ABSTRACT
Post-translational modifications, such as phosphorylation, ubiquitination and acetylation, play crucial roles in the regulation of autophagy. Acetylation has emerged as an important regulatory mechanism for autophagy. Acetylation regulates autophagy initiation and autophagosome formation by targeting core components of the ULK1 complex, the BECN1-PIK3C3 complex, and the LC3 lipidation system. Recent studies have shown that acetylation occurs on the key proteins participating in autophagic cargo assembly and autophagosome-lysosome fusion, such as SQSTM1/p62 and STX17. In addition, acetylation controls autophagy at the transcriptional level by targeting histones and the transcription factor TFEB. Here, we review the current knowledge on acetylation of autophagy proteins and their regulations and functions in the autophagy pathway with focus on recent findings.
Abbreviations : ACAT1: acetyl-CoA acetyltransferase 1; ACSS2: acyl-CoA synthetase short chain family member 2; AMPK: AMP-activated protein kinase; ATG: autophagy-related; CALCOCO2/NDP52: calcium binding and coiled-coil domain 2; CCAR2/DBC1: cell cycle and apoptosis regulator 2; BECN1: beclin 1; CMA: chaperone-mediated autophagy; CREBBP/CBP: CREB binding protein; EP300/p300: E1A binding protein p300; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; GSK3: glycogen synthase kinase 3; HDAC6: histone deacetylase 6; HSPA8/HSC70: heat shock protein family A (Hsp70) member 8; KAT2A/GCN5: lysine acetyltransferase 2A; KAT2B/PCAF: lysine acetyltransferase 2B; KAT5/TIP60: lysine acetyltransferase 5; KAT8/MOF: lysine acetyltransferase 8; LAMP2A: lysosomal associated membrane protein 2A; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; MTOR: mechanistic target of rapamycin kinase; NBR1: NBR1 autophagy cargo receptor; OPTN: optineurin; PD: Parkinson disease; PE: phosphatidylethanolamine; PIK3C3/VPS34: phosphatidylinositol 3-kinase catalytic subunit type 3; PKM2: pyruvate kinase M1/2; PtdIns3P: phosphatidylinositol-3-phosphate; PTM: post-translational modification; RB1CC1/FIP200: RB1 inducible coiled-coil 1; RUBCN/Rubicon: rubicon autophagy regulator; RUBCNL/Pacer: rubicon like autophagy enhancer; SIRT1: sirtuin 1; SNAP29: synaptosome associated protein 29; SNARE: soluble N-ethylamide-sensitive factor attachment protein receptor; SQSTM1/p62: sequestosome 1; STX17: syntaxin 17; TFEB: transcription factor EB; TP53/p53: tumor protein p53; TP53INP2/DOR: tumor protein p53 inducible nuclear protein 2; UBA: ubiquitin-associated; ULK1: unc-51 like autophagy activating kinase 1; VAMP8: vesicle associated membrane protein 8; WIPI2: WD repeat domain, phosphoinositide interacting 2.
KEYWORDS: Acetylation, acetyltransferase, autophagy, deacetylase, post-translational modification
Introduction
Protein acetylation regulates various biological processes, such as glycolysis, lipid synthesis, DNA damage repair, and cell cycle progression, by modulating stability, interaction with binding partners, enzymatic activity, or subcellular localization of the targets [1,2]. The acetylation state of proteins is determined by the opposing actions of acetyltransferases and deacetylases. The acetyltransferases are responsible for the transfer of the acetyl group from acetyl-CoA to the lysine residues of the substrates. The deacetylases are responsible for the removal of the acetyl group from the modified lysine residues.
Protein acetylation has emerged as a pivotal regulatory mechanism for autophagy [3–12]. Many proteins functioning at different stages of the autophagy pathway have been identified as targets of acetylation [3–12]. In this review, we will discuss the functions and mechanisms of acetylation and corresponding acetyltransferases and deacetylases in the regulation of autophagy mainly focusing on those involved in autophagy initiation, MAP1LC3/LC3 (microtubule associated protein 1 light chain 3) conjugation system, autophagic cargo assembly, autophagosome-lysosome fusion and transcriptional control of autophagy-related genes [3–12].
Acetylation of autophagy initiation machinery
Autophagy induction occurs when ULK1 (unc-51 like autophagy activating kinase 1) is activated in response to starvation [13,14]. ULK1 forms a protein complex with several partners, including RB1CC1/FIP200 (RB1 inducible coiled-coil 1), ATG13 (autophagy related 13) and ATG101, to regulate autophagy initiation [15–19]. The activity of ULK1 complex is regulated by MTOR (mechanistic target of rapamycin kinase) complex 1 (MTORC1) and AMP-activated protein kinase (AMPK) [20]. The ULK1 complex activates the BECN1 (beclin 1)-PIK3C3/VPS34 (phosphatidylinositol 3-kinase catalytic subunit type 3) complex that catalyzes the local synthesis of phosphatidylinositol-3-phosphate (PtdIns3P) [21]. PtdIns3P recruits the downstream effector WIPI2 (WD repeat domain, phosphoinositide interacting 2) to promote phagophore formation and expansion [22].
Acetylation occurs on ULK1, PIK3C3 and BECN1 [5,7,23] (Figure 1, Table 1). During deprivation of growth factors, GSK3 (glycogen synthase kinase 3) phosphorylates and activates the acetyltransferase KAT5/TIP60 (lysine acetyltransferase 5) [5]. KAT5/TIP60 acetylates ULK1 at K162 and K606 and thereby activates ULK1 in response to serum deprivation [5] (Figure 1, Table 1). KAT5/TIP60-mediated acetylation of ULK1 activates ULK1 independently of MTORC1- and AMPK-mediated phosphorylations of ULK1 [5].
Figure 1.
