Abstract
Background and Aims
Invasive plants often colonize wide-ranging geographical areas with various local microenvironments. The specific roles of epigenetic and genetic variation during such expansion are still unclear. Chenopodium ambrosioides is a well-known invasive alien species in China that can thrive in metalliferous habitats. This study aims to comprehensively understand the effects of genetic and epigenetic variation on the successful invasion of C. ambrosioides.
Methods
We sampled 367 individuals from 21 heavy metal-contaminated and uncontaminated sites with a wide geographical distribution in regions of China. We obtained environmental factors of these sampling sites, including 13 meteorological factors and the contents of four heavy metals in soils. Microsatellite markers were used to investigate the demographic history of C. ambrosioides populations in China. We also analysed the effect of epigenetic variation on metalliferous microhabitat adaptation using methylation-sensitive amplified polymorphism (MSAP) markers. A common garden experiment was conducted to compare heritable phenotypic variations among populations.
Key Results
Two distinct genetic clusters that diverged thousands of years ago were identified, suggesting that the eastern and south-western C. ambrosioides populations in China may have originated from independent introduction events without recombination. Genetic variation was shown to be a dominant determinant of phenotypic differentiation relative to epigenetic variation, and further affected the geographical distribution pattern of invasive C. ambrosioides. The global DNA unmethylation level was reduced in metalliferous habitats. Dozens of methylated loci were significantly associated with the heavy metal accumulation trait of C. ambrosioides and may contribute to coping with metalliferous microenvironments.
Conclusions
Our study of C. ambrosioides highlighted the dominant roles of genetic variation in large geographical range expansion and epigenetic variation in local metalliferous habitat adaptation.
Keywords: Biological invasion, heavy metals, DNA methylation, epigenetics, Chenopodium ambrosioides, phenotypic variation
INTRODUCTION
Biological invasion has become a severe problem worldwide. Exploring the evolutionary processes of alien species during establishment and range expansion is critical for understanding successful invasion (Banerjee et al., 2019). Genetic diversity is commonly thought to play a role in successful invasion by facilitating the efficacy of selection (Crawford and Whitney, 2010; van Kleunen et al., 2015; Vilatersana et al., 2016; van Boheemen et al., 2017). However, demographic bottlenecks often exist in invasive populations due to loss of a number of individuals, and lead to a loss of genetic variation (Estoup et al., 2016). Some genetically depleted populations may still produce phenotypic variation in response to heterogeneous environments and help species to establish in a new environment (Bossdorf et al., 2008; Hagenblad et al., 2015; Spens and Douhovnikoff, 2016; Banerjee et al., 2019). The existence of these invasive plants with limited genetic diversity indicates that we need to have a deeper understanding of the environmental adaptation mechanism of invasive species (Richards et al., 2012; Chen et al., 2015).
Epigenetic modifications have been reported to regulate gene expression and phenotypic variations (Niederhuth and Schmitz, 2014; Wibowo et al., 2016; Chen et al., 2022). Increasing numbers of studies point to the importance of epigenetic variation for the successful invasion of plants with low genetic diversity (Chwedorzewska and Bednarek, 2012; Richards et al., 2012). When casual/introduced aliens or invasive aliens are exposed to environmental stresses, epigenetic modifications can be induced, which lead to phenotypic plasticity in plant traits independent of genetic variation (Banerjee et al., 2019). The studies on epigenetic variation conducted to date have mainly focused on invasive species with limited genetic diversity. However, some invasive species, especially those that have undergone multiple invasions, possess relatively high genetic diversity (Meimberg et al., 2010). A gap remains in the comprehensive understanding of the respective effects of genetic and epigenetic variation on the invasion success of species undergoing multiple invasions or introductions. In the process of expansion, invasive plants often colonize a wide range of geographical areas with different environmental characteristics. Both soil and climate factors may affect the phenotypic variation of plants (Chen et al., 2020, 2022). Additionally, invasive plants are frequently exposed to various biotic and abiotic stresses that they encounter in local microenvironments. However, the specific roles played by epigenetic and genetic variation in large geographical area expansion and local habitat adaptation are largely unknown.
Interestingly, many invasive plants, such as Chenopodium ambrosioides L. (Zhang et al., 2012) and Phytolacca americana L. (Tie et al., 2005; Chen et al., 2015), have been found to thrive in metalliferous habitats. Recently, an increasing number of studies have highlighted the role of epigenetic modifications in response to heavy metal stress (Kumar et al., 2012; Ueda and Seki, 2020; Jing et al., 2022). Various genes which have been reported to be involved in metal exclusion or detoxification are potentially regulated by a reversible epigenetic mechanism (Chaudhary et al., 2016; Gallo-Franco et al., 2020). Phenotypic plasticity induced by epigenetic variation may promote the rapid adaptation of invasive plants to a range of heavy metal-contaminated environments. However, the heavy metal adaptation mechanisms of invasive plants during expansion into new environments have not been completely elucidated.
Chenopodium ambrosioides is a tetraploid perennial herb native to tropical America (Kolano et al., 2012). At the beginning of the 18th century, C. ambrosioides was introduced randomly or deliberately to the rest of the world and widely adopted as an anthelmintic remedy (Morton, 1981; Kliks, 1985). In China, C. ambrosioides is well known as a common invasive alien species and distributed in most regions of southern China (Ma, 2013). Previous studies have reported that C. ambrosioides is a manganese/lead hyperaccumulator that can flourish in heavy metal-contaminated habitats and accumulate large amounts of heavy metals in its aerial tissues (Zhang et al., 2012; Rivera-Becerril et al., 2013). In this study, we assessed the epigenetic and genetic patterns of invasive C. ambrosioides populations distributed in a large geographical region of China with different levels of heavy metal contamination. By integrating genetic, epigenetic and phenotypic data, we investigated (1) the demographic history of invasive C. ambrosioides populations in China; (2) the role of genetic and epigenetic variation in geographical distribution pattern; and (3) the effect of epigenetic variation on metalliferous microhabitat adaptation. Our study revealed the specific roles of genetic variation at large geographic scale and epigenetic variation at local metalliferous habitat scale.
MATERIALS AND METHODS
Plant materials
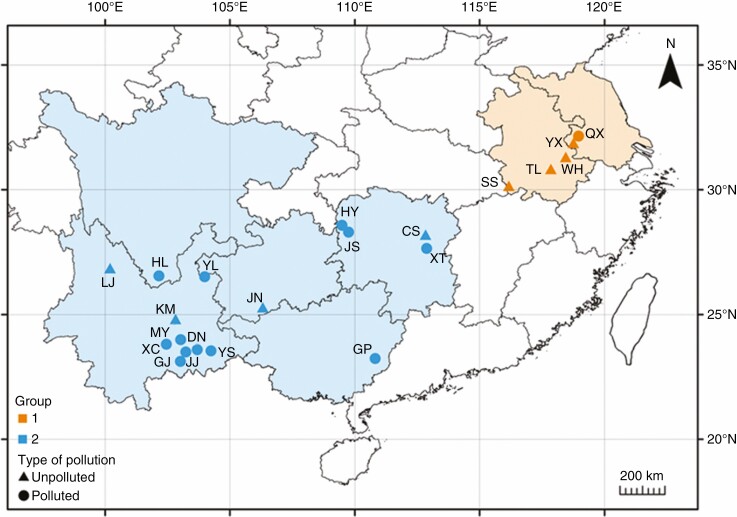
In August 2015, we obtained 21 C. ambrosioides populations from heavy metal-contaminated and uncontaminated sites in eastern and south-western China (Fig. 1; Supplementary data Table S1). At each sampling sites, about 13–18 current-year plants at least 15 m apart from each other were selected as a population, and their leaves, seeds and soil samples were collected. The longitude and latitude information of the sampling sites was recorded. Typical mature leaves at the middle of the stem were collected. The leaf tissues were carefully washed in tap water and rinsed in deionized water. A portion of the leaf tissues was ground in liquid nitrogen, and genomic DNA was extracted using DNA Plantzol Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. The remaining leaf tissues were dried at 80 °C to a constant weight for heavy metal analysis. Soil samples were collected within 5 cm of the rhizosphere of C. ambrosioides in the 0–20 cm depth range below the surface. The soil samples were air-dried at room temperature and further dried at 105 °C for 6 h before heavy metal analysis. Seeds were collected for common garden experiments. It should be noted that we failed to collect enough seeds from the SS site (Susong County, Anhui Province), thus this population was not included in subsequent common garden experiments.
Fig. 1.
Sampling locations of Chenopodium ambrosioides populations. Orange and blue represent groups 1 and 2, respectively. The circles and triangles represent populations collected from heavy metal-polluted and unpolluted areas, respectively. The detailed information of locality, habitat, longitude and latitude of each population is shown in Supplementary data Table S1.
Molecular data collection
Potential simple sequence repeats (SSRs) were searched in the genome sequences of C. ambrosioides (SRA accession: SRR8132837) using the MISA tool (http://pgrc.ipk-gatersleben.de/misa/misa.html), and SSR primer pairs were designed to generate PCR products ranging in size from 100 to 300 bp using Primer3 (https://probes.pw.usda.gov/cgi-bin/batchprimer3/batchprimer3.cgi). After evaluating the polymorphism potential of the designed SSR markers using 12 C. ambrosioides individuals, 12 polymorphic SSR loci and 367 individuals collected from 21 populations were chosen to assess the genetic diversity levels and population structure of C. ambrosioides (Supplementary data Table S7). The 5ʹ end of the forward primer of each pair was labelled with a fluorescent dye (6-FAM, HEX or TAMRA). Fragments were separated on an ABI 3730xl DNA Analyzer (Applied Biosystems, Forster City, CA, USA) after PCR amplification, and alleles were manually scored using GeneMaker Software version (ver 2.2.0; SoftGenetics, State College, PA, USA) with GeneScan 500 LIZ as an internal size standard.
