Abstract
Vitiligo is impacted by environmental triggers. We studied the contribution of the microbiome in FH mice, where depigmentation is mediated by tyrosinase-reactive T cells. Mice received oral antibiotics and were monitored for depigmentation. The microbiome was studied in fecal and skin samples by 16S rRNA analysis. Resulting T-cell distributions were evaluated. In untreated mice, pigment loss did not expand to the pelage, whereas mice in the ampicillin group were about 1/3 depigmented at 30 weeks. Contrary to models of autoimmunity that are less dependent on IFN-γ, ampicillin but not neomycin treatment correlated with accelerated disease and reduced bacteria in fecal pellets. Modified cytokine patterns in tissue and serum suggest a response that transcends the gut. Ampicillin-induced depigmentation was accompanied by gut but not skin dysbiosis, and reduced T cells in both sites. Neomycin induced a redistribution of gut T cells and accumulation of skin Tregs. This treatment spurred a Bacteroides-dominated population of fecal bacteria. Reduced diversity is prominent especially after ampicillin treatment, when the gut is dominated by Pseudomonas species. In line with current concepts relating the microbiome and the immune system, we predict that dietary measures might promote skin health and delay vitiligo onset.
Keywords: vitiligo, microbiome, Pseudomonas, T cells
INTRODUCTION
Vitiligo patients exhibit progressive skin depigmentation in response to environmental triggers. Depigmentation follows loss of melanocytes mediated by skin-infiltrating, cytotoxic T cells. Such T cells, found in genetically predisposed individuals, mediate responses towards melanosomal proteins (Eby et al., 2018, Palermo et al., 2005). Hereditary factors are increasingly well understood (Jin et al., 2016), and support an autoimmune disease etiology.
While environmental triggers have been correlated with induction of vitiligo, the mechanisms underlying their roles in pathogenesis (Jeon et al., 2014), or how these impact T-cell function (Garzorz et al., 2015), are not well understood. It is thought that chemical, emotional, and hormonal stressors trigger vitiligo through cellular stress pathways (Manga et al., 2016, Patel et al., 2017). Importantly, some patients list infection or antibiotics as etiologic factors, and H. pylori was implicated in vitiligo (Magen and Delgado, 2014), leading us to hypothesize that dysbiosis was a trigger for development of vitiligo.
Contributions of cutaneous bacteria to disease are well-acknowledged, including P. acnes to acne (Liu et al., 2015), and S. aureus to psoriasis (Chang et al., 2018) and atopic dermatitis (Seddon and Hughes, 2018). A breach of skin integrity offers microbes a port of entry to trigger innate, humoral, or cell-mediated immune responses (Prescott et al., 2017). Importantly, depigmented vitiligo lesions harbor markedly different microbes, suggesting an impact on intact skin as well (Ganju et al., 2016). Yet, bacteria in distant tissue sites may also be involved. Gut dysbiosis influences systemic immunity and has been implicated in a number of autoimmune conditions (Honda and Littman, 2016). While microbial diversity helps maintain immune homeostasis, individual species might elicit pathogenic responses (Duvallet et al., 2017) as exemplified by Ro60-producing commensal bacteria in lupus (Greiling et al., 2018).
Gut health was likewise shown to influence melanoma progression, and in the context of immune responses against melanocytes, melanoma and vitiligo represent different ends of a spectrum. Where the objective in melanoma is to enhance immune responses, in vitiligo the objective is to dampen them. In melanoma, Bifidobacterium promoted anti-tumor immunity in mice (Sivan et al., 2015) and was associated with greater anti-PD-1 responses in patients (Matson et al., 2018). Therefore, some bacterial strains might worsen vitiligo, as they enhance immunity to melanoma.
The mechanism linking bacterial colonization and immune responses is a subject of intense investigation. Humoral responses can result from bacterial infections, wherein antibodies target bacteria. Yet antibody responses to particular bacterial strains have also been shown to elicit autoimmunity via molecular mimicry to host antigens (Proal et al., 2013). One example was demonstrated in a seminal study of neomycin-induced segmented filamentous bacteria (SFB) contribution to arthritic ankle thickening in mice (Wu et al., 2010). This was also shown to occur in human autoimmune arthritis as a result of increased abundance of B. adolescentis within small intestine lamina propria (Tan et al., 2016).
Microbial peptides may also elicit CD8 T cell responses, though these responses are less understood (Tai et al., 2016). A better understanding of microbes impacting T-cell activation might reveal whether gut health impacts vitiligo development. To address this, we bred FH mice with progressive vitiligo. In these TCR transgenic mice, gradual depigmentation is mediated by cutaneous CD8 T cells (Gregg et al., 2010). Depigmentation follows recognition of murine tyrosinase269–276 (FMDGTMSQV) presented by HLA-A2D (Colella et al., 2000). Lesions develop symmetrically as in human vitiligo, and depigmentation likewise depends on CXCR3 and IFN-γ (Rashighi et al., 2014). Pups were subjected to antibiotics and subsequent changes to the microbiome and associated depigmentation were followed. Fecal specimens and intestinal, serum, spleen and skin samples were collected. Fecal and skin specimens were analyzed by 16SrRNA gene sequencing. T-cell distribution was followed in the skin and gut, and FACS and cytokine arrays were performed. Our studies indicate that antibiotic treatment and subsequent microbial dysbiosis impacts melanocyte-reactive T-cell function, suggesting dietary changes might likewise influence disease kinetics (Garcia-Larsen et al., 2018).
