ABSTRACT
Macroautophagy/autophagy, a highly conserved catabolic pathway that maintains proper cellular homeostasis is stringently regulated by numerous autophagy-related (Atg) proteins. Many studies have investigated autophagy regulation at the transcriptional level; however, relatively little is known about translational control. Here, we report the upstream open reading frame (uORF)-mediated translational control of multiple Atg proteins in Saccharomyces cerevisiae and in human cells. The translation of several essential autophagy regulators in yeast, including Atg13, is suppressed by canonical uORFs under nutrient-rich conditions, and is activated during nitrogen-starvation conditions. We also found that the predicted human ATG4B and ATG12 non-canonical uORFs suppress downstream coding sequence translation. These results demonstrate that uORF-mediated translational control is a widely used mechanism among ATG genes from yeast to human and suggest a model for how some ATG genes bypass the general translational suppression that occurs under stress conditions to maintain a proper level of autophagy.
Abbreviations: 5’ UTR, 5’ untranslated region; Atg, autophagy-related; CDS, coding sequence; Cvt, cytoplasm-to-vacuole targeting; HBSS, Hanks’ balanced salt solution; PA, protein A; PE, phosphati-dylethanolamine; PIC, preinitiation complex; PtdIns3K, phosphatidylinositol 3-kinase; qRT-PCR, quantitative reverse transcription PCR; Ubl, ubiquitin-like; uORF, upstream open reading frame; WT, wild-type.
KEYWORDS: Autophagy, human, lysosome, stress, translational regulation, vacuole, yeast
Introduction
Macroautophagy (hereafter referred to as autophagy) is an evolutionarily conserved cellular process in which intracellular components and dysfunctional organelles are delivered to lysosomes/vacuoles for degradation and recycling [1]. Aberrant autophagy has been implicated in a series of diseases, including tumorigenesis, neurodegeneration, and metabolic diseases [2,3]. To date, more than 40 autophagy-related (ATG) genes has been identified as components of the autophagy pathway in yeast [4]. As a highly conserved process among eukaryotes, most of the protein products of these ATG genes have homologs or functional counterparts in human. Under nutrient-rich conditions, autophagy is preserved at a relatively low level to work as a cellular quality control mechanism. Upon stress conditions, such as nutrient deprivation, high-level autophagy is induced to effectively promote bioenergetic homeostasis and maintain cell survival. Active autophagy is largely dependent on proper expression of ATG genes. Previous studies have demonstrated many transcriptional activators and repressors in autophagy regulation [5]; however, little is known about how cells efficiently regulate autophagy at the level of translation.
Of note, during stress conditions, such as nutrient limitation, translational activity of most genes is downregulated to save metabolic energy and avoid the generation of dysfunctional proteins. The global translational downregulation of genes upon stress induction is achieved through Sui2/eIF2α phosphorylation, which affects formation of the 43S preinitiation complex involved in eukaryotic translation initiation [6]. Though cellular stress leads to globally decreased translation of most genes, a small number of mRNAs display upregulated translational activity. For example, many ATG genes are reported to bypass the global translational suppression induced by Sui2/eIF2α phosphorylation during nutrient deprivation [7]. Recently, two studies from our lab showed that Dhh1, a DExD/H-box RNA helicase, and Psp2, an RGG motif protein, are engaged in efficient translational regulation of Atg1 and Atg13, two important components of the Atg1 kinase complex, the key autophagy initiator [8,9]. In addition, translation of ATG5, ATG7, ATG12, ATG16L1, and BECN1 are regulated by some RNA-binding proteins such as ELAVL1/HuR and ELAVL4/HuD, HNRNPA1/hnRPA1, and ZFP36/TTP [10–14]. However, it is not clear how other ATG genes get upregulated as part of the translational reprogramming that occurs under stress conditions.
Upstream open reading frames (uORFs) are short coding sequences with start codons located in the 5' untranslated region (5' UTR) of eukaryotic mRNAs [15]. Though most translation events occur via the main ORF of mRNAs, many translation events also occur within the 5' UTR of a gene, including those that initiate at uORFs. The stop codon of a uORF is typically located before the stop codon of the protein coding sequence (CDS). Therefore, a uORF may either end before the start of the CDS or may partially overlap with the CDS depending on the location of its associated stop codon. It is widely accepted that in most cases, uORFs act as important cis-regulatory elements to contrtol translation of the corresponding CDS, especially upon amino acid starvation [16]. Nearly half of the mRNA transcripts in yeast and human possess uORFs [15,17]. However, not all uORFs are active in translation regulation, and the mechanism for how uORFs modify CDS translation varies on a case-by-case basis. Currently, proposed models for how uORFs alter CDS translation include leaky scanning, translation reinitiation and ribosome stalling [18–20]. In addition, it is proposed that the peptide produced by a uORF may have biological functions [21]. As uORFs of many genes, such as GCN4 in yeast and ATF4 in mammals [19,22], are closely correlated with stress-activated gene upregulation, we raised the question of whether uORFs play a role in downregulating ATG gene expression to help maintain a basal level of autophagy under nutrient-rich conditions; bypass of the uORFs would allow increased gene expression when cells are undergoing global translation suppression during stress.
In this paper, we report that uORFs are involved in the translational control of a multitude of ATG genes in both S. cerevisiae and human cells. We first demonstrate that an ATG13 uORF acts as a cis-acting element, the bypass of which facilitates translational expression of Atg13 in response to stress-induced eIF2 phosphorylation in yeast. The translational event of Atg13 mediated by the uORF is dependent on the surrounding contexts of the uORF start codon, and on the Gcn2 kinase. We also find that at least four uORFs of yeast ATG5 differentially contribute to ATG5 translational control; bypass of the ATG5 and ATG13 uORFs results in increased autophagy activity. Yeast ATG12 and yeast ATG19 possess at least one active uORF, to regulate downstream CDS translation. In addition to the aforementioned ATG genes in yeast, a series of genes encoding the core autophagy machinery with potential active uORFs in human cells are delineated here. We determine that the predicted active non-canonical uORFs of ATG4B and ATG12 strongly impede CDS translation in human cells. Altogether, our results show that uORFs have a widespread impact on translational control of ATG genes to mediate stress-induced autophagy.
Results
An inhibitory ATG13 uORF mediates Atg13 expression in response to nitrogen starvation
Ribosome profiling, also known as Ribo-seq, utilizes high-throughput sequencing to identify short mRNA fragments that are protected by 80S ribosomes, and therefore provides extensive information on the translational events governed by the translating ribosomes and the corresponding translational efficiency [23]. Several genome-wide translation analyses by ribosome profiling provide a list of genes that may have potential active uORFs mediating downstream CDS translational efficiency or encoding small peptides that may have biological functions [17,24,25]. A previous ribosome profiling study in yeast by Ingolia et al. [17] hinted that uORFs of many genes may have a functional role in translational responses to acute amino acid starvation. Because amino acid deprivation is an important upstream signal that stimulates autophagy, we hypothesized that uORFs are involved in the translational control of autophagy.
To investigate the role of uORFs in core autophagy machinery in yeast, we initiated our analysis by extracting ATG genes from the subset of genes predicted to have active uORFs with the canonical start codon AUG based on ribosome profiling [17]. We found that ATG5, ATG12, ATG13 and ATG19 are highly likely to possess active uORFs (Table S1). However, no previous reports have associated uORFs with ATG genes. In addition, although uORFs are prominently more active in translation than other regions of the 5' UTR, their translational efficiencies are not comparable to the corresponding CDS. Indeed, some putative translational signals identified by ribosome profiling cannot be experimentally validated and might be false positives. Moreover, the functional activity of uORFs has been demonstrated for only a limited number of genes. Therefore, the reliability of the identification of uORFs and the mechanism of how the uORFs in specific genes regulate gene expression requires experimental evaluation.
Atg13 is an essential component of the Atg1 kinase complex that plays an irreplaceable role in autophagy initiation and in sensing upstream nutrient signals [26]. ATG13 is predicted to possess a canonical uORF that initiates at an AUG codon in the 5' UTR and ends inside the downstream ATG13 CDS (Figure 1A). This ATG13 uORF encodes a 46-amino-acid polypeptide. To determine whether this canonical ATG13 uORF plays a role in Atg13 translation, we constructed plasmids expressing either wild-type (WT) Atg13 tagged with the protein A (PA) epitope or a mutant version in which we mutated the AUG start codon of the ATG13 uORF to UUG to prevent ATG13 uORF translation. We then measured plasmid-based protein expression of Atg13-PA in atg13Δ cells. The ATG13 uORF mutation resulted in a fold-change of approximately 2.3 in Atg13-PA protein level under nutrient-rich conditions (Figure 1B, 1C). Although nitrogen starvation stimulated Atg13-PA upregulation in WT cells, Atg13-PA levels were not significantly changed following 2 h of nitrogen starvation in cells with the ATG13 uORF mutation. These results together demonstrate that the ATG13 uORF is involved in controlling expression of ATG13 in a nutrient-dependent manner.
Figure 1.
