ABSTRACT
The evolutionary plant-pathogen arms race has equipped plants with the immune system that can defend against pathogens. Pattern-triggered immunity and effector-triggered immunity are two major branches of innate immunity that share immune responses, including oxidative bursts, transcriptional reprogramming, and cell wall modifications such as lignin deposition. In a previous study, we reported that lignin rapidly accumulates in pathogen-infected Arabidopsis leaves and acts as a mechanical barrier, spatially restricting pathogens and cell death. Lignin deposition into the cell wall is a three-step process: monolignol biosynthesis, transport, and polymerization. While monolignol biosynthesis and polymerization are relatively well understood, the mechanism of monolignol transport remains unclear. In this study, we show that macroautophagy/autophagy modulates pathogen-induced lignin formation. Lignification and other immune responses were impaired in autophagy-defective atg (autophagy-related) mutants. In microscopy analyses, monolignols formed punctate structures in response to pathogen infection and colocalized with autophagic vesicles. Furthermore, autophagic activity and lignin accumulation were both enhanced in dnd1 (defense, no death 1) mutant with elevated disease resistance but no cell death and crossing dnd1-1 with atg mutants resulted in a lignin deficit, further supporting that lignin formation requires autophagy. Collectively, these findings demonstrate that lignification, particularly monolignol transport, is achieved through autophagic membrane trafficking in plant immunity.Abbreviations: ABC transporter: ATP-binding cassette transporter; ACD2/AT4G37000: accelerated cell death 2; ATG: autophagy-related; C3′H/AT2G40890: p-coumaroyl shikimate 3-hydroxylase; C4H/AT2G30490: cinnamate 4-hydroxylase; CA: coniferyl alcohol; CaMV: cauliflower mosaic virus; CASP: Casparian strip membrane domain protein; CASPL: CASP-like protein; CBB: Coomassie Brilliant Blue; CCoAOMT1/AT4G34050: caffeoyl-CoA O-methyltransferase 1; CCR1/AT1G15950: cinnamoyl-CoA reductase 1; CFU: colony-forming unit; COMT1/AT5G54160: caffeic acid O-methyltransferase 1; Con A: concanamycin A; DMAC: dimethylaminocoumarin; DND1/AT5G15410: defense, no death 1; CNGC2: cyclic nucleotide-gated channel 2; ER: endoplasmic reticulum; ESB1/AT2G28670/DIR10: enhanced suberin 1; ETI: effector-triggered immunity; EV: extracellular vesicle; F5H/AT4G36220: ferulate-5-hydroxylase; Fluo-3 AM: Fluo-3 acetoxymethyl ester; GFP: green fluorescent protein; HCT/AT5G48930: p-hydroxycinnamoyl-CoA:quinate/shikimate p-hydroxycinnamoyltransferase; HR: hypersensitive response; LAC: laccase; LTG: LysoTracker Green; LSD1/AT4G200380: lesion stimulating disease 1; PAL1/AT2G37040: phenylalanine ammonia-lyase 1; PAMP: pathogen-associated molecular patterns; PCD: programmed cell death; PE: phosphatidylethanolamine; PRX: peroxidase; Pst DC3000: Pseudomonas syringe pv. tomato DC3000; PTI: pattern-triggered immunity; SA: salicylic acid; SD: standard deviation; SID2/AT1G7410: SA induction-deficient 2; UGT: UDP-glucosyltransferase; UPLC: ultraperformance liquid chromatography; UPS: unconventional protein secretion; V-ATPase: vacuolar-type H+-translocating ATPase
KEYWORDS: Arabidopsis, autophagy, cell death, dnd1, immunity, lignin, monolignol transport
Introduction
Phenolic polymer lignin is an essential cell wall component required for plant growth and development [1]. Lignin provides structural rigidity and impermeability to the cell wall, enabling plants to stand upright and transport water over long distances via the vascular system. Lignin polymers are mainly composed of monolignols, p-coumaryl, coniferyl, and sinapyl alcohols, which are oxidatively coupled to p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) lignin units, respectively [2,3]. Monolignol composition and lignin structure vary among plant species, tissues, cells, and even individual cell wall layers, which impacts plant growth, development, and environmental responses [4,5]. Lignin is also critical for innate immunity, providing physical protection to plants against invading pathogens [6,7]. Lignin biogenesis is rapidly induced through transcriptional regulation in two modes of the innate immune system: pattern-triggered immunity (PTI), a basal immunity induced by the recognition of pathogen-associated molecular patterns (PAMPs), and effector-triggered immunity [8], a more robust immune response induced by the recognition of effectors injected by pathogens into plant cells [8–12]. ETI is frequently accompanied by programmed cell death (PCD), termed the hypersensitive response (HR) [13]. Recently, we showed that lignin is readily formed in response to pathogen infection, as an ETI response, to spatially restrict pathogens to the infection site, thus conferring disease resistance [11].
Lignin deposition into the cell wall is a three-step process consisting of monolignol biosynthesis, transport, and polymerization. Monolignols are generated by the phenylpropanoid pathway in the cytosol, and its constituent enzymes have been extensively studied [14–16]. After biosynthesis, monolignols are exported to the cell wall and polymerized via cell wall-localized oxidation systems, which include oxidative enzymes laccases (LACs) and peroxidases (PRXs) [17]. Although LACs and PRXs belong to large gene families and therefore have redundant activities, several lac and prx mutants displayed lignin deficiency in vascular tissues [18–21]. In particular, PRX64/AT5G42180, together with other factors such as the NADPH oxidase RBOHF/AT1G64060, ESB1/AT2G28670/DIR10 (enhanced suberin 1), and Casparian strip membrane domain proteins (CASPs), was required for localized lignin deposition and subsequent formation of the Casparian strip, a diffusion barrier in root endodermal cells [22–25].
Compared to monolignol biosynthesis and polymerization, the mechanism of monolignol export is poorly understood, while four possible models have been proposed [26–28]. Hydrophobic monolignols could be transported by simple diffusion across the plasma membrane or by Golgi-derived vesicle trafficking. A recent study showed that monolignols can diffuse across membranes in a process driven by a polymerization-induced concentration gradient [29]. Alternatively, monolignols are glycosylated and stored as monolignol glucosides in the vacuole. In gymnosperms, the stored glucoconjugates may be transported to the cell wall and used for polymerization [16,30]. However, mutations in Arabidopsis UGT (UDP-glucosyltransferase) genes, encoding monolignol glycosylating enzymes, do not change or rather increased the lignin content, suggesting that monolignol glycosylation may function to sequester toxic monolignols into the vacuole, but not to export monolignols for lignification in angiosperms [31–33]. In addition, it has been demonstrated that monolignol transport is, at least partially, an ATP-dependent active process involving ATP-binding cassette (ABC) transporters [34,35]. ABC transporters form a large family of transmembrane proteins that function in the transport of diverse molecules, such as metabolites, ions, hormones, and lipids [36,37]. The Arabidopsis ABC transporter AtABCG29/AT3G16340 was identified as a p-coumaryl alcohol transporter targeted to the plasma membrane of the root endodermis and vascular tissues [38]. When expressed in yeasts, AtABCG29 increased p-coumaryl alcohol efflux, reducing its toxic effect, and the isolated microsomes showed p-coumaryl alcohol transport activity. Furthermore, a group of ABCG genes was co-expressed with MYB58/AT1G16490, a transcription factor that controls the expression of monolignol biosynthetic genes in differentiating Arabidopsis tracheary element cell cultures, further suggesting the potential involvement of ABC transporters in lignification [39].
Autophagy is a highly conserved catabolic process in eukaryotes, in which intracellular components and organelles are sequestered by double membrane-bound autophagosomes and delivered to the vacuole or lysosome for degradation and recycling [40,41]. Dozens of ATG (autophagy-related) proteins, identified in yeast, animals, and plants, comprise the core autophagy machinery and drive autophagosome biogenesis [42,43]. Genetic and molecular studies have demonstrated that autophagy maintains cellular homeostasis and plays important roles in development, senescence, and responses to abiotic and biotic stresses in plants [44,45]. Autophagy is readily induced in plants by pathogen infection and positively or negatively regulates disease resistance, depending on the lifestyle of invading pathogens [46–50]. Notably, autophagy plays both pro-survival and pro-death roles in regulating ETI-associated HR PCD. Silencing or knockout of ATG genes in tobacco and Arabidopsis led to the spread of chlorotic cell death beyond the infection site, supporting the pro-survival activity of autophagy [49,51,52]. Meanwhile, autophagy functions as a pro-death mechanism, as demonstrated by decreased HR cell death in atg mutants [46]. Constitutive activation of the Rab GTPase RabG3b, an autophagic component, promoted HR PCD through autophagic cell death [53]. Despite considerable evidence for the role of autophagy in cell death and immunity, the underlying mechanism remains unclear.
We previously demonstrated that lignin deposition is rapidly induced during incompatible plant-pathogen interactions and confers disease resistance in Arabidopsis [10,11]. Lignin deficiency resulted in impaired immune responses, i.e., increased susceptibility and the spread of pathogens and HR cell death to the nearby uninfected region, suggesting that the lignin-deposited structure functions as a physical barrier, spatially limiting pathogens and HR PCD to the infection site. In this study, we report that autophagy is a key regulator of pathogen-induced lignification. The observed immune defects caused by lignin deficiency were phenocopied in atg mutants. In pathogen-infected cells, monolignols formed punctate structures in a manner dependent on autophagic activation and colocalized to autophagic vesicles. Here we propose that autophagy mediates pathogen-induced lignification by regulating monolignol transport.
Results
Autophagy is required for pathogen-induced lignification in ETI
We previously reported that pathogen-induced lignification is important for innate immunity [10,11]. HR PCD was triggered and localized at the infection site in response to ETI-eliciting avirulent bacterial pathogens, such as Pseudomonas syringae pv. tomato (Pst) DC3000 (AvrRpm1), but spread across entire infected leaves of lignin-deficient knockout mutants palQ and myb15 for PAL (phenylalanine ammonia-lyase) and MYB transcription factor MYB15/AT3G23250, respectively. PAL catalyzes the conversion of phenylalanine to cinnamic acid, the first committed step in the lignin biosynthetic pathway, and MYB15 mediates the expression of lignin biosynthetic genes [10,11]. Notably, this cell death phenotype was similar to spreading HR cell death observed in atg mutants during innate immune responses [49,53]. This led us to suspect that pathogen-induced lignification may be impaired in atg mutants. To examine this, autophagy-defective atg5-1 and atg7-2 mutants were inoculated with Pst DC3000 (AvrRpm1) and assessed for lignin formation. HR cell death gradually spread over the infection site and lignin deposition was defective in atg5-1 and atg7-2 mutants, as visualized by phloroglucinol staining and quantified by the acetyl bromide assay (Figure 1a-d). We previously demonstrated that lignin deposition enhances disease resistance and prevents the spread of invading pathogens [11]. Consistently, the growth of Pst DC3000 (AvrRpm1) was higher in atg mutants than in wild-type plants (Figure 1e). In addition, green fluorescent protein (GFP)-labeled Pst DC3000 (AvrRpt2) was inoculated into wild-type and atg leaves, and its localization was monitored. In line with previous results, fluorescent bacteria were restricted to the infected site in wild-type plants but moved to the nearby uninfected site in atg mutants (Figure 1f).
Figure 1.