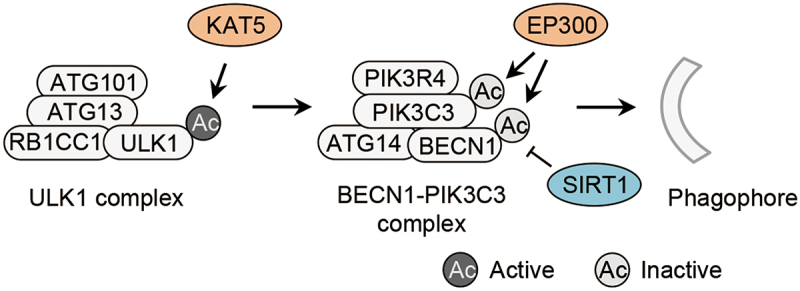
Acetylation of autophagy initiation machinery. Acetylation of ULK1 by the acetyltranferase KAT5/TIP60 activates the ULK1 complex, and acetylation of PIK3C3 and BECN1 regulated by the acetyltransferase EP300 and the deacetylase SIRT1 inhibits the BECN1-PIK3C3 complex, both of which control the initiation of autophagy. Ac, acetylation; ATG, autophagy-related; BECN1, beclin 1; EP300/p300, E1A binding protein p300; KAT5/TIP60, lysine acetyltransferase 5; RB1CC1/FIP200, RB1 inducible coiled-coil 1; SIRT1, sirtuin 1; ULK1, unc-51 like autophagy activating kinase 1; PIK3C3/VPS34, phosphatidylinositol 3-kinase catalytic subunit type 3; PIK3R4, phosphoinositide-3-kinase regulatory subunit 4.
Table 1.
Acetyltransferases and deacetylases that modify autophagy-related proteins*.
| Acetyltransferases |
Target proteins |
Acetylation sites |
Deacetylases |
Effects on autophagy |
Underlying mechanisms |
References |
| EP300 | BECN1 | K430, K437 | SIRT1 | Inhibits autophagy initiation | Decreases PIK3C3 activity | [23] |
| PIK3C3 | K29, K771, K781 | Unknown | Inhibits autophagy initiation | Decreases PIK3C3 activity | [7] | |
| EP300-CREBBP | LC3 | K49, K51 | SIRT1 | Inhibits LC3 lipidation | Inhibits the redistribution of nuclear LC3 into cytoplasm and the binding of LC3 to ATG7 | [3,4,32] |
| ATG5 | Unknown | SIRT1 | Unknown | Unknown | [3,32] | |
| ATG7 | Unknown | SIRT1 | Unknown | Unknown | [3,32] | |
| ATG12 | Unknown | Unknown | Unknown | Unknown | [32] | |
| CREBBP | STX17 | K219, K223 | HDAC2 | Inhibits autophagosome -lysosome fusion | Inhibits the interaction between STX17 and SNAP29 | [6] |
| KAT5/TIP60/Esa1 | ULK1 | K162, K606 | Unknown | Promotes autophagy initiation | Increases the activity of ULK1 complex | [5] |
| Atg3 | K19, K48, K183 | Rpd3 | Promotes Atg8 lipidation | Increases the interaction between Atg3 and Atg8 | [8] | |
| SQSTM1 | K420, K435 | HDAC6 | Promotes cargo assembly | Promotes SQSTM1 binding to ubiquitin and ubiquitinated substrates | [9] | |
| RUBCNL | K483, K523, K533, K573, K633 | Unknown | Promotes autophagosome-lysosome fusion | Promotes the interaction of RUBCNL with HOPS complex and STX17 | [52] | |
| KAT2A/GCN5 | ATG7 | K338 | Unknown | Inhibits LC3 lipidation | Decreases the activity of ATG7 | [34] |
| TFEB | K116, K274, K279 | SIRT1 (K116) | Inhibits transcriptional regulation of autophagy | Inactivates the transcriptional activity of TFEB | [10,11] | |
| ACAT1 | TFEB | K91, K103, K116, K430 | SIRT1 (K116) | Promotes transcriptional regulation of autophagy | Activates the transcriptional activity of TFEB | [10,12] |
| KAT8/MOF | Histone H4 | K16 | SIRT1 | Inhibits transcriptional regulation of autophagy | Inhibits the transcription of autophagy-related genes | [62] |
*Many of the listed sites need further rigorous validation to be defined as target sites of the acetyltransferases and deacetylases
The lipid kinase PIK3C3/VPS34, the core subunit of the BECN1-PIK3C3 complex, is acetylated at K29, K771 and K781 by the acetyltransferase EP300/p300 (E1A binding protein p300) [7] (Figure 1, Table 1). The acetylation at K29 suppresses the interaction between PIK3C3 and BECN1, whereas the acetylation at K771 diminishes the affinity of PIK3C3 for its substrate phosphatidylinositol (PI) [7]. Another study has shown that EP300-mediated acetylation of BECN1 at K430 and K437 promotes the interaction between BECN1 and RUBCN/Rubicon (rubicon autophagy regulator), a negative regulator of PIK3C3 [23] (Figure 1, Table 1). Thus, the acetylation of BECN1 K430 and K437 suppresses PIK3C3 activity [23]. By acetylating both PIK3C3 and BECN1, EP300 might potently suppress PIK3C3 activity and autophagy induction. SIRT1 (sirtuin 1) acts as the major deacetylase for BECN1 K430 and K437 [23] (Figure 1, Table 1). PIK3C3 and BECN1 form multiple protein complexes participating not only in autophagy but also in other membrane processes [24–27]. Thus, EP300-mediated acetylation of PIK3C3 and BECN1 and SIRT1-mediated deacetylation of BECN1 might regulate diverse membrane processes in cells.