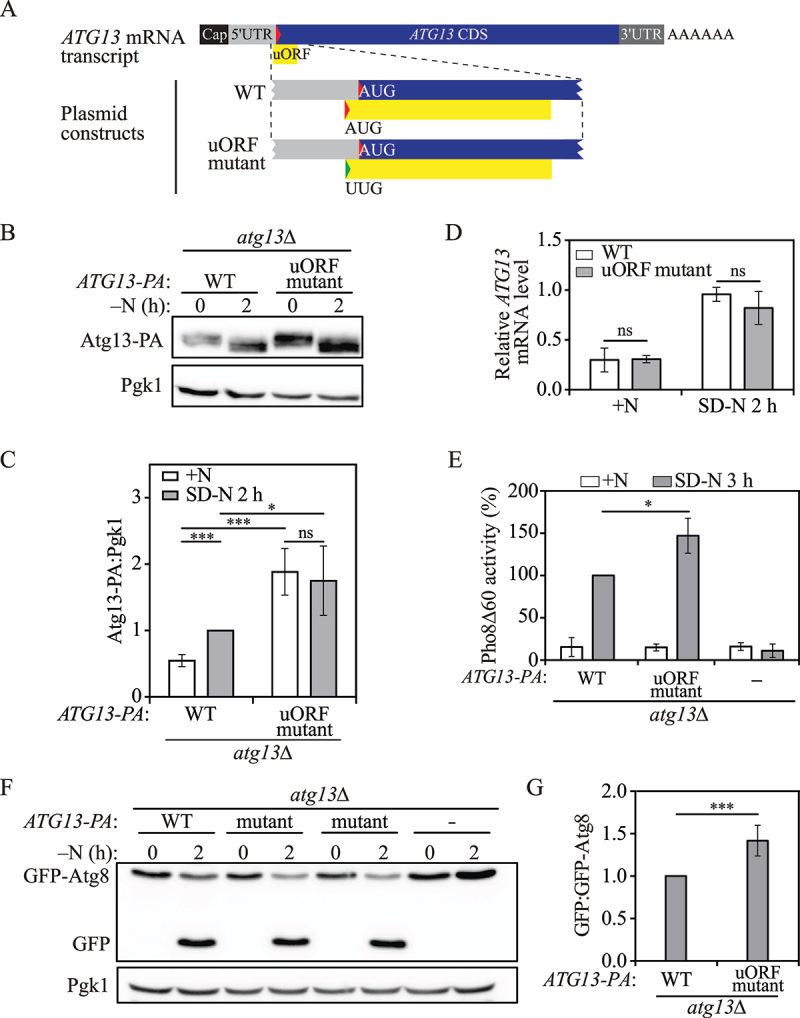
An ATG13 uORF regulates autophagy under nitrogen-starvation conditions in yeast. (A) Schematic representation of ATG13 mRNA transcript with a potential functional uORF identified by ribosome profiling (yellow box). This ATG13 uORF has a start codon (AUG) in the 5’ UTR, and in-frame stop codon preceding the end of the ATG13 CDS. The mutation made in the start codon of the ATG13 uORF in the plasmid construct is shown as indicated (represented by the change from a red triangle to a green triangle). (B) Atg13-PA levels were measured by western blot in WT and ATG13 uORF mutant cells under growing conditions and after 2 h of nitrogen-starvation conditions. Pgk1 was used as a loading control. (C) The ratio of Atg13-PA to Pgk1 was quantified. Mean±SD of n = 3 independent experiments are shown as indicated. Student’s t-test; ns: not significant, **: p < 0.01, ***: p < 0.005. (D) ATG13-PA mRNA levels were determined in WT and ATG13 uORF mutant cells under growing and 1-h nitrogen-starvation conditions by RT-qPCR. Mean±SD of n = 3 independent experiments are shown as indicated. ANOVA; ns: not significant. (E) Autophagy activity was measured with the Pho8Δ60 assay in WT, ATG13 uORF mutant, and atg13Δ strains under growing conditions and after 3 h of nitrogen starvation. Mean±SD of n = 3 independent experiments are shown as indicated. Student’s t-test; *: p < 0.05. (F) Autophagy was measured using the GFP-Atg8 processing assay in WT, ATG13 uORF mutant and atg13Δ strains under growing conditions and after 2 h of nitrogen starvation; a representative image is shown. (G) The ratio of free GFP to GFP-Atg8 after 2 h of nitrogen starvation was quantified. Mean±SD of n = 3 independent experiments are shown as indicated. Student’s t-test; ***: p < 0.005. SD-N, synthetic minimal medium lacking nitrogen; +N (YPD), yeast extract-peptone-dextrose; WT, wild type; PA, protein A.
Atg13 protein expression is controlled transcriptionally and post-transcriptionally. To exclude the potential effects of the ATG13 uORF on ATG13 transcription or mRNA stability, we measured the level of ATG13 mRNA in WT cells and ATG13 uORF mutant cells by real-time quantitative reverse transcription PCR (qRT-PCR). We found that the ATG13 mRNA level was highly upregulated upon nitrogen starvation as reported previously; however, there was no significant difference in the ATG13 mRNA levels between WT cells and the cells with the ATG13 uORF mutation (Figure 1D), demonstrating that the effect of the ATG13 uORF on ATG13 expression occurs most likely at the translational level.
The ATG13 uORF is an inhibitory regulator for autophagy
Because the ATG13 uORF mutation led to an elevated level of Atg13-PA under nutrient-rich conditions relative to the WT, we speculated that the inhibitory uORF might therefore act as a regulator of autophagy. To investigate autophagy activity, we performed the quantitative Pho8Δ60 assay, in which we generated a mutant of the vacuolar phosphatase Pho8 that lacks the first 60 amino acids of its N terminus [27]. The mutant Pho8 can be delivered to the vacuole only through non-selective autophagy, which leads to the cleavage of its propeptide by vacuolar hydrolases and subsequent activation of its phosphatase activity. Hence, phosphatase activity in the Pho8Δ60 assay demonstrates autophagy flux in the cells. We observed a significant increase in autophagy activity in ATG13 uORF mutant cells compared with WT cells after 3 h of nitrogen starvation (Figure 1E), suggesting that the elevated level of Atg13 allowed cells to initiate a more rapid autophagic response once shifted to nitrogen-starvation conditions.
To extend our analysis, we carried out the GFP-Atg8 processing assay. Atg8 is a ubiquitin-like protein required for autophagosome formation. Upon autophagy induction, the phosphatidylethanolamine (PE)-conjugated form of Atg8 initially resides on both sides of the phagophore; Atg8–PE on the outer surface of the mature autophagosome is recycled back into the cytosol, whereas the population inside the autophagosome is delivered to the vacuole and degraded. As GFP is relatively resistant to vacuolar hydrolases, the amount of free GFP generated by the GFP-Atg8 processing assay can be used to quantify autophagic delivery to the vacuole [28]. Consistent with the Pho8Δ60 assay results, we found that mutation of the ATG13 uORF resulted in increased autophagy activity after 2 h of nitrogen starvation, as observed by the ratio of free GFP to GFP-Atg8 (Figure 1F, 1G).
Moreover, we tested Atg1 protein levels in WT and ATG13 uORF mutant cells. Atg1 is a serine/threonine kinase that is essential for autophagosome formation. Atg13 physically interacts with Atg1, and the phosphorylation level of Atg13 affects Atg1 kinase activity and subsequent autophagy induction [29]. We found that Atg1 levels were not significantly changed by the ATG13 uORF mutation in either nutrient-rich conditions or after 2 h of nitrogen starvation (Figure S1), excluding the possibility that the increased autophagy activity in ATG13 uORF mutant cells is due to its impact on Atg1 protein levels. We also asked whether the observed increase in autophagy could be explained by enhanced Atg13 protein levels. We found that overexpression of Atg13-PA under the control of the CUP1 promoter resulted in a higher level of autophagy as shown by Pho8Δ60 activity (Figure S2), indicating that elevated Atg13 levels contribute to autophagy activation. Collectively, our results indicate that the translational upregulation of ATG13 mediated by bypassing the uORF promotes autophagy.
Translational control of ATG13 by its uORF is dependent on a partial Kozak sequence and Gcn2 kinase
Next, we asked how the identified ATG13 uORF regulates Atg13 translation. It is commonly acknowledged that when uORFs show a repressive effect in CDS translation, it is presumably because the translation of uORFs can occur at the expense of reducing translation initiation of the downstream CDS [30]. During translation, the GTP-bound eIF2 complex binds the initiator methionyl-tRNA (Met-tRNAiMet) to form the ternary complex eIF2-GTP-Met-tRNAiMet, and subsequently interacts with the 40S ribosomal subunit, Tif11/eIF1A and the eIF3 complex to assemble into the 43S preinitiation complex (PIC) [31]. Starting from the 5' ends of mRNAs, the PIC scans for a start codon, which is usually the first AUG base pairing with Met-tRNAiMet, in the 5' to 3' direction. Once the start codon is identified, GTP is hydrolyzed by eIF2, causing the eIF proteins to dissociate from the PIC, which allows the joining of the 60S ribosomal subunit to form an 80S ribosome; translation elongation then begins with the decoding of the second codon [31].
In addition to the activity of the PIC, the context of nucleotides of the start codons, such as the Kozak sequence, plays an important role to strengthen or weaken start codon recognition. An optimal Kozak consensus sequence is defined as GCCA/GCCAUGG, where the A of the AUG start codon (bold) is designated +1, a purine is present at position −3 (underlined) and a guanosine is present at +4 (underlined) [32]. This optimal Kozak sequence provides stabilizing interactions with the PIC, and hence dictates a strong recognition context for the start codon. The correct AUG for the ATG13 CDS follows these rules for an optimal Kozak sequence (Figure 2A). In contrast, the ATG13 uORF possesses a relatively weak Kozak sequence with an A at position +4. Hence, we hypothesized a leaky scanning model for uORF-mediated Atg13 translation. In the leaky scanning model, AUG start codons within uORFs that are present within a weak Kozak sequence may be recognized, starting the translation of the uORF, or “scanned through” meaning this start codon is bypassed. In the latter situation, translation efficiency of the downstream CDS is elevated.
Figure 2.
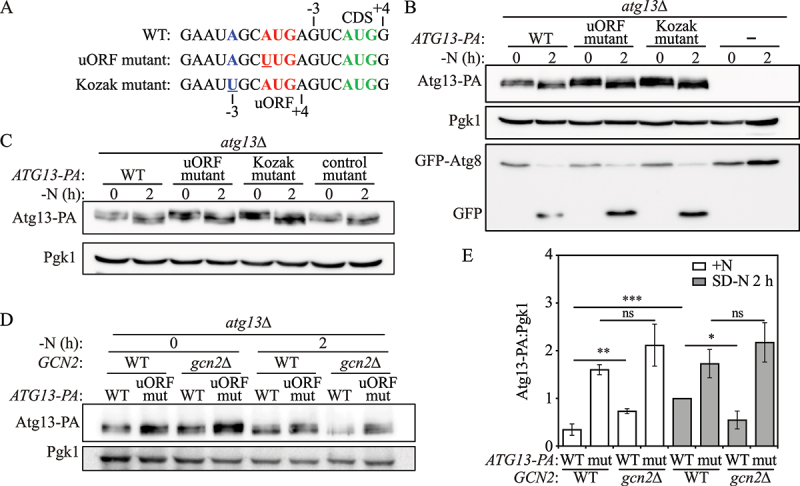
uORF-mediated Atg13 translation is Gcn2 dependent. (A) The mutations made in the Kozak mutant and uORF mutant cells are shown as indicated; mutated regions are presented in blue and red colors and mutated nucleotides are underlined. The correct AUG start codon of the ATG13 CDS is presented in green. (B) Atg13-PA protein levels and autophagy activity were measured by western blot and GFP-Atg8 processing in atg13Δ strains with empty vector or vectors expressing WT Atg13-PA, an ATG13 uORF mutant, or a Kozak mutant under growing and 2-h nitrogen-starvation conditions. (C) Atg13-PA protein levels were measured in atg13Δ strains with vectors expressing WT Atg13-PA, an ATG13 uORF mutant, a Kozak mutant, or a control mutant under growing and 2-h nitrogen-starvation conditions. (D)Atg13-PA protein levels were measured by western blot in either atg13Δ or atg13Δ gcn2Δ cells expressing WT Atg13-PA or an ATG13 uORF mutant under growing and 2-h nitrogen-starvation conditions; a representative image is shown. (E) The ratio of Atg13-PA to Pgk1 was quantified. Mean±SD of n = 3 independent experiments are shown as indicated. Student’s t-test; *: p < 0.05, **: p < 0.01, ***: p < 0.005, ns: not significant. SD-N, synthetic minimal medium lacking nitrogen; +N (YPD), yeast extract-peptone-dextrose; WT, wild type; PA, protein A.
To test whether the initiation context of the uORF start codon affects Atg13 translation, we constructed strains with either 1) a mutated initiation sequence for the ATG13 uORF, converting A to U in the AUG codon, or 2) a mutation in the Kozak sequence converting A to U at position −3 (Figure 2A). We found that the mutation in the Kozak sequence led to increased Atg13-PA translation similar to the level of a direct mutation in the ATG13 uORF start codon (Figure 2B). Moreover, we measured GFP-Atg8 processing to determine the effect of the mutations on autophagy activity. We found that Kozak mutant cells displayed increased levels of autophagy to a similar extent as uORF mutant cells (Figure 2B). As a control, we mutated an AGG codon to UUG in the 5' UTR of the ATG13 mRNA transcript that is 46 nucleotides upstream of the uORF start codon. The control mutant showed similar Atg13-PA protein levels as WT cells (Figure 2C). These results suggest that the partial Kozak sequence for the start codon of the ATG13 uORF is essential for regulating uORF-mediated Atg13 translation.