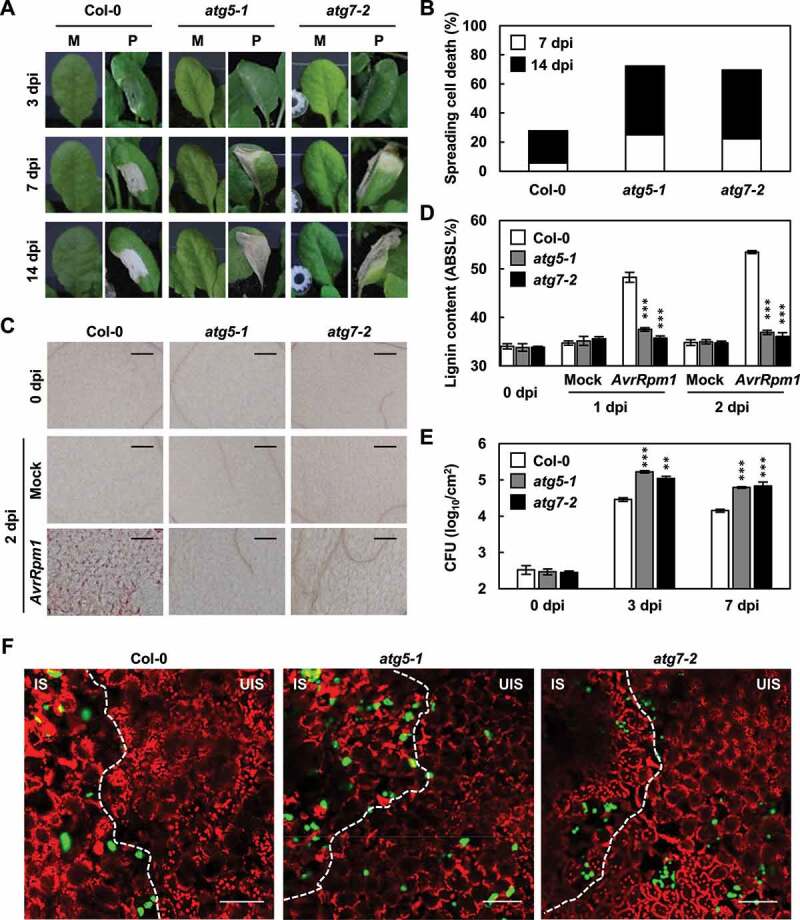
Autophagy positively regulates lignin accumulation and immune responses. (a) Cell death phenotypes of wild-type and atg leaves treated with mock and Pst DC3000 (AvrRpm1). M, mock; P, Pst DC3000 (AvrRpm1). (b) Quantification of leaves (n ≥ 30) with spreading cell death as in (a). (c) Phloroglucinol staining of wild-type and atg leaves treated with mock and Pst DC3000 (AvrRpm1). Scale bars: 100 μm. (d) Quantification of lignin content in wild-type and atg leaves treated with mock and Pst DC3000 (AvrRpm1). Data represent means ± SD (n = 4; 3–9 leaves each). (e) Measurements of Pst DC3000 (AvrRpm1) growth in wild-type and atg plants. Data represent means ± SD (n = 3). (f) Colonization patterns of GFP-Pst DC3000 (AvrRpt2) in wild-type and atg plants at 2 dpi. Scale bars: 100 μm. Six-week-old plants were inoculated with mock (10 mM MgCl2) and Pst DC3000 (AvrRpm1) at 2 × 105 CFU/ml for growth assays and at 108 CFU/ml for other experiments. Asterisks indicate significant differences from the respective Col-0 (t test; **P < 0.01; ***P < 0.001) in (d and e). AvrRpm1, Pst DC3000 (AvrRpm1); dpi, days post-inoculation; IS, infected site; UIS, uninfected site.
It was previously shown that spreading HR cell death in atg mutants is an age-related phenotype [46,52,54]. The spread of chlorotic cell death was prominent in 7-8-week old atg plants, but not clearly observed in 3-4-week-old atg plants. Since we used “old” (6-week-old) plants, infection experiments were conducted with “young” (4-week-old) plants to investigate whether the observed immune responses are age-dependent. Consistent with previous observations, the incidence of spreading cell death was largely decreased in Pst DC3000 (AvrRpm1)-infected young atg plants (Fig. S1). However, lignin formation showed little difference and was defective in both old and young atg plants. Increased susceptibility and the spread of pathogens to the uninfected site were also observed in young atg plants. These results suggest that pathogen-induced lignification is dependent on autophagy but is not affected by plant age.
Autophagy is implicated in monolignol transport required for pathogen-induced lignification
With these results, we raised the question as to how autophagy is implicated in pathogen-induced lignification. Considering that lignin biosynthesis is a three-step process composed of monolignol biosynthesis, transport, and polymerization, autophagy, a membrane trafficking process, may be involved in monolignol transport. We first checked whether the expression of monolignol biosynthetic genes is altered in atg mutants. Quantitative real-time RT-PCR analysis was performed to determine the expression of genes encoding monolignol biosynthetic enzymes, including PAL1, C4H (cinnamate 4-hydroxylase), HCT (p-hydroxycinnamoyl-CoA:quinate/shikimate p-hydroxycinnamoyltransferase), C3′H (p-coumaroyl shikimate 3-hydroxylase), CCoAOMT1 (caffeoyl-CoA O-methyltransferase 1), CCR1 (cinnamoyl-CoA reductase 1), F5H (ferulate-5-hydroxylase), and COMT1 (caffeic acid O-methyltransferase 1). The expression of monolignol biosynthetic genes was increased in response to Pst DC3000 (AvrRpm1) treatment, but did not differ between wild-type and atg mutants (Fig. S2). To examine whether monolignol biosynthesis is altered in atg mutants, soluble metabolites were extracted from wild-type and atg plants before and after Pst DC3000 (AvrRpm1) inoculation and analyzed for monolignol content by ultraperformance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) analysis (Fig. S3 and Table S2). Pathogen treatment substantially increased monolignol production in both wild-type and atg mutants. These results suggest that autophagy is not related to monolignol biosynthesis but to subsequent processes.
Next, to determine the correlation between autophagy and monolignol transport and polymerization, we investigated whether lignin deposition is restored in atg mutants by external application of monolignols. Recovery of lignification by monolignol application would imply a defect in monolignol transport, but not in the polymerization process in atg mutants. Therefore, atg5-1 and atg7-1 mutants were treated with coniferyl alcohol (CA), the monolignol composing G lignin units, prior to bacterial infiltration. G lignin has been shown to mainly contribute to disease resistance [8]. Pre-treatment of atg mutants with CA resulted in lignin accumulation up to approximately 86% of that in wild-type plants infected with Pst DC3000 (AvrRpm1) (Figure 2a). Other immune defects were concomitantly rescued in CA-treated atg mutants: bacterial growth and the spread of cell death were reduced, and pathogens were restricted to the infection site (Figure 2b-e). These results suggest that autophagy is implicated in monolignol transport during the ETI response.
Figure 2.
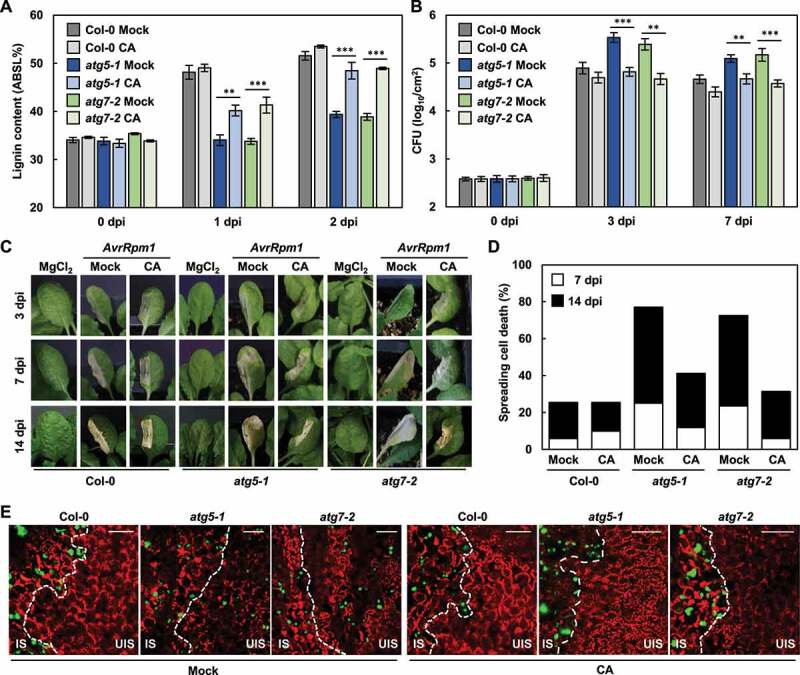
Immune defects in atg mutants are rescued by external supply of monolignols. (a) Quantification of lignin content in wild-type and atg leaves after mock and CA pre-treatments and Pst DC3000 (AvrRpm1) inoculation. Data represent means ± SD (n = 4; 3–9 leaves each). (b) Measurements of Pst DC3000 (AvrRpm1) growth in wild-type and atg plants. Data represent means ± SD (n = 3). (c) Cell death phenotypes of wild-type and atg leaves after mock and CA pre-treatments and Pst DC3000 (AvrRpm1) inoculation. (d) Quantification of leaves (n ≥ 30) with spreading cell death as in (c). (e) Colonization patterns of GFP-Pst DC3000 (AvrRpt2) in wild-type and atg plants after mock and CA pre-treatments at 2 dpi. Scale bars: 100 μm. Six-week-old plants were pre-treated with mock (0.1% DMSO) and CA (50 μM) for 1 h and inoculated with Pst DC3000 (AvrRpm1) at 2 × 105 CFU/ml for growth assays and at 108 CFU/ml for other experiments. Asterisks indicate significant differences in (a and b) (t test; **P < 0.01; ***P < 0.001). AvrRpm1, Pst DC3000 (AvrRpm1); CA, coniferyl alcohol; dpi, days post-inoculation; IS, infected site; UIS, uninfected site.
Additionally, we asked whether trafficking of secretory and plasma membrane proteins involved in monolignol polymerization may also be modulated by autophagy. Since CASP-like proteins (CASPLs) CASPL1D1/AT4G15610 and CASPL4D1/AT2G39530, supposedly four-membrane-span proteins, are required for pathogen-induced lignin formation [11], their subcellular localization was determined in protoplasts by transiently expressing CASPL1D1 and CASPL4D1 fused with mCherry and Citrine at the C-terminus, respectively (Fig. S4). CASPL1D1 and CASPL4D1 were localized to the plasma membrane in atg mutants as in wild-type cells, suggesting that their localization to the plasma membrane is autophagy-independent.
Monolignols colocalize with autophagic vesicles in cells upon pathogen infection
It was previously shown that when incubated with protoplasts, fluorescence-tagged monolignols penetrate the plasma membrane and diffuse into the cytoplasm, and therefore can be used as imaging probes [55,56]. Considering that autophagy may regulate monolignol transport, we investigated whether monolignols colocalize with autophagic vesicles. To locate monolignols and autophagosome structures in cells, we used fluorescence-tagged monolignol, dimethylaminocoumarin (DMAC)-tagged CA (DMAC-CA), and transgenic Col-0 plants overexpressing GFP-ATG8a fusion proteins (35S:GFP-ATG8a). Protoplasts were isolated from 35S:GFP-ATG8a plants before and after Pst DC3000 (AvrRpm1) infection and incubated with CA or DMAC-CA. GFP-ATG8a and DMAC-CA fluorescence diffused in the cytoplasm of cells from un-treated 35S:GFP-ATG8a plants. In contrast, protoplasts from Pst DC3000 (AvrRpm1)-treated plants formed GFP-ATG8a-labeled autophagic vesicles and concurrently DMAC-CA-associated aggregates or punctate structures (Figure 3). DMAC-CA puncta entirely colocalized with autophagic vesicles in pathogen-activated protoplasts, but autophagosomes did not all overlap with DMAC-CA structures. Furthermore, we monitored DMAC-CA monolignols and GFP-ATG8a autophagic vesicles by live cell imaging and found that they move together in protoplasts, further verifying their colocalization (Movie S1).
Figure 3.