RESULTS
Antibiotic treatment impacts vitiligo development.
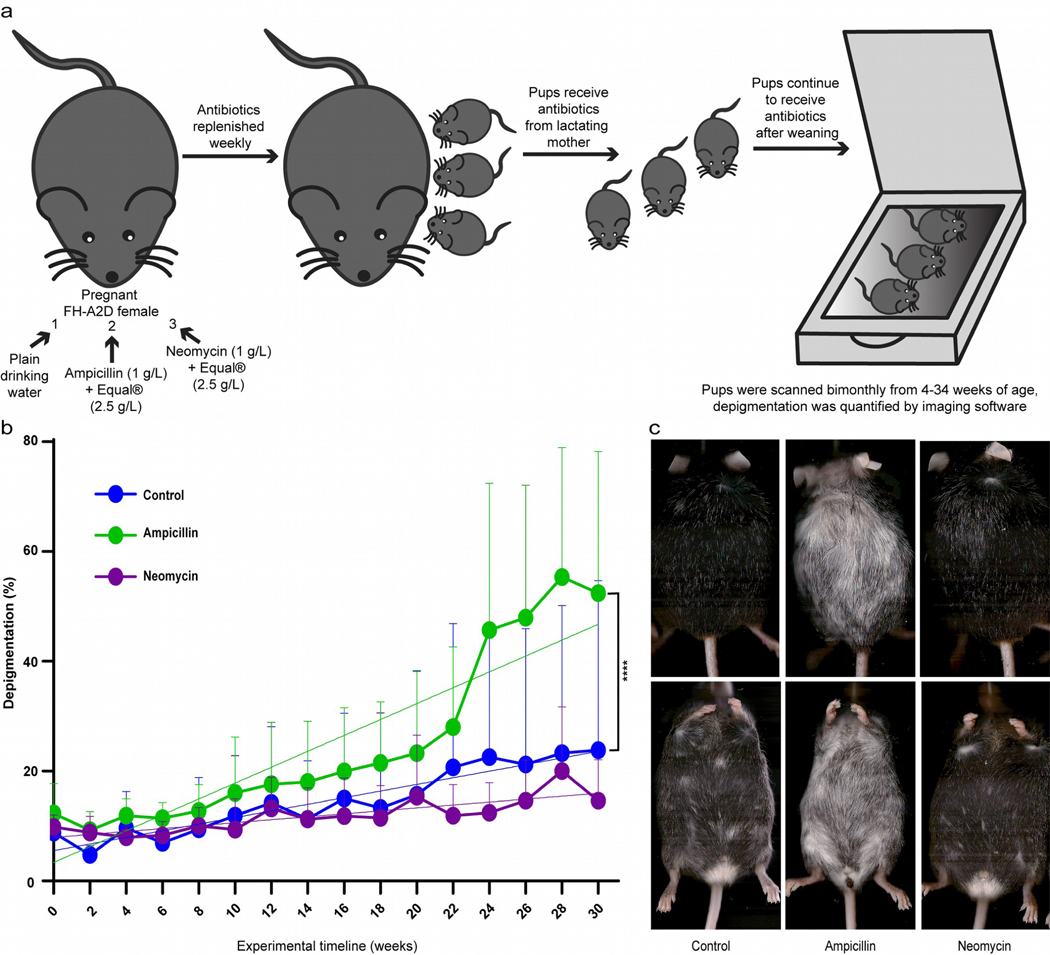
FH mice were treated and followed as in Fig. 1a. Testing the hypothesis H0 that slopes are equal and disease development is unrelated to antibiotic exposure, pups in fact developed vitiligo with different kinetics among treatments (Fig. 1b). Ampicillin accelerated vitiligo development (P<0.0001) whereas mice treated with neomycin trended towards delayed vitiligo (P=0.07). Examples in Fig. 1c include total depigmentation in an ampicillin-treated animal. Untreated animals (N=10) depigment at 0.6 +/− 0.16 %/wk, or 0.27 +/− 0.06% per week in neomycin treated mice (N=8). Meanwhile, vitiligo accelerated 2.4 fold to 1.45 +/− 0.14% in presence of ampicillin (N=10). Similar to human patients, depigmentation was discontinuous. Melanocytes were mostly lost in all treatment groups at end point, while faint TRP-2 expression was still observed in controls and melanized hairs remained in both neomycin and control groups (Fig. S1). Thus microbes might affect autoimmune responses to melanocytes. As long-term antibiotic use has been associated with increased reactive oxygen species (ROS) production and melanocyte stress in vitiligo has been associated with ROS, we also assessed oxidative stress in antibiotic treated FH mice. Increased depigmentation in ampicillin-treated mice was not ascribable to increased intestinal oxidative stress when comparing 4-hydroxy-2-nonenal and HSP70 expression in colon tissue (Fig. S2). Thus, we pursued the microbial dysbiosis as a potential contributing factor. As antibiotics can affect the microbiome in different sites, we next determined the microbial composition in both skin and gut.
Fig. 1. Systemic antibiotics alter vitiligo development.
(a) Experimental outline, showing triage into 3 groups treated with normal drinking water (control, N=10), or neomycin (N=8) or ampicillin (N=10). Treatment continued until 35 weeks of age. (b) Depigmentation was measured by scanning and image analysis every 2 weeks, starting at 4 weeks old. Antibiotic treatments resulted in significantly altered depigmentation at P<0.0001 for H0: slopes are equal. The slopes are 0.27 +/− 0.06 for neomycin, 0.6 +/− 0.16 for control mice and 1.45 +/−0.14 for ampicillin treated mice over the period of study. (c) Examples of terminal mouse scans. (****p<0.0001).
Systemic antibiotics primarily limit microbial diversity in the gut.
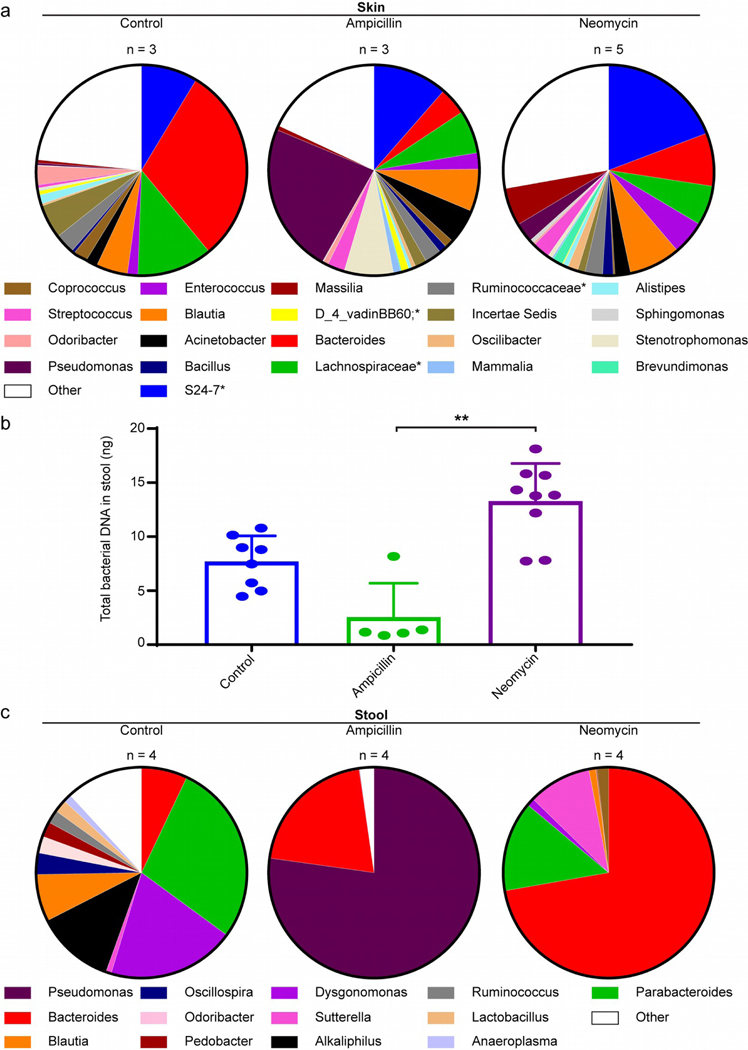
Next, we investigated if and where the microbiome was impacted. We isolated DNA from available skin samples and amplified 16S rRNA-encoding genes using degenerate primers. Depigmentation curves are in Fig. S3. Relative percentual abundance of microbial sequences is displayed under Fig. 2a for skin samples from controls (N=3), neomycin (N=5) and ampicillin-treated mice (N=3). Minimal differences among groups are overshadowed by differences within groups in Fig. S4a, except reduced Incertae Sedis (P=0.004) in skin from neomycin-treated mice. As its abundance diminished from 5.1% in controls to 1.1% among neomycin-treated mice without affecting depigmentation, no clinical impact could be assigned to this difference. Antibiotic-infused drinking water had little impact on cutaneous colonization, suggesting that autoimmune T cells attacking the skin are instead guided by more distant microbes. This prompted an evaluation of gut speciation and bacterial abundance for control and antibiotic treated mice (N=4 per group) to look for microbial species aligning with depigmentation. We compared bacterial abundance in excrements from control (N=8), ampicillin (N=5) and neomycin (N=9) treatment groups (Fig. 2b). Standard deviations apply to inter-group variation. Results trended towards reduced DNA content in ampicillin-treated mice and increased DNA content in neomycin-treated mice. Stool from neomycin treated mice contained 5-fold more DNA than ampicillin treated mice (P=0.0002). Statistical comparisons made are mentioned only when significant (P<0.05). For both antibiotics, microbial diversity was affected (Fig. 2c), resulting in reduced relative prevalence of Alkaliphilus (both P<0.0001), Dysgonomonas (both P<0.0001), Odoribacter (both P=0.0009), Oscillospira (both P=0.006), Pedobacter (P<0.0001 for ampicillin and P=0.0001 for neomycin) and Ruminococcus (P=0.003 for both) and for the ampicillin group only, Parabacteroides (P<0.0001). Sutterella species were reduced in ampicillin, and increased in neomycin-treated mice (both P=0.0003). After ampicillin treatment, the gut was primarily inhabited by Pseudomonas whereas neomycin favored Bacteroides outgrowth (P<0.0001). These differences are more consistent than in skin (Fig. S4b). Dominant species were Pseudomonas aeruginosa and Bacteroides vulgatus, respectively (Fig. S5). This suggests that specific species align with, and might launch or inhibit immune responses.