Acetylation of LC3 conjugation machinery
The conversion of soluble LC3 into membrane-bound LC3–phosphatidylethanolamine (PE) is an important event in autophagy. Membrane-bound LC3–PE controls several pivotal processes in autophagy, including growth and expansion of the phagophore, recruitment of cargoes, and fusion of autophagosomes and lysosomes [28–30]. The formation of LC3–PE relies on the ubiquitination-like conjugation system by which soluble LC3 is conjugated with PE under the assistance of the E1-like enzyme ATG7, the E2-like enzyme ATG3, and the E3-like ATG12–ATG5-ATG16L1 complex [22,31].
The core components involved in LC3 lipidation system are targeted by protein acetylation [3,4,8,32–34] (Figure 2, Table 1). LC3 (including other Atg8-family proteins) can be acetylated by either EP300 or its closely related CREBBP (CREB binding protein) at K49 and K51 [4,32]. The acetylation of LC3 at both sites plays a negative role in autophagy by suppressing the redistribution of nuclear LC3 into cytoplasm [4]. In starved cells, the deacetylated nuclear LC3 is transported to the cytoplasm by the nuclear protein TP53INP2/DOR (tumor protein p53 inducible nuclear protein 2) [4]. Interestingly, MTORC1 seems to be a major upstream regulator determining the distribution of TP53INP2 in the nucleus, including the nucleolus, and in the cytoplasm [35–37]. The acetylation of LC3 at both sites also suppresses its binding to ATG7, thus resulting in reduction of LC3–PE formation [4]. In addition, the acetylation of LC3 at both sites is also shown to stabilize LC3 by inhibiting the proteasome-dependent degradation of LC3 [38]. Atg3 is acetylated at K19, K48 and K183 by Esa1 (KAT5/TIP60 in mammalian cells) in yeast [8]. Contrary to the LC3 acetylation, the Atg3 acetylation at K19 and K48 enhances autophagy by promoting its interaction with Atg8 (the yeast homolog of LC3) and facilitating Atg8 lipidation [8].
Figure 2.

Acetylation of LC3 conjugation machinery. Acetylation of Atg3 controlled by the acetyltransfease Esa1 (KAT5/TIP60 in mammalian cells) and the deacetylase Rpd3 promotes Atg8 lipidation in yeast. Acetylation of ATG5, ATG7, ATG12 and LC3 is mediated by the acetyltransfease EP300-CREBBP and deacetylation of ATG5, ATG7 and LC3 is mediated by the deacetylase SIRT1. In addition, the acetyltransfease KAT2A/GCN5 also acetylates ATG7. In particular, deacetylation promotes the cytoplasmic translocation of nuclear LC3 under the assistance of the nuclear protein TP53INP2 and stimulates LC3 lipidation in the cytoplasm. CREBBP/CBP, CREB binding protein, KAT2A/GCN5, lysine acetyltransferase 2A; MAP1LC3/LC3, microtubule-associated protein 1 light chain 3; TP53INP2, tumor protein p53 inducible nuclear protein 2.
ATG5, ATG7, and ATG12 are acetylated by both EP300 and CREBBP [32]. In addition, ATG7 is also acetylated at K338 by KAT2A/GCN5 (lysine acetyltransferase 2A) [34]. How acetylation regulates the autophagic functions of ATG5, ATG7 and ATG12 is unclear. Some studies have shown that ATG5 and ATG7 regulate DNA damage response and the transcriptional activity of TP53/p53 (tumor protein p53), respectively [39,40]. Thus, it is possible that the acetylated forms of ATG5 and ATG7 could regulate autophagy-independent processes, such as the DNA damage response.
SIRT1 acts as the major deacetylase that removes the acetyl group from the acetylated lysine residues of ATG5, ATG7 and LC3 [3]. Rpd3 (a histone deacetylase) is responsible for the deacetylation of Atg3 at K19, K48 and K183 in yeast [8]. The deacetylase of ATG3 in mammalian cell remains unclear. Of note, ATG5, ATG7, ATG12 and LC3 undergo deacetylation while Atg3 undergoes acetylation upon autophagy induction [3,4,8,32], indicating that EP300-CREBBP and KAT5/TIP60 might have opposing effects on autophagy.
Acetylation of autophagic cargo assembly proteins
A group of proteins called autophagy receptor proteins play roles in recruiting specific types of cargoes to phagophores [41]. SQSTM1/p62 (sequestosome 1) recruits ubiquitinated proteins via its ubiquitin-associated (UBA) domain [42,43]. The UBA domain of SQSTM1 can undergo intermolecular dimerization, which reduces the affinity of the domain toward ubiquitinated substrates [44,45]. Various post-translational modifications (PTMs), including phosphorylation and ubiquitination, occur on the UBA domain of SQSTM1 and phosphorylation has been shown to disrupt the dimerization [46,47].
KAT5/TIP60 acetylates SQSTM1 at K420 and K435 and facilitates its autophagic degradation in starved cells [9] (Figure 3, Table 1). The acetylation occurs on the UBA domain of SQSTM1 and disrupts the dimerization. This enhances SQSTM1 binding to ubiquitinated substrates and promotes SQSTM1-mediated autophagic cargo assembly [9]. Given that the UBA or UBA-like domain is also present in other autophagy receptors, such as NBR1 (NBR1 autophagy cargo receptor), OPTN (optineurin) and CALCOCO2/NDP52 (calcium binding and coiled-coil domain 2) [48], it is possible that KAT5/TIP60-mediated acetylation might be a general mechanism adopted by the autophagy receptors to strengthen their affinity for ubiquitinated substrates.
Figure 3.
