Abstract
Background and Aims
Although plastid genes are widely used in phylogenetic studies, signals of positive selection have been scarcely investigated in the grape family. The plastomes from 91 accessions of Vitaceae were examined to understand the extent to which positive selection is present and to identify which genes are involved. Moreover, the changes through time of genes under episodic positive selection were investigated and the hypothesis of an adaptive process following the Cretaceous–Palaeogene (K/Pg) transition about 66 million years ago was tested.
Methods
Different codon-substitution models were used to assess pervasive and episodic positive selection events on 70 candidate plastid genes. Divergence times between lineages were estimated and stochastic character mapping analysis was used to simulate variation over time of the genes found to be under episodic positive selection.
Key Results
A total of 20 plastid genes (29 %) showed positive selection. Among them, 14 genes showed pervasive signatures of positive selection and nine genes showed episodic signatures of positive selection. In particular, four of the nine genes (psbK, rpl20, rpoB, rps11) exhibited a similar pattern showing an increase in the rate of variation close to the K/Pg transition.
Conclusion
Multiple analyses have shown that the grape family has experienced ancient and recent positive selection events and that the targeted genes are involved in essential functions such as photosynthesis, self-replication and metabolism. Our results are consistent with the idea that the K/Pg transition has favoured an increased rate of change in some genes. Intense environmental perturbations have influenced the rapid diversification of certain lineages, and new mutations arising on some plastid genes may have been fixed by natural selection over the course of many generations.
Keywords: Adaptive evolution, Cretaceous–Palaeogene, mass extinction, episodic selection, grape, plastid genes, plastomes, positive selection, Vitis vinifera, Vitaceae
INTRODUCTION
Knowing how natural selection has affected the evolution of species over time is one of the most relevant and fascinating topics in evolutionary biology. Nevertheless, to date, we still know little about the forces that may have operated in the past shaping present lineages (Dobzhansky, 1973; Zhong et al., 2015). For the last 100 million years (Myr), genomes have been widely shaped by environmental perturbations (Soltis and Soltis, 2016; 2021). In particular, ancient whole-genome duplication (WGD) events seem to be related to the Cretaceous–Palaeogene (K/Pg) mass extinction [66 million years ago (Ma)] caused by the impact of an asteroid in Mexico or volcanic activity (Vellekoop et al., 2014; Chiarenza et al., 2020). Although the K/Pg mass extinction is not considered the main cause of extinction of plants, it may have influenced the evolution and diversification of taxa (Cui et al., 2019; Koenen et al., 2021). Dust clouds and sulphur aerosols are thought to have prevented solar radiation, producing a rapid global cooling, the so-called ‘impact winter’. Polyploidization events appear to have contributed to the adaptation of several plants to changing environments (Fawcett et al., 2009; Vanneste et al., 2014; Cai et al., 2019). Apart from chromosomal duplications that show important indications in favour of an adaptive process (Van de Peer et al., 2017), single genes could also have been affected by environment perturbations and newly arising mutations might have been selected.
While in the past attention has been paid to understanding the negative pressure that operated on genomes, today there is a growing interest in identifying positive selection (Nielsen, 2005; Murrell et al., 2012). For example, the identification of positively selected genes (PSGs) has proved a powerful tool in evolutionary studies focusing on adaptation to climate change (Anderson and Song, 2020). Theoretical and applied papers have proposed various methods for estimating Darwinian selection (Kimura, 1968; Nei and Kumar, 2000). In contrast to the approach to population genetics which depends on assumptions concerning the demographic history of populations, comparative approaches involve data from different taxa and have been developed to detect selection in the past (Nei and Kumar, 2000; Nielsen, 2005). These phylogenetic methods detect positive and negative selection by estimating the ratio of non-synonymous to synonymous substitution rates (ω) and test for significant deviations from the neutral expectation of ω = 1 (Nielsen and Yang, 1998). Recently, specific tests have been developed to identify pervasive and episodic positive selection (Weaver et al., 2018). While the former methods imply that the identified sites are under continuous change, the latter indicate that the sites have changed, and that they will be maintained in the lineage providing an adaptive advantage.
Largely used in phylogenetic studies due to the ease with which the DNA can be sequenced, plastid genes have usually been considered to evolve as a single locus, and the mechanisms of evolution have not been greatly studied in recent years (Doyle, 1992; Gonçalves et al., 2019). A growing body of evidence suggests that positive selection in plastid genes is much more common than previously believed (Kapralov and Filatov, 2007). Changes in photosystem genes to adapt to changing climatic conditions have been reported in several angiosperm families (Bock et al., 2014), although most studies focused on a limited number of sequences. High-throughput sequencing technologies have recently offered an extraordinary opportunity to access complete plastid genomes and to explore the evolution of organelles (Sabater, 2018). For example, widely diffused signs of adaptive evolution at the molecular level have been found across the plastomes in Poaceae (Gao et al., 2019b). Different studies have shown that about one-third of the current plastid genes in grasses display signs of pervasive positive selection and that several episodic positive selection events probably occurred in the family Poaceae in the past (Zhong et al., 2009; Piot et al., 2018). Although, so far, few efforts have been made to explore how and when the plastid genes adapted to climate variations, recent phylogenetic methods have opened new ways to elucidate these pending issues.
The grape family (Vitaceae) is spread throughout temperate, tropical and subtropical areas and it began to diversify during the Late Cretaceous (Magallón and Castillo, 2009; Manchester et al., 2013). Taxa in the family are characterized by leaf-opposed tendrils used to climb trees (Gerrath and Posluszny, 1988; Zhang et al., 2015a), but throughout their evolution, different genera have adapted to occupy specific habitats by modifying their own habits (vines, lianas, trees, herbs and stem succulents). While Vitis, Ampelopsis and Parthenocissus adapted mostly to the cool climate in the Neogene (Zachos et al., 2001; Chen, 2009; Ma et al., 2016; Ickert-Bond et al., 2018), the genera Cyphostemma, Cayratia, Tetrastigma and Cissus include several species that show Crassulacean acid metabolism (CAM) and succulence (Chen, 2009), features which have occurred in many plant families adapted to drought (Heyduk et al., 2019). Phylogenetically related to the non-climbing monogeneric Leeaceae (Lu, et al., 2018) the grape family includes the grapevine (Vitis vinifera L.), one of the most economically important crops in the world (Grassi and De Lorenzis, 2021). In recent years, several wild grapes have been well documented as important genetic resources showing resistance genes against environmental stresses and diseases as well as important sources of molecules beneficial to human health (Szekeres et al., 2011; Lekshmi et al., 2015; Méndez-López et al., 2020). As a consequence of the development of new molecular biology techniques and the increasing interest in wild relatives, the phylogeny of the grape family based on plastid DNA has improved greatly over the last decade allowing the identification of five major clades in the family: the Ampelocissus–Vitis–Nothocissus-Pterisanthes clade, the Parthenocissus–Yua clade, the Cayratia–Cyphostemma–Tetrastigma clade, the Cissus clade, and the Ampelopsis–Rhoicissus–Clematicissus clade (Liu et al., 2013, 2016; Zhang et al., 2015b; Lu et al., 2018). Positive selection in grape plastomes has been investigated only in a few taxa (Yue et al., 2010; Raman and Park, 2016; Yu et al., 2019), but the substitution rate heterogeneity observed in non-coding sequences suggests that different lineages may have accelerated or slowed down during their evolution (Zecca et al., 2020a). Moreover, the phylogenetic discordances observed between phylogenetic trees based on coding and non-coding plastid regions suggest that selection forces may have influenced the evolution of some genes (Zecca et al., 2020a).
In this work, different codon-substitution models have been used to assess how positive selective forces have shaped the evolutionary history of plastomes within the grape family. In particular, analysing data from 91 accessions of Vitaceae we aimed: (1) to understand the extent to which positive selection is present in plastomes; (2) to identify genes that are under positive selection, distinguishing between pervasive and episodic positive selection; (3) to investigate changes over time of genes under episodic positive selection and, based on those changes, to test the hypothesis of an adaptive process following the K/Pg transition about 66 Ma (Van de Peer et al., 2017; Cai et al., 2019) within the grape family (hereinafter we refer to it as the ‘Cretaceous–Palaeogene hypothesis’). In discussing our results, we referred to the Geologic Time Scale proposed by the Geological Society of America (Walker et al., 2018).
MATERIALS AND METHODS
Data collection
All the plastid genome sequences of Vitis (59), Ampelopsis (3) and Tetrastigma (1) species were downloaded from the public database available at the National Center for Biotechnology Information (NCBI GenBank). Furthermore, a plastid alignment of 28 Vitaceae accessions originally published by Zhang et al. (2015b) was downloaded from the Dryad database (https://doi.org/10.5061/dryad.d15v3) including Vitis (4), Pterisanthes (1), Ampelocissus (1), Parthenocissus (2), Rhocissus (1), Ampelopsis (2), Nekemias (1), Cissus (6), Tetrastigma (4), Cyphostemma (4), Cayratia (1) and Leea (1). Overall, 73 genes were identified in single-copy regions using the reference plastome of V. vinifera (NC_007957.1).
Data preparation and diversity analysis
Multiple sequence alignment was performed for each gene separately with the online version of MAFFT v.7 and successively all introns were eliminated. The ndhF gene was excluded owing to uncertain alignment due to large indels whereas petB was excluded owing to identity with mitochondrial DNA. Parsimony-informative sites (P), nucleotide diversity (π) and haplotype diversity (HD) were calculated in 71 genes using the program DnaSP v.6.0.7 (http://www.ub.edu/dnasp/) (Supplementary Data Table S1). Alignment of 71 plastid genes is available in File S1.
To detect the protein-coding genes under selection, each gene was prepared using Codon Alignment v.2.1.0 (https://www.hiv.lanl.gov) and all stop codons were deleted using the CleanStopCodons tool implemented in HyPhy software (Pond and Muse, 2005). Since the EasyCodeML software requires a phylogram as the input file, we inferred a maximum-likelihood (ML) tree for all datasets produced using RaxML-HPC BlackBox (Stamatakis, 2014, p. 201) performed under the GTRGAMMA model of substitution. Analyses were performed on the CIPRES Science Gateway (Miller et al., 2010). The phylogenetic trees obtained are available in Supplementary Data File S2. Successively, each gene was analysed independently to search for signs of positive selection, as explained below.