Fig. 2. Antibiotics affect microbial diversity in the gut and not in the skin.
(a) Isolates from total skin homogenates at 35 weeks were pre-enriched for microbial DNA before completing 16S rRNA analysis. Microbial diversity is maintained in the skin after antibiotic treatment. (b) Relative bacterial DNA content relative to reference strain R. productus determined by qPCR in samples from 3 control, 5 ampicillin-treated or 9 neomycin treated mice. The amounts significantly differed between ampicillin and neomycin groups, with concentrations trending towards a decrease and increase compared to controls, respectively. (c) Fecal DNA analysis reveals marked skewing of the microbiome. Ampicillin treatment is associated with enrichment of Pseudomonas species in the gut, whereas neomycin treatment enriched for the Bacterioides genus. (*Genus not identifiable).
Reduced microbial diversity in the gut correlates with T-cell redistribution.
The consequences of antibiotics for vitiligo development oppose those reported for autoimmune arthritis (Wu et al., 2010), suggesting that individual antibiotics favor different subsets of lymphocytes, resulting in neomycin-induced B cell activity in autoimmune arthritis (Wu et al., 2010) and ampicillin-supported T cell-mediated cytotoxicity in vitiligo (Le Poole and Luiten, 2008), quantified here. Examples in Fig. 3a–c demonstrate extensive depigmentation in response to ampicillin while T-cell abundance dwindled by 68% (P=0.047 and 0.014 compared to control and neomycin, respectively; N=3) (Fig. 3g). Remaining T cells were sparsely distributed over non-pigmented hair follicles, quantified in Fig. S6. Moreover, Tregs (Fig. 3d–f) were 16-fold more abundant (P=0.02) in skin after neomycin treatment compared to untreated mice (Fig. 3h), and may help control vitiligo. This might explain skin T-cell abundance in neomycin-treated mice, with pigmentation largely intact. We also evaluated gut T cells in sections with greatest microbial diversity. Initial screening (Fig. S7) revealed differences in T-cell infiltration primarily in the ileum. At N=3, CD4 T cells were 51% reduced (P=0.04) by neomycin and 83% (P=0.004) by ampicillin treatment (Fig. 4a). CD4 T cells were peripherally located within villi of control mice (Fig. 4b), but central in villi after ampicillin (Fig. 4c) and especially neomycin treatment (Fig. 4d). Cytotoxic T cells in Fig. 4e were likewise 65% reduced (P=0.001) by neomycin, and 92% (P=0.0002) by ampicillin treatment. Their distribution is affected similar to CD4 T cells (Fig. 4f–h) making less contact with gut contents after neomycin treatment. Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) revealed that apoptosis was not increased in the ileum of ampicillin treated mice (Fig. S8). Overall, antibiotics impact gut T-cell distribution and abundance, suggesting that viability or mobility is affected, with possible relocation to sites of antigen expression.
Fig. 3. Ampicillin mediates T-cell depletion from the skin.
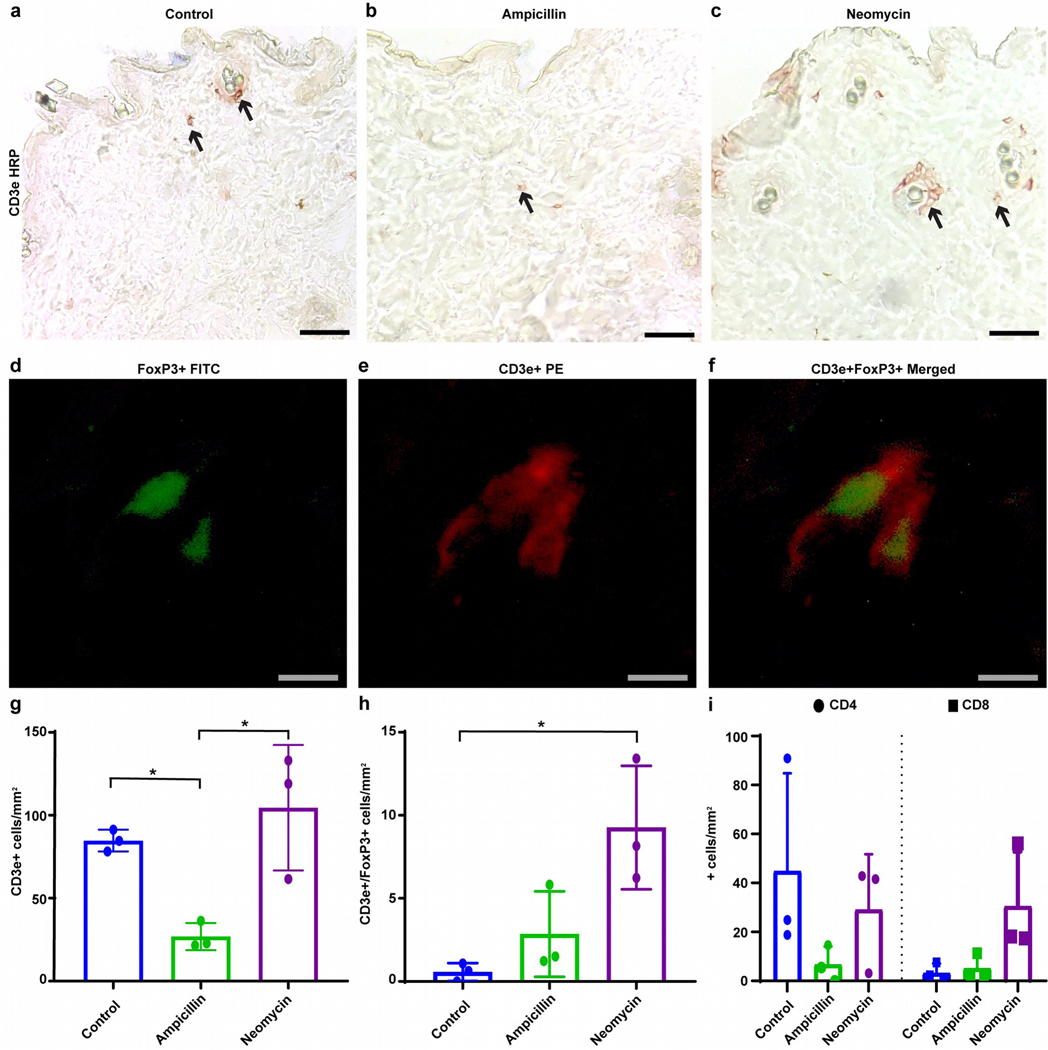
Skin tissue was evaluated for T-cell infiltration using antibodies to CD3e, showing examples of (a) control skin, and skin from (b) ampicillin and (c) neomycin treated mice. Representative staining for Treg in skin used to quantify positive (d) FoxP3 cells in FITC (green), (e) CD3e cells in PE (red), (f) and merged FoxP3 (green) and CD3e (red) cells are shown. Skin quantifications ± SD (N=3 per group) from control or ampicillin or neomycin treated mice for (g) CD3e, (h) FoxP3/CD3e, and (i) CD4 and CD8 are shown. (Scale bar = (a-c) 50μm, (d-f) 10μm, *p<0.05).
Fig. 4. Antibiotic treatment redistributes T cells in the gut.