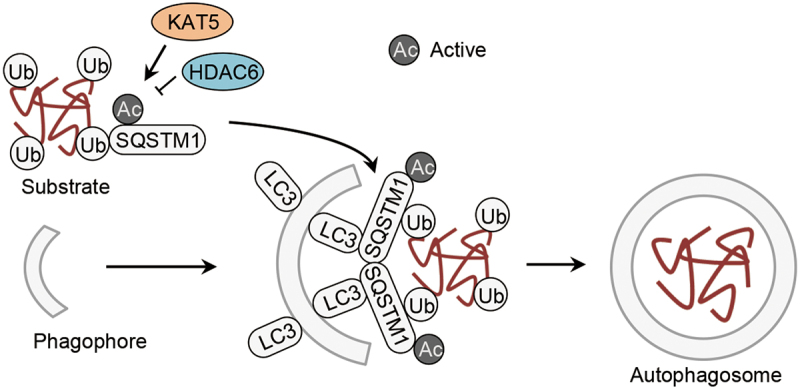
Acetylation of autophagic cargo assembly proteins. Acetylation of SQSTM1/p62 controlled by the acetyltransfease KAT5/TIP60 and the deacetylase HDAC6 facilitates autophagic cargo assembly. HDAC6, histone deacetylase 6; SQSTM1/p62, sequestosome 1; Ub, ubiquitin.
Acetylation of autophagosome-lysosome fusion machinery
The fusion between autophagosome and lysosome is required for the degradation of autophagy substrates. Various components, including RAB GTPases, soluble N-ethylamide-sensitive factor attachment protein receptors (SNAREs) and tethering factors, regulate autophagosome-lysosome fusion [49–51].
Acetylation controls autophagosome-lysosome fusion by targeting STX17 (syntaxin 17), an autophagosome-localized SNARE, and RUBCNL/Pacer (rubicon like autophagy enhancer) [6,52] (Figure 4, Table 1). In starved cells, the acetylation of STX17 at K219 and K223 is decreased due to the inactivation of its acetyltransferase CREBBP [6]. The deacetylation state of STX17 promotes the recruitment of SNAP29 (synaptosome associated protein 29) from the cytosol by enhancing their interaction [6]. The STX17-SNAP29 complex interacts with VAMP8 (vesicle associated membrane protein 8) at the lysosome to form the trimeric SNARE complex consisting of STX17, SNAP29, ‘and’ VAMP8, which is required for the autophagosome-lysosome fusion [6,49]. Conversely, the acetyltransferase KAT5/TIP60-mediated acetylation of RUBCNL promotes its interaction with the homotypic fusion and vacuole protein sorting (HOPS) complex to facilitate the autophagosome-lysosome fusion [52]. Thus, EP300-CREBBP and KAT5/TIP60 play opposing roles in the regulation of autophagosome-lysosome fusion as they do in the regulation of autophagy initiation.
Figure 4.

Acetylation of autophagosome-lysosome fusion machinery. Acetylation of RUBCNL/Pacer by the acetyltransferase KAT5/TIP60, and acetylation of STX17 controlled by the acetyltransferase CREBBP and the deacetylase HDAC2, promotes and inhibits autophagosome-lysosome fusion, respectively. In addition, the deacetylase HDAC6 is recruited to the lysosome by ATP13A2 to deacetylate CTTN to promote autophagosome-lysosome fusion. ATP13A2, ATPase cation transporting 13A2; CREBBP/CBP, CREB binding protein; CTTN, cortactin; HDAC2, histone deacetylase 2; HDAC6, histone deacetylase 6; HOPS, homotypic fusion and vacuole protein sorting; RUBCNL/Pacer, rubicon like autophagy enhancer; SNAP29, synaptosome associated protein 29; STX17, syntaxin 17; VAMP8, vesicle associated membrane protein 8.
HDAC6 (histone deacetylase 6) facilitates the autophagosome-lysosome fusion in selective autophagy [53]. Polyamine-transporting ATP13A2 (ATPase cation transporting 13A2) recruits HDAC6 to the lysosome where HDAC6 deacetylates CTTN (cortactin), a protein that acts as a potent promoter of the autophagosome-lysosome fusion [54] (Figure 4). Mutations of ATP13A2 have been found in Parkinson disease (PD) patients [55,56]. Those mutations prevent ATP13A2 to facilitate the autophagosome-lysosome fusion, thus leading to the accumulation of protein aggregates and damaged mitochondria [54]. This suggests that dysregulation of HDAC6-mediated autophagosome-lysosome fusion might be involved in the pathogenesis of PD.
While both EP300 and CREBBP participate in autophagosome formation, only CREBBP is involved in the regulation of autophagosome-lysosome fusion. Elucidating the molecular basis underlying the preference of EP300 and CREBBP for their substrates would help understand the functional difference between EP300 and CREBBP in various biological processes, including autophagy.
Transcriptional control of autophagy-related genes by acetylation
Autophagy is regulated at the transcriptional level [57,58]. As a member of MiT/TFE family, TFEB (transcription factor EB) enhances the expression of autophagy- and lysosome-related genes, such as MAP1LC3B, SQSTM1, LAMP1 and CTSD [58]. TFEB is regulated by multiple protein kinases, including MTORC1 [59–61]. MTORC1-mediated phosphorylation prevents the nuclear translocation of TFEB and blocks its transcriptional activity [61].
Several studies have shown that acetylation regulates TFEB activity [10–12] (Figure 5, Table 1). TFEB is acetylated by the acetyltransferases ACAT1 (acetyl-CoA acetyltransferase 1) and KAT2A/GCN5 [11,12]. Interestingly, ACAT1 and KAT2A/GCN5 have the opposing effects on the transcriptional activity of TFEB via acetylation [11,12]. ACAT1 acetylates TFEB at K91, K103, K116 and K430 and promotes its transcriptional activity, whereas KAT2A/GCN5 acetylates TFEB at K116, K274 and K279 and suppresses its transcription activity [11,12]. Different intracellular or environmental cues might stimulate the distinct acetyltransferases to control the activity of TFEB.