We screened 367 individuals from 21 populations (using the same samples employed in SSR analysis) via the methylation-sensitive AFLP method (MSAP, or MS-AFLP, Reyna-López et al., 1997; Chen et al., 2020) to analyse the epigenetic variation of C. ambrosioides. Genomic DNA was double digested with EcoRI/MspI or EcoRI/HpaII. MspI and HpaII are isoschizomers with different sensitivities to cytosine methylation at the same tetranucleotide restriction site (5ʹ-CCGG). Cleavage by HpaII is blocked when the inner or outer cytosine is methylated on both strands, whereas cleavage by MspI is blocked when the outer cytosine is hemimethylated or fully methylated (Reyna-López et al., 1997). After pre-amplification, nine fluorescently labelled primer pairs were used for selective amplification (Supplementary data Table S9). Subsequently, fragments were separated on an ABI 3730xl DNA analyser, and fragments with lengths of 69–450 bp were identified using GENEMARKER ver. 2.2.0 (SoftGenetics). We further transformed the epigenetic fragments into a binary character matrix, with ‘0’ indicating absence and ‘1’ indicating presence. Four types of methylation were defined according to whether a fragment was present in MspI and HpaII: type 1, no methylation, present in both MspI and HpaII (1, 1); type 2, hemimethylation of internal or external cytosines, present in HpaII but absent in MspI (1, 0); type 3, full methylation of internal cytosines, present in MspI but absent in HpaII (0, 1); and type 4, either full methylation of cytosines or sequence polymorphism, absent in both MspI and HpaII (0, 0) (Schulz et al., 2013). Types 2 and 3 (type 2 + 3) were treated as total methylation. Then, an MSAP profile was constructed to compute epigenetic parameters according to two methylation states: unmethylated (type 1 coded as 0) and methylated (type 2 and type 3 coded as 1), and type 4 was treated as missing data (Foust et al., 2016; Chen et al., 2020).
Heavy metal analysis
Dried plant samples collected in fields were ground and digested with concentrated HNO3 and HClO4 (87:13, v/v). Dried soil samples were digested with a mixture of concentrated HNO3:HClO4 (4:1, v/v). The total concentrations of four types of heavy metals (Mn, Zn, Cd and Pb) in the solutions were determined by inductively coupled plasma atomic emission spectrometry (ICP-AES; Optima 3300 DV, Perkin-Elmer, Waltham, MA, USA) (Supplementary data Tables S3 and S4). Unpolluted habitats were defined as soils with Cd concentrations <1 mg kg−1 and Zn and Pb concentrations <500 mg kg−1 based on environmental quality standards for soils in China (SEPAC, 1995). Because no equivalent Chinese standard exists for Mn, the maximum soil Mn level for an unpolluted habitat was defined as 1800 mg kg−1 according to soil quality standards for habitat and agriculture in Thailand (PCD, 1995). In addition, the Nemerow integrated pollution index (NIPI) of toxic elements in soil was determined to quantify the ecological pollution levels of 21 sites (Supplementary data Table S5). We calculated NIPI using the following formula: Pi = {[(Piave)2 + (Pimax)2]/2}1/2, where Pi is the NIPI, Piave is the average pollution index of element i, and Pimax is the highest pollution index of element i (Ma et al., 2016).
Meteorological data
We collected meteorological data of counties covering all 21 sampling sites for the whole of 2015 from the National Climate Centre of China (http://ncc-cma.net/cn/). The obtained parameters were as follows: altitude (ALT, m), 20–20 time precipitation (PRE_Time, mm), average wind speed in 2 min (WS_2 min, m s–1), average annual wind speed (AAWS, m s–1), maximum wind speed (MWS, m s–1), annual gale days (GD_Ann, d), sunshine hours (SSH, h), average air pressure (AP_Avg, hpa), average temperature (TEM_Avg, °C), average ground surface temperature (GST_Avg, °C), accumulated temperature (ATEM, d·°C), average relative humidity (RH_Avg, %) and minimum average humidity (HRMin_Avg, %) (Supplementary data Table S6).
Phenotypic analysis
The common garden experiment was conducted to compare heritable phenotypic variations among populations. The seeds were collected from 4–5 individuals of 20 populations of C. ambrosioides (except for SS). The common garden experiment was conducted in a greenhouse operated by Nanjing Agricultural University [average temperature 30/25 °C (day/night); relative humidity 60–80 %; photoperiod 14/10 h (day/night)]. The seeds were germinated in a mixture of perlite and vermiculite on plastic plates. After germination, the seedlings were transplanted to plastic buckets containing 1/8 strength Hoagland nutrient solution and cultivated for 14 d. Thereafter, the seedlings were cultivated in full-strength Hoagland nutrient solution. We renewed the Hoagland nutrient solution every other day. After 30 d, the seedlings were harvested. We measured root and shoot lengths (RL and SL) and root and shoot fresh weights (RFW and SFW), and counted blade numbers (BN) and lateral branch numbers (LBN). Leaf area (LA) was measured with an LI-3000C leaf area meter (LI-COR, Lincoln, NE, USA). The relative chlorophyll content (Chl) was determined by measuring the transmittance of leaves in two wavelength regions (650 and 940 nm) with a SPAD-502 meter (Konica Minolta, Osaka, Japan). The net photosynthetic rate (Pn), transpiration rate (Tr), stomatal conductance (Gs), and intercellular carbon dioxide (Ci) were measured with an LI-6400 portable photosynthesis system (LI-COR) (Schories and Mehlig, 2000). The light intensity was 1000 μmol m–2 s–1, and the area of the leaf chamber was 3 × 2 cm2 (Supplementary data Table S2). Principal co-ordinate analysis (PCoA) based on the phenotypic data was performed using the ‘vegan’ function in R. PCoA can visually display the similarity or difference of plant samples based on phenotypic traits. In this study, a Euclidean distance matrix was constructed based on the value of all 12 phenotypic traits, and a series of eigenvalues were sorted. The most important eigenvalues in the top two (PCo1 and PCo2) which interpreted 69.42 % total variability were selected and displayed in the co-ordinate system.
Genetic variation analysis
The number of alleles (NA), observed heterozygosity (HO), expected heterozygosity (HE) and polymorphism information content (PIC) were calculated for each SSR locus using CERVUS (ver. 3.0.7, Kalinowski et al., 2007) (Supplementary data Table S8). Deviations from Hardy–Weinberg equilibrium were analysed using GENEPOP (ver. 4.7.5, Rousset, 2008). Null allele frequencies were estimated using the software FreeNa (Chapuis and Estoup, 2007) with the number of replicates fixed to 25 000. Chenopodium ambrosioides is a tetraploid species (Kolano et al., 2012). The null allele is often suggested to be included as an estimator of allele frequencies in polyploid organisms (De Silva et al., 2005; Dufresne et al., 2014). In this study, we removed the locus of CHAM96, whose null allele frequency was particularly high (0.246), from our subsequent analysis (Supplementary data Table S8). For each population of C. ambrosioides, Shannon’s diversity index (Hʹ) was calculated with GenAlEx software (ver. 6.5).
We performed Bayesian clustering to characterize population structure using STRUCTURE software (ver. 2.3.4). The admixture model with independent allele frequencies was used; ten replications were performed for each K (range: K = 1–21) and the optimal K was estimated based on the parameter ΔK (Evanno, 2005). We also inferred population structure including the spatial information (longitude and latitude) by GENELAND (ver. 4.9.2) (Guillot et al., 2005). The spatial and correlated allele frequencies model was used; ten replications were simulated and the optimal number of population was chosen by the simulation with the highest average posterior probability. Subsequently, we estimated genetic structure via PCoA based on Nei’s distance matrix using GenAlEx software. Neighbor–Joining (NJ) analysis (bootstraps = 1000) was conducted in PHYLIP software (ver. 3.69; Felsenstein, 1993) based on Nei’s distance calculated by PowerMarker (ver. 3.25) (Liu and Muse, 2005) to reveal the genetic relationships among populations. Then, analysis of molecular variance (AMOVA) was performed to assess the overall genetic differences among C. ambrosioides populations (ARLEQUIN software ver. 3.5). We also conducted a hierarchical AMOVA of C. ambrosioides based on the optimum genetic cluster of populations in STRUCTURE.
Contemporary migration rate (past few generations; <5 generations) was estimated using a Bayesian approach in BAYESASS (ver. 3.04; Wilson and Rannala, 2003). The BAYESASS program estimates contemporary migration rates (mc, fraction of immigrant individuals) according to the proportion of individuals in each population sample that are assigned to other populations with high probability. This method relaxes some equilibrium assumptions, which allows deviation from Hardy–Weinberg equilibrium but assumes linkage disequilibrium. Each Markov chain Monte Carlo (MCMC) run was performed with a burn-in of the first 1 000 000 steps of 10 000 000 total iterations. Model convergence was assessed via the comparison of the posterior probability densities of inbreeding coefficients and allele frequencies across 100 replicate runs, each with a different initial seed. We adjusted the mixing parameters to ensure that the posterior acceptance rates for each parameter were between 20 % and 60 % according to Wilson and Rannala (2003). The mean value and 95 % confidence intervals of mc are reported in the Results. Historical gene flow (much longer period of time, approx. 4Ne generations in the past) was estimated with Migrate-n (ver. 3.7.2, Beerli and Felsenstein, 2001). The Migrate-n program estimates the parameters θ (θ = 4 Neμ, where μ is the mutation rate per generation and is estimated for nSSRs as 10–3; Udupa and Baum, 2001) and M (mutation-scaled rates of migration, M = mh/μ, where mh is the historical migration rate per generation). The Migrate-n was conducted with the Brownian motion microsatellite model to estimate the parameters using either a Bayesian inference or maximum likelihood framework. We ran Migrate-n with a full migration matrix model between 21 populations of C. ambrosioides. An exponential prior distribution was set for both M (0–1000, mean = 500, delta = 100) and θ (0.0–0.10, mean = 0.05, delta = 0.01) with the historical migration model. The prior distribution of the parameters was set for both M (0–1000, mean = 500, delta = 100) and θ (0.0–0.10, mean = 0.05, delta = 0.01). Following a burn-in of 10 000 iterations, each run visited a total of 500 000 parameter values and recorded 5000 genealogies at a sampling increment of 100. We used a static heating scheme at four temperatures (1, 1.5, 3 and 6 °C) to efficiently search the genealogy space. We present the mean and 95 % confidence intervals for mh in the Results. We further estimated the gene flow value (Nm) as follows: Nm = θ × M/4, and θ and M were calculated with Migrate-n (Beerli and Felsenstein, 1999).