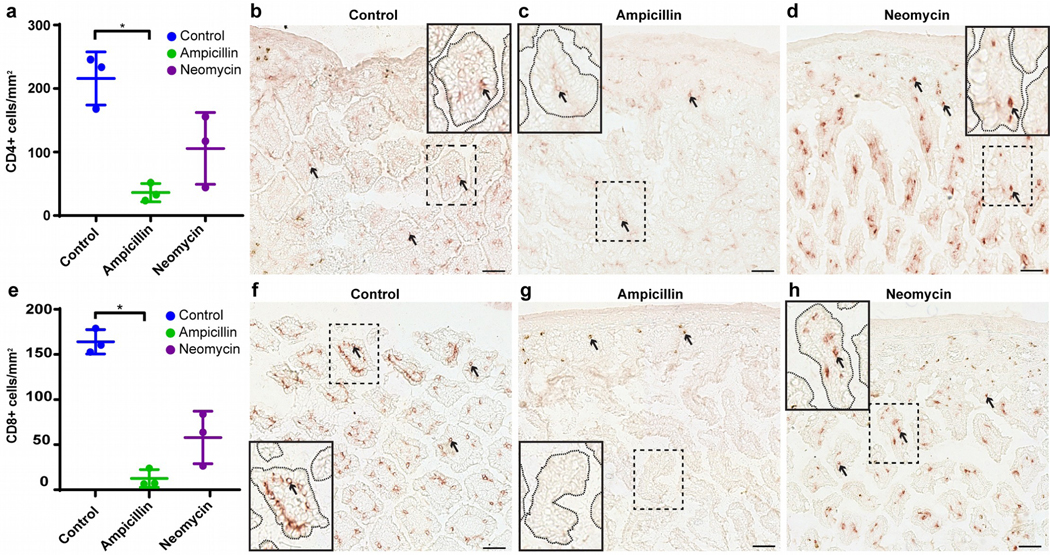
(a) CD4+ T cells are significantly reduced in available ileum samples of ampicillin-treated mice (N=3). Examples are from (b) control, (c) ampicillin and (d) neomycin treated mice, respectively. (e) Meanwhile, CD8+ T cells are significantly reduced (N=3) compared to (f) control tissue by (g) ampicillin but not (h) neomycin treatment. Inserts show marked redistribution of CD4+ and CD8+ T cells within the villi. Arrows: examples of stained T cells. (Scale bar = 50μm (b-d, f-h), *p<0.05).
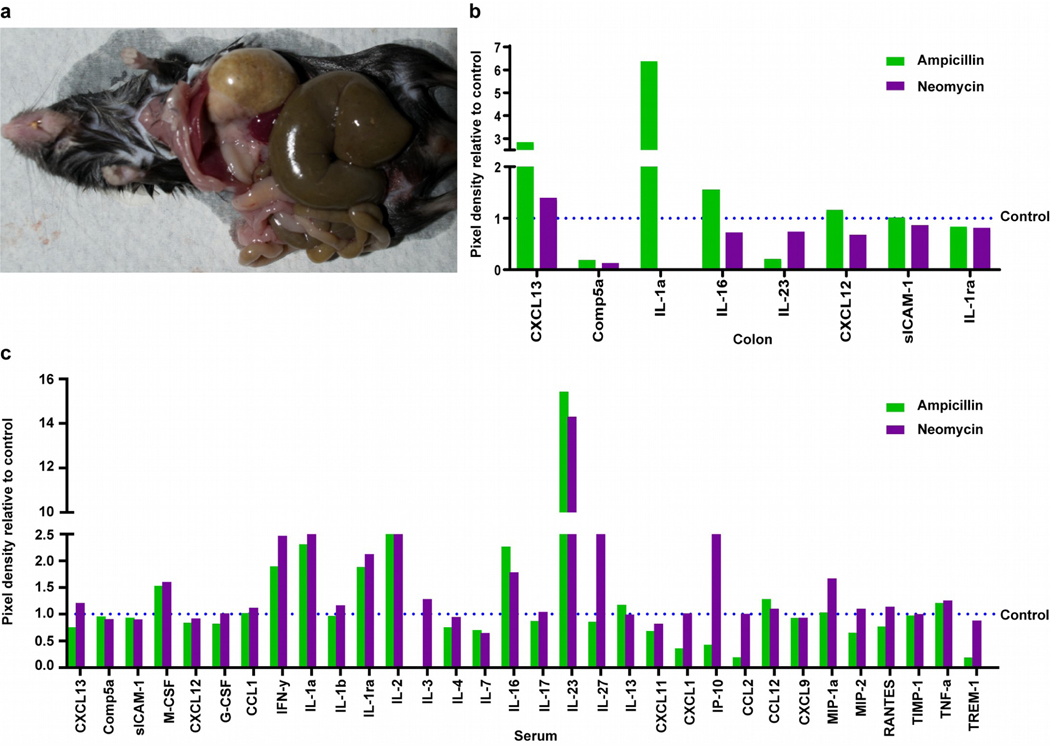
Prolonged oral ampicillin treatment affects systemic immune molecules.
Oral antibiotics affected vitiligo development, suggesting bacteria might signal to tissues distal from the gut. Indeed, ampicillin-exposed mice exhibited distended abdomens with expansion of the GI tract, particularly the cecum (Fig. 5a), suggestive of altered permeability. To identify immune status in the gut and other tissues, we first assessed cytokine production in the gut by microarray displayed in Fig. 5b. Antibiotics reduced complement 5A levels by 82% for ampicillin, and 88% for neomycin treatment. IL-23 appeared selectively reduced by 80% in ampicillin-treated samples while CXCL13 was 2.8 fold greater. The greatest change was in IL-1α, increased 6.3 fold in ampicillin and absent in neomycin treated mice, distinguishing responses to either antibiotic. To evaluate systemic changes in immune responsiveness following prolonged antibiotics, we performed the array on pooled serum cytokines (Fig. 5c). Antibiotic treatment was associated with a <2.5 fold upregulation of M-CSF, IL-16, IL-1α, IL-1ra, IFN-γ, and IL-2. Moreover, a ~15-fold increase in IL-23 was observed. Antibiotics thus appeared to elicit inflammatory cytokine responses. Ampicillin-enhanced depigmentation might however be better explained by cytokines differentially released after either antibiotic, including abolished expression of IL-3 possibly impacting pDC activation, 65% reduced neuroprotective CXCL1, 80% reduced Th2 cytokine CCL-2, 75% reduced innate pro-inflammatory TREM-1, or by vitiligo biomarker CXCL-10 reduced by ampicillin yet 2.5 fold increased by neomycin. Thus accelerated depigmentation accompanies greater inflammatory and reduced type 2 cytokines.
Fig.5. Gut dysbiosis is accompanied by tissue damage and altered cytokine responses in the gut.
(a) Enlarged cecum, liver and stomach in an ampicillin treated mouse, reflecting tissue damage. (b) A sandwich array performed to identify cytokines up and downregulated in 4 pooled colon tissue homogenates per group. The dotted line represents the relative value for control samples. (c) Array performed on serum to evaluate systemic changes (4 pooled samples/ group), with less IL-3, CXCL1, CXCL10 (=IP-10), CCL2 and TREM-1, and more IL-27 and CXCL10 in serum from ampicillin versus neomycin samples, respectively. (**p<0.01).
T-cell functionality is impacted by antibiotic treatment.