Positive selection analyses – site model tests
The site model (SM) test was performed using the CODEML algorithm (Yang, 1997) implemented in EasyCodeML (Gao et al., 2019a). Site models use a statistical distribution to account for variation of the ω ratio among codons as described by Nielsen and Yang (1998). A ratio (ω) of the non-synonymous substitution rate (Ka) to the synonymous substitutions rate (Ks) greater than 1 suggests that a gene is positively selected (Yang and Nielsen, 2002). Codon substitution models were investigated under the preset mode and two likelihood ratio tests (LRTs) were performed to detect positively selected sites: M7 vs. M8 and M8a vs. M8. The M8–M7 comparison offers a very stringent test of positive selection, but the M8–M8a comparison yields fewer false positives (Gao et al., 2019a). To minimize the possibility of stumbling in local optima, we ran the M8 model with different starting ω values (ω = 2, 1 and 0.5) as suggested by Anisimova et al. (2002). We selected the clean data option for the accD, ycf1, rpl22, psbT and rpoC2 genes due to the presence of indels. Positive selection was reported if the estimated ω parameter was greater than 1 and the LRT was significant (P < 0.05). Since a variety of approaches showing different power and limitations have been proposed to identify codons under positive selection in an alignment, we chose to perform the Bayes empirical Bayes (BEB) analysis and the FUBAR analysis implemented respectively in EasyCodeML and the DATAMONKEY web interface (Yang et al., 2005; Murrell et al., 2013; Weaver et al., 2018). The BEB method, calculating posterior probabilities for site classes, was implemented for the M8 model. FUBAR uses a Bayesian approach to infer non-synonymous and synonymous substitution rates on a per-site basis for a given coding alignment. FUBAR was run using the following default option: number of Markov chain Monte Carlo (MCMC) chains to run = 5, length of each chain = 2000 000, burn-in = 1000 000, samples drawn from each chain = 100; advanced options: number of grid points = 50; concentration parameter of the Dirichlet prior = 0.5. For each codon, positive selection was reported when both BEB and FUBAR analysis showed P > 0.9.
Positive selection analyses – branch site model tests
We used the aBSREL method (adaptive branch-site random effects likelihood) implemented in the DATAMONKEY web server to identify those branches that have high values of ω (Smith et al., 2015). Specifically, aBSREL uses AICc (small sample Akaike’s information criterion) to infer the optimal number of ω rate classes for each branch. aBSREL tests, for each branch in the phylogeny, whether a proportion of sites have evolved under positive selection but it does not test for selection at specific sites. aBSREL requires no prior knowledge of the lineages of interest and an LRT was performed at each branch comparing the full model to a null model where branches do not have rate classes of ω > 1. First, we performed an exploratory analysis in which all branches of the phylogeny were tested. Then, we verified specific hypotheses, based on the results of the exploratory analysis. Branches of interest were selected and tested for positive selection. If more branches were tested simultaneously, the Holm–Bonferroni procedure implemented in the software was applied. Positive selection was reported if the LRT was significant (P < 0.05). A branch site model (BSM) test implemented in EasyCodeML was performed to verify signs of episodic selection previously observed by aBSREL analysis. The MLs obtained by the BSM, which allows positive selection along specified branches (Model A), were compared with those obtained by the null model (Model Anull), which allows neutral evolution and negative selection (Zhang et al., 2005). BSM allows us to identify signs of positive selection in each gene but no information is given for the single codons, and thus if the LRT yielded significant results, Bayesian and likelihood methods are then used to identify amino acid residues that have potentially evolved under selection. We performed the BEB and the fixed effects likelihood (FEL) analyses (Kosakovsky Pond and Frost, 2005; Yang et al., 2005) implemented respectively in the EasyCodeML and the DATAMONKEY web servers in order to identify the codons under positive selection. The BEB method calculating posterior probabilities for site classes is implemented for the branch-site model A. FEL fits an MG94xREV model to each codon site to infer site-specific non-synonymous and synonymous substitution rates. Positive selection of each codon estimated by BEB analysis was reported for P > 0.9 whereas for FEL analysis we used a P-value of 0.05 as a cut-off.
RELAX analysis
All branches that showed positive selection in aBSREL and BSM analysis were successively tested using the RELAX method (Wertheim et al., 2015) available at the DATAMONKEY web server. RELAX, which includes a selection intensity parameter (k), is a tool that shows whether a branch is under relaxed or intensified selective strength. RELAX requires the branch (tested branch) that will be compared against reference branches to be specified a priori. All branches not specified a priori as a tested branch were used as reference branches. A significant k > 1 shows that the selection strength has been intensified, whereas a significant k < 1 indicates that the strength of the selection has been relaxed.
Divergence time estimation
Correct estimation of the time of divergence during evolution is a prerequisite to know when the positive selection occurred along lineages. Since positive selection can affect the estimate of divergence between lineages, we chose to produce a chronogram based only on the plastid non-coding DNA regions. Divergence times between lineages were estimated using BEAST v.1.10.2 (Suchard et al., 2018) implemented on the CIPRES Science Gateway. The dataset used in molecular dating analysis consisted of 76 spacers and 19 introns, and a detailed description of DNA regions used is available in Zecca et al. (2020a). Due to the considerable computation time, we analysed the DNA regions selected without partitioning and a fixed ML topology was used (Supplementary Data Fig. S1). Five hundred independent ML searches were performed under the GTRGAMMA model of substitution, starting from 500 randomized maximum-parsimony trees in RaxML-HPC2 v.8.2.12 (Stamatakis, 2014) and executing 1000 rapid bootstrap replicates. Three independent MCMC runs were conducted in BEAST v.1.10.2 using the GTR+G+I model (nucleotide base frequencies are estimated empirically from the data) as estimated by the jModelTest2 (Darriba et al., 2012), the Fixed Local Clock (Yoder and Yang, 2000) and the Yule pure birth tree model (Udny Yule, 1925). Although the Palaeobiological Database (https://paleobiodb.org) contains several fossils attributed to the grape family, several records appear not to be suitable for our study because they belong to taxa with ambiguous phylogenetic positions or are uncertain in the classification (Manchester et al., 2013). In recent years some studies have focused on the estimation of divergence times using both fossils and local clock models (Koenen et al., 2021). Moreover, Zecca et al. (2020a) showed that evolutionary rates of non-coding DNA in grape plastomes vary across lineages, supporting the use of clock partitions on different clades. Thus, after a preliminary survey to compare the main calibration strategies applied in different papers, we chose to adopt fossil calibrations combined with five fixed local clock models as described in the calibration strategy in Supplementary Data Table S2. Bayesian analyses were run for 80–100 × 106 generations, with all parameters sampled every 50 000 generations. LogCombiner v.1.10 was used to combine the independent runs in a single trace after a 5 % burn-in. Stationarity of each chain was assessed using Tracer v.1.6 (http://beast.community/tracer) on the basis of the effective sample size (only ESS values >200 were accepted).
Investigating trait changes through time
Genes found to be under positive selection along specific branches (i.e. those identified by both aBSREL and BSM methods) were further analysed as discrete traits. For each gene, its character states were defined by the sequence of amino acids found corresponding to the codons that resulted under episodic positive selection in either BEB or FEL analysis. Character states were named using the appropriate combinations of the amino acid one-letter code and were assigned to the studied species. An example of character states encoding is given in Fig. 1 and the character data matrix used in the following analyses is available in Supplementary Data File S3. We applied the fitDiscrete function from the R package ‘geiger’ (Pennell et al., 2014; R Core Team, 2021) to fit the equal-rates (ER), the symmetrical (SYM) and the all-rates-different (ARD) likelihood models for discrete character evolution to the chronogram obtained from BEAST using the non-coding regions (hereafter called ‘reference tree’). Estimated models were ranked based on their Akaike weights computed using the function aicw. For each gene, the model that received the highest support was used to perform stochastic character mapping on the reference phylogenetic tree. When different models received identical support, the simplest was chosen. Simulated stochastic character maps were obtained using the make.simmap function from the R package ‘phytools’ (Revell, 2012), with the following additional settings: nsim = 1000, Q = ‘empirical’, pi = ‘estimated’. Sampled stochastic map character histories were used to generate ‘changes through time’ plots (‘phytools’ function ctt; segments = 45) showing the mean number of changes and the mean rate of changes per unit time. Four genes showed similar patterns in the mean rate of changes per unit time. Information from these genes was pulled together to derive a new combined (multistate) character, whose states were obtained by concatenating the character states found at the four genes. The combined character was further employed to perform stochastic character mapping analysis and to generate a new ‘changes through time’ plot, following the same procedure explained above. The transition matrix computed during stochastic mapping of the combined character and the reference tree was used to simulate trait changes through time under a null model of a constant rate of character evolution through time, using the function sim.multiCtt (nsim = 1000).
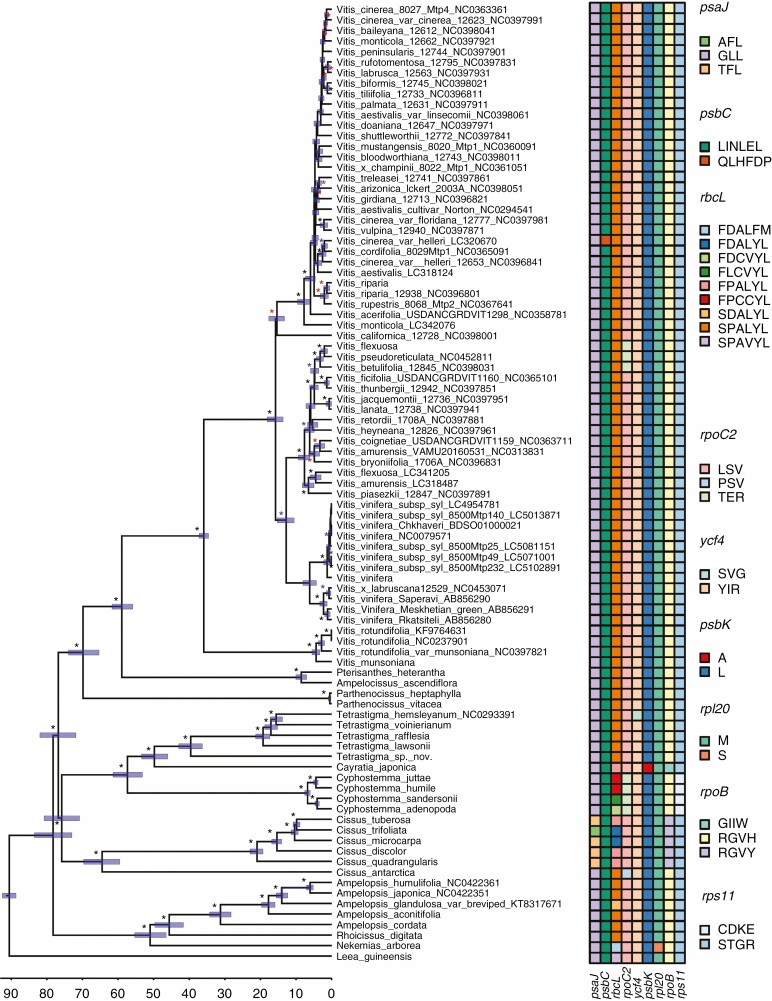
Fig. 1.