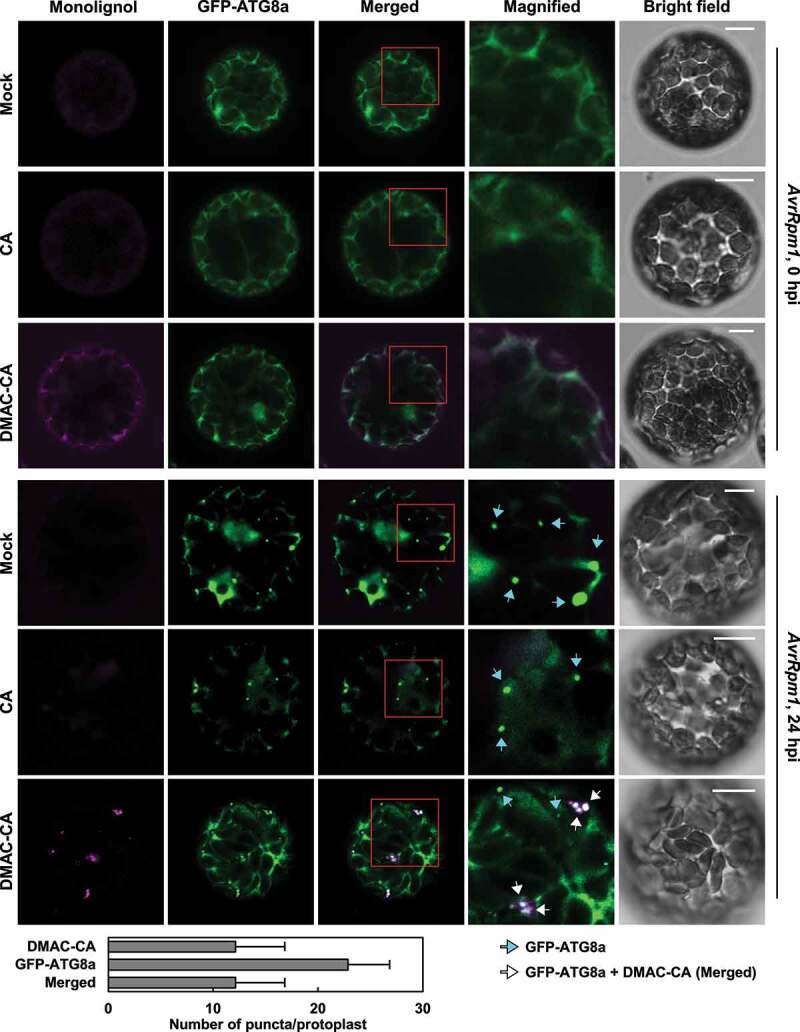
Monolignols colocalize with autophagic vesicles in protoplasts upon pathogen infection. Representative microscopic images of monolignol (DMAC-CA) and autophagic (GFP-ATG8a) structures in GFP-ATG8a-transgenic protoplasts after Pst DC3000 (AvrRpm1) infection. Five-week-old 35S:GFP-ATG8a plants were treated with Pst DC3000 (AvrRpm1) at 106 CFU/ml for 24 h and subjected to protoplast isolation. The isolated protoplasts were incubated with mock (0.1% DMSO), CA (50 μM), and DMAC-CA (50 μM) for 4 h. Images of boxed areas are magnified in the right panel. Cyan arrows indicate autophagic puncta (GFP-ATG8a) only and white arrows indicate colocalization of monolignol (DMAC-CA) and autophagic (GFP-ATG8a) structures. The graph shows quantification of monolignol (DMAC-CA) and autophagic (GFP-ATG8a) puncta at 24 hpi. Data represent means ± SD (n = 7). Scale bars: 10 μm. CA, coniferyl alcohol; DMAC-CA, dimethylaminocoumarin-coniferyl alcohol; AvrRpm1, Pst DC3000 (AvrRpm1); hpi, hours post-inoculation.
We examined whether colocalization of DMAC-CA monolignols with autophagic vesicles is also observed in plants. 35S:GFP-ATG8a plants were infiltrated with DMAC-CA before pathogen infection and DMAC-CA diffusion into cells was observed. Likewise, DMAC-CA and GFP-ATG8a positive structures were generated and colocalized in the cytoplasm in response to Pst DC3000 (AvrRpm1) treatment (Figure 4a). However, these punctate structures did not appear in the atg mutant background (Figure 4b), indicating that GFP-ATG8a-associated structures are canonical autophagic vesicles and that the formation of monolignol puncta depends on autophagic activation. Live cell imaging further revealed co-movement of DMAC-CA monolignols and GFP-ATG8a autophagic vesicles in cells (Movie S2). Intriguingly, vesicles moved along cytoskeletal elements, some of which appeared to be transported towards the plasma membrane and even out of the cell (Movie S2 and Figure 4c). These results together suggest that monolignols are selected as cargo for autophagosomes and delivered to the cell wall via the autophagic process.
Figure 4.
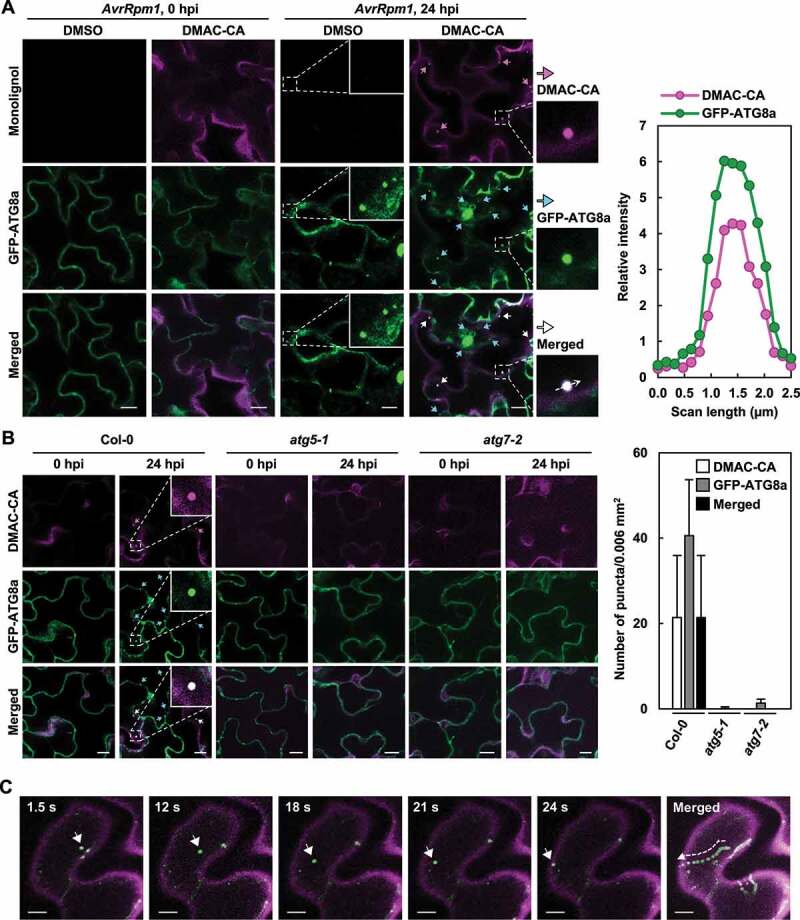
Monolignols colocalize with autophagic vesicles in plants upon pathogen infection. (a) Representative microscopic images of monolignol (DMAC-CA) and autophagic (GFP-ATG8a) structures in GFP-ATG8a-transgenic leaves after Pst DC3000 (AvrRpm1) infection. The graph shows relative intensity profiles of monolignol (DMAC-CA) and autophagic (GFP-ATG8a) signals across transect in the merged image. (b) Representative microscopic images of monolignol (DMAC-CA) and autophagic (GFP-ATG8a) structures in GFP-ATG8a-transgenic wild-type and atg leaves after Pst DC3000 (AvrRpm1) infection. The graph shows quantification of monolignol (DMAC-CA) and autophagic (GFP-ATG8a) puncta at 24 hpi. Data represent means ± SD (n = 6). (c) Time-lapse tracking of monolignol (DMAC-CA) and autophagic (GFP-ATG8a) structures captured in Movie S2. Time in seconds in the movie is shown at the top of images. A representative monolignol (DMAC-CA) and autophagic (GFP-ATG8a) structure (arrowhead) co-migrates in successive time-lapse images. Six-week-old plants were pre-treated with mock (0.1% DMSO) and DMAC-CA (50 μM) for 1 h, and treated with Pst DC3000 (AvrRpm1) at 106 CFU/ml for 24 h. In (a and b), monolignol (DMAC-CA) and autophagic (GFP-ATG8a) structures are indicated by arrows and the representative structures are shown at higher magnification. Scale bars: 10 μm. AvrRpm1, Pst DC3000 (AvrRpm1); DMAC-CA, dimethylaminocoumarin-coniferyl alcohol; hpi, hours post-inoculation.
We also speculated that autophagic vesicles would be released to the apoplast if autophagy is implicated in monolignol transport to the cell wall. Therefore, extracellular vesicles (EVs) were isolated from the apoplastic wash fluid of 35S:GFP-ATG8a leaves using vacuum infiltration and ultracentrifugation [57,58]. Microscopic analysis revealed that GFP-ATG8a-associated EVs accumulate in the apoplast following Pst DC3000 (AvrRpm1) infection (Figure 5a). In addition, apoplastic wash fluids and EVs were isolated from wild-type leaves and analyzed by immunoblotting with antibodies against ATG8a, the secreted protein PR1 (pathogenesis-related 1), and the cytosolic proteins GST (glutathione S-transferase) and actin (Figure 5b). The expression of ATG8a and PR1 proteins was increased by pathogen treatment, and they both, but not cytosolic GST and actin, were detected in apoplastic wash fluids. Of the tested proteins, ATG8a was associated with EVs isolated from pathogen-infected leaves. These results further support that monolignol transport is mediated by the autophagic pathway.
Figure 5.
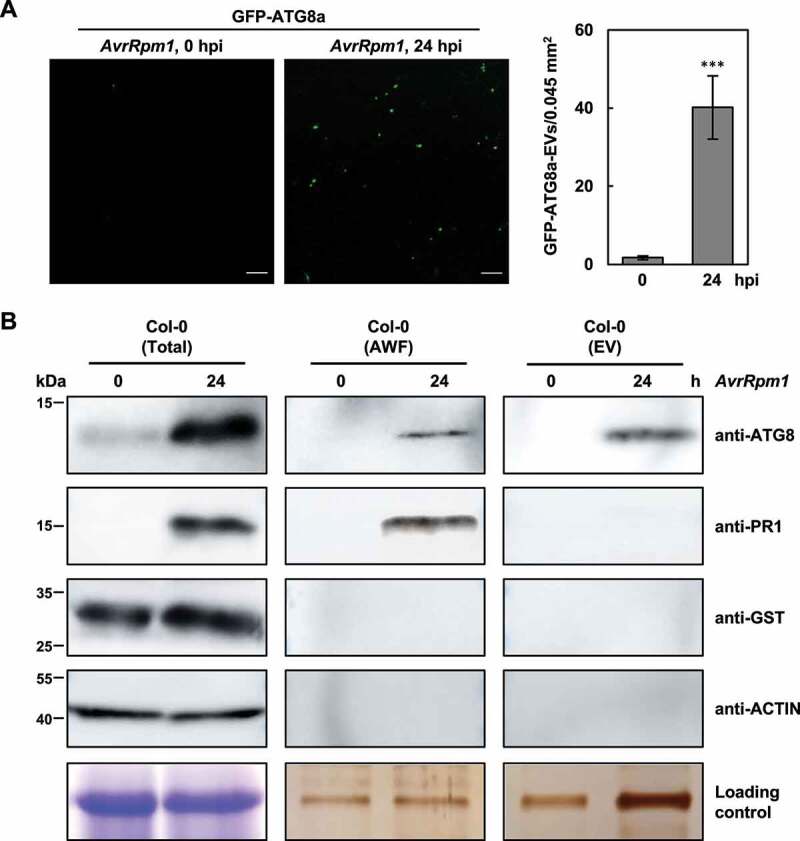
Autophagic vesicles are released to the apoplast upon pathogen infection. (a) Confocal microscopic images of extracellular vesicles (EVs) isolated from the apoplastic wash fluid of 35S:GFP-ATG8a leaves before and after Pst DC3000 (AvrRpm1) infection. Fluorescent spots, corresponding to GFP-ATG8a-labeled EVs, were quantified as shown in the graph. Data represent means ± SD (n = 8). Asterisks indicate significant difference from 0 hpi (t test; ***P < 0.001). Scale bars: 20 μm. (b) Western blot analysis of the apoplastic wash fluid (AWF) and EVs isolated from untreated or Pst DC3000 (AvrRpm1)-treated wild-type leaves with anti-ATG8a, anti-PR1, anti-GST, and anti-actin antibodies. Total proteins from two to three leaves were directly extracted in SDS sample buffer and used as a positive control. Coomassie Brilliant Blue staining for total proteins and silver staining for AWF and EVs served as loading controls. Six-week-old plants were treated with Pst DC3000 (AvrRpm1) at 106 CFU/ml for 24 h. AvrRpm1, Pst DC3000 (AvrRpm1); hpi, hours post-inoculation.
Lignification and autophagy activity are enhanced in HR-suppressed dnd1 mutant
Our previous [10,11] and present results show that lignin deposition is required to restrict HR PCD. This prompted us to measure the lignin content in known cell death-defective mutants, i.e., acd2-2 (accelerated cell death 2), lsd1-2 (lesion stimulating disease 1), and dnd1-1 (defense, no death 1) mutants [53,59–61]. While HR cell death was expanded in acd2-2 and lsd1-1 mutants as in atg5-1, but suppressed in dnd1-1 as previously reported, the lignin content was decreased in acd2-2, lsd1-2, and atg5-1, but increased in dnd1-1 mutants compared to wild-type in response to Pst DC3000 (AvrRpm1) treatment (Figure 6a,b). Notably, lignin constitutively accumulated in dnd1-1 plants. These results imply that lignification is correlated with the restriction of HR cell death.
Figure 6.