We used Wilcoxon’s signed rank test vs. the mode-shift test implemented in BOTTLENECK 1.2.1 (Piry et al., 1999) to detect population declines over historical vs. contemporary time scales (approx. 4Ne generations in the past vs. the past few generations; Luikart and Cornuet, 1998). We performed 10 000 simulations under the SMM (strict stepwise mutation model) and TPM (the two-phase model with 95 % single-step mutations and 5 % multiple-step mutations), with a variance of 12 as recommended by Piry et al. (1999). Significant P-values were taken as evidence of bottlenecks.
We assessed the demographic history of divergence between populations by approximate Bayesian computation (ABC) implemented in DIYABC (ver. 2.1; Cornuet et al., 2008, 2014). We conceptualized two ABC models, which were used to estimate the divergence time (ABC1) and changes in the effective population size (ABC2) separately. In the ABC1 model, our analysis divided the 21 populations into two groups based on genetic clustering analysis conducted in STRUCTURE, and constructed a total of three scenarios to estimate the divergence time between the two population groups. Scenario 1 considered group 1 as the first invasive population, with colonization via eastern China, followed by the divergence of group 2 from group 1 at time t; scenario 2 considered group 2 as the first invasive population, with colonization via south-eastern China, followed by the divergence of group 1 from group 2; and scenario 3 considered both groups to have split simultaneously from one ancestral population at time t. The training sets included a total of 1 500 000 simulated datasets. In the ABC2 model, three simple population demographic scenarios were tested to examine the changes in effective population size for different groups based on STRUCTURE clustering (group 1, group 2 and total). Scenario 1 was a bottleneck model (Na > N; the effective population size changed from Na to N at time t); scenario 2 was an expansion model (Na < N; the effective population size changed from Na to N at time t); and scenario 3 was conceptualized as a constant model (Na = N; the effective population size was constant at N from the present to the past). The parameters t, N1, N2 and Na were set to 30 000, 20 000, 20 000 and 80 000, respectively. Default values were used for the remaining parameters. To test the best scenario for ABC1, we applied both logistic regression and a direct method to the 1 % of the simulated datasets closest to the observed datasets to estimate the relative posterior probability of the scenarios. For ABC2, we predicted the best scenario by estimating its posterior probability through ABC.
Epigenetic variation analysis
The frequency of each methylation type was calculated using the msap package in R (Schulz et al., 2013). Hʹ for epigenetic data was evaluated with GenAlEx software (ver. 6.5; Peakall and Smouse, 2006). NJ analysis (bootstraps = 1000) was performed based on epigenetic Nei’s distance using PHYLIP software (ver. 3.69; Felsenstein, 1993). Nei’s distance for epigenetic data was calculated with AFLPsurv software (ver. 1.0; Vekemans, 2002). We further estimated epigenetic structure based on the PCoA performed in GenAlEx software. Subsequently, both AMOVA and hierarchical AMOVA were used to assess the epigenetic differences among C. ambrosioides populations (ARLEQUIN ver. 3.5; Excoffier and Lischer, 2010). The grouping in hierarchical AMOVA was based on the best genetic cluster of populations in STRUCTURE.
The pairwise epigenetic and genetic differentiation coefficient (FST) was calculated with GenAlEx software. Then, we detected the correlations between the FST values of genetic and epigenetic differentiation using Mantel test in GenAlEx. A simple linear regression model was used to explore patterns of spatial autocorrelation with genetic or epigenetic variation by calculating the correlations between geographic distance (log-transformed) and genetic or epigenetic differentiation (FST/1 – FST) (Diniz-Filho et al., 2013). These analyses were performed separately on the populations belonging to groups 1 and 2 (based on cluster results obtained in STRUCTURE) and among all populations. The simple linear regression of pairwise comparisons was performed with the ‘lm’ function in R.
We used distance-based redundancy analysis (dbRDA) to further evaluate the correlation between epigenetic and genetic variation. The dbRDA was performed with the ‘capscale’ function of the vegan package in R (Oksanen et al., 2019) using the following formula: capscale (x~y), where x = the Euclidean distance matrix for MSAP as the dependent variable, and y = the data of 11 microsatellite fragments, which were normalized with min–max standardization as predictors of epigenetic variation. Additionally, we used dbRDA to explore the effect of environmental factors on phenotypic, genetic and epigenetic variation. Two types of environmental factors were considered, namely 13 meteorological factors and the contents of four metals in soils, which were analysed separately. The relationships between phenotypic variation and genetic or epigenetic variation were also analysed using dbRDA. The phenotypic factors used in dbRDA were determined in common garden experiments and included RL, SL, RFW, SFW, BN, LBN, LA, Chl, Pn, Tr, Gs and Ci.
To investigate the association between leaf heavy metal contents and methylated loci (unmethylated loci and total methylated loci), we performed epigenome-wide association analysis (EWAS) using a mixed linear model (MLM) in TASSEL (ver. 5.2.52) (Bradbury et al., 2007). Both the kinship matrix and P-matrix were generated in TASSEL to fit the MLM using unmethylated loci or total methylated loci to account for multiple levels of relatedness (Zhao et al., 2007). Here, we used a P-matrix based on principal component analysis (PCA) rather than a Q-matrix based on the STRUCTURE algorithm to summarize genome-wide patterns of relatedness because the STRUCTURE algorithm was not suitable for estimating population epigenetic structure. The heavy metal content was determined as the sum of the contents of four heavy metals (Cd, Mn, Pb and Zn) after normalization with min–max standardization. The significance of locus–trait associations was defined according to P < 0.05 or 0.01. Manhattan plots were generated using the qqman package in R (Turner, 2018).
Statistical analysis
Data were statistically analysed using the software SPSS 22.0. The Student’s t-test was used to compare the datasets (i.e. phenotypic traits, Shannon’s diversity and methylation frequency) between two groups, and significant differences among means were determined at P < 0.05.
RESULTS
Phenotypic variations of invasive C. ambrosioides populations
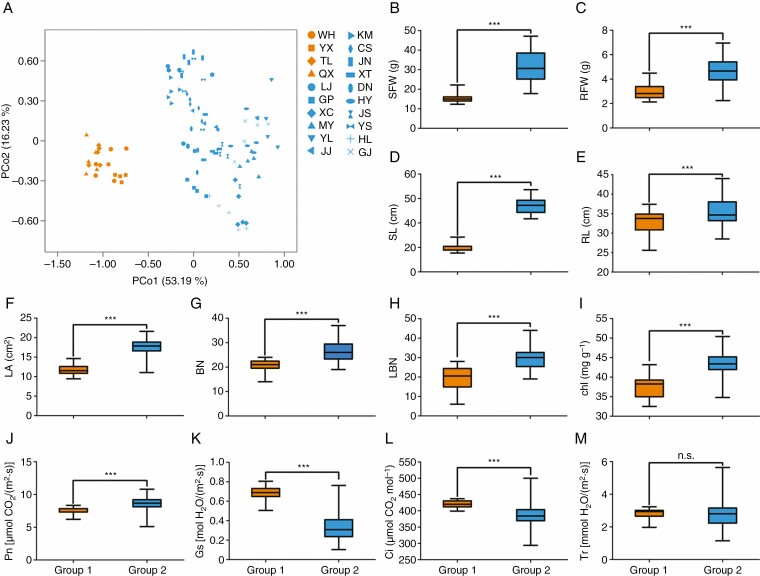
We compared heritable phenotypic variations among invasive C. ambrosioides populations by a common garden experiment. The PCoA showed that the first two principal co-ordinates (PCo1 and PCo2) explained 69.42 % of the overall phenotypic variation, which could divided the invasive C. ambrosioides population in China into two distinct groups (Fig. 2A). Group 1 included four populations mainly distributed in eastern China, whereas group 2 included the remaining 16 populations distributed in south-western China (Figs 1 and 2A). The values of fresh weights of shoot and root (SFW and RFW), lengths of shoot and root (SL and RL), leaf area (LA) and blade and lateral branch numbers (BN and LBN) were significantly higher in group 2 than in group 1(P < 0.001, Fig. 2B–H). Both the relative chlorophyll content (Chl) and the net photosynthetic rate (Pn) were significantly higher in group2 (P < 0.001), and the values of stomatal conductance (Gs) and intercellular carbon dioxide (Ci) were significantly higher in group 1 (P < 0.001, Fig. 2I–M; Supplementary data Table S2).
Fig. 2.
Phenotypic index of C. ambrosioides in the common garden. (A) Principal co-ordinate analysis (PCoA) of C. ambrosioides based on phenotypic data. PCo1 and PCo2 represent the first two principal co-ordinates of the dataset, and each principal co-ordinate explains 53.19 % and 16.23 %, respectively, of total phenotypic variation. Comparison of C. ambrosioides based on SFW (B), RFW (C), SL (D), RL (E), LA (F), BN (G), LBN (H), Chl (I), Pn (J), Gs (K), Ci (L) and Tr (M). Significant differences among means were determined using the Student’s t-test. P stands for significance test of correlation which values are indicted as: n.s., P > 0.05; ***P < 0.001.