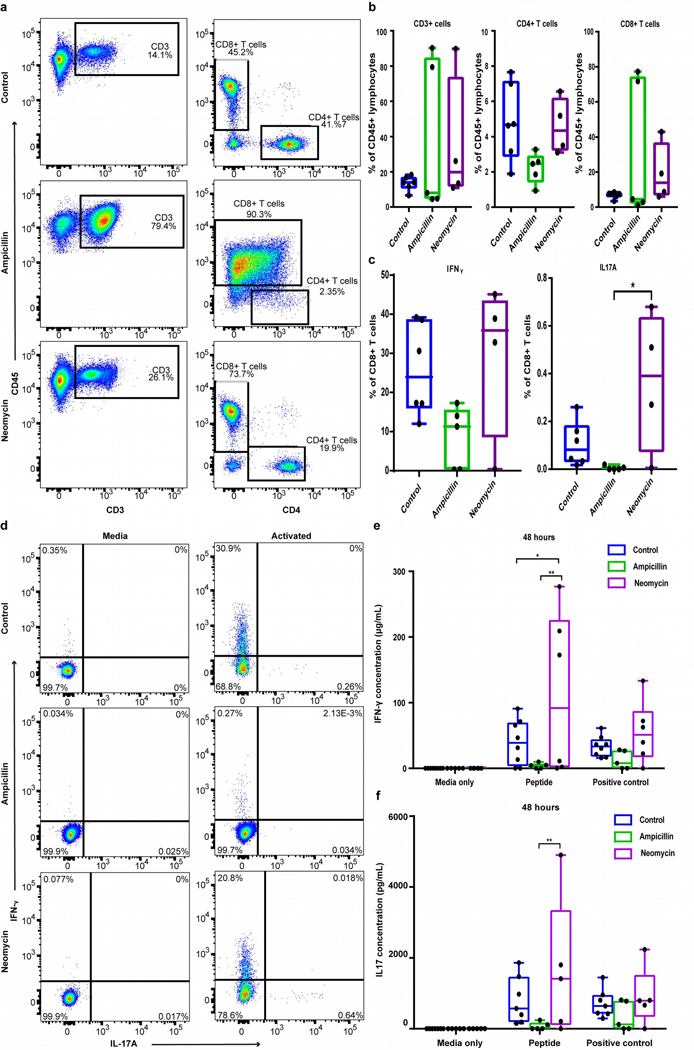
Given the role of T cells in depigmentation and altered intestinal cytokine responses, antibiotic treatment might alter systemic T-cell functionality. We evaluated this by multiparameter FACS analysis (gating shown in Fig. S9). Unstimulated splenocytes showed T cells representing up to 90% of CD45+ leukocytes in some antibiotic-treated mice (Fig. 6a). This was driven by increased CD8+ T cells in those animals, which in the ampicillin-treated mice co-expressed CD4, suggestive of chronic activation (Nascimbeni et al., 2004). Overall, a trend towards proportionately fewer CD4 T cells appeared in ampicillin-treated mice (Fig. 6b). After non-specific stimulation, CD8+ T cells from ampicillin-treated mice appeared refractory, trending towards 65% less IFN-γ-producing cells accompanied by elimination of IL-17A-producing cells compared to neomycin treatment (p=0.014; Fig. 6c). Similar patterns were observed in response to murine tyrosinase269–276 or CD3/CD28 T cell activator beads. Minimal IFN-γ was found after ampicillin treatment, while neomycin enhanced IFN-γ compared to ampicillin-treated (p=0.004) and control mice (p=0.04; Fig. 6d, e). IL-17 was elevated in neomycin- versus ampicillin-treated mice (p=0.004; Fig. 6d, f). Smaller differences appeared after 24 hours (not shown). As healthy FH T cells respond with robust cytokine release, these data point to dysfunctional and exhausted T cells resulting from ampicillin treatment following enhanced depigmentation in these mice.
Fig. 6. Antibiotic treatment skews T-cell distribution and functionality.
(a) Example FACS plots and (b) bar graphs of total T cells, CD4+ and CD8+ T cells among CD45+ splenocytes. (c) IFN-γ and IL-17A were measured in CD8+ T cells from antibiotic-treated or control mice after 5 hrs incubation with or without leukocyte activation cocktail (LAC). (d) IFN-γ and IL-17A+ cells in live CD8+ T cells among LAC or media-treated controls cells. (e) IFN-γ and (f) IL-17 were measured by ELISA in splenocyte supernatants from antibiotic-treated and control mice stimulated with media, Tyrosinase369–377 (30μg/ml) or CD3/CD28 activator beads for 48 hrs. (*p<0.05, **p<0.01).
DISCUSSION
Applying antibiotics and manipulating the microbiome profoundly affected vitiligo in our mice. Though similar observations were reported in antibody-mediated autoimmunity (Lopez et al., 2016), humoral responses might directly target bacteria whereas cytotoxic T cells cannot. A mechanism whereby loss of bacterial diversity results in accelerated cytotoxic T-cell responses to self- antigens is less well understood. The influence of microbes on immunity and health is attracting attention (Ash and Mueller, 2016), with antibiotics offering a valuable investigative tool (Blaser, 2016). Studies in mice are justified herein by similarity to the human gut microbiome (Krych et al., 2013), with specific attention to the ileum where bacterial adherence and T-cell activation have the greatest impact (Schnupf et al., 2017).
Another question is how gut microbes impact immune responses in distant sites such as skin (Salem et al., 2018). Responses might rather be due to skin commensals, which can impact non-classical MHC I-restricted T cells (Linehan et al., 2018). Ampicillin can induce ROS formation in bacteria, and even human cells which might contribute to gut permeability (Kalghatgi et al., 2013). ROS formation is relevant to vitiligo, where increased cytokine production correlates with increased ROS and reduced antioxidants (Mitra et al., 2017). And vitiligo patient studies indicate that depigmentation is associated with some loss of bacterial diversity and lesional under-representation of Corynebacteriaceae, whereas the classes of Gammaproteobacteria and Flavobacteria are overrepresented (Ganju et al., 2016). Non-lesional skin showed similarity to healthy controls (Grice et al., 2008). These findings incentivize further studies into a role for microbes in vitiligo development.
There is precedent for gut microbes to associate with cutaneous autoimmunity, as in psoriasis (Byrd et al., 2018). There is also evidence that antibiotic treatment reduces gut bacteria and increases shedding of bacterial components (Vazquez-Mendoza et al., 2018). In light of these studies, we propose that ampicillin use allows for outgrowth of proinflammatory gut strains and/or provides antigens that can be taken up by gut dendritic cells to activate T cells against melanocytes. Furthermore, our mice displayed inflammatory cytokine patterns in the gut. This might help activate DCs and T cells to induce autoreactive immune responses (Wekerle, 2017).
This would imply that bacterial homogenates might share antigenic epitopes with self-antigens (Li et al., 2018), such as Ro60 ortholog producing commensal bacteria (Greiling et al., 2018). Indeed, after antibiotic treatment, vitiligo ensued only in mice expressing the tyrosinase-reactive T-cell receptor transgene and HLA-A2D, suggesting that vitiligo develops only if self-reactive T cells can respond to peptide-HLA complexes. Meanwhile, unresponsive T cells found in some ampicillin-treated mice suggest that melanocyte-responsive T cells became exhausted or apoptotic after melanocyte elimination (Ekmekciu et al., 2017).
Immune responses in vitiligo are similar to alopecia areata. There, gut permeability was linked to peripheral inflammation and tissue destruction (Borde and Astrand, 2018). In fact, some patients exhibited improvement in their condition following treatment for Clostridium difficile (Rebello et al., 2017). C. difficile infections correlate with reduced microbial diversity and greater host DNA content in stool, suggestive of reduced integrity of the gut lining (Vincent et al., 2015). Meanwhile, in mice monocolonized with Lactobacillus murinus, resulting biotin depletion was held responsible for alopecia development (Hayashi et al., 2017).
Neomycin treatment might offer some protection from vitiligo. Interestingly, prenatal neomycin is likewise associated with protection from diabetes development in NOD mice (Hu et al., 2015). We observed marked redistribution of T cells in the ileum of neomycin treated mice. Moreover, Tregs were overabundant in pre-vitiliginous skin. As tolerogenic differentiation of T cells can be supported by the gut microbiota (Lui et al., 2016), Treg differentiation might be favored by neomycin. This is certainly desirable in vitiligo to counter the contribution of IFNγ (Le Poole and Mehrotra, 2017). Of note, increased abundance of Treg coincides with reduced abundance of cutaneous Incertae Sedis. Meanwhile mucosal-associated invariant T cells (MAIT cells) could play a role in connecting gut dysbiosis and autoimmunity, as these cells respond to bacterial metabolites while mounting both antimicrobial and effector T cell functions, as suggested for several autoimmune conditions including multiple sclerosis (Buscarinu et al., 2018).