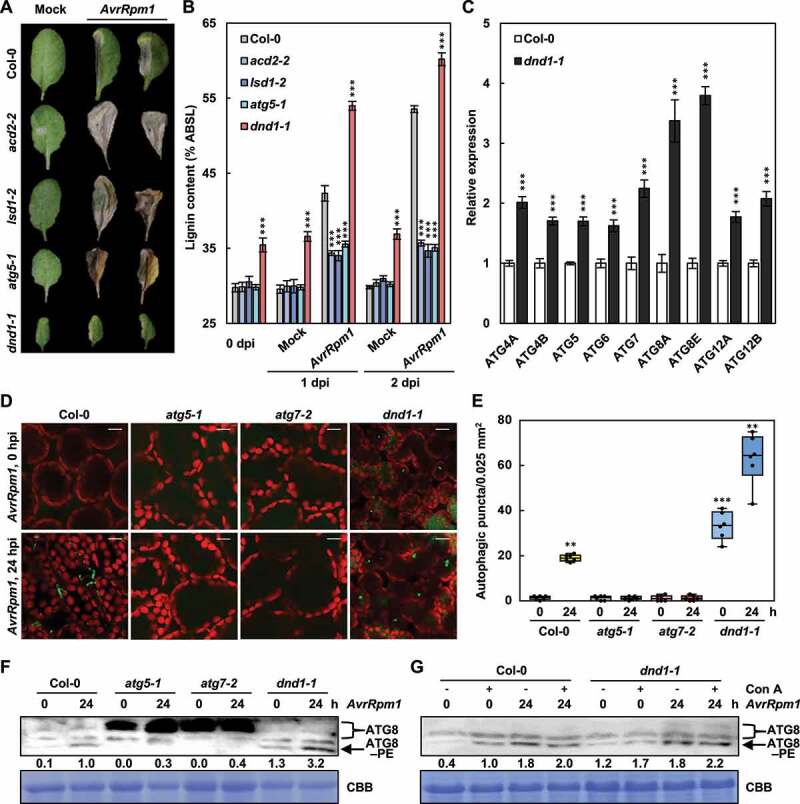
Lignification and autophagy are enhanced in defense, no death 1 (dnd1) mutant. (a) Cell death phenotypes of wild-type and cell death-defective mutant leaves treated with mock and Pst DC3000 (AvrRpm1) for 7 days. (b) Quantification of lignin content in wild-type and cell death-defective mutant leaves treated with mock and Pst DC3000 (AvrRpm1). Data represent means ± SD (n = 4; 3–9 leaves each). (c) Quantitative real-time RT-PCR analysis of ATG genes in wild-type and dnd1-1 plants. Data represent means ± SD (n = 4). (d) LTG staining of autophagic structures in wild-type and cell death-defective mutant leaves after Pst DC3000 (AvrRpm1) infection. Scale bars: 20 μm. (e) Quantification of LTG-stained autophagic structures in (d). Autophagic puncta were counted in areas of 0.025 mm2. Data represent means ± SD (n = 6). (f) ATG8 lipidation analysis in wild-type and dnd1-1 plants after Pst DC3000 (AvrRpm1) infection. Total proteins stained by Coomassie Brilliant Blue serve as a loading control. Relative intensities of ATG8–PE normalized to the loading control are shown as numerical values. (g) ATG8 lipidation analysis in wild-type and dnd1-1 plants after mock and Con A pre-treatments and Pst DC3000 (AvrRpm1) inoculation. Total proteins stained by Coomassie Brilliant Blue serve as a loading control. Relative intensities of ATG8–PE normalized to the loading control are shown as numerical values. Six-week-old plants were pre-treated with mock (0.1% DMSO) and Con A (5 μM) for 8 h in (g) and inoculated with Pst DC3000 (AvrRpm1) at 108 CFU/ml in (a and b) and at 106 CFU/ml in (d-g). Asterisks indicate significant differences from the respective Col-0 in (b and c) and from the time point 0 h in (e) (t test; **P < 0.01; ***P < 0.001). AvrRpm1, Pst DC3000 (AvrRpm1); dpi, days post-inoculation; hpi, hours post-inoculation; ATG8–PE, ATG8–phosphatidylethanolamine; Con A, concanamycin A; CBB, Coomassie Brilliant Blue.
Over-accumulation of lignin in dnd1-1 mutant raised the question as to whether linin formation in dnd1-1 plants is also dependent on autophagy. DND1 encodes CNGC2 (cyclic nucleotide-gated channel 2), a Ca2+ influx channel that regulates Ca2+-based PTI [62,63]. First, the expression of ATG genes was analyzed in wild-type and dnd1-1 plants. The transcript levels of all tested ATG genes were higher in dnd1-1 than in wild-type plants (Figure 6c). Next, the level of autophagic activation in Pst DC3000 (AvrRpm1)-inoculated wild-type, atg5-1, atg7-1, and dnd1-1 plants was determined using LysoTracker Green (LTG), which stains acidic organelles including autophagosome structures [53]. The formation of LTG-stained punctate structures was induced in wild-type leaves, but was constitutive and further increased by pathogen treatment in dnd1-1 mutant (Figure 6d,e). We often observed enlarged autophagosomes after Pst DC3000 (AvrRpm1) infection in dnd1-1 leaves. This may reflect an increase in autophagy activity, which is modulated by the size and number of autophagosomes [42,64]. LTG-stained puncta were not detected in atg mutants, implying that these structures correspond to autophagic vesicles. To further examine the induction of autophagy, we analyzed the formation of the ATG8-phosphatidylethanolamine (PE) conjugate as a marker for autophagic activation by western blotting using an anti-ATG8a antibody. Consistently, both constitutive and pathogen-induced ATG8–PE accumulation was higher in dnd1-1 than in wild-type plants (Figure 6f). Autophagic flux represents the dynamic process of autophagy, encompassing autophagosome formation, fusion with lysosomes/vacuoles, and subsequent degradation of autolysosomal contents. Therefore, measuring autophagic flux is important because increases in autophagosome or ATG8–PE formation may be explained either by autophagic induction or by impairment of the last steps, such as fusion with lysosomes/vacuoles and subsequent breakdown. Concanamycin A (Con A) is a vacuolar ATPase inhibitor that inhibits vacuolar hydrolases and is therefore frequently used to monitor autophagic flux [65]. Con A treatment led to the increase in ATG8–PE production in both wild-type and dnd1-1 plants before and after Pst DC3000 (AvrRpm1) infection (Figure 6g). These results together suggest that dnd1-1 mutant has enhanced activity but normal flux of autophagy.
Lignin accumulation depends on autophagy in dnd1 mutant
To assess whether linin formation depends on autophagy in dnd1-1 plants, we crossed dnd1-1 with atg5-1 and atg7-2 mutants (Fig. S5). The dwarf phenotype of dnd1-1 mutant was retained and early senescence in atg mutants was accelerated in dnd1-1 atg5-1 and dnd1-1 atg7-2 double mutants (Fig. S5A) [52,61]. As expected, the enhanced autophagic activity in dnd1-1 mutant was abolished in dnd1-1 atg5-1 and dnd1-1 atg7-2 mutants, as determined by LTG staining of autophagic structures and immunoblotting of ATG8–PE adducts (Fig. S5B,C).
These single and double mutants were then subjected to lignin quantification. Both constitutive and AvrRpm1-induced lignin accumulation in dnd1-1 mutant was eliminated in dnd1-1 atg5-1 and dnd1-1 atg7-2 mutants, as in atg5-1 and atg7-2 mutants (Figure 7a). It was previously shown that dnd1 mutant has elevated disease resistance to both avirulent and virulent pathogens [61,62]. Consistently, the growth of Pst DC3000 (AvrRpm1) was significantly decreased in dnd1-1, compared to wild-type plants, but increased in dnd1-1 atg5-1 and dnd1-1 atg7-2 mutants to the level in atg mutants (Figure 7b). Noticeably, AvrRpm1-elicited HR cell death was not only restored but also unrestricted and spread in dnd1-1 atg5-1 and dnd1-1 atg7-2 mutants, as in atg mutants (Figure 7c-f). These results demonstrate that excessive lignification is due to enhanced autophagic activity in dnd1 mutant, and therefore, autophagy is essential for immunity-associated lignification.
Figure 7.
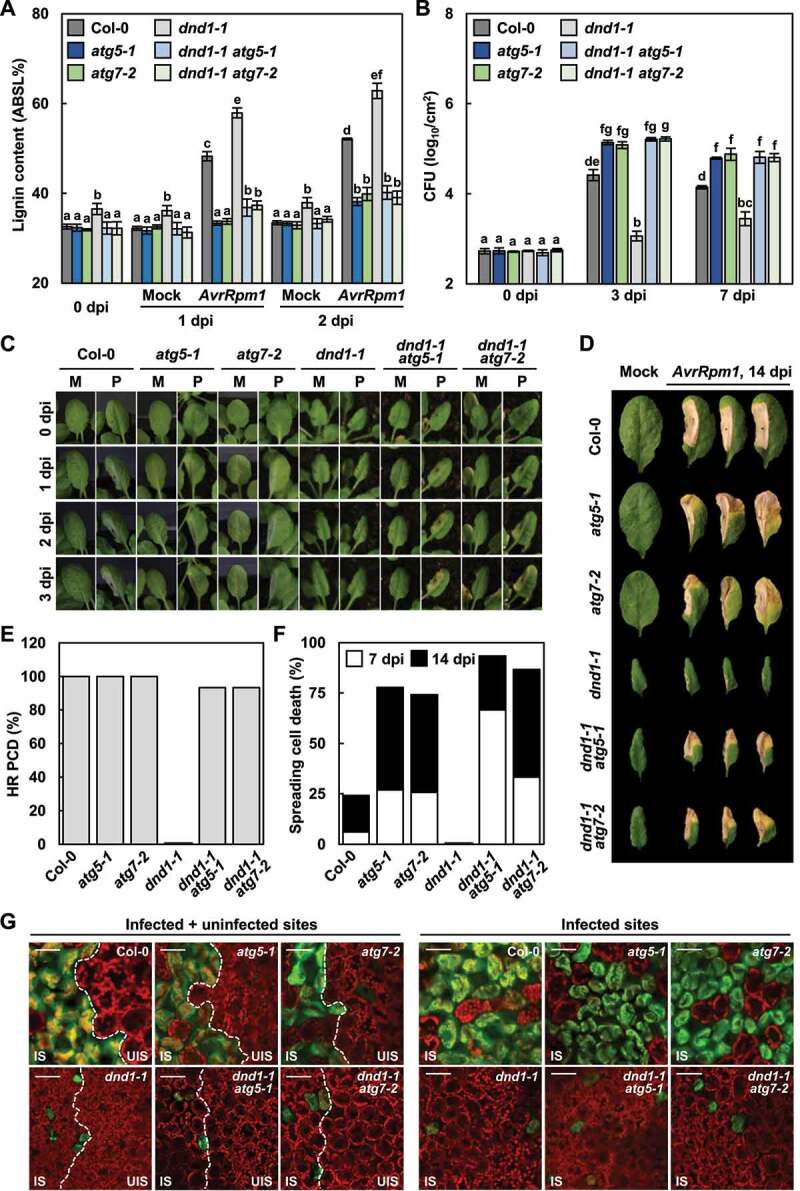
Lignin accumulation depends on autophagy in defense, no death 1 (dnd1) mutant. (a) Quantification of lignin content in wild-type, dnd1, atg, and dnd1 atg leaves treated with mock and Pst DC3000 (AvrRpm1). Data represent means ± SD (n = 4; 3–9 leaves each). (b) Measurements of Pst DC3000 (AvrRpm1) growth in wild-type, dnd1, atg, and dnd1 atg plants. Data represent means ± SD (n = 3). (c) Cell death phenotypes of wild-type, dnd1, atg, and dnd1 atg leaves treated with mock and Pst DC3000 (AvrRpm1) for 1–3 days. M, mock; P, Pst DC3000 (AvrRpm1). (d) Cell death phenotypes of wild-type, dnd1, atg, and dnd1 atg leaves treated with mock and Pst DC3000 (AvrRpm1) for 14 days. (e) Quantification of leaves (n ≥ 30) with HR cell death as in (c). (f) Quantification of leaves (n ≥ 30) with spreading cell death as in (d). (g) Fluo-3 AM staining of Ca2+ ions in Pst DC3000 (AvrRpm1)-inoculated wild-type, dnd1, atg, and dnd1 atg leaves. Fluo-3 AM emits green fluorescence after Ca2+ binding. Scale bars: 50 μm. Six-week-old plants were inoculated with mock (10 mM MgCl2) and Pst DC3000 (AvrRpm1) at 2 × 105 CFU/ml for growth assays and at 108 CFU/ml for other experiments. Different letters indicate significant differences in (a and b) (Tukeys HSD test; P < 0.05). AvrRpm1, Pst DC3000 (AvrRpm1); dpi, days post-inoculation; IS, infected site; UIS, uninfected site.