Genetic and epigenetic structure and diversity of invasive C. ambrosioides populations
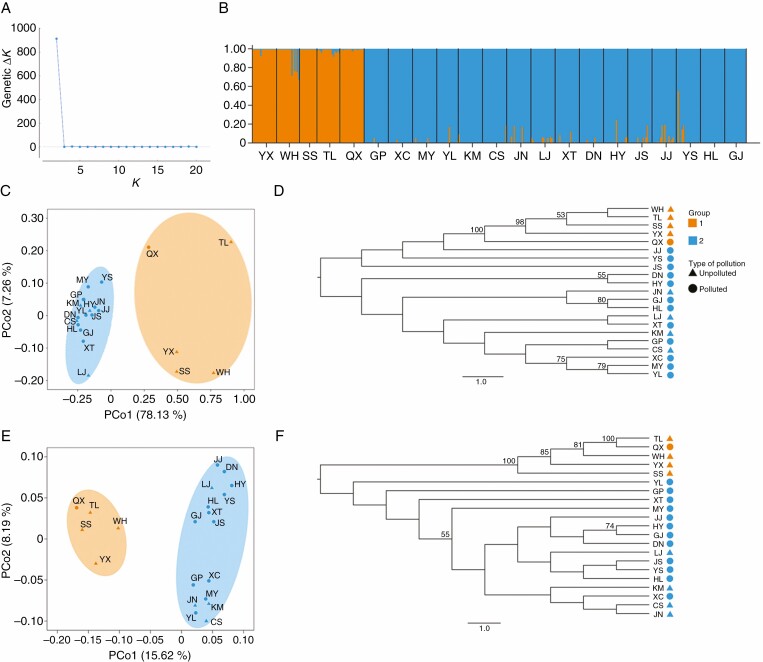
We further analysed the genetic and epigenetic structures of invasive C. ambrosioides. In the STRUCTURE analysis, the 21 populations of C. ambrosioides could be optimally clustered into two distinct genetic groups based on the ΔK criterion (Fig. 3A). The grouping of populations based on genetic variation was consistent with that of phenotypic variation. Group 1 (orange) included five populations distributed in eastern China (including SS), whereas group 2 (blue) included the remaining 16 populations distributed in south-western China (Figs 1 and 3B). When we inferred population structure including the spatial information using GENELAND analysis, the populations were grouped under four optimum clusters. Sixteen populations distributed in south-western China belong to a cluster which was the same as group 2 in the STRUCTURE analysis, whereas the five populations in eastern China were divided into three clusters (Supplementary data Fig. S1). The PCoA showed that the first two principal co-ordinates accounted for 85.39 % of the genetic variation in the C. ambrosioides populations, which indicated a deep genetic division between the eastern (group 1) and south-western (group 2) populations (Fig. 3C). In the NJ tree based on genetic data, the eastern and south-western populations were grouped into two distinct clades (Fig. 3D). Similarly, PCoA and NJ analysis based on epigenetic data grouped the eastern and south-western populations into two clusters (group 1 and group 2, Fig. 3E, F). It is worth noting that the first two principal co-ordinates in the epigenetic PCoA accounted for only 23.81 % of the epigenetic variation observed in the C. ambrosioides populations, which was much lower than that in the genetic PCoA (85.39 %). The results indicated that the genetic division between the eastern and south-western populations was much deeper than the epigenetic division. Accordingly, the AMOVA results showed that more genetic variation (44.31 %) than epigenetic variation (16.67 %) was explained by the differences among populations (Table 1). Hierarchical AMOVA showed that 50.14 % of the genetic variation was distributed between the eastern and south-western groups (group 1 and group 2), whereas only 12.34 % of the epigenetic variation was distributed between groups (Table 1). Unlike the genetic variation, most of the epigenetic variation (76.83 %) could be attributed to the variation within populations.
Fig. 3,
Population structure of C. ambrosioides-based genetic and epigenetic data. (A) ΔK plot in STRUCTURE analysis. (B) Individual assignment to each STRUCTURE cluster for K = 2 based on genetic data. Each individual is represented by a thin vertical line. (C–F) PCoA and NJ analysis of C. ambrosioides based on the SSR profile (C, D) and MSAP profile (E, F). PCo1 and PCo2 represent the first two principal co-ordinates of the dataset, which explain 85.39 % of total genetic variation and 23.81 % of total epigenetic variation.
Table 1.
(A) Genetic and epigenetic AMOVA for 21 populations of C. ambrosioides analysed. (B) Three-level hierarchical analysis of epigenetic and genetic molecular variation (AMOVA) for C. ambrosioides, and the groups divided based on the best genetic cluster of populations in STRUCTURE
| Source of variation | d.f. | Variance components | Percentage of variation | F-statistics |
|---|---|---|---|---|
| (A) AMOVA | ||||
| Genetic variation | ||||
| Among populations | 20 | 1.43 | 44.31 | F ST = 0.443 |
| Within populations | 713 | 1.80 | 55.69 | |
| Epigenetic variation | ||||
| Among populations | 20 | 47.72 | 16.67 | F ST = 0.167 |
| Within populations | 346 | 238.52 | 83.33 | |
| (B) Hierarchical AMOVA | ||||
| Genetic variation | ||||
| Between group 1 and group 2 | 1 | 2.37 | 50.14 | F CT = 0.501 |
| Among populations within group | 19 | 0.56 | 11.83 | F SC = 0.237 |
| Within populations | 713 | 1.80 | 38.04 | F ST = 0.620 |
| Epigenetic variation | ||||
| Between group 1 and group 2 | 1 | 38.31 | 12.34 | F CT = 0.123 |
| Among populations within group | 19 | 33.63 | 10.83 | F SC = 0.124 |
| Within populations | 346 | 238.52 | 76.83 | F ST = 0.232 |
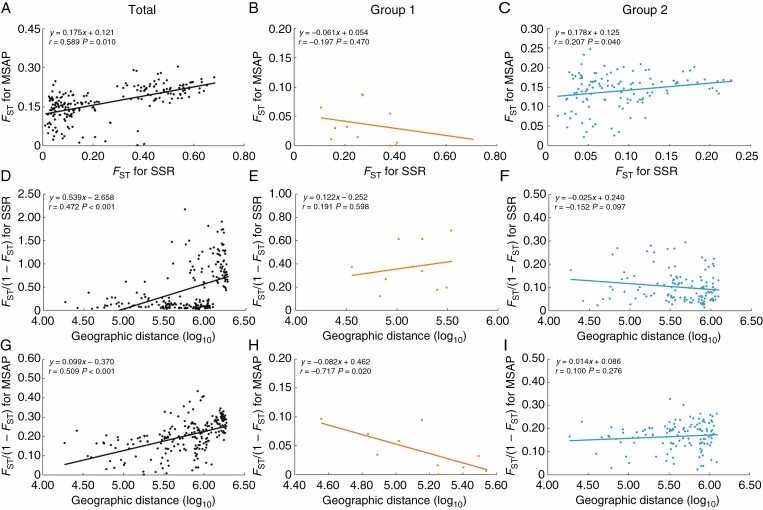
We found that the genetic FST values were significantly related to the epigenetic FST values among all 21 populations of C. ambrosioides (total: r = 0.589, P = 0.010) or within south-western populations (group 2: r = 0.207, P = 0.040), but no such relationship was detected within the eastern populations (group 1, Fig. 4A–C). We further analysed the correlations between geographic differentiation and genetic/epigenetic differentiation. Positive spatial autocorrelation patterns were found for both the genetic (r = 0.472, P < 0.001) and epigenetic (r = 0.509, P < 0.001) variation among the 21 populations (Fig. 4D, G). However, we did not detect such a linear positive correlation for either genetic or epigenetic variation when the populations within group 1 or 2 were analysed separately (Fig. 4E, F, H, I).
Fig. 4.
Correlations between genetic and epigenetic variations (measured by FST) among C. ambrosioides populations in total (A), group 1 (B) and group 2 (C). Spatial autocorrelation pattern detected for genetic (D–F) and epigenetic (G–I) variations (measured by FST/1 – FST) among populations in total (D, G), group 1 (E, H) and group 2 (F, I). Geographic distance is log-transformed. r = correlation coefficient, P = significance of correlation.
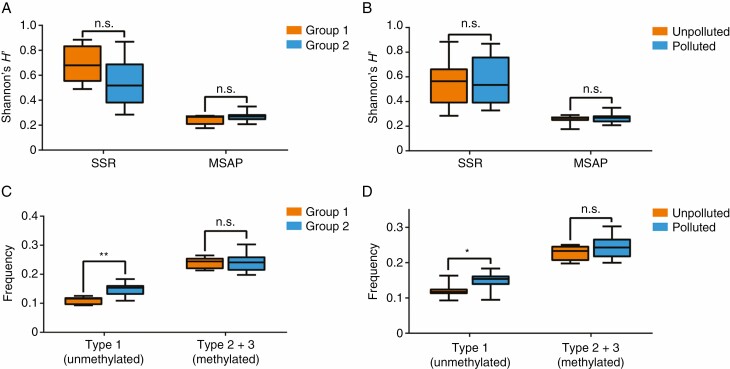
We then analysed genetic and epigenetic diversity. The analysis of Shannon’s diversity showed that the epigenetic diversity was generally lower than the genetic diversity in both the eastern and south-western populations (Supplementary data Table S1). Interestingly, the mean epigenetic diversity was slightly lower in the eastern populations than in the south-western populations (HʹMSAP: 0.2446 ± 0.0192 compared with 0.2711 ± 0.0090; Supplementary data Table S1; Fig. 5A), while genetic diversity was higher in the eastern populations than in the south-western populations (HʹSSR: 0.6906 ± 0.0694 compared with 0.5277 ± 0.0460; Supplementary data Table S1; Fig. 5A). No significant difference in either epigenetic or genetic diversity was detected between the heavy metal-polluted and unpolluted populations (Fig. 5B). We also analysed the frequency of each methylation type in both eastern and south-western populations. The frequency of unmethylated loci (type 1) was significantly higher in the south-western populations than in the eastern populations (Fig. 5C; Supplementary data Table S10). Considering that the eastern populations were mainly unpolluted (except for QX), we also compared the frequency of each methylation type between the heavy metal-polluted and unpolluted populations. We found that the frequency of unmethylated loci (type 1) was higher in the metal-polluted populations than in the unpolluted populations (Fig. 5D; Supplementary data Table S10).