Serum cytokine profiles would indicate that antibiotics, particularly ampicillin, support inflammation. Of particular interest are the opposing changes in CXCL10, a chemokine central to cutaneous T-cell recruitment (Rashighi et al., 2014), melanocyte viability (Tulic et al., 2019) and depigmentation. Its levels were upregulated in neomycin treated, and downregulated in ampicillin treated mice. Though CXCL10 levels do not reflect depigmentation levels, they might align with ongoing autoreactivity by rerouting T cells to sites other than skin.
The presence of Pseudomonas aeruginosa correlated with accelerated disease. This species is also more frequent in ulcerative colitis (Walujkar et al., 2018). P. aeruginosa has been detected in the circulation and is an opportunistic pathogen. Its overrepresentation might contribute to gut permeability after ampicillin treatment (Yu and Martin, 2000). Bacteroides species were associated with delayed autoimmune responses to neomycin treatment. Bacteroides gut colonization can help prevent inflammatory bowel disease (Samborski and Grzymislawski, 2015).
Diversity is likewise important for anti-tumor responses. An interesting dilemma exists wherein a healthy gut promotes anti-melanoma responses, as does vitiligo. This suggests that particular species might be enriched in vitiligo development, while promoting vitiligo development and activation of anti-melanoma T cells. Therefore, our findings might inform future melanoma treatment. The use of probiotics could prove therapeutic, yet the preferred probiotic preparation might vary between both conditions. For example, Bifidobacterium adolescentis is an off-the-shelf probiotic equivalent to murine segmented filamentous bacteria (Tan et al., 2016). The latter were however shown to drive pathogenic Th17 responses and accelerate arthritis (Wu et al., 2010). Yet Th17 can also drive desirable anti-melanoma responses (Bowers et al., 2017). An exopolysaccharide preparation from probiotic Bacillus subtilis can however potentially limit CD8+ T-cell activation by inducing the development of M2 macrophages (Paynich et al., 2017), which might benefit vitiligo patients.
As nutrition, gut health and immune responses are intimately linked (Kau et al., 2011), our findings suggest that diets favoring a healthy gut might delay vitiligo onset or expansion (Grimes and Nashawati, 2017), and may prompt studies into the impact of nutrients on depigmentation. Similarly, fecal transplants, a popular remedy for dysbiosis, could prove therapeutic in vitiligo (Khanna, 2018).
In sum, vitiligo develops in individuals with melanocyte-reactive T cells that have been activated against self through some triggering event. We hypothesize that gut dysbiosis is one such trigger, and here we provide some evidence that altered gut microbiota can drive or delay depigmentation in mice depending upon which species are present.
MATERIALS AND METHODS
Antibiotic Administration
FH-A2D mice (Gregg et al., 2010) were maintained under protocols approved by Loyola’s Institutional Animal Care and Use Committee following the Guide. Mice were administered antibiotics upon pregnancy and litters were maintained on ampicillin or neomycin, or left untreated (n = 10 for control (6♂, 4♀) and ampicillin (5♂, 5♀), n = 8 for neomycin (5♂, 3♀) groups). Antibiotics were administered as 1 g/L of ampicillin sodium salt (Sigma-Aldrich, St. Louis, MO) or 1 g/L of neomycin sulfate (Fisher BioReagents, Pittsburgh, PA), combined with 2.5 g/L of Equal® Sweetener in drinking water (Wu et al., 2010). Pups continued antibiotic treatment via lactation and post-weaning. Water was exchanged weekly, keeping stocks at −20°C for up to one month.
Measuring Depigmentation
From 4 weeks of age, pups were scanned biweekly to monitor depigmentation. Depigmentation was measured by flatbed scanning (Hewlett-Packard, Palo Alto, CA) under isoflurane anesthesia. Using Adobe Photoshop software (Adobe Systems, San Jose, CA) luminosity was measured (Denman et al., 2008) as explained in the supplement.
Immunostaining
Ileum, colon and abdominal skin samples were frozen in Optimal Cutting Temperature Compound (Sakura Finetek, Torrance, CA). Eight μm cryosections were cut (Leica, Wetzlar, Germany). Total T cells were detected by pan-T-cell marker CD3. For CD3, CD4, CD8a, and TRP-2 staining, sections were acetone-fixed. For FoxP3/CD3 staining, fixed sections were permeabilized using True-Nuclear Transcription factor buffer (BioLegend, San Diego, CA). Sections were treated with SuperBlock (ScyTek Laboratories, Logan, UT). TUNEL staining was performed following manufacturer’s instructions of the ApopTag Detection Kit (Sigma-Aldrich). Staining protocols and antibodies are in the supplemental file. Sections were imaged on a Revolve microscope (Echo Laboratories, San Diego, CA) or TissueFAXS PLUS imaging system (TissueGnostics). Cells were quantified using Adobe Photoshop software.
Immunoblotting
Proteins from snap frozen colon samples were combined with Laemmli sample buffer (Bio-Rad Laboratories, Hercules, CA) and 2-Mercaptoethanol (Sigma-Aldrich) and denatured before loading onto a 4–20% Mini-PROTEAN TGX gel (Bio-Rad), and transferred to a polyvinylidene difluoride (PVDF) membrane (Sigma-Aldrich) for immunostaining outlined in the supplement.
Bacterial DNA Isolation and analysis
Fecal samples were collected from available mice scanned for 8 to 26 weeks. Collection included mice of comparable ages to represent each treatment group. These and abdominal skin samples were stored at −80°C until use. DNA was collected from fecal samples using QIAmp DNA stool mini kits (Qiagen, Hilden, Germany). Skin DNA was purified using DNAeasy Blood and Tissue Kits (Qiagen), following manufacturer’s instructions. (Eu)bacterial primers (Barman et al., 2008) were used to estimate bacterial content in fecal DNA (selected from randomized mice independent of depigmentation) by qRT-PCR (Applied Biosystems, Foster City, CA) using reference strain R. productus for ΔΔCt analysis. DNA samples from feces and skin were submitted for 16S rRNA sequencing at UIC’s Research Informatics Core (RIC), performing basic processing, annotation and quantification of amplicon sequence data using a primer pair consisting of 515 forward: ACACTGACGACATGGTTCTACAGTGYCAGCMGCCGCGGTAA and 806 reverse: TACGGTAGCAGAGACTTGGTCTGGACTACNVGGGTWTCTAAT. Sequencing readouts were analyzed using BaseSpace online software (Illumina, San Diego, CA). A detailed protocol for 16S rRNA sequencing can be found in the supplement. See the data availability statement for location of raw data.
In vitro splenocyte stimulation
Cryopreserved splenocytes were thawed into RMPI 1640 with 10% FBS, 1% antibiotics, 50μM 2-mercaptoethanol and 20μg/ml DNAseI (Thermo Fisher Scientific). A million splenocytes were stimulated for 5, 24 or 48 hours with or without 30μg/ml murine tyrosinase369–377 (GenScript), Leukocyte activation cocktail (BD Biosciences; 5 hours only) or 3:1 Dynabeads Mouse T-Activator CD3/CD28 (Thermo Fisher Scientific) for 24 and 48 hours, adding brefeldin A for the last 5. Supernatants were used in ELISAs. Cells were prepared for flow cytometry.
Flow cytometry
Before surface staining, cells were incubated with mouse Fc Block (Biolegend) and LIVE/DEAD Fixable Near IR Dead Cell dye (Thermo Fisher Scientific) per manufacturer’s instructions. Antibodies are in the supplement. Data were acquired on a BD FACSymphony flow cytometer, and analyzed using FlowJo v10.3.0 (FlowJo LLC, OR, USA).
Cytokine ELISAs
Supernatants from 24 and 48hr splenocyte stimulations were used in murine IFN-γ and IL-17 Quantikine ELISA kits (R & D Systems, MN, USA) per manufacturer’s instructions. Data were acquired on a Synergy HT reader (Biotek, VT, USA) equipped with Gen5 v1.08 (Biotek, VT, USA) and analyzed using Prism v7.03 (GraphPad Software, CA, USA).