While lignin deficiency accounted for defects in immune responses, the recovery of HR PCD in dnd1-1 atg mutants was unexpected. Previous reports demonstrated that DND1/CNGC2 functions as a Ca2+ channel and triggers Ca2+ influx in immunity [63] and that the cytoplasmic Ca2+ increase is required for HR cell death [66,67]. Accordingly, we determined intracellular Ca2+ changes using the fluorescent Ca2+ indicator, Fluo-3 acetoxymethyl ester (Fluo-3 AM) [68]. In line with previous findings, the fluorescent signal of Ca2+ was largely increased in wild-type cells, but not in dnd1-1 in response to Pst DC3000 (AvrRpm1) infection (Figure 7g). However, the Ca2+ level was little elevated in dnd1 atg mutants, in which HR cell death was restored. The mechanism by which dnd1 atg mutants with low Ca2+ production develop HR PCD remains to be elucidated.
Autophagic induction is salicylic acid-dependent in dnd1 mutant
It was previously reported that autophagy is induced by salicylic acid (SA) signaling [52]. In line with this finding, SA treatment activated autophagy in wild-type plants, as shown by the increased ATG8–PE formation (Figure 8a). Given that SA levels are constitutively elevated in dnd1 mutant [61], we investigated whether autophagy induction is due to SA accumulation in dnd1-1 plants. SID2/AT1G7410 (SA induction-deficient 2) encodes isochorismate synthase, which is required for SA biosynthesis [69]. Therefore, we used dnd1-1 sid2-1 mutant [70] to assess SA dependence of autophagy. AvrRpm1-induced autophagy was analyzed in dnd1-1 sid2-1 mutant by ATG expression analysis, LTG staining, and ATG8 immunoblotting. ATG expression was promoted in dnd1-1 but decreased in dnd1-1 sid2-1 mutant to the levels in wild-type plants (Figure 8b). ATG8–PE forms and abundant autophagic structures seen in dnd1-1 were substantially reduced in dnd1-1 sid2-1 mutant both before and after pathogen infection (Figure 8c-e). Consistently, pathogen-induced lignification was defective in dnd1-1 sid2-1 mutant (Figure 8f). These results indicate that autophagy and lignification are induced by SA signaling during immune responses.
Figure 8.
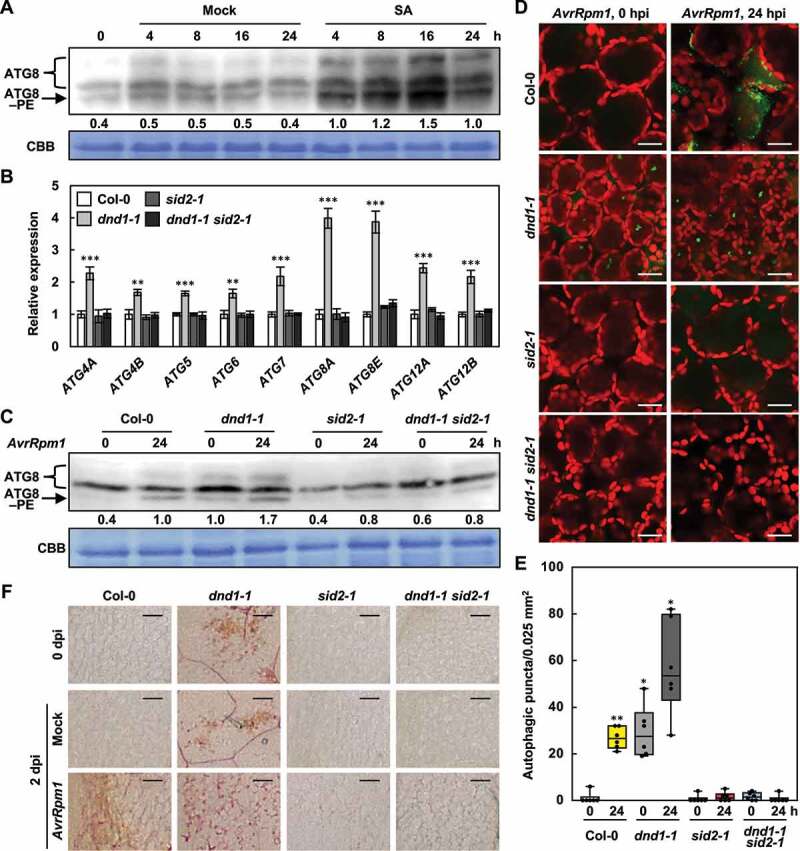
Autophagy and lignification are induced by SA signaling. (a) ATG8 lipidation analysis in wild-type plants treated with mock (water) and SA (1 mM). Total proteins stained by Coomassie Brilliant Blue serve as a loading control. Relative intensities of ATG8–PE normalized to the loading control are shown as numerical values. (b) Quantitative real-time RT-PCR analysis of ATG genes in wild-type, dnd1, sid2, and dnd1 sid2 plants. Data represent means ± SD (n = 4). (c) ATG8 lipidation analysis in wild-type, dnd1, sid2, and dnd1 sid2 plants treated with Pst DC3000 (AvrRpm1). Total proteins stained by Coomassie Brilliant Blue serve as a loading control. Relative intensities of ATG8–PE normalized to the loading control are shown as numerical values. (d) LTG staining of autophagic structures in wild-type, dnd1, sid2, and dnd1 sid2 leaves after Pst DC3000 (AvrRpm1) infection. Scale bars: 20 μm. (e) Quantification of LTG-stained autophagic structures in (d). Autophagic puncta were counted in areas of 0.025 mm2. Data represent means ± SD (n = 6). (f) Phloroglucinol staining of wild-type, dnd1, sid2, and dnd1 sid2 leaves infiltrated with mock and Pst DC3000 (AvrRpm1). Scale bars: 100 μm. Six-week-old plants were inoculated with mock (10 mM MgCl2) and Pst DC3000 (AvrRpm1) at 106 CFU/ml in (a, c-e) and at 108 CFU/ml in (f). Asterisks indicate significant differences from the respective Col-0 in (b) and from the time point 0 h of Col-0 in (e) (t test; *P < 0.05; **P < 0.01; ***P < 0.001). SA, salicylic acid; ATG8–PE, ATG8–phosphatidylethanolamine; CBB, Coomassie Brilliant Blue; AvrRpm1, Pst DC3000 (AvrRpm1); hpi, hours post-inoculation; dpi, days post-inoculation.
Discussion
In this work, we demonstrated that autophagy is essential for pathogen-induced lignin deposition. Autophagy-deficient mutants atg5-1 and atg7-2 displayed reduced lignin accumulation, resulting in immune defects, i.e., the spread of invading bacterial pathogens and HR PCD and the increased pathogen growth. We also found that monolignols are transported via the autophagic pathway, as revealed by co-localization and co-movement of fluorescence-tagged monolignol and autophagic structures in immunity-activated cells. Studies of dnd1-1 mutant with enhanced lignification and autophagic activation provide additional evidence that immunity-associated lignin production requires autophagy in an SA-dependent manner. We propose that autophagy functions as a membrane trafficking pathway to facilitate monolignol transport, and therefore, lignin formation in plant immunity (Figure 9).
Figure 9.
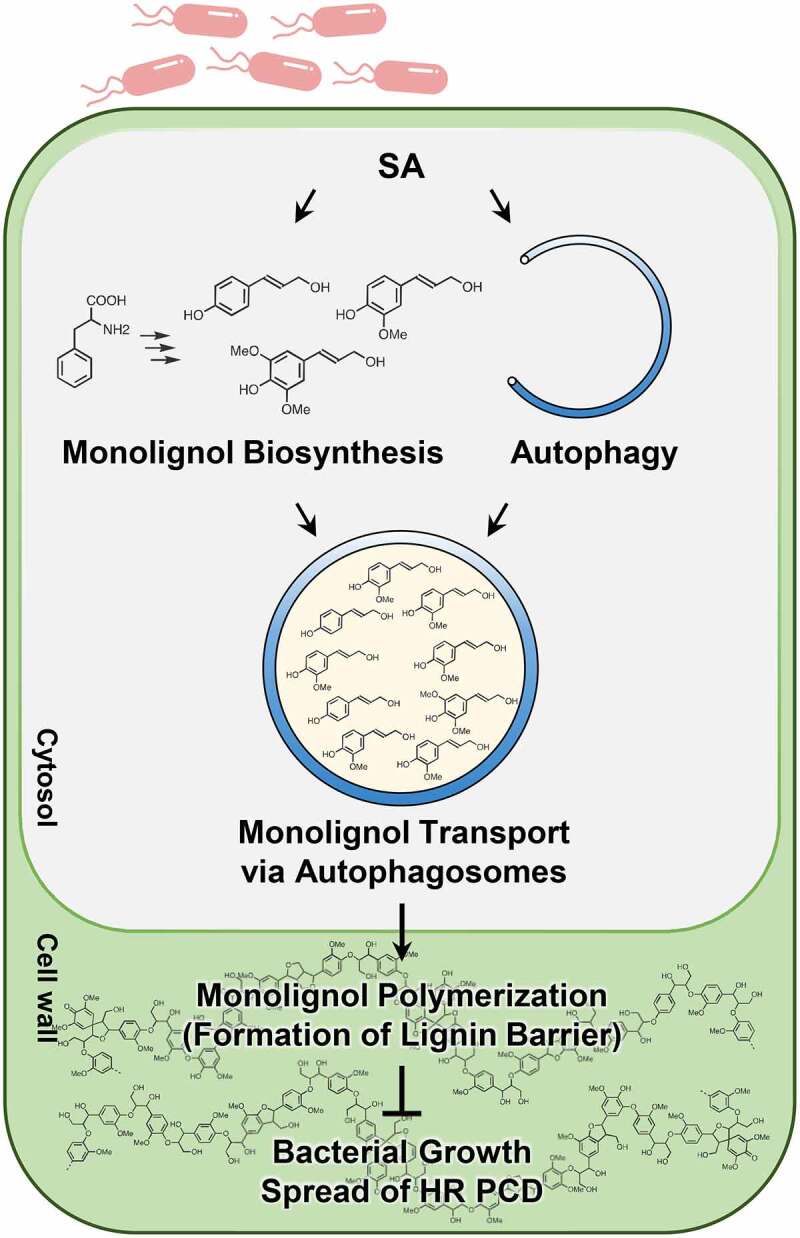
Model for the role of autophagy in pathogen-induced lignification. Avirulent pathogen infection activates SA signaling, resulting in the induction of monolignol biosynthesis and autophagy. Monolignols may be selected as cargo of autophagosomes in the cytosol and transported to the cell wall through the autophagic trafficking process. In this way, autophagy leads to lignification, which in turn enhances disease resistance and prevents the spread of HR PCD.
Compared to well-documented studies of monolignol biosynthesis and polymerization, the transport mechanism of monolignols has remained elusive. Previous in vitro uptake assays demonstrated that monolignols are transported via plasma membrane-derived vesicles, and this process is inhibited either by excluding ATP or by including transporter inhibitors, suggesting the involvement of ABC transporters in monolignol transport [35]. A subsequent study suggested that ABCG29, an Arabidopsis ABC transporter, pumps p-coumaryl alcohol across the plasma membrane of the root endodermis and vascular tissues [38]. ABCG29 expression rendered yeast cells the selective transporter activity for p-coumaryl alcohol but not for other lignin monomers. This finding raised questions on how other monolignols, coniferyl and sinapyl alcohols, are transported and whether they are transported by other ABC transporters or by other transport mechanisms. In fact, studies in differentiating xylem of some woody species (hybrid poplar, Japanese cypress, and bamboo) suggested that transport of monolignols could be also mediated by a proton gradient formed by vacuolar-type H+-ATPase (V-ATPase) [71,72]. Given that ABC transporters constitute a large family, ABC transporter members may have distinct specificities for monolignol types. Alternatively, ABC transporters may be responsible for part of monolignol transport, functioning only in certain cells and tissues or in particular developmental stages and conditions.
The classical functions of autophagy include nutrient recycling, degradation of damaged organelles and proteins, and regulation of PCD [44,45]. In contrast to canonical degradative autophagy, growing evidence indicates that autophagy participates in cellular secretion [73,74]. In eukaryotic cells, most secreted proteins have the amino-terminal signal sequences required for targeting to the endoplasmic reticulum (ER) and move through the ER and Golgi-dependent conventional secretory pathway [75]. However, numerous secreted proteins lack signal peptides and are delivered to the extracellular space through alternative routes, collectively termed unconventional protein secretion (UPS) pathways [76,77]. A main type of UPS routes is secretory autophagy that facilitates unconventional secretion of a variety of cytosolic cargo [73,74]. Among the secretory autophagy cargo are interleukins, cytosolic proteins, protein aggregates (SNCA/α-synuclein, amyloid-β), and granule contents [73,74,78]. Autophagy functions as part of metabolic pathways and the impairment of autophagy is implicated in metabolic diseases [79]. Some evidence suggests that secretory autophagy has a role in the extracellular release of metabolites. Cancer-associated fibroblasts execute autophagy-dependent secretion of metabolites such as amino acids, fatty acids, ketone, lactate, and alanine to fuel tumor cells [80–82]. In addition, autophagy governs ATP secretion, which is instrumental for melanoma cell migration and invasion [83].