Fig. 5.
Magnitudes of genetic and epigenetic variation among C. ambrosioides populations based on SSR and MSAP. Shannon’s diversity in groups 1 and 2 (A) and in heavy metal-polluted and unpolluted areas (B); frequency of two types of methylation, including type 1 (no methylation) and type 2 + 3 (total methylation), between groups 1 and 2 (C) and between heavy metal-polluted and unpolluted areas (D). Boxplots indicate medians, 25th and 75th percentiles and minimum and maximum values. Significance based on the Student’s t-test is indicated as n.s. P > 0.05; *P < 0.05; **P < 0.01.
Demographic history of invasive C. ambrosioides populations
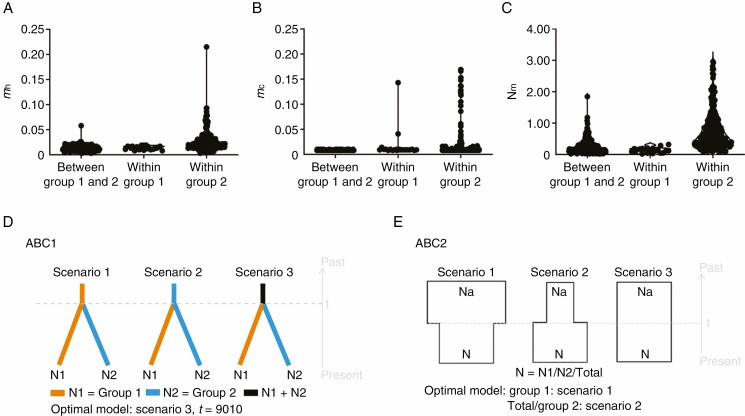
Both the Migrate-n and BAYESASS results showed extremely low levels of migration rates between groups 1 and 2 (mh or mc < 0.05, and their 95 % confidence intervals overlapped zero; Fig. 6A, B; Supplementary data Table S11). The Migrate-n results showed low levels of historical migration rates (mh) among most populations within group 1 or 2 (mh < 0.1), except a moderate migration rate (mh = 0.215) from population HL to XT within group 2 (Fig. 6A; Supplementary data Table S11). The BAYESASS results also showed extremely low levels of contemporary migration rates (mc < 0.05) among almost all populations within group 1, while a moderate migration rate (0.1 < mc < 0.2) was detected among some populations within group 2 (Fig. 6B; Supplementary data Table S11). Limited gene flow values (almost all Nm < 1) was observed between groups 1 and 2 and among populations within group 1, while relatively high Nm values (Nm > 1) were observed among some populations within group 2 (Fig. 6C). Wilcoxon’s test revealed that five populations in group 2 (MY, XT, HY, JS and JJ) had a historical bottleneck either in the SMM or the TPM model (P < 0.05; Supplementary data Table S12), while none of the populations in group 1 experienced a historical bottleneck. In the mode-shift test, all populations except population HL showed an L-shaped distribution of alleles, providing no evidence of a recent bottleneck (Supplementary data Table S12).
Fig. 6.
Violin plot of gene flow (Nm) (A), historical migration rates (mh) (B) and contemporary migration rates (mc) (C) based on all 21 populations of C. ambrosioides. (D and E) Conceptual models for assessing the demographic history of C. ambrosioides by approximate Bayesian computation (ABC). ABC1 (D), the three scenarios tested to estimate the divergence time between two population groups based on genetic clustering in STRUCTURE analysis. ABC2 (E), the three scenarios tested to assess the changes in effective population size. N represents the effective population size of the corresponding population group during the relevant time period, and t represents the time scale measured in the number of generations.
The DIYABC analyses performed to estimate the timing of the divergence between the two cluster groups (ABC1) indicated that scenario 3 (Fig. 6D; Supplementary data Table S13) showed the highest posterior probabilities in both the direct (0.4400, 95 % CI = 0.0049–0.8751) and logistic regression comparisons (0.3861, 95 % CI = 0.3448–0.4274). In scenario 3, we hypothesized the simultaneous divergence of the two groups from one ancestral population 9010 (95 % CI = 1530–27 800) generations ago (Supplementary data Table S14). Based on an estimate of the generation time of C. ambrosioides of approx. 1 year based on our field observations, the divergence times of C. ambrosioides were converted into an absolute time of 9010 years. The analyses for changes in the effective population size (ABC2) indicated that scenario 2 (expansion model) showed the highest posterior probability value both across all 21 populations and in group 2 alone (Fig. 6E; Supplementary data Table S15). However, the best model of scenario 1 (bottleneck model) was detected across the populations of group 1. The median values of t (time scale measured in the number of generations) were 1490 (95 % CI = 40.1–21 800; for the total), 564 (95 % CI = 19.9–15 200; for group 2) and 14 200 (95 % CI = 2770–28 900; for group 1) generations ago (Supplementary data Table S13), corresponding to 1490, 564 and 14 200 years, respectively
Relationships among epigenetic, genetic, phenotypic and environmental variation
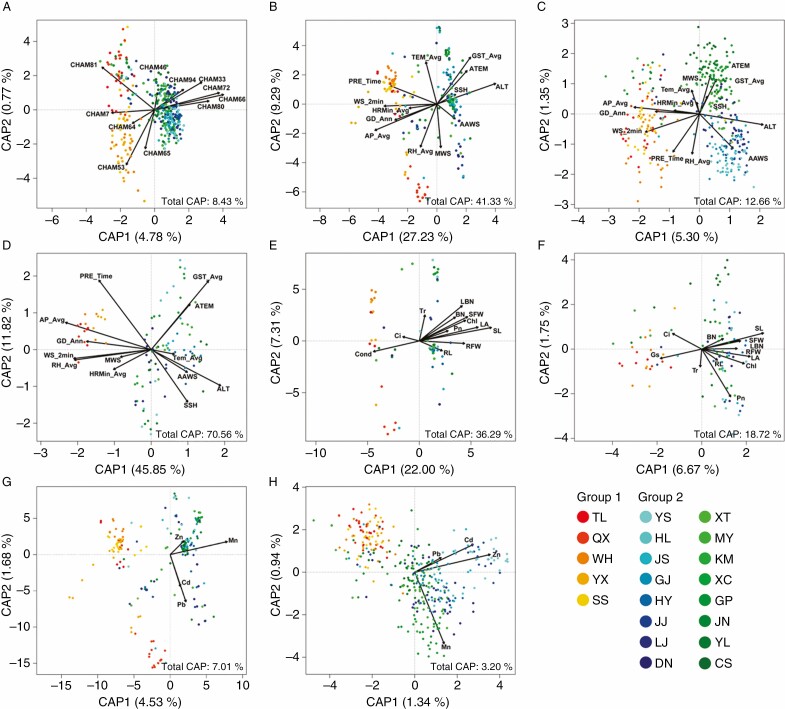
We analysed the relationships between epigenetic and genetic variation by dbRDA, which showed that the genetic variation based on 11 polymorphic SSR loci could explain 8.43 % of the epigenetic variation observed in C. ambrosioides (Fig. 7A). Epigenetic variation was significantly correlated with all 11 SSR loci (Supplementary data Table S16).
Fig. 7.
The relationships among genetic, epigenetic, environmental and phenotypic variation determined using distance-based redundancy analyses (dbRDA). Two types of environmental factors were used, comprising 13 meteorological factors and the contents of four metals in soils. (A) Epigenetic variation in C. ambrosioides populations determined using genetic data as predictors; (B) genetic, (C) epigenetic and (D) phenotypic variations in C. ambrosioides populations determined using meteorological factors as predictors; (E) genetic and (F) epigenetic variation in C. ambrosioides populations determined using phenotypic data as predictors; (G) genetic and (H) epigenetic variation in C. ambrosioides populations determined using four heavy metals in soil as predictors. The meteorological data include altitude (ALT), 20–20 time precipitation (PRE_Time), 2 min average wind speed (WS_2 min), average annual wind speed (AAWS), maximum wind speed (MWS), annual gale days (GD_Ann), sunshine hours (SSH), average air pressure (AP_Avg), average temperature (TEM_Avg), average ground surface temperature (GST_Avg), accumulated temperature (ATEM), average relative humidity (RH_Avg) and minimum average humidity (MRH_Avg). The phenotypic data included root length (RL), shoot length (SL), root fresh weight (RFW), shoot fresh weight (SFW), blade number (BN), lateral branch number (LBN), leaf area (LA), relative chlorophyll content (Chl), net photosynthetic rate (Pn), transpiration rate (Tr), stomatal conductance (Gs) and intercellular carbon dioxide (Ci). The four heavy metals are Zn, Mn, Pb and Cd.
We compared meteorological factors between the eastern and south-western populations, and found that the south-western region showed a higher ALT and a lower WS_2 min, GD_Ann and AP_Avg (Supplementary data Table S6). Then, we further analysed the effects of meteorological factors on genetic or epigenetic variation by dbRDA. The 13 meteorological descriptors explained 41.33 % of the genetic variation but only explained 12.66 % of the epigenetic variation in the C. ambrosioides populations (Fig. 7B, C). In addition, these meteorological descriptors explained 70.56 % of the phenotypic variation (Fig. 7D). Most meteorological factors were significantly correlated with phenotypic, genetic and epigenetic variation (Supplementary data Table S16). Among these meteorological factors, ALT, WS_2 min, GD_Ann and AP_Avg were significantly different in the eastern and south-western regions (Supplementary data Table S6). We also analysed the effects of heavy metal in soils on genetic or epigenetic variation by dbRDA. Compared with meteorological factors, soil heavy metal content had less influence on the overall genetic and epigenetic variation (Fig. 7G, H). The determination coefficient between the contents of Mn, Zn and Cd in soil and epigenetic variation (0.3184, 0.3872 and 0.3207) was greater than that of genetic variation (0.1097, 0.0104 and 0.0350) (Supplementary data Table S16).