Cytokine Array Analysis
Cryopreserved colon was homogenized using the BeadBug Microtube Homogenizer and 1.5 mm Triple-Pure High Impact Zirconium pre-filled tubes (Benchmark Scientific, Sayreville, NJ) with T-PER tissue protein extraction reagent (ThermoFisher Scientific) and protease inhibitor cocktail (ThermoFisher Scientific). Colon proteins were quantified using the Pierce BCA Protein Assay Kit per manufacturer’s protocol (ThermoFisher Scientific), and analyzed using Proteome Profiler Mouse Cytokine Array Kits (R & D Systems, Minneapolis, MN) per manufacturer’s protocol. Twentyfive μL of serum per mouse (N=4) was pooled by treatment. Membranes were imaged on a LAS-3000 luminescent imager (Fujifilm, Tokyo, Japan) and analyzed using ImageJ software.
Statistical analysis
The data were evaluated by one way analysis of variance accounting for different variances across the treatment groups, with post-hoc Tukey-Kramer comparisons. Statistical significance is represented as *p<0.05, ** p<0.01, *** p<0.001 or ****p<0.0001.
Supplementary Material
ACKNOWLEDGEMENTS
Research was supported in part by NIH RO1 AR057643 to CLP, and by Cancer Center Support Grant (NCI CA060553) to Northwestern’s Flow Cytometry and Imaging Cores.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
CRediT STATEMENT
Conceptualization, I.C.L.P. and K.K.; Methodology, I.C.L.P. and K.K.; Formal Analysis, A.W.R.; Investigation, E.R.D., C.C., Z.M., and S.A.; Resources, V.H.E.; Writing – Original Draft, I.C.L.P; Writing – Review & Editing, I.C.L.P, V.H.E., E.R.D., and C.C.; Visualization, E.R.D. and C.C.; Supervision, I.C.L.P.; Project Administration, I.C.L.P.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DATA AVAILABILITY STATEMENT
Metagenome datasets have been made available on NCBI’s Sequence Read Archive via BioProject accession number: PRJNA559873.
REFERENCES
- Ash C, Mueller K. Manipulating the Microbiota. Science (New York, NY) 2016;352(6285):530–1. [DOI] [PubMed] [Google Scholar]
- Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, et al. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infection and immunity 2008;76(3):907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser MJ. Antibiotic use and its consequences for the normal microbiome. Science (New York, NY) 2016;352(6285):544–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde A, Astrand A. Alopecia areata and the gut-the link opens up for novel therapeutic interventions. Expert opinion on therapeutic targets 2018;22(6):503–11. [DOI] [PubMed] [Google Scholar]
- Bowers JS, Nelson MH, Majchrzak K, Bailey SR, Rohrer B, Kaiser AD, et al. Th17 cells are refractory to senescence and retain robust antitumor activity after long-term ex vivo expansion. JCI insight 2017;2(5):e90772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscarinu MC, Romano S, Mechelli R, Pizzolato Umeton R, Ferraldeschi M, Fornasiero A, et al. Intestinal Permeability in Relapsing-Remitting Multiple Sclerosis. Neurotherapeutics : the journal of the American Society for Experimental NeuroTherapeutics 2018;15(1):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nature reviews Microbiology 2018;16(3):143–55. [DOI] [PubMed] [Google Scholar]
- Chang HW, Yan D, Singh R, Liu J, Lu X, Ucmak D, et al. Alteration of the cutaneous microbiome in psoriasis and potential role in Th17 polarization. Microbiome 2018;6(1):154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colella TA, Bullock TN, Russell LB, Mullins DW, Overwijk WW, Luckey CJ, et al. Self-tolerance to the murine homologue of a tyrosinase-derived melanoma antigen: implications for tumor immunotherapy. The Journal of experimental medicine 2000;191(7):1221–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denman CJ, McCracken J, Hariharan V, Klarquist J, Oyarbide-Valencia K, Guevara-Patino JA, et al. HSP70i accelerates depigmentation in a mouse model of autoimmune vitiligo. The Journal of investigative dermatology 2008;128(8):2041–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvallet C, Gibbons SM, Gurry T, Irizarry RA, Alm EJ. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nature communications 2017;8(1):1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eby JM, Smith AR, Riley TP, Cosgrove C, Ankney CM, Henning SW, et al. Molecular properties of gp100-reactive T-cell receptors drive the cytokine profile and antitumor efficacy of transgenic host T cells. Pigment cell & melanoma research 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekmekciu I, von Klitzing E, Fiebiger U, Escher U, Neumann C, Bacher P, et al. Immune Responses to Broad-Spectrum Antibiotic Treatment and Fecal Microbiota Transplantation in Mice. Frontiers in immunology 2017;8:397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganju P, Nagpal S, Mohammed MH, Nishal Kumar P, Pandey R, Natarajan VT, et al. Microbial community profiling shows dysbiosis in the lesional skin of Vitiligo subjects. Scientific reports 2016;6:18761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Larsen V, Ierodiakonou D, Jarrold K, Cunha S, Chivinge J, Robinson Z, et al. Diet during pregnancy and infancy and risk of allergic or autoimmune disease: A systematic review and meta-analysis. PLoS medicine 2018;15(2):e1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzorz N, Alsisi M, Todorova A, Atenhan A, Thomas J, Lauffer F, et al. Dissecting susceptibility from exogenous triggers: the model of alopecia areata and associated inflammatory skin diseases. Journal of the European Academy of Dermatology and Venereology : JEADV 2015;29(12):2429–35. [DOI] [PubMed] [Google Scholar]
- Gregg RK, Nichols L, Chen Y, Lu B, Engelhard VH. Mechanisms of spatial and temporal development of autoimmune vitiligo in tyrosinase-specific TCR transgenic mice. Journal of immunology (Baltimore, Md : 1950) 2010;184(4):1909–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiling TM, Dehner C, Chen X, Hughes K, Iniguez AJ, Boccitto M, et al. Commensal orthologs of the human autoantigen Ro60 as triggers of autoimmunity in lupus. Sci Transl Med 2018;10(434). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Renaud G, Young AC, Bouffard GG, Blakesley RW, et al. A diversity profile of the human skin microbiota. Genome research 2008;18(7):1043–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes PE, Nashawati R. The Role of Diet and Supplements in Vitiligo Management. Dermatologic clinics 2017;35(2):235–43. [DOI] [PubMed] [Google Scholar]
- Hayashi A, Mikami Y, Miyamoto, Kamada N, Sato T, Mizuno S, et al. Intestinal Dysbiosis and Biotin Deprivation Induce Alopecia through Overgrowth of Lactobacillus murinus in Mice. Cell reports 2017;20(7):1513–24. [DOI] [PubMed] [Google Scholar]
- Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature 2016;535(7610):75–84. [DOI] [PubMed] [Google Scholar]
- Hu Y, Peng J, Tai N, Hu C, Zhang X, Wong FS, et al. Maternal Antibiotic Treatment Protects Offspring from Diabetes Development in Nonobese Diabetic Mice by Generation of Tolerogenic APCs. Journal of immunology (Baltimore, Md : 1950) 2015;195(9):4176–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon IK, Park CJ, Lee MH, Lee DY, Kang HY, Hann SK, et al. A Multicenter Collaborative Study by the Korean Society of Vitiligo about Patients’ Occupations and the Provoking Factors of Vitiligo. Annals of dermatology 2014;26(3):349–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Andersen G, Yorgov D, Ferrara TM, Ben S, Brownson KM, et al. Genome-wide association studies of autoimmune vitiligo identify 23 new risk loci and highlight key pathways and regulatory variants. Nature genetics 2016;48(11):1418–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalghatgi S, Spina CS, Costello JC, Liesa M, Morones-Ramirez JR, Slomovic S, et al. Bactericidal antibiotics induce mitochondrial dysfunction and oxidative damage in Mammalian cells. Sci Transl Med 2013;5(192):192ra85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature 2011;474(7351):327–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna S. Microbiota Replacement Therapies: Innovation in Gastrointestinal Care. Clinical pharmacology and therapeutics 2018;103(1):102–11. [DOI] [PubMed] [Google Scholar]