Divergence times of the main lineages of Vitaceae based on non-coding plastid DNA and estimated using BEAST 1.10.2. Black, blue and red asterisks represent bootstrap values >90, >75 and >50 respectively. The scale at the bottom of the figure is reported in millions of years before the present while blue bars represent the associated credibility interval (95 % HPD). The character data matrix drawn next to the tips of the tree shows the distribution of character states across the nine genes identified by both aBSREL and BSM methods (columns) and the species analysed (rows). For any given gene, the character states were defined by the sequence of amino acids found corresponding to the codons that were under episodic positive selection. Cells in the matrix were coloured based on the character states identified in each gene and character states were named using the appropriate combinations of the amino acid one-letter code. For example, in the case of the psaJ gene, three codons were under positive selection, resulting in three different character states: AFL (i.e. first codon: alanine; second codon: phenylalanine; third codon: leucine), GLL (i.e. first codon: glycine; second codon: leucine; third codon: leucine) and TFL (i.e. first codon: threonine; second codon: phenylalanine; third codon: leucine). In the case of a synonymous mutation, the same amino acid was used to encode character states, as in the case of the third codon of the psaJ gene. Physical positions of amino acids under selection relative to their alignment are given in Table 2. Gene names and colours used to represent different character states in each gene are given in the figure.
In addition, we applied the ‘phytools’ functions make.era.map and fitmultiMk to the combined character matrix to generate and fit models in which the rate of change of the composite trait varied between different times on the reference tree. Temporal boundaries were chosen based on: (1) the ‘Cretaceous–Palaeogene hypothesis’), and (2) the observed trend in rate variation over time as shown in the ‘changes through time’ plots, combined with an ML search procedure (see Results). The LRT was used to compare different heterogeneous rate models (i.e. multi-rate models) against the null model (i.e. constant rate along the tree).
Amino acid substitution rates across different lineages of Vitaceae
BEAST v.2.6.3 was used to explore the variation in substitution rates across the lineages of Vitaceae using the amino acidic alignment based on the nine proteins selected by aBSREL and BSM analyses. To reduce the calculation time, the nine proteins selected were analysed without partitioning. Three independent MCMC runs were conducted using the JTT+G+I model as estimated by Prottest 3.4 (Abascal et al., 2005), the Random Local Clock (Drummond and Suchard, 2010) and the Yule pure birth tree model (Udny Yule, 1925). The clock rate was set to 1 (substitutions/site/years) to estimate relative branch lengths. Bayesian analyses were run for 170–280 × 106 generations, with parameters sampled every 5000 generations and trees sampled every 20 000 generations.
RESULTS
Diversity analysis
A complete list of all 71 plastid genes analysed in this study, including total nucleotide sites (TS), variable sites (VS), parsimony-informative sites (P), nucleotide diversity (π) and haplotype diversity (HD), is given in Supplementary Data Table S1. Notably, the ycf1 gene showed the largest number of variable and parsimony-informative sites and the highest values of nucleotide and haplotype diversity among the plastid genes analysed (Table S1). Moreover, premature stop codons were found in V. mustagensis and V. acerifolia, suggesting a loss of gene functions in these species. Our findings are consistent with the idea that the ycf1 gene underwent a pseudogenization process in the family Vitaceae (albeit to varying degrees in different lineages). Consequently, we decided to exclude the ycf1 gene from the positive selection analyses.
Pervasive positive selection
Fourteen out of 70 plastid genes showed pervasive signs of positive selection according to the results of the SM test. PSGs and their relative ω values, together with amino acid residues under selection as identified by FUBAR and BEB analysis, are summarized in Table 1. Regardless of the codon-specific test used to perform the analysis, the rbcL gene showed the largest number of codons targeted by positive selection of all the PSGs previously identified by the SM test.
Table 1.
Summary of results of the SM test. BEB and FUBAR were applied to identify the codons targeted by positive selection. PSG = positively selected genes; ω = ratio of non-synonymous (Ka) to synonymous substitution rate (Ks); L = likelihood; M8 vs. M7 and M8a are the models tested as described in the Materials and Methods. Positive selection results were reported if the estimated ω was >1 and the likelihood ratio test was significant (P < 0.05). Positive selection of each codon estimated by BEB and FUBAR analysis is reported if P > 0.9. Position in the alignment, letter of amino acid residue and probability are reported for each codon selected
| PSG | Ln L(M8) | ω(M8) | Ln L(M7) | P (M7–M8) | BEB P > 0.9 | FUBAR P > 0.9 |
|---|---|---|---|---|---|---|
| Ln L(M8a) | P (M8a–M8) | |||||
| accD | −4465.1820 | 4.144 | −4480.7112 | 1.8 × 10−7 | 71 Q 0.99, 94 D 0.90, 104 V 0.96, 197 H 0.94, 208 G 0.96, 217 L 0.96, 323 P 0.92, 490 K 0.97 | 71 Q 0.98, 94 D 0.96, 283 L 96, 490 K 0.95 |
| −4478.1102 | 3.7 × 10−7 | |||||
| atpA | −3964.9188 | 51.867 | −4035.7022 | <10−10 | 488 T 0.99, 500 R 0.98, | 459 A 0.97, 465 L 0.97, 488 T 0.98, 500 R 0.95 |
| −4024.0715 | <10−10 | |||||
| matK | −5187.2953 | 3.723 | −5193.9524 | 1.2 × 10−3 | 118 A 0.91, 349 A 0.91 | 88 Y 0.91, 168 A 0.92, 349 A 0.95, 482 L 0.91 |
| −5174.2554 | 3.3 × 10−3 | |||||
| ndhH | −2876.0302 | 5.393 | −2886.9280 | 1.8 × 10−5 | 269 V 1.00 | 269 V 1.00 |
| −2879.5367 | 8.3 × 10−3 | |||||
| psaA | −4841.1805 | 2.513 | −4865.7854 | <10−10 | 19 I 0.94,152 N 0.98, 209 S 0.94, 626 L 0.96 | 19 I 0.93, 152 N 0.98, 209 S 0.98, 626 L 0.98 |
| −4844.5873 | 9.0 × 10−3 | |||||
| psaB | −4551.2542 | 1.365 | −4561.0552 | 5.5 × 10−5 | 273 I 0.92 | 273 I 0.93, 245 G 0.93 |
| −4547.3340 | 5.1 × 10−3 | |||||
| psaI | −363.8115 | 11.351 | −375.7080 | 6.8 × 10−6 | 3 T 0.90, 4 L 0.96, 7 P 0.99, 8 S 1.00, 10 L 1.00, 15 G 0.99 | 4 L 0.96 |
| −331.8516 | <10−9 | |||||
| psbA | −2111.5589 | 2.004 | −2128.1153 | 6.5 × 10−8 | 155 A 0.98, 351 I 0.92 | 155 A 0.99, 235 A 0.92 |
| −2105.3722 | 4.3 × 10−4 | |||||
| psbD | −1983.7730 | 3.569 | −1997.1252 | 1.6 × 10−6 | 146 A 0.97 | 146 A 0.97 |
| −1985.9045 | 3.9 × 10−2 | |||||
| psbK | −461.9339 | 2.725 | −466.2367 | 1.4 × 10−2 | 47 L 1.00 | 22 N 0.93 |
| −455.8277 | 4.7 × 10−4 | |||||
| rbcL | −4018.5596 | 4.215 | −4063.6618 | <10−10 | 28 E 0.98, 30 E 0.97, 33 P 1.00, 86 H 1.00, 91 A 0.99, 97 F 0.99, 140 I 0.95, 142 P 1.00, 145 S 1.00, 219 L 0.99, 225 I 0.99, 251 I 1.00, 255 V 1.00, 256 F 0.91, 279 S 1.00, 309 M 1.00, 353 F 0.92, 461 V 0.95, 470 P 1.00 | 10 S 0.98, 30 E 0.92, 33 P 0.93, 86 H 1.00, 91 A 0.99, 95 S 0.94, 140 I 0.94, 142 P 1.00, 143 A 0.97, 145 S 1.00, 219 L 0.99, 225 I 0.99, 251 I 0.95, 255 V 0.99, 256 F 0.95, 309 M 0.95, 320 L 0.91, 354 V 0.94, 461 V 0.98, 470 P 0.99 |
| −4089.8081 | <10−10 | |||||
| rpoA | −2567.0870 | 5.590 | −2572.4374 | 4.7 × 10−3 | 155 R 0.91, 249 S 0.97, 316 R 0.90 | 7 T 0.93, 105 G 0.94, 249 S 0.90, 272 L 0.91, 316 R 0.94 |
| −2556.2576 | 3.2 × 10−6 | |||||
| rpoC2 | −1521.3402 | 3.675 | −11529.2829 | 3.5 × 10−4 | 434 F 0.96, 507 L 0.93, 508 S 0.96, 509 V 0.93, 1008 A 0.91, 1370 M 0.91 | 434 F 0.99, 509 V 0.95, 758 K 0.91,766 L 0.91 |
| −11217.1235 | < 10−10 | |||||
| rps19 | −812.0943 | 11.835 | −817.2284 | 5.9 × 10−3 | 29 W 0.92 | 24 W 0.92, 29 W 0.95 |
| -806.7812 | 1.1 × 10−3 |
Episodic positive selection
The aBSREL model was used for the recognition of episodic diversifying selection in the phylogenetic trees and then confirmed using the BSM test. aBSREL found evidence of episodic positive selection in 21 genes (Supplementary Data Table S3). The psaJ, psbC, psbK, rbcL, rpl20, rpoB, rpoC2, rps11 and ycf4 genes were also confirmed by the BSM test and may be considered suitable candidates as being responsible for functional adaptations. Sites under selection identified by FEL and BEB analysis are summarized in Table 2. We used the RELAX method to test whether the positive selection is intensified or relaxed in all nine genes previously identified by the BSM test. The value of the K selection intensity parameter for each branch affected by episodic diversifying selection is reported in Table S4. The psbC, rpoB, rpoC2, rps11, ycf4 and rbcL genes showed intensification signals.
Table 2.