Then, we analysed the relationship between phenotypic variation and genetic or epigenetic variation by dbRDA. Similar to meteorological descriptors, the 12 phenotypic factors explained 36.29 % of the genetic variation but only 18.72 % of the epigenetic variation in the C. ambrosioides populations (Fig. 7E, F). Most phenotypic factors were significantly different between the eastern and south-western regions (Supplementary data Table S2).
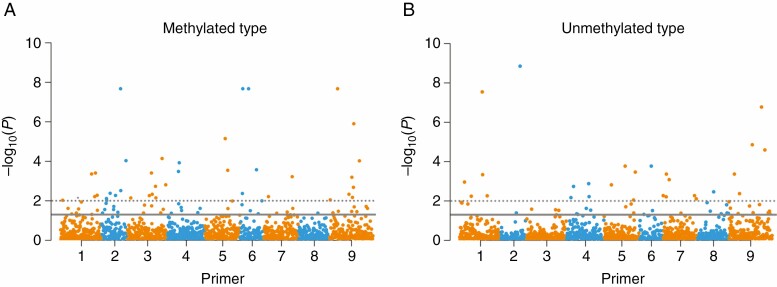
Finally, we performed EWAS to examine the association between leaf heavy metal contents and methylated loci, as we found that the frequency of unmethylated loci (type 1) was significantly higher in the heavy metal-polluted populations. A total of 79 methylated loci (type 2 + 3) and 60 unmethylated loci (type 1) were associated with the heavy metal contents of leaves (P < 0.05, Fig. 8A, B).
Fig. 8.
Manhattan plots of total methylated (A) and unmethylated loci (B) significantly associated with leaf heavy metal contents of C. ambrosioides. The x-axis represents different loci amplified with eight MSAP primer pairs (Supplementary data Table S9), and the y-axis represents –log10 (P-values). The solid line indicates P < 0.05, and the dashed line indicates P < 0.01.
DISCUSSION
Demographic history of invasive C. ambrosioides populations in China
Multiple introductions play important roles in the success of biological invasions, as they are likely to include independent genotypes and provide sufficient variation for local adaptation (Kolbe et al., 2004; Lavergne and Molofsky, 2007; Meimberg et al., 2010). Here, we assessed the genetic structure and patterns of introduction among 21 invasive C. ambrosioides populations in China. The occurrence of two independent introductions into China was detected. First, two distinguishable genotype lineages were identified among the C. ambrosioides populations in China by STRUCTURE analysis (Fig. 3B). According to DIYABC analyses, the approximate divergence time between the two different lineages was estimated to be 9010 years ago (Supplementary data Table S14). However, C. ambrosioides is native to tropical America and was exported to the rest of the world at the beginning of the 18th century (Morton, 1981); the first record of C. ambrosioides in China comes from 1919 (Herbarium of Biology, Peking University; Voucher: PEY0008776). Thus, the polymorphisms distinguishing the lineages of C. ambrosioides in China evolved independently in its native range, prior to its recorded introduction into China. Furthermore, we found a geographic pattern of the occurrence of the two different lineages (Figs 1 and 3B). One genotype lineage was predominantly distributed in eastern China (group 1), and the other was found in south-western China (group 2). No admixture genotype shared between the populations of groups 1 and 2 was detected. The earliest records of C. ambrosioides occurrence in both eastern and south-western China come from the 1930s (Herbarium, Institute of Botany, Chinese Academy of Sciences; Vouchers: 00510268 and 00510368). This obvious east–south-west split of genotypes indicates independent C. ambrosioides invasion events in China. Since we did not sample populations from tropical America, the sources of these two Chinese lineages in their native areas are not known. In future studies, more native populations of C. ambrosioides should be sampled to further elucidate their sources and pathways of introduction.
According to our contemporary and historical migration analyses, gene flow between the eastern and south-western groups of C. ambrosioides is limited (Fig. 6A–C). In addition, limited gene flow was also observed among populations within group 1. Consistent with the results of gene flow, GENELAND analysis showed that the populations in eastern China were further divided into three clusters (Supplementary data Fig. S1). Previous studies have reported that the introduction of multiple genotypes could increase the likelihood of establishment in the introduced range, and further genetic admixture among different introductions could increase the efficacy of selection in a new range (Roman and Darling, 2007; Rius and Darling, 2014; van Kleunen et al., 2015). The geographical splitting of distinct genotypes in multiple introductions of C. ambrosioides could be regarded as an early stage in range expansion. Subsequent recombination between different genotypes might be expected to facilitate invasion in the future (Meimberg et al., 2010; van Boheemen et al., 2017).
Roles of genetic and epigenetic variation in C. ambrosioides invasion
We found phenotypic variations between the eastern and south-western groups of C. ambrosioides (Fig. 2A). Phenotypic variation plays a key role in the successful invasion of exotic plant species (Hagenblad et al., 2015). Both genetic and epigenetic variation can contribute to phenotypic variation (Scoville et al., 2011; Herrera and Bazaga, 2013; Vilatersana et al., 2016; van Boheemen et al., 2017). In this study, the roles of genetic and epigenetic variation in C. ambrosioides invasion were comprehensively evaluated.
Roles of genetic and epigenetic variation in the geographical distribution pattern of invasive C. ambrosioides
Although a spatial autocorrelation pattern was detected for both genetic and epigenetic variation throughout the range of the invasive populations (Fig. 4B, C), a higher proportion of genetic variation than epigenetic variation was found between eastern and south-western populations. Meanwhile, we found that even though the genetic and epigenetic pairwise FST values were significantly related (Fig. 4A), genetic variation had a greater effect on phenotypic variation than epigenetic variation (Fig. 7E, F). Thus, we deduced that the obvious geographical split of the two independent introductions is mainly due to phenotypic differentiation, which has a greater genetic basis than an epigenetic basis.
In invasive C. ambrosioides populations, climatic variation significantly contributed to the observed phenotypic differentiation (Fig. 7D). One possible explanation for this finding is that different phenotypic traits of the eastern and south-western populations may improve their ability to adapt to the different climates of their distinct distribution regions. Although both genetic and epigenetic variation were found to be significantly affected by climatic factors, climatic factors showed a stronger effect on genetic differentiation (Fig. 7B, C). Therefore, genetic variation is a more dominant determinant of the expansion of C. ambrosioides into geographically and meteorologically different regions than epigenetic variation. Genetic variation facilitating range expansion by improving the fitness of invasive populations has also been reported in other invasive plants, such as Senecio pterophorus and Ambrosia artemisiifolia (Vilatersana et al., 2016; van Boheemen et al., 2017).
Effect of epigenetic variation on metalliferous microhabitat adaptation
Chenopodium ambrosioides has been reported to be a manganese/lead hyperaccumulator that thrives in heavy metal-contaminated habitats (Zhang et al., 2012; Rivera-Becerril et al., 2013). Here, we used a series of invasive populations from both metalliferous and non-metalliferous habitats to investigate the molecular basis of C. ambrosioides adaptation to heavy metal stresses in local microhabitats. Our comparison of genetic diversity between heavy metal-polluted and unpolluted populations of C. ambrosioides did not reveal any significant differences (Supplementary data Table S1; Fig. 4E). In addition, no evidence of an intralineage genotype associated with either metalliferous or non-metalliferous habitats was found (Fig. 3B). Although several studies have indicated the relationship between population genetic variation and heavy metal tolerance in hyperaccumulators (Verbruggen et al., 2009; Wójcik et al., 2013), the characterization of the genetic variation of C. ambrosioides from metalliferous and non-metalliferous habitats failed to reveal adaptive variation, so we further performed an epigenome-wide analysis. Epigenetic modifications in gene expression have been recognized as an important mechanism underlying the plastic responses of plant traits to heterogeneous environments (Herrera and Bazaga, 2013; Chen et al., 2022). Our hierarchical AMOVAs showed that the proportion of epigenetic variation within the C. ambrosioides populations (76.83 %) was much greater than the proportion of genetic variation (38.04 %) (Table 1). Furthermore, the correlation between genetic and epigenetic variation disappeared within the eastern lineages (Fig. 4B). Thus, the potential for each lineage of C. ambrosioides to adapt to complicated microenvironments through specific epigenetic modifications should be considered. We found that the determination coefficient of soil heavy metal content on epigenetic variation was greater than that of genetic variation (Supplementary data Table S16). The frequency of unmethylated loci was much higher in the heavy metal-polluted populations than in the unpolluted populations (Supplementary data Table S10; Fig. 4G). Changes in global DNA methylation levels under heavy metal stress have similarly been detected in other species (Ou et al., 2012; Feng et al., 2016; Fan et al., 2020). In addition, recent studies have reported that specific methylation changes can be induced by heavy metal stress and display transgenerational inheritance over many generations (Cong et al., 2019; Fan et al., 2020; Jing et al., 2022). For example, Feng et al. (2016) identified specific differentially methylated regions induced by heavy metal stress in rice, which were closely associated with transcriptional differences in stress-response genes involved in metal transport, metabolic processes and transcriptional regulation. Our EWAS identified dozens of total methylated and unmethylated loci that were significantly associated with heavy metal contents in the leaves of C. ambrosioides (Fig. 7). Together, our results suggest that epigenetic variation plays an important role in the metal accumulation trait of C. ambrosioides, which allows it to cope with high heavy metal concentrations in the microenvironment.