- Krych L, Hansen CH, Hansen AK, van den Berg FW, Nielsen DS. Quantitatively different, yet qualitatively alike: a meta-analysis of the mouse core gut microbiome with a view towards the human gut microbiome. PloS one 2013;8(5):e62578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Poole IC, Luiten RM. Autoimmune etiology of generalized vitiligo. Curr Dir Autoimmun 2008;10:227–43. [DOI] [PubMed] [Google Scholar]
- Le Poole IC, Mehrotra S. Replenishing Regulatory T Cells to Halt Depigmentation in Vitiligo. The journal of investigative dermatology Symposium proceedings 2017;18(2):S38–s45. [DOI] [PubMed] [Google Scholar]
- Li B, Selmi C, Tang R, Gershwin ME, Ma X. The microbiome and autoimmunity: a paradigm from the gut-liver axis. Cellular & molecular immunology 2018;15(6):595–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linehan JL, Harrison OJ, Han SJ, Byrd AL, Vujkovic-Cvijin I, Villarino AV, et al. Non-classical Immunity Controls Microbiota Impact on Skin Immunity and Tissue Repair. Cell 2018;172(4):784–96.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PF, Hsieh YD, Lin YC, Two A, Shu CW, Huang CM. Propionibacterium acnes in the pathogenesis and immunotherapy of acne vulgaris. Current drug metabolism 2015;16(4):245–54. [DOI] [PubMed] [Google Scholar]
- Lopez P, Sanchez B, Margolles A, Suarez A. Intestinal dysbiosis in systemic lupus erythematosus: cause or consequence? Current opinion in rheumatology 2016;28(5):515–22. [DOI] [PubMed] [Google Scholar]
- Lui JB, McGinn LS, Chen Z. Gut microbiota amplifies host-intrinsic conversion from the CD8 T cell lineage to CD4 T cells for induction of mucosal immune tolerance. Gut microbes 2016;7(1):40–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magen E, Delgado JS. Helicobacter pylori and skin autoimmune diseases. World journal of gastroenterology 2014;20(6):1510–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manga P, Elbuluk N, Orlow SJ. Recent advances in understanding vitiligo. F1000Research 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science (New York, NY) 2018;359(6371):104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S, De Sarkar S, Pradhan A, Pati AK, Pradhan R, Mondal D, et al. Levels of oxidative damage and proinflammatory cytokines are enhanced in patients with active vitiligo. Free radical research 2017;51(11–12):986–94. [DOI] [PubMed] [Google Scholar]
- Nascimbeni M, Shin EC, Chiriboga L, Kleiner DE, Rehermann B. Peripheral CD4(+)CD8(+) T cells are differentiated effector memory cells with antiviral functions. Blood 2004;104(2):478–86. [DOI] [PubMed] [Google Scholar]
- Palermo B, Garbelli S, Mantovani S, Scoccia E, Da Prada GA, Bernabei P, et al. Qualitative difference between the cytotoxic T lymphocyte responses to melanocyte antigens in melanoma and vitiligo. European journal of immunology 2005;35(11):3153–62. [DOI] [PubMed] [Google Scholar]
- Patel S, Rauf A, Khan H, Meher BR, Hassan SSU. A holistic review on the autoimmune disease vitiligo with emphasis on the causal factors. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie 2017;92:501–8. [DOI] [PubMed] [Google Scholar]
- Paynich ML, Jones-Burrage SE, Knight KL. Exopolysaccharide from Bacillus subtilis Induces Anti-Inflammatory M2 Macrophages That Prevent T Cell-Mediated Disease. Journal of immunology (Baltimore, Md : 1950) 2017;198(7):2689–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott SL, Larcombe DL, Logan AC, West C, Burks W, Caraballo L, et al. The skin microbiome: impact of modern environments on skin ecology, barrier integrity, and systemic immune programming. The World Allergy Organization journal 2017;10(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proal AD, Albert PJ, Marshall TG. The human microbiome and autoimmunity. Current opinion in rheumatology 2013;25(2):234–40. [DOI] [PubMed] [Google Scholar]
- Rashighi M, Agarwal P, Richmond JM, Harris TH, Dresser K, Su MW, et al. CXCL10 is critical for the progression and maintenance of depigmentation in a mouse model of vitiligo. Sci Transl Med 2014;6(223):223ra23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebello D, Wang E, Yen E, Lio PA, Kelly CR. Hair Growth in Two Alopecia Patients after Fecal Microbiota Transplant. ACG case reports journal 2017;4:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem I, Ramser A, Isham N, Ghannoum MA. The Gut Microbiome as a Major Regulator of the Gut-Skin Axis. Frontiers in microbiology 2018;9:1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samborski P, Grzymislawski M. The Role of HSP70 Heat Shock Proteins in the Pathogenesis and Treatment of Inflammatory Bowel Diseases. Advances in clinical and experimental medicine : official organ Wroclaw Medical University 2015;24(3):525–30. [DOI] [PubMed] [Google Scholar]
- Schnupf P, Gaboriau-Routhiau V, Sansonetti PJ, Cerf-Bensussan N. Segmented filamentous bacteria, Th17 inducers and helpers in a hostile world. Current opinion in microbiology 2017;35:100–9. [DOI] [PubMed] [Google Scholar]
- Seddon O, Hughes H. Staphylococcus aureus and atopic dermatitis: a complex relationship. The British journal of dermatology 2018;178(6):1234. [DOI] [PubMed] [Google Scholar]
- Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science (New York, NY) 2015;350(6264):1084–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai N, Peng J, Liu F, Gulden E, Hu Y, Zhang X, et al. Microbial antigen mimics activate diabetogenic CD8 T cells in NOD mice. The Journal of experimental medicine 2016;213(10):2129–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan TG, Sefik E, Geva-Zatorsky N, Kua L, Naskar D, Teng F, et al. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proceedings of the National Academy of Sciences of the United States of America 2016;113(50):E8141–e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulic MK, Cavazza E, Cheli Y, Jacquel A, Luci C, Cardot-Leccia N, et al. Innate lymphocyte-induced CXCR3B-mediated melanocyte apoptosis is a potential initiator of T-cell autoreactivity in vitiligo. Nature communications 2019;10(1):2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Mendoza P, Elghandour MMM, Alaba PA, Sanchez-Aparicio P, Alonso-Fresan MU, Barbabosa-Pliego A, et al. Antimicrobial and bactericidal impacts of Bacillus amyloliquefaciens CECT 5940 on fecal shedding of pathogenic bacteria in dairy calves and adult dogs. Microbial pathogenesis 2018;114:458–63. [DOI] [PubMed] [Google Scholar]
- Vincent C, Mehrotra S, Loo VG, Dewar K, Manges AR. Excretion of Host DNA in Feces Is Associated with Risk of Clostridium difficile Infection. Journal of immunology research 2015;2015:246203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walujkar SA, Kumbhare SV, Marathe NP, Patangia DV, Lawate PS, Bharadwaj RS, et al. Molecular profiling of mucosal tissue associated microbiota in patients manifesting acute exacerbations and remission stage of ulcerative colitis. World journal of microbiology & biotechnology 2018;34(6):76. [DOI] [PubMed] [Google Scholar]
- Wekerle H. Brain Autoimmunity and Intestinal Microbiota: 100 Trillion Game Changers. Trends Immunol 2017;38(7):483–97. [DOI] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 2010;32(6):815–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Martin CM. Increased gut permeability and bacterial translocation in Pseudomonas pneumonia-induced sepsis. Critical care medicine 2000;28(7):2573–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Metagenome datasets have been made available on NCBI’s Sequence Read Archive via BioProject accession number: PRJNA559873.