google Scholar]
- 165.Reyes-Ramirez A, Ibarra JE. 2008. Plasmid patterns of Bacillus thuringiensis type strains. Appl Environ Microbiol 74:125–129. [PubMed] 10.1128/AEM.02133-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gonzalez JM, Jr, Brown BJ, Carlton BC. 1982. Transfer of Bacillus thuringiensis plasmids coding for delta-endotoxin among strains of B. thuringiensis and B. cereus. Proc Natl Acad Sci USA 79:6951–6955. [PubMed] 10.1073/pnas.79.22.6951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gammon K, Jones GW, Hope SJ, de Oliveira CM, Regis L, Silva Filha MH, Dancer BN, Berry C. 2006. Conjugal transfer of a toxin-coding megaplasmid from Bacillus thuringiensis subsp. israelensis to mosquitocidal strains of Bacillus sphaericus. Appl Environ Microbiol 72:1766–1770. [PubMed] 10.1128/AEM.72.3.1766-1770.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wang P, Zhang C, Zhu Y, Deng Y, Guo S, Peng D, Ruan L, Sun M. 2013. The resolution and regeneration of a cointegrate plasmid reveals a model for plasmid evolution mediated by conjugation and oriT site-specific recombination. Environ Microbiol 15:3305–3318. [PubMed] 10.1111/1462-2920.12177 [DOI] [PubMed] [Google Scholar]
- 169.Hu X, Hansen BM, Yuan Z, Johansen JE, Eilenberg J, Hendriksen NB, Smidt L, Jensen GB. 2005. Transfer and expression of the mosquitocidal plasmid pBtoxis in Bacillus cereus group strains. FEMS Microbiol Lett 245:239–247. [PubMed] 10.1016/j.femsle.2005.03.012 [DOI] [PubMed] [Google Scholar]
- 170.Huang T, Liu J, Song F, Shu C, Qiu J, Guan X, Huang D, Zhang J. 2004. Identification, distribution pattern of IS231 elements in Bacillus thuringiensis and their phylogenetic analysis. FEMS Microbiol Lett 241:27–32. [PubMed] 10.1016/j.femsle.2004.09.037 [DOI] [PubMed] [Google Scholar]
- 171.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–W469. [PubMed] 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Dereeper A, Audic S, Claverie JM, Blanc G. 2010. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol 10:8. [PubMed] 10.1186/1471-2148-10-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yonogi S, Matsuda S, Kawai T, Yoda T, Harada T, Kumeda Y, Gotoh K, Hiyoshi H, Nakamura S, Kodama T, Ilda T. 2014. BEC, a novel enterotoxin of Clostridium perfringens found in human clinical isolates from acute gastroenteritis outbreaks. Infect Immun 82:2390-2399. [PubMed] 10.1128/IAI.01759-14 [DOI] [PMC free article] [PubMed] [Google Scholar]