Conclusion
In this study, we found that the eastern and south-western populations of C. ambrosioides in China may have originated from independent invasion events without recombination. Genetic variation is a more dominant determinant of the phenotypic differentiation and geographical distribution pattern of invasive C. ambrosioides than epigenetic variation. Additionally, the global DNA methylation level and specific methylated loci contribute to the adaptation of invasive C. ambrosioides to local metalliferous habitats.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Figure S1: results of GENELAND analysis of 21 populations of Chenopodium ambrosioides. Table S1: the locality, habitat, longitude and latitude, sample size, genetic and epigenetic diversity index of the 21 populations of Chenopodium ambrosioides. Table S2: phenotypic data of C. ambrosioides populations in the common garden experiment. Table S3: heavy metal content in soil of C. ambrosioides sampling sites. Table S4: heavy metal content in leaves of C. ambrosioides populations. Table S5: Nemerow integrated pollution index of C. ambrosioides populations. Table S6: meteorological data across C. ambrosioides sampling sites. Table S7: characterization of 12 polymorphic genomic SSR markers for C. ambrosioides populations. Table S8: polymorphism information of SSR loci. Table S9: adapters and primers used for MSAP epigenotyping. Table S10: methylation frequency of 21 wild populations of C. ambrosioides. Table S11: mean contemporary migration rate estimated from BAYEASS and historical migration rate estimated from Migrate-n across the 21 C. ambrosioides populations. Table S12: bottleneck analysis for 21 populations of C. ambrosioides. Table S13: posterior probability of each of the three scenarios for C. ambrosioides, and their 95 % confidence interval based on the logistic and direct estimate. Table S14: estimated divergence parameters for the population groups of C. ambrosioides. Table S15: posterior probability and their 95 % confidence interval of each of three scenarios for C. ambrosioides, and the population groups to evaluate effective population size changes. Table S16: the results of correlating predictor factor variation to epigenetic, genetic and phenotypic variation in dbRDA analysis.
Contributor Information
Hanchao Zhang, College of Life Sciences, Nanjing Agricultural University, Nanjing, Jiangsu, PR China.
Yongwei Tang, College of Life Sciences, Nanjing Agricultural University, Nanjing, Jiangsu, PR China.
Quanyuan Li, College of Life Sciences, Nanjing Agricultural University, Nanjing, Jiangsu, PR China.
Shangjun Zhao, College of Life Sciences, Nanjing Agricultural University, Nanjing, Jiangsu, PR China.
Zhou Zhang, College of Life Sciences, Nanjing Agricultural University, Nanjing, Jiangsu, PR China.
Yahua Chen, College of Life Sciences, Nanjing Agricultural University, Nanjing, Jiangsu, PR China; Jiangsu Collaborative Innovation Centre for Solid Organic Waste Resource Utilization, Nanjing Agricultural University, Nanjing, Jiangsu, PR China.
Zhenguo Shen, College of Life Sciences, Nanjing Agricultural University, Nanjing, Jiangsu, PR China; Jiangsu Collaborative Innovation Centre for Solid Organic Waste Resource Utilization, Nanjing Agricultural University, Nanjing, Jiangsu, PR China.
Chen Chen, College of Life Sciences, Nanjing Agricultural University, Nanjing, Jiangsu, PR China; Jiangsu Collaborative Innovation Centre for Solid Organic Waste Resource Utilization, Nanjing Agricultural University, Nanjing, Jiangsu, PR China.
FUNDING
This work was supported by grants from the National Natural Science Foundation of China (31770404), the Fundamental Research Funds for the Central Universities (SKY202101) and the China Agriculture Research System (CARS-10-B24).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
LITERATURE CITED
- Banerjee AK, Guo W, Huang Y. 2019. Genetic and epigenetic regulation of phenotypic variation in invasive plants – linking research trends towards a unified framework. NeoBiota 49: 77–103. doi: 10.3897/neobiota.49.33723. [DOI] [Google Scholar]
- Beerli P, Felsenstein J. 1999. Maximum-likelihood estimation of migration rates and effective population numbers in two populations using a coalescent approach. Genetics 152: 763–773. doi: 10.1093/genetics/152.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli P, Felsenstein J. 2001. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proceedings of the National Academy of Sciences, USA 98: 4563–4568. doi: 10.1073/pnas.081068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boheemen LA, Lombaert E, Nurkowski KA, Gauffre B, Rieseberg LH, Hodgins KA. 2017. Multiple introductions, admixture and bridgehead invasion characterize the introduction history of Ambrosia artemisiifolia in Europe and Australia. Molecular Ecology 26: 5421–5434. doi: 10.1111/mec.14293. [DOI] [PubMed] [Google Scholar]
- Bossdorf O, Richards CL, Pigliucci M. 2008. Epigenetics for ecologists. Ecology Letters 11: 106–115. [DOI] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Chapuis MP, Estoup A. 2007. Microsatellite null alleles and estimation of population differentiation. Molecular Biology and Evolution 24: 621–631. doi: 10.1093/molbev/msl191. [DOI] [PubMed] [Google Scholar]
- Chaudhary K, Jan S, Khan S. 2016. Heavy metal ATPase (HMA2, HMA3, and HMA4) genes in hyperaccumulation mechanism of heavy metals. In: Ahmad P, ed. Plant metal interaction: emerging remediation techniques. Amsterdam: Elsevier, 545–556. [Google Scholar]
- Chen C, Zhang H, Wang A, Lu M, Shen Z, Lian C. 2015. Phenotypic plasticity accounts for most of the variation in leaf manganese concentrations in Phytolacca americana growing in manganese-contaminated environments. Plant and Soil 396: 215–227. doi: 10.1007/s11104-015-2581-7. [DOI] [Google Scholar]
- Chen C, Zheng Z, Bao Y, et al. 2020. Comparisons of natural and cultivated populations of Corydalis yanhusuo indicate divergent patterns of genetic and epigenetic variation. Frontiers in Plant Science 11: 985. doi: 10.3389/fpls.2020.00985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Wang M, Zhu J, et al. 2022. Long-term effect of epigenetic modification in plant–microbe interactions: modification of DNA methylation induced by plant growth-promoting bacteria mediates promotion process. Microbiome 10: 36. doi: 10.1186/s40168-022-01236-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chwedorzewska KJ, Bednarek PT. 2012. Genetic and epigenetic variation in a cosmopolitan grass Poa annua from Antarctic and Polish populations. Polish Polar Research 33: 63–80. doi: 10.2478/v10183-012-0004-5. [DOI] [Google Scholar]
- Cong W, Miao Y, Xu L, et al. 2019. Transgenerational memory of gene expression changes induced by heavy metal stress in rice (Oryza sativa L.). BMC Plant Biology 19: 282. doi: 10.1186/s12870-019-1887-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornuet JM, Santos F, Beaumont MA, et al. 2008. Inferring population history with DIY ABC: a user-friendly approach to approximate Bayesian computation. Bioinformatics 24: 2713–2719. doi: 10.1093/bioinformatics/btn514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornuet JM, Pudlo P, Veyssier J, et al. 2014. DIYABC v2.0: a software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 30: 1187–1189. doi: 10.1093/bioinformatics/btt763. [DOI] [PubMed] [Google Scholar]
- Crawford KM, Whitney KD. 2010. Population genetic diversity influences colonization success. Molecular Ecology 19: 1253–1263. doi: 10.1111/j.1365-294X.2010.04550.x. [DOI] [PubMed] [Google Scholar]
- De Silva HN, Hall AJ, Rikkerink E, McNeilage MA, Fraser LG. 2005. Estimation of allele frequencies in polyploids under certain patterns of inheritance. Heredity 95: 327–334. doi: 10.1038/sj.hdy.6800728. [DOI] [PubMed] [Google Scholar]
- Diniz-Filho JAF, Soares TN, Lima JS, et al. 2013. Mantel test in population genetics. Genetics and Molecular Biology 36: 475–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne F, Stift M, Vergilino R, Mable BK. 2014. Recent progress and challenges in population genetics of polyploid organisms: an overview of current state-of-the-art molecular and statistical tools. Molecular Ecology 23: 40–69. doi: 10.1111/mec.12581. [DOI] [PubMed] [Google Scholar]
- Estoup A, Ravigné V, Hufbauer R, Vitalis R, Gautier M, Facon B. 2016. Is there a genetic paradox of biological invasion? Annual Review of Ecology, Evolution, and Systematics 47: 51–72. doi: 10.1146/annurev-ecolsys-121415-032116. [DOI] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14: 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HE. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10: 564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Fan SK, Ye JY, Zhang LL, et al. 2020. Inhibition of DNA demethylation enhances plant tolerance to cadmium toxicity by improving iron nutrition. Plant, Cell & Environment 43: 275–291. doi: 10.1111/pce.13670. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 1993. PHYLIP (phylogenetic inference package) version 3.6. Department of Genetics, University of Washington, Seattle.