Summary of results of aBSREL and BSM analyses. BEB and FEL analyses were applied to identify the codons targeted by positive selection. PSG = positively selected genes; Branch = stem of the specified species or clade; likelihood ratio test (LRT) is reported if positive selection is significant (P < 0.05). Positive selection of each codon estimated by BEB and FEL analysis is reported if P > 0.9 and P < 0.05 respectively. Position in the alignment, letter of amino acid residue and probability are reported for each codon selected
| PSG | Branch | aBSREL | BSM | BEB P > 0.9 | FEL P < 0.05 | ||
|---|---|---|---|---|---|---|---|
| LRT | P | LRT | P | ||||
| psaJ |
Cissus tuberosa
Cissus microcarpa Cissus discolor Cissus trifoliata Cissus quadrangularis |
6.723 | 1.2 × 10−2 | 12.318 | 4.5 × 10−4 | 20 G 0.98, 32 L 0.92, 37 L 0.91 | 20 G 2.8 × 10−2 |
| psbC | Vitis cinerea var helleri | 19.395 | <10−5 | 6.170 | 1.3 × 10−2 | 42 L 1.00, 43 I 0.97, 44 N 0.97, 45 L 0.97, 78 E 0.96, 95 L 0.96 | 42 L < 10−4, 43 I 2.0 × 10−3, 44 N 1.0 × 10−2, 45 L 3.0 × 10−3, 78 E 5.0 × 10−3, 95 L 3.2 × 10−2 |
| psbK | Cayratia japonica | 10.439 | 1.2 × 10−3 | 12.708 | 3.6 × 10−4 | 7 L 0.98 | 7 L 1.0 × 10−3 |
| rbcL | Nekemias arborea | 16.437 | 1.0 × 10−4 | 8.705 | 3.1 × 10−3 | 371 L 0.95 | 10 S 1.0 × 10−3, 33 P 1.0 × 10−3, 363 Y 3.1 × 10−2, 371 L 9.0 × 10−3 |
|
Cyphostemma sandersonii
Cyphostemma adenopoda Cyphostemma juttae Cyphostemma humile |
20.947 | <10−5 | 5.919 | 1.5 × 10−2 | 99 A 0.925 | 99 A 2.0 × 10−2, 219 L 4.3 × 10−2 | |
| rpl20 | Nekemias arborea | 7.4669 | 8.4 × 10−3 | 6.635 | 1.0 × 10−2 | 110 M 0.96 | – |
| rpoB | Cayratia japonica | 12.196 | 8.0 × 10−4 | 14.880 | 1.1 × 10−4 | 5 G 0.93 | 3 R 2.0 × 10−3, 5 G 3.0 × 10−3, 145 V 1.8 × 10−2, 446 H 1.1 × 10−2 |
| rpoC2 |
Vitis betulifolia
Vitis pseudoreticulata Vitis flexuosa |
72.219 | <10−5 | 52.775 | <10−10 | 508 S 0.93 | 507 L < 10−4, 508 S < 10−4, 509 V < 10−4 |
| rps11 |
Cyphostemma sandersonii
Cyphostemma adenopoda Cyphostemma juttae Cyphostemma humile |
7.214 | 9.6 × 10−3 | 14.525 | 1.4 × 10−4 | 54 S 0.90, 74 T 0.99,77 G 0.99, 81 R 1.00 | 54 S 3.1 × 10−2, 74 T 2.2 × 10−2, 77 G 2.7 × 10−2, 81 R 8.0 × 10−3 |
| ycf4 | Tetrastigma hemsleyanum | 7.20 | 9.6 × 10−3 | 7.21 | 9.6 × 10−3 | 145 Y 0.99 | 145 Y 1.0 × 10−3, 148 I 2.2 × 10−2,149 R 2.9 × 10−2 |
Estimation of divergence times
Divergence time analysis was performed using non-coding DNA and the dataset shows a total of 43 894 nucleotides, 10 920 variable nucleotides and 5878 parsimony-informative sites. The 4560 trees obtained by three independent MCMC runs using BEAST v.1.10.2 were combined to produce the maximum clade credibility tree. Bayesian estimates of divergence times are presented in Fig. 1 and the phylogenetic trees obtained are available in Supplementary Data File S2. The clade that includes Ampelocissus–Pterisanthes–Vitis is estimated to have diverged from Parthenocissus in the late Cretaceous 69.85 Ma [95 % highest posterior density (HPD): 73.91–65.36 Ma) whereas the clade that includes Tetrastigma–Cayratia–Cyphostemma was estimated to have diverged from the Cissus clade at 75.80 Ma (95 % HPD: 80.77–70.82 Ma). Overall, our divergence time estimates of the major clades are close to those inferred by Wen et al. (2018) and Zecca et al. (2020a).
Changes through time plots and epoch model tests
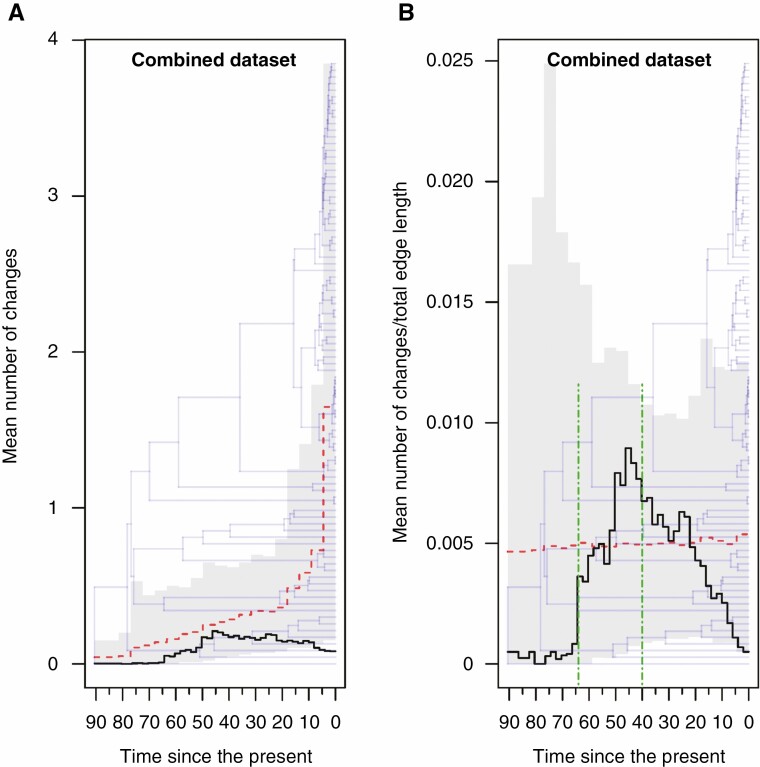
The ER model for discrete character evolution was selected for all genes except psaJ (SYM) and psbC (ARD). Changes through time plots generated from sampled stochastic character maps for each gene are shown in Supplementary Data Fig. S2 and Fig. 2. Four out of nine genes showed similar patterns in mean rate of change per unit time (psbK, rpl20, rpoB, rps11), whereas the remaining genes showed different trends. The ER model provided a better fit to the data even with the combined character. Figure 3 shows the simulated evolution of the combined character obtained by concatenating the character states found at the four genes, mapped along the branches of the reference chronogram. The ‘changes through time’ plots showing the sampled mean number of changes (left panel) and the sampled mean rate of change per unit time (right panel) for the combined character are shown in Fig. 4 together with the expected means and their expected 95 % distributions under the null model. Moving from the past to the present, the graph representing the rate variation over time shows a very low rate for more than 20 Myr, followed by a rapid increase in the rate of change, succeeded in turn by a prolonged decrease until recent times. We then formally tested whether epoch models that permit a heterogeneous rate of trait evolution between different time periods fit our data better than the single rate model. The Cretaceous–Palaeogene hypothesis suggests that events that mark the end of the Cretaceous Period and the beginning of the Palaeogene Period boosted adaptive evolution in different organisms during the first part of the Cenozoic Era. Accordingly, we fitted a two-epoch (i.e. two-rates) model setting the temporal boundary between epochs at 66 Ma. The LRT favoured the two-epoch model over the single rate model (d.f. = 1, χ2 = 3.8681, P = 0.0492). Our previous results based on stochastic mapping analysis seemed to point to a more complex pattern and therefore we fitted a three-epoch (i.e. three-rate) model assuming two epoch boundaries (i.e. two rate transition boundaries). As shown in Fig. 4, our results identified an abrupt increase in the rate of variation at 65–64 Ma, suggesting these dates as good candidates for the oldest transition time. The placement of the second boundary was less obvious. Therefore, to determine both epoch boundaries we applied the following procedure; the first-rate shift was fixed at either 65 or 64 Ma while the second shift was allowed to vary between 50 and 20 Ma with a step size of 1 Myr, resulting in a total of 62 different combinations of temporal boundaries. All generated models were fitted using the same procedure explained above and the ML model was taken as the final best model to test against the null model (Supplementary Data Fig. S3). At the end of the ML search procedure, the final ML model (log likelihood = −15.223) had transition times fixed at 64 and 40 Ma, respectively. Again, the LRT favoured the multi-regime model over the single rate model (d.f. = 2, χ2 = 6.4782, P = 0.0392).
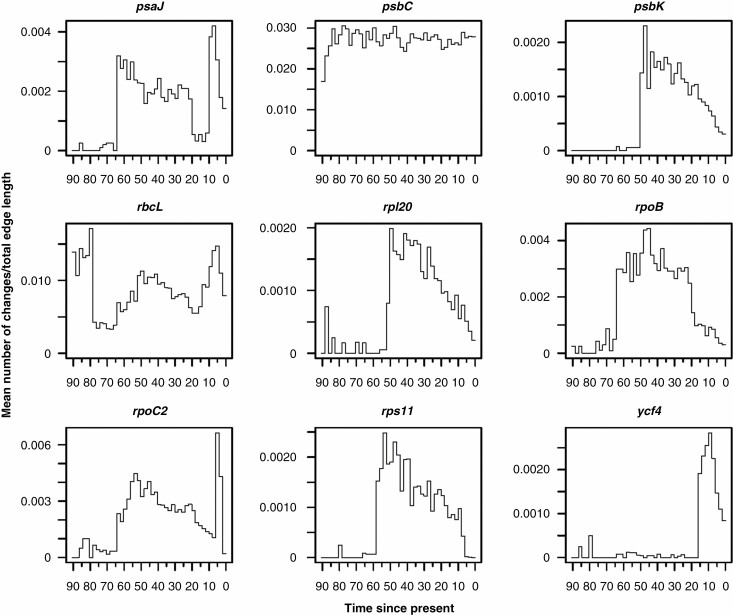
Fig. 2.
Changes through time plots generated from sampled stochastic character maps for the nine genes found to be under episodic positive selection by both aBSREL and BSM methods. For each gene the mean rate of changes per unit time is shown. Gene names are given above plots and time is expressed in millions of years since the present.
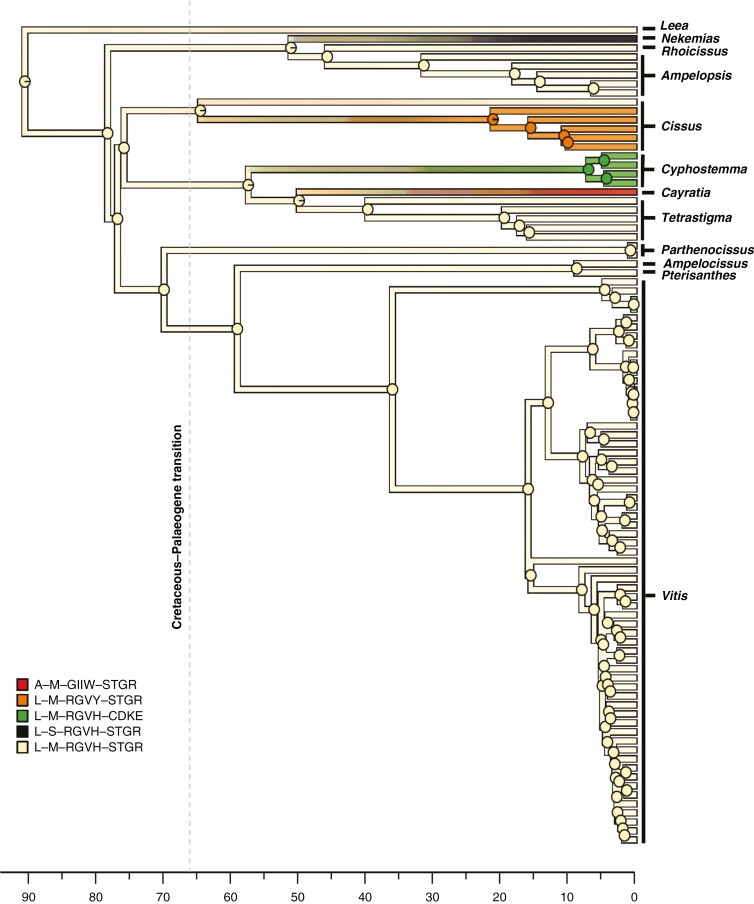
Fig. 3.