Although the partial Kozak sequence for the ATG13 uORF affects Atg13 translation, suggesting a leaky scanning mechanism, we also examined the role of Gcn2, which is typically involved in translation reinitiation. Previous studies showed that upon amino acid starvation, the protein kinase Gcn2 phosphorylates Sui2/eIF2α, and hence inhibits the formation of new ternary complexes, thus impeding the reinitiation of translation at the CDS after ribosome termination [6,33]. To further explore the mechanism of the ATG13 uORF, we deleted GCN2 in WT and ATG13 uORF mutant cells expressing Atg13-PA.
In the cells expressing WT Atg13-PA, 2-h nitrogen starvation led to the expected increase in Atg13-PA levels; however, Gcn2 deficiency led to diminished Atg13-PA protein levels under 2-h nitrogen starvation conditions (Figure 2D, 2E, Figure S3), indicating that Gcn2 mediates translational expression of Atg13 under these conditions. In contrast, Atg13-PA protein levels were not significantly affected by GCN2 knockout in ATG13 uORF mutant cells under identical nitrogen-starvation conditions, suggesting that the bypass of uORF-mediated Atg13 translational regulation in WT cells upon nitrogen starvation was Gcn2 dependent. These results together indicate that the ATG13 uORF regulates Atg13 expression through a Gcn2-dependent mechanism.
Multiple inhibitory ATG5 uORFs mediates Atg5 translation and autophagy regulation
Atg5 is an essential protein involved in the process of phagophore expansion and autophagosome formation [34,35]. During autophagy, Atg5 is covalently conjugated with Atg12 [35]. The resulting irreversible Atg12–Atg5 conjugate functions as an E3-like enzyme to facilitate Atg8 lipidation [36]. ATG5 is predicted to have five potential canonical uORFs (Table S1). We investigated four of these putative uORFs, which have been shown to actively bind to translating ribosomes. All these uORFs have both start and stop codons located inside the 5' UTR of ATG5 (Figure 3A). The most upstream of these sites, uORF1, encodes a 13 amino-acid polypeptide and its corresponding stop codon is 33 nucleotides ahead of the downstream ATG5 CDS. uORF2 encodes a 16 amino-acid polypeptide and its corresponding stop codon is 8 nucleotides ahead of the downstream ATG5 CDS. uORF3 and uORF4 share the same stop codon and encode extremely small peptides with a length of 2 or 1 amino acids, respectively.
Figure 3.
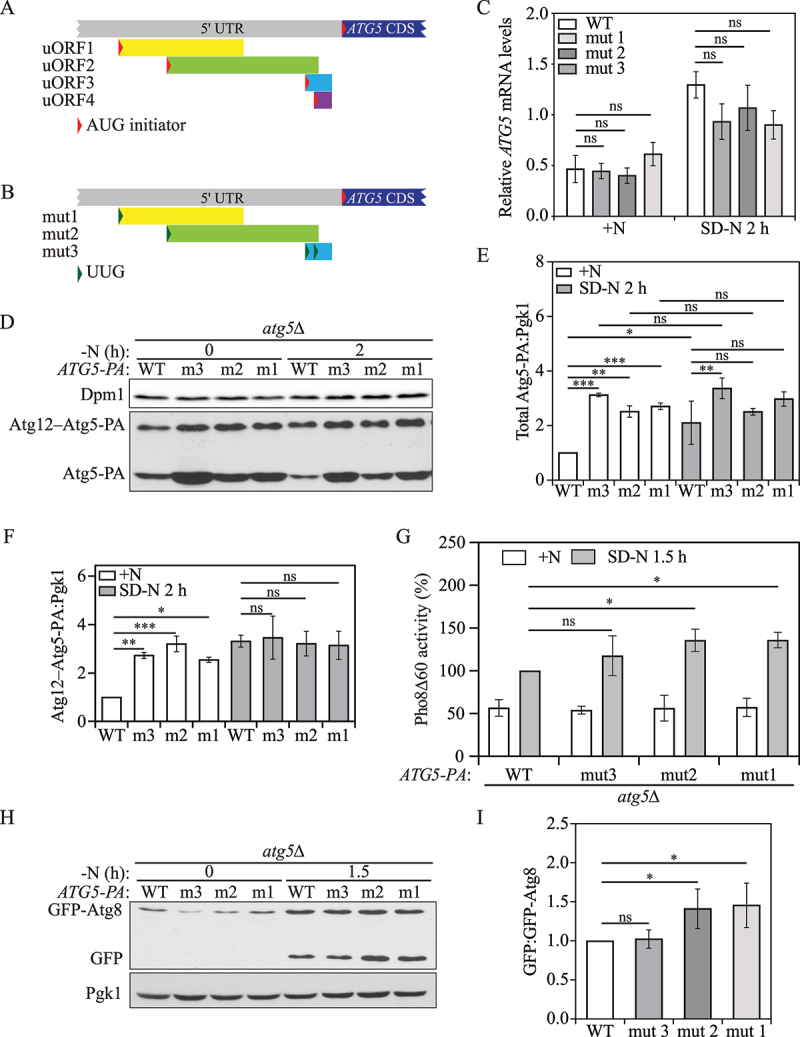
Multiple ATG5 uORFs mediate Atg5 translation and autophagy in yeast. (A) Schematic representation of partial ATG5 mRNA transcript with 4 potential functional uORFs identified by ribosome profiling (represented by yellow, blue, green, and purple boxes). Each ATG5 uORF has a start codon (AUG) in the 5’ UTR, and a stop codon preceding the ATG5 CDS. (B) The mutations made in the start codon of ATG5 uORFs in the plasmid constructs are shown as indicated (represented as a change from a red triangle to a green triangle). (C) ATG5-PA mRNA levels were determined in WT and ATG5 uORF mutant cells under growing and 1-h nitrogen-starvation conditions by RT-qPCR. Mean±SD of n = 3 independent experiments are shown as indicated. ANOVA; ns: not significant. (D) Atg5-PA levels were measured by western blot in WT and ATG5 uORF mutant cells under growing conditions and after 2 h of nitrogen starvation. Pgk1 was used as a loading control. (E) The ratio of total Atg5-PA to Pgk1 was quantified. Mean±SD of n = 3 independent experiments are shown as indicated. ANOVA; ns: not significant, *: p < 0.05, **: p < 0.01, ***: p < 0.005. (F) The ratio of Atg12–Atg5-PA to Pgk1 was quantified. Mean±SD of n = 3 independent experiments are shown as indicated. ANOVA; ns: not significant, *: p < 0.05, **: p < 0.01, ***: p < 0.005. (G) Autophagy activity was measured using the Pho8Δ60 assay in WT, and ATG5 uORF mutant cells under growing conditions and after 3 h of nitrogen starvation. Mean±SD of n = 3 independent experiments are shown as indicated. ANOVA; *: p < 0.05, ns: not significant. (H) Autophagy was measured with the GFP-Atg8 processing assay in WT, and ATG5 uORF mutant strains under growing conditions and after 1.5 h of nitrogen starvation; a representative image is shown. (I) The ratio of free GFP to GFP-Atg8 after 1.5 h of nitrogen starvation was quantified. Mean±SD of n = 3 independent experiments are shown as indicated. ANOVA; *: p < 0.05. SD-N, synthetic minimal medium lacking nitrogen; +N (YPD), yeast extract-peptone-dextrose; WT, wild type; PA, protein A.
To study the roles of each individual uORF in Atg5 translation, we constructed plasmids that express either WT Atg5-PA or a mutant version by switching the start codons of uORF1, uORF2 or both uORF3 and uORF4 from AUG to UUG (Figure 3B); uORF3 and uORF4 were mutated together because they partially overlap and share the same stop codon. These plasmids were then transformed into atg5Δ cells. We named the cells with mutated uORF1 and mutated uORF2 “mut 1” and “mut 2”, respectively, whereas those expressing mutated uORF3 and uORF4 were named “mut 3”.
To exclude the possibility that these ATG5 uORFs have a role in ATG5 transcription or mRNA stability, we first performed RT-qPCR. No significant difference in ATG5 mRNA levels was detected among the WT and mutant strains under both growing and nitrogen-starvation conditions (Figure 3C). Next, we measured Atg5-PA levels by western blot. Atg5-PA has two forms on the blot: the free form of Atg5-PA and the Atg12–Atg5-PA conjugate. We first focused on total Atg5-PA (free Atg5-PA+Atg12–Atg5-PA) levels to explore the role of the ATG5 uORFs on Atg5 translation. All three mutant cells showed increased total Atg5-PA levels under growing conditions compared with WT cells (Figure 3D, 3E). Although total Atg5-PA levels of WT cells were significantly increased after 2 h of nitrogen starvation, we did not find significant changes of the total Atg5-PA levels between growing and starvation conditions in any of the mutant cells. These results suggest that these four predicted uORFs are involved in downregulating ATG5 under growing conditions.
Moreover, no significant difference was observed following 2 h of starvation in strains with either uORF1 or uORF2 mutation relative to the WT (Figure 3D, 3E), suggesting that the disruption of either ATG5 uORF1 or uORF2 eliminated the entire inhibitory role of ATG5 uORFs in the regulatory mechanism of translation. In contrast, double mutations in uORF3 and uORF4 resulted in higher levels of Atg5-PA compared with the other two mutants. This difference may reflect the relative position of uORF3 and uORF4; the corresponding stop codon ends just 3 nucleotides before the start codon of the ATG5 CDS, and the short distance would provide less time for the 40S ribosome to reassemble factors needed for reinitiation after translation termination at either of these uORFs.
The formation of the Atg12–Atg5 conjugate is essential for autophagy [37]. We found that Atg12–Atg5-PA levels in mut 1, mut 2 and mut 3 resulted in fold-changes of 2.6-, 3.2-, and 2.7 relative to WT cells in nutrient-rich conditions (Figure 3D, 3F). The Atg12–Atg5 levels in WT cells quickly increased to that of the three mutant strains after 2 h of starvation, at which time the conjugation appeared to be saturated. Next, we examined the effect of the uORF mutants on autophagy activity using the Pho8Δ60 assay. None of the three mutants showed a significant difference in autophagy activity compared with the WT after 3 h of starvation (Figure S4), presumably because the Atg12–Atg5-PA levels in WT cells reached the saturation point within 2 h of starvation (Figure 3D, 3F). To further examine our hypothesis, we took advantage of the shorter time course that is possible with the GFP-Atg8 processing assay. In contrast to the results seen with the Pho8∆60 assay, we found an ~40% increase in autophagy activity in both the uORF1 and uORF2 mutant strains following 1.5 h of starvation (Figure 3H, 3I); there was no detectable increase in the strains containing double mutations in uORF3 and uORF4. To reconcile these differences, we extended our analysis by performing a short-term Pho8∆60 assay with a time course closer to that of the GFP-Atg8 processing assay. We found that consistent with the latter, after 1.5 h of starvation, the uORF1 and uORF2 mutant strains showed elevated autophagy activity (Figure 3G). These data collectively suggest that ATG5 uORF1 and uORF2 play a predominant role in autophagy regulation compared with uORF3 and uORF4, even though the latter have a more pronounced effect on the level of free Atg5.