Autophagy is critical for metabolism and nutrient allocation in plant growth and stress responses and has been implicated in trafficking of metabolites in plants [84,85]. Anthocyanins, secondary metabolites, are transported from the cytosol to the vacuole in an autophagy-dependent manner. Anthocyanins colocalized with ER-derived vesicle-like structures and anthocyanin accumulation was reduced in atg mutants, suggesting that anthocyanins can be selected as autophagic cargo [86,87]. A different autophagy-dependent mechanism has also been suggested, based on the observation that anthocyanin aggregates are in close contact with the vacuolar surface and directly engulfed by the vacuolar membrane in a microautophagy-like process [88]. An earlier study reported that anthocyanins are located in the cell wall, and approximately 30% of flavonoids accumulate in the cell wall of a flower petal [89]. In other instances, vesicles participated in the transport of phytochemicals and defense-related compounds to the cell surface [90,91]. Moreover, phytochemicals accumulated in the cell wall through the Golgi-independent trafficking process. Therefore, it would be worthwhile to investigate whether the extracellular transport of flavonoids and metabolic compounds is mediated by autophagic pathways.
EXO70B1, an Arabidopsis exocyst subunit, has been engaged in autophagy-dependent anthocyanin transport to the vacuole [84]. The exocyst is an octameric vesicle tethering complex involved in the tethering and targeting of vesicles to the plasma membrane prior to vesicle fusion, and is composed of eight subunits, SEC3, SEC5, SEC8, SEC10, SEC15, EXO70, and EXO84 [92]. EXO70B1-positive compartments colocalized with autophagic bodies and anthocyanin content was decreased in exo70B1 mutants. Furthermore, exo70B1 mutants developed spontaneous leaf lesions mediated by SA accumulation, phenotypes associated with atg mutants [52,84]. SA may be transported via the autophagy-dependent pathway, and SA accumulation in exo70B1 and atg mutants may be due to defects in this pathway [93,94]. This leads to the speculation that SA and other metabolites, such as monolignols, are selected as cargo for autophagosomes and are released out of cells through the autophagic trafficking process. A subsequent question is whether and how metabolites can be enclosed in autophagosomes as cargo. The phenylpropanoid pathway produces a wide array of secondary metabolites, such as monolignols and flavonoids, which are catalyzed by numerous enzymes [95]. Growing evidence shows that these metabolic enzymes are organized into multienzyme complexes called “metabolons” bound to the surface of the ER [96–98], leading to compartmentalization of metabolite synthesis within the cell. It is conceivable that monolignol concentrates formed around the ER surface are directly engulfed by phagophores or early autophagic structures, particularly because the ER has been revealed as a membrane source for autophagosome formation [99,100]. This would be an efficient way to concentrate and release a large amount of monolignols, which in turn allows for the rapid establishment of a lignin barrier against invading pathogens. Further studies are needed to determine how monolignols are selectively packaged into autophagosomes and how the switch between the degradative and secretory pathways is regulated.
In this study, an intriguing observation was over-accumulation of lignin in dnd1 mutant that was previously isolated as a mutant line with no HR cell death but increased resistance to avirulent P. syringae pathogens [61]. We found that dnd1 mutant has enhanced autophagic activity, and when crossed with atg mutants, failed to accumulate lignin and restrict bacterial growth. These results suggest that both enhanced lignification and disease resistance are attributed to autophagy in dnd1 plants. Furthermore, autophagic induction was SA-dependent in dnd1 mutant, as revealed by a great reduction of autophagic activity in dnd1 sid2 mutant. These results led us to propose that lignin formation requires autophagy, which is activated by the SA pathway during plant-pathogen interactions (Figure 9). Meanwhile, we observed that HR cell death is restored in dnd1 atg mutant plants. DND1 encodes a cyclic nucleotide-gated channel CNGC2 [62] that functions as a Ca2+ influx channel [63,101]. Ca2+ signaling is critical for the regulation of plant immunity [102,103]. PTI signaling events include a rapid and transient rise in cytosolic Ca2+, and DND1/CNGC2 has been implicated in Ca2+-dependent PTI [63]. HR PCD, a hallmark of ETI, is associated with sustained Ca2+ elevation and is inhibited by Ca2+ channel blockers [67]. While the loss-of-HR phenotype of dnd1 plants supports the importance of Ca2+ signaling in HR, how HR cell death is recovered in dnd1 atg mutants in which Ca2+ elevation does not occur in response to pathogen infection remains unclear. Because PAMP-triggered Ca2+ signals and immune responses were not sufficiently removed in dnd1/cngc2 mutants [63], it is conceivable that other types of channels or Ca2+-independent pathways may function in place of CNGC2 for the ETI process.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana (Col-0) plants were grown at 23°C under long-day conditions in a 16-h light/8-h dark cycle for general growth and under short-day conditions in a 8-h light/16-h dark cycle for pathogen infection. The mutant lines used in this study are atg5-1 (SAIL_129_B07), atg7-2 (GABI_655B06), acd2-2 (CS3734), lsd1-2 (CS68734), dnd1-1 (CS6523), sid2-1 [104], dnd1-1 sid2-1 [70], GFP-ATG8a atg5-1 [65], and GFP-ATG8a atg7-2 [65]. Double mutants were generated by crossing dnd1-1 with atg5-1 and atg7-2 and identified by genotyping with the MboI derived cleaved amplified polymorphic sequences (dCAPS) marker [105] and gene-specific primers (Table S1).
Plant treatments
The bacterial pathogen Pst DC3000 (AvrRpm1) was grown at 28°C on Kings B agar media containing 50 μg/ml kanamycin (Duchefa, K0126) and 100 μg/ml rifampicin (Duchefa, R0146). Bacteria were suspended in 10 mM MgCl2 and syringe-infiltrated into leaves at 1 × 108 colony-forming unit (CFU)/ml (OD600 = 0.1) for HR PCD development, at 2 × 105 CFU/ml (OD600 = 0.0002) for bacterial growth, and at 1 × 106 CFU/ml (OD600 = 0.001) for ATG8 lipidation and microscopic analysis. CA (Sigma-Aldrich, 223,735) and DMAC-CA [55,56] were dissolved in 100% dimethyl sulfoxide (DMSO; Duchefa, D1370) to make 50 mM stock solutions. Protoplast suspensions were treated with CA and DMAC-CA at a final concentration of 50 μM and incubated for 4 h. Arabidopsis leaves were infiltrated with CA (50 μM) and DMAC-CA (50 μM) for 1 h, with Con A (5 μM; BioVision, B2091) for 8 h, and with SA (1 mM; Duchefa, S1367) for 4–24 h, as indicated. Chemical-treated leaves were air-dried before pathogen inoculation.
Bacterial growth assay
Bacterial growth was determined, as previously described [11]. Leaves of 4- or 6-week-old plants were syringe-inoculated with 2 × 105 CFU/ml of Pst DC3000 (AvrRpm1). Leaf discs (3 mm in diameter) from infiltrated leaves were pooled and ground in 10 mM MgCl2. To determine CFU, serial dilutions were plated on Kings B agar medium containing 50 μg/ml kanamycin (Duchefa, K0126) and 100 μg/ml rifampicin (Duchefa, R0146). Experiments were repeated 3–5 times with biologically independent samples.
Gene expression analysis
Total RNAs were extracted from Arabidopsis leaves using TRIsure reagent (Bioline, BIO-38033) and reverse-transcribed into cDNAs using the PrimeScriptTM RT reagent Kit (TaKaRa, RR037A). Quantitative real-time PCR was performed on a LightCycler 480 system (Roche, Mannheim, Germany) using the KAPA SYBR FAST qPCR master mix (Kapa Biosystems, KK4611) with gene-specific primers (Table S1). For transcript normalization, Actin2/AT3G18780 was used as a reference gene. Data were analyzed using LC480Conversion and LinRegPCR software (Heart Failure Research Center). Experiments were repeated 3–5 times with biologically independent samples.
Phloroglucinol staining
Lignin was stained with phloroglucinol, as previously described [11]. Arabidopsis leaves were dehydrated in 100% ethanol overnight, gradually rehydrated in a graded series of ethanol (75%, 50%, and 25%) and water for 30 min each, and then stained with 3% phloroglucinol (Sigma-Aldrich, P3502) dissolved in 30% HCl for 1 min. The stained leaves were observed under an optical microscope (Leica EZ4E, Leica Microsystems, Germany).
Lignin quantification
Lignin content was determined by the acetyl bromide-based method, as previously described [11]. Arabidopsis leaves were frozen and ground in liquid nitrogen. The dried samples (3 ± 1 mg, accurately weighed) were washed serially with 70% ethanol, chloroform:methanol (1:1 v:v), and acetone. The washed pellets were completely dried at 45°C and digested with 25% acetyl bromide in acetic acid at 70°C for 1 h by vortexing. Samples were cooled on ice and centrifuged for 5 min at 16,000 g. The supernatants (100 μl) were transferred to glass tubes and mixed with 400 μl of 2 M NaOH, 70 μl of 0.5 M hydroxylamine hydrochloride (Sigma-Aldrich, 255,580), and 430 μl of acetic acid. The prepared solutions were transferred to 96-well microplates, and the absorbance was measured using a microplate reader (SpectraMax i3x, Molecular Devices, San Jose, CA, USA) at 280 nm. The content of acetyl bromide soluble lignin (% ABSL) was calculated using Beers Law [106]. The extinction coefficient used for Arabidopsis was 15.69 l/g cm [107].
Monolignol quantification
Monolignol content was analyzed, according to a previously described method with minor modifications [108]. Briefly, Arabidopsis leaves were frozen and ground in liquid nitrogen. Leaf powder (200 mg) was homogenized with 1 ml of 75% methanol containing the internal standard (1 μg/ml, p-fluoro-DL-phenylalanine) and incubated at 65°C for 30 min. The samples were then centrifuged at 16,000 g for 15 min. The supernatants were dried in a centrifugal evaporator at room temperature, and the resultant pellets were dissolved in 60 μl of 75% methanol. Subsequently, 10 µl of the prepared samples were injected into the UPLC-MS/MS system. The chromatographic system consisted of a Waters Acquity UPLC system (Waters, Milford, MA, USA) coupled with a quadrupole time-of-flight (Q-TOF) mass spectrometer (Synapt G2-Si, Waters, Milford, MA, USA). Chromatographic separation was carried out on an Acquity UPLC BEH C8 column (2.1 mm x 100 mm, 1.7 µm). The mobile phase (delivered at 0.3 ml/min) consisted of solvent A (2.5 mM ammonium acetate in water adjusted to pH 5.3 with glacial acetic acid) and solvent B (98% acetonitrile, 2% water, 0.02% formic acid). Mass profiling was performed in negative scan mode with mass-to-charge-ratio (m/z) of 50 to 600, and the mass spectrometry parameters were set as follows: capillary voltage, 2,000 V; cone voltage, 20 V; source temperature, 110°C; desolvation temperature, 550°C; desolvation gas flow, 900 l/h. To ensure the accuracy of the measured mass, leucine-encephalin (m/z 554.2615 in the negative mode) was used as a reference lock-mass compound. Quantitative analysis of metabolites was performed in the multiple reaction monitoring (MRM) mode, and the detailed MRM conditions for each analyte are listed in Table S2. The data were processed using MassLynxTM 4.1 software.
EV isolation
EVs were isolated from the apoplastic wash fluids of leaves, as previously described [57,58]. Plant leaves were vacuum-infiltrated with isolation buffer (20 mM MES-KOH, pH 5.7, 0.1 M NaCl, 2 mM CaCl2) and centrifuged for 10 min at 900 g. The collected fluids were centrifuged for 30 min at 2,000 g to remove cell debris, filtered through a 0.45-μm filter, and further cleaned by centrifugation for 30 min at 10,000 g. The resulting supernatants were used as apoplastic wash fluids in immunoblotting. To isolate EVs, the apoplastic wash fluids were transferred to ultracentrifuge tubes and centrifuged for 1 h at 100,000 g. The pellets were washed, resuspended in isolation buffer, and used as EVs in microscopic analysis and immunoblotting.