Simulated evolution of the multi-state discrete character obtained by combining data from four plastid genes (i.e. the combined dataset) mapped along the branches of the reference chronogram inferred by BEAST. State transitions along the tree edges and marginal posterior probabilities of ancestral states in internal nodes are based on stochastic mapping simulations. For the psbK, rpl20, rpoB and rps11 genes, the character states of each single gene were defined by the sequence of amino acids found corresponding to the codons that were under episodic positive selection and character states were named using the appropriate combinations of the amino acid one-letter code as explained in the main text and in Fig. 1. The combined character states were obtained by concatenating the character states found at the four genes. Concatenation was carried out in the following order: psbK–rpl20–rpoB–rps11. Overall, five combined states were found. Acronyms of the five combined states and colours used to represent them are given in the figure. The dashed vertical grey line represents the Cretaceous–Palaeogene transition at 66 Ma. Time is measured in millions of years since the present.
Fig. 4.
Changes through time plots generated from sampled stochastic character maps for the combined character (i.e.information from the psaK, rpl20, rpoB and rps11 genes). Time is measured in millions of years since the present. (A) The sampled mean number of changes per unit time (black line), the expected mean (red dashed line) and the expected 95 % distributions under the null model (grey shading) are superimposed on the reference chronogram obtained from BEAST. (B) The sampled mean rate of changes per unit time (black line), the expected mean (red dashed line) and the expected 95 % distributions under the null model (grey shading) are superimposed on the reference chronogram obtained from BEAST. The two epoch boundaries applied in the tested ML three-epochs model (see main text for details) are indicated by dot dash vertical green lines drawn at 64 and 40 Ma, respectively.
Substitution rate heterogeneity in Vitaceae
The amino acid matrix obtained from the nine PSGs identified in both the aBSREL and BSM tests shows 3983 residues, of which 584 were variable and 283 parsimony-informative. The inferred maximum clade credibility tree is shown in Supplementary Data Fig. S4 and in Fig. S5 with the median substitution rates and their 95 % HPD intervals displayed on branches. Our findings revealed a substantial variation in the relative substitution rates across different lineages, ranging from 0.11 in V. vinifera to 3.27 in Cissus trifloliata. Notably, Leea, Cyphostemma and Cissus lineages evolve faster than other lineages of Vitaceae.
DISCUSSION
To the best of our knowledge, in this paper we present the first broad analysis of positive selection across many lineages of the family Vitaceae using genome-wide plastome data. We integrated different methods to explore genomic data and this has proven to be a robust and effective approach to identify genes targeted by positive selection. Multiple analyses have shown that Vitaceae have experienced several positive selection events over 90 Myr of evolution and that the targeted genes are involved in essential functions such as photosynthesis, self-replication and metabolism. In particular, we found evidence of both pervasive and episodic positive selection. In the following, we discuss our outcome from this dual perspective and suggest their implications for future work.
Pervasive positive selection in the family Vitaceae
We identified 14 genes targeted by pervasive positive selection mainly located in the large single-copy region (Table 1). Eight genes are related to light-dependent proteins of the photosynthetic light reactions: psaA, psaB and psaI encoding photosystem I proteins; psbA, psbD and psbK encoding photosystem II proteins; and atpA encoding the ATP synthase and ndhH encoding the NADH-dehydrogenase genes. Three genes are related to proteins for transcription and post-transcriptional modifications: matK encoding the maturase K; and rpoA and rpoC2 encoding plastid polymerases. The remaining genes were rps19, which encodes a small ribosomal protein, accD involved in lipid acid synthesis and rbcL that encodes the large subunit of Rubisco related to photosynthetic dark reactions (Wicke et al., 2011).
Contrary to our results, the Ka/Ks ratio obtained by Raman and Park (2016) was <1 for all genes, except for rpl22 and rps19. The observed discrepancy between results can be explained as being due to the different number of accessions analysed in the two studies. In contrast to Raman and Park (2016), we examined a large dataset including different genera and species of Vitaceae. Sampling from many different lineages has been shown to improve the power and the accuracy of PSG prediction (Anisimova et al., 2002), producing more reliable results. In particular, we found widespread signs of positive selection in the rbcL gene (coding for the large subunit of Rubisco) in which 19 and 20 positively selected residues were detected by BEB and FUBAR analyses, respectively. Other authors have suggested that Rubisco could have evolved to improve its efficiency according to the availability of CO2 and that changes on some residues have effectively influenced the photosynthetic performance in some plants, facilitating adaptation and colonization of various habitats (Kapralov and Filatov, 2007; Hao et al., 2010; Galmes et al., 2014). In recent studies, using phylogenetic analysis at diverse taxonomic levels, different authors have shown that positive selection signs on particular amino acid residues were due to adaptation to dry or wet environments in Hawaiian endemic plants belonging to the genus Schiedea (Caryophyllaceae) and to aquatic or terrestrial habitats in the genus Potamogeton (Potamogetonaceae) (Kapralov and Filatov, 2006; Iida et al., 2009). Moreover, it has been shown that the adaptive evolution of Rubisco kinetics is linked to changes in intracellular concentrations of CO2 in both C3 and C4 plants (Kapralov and Filatov, 2007; Kapralov et al., 2011; Hermida-Carrera et al., 2020). Some of the amino acid residues of the rbcL gene found as the targets of positive selection in this study have been previously identified to have functional roles in different land plants. For example, site 225 (Leu/Ile) contributes to the linkage between large and small subunits of Rubisco (Makowski et al., 2008; Sen et al., 2011). It is known that the ancestral methionine at site 309 can be replaced in some plants by the smaller, more hydrophobic isoleucine which acts as a catalytic switch, making the Rubisco faster but less CO2-specific (Kapralov et al., 2011, 2012; Whitney et al., 2011; Galmes et al., 2014). Furthermore, four positively selected sites we identified (91, 142, 225 and 255) coincide with those reported in a large study that included several CAM species from the families Orchidaceae and Bromeliaceae (Hermida-Carrera et al., 2020). CAM is an adaptive mechanism found in drought-adapted plants in arid and semi-arid ecosystems that allows the efficiency of water and nutrient assimilation to be improved (Drennan and Nobel, 2000). Although most species are climbing, some genera of Vitaceae have adapted well to xerophytic habitats by modifying their habit, such as several African grapes that show erect growth, succulent stems and CAM metabolism (Chen, 2009). For example, the principal centres of diversification of Cyphostemma are in eastern Africa and Madagascar where arid conditions have dominated over the past 65 Myr (Hearn et al., 2018). Since several species, especially those included in Cissus and Cyphostemma, show phenotypes adapted to specific environments (De Santo et al., 1983; Olivares et al., 1984; Chen, 2009; Hearn et al., 2018), we suggest that continuous changes observed in the rbcL gene may have improved the catalytic performance of Rubisco activity in response to varying environmental conditions. However, a larger sampling of plants characterized by well-defined phenotypes (including CAM traits) is necessary to test specific hypotheses of adaptivity.
Recent episodic positive selection
Adaptive evolution has been reported to be generally more frequent in recent and rapid evolutionary radiations than in ancient lineages (Pease et al., 2016; Nevado et al., 2019). Nonetheless, in our survey we found several examples of both ancient and recent branches targeted by positive selection (Supplementary Data Table S3). We limit our discussion to those branches that were identified by both aBSREL and BSM methods (Table 2).
We found examples of recent branches targeted by positive selection in both American and Asian Vitis species (Table 2). Vitis cinerea var. helleri is endemic to central Texas and it is well adapted to the calcareous mountains of the Edwards Plateau. This agronomically important species has been widely used to produce rootstock given its tolerance to dryness, salinity and high limestone content in soils and due to its resistance to phylloxera (Pittcock, 1997). Our results showed six codons targeted by positive selection in the psbC gene, a fundamental component of the core complex of photosystem II. Vitis cinerea is a highly variable, probably not monophyletic taxon, consisting of at least four varieties distinguishable by several morphological traits (Moore et al., 2016; Wen et al., 2018; Zecca et al., 2020b). A recent anatomical study has shown a well-developed compact palisade layer on the leaves of V. cinerea var. helleri, typical of species adapted to high light environments (Ickert-Bond et al., 2018). The Edwards Plateau is an ecological region characterized by a transition zone from mesic to arid climates subject to periodic drought and erratic precipitations (Griffith et al., 2007). During the glacial phases of the Pleistocene, mean annual temperature in central Texas was below 5–10 °C and the Edwards Plateau was a mesic grassland with few trees. With increasing temperatures, the grassland transformed into open woodland dominated by several species of trees (Seersholm et al., 2020). Thus, adaptation to specific habitats may have driven the evolution of the psbC gene and positive diversifying selection may have affected the entire species or only local populations.
The stem of the clade consisting of V. betulifolia, V. pseudoreticulata and V. flexuosa also showed some codons of the rpoC2 gene under intensive positive selection (see Table 2). Molecular dating analysis suggests that this clade could have split in relatively recent times (Fig. 1). Vitis betulifolia is a wild species distributed in central China that can grow anywhere from forest to mountains up to 3600 m and it overlaps with the other two species in the regions of Henan, Hubei and Hunan (http://www.efloras.org/index.aspx; Weckerle et al., 2006; Tröndle et al., 2010). Vitis flexuosa has a large native range in tropical and temperate Asia and it is more tolerant to abiotic stress than cultivated grapes (Moon et al., 2017; Lee et al., 2021). Vitis pseudoreticulata is a wild grape growing in southern China characterized by high resistance to moisture, anthracnose and white rot (Ma et al., 2019). Previous studies have reported a positive selection for the rpoC2 gene in Oryza species (Poaceae) adapted to either shady or sunny environments (Gao et al., 2019b) and in the genus Cardamine (Brassicaceae), possibly because of adaptation to high-altitude environments (Hu et al., 2015). Similar adaptations to different light regimes and altitudes could also explain our findings in V. betulifolia, V. pseudoreticulata and V. flexuosa.