Atg12 and Atg19 are translationally regulated by uORFs
Based on the above findings for Atg5 and Atg13, we decided to examine the potential roles of the predicted canonical uORFs of ATG12 and ATG19, to determine whether they may also play an inhibitory role in translation regulation. As mentioned above, Atg12 is a component of the Atg12–Atg5 conjugate, the formation of which is essential for autophagosome formation [36]. Atg19 is a receptor protein for the cytoplasm-to-vacuole targeting (Cvt) pathway, a type of selective autophagy that specifically delivers certain resident hydrolases into the yeast vacuole [38]. The predicted ATG12 uORF encodes a 3-amino-acid peptide, and its stop codon is inside the downstream CDS (Figure 4A). The ATG19 5' UTR contains three putative uORFs (Table S1). We studied the only ATG19 uORF that contains a suboptimal Kozak sequence surrounding its start codon. A previous study that exploits computer simulations to model scanning and ribosomal translational elongation behavior showed that the uORFs with a greater length are more likely to display inhibitory effects of downstream CDS translation [39]. The potential ATG19 uORF is relatively longer with a length of 15 amino acids and ends 48 nucleotides before the ATG19 CDS. We constructed plasmids expressing WT Atg12-PA or WT Atg19-PA, or their uORF mutant versions by changing the corresponding uORF start codons from AUG to UUG. These plasmids were transformed into atg12Δ or atg19Δ cells, respectively.
Figure 4.
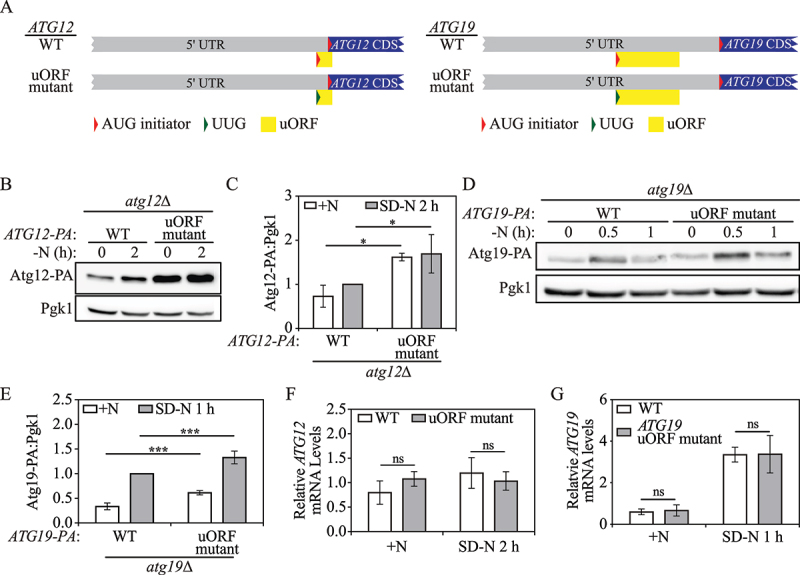
Yeast Atg12 and Atg19 translation are mediated by a single inhibitory uORF in the corresponding 5’ UTR. (A) Schematic representation of partial ATG12 or ATG19 mRNA transcripts with a potential functional uORF identified by ribosome profiling (yellow box). The ATG12 uORF has a start codon (AUG) in the 5’ UTR, and an in-frame stop codon preceding the end of the ATG12 CDS. The ATG19 uORF has a start codon (AUG) in the 5’ UTR, and a stop codon preceding the ATG19 CDS. The mutations made in the start codon of the ATG12 or ATG19 uORF in the plasmid constructs are shown as indicated (represented as a change from a red triangle to a green triangle). (B) Atg12-PA levels were measured by western blot in WT and ATG12 uORF mutant cells under growing conditions and after 2 h of nitrogen starvation. Pgk1 was used as a loading control. (C) The ratio of Atg12-PA to Pgk1 was quantified. Mean±SD of n = 3 independent experiments are shown as indicated. ANOVA; *: p < 0.05. (D) Atg19-PA levels were measured by western blot in WT and ATG19 uORF mutant cells under growing conditions and after 0.5 h and 1 h of nitrogen starvation. (E) The ratio of total Atg19-PA to Pgk1 under growing and 1-h nitrogen-starvation conditions was quantified. Mean±SD of n = 4 independent experiments are shown as indicated. ANOVA; ***: p < 0.005. (F) ATG12-PA mRNA levels were determined in WT and ATG12 uORF mutant cells under growing and 2-h nitrogen-starvation conditions by RT-qPCR. Mean±SD of n = 3 independent experiments are shown as indicated. ANOVA; ns: not significant. (G) ATG19-PA mRNA levels were determined in WT and ATG19 uORF mutant cells under growing and 1-h nitrogen-starvation conditions by RT-qPCR. Mean±SD of n = 3 independent experiments are shown as indicated. ANOVA; ns: not significant. SD-N, synthetic minimal medium lacking nitrogen; +N (YPD), yeast extract-peptone-dextrose; WT, wild type; PA, protein A.
To examine the role of the ATG12 and ATG19 uORFs on translation, we measured protein levels by western blot. ATG12 uORF mutant cells displayed a fold-change of 2.3 and 1.7 in Atg12-PA levels compared with WT cells in nutrient-rich conditions and after 2 h of nitrogen starvation, respectively (Figure 4B, 4C), implying that the ATG12 uORF controls Atg12-PA levels under both conditions. ATG19 uORF mutant cells demonstrated a fold-change of 1.9 and 1.3 in Atg19-PA levels compared with WT cells in nutrient-rich and nitrogen starvation conditions, respectively (Figure 4D, 4E), indicating a similar regulatory role as seen with the ATG12 uORF. To determine if the ATG12 and ATG19 uORFs affect corresponding mRNA stability, we measured mRNA levels by RT-qPCR. Under either nutrient-replete or nitrogen-starvation conditions, the mutations of the uORF start codons did not significantly change ATG12 or ATG19 mRNA levels (Figure 4F, 4G). These data suggest that the ATG12 and ATG19 uORFs repress their corresponding CDS translation.
More than 20 ATG genes have potential active uORFs (canonical and non-canonical) in human
Next, we asked whether uORFs play a role in the translational control of ATG genes in more-complex eukaryotes. Recently, several genome-wide ribosome profiling studies were performed in human cells [25,40,41]. Active uORFs widely exist in the human genome; however, the precise number and roles of the uORFs are unknown. A previous study by McGillivray et al. exploited machine learning algorithm training based on uORFs labeled within ribosome profiling experiments to predict potential active uORFs that are bound by ribosomes in human cells [25].
To gain a comprehensive scope of uORF-mediated translation regulation of ATG genes in human, we extracted the genes encoding core autophagy machinery from the list of genes with potential active uORFs predicted in the study by McGillivray et al. [25]. The set of core ATG genes used here encodes 5 groups of proteins, including 1) the ULK1 kinase complex, composed of ULK1, ULK2, RB1CC1/FIP200, ATG13, and ATG101; 2) the class III phosphatidylinositol 3-kinase (PtdIns3K) complex, which is composed of BECN1/Beclin 1, PIK3C3/VPS34, PIK3R4/VPS15, ATG14, NRBF2, and UVRAG; 3) the ATG9 trafficking system, which is composed of ATG2A, ATG2B, WIPI1, WIPI2, WDR45/WIPI4 and the transmembrane protein ATG9A; 4) the Atg8-family ubiquitin-like (Ubl) conjugation system, composed of ATG4A, ATG4B, ATG4C, ATG4D, ATG3, MAP1LC3A, MAP1LC3B, MAP1LC3C, GABARAP, and GABARAPL2; 5) the ATG12 Ubl conjugation system, composed of ATG5, ATG12, ATG16L1, ATG7, and ATG10 [3,42]. We provide the list of uORFs predicted to be active for ATG genes in Table S2. We found that among the 32 chosen genes, 18 of them possess one or more canonical uORFs with an AUG start codon (Figure 5A). We noticed that ATG5 had 3 potential AUG-initiated uORFs, whereas ATG12, and ATG13 did not display the existence of uORFs with AUG start codons.
Figure 5.
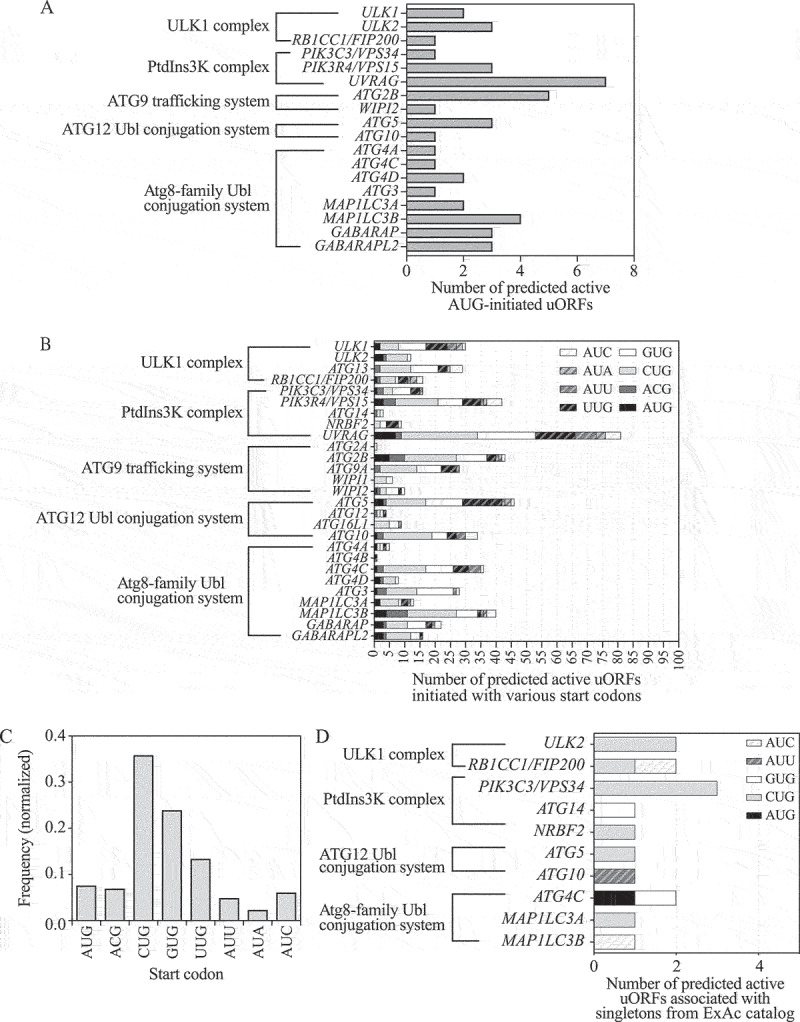
More than 20 human ATG genes are predicted to have functional uORFs. (A) The number of active canonical uORFs initiated with an AUG start codon for ATG genes predicted by a machine learning algorithm [25]. ATG genes are classified into 5 groups, including those encoding components of the ULK1 complex, the PtdIns3K complex, the ATG9 trafficking system, the ATG12 Ubl conjugation system, and the Atg8-family Ubl conjugation system. ATG genes with no active canonical uORFs are not shown. (B) Composition analysis of uORFs (initiated with AUG or near-cognate start codons) in ATG genes predicted by a machine learning algorithm [25]. ATG genes with no active uORFs are not shown. (C) The frequency of AUG start codons and near-cognate start codons of predicted positive uORFs in all ATG genes. (D) Ultra-rare variants (singletons) from the Exome Aggregation Consortium (ExAC) catalog v.1 that are associated with predicted active uORFs of ATG genes are counted [25,46]. The composition of uORFs (initiated with AUG and near-cognate codons) that are associated with singleton variants are analyzed. PtdIns3K, phosphatidylinositol 3-kinase; Ubl, ubiquitin-like.