Immunoblot analysis
Proteins in the apoplastic wash fluid were precipitated with 20% trichloroacetic acid (w/v) and washed three times with acetone. The apoplastic protein pellets and collected EVs were resuspended in 6x SDS sample buffer (375 mM Tris-HCl, pH 6.8, 50% glycerol, 9% SDS, 1.2 M β-mercaptoethanol, 0.03% bromophenol blue). The samples were boiled for 10 min and clarified via centrifugation. Proteins were separated on 12% SDS polyacrylamide gels and transferred onto polyvinylidene fluoride membranes. The membranes were incubated with anti-ATG8a (Abcam, ab77003), anti-GST (Abcam, ab111947), anti-actin (Abcam, ab197345), and anti-PR1 (Agrisera, AS10 687) antibodies. Antibody-bound proteins were detected by incubation with secondary antibodies conjugated to horseradish peroxidase using an enhanced chemiluminescence (ECL) system (Amersham Biosciences).
Microscopy analysis
LTG staining was performed, as previously described [53]. Leaves were infiltrated with 1 μM LTG DND-26 (Invitrogen, L7526) and incubated for 1 h. Ca2+ ions were visualized by the fluorescent Ca2+ indicator Fluo-3 AM (ThermoFisher, F14218), as previously described [109]. Leaves were stained with 20 μM Fluo-3 AM in 20 μM MES-KOH (pH 5.7) for 1 h at 4°C, followed by washing twice with 20 μM MES-KOH (pH 5.7). Images were acquired using a confocal microscope (LSM 700, Carl Zeiss, Jena, Germany). Excitation and emission were set at 488 and 515 nm for LTG and Fluo-3 AM, 639 and 710 nm for chlorophyll, and 488 and 510 nm for GFP. To quantify the number of puncta, the images were analyzed by ImageJ (http://rsb.info.nih.gov/ij/) and manual counting.
Protoplast isolation and transfection
Protoplasts were isolated from rosette leaves of 5-week-old Col-0 plants, as previously described [110]. Leaves were sliced and incubated for 3 h in an enzyme solution (20 mM MES-KOH, pH 5.7, 1% cellulase R-10 [Yakult], 0.4% macerozyme R-10 [Yakult], 0.4 M D-mannitol, 20 mM KCl, 10 mM CaCl2, 0.1% BSA [MP Biomedicals, 160,069]). Protoplasts were isolated by filtering through a 70-μm cell strainer and centrifugation. The isolated protoplasts were resuspended in W5 solution (1.5 mM MES-KOH, pH 5.6, 154 mM NaCl, 125 mM CaCl2, 5 mM KCl). Protoplast transfection was carried out by a polyethylene glycol-mediated method [110].
Subcellular localization analysis
For CASPL1D1-mCherry and CASPL4D1-Citrine, the CASPL1D1 and CASPL4D1 coding regions were fused to the N-termini of mCherry and Citrine, respectively. The constructs were placed under the control of the CaMV 35S promoter and were introduced into protoplasts. For DMAC-CA and GFP-ATG8a, protoplasts or leaves of 35S:GFP-ATG8a plants were treated with 50 μM DMAC-CA. Images were acquired using a confocal microscope (LSM 800, Carl Zeiss, Jena, Germany). Excitation and emission were set at 555 and 630 nm for mCherry, 515 and 530 nm for Citrine, 405 and 490 nm for DMAC-CA, and 488 and 510 nm for GFP.
ATG8 lipidation assay
Immunoblotting for an ATG8 lipidation assay was performed, as previously described [53]. Total proteins were extracted from 5-6-week-old plants grown under short-day conditions. Equal amounts of proteins were subjected to 15% SDS-PAGE in the presence of 6 M urea and transferred to polyvinylidene fluoride membranes in a semi-dry transfer system (Bio-Rad). The blots were probed with the ATG8a antibody (Abcam, ab77003) overnight at 4°C. Following incubation with a horseradish peroxidase-conjugated secondary antibody (Cytiva, NA934), antibody-bound proteins were detected using Amersham Imager 680 (GE Healthcare, Marlborough, MA, USA).
Statistical analysis
Statistical analyses were performed using GraphPad Prism (v. 8.0). The data were expressed as mean ± standard deviation [86]. Significant differences between experimental groups were evaluated by one-way ANOVA with Tukeys HSD test or unpaired Students t test for multiple comparisons or single comparisons, respectively. Statistical significance was set at P < 0.05. Detailed information about statistical analysis is described in the figure legends. All experiments were repeated 3-5 times with similar results.
Supplementary Material
Acknowledgments
We thank Mikael Brosché (University of Helsinki) for dnd1-1 sid2-1 seeds and Taijoon Chung (Pusan National University) for GFP-ATG8a atg5-1 and GFP-ATG8a atg7-2 seeds.
Funding Statement
This work was supported by a Korea University grant and National Research Foundation of Korea (NRF) grants (2018R1A5A1023599, 2021R1A2C1003213, 2021R1A6A1A10045235, 2021R1A6A3A01088290) from the Korean government (MSIP). YT acknowledges support from the Japan Society for the Promotion of Science (JSPS KAKENHI, JP#20H03044).
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15548627.2022.2085496.
References
- [1].Vanholme R, Demedts B, Morreel K, et al. Lignin biosynthesis and structure. Plant Physiol. 2010;153(3):895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Boerjan W, Ralph J, Baucher M.. Lignin biosynthesis. Annu Rev Plant Biol. 2003;54(1):519–546. [DOI] [PubMed] [Google Scholar]
- [3].Ralph J, Lundquist K, Brunow G, et al. Lignins: natural polymers from oxidative coupling of 4-hydroxyphenyl-propanoids. Phytochem Rev. 2004;3(1):29–60. [Google Scholar]
- [4].Voxeur A, Wang Y, Sibout R. Lignification: different mechanisms for a versatile polymer. Curr Opin Plant Biol. 2015;23:83–90. [DOI] [PubMed] [Google Scholar]
- [5].Wang Y, Chantreau M, Sibout R, et al. Plant cell wall lignification and monolignol metabolism. Front Plant Sci. 2013;4:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Malinovsky FG, Fangel JU, Willats WG. The role of the cell wall in plant immunity. Front Plant Sci. 2014;5:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nicholson RL, Hammerschmidt R. Phenolic-compounds and their role in disease resistance. Annu Rev Phytopathol. 1992;30(1):369–389. [Google Scholar]
- [8].Chezem WR, Memon A, Li FS, et al. SG2-type R2R3-MYB transcription factor MYB15 controls defense-induced lignification and basal immunity in Arabidopsis. Plant Cell. 2017;29(8):1907–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cui H, Tsuda K, Parker JE. Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol. 2015;66(1):487–511. [DOI] [PubMed] [Google Scholar]
- [10].Kim SH, Lam PY, Lee MH, et al. The Arabidopsis R2R3 MYB transcription factor MYB15 is a key regulator of lignin biosynthesis in effector-triggered immunity. Front Plant Sci. 2020;11:1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lee MH, Jeon HS, Kim SH, et al. Lignin-based barrier restricts pathogens to the infection site and confers resistance in plants. EMBO J. 2019;38(23):e101948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zipfel C. Plant pattern-recognition receptors. Trends Immunol. 2014;35(7):345–351. [DOI] [PubMed] [Google Scholar]
- [13].Coll NS, Epple P, Dangl JL. Programmed cell death in the plant immune system. Cell Death Differ. 2011;18(8):1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bonawitz ND, Chapple C. The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu Rev Genet. 2010;44(1):337–363. [DOI] [PubMed] [Google Scholar]
- [15].Vanholme R, De Meester B, Ralph J, et al. Lignin biosynthesis and its integration into metabolism. Curr Opin Biotechnol. 2019;56:230–239. [DOI] [PubMed] [Google Scholar]
- [16].Whetten R, Sederoff R. Lignin biosynthesis. Plant Cell. 1995;7(7):1001–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tobimatsu Y, Schuetz M. Lignin polymerization: how do plants manage the chemistry so well? Curr Opin Biotechnol. 2019;56:75–81. [DOI] [PubMed] [Google Scholar]
- [18].Berthet S, Demont-Caulet N, Pollet B, et al. Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell. 2011;23(3):1124–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Herrero J, Fernandez-Perez F, Yebra T, et al. Bioinformatic and functional characterization of the basic peroxidase 72 from Arabidopsis thaliana involved in lignin biosynthesis. Planta. 2013;237(6):1599–1612. [DOI] [PubMed] [Google Scholar]
- [20].Shigeto J, Itoh Y, Hirao S, et al. Simultaneously disrupting AtPrx2, AtPrx25 and AtPrx71 alters lignin content and structure in Arabidopsis stem. J Integr Plant Biol. 2015;57(4):349–356. [DOI] [PubMed] [Google Scholar]
- [21].Zhao Q, Nakashima J, Chen F, et al. LACCASE Is necessary and nonredundant with PEROXIDASE for lignin polymerization during vascular development in Arabidopsis. Plant Cell. 2013;25(10):3976–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hosmani PS, Kamiya T, Danku J, et al. Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root. Proc Natl Acad Sci U S A. 2013;110(35):14498–14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lee Y, Rubio MC, Alassimone J, et al. A mechanism for localized lignin deposition in the endodermis. Cell. 2013;153(2):402–412. [DOI] [PubMed] [Google Scholar]
- [24].Naseer S, Lee Y, Lapierre C, et al. Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc Natl Acad Sci U S A. 2012;109(25):10101–10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Roppolo D, De Rybel B, Denervaud Tendon V, et al. A novel protein family mediates Casparian strip formation in the endodermis. Nature. 2011;473(7347):380–383. [DOI] [PubMed] [Google Scholar]
- [26].Liu CJ. Deciphering the enigma of lignification: precursor transport, oxidation, and the topochemistry of lignin assembly. Mol Plant. 2012;5(2):304–317. [DOI] [PubMed] [Google Scholar]
- [27].Liu CJ, Miao YC, Zhang KW. Sequestration and transport of lignin monomeric precursors. Molecules. 2011;16(1):710–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Perkins M, Smith RA, Samuels L. The transport of monomers during lignification in plants: anything goes but how? Curr Opin Biotechnol. 2019;56:69–74. [DOI] [PubMed] [Google Scholar]
- [29].Perkins ML, Schuetz M, Unda F, et al. Monolignol export by diffusion down a polymerization-induced concentration gradient. Plant Cell. 2022;34(5):2080–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Aoki D, Hanaya Y, Akita T, et al. Distribution of coniferin in freeze-fixed stem of Ginkgo biloba L. by cryo-TOF-SIMS/SEM. Sci Rep. 2016;6(1):31525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lanot A, Hodge D, Jackson RG, et al. The glucosyltransferase UGT72E2 is responsible for monolignol 4-O-glucoside production in Arabidopsis thaliana. Plant J. 2006;48(2):286–295. [DOI] [PubMed] [Google Scholar]
- [32].Le Roy J, Huss B, Creach A, et al. Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front Plant Sci. 2016;7:735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lin JS, Huang XX, Li Q, et al. UDP-glycosyltransferase 72B1 catalyzes the glucose conjugation of monolignols and is essential for the normal cell wall lignification in Arabidopsis thaliana. Plant J. 2016;88(1):26–42. [DOI] [PubMed] [Google Scholar]
- [34].Ehlting J, Mattheus N, Aeschliman DS, et al. Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J. 2005;42(5):618–640. [DOI] [PubMed] [Google Scholar]
- [35].Miao YC, Liu CJ. ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proc Natl Acad Sci U S A. 2010;107(52):22728–22733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tht D, Martinoia E, Lee Y. Functions of ABC transporters in plant growth and development. Curr Opin Plant Biol. 2018;41:32–38. [DOI] [PubMed] [Google Scholar]
- [37].Rea PA. Plant ATP-binding cassette transporters. Annu Rev Plant Biol. 2007;58(1):347–375. [DOI] [PubMed] [Google Scholar]
- [38].Alejandro S, Lee Y, Tohge T, et al. AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr Biol. 2012;22(13):1207–1212. [DOI] [PubMed] [Google Scholar]
- [39].Takeuchi M, Kegasa T, Watanabe A, et al. Expression analysis of transporter genes for screening candidate monolignol transporters using Arabidopsis thaliana cell suspensions during tracheary element differentiation. J Plant Res. 2018;131(2):297–305. [DOI] [PubMed] [Google Scholar]
- [40].Chung T. See how I eat my greens-autophagy in plant cells. J Plant Biol. 2011;54(6):339–350. [Google Scholar]
- [41].Marshall RS, Vierstra RD. Autophagy: the master of bulk and selective recycling. Annu Rev Plant Biol. 2018;69(1):173–208. [DOI] [PubMed] [Google Scholar]
- [42].Feng Y, He D, Yao Z, et al. The machinery of macroautophagy. Cell Res. 2014;24(1):24–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Suzuki H, Osawa T, Fujioka Y, et al. Structural biology of the core autophagy machinery. Curr Opin Struct Biol. 2017;43:10–17. [DOI] [PubMed] [Google Scholar]
- [44].Qi H, Xia FN, Xiao S. Autophagy in plants: physiological roles and post-translational regulation. J Integr Plant Biol. 2021;63(1):161–179. [DOI] [PubMed] [Google Scholar]