Interestingly, RELAX analysis identified both psbC and rpoC2 genes as subject to intensified selection along the branches leading to V. cinerea var. helleri and V. betulifolia, V. flexuosa and V. pseudoreticulata, respectively (Supplementary Data Table S4). These findings suggest that the environmental constraints that promoted adaptive evolution in these species may still be ongoing. Once phylogenetic approaches have identified positive selection, specific methods should be applied to further explore adaptive traits within populations (Rees et al., 2020). Events of rapid adaptive evolution may ease the colonization of a wide variety of environments and some authors have proposed that the introduction of novel and beneficial alleles also has repercussions for speciation events and endurance of taxa (Blanquart et al., 2013; Nevado et al., 2019). Therefore, investigating the local adaptation events at a population level can reveal important information about the function and origin of alleles. However, it has been shown that beneficial alleles can only partially arise from de novo mutations and that in certain circumstances they can be introduced by admixture with other species (Pease et al., 2016). As recent studies have shown that extensive admixture is more common than thought in ancient and modern Vitis species (Ma et al., 2018; Zecca et al., 2020b), further studies of V. cinerea var. helleri, V. betulifolia, V. flexuosa and V. pseudoreticulata populations are highly recommended. These investigations are of crucial importance especially for Chinese wild grapes, because although these species have enormous economic potential for breeding, there is still little information available about their distribution, evolution and agronomic traits (Wan et al., 2008; Zecca et al., 2012; Wang et al., 2014; Ma et al., 2018). Overall, these findings highlight how positive selection can affect biodiversity and the survival capacity of species, as well as having important repercussions for the selection of agronomic traits.
Ancient episodic positive selection and the Cretaceous–Palaeogene transition hypothesis
Adaptive evolution has certainly contributed to shaping the past and the current diversity at the level of species, genomes and genes. However, signs of this are not always obvious. Probably the best-known example is related to the mass extinction that occurred about 66 Ma. In the wake of the K/Pg mass extinction, many surviving organisms began to diversify into the niches formerly occupied by other species. The intense environmental perturbations following the Cretaceous–Palaeogene transition further favoured periods of rapid diversification. Over time, the combination of new niches available for colonization, changing environments, new emerging traits and natural selection has promoted adaptive radiations in different taxa. Plants partially diverge from this general context because the K/Pg mass extinction is not believed to have been the main cause of plant extinction (Koenen et al., 2021). Nonetheless, the Cretaceous–Palaeogene transition may still have influenced the evolution and the diversification of several plant lineages, increasing origination rates in angiosperms and contributing to their current ecological and evolutionary dominance (Silvestro et al., 2015; Cui et al., 2019).
Here we focused our attention on the gene level by analysing variations over time of the nine plastid genes that we found to be under positive selection in both aBSREL and BSM analyses (Table 2). Our general expectation was to observe different patterns of variation for different genes. This was partially met because five of the nine genes analysed showed distinct trends (Supplementary Data Fig. S2 and Fig. 2). Contrary to our expectation, however, the psbK, rpl20, rpoB and rps11 genes presented similar patterns of variation over time, showing a rapid acceleration in the mean rate of change during the first part of the Palaeogene, followed by a prolonged slowdown until recent times (Fig. 2). The same signal was also seen when these genes were combined in a new multistate character (Fig. 4, right panel) showing a sudden increase in the mean rate of change at about 65–64 Ma.
Our findings show a certain compatibility with the ‘Cretaceous–Palaeogene hypothesis’ and allow us to confirm the occurrence of heterogeneous rates over time. Changing environments may have promoted variation, and new favourable mutations arising on the psbK, rpl20, rpoB and rps11 genes may have been fixed by natural selection over the course of many generations. Although the two-epoch model identified 66 Ma (i.e. the estimated age of the Cretaceous–Palaeogene transition) as a significant time boundary between different rate regimes, it seems reasonable that the process actually started later than 66 Ma and that it took a certain lapse of time to leave a detectable imprint on genes (Fig. 3). Thus, it is not surprising that the three-epoch model identified 64 Ma as the most likely date for the first sign of a steep increase in the mean rate of change. The second shift was instead placed at 40 Ma, far in time from the first-rate transition. It is worthwhile noting here that the acceleration in the rates of change persisted for millions of years before slowing down definitively. Therefore, even though the Cretaceous–Palaeogene upheavals offered the trigger for this steep increase in variation, it is plausible that other subsequent historical events and evolutionary constraints helped keep this process active on these genes for millions of years. Remarkable, between the Palaeocene–Eocene Thermal Maximum (56 Ma) and the end of the Early Eocene Climate Optimum (EECO, 47 Ma) a pronounced warming trend took place, reaching temperatures that were more than 10 °C higher than they are today (Westerhold et al., 2020). The EECO was followed by a 13-Myr-long trend toward cooler conditions interspersed with transient warming events including the Middle Eocene Climatic Optimum (~40 Ma), after which the decrease in temperature became more marked culminating in the Eocene–Oligocene Transition (EOT, 34 Ma) (Westerhold et al., 2020). These climatic trends fit well with the observed pattern in rate variations, but testing the impact of these climate variations is beyond the scope of this paper. Further studies will help to clarify this issue.
Overall, our results are consistent with the idea that the events linked to the dramatic K/Pg mass extinction favoured an increased rate of change in some plastid genes that were subsequently targeted by adaptive selection in several genera of the family Vitaceae, such as Cayratia, Cyphostemma and Nekemias (Table 2). Interestingly, albeit with different general patterns, the psaJ and rpoC2 genes and partially the rbcL gene also showed an acceleration in the rate of change across the Cretaceous–Palaeogene transition (Fig. 2), suggesting that other genes could have shared a similar fate in additional lineages of Vitaceae. Finally, it is important to note that other more recent events have also certainly contributed to shaping the evolutionary history of Vitaceae. The sudden increase in the rate of change followed by a rapid decline shown by the psaJ, rpoC2, rbcL and ycf4 genes in the last few million years could suggest further episodes of adaptive evolution.
From the latter part of the Oligocene (~26–27 Ma) a global warming trend began (Zachos et al., 2001; Westerhold et al., 2020) which peaked in the Miocene Climatic Optimum (MCO, ~17–14 Ma) (Flower and Kennett 1995; Westerhold et al. 2020). This general trend was interrupted by some brief periods of glaciation, characterized by abrupt changes in temperature, which contrast with the background trend and that were usually accompanied by a major perturbation in the global carbon cycle (Zachos et al., 2001). In particular, a brief but deep (~200 kyr) glacial maximum occurred at ~20 Ma (Paul et al., 2000). This event, referred to as Mi-1, was followed by a series of intermittent but smaller glaciations and was accompanied by accelerated rates of turnover and speciation in certain groups of biotas (Zachos et al., 2001). The MCO was followed by a cooling phase interspersed with short warmer phases, such as the Tortonian Thermal Maximum and the mid-Pliocene Warm Period, until the establishment of a major glacial event at 3.3 Ma (mid-Pliocene M2 glacial; Westerhold et al., 2020). All these global climatic fluctuations might be related to the observed spike in the rate of change at 20–5 Ma in the psaJ, rpoC2, rbcL and ycf4 genes. Nonetheless, we cannot exclude that local conditions may have played a crucial role as well. Thus, further studies will be needed to clarify the underlying causes involved.
Substitution rates, positive selection and phylogeny in Vitaceae
We observed accelerated relative substation rates in Cissus, Cyphostemma and Leea lineages when the nine proteins positively selected by both aBSREL and BSM tests were analysed (Supplementary Data Figs S4 and S5). Increases in substitution rates might have triggered the ability of ancestral lineages to adapt, also influencing their descendants. For example, high substitution rates are often associated with high temperatures and some authors have proposed that the environmental energy has driven evolutionary rates and the diversification in flowering plants (Davies et al., 2004; Wright et al., 2006). We suggest that accelerated evolutionary diversification in some photosynthetic genes may have facilitated the adaptation of certain lineages of Vitaceae to specific habitats, as previously proposed for other plants (Kapralov and Filatov, 2006; Iida et al., 2009). High rates in Cissus, Cyphostemma and Leea lineages were also found by Zecca et al. (2020a), exploring the non-coding regions of plastomes. The authors observed that there exists heterogeneity among lineages and that different habits are probably linked to different substitution rates. Together, all these findings provide the basis for future studies aimed at understanding what factors have driven the evolution of plastomes in Vitaceae. We cannot state here whether the forces that acted on genes were the same as acted on spacers and introns. However, recent studies have shown that non-coding DNA regions can also be subject to significant levels of positive selection (Williamson et al., 2014).
The fact that external forces can influence the evolution of genes and non-coding DNA can have implications in the field of species delimitation/identification. High levels of positive selection may affect the reconstructive ability of phylogenetic methods and their ability to estimate divergence times between lineages correctly. Normally, positive selection should be tested beforehand, and positively selected markers should be avoided in phylogenetic studies. We recommend that at least codons evolving under positive selection should be excluded to improve the analysis when coding DNA is used. Our work offers a first step in this direction, providing useful information on positively selected plastid genes and codons. Methods also exist to detect selection on non-coding DNA, albeit they are not widely used because of their many limitations and difficulties in formulating appropriate and powerful models (Zhen and Andolfatto, 2012). Pending the development of new methods, we recommend using a large number of non-coding markers to reduce the impact of the fraction of functional non-coding DNA.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1. List of plastid genes analysed. Table S2. Calibration strategy. Table S3. Summary of results obtained by aBSREL analysis. Table S4. Genes under relaxed or intensified selective strength. Figure S1. Best maximum likelihood tree. Figure S2. Changes through time plots. Figure S3. Log-likelihood values. Figures S4 and S5. Maximum clade credibility trees. File S1. All genes analysed in this work. File S2. All trees obtained in this work, reported in Newick or Nexus format. File S3. Character states encoding.
Contributor Information
Giovanni Zecca, University of Milan-Bicocca, Department of Biotechnology and Bioscience, Piazza della Scienza 2, 20126, Milano, Italy.
Davide Panzeri, University of Milan-Bicocca, Department of Biotechnology and Bioscience, Piazza della Scienza 2, 20126, Milano, Italy.
Fabrizio Grassi, University of Milan-Bicocca, Department of Biotechnology and Bioscience, Piazza della Scienza 2, 20126, Milano, Italy; NBFC, National Biodiversity Future Center, Palermo 90133, Italy.