Several prior studies revealed that in addition to canonical AUG start codons, numerous uORFs initiate with non-AUG start codons, including ACG, AUU, AUA, AUC, CUG, GUG and UUG [43,44]. We therefore decided to examine whether ATG12, ATG13 and other human ATG genes exploit non-canonical uORFs, which initiate with a non-AUG start codon, to regulate their translation. Hence, we determined the composition of potential active uORFs (including uORFs that started with AUG and near-cognate start codons) of ATG genes. We found that 27 out of 32 human ATG genes have at least one predicted functional uORF (Figure 5B). ATG5, ATG12, and ATG13 have 46, 4 and 29 uORFs, respectively. AUG has been identified as the most prevalent uORF start codon in the genome [25]. However, our analysis of the data from McGillivray et al. [25] indicated that uORFs for ATG genes exhibit translation initiation mostly at CUG and GUG (Figure 5C). CUG and GUG are more likely to be scanned through/bypassed during start codon recognition, suggesting that leaky scanning is a common feature in the repressive mechanism for uORF-mediated translational control of ATG genes.
Identification of singleton variants provides tremendous information for human phenotypic variation and disease susceptibility [45]. We scanned singleton variants on start codons of predicted uORFs of ATG genes from the ExAc catalog [25,46]. We found that 10 genes encoding core autophagy machinery, including ULK2, RB1CC1/FIP200, PIK3C3/VPS34, ATG14, NRBF2, ATG5, ATG10, ATG4C, MAP1LC3A and MAP1LC3B, are associated with altered start codons of predicted active uORFs at a low allele mutation frequency (from 8.24e−06 to 1.04e−05) (Table S3, Figure 5D). These data imply that there is a high possibility that the predicted uORFs in these ATG genes are active and functionally important, because a lower frequency of singletons is often due to heavy selection pressure of regions with essential functions [47].
Non-canonical uORFs regulate starvation-induced ATG4B and ATG12 translation in human
Among the four members of the ATG4 family (ATG4A, ATG4B, ATG4C, and ATG4D), the role of ATG4B is best characterized. ATG4B is a crucial cysteine protease that proteolytically cleaves Atg8-family proteins (LC3 and GABARAP subfamilies) to their active forms and promotes efficient autophagosome biogenesis [48]. We noticed that ATG4B is predicted to have only one active non-canonical uORF that initiates with a UUG codon and partially overlaps with the downstream CDS (Figure 6A). To validate the function of this single uORF in ATG4B expression regulation, we generated constructs with a luciferase reporter coding sequence downstream of the ATG4B 5' UTR containing either the WT predicted active ATG4B uORF or a mutant version with the start codon changed from UUG to AAG. We chose AAG because a previous study suggested that AAG is employed as the start codon of uORFs with the least probability of function [49]. The constructs containing WT ATG4B 5'UTR-LUC and uORF mutant ATG4B 5'UTR-LUC were transfected into 293 FT cells.
Figure 6.
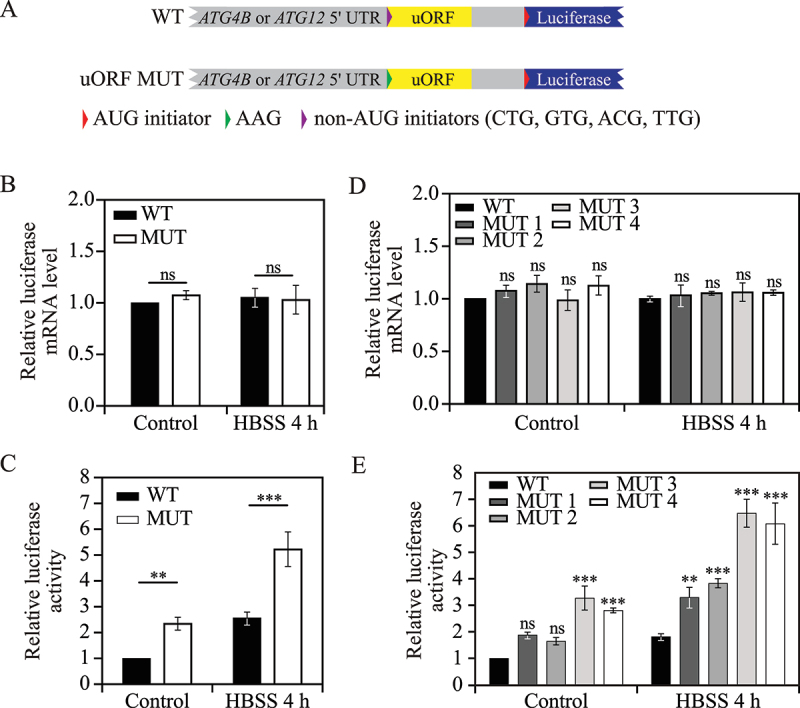
Non-canonical uORFs regulate ATG4B and ATG12 translation in human. (A) Schematic representation of WT and mutant human ATG12 or ATG4B 5'UTR-LUC constructs. ATG12 or ATG4B 5’ UTR-containing uORFs (yellow box) with non-AUG start codons (purple triangle) were cloned upstream of the luciferase CDS in WT constructs. The start codons of the uORFs were changed to AAG (green triangle) in the mutant constructs. (B) Relative luciferase mRNA levels were measured in 293 FT cells transfected with WT and uORF mutant ATG4B 5'UTR-LUC constructs under control (nutrient-rich medium) and 4-h HBSS starvation conditions. Mean±SD of n = 3 independent experiments are shown as indicated. ANOVA; ns: not significant. (C) Relative luciferase activities were measured in 293 FT cells transfected with WT and uORF mutant ATG4B 5'UTR-LUC constructs under control (nutrient-rich medium) and 4-h HBSS starvation conditions. Mean±SD of n = 3 independent experiments are shown as indicated. ANOVA; **: p < 0.01, ***: p < 0.005. (D) Relative luciferase mRNA levels were measured in 293 FT cells transfected with WT and uORF mutant (MUT 1 to MUT 4) ATG12 5'UTR-LUC constructs as in (B). Mean±SD of n = 3 independent experiments are shown as indicated. ANOVA; ns: not significant. (E) Relative luciferase activities were measured in 293 FT cells transfected with WT and uORF mutant (MUT 1 to MUT 4) ATG4B 5'UTR-LUC constructs as in (C). Mean±SD of n = 3 independent experiments are shown as indicated. ANOVA; **: p < 0.01, ***: p < 0.005. WT, wild type; HBSS, Hanks’ balanced salt solution.
To investigate whether and how the ATG4B uORF affects protein translation upon stress, we determined the luciferase activity and luciferase mRNA levels under nutrient-rich conditions or after 4 h of Hanks’ balanced salt solution (HBSS)-mediated starvation. The luciferase mRNA level was not affected by mutation of the ATG4B uORF during either growing or starvation conditions (Figure 6B), suggesting the ATG4B uORF does not affect mRNA stability. However, the cells with the mutant construct displayed a fold-increase of 2.4 and 2.0 in luciferase activity compared with WT cells under nutrient-rich and HBSS starvation conditions, respectively (Figure 6C). These results indicate that the ATG4B uORF inhibits downstream ORF translation under both conditions.
Although human ATG12 is highly conserved down to yeast Atg12 in terms of structure and function [50,51], human ATG12 mRNA was not predicted to possess a canonical uORF as seen with yeast ATG12. Instead, ATG12 had four predicted non-canonical uORFs, all of which initiate with non-AUG codons, in the order of CUG, GUG, ACG, and UUG from 5' to 3' (Figure 6A). We named the four non-canonical uORFs as uORF1, uORF2, uORF3 and uORF4 based on the order of start codon positions from 5' to 3'. To determine the role of each ATG12 uORF in translational regulation, we constructed a WT ATG12 5' UTR-LUC plasmid that contains the luciferase reporter coding sequence downstream of the ATG12 5' UTR. We then generated a set of mutant plasmids by mutating the start codon of each predicted uORF to AAG. These constructs were again transfected into 293 FT cells.
We first measured luciferase mRNA levels by RT-qPCR. Luciferase mRNA levels were not affected by mutation in any of the ATG12 uORFs during either growing or 4-h HBSS starvation conditions (Figure 6D), demonstrating no effect of ATG12 uORFs on mRNA stability. Next, we examined luciferase activity as a readout of translation activity. During growing conditions, mutations in uORF1 or uORF2 did not significantly change luciferase activity, whereas mutations in uORF3 and uORF4 resulted in a fold-increase of 3.27 and 2.81, respectively (Figure 6E). Upon 4 h of HBSS starvation, we observed a fold-increase of 1.82, 2.11, 3.57 and 3.35 in luciferase activity in the cells with mutations in the start codons of uORF1 to uORF4, respectively. Collectively, these results indicate that the ATG12 uORF3 and uORF4 are predominant cis regulators for downstream ORF translation, although ATG12 uORF1 and uORF2 displayed a mild inhibitory effect on translation.
Discussion
Though substantial effort has been made to delineate the mechanisms for how ATG genes are regulated transcriptionally and post-translationally, little is known about how ATG genes bypass the general translation suppression that occurs during stress conditions to maintain the activation of autophagy. In this study, we proposed and validated the hypothesis that uORFs act as an important regulatory element for the translation of ATG genes, especially in response to starvation, controlling proteins that participate in different stages of autophagy, including initiation, phagophore expansion, and cargo selection.