Figure 5.

Transcriptional control of autophagy-related genes by acetylation. At the transcriptional level, acetylation of TFEB by the acetyltransferases ACAT1 and KAT2A/GCN5 activates and inactivates the transcriptional activity of TFEB, respectively. Of note, acetylation of histone H4, mediated by the acetyltransferase KAT5/TIP60 or controlled by the acetyltransferase KAT8/MOF and the deacetylase SIRT1, promotes and inhibits the transcription of autophagy- and lysosome-related genes, respectively. In addition, upon energy depletion, AMPK phosphorylates ACSS2 and drives its nuclear translocation, where ACSS2 binds to TFEB and produces acetyl-CoA from acetate to stimulate histone acetylation locally, leading to the transcription of autophagy- and lysosome-related genes. ACAT1, acetyl-CoA acetyltransferase 1; ACSS2, acyl-CoA synthetase short chain family member 2; AMPK, AMP-activated protein kinase; KAT2A/GCN5, lysine acetyltransferase 2A; KAT8/MOF, lysine acetyltransferase 8; MTORC1, mechanistic target of rapamycin kinase complex 1; TFEB, transcription factor EB.
Acetylation of histones is also closely linked to the transcription of autophagy-related genes [62,63] (Figure 5, Table 1). The transcription of autophagy-related genes is repressed by the acetyltransferase KAT8/MOF (lysine acetyltransferase 8) and activated by the acetyltransferase KAT5/TIP60 [62,63]. KAT8/MOF acetylates histone H4 K16 to repress the transcription of autophagy-related genes, such as MAP1LC3B, NBR1 and ULK1, and the acetylation is reversed by SIRT1 [62]. KAT5/TIP60 acetylates histone H4 to activate the transcription of autophagy-related genes, such as MAP1LC3B, GABARAP, SQSTM1 and ULK1 [63]. The acetylation sites remain unknown.
Upon glucose starvation, AMPK-dependent phosphorylation drives the nuclear translocation of ACSS2 (acyl-CoA synthetase short chain family member 2) [64]. Nuclear ACSS2 then binds to TFEB and utilizes acetate, which is generated from histone deacetylation, to locally produce acetyl-CoA for histone acetylation in the promoter regions of TFEB target genes, leading to the transcription of autophagy- and lysosome-related genes [64]. Although intracellular acetyl-CoA level is downregulated in cells cultured in serum-free Hanks’ balanced salt solution (HBSS) [65], ACSS2-mediated local production of acetyl-CoA helps maintain histone acetylation in the promoter regions of specific genes [64]. The acetyltransferase that mediates the histone acetylation in the promoter regions of TFEB-regulated genes remains unknown.
The activity of KAT2A toward the acetylation of TFEB is decreased upon treatment of nutrient-free culture medium or torin 1 (MTOR inhibitor) [11], suggesting that MTOR might act as an upstream regulator of KAT2A. Mechanistically, KAT2A-mediated acetylation suppresses the transcriptional activity of TFEB through disrupting its dimerization, which decreases its affinity for DNA [11]. Thus, MTOR might control not only the subcellular localization but also the DNA affinity of TFEB. The molecular mechanism underlying MTOR-dependent activation of KAT2A remains unclear and needs further investigation.
Regulation of autophagy-related acetyltransferases and deacetylases
The acetylation states of the autophagy-related proteins are controlled by signaling molecules responding to cellular nutrients and growth factors. As the major deacetylase functioning in autophagy, SIRT1 can be activated through an AMPK-dependent manner upon glucose deprivation [66]. AMPK-mediated phosphorylation of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) triggers its nuclear translocation [66]. Nuclear GAPDH then binds and activates SIRT1 by displacing CCAR2/DBC1 (cell cycle and apoptosis regulator 2) [66], the major repressor protein of SIRT1 [67,68]. Activated SIRT1 then deacetylates nuclear LC3 to facilitate autophagy initiation [3,4,66]. However, whether deacetylation of other ATG proteins by SIRT1 is also nuclear events remain unknown.
AMPK regulates the acetyltransferase activity of EP300 [69,70]. AMPK-mediated phosphorylation of EP300 at the N-terminal region blocks its acetyltransferase activity [69,70]. EP300 activity is regulated by intracellular acetyl-CoA [65]. Incubation of the cells with serum-free HBSS causes depletion of intracellular acetyl-CoA and inhibits the autoacetylation of EP300, leading to EP300 inactivation and autophagy induction [65]. Amino acid deprivation promotes the nuclear translocation of EP300 and affects the accessibility of EP300 to its nuclear and cytoplasmic substrates, such as TP53 and ATG7, whereby enhancing the acetylation of TP53 and decreasing the acetylation of ATG7 and leading to TP53 activation and autophagy induction [71]. In addition, MTORC1 regulates the activity of EP300-CREBBP [72]. MTORC1 phosphorylates EP300 at S2271, S2279, S2291 and S2315, thereby abolishing its intramolecular inhibition and resulting in its activation [72–74]. Thus, MTORC1 can suppress autophagy via activation of EP300-CREBBP. Several natural compounds, such as spermidine and curcumin, have been shown to directly bind to EP300-CREBBP and inhibit the acetyltransferase activity, leading to activation of autophagy [75–77]. Notably, genetic or pharmacological inhibition of EP300-CREBBP is sufficient to activate autophagy [32,75–77].
EP300-CREBBP and KAT5/TIP60 play the opposing roles as the major acetyltransferases in many different stages of autophagy. How their acetyltransferase activities are coordinately regulated during the autophagy process needs more investigation. Regarding KAT5/TIP60, which enhances autophagy [5], it remains elusive how KAT5/TIP60 can maintain its high acetyltransferase activity under growth factor deprivation, the condition that can lower the intracellular acetyl-CoA level.