LITERATURE CITED
- Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Anderson JT, Song B-H, Song B-H. 2020. Plant adaptation to climate change—Where are we? Journal of Systematics and Evolution 58: 533–545. doi: 10.1111/jse.12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M, Bielawski JP, Yang Z. 2002. Accuracy and power of Bayes prediction of amino acid sites under positive selection. Molecular Biology and Evolution 19: 950–958. doi: 10.1093/oxfordjournals.molbev.a004152. [DOI] [PubMed] [Google Scholar]
- Blanquart F, Kaltz O, Nuismer SL, Gandon S. 2013. A practical guide to measuring local adaptation. Ecology Letters 16: 1195–1205. doi: 10.1111/ele.12150. [DOI] [PubMed] [Google Scholar]
- Bock DG, Andrew RL, Rieseberg LH. 2014. On the adaptive value of cytoplasmic genomes in plants. Molecular Ecology 23: 4899–4911. doi: 10.1111/mec.12920. [DOI] [PubMed] [Google Scholar]
- Cai L, Xi Z, Amorim AM, et al. 2019. Widespread ancient whole-genome duplications in Malpighiales coincide with Eocene global climatic upheaval. New Phytologist 221: 565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IJU. 2009. History of Vitaceae inferred from morphology-based phylogeny and the fossil record of seeds. Dissertation. University of Florida. [Google Scholar]
- Chiarenza AA, Farnsworth A, Mannion PD, et al. 2020. Asteroid impact, not volcanism, caused the end-Cretaceous dinosaur extinction. Proceedings of the National Academy of Sciences 117: 17084–17093. doi: 10.1073/pnas.2006087117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y-M, Wang W, Ferguson DK, Yang J, Wang Y-F. 2019. Fossil evidence reveals how plants responded to cooling during the Cretaceous–Paleogene transition. BMC Plant Biology 19: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772–772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies TJ, Savolainen V, Chase MW, Moat J, Barraclough TG. 2004. Environmental energy and evolutionary rates in flowering plants. Proceedings of the Royal Society of London. Series B: Biological Sciences 271: 2195–2200. doi: 10.1098/rspb.2004.2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santo AV, Alfani A, Russo G, Fioretto A. 1983. Relationship between CAM and succulence in some species of Vitaceae and Piperaceae. Botanical Gazette 144: 342–346. doi: 10.1086/337382. [DOI] [Google Scholar]
- Dobzhansky T. 1973. Nothing in biology makes sense except in the light of evolution. The American Biology Teacher 35: 125–129. doi: 10.2307/4444260. [DOI] [Google Scholar]
- Doyle JJ. 1992. Gene trees and species trees: molecular systematics as one-character taxonomy. Systematic Botany 17: 144–163. [Google Scholar]
- Drennan PM, Nobel PS. 2000. Responses of CAM species to increasing atmospheric CO2 concentrations. Plant, Cell & Environment 23: 767–781. [Google Scholar]
- Drummond AJ, Suchard MA. 2010. Bayesian random local clocks, or one rate to rule them all. BMC Biology 8: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett JA, Maere S, Van De Peer Y. 2009. Plants with double genomes might have had a better chance to survive the Cretaceous–Tertiary extinction event. Proceedings of the National Academy of Sciences 106: 5737–5742. doi: 10.1073/pnas.0900906106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower BP, Kennett JP, Kennett JP. 1995. Middle Miocene deepwater paleoceanography in the southwest Pacific: Relations with East Antarctic Ice Sheet development. Paleoceanography 10: 1095–1112. doi: 10.1029/95pa02022. [DOI] [Google Scholar]
- Galmes J, Andralojc PJ, Kapralov MV, et al. 2014. Environmentally driven evolution of Rubisco and improved photosynthesis and growth within the C 3 genus Limonium (Plumbaginaceae). New Phytologist 203: 989–999. doi: 10.1111/nph.12858. [DOI] [PubMed] [Google Scholar]
- Gao F, Chen C, Arab DA, Du Z, He Y, Ho SY. 2019a. EasyCodeML: A visual tool for analysis of selection using CodeML. Ecology and Evolution 9: 3891–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao LZ, Liu YL, Zhang D, et al. 2019b. Evolution of Oryza chloroplast genomes promoted adaptation to diverse ecological habitats. Communications Biology 2: 278. doi: 10.1038/s42003-019-0531-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrath JM, Posluszny U, Posluszny U. 1988. Morphological and anatomical development in the Vitaceae. I. Vegetative development in Vitis riparia. Canadian Journal of Botany 66: 209–224. doi: 10.1139/b88-037. [DOI] [Google Scholar]
- Gonçalves DJ, Simpson BB, Ortiz EM, Shimizu GH, Jansen RK. 2019. Incongruence between gene trees and species trees and phylogenetic signal variation in plastid genes. Molecular Phylogenetics and Evolution 138: 219–232. doi: 10.1016/j.ympev.2019.05.022. [DOI] [PubMed] [Google Scholar]
- Grassi FD, De Lorenzis G. 2021. Back to the origins: background and perspectives of grapevine domestication. International Journal of Molecular Sciences 22: 4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith G, Bryce S, Omernik J, Rogers A. 2007. Ecoregions of Texas. Austin, TX:Texas Commission on Environmental Quality. [Google Scholar]
- Hao DC, Mu J, Xiao PG. 2010. Molecular evolution and positive Darwinian selection of the gymnosperm photosynthetic Rubisco enzyme. Botanical Studies 51: 491–510. [Google Scholar]
- Hearn DJ, Evans M, Wolf B, McGinty M, Wen J. 2018. Dispersal is associated with morphological innovation, but not increased diversification, in Cyphostemma (Vitaceae). Journal of Systematics and Evolution 56: 340–359. doi: 10.1111/jse.12417. [DOI] [Google Scholar]
- Hermida-Carrera C, Fares MA, Font-Carrascosa M, et al. 2020. Exploring molecular evolution of rubisco in C 3 and CAM Orchidaceae and Bromeliaceae. BMC Evolutionary Biology 20: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyduk K, Moreno-Villena JJ, Gilman I, et al. 2019. The genetics of convergent evolution: insights from plant photosynthesis. Nature Reviews Genetics 20: 485–493. [DOI] [PubMed] [Google Scholar]
- Hu S, Sablok G, Wang B, et al. 2015. Plastome organization and evolution of chloroplast genes in Cardamine species adapted to contrasting habitats. BMC Genomics 16: 306. doi: 10.1186/s12864-015-1498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ickert-Bond SM, Harris AJ, Lutz S, Wen J. 2018. A detailed study of leaf micromorphology and anatomy of New World Vitis L. subgenus Vitis within a phylogenetic and ecological framework reveals evolutionary convergence. Journal of Systematics and Evolution 56: 309–330. [Google Scholar]
- Iida S, Miyagi A, Aoki S, Ito M, Kadono Y, Kosuge K. 2009. Molecular adaptation of rbcL in the heterophyllous aquatic plant Potamogeton. PLoS One 4: e4633. doi: 10.1371/journal.pone.0004633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapralov MV, Filatov DA, Filatov DA. 2006. Molecular adaptation during adaptive radiation in the Hawaiian endemic genus Schiedea. PLoS One 1: e8. doi: 10.1371/journal.pone.0000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapralov MV, Filatov DA, Filatov DA. 2007. Widespread positive selection in the photosynthetic Rubisco enzyme. BMC Evolutionary Biology 7: 73. doi: 10.1186/1471-2148-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapralov MV, Kubien DS, Andersson I, Filatov DA. 2011. Changes in Rubisco kinetics during the evolution of C4 photosynthesis in Flaveria (Asteraceae) are associated with positive selection on genes encoding the enzyme. Molecular Biology and Evolution 28: 1491–1503. doi: 10.1093/molbev/msq335. [DOI] [PubMed] [Google Scholar]
- Kapralov MV, Smith JAC, Filatov DA. 2012. Rubisco evolution in C4 eudicots: an analysis of Amaranthaceae sensu lato. PLoS One 7: e52974. doi: 10.1371/journal.pone.0052974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. 1968. Evolutionary rate at the molecular level. Nature 217: 624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- Koenen EJ, Ojeda DI, Bakker FT, et al. 2021. The origin of the legumes is a complex paleopolyploid phylogenomic tangle closely associated with the Cretaceous–Paleogene (K–Pg) mass extinction event. Systematic Biology 70: 508–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW, Frost SD. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Molecular Biology and Evolution 22: 1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Lee JH, Kim SA, Ahn SY, Yun HK. 2021. Heat shock transcriptional factor genes (VfHSFs) of Vitis flexuosa respond differentially to high temperature in grapevines. Horticulture, Environment and Biotechnology 62: 87–97. [Google Scholar]
- Lekshmi RK, Rajesh R, Mini S. 2015. Ethyl acetate fraction of Cissus quadrangularis stem ameliorates hyperglycaemia-mediated oxidative stress and suppresses inflammatory response in nicotinamide/streptozotocin induced type 2 diabetic rats. Phytomedicine 22: 952–960. doi: 10.1016/j.phymed.2015.06.014. [DOI] [PubMed] [Google Scholar]
- Liu XQ, Ickert-Bond SM, Chen LQ, Wen J. 2013. Molecular phylogeny of Cissus L. of Vitaceae (the grape family) and evolution of its pantropical intercontinental disjunctions. Molecular Phylogenetics and Evolution 66: 43–53. doi: 10.1016/j.ympev.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Liu XQ, Ickert-Bond SM, Nie ZL, Zhou Z, Chen LQ, Wen J. 2016. Phylogeny of the Ampelocissus–Vitis clade in Vitaceae supports the New World origin of the grape genus. Molecular Phylogenetics and Evolution 95: 217–228. doi: 10.1016/j.ympev.2015.10.013. [DOI] [PubMed] [Google Scholar]
- Lu L, Cox CJ, Mathews S, Wang W, Wen J, Chen Z. 2018. Optimal data partitioning, multispecies coalescent and Bayesian concordance analyses resolve early divergences of the grape family (Vitaceae). Cladistics 34: 57–77. doi: 10.1111/cla.12191. [DOI] [PubMed] [Google Scholar]
- Ma ZY, Wen J, Ickert-Bond SM, Chen LQ, Liu XQ. 2016. Morphology, structure, and ontogeny of trichomes of the grape genus (Vitis, Vitaceae). Frontiers in Plant Science 7: 704. doi: 10.3389/fpls.2016.00704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma ZY, Wen J, Tian J-P, Jamal A, Chen LQ, Liu XQ. 2018. Testing reticulate evolution of four Vitis species from East Asia using restriction-site associated DNA sequencing. Journal of Systematics and Evolution 56: 331–339. doi: 10.1111/jse.12444. [DOI] [Google Scholar]
- Ma C, Fu P, Wang L, et al. 2019. The complete chloroplast genome sequence of Vitis pseudoreticulata. Mitochondrial DNA Part B 4: 3630–3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magallón S, Castillo A, Castillo A. 2009. Angiosperm diversification through time. American Journal of Botany 96: 349–365. doi: 10.3732/ajb.0800060. [DOI] [PubMed] [Google Scholar]
- Makowski M, Sobolewski E, Czaplewski C, O\ldziej S, Liwo A, Scheraga HA. 2008. Simple physics-based analytical formulas for the potentials of mean force for the interaction of amino acid side chains in water. IV. Pairs of different hydrophobic side chains. The Journal of Physical Chemistry. B 112: 11385–11395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchester SR, Kapgate DK, Wen J. 2013. Oldest fruits of the grape family (Vitaceae) from the Late Cretaceous Deccan Cherts of India. American Journal of Botany 100: 1849–1859. doi: 10.3732/ajb.1300008. [DOI] [PubMed] [Google Scholar]
- Méndez-López LF, Garza-González E, Ríos MY, et al. 2020. Metabolic profile and evaluation of biological activities of extracts from the stems of Cissus trifoliata. International Journal of Molecular Sciences 21: 930. doi: 10.3390/ijms21030930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M, Pfeiffer WT, Schwartz T. 2010. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In: Gateway Computing Environments Workshop (GCE), New Orleands, LA, 1–8. doi: 10.1109/GCE.2010.5676129 [DOI] [Google Scholar]
- Moon JS, Hur YY, Jung S-M, et al. 2017. Transcript profiling of native Korean grapevine species Vitis flexuosa exposed to dehydration and rehydration treatment. Horticulture, Environment and Biotechnology 58: 66–77. doi: 10.1007/s13580-017-0064-x. [DOI] [Google Scholar]