We first hypothesized that the canonical ATG13 uORF mediates Atg13 translation through a leaky-scanning mechanism and showed that the suboptimal Kozak sequence for the ATG13 uORF is involved in regulation of Atg13 translation. However, the +4 position in the Kozak sequence may not play as much of a role as the −3 position [52]. Thus, depending on the relative importance of the −3 and +4 positions, if we consider the ATG13 uORF to have an optimal Kozak sequence, the likelihood of a leaky-scanning mechanism is decreased. Accordingly, we also considered whether the ATG13 uORF exploits a translation reinitiation mechanism. The reinitiation model was proposed by Kozak and suggests that when there is a relatively long distance between a uORF and the downstream ORF, the ribosomes are able to reassemble along with initiation factors and initiate the translation of the downstream ORF [32]. Our data fit either mechanism, but we think there is a low probability that this ATG13 uORF exploits a reinitiation mechanism; reinitiation is found to be highly inefficient when the two ORFs are located very close together, within 8 nucleotides, or overlap with each other [53]. In addition, the reinitiation model is based on the presence of a termination site downstream of the uORF, but upstream of the canonical ORF. Considering that the identified ATG13 uORF in this study ends inside the ATG13 CDS, we think that the reinitiation model is not likely to be used. The main piece of data that would seem to argue against the leaky-scanning model is the finding that Gcn2 is required to bypass the uORF-mediated decrease in translation. However, the general regulation of ATG gene expression is typically opposite that of most other genes. That is, under starvation conditions approximately 98% of the genes in a cell are downregulated, whereas ATG genes are upregulated. Along these lines, factors that promote the expression and translation of genes in growing conditions, such as TOR, often act in the opposite manner for ATG genes. Although we do not know the precise mechanism, we hypothesize that Gcn2 may be acting in this way, participating in the downregulation of most genes under starvation conditions, while facilitating the expression of ATG genes. Thus, we hypothesize that the decreased availability of ternary complexes mediated by Gcn2 upon starvation contributes to the leaky scanning mechanism, by decreasing the efficiency of start codon recognition for the ATG13 uORF compared to that for the ATG13 CDS. Similarly, the uORFs for ATG5, ATG12 and ATG19 have weak Kozak sequences or completely lack a canonical sequence, in line with a leaky-scanning mechanism. At present, we cannot make a definitive conclusion as to which mechanism, leaky scanning or translation reinitiation, is used in the context of the ATG uORFs, but overall our results favor the former.
We showed that uORF-mediated Atg13 translation is dependent on Gcn2 kinase activity in yeast. It has been reported that many human genes, such as ATF4 and ATF5, exploit uORFs to regulate translation by EIF2 phosphorylation [22,54]. In human cells, there are four EIF2A/eIF2α kinases (EIF2AK1/HRI, EIF2AK2/PKR, EIF2AK3/PERK, and EIF2AK4/GCN2) involved in the integrated stress response [55]. These kinases are activated in response to various types of stress. Among all the human EIF2A kinases, EIF2AK4/GCN2 and EIF2AK3/PERK are altered upon nutrient deprivation and ER stress, respectively. uORFs regulate ATF4 translation in response to EIF2 phosphorylation during ER stress. Because the upregulation of the ATG proteins with the omission of the uORFs seem to be most consistent with the translational control of Gcn4 in yeast or ATF4 in human, we think it is also possible and worth investigating whether activation of EIF2AK3/PERK by ER stress may have a potential role in uORF-mediated translational control of ATG genes in human cells.
We identified inhibitory effects of canonical uORFs on downstream CDS translation of ATG5, ATG12, ATG13 and ATG19 during both nutrient-replete and -depleted conditions; however, we are aware that the underlying translation regulation mechanism might be at least in part different from what we propose here. For example, the protein levels of Atg13 and Atg19 are both upregulated upon starvation. We did not observe a significant increase of Atg13 protein levels in the ATG13 uORF mutant cells upon starvation compared to growing conditions, whereas in the ATG19 uORF mutant cells, Atg19 protein levels were markedly enhanced upon starvation, suggesting that the identified ATG19 uORF may have less impact on its CDS translation compared with the identified ATG13 uORF. There are multiple possibilities to explain these observations. For example, ATG19 mRNA may have more actively translated uORFs that were not identified by ribosome profiling due to the low sensitivity of translational signal capture, and thus the upregulation of the Atg19 protein by a starvation stimulus may not be fully eliminated by simply mutating one uORF. It is also possible that there exists a more dominant but currently unknown mechanism for Atg19 translational control, such as RNA-binding proteins and trans-acting regulators like microRNAs [56,57]. A previous study reported that Dhh1, a DExD/H-box RNA helicase, is required for efficient translation of some Atg proteins such as Atg1 and Atg13 [8]. RNA-binding proteins, such as ELAVL1, ELAVL4, HNRNPA1 and ZFP36, are involved in translation regulation of ATG5, ATG16, ATG7 and BECN1 in various human cell settings [10–14]. In addition, EIF5A is required for efficient ATG3 translation [58]. It will be interesting to further explore the mechanism by which some ATG genes regulate translation via a cis-acting uORF.
We also note that we observed that ATG19 uORF mutant cells demonstrated a fold-change of 1.9 and 1.3 in Atg19-PA levels compared with WT cells in nutrient-rich and nitrogen-starvation conditions, respectively. However, an analysis of prApe1 maturation to monitor activity of the Cvt pathway did not show a difference between WT and ATG19 uORF mutant cells (data not shown). Atg19 interacts with Atg11 prior to binding Atg8; thus, increasing the expression of Atg19 and not Atg11 may not be adequate to induce a significant change in the delivery of prApe1 to the vacuole via the Cvt pathway.
Mutations in either ATG5 uORF1 or uORF2 resulted in enhanced total Atg5 and Atg12–Atg5 levels, as well as a consequent increase in autophagy. Intriguingly, although we observed a significant increase in these levels in cells containing mutations in ATG5 uORF3 and uORF4, autophagy activity monitored by the GFP-Atg8 processing and Pho8∆60 assays was not consequently enhanced. uORF3 encodes a dipeptide, and the stop codon of uORF3 and uORF4 is only 3 nucleotides upstream of the ATG5 CDS. It is possible that besides having a role in impeding the translation of the downstream CDS, the dipeptide generated by uORF3 is biologically functional and involved in autophagy activation. Due to technical limitations, we failed to directly overexpress or detect the uORF micro-peptide; however, a recent report showed that it is common that micro-peptides generated by uORFs are able to stabilize protein complexes together with CDS proteins [59]. Hence, the precise role of yeast ATG5 uORF3 and uORF4 in autophagy regulation is yet to be uncovered. Furthermore, canonical uORFs are more conserved compared with non-canonical ones in general [25]. Therefore, it would be interesting to determine whether canonical uORFs of human ATG5 display similar unknown features as the uORF3 of yeast ATG5.
A previous study showed that the translational enhancement of ATG5, ATG12 and ATG16L1 upon starvation is mediated by the interaction between the RNA binding protein ELAVL1/HuA and the 3' UTR of the corresponding mRNAs in human cells [60]. In addition, three canonical uORFs and many non-canonical uORFs of ATG5 are predicted to be translationally active in human cells in our study, suggesting a very complex mechanism underlying ATG5 translation regulation and consequent autophagy induction upon stress. The detailed landscape requires further investigation.
Many studies indicated that gain and loss of uORFs frequently occur in the human population [15,61]. A subset of EIF2A-targeted uORFs regulate oncogenic mRNAs during early stages of tumorigenesis [62]. In addition to the scanning singleton variants associated with uORFs on ATG genes from the ExAc catalog (Figure 5), we also investigated if there are recurrent somatic mutations in cancers (obtained from the COSMIC database) occurring on predicted active uORFs of ATG genes in human [25,63]. Although 15 uORFs of ATG genes are linked with singleton variants in the ExAc catalog, we did not find any mutations in uORFs of ATG genes associated with cancers from COSMIC. The identification of uORFs of ATG genes affected by singleton variants from the ExAC catalog suggests that these uORFs play an essential role in regulating autophagy as it reflects considerable selection pressure. We were not surprised to find that no uORFs of ATG genes are recurrently associated with cancers. Although autophagy is broadly recognized to play a bidirectional role in tumor suppression and cancer cell growth promotion, mutations of core ATG genes in cancer are not common [64]. The escape of genetic mutations in uORFs of ATG genes is consistent with this phenomenon; this escape presumably occurs because proper autophagy is essential for maintaining nuclear and mitochondrial stability [3]. In addition, uORF mutations may cause excessive autophagy that are harmful for cell survival.
In summary, we found that canonical uORFs on the mRNA transcripts of multiple yeast ATG genes, including ATG5, ATG12, ATG13, and ATG19, inhibit downstream CDS translation under nutrient-rich conditions. The bypass of these uORFs plays an essential role in the translational upregulation of these ATG genes upon nitrogen starvation. ATG13 uORF translation is largely dependent on its Kozak sequence as well as Gcn2 kinase activity. We showed that the defined ATG5 uORF1 and uORF2 are essential for Atg5 translational control to regulate autophagy activation. Furthermore, we elucidated that non-canonical uORFs of human ATG4B and ATG12 play an inhibitory role of downstream CDS translation by reducing translation efficiency. Functional activity of uORFs has been demonstrated for only a limited number of genes, such as GCN4 in yeast and ATF4 in human; we extended the examples to ATG genes, the genes encoding components of autophagy, a critical mechanism for cells to maintain survival and promote bioenergetic homeostasis in response to stress. Furthermore, there are currently no reports associating uORFs with the translational control of ATG genes. Thus, for the first time, we showed that a large subset of ATG genes is regulated by a specific uORF-mediated translational mechanism to modulate autophagy activity.
Materials and Methods
Yeast strains, media and growth conditions
Yeast strains used in this paper are listed in Table S4. For nutrient-rich conditions, yeast cells were grown in YPD medium (1% [wt:vol] yeast extract, 2% [wt:vol] peptone, 2% [wt:vol] glucose). Autophagy was induced by shifting mid-log phase yeast cells from rich medium to nitrogen-starvation medium SD-N (0.17% yeast nitrogen base without ammonium sulfate or amino acids, 2% [wt:vol] glucose) for the indicated times.
Key Reagents
Key Reagents used in the study are listed in Table 1.
Table 1.
Key reagents used in these studies.