Future perspectives
Although acetyltransferases and deacetylases have been intensively studied in their regulation of macroautophagy, their roles in other types of autophagy remain poorly understood. Deacetylation of mitochondrial proteins has been shown to initiate mitophagy [78,79], suggesting that protein acetylation is involved in the regulation of selective autophagy. However, protein acetylation-based signaling pathways in the regulation of selective autophagy, such as mitophagy, reticulophagy or xenophagy, remain largely unknown.
Several recent studies have shown that acetylation helps target the substrates for degradation through chaperone-mediated autophagy (CMA) [80–82]. Acetyltransferase KAT2B/PCAF (lysine acetyltransferase 2B) acetylates PKM2 (pyruvate kinase M1/2) in cells cultured at a high glucose concentration, which promotes the interaction between PKM2 and HSPA8/HSC70 (heat shock protein family A (Hsp70) member 8) [82], the chaperone for CMA. Thus, KAT2B-mediated acetylation can lead to the lysosomal degradation of PKM2 by CMA [82]. Whether the core machinery of CMA, such as HSPA8 or LAMP2A (lysosomal associated membrane protein 2A), is regulated by acetylation is unknown. Interestingly, EP300-CREBBP undergoes CMA-dependent degradation in cells treated with a chemotherapy drug that interferes with nucleic acid synthesis [83]. Thus, CMA-dependent degradation appears to engage extensively in acetylation-dependent cellular events in response to different cellular conditions.
Considering that many other types of PTMs are involved in autophagy regulation [84–87], it will be interesting to explore the potential crosstalk between acetylation and other types of PTMs during the autophagy process. Notably, dysregulation of autophagy has been intensively studied in a plethora of human diseases, including cancer and neurodegeneration [88–90]. Whether protein acetylation contributes to the dysregulation of autophagy in pathological conditions remains largely unknown. Given that protein acetylation integrates various intracellular and environmental cues to tightly control autophagy, it is highly plausible that acetylation of autophagy proteins might be dysregulated and contribute to pathologies of many human diseases.
Acknowledgments
We are grateful to Dr. Do-Hyung Kim (University of Minnesota, Minneapolis, US) for his constructive comments and careful editing work on the manuscript. We thank all members of Dr. Wan’s lab for helpful discussions.
Funding Statement
This work was supported by the National Natural Science Foundation of China (31970694, 31701213), the Young Elite Scientists Sponsorship Program by China Association for Science and Technology (CAST) (2019QNRC001), the Chao Kuang Piu High-tech Development Fund (2020QN024), and the Training Program for Excellent Young Innovators of Changsha (kq2106066).
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- [1].Narita T, Weinert BT, Choudhary C.. Functions and mechanisms of non-histone protein acetylation. Nat Rev Mol Cell Biol. 2019;20:156–174. [DOI] [PubMed] [Google Scholar]
- [2].Xia C, Tao Y, Li MS, et al. Protein acetylation and deacetylation: An important regulatory modification in gene transcription. Exp Ther Med. 2020;20:2923–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lee IH, Cao L, Mostoslavsky R, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105:3374–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Huang R, Xu Y, Wan W, et al. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol Cell. 2015;57:456–466. [DOI] [PubMed] [Google Scholar]
- [5].Lin SY, Li TY, Liu Q, et al. GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science. 2012;336:477–481. [DOI] [PubMed] [Google Scholar]
- [6].Shen Q, Shi Y, Liu J, et al. Acetylation of STX17 (syntaxin 17) controls autophagosome maturation. Autophagy. 2021;17:1157–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Su H, Yang F, Wang Q, et al. VPS34 acetylation controls its lipid kinase activity and the initiation of canonical and non-canonical autophagy. Mol Cell. 2017;67:907–921 e907. [DOI] [PubMed] [Google Scholar]
- [8].Yi C, Ma M, Ran L, et al. Function and molecular mechanism of acetylation in autophagy regulation. Science. 2012;336:474–477. [DOI] [PubMed] [Google Scholar]
- [9].You Z, Jiang WX, Qin LY, et al. Requirement for p62 acetylation in the aggregation of ubiquitylated proteins under nutrient stress. Nature Commun. 2019;10:5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bao J, Zheng L, Zhang Q, et al. Deacetylation of TFEB promotes fibrillar Abeta degradation by upregulating lysosomal biogenesis in microglia. Protein Cell. 2016;7:417–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wang Y, Huang Y, Liu J, et al. Acetyltransferase GCN5 regulates autophagy and lysosome biogenesis by targeting TFEB. EMBO Rep. 2020;21:e48335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhang J, Wang J, Zhou Z, et al. Importance of TFEB acetylation in control of its transcriptional activity and lysosomal function in response to histone deacetylase inhibitors. Autophagy. 2018;14:1043–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chan EYW, Kir S, Tooze SA. SiRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem. 2007;282:25464–25474. [DOI] [PubMed] [Google Scholar]
- [14].Matsuura A, Tsukada M, Wada Y, et al. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. [DOI] [PubMed] [Google Scholar]
- [15].Chang YY, Neufeld TP. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell. 2009;20:2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ganley IG, Lam DH, Wang JR, et al. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hosokawa N, Hara T, Kaizuka T, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hosokawa N, Sasaki T, Iemura S, et al. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 2009;5:973–979. [DOI] [PubMed] [Google Scholar]