- [45].Wang P, Mugume Y, Bassham DC. New advances in autophagy in plants: regulation, selectivity and function. Sem Cell Dev Biol. 2018;80:113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hofius D, Schultz-Larsen T, Joensen J, et al. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell. 2009;137(4):773–783. [DOI] [PubMed] [Google Scholar]
- [47].Lai Z, Wang F, Zheng Z, et al. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 2011;66(6):953–968. [DOI] [PubMed] [Google Scholar]
- [48].Lenz HD, Haller E, Melzer E, et al. Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. 2011;66(5):818–830. [DOI] [PubMed] [Google Scholar]
- [49].Patel S, Dinesh-Kumar SP. Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy. 2008;4(1):20–27. [DOI] [PubMed] [Google Scholar]
- [50].Wang Y, Nishimura MT, Zhao T, et al. ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J. 2011;68(1):74–87. [DOI] [PubMed] [Google Scholar]
- [51].Liu Y, Schiff M, Czymmek K, et al. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121(4):567–577. [DOI] [PubMed] [Google Scholar]
- [52].Yoshimoto K, Jikumaru Y, Kamiya Y, et al. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell. 2009;21(9):2914–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kwon SI, Cho HJ, Kim SR, et al. The Rab GTPase RabG3b positively regulates autophagy and immunity-associated hypersensitive cell death in Arabidopsis. Plant Physiol. 2013;161(4):1722–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Munch D, Rodriguez E, Bressendorff S, et al. Autophagy deficiency leads to accumulation of ubiquitinated proteins, ER stress, and cell death in Arabidopsis. Autophagy. 2014;10(9):1579–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Tobimatsu Y, Davidson CL, Grabber JH, et al. Fluorescence-tagged monolignols: synthesis, and application to studying in vitro lignification. Biomacromolecules. 2011;12(5):1752–1761. [DOI] [PubMed] [Google Scholar]
- [56].Tobimatsu Y, Wagner A, Donaldson L, et al. Visualization of plant cell wall lignification using fluorescence-tagged monolignols. Plant J. 2013;76(3):357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].He B, Cai Q, Qiao L, et al. RNA-binding proteins contribute to small RNA loading in plant extracellular vesicles. Nat Plants. 2021;7(3):342–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Rutter BD, Innes RW. Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol. 2017;173(1):728–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Greenberg JT, Guo A, Klessig DF, et al. Programmed cell death in plants: a pathogen-triggered response activated coordinately with multiple defense functions. Cell. 1994;77(4):551–563. [DOI] [PubMed] [Google Scholar]
- [60].Munch D, Teh OK, Malinovsky FG, et al. Retromer contributes to immunity-associated cell death in Arabidopsis. Plant Cell. 2015;27(2):463–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Yu IC, Parker J, Bent AF. Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc Natl Acad Sci U S A. 1998;95(13):7819–7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Clough SJ, Fengler KA, Yu IC, et al. The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc Natl Acad Sci U S A. 2000;97(16):9323–9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Tian W, Hou C, Ren Z, et al. A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature. 2019;572(7767):131–135. [DOI] [PubMed] [Google Scholar]
- [64].Jin M, Klionsky DJ. Regulation of autophagy: modulation of the size and number of autophagosomes. FEBS Lett. 2014;588(15):2457–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Shin KD, Lee HN, Chung T. A revised assay for monitoring autophagic flux in Arabidopsis thaliana reveals involvement of AUTOPHAGY-RELATED9 in autophagy. Mol Cells. 2014;37(5):399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Ma W, Berkowitz GA. The grateful dead: calcium and cell death in plant innate immunity. Cell Microbiol. 2007;9(11):2571–2585. [DOI] [PubMed] [Google Scholar]
- [67].Ren H, Zhao X, Li W, et al. Calcium signaling in plant programmed cell death. Cells. 2021;10(5):1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Gee KR, Brown KA, Chen WNU, et al. Chemical and physiological characterization of fluo-4 Ca2+-indicator dyes. Cell Calcium. 2000;27(2):97–106. [DOI] [PubMed] [Google Scholar]
- [69].Wildermuth MC, Dewdney J, Wu G, et al. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414(6863):562–565. [DOI] [PubMed] [Google Scholar]
- [70].Xu E, Brosche M. Salicylic acid signaling inhibits apoplastic reactive oxygen species signaling. BMC Plant Biol. 2014;14(1):155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Tsuyama T, Kawai R, Shitan N, et al. Proton-dependent coniferin transport, a common major transport event in differentiating xylem tissue of woody plants. Plant Physiol. 2013;162(2):918–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Tsuyama T, Matsushita Y, Fukushima K, et al. Proton gradient-dependent transport of p-glucocoumaryl alcohol in differentiating xylem of woody plants. Sci Rep. 2019;9(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].New J, Thomas SM. Autophagy-dependent secretion: mechanism, factors secreted, and disease implications. Autophagy. 2019;15(10):1682–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Ponpuak M, Mandell MA, Kimura T, et al. Secretory autophagy. Curr Opin Cell Biol. 2015;35:106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Wang XF, Chung KP, Lin WL, et al. Protein secretion in plants: conventional and unconventional pathways and new techniques. J Exp Bot. 2018;69(1):21–37. [DOI] [PubMed] [Google Scholar]
- [76].Malhotra V. Unconventional protein secretion: an evolving mechanism. EMBO J. 2013;32(12):1660–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Rabouille C. Pathways of unconventional protein secretion. Trends Cell Biol. 2017;27(3):230–240. [DOI] [PubMed] [Google Scholar]
- [78].Cavalli G, Cenci S. Autophagy and protein secretion. J Mol Biol. 2020;432(8):2525–2545. [DOI] [PubMed] [Google Scholar]
- [79].Gonzalez CD, Resnik R, Vaccaro MI. Secretory autophagy and its relevance in metabolic and degenerative disease. Front Endocrinol. 2020;11:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Guido C, Whitaker-Menezes D, Capparelli C, et al. Metabolic reprogramming of cancer-associated fibroblasts by TGF-beta drives tumor growth: connecting TGF-beta signaling with “Warburg-like” cancer metabolism and L-lactate production. Cell Cycle. 2012;11(16):3019–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Martinez-Outschoorn UE, Lisanti MP, Sotgia F. Catabolic cancer-associated fibroblasts transfer energy and biomass to anabolic cancer cells, fueling tumor growth. Sem Cancer Biol. 2014;25:47–60. [DOI] [PubMed] [Google Scholar]
- [82].Sousa CM, Biancur DE, Wang X, et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature. 2016;536(7617):479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Martin S, Dudek-Peric AM, Garg AD, et al. An autophagy-driven pathway of ATP secretion supports the aggressive phenotype of BRAF(V600E) inhibitor-resistant metastatic melanoma cells. Autophagy. 2017;13(9):1512–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Kulich I, Pecenkova T, Sekeres J, et al. Arabidopsis exocyst subcomplex containing subunit EXO70B1 is involved in autophagy-related transport to the vacuole. Traffic. 2013;14(11):1155–1165. [DOI] [PubMed] [Google Scholar]
- [85].Michaeli S, Galili G, Genschik P, et al. Autophagy in plants–what’s new on the menu? Trends Plant Sci. 2016;21(2):134–144. [DOI] [PubMed] [Google Scholar]
- [86].Pourcel L, Irani NG, Lu Y, et al. The formation of anthocyanic vacuolar inclusions in Arabidopsis thaliana and implications for the sequestration of anthocyanin pigments. Mol Plant. 2010;3(1):78–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Poustka F, Irani NG, Feller A, et al. A trafficking pathway for anthocyanins overlaps with the endoplasmic reticulum-to-vacuole protein-sorting route in Arabidopsis and contributes to the formation of vacuolar inclusions. Plant Physiol. 2007;145(4):1323–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Chanoca A, Kovinich N, Burkel B, et al. Anthocyanin vacuolar inclusions form by a microautophagy mechanism. Plant Cell. 2015;27(9):2545–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Markham KR, Ryan KG, Gould KS, et al. Cell wall sited flavonoids in lisianthus flower petals. Phytochemistry. 2000;54(7):681–687. [DOI] [PubMed] [Google Scholar]
- [90].Lin Y, Irani NG, Grotewold E. Sub-cellular trafficking of phytochemicals explored using auto-fluorescent compounds in maize cells. BMC Plant Biol. 2003;3(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Meyer D, Pajonk S, Micali C, et al. Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J. 2009;57(6):986–999. [DOI] [PubMed] [Google Scholar]
- [92].Heider MR, Munson M. Exorcising the exocyst complex. Traffic. 2012;13(7):898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Kulich I, Zarsky V. Autophagy-related direct membrane import from ER/cytoplasm into the vacuole or apoplast: a hidden gateway also for secondary metabolites and phytohormones? Int J Mol Sci. 2014;15(5):7462–7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Pečenková T, Marković V, Sabol P, et al. Exocyst and autophagy-related membrane trafficking in plants. J Exp Bot. 2018;69(1):47–57. [DOI] [PubMed] [Google Scholar]
- [95].Fraser CM, Chapple C. The phenylpropanoid pathway in Arabidopsis. Arabidopsis Book. 2011;9:e0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Bassard JE, Richert L, Geerinck J, et al. Protein-protein and protein-membrane associations in the lignin pathway. Plant Cell. 2012;24(11):4465–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Chen HC, Li QZ, Shuford CM, et al. Membrane protein complexes catalyze both 4-and 3-hydroxylation of cinnamic acid derivatives in monolignol biosynthesis. Proc Natl Acad Sci U S A. 2011;108(52):21253–21258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Gou M, Ran X, Martin DW, et al. The scaffold proteins of lignin biosynthetic cytochrome P450 enzymes. Nat Plants. 2018;4(5):299–310 [DOI] [PubMed] [Google Scholar]
- [99].Hayashi-Nishino M, Fujita N, Noda T, et al. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nature Cell Biol. 2009;11(12):1433–1437. [DOI] [PubMed] [Google Scholar]
- [100].Kotani T, Kirisako H, Koizumi M, et al. The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc Natl Acad Sci U S A. 2018;115(41):10363–10368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Leng Q, Mercier RW, Yao W, et al. Cloning and first functional characterization of a plant cyclic nucleotide-gated cation channel. Plant Physiol. 1999;121(3):753–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Cheval C, Aldon D, Galaud JP, et al. Calcium/calmodulin-mediated regulation of plant immunity. Biochim Biophys Acta. 2013;1833(7):1766–1771. [DOI] [PubMed] [Google Scholar]
- [103].Tian W, Wang C, Gao Q, et al. Calcium spikes, waves and oscillations in plant development and biotic interactions. Nat Plants. 2020;6(7):750–759. [DOI] [PubMed] [Google Scholar]
- [104].Adie BAT, Perez-Perez J, Perez-Perez MM, et al. ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell. 2007;19(5):1665–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Genger RK, Jurkowski GI, McDowell JM, et al. Signaling pathways that regulate the enhanced disease resistance of Arabidopsis “Defense, No Death” mutants. Mol Plant Microbe Interact. 2008;21(10):1285–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Kapp N, Barnes WJ, Richard TL, et al. Imaging with the fluorogenic dye Basic Fuchsin reveals subcellular patterning and ecotype variation of lignification in Brachypodium distachyon. J Exp Bot. 2015;66(14):4295–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Foster CE, Martin TM, Pauly M. Comprehensive compositional analysis of plant cell walls (Lignocellulosic biomass) part I: lignin. J Vis Exp. 2010;37:e1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Jaini R, Wang P, Dudareva N, et al. Targeted metabolomics of the phenylpropanoid pathway in Arabidopsis thaliana using reversed phase liquid chromatography coupled with tandem mass spectrometry. Phytochem Analysis. 2017;28(4):267–276. [DOI] [PubMed] [Google Scholar]
- [109].Li W, Xu F, Chen S, et al. A comparative study on Ca content and distribution in two Gesneriaceae species reveals distinctive mechanisms to cope with high rhizospheric soluble calcium. Front Plant Sci. 2014;5:647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2(7):1565–1572. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