| REAGENT or RESOURCE |
SOURCE |
IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-YFP | Clontech | 632381; RRID: AB_2313808 |
| Anti-peroxidase (anti-protein A) | Jackson ImmunoResearch | 323–005-024; RRID:AB_2315781 |
| Anti-Pgk1 | Dr. Jeremy Thorner | N/A |
| Anti-Atg1 | In house | N/A |
| Goat anti-rabbit IgG, HRP | Fisher Scientific | ICN55676; RRID:AB_2334589 |
| Rabbit anti-mouse IgG, HRP |
Jackson ImmunoResearch |
315–035-003; RRID:AB_2340061 |
| Chemicals, peptides, and recombinant proteins | ||
| Power SYBR Green PCR Master Mix | Applied Biosystems/ThermoFisher Scientific | ILT4367659 |
| p-Nitrophenyl phosphate | Sigma-Aldrich | N4645 |
| Complete EDTA-free protease inhibitor cocktail | Roche | 05056489001 |
| TURBO DNA-free kit | Fisher Scientific | AM1907 |
| RNasin® PLUS Rnase inhibitor | Fisher Scientific | PRN2615 |
| Lipofectamine 3000 |
Invitrogen |
L3000001 |
| Critical commercial assays | ||
| BCA Protein assay | Fisher Scientific | PI23223, PI23224 |
| Dual Luciferase Assay System | Promega | E1910 |
| NucleoSpin® RNA II | Clontech | 740955.50 |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems/ThermoFisher Scientific | ILT4368814 |
| DNA Gel Purification Kit | Qiagen | 28706X4 |
| Q5® Site-Directed Mutagenesis Kit |
New England BioLabs |
E0554S |
| Experimental models: Cell lines | ||
| Human: HEK293-FT | Thermo Fisher Scientific | R70007 |
| Human: HeLa |
American Type Culture Collection |
CCL-2 |
| Experimental models: Organisms/strains | ||
|
S. cerevisiae: strain background: SEY6210 |
See Table S4 |
N/A |
| Oligonucleotides | ||
| Primers for RT-qPCR in yeast | [66] | N/A |
| Primers for RT-qPCR in human | See Table S5 | N/A |
| Primers for plasmid cloning and strains construction |
See Table S6 |
N/A |
| Recombinant DNA | ||
| pRS406-ATG13 5'UTR-ATG13-PA | This study | N/A |
| pRS406-ATG135'UTRmut-ATG13-PA | This study | N/A |
| pRS406-ATG12 5'UTR-ATG12-PA | This study | N/A |
| pRS406-ATG125'UTRmut-ATG12-PA | This study | N/A |
| pRS406-ATG19 5’ UTR-ATG19-PA | This study | N/A |
| pRS406-ATG195'UTRmut-ATG19-PA | This study | N/A |
| pRS416-ATG5 5'UTR-ATG5-PA | This study | N/A |
| pRS416-ATG55'UTRmut1-ATG5-PA | This study | N/A |
| pRS416-ATG55'UTRmut2-ATG5-PA | This study | N/A |
| pRS416-ATG55'UTRmut3-ATG5-PA | This study | N/A |
| pCMV-ATG4B 5'UTR-LUC | This study | N/A |
| pCMV-ATG4B5'UTRmut-LUC | This study | N/A |
| pCMV-ATG12 5'UTR-LUC | This study | N/A |
| pCMV-ATG125'UTRmut1-LUC | This study | N/A |
| pCMV-ATG125'UTRmut2-LUC | This study | N/A |
| pCMV-ATG125'UTRmut3-LUC | This study | N/A |
| pCMV-ATG125'UTRmut4-LUC | This study | N/A |
|
pRL-SV40P |
[67] |
Addgene, 27163; deposited by Ron Prywes |
| Software and algorithms | ||
| CFX Manager Software | Bio-Rad | N/A |
| R studio | R studio® | https://www.rstudio.com/ |
| Bio-Rad Image Lab Software | Bio-Rad | |
| GraphPad Prism 9 | GraphPad | https://www.graphpad.com/ |
Cell culture and HBSS starvation
The 293 FT (R70007) and HeLa (CCL-2) cell lines were obtained from Thermo Fisher Scientific and the American Type Culture Collection, respectively. These cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Thermo Fisher Scientific, 11,995,073) supplemented with 10% heat-inactivated fetal bovine serum (Thermo Fisher Scientific, A3840001) and 1% penicillin and streptomycin (Thermo Fisher Scientific, 15,070–063) at 37°C, 95% humidity, and 5% CO2. Hanks’ balanced salt solution (HBSS) was obtained from Sigma-Aldrich (55037C). Starvation was induced by changing the DMEM medium to HBSS for 4 h. Cell line identity was validated by short tandem repeat profiling, and routine mycoplasma testing was negative for contamination.
Luciferase reporter assay
293 FT cells were seeded at a density of 5 × 104 cells per well in a 24-well plate and grown to 50% confluency. One μg of the 5' UTR-Firefly luciferase reporter construct (Addgene, 45968; deposited by Dr. Ralf Kuehn) and 100 ng of Renilla luciferase reporter construct (Addgene, 27163; deposited by Dr. Ron Prywes) were transfected into 293 FT cells using Lipofectamine 3000 (Invitrogen, L3000001). After 24 h, cells were treated with HBSS for an additional 4 h, and then Firefly and Renilla luciferase activities were determined using a Dual Luciferase Assay System (Promega, E1910). Firefly luciferase signals were normalized to Renilla luciferase internal control signals in each experiment.
RT-qPCR
For yeast, the cells were cultured in YPD medium to mid-log phase, and an aliquot was collected as the nutrient-rich sample. The remaining cells were shifted to SD-N medium for autophagy induction. Cells were then collected and frozen in liquid nitrogen. Total RNA was extracted using an RNA extraction kit (NucleoSpin RNA II; Clontech, 740955.50). Reverse transcription was carried out using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems/ThermoFisher Scientific, ILT4368814). RT-qPCR was performed using the Power SYBR Green PCR Master Mix (Applied Biosystems/ThermoFisher Scientific, ILT4367659). The relative abundance of reference mRNAs and normalization for different total RNA amounts was done as described previously [65,66].
For human cells, 293 FT cells were seeded at a density of 5 × 104 cells per well in a 24-well plate and grown to 50% confluency. One μg of the 5'UTR-Firefly luciferase reporter construct and 100 ng of Renilla luciferase reporter construct [67] were transfected into 293 FT cells using Lipofectamine 3000. After 24 h, cells were treated with HBSS for an additional 4 h. mRNA levels were determined by real-time PCR analysis.
Western blot
Western blot was performed as described previously [8,68]. The antiserum to Pgk1 was generously provided by Dr. Jeremy Thorner (University of California, Berkeley). Antibody to YFP (Clontech, 632381) was used to detect GFP-tagged proteins. Atg13-PA, Atg5-PA, Atg12-PA, and Atg19-PA were detected with antibody to PA (Jackson ImmunoResearch, 323–005-024). The endogenous antibody to Atg1 was described previously [69]. Western blot bands were quantified by Bio-Rad Image Lab Software. Lanes/bands were manually selected and added for lane file analysis. An analysis table showing band intensities was used to calculate relative protein expression levels. The bands used are described in the figure legends.
Plasmids
Plasmids used in the yeast cells were generated as follows: The pRS406-ATG13 5'UTR-ATG13-PA plasmid was made by fast-cloning from the integrating vector pRS406 as described previously [8]. The DNA fragment containing 600 base pairs (bps) upstream of the start codon of ATG13, the ORF encoding ATG13-PA, and 423 bps downstream of the ATG13 CDS was amplified by PCR from the genomic DNA of a yeast strain in which the ATG13 gene was C-terminally-tagged with one copy of PA followed by the ATG13 endogenous 3' UTR. pRS406-ATG13 5'UTRmut-ATG13-PA, the plasmid with the ATG13 uORF mutation, was generated by site-directed mutagenesis based on the pRS406-ATG13 5'UTR-ATG13-PA plasmid. Similarly, the pRS406-ATG12 5'UTR-ATG12-PA and pRS406-ATG19 5'UTR-ATG19-PA plasmids were generated by fast-cloning from the pRS406 vector. The plasmids with uORF mutations, pRS406-ATG19 5'UTRmut-ATG19-PA and pRS406-ATG12 5'UTRmut-ATG12-PA were mutated by site-directed mutagenesis. These integrating plasmids were linearized using StuI before being integrated into the corresponding strains. The pRS416-ATG5 5'UTR-ATG5-PA plasmid was generated by fast-cloning from the centromeric vector pRS416. uORF mutation versions of the pRS416-ATG5 5'UTR-ATG5-PA plasmid, including pRS416-ATG5 5'UTRmut1-ATG5-PA, pRS416-ATG5 5'UTRmut2-ATG5-PA, and pRS416-ATG5 5'UTRmut3-ATG5-PA were generated by site-directed mutagenesis.
Plasmids used in the human cells were generated as follows: 518 nucleotides of the 5' UTR of ATG4B mRNA and 315 nucleotides of the 5' UTR of ATG12 mRNA containing the wild-type uORFs were PCR-amplified using DNA polymerase on genomic DNA derived from the HeLa cell line, together with PCR primers. PCR-amplified 5' UTRs were purified using a DNA Gel Purification Kit (Qiagen, 28706X4), digested with KpnI and MluI, and inserted into the KpnI and MluI sites of a Firefly luciferase reporter system using T4 DNA ligase (New England BioLabs, M0202S). Site directed mutagenesis of uORFs were performed using a kit (New England BioLabs, E0554S) with PCR primers. The sequence of constructs was confirmed by dideoxy sequencing.
Identification of putative uORFs of ATG genes
The following published datasets were analyzed in this study: (1) All possible canonical uORFs annotated by ribosome profiling performed by Ingolia et al. [17]; (2) canonical and noncanonical uORFs catalog predicted by a machine learning algorithm from the study by McGillivray et al. [25]. Potential translational active uORFs associated with core ATG genes were extracted and further analyzed based on their position, type, and start codon usage, etc. as shown in Tables S1 and S2.
Autophagy assays
The Pho8Δ60 assay and GFP-Atg8 processing assay were performed as described previously [29,70].
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 9. Statistical significance was determined from at least 3 independent experiments using either Student’s t-test or ANOVA. *: p < 0.05; **: p < 0.01; ***: p < 0.005. Number of independent experiments (n), statistical test utilized, dispersion of measurements and significance are described in the figure legends.
Supplementary Material
Funding Statement
This work was supported by the National Institute of General Medical Sciences [GM131919] and the National Cancer Institute [R01CA160417].