- [19].Jung CH, Jun CB, Ro SH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kim J, Kundu M, Viollet B, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Russell RC, Tian Y, Yuan H, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Dooley HC, Razi M, Polson HE, et al. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cell. 2014;55:238–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sun T, Li X, Zhang P, et al. Acetylation of Beclin 1 inhibits autophagosome maturation and promotes tumour growth. Nat Commun. 2015;6:7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Itakura E, Kishi C, Inoue K, et al. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kihara A, Noda T, Ishihara N, et al. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sun QM, Fan WL, Chen KL, et al. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 2008;105:19211–19216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex–at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20:355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kumar S, Jain A, Farzam F, et al. Mechanism of Stx17 recruitment to autophagosomes via IRGM and mammalian Atg8 proteins. J Cell Biol. 2018;217:997–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. [DOI] [PubMed] [Google Scholar]
- [30].Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. [DOI] [PubMed] [Google Scholar]
- [31].Ichimura Y, Kirisako T, Takao T, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. [DOI] [PubMed] [Google Scholar]
- [32].Lee IH, Finkel T. Regulation of autophagy by the p300 acetyltransferase. J Biol Chem. 2009;284:6322–6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Xue SJ, Mao FX, Hu DB, et al. Acetylation of BmAtg8 inhibits starvation-induced autophagy initiation. Mol Cell Biochem. 2019;457:73–81. [DOI] [PubMed] [Google Scholar]
- [34].Zhang SL, Liang ML, Naqvi NI, et al. Phototrophy and starvation-based induction of autophagy upon removal of Gcn5-catalyzed acetylation of Atg7 in magnaporthe oryzae. Autophagy. 2017;13:1318–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Nowak J, Archange C, Tardivel-Lacombe J, et al. The TP53INP2 protein is required for autophagy in mammalian cells. Mol Biol Cell. 2009;20:870–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Xu Y, Wan W. The bifunctional role of TP53INP2 in transcription and autophagy. Autophagy. 2020;16:1341–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Xu Y, Wan W, Shou X, et al. TP53INP2/DOR, a mediator of cell autophagy, promotes rDNA transcription via facilitating the assembly of the POLR1/RNA polymerase I preinitiation complex at rDNA promoters. Autophagy. 2016;12:1118–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Song TT, Su HF, Yin W, et al. Acetylation modulates LC3 stability and cargo recognition. FEBS Lett. 2019;593:414–422. [DOI] [PubMed] [Google Scholar]
- [39].Lee IH, Kawai Y, Fergusson MM, et al. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science. 2012;336:225–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Maskey D, Yousefi S, Schmid I, et al. ATG5 is induced by DNA-damaging agents and promotes mitotic catastrophe independent of autophagy. Nature Commun. 2013;4:2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bjorkoy G, Lamark T, Brech A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Donaldson KM, Li W, Ching KA, et al. Ubiquitin-mediated sequestration of normal cellular proteins into polyglutamine aggregates. Proc Natl Acad Sci U S A. 2003;100:8892–8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Isogai S, Morimoto D, Arita K, et al. Crystal structure of the ubiquitin-associated (UBA) domain of p62 and its interaction with ubiquitin. J Biol Chem. 2011;286:31864–31874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Raasi S, Varadan R, Fushman D, et al. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;12:708–714. [DOI] [PubMed] [Google Scholar]
- [46].Lim J, Lachenmayer ML, Wu S, et al. Proteotoxic stress induces phosphorylation of p62/SQSTM1 by ULK1 to regulate selective autophagic clearance of protein aggregates. PLoS Genet. 2015;11:e1004987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Peng H, Yang J, Li G, et al. Ubiquitylation of p62/sequestosome1 activates its autophagy receptor function and controls selective autophagy upon ubiquitin stress. Cell Res. 2017;27:657–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kirkin V, Rogov VV. A diversity of selective autophagy receptors determines the specificity of the autophagy pathway. Mol Cell. 2019;76:268–285. [DOI] [PubMed] [Google Scholar]
- [49].Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269. [DOI] [PubMed] [Google Scholar]
- [50].Jiang P, Nishimura T, Sakamaki Y, et al. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol Biol Cell. 2014;25:1327–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wang Z, Miao G, Xue X, et al. The Vici syndrome protein EPG5 is a Rab7 effector that determines the fusion specificity of autophagosomes with late endosomes/lysosomes. Mol Cell. 2016;63:781–795. [DOI] [PubMed] [Google Scholar]
- [52].Cheng X, Ma X, Zhu Q, et al. Pacer is a mediator of mTORC1 and GSK3-TIP60 signaling in regulation of autophagosome maturation and lipid metabolism. Mol Cell. 2019;73:788–802 e787. [DOI] [PubMed] [Google Scholar]
- [53].Lee JY, Koga H, Kawaguchi Y, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010;29:969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Wang RX, Tan JQ, Chen TT, et al. ATP13A2 facilitates HDAC6 recruitment to lysosome to promote autophagosome-lysosome fusion. J Cell Biol. 2019;218:267–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Park JS, Blair NF, Sue CM. The role of ATP13A2 in Parkinson’s disease: Clinical phenotypes and molecular mechanisms. Mov Disord. 2015;30:770–779. [DOI] [PubMed] [Google Scholar]
- [56].Rochet JC. New insights into lysosomal dysfunction in Parkinson’s disease: an emerging role for ATP13A2. Mov Disord. 2012;27:1092. [DOI] [PubMed] [Google Scholar]