- Moore MO, Wen J; Flora of North America Editorial Committee. 2016. Vitaceae. Flora of North America North of Mexico 12: 3–13. [Google Scholar]
- Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. 2012. Detecting individual sites subject to episodic diversifying selection. PLoS Genetics 8: e1002764. doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B, Moola S, Mabona A, et al. 2013. FUBAR: a fast, unconstrained Bayesian approximation for inferring selection. Molecular Biology and Evolution 30: 1196–1205. doi: 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. Oxford: Oxford university press. [Google Scholar]
- Nevado B, Wong EL, Osborne OG, Filatov DA. 2019. Adaptive evolution is common in rapid evolutionary radiations. Current Biology 29: 3081–3086.e5. doi: 10.1016/j.cub.2019.07.059. [DOI] [PubMed] [Google Scholar]
- Nielsen R. 2005. Molecular signatures of natural selection. Annual Review Genetics 39: 197–218. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Yang Z, Yang Z. 1998. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148: 929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares E, Urich R, Montes G, Coronel I, Herrera A. 1984. Occurrence of Crassulacean acid metabolism in Cissus trifoliata L. (Vitaceae). Oecologia 61: 358–362. doi: 10.1007/BF00379635. [DOI] [PubMed] [Google Scholar]
- Paul HA, Zachos JC, Flower BP, Tripati A. 2000. Orbitally induced climate and geochemical variability across the Oligocene/Miocene boundary. Paleoceanography 15: 471–485. doi: 10.1029/1999pa000443. [DOI] [Google Scholar]
- Pease JB, Haak DC, Hahn MW, Moyle LC. 2016. Phylogenomics reveals three sources of adaptive variation during a rapid radiation. PLoS Biology 14: e1002379. doi: 10.1371/journal.pbio.1002379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell MW, Eastman JM, Slater GJ, et al. 2014. geiger v2. 0: an expanded suite of methods for fitting macroevolutionary models to phylogenetic trees. Bioinformatics 30: 2216–2218. doi: 10.1093/bioinformatics/btu181. [DOI] [PubMed] [Google Scholar]
- Piot A, Hackel J, Christin P-A, Besnard G. 2018. One-third of the plastid genes evolved under positive selection in PACMAD grasses. Planta 247: 255–266. doi: 10.1007/s00425-017-2781-x. [DOI] [PubMed] [Google Scholar]
- Pittcock JK. 1997. Molecular marker characterization of selected grape species found in Texas and New Mexico. Dissertation. Texas Tech University. [Google Scholar]
- Pond SLK, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies In: Nielsen R (ed) Statistical methods in molecular evolution. Berlin: Springer, 125–181. [Google Scholar]
- R Core Team. 2021. R: A language and environment for statistical computing. Vienna:R Foundation for Statistical Computing. https://www.R-project.org/. [Google Scholar]
- Raman G, Park SJ, Park S. 2016. The complete chloroplast genome sequence of Ampelopsis: gene organization, comparative analysis, and phylogenetic relationships to other angiosperms. Frontiers in Plant Science 7: 341. doi: 10.3389/fpls.2016.00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees JS, Castellano S, Andrés AM. 2020. The genomics of human local adaptation. Trends in Genetics 36: 415–428. doi: 10.1016/j.tig.2020.03.006. [DOI] [PubMed] [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution 3: 217–223. [Google Scholar]
- Sabater B. 2018. Evolution and function of the chloroplast. Current investigations and perspectives. International Journal of Molecular Sciences 19: 3095. doi: 10.3390/ijms19103095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seersholm FV, Werndly DJ, Grealy A, et al. 2020. Rapid range shifts and megafaunal extinctions associated with late Pleistocene climate change. Nature Communication 11: 2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen L, Fares MA, Liang B, et al. 2011. Molecular evolution of rbcL in three gymnosperm families: identifying adaptive and coevolutionary patterns. Biology Direct 6: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestro D, Cascales-Miñana B, Bacon CD, Antonelli A. 2015. Revisiting the origin and diversification of vascular plants through a comprehensive Bayesian analysis of the fossil record. New Phytologist 207: 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD, Wertheim JO, Weaver S, Murrell B, Scheffler K, Kosakovsky Pond SL. 2015. Less is more: an adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Molecular Biology and Evolution 32: 1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE. 2016. Ancient WGD events as drivers of key innovations in angiosperms. Current Opinion in Plant Biology 30: 159–165. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE. 2021. Plant genomes: Markers of evolutionary history and drivers of evolutionary change. Plants, People, Planet, 3: 74–82. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. 2018. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evolution 4: vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres T, Saiko P, Fritzer-Szekeres M, Djavan B, Jäger W. 2011. Chemopreventive effects of resveratrol and resveratrol derivatives. Annals of the New York Academy of Sciences 1215: 89–95. [DOI] [PubMed] [Google Scholar]
- Tröndle D, Schröder S, Kassemeyer H-H, Kiefer C, Koch MA, Nick P. 2010. Molecular phylogeny of the genus Vitis (Vitaceae) based on plastid markers. American Journal of Botany 97: 1168–1178. [DOI] [PubMed] [Google Scholar]
- Udny Yule G. 1925. A mathematical theory of evolution, based on the conclusions of Dr. JC Willis, FRS. Philosophical Transactions of the Royal Society of London, Series B 213: 21–87. [Google Scholar]
- Van de Peer Y, Mizrachi E, Marchal K. 2017. The evolutionary significance of polyploidy. Nature Reviews Genetics 18: 411–424. [DOI] [PubMed] [Google Scholar]
- Vanneste K, Baele G, Maere S, Van de Peer Y. 2014. Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous–Paleogene boundary. Genome Research 24: 1334–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellekoop J, Sluijs A, Smit J, et al. 2014. Rapid short-term cooling following the Chicxulub impact at the Cretaceous–Paleogene boundary. Proceedings of the National Academy of Sciences 111: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JD, Geissman JW, Bowring SA, Babcock LE, compilers. 2018. Geologic Time Scale v. 5.0. Boulder: Geological Society of America, doi: 10.1130/2018.CTS005R3C. [DOI] [Google Scholar]
- Wan Y, Schwaninger H, Li D, Simon CJ, Wang Y, He P. 2008. The eco-geographic distribution of wild grape germplasm in China. Vitis-Geilweilerhof 47: 77. [Google Scholar]
- Wang L, Wei J, Zou Y, et al. 2014. Molecular characteristics and biochemical functions of VpPR10s from Vitis pseudoreticulata associated with biotic and abiotic stresses. International Journal of Molecular Sciences 15: 19162–19182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S, Shank SD, Spielman SJ, Li M, Muse SV, Kosakovsky Pond SL. 2018. Datamonkey 2.0: a modern web application for characterizing selective and other evolutionary processes. Molecular Biology and Evolution 35: 773–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckerle CS, Huber FK, Yongping Y, Weibang S. 2006. Plant knowledge of the Shuhi in the Hengduan Mountains, southwest China. Economic Botany 60: 3–23. [Google Scholar]
- Wen J, Harris AJ, Kalburgi Y, et al. 2018. Chloroplast phylogenomics of the New World grape species (Vitis, Vitaceae). Journal of Systematics and Evolution 56: 297–308. [Google Scholar]
- Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K. 2015. RELAX: detecting relaxed selection in a phylogenetic framework. Molecular Biology and Evolution 32: 820–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerhold T, Marwan N, Drury AJ, et al. 2020. An astronomically dated record of Earth’s climate and its predictability over the last 66 million years. Science 369: 1383–1387. [DOI] [PubMed] [Google Scholar]
- Wicke S, Schneeweiss GM, dePamphilis CW, et al. 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Molecular Biology 76: 273–297. doi: 10.1007/s11103-011-9762-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Sharwood RE, Orr D, White SJ, Alonso H, Galmés J. 2011. Isoleucine 309 acts as a C4 catalytic switch that increases ribulose-1, 5-bisphosphate carboxylase/oxygenase (rubisco) carboxylation rate in Flaveria. Proceedings of the National Academy of Sciences 108: 14688–14693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson RJ, Josephs EB, Platts AE, et al. 2014. Evidence for widespread positive and negative selection in coding and conserved noncoding regions of Capsella grandiflora. PLoS Genetics 10: e1004622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S, Keeling J, Gillman L. 2006. The road from Santa Rosalia: a faster tempo of evolution in tropical climates. Proceedings of the National Academy of Sciences 103: 7718–7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Computer Applications in the Biosciences 13: 555–556. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Molecular Biology and Evolution 19: 908–917. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wong WS, Nielsen R. 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Molecular Biology and Evolution 22: 1107–1118. [DOI] [PubMed] [Google Scholar]
- Yoder AD, Yang Z. 2000. Estimation of primate speciation dates using local molecular clocks. Molecular Biology and Evolution 17: 1081–1090. [DOI] [PubMed] [Google Scholar]
- Yu X, Tan W, Zhang H, Gao H, Wang W, Tian X. 2019. Complete chloroplast genomes of Ampelopsis humulifolia and Ampelopsis japonica: molecular structure, comparative analysis, and phylogenetic analysis. Plants 8: 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J-X, Li J, Wang D, Araki H, Tian D, Yang S. 2010. Genome-wide investigation reveals high evolutionary rates in annual model plants. BMC Plant Biology 10: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachos J, Pagani M, Sloan L, Thomas E, Billups K. 2001. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 292: 686–693. [DOI] [PubMed] [Google Scholar]
- Zecca G, Abbott JR, Sun W-B, Spada A, Sala F, Grassi F. 2012. The timing and the mode of evolution of wild grapes (Vitis). Molecular Phylogenetics and Evolution 62: 736–747. [DOI] [PubMed] [Google Scholar]
- Zecca G, Grassi F, Tabidze V, et al. 2020a. Dates and rates in grape’s plastomes: evolution in slow motion. Current Genetics 66: 123–140. [DOI] [PubMed] [Google Scholar]
- Zecca G, Labra M, Grassi F. 2020b. Untangling the evolution of American wild grapes: Admixed species and how to find them. Frontiers in Plant Science 10: 1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Molecular Biology and Evolution 22: 2472–2479. [DOI] [PubMed] [Google Scholar]
- Zhang N, Wen J, Zimmer EA. 2015a. Expression patterns of AP1, FUL, FT and LEAFY orthologs in Vitaceae support the homology of tendrils and inflorescences throughout the grape family. Journal of Systematics and Evolution 53: 469–476. [Google Scholar]
- Zhang N, Wen J, Zimmer EA. 2015b. Congruent deep relationships in the grape family (Vitaceae) based on sequences of chloroplast genomes and mitochondrial genes via genome skimming. PLoS One 10: e0144701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Y, Andolfatto P. 2012. Methods to detect selection on noncoding DNA. Evolutionary Genomics 2: 141–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Yonezawa T, Zhong Y, Hasegawa M. 2009. Episodic evolution and adaptation of chloroplast genomes in ancestral grasses. PLoS One 4: e5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Sun L, Penny D. 2015. The origin of land plants: a phylogenomic perspective. Evolutionary Bioinformatics 11: EBO–S29089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.