- Feng SJ, Liu XS, Tao H, et al. 2016. Variation of DNA methylation patterns associated with gene expression in rice (Oryza sativa) exposed to cadmium. Plant, Cell & Environment 39: 2629–2649. doi: 10.1111/pce.12793. [DOI] [PubMed] [Google Scholar]
- Foust CM, Preite V, Schrey AW, et al. 2016. Genetic and epigenetic differences associated with environmental gradients in replicate populations of two salt marsh perennials. Molecular Ecology 25: 1639–1652. doi: 10.1111/mec.13522. [DOI] [PubMed] [Google Scholar]
- Gallo-Franco JJ, Sosa CC, Ghneim-Herrera T, Quimbaya M. 2020. Epigenetic control of plant response to heavy metal stress: a new view on aluminum tolerance. Frontiers in Plant Science 11: 602625. doi: 10.3389/fpls.2020.602625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot G, Mortier F, Estoup A. 2005. GENELAND: a computer package for landscape genetics. Molecular Ecology Notes 5: 712–715. doi: 10.1111/j.1471-8286.2005.01031.x. [DOI] [Google Scholar]
- Hagenblad J, Hülskötter J, Acharya KP, et al. 2015. Low genetic diversity despite multiple introductions of the invasive plant species Impatiens glandulifera in Europe. BMC Genetics 16: 103. doi: 10.1186/s12863-015-0242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera CM, Bazaga P. 2013. Epigenetic correlates of plant phenotypic plasticity: DNA methylation differs between prickly and nonprickly leaves in heterophyllous Ilex aquifolium (Aquifoliaceae) trees. Botanical Journal of the Linnean Society 171: 441–452. [Google Scholar]
- Jing M, Zhang H, Wei M, et al. 2022. Reactive oxygen species partly mediate DNA methylation in responses to different heavy metals in pokeweed. Frontiers in Plant Science 13: 845108. doi: 10.3389/fpls.2022.845108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski ST, Taper ML, Marshall TC. 2007. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Molecular Ecology 16: 1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. [DOI] [PubMed] [Google Scholar]
- van Kleunen M, Röckle M, Stift M. 2015. Admixture between native and invasive populations may increase invasiveness of Mimulus guttatus. Proceedings of the Royal Society B: Biological Sciences 282: 20151487. doi: 10.1098/rspb.2015.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliks MM. 1985. Studies on the traditional herbal anthelmintic Chenopodium ambrosioides L.: ethnopharmacological evaluation and clinical field trials. Social Science & Medicine 21: 879–886. [DOI] [PubMed] [Google Scholar]
- Kolano B, Tomczak H, Molewska R, Jellen EN, Maluszynska J. 2012. Distribution of 5S and 35S rRNA gene sites in 34 Chenopodium species (Amaranthaceae). Botanical Journal of the Linnean Society 170: 220–231. doi: 10.1111/j.1095-8339.2012.01286.x. [DOI] [Google Scholar]
- Kolbe JJ, Glor RE, Schettino LRG, Lara AC, Larson A, Losos JB. 2004. Genetic variation increases during biological invasion by a Cuban lizard. Nature 431: 177–181. [DOI] [PubMed] [Google Scholar]
- Kumar M, Bijo AJ, Baghel RS, Reddy CRK, Jha B. 2012. Selenium and spermine alleviate cadmium induced toxicity in the red seaweed Gracilaria dura by regulating antioxidants and DNA methylation. Plant Physiology and Biochemistry 51: 129–138. doi: 10.1016/j.plaphy.2011.10.016. [DOI] [PubMed] [Google Scholar]
- Lavergne S, Molofsky J. 2007. Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proceedings of the National Academy of Sciences, USA 104: 3883–3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Muse SV. 2005. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21: 2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- Luikart G, Cornuet JM. 1998. Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conservation Biology 12: 228–237. [Google Scholar]
- Ma JS. 2013. The checklist of the Chinese invasive plants. Beijing: Higher Education Press, 25–27. [Google Scholar]
- Ma L, Yang ZG, Li L, Wang L. 2016. Source identification and risk assessment of heavy metal contaminations in urban soils of Changsha, a mine-impacted city in Southern China. Environmental Science and Pollution Research 23: 17058–17066. doi: 10.1007/s11356-016-6890-z. [DOI] [PubMed] [Google Scholar]
- Meimberg H, Milan NF, Karatassiou M, Espeland EK, McKay JK, Rice KJ. 2010. Patterns of introduction and adaptation during the invasion of Aegilops triuncialis (Poaceae) into Californian serpentine soils. Molecular Ecology 19: 5308–5319. doi: 10.1111/j.1365-294X.2010.04875.x. [DOI] [PubMed] [Google Scholar]
- Morton JF. 1981. Atlas of medicinal plants of Middle America: Bahamas to Yucatan. Springfield, IL: Charles C. Thomas. [Google Scholar]
- Niederhuth CE, Schmitz RJ. 2014. Covering your bases: inheritance of DNA methylation in plant genomes. Molecular Plant 7: 472–480. doi: 10.1093/mp/sst165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Friendly M, et al. 2019. Vegan community ecology package: ordination methods, diversity analysis and other functions for community and vegetation ecologists. R package v.2.5-6. http://CRAN.R-project.org/package=vegan; last accessed 20 December 2021.
- Ou X, Zhang YH, Xu CM, et al. 2012. Transgenerational inheritance of modified DNA methylation patterns and enhanced tolerance induced by heavy metal stress in rice (Oryza sativa L.). PLoS One 7: e41143. doi: 10.1371/journal.pone.0041143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PCD (Pollution Control Department, Thailand). 1995. Notification of National Environmental Board No. 10, B.E 2538 (1995) under the Enhancement and Conservation of National Environmental Quality Act B.E.2535 (1992), published in the Royal Government Gazette No. 112 Part 52 dated May 25, B.E.2538. https://www.pcd.go.th/
- Peakall R, Smouse PE. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes 6: 288–295. doi: 10.1111/j.1471-8286.2005.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piry S, Luikart G, Cornuet JM. 1999. BOTTLENECK: a computer program for detecting recent reductions in the effective size using allele frequency data. Journal of Heredity 90: 502–503. doi: 10.1093/jhered/90.4.502. [DOI] [Google Scholar]
- Reyna-López GE, Simpson J, Ruiz-Herrera J. 1997. Differences in DNA methylation patterns are detectable during the dimorphic transition of fungi by amplification of restriction polymorphisms. Molecular and General Genetics 253: 703–710. [DOI] [PubMed] [Google Scholar]
- Richards CL, Schrey AW, Pigliucci M. 2012. Invasion of diverse habitats by few Japanese knotweed genotypes is correlated with epigenetic differentiation. Ecology Letters 15: 1016–1025. doi: 10.1111/j.1461-0248.2012.01824.x. [DOI] [PubMed] [Google Scholar]
- Rius M, Darling JA. 2014. How important is intraspecific genetic admixture to the success of colonising populations? Trends in Ecology and Evolution 29: 233–242. doi: 10.1016/j.tree.2014.02.003. [DOI] [PubMed] [Google Scholar]
- Rivera-Becerril F, Juárez-Vázquez LV, Hernández-Cervantes SC, et al. 2013. Impacts of manganese mining activity on the environment: interactions among soil, plants, and arbuscular mycorrhiza. Archives of Environmental Contamination and Toxicology 64: 219–227. doi: 10.1007/s00244-012-9827-7. [DOI] [PubMed] [Google Scholar]
- Roman J, Darling JA. 2007. Paradox lost: genetic diversity and the success of aquatic invasions. Trends in Ecology & Evolution 22: 454–464. doi: 10.1016/j.tree.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Rousset F. 2008. genepop’007: a complete re‐implementation of the genepop software for Windows and Linux. Molecular Ecology Resources 8: 103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Schories D, Mehlig U. 2000. CO2 gas exchange of benthic microalgae during exposure to air: a technique for the rapid assessment of primary production. Wetlands Ecology and Management 8: 273–280. [Google Scholar]
- Schulz B, Eckstein RL, Durka W. 2013. Scoring and analysis of methylation‐sensitive amplification polymorphisms for epigenetic population studies. Molecular Ecology Resources 13: 642–653. doi: 10.1111/1755-0998.12100. [DOI] [PubMed] [Google Scholar]
- Scoville AG, Barnett LL, Bodbyl‐Roels S, Kelly JK, Hileman LC. 2011. Differential regulation of a MYB transcription factor is correlated with transgenerational epigenetic inheritance of trichome density in Mimulus guttatus. New Phytologist 191: 251–263. doi: 10.1111/j.1469-8137.2011.03656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEPAC (State Environmental Protection Administration of China). 1995. Environmental Quality Standard for Soils (GB15618-1995). Beijing: Standards Press of China. [Google Scholar]
- Spens AE, Douhovnikoff V. 2016. Epigenetic variation within Phragmites australis among lineages, genotypes, and ramets. Biological Invasions 18: 2457–2462. doi: 10.1007/s10530-016-1223-1. [DOI] [Google Scholar]
- Tie BQ, Yuan M, Tang MZ. 2005. Phytolacca americana L.: a new manganese accumulator plant. Journal of Agro-Environment Science 24: 340–343. [Google Scholar]
- Turner. 2018. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. Journal of Open Source Software 3: 731. doi: 10.21105/joss.00731. [DOI] [Google Scholar]
- Udupa S, Baum M. 2001. High mutation rate and mutational bias at (TAA)n microsatellite loci in chickpea (Cicer arietinum L.). Molecular Genetics and Genomics 265: 1097–1103. doi: 10.1007/s004380100508. [DOI] [PubMed] [Google Scholar]
- Ueda M, Seki M. 2020. Histone modifications form epigenetic regulatory networks to regulate abiotic stress response. Plant Physiology 182: 15–26. doi: 10.1104/pp.19.00988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vekemans X. 2002. AFLP-SURV: a program for genetic diversity analysis with AFLP (and RAPD) population data. Distributed by the author, version 1.0. Laboratoire de Génétique et Ecologie Végétale, Université Libre de Bruxelles, Bruxelles, Belgium. [Google Scholar]
- Verbruggen N, Hermans C, Schat H. 2009. Molecular mechanisms of metal hyperaccumulation in plants. New Phytologist 181: 759–776. doi: 10.1111/j.1469-8137.2008.02748.x. [DOI] [PubMed] [Google Scholar]
- Vilatersana R, Sanz M, Galian A, Castells E. 2016. The invasion of Senecio pterophorus across continents: multiple, independent introductions, admixture and hybridization. Biological Invasions 18: 2045–2065. doi: 10.1007/s10530-016-1150-1. [DOI] [Google Scholar]
- Wibowo A, Becker C, Marconi G, et al. 2016. Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity. Elife 5: e13546. doi: 10.7554/eLife.13546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GA, Rannala B. 2003. Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163: 1177–1191. doi: 10.1093/genetics/163.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wójcik M, Dresler S, Jawor E, Kowalczyk K, Tukiendorf A. 2013. Morphological, physiological, and genetic variation between metallicolous and nonmetallicolous populations of Dianthus carthusianorum. Chemosphere 90: 1249–1257. doi: 10.1016/j.chemosphere.2012.09.068. [DOI] [PubMed] [Google Scholar]
- Zhang WH, Huang Z, He LY, Sheng XF. 2012. Assessment of bacterial communities and characterization of lead-resistant bacteria in the rhizosphere soils of metal-tolerant Chenopodium ambrosioides grown on lead–zinc mine tailings. Chemosphere 87: 1171–1178. [DOI] [PubMed] [Google Scholar]
- Zhao KY, Aranzana MJ, Kim S, et al. 2007. An Arabidopsis example of association mapping in structured samples. PLoS Genetics 3: e4. doi: 10.1371/journal.pgen.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.