Disclosure statement
The authors declare no competing interests.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Yin Z, Popelka H, Lei Y, et al. The Roles of Ubiquitin in Mediating Autophagy. Cells. 2020;9(9):2025. DOI: 10.3390/cells9092025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wen X, Yang Y, Klionsky DJ.. Moments in autophagy and disease: past and present. Mol Aspects Med. 2021 April 28; 82:100966. 10.1016/j.mam.2021.100966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Yang Y, Klionsky DJ. Autophagy and disease: unanswered questions.Cell Death Differ. 2020. [2020 March 01];27(3):858–871.DOI: 10.1038/s41418-019-0480-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Parzych KR, Ariosa A, Mari M, et al. A newly characterized vacuolar serine carboxypeptidase, Atg42/Ybr139w, is required for normal vacuole function and the terminal steps of autophagy in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2018;29(9):1089–1099. DOI: 10.1091/mbc.E17-08-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Feng Y, Yao Z, Klionsky DJ. How to control self-digestion: transcriptional, post-transcriptional, and post-translational regulation of autophagy.Trends Cell Biol. 2015. [2015 June 01];25(6):354–363.DOI: 10.1016/j.tcb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chikashige Y, Kato H, Thornton M, et al. Gcn2 eIF2α kinase mediates combinatorial translational regulation through nucleotide motifs and uORFs in target mRNAs. Nucleic Acids Res. 2020;48(16):8977–8992. DOI: 10.1093/nar/gkaa608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zou K, Ouyang Q, Li H, et al. A global characterization of the translational and transcriptional programs induced by methionine restriction through ribosome profiling and RNA-seq. BMC Genomics. 2017 February 17 18(1):189. 10.1186/s12864-017-3483-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Liu X, Yao Z, Jin M, et al. Dhh1 promotes autophagy-related protein translation during nitrogen starvation. PLoS Biol. 2019;17(4):e3000219. DOI: 10.1371/journal.pbio.3000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Yin Z, Liu X, Ariosa A, et al. Psp2, a novel regulator of autophagy that promotes autophagy-related protein translation. Cell Res. 2019 December 01 29(12):994–1008. 10.1038/s41422-019-0246-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Acevo-Rodríguez PS, Maldonado G, Castro-Obregón S, et al. Autophagy regulation by the translation machinery and its implications in cancer [Review]. Front Oncol. 2020. March 13;10(322). DOI: 10.3389/fonc.2020.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kim C, Kim W, Lee H, et al. The RNA-binding protein HuD regulates autophagosome formation in pancreatic β cells by promoting autophagy-related gene 5 expression. J Biol Chem. 2014;289(1):112–121. DOI: 10.1074/jbc.M113.474700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ji E, Kim C, Kang H, et al. RNA binding protein HuR promotes autophagosome formation by regulating expression of autophagy-related proteins 5, 12, and 16 in human hepatocellular carcinoma cells. Mol Cell Biol. 2019;39(6):e00508–18. DOI: 10.1128/MCB.00508-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Palanisamy K, Tsai TH, Yu TM, et al. RNA‐binding protein, human antigen R regulates hypoxia‐induced autophagy by targeting ATG7/ATG16L1 expressions and autophagosome formation. J Cell Physiol. 2019;234(5):7448–7458. DOI: 10.1002/jcp.27502. [DOI] [PubMed] [Google Scholar]
- [14].Zhang Z, Guo M, Li Y, et al. RNA-binding protein ZFP36/TTP protects against ferroptosis by regulating autophagy signaling pathway in hepatic stellate cells. Autophagy. 2020;16(8):1482–1505. DOI: 10.1080/15548627.2019.1687985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proceedings of the National Academy of Sciences. 2009;106(18):7507–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhang H, Wang Y, Lu J. Function and evolution of upstream ORFs in eukaryotes. Trends Biochem Sci. 2019;44(9):782–794. [DOI] [PubMed] [Google Scholar]
- [17].Ingolia NT, Ghaemmaghami S, Newman JRS, et al. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–223. DOI: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Fernandez J, Yaman I, Huang C, et al. Ribosome stalling regulates IRES-mediated translation in eukaryotes, a parallel to prokaryotic attenuation. Mol Cell. 2005. [2005 February 04];17(3):405–416. DOI: 10.1016/j.molcel.2004.12.024. [DOI] [PubMed] [Google Scholar]
- [19].Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. [DOI] [PubMed] [Google Scholar]
- [20].Munzarová V, Pánek J, Gunišová S, et al. Translation Reinitiation Relies on the Interaction between eIF3a/TIF32 and Progressively Folded cis-Acting mRNA Elements Preceding Short uORFs. PLoS Genet. 2011;7(7):e1002137. DOI: 10.1371/journal.pgen.1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Andrews SJ, Rothnagel JA. Emerging evidence for functional peptides encoded by short open reading frames.Nat Rev Genet. 2014. [2014 March 01];15(3):193–204.DOI: 10.1038/nrg3520. [DOI] [PubMed] [Google Scholar]
- [22].Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(31):11269–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ingolia NT, Brar GA, Rouskin S, et al. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012 August 01 7(8):1534–1550. 10.1038/nprot.2012.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chew G-L, Pauli A, Schier AF. Conservation of uORF repressiveness and sequence features in mouse, human and zebrafish.Nat Commun. 2016. [2016 May 24];7(1):11663.DOI: 10.1038/ncomms11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].McGillivray P, Ault R, Pawashe M, et al. A comprehensive catalog of predicted functional upstream open reading frames in humans. Nucleic Acids Res. 2018;46(7):3326–3338. DOI: 10.1093/nar/gky188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Reggiori F, Tucker KA, Stromhaug PE, et al. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004. Jan;6(1):79–90.DOI: 10.1016/S1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- [27].Klionsky DJ, Abdel-Aziz AK, Abdelfatah S, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)1. Autophagy. [2021 January 02];17(1):1–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Cheong H, Klionsky DJ. Biochemical methods to monitor autophagy-related processes in yeast. Methods Enzymol. 2008;451:1–26. [DOI] [PubMed] [Google Scholar]
- [29].Memisoglu G, Eapen VV, Yang Y, et al. PP2C phosphatases promote autophagy by dephosphorylation of the Atg1 complex. Proceedings of the National Academy of Sciences. 2019:201817078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hinnebusch AG, Ivanov IP, Sonenberg N. Translational control by 5’-untranslated regions of eukaryotic mRNAs. Science. 2016 Jun 17; 352(6292):1413–1416. 10.1126/science.aad9868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009 Feb 20; 136(4):731–745. 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31; 44(2):283–292. 10.1016/0092-8674(86)90762-2 [DOI] [PubMed] [Google Scholar]
- [33].Barbosa C, Peixeiro I, Romão L. Gene expression regulation by upstream open reading frames and human disease. PLoS Genet. 2013;9(8):e1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kim M, Sandford E, Gatica D, et al. Mutation in ATG5 reduces autophagy and leads to ataxia with developmental delay. Elife. 2016 Jan 26;5: 10.7554/eLife.12245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mizushima N, Noda T, Yoshimori T, et al. A protein conjugation system essential for autophagy. Nature. 1998 September 1 395(6700):395–398. 10.1038/26506 [DOI] [PubMed] [Google Scholar]
- [36].Walczak M, Martens S. Dissecting the role of the Atg12-Atg5-Atg16 complex during autophagosome formation. Autophagy. 2013. Mar;9(3):424–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hanada T, Noda NN, Satomi Y, et al. The Atg12-Atg5 Conjugate Has a Novel E3-like Activity for Protein Lipidation in Autophagy*. J Biol Chem. 2007 December 28 282(52):37298–37302. 10.1074/jbc.C700195200 [DOI] [PubMed] [Google Scholar]
- [38].Scott SV, Guan J, Hutchins MU, et al. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol Cell. 2001. Jun;7(6):1131–1141.DOI: 10.1016/S1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Andreev DE, Arnold M, Kiniry SJ, et al. TASEP modelling provides a parsimonious explanation for the ability of a single uORF to derepress translation during the integrated stress response. Elife. 2018;7:e32563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Fritsch C, Herrmann A, Nothnagel M, et al. Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting. Genome Res. 2012;22(11):2208–2218. DOI: 10.1101/gr.139568.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lee S, Liu B, Lee S, et al. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proceedings of the National Academy of Sciences. 2012;109(37):E2424–E2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Feng Y, He D, Yao Z, et al. The machinery of macroautophagy. Cell Res. 2014 January 1 24(1):24–41. 10.1038/cr.2013.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kearse MG, Wilusz JE. Non-AUG translation: a new start for protein synthesis in eukaryotes. Genes Dev. 2017;31(17):1717–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Spealman P, Naik AW, May GE, et al. Conserved non-AUG uORFs revealed by a novel regression analysis of ribosome profiling data. Genome Res. 2018. Feb;28(2):214–222.DOI: 10.1101/gr.221507.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Tennessen JA, Bigham AW, O’Connor TD, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337(6090):64–69. DOI: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016 July 1 536(7616):285–291. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ke X, Taylor MS, Cardon LR. Singleton SNPs in the human genome and implications for genome-wide association studies. Eur J Hum Genet. 2008 March 1; 16(4):506–515. 10.1038/sj.ejhg.5201987 [DOI] [PubMed] [Google Scholar]
- [48].Li M, Hou Y, Wang J, et al. Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J Biol Chem. 2011 Mar 4 286(9):7327–7338. 10.1074/jbc.M110.199059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Michel AM, Andreev DE, Baranov PV. Computational approach for calculating the probability of eukaryotic translation initiation from ribo-seq data that takes into account leaky scanning. BMC Bioinformatics. 2014 November 21; 15(1):380. 10.1186/s12859-014-0380-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Noda NN, Fujioka Y, Hanada T, et al. Structure of the Atg12–Atg5 conjugate reveals a platform for stimulating Atg8–PE conjugation. EMBO Rep. 2013;14(2):206–211. DOI: 10.1038/embor.2012.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Otomo C, Metlagel Z, Takaesu G, et al. Structure of the human ATG12~ATG5 conjugate required for LC3 lipidation in autophagy. Nat Struct Mol Biol. 2013 January 1 20(1):59–66. 10.1038/nsmb.2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Xia X, Dermitzakis E. The +4G site in kozak consensus is not related to the efficiency of translation initiation. PLOS ONE. 2007;2(2):e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol Cell Biol. 1987;7(10):3438–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Watatani Y, Ichikawa K, Nakanishi N, et al. Stress-induced translation of ATF5 mRNA is regulated by the 5’-untranslated region. J Biol Chem. 2008 Feb 1 283(5):2543–2553. 10.1074/jbc.M707781200 [DOI] [PubMed] [Google Scholar]
- [55].Andreev DE, O’Connor PB, Fahey C, et al. Translation of 5’ leaders is pervasive in genes resistant to eIF2 repression. Elife. 2015 Jan 26;4:e03971. 10.7554/eLife.03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Babitzke P, Baker CS, Romeo T. Regulation of translation initiation by RNA binding proteins. Annu Rev Microbiol. 2009;63:27–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Cottrell KA, Szczesny P, Djuranovic S. Translation efficiency is a determinant of the magnitude of miRNA-mediated repression. Sci Rep. 2017 November 02; 7(1):14884. 10.1038/s41598-017-13851-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lubas M, Harder LM, Kumsta C, et al. eIF5A is required for autophagy by mediating ATG3 translation. EMBO Rep. 2018. Jun;19(6). doi: 10.15252/embr.201846072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Chen J, Brunner A-D, Cogan JZ, et al. Pervasive functional translation of noncanonical human open reading frames. Science. 2020;367(6482):1140–1146. DOI: 10.1126/science.aay0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ji E, Kim C, Kang H, et al. RNA binding protein hur promotes autophagosome formation by regulating expression of autophagy-related proteins 5, 12, and 16 in human hepatocellular carcinoma cells. mol cell biol. 2019 mar 15;39:(6):e00508–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zhang H, Dou S, He F, et al. Genome-wide maps of ribosomal occupancy provide insights into adaptive evolution and regulatory roles of uORFs during Drosophila development. PLoS Biol. 2018;16(7):e2003903. DOI: 10.1371/journal.pbio.2003903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Sendoel A, Dunn JG, Rodriguez EH, et al. Translation from unconventional 5’ start sites drives tumour initiation. Nature. 2017;541(7638):494–499. DOI: 10.1038/nature21036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Forbes SA, Beare D, Boutselakis H, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2016;45(D1):D777–D783. DOI: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016 Sep 1; 30(17):1913–1930. 10.1101/gad.287524.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Gatica D, Hu G, Liu X, et al. The Pat1-Lsm complex stabilizes ATG mRNA during nitrogen starvation-induced autophagy. Mol Cell. 2019 January 17 73(2):314–324.e4. 10.1016/j.molcel.2018.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Hu G, McQuiston T, Bernard A, et al. A conserved mechanism of TOR-dependent RCK-mediated mRNA degradation regulates autophagy. Nat Cell Biol. 2015 July 1 17(7):930–942. 10.1038/ncb3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Chen X, Prywes R. Serum-induced expression of the cdc25A gene by relief of E2F-mediated repression. Mol Cell Biol. 1999. Jul;19(7):4695–4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Feng Y, Ariosa AR, Yang Y, et al. Downregulation of autophagy by Met30-mediated Atg9 ubiquitination. Proceedings of the National Academy of Sciences. 2021;118(1):e2005539118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Abeliovich H, Zhang C, William A, et al. Chemical genetic analysis of apg1 reveals a non-kinase role in the induction of autophagy. Mol Biol Cell. 2003;14(2):477–490. DOI: 10.1091/mbc.e02-07-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Feng Y, Backues SK, Baba M, et al. Phosphorylation of Atg9 regulates movement to the phagophore assembly site and the rate of autophagosome formation. Autophagy. 2016 April 2 12(4):648–658. 10.1080/15548627.2016.1157237 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