- [57].Seok S, Fu T, Choi SE, et al. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature. 2014;516:108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Settembre C, Di Malta C, Polito VA, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ferron M, Settembre C, Shimazu J, et al. A RANKL-PKCbeta-TFEB signaling cascade is necessary for lysosomal biogenesis in osteoclasts. Genes Dev. 2013;27:955–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Palmieri M, Pal R, Sardiello M. AKT modulates the autophagy-lysosome pathway via TFEB. Cell Cycle. 2017;16:1237–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Settembre C, Zoncu R, Medina DL, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Fullgrabe J, Lynch-Day MA, Heldring N, et al. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature. 2013;500:468–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Yu YS, Shin HR, Kim D, et al. Pontin arginine methylation by CARM1 is crucial for epigenetic regulation of autophagy. Nature Commun. 2020;11:6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Li XJ, Yu WL, Qian X, et al. Nucleus-translocated ACSS2 promotes gene transcription for lysosomal biogenesis and autophagy. Mol Cell. 2017;66:684–697 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Marino G, Pietrocola F, Eisenberg T, et al. Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol Cell. 2014;53:710–725. [DOI] [PubMed] [Google Scholar]
- [66].Chang CM, Su H, Zhang DH, et al. AMPK-dependent phosphorylation of GAPDH triggers Sirt1 activation and is necessary for autophagy upon glucose starvation. Mol Cell. 2015;60:930–940. [DOI] [PubMed] [Google Scholar]
- [67].Kim JE, Chen JJ, Lou ZK. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. [DOI] [PubMed] [Google Scholar]
- [68].Zhao WH, Kruse JP, Tang Y, et al. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Yang WB, Hong YH, Shen XQ, et al. Regulation of transcription by AMP-activated protein kinase: phosphorylation of p300 blocks its interaction with nuclear receptors. J Biol Chem. 2001;276:38341–38344. [DOI] [PubMed] [Google Scholar]
- [70].Zhang Y, Qiu J, Wang XM, et al. AMP-activated protein kinase suppresses endothelial cell inflammation through phosphorylation of transcriptional coactivator p300. Arterioscler Thromb Vasc Biol. 2011;31:2897–2908. 2011. [DOI] [PubMed] [Google Scholar]
- [71].Sebti S, Prebois C, Perez-Gracia E, et al. BAT3 modulates p300-dependent acetylation of p53 and autophagy-related protein 7 (ATG7) during autophagy. Proc Natl Acad Sci U S A. 2014;111:4115–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Wan W, You Z, Xu Y, et al. mTORC1 phosphorylates acetyltransferase p300 to regulate autophagy and lipogenesis. Mol Cell. 2017;68:323–335 e326. [DOI] [PubMed] [Google Scholar]
- [73].Delvecchio M, Gaucher J, Aguilar-Gurrieri C, et al. Structure of the p300 catalytic core and implications for chromatin targeting and HAT regulation. Nat Struct Mol Biol. 2013;20:1040–1046. [DOI] [PubMed] [Google Scholar]
- [74].Thompson PR, Wang DX, Wang L, et al. Regulation of the p300 HAT domain via a novel activation loop. Nat Struct Mol Biol. 2004;11:308–315. 2004. [DOI] [PubMed] [Google Scholar]
- [75].Marcu MG, Jung YJ, Lee S, et al. Curcumin is an inhibitor of p300 histone acetylatransferase. Med Chem. 2006;2:169–174. [DOI] [PubMed] [Google Scholar]
- [76].Pietrocola F, Lachkar S, Enot DP, et al. Spermidine induces autophagy by inhibiting the acetyltransferase EP300. Cell Death Differ. 2015;22:509–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Xu YF, Wu YS, Wang L, et al. Identification of curcumin as a novel natural inhibitor of rDNA transcription. Cell Cycle. 2020;19:3362–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Scott I, Webster BR, Chan CK, et al. GCN5-like protein 1 (GCN5L1) controls mitochondrial content through coordinated regulation of mitochondrial biogenesis and mitophagy. J Biol Chem. 2014;289:2864–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Webster BR, Scott I, Han K, et al. Restricted mitochondrial protein acetylation initiates mitochondrial autophagy. J Cell Sci. 2013;126:4843–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Bonhoure A, Vallentin A, Martin M, et al. Acetylation of translationally controlled tumor protein promotes its degradation through chaperone-mediated autophagy. Eur J Cell Biol. 2017;96:83–98. [DOI] [PubMed] [Google Scholar]
- [81].Huang HC, Liu RJ, Huang YH, et al. Acetylation-mediated degradation of HSD17B4 regulates the progression of prostate cancer. Aging (Albany NY). 2020;12:14699–14717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Lv L, Li D, Zhao D, et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell. 2011;42:719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Du CZ, Huang DD, Peng YF, et al. 5-Fluorouracil targets histone acetyltransferases p300/CBP in the treatment of colorectal cancer. Cancer Lett. 2017;400:183–193. [DOI] [PubMed] [Google Scholar]
- [84].Kim YM, Jung CH, Seo M, et al. mTORC1 phosphorylates UVRAG to negatively regulate autophagosome and endosome maturation. Vol. 57. Mol Cell; 2015. p. 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Kwon YT, Ciechanover A. The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem Sci. 2017;42:873–886. [DOI] [PubMed] [Google Scholar]
- [86].Wan W, Liu W. MTORC1 regulates autophagic membrane growth by targeting WIPI2. Autophagy. 2019;15:742–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Wan W, You Z, Zhou L, et al. mTORC1-regulated and HUWE1-mediated WIPI2 degradation controls autophagy flux. Mol Cell. 2018;72:303–315 e306. [DOI] [PubMed] [Google Scholar]
- [88].Collier JJ, Guissart C, Olahova M, et al. Developmental consequences of defective ATG7-mediated autophagy in humans. New Engl J Med. 2021;384:2406–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Mizushima N, Levine B. Autophagy in human diseases. New Engl J Med. 2020;383:1564–1576. [DOI] [PubMed] [Google Scholar]
- [90].Yang Y, Klionsky DJ. Autophagy and disease: unanswered questions. Cell Death Differ. 2020;27:858–871. [DOI] [PMC free article] [PubMed] [Google Scholar